Graphical Abstract
Keywords: ketogenic diet, ketone bodies, β-hydroxybutyrate, ketone ester, microbiome, bifidobacteria, Th17 cells, intestinal immunity, adipose tissue
SUMMARY
Very low-carbohydrate, high-fat ketogenic diets (KDs) induce a pronounced shift in metabolic fuel utilization that elevates circulating ketone bodies; however, the consequences of these compounds for host-microbiome interactions remain unknown. Here, we show that KDs alter the human and mouse gut microbiota in a manner distinct from high-fat diets (HFDs). Metagenomic and metabolomic analyses of stool samples from an 8-week inpatient study revealed marked shifts in gut microbial community structure and function during the KD. Gradient diet experiments in mice confirmed the unique impact of KDs relative to HFDs with a reproducible depletion of bifidobacteria. In vitro and in vivo experiments showed that ketone bodies selectively inhibited bifidobacterial growth. Finally, mono-colonizations and human microbiome transplantations into germ-free mice revealed that the KD-associated gut microbiota reduces the levels of intestinal pro-inflammatory Th17 cells. Together, these results highlight the importance of trans-kingdom chemical dialogues for mediating the host response to dietary interventions.
INTRODUCTION
Very low carbohydrate (CHO), high-fat ketogenic diets (KDs) are used for multiple diseases, but their metabolic and immune consequences remain unclear (Hall and Chung, 2018). We hypothesized that KDs could act in part via the gut microbiota. The gut microbiota is highly sensitive to dietary intake (Carmody et al., 2015) and plays a central role in coordinating host metabolism (Cani et al., 2019) and immunity (Honda and Littman, 2016). Work in rodents (Cani et al., 2007; Olson et al., 2018) and humans (David et al., 2014; Mardinoglu et al., 2018) shows that KDs alter the structure and function of the gut microbiota in states of health and disease. Yet multiple fundamental questions remain unaddressed. In humans, the published studies have been confounded by other changes in caloric and macronutrient intake (David et al., 2014; Mardinoglu et al., 2018). The mechanisms through which KDs alter gut bacterial abundance are unknown as are the downstream consequences of these changes for host metabolism and immunity. Here, we dissect these questions through the combined use of controlled dietary interventions in human subjects and follow-on studies in mice. Together, our results emphasize the importance of considering the chemical dialogues between host and microbial cells to gain a more comprehensive view of the mechanistic links between diet and host physiology.
RESULTS
Ketogenic diets reproducibly alter the human gut microbiome
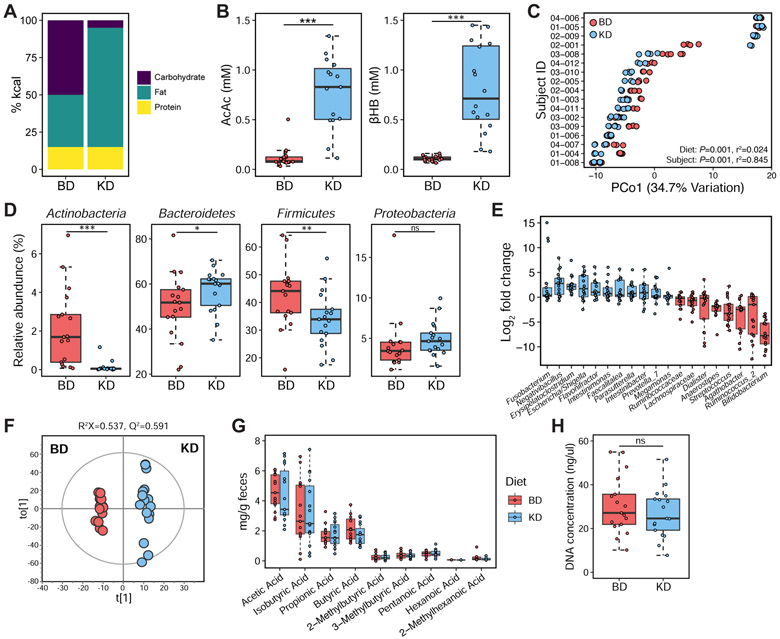
We conducted an inpatient crossover study in 17 adult overweight or class I obese non-diabetic men. Participants were first fed a baseline diet for 4 weeks (BD, 50% CHO, 15% Protein, 35% Fat) followed by a 4-week KD (5% CHO, 15% Protein, 80% Fat) (Figure 1A), resulting in elevated circulating plasma ketone bodies (Figure 1B). Five daily stool samples were collected in the final week of each diet period. Subjects were in a similar state of mild negative energy balance during both stool collection periods (Hall et al., 2016). 16S rRNA gene sequencing (16S-seq) revealed significant shifts in the gut microbiota between the two diets (Figures 1C and S1A; Table S1), including an altered relative abundance of the Actinobacteria, Bacteroidetes, and Firmicutes phyla (Figure 1D). 19 genera were significantly different with Bifidobacterium showing the greatest decrease on the KD (Figure 1E). Metagenomic sequencing of paired stool samples from the two diet arms confirmed the reduction of several bifidobacterial species on KD (Figures 2A and 2B; Table S2). Global profiling of the fecal metabolome by 1H NMR revealed a significant separation of fecal metabolite profiles between BD and KD stages (Figure 1F and S1B; Table S3). We did not observe any significant differences in the short-chain fatty acid levels (Figure 1G) or bacterial load (Figure 1H) between the two diet arms.
Figure 1. A ketogenic diet alters the human gut microbial community structure relative to an isocaloric control diet.
(A) Daily diet composition during the baseline diet (BD) and ketogenic diet (KD) stages respectively, represented in percent calories.
(B) Plasma levels of the ketone bodies acetoacetate (AcAc) and βHB (βHB) significantly increased during KD across all 17 study participants (***P < 0.001; paired t-test).
(C) Reproducible shifts in gut microbial community structure are observed across study participants on the first principle coordinate (PCo1) of PhILR Euclidean distances between the two diet stages (P = 0.001, r2 = 0.024, ADONIS with Subject ID as stratum), despite significant inter-individual variation in community structure between subjects (P = 0.001, r2 = 0.845, ADONIS testing for Subject ID).
(D) KD alters the relative abundance of the major phyla in the human gut microbiota (*P < 0.05, **P < 0.01, ***P < 0.001, ns = not significant; paired Wilcoxon test).
(E) Fold change of genera whose abundances were significantly altered by KD (FDR < 0.05, DESeq2).
(F) OPLS-DA score plot from 1H NMR spectra of fecal metabolite profiles between BD and KD diet stages across 17 study participants. The OPLS-DA model was validated with a 7-fold cross validation method; R2 value represents predictive power (R2>0.5 suggests the model is robust) and Q2>0.4 value suggests the model is valid.
(G) GC-MS analysis of human fecal short-chain fatty acids (SCFAs) did not reveal differences in SCFA concentration between BD (n = 15 samples) and KD (n = 15 samples) diet stages. Statistical analyses performed using paired t-tests for each SCFA.
(H) Fecal DNA content is not significantly different between the two diet stages (ns = not significant; paired t-test).
Data are presented as mean±SEM. Data points represent single samples (C, F-G) or the average of samples per study participant on each diet stage (D-E, H). See also Figure S1, Tables S1 and S3.
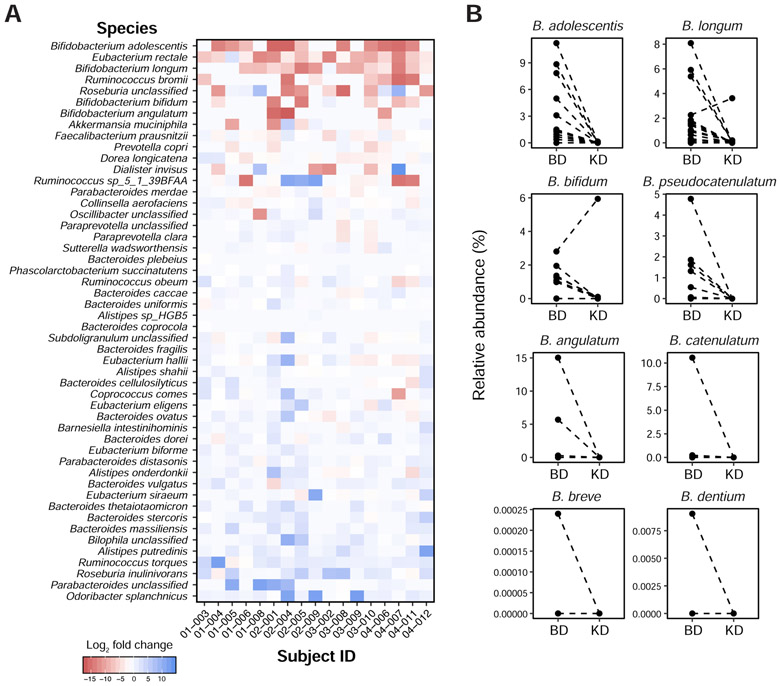
Figure 2. Metagenomic sequencing reveals decreased abundance of bifidobacterial species on the ketogenic diet in our human cohort.
(A) Heatmap showing fold change of the 50 most abundant species, comparing the ketogenic diet (KD) to baseline diet (BD) stages across all 17 subjects in our human study. Bacterial species are ordered by the magnitude and direction of the average fold change, from the most negative (top) to positive (bottom).
(B) The ketogenic diet results in consistent reductions in the abundance of bifidobacterial species in our human cohort. Each line connects paired samples between the two diet arms from each study subject, for bifidobacterial species detected in a study subject’s gut microbiota.
See also Table S2.
Ketogenic diets alter the gut microbiome in a manner distinct from high-fat diets
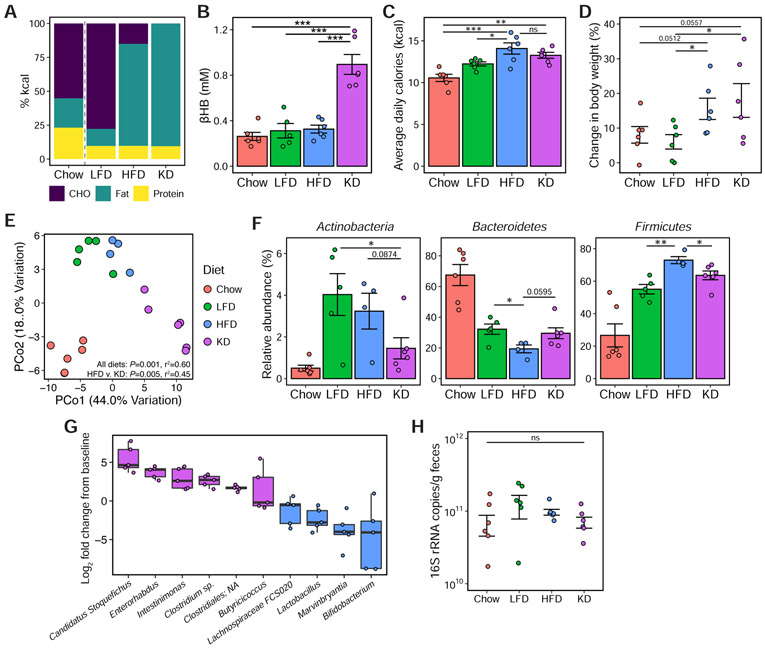
HFDs reproducibly increase the Firmicutes at the expense of the Bacteroidetes (Bisanz et al., 2019), yet our results indicate that KDs have the opposite effect. To address this discrepancy, we fed C57BL/6J mice 3 matched diets for 3 weeks (Figure 3A): low-fat diet (LFD, 78/12/10 CHO/fat/protein); high-fat diet (HFD, 15/75/10); and KD (0/90/10). The KD raised plasma levels of the ketone body β-hydroxybutyrate (βHB) compared to the other diets (Figure 3B). Mice fed the HFD and KD consumed more calories (Figure 3C) and gained more body weight (Figure 3D) compared to mice fed chow or LFD.
Figure 3. The impact of a ketogenic diet on the gut microbiota is distinct from a high-fat diet.
(A) Macronutrient composition (in percent calories) of the chow diet and three semi-purified diets: low-fat diet (LFD), high-fat diet (HFD) and ketogenic diet (KD).
(B) Circulating βHB levels in mice on diets for 3 weeks (n = 6 mice/group, ***P < 0.001; one-way ANOVA with Tukey’s test).
(C) Overall mean caloric intake represented as kcal per mouse per day over 3 weeks of dietary intervention (n = 6 mice/group, *P < 0.05, **P < 0.01, ***P < 0.001, ns = not significant; one-way ANOVA with Tukey’s test).
(D) Percent change in body weight compared to baseline after 3 weeks of dietary intervention (n = 6 mice/group, *P < 0.05; Kruskal-Wallis with Dunn’s test).
(E) Principal coordinate analysis of PhILR Euclidean distances reveal significant effect of diet on community composition (P = 0.001, r2 = 0.60; ADONIS) and also significant differences in community composition comparing HFD to KD (P = 0.005, r2 = 0.45; ADONIS).
(F) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets (n = 4-6 mice/group, *P < 0.05, **P < 0.01; Kruskal-Wallis with Dunn’s test).
(G) Fold change from baseline of genera whose abundances were significantly different between HFD and KD (FDR < 0.1, DESeq2), in mice fed KD (n = 6 mice).
(H) Fecal bacterial DNA content, expressed as 16S rRNA gene copies/ gram wet weight, of mice fed the respective diets is not significantly different (n = 6 mice/group, ns = not significant; one-way ANOVA with Tukey’s test).
See also Figures S2 and S3, and Table S4.
16S-seq revealed a significant effect of diet on the gut microbiota (Figure 3E). The HFD increased the relative abundance of the Firmicutes phylum at the expense of the Bacteroidetes; however, the KD reversed this trend (Figure 3F). The KD also decreased the relative abundance of Actinobacteria (Figure 3F). Comparing HFD and KD, we found 10 differentially abundant genera (Figure 3G) with Bifidobacterium showing the greatest decrease on the KD. Total colonization was comparable between diets (Figure 3H). The microbial shifts were stable and reproducible over 10 weeks of dietary intervention (Figures S2A-C) in animals at an independent mouse facility and with longer-term feeding (Newman et al., 2017a) over a period of up to 11 months (Figures S2H,I). Caloric intake, body weight, and adiposity were significantly higher on HFD and KD compared to LFD (Figures S2D-F). Mice in the KD group gained significantly less weight compared to HFD (Figure S2E) despite no significant differences in gonadal fat or liver weights (Figures S2F,G).
We replicated these findings with an alternative CHO source enriched for plant polysaccharides (Figure S3A; Table S4). Mice were sequentially fed these diets for 3 days each in order of decreasing dietary CHO. The gut microbiota shifted within a day following the chow-LFD and HFD-KD transitions (Figure S3B). Bacteroidetes decreased in response to CHO restriction with a corresponding increase in the Firmicutes (Figure S3C). Actinobacteria had a non-linear response to CHO which was also detectable at the genus level (Figure S3D). These results were consistent with a 3-week dietary intervention wherein mice were split into separate diets (Figure S3E) and experiments without added resistant starch (Figure 3F). The non-linear relationship between diet and the gut microbiota suggests that there may be a tipping point at which the microbiota responds differently to macronutrient intake.
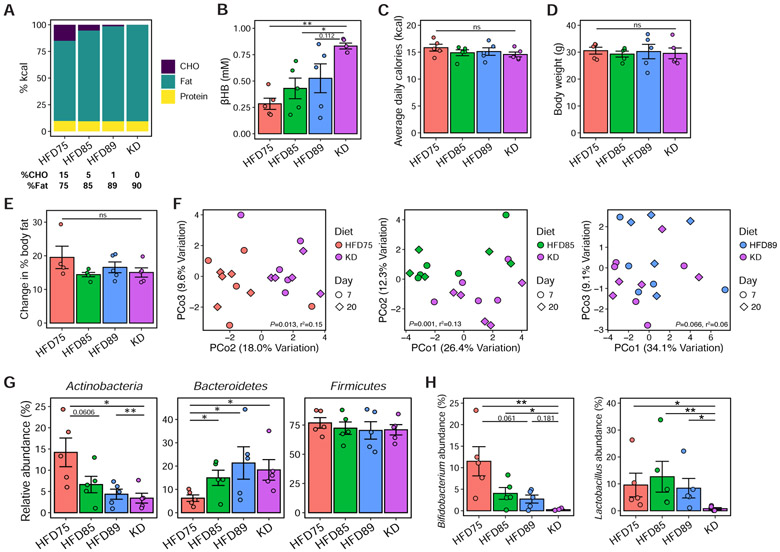
To more precisely identify this tipping point we tested diets containing CHO levels (Figure 4A) between the original HFD (HFD75; 15% CHO) and KD (0% CHO): HFD85 (5/85/10 CHO/fat/protein) and HFD89 (1/89/10). Circulating ketone body levels increased as dietary CHO decreased (Figure 4B). There were no significant differences in caloric intake, body weight, or adiposity (Figures 4C-E). The gut microbiota was significantly altered between HFD75/HFD85 and KD, but not between HFD89 and KD (Figure 4F). Bacteroidetes increased on all three diets compared to HFD75, with a progressive decrease in Actinobacteria with decreasing CHO (Figure 4G). We detected two differentially abundant genera between HFD89 and KD (Figure 4H). Bifidobacterium showed a progressive decline with decreasing CHO (Figure 4H), reflecting the overall Actinobacteria phylum (Figure 4G). Lactobacillus was significantly decreased on the KD (Figure 4H) despite no overall changes in the Firmicutes phylum (Figure 4G). These results suggest that partial induction of ketogenesis is sufficient to alter the gut microbiota despite the continued intake of CHO.
Figure 4. Modest changes in dietary carbohydrate associated with partial induction of ketogenesis is sufficient to alter gut microbial composition.
(A) Macronutrient composition (in percent calories) of four semi-purified diets in order of decreasing CHO content from 15% (HFD75) to 0% (KD). The CHO and fat content of each diet are shown below the barplots, with protein content matched at 10% kcal across all diets.
(B) Circulating βHB levels in mice fed the respective diets for 3 weeks (n = 5 mice/group, *P < 0.05, **P < 0.01; one-way ANOVA with Tukey’s test).
(C) Overall mean caloric intake, represented as kcal per mouse per day over 3 weeks of dietary intervention, was not significantly different between diet groups (n = 5 mice/group, ns = not significant; one-way ANOVA).
(D) Body weights of mice after 3 weeks of dietary intervention were not significantly different between diet groups (n = 5 mice/group, ns = not significant; one-way ANOVA).
(E) Changes in percent body fat, measured using EchoMRI at baseline and after 3 weeks of dietary intervention, were not significantly different between diet groups (n = 5 mice/group, ns = not significant; one-way ANOVA).
(F) Principal coordinate analyses of PhILR Euclidean distances reveal significant differences in community composition comparing HFD75 to KD (P = 0.013, r2 = 0.15) and HFD85 to KD (P = 0.001, r2 = 0.13) but not HFD89 to KD (P = 0.066, r2 = 0.06). Each data point represents a single fecal sample, with the color and shape of the symbols representing diet group and day of fecal collection respectively (n = 5 mice/group; ADONIS testing for effect of diet and accounting for within-mouse replicates).
(G) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets for 3 weeks (n = 5 mice/group, *P < 0.05, **P < 0.01; Kruskal-Wallis with Dunn’s test).
(H) Relative abundances of two genera (Bifidobacterium and Lactobacillus) that are significantly different comparing HFD89 to KD (FDR<0.05, DESeq2) are shown across diet groups (n = 5 mice/group, *P < 0.05, **P < 0.01; Kruskal-Wallis with Dunn’s test). Data are presented as mean±SEM. Each data point represents an individual, singly-housed animal (B-E, G-H).
Ketone bodies directly inhibit the growth of gut bacteria
Multiple mechanisms could contribute to differences in the gut microbiota between the HFD and KD. Prior work showed that mucus foraging increases in mice fed polysaccharide-deficient diets (Desai et al., 2016; Earle et al., 2015); however, in our experiments the mucus layer was maintained even in the absence of dietary CHO (Figure S4A-C) and Muc2-deficient mice lacking the major colonic mucin still exhibited similar gut microbiota shifts on the KD (Figure S4D-F). We also considered bile acids due to their role in fat emulsification; however, we did not observe a significant difference in the overall bile acid pool in human fecal samples (Figure S5A; Table S5) or mouse cecal contents (Figure S5B; Table S5) between diets. Finally, we focused on the ketone bodies that are indicative of increased lipid oxidation in response to CHO restriction. While the liver is the primary ketogenic organ, intestinal epithelial cells are also capable of producing ketone bodies (Puchalska and Crawford, 2017) and βHB increases in intestinal tissue on a KD (Tognini et al., 2017).
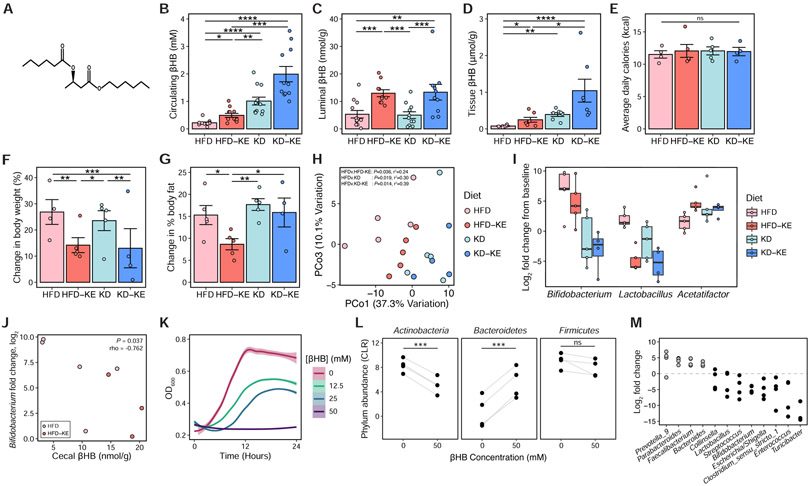
To investigate the impact of ketone bodies on the gut microbiota in the absence of CHO restriction, we supplemented the HFD with a synthetic ketone ester (KE; Figure 5A), mimicking intestinal production of βHB. As a positive control, we also included the KD with or without KEs. In two independent experiments, KE supplementation significantly raised circulating levels of βHB over baseline on the HFD, albeit to a lower level than KD consumption for 3 weeks (Figure 5B). KE supplementation to either a HFD or KD significantly increased βHB concentrations in the intestinal lumen (Figure 5C). We also observed elevated βHB in intestinal tissue (Figure 5D). Caloric intake was similar across groups (Figure 5E); however, mice fed KEs gained significantly less body weight (Figure 5F). Mice fed the HFD-KE also gained significantly less adiposity than HFD controls (Figure 5G). 16S-seq revealed that KE supplementation significantly altered the gut microbiota on a HFD (Figure 5H). Three genera were consistently altered in response to dietary and pharmacological induction of ketosis (Figure 5I). Of these three, only Bifidobacterium had a significant negative correlation with βHB in the intestinal lumen (Figure 5J) and also decreased in abundance on the KD in both humans and mice (Figure 1E and 3G).
Figure 5. Ketone bodies directly inhibit gut bacterial growth.
(A) Chemical structure of the ketone ester (KE) comprising βHB linked to two 6-carbon medium chain fatty acids which are metabolized to βHB.
(B) Circulating βHB levels in mice fed HFD, KE-supplemented HFD (HFD-KE), KD, or KE-supplemented KD (KD-KE) respectively for 3 weeks (n = 11-12 mice/group from two independent experiments, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Kruskal-Wallis with Dunn’s test).
(C) βHB concentrations in cecal contents of mice fed the respective diets for 3 weeks (n = 11-12 mice/group from two independent experiments, **P < 0.01, ***P < 0.001; Kruskal-Wallis with Dunn’s test).
(D) βHB concentrations in colon tissues of mice fed the respective diets for 3 weeks (n = 7 mice/group, *P < 0.05, **P < 0.01, ****P < 0.0001; Kruskal-Wallis with Dunn’s test).
(E) Overall mean caloric intake, represented as kcal per mouse per day over 3 weeks of dietary intervention, was not significantly different between groups (n = 4-5 mice/group, ns = not significant; Kruskal-Wallis test).
(F) Percent change in body weight from baseline after 3 weeks of dietary intervention was significantly lower in mice fed the KE-supplemented diets (HFD-KE and KD-KE) compared to either HFD or KD (n = 4-5 mice/group, *P < 0.05, **P < 0.01, ***P < 0.001; Kruskal-Wallis with Dunn’s test).
(G) Change in percent body fat, measured using EchoMRI at baseline and after 3 weeks, was significantly lower in mice fed HFD-KE compared to other diets (n = 4-5 mice/group, *P < 0.05, **P < 0.01; Kruskal-Wallis with Dunn’s test).
(H) Principal coordinates analysis of PhILR Euclidean distances reveals distinct clustering of microbial communities between diet groups (P = 0.001, r2 = 0.41, ADONIS), and microbial communities on the HFD diet are significantly different from each of the other diet groups (P and r2 values are indicated on the PCoA plot, ADONIS).
(I) Fold change of genera that are consistently altered in comparisons between HFD vs. KD and HFD vs. HFD-KE (FDR < 0.2, linear mixed effects model with MouseID as random effect), compared to baseline in the microbial communities of mice fed the respective diets (n = 4-5 mice/group).
(J) Negative correlation between cecal βHB levels and Bifidobacterium fold change in mice fed HFD or HFD-KE for 3 weeks (P = 0.037, rho = −0.762; Spearman’s rank correlation).
(K) Growth of fecal suspensions from four healthy donors measured under anaerobic conditions with different concentrations of βHB (12.5, 25 and 50mM) or vehicle control (0mM) over 24 hours. The average growth curves among the four donors, each grown in quadruplicates per concentration, are shown with shaded areas representing standard error (using stat_smooth function in R) and optical density (600 nm) measured at 15 min intervals over 24 hours.
(L) Centered log2-ratio (CLR) normalized abundances of the major phyla in four ex vivo communities incubated with 50 mM βHB or vehicle control (0 mM) for 16 hours. Each data point represents the average value from quadruplicates for a single donor community. Each line connects the microbial communities from the same donor, incubated with βHB or vehicle control (***P < 0.001, ns = not significant; paired t-test).
(M) Fold change of genera whose abundances were significantly altered by 50mM βHB compared to vehicle control (FDR < 0.05, DESeq2) at 16 hours. Each data point represents the average value from quadruplicates for a single donor community.
Data are presented as mean±SEM. Each data point represents an individual, singly-housed animal (B-G). See also Figure S6.
βHB had similar impacts on the human gut microbiota. We incubated stool samples from 4 human donors fed a BD with three different concentrations of βHB (12.5, 25 and 50 mM) or vehicle controls. βHB inhibited the growth of these ex vivo communities in a dose-dependent manner (Figure 5K) and 16S-seq revealed broad changes in the microbial communities that were evident at the phylum level (Figure 5L) in the presence of 50 mM βHB despite no overt growth (Figure 5K). We observed significantly lower abundance of Actinobacteria and increased Bacteroidetes comparing 50 mM βHB to vehicle control. We also found a total of 12 genera in the ex vivo communities that were significantly altered in the presence of βHB that included decreased abundance of Bifidobacterium and Lactobacillus (Figure 5M). These results are consistent with our in vivo findings above where we observed the selective restriction of both genera in the gut microbial communities of mice fed the ketone ester and extend the relevance of our findings to the human gut microbiota.
βHB directly limits Bifidobacterium growth in vitro. We isolated a strain of Bifidobacterium adolescentis (strain BD1) — the most differentially abundant species of Bifidobacterium in our human cohort (Figure 2A) — from human stool samples. βHB inhibited the in vitro growth of B. adolescentis BD1 in a dose-dependent manner (Figure S6A) and was bacteriostatic at the highest concentration tested (Figure S6B). Butyrate, a short-chain fatty acid that is chemically similar to βHB, produced similar growth-inhibitory effects (Figure S6C). Neutralization of βHB and butyrate prior to addition to the growth media abrogated their growth inhibitory effects (Figures S6A and S6C). Acidification of the growth media to match the pH of media at known concentrations of βHB recapitulated the inhibition of B. adolescentis growth (Figure S6D), suggesting that βHB prevents the in vitro growth of B. adolescentis through a pH-dependent mechanism.
To assess the specificity of βHB, we tested it against a panel of 35 gut bacterial isolates from 6 different phyla (Figure S6E,F) including 4 bifidobacterial strains isolated from our human cohort: 2 strains of Bifidobacterium adolescentis (strain BD1 and BD2); Bifidobacterium longum (strain BD3); and Bifidobacterium angulatum (strain BD4). For most isolates, the concentration required to prevent ≥90% growth (MIC90) was between 25-50 mM, with the exception of isolates in the genera Eggerthella and Lactobacillus that were insensitive to growth inhibition by βHB at the highest concentration tested. We also detected growth-promoting effects of βHB on select bacteria (Figures S6E). Contrary to our in vivo (Figure 5I) and ex vivo (Figure 5M) studies, we did not detect a selective effect of βHB against Bifidobacterium. The Bacteroidetes tended to be more sensitive to growth inhibition by βHB relative to the other phyla (Figure S6F), in contrast to the increased relative abundance of Bacteroidetes observed during KD consumption in humans (Figure 1D), mice (Figures 3F,4G), and ex vivo incubations (Figure 5L). Thus, the target specificity of βHB for Bifidobacterium observed in vivo could be driven by inter-species interactions and/or environmental factors.
Consistent with the latter hypothesis, the βHB concentrations used to achieve MIC90 in bacterial monocultures are roughly the same order of magnitude as circulating βHB levels in humans — where consistent serum βHB levels above 2 mM are reached with a ketogenic diet (Newman and Verdin, 2014) — but exceed the estimated physiological range in the intestinal lumen. Our data suggests that the βHB concentration in bulk cecal contents of mice fed a ketogenic diet is ~5 nmol/g (Figure 5C), which is multiple orders of magnitude lower than the concentrations tested in our in vitro assays (12.5-50 mM). Nevertheless, we found that the concentration of βHB in the intestinal tissue is roughly three orders of magnitude higher compared to luminal contents (Figure 5D), which could suggest that bacteria residing close to the intestinal epithelium may be exposed to relatively high local concentrations of βHB similar to the concentrations tested in the in vitro assays. Further investigations are needed to elucidate the mechanism of action of βHB on bifidobacteria within the context of complex microbial communities and gastrointestinal physiology.
Downstream consequences for host pathophysiology
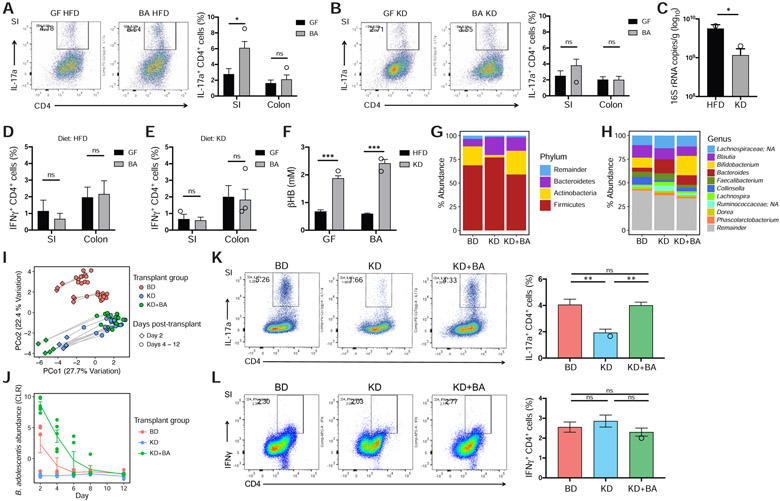
Human gut bifidobacteria, including B. adolescentis, induce pro-inflammatory intestinal Th17 cells in mono-colonized mice (Tan et al., 2016). Mono-colonization of germ-free mice with B. adolescentis BD1 resulted in a significant increase in CD4+ IL-17a+ (Th17) cells in the small (but not large) intestines of mice (Figures 6A and S7A) that were fed a HFD over the course of the experiment. Consumption of a KD prevented the induction of intestinal Th17 cells in mice mono-colonized with B. adolescentis (Figure 6B), consistent with a significant decrease in B. adolescentis colonization in the ileum compared to HFD-fed mice (Figure 6C). CD4+ IFNγ+ (Th1) cell levels in the intestines were unaffected by B. adolescentis mono-colonization on either diet (Figures 6D,E), consistent with the previous report showing preferential induction of Th17 cells in the small intestine by B. adolescentis (Tan et al., 2016). These data suggest that the KD, in part due to the host production of ketone bodies (Figure 6F), mediates the lack of intestinal Th17 induction in these mice by reducing colonization levels of B. adolescentis.
Figure 6. The KD-associated microbiota reduces gut Th17 cell accumulation in gnotobiotic mice.
(A) B. adolescentis BD1 (BA) induces a robust Th17 population in the small intestine (SI) but not large intestine (colon) of mice fed HFD. Th17 populations are measured as IL-17a+ CD4+ live cell percentages (see gating strategy in Figure S7A). Black and grey bars represent germ-free (GF) and BA-monocolonized mice respectively (n = 4 mice/group, *P < 0.05, ns = not significant; Welch’s t-test). Representative flow cytometry plots of the IL-17a+ CD4+ live cell populations are shown to the left.
(B) BA-monocolonized mice fed KD do not show intestinal Th17 cell induction compared to germ-free controls fed KD, in the SI and colon (n = 4 mice/group, ns = not significant; Welch’s t-test). Representative flow cytometry plots of the IL-17a+ CD4+ live cell populations are shown to the left.
(C) Bacterial DNA content, expressed as 16s rRNA gene copies/ gram wet weight in the ileal contents from BA-monocolonized mice, is significantly lower in mice fed KD compared to HFD (n = 3-4 mice/group, *P < 0.05; Welch’s t-test).
(D, E) No significant difference in Th1 populations in the SI and colon between germ-free (GF, black bars) and BA-monocolonized (BA, grey bars) mice fed (D) HFD and (E) KD respectively. Th1 populations are measured as IFNγ+ CD4+ live cell percentages (n = 3-4 mice/group, ns = not significant; Welch’s t-test).
(F) Circulating βHB levels are significantly elevated in both GF and BA-monocolonized mice fed KD compared to HFD. Black and grey bars represent HFD and KD respectively (n = 4 mice/group, ***P < 0.001; Welch’s t-test).
(G, H) Community composition at (G) phylum and (H) genus level of donor fecal samples used for gnotobiotic transplant, pooled from the same four individuals during the baseline diet (BD) and ketogenic diet (KD) stages respectively, and a third sample comprising the KD donor sample supplemented with B. adolescentis BD1 (KD+BA).
(I) Fecal microbiota of mice colonized with the donor samples shown in panels G and H, sampled every two days with sacrifice occurring 12 days post-transplantation (n = 5-6 mice/group).
(J) Abundance of B. adolescentis in the fecal microbiota of mice in the three transplant groups, sampled over the course of the experiment (n = 5-6 mice/group). BA abundance is significantly different between transplant groups (Pgroup = 1.8e-5; PKD+BAv.KD < 0.001; PKD+BAv.BD < 0.01; PBDv.KD = 0.053). Statistical analysis carried out using a linear mixed effects model with MouseID as random effect and Tukey’s test.
(K) Mice that received the KD microbiota showed significantly lower levels of intestinal Th17 cells than either BD or KD+BA transplant groups. Tissues were harvested on day 12 post-transplantation. Representative flow cytometry plots of the IL-17a+ CD4+ live cell populations are shown to the left (n = 4-6 mice/group, **P < 0.01, ns = not significant; one-way ANOVA with Tukey’s test).
(L) Th1 cell populations in the small intestine are not significantly different between transplant groups. Representative flow cytometry plots of the IFNγ+ CD4+ live cell populations are shown to the left (n = 4-6 mice/group, ns = not significant; one-way ANOVA with Tukey’s test).
Data are presented as mean±SEM. Each data point represents an individual, singly-housed animal (A-F, K-L).
We next wanted to investigate if changes in Bifidobacterium relative abundance in the context of an intact human gut microbiota would translate to differences in intestinal Th17 cell accumulation. We inoculated germ-free mice with fecal microbiota from human donors collected during the BD or KD stages and the Bifidobacterium-depleted KD microbiota supplemented with B. adolescentis BD1 (KD+BA). Supplementation recapitulated Bifidobacterium abundance in the BD microbiota at both the phylum (Figure 6G) and genus (Figure 6H) levels. 16S-seq of fecal samples showed that distinct microbial communities were established between BD and both KD microbiota groups (Figure 6I). Bifidobacterium abundance was significantly lower in the KD group compared with either BD or KD+BA groups (Figure 6J). Mice that received the KD microbiota showed significantly lower intestinal Th17 frequency compared to either BD or KD+BA groups which were detectable on day 12 post-transplantation (Figure 6K). There were no significant differences in the Th1 cell population (Figure 6L). These results demonstrate that changes in the abundance of Bifidobacterium in complex human gut microbial communities can have a significant impact on intestinal Th17 accumulation.
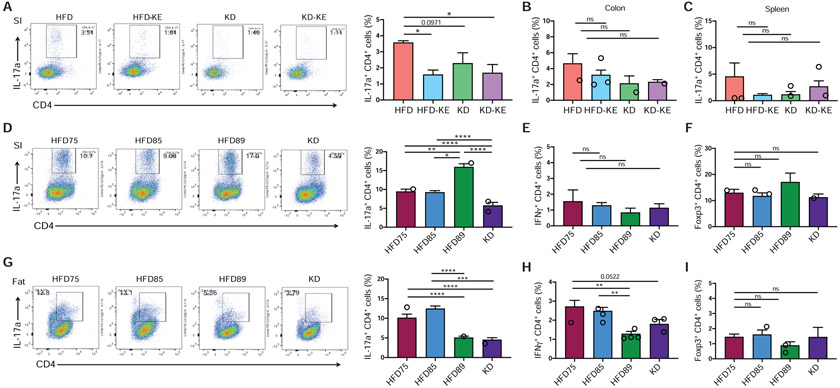
Given that human fecal transplantation of BD vs KD-associated microbiota was sufficient to reveal differential accumulation of Th17 cells in the gut, we hypothesized that differences in intestinal Th17 frequency should also be detectable in our diet experiments with conventional mice. Consistent with this hypothesis, conventional mice fed a KD exhibited lower levels of Th17 cells in the small intestine (SI) compared to mice fed a HFD (Figures 7A and 7D). Similarly, mice fed the KE-supplemented HFD, which reduced Bifidobacterium abundance in the gut microbiota relative to HFD (Figure 5I), led to lower Th17 cell accumulation in the small intestines (Figure 7A). There were no significant differences in Th17 cell levels in the large intestines (Figure 7B) or spleen (Figure 7C). Immunophenotyping of other immune cell populations in the intestine and spleen (Figure S7B) did not reveal consistent changes comparing HFD-KE and KD to HFD; the observed immune changes were specific to small intestinal Th17 cells.
Figure 7. The ketogenic diet and ketone ester feeding reduces intestinal Th17 cell accumulation in conventional mice.
(A) SPF mice fed either KD or ketone ester (KE)-supplemented diets show significantly lower frequencies of small intestinal Th17 cells compared to mice fed HFD for 3 weeks (n = 4-5 mice/group, *P < 0.05; one-way ANOVA with Tukey’s test). Representative flow cytometry plots of the IL-17a+ CD4+ live cell populations are shown to the left.
(B, C) No significant differences in Th17 cell levels in the (B) colon and (C) spleen of mice fed the respective diets for 3 weeks (n = 4-5 mice/group, ns = not significant; one-way ANOVA with Tukey’s test).
(D) SPF mice fed KD show significantly lower frequencies of small intestinal Th17 cells compared to mice fed similar high-fat, low-CHO diets shown in Figure 4A for 3 weeks (n = 4-5 mice/group, *P < 0.05, **P < 0.01, ****P < 0.0001; one-way ANOVA with Tukey’s test). Representative flow cytometry plots of the IL-17a+ CD4+ live cell populations are shown to the left.
(E, F) No significant differences in (E) IFNγ+ CD4+ Th1 and (F) Foxp3+ CD4+ Treg cell populations in the small intestine between groups after 3 weeks of dietary intervention (n = 4-5 mice/group, ns = not significant; one-way ANOVA with Tukey’s test).
(G) Th17 cell populations in the epididymal white adipose tissue of mice fed the respective diets for 3 weeks. Representative flow cytometry plots of the IL-17a+ CD4+ live cell populations in epididymal fat are shown to the left (n = 5 mice/group, ***P < 0.001, ****P < 0.0001; one-way ANOVA with Tukey’s test).
(H) IFNγ+ CD4+ (Th1) cell populations in the epididymal white adipose tissue of mice fed the respective diets for 3 weeks (n = 5 mice/group, **P < 0.01; one-way ANOVA with Tukey’s test).
(I) Foxp3+ CD4+ (Treg) cell populations in the epididymal white adipose tissue are not significantly different between groups after 3 weeks of dietary intervention (n = 5 mice/group, ns = not significant; one-way ANOVA with Tukey’s test).
Data are presented as mean±SEM. Each data point represents an individual, singly-housed animal. See also Figure S7.
Differences in SI Th17 cell accumulation could also be observed with small changes in dietary CHO intake (Figure 7D). Intestinal Th17 frequency was significantly lower in mice fed KD compared to similar high-fat, low-CHO diets (Figure 4A), consistent with a significant decrease in Bifidobacterium abundance on the KD (Figure 4H). The observed intestinal Th17 cell levels are not directly proportional to Bifidobacterium abundance since we observed a significant decrease in intestinal Th17 cell frequency only on the KD (Figure 7D) despite the progressive decline in Bifidobacterium abundance with decreasing dietary CHO (Figure 4H). Furthermore, HFD89 (1% CHO) resulted in a significant increase in Th17 cells relative to HFD85 (5% CHO). These results suggest that there may be an abundance threshold that is required for bifidobacteria in the gut to induce Th17 cell responses in the lamina propria or that other host or microbial factors may contribute to the observed Th17 differences. There were no significant changes in other immune cells subsets including Th1 (Figure 7E) and Treg (Figure 7F) cell populations.
Given increasing evidence for immunological crosstalk between the gut and other tissue compartments (Lee et al., 2018; Powell et al., 2017), we next asked if the observed differences in Th17 responses in the gut could be detected more systemically. Although we did not observe any differences in Th17 frequency in the spleen, we detected a significant drop in Th17 cell accumulation in visceral adipose tissue on both HFD89 and KD compared to the other diets (Figure 7G) with a similar pattern for the Th1 cell population (Figure 7H) and no differences in the Treg cell population (Figure 7I). These results complement recent findings which highlighted the effects of a ketogenic diet on immune cells in the visceral adipose tissue (Goldberg et al., 2020) and raises the intriguing question of whether the observed changes in adipose Th17 cells are related to gut microbial-induced changes in the intestinal immune environment or due to circulating βHB which may directly modulate immune cell function in the adipose tissue (Puchalska and Crawford, 2017). Our findings highlight how very low-CHO diets (0-1%) are distinctive from low-CHO diets (5-15%) in both gut microbial and immune responses.
DISCUSSION
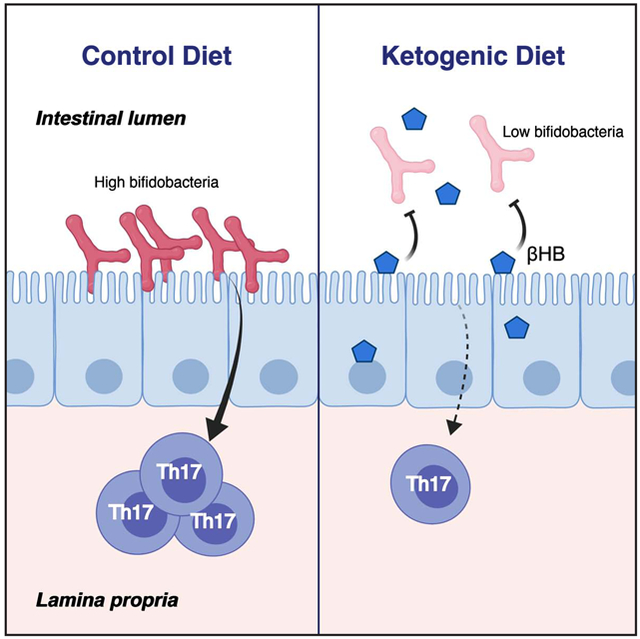
Taken together, these results suggest that diet-induced changes in host metabolites alter the gut microbiota with downstream consequences for immune cells. The impact of the KD on the gut microbiota is distinctive from HFDs, due in part to the concomitant host production of ketone bodies. Inhibition of bifidobacterial growth by ketone bodies results in KD-associated decreases in intestinal Th17 cell levels and possibly also adipose tissues. Given the links between obesity and chronic low-grade inflammation (Hotamisligil, 2017), decreased levels of pro-inflammatory Th17 cells in both gut and adipose tissues on a KD may be a potential mechanism contributing to the greater efficacy of KD in improving some aspects of metabolic syndrome such as glycemic control (Sainsbury et al., 2018; Saslow et al., 2017) and reductions in body fat (Mardinoglu et al., 2018; Volek et al., 2009).
Aside from reported metabolic effects in clinical trials, KDs have been used as dietary treatment for intractable epilepsy since the 1920s and recent studies have found significantly positive outcomes with the use of KDs for treatment of refractory epilepsy in children and adults (Kverneland et al., 2015; Liu et al., 2018; Neal et al., 2008) where anti-epileptic drugs have failed. However, the mechanisms mediating the efficacy of KDs as treatment for epilepsy remain unknown. A recent paper provided intriguing evidence for the role of the gut microbiota in mediating anti-seizure effects of KD in mouse models of refractory epilepsy (Olson et al., 2018). Interestingly, another recent paper showed increased proportion of circulating Th17 cells in patients with intractable epilepsy compared to control individuals that was partially reversed following KD consumption (Ni et al., 2016), highlighting the potential role of Th17 immune responses in the pathogenesis of epilepsy, and consistent with our data showing decreased Th17 cell frequency in both the gut and adipose tissues with KD consumption. These observations together provide the premise for future investigations into the potential role of Th17 immune responses as a mechanism underlying the efficacy of KD as a treatment for refractory epilepsy and potentially other diseases associated with increased Th17 activation such as autoimmune diseases. More research is required to better understand the long-term consequences of KDs in humans in both health and disease.
Our study highlights the role of the gut microbiota in mediating changes in intestinal Th17 cell responses to KD consumption. We focused on Bifidobacterium which had a reproducible decrease in abundance during KD consumption in both humans and mice and also with feeding of a synthetic KE in mice. Changes in Bifidobacterium abundance resulted in a significant alteration to intestinal Th17 cell populations. The impact of diet and the gut microbiota on Th17 cells may be beneficial or detrimental depending on the context, warranting comparative analyses of these complex interactions in the context of inflammatory diseases (Tan et al., 2016), defense against enteric pathogens (Honda and Littman, 2016), and cancer immunotherapy (Matson et al., 2018), among others. Further investigation is also needed to explore the potential consequences of changes in other members of the gut microbiota observed with KD consumption. Lactobacillus were decreased in response to both the KD and KE supplementation in mice. In humans, both Fusobacteria and Escherichia were significantly enriched on the KD, which have both been implicated in predisposition to colorectal cancer (Dalmasso et al., 2014; Kostic et al., 2013). These results suggest that the short- and long-term impact of KDs on host health and disease likely depends upon a complex series of beneficial, neutral, and detrimental shifts in microbial ecology.
βHB has anti-inflammatory effects due to its ability to inhibit the NLRP3 inflammasome (Youm et al., 2015). Production of βHB during a KD could directly impact immune responses in addition to altering microbial mediated immunomodulation. Nevertheless, our current study provides evidence for a causal contribution of the gut microbiota in mediating the immune responses observed with KD consumption, as the transplantation of fecal microbiota from human donors fed KD versus a baseline diet into germ-free mice was sufficient to reveal differences in intestinal Th17 cell accumulation. Our data supports the contribution of select gut bacterial changes, specifically Bifidobacterium, in modulating the intestinal Th17 cell population, consistent with previous work which showed that Bifidobacterium robustly induced intestinal Th17 cells (Tan et al., 2016).
Supplementation of oral ketone esters to a HFD suggests that KD-induced microbiota shifts are mediated in part through host production of ketone bodies. In the context of the KD, both CHO restriction and βHB may contribute to the marked reductions in Bifidobacterium and are not mutually exclusive. We attempted to dissect the relative importance of CHO versus βHB though our fine dietary gradient experiments and KE supplementation. The decline in Bifidobacterium abundance was associated with the induction of ketogenesis and not the complete lack of CHO, providing additional support for the importance of ketone bodies. However, we also observed low Bifidobacterium levels in mice fed a low-fat, high-plant-polysaccharide chow diet, warranting additional studies.
Further investigations are also needed to dissect the relative contribution of hepatic versus intestinal ketogenesis to the observed gut microbial changes. While local production of ketone bodies in the gut epithelium is likely to be more relevant to the gut microbiome, it is possible that elevations of circulating βHB from hepatic production during KD could also directly or indirectly shape the gut microbiota. Comparing the relative impact of intestinal versus hepatic ketogenesis on the gut microbiome might provide some insights into the physiological roles of ketone body production in the gut epithelium (Cheng et al., 2019; Miyamoto et al., 2019).
Overall, this work extends important previous findings from our laboratory (Carmody et al., 2015; David et al., 2014) and others (Tan et al., 2016), providing evidence that the impact of diet on the gut microbiota is not only translatable from animal models to humans, but also appears to play a causal role in mediating host immune responses to diet. Continued progress in elucidating the mechanistic basis for these observations could help inform more personalized approaches to utilize dietary interventions for the prevention and treatment of human disease.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter Turnbaugh (peter.turnbaugh@ucsf.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All sequencing datasets generated in this study are deposited in the NCBI Sequence Read Archive under BioProject PRJNA529487.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Study Design
The human study was approved by the Institutional Review Boards of the National Institute of Diabetes and Digestive and Kidney Diseases (NCT01967563), the Pennington Biomedical Research Center (2013-3-PBRC), Columbia University Medical Center (IRB-AAAL7113), and the Translational Research Institute for Metabolism and Diabetes (IRB 493675). Written informed consent was obtained from each participant.
Seventeen men between the ages of 18 - 50 years with body mass index between 25 - 35 kg/m2 were admitted to metabolic wards (5 subjects at NIH, and 4 subjects at each of the three other sites) and consumed a baseline diet for 4 weeks followed by an isocaloric ketogenic diet for another 4 weeks. Details on the study design, subject recruitment, and diets have been previously reported (Hall et al., 2016). Fecal samples were collected daily in the final week of each diet stage and stored at −80°C. Participants underwent weekly fasting blood draws for assessment of ketosis. Plasma ketone levels in the final week of each diet stage is reported here.
Mice
All animal experiments were conducted under protocols (AN170378 and AN170098) approved by the UCSF Institutional Animal Care and Use Committee. SPF or germ-free C57BL/6J mice were either born at UCSF or purchased from Jackson Laboratory. Germ-free and gnotobiotic mice were maintained within the UCSF Gnotobiotic Core Facility. Muc2−/− mice on C57BL6/J background were generated as previously described (Velcich et al., 2002) and obtained from Eugene Chang (University of Chicago). Littermates were randomly assigned to experimental groups. Unless otherwise noted, experiments were performed using wild-type C57BL/6J mice of both sexes between age 5 - 10 weeks at the start of experiment and were singly-housed during experiment.
METHOD DETAILS
Diets
All diets were provided ad libitum unless otherwise noted. Semi-purified diets were purchased from Envigo, and the ingredients used in these diets are listed in Table S4. Standard chow diet (Lab Diet 5058) was used in the SPF facility, while standard autoclaved chow diet (Lab Diet 5021) was used in the gnotobiotic facility. All diets used for gnotobiotic experiments were either autoclaved or irradiated to ensure sterility.
Bacteria Culturing Conditions
All bacteria were cultured at 37°C in an anaerobic chamber (Coy Laboratory Products) with an atmosphere composed of 2-3% H2, 20% CO2, and the balance N2. Culture media was composed of brain heart infusion (BHI) media supplemented with L-cysteine-HCl (0.05%, w/v), resazurin (0.0001%, w/v), hemin (5 μg/mL) and Vitamin K (1 μg/mL), referred to herein as BHI+ and allowed to equilibrate in the anaerobic environment prior to use.
Isolation of Bifidobacteria
Selective isolation of bifidobacteria was carried out by plating diluted human fecal samples on TOS-propionate agar (Sigma) containing lithium-mupirocin supplement (Sigma), followed by incubation at 37°C overnight in an anaerobic Coy chamber. Single colonies were randomly selected and identified by Sanger sequencing of the full-length 16S rRNA gene. A single colony identified as B. adolescentis (strain BD1) was inoculated into fresh BHI media and incubated anaerobically for 24 hours, mixed with autoclaved glycerol and stored in a cryovial at −80°C until further use.
Bacterial Growth Curves
The compounds used for bacterial growth curve assays are DL-β-Hydroxybutyric acid (Sigma 166898) and butyric acid (Sigma B103500). All compounds were dissolved in water and filter-sterilized to make 1M stock solutions. Two-fold serial dilutions of all compounds were freshly prepared prior to the start of growth curve assays. 10 μL of each dilution or BHI+ media (for growth controls) were added in triplicate to wells of 96-well plates containing 40 μL of BHI+ media. Fresh cultures of bacteria were cultivated in BHI+ media at 37°C for 24 hours under anaerobic conditions, diluted to OD 0.001, then 50 μL of the diluted cultures were added to each well of 96-well plates except for sterile controls for which 50 μL of BHI+ media were added, making a total volume of 100 μL in each well. Plates were sealed with tape to minimize evaporation and incubated at 37°C for 24 hours with continuous shaking. Growth curves were acquired by tracking optical density (OD) at 600 nm with a microplate spectrophotometer (EON, BioTek Instruments), with measurements taken every 15 min.
Ex Vivo Incubation of Human Stool Samples
An aliquot of frozen stool sample from each human donor was resuspended in 10 volumes (by weight) of BHI+ media in an anaerobic Coy chamber, vortexed for 1 min and left to settle for 5 min, and the clarified supernatant was then aliquoted for evaluation of ex vivo growth. 1:100 dilution of each fecal slurry was inoculated into BHI+ media containing different concentrations of DL-β-Hydroxybutyric acid (Sigma 166898) in quadruplicates per condition. Growth curves were acquired by measuring optical density (600 nm) every 15 min for 24 hours with continuous shaking.
Gnotobiotic Colonization
B. adolescentis BD1 was inoculated into fresh BHI+ media and incubated at 37°C for 24 hours in an anaerobic Coy chamber. A single 200 μL aliquot of the B. adolescentis culture (1.3 x 108 cfu/mL) or sterile BHI+ media as vehicle control was administered by oral gavage into each germ-free mouse within its gnotobiotic isolator. Mice were switched from standard chow diet to either HFD or KD on the same day as colonization, and maintained on their respective diets during the 12-day experiment.
Fecal Microbiota Transplants
Four human donors were selected for fecal microbiota transplants, one from each study site. Frozen fecal samples from these human donors during BD and KD diet stages respectively were resuspended in 10 volumes (by weight) of BHI+ media in an anaerobic Coy chamber. Each diluted sample was vortexed for 1 min and left to settle for 5 min, and equal volumes of the clarified supernatants were pooled for gavage (one pool from four donor fecal samples collected during BD and a second pool containing another four samples collected from the same donors during KD). The KD microbiota pool was split into two volumes, and a third pool was prepared by spiking in fresh culture of B. adolescentis to one volume of the KD pool. A single 200 μL aliquot of each pooled sample was administered by oral gavage into each germ-free mouse within its gnotobiotic isolator. All mice were maintained on a standard autoclaved chow diet over the 12-day experiment.
DNA Extraction
Human stool samples were homogenized with bead beating for 10 min on a vortex fitted with tube holder (MoBio, 13000-V1), using beads of mixed size and material (Lysing Matrix E 2mL Tube, MP Biomedicals) and the lysis solution provided in the PowerSoil bacterial DNA extraction kit (MoBio), and DNA was subsequently purified according to the manufacturer’s instructions.
All other samples were homogenized with bead beating for 5 min (Mini-Beadbeater-96, BioSpec) using the ZR BashingBead lysis matrix containing 0.1 and 0.5 mm beads (ZR-96 BashingBead Lysis Rack, Zymo Research) and the lysis solution provided in the ZymoBIOMICS 96 MagBead DNA Kit (Zymo Research). The samples were then centrifuged for 5 min at 3,000 g and the supernatant was transferred to 1mL deep-well plates. The DNA was then purified using the ZymoBIOMICS 96 MagBead DNA Kit (Zymo Research) according to the manufacturer's instructions.
qPCR Assessment of Bacterial Loads
DNA was extracted as described above. Quantification against a standard curve of purified genomic DNA from Escherichia coli was performed by quantitative PCR of the V6 region using BioRad iTaq Universal Probes Supermix in triplicate 10 μL reactions using 200 nM of the following oligonucleotides: 5’-TGGAGCATGTGGTTTAATTCGA-3’, 5’-TGCGGGACTTAACCCAACA-3’, 5’-[Cy5]CACGAGCTGACGACARCCATGCA[BHQ3] -3’. Reactions were performed on a BioRad CFX384 real-time thermocycler with the following cycle parameters: 95°C for 5 min followed by 40 cycles of 95°C for 5 sec, 60°C for 15 sec.
16S rRNA Sequencing and Analysis
DNA was isolated from human and mouse samples as described above. For human samples, 16S rRNA gene PCR was carried out using GoLay-barcoded 515F/806R primers (Caporaso et al., 2012) targeting the V4 region of the 16S rRNA gene according to the methods of the Earth Microbiome Project (earthmicrobiome.org). Amplicons were quantified with PicoGreen (Quant-It dsDNA, Life Technologies) and pooled at equimolar concentrations. For other samples, 16S rRNA gene PCR was carried out as per reference protocol and primers (Gohl et al., 2016). Amplicons were pooled and normalized using the SequalPrep Normalization Plate Kit (Invitrogen). For both mouse and human samples, aliquots of the pools were then column (MinElute PCR Purification Kit, Qiagen) and gel purified (QIAquick Gel Extraction Kit, Qiagen). Libraries were then quantified and sequenced with a 600 cycle MiSeq Reagent Kit (250x250; Illumina) with ~15% PhiX.
Sequencing reads were demultiplexed using QIIME before denoising and processing with DADA2 (Callahan et al., 2016). Taxonomy was assigned using the DADA2 implementation of the RDP classifier (Wang et al., 2007) using the DADA2 formatted training sets for SILVA123 (benjjneb.github.io/dada2/assign.html). A phylogenetic tree was constructed using FastTree (Price et al., 2010) with midpoint rooting. Sequence variants were filtered such that they were present in more than one sample with at least a total of 10 reads. The PhILR Euclidean distance was calculated by first carrying out the phylogenetic isometric log ratio transformation (philr, PhILR (Silverman et al., 2017)) followed by calculating the Euclidean distance (vegdist, Vegan (Dixon, 2003)). Principal coordinates analysis was carried out using the pcoa function of APE (Paradis et al., 2004). ADONIS calculations were carried out (adonis, Vegan) with 999 replications on each distance metric. The Wald test in the DESeq2 package (Love et al., 2014) was used to analyze differential abundances on count data, using features that represented at least 0.05% of total sequencing reads. Centered log2-ratio (CLR) normalized abundances were calculated as Aclr=[log2(A1/ga), log2(A2/ga),… log2(An/ga),], where A is a vector of read counts with a prior of 0.5 added and ga is the geometric mean of all values of A. Time-course analyses were carried out using linear mixed effects models (lmerTest, (Kuznetsova et al., 2017)) using mouse as a random effect to account for repeated sampling across time. Corrections for multiple hypothesis testing to false discovery rate (FDR) using the Benjamin-Hochberg method (Benjamini and Hochberg, 1995) were performed where applicable.
Metagenomic Shotgun Sequencing and Analysis
Fecal DNA from human donors was isolated as described above. Whole-genome shotgun libraries were prepared using the Nextera XT DNA Library Prep Kit (Illumina). Paired ends of the libraries were sequenced on the Illumina HiSeq 2500 (Rapid Run mode) or NovaSeq 6000 platform. Raw Illumina reads underwent quality trimming and adaptor removal using Trimmomatic (Bolger et al., 2014) and host read removal by mapping to the human genome (GRCh38) with Bowtie2 (Langmead and Salzberg, 2012). Taxonomic profiling and annotation was performed using MetaPhlAn2 (Segata et al., 2012).
Lamina Propria Lymphocyte Isolation
Lamina propria lymphocytes (LPLs) were isolated with slight modifications of previously described techniques (Atarashi et al., 2011; Kubinak et al., 2015; Round et al., 2011). In brief, small intestinal and colonic tissues were splayed longitudinally with mucus removed by scraping then stored in complete RPMI (10% fetal bovine serum, 100 units per mL penicillin and streptomycin, β-mercaptoethanol, glutamate, sodium pyruvate, HEPES and non-essential amino acids). Supernatants were discarded after filtering through a 100 μM filter, and remaining tissue incubated in 1X HBSS (without Ca2+and Mg2+) containing 5 mM EDTA (Promega) and 1 mM DL-Dithiothreitol (DTT) (Bioplus chemicals) for 45 min at 37°C on a shaker. Supernatants were filtered through a 100μM filter and discarded, and remaining tissue was incubated for 45 min (colon) or 35 min (SI) at 37°C on a shaker in a solution containing 1X HBSS containing 5% (v/v) fetal bovine serum (GIBCO heat inactivated), 1 U/mL Dispase (Sigma), 0.5 mg/mL Collagenase VIII (Sigma), and 20 μg/mL DNaseI (Sigma). The supernatant was filtered over a 40 μm cell strainer into cold sterile 1X PBS. Cells were subjected to a Percoll (VWR) gradient (40%/80% [v/v] gradient) and spun at 2000 RPM for 20 min with no brake and no acceleration. Cells at the interface were collected, washed with PBS, and prepared for flow cytometry analysis.
Adipose Lymphocyte Isolation
Epididymal fat pads were dissected from the same side of mice following euthanasia and temporarily stored in 3 mL of cold Ham’s F-10 medium prior to digestion. The tissues were then finely minced with scissors and digested in 3 mL of 2X digestion buffer (2 mg/mL at 250U/mg Collagenase type I, Worthington, and 30 mg/mL bovine serum albumin in Ham’s F-10 medium) for 30 min at 37°C on a shaker, vortexing every 15 min. Cells were spun down at 1600 RPM for 7 min, resuspended in 10 mL of cold sterile 1X PBS, and filtered through a 40 μm cell strainer. Cells were spun down again, resuspended in PBS and prepared for flow cytometry analysis.
Flow Cytometry
LPLs were isolated from the small intestinal and colonic lamina propria as described above. Fat lymphocytes were isolated as described above. Spleen cells were prepped through gentle mashing with a syringe plunger in 1 mL 1X PBS. Spleen cells were treated with 1X RBC Lysis Buffer (Biolegend) to lyse and remove red blood cells. For extracellular stains, surface staining was done in staining buffer: 1X HBSS (Corning) supplemented with 10 mM HEPES (Fisher Scientific), 2 mM EDTA (Invitrogen), and 0.5% (v/v) fetal bovine serum (heat inactivated) for 20 min at 4°C. Cells were then washed twice in staining buffer then assayed via flow cytometry. The following antibodies were used: anti-CD3 (17A2, Fisher Scientific), anti-CD4 (GK1.5, Biolegend), anti-CD8a (53.67, Biolegend), anti-CD44 (IM-7, VWR), anti-CD62L (MEL-14, Fisher Scientific), anti-CD11c (N418, Biolegend), anti-CD11b (M1/70, VWR), anti-MHCII (M5/114.15.2, Biolegend), anti-F4/80 (BM8, Biolegend), anti-LygC (HK1.4, Biolegend), anti-Ly6G (1A8, Biolegend), anti-B220 (RA3-6B2, Fisher Scientific), anti-IgD (11.26c.2a, VWR), anti-IgA (NA, Fisher Scientific), anti-IgM (RMM-1, VWR). For intracellular staining, cells were first stimulated with ionomycin (1000 ng/mL), PMA (50 ng/mL), and Golgi Plug (1 μL/sample) (BD Bioscience) overnight or 4-6 hours at 37°C. Alternatively, cells were stimulated with a cell stimulation cocktail (Fisher Scientific) containing PMA and ionomycin according to the manufacturer's instructions, and Golgi plug was again added. Stimulated cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher) according to the manufacturer’s instructions. Cells were surface stained with anti-CD3 (17A2, Invitrogen), anti-CD4 (GK1.5, Biolegend), washed, and then fixed/permeabilized in 100 μL Perm/Fix buffer (BD Bioscience). Cells were washed twice in Perm/Wash buffer (BD Bioscience) and then stained for intracellular cytokines: anti-Rorγt (B2D, Invitrogen), anti-IFNγ (XMG1.2, Millipore), anti-IL17a (ebio17B7, Invitrogen), anti-Foxp3 (150D, Biolegend). Cells were washed in Perm/Wash buffer twice and then re-suspended in staining buffer for flow cytometry analysis. Gating of cell populations was done using isotype and single stain controls. These data were collected with a BD LSR Fortessa and analyzed with FlowJo software (Version 10.6.1).
β-hydroxybutyrate (βHB) Quantitation by UPLC-Orbitrap-MS
For measurements of circulating β-hydroxybutyrate (βHB), blood was obtained from mice by cardiac puncture immediately following euthanasia in the day between 9am till noon. Blood was collected into collection tubes coated with K2-EDTA (Terumo T-MQK) or with serum separator (BD Microtainer 365978), centrifuged at 13,000 RPM for 10 min at 4°C to separate the plasma or serum, and frozen at −20°C until use. βHB levels were quantified using the LiquiColor β-Hydroxybutyrate colorimetric test (StanBio) according to the manufacturer’s instructions. Baseline absorbance of the sample-enzyme mixture prior to adding catalyst were subtracted from the final absorbance values in order to account for any sample hemolysis, with 2-3 replicates run per sample.
For measurements of βHB in intestinal contents and tissues, cecal content and colon tissue samples (25 mg) were extracted as previously described (Tian et al., 2018, 2019). Briefly, samples were extracted with 1 mL of ice-cold methanol [80% v/v, 0.1% formic acid] (for cecal content) or 0.5 ml of ice cold methanol [80% v/v] (for colon tissue), followed by homogenization. After centrifugation, the supernatants were dried in vacuum and reconstituted in 100 μL of methanol (3% v/v). Quantitative analysis was performed by the LC-MS system consisting of a Dionex Ultimate 3000 quaternary HPLC pump, a Dionex 3000 column compartment, a Dionex 3000 autosampler, and an Exactive plus orbitrap mass spectrometer controlled by Xcalibur 2.2 software (all from Thermo Fisher Scientific, Waltham, MA). Analytes were detected by multiple reaction monitoring and normalized by a 13C stable isotope labeled internal standard (Cambridge Isotope Laboratories, Inc., Tewksbury, MA). The results were quantified using a standard curve with concentrations ranging from 0.1 μM to 50 μM.
Ketone Ester Synthesis
The synthesis method for the hexanoyl hexyl β-hydroxybutyrate (C6x2-βHB) ester was developed by Scott Ulrich of Ithaca College (Ithaca, New York, USA) as previously described (Newman et al., 2017b), and the ester was then custom ordered in bulk from AK Scientific (Union City, CA). The βHB moiety of the ester is chiral, and the compound was synthesized as an enantiomeric mixture. Briefly, hexyl β-hydroxybutyrate was prepared by suspending sodium β-hydroxybutyrate in dry dimethylformamide and reacting with 1-bromohexane, followed by hexyl acyl chloride. The identity of the final product was confirmed by 1H NMR and gas chromatograph mass spectrometry (GC/MS), with 97.5% purity on GC.
NMR Global Metabolomics
NMR spectroscopy was performed at 298K on a Bruker Avance III 600-MHz spectrometer configured with a 5 mm inverse cryogenic probe (Bruker Biospin, Germany) as previously described (Cai et al., 2017). 50 mg of fresh human feces were extracted with 1 mL of phosphate buffer (K2HPO4/NaH2PO4, 0.1 M, pH 7.4, 50% v/v D2O) containing 0.005% sodium 3-(trimethylsilyl) [2,2,3,3-2H4] propionate (TSP-d4) as a chemical shift reference (δ 0.00). Samples were freeze-thawed three times with liquid nitrogen and water bath for thorough extraction, then homogenized (6500 rpm, 1 cycle, 60s) and centrifuged (11 180g, 4 °C, 10 min). The supernatants were transferred to a new 2 mL tube. Additional 600 μL of PBS was added to the pellets, followed by the same extraction procedure described above. Combined fecal extracts were centrifuged (11,180g, 4°C, 10 min), 600 μL supernatants were transferred to a 5 mm NMR tube (Norell, Morganton, NC) for NMR spectroscopy analysis. A standard one-dimensional NOESY pulse sequence noesypr1d (recycle delay-90°-t1-90°-tm-90°-acquisition) is used with a 90 pulse length of approximately 10s (−9.6 dbW) and 64 transients are recorded into 32k data points with a spectral width of 9.6 KHz.
NMR spectra were processed as previously described (Cai et al., 2017). First, all spectra quality was improved with topspin 3.0 (Bruker Biospin, Germany) for phase and baseline correction and chemical shift calibration. Then AMIX software (version: 3.9.14, Brucker Biospin, Germany) was used for bucketing (bucket width 0.004 ppm), interference signal removing (water and polyethylene glycol), and scaling (total intensity). The following multivariate data analysis was performed with SIMCA 13 (Umetrics, Sweden). Orthogonal projection to latent structure-discriminant analysis (OPLS-DA) was performed with UV scaling, and the model was validated with a 7-fold cross validation method. R2X and Q2 values generated from the method represent the predictive power and validity of the models, respectively.
Targeted Bile Acid Quantitation by UPLC-MS/MS
Bile acids quantitation in mouse cecal content, serum and human feces was performed with an ACQUITY UPLC using a BEH C8 column (1,7 μm, 100 mm x 2.1 mm) coupled with a Xevo TQ-S mass spectrometer equipped with an electrospray ionization (ESI) source operating in negative mode (All Waters, Milford, MA) as previously described (Sarafian et al., 2015). Selected ion monitoring (SIR) for non-conjugated bile acids and multiple reaction monitoring (MRM) for conjugated bile acids were used. 50 mg of cecal content/feces was pre-weighed, mixed with 1 mL of pre-cooled methanol containing 0.5 μM stable-isotope-labeled bile acids (internal standards) and 1.0 mm diameter zirconia/silica beads (BioSpec, Bartlesville, OK), followed by thorough homogenization and centrifugation. Supernatant was transferred to an autosampler vial for analysis. 100 μL of serum was extracted by adding 200 μL pre-cooled methanol containing 0.5 μM internal standard bile acids. Following centrifugation, the supernatant of the extract was transferred to an autosampler vial for quantitation. Calibration curves of individual bile acids were drafted with bile acid standards for quantitation of the biological abundance of bile acids.
Targeted SCFAs quantitation by GC-MS
Targeted analysis of short-chain fatty acids (SCFAs) in human feces was performed with an Agilent 7890A gas chromatograph coupled with an Agilent 5975 mass spectrometer (Agilent Technologies Santa Clara, CA) using a propyl esterification method as previously described (Cai et al., 2017). 50 mg of human fecal samples were pre-weighed, mixed with 1 mL of 0.005 M NaOH containing 10 μg/mL caproic acid-6,6,6-d3 (internal standard) and 1.0 mm diameter zirconia/silica beads (BioSpec, Bartlesville, OK). The mixture was thoroughly homogenized and centrifuged (13200g, 4°C, 20 min). 500 μL of supernatant was transferred to a 20 mL glass scintillation vial. 500 μL of 1-propanol/pyridine (v/v=3/2) solvent was added into the vial, followed by a slow adding of an aliquot of 100 μL of esterification reagent propyl chloroformate. After a brief vortex of the mixture for 1 minute, samples were derivatized at 60 °C for 1 hour. After the derivatization, samples were extracted with hexane in a two-step procedure (300 μL + 200 μL ) as described (Zheng et al., 2013). First, 300 μL of hexane was added to the sample, briefly vortexed and centrifuged (2000g, 4°C, 5 min), and 300 μL of the upper layer was transferred to a glass autosampler vial. Second, an additional 200 μL of hexane was added to the sample, vortexed, centrifuged, and the 200 μL upper layer was transferred to the glass autosampler vial. A combination of 500 μL of extracts were obtained for GC-MS analysis. A calibration curve of each SCFA was generated with series dilution of the SCFA standard for absolute quantitation of the biological concentration of SCFAs in human feces samples.
RNA Extraction and qPCR Analysis for Gene Expression
Total RNA was isolated from colonic tissues using the Direct-zol-96 MagBead kit (Zymo Research) as per the manufacturer’s instructions. For qPCR analysis, cDNA was synthesized using iScript Reverse Transcription Supermix (BioRad), and qPCR was performed using a commercially available TaqMan Gene Expression Assay for Muc2 (Assay ID: Mm01276694_g1) with iTaq Universal Probes Supermix (BioRad) on a BioRad CFX384 real-time thermocycler. Samples were run in triplicates and normalized to the B2m housekeeping gene (Taqman Gene Expression Assay ID: Mm00437762_m1).
Intestinal Tissue Collection, Fixation and Processing
Sections of the distal colon, closest to the rectum and containing a fecal pellet, were collected from mice following euthanasia and placed in cassettes. Tissues were fixed in methacarn solution and processed as previously described (Earle et al., 2015; Johansson and Hansson, 2012). Colon samples within cassettes were then submerged in melted paraffin at 68°C for 1 hr, removed, and kept at room temperature until sectioning. Paraffin blocks were cut into 4-μm-thick sections and deparaffinized for FISH staining.
Fluorescence In Situ Hybridization (FISH)
FISH was performed as previously described (Earle et al., 2015; Johansson and Hansson, 2012). Briefly, deparaffinized sections were incubated with the universal bacterial EUB338 probe (Amann et al., 1990) diluted to 10ng/μL in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.4], 0.01% sodium dodecyl sulfate, 10% formamide) at 50°C for 20 min in the dark and washed three times in PBS. Samples were then incubated with 10μg/mL DAPI (EMD Millipore) and 40μg/mL FITC-UEA1 (Sigma) in the dark for 1 hr at 4°C, washed three times in PBS, allowed to dry, and mounted in Vectashield mounting medium (Vector Labs).
Imaging and Quantitation of Mucus Layer in Colon Sections
Images were acquired on a Zeiss LSM 880 or Leica SP5 confocal microscope. Samples were imaged with a 63x oil-immersion objective. For quantification of mucus thickness, images were acquired with a frame size of 1912 X 1912 and 16-bit depth on the Zeiss LSM 880 microscope, with tile scans acquired using a mechanical stage and overlapping images (3 x 5) stitched using the ZEN Imaging Software (black edition). Mucus thickness was then quantified using the analysis platform BacSpace (Earle et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
Where applicable, statistical analysis was performed using either R version 3.5 or GraphPad Prism 8. ANOVA with Tukey’s post hoc analysis was used for parametric analysis of variance between groups, and two-tailed paired Student’s t test or unpaired Welch’s t test was used for pairwise comparisons. The Kruskal-Wallis with Dunn’s test was used for non-parametric analysis of variance between groups, and a two-tailed Wilcoxon rank-sum test or Wilcoxon signed-rank test was used for pairwise comparisons. For longitudinal data analyses, linear mixed effects models were performed using lmerTest (Kuznetsova et al., 2017) with multiple testing corrections via the ghlt function of multcomp using TukeyHSD. Analysis and data visualization of microbial populations were carried out in R as described above. Data are presented as mean±SEM unless otherwise noted.
Supplementary Material
Figure S1. The ketogenic diet reproducibly alters the structure and function of the human gut microbiota, related to Figure 1.
(A) Principal coordinate analysis of PhILR Euclidean distances reveal distinct clustering of gut microbial communities between baseline diet (BD) and ketogenic diet (KD) stages across study participants, related to Figure 1C. P values were determined using ADONIS tests for the effect of diet within each subject.
(B) Representative 600 MHz NMR spectra of feces from baseline diet (BD) and ketogenic diet (KD) stages with metabolites identification, related to Figure 1F. The regions of δ 5.2-9.0 were vertically expanded 16 times. Keys: 1,Butyrate; 2,Branched-chain amino acids; 3,Propionate; 4,Lactate; 5,Threonine; 6,Alanine; 7,n-Butyrate; 8,Lysine; 9,Arginine; 10,Acetate; 11,Proline; 12,Succinate; 13,Taurine; 14,β-Glucose; 15,Glycine; 16,α-Glucose; 17,Uracil; 18,Fumarate; 19,Tyrosine; 20,Histidine; 21,Phenylalanine; 22,Tryptophan; 23,Xanthine; 24,Hypoxanthine; 25,Formate; 26,Nicotinurate; 27,Uridine; 28,Polyethylene glycol (PEG).
Figure S2. The ketogenic diet results in stable and reproducible shifts in gut microbial composition, related to Figure 3.
(A) Circulating βHB levels of mice fed a low-fat diet (LFD), high-fat diet (HFD) or ketogenic diet (KD) for 10 weeks (n = 8 mice/group, ***P < 0.001; One-way ANOVA with Tukey’s test).
(B) Principal coordinate analysis of PhILR Euclidean distances reveal distinct clustering of KD samples and a significant effect of diet on community composition (P = 0.001, r2= 0.31, ADONIS).
(C) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets, averaged from weekly samples. (n = 8 mice/group with mice housed in pairs, *P < 0.05, **P < 0.01, ***P < 0.001; Kruskal-Wallis with Dunn’s test).
(D) Overall mean caloric intake represented as kcal per mouse per day on the respective diets. Food intake was measured weekly over 11 weeks (n = 8 mice/group with mice housed in pairs, **P < 0.01, ***P < 0.001, ns = not significant; One-way ANOVA with Tukey’s test).
(E) Body weight trajectories of mice fed the respective diets over 11 weeks (n = 8 mice/group). Asterisks denote significant differences from LFD and HFD respectively (PLFDv.HFD = 6.5e-5; PLFDv.KD = 0.049; PHFDv.KD = 0.049). Statistical analysis carried out using linear mixed effects model with MouseID as random effect and Tukey’s test.
(F) Gonadal fat mass in mice fed the respective diets for 11 weeks (n = 8 mice/group, **P < 0.01, ****P < 0.0001; One-way ANOVA with Tukey’s test).
(G) Liver mass in mice fed the respective diets for 11 weeks (n = 8 mice/group, *P < 0.05, ns = not significant; One-way ANOVA with Tukey’s test).
(H) Principal coordinate analysis of fecal samples collected at a single time point from a healthspan and longevity cohort (Newman et al., 2017a) reveals a significant effect of diet on community composition (P = 0.001, r2= 0.33, ADONIS) with long-term feeding of the respective diets (range 28 - 311 days at the time of sample collection). Each data point represents an individual animal (n = 21 - 35 mice/group with 3-5 mice housed per cage).
(I) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets long-term, related to panel H (n = 21 - 35 mice/group, *P < 0.05, ***P < 0.01; Kruskal-Wallis with Dunn’s test). Data are presented as mean±SEM.
Figure S3. The effect of a ketogenic diet on the gut microbiota is replicable with an alternative carbohydrate source enriched for plant polysaccharides, related to Figure 3.
(A) Macronutrient composition (in percent calories) of the chow diet and a series of semi-purified diets containing resistant starch and varying CHO and fat content: low-fat diet (LFD), 45%-fat high-fat diet (HFD45), 75%-fat high-fat diet (HFD75), and ketogenic diet (KD).
(B) Rapid shifts in gut microbial composition are observed within a day of feeding of each diet. Mice were fed each diet for 3 days in a sequential order: Chow → LFD → HFD45 → HFD75 → KD, and dashed vertical lines represent the switch of diets. Axis 1 of principal coordinate analysis (PCo1) on PhILR Euclidean distances is shown over the 15-day study period (n = 4 mice).
(C) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets over 15 days reveal both linear (Bacteroidetes and Firmicutes) and non-linear (Actinobacteria) dose responses to dietary CHO (n = 4 mice).
(D) Abundance of Bifidobacterium in the murine fecal microbiota over study period, showing decrease on KD to a level similar to that of chow diet (n = 4 mice).
(E) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets over 3 weeks in a separate experiment (n = 4 mice/group, *P < 0.05; Kruskal-Wallis with Dunn’s test). Data are presented as mean±SEM. Each data point represents an individual, singly-housed animal.
Figure S4. A ketogenic diet (KD) maintains a robust mucus layer despite the lack of fermentable carbohydrates and gut microbial shift on KD is independent of colonic mucin, related to Figures 2 and 3.
(A) Representative images of distal colon sections from SPF mice fed chow, low-fat diet (LFD), high-fat diet (HFD), or ketogenic diet (KD). Sections were stained with DAPI (epithelium, blue), UEA1 lectin (mucus, green) and EUB338 universal bacterial FISH probe (red). Images were further processed using BacSpace (Earle et al., 2015) and the computed luminal debris appears in yellow.
(B) Average mucus widths in distal colons of SPF mice fed the respective diets after 3 weeks, quantified using BacSpace (Earle et al., 2015) (n = 3 - 5 mice/group, ns = not significant; one-way ANOVA).
(C) Quantitative PCR of colonic Muc2 transcripts reveal no significant difference in Muc2 relative gene expression (n = 3 - 5 mice/group, ns = not significant; one-way ANOVA).
(D) Representative images of distal colon sections from wild-type (WT) and Muc2−/− mice respectively, showing lack of goblet cells and mucus layer in Muc2−/− mice with bacteria in close contact with gut epithelium. Sections were stained with DAPI (epithelium, blue), UEA1 lectin (mucus, green) and EUB338 universal bacterial FISH probe (red).
(E) Principal coordinate analysis of PhILR Euclidean distances reveal significant clustering of microbial communities in WT and Muc2−/− littermates by diet (P = 0.001, r2= 0.41, ADONIS) but not genotype (P = 0.076, r2= 0.10, ADONIS); n = 3-4 mice per genotype-diet group.
(F) Average fold difference of genera whose abundances are significantly different between HFD and KD (FDR < 0.1, DESeq2), in WT and Muc2−/− mice respectively (n = 3-4 mice per genotype-diet group). Data are presented as mean±SEM in B and C. Each data point represents an individual, singly-housed animal in B, C and E.
Figure S5. Overall bile acid pool is not significantly altered by a ketogenic diet in humans and mice, related to Figures 1 and 3.
(A, B) Concentrations of total, free, and conjugated bile acids (BA) are not significantly different in (A) fecal samples from study participants during the baseline diet (BD) and ketogenic (KD) diet stages, or (B) cecal contents from mice fed a high-fat diet (HFD) or ketogenic diet (KD). Data points represent paired stool samples from study participants during the two diet arms (A) or individual mice (B). Statistical analyses performed using paired (A) or unpaired (B) t tests, ns = not significant.
Figure S6. Growth responses of B. adolescentis and other human gut bacteria to βHB in vitro, related to Figure 5.
(A) Growth curves of B. adolescentis BD1 grown under anaerobic conditions with different concentrations of βHB (left) or neutralized βHB (right), compared to positive control grown without βHB (red).
(B) Colony-forming units (CFUs/ml) in B. adolescentis BD1 cultures prior to and at 3 hrs after addition of 50 mM β-hydroxybutyrate (βHB) or vehicle control (BHI media) at mid-exponential phase. Mean and SEM of quadruplicate cultures shown (**P < 0.01, ns = not significant; Welch’s t-test).
(C) Growth curves of B. adolescentis BD1 grown under anaerobic conditions with different concentrations of butyrate (left) or neutralized butyrate (right), compared to positive control grown without butyrate (red).
(D) Growth curves of B. adolescentis BD1 grown under anaerobic conditions in acidified media matching pH of the respective βHB concentrations in panel A, compared to growth media at pH 7 (positive control, red). Growth curves in A,C,D show mean±SEM of triplicate cultures, with optical density (600 nm) measured at 15 min intervals over 24 hours.
(E) A diverse panel of 35 gut bacterial isolates were incubated with varying concentrations of βHB, and a maximum likelihood phylogenetic tree using 16S rRNA gene sequences was constructed. Heatmap shows fold change of carrying capacity relative to media control at the respective βHB concentrations. V, Verrucomicrobia; P, Proteobacteria; F, Fusobacteria.
(F) The minimum inhibitory concentration required to prevent ≥90% growth (MIC90) of bacterial isolates spanning four major phyla. Bacteroidetes tended to be sensitive relative to the other phyla when tested in vitro (*P < 0.05; Kruskal-Wallis with Dunn’s test). Mean±SEM shown.
(G) Representative growth curves of selected isolates grown under anaerobic conditions with different concentrations of βHB. Growth curves show mean±SEM of triplicate cultures, with optical density (600 nm) measured at 30 min intervals over 24 hours.
Figure S7. Immune changes driven by ketone ester supplementation and ketogenic diets, related to Figures 5 - 7.
(A) Gating strategy for Th17 cell populations.
(B) Flow cytometry analysis of immune cell populations in the small intestine (top panel), large intestine (middle panel) and spleen (bottom panel) of mice fed either a high-fat diet (HFD), ketone ester (KE)-supplemented HFD (HFD-KE) or ketogenic diet (KD) for 3 weeks. Th17 levels are measured as IL-17a+ CD4+ live cells, Th1 levels are measured as IFNγ+ CD4+ live cells, and Treg levels are measured as Foxp3+ CD4+ live cells. The CD62LloCD44hi populations were gated on CD3+ CD4+ cells. Frequencies of the immune cell populations are presented relative to the HFD group which is set to 1 with mean and SEM shown (n = 5-6 mice/group, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; one-way ANOVA with Tukey’s test).
Table S1. List of samples subjected to 16S rRNA gene sequencing, related to Figures 1, 3-6, and S1 - S4.
Table S2. List of human fecal samples subjected to shotgun metagenomic sequencing, related to Figure 2.
Table S3. Relative quantification (relative to total signal intensity) of fecal metabolites by 1H NMR in study participants fed the baseline diet (BD) and ketogenic diet (KD) in our human cohort, related to Figures 1F and S2.
Table S5. Bile acid measurements of fecal samples from human participants fed the baseline diet (BD) or ketogenic diet (KD), and cecal contents from mice fed a high-fat diet (HFD) or ketogenic diet (KD), related to Figures 1 and 3.
Highlights:
Ketogenic diets (KDs) alter the gut microbiota in a manner distinct from high-fat diets
Gut microbial shifts on KDs are driven in part through host production of ketone bodies
β-hydroxybutyrate selectively inhibits bifidobacterial growth
The KD-associated gut microbiota reduces levels of intestinal Th17 cells
Ketogenic diets differ from high fat diets in that they alter the gut microbiome to affect the level of gut Th17 cells
ACKNOWLEDGEMENTS
We thank Jordan Bisanz and the rest of the Turnbaugh lab for advice and feedback, the UCSF Gnotobiotic Core staff (Jolie Ma and Kimberly Ly), KC Huang (Stanford) for help with microscopy, and the CZ Biohub Sequencing Platform. This work was supported by the Nutrition Science Initiative, the NIH (R01HL122593; R21CA227232; P30DK098722), the UCSF SSEW Initiative, and the Searle Scholars Program (SSP-2016-1352). QYA was supported by A*STAR. ADP is supported by the PA Dept of Health using Tobacco CURE funds. PJT holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, is a Chan Zuckerberg Biohub investigator, and is a Nadia’s Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42-16).
Footnotes
DECLARATION OF INTERESTS
JCN and EV are co-founders with equity interest of BHB Therapeutics Ltd, which is developing products related to ketone bodies. PJT is on the scientific advisory boards for Kaleido, Pendulum, Seres, and SNIPRbiome; there is no direct overlap between the current study and these consulting duties. All other authors have no relevant declarations.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, and Stahl DA (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol 56, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol 57, 289–300. [Google Scholar]
- Bisanz JE, Upadhyay V, Turnbaugh JA, Ly K, and Turnbaugh PJ (2019). Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe 26, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang J, Tian Y, Zhang L, Hatzakis E, Krausz KW, Smith PB, Gonzalez FJ, and Patterson AD (2017). Orthogonal comparison of GC-MS and 1H NMR spectroscopy for short chain fatty acid quantitation. Anal. Chem 89, 7900–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. [DOI] [PubMed] [Google Scholar]
- Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, and Everard A (2019). Microbial regulation of organismal energy homeostasis. Nature Metabolism 1, 34–46. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, and Turnbaugh PJ. (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-W, Biton M, Haber AL, Gunduz N, Eng G, Gaynor LT, Tripathi S, Calibasi-Kocal G, Rickelt S, Butty VL, et al. (2019). Ketone body signaling mediates intestinal stem cell homeostasis and adaptation to diet. Cell 178, 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A, and Bonnet R (2014). The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes 5, 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci 14, 927–930. [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, and Sonnenburg JL (2015). Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, et al. (2016). Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol 34, 942–949. [DOI] [PubMed] [Google Scholar]
- Goldberg EL, Shchukina I, Asher JL, Sidorov S, Artyomov MN, and Dixit VD (2020). Ketogenesis activates metabolically protective γδ T cells in visceral adipose tissue. Nature Metabolism 2, 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KD, and Chung ST (2018). Low-carbohydrate diets for the treatment of obesity and type 2 diabetes. Curr. Opin. Clin. Nutr. Metab. Care 21, 308–312. [DOI] [PubMed] [Google Scholar]
- Hall KD, Chen KY, Guo J, Lam YY, Leibel RL, Mayer LE, Reitman ML, Rosenbaum M, Smith SR, Walsh BT, et al. (2016). Energy expenditure and body composition changes after an isocaloric ketogenic diet in overweight and obese men. Am. J. Clin. Nutr 104, 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS (2017). Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185. [DOI] [PubMed] [Google Scholar]
- Johansson MEV, and Hansson GC (2012). Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH In Mucins: Methods and Protocols, McGuckin MA, and Thornton DJ, eds. (Totowa, NJ: Humana Press; ), pp. 229–235. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM, and Round JL (2015). MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff P, and Christensen R (2017). lmerTest Package: tests in linear mixed effects models. Journal of Statistical Software 82, 1–26. [Google Scholar]
- Kverneland M, Selmer KK, Nakken KO, Iversen PO, and Taubøll E (2015). A prospective study of the modified Atkins diet for adults with idiopathic generalized epilepsy. Epilepsy Behav. 53, 197–201. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Wollam J, and Olefsky JM (2018). An integrated view of immunometabolism. Cell 172, 22–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yang Y, Wang Y, Tang H, Zhang F, Zhang Y, and Zhao Y (2018). Ketogenic diet for treatment of intractable epilepsy in adults: A meta-analysis of observational studies. Epilepsia Open 3, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A, Wu H, Bjornson E, Zhang C, Hakkarainen A, Räsänen SM, Lee S, Mancina RM, Bergentall M, Pietiläinen KH, et al. (2018). An integrated understanding of the rapid metabolic benefits of a carbohydrate-restricted diet on hepatic steatosis in humans. Cell Metab. 27, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto J, Ohue-Kitano R, Mukouyama H, Nishida A, Watanabe K, Igarashi M, Irie J, Tsujimoto G, Satoh-Asahara N, Itoh H, et al. (2019). Ketone body receptor GPR43 regulates lipid metabolism under ketogenic conditions. Proc. Natl. Acad. Sci. USA 116, 23813–23821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal EG, Chaffe H, Schwartz RH, Lawson MS, Edwards N, Fitzsimmons G, Whitney A, and Cross JH (2008). The ketogenic diet for the treatment of childhood epilepsy: a randomised controlled trial. Lancet Neurol. 7, 500–506. [DOI] [PubMed] [Google Scholar]
- Newman JC, and Verdin E (2014). Ketone bodies as signaling metabolites. Trends Endocrinol. Metab 25, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Covarrubias AJ, Zhao M, Yu X, Gut P, Ng C-P, Huang Y, Haldar S, and Verdin E (2017a). Ketogenic diet reduces midlife mortality and improves memory in aging mice. Cell Metab. 26, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Kroll F, Ulrich S, Palop JJ, and Verdin E (2017b). Ketogenic diet or BHB improves epileptiform spikes, memory, survival in Alzheimer’s model. bioRxiv. 10.1101/136226 [DOI]
- Ni F-F, Li C-R, Liao J-X, Wang G-B, Lin S-F, Xia Y, and Wen J-L (2016). The effects of ketogenic diet on the Th17/Treg cells imbalance in patients with intractable childhood epilepsy. Seizure 38, 17–22. [DOI] [PubMed] [Google Scholar]
- Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, and Hsiao EY (2018). The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 173, 1728–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, and Strimmer K (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Powell N, Walker MM, and Talley NJ (2017). The mucosal immune system: master regulator of bidirectional gut-brain communications. Nat. Rev. Gastroenterol. Hepatol 14, 143–159. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalska P, and Crawford PA (2017). Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 25, 262–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, and Mazmanian SK (2011).The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury E, Kizirian NV, Partridge SR, Gill T, Colagiuri S, and Gibson AA (2018). Effect of dietary carbohydrate restriction on glycemic control in adults with diabetes: A systematic review and meta-analysis. Diabetes Res. Clin. Pract 139, 239–252. [DOI] [PubMed] [Google Scholar]
- Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJW, Patel VC, Dumas M-E, Holmes E, and Nicholson JK (2015). Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal. Chem 87, 9662–9670. [DOI] [PubMed] [Google Scholar]
- Saslow LR, Daubenmier JJ, Moskowitz JT, Kim S, Murphy EJ, Phinney SD, Ploutz-Snyder R, Goldman V, Cox RM, Mason AE, et al. (2017). Twelve-month outcomes of a randomized trial of a moderate-carbohydrate versus very low-carbohydrate diet in overweight adults with type 2 diabetes mellitus or prediabetes. Nutr. Diabetes 7, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, and Huttenhower C (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JD, Washburne AD, Mukherjee S, and David LA (2017). A phylogenetic transform enhances analysis of compositional microbiota data. Elife 6, e21887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu H-JJ, et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 113, E8141–E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Nichols RG, Roy P, Gui W, Smith PB, Zhang J, Lin Y, Weaver V, Cai J, Patterson AD, et al. (2018). Prebiotic effects of white button mushroom (Agaricus bisporus) feeding on succinate and intestinal gluconeogenesis in C57BL/6 mice. J. Funct. Foods 45, 223–232. [Google Scholar]
- Tian Y, Cai J, Gui W, Nichols RG, Koo I, Zhang J, Anitha M, and Patterson AD (2019). Berberine directly affects the gut microbiota to promote intestinal farnesoid X receptor activation. Drug Metab. Dispos 47, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, Baldi P, and Sassone-Corsi P (2017). Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 26, 523–538.e5. [DOI] [PubMed] [Google Scholar]
- Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, and Augenlicht L (2002). Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729. [DOI] [PubMed] [Google Scholar]
- Volek JS, Phinney SD, Forsythe CE, Quann EE, Wood RJ, Puglisi MJ, Kraemer WJ, Bibus DM, Fernandez ML, and Feinman RD (2009). Carbohydrate restriction has a more favorable impact on the metabolic syndrome than a low fat diet. Lipids 44, 297–309. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, and Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm Y-H, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, D’Agostino D, Planavsky N, Lupfer C, Kanneganti TD, et al. (2015). The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med 21, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Qiu Y, Zhong W, Baxter S, Su M, Li Q, Xie G, Ore BM, Qiao S, Spencer MD, et al. (2013). A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 9, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The ketogenic diet reproducibly alters the structure and function of the human gut microbiota, related to Figure 1.
(A) Principal coordinate analysis of PhILR Euclidean distances reveal distinct clustering of gut microbial communities between baseline diet (BD) and ketogenic diet (KD) stages across study participants, related to Figure 1C. P values were determined using ADONIS tests for the effect of diet within each subject.
(B) Representative 600 MHz NMR spectra of feces from baseline diet (BD) and ketogenic diet (KD) stages with metabolites identification, related to Figure 1F. The regions of δ 5.2-9.0 were vertically expanded 16 times. Keys: 1,Butyrate; 2,Branched-chain amino acids; 3,Propionate; 4,Lactate; 5,Threonine; 6,Alanine; 7,n-Butyrate; 8,Lysine; 9,Arginine; 10,Acetate; 11,Proline; 12,Succinate; 13,Taurine; 14,β-Glucose; 15,Glycine; 16,α-Glucose; 17,Uracil; 18,Fumarate; 19,Tyrosine; 20,Histidine; 21,Phenylalanine; 22,Tryptophan; 23,Xanthine; 24,Hypoxanthine; 25,Formate; 26,Nicotinurate; 27,Uridine; 28,Polyethylene glycol (PEG).
Figure S2. The ketogenic diet results in stable and reproducible shifts in gut microbial composition, related to Figure 3.
(A) Circulating βHB levels of mice fed a low-fat diet (LFD), high-fat diet (HFD) or ketogenic diet (KD) for 10 weeks (n = 8 mice/group, ***P < 0.001; One-way ANOVA with Tukey’s test).
(B) Principal coordinate analysis of PhILR Euclidean distances reveal distinct clustering of KD samples and a significant effect of diet on community composition (P = 0.001, r2= 0.31, ADONIS).
(C) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets, averaged from weekly samples. (n = 8 mice/group with mice housed in pairs, *P < 0.05, **P < 0.01, ***P < 0.001; Kruskal-Wallis with Dunn’s test).
(D) Overall mean caloric intake represented as kcal per mouse per day on the respective diets. Food intake was measured weekly over 11 weeks (n = 8 mice/group with mice housed in pairs, **P < 0.01, ***P < 0.001, ns = not significant; One-way ANOVA with Tukey’s test).
(E) Body weight trajectories of mice fed the respective diets over 11 weeks (n = 8 mice/group). Asterisks denote significant differences from LFD and HFD respectively (PLFDv.HFD = 6.5e-5; PLFDv.KD = 0.049; PHFDv.KD = 0.049). Statistical analysis carried out using linear mixed effects model with MouseID as random effect and Tukey’s test.
(F) Gonadal fat mass in mice fed the respective diets for 11 weeks (n = 8 mice/group, **P < 0.01, ****P < 0.0001; One-way ANOVA with Tukey’s test).
(G) Liver mass in mice fed the respective diets for 11 weeks (n = 8 mice/group, *P < 0.05, ns = not significant; One-way ANOVA with Tukey’s test).
(H) Principal coordinate analysis of fecal samples collected at a single time point from a healthspan and longevity cohort (Newman et al., 2017a) reveals a significant effect of diet on community composition (P = 0.001, r2= 0.33, ADONIS) with long-term feeding of the respective diets (range 28 - 311 days at the time of sample collection). Each data point represents an individual animal (n = 21 - 35 mice/group with 3-5 mice housed per cage).
(I) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets long-term, related to panel H (n = 21 - 35 mice/group, *P < 0.05, ***P < 0.01; Kruskal-Wallis with Dunn’s test). Data are presented as mean±SEM.
Figure S3. The effect of a ketogenic diet on the gut microbiota is replicable with an alternative carbohydrate source enriched for plant polysaccharides, related to Figure 3.
(A) Macronutrient composition (in percent calories) of the chow diet and a series of semi-purified diets containing resistant starch and varying CHO and fat content: low-fat diet (LFD), 45%-fat high-fat diet (HFD45), 75%-fat high-fat diet (HFD75), and ketogenic diet (KD).
(B) Rapid shifts in gut microbial composition are observed within a day of feeding of each diet. Mice were fed each diet for 3 days in a sequential order: Chow → LFD → HFD45 → HFD75 → KD, and dashed vertical lines represent the switch of diets. Axis 1 of principal coordinate analysis (PCo1) on PhILR Euclidean distances is shown over the 15-day study period (n = 4 mice).
(C) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets over 15 days reveal both linear (Bacteroidetes and Firmicutes) and non-linear (Actinobacteria) dose responses to dietary CHO (n = 4 mice).
(D) Abundance of Bifidobacterium in the murine fecal microbiota over study period, showing decrease on KD to a level similar to that of chow diet (n = 4 mice).
(E) Relative abundance of the major phyla in the microbial communities of mice fed the respective diets over 3 weeks in a separate experiment (n = 4 mice/group, *P < 0.05; Kruskal-Wallis with Dunn’s test). Data are presented as mean±SEM. Each data point represents an individual, singly-housed animal.
Figure S4. A ketogenic diet (KD) maintains a robust mucus layer despite the lack of fermentable carbohydrates and gut microbial shift on KD is independent of colonic mucin, related to Figures 2 and 3.
(A) Representative images of distal colon sections from SPF mice fed chow, low-fat diet (LFD), high-fat diet (HFD), or ketogenic diet (KD). Sections were stained with DAPI (epithelium, blue), UEA1 lectin (mucus, green) and EUB338 universal bacterial FISH probe (red). Images were further processed using BacSpace (Earle et al., 2015) and the computed luminal debris appears in yellow.
(B) Average mucus widths in distal colons of SPF mice fed the respective diets after 3 weeks, quantified using BacSpace (Earle et al., 2015) (n = 3 - 5 mice/group, ns = not significant; one-way ANOVA).
(C) Quantitative PCR of colonic Muc2 transcripts reveal no significant difference in Muc2 relative gene expression (n = 3 - 5 mice/group, ns = not significant; one-way ANOVA).
(D) Representative images of distal colon sections from wild-type (WT) and Muc2−/− mice respectively, showing lack of goblet cells and mucus layer in Muc2−/− mice with bacteria in close contact with gut epithelium. Sections were stained with DAPI (epithelium, blue), UEA1 lectin (mucus, green) and EUB338 universal bacterial FISH probe (red).
(E) Principal coordinate analysis of PhILR Euclidean distances reveal significant clustering of microbial communities in WT and Muc2−/− littermates by diet (P = 0.001, r2= 0.41, ADONIS) but not genotype (P = 0.076, r2= 0.10, ADONIS); n = 3-4 mice per genotype-diet group.
(F) Average fold difference of genera whose abundances are significantly different between HFD and KD (FDR < 0.1, DESeq2), in WT and Muc2−/− mice respectively (n = 3-4 mice per genotype-diet group). Data are presented as mean±SEM in B and C. Each data point represents an individual, singly-housed animal in B, C and E.
Figure S5. Overall bile acid pool is not significantly altered by a ketogenic diet in humans and mice, related to Figures 1 and 3.
(A, B) Concentrations of total, free, and conjugated bile acids (BA) are not significantly different in (A) fecal samples from study participants during the baseline diet (BD) and ketogenic (KD) diet stages, or (B) cecal contents from mice fed a high-fat diet (HFD) or ketogenic diet (KD). Data points represent paired stool samples from study participants during the two diet arms (A) or individual mice (B). Statistical analyses performed using paired (A) or unpaired (B) t tests, ns = not significant.
Figure S6. Growth responses of B. adolescentis and other human gut bacteria to βHB in vitro, related to Figure 5.
(A) Growth curves of B. adolescentis BD1 grown under anaerobic conditions with different concentrations of βHB (left) or neutralized βHB (right), compared to positive control grown without βHB (red).
(B) Colony-forming units (CFUs/ml) in B. adolescentis BD1 cultures prior to and at 3 hrs after addition of 50 mM β-hydroxybutyrate (βHB) or vehicle control (BHI media) at mid-exponential phase. Mean and SEM of quadruplicate cultures shown (**P < 0.01, ns = not significant; Welch’s t-test).
(C) Growth curves of B. adolescentis BD1 grown under anaerobic conditions with different concentrations of butyrate (left) or neutralized butyrate (right), compared to positive control grown without butyrate (red).
(D) Growth curves of B. adolescentis BD1 grown under anaerobic conditions in acidified media matching pH of the respective βHB concentrations in panel A, compared to growth media at pH 7 (positive control, red). Growth curves in A,C,D show mean±SEM of triplicate cultures, with optical density (600 nm) measured at 15 min intervals over 24 hours.
(E) A diverse panel of 35 gut bacterial isolates were incubated with varying concentrations of βHB, and a maximum likelihood phylogenetic tree using 16S rRNA gene sequences was constructed. Heatmap shows fold change of carrying capacity relative to media control at the respective βHB concentrations. V, Verrucomicrobia; P, Proteobacteria; F, Fusobacteria.
(F) The minimum inhibitory concentration required to prevent ≥90% growth (MIC90) of bacterial isolates spanning four major phyla. Bacteroidetes tended to be sensitive relative to the other phyla when tested in vitro (*P < 0.05; Kruskal-Wallis with Dunn’s test). Mean±SEM shown.
(G) Representative growth curves of selected isolates grown under anaerobic conditions with different concentrations of βHB. Growth curves show mean±SEM of triplicate cultures, with optical density (600 nm) measured at 30 min intervals over 24 hours.
Figure S7. Immune changes driven by ketone ester supplementation and ketogenic diets, related to Figures 5 - 7.
(A) Gating strategy for Th17 cell populations.
(B) Flow cytometry analysis of immune cell populations in the small intestine (top panel), large intestine (middle panel) and spleen (bottom panel) of mice fed either a high-fat diet (HFD), ketone ester (KE)-supplemented HFD (HFD-KE) or ketogenic diet (KD) for 3 weeks. Th17 levels are measured as IL-17a+ CD4+ live cells, Th1 levels are measured as IFNγ+ CD4+ live cells, and Treg levels are measured as Foxp3+ CD4+ live cells. The CD62LloCD44hi populations were gated on CD3+ CD4+ cells. Frequencies of the immune cell populations are presented relative to the HFD group which is set to 1 with mean and SEM shown (n = 5-6 mice/group, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; one-way ANOVA with Tukey’s test).
Table S1. List of samples subjected to 16S rRNA gene sequencing, related to Figures 1, 3-6, and S1 - S4.
Table S2. List of human fecal samples subjected to shotgun metagenomic sequencing, related to Figure 2.
Table S3. Relative quantification (relative to total signal intensity) of fecal metabolites by 1H NMR in study participants fed the baseline diet (BD) and ketogenic diet (KD) in our human cohort, related to Figures 1F and S2.
Table S5. Bile acid measurements of fecal samples from human participants fed the baseline diet (BD) or ketogenic diet (KD), and cecal contents from mice fed a high-fat diet (HFD) or ketogenic diet (KD), related to Figures 1 and 3.
Data Availability Statement
All sequencing datasets generated in this study are deposited in the NCBI Sequence Read Archive under BioProject PRJNA529487.