Abstract
We present the case of a 71-year-old man who, despite becoming asymptomatic after having some mild symptoms of COVID-19, had SARS-CoV-2 RNA detected for 37 days after onset, from his concentrated and purified saliva specimens using sugar chain-immobilized gold nanoparticles. It was suggested that the early morning saliva specimens were more likely to show positive results than those obtained later in the day.
Keywords: SARS-CoV-2, COVID-19, Saliva, RT-PCR, Sugar chain-immobilized magnetic gold nanoparticle, Virus shedding
1. Introduction
COVID-19 was reported in Wuhan, China on December 31, 2019, and spread around the world quickly. On March 11, 2020, the WHO declared it to be a pandemic [1]. The current definitive diagnosis of COVID-19 is mainly performed using real-time reverse transcription polymerase chain reaction (RT-PCR) from lower respiratory tract specimens or nasopharyngeal specimens [2]. Virus monitoring is performed on confirmed cases, and hospital discharge requires two days’ consecutive negative confirmation of nasopharyngeal specimens [3]. Despite this monitoring standard being widely used globally, it has the following disadvantages: healthcare workers being exposed to the virus during specimen collection, thus creating a need for personal protective equipment (PPE) despite the current shortage of medical resources, and the performance of uncomfortable or invasive procedures on patients. To overcome these disadvantages, we came up with an RT-PCR test using saliva specimens, which were pre-treated with sugar chain-immobilized magnetic gold nanoparticles (SMGNP) to concentrate and purify virus particles at a rate of 5 min for one specimen. In evaluating RT-PCR using saliva specimens, we found the appropriate timing to collect saliva specimens, and present a case in which viral RNA was detected in saliva specimens for 37 days after onset. Our report contributes to knowledge of virus shedding and alternative testing methods.
2. Case report
On February 12, 2020, a Japanese man aged 71 years with only a history of allergic rhinitis was transported to our hospital from a cruise ship with an outbreak of COVID-19, anchored in Yokohama for quarantine. He had been on the cruise ship since January 20, 2020. He complained of body aches on February 5. On February 7, his temperature reached 37.5 °C. RT-PCR was performed, and on February 9, COVID-19 was confirmed. When he came to our hospital, his vital signs were within normal range and his laboratory results were quite normal. He had a dry cough and nasal discharge but his functions were otherwise normal. He was hospitalized for follow-up and confirmation of a RT-PCR negative result for SARS-CoV-2. On February 13, we received his written informed consent to participate in a study to establish an alternative and rapid diagnostic method using saliva specimens. This study was approved by the Institutional Review Board of Hamamatsu Medical Center (2019-122) based on the Ethical Guidelines for Medical Research Targeting Humans, provided by the Japanese Ministry of Health, Labor and Welfare. Saliva specimens were collected on the same day as the oropharyngeal or nasopharyngeal specimens submitted to the National Institute of Infectious Diseases (NIID) for viral monitoring. To determine the best time for obtaining the saliva specimens, daytime saliva specimens (DSS) were collected until March 1 and early morning saliva specimens (EMSS) were collected from March 3. We gave him a collection container marked with a 600-μL line the day before his submitting saliva specimens. Saliva specimen collections were carried out by him alone, spitting saliva up to the marked line, which was confirmed by the nurse especially in the case of EMSS. We concentrated and purified virus particles from 600μL of his saliva specimens using SMGNP, and extracted the RNA. SMGNP is composed of iron and gold of about 5 nm size, immobilized with sugar chain (sulfated oligosaccharide), to which the virus binds. The following procedure was used according to the previously established method with modification [[4], [5], [6]]. When SMGNPs are added to the viral solution, SMGNPs adsorbs on the surface of the viral particles via the sugar chain to capture the viruses. A secondary solution of magnetic micro-particles (MMPs, size: about 1 μm) composed of iron were added to the solution to collect the SMGNP-captured viruses. Then, magnetic separation was carried out to obtain the SMGNP-virus-MMP complex, in which viral particles were separated from the viral solution. Finally by adding a detergent (0.1% sodium lauryl sulfate aqueous solution) to the complex, the viral RNA was eluted. Since the viral particles were purified during the separation step, it was possible to directly apply the RT-PCR without further purification. The extracted RNA was cryopreserved (from February 13 to March 1, 2020) or refrigerated (from March 3 to 20), and sent to Kagoshima University to perform RT-PCR (intercalation method) using Centers for Disease Control and Prevention (CDC)-proposed primer sets with a slight modification (Forward primer: GACCCCAAAATCAGCGAAATG, Reverse primer: ATGTTGAGTGAGAGCGGTG) [7]. The assays at Kagoshima University (KU) were all done without any information about the NIID RT-PCR results.
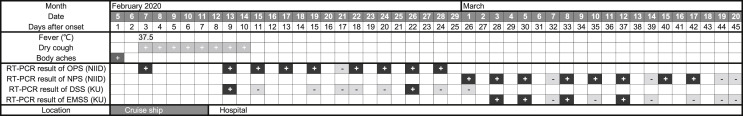
Although the patient looked healthy and had only body aches, a one-day-fever, and dry cough lasting several days, he had positive RT-PCR from NIID until day 42 after onset. His EMSS was positive up to day 37, and changed to negative on day 39. On day 45, he received 2 days’ consecutive negative RT-PCR NIID results based on his nasopharyngeal specimens, and was discharged in good health (Fig. 1 ).
Fig. 1.
Symptoms and RT-PCR results of the patient with COVID-19 from onset to discharge. Two saliva specimens on day13 (February 17) and day 40 (March 15) had not been submitted. Abbreviations; OPS: oropharyngeal specimens; NPS: nasopharyngeal specimens; DSS: daytime saliva specimens; EMSS: early morning saliva specimens; NIID: The National Institute of Infectious Diseases; KU: Kagoshima University.
3. Discussion
The WHO has claimed that virus shedding patterns are not yet well understood and further investigations are needed to better understand the timing, compartmentalization and quantity of viral shedding to inform optimal specimen collection [2]. No mention was made of saliva specimens. The use of saliva specimens for diagnosis or virus monitoring has several advantages: reducing virus exposure to healthcare workers, saving PPE and collection time, and being non-invasive.
The detection of SARS-CoV-2 in saliva has been reported [8,9]. The study also reported using saliva collected in the early morning, but it was not stated whether these samples were compared with specimens collected later in the day [9]. In our case, saliva specimens collected during the day had a lower rate of positive concordance when compared to NIID results. For DSS, the sensitivity was 25.0% (2/8) and the specificity was 100% (1/1) based on NIID results. In contrast, when the EMSS were used, the results came close to matching the NIID results of RT-PCR performed on the nasopharyngeal specimens (Fig. 1). The number of the EMSS was small, but the sensitivity based on NIID results was 66.7% (4/6) and the specificity was 100% (4/4).
There may be several factors affecting the different detection rates relating to the time of saliva specimens collection. The first factor is the difference in the amount of virus in the saliva specimens. During sleep, the cessation of salivary outflow can result in a decrease in oral viral clearance, resulting in an increase in viral load in salivary specimens in early morning saliva [10]. In addition, early morning saliva may be more likely to be contaminated with sputum, resulting in an increased viral load in the saliva specimens [9]. The second factor is the volume of the saliva specimens. If the viruses are uniformly present in the saliva, a higher volume of saliva would increase the amount of virus in the concentrated specimen. However, since we uniformly used 600μL saliva specimens for the SMGNP concentration and RNA extraction, the second factor can be ignored. The third factor is the variation of inhibitory agents on RT-PCR reaction in saliva specimens at different collection times. Known inhibitors for PCR include organic compounds, hemoglobin, protein, IgG, food, and calcium [11]. Detection sensitivity would be reduced if these substances are present in the collected saliva specimens. For example, hemoglobin is known to be detected in saliva after tooth brushing [12], and PCR inhibition can occur in saliva specimens after tooth brushing. In general, a variety of PCR inhibitors are found in foods [11]. In post-meal saliva specimens, food-derived inhibitors may be present, resulting in a reduced detection rate. Based on the above considerations, EMSS collected before breakfast and tooth brushing have the potential to contain more virus and fewer inhibitory agents than DSS. The finding of this manuscript may support the validity of using EMSS, as suggested in another study [9].
The results of the last four saliva specimens point towards an appropriate collection method. It can be suggested that virus monitoring after definitive diagnosis should be performed with EMSS concentrated and purified using SMGNP, and then performed with a nasopharyngeal specimen after the EMSS produces negative results.
For the virus shedding period, viral RNA was detected up to 25 days after symptom onset in previous reports [9]. For specimens other than saliva, it has been reported that virus RNA was detected up to 37 days after onset [13]. In our case, viral RNA was confirmed for a longer period than these reports. Kelvin Kai-Wang To et al. reported that the higher the initial viral load, the longer the detection period, and that older individuals tended to have higher peak viral loads [9]. Alraddadi BM et al. reported that in MERS-CoV person with allergic rhinitis had a relative risk of 2.21 for infection [14]. Our case was elderly and had allergic rhinitis, which may be the reason why the viral RNA was confirmed over a longer period of time. In COVID-19 cases with allergic rhinitis, it is necessary to verify whether the virus excretion period is prolonged.
One of the prospects for saliva as a diagnostic specimen is its application to rapid antigen testing. Previous studies have estimated that coronavirus levels in saliva specimens are as low as 1 in 10–1000 compared to nasopharyngeal or lower respiratory tract specimens [9,15]. The detection sensitivity of rapid antigen testing has been reported to be between 105 and 107 copies/μL for influenza diagnosis, which is 102 to 10⁴ times less sensitive than PCR [16]. Therefore, rapid antigen testing using saliva specimens is considered impractical due to its low sensitivity.
Our case presented: First, when using saliva specimens for virus monitoring, early morning specimens should be used. Second, EMSS concentrated and purified using SMGNP may be an alternative method for virus monitoring, when followed up with nasopharyngeal specimens. Third, it is possible that the virus can be detected in saliva for 37 days after onset, even after the patient becomes asymptomatic.
There are some limitations to our case. Firstly, the comparison standards for DSS and EMSS are different. In order to compare the sensitivity and specificity, the same standard test must be compared. However, the specimens submitted to NIID were changed in the middle of the process, and most of the DSS specimens were compared to oropharyngeal specimens (8/9), and all of the EMSS specimens were compared to nasopharyngeal specimens (10/10). When comparing nasopharyngeal specimens and oropharyngeal specimens, it has been reported that nasopharyngeal specimens may have a higher viral load [17]. It has also been reported that the viral load tends to decrease in excretion over time from onset [9]. In other words, EMSS was compared to a more sensitive standard test with a lower viral load than DSS was. Therefore, we can state that the efficacy of EMSS is not overestimated in comparison to DSS. Second, 4–6 days elapsed between virus extraction and RT-PCR of the saliva specimens, which may have resulted in RNA disruption and reduced sensitivity compared to NIID results obtained by RT-PCR on the day of specimen collection or the next day.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Hamamatsu Medical Center (2019-122) based on the Ethical Guidelines for Medical Research Targeting Humans provided by the Japanese Ministry of Health, Labor and Welfare.
Consent for publication
We received the patient's written informed consent. A copy of the written consent is available for review by the Editor of this journal.
Availability of data and materials
The datasets used for the current study are available from the corresponding author on reasonable request.
Funding
This research received no external funding.
Declaration of Competing interest
The authors: No reported conflict of interest except Dr. Suda. Dr. Suda reports other from SUDx-Biotec Corporation, during the conduct of the study; grants from Japan Ministry of Agriculture, Forestry and Fisheries, grants from Japan Agency for Medical Research and Development, outside the submitted work; In addition, Dr. Suda has a US patent #9464281 licensed and Dr. Suda is also a president of SUDx-Biotec Corporation. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
The Individuals included in acknowledgements: Bo reported as an employee of Kagoshima University with a remit to support the internationalization of the university, she received no payment or services for her contribution to this paper, which was limited to proofreading for grammatical errors and providing stylistic suggestions. Arima reported she is an employee of SUDx-Biotec Corporation. Regarding this paper, she helped the PCR assay, however, did not receive any additional salary. All individuals included in acknowledgements have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Acknowledgments
We thank Eriko Arima and Bo Causer for support in preparing RT-PCR assay and English check, respectively.
Footnotes
Yasuhisa Tajima designed the study, and drafted the work. Yasuo Suda developed RT-PCR for SARS-CoV-2 using saliva specimens and did all RT-PCR using saliva specimens. Yasuo Suda and Kunio Yano revised the work critically for important intellectual content. All authors contributed to the acquisition, analysis, or interpretation of data for the work. All authors have given the final approval of the version to be published and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy of any part of the work are appropriately investigated and resolved.
References
- 1.The World Health Organization Novel coronavirus—China. https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china from:
- 2.The World Health Organization Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Interim guidance 2 March 2020. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 from:
- 3.ECDC Novel coronavirus (SARS-CoV-2) Discharge criteria for confirmed COVID-19 cases – when is it safe to discharge COVID-19 cases from the hospital or end home isolation? https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-Discharge-criteria.pdf from:
- 4.Suda Y., Nagatomo M., Yokoyama R., Ohzono M., Aoyama K., Zhang X., et al. Highly sensitive detection of influenza virus in saliva by real-time PCR method using sugar chain-immobilized gold nanoparticles; application to clinical studies. Biotechnology Reports. 2015;(7):64–71. doi: 10.1016/j.btre.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saksono B., Dewi B.E., Nainggolan L., Suda Y. A highly sensitive diagnostic system for detecting dengue viruses using the interaction between a sulfated sugar chain and a virion. PloS One. 2015 May 26;10(5):e0123981. doi: 10.1371/journal.pone.0123981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suda Y, Wakao M, Kodama T. Method for concentrating viruses, method for concentrating cells or bacteria, and magnetic composite; United States Patent, 2016 Oct 11:US 9,464,281 B2.
- 7.Research C.D.C. Use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primer and probe information from. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- 8.To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 Feb 12 doi: 10.1093/cid/ciaa149. pii: ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020 May;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. pii: S1473-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphrey S.P., Williamson R.T. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001 Feb;85(2):162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 11.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol. 2012 Nov;113(5):1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 12.Hofman Lindsay F. Human saliva as a diagnostic specimen. J Nutr. 2001 May;131(5) doi: 10.1093/jn/131.5.1621S. 1621S-5S. [DOI] [PubMed] [Google Scholar]
- 13.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 Mar 28;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. pii: S0140-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alraddadi B.M., Al-Salmi H.S., Jacobs-Slifka K., Slayton R.B., Estivariz C.F., Geller A.I., et al. Risk factors for Middle East respiratory syndrome coronavirus infection among healthcare personnel. Emerg Infect Dis. 2016 Nov;22(11):1915–1920. doi: 10.3201/eid2211.160920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drosten C., Chiu L.L., Panning M., Leong H.N., Preiser W., Tam J.S., et al. Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol. 2004 May;42(5):2043–2047. doi: 10.1128/JCM.42.5.2043-2047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsushima Y., Uno N., Sasaki D., Morinaga Y., Hasegawa H., Yanagihara K. Quantitative RT-PCR evaluation of a rapid influenza antigen test for efficient diagnosis of influenza virus infection. J Virol Methods. 2015 Feb;212:76–79. doi: 10.1016/j.jviromet.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Zou L., Ruan F., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020 Mar 19;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used for the current study are available from the corresponding author on reasonable request.