SUMMARY
We perform a CRISPR-Cas9 genome-wide screen in glioblastoma stem cells and identify integrin αvβ5 as an internalization factor for Zika virus (ZIKV). Expression of αvβ5 is correlated with ZIKV susceptibility in various cells and tropism in developing human cerebral cortex. A blocking antibody against integrin αvβ5, but not αvβ3, efficiently inhibits ZIKV infection. ZIKV binds to cells but fails to internalize when treated with integrin αvβ5-blocking antibody. αvβ5 directly binds to ZIKV virions and activates focal adhesion kinase, which is required for ZIKV infection. Finally, αvβ5 blocking antibody or two inhibitors, SB273005 and cilengitide, reduces ZIKV infection and alleviates ZIKV-induced pathology in human neural stem cells and in mouse brain. Altogether, our findings identify integrin αvβ5 as an internalization factor for ZIKV, providing a promising therapeutic target, as well as two drug candidates for prophylactic use or treatments for ZIKV infections.
Graphical Abstract

In Brief
Wang et al. show that Zika virus (ZIKV) uses integrin αvβ5 to infect neural stem cells. ZIKV infection can be inhibited by αvβ5 blocking antibody or inhibitors, SB273005 and cilengitide, in human neural stem cells and in mouse brain, providing drug candidates for prophylactic use or treatments for ZIKV infections.
INTRODUCTION
Zika virus (ZIKV) is a re-emerging arbovirus belonging to the Flavivirus genus that includes other mosquito-borne human pathogens such as dengue virus (DENV1–4), West Nile virus (WNV), yellow fever virus (YFV), and Japanese encephalitis virus (JEV), among others (Lazear and Diamond, 2016). One half of people on Earth are at risk for ZIKV infection, and there is no safe and effective treatment or vaccine. ZIKV infection is associated with severe fetal abnormalities, including microcephaly, hydranencephaly, and intrauterine fetal growth restriction (Brasil et al., 2016; Noronha et al., 2016; Sarno et al., 2016). In vitro and in vivo studies have shown that ZIKV preferentially infects neural stem/progenitor cells and immature neurons in the developing brain and dysregulates various cellular processes (Cugola et al., 2016; Dang et al., 2016; Li et al., 2016; Tang et al., 2016). These processes are thought to directly cause microcephaly and other brain abnormalities in infants infected in utero. The molecular mechanisms by which ZIKV dysregulates critical human neural stem cell (hNSC) functions are not well understood.
ZIKV is a mosquito-borne flavivirus originally discovered in 1947 (Driggers et al., 2016) that had caused sporadic disease in Africa and Asia. Recent outbreaks occurred in 2007 in Micronesia and in 2013 in French Polynesia (Broutet et al., 2016). The Brazilian outbreak of ZIKV in 2015–2016 has raised alarms about enhanced viral pathogenicity and expansion of its global range. ZIKV has a single positive (+) strand RNA genome coding for a single polyprotein, which is cleaved by viral and host proteases to produce three structural and seven nonstructural proteins (Miner and Diamond, 2017).
A number of genome-wide CRISPR screens have been performed in flavivirus infection models and have begun to illuminate our understanding of host pathways important in the life cycle of flaviviruses. Two CRISPR screens against WNV infection have been performed in human cells and identified members of the endoplasmic reticulum membrane complex (EMC) and endoplasmic reticulum-associated signal peptidase complex (SPCS) (Ma et al., 2015). A CRISPR screen against Dengue virus (DENV) and Hepatitis C virus (HCV) again confirmed the importance of endoplasmic reticulum (ER) protein complexes in the replication of flaviviruses (Marceau et al., 2016). Another study evaluated two different genome-wide RNAi pools in DENV infection, conducted a CRISPR screen against ZIKV infection in HeLa cells, and also confirmed the importance of the EMC complex in DENV and ZIKV infection (Savidis et al., 2016). Recently, two CRISPR screens were performed to identify ZIKV dependency factors in neural progenitor cells (Li et al., 2019; Wells et al., 2018). These screens identified heparan sulfation, endocytosis, ER processing, and Golgi and interferon functions (Li et al., 2019) as well as vacuolar ATPase in addition to heparan sulfation and oligomeric Golgi complex as ZIKV-dependent factors (Wells et al., 2018).
Integrins, a family of 24 heterodimers consisting of α and β subunits, are transmembrane adhesion receptors that are key components of cell signaling mechanisms involved in cancer progression and metastasis (Hynes, 2002). Specific ligands bind and cluster integrins to regulate vehicle trafficking and transduce both outside-in and inside-out signaling events (Hynes, 2002). In one of the outside-in signaling mechanisms of integrins, focal adhesion kinase (FAK) is phosphorylated and activated to recruit additional kinases and induce complex signaling cascade to regulate cell survival, proliferation, and migration (Mitra and Schlaepfer, 2006). Therefore, FAK inhibitors have been developed to control migration, invasion, and metastasis of various tumors. Several viruses had been reported to use integrins as receptors or co-receptors, including adenovirus (Summerford et al., 1999; Wickham et al., 1993), Kaposi’s sarcoma-associated herpesvirus (KSHV) (Akula et al., 2002), foot-and-mouth disease virus (Jackson et al., 2004), echovirus (Bergelson et al., 1992), human metapneumovirus (Cseke et al., 2009), hantavirus (Larson et al., 2005), human parechovirus (Triantafilou et al., 2000), reovirus (Maginnis et al., 2006), and rotavirus (Guerrero et al., 2000).
AXL receptor had been initially described as an entry receptor for ZIKV (Hamel et al., 2015; Meertens et al., 2017; Nowakowski et al., 2016; Retallack et al., 2016; Savidis et al., 2016). However, some reports argued that AXL was not essential for ZIKV entry in human NSCs and organoids (Wells et al., 2016). In addition, AXL was not required for ZIKV infection in mice (Hastings et al., 2017; Li et al., 2017). ZIKV utilizes AXL receptor for infection in human fetal endothelial cells and AXL promotes virus replication by antagonizing type I interferon signaling in astrocytes (Chen et al., 2018a; Richard et al., 2017; Tabata et al., 2016). Despite this progress, however, the determinants of cell and tissue tropism of ZIKV, the entry receptors or co-receptors, and the host factors required for ZIKV replication in the CNS are still largely unknown (Miner and Diamond, 2017). In order to address these questions, we performed the a CRISPR-Cas9 genome-wide screen in glioblastoma stem cells (GSCs) to identify factors determining ZIKV neurotropism. In parallel, to distinguish neuro-specific host factors from other cell types, a genome-wide CRISPR screen was performed in HEK293FT cells. Our studies identified a unique GSCs-specific list of 92 genes that linked to ZIKV infection in GSCs. Integrin αvβ5, but not αvβ3, was identified as an entry factor mediating ZIKV internalization and ZIKV virions physically interacted with integrin αvβ5. The expression of ITGB5 correlated with ZIKV susceptibility in various cell lines and primary cell types in CNS. Receptor blocking antibodies as well as two αvβ5 pharmaceutical inhibitors blocked ZIKV infection and significantly reduced ZIKV-induced pathology in hNSC and mouse brains.
RESULTS
Genome-wide CRISPR Knockout Screen in GSCs Identifies ZIKV Dependency Factors
Here, we performed a genome-wide pooled CRISPR knockout screen to gain insight into host dependency factors of ZIKV infection in human neural stem-like cells. We used a genome-wide CRISPR library generated by Root lab (targeting 19,114 coding genes, 4 single guide RNAs [sgRNAs]/gene), named Brunello (Doench et al., 2016). As illustrated in Figure 1A, human glioblastoma stem cells (GSCs) were transduced with the lentiviral Brunello CRISPR library (400 million cells at MOI = 0.3), selected with puromycin for 7 days for stable viral integration and completion of genome editing. GSCs (TS576) are highly permissive to ZIKV (Zhu et al., 2017). Stemness of the cells was confirmed with high expression of stem cell marker SOX2, and lack of differentiation maker GFAP expression (Figures 1B and S1A). GSCs shares similarities with neural stem cells and stemness markers are constantly detected during the culture. We infected cells with ZIKV-ZsGreen reporter virus. This virus was constructed based on the BeH819015 strain (GI: 975885966), a representative of the most prominent group of ZIKV isolates from South America (Mutso et al., 2017). A robust ZsGreen signal was observed indicating efficient viral infection (Figures 1B and S1B). When infected at a lower MOI (MOI = 0.01), viral replication was significant increased at 5 and 6 days, and all cells were infected with ZIKV at 7 days. When infected at a higher MOI (MOI = 0.5), the viral infection significantly increased in 3 days and 4 days, reaching approximately full infection 5 days post-infection (Figure 1C). Cell death was apparent at day 3, when around 80% of cells were died from virus-induced cytopathic effect (Figure S1C). Stemness of cells was not changed by ZIKV infection (Figure S1D).
Figure 1. Identification of ZIKV Dependency Factors in Glioblastoma Stem Cells (GSCs) by a Genome-wide CRISPR Knockout Screen.
(A) Schematic representation of the genome-wide CRISPR/Cas9 screen in GSCs. Briefly, GSC cells were transduced with Brunello sgRNA pooled library and selected by puromycin treatment. Transduced cells were mock-infected or infected with ZIKV ZsGreen strain (ZIKV-ZsGreen) at an MOI of 0.01 for 7 days. ZsGreen negative cells were sorted out and cultured for 2 weeks in the presence of ZIKV inhibitor, NITD008. Expanded cells were re-infected with ZIKV for 1 week and sorted for resistant cells. Genomic DNA was harvested, and sgRNA sequencing libraries were prepared by nested PCR and quantified on Illumina NextSeq.
(B) Characterization of GSC (TS576) stemness. GSCs are shown to highly express stem cell marker SOX2 and lack of differentiation maker GFAP expression. n = 3.
(C) GSCs are highly permissive for ZIKV-ZsGreen infection. GSCs were infected with ZIKV-ZsGreen at an MOI of 0.5 for 5 days. n = 3.
(D) Replication curve of ZIKV-ZsGreen in GSCs infected at an MOI of 0.5 or 0.01. ZsGreen-positive cells were quantified by flow cytometry. n = 3.
(E) Identification of sgRNAs consistently enriched in ZsGreen-negative versus uninfected cells. STARS analysis was performed, and 271 genes are shown for illustration. Previously identified genes are highlighted in blue. A few of newly identified genes in our study are highlighted in red and orange.
(F) Enrichment of specific sgRNAs identified by STARS analysis is enhanced after expansion and second sorting. Known targets that have been identified were highlighted by red asterisk. Color key represents fold change.
We utilized two rounds of sorting to enrich ZIKV resistant cells to identify host genes important in ZIKV infection. We challenged 75 million of the transduced and selected cells with ZIKV at a low MOI (MOI = 0.01) for 7 days. The other 40 million cells were mock infected as control (Figure 1A). After 7 days of infection, ZsGreen-negative cells were sorted out with a very restricted gating strategy (Figures S1E and S1F). Half of the cells were collected and another half were retained for a second round of infection and sorting. A ZIKV NS5 inhibitor (NITD008) was used to reduce the cytopathic effect of ZIKV and allowed cells to expand for 2 weeks. The expanded cells were re-infected with ZIKV-ZsGreen (MOI = 0.01) for 7 days and sorted for ZsGreen negative cells. Genomic DNA was extracted from harvested cells and sgRNAs were PCR-amplified and deep sequenced on an Illumina NextSeq (Figure 1A). Normalized reads of ZIKV treated cells were compared to mock infection cells. The frequency of the amplified CRISPR plasmid and mock group sequencing reached >95% of the entire library, confirming that it was a genome-wide screen (Figures S1G and S1H).
The use of multiple sgRNAs targeting each gene in pooled screening allows for further discrimination of valid hits by identifying genes in which multiple sgRNAs are enriched in the infection-resistant population. We used the STARS algorithm to rank screening hits with consistent enrichment among multiple sgRNAs in late ZIKV-infected cells compared to mock infected cells of the same duration (Doench et al., 2016). STARS analysis was utilized to score and rank the sgRNAs that were enriched in 1st and 2nd round screen compared with mock group. We excluded the genes whose sgRNAs had low sequencing coverage (<5 reads per million). We obtained a final list of 271 candidate genes (Table S1) with at least 2 separate sgRNAs that were enriched (Figures 1E and S1I). For illustration purposes, Figure 1F shows 11 gene targets with significant fold change and further enrichment compared to the first infection. Our hits, including SSR3, STT3A, MMGT1, and SSR2, EMC2, EMC3, EMC6 are consistent with the prior screens in flaviviruses (Ma et al., 2015; Marceau et al., 2016; Zhang et al., 2016) and were also top scoring hits in the ZIKV HeLa screen (Savidis et al., 2016).
We are interested in identifying unique genes that determine ZIKV susceptibility in GSCs. Thus, another screen was conducted in the HEK293FT cell line. HEK293FT cells were transduced with the CRISPR library (MOI = 0.3), selected with puromycin for 7 days. Then 30 million of the transduced and selected cells were challenged with ZIKV or mock infection. Cells were harvested at two time points, early (4 days post ZIKV infection) and late (7 days post ZIKV infection) time points of infection as well as the uninfected mock control. Genomic DNA was extracted from harvested cells and sgRNAs were amplified and deep sequenced. Normalized reads of ZIKV treated cells were compared to mock infection cells (Figures S2A-S2C). We examined the list of enrichment for genes (Table S2) in which multiple sgRNAs also had higher fold change in both early and late infection time points. This generated a gene list (in total 159 genes) with at least 2 separate sgRNAs that were not only enriched both at early and late time points but also enriched to a greater extent at the later time point than at the early time point (Figures S2D and S2E). We then excluded the genes whose sgRNAs had low sequencing coverage (<5 reads per million in both replicates) and obtained a final list of 101 candidate genes (Table S2). Top hits are involved in ER membrane complex and endocytosis that is required for efficient viral replication or internalization (Figures S2F and S2G). We validated the function of the two top hits (MMGT1 and EMC6) and two endocytosis-related genes (COPB2 and DNM2) by performing sgRNA knockout (KO) (2 sgRNAs per target) (Figure S2H). KO of COPB2, DNM2, significantly impaired the ability of ZIKV to productively infect target cells (Figure S2H). We also validated 4 additional genes in our candidate list (STOM, RPL23, SV2C, TMTC3) whose KO by CRISPR/Cas9 also inhibited ZIKV replication (Figure S2H). These results demonstrate that we have successfully identified 293FT-specific genes involved in ZIKV infection.
Identification of ZIKV Dependency Factors Specific for GSCs and Their Validation in ZIKV Infection
To identify the factors that contribute to ZIKV infection in GSCs, we compared CRISPR screen data in GSCs with other screens performed in 293FT and HeLa cells (Savidis et al., 2016). The Venn diagram comparing our hits in GSCs and 293FT, and from the previous HeLa screen is shown in Figure 2A. Ten common genes identified belong to ER translocation complex, ERAD pathway and oligosaccharyltransferase complex which were characterized in previous study for flavivirus infection (Marceau et al., 2016). Strikingly, 92 hits were unique to our GSCs cells screen and have not been previously linked to ZIKV infection (Figure 2A). Next, we selected a few of the top hits for further functional validation. TS576 cells were transduced with short hairpin RNA (shRNA) lentiviral particles to knockdown target genes. As shown in Figure S3A, the mRNA of the 11 targets were significantly decreased after shRNA transduction. Importantly, ZIKV infection was inhibited when we silenced the expression of eleven genes including: CENPH, ITGB5, MYLPF, HOMER1, BAALC, GABBR2, EPHA10, PTPN2, GCNT7, TRAM1, and TMEM41B. Among them, ITGB5, an integrin family member dramatically decreased virus replication by 86%. Knockdown of two other neural genes, HOMER1 and BAALC, also significantly reduced virus replication (Figures 2B and S3B), suggesting that these factors contribute to ZIKV neurotropism. TMEM41B that has been reported to be essential for DENV infection (Marceau et al., 2016) was also involved in ZIKV infection. GCNT7, a glucosaminyl transferase, which might participate in glycosylation of ZIKV envelope in Golgi apparatus affecting the budding process as well as infectivity of virions. Altogether, our results have identified 92 unique host dependency factors in GSCs and confirmed that silencing of 11 targets efficiently reduced virus replication.
Figure 2. Identification of GSC-Specific ZIKV Dependency Factors.
(A) Comparison of targets from three genome-wide screens in GSC and 293 FT (this study) and HeLa cells (Savidis et al., 2016). Ten common targets are shown in the green box (right). A representative list of GSC-specific targets is shown in blue box.
(B) Eleven unique targets are essential for ZIKV infection in GSCs. GSCs were transduced with lentiviruses expressing non-targeting control (NC) shRNA or pooled shRNAs for indicated genes. After 2 days, cells were infected with ZIKV ZsGreen at an MOI of 0.01 for 3 days. Cells were harvested to extract the RNA, and RT-PCR was performed to quantify the expression of ZIKV NS3 gene relative to GAPDH. Mean ± SD of n = 3. *p < 0.05, **p < 0.01 by Student’s t test.
(C) ITGB5 KO efficiency in TS576. TS576 cells were transduced with non-targeting sgRNA (NTC) or four separate sgRNAs targeting ITGB5. Cells were selected by 1 μg/mL puromycin for 7 days. The expression of αvβ5 on cell surface was monitored by flow cytometry. Blue shading represents cell staining with isotype antibody, red shading represents NTC cells staining with αvβ5 antibody, golden shading represents four ITGB5-edited cells staining with αvβ5 antibody. One representative of three independent experiments is shown.
(D) ITGB5 KO dramatically reduced ZIKV infection. NTC- or ITGB5-edited cells were infected with ZIKV-ZsGreen at MOI = 0.05 for 72 h. The percentage of infected cells were measured by flow cytometry. Mean ± SD of n = 3. ****p < 0.0001 by Student’s t test.
(E) αvβ5 expressions is restored by trans-complementation of ITGB5. The expressions of αvβ5 was characterized in control or ITGB5 KO TS576 cells transduced with empty vectors (e.v.) or rescue vectors (sg1_ β5 or sg2_β5) for 2 days. The expression of αvβ5 on cell surface was monitored by flow cytometry. Golden shading represents cells staining with isotype antibody, blue shading represents cells transduced with empty vectors (e.v.) staining with αvβ5 antibody, red shading represents cells transduced with rescuing vectors (sg1_ β5 or sg2_β5) staining with αvβ5 antibody. A representative of three independent experiments is shown.
(F) Trans-complementation of ITGB5 in ITGB5 KO TS576 cells restored ZIKV infectivity. The control and ITGB5 KO TS576 cells expressing e.v. or resistant ITGB5 were infected with ZIKV-ZsGreen at MOI = 0.05 for 72 h. The percentage of infected cells were measured by flow cytometry. Mean ± SD of n = 3. **p < 0.01, ***p < 0.001, ****p < 0.0001 by Student’s t test.
Various integrin family members had been identified as receptors for diverse viruses including flaviviruses (Stewart and Nemerow, 2007). To further investigate the role of ITGB5 in ZIKV infection, we generated ITGB5 KO cells using four distinct sgRNAs. Because integrin β5 forms and functions as heterodimers only with integrin αv subunit (Ley et al., 2016), we determined the expressions of ITGB5 by surface staining of αvβ5. As shown in Figure 2C, all of the four sgRNAs could ablate αvβ5 expressions on the cell membrane. Consistent with the screening and shRNA results, ITGB5 KO showed a robust resistance to infection by ZIKV-ZsGreen (Figures 2D, S3C, and S3D). To exclude the off-target effects of the sgRNAs, a cDNA rescue experiment was performed. We designed rescuing ITGB5 vectors lacking either sgRNA1 or 2 target sites to rescue the expression of ITGB5 in KO cells (Figure S3E). The rescuing vectors were delivered into TS576 control or ITGB5 KO cells. αvβ5 was confirmed to be overexpressed and rescued in control and KO cells, respectively. Compared to sgRNA 1 resistant construct (sg1_ β5), sgRNA 2 resistant construct (sg2_ β5) reached a higher rescued expression of αvβ5 (Figure 2E), probably due to the difference in the codon preference in the muted sites leading to the difference in the translational efficiency. To determine whether the ectopic expression of integrin β5 will enhance ZIKV infection, we infected TS576 cells overexpressing sgRNA resistant ITGB5 with ZIKV and analyzed viral replication by quantifying green cells. We found that ZIKV infection was increased in TS576 cells expressing both sgRNA-resistant vectors (Figure 2F), demonstrating that the trans-complementation of ITGB5 in ITGB5 KO cells could restore viral infectivity. Consistent with the β5 expression by two sgRNA-resistant vectors observed in Figure 2E, sg2_ β5 vector rendered TS576 cells more susceptible to ZIKV infection as it led to higher β5 expression (Figure 2F). These results show that ITGB5 is essential for ZIKV infection.
Integrin αvβ5 Mediates Internalization of ZIKV
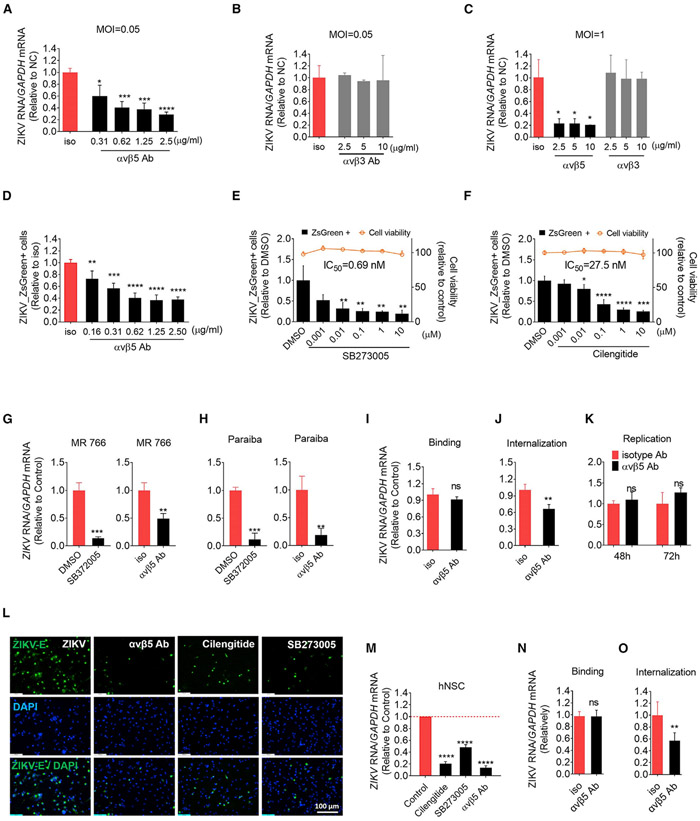
WNV and Japanese encephalitis virus use integrin αvβ3, but not αvβ5, for viral entry (Chu and Ng, 2004; Cui et al., 2018). In our CRISPR screen, ITGB5, but not ITGB3, was enriched in ZIKV infection resistant cells. Here, anti-integrin αvβ5 antibody was utilized to block its function to determine the role of integrin β5 in ZIKV infection. The results showed that pre-treatment of anti-integrin αvβ5 blocking antibody significantly inhibited ZIKV infection in dose-dependent manner at low and high MOIs conditions compared with isotype antibody treatment (Figures 3A and 3C). The flow data analysis also showed that this blocking antibody reduced the percentage of ZIKV-positive cells in a dose-dependent manner (Figure 3D). The released viral RNA in supernatant was also decreased by the blocking antibody (Figure S4A). To determine the specificity of the roles of αvβ5 in ZIKV infection, anti-integrin αvβ3 blocking antibody was included. We observed that αvβ3 was expressed at the surface of TS576 cells (Figure S4B). Treatment of αvβ3 antibody did not inhibit ZIKV infection at different MOIs even at higher concentrations (Figures 3B, 3C, and S4C). These results suggest that unlike other members of the flavivirus family, ZIKV prefers to utilize αvβ5, but not αvβ3, for its infection. Considering that integrin-ligand binding is dependent on cations (Summerford et al., 1999; Wickham et al., 1993; Zhang and Chen, 2012), we treated the cells with 5 mM EDTA during ZIKV infection. Our results showed that ZIKV infection was significantly reduced by the treatment of EDTA, further supporting the notion that αvβ5 function is essential for ZIKV infection (Figures S4D-S4F).
Figure 3. Integrin β5 Is Identified as an Internalization Factor for ZIKV and Contributes to Its Neurotropism.
(A–D) αvβ5, but not αvβ3, is essential for ZIKV infection. TS576 cells were treated with various concentrations of anti-integrin αvβ5, αvβ3, or isotype antibody for 1 h and infected with ZIKV with MOI = 0.05 (A, B, and D) or MOI = 1 (C). At 72 h (A, B, and D) or 48 h (C) post-infection, cellular ZIKV RNA was quantified by qPCR and the percentage of infected cells were measured by flow cytometry (D). Mean ± SD of n = 3. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, by Student’s t test.
(E) αvβ5 inhibitor, SB273005 represses ZIKV infection. TS576 cells were pre-treated with indicated concentrations of SB273005 or DMSO for 1 h and infected with ZIKV with MOI = 0.05 for 72 h. The percentage of infected cells were measured by flow cytometry. Cell viability was measured in TS576 cells treated with indicated concentration of SB273005 for 3 days. Mean ± SD of n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test.
(F) αvβ5 inhibitor, cilengitide, represses ZIKV infection. ZIKV infection assay was performed as (E) with indicated concentrations of cilengitide. Cell viability was measured in TS576 cells treated with indicated concentration of cilengitide for 3 days. Mean ± SD of n = 3. *p < 0.05, ****p < 0.0001 by Student’s t test.
(G and H) αvβ5 is a broad dependent factor for different ZIKV strains. TS576 was pretreated with antibody or drug as (E) and then infected with MR766 (G) or Paraiba strain (H) at MOI 0.5 for 48 h. Cellular ZIKV RNA was quantified by qPCR. Mean ± SD of n = 3. **p < 0.01, ***p < 0.001 by Student’s t test.
(I and J) αvβ5 is essential for ZIKV endocytosis, but not for binding in TS576 cells. (I) TS576 cells were treated with 2.5 μg/mL anti-integrin αvβ5 or isotype antibody for 1 h and then challenged with ZIKV with MOI = 20 at 4°C for 2 h. Cells were washed, and total RNA was extracted and quantified. (J) Cells were treated as in (I) and then after washing, infected cells were transferred to 37°C for 4 h, and un-internalized virions were removed by Accumax. Total cellular RNA was extracted, and ZIKV RNA was quantified by qPCR. Mean ± SD of n = 3. **p < 0.01 by Student’s t test. ns, not significant.
(K) Inhibiting αvβ5 does not affect ZIKV replication. TS576 was transfected with 500 ng ZIKV-ZsGreen replicon RNA and then pretreated with 2.5 μg/mL αvβ5 antibody or isotype antibody. 48 or 72 h after transfection, cells were collected for RNA quantification of ZIKV. Mean ± SD of n = 3. ns, not significant by Student’s t test.
(L and M) αvβ5 is essential for ZIKV infection in primary human fetal brain-derived neural stem cells (hfNSC). hfNSC was pretreated with 2.5 μg/mL αvβ5 antibody or isotype antibody, DMSO, or 2.5 μM SB273005 or 2.5 μM cilengitide for 1 h, and then challenged with MR766 at MOI = 1 for 72 h. (L) ZIKV E protein was stained by immunofluorescence assay. (M) Total cellular RNA was extracted and ZIKV RNA was quantified by RT-PCR. Mean ± SD of n = 3. ****p < 0.0001 by Student’s t test.
(N and O) αvβ5 is essential for ZIKV endocytosis, but not for binding in hfNSC. (N) hfNSC were treated with 2.5 μg/mL anti-integrin αvβ5 or isotype antibody for 1 h and then challenged with MR766 with MOI = 20 at 4°C for 2 h. Cells were washed, and total RNA was extracted and quantified. (O) Cells were treated as in (I) and then after washing, infected cells were transferred to 37°C for 4 h, and un-internalized virions were removed by Accumax. Total cellular RNA was extracted and ZIKV RNA was quantified by qPCR. Mean ± SD of n = 3. **p < 0.01 by Student’s t test. ns, not significant.
Because integrins are promising drug targets for cancer therapy, there are a number of integrin inhibitors in clinical trials to treat a variety of cancers (Ley et al., 2016). We selected two αvβ5 integrin inhibitors, SB273005 and cilengitide, to validate αvβ5 integrin function and also to evaluate their potential for anti-ZIKV drug development. Our results showed that treatment with these inhibitors significantly blocked ZIKV infection in TS576 cells in a dose-dependent manner (Figures 3E, 3F, and S4G-S4I). Strikingly, both SB273005 and cilengitide showed significant anti-ZIKV activity at nanomolar concentrations. The half-maximal inhibitory concentration (IC50) of SB273005 and cilengitide were 0.69 nM and 27.5 nM, respectively (Figures 3E and 3F). In addition, the IC50s were quite similar to their half-maximal binding concentrations to αvβ5 integrin in vitro (SB273005, 0.3 nM; cilengitide, 37 nM) (Liu et al., 2008; Miller et al., 2000). These inhibitors did not exhibit any cytotoxicity, even at 10 μM in TS576 (Figures 3E and 3F). These results further validate that αvβ5 integrin is a key host factor for ZIKV infection and provide two promising drug candidates targeting αvβ5 integrin for ZIKV infections.
Although ZIKV-ZsGreen (BeH819015) is a representative of the most prominent group isolated from South America (Mutso et al., 2017), we decided to verify the roles of αvβ5 in infections of another Asian lineage strain Paraiba and an African lineage strain MR766. Similar to ZIKV-ZsGreen results, treatment of the both blocking antibody and SB273005 significantly reduced virus infection in TS576 (Figures 3G and 3H). Altogether, these results demonstrate that αvβ5 plays a key role in infections of different ZIKV strains.
Because integrin αvβ5 is a transmembrane protein and blocking its ligands-binding domains inhibits ZIKV infection, it suggests that integrin αvβ5 promotes ZIKV entry. To test this hypothesis, we performed virus binding and internalization assays. TS576 cells were treated with anti-integrin αvβ5 or isotype antibody for 1 h and then challenged with ZIKV at 4°C for 2 h. For binding assay, cells were washed, and total RNA was extracted and quantified. For internalization assay, cells were washed and transferred to 37°C for 4 h, and un-internalized virions were removed by Accumax. Total cellular RNA was extracted and ZIKV RNA was quantified by qPCR. These results showed that at 4°C, ZIKV binding to cells was not affected by treatment of the blocking antibody (Figure 3I). For virus internalization, less virus RNA was detected within the cells by pre-treatment of anti-integrin αvβ5 antibody (Figure 3J). To exclude the possibility that αvβ5 is involved in viral replication, we transfected in vitro-transcribed ZIKV replicon into TS576 cells (Mutso et al., 2017). No difference of ZIKV RNA replication was detected after treatment with the blocking antibody (Figure 3K). These results show that integrin αvβ5 mediates virus internalization but not binding.
Next, to evaluate physiological significance of αvβ5, we investigated the role of integrin αvβ5 in primary human neural stem cells (hNSCs), which ZIKV preferentially infect in vitro and in vivo (Dang et al., 2016; Tang et al., 2016). hNSCs were characterized by analyzing Nestin expression (Figure S4J). hNSCs were treated with either αvβ5 blocking antibody or two inhibitors followed by ZIKV infection. Consistent with the results obtained in TS576 cells (Figures 3A, 3D, 3F, and 3G), anti-αvβ5 blocking antibody, cilengitide, and SB273005 efficiently inhibited ZIKV infection in hNSCs (Figures 3L and 3M). Similarly, αvβ5 blocking antibody exerted no effect on viral binding but significantly reduced virus internalization in hNSCs (Figures 3N and 3O). This suggests that integrin αvβ5 acts as an internalization factor for ZIKV both in TS576 and hNSCs.
ZIKV Virions Directly Interact with Integrin αvβ5, and FAK Activation Is Essential for ZIKV Infection
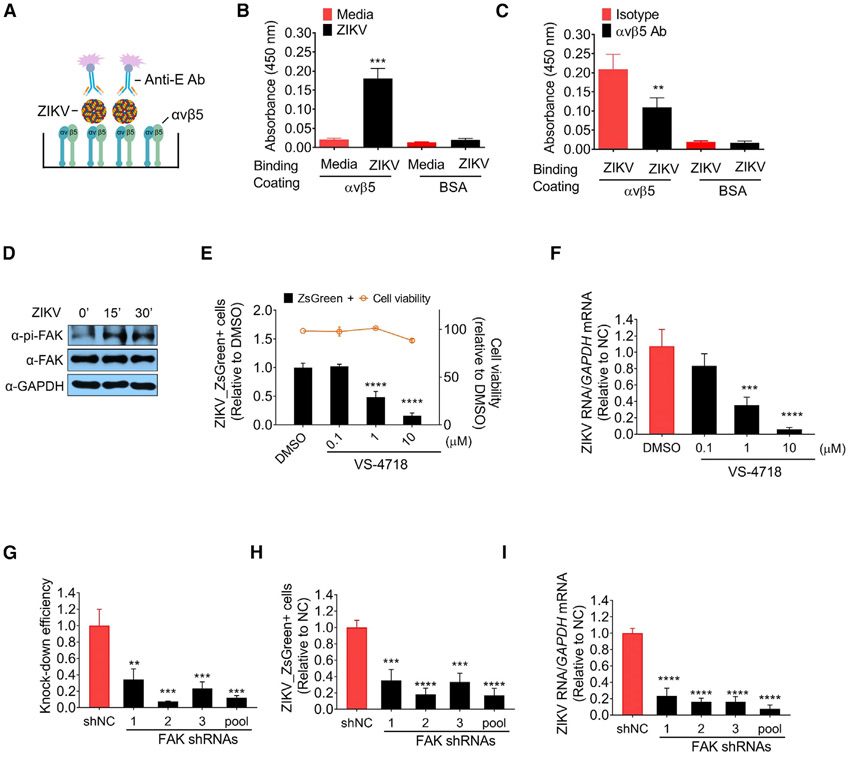
To determine whether ZIKV virions directly bind integrin αvβ5, we performed ZIKV-αvβ5 ELISA experiments by coating purified integrin αvβ5 on plates and incubating with ZIKV virions. The principle of this assay is shown in Figure 4A. We found that ZIKV virions specifically bind with integrin αvβ5 but not with BSA (Figure 4B). Furthermore, the binding of ZIKV-αvβ5 was inhibited by αvβ5 blocking antibody, while isotype antibody showed no effects on ZIKV and integrin interactions (Figure 4C). Along with the results that αvβ5 blocking antibody reduced ZIKV internalization in both TS576 and primary NSCs (Figures 3J and 3O), the physical interaction of ZIKV-αvβ5 is essential for ZIKV infection. Together, these results demonstrate that ZIKV virions physically interact with integrin αvβ5, and this interaction is essential for ZIKV infection.
Figure 4. ZIKV Virions Directly Interact with Integrin αvβ5 and FAK Activation Is Essential for ZIKV Infection.
(A) Principle of the ZIKV-integrin ELISA assay. Purified integrin αvβ5 was coated on 96-well ELISA plate and incubated with MR766. Bound virus was detected by biotin-conjugated anit-E antibody and HRP-streptavidin.
(B) ZIKV virions interacted with integrin αvβ5. Purified integrin αvβ5 or BSA was coated on 96-well plate overnight at 4°C and incubated with MR766 or media for 2 h at 4°C. Bound virus was detected by biotin-conjugated anit-E antibody and HRP-streptavidin. Absorbance (450 nm) was measured for each well to quantify the bound virions. Mean ± SD of n = 3. ***p < 0.001 by Student’s t test.
(C) ZIKV binding to integrin αvβ5 is neutralized by αvβ5 blocking antibodies. Purified integrin αvβ5 or BSA was coated on 96-well plate overnight at 4°C and incubated with αvβ5 blocking antibody (P1F6, 5 μg/mL) at 37°C for 30 min, and then incubated with MR766 or media for 2 h at 4°C. Bound virus was detected by biotin-conjugated anit-E antibody and HRP-streptavidin. Absorbance (450 nm) was measured for each well to quantify the bound virions. Mean ± SD of n = 3. **p < 0.01 by Student’s t test.
(D) FAK is activated by ZIKV infection. Western blot analysis of phosphorylated FAK (pi-FAK) or FAK was performed in mock or ZIKV (MOI = 50)-infected TS576 cells for indicated time points. GAPDH was shown as loading control. One representative of three independent experiments is shown.
(E and F) FAK inhibitor, VS-4718 represses ZIKV infection. TS576 cells were pre-treated with indicated concentrations of VS-4718 or DMSO for 1 h and infected with ZIKV with MOI = 0.05 for 72 h. The percentage of infected cells were measured by flow cytometry. Cell viability was measured in TS576 cells treated with indicated concentration of VS-4718 for 3 days (E). ZIKV RNA was quantified by RT-PCR (F). Mean ± SD of n = 3. ***p < 0.001, ****p < 0.0001 by Student’s t test.
(G–I) Knock down of FAK inhibits ZIKV infection. TS576 cells were transduced with three different shRNAs or shRNA pool for 2 days and then infected with ZIKV with MOI = 0.05 for 72 h. The knock down efficiency was determined by measuring the expressions of FAK using RT-PCR (G). The percentage of infected cells were measured by flow cytometry (H). ZIKV RNA was quantified by RT-PCR (I). Mean ± SD of n = 3. **p < 0.01, ***p < 0.001, ****p < 0.0001 by Student’s t test.
Despite the significance of integrins in cancer and other disease, the complete understanding of integrin-specific signalosomes is still not available. Canonical integrin signaling involves FAK phosphorylation at Y397 that leads to signaling cascades regulating cell survival, proliferation, adhesion, migration, and invasion (Desgrosellier and Cheresh, 2010; Tancioni et al., 2014). To investigate the mechanism of integrin-mediated signaling by ZIKV, we probed the activity of FAK by ZIKV infection. We infected TS576 cells with ZIKV and analyzed FAK phosphorylation at different time points. We detected a peak induction of activated FAK (pi-FAK-Y397) within 15 min after infection (Figure 4D). Next, we determined the functional relevance of FAK phosphorylation in ZIKV infection by two methods: (1) pharmacological inhibition of FAK phosphorylation by VS-4718 (Serrels et al., 2015; Tancioni et al., 2014; Figures 4E and 4F), and (2) silencing of FAK by three separate shRNAs as well pooled shRNAs (Figures 4G-4I). Both of these approaches to inhibit FAK function resulted in a significant repression of ZIKV infection (Figures 4G–4I). Altogether, these results show that ZIKV-integrin interactions involve FAK-mediated signaling pathway, and inhibition of FAK provides another strategy to inhibit ZIKV infection. Future studies could elucidate how FAK activation reprograms hNSCs transcriptional and proteomic networks to modulate ZIKV infection.
αvβ5 Expression Correlates with ZIKV Susceptibility and Contributes to ZIKV Neurotropism
To gain insight into the correlation of αvβ5 with viral neurotropism, we next investigated the correlation between αvβ5 expression and ZIKV susceptibility in multiple cell types including GSCs (TS576), microglia, 293FT, H9, and Jurkat. High expression levels of αvβ5 were detected in TS576 and a microglia cell line, which ZIKV preferentially infect in vitro and in vivo (Chen et al., 2018b; Lum et al., 2017; Zhu et al., 2017). 293FT cells were less permissive to ZIKV infection and showed a relative low expression of αvβ5 (Figures 5A and 5B). Consistent with previous findings (Chan et al., 2016), lymphocytes were not susceptible to ZIKV infection. The result showed that integrin αvβ5 was undetectable in H9 and Jurkat cells (Figure 5A). The susceptibility of these cells to ZIKV infection was monitored by quantifying infected cell ratios (Figure 4B). The qPCR results reconfirmed that mRNA levels of ITGB5 and ZIKV RNA replication were consistent with the flow analysis data (Figure S5A).
Figure 5. Integrin β5 (ITGB5) Expression Correlates with ZIKV Susceptibility and Contributes to ZIKV Neurotropism.
(A) Surface level of integrin αvβ5 on TS576, microglia, 293FT, Jurkat, and H9 cells were monitored by flow cytometry. Blue shading represents cell staining with a control isotype antibody, red shading represents cell staining with αvβ5 antibody. A representative of three independent experiments is shown.
(B) The indicated cells were challenged with ZIKV-ZsGreen at an MOI of 1 for 2 days. Infected cells were shown as ZsGreen-positive cell and were quantified by flow cytometry. Mean ± SD of n = 3.
(C and D) Single-cell transcriptomics data from developing human cerebral cortex were extracted from Nowakowski et al. (2016). Violin plots showing distribution of expression levels of ITGB5 (C) and SOX2 (D) across single cells of each respective cell type including neuron, interneuron, radial glia, astrocyte, microglia, endothelial, and IPC are presented.
(E) Overexpression of integrin β5 in Jurkat cells. Surface levels of integrin αvβ5 on Jurkat cells were measured by flow cytometry. Blue shading represents empty vector transduced cells, red shading represents β5-overexpression vector transduced cells. Both cells were stained with αvβ5 antibody. A representative of three independent experiments is shown.
(F–I) Expression of integrin β5 converts Jurkat cells susceptible to ZIKV infection. Empty vector (e.v.) or β5-overexpression vector transduced cells were infected with ZIKV-ZsGreen (F–H) at MOI = 5 for 72 h. (F) Green fluorescence field and bright field were shown to illustrate the ZIKV-infected cells. (G) The ZsGreen-positive cells were quantified by flow cytometry. Mean ± SD of n = 3. (H) Total cellular RNA was extracted, and ZIKV RNA was quantified by RT-PCR. Mean ± SD of n = 3. (I) Empty vector or β5-overexpression vector transduced cells were infected with MR766 at MOI = 5 for 48 h. Total cellular RNA was extracted and ZIKV RNA was quantified by RT-PCR. Mean ± SD of n = 3. ***p < 0.001 by Student’s t test.
Because ZIKV is a neurotropic flavivirus that preferentially infects radial glia, astrocyte, microglia, and endothelial cells, but not neurons in fetal brain (Miner and Diamond, 2017; Retallack et al., 2016), we analyzed single-cell transcriptomics data of different cell types isolated from developing human cerebral cortex (Nowakowski et al., 2016). The expression profiles showed that ITGB5 expression was enriched in radial glia, astrocyte, microglia, and endothelial cells, but not neurons and interneurons (Figure 5C), which exactly matched the cell tropism of ZIKV (Miner and Diamond, 2017). Furthermore, single-cell RNA (scRNA) sequence analysis showed that ITGB5 was highly expressed in outer radial glia but not in interneurons, neurons, or intermediate progenitor cells (IPC) (Pollen et al., 2015). Immunolabeling data also showed that ITGB5 was a marker for outer radial glia because it perfectly matched with the expression of radial glia marker PAX6 (Pollen et al., 2015). In addition, SOX2, a stemness marker, and AXL, a potential receptor for ZIKV, showed much higher expression in those cell types vulnerable to ZIKV infection (Figures 5D and S5B; Meertens et al., 2017). Meanwhile, ITGB3 was absent in most cell types and only showed slight expression in microglia (Figure S5C).
To further validate this correlation between ITGB5 expression and ZIKV susceptibility, we performed ectopic expression of integrin αvβ5 in Jurkat cells (non-permissive cell) and analyzed ZIKV infection (Figure 5E). Strikingly, compared with control vector, overexpression of αvβ5 converted non-permissive Jurkat cells into cells that were susceptible to ZIKV infection. The ZsGreen-positive cell ratios (Figures 5F and 5G) as well as viral genomic RNA replication (Figure 5H) dramatically increased in αvβ5-overexpressed cells. These findings were also consistent using another ZIKV strain MR766 (Figure 5I). Together with the results that ITGB5 KO dramatically decreased ZIKV infection (Figure 2D), these data support the notion of a strong correlation between αvβ5 expression and ZIKV susceptibility.
Inhibition of Integrin αvβ5 Alleviates ZIKV-Induced Pathology in hNSCs and Mouse Brain
To further investigate the physiological significance of integrin αvβ5 during ZIKV infection, we analyzed the function of ZIKV-integrin in hNSCs and in mouse brain. Because ZIKV prefers to infect NSCs leading to depletion of NSCs and impaired neurogenesis during brain development (Dang et al., 2016; Tang et al., 2016), hNSC neurospheres had been utilized to recapitulate in vivo neurogenesis phenotypes for ZIKV infection (Dang et al., 2016). We produced hNSC-derived neurospheres and determined the effect of ZIKV infection and integrin inhibition. Compared with the uninfected group, ZIKV infection resulted in a significant decrease in overall neurosphere size (Figure 6A). The treatment of αvβ5 blocking antibody, cilengitide, and SB273005 significantly rescued the destruction of neurospheres caused by ZIKV infection (Figures 6A and 6B). In addition, ZIKV infections cause apoptosis and induce cell death of hNSC (Dang et al., 2016; Tang et al., 2016). Consistent with the neurosphere size results, ZIKV infection dramatically reduced cell viability and treatment of cilengitide, αvβ5 blocking Ab, or SB273005 reversed this effect in both neurospheres produced from human-induced pluripotent stem cell (iPSC)-derived NSCs (ihNSC) (Figure 6C) and human fetal-derived primary NSCs (hfNSC) (Figure S6A). In addition, cilengitide reduced ZIKV-induced apoptosis in hfNSC (Figure S6B). Altogether, these results show that ZIKV-integrin interactions are required in 3D neurosphere models of hNSCs and blocking integrins ameliorates ZIKV-mediated cell death in hNSCs.
Figure 6. Inhibition of Integrin αvβ5 Alleviates ZIKV-Induced Pathology in hNSCs and Mouse Brain.
(A and B) Inhibition of integrin αvβ5 ameliorates ZIKV-induced malformation of 3D NSC neurospheres. The neurospheres of iPSC-derived NSCs were mock-infected or infected with MR766 at MOI = 1. The infected cells were mock-treated or treated with 2.5 μM cilengitide or 2.5 μg/mL αvβ5 blocking antibody or 2.5 μM SB273005. (A) 72 h post-infection, bright field images of neurospheres are shown. (B) Neurosphere size was quantified by ImageJ and the size distribution is shown. Box and whisker plots show 10th–90th percentiles. N >50 neurospheres per group. **p < 0.01, *p < 0.05 by Student’s t test.
(C) Inhibition of integrin αvβ5 reversed ZIKV-induced cell death of hNSCs. The neurospheres of iPSC-derived NSCs were mock-infected or infected with MR766 at MOI = 1. The infected cells were mock-treated or treated with 2.5 μM cilengitide or 2.5 μg/mL αvβ5 blocking antibody or 2.5 μM SB273005. Cell viability was measured 48h post-infection. Mean ± SD of n = 4. ***p < 0.001 by Student’s t test.
(D) Cilengitide significantly decreases ZIKV infection in mouse brain. C57BL/6 mice (4- to 5-week-old) were administered with IFNAR1 monoclonal antibody (mAb) (MAR1-5A3) before 1-day prior infection. Then the mice were challenged with MR766 (2 × 104 plaque-forming unit [PFU]/mouse) by intraperitoneal (i.p.) injection. Mice were treated with integrin inhibitor (cilengitide) (20 mg/kg body weight) daily by i.p. PBS was used as a vehicle control. Mice were sacrificed and brain was collected and analyzed using RT-PCR. Mean ± SD of n = 4. ****p < 0.0001 by Student’s t test.
(E and F) Cilengitide reduces the ZIKV infection in the cortical region of mouse brain. Mice were treated as in (D). (E) ZIK-infected neural stem cell marker (Nestin)-positive cells were quantified as described earlier (Youssef et al., 2019; Zhang et al., 2019). (F) Images showing immunostaining of neural stem cell marker (Nestin) (red) with ZIKV envelope antigen antibody (ZIKV-E) cells (green) in the cortex region. MZ, marginal zone; CP, cortical plate; SP, subplate zone; IZ, intermediate zone. Representative of 2 mice in each group. *p < 0.05, **p < 0.01 by Student’s t test.
Because cilengitide exhibited robust inhibition of ZIKV infection in cellular and neurosphere models (Figures 3F, 3L, 3M, and 6A-6C), we sought to determine its effects to inhibit ZIKV in vivo. C57BL/6 mice were administered with IFNAR1 blocking Ab before ZIKV infection and treated daily with cilengitide (20 mg/kg body weight) or PBS only. Animals were euthanized at 6 days post-infection, and viral levels were analyzed by qRT-PCR or by immunohistochemistry. At the whole brain level, we found that cilengitide treatment significantly suppressed levels of ZIKV RNA (Figure 6D). Immunohistochemical staining revealed efficient infection of ZIKV in hNSC as observed by colocalization of ZIKV envelop protein with NSC marker Nestin in the cortex (Figures 6E, 6F, S6C, and S6D) and the dentate gyrus region of the hippocampus (Figures S6C and S6D). Treatment of cilengitide reduced ZIKV infection in NSCs in the both cortical and hippocampal regions of mouse brain. All in all, these results established that inhibition of integrin αvβ5 could ameliorate ZIKV-induced malformation of 3D NSC neurosphere, reverse ZIKV-induced cell death, and decrease ZIKV infection in neural stem cells from both cortical and hippocampal region of mice brains.
DISCUSSION
To systemically investigate the key factors essential for ZIKV infection, Savidis et al. (2016) performed a genome-wide CRISPR screen in HeLa cells. EMC was reconfirmed to be involved in ZIKV and also other falvivirus family members’ replication at an early stage (Ma et al., 2015; Savidis et al., 2016). However, because ZIKV neurotropism is different from other members of Flaviviridae such as WNV and JEV, unique factors should determine this specificity (Laureti et al., 2018). Chavali et al. (2017) reported that Musashi-1, a neural precursor protein, directly interacted with ZIKV genome and also involved in the development of microcephaly. Another orthogonal proteomics survey in neural progenitor cells revealed hundreds of interacting proteins in which many of them were associated with neuronal development and retinal defects (Scaturro et al., 2018). These findings provided a clue that investigation in neural cells could be important in elucidating ZIKV neurotropism. Therefore, we performed a genome-wide CRISPR screen in GSCs. Because our screening strategy required stem cell expansion, cell sorting, and two rounds of infection for enrichment, primary NSCs could not be maintained for these long-term studies. GSCs can be preferentially infected by ZIKV in vitro and in vivo, thus they can provide a good model system to study ZIKV neurotropism (Chen et al., 2018b; Goffart et al., 2013; Pollen et al., 2015; Zhu et al., 2017). Comparative analysis of our results obtained from GSCs and 293FT cells screens and the results from Savidis et al. (2016) in HeLa cells resulted in identification of a number of neural genes (Figure 2). Among them, HOMER1 and BAALC were further confirmed to be involved in ZIKV infection. HOMER1 is a postsynaptic scaffold protein that might interact with metabotropic glutamate receptors to control intracellular calcium signaling to affect ZIKV infection (Luo et al., 2012). BAALC is highly expressed in various neural tissues and might control neural progenitor differentiation and proliferation (Moodbidri and Shirsat, 2006; Tanner et al., 2001). The underlying mechanism of these factors involved in ZIKV infection needs further investigation.
ITGB5 was one of the top ten targets that was specifically present in our GSCs screen (Figures 1F and 2A). The ITGB5 KO and rescue experiments demonstrated its essential role in ZIKV infection (Figures 2C-2F). Furthermore, ZIKV virions directly interacted with integrin αvβ5 (Figures 4A-4C). In both TS576 and hNSCs, viral internalization was inhibited by blocking integrin αvβ5 (Figures 3J and 3O). In addition, there was a strong correlation between ITGB5 expression and ZIKV susceptibility in various cell lines (Figures 5A and 5B). Strikingly, ITGB5 was specially and highly expressed in outer radial glia, which are the neural stem cells of the neocortex, explaining ZIKV tropism for hNSC (Figure 5C; Pollen et al., 2015). Importantly, ectopic expression of ITGB5 in non-permissive Jurkat cells renders the cells susceptible to ZIKV infection. Altogether, these results suggest that integrin αvβ5 is an essential internalization factor for ZIKV. ITGB5 was neither identified as a top hit nor selected for further validation in ZIKV infection screens (Li et al., 2019; Wells et al., 2018). The reasons could be at least 2-fold. First, we utilized reporter virus, which is a straightforward readout and decreased the background. Second, we used 3D neurosphere of GSC for a screen that was a more physiologically relevant model than the other stem cell culture conditions (Dang et al., 2016; Garcez et al., 2016). Future studies employing αvβ5 and interferon (IFN) knockout animal models in the context of flavivirus infections could further validate the functional requirements of this factor for ZIKV infection.
Integrins have been demonstrated to promote various viral entry (Stewart and Nemerow, 2007). For some flaviviruses, such as WNV and JEV, integrin β3, but not β5, mediates binding and internalization (Chu and Ng, 2004). ITGB5 was significantly expressed in neural cells compared to ITGB3, especially in cells that are susceptible for ZIKV infection (Figures 5C and 5D). This may help elucidate the uniqueness of ZIKV neurotropism compared with WNV and JEV (Olmo et al., 2017). Interestingly, integrin αvβ5 was shown to mediate adeno-associated virus type 2 internalization but not binding, similar to our findings for ZIKV (Summerford et al., 1999). This suggests integrin αvβ5 might have similar functions in the two viruses from different families. In addition, integrin αvβ5, as a mediator of host signalosome, might be activated by ZIKV and trigger signal pathways to rearrange cytoskeleton and recruit components to assemble endocytic vesicles. The outside-in signal might also contribute to a permissive initial infection (Hussein et al., 2015). Investigations of ZIKV-integrin interactions not only contribute to furthering our understanding of ZIKV neurotropism and pathogenesis, but also reveal promising therapeutic targets for ZIKV infection.
Integrins are promising drug targets for cancers and other pathological disorders (Arruda Macêdo et al., 2015). Three integrins had been exploited as therapeutic targets by designing blocking antibodies, short peptides, and small molecules to interfere with their functions (Ley et al., 2016). In our study, integrin αvβ5 inhibitors SB273005 and cilengitide showed remarkable anti-ZIKV activity with IC50 of 0.69 nM and 27.5 nM, respectively. These IC50 values are quite close to their binding IC50 to integrin αvβ5 in vitro (Badger et al., 2001; Miller et al., 2000). Importantly, inhibition of integrin αvβ5 by these two inhibitors and a blocking antibody ameliorated ZIKV-induced malformation of NSC 3D neurospheres and reversed ZIKV-induced cell death. Furthermore, as a proof of concept study, cilengitide significantly reduced ZIKV infection of neural stem cells in both cortical and hippocampal region of mouse brains. These findings revealed that integrin αvβ5 is important for ZIKV infection in vivo. Because cilengitide was not directly injected in the brains of animals or formulated for CNS delivery, overall observed effects of cilengitide on ZIKV inhibition in vivo were not dramatic when the whole brains were analyzed. Nonetheless, these results validate the role of integrin in ZIKV infection in mouse and provide a proof of concept for integrin blockade in controlling viral infections. Because SB273005 and cilengitide have been tested in dozens of clinical trials and are available in various formulations, these drugs can potentially be used as prophylactics or treatments for ZIKV infections. These compounds also provide promising lead molecules for further antiviral drug development.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tariq Rana (trana@ucsd.edu).
All unique/stable reagents generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All studies were conducted in accordance with approved IRB protocols by the University of California, San Diego. All animal work was approved by the Institutional Review Board at the University of California, San Diego and was performed in accordance with Institutional Animal Care and Use Committee guidelines.
Cell lines and Culturing Conditions
Glioblastoma stem cells (GSCs, TS576) were cultured as described previously (Benitez et al., 2018; Inda et al., 2010). In brief, TS576 were cultured in DMEM/F12 medium supplemented with 1:100 B27 without vitamin A, EGF (20ng/ml), FGF (10ng/ml) and penicillin-streptomycin (100IU/ml) at 37°C. H9 and Jurkat were cultured in RPMI1640 medium with 10% FBS at 37°C. 293FT, Vero and microglia cells were cultured in D10 medium (DMEM supplemented with 10% FBS). Human iPSC derived neural stem cells (hiNSC) were obtained from Dr. Frank Furnari Lab at UCSD and hiNSC were cultured in DMEM/F12 with 2mM Glutamax, 1% N2 supplement, 1% B27 supplement, 50 mM ascorbic acid, 3 μM CHIR99021, 0.5μM Purmorphamine and 1% Pen/Strep on Matrigel coated plates at 37°C. Primary human fetal NSCs (hfNSC) (Thermo Fisher, A15654) were cultured in DMEM/F12 with 2mM Glutamax, 2% StemPro supplement, EGF (20ng/ml), bFGF (20ng/ml), 6 U/ml heparin, 200μM ascorbic acid and 1% Pen/Strep on Matrigel coated plates at 37°C. Neurospheres were formed by generating single cell suspensions using Stempro accutase and seeding in untreated plates. hfNSC was isolated from a male donor and authenticated by evaluating differentiation potential to neuron, glial cell and the expression of markers of Nestin, SOX2, HLA–DR and CD11b.
ZIKV strain, virus stock preparation and viral titer measurements
ZIKV-ZsGreen reporter virus was rescued as described previously (Mutso et al., 2017). In brief, ZIKV-ZsGreen infectious RNA was transfected into Vero cells and supernatant containing rescued virus were collected at 5 days post-transfection. Prototype MR766 ZIKV virus and Paraiba strain was produced in Vero cells after inoculum with stock virus at MOI = 1 in E-MEM 10% FBS medium. 24h later, media was removed, and the cells were washed with PBS. DMEM/F12 medium with B27 and growth factors were added. Supernatant was harvested after 36 hours. Media was replaced 24 hr after inoculation and viral supernatant was collected at 48 hr post-inoculation. Viral titer was assessed using plaque assay on Vero cells.
Mice handling
C57BL/6J mice (4 to 5 week old, male) were purchased from MMRRC Jackson Laboratories and housed according to regulatory standards approved by the Institute Review Board of the University of California, San Diego.
METHOD DETAILS
Pooled library amplification and viral production
Brunello library (Cat# 73179) and GeCKO v2 A library (cat# 1000000048) pooled plasmid (lentiCRISPRv2) was obtained from addgene and amplified per recommended protocols via electroporation in lucigen endura electrocompetent cells. 293FT cells were harvested with DMEM and co-transfected with 12ug of the plasmid library, 9ug psPAX2 vector, and 6ug pMD2.G vector into 10cm plates using 48uL of PLUS reagent (Invitrogen 11514-015) and 60uL of lipofectamine in Opti-MEM media. Media was aspirated then after 6 hours and replaced with fresh D10 media. 24h later, D10 media was removed and the cells were washed with PBS. DMEM/F12 medium with B27 and growth factors were added. Supernatant was harvested after 24 h and centrifuged at 3,000 rpm at 4°C for 10 minutes and then filtered through 0.45um low protein binding membrane (Millipore) and concentrated using Amicon Ultra-15 Centrifugal Filter (Millipore) for 40 minutes at 4°C at 4,000 rpm. Virus was then aliquoted and frozen at −80°C.
Pooled Library Transduction, Zika Virus Challenge, and resistant cell isolation
Functional virus titer was obtained by measuring puromycin resistance after transduction via spin infection as described in Zhang lab papers (Joung et al., 2017; Shalem et al., 2015). A titer resulting in 30% of cells surviving puromycin selection has been calculated to correspond to a MOI of 0.3. Four hundred million TS576 cells was transduced with library to reach a coverage of 500 for each group (~500 cells for the same sgRNA, the library contains 77,441 sgRNAs). Transduced cells were selected with puromycin at 1 μg/ml for 7 days and split every 4 days. For ZIKV infection group, a total of 75 million cells were infected at MOI = 0.01 for 7days. ZsGreen negative cells were sorted out, half of the cells were split out for sequencing. The other half was kept in culture and expanded for 2 weeks in the presence of ZIKV inhibitor, NITD008. Expanded cells were re-infected with ZIKV at an MOI of 0.01 for 1 week and sorted for resistant cells. These cells were collected for sequencing.
sgRNA Library Quantification by Deep Sequencing
Genomic DNA was extracted from cells using the salt-precipitation protocol as described in Chen et al. (2015). The sgRNA library readout was performed following described protocols using a two-step nested PCR process in which PCR1 amplifies the lentiviral sequence containing the 20bp sgRNA cassette followed by PCR2 which attaches illumina sequencing adapters and barcodes. Primer sequences were obtained from the Zhang lab online resource (http://genome-engineering.org/gecko/) using v2Adaptor_F and v2Adapter_R for PCR1 and Primers F01-F06 and R01-R02 for PCR2 (Joung et al., 2017). All PCR was performed using Herculase II Fusion DNA Polymerase (Agilent). Enough PCR1 reactions were performed to maintain full library representation and PCR2 was performed with one reaction per 104 constructs (7 reactions per sample). 10ug of genomic DNA was used per PCR1 reaction. After pooling PCR1 product, 10uL of pooled PCR1 product was used for each PCR2 reaction. PCR products were purified and quantified with Qubit and/or bioanalyzer and diluted libraries were sequenced on Illumina NextSeq.
ZIKV infection in mice
C57BL/6J mice (4 to 5 week old, male, from Jackson Laboratories) were administered as a single dose with 2mg of an IFNAR1 mAb (MAR1-5A3) before one-day infection of ZIKV and 0.5mg of IFNAR1 mAb at second and fourth day after ZIKV infection by intraperitoneal (i.p.) injection as previously described (Gorman et al., 2018). Mice were treated with cilengitide (20 mg/kg body weight) daily for 6 days by i.p. and were infected with ZIKV MR766 (2x104 PFU/mouse) in 200ul volume through i.p. injection. Mice were sacrificed and brain was collected and analyzed by RT-qPCR or by immunohistochemistry.
Data processing and analysis
Sequencing reads were de-multiplexed and adapters trimmed using cutadapt (https://cutadapt.readthedocs.io/en/stable/, https://doi.org/10.14806/ej.17.1.200) leaving only the 20bp sgRNA spacer sequences. The sgRNA spacer sequences were then mapped to the Brunello or GeCKO library using bowtie (Langmead and Salzberg, 2012) allowing a maximum of one mismatch and allowing only uniquely aligning reads. Only sgRNA spacers with multiple reads were analyzed (sgRNA spacer with only a single read were filtered out). Normalized read counts were obtained by normalizing to total read count per sample (normalized reads per sgRNA = reads per sgRNA/total reads for all sgRNAs in sample x 106+1). Normalized reads were used for input in STARS software (https://portals.broadinstitute.org/gpp/public/dir/download?dirpath=software&filename=STARS_v1.3.zip) following instruction (Doench et al., 2016).
Lentivirus production and transduction
shRNA vectors were obtained from the Functional Genomics Center of La Jolla institute of allergy and immunology, the sequences are listed in Table S3. For knockout of ITGB5, four sgRNA oligos targeting ITGB5 were designed and inserted into sgRNA expressing vector (lentiCRISPR v2, addgene 52961). ITGB5 was cloned into a pLVX plasmid for ITGB5 overexpression experiments. Two rescue ITGB5 vectors were generated by point mutation kit (NEB). All the cloning primers were listed in Table S3. Lentiviral particles were prepared by co-transfection of shRNA, sgRNA or overexpression vectors (1.8 μg), psPAX.2 (1.2 μg) and pMD2.G (0.6 μg) vectors in 0.8 million 293FT cells. Lipofectamine 2000 and Opti-MEM were used according to manufacturer’s instruction. Six hours post transfection, supernatant was removed and D10 medium was added. 24h later, D10 media was removed and the cells were washed with PBS. DMEM/F12 medium with B27 and growth factors were added. Supernatant was harvested after 24 hours and filtered at 0.22 μm and added to target TS576. After 12 hours lentivirus was removed and replaced with fresh medium.
Immunohistochemistry
Brains were removed and post-fixed in 10% paraformaldehyde overnight at 4°C followed by cryopreservation in 10, 20, 30% (w/v) sucrose in PBS. Serial coronal sections of 20-30 μm thickness encompassing the neurogenic regions such as hippocampus and sub-ventricular zone (SVZ) were cut using a freezing cryostat (Leica Biosystems, CM3050s). In order to examine the ZIKV infection, double immunofluorescence analysis was carried out in the cortex and hippocampus. Coronal sections beginning at bregma −1.50 to −3.50 mm through the dorsal hippocampus encompassing the dentate gyrus region and cortex were co-labeled with Nestin/ZIKV-E. Free-floating sections were washed and carried out antigen retrieval with citrate buffer (pH 6.2) and blocked with 3% NGS, 0.1% Triton X-100, and 0.5% BSA for 2 h. Sections were then incubated in mouse anti-ZIKV-E/anti-flavivirus group antigen antibody 4G2 (1:500), chicken anti-Nestin (1:500) for 24 h at 4°C. Secondary antibodies used included anti-mouse and anti-chicken Alexa Fluor 488 (1:400); and anti-mouse and anti-chicken Alexa Fluor 594 (1:400). Sections were mounted with DAPI containing Hard Set anti-fade mounting medium (Vectashield, Vector Laboratories, CA, USA) and stored in the dark at 4°C. Images from the stained slides were acquired for fluorescence co-labeling under inverted Leica fluorescence microscope (DMI 3000B) and Keyence All in one fluorescence microscope BZ-X800. The images were analyzed and background corrected by ImageJ software as described earlier (Youssef et al., 2019; Zhang et al., 2019) and BZ-X analyzer as described in user manual. For quantification, cell counting in cortical and hippocampal region for Nestin/ZIKV-E double positive cells were performed in coronal sections beginning at bregma −1.50 to −3.50 mm. Nestin/ZIKV-E double positive cells, in at least 5 sections in each group, were analyzed and cells were identified by their DAPI labeled nuclei.
ZIKV infection experiment in presence of blocking antibodies or inhibitors
TS576 cells were treated with antibodies or inhibitors for 1h and infected with ZIKV-ZsGreen of MOI = 0.05 or 1. Cells were collected 48h or 72h post-infection. For RNA quantification, cells were lysed in TRIzol and total RNA was extracted. 1ug RNA of each sample was used to generate cDNA with a reverse transcription kit. Viral RNA was quantified relative to cellular GAPDH mRNA by real-time qPCR. For flow analysis, cells were dissociated with accumax and fixed in 2% PFA with PBS. The samples were filtered and subject for flow analysis using BD ACCURI C6 Flow Cytometer. Data analysis was performed using FlowJo v10 software (FlowJo LLC). For cell viability assays, cells seeding in 96-well plate were mixed with Celltiter-Glo reagent (Promega, G7572) according to the manufacturer’s protocol, and luminescence intensity was measured to quantify the cell viability. The cytotoxicity of inhibitors and cell viability of neurospheres were also determined using this method.
ZIKV binding and internalization assays
TS576 cells or hNSCs were treated with 2.5ug/ml anti-integrin αvβ3, αvβ5 or isotype antibody for 1h and incubated with ZIKV of MOI = 20 at 4 degrees for two hours. For the binding assays, cells were then washed with chilled PBS for three times, and cells were lysed using TRIzol and binding viral RNA was quantified by qPCR. For the internalization assay, cells were incubated in 37 degrees for 4h. And further washed with PBS for three times. Pellets were lysed by TRIzol and total RNA was extracted to quantify internalized viral RNA.
ELISA Binding
To detect ZIKV-αvβ5 direct interaction, integrin αvβ5 or BSA were coated (400ng/well) in TBS supplemented with 10mM Ca2+, 10mM Mg2+ and 1mM Mn2+ overnight. The wells were blocked with 2% BSA in TBS for 2h at 37°C. MR766 virions (107 PFU/well) were added and incubated at 4°C for 2h. Unbound virions were removed by washing with TBS for four times and 0.5 μg/ml biotinylated 4G2 (Novus, NBP2-52709B) and HRP-Streptavidin were added to detect the binding virions. TMB substrate was incubated and the intensity at 450nm was measured.
Immunofluorescence staining of NSCs and GSCs
NSCs or GSCs were washed with PBS and then fixed with 4% paraformaldehyde for 1 hour. Fixed cells were blocked in 5% BSA in PBS for 1 hours, washed three times with PBS+0.1% Triton X-100 (PBST), and incubated with anti-flavivirus group antigen antibody 4G2 (1:500) or SOX2 (1:500), or GFAP (1:500) at 4°C overnight. GSCs were then washed 3 times with PBST and incubated with FITC-conjugated secondary antibody for 1 hour. After 3 washes in PBST, DAPI was added and images were acquired. All images were acquired on a Leica DMI 3000B.
QUANTIFICATION AND STATISTICAL ANALYSIS
Graphs were plotted using Prism 8.0.2 (GraphPad Inc.) Differences between groups were analyzed using Student’s t test (paired, two-sided) if not stated in the figure legends otherwise. Data are presented as the means ± standard deviation (SD), with a p value < 0.05 considered statistically significant. The values of n refers to the number of mice used in the mice study part. In the other parts, it refers to the number of dependent experiments.
DATA AND CODE AVAILABILITY
Raw and processed data are provided in the Gene Expression Omnibus (accession number GSE139142 for sgRNA library deep sequencing).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| αvβ5 blocking antibody | Millipore | Cat# MAB1961Z, RRID:AB_94466 |
| αvβ3 blocking antibody | Millipore | Cat# MAB1976Z, RRID:AB_11212117 |
| AF488-conjugated anti-αvβ5 | BioLegend | Cat# 920010, RRID:AB_2629782 |
| AF488-conjugated mouse IgG1 | BioLegend | Cat# 400132, RRID:AB_400129 |
| Anti-phosphorylated FAK | Abcam | Cat# ab81298, RRID:AB_1640500 |
| Anti-FAK | Abcam | Cat# ab131435, RRID:AB_11154758 |
| anti-ZIKV-E | Millipore | Cat# MAB10216, RRID:AB_827205 |
| Anti-Nestin | Abcam | Cat# ab134017, RRID:AB_2753197 |
| Anti-Sox2 | Abcam | Cat# ab97959, RRID:AB_2341193 |
| Anti-cleaved caspase 3 | Sigma | Cat# C8487, RRID:AB_476884 |
| Anti-GFAP | Sigma | Cat# G3893, RRID:AB_477010 |
| Bacterial and Virus Strains | ||
| MR766 | National Institutes of Health | LC002520.1 |
| Paraiba | Stevenson Laboratory | KX280026.1 |
| ZsGreen strain | Mutso et al., 2017 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SB273005 | Selleck Chemicals | Cat# S7540 |
| cilengitide | Sigma | Cat# SML1594 |
| VS-4718 | Selleck Chemicals | Cat# S7653 |
| NITD008 | R&D | Cat# 6045/1 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE139142 |
| Experimental Models: Cell Lines | ||
| TS576 | Dr. Frank Furnari Lab | N/A |
| Human iPSC derived neural stem cells (hiNSC) | Dr. Frank Furnari Lab | N/A |
| H9 | ATCC | HTB-176 |
| Jurkat | NIH AIDS Reagent Program | Cat# 177 |
| 293FT | ATCC | CRL 11268 |
| human fetal NSCs (hfNSC) | Thermo Fisher | A15654 |
| Vero | ATCC | CCL-81 |
| microglia | Dr Jonathan Karn Lab | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse model: C57BL/6J | Jackson Labs | N/A |
| Oligonucleotides | ||
| Primers for crispr screening, shRNA, RT-PCR, and vector, see Table S3 | This paper | N/A |
| Recombinant DNA | ||
| ITGB5_sgRNA1 | This paper | N/A |
| ITGB5_sgRNA2 | This paper | N/A |
| ITGB5_sgRNA3 | This paper | N/A |
| ITGB5_sgRNA4 | This paper | N/A |
| plvx-ITGB5 | This paper | N/A |
| ITGB5_sg 1 mut | This paper | N/A |
| ITGB5_sg 2 mut | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 8 | Software | RRID: SCR_002798 https://www.graphpad.com/ |
| ImageJ | Software | ImageJ; RRID: SCR_003070 https://imagej.nih.gov/ij/download.html |
| FlowJo | Software | Version 10 https://www.flowjo.com/solutions/flowjo/downloads |
Highlights.
CRISPR screens identify 92 GSC-specific factors linked to ZIKV infection
Integrin αvβ5 mediates ZIKV internalization into hNSCs providing two drug candidates
Integrin αvβ5 expression correlates with ZIKV susceptibility and neurotropism
Inhibition of αvβ5 alleviates ZIKV-induced pathology in hNSCs and in mouse brain
ACKNOWLEDGMENTS
We thank Dr. Andres Merits at University of Tartu, Tartu, Estonia, for providing ZIKV-ZsGreen and ZIKV-replicon clones. We thank Michael Diamond, David Cheresh, and David Schlaepfer for sharing their insight into the integrin and ZIKV receptors mechanisms. We also thank the support from Dr. Kristen Jepsen of IGM at UCSD for her help with the HT-seq and Dr. Celsa Spina and Tara Rambaldo at UCSD CFAR for cell sorting. We thank members of the Rana lab for helpful discussions and advice. This work was supported in part by grants from the NIH (AI125103, CA177322, DA039562, DA046171, and DA049524).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.020.
DECLARATION OF INTERESTS
T.M.R. is a founder of ViRx Pharmaceuticals and has an equity interest in the company. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
REFERENCES
- Akula SM, Pramod NP, Wang FZ, and Chandran B (2002). Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108, 407–419. [DOI] [PubMed] [Google Scholar]
- Arruda Macêdo JK, Fox JW, and de Souza Castro M (2015). Disintegrins from snake venoms and their applications in cancer research and therapy. Curr. Protein Pept. Sci 16, 532–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger AM, Blake S, Kapadia R, Sarkar S, Levin J, Swift BA, Hoffman SJ, Stroup GB, Miller WH, Gowen M, and Lark MW (2001). Disease-modifying activity of SB 273005, an orally active, nonpeptide alphavbeta3 (vitronectin receptor) antagonist, in rat adjuvant-induced arthritis. Arthritis Rheum. 44, 128–137. [DOI] [PubMed] [Google Scholar]
- Benitez JA, Ma J, D’Antonio M, Boyer A, Camargo MF, Zanca C, Kelly S, Khodadadi-Jamayran A, Jameson NM, Andersen M, et al. (2018). Publisher Correction: PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat. Commun 9, 16217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson JM, Shepley MP, Chan BM, Hemler ME, and Finberg RW (1992). Identification of the integrin VLA-2 as a receptor for echovirus 1. Science 255, 1718–1720. [DOI] [PubMed] [Google Scholar]
- Brasil P, Pereira JP Jr., Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, et al. (2016). Zika Virus Infection in Pregnant Women in Rio de Janeiro. N. Engl. J. Med 375, 2321–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutet N, Krauer F, Riesen M, Khalakdina A, Almiron M, Aldighieri S, Espinal M, Low N, and Dye C (2016). Zika Virus as a Cause of Neurologic Disorders. N. Engl. J. Med 374, 1506–1509. [DOI] [PubMed] [Google Scholar]
- Chan JF, Yip CC, Tsang JO, Tee KM, Cai JP, Chik KK, Zhu Z, Chan CC, Choi GK, Sridhar S, et al. (2016). Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg. Microbes Infect 5, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali PL, Stojic L, Meredith LW, Joseph N, Nahorski MS, Sanford TJ, Sweeney TR, Krishna BA, Hosmillo M, Firth AE, et al. (2017). Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science 357, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang YF, Yang Y, Zou P, Chen J, He Y, Shui SL, Cui YR, Bai R, Liang YJ, et al. (2018a). AXL promotes Zika virus infection in astrocytes by antagonizing type I interferon signalling. Nat. Microbiol 3, 302–309. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wu J, Ye Q, Ma F, Zhu Q, Wu Y, Shan C, Xie X, Li D, Zhan X, et al. (2018b). Treatment of Human Glioblastoma with a Live Attenuated Zika Virus Vaccine Candidate. MBio 9, e01683–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. (2015). Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JJ, and Ng ML (2004). Interaction of West Nile virus with alpha v beta 3 integrin mediates virus entry into cells. J. Biol. Chem 279, 54533–54541. [DOI] [PubMed] [Google Scholar]
- Cseke G, Maginnis MS, Cox RG, Tollefson SJ, Podsiad AB, Wright DW, Dermody TS, and Williams JV (2009). Integrin alphavbeta1 promotes infection by human metapneumovirus. Proc. Natl. Acad. Sci. USA 106, 1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wu Y, Fan D, Gao N, Ming Y, Wang P, and An J (2018). Peptides P4 and P7 derived from E protein inhibit entry of dengue virus serotype 2 via interacting with β3 integrin. Antiviral Res. 155, 20–27. [DOI] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, and Rana TM (2016). Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell 19, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier JS, and Cheresh DA (2010). Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, et al. (2016). Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N. Engl. J. Med 374, 2142–2151. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, and Rehen SK (2016). Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818. [DOI] [PubMed] [Google Scholar]
- Goffart N, Kroonen J, and Rogister B (2013). Glioblastoma-initiating cells: relationship with neural stem cells and the micro-environment. Cancers (Basel) 5, 1049–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH 3rd., et al. (2018). An Immunocompetent Mouse Model of Zika Virus Infection. Cell Host Microbe 23, 672–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero CA, Méndez E, Zárate S, Isa P, López S, and Arias CF (2000). Integrin alpha(v)beta(3) mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97, 14644–14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, et al. (2015). Biology of Zika Virus Infection in Human Skin Cells. J. Virol 89, 8880–8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings AK, Yockey LJ, Jagger BW, Hwang J, Uraki R, Gaitsch HF, Parnell LA, Cao B, Mysorekar IU, Rothlin CV, et al. (2017). TAM Receptors Are Not Required for Zika Virus Infection in Mice. Cell Rep. 19, 558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein HA, Walker LR, Abdel-Raouf UM, Desouky SA, Montasser AK, and Akula SM (2015). Beyond RGD: virus interactions with integrins. Arch. Virol 160, 2669–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. (2010). Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 24, 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Clark S, Berryman S, Burman A, Cambier S, Mu D, Nishimura S, and King AM (2004). Integrin alphavbeta8 functions as a receptor for foot-and-mouth disease virus: role of the beta-chain cytodomain in integrin-mediated infection. J. Virol 78, 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, and Zhang F (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc 12, 828–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RS, Brown DC, Ye C, and Hjelle B (2005). Peptide antagonists that inhibit Sin Nombre virus and hantaan virus entry through the beta3-integrin receptor. J. Virol 79, 7319–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureti M, Narayanan D, Rodriguez-Andres J, Fazakerley JK, and Kedzierski L (2018). Flavivirus Receptors: Diversity, Identity, and Cell Entry. Front. Immunol 9, 2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, and Diamond MS (2016). Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol 90, 4864–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K, Rivera-Nieves J, Sandborn WJ, and Shattil S (2016). Integrin-based therapeutics: biological basis, clinical use and new drugs. Nat. Rev. Drug Discov 15, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, and Xu Z (2016). Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 19, 120–126. [DOI] [PubMed] [Google Scholar]
- Li F, Wang PR, Qu LB, Yi CH, Zhang FC, Tang XP, Zhang LG, and Chen L (2017). AXL is not essential for Zika virus infection in the mouse brain. Emerg. Microbes Infect 6, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Muffat J, Omer Javed A, Keys HR, Lungjangwa T, Bosch I, Khan M, Virgilio MC, Gehrke L, Sabatini DM, and Jaenisch R (2019). Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc. Natl. Acad. Sci. USA 116, 9527–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang F, and Chen X (2008). Integrin alpha(v)beta(3)-Targeted Cancer Therapy. Drug Dev. Res 69, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum FM, Low DK, Fan Y, Tan JJ, Lee B, Chan JK, Rénia L, Ginhoux F, and Ng LF (2017). Zika Virus Infects Human Fetal Brain Microglia and Induces Inflammation. Clin. Infect. Dis 64, 914–920. [DOI] [PubMed] [Google Scholar]
- Luo P, Li X, Fei Z, and Poon W (2012). Scaffold protein Homer 1: implications for neurological diseases. Neurochem. Int 61, 731–738. [DOI] [PubMed] [Google Scholar]
- Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi JG, Shi G, Qi L, et al. (2015). A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell Rep. 12, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maginnis MS, Forrest JC, Kopecky-Bromberg SA, Dickeson SK, Santoro SA, Zutter MM, Nemerow GR, Bergelson JM, and Dermody TS (2006). Beta1 integrin mediates internalization of mammalian reovirus. J. Virol 80, 2760–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, and Carette JE (2016). Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau VM, et al. (2017). Axl Mediates ZIKA Virus Entry in Human Glial Cells and Modulates Innate Immune Responses. Cell Rep. 18, 324–333. [DOI] [PubMed] [Google Scholar]
- Miller WH, Alberts DP, Bhatnagar PK, Bondinell WE, Callahan JF, Calvo RR, Cousins RD, Erhard KF, Heerding DA, Keenan RM, et al. (2000). Discovery of orally active nonpeptide vitronectin receptor antagonists based on a 2-benzazepine Gly-Asp mimetic. J. Med. Chem 43, 22–26. [DOI] [PubMed] [Google Scholar]
- Miner JJ, and Diamond MS (2017). Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe 21, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, and Schlaepfer DD (2006). Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol 18, 516–523. [DOI] [PubMed] [Google Scholar]
- Moodbidri MS, and Shirsat NV (2006). Induction of BAALC and down regulation of RAMP3 in astrocytes treated with differentiation inducers. Cell Biol. Int 30, 210–213. [DOI] [PubMed] [Google Scholar]
- Mutso M, Saul S, Rausalu K, Susova O, Žusinaite E, Mahalingam S, and Merits A (2017). Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J. Gen. Virol 98, 2712–2724. [DOI] [PubMed] [Google Scholar]
- Noronha Ld., Zanluca C, Azevedo ML, Luz KG, and Santos CN (2016). Zika virus damages the human placental barrier and presents marked fetal neurotropism. Mem. Inst. Oswaldo Cruz 111, 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, and Kriegstein AR (2016). Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell 18, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo IG, Carvalho TG, Costa VV, Alves-Silva J, Ferrari CZ, Izidoro-Toledo TC, da Silva JF, Teixeira AL, Souza DG, Marques JT, et al. (2017). Zika Virus Promotes Neuronal Cell Death in a Non-Cell Autonomous Manner by Triggering the Release of Neurotoxic Factors. Front. Immunol 8, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, et al. (2016). Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. USA 113, 14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AS, Shim BS, Kwon YC, Zhang R, Otsuka Y, Schmitt K, Berri F, Diamond MS, and Choe H (2017). AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc. Natl. Acad. Sci. USA 114, 2024–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno M, Sacramento GA, Khouri R, do Rosário MS, Costa F, Archanjo G, Santos LA, Nery N Jr., Vasilakis N, Ko AI, and de Almeida AR (2016). Zika Virus Infection and Stillbirths: A Case of Hydrops Fetalis, Hydranencephaly and Fetal Demise. PLoS Negl. Trop. Dis 10, e0004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, et al. (2016). Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep. 16, 232–246. [DOI] [PubMed] [Google Scholar]
- Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, Bartenschlager R, Götz M, and Pichlmair A (2018). An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561, 253–257. [DOI] [PubMed] [Google Scholar]
- Serrels A, Lund T, Serrels B, Byron A, McPherson RC, von Kriegsheim A, Gómez-Cuadrado L, Canel M, Muir M, Ring JE, et al. (2015). Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell 163, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, and Zhang F (2015). High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PL, and Nemerow GR (2007). Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 15, 500–507. [DOI] [PubMed] [Google Scholar]
- Summerford C, Bartlett JS, and Samulski RJ (1999). AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med 5, 78–82. [DOI] [PubMed] [Google Scholar]
- Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, and Pereira L (2016). Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe 20, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancioni I, Uryu S, Sulzmaier FJ, Shah NR, Lawson C, Miller NL, Jean C, Chen XL, Ward KK, and Schlaepfer DD (2014). FAK Inhibition disrupts a β5 integrin signaling axis controlling anchorage-independent ovarian carcinoma growth. Mol. Cancer Ther 13, 2050–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. (2016). Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 18, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner SM, Austin JL, Leone G, Rush LJ, Plass C, Heinonen K, Mrózek K, Sill H, Knuutila S, Kolitz JE, et al. (2001). BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc. Natl. Acad. Sci. USA 98, 13901–13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Triantafilou M, Takada Y, and Fernandez N (2000). Human parechovirus 1 utilizes integrins alphavbeta3 and alphavbeta1 as receptors. J. Virol 74, 5856–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, Kommineni S, Bilican B, Klim JR, Hill EJ, et al. (2016). Genetic Ablation of AXL Does Not Protect Human Neural Progenitor Cells and Cerebral Organoids from Zika Virus Infection. Cell Stem Cell 19, 703–708. [DOI] [PubMed] [Google Scholar]
- Wells MF, Salick MR, Piccioni F, Hill EJ, Mitchell JM, Worringer KA, Raymond JJ, Kommineni S, Chan K, Ho D, et al. (2018). Genome-wide screens in accelerated human stem cell-derived neural progenitor cells identify Zika virus host factors and drivers of proliferation. bioRxiv. 10.1101/476440. [DOI] [Google Scholar]
- Wickham TJ, Mathias P, Cheresh DA, and Nemerow GR (1993). Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73, 309–319. [DOI] [PubMed] [Google Scholar]
- Youssef M, Atsak P, Cardenas J, Kosmidis S, Leonardo ED, and Dranovsky A (2019). Early life stress delays hippocampal development and diminishes the adult stem cell pool in mice. Sci. Rep 9, 4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, and Chen J (2012). The regulation of integrin function by divalent cations. Cell Adhes. Migr 6, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, et al. (2016). A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Tan YW, Yam WK, Tu H, Qiu L, Tan EK, Chu JJH, and Zeng L (2019). In utero infection of Zika virus leads to abnormal central nervous system development in mice. Sci. Rep 9, 7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gorman MJ, McKenzie LD, Chai JN, Hubert CG, Prager BC, Fernandez E, Richner JM, Zhang R, Shan C, et al. (2017). Zika virus has oncolytic activity against glioblastoma stem cells. J. Exp. Med 214, 2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data are provided in the Gene Expression Omnibus (accession number GSE139142 for sgRNA library deep sequencing).