Significance
G-protein–coupled receptors (GPCRs) are the target of approximately 34% of the current medications. However, their in vivo assessment remains arduous, mainly due to their structural properties, their low abundance, and the lack of highly specific antibodies. Using proteomics and unique transgenic mouse models, we reveal an in vivo GPCR interactome. We applied this approach to the δ-opioid receptor (DOPr) and identified several interactors from brain tissues. Since this procedure can be transposed to any other receptors, our work provides leads for the discovery of molecular mechanisms regulating GPCR functions in vivo.
Keywords: G-protein–coupled receptors, δ-opioid receptor, mass spectrometry, mouse model, GPCR interactome
Abstract
With over 30% of current medications targeting this family of proteins, G-protein–coupled receptors (GPCRs) remain invaluable therapeutic targets. However, due to their unique physicochemical properties, their low abundance, and the lack of highly specific antibodies, GPCRs are still challenging to study in vivo. To overcome these limitations, we combined here transgenic mouse models and proteomic analyses in order to resolve the interactome of the δ-opioid receptor (DOPr) in its native in vivo environment. Given its analgesic properties and milder undesired effects than most clinically prescribed opioids, DOPr is a promising alternative therapeutic target for chronic pain management. However, the molecular and cellular mechanisms regulating its signaling and trafficking remain poorly characterized. We thus performed liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses on brain homogenates of our newly generated knockin mouse expressing a FLAG-tagged version of DOPr and revealed several endogenous DOPr interactors involved in protein folding, trafficking, and signal transduction. The interactions with a few identified partners such as VPS41, ARF6, Rabaptin-5, and Rab10 were validated. We report an approach to characterize in vivo interacting proteins of GPCRs, the largest family of membrane receptors with crucial implications in virtually all physiological systems.
With over 800 members, G-protein–coupled receptors (GPCRs) form the largest human membrane protein family and are characterized by their unique structure exhibiting an extracellular N terminus domain, seven hydrophobic transmembrane α-helices, alternating intracellular and extracellular loops, and a cytosolic C terminus. Approximately 34% of all drugs actually approved by the US Food and Drug Administration specifically target GPCRs, highlighting their significant pharmacological relevance (1). Following their synthesis in the endoplasmic reticulum, GPCRs undergo several posttranslational modifications ensuring their proper maturation and folding. Once they pass all of the quality control steps leading to their sequential delivery to the Golgi apparatus and the trans-Golgi network (TGN), GPCRs are exported to the plasma membrane where they can be activated by their respective ligands, and thereby elicit specific physiological responses. In order to achieve complete maturation and correct localization, trafficking of GPCRs along the biosynthetic pathway needs to be tightly regulated by a myriad of proteins executing well-defined and specific functions such as chaperones, transport vesicles, and escort proteins (reviewed in ref. 2).
Belonging to the rhodopsin-like class A family of GPCRs, the δ-opioid receptor (DOPr) induces analgesic effects and attenuates pain hypersensitivities in several chronic pain models including neuropathic, inflammatory, diabetic, and cancer pain (3, 4), while producing considerably less undesired effects than most clinical opioid therapeutics (5–7). Moreover, DOPr activation has been associated with anxiolytic, antidepressant, as well as cardioprotective and neuroprotective effects, thus rendering it an attractive therapeutic target for chronic pain management (8). Interestingly, we and others have previously observed a correlation between the translocation of DOPr from the intracellular compartments to the plasma membrane and the extent to which DOPr agonists mediated analgesic effects (9–11). However, the in vivo mechanisms and regulatory proteins involved in DOPr trafficking and signaling remain largely unknown and a thorough understanding of these cellular processes is crucial in order to develop potent and better-tolerated analgesics.
Due to their low expression levels and the lack of specific antibodies, the study of protein–protein interactions of endogenous GPCRs still represents a challenging task (12, 13). Therefore, in order to study the molecular and cellular mechanisms involved in DOPr regulation in vivo, we developed and characterized a transgenic knockin (KI) mouse expressing a FLAG-tagged version of the DOPr replacing the endogenous receptor. Our findings show that the FLAG-DOPr from KI animals is expressed at similar levels and in the same brain areas as the wild-type (WT) receptor, allowing us to identify endogenous interacting proteins of DOPr in the central nervous system (CNS) by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis on immunoprecipitated FLAG-DOPr. A distinguishing feature of this in vivo proteomic analysis is that the GPCR is expressed under the control of its endogenous promoter. We provide a list of potential DOPr-interacting partners identified using our newly generated FLAG-DOPr-KI mouse and validate the functional implication of one of the identified partners, Rab10, on the trafficking of DOPr. Our findings open a path to the study of protein–protein interactions and signal transduction of endogenous GPCRs occurring in vivo, notably the DOPr in pain-related pathways.
Results
Generation of FLAG-DOPr Knockout and Knockin Mice.
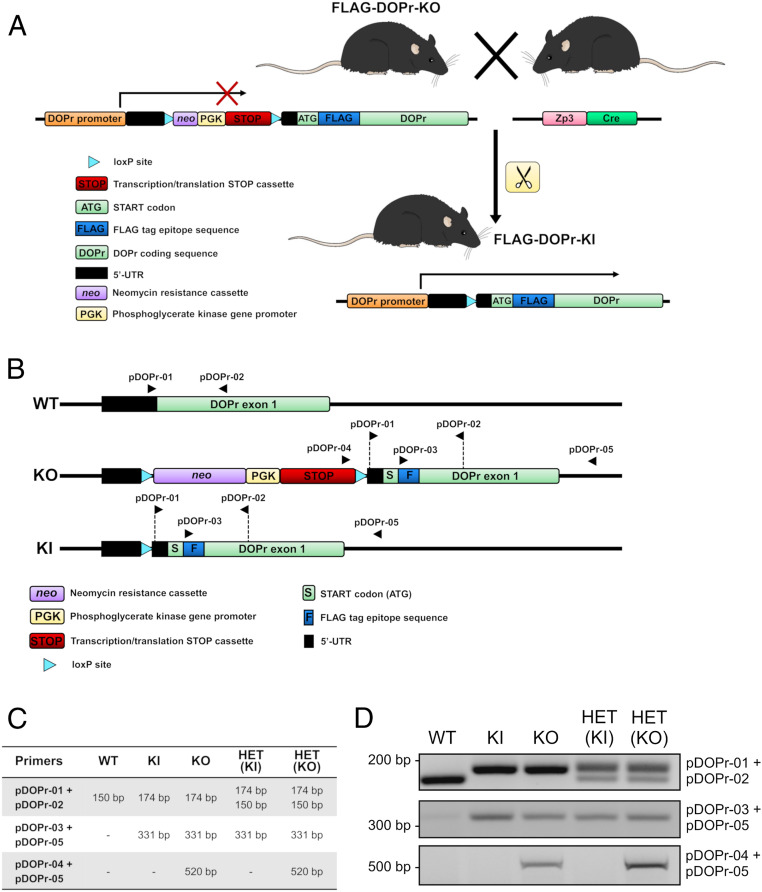
For the purpose of this study, we generated a KI mouse in which the endogenous WT receptor is being replaced by a N-terminal flagged version of DOPr. Using a classical homologous recombination-based approach, a sequence encoding the FLAG-tag epitope was inserted into the open-reading frame, following the initiation codon at the 5′-end of DOPr first exon, along with a translational neomycin STOP cassette in the 5′-untranslated region of the OPRD1 gene. Mice expressing these genotypic characteristics are henceforth referred to as FLAG-DOPr-KO or KO (Fig. 1A). The KI mouse line (or FLAG-DOPr-KI) was then generated by breeding KO mice with Zp3-Cre mice, resulting in the excision of the sequence comprised between the two loxP sites, and thereby allowing the expression of the FLAG-tagged DOPr in tissues that would normally express the WT DOPr (Fig. 1A). Genotypes were confirmed by PCR using various combinations of primers (Fig. 1 B–D), and sequencing of the relevant regions of the genomic DNA from KO and KI mice was also performed in order to certify the integrity of each sequence.
Fig. 1.
Generation of FLAG-DOPr-KI mice. (A) Schematic representation of the generation of FLAG-DOPr-KI mice obtained by breeding FLAG-DOPr-KO mice with Zp3-Cre mice. In KO mice, a sequence encoding the FLAG tag epitope was introduced immediately after the START codon, in the 5′-end of the DOPr coding sequence. A translational STOP cassette flanked by two loxP sites was also inserted in the 5′-untranslated region (5′-UTR) of the OPRD1 gene to disable the expression of FLAG-DOPr. Breeding with Zp3-Cre mice enables the excision of the STOP cassette, therefore allowing the expression of FLAG-DOPr instead of the endogenous DOPr in all tissues normally expressing DOPr. Genotypes were confirmed by PCR using various combinations of primers (B–D). For each genotype, primer locations are indicated with black arrowheads (B), and the expected lengths of the fragments generated by PCR are shown (C). The complete sequence of each primer can be found in SI Appendix, Table S5. (D) The PCR amplification using pDOPr-01 and 02 (Upper) distinguishes between WT (150 bp), homozygous transgenic (174 bp), and heterozygous (HET) (150 and 174 bp) mice, while the amplification using the specific sequence encoding the FLAG tag epitope resulted in a band of 331 bp for all genotypes except WT (pDOPr-03 and 05; Middle). The presence of an intact STOP cassette is shown by a band of 520 bp in KO and HET(KO) mice (pDOPr-04 and 05, Bottom).
Expression and Distribution of FLAG-DOPr in KI Mice.
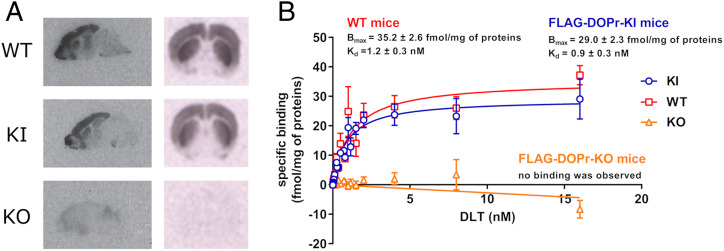
To examine the expression pattern of FLAG-DOPr in the brain of KI mice, binding assays were carried out on brain slices using 125I-Deltorphin I (125I-DLT), a selective DOPr agonist. The results showed that FLAG-DOPr was predominantly expressed in the forebrain, which includes the cortex, the olfactory bulb, the amygdala, and the striatum, which corresponds to the distribution pattern observed for the endogenous DOPr in WT mice (Fig. 2A). The specificity of DOPr labeling and localization is supported by the absence of 125I-DLT binding in brain slices from KO mice (Fig. 2A). Additional coronal brain sections of WT, KI, and KO are shown in SI Appendix, Fig. S1A. The affinity (Kd) and the Bmax of FLAG-DOPr expressed in KI mice were then measured by saturation binding assays using brain membrane preparations from WT, KO, and KI mice (Fig. 2B). For KI mice, the values obtained for Kd and Bmax were 0.9 ± 0.3 nM and 29.0 ± 2.3 fmol/mg of proteins, respectively, which was not significantly different from WT animals (Kd = 1.2 ± 0.3 nM and Bmax = 35.2 ± 2.6 fmol/mg of proteins). No specific binding was detected in KO mice. Specificity of DOPr KO was also assessed by comparing the binding patterns of 125I-DLT and 125I-DAMGO, a selective μ-opioid receptor (MOPr) agonist, in mouse brain sections. The results showed that 125I-DLT–specific binding was absent in KO mice, whereas binding of 125I-DAMGO to MOPr remained unchanged when WT and KO mice were compared (SI Appendix, Fig. S1 B and C).
Fig. 2.
Expression and distribution of FLAG-DOPr in the CNS of KI mice. (A) Autoradiography of 125I-DLT binding on sagittal and coronal brain sections of WT, KI, and KO mice. (B) Pharmacological properties (Kd and Bmax) of DOPr (either WT or FLAG-DOPr) expressed in WT (red squares), KI (blue circles), and KO mice (orange triangles) were evaluated with saturation binding assays on brain membranes using 125I-DLT. In the absence of residual specific binding in KO mice, Kd and Bmax were not calculated. Results are the mean ± SEM of four independent experiments, each performed in triplicates.
Characterization of FLAG-DOPr Activation in KI Mice.
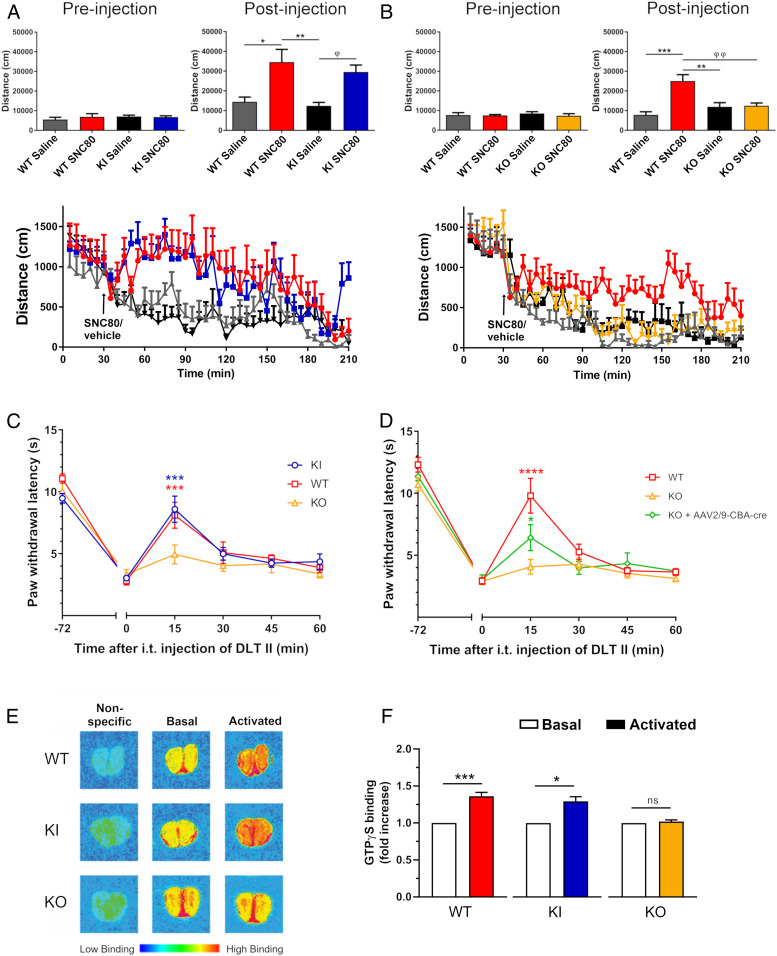
To assess in vivo functional and physiological activation of FLAG-DOPr in KI mice, we first examined the effects of DOPr activation in locomotor and nociceptive tests. Previous studies reported an increase in locomotor activity following subcutaneous (s.c.) administration of 10 mg/kg of the DOPr agonist SNC80 (14, 15). Locomotor activity of WT, KI, and KO animals was therefore evaluated by measuring the total distance traveled following administration of SNC80. The selective DOPr agonist induced locomotor hyperactivity in WT and KI mice to a similar extent (Fig. 3A) but not in KO mice (Fig. 3B). Furthermore, since DOPr activation is associated with antinociceptive properties in mice (3, 4, 8), we characterized the nociceptive behavioral response to thermal stimuli. Using the complete Freund’s adjuvant (CFA) chronic pain model, our data showed similar thermal antihyperalgesic effects in WT and KI animals following intrathecal administration of 1 µg of Deltorphin II (DLT II) (Fig. 3C). Conversely, spinal administration of DLT II in KO mice had no effect (Fig. 3C). Another important feature of this KO mouse line resides in the fact that the translational STOP cassette is flanked by two loxP sites, thereby allowing the excision of the sequence using the Cre/LoxP system (Fig. 1A). In this model, the Cre-driven removal of the translational STOP cassette makes it possible to rescue the expression of DOPr in a tissue- and/or cell-specific manner. We have previously demonstrated that the recombinant adenoassociated virus rAAV2/9-CBA-Cre-GFP predominantly targeted dorsal root ganglia (DRGs) following intrathecal administration (16). Interestingly, when administered to KO mice, our results showed that the antihyperalgesic effects of DLT II were partially reinstated 6 wk after the viral infection, supporting a role for DOPr expressed in primary afferents in the control of pain induced by a thermal stimulus (Fig. 3D).
Fig. 3.
Functional activation of FLAG-DOPr in KI mice. SNC80 (10 mg/kg; s.c.)-induced locomotor activity was assessed in WT, (A) KI (WT, n = 5; KI, n = 6; *P = 0.0102, **P = 0.0032, and φP = 0.0170; one-way ANOVA followed by Tukey’s post hoc test), and (B) KO mice (WT, n = 5–7; KO, n = 6; **P = 0.0038, ***P = 0.0004, and φφP = 0.0056; one-way ANOVA followed by Tukey’s post hoc test). DLT II (1 µg; i.t.)-induced antihyperalgesic effects was evaluated in WT, KO, and (C) KI mice using the Hargreaves test and the CFA model of inflammation (WT, n = 11 and ***P = 0.0005; KI, n = 7 and ***P = 0.0004; two-way ANOVA followed by Tukey’s post hoc test compared to KO [n = 11]) or (D) KO mice intrathecally injected with the recombinant adenoassociated virus AAV2/9-CBA-Cre (WT, n = 11 and ****P < 0.0001; KO + AAV2/9-CBA-Cre, n = 8 and *P = 0.0330; two-way ANOVA followed by Tukey’s post hoc test compared to KO [n = 8]). (E and F) The integrity of the G-protein coupling of DOPr (and FLAG-DOPr) was evaluated using the 35S-GTPγS binding assay on coronal sections of olfactory bulb (20 µm) from WT, KI, and KO mice. 35S-GTPγS binding density is shown in color scale (E). Quantification of binding signal was carried out on olfactory bulb sections (WT, n = 7 and ***P = 0.0004; KO, n = 4 and P = 0.1457; KI, n = 4 and *P = 0.0182; one-sample Wilcoxon test) from three independent experiments. Data are expressed as fold increase in GTPγS binding for nontreated (basal) and DLT II-treated sections (activated). Nonspecific binding was assessed in the presence of 10 µM GTPγS. n.s., not significant.
Furthermore, the function of DOPr was also determined by 35S-GTPγS binding experiments on olfactory bulb sections of WT, KI, and KO mice following stimulation of DOPr with DLT II, as we described previously (17). As shown in Fig. 3E, nonspecific and basal activities were not different between WT, KI, and KO mice. However, incubation of olfactory bulb sections with 10 µM DLT II led to a significant increase in 35S-GTPγS binding in brain slices of WT and KI mice, but not in KO animals, where activity was similar to the basal level (Fig. 3 E and F). Altogether, these results indicate that FLAG-DOPr expressed in KI mice exhibits pharmacological properties, expression levels, and distribution patterns, as well as physiological functions that match those of the WT endogenous receptor. This establishes the FLAG-DOPr-KI mouse as a physiologically relevant model to study the regulation of the FLAG-tagged receptor in vivo under endogenous control of expression, as well as a useful tool to study DOPr function in distinct cell populations.
Identification of Potential Interaction Partners of DOPr by Liquid Chromatography–Tandem Mass Spectrometry Analysis.
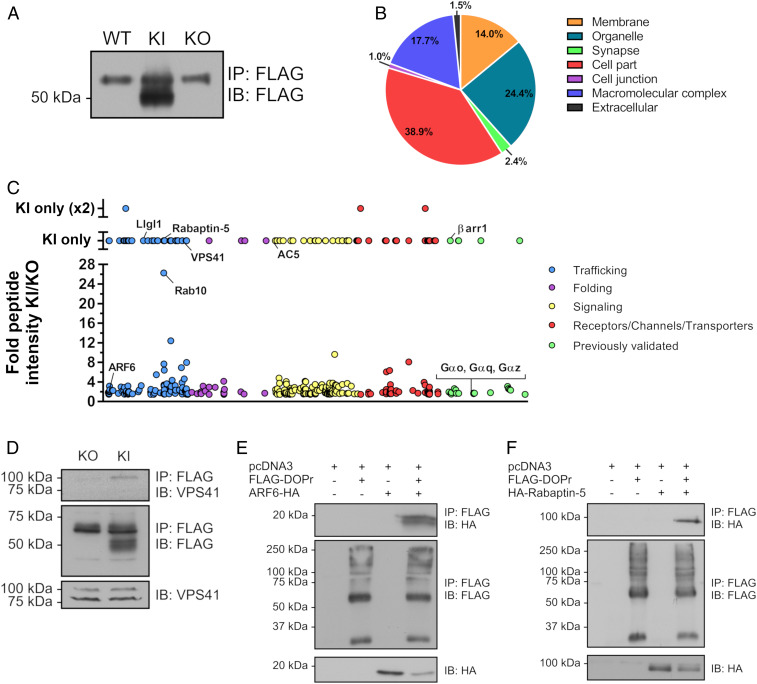
As mentioned above, despite their therapeutic potential, the proteins interacting with GPCRs in vivo remain to be identified. In this regard, we sought to identify novel in vivo DOPr-interacting partners using our FLAG-DOPr-KI mouse model. Using anti-FLAG M2 monoclonal antibodies coupled to magnetic beads, we immunoprecipitated FLAG-DOPr from homogenates of mouse brains. As shown in Fig. 4A, FLAG-DOPr (∼50 kDa) was successfully immunoprecipitated from KI mouse forebrain lysates but was not detected in the immunoprecipitates of WT and KO mice brain homogenates. The upper band (∼60 kDa) is thought to represent nonspecific antibody binding since it was also observed in the WT and KO lanes. For the identification of proteins that interact with DOPr, immunoprecipitation of FLAG-DOPr from brain lysates of five KI mice was performed, followed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. As a control, the same procedure was simultaneously carried out using brains from KO mice. Candidate interacting proteins were considered as positive hits when the ratio of peptide intensity KI/KO displayed at least a 1.5-fold increase. This protocol was performed twice, and the positive hits from both LC-MS/MS analyses were combined. Using the PANTHER Classification System, the positive hits were clustered into seven different cellular components (Fig. 4B). A large proportion of the identified interacting proteins were associated with the compartment named cell part (38.9%), which includes predominantly cytoplasmic and vesicle-associated proteins, but also with membranes (14.0%) and various organelles (24.4%). Candidates of interest were further classified according to their reported functions (Fig. 4C), covering intracellular trafficking (SI Appendix, Table S1), protein folding (SI Appendix, Table S2), signal transduction (SI Appendix, Table S3), and proteins belonging to the receptors/transporters/channels family (SI Appendix, Table S4).
Fig. 4.
Identification of potential DOPr-interacting proteins from brains of FLAG-DOPr-KI mice. (A) Immunoprecipitation of FLAG-DOPr (∼50-kDa band) was performed on forebrain lysates from WT, KI, and KO mice using a M2 mouse monoclonal anti-FLAG antibody immobilized on magnetic beads. Immunoblotting was carried out using a rabbit polyclonal anti-FLAG antibody. Following LC-MS/MS analysis of immunoprecipitated FLAG-DOPr from the forebrain of KI mice, identified DOPr-interacting proteins were classified according to their cellular localization using the PANTHER Classification System online tool (B) and reported molecular functions covering intracellular trafficking (blue), folding (purple), signal transduction (yellow), and proteins belonging to the receptor/channel/transporter families (red) (C). Previously reported DOPr interactors are identified in green (C). In order to validate an endogenous interaction, immunoprecipitation of FLAG-DOPr from brain lysates of KO and KI mice was carried out using a M2 mouse monoclonal anti-FLAG antibody immobilized on magnetic beads and immunoblotting was performed using a rabbit polyclonal anti-VPS41 antibody (D). Further validation of DOPr-interacting proteins was assessed in lysates of HEK293 cells transiently expressing the human FLAG-DOPr and (E) ARF6-HA or (F) HA-Rabaptin-5 using a M2 mouse monoclonal anti-FLAG antibody. Immunoblotting of the receptor was carried out with FLAG-specific rabbit polyclonal antibody (D–F), and ARF6-HA and HA-Rabaptin-5 were immunoblotted using HA-specific HRP-conjugated antibodies. (A and D–F) Blots shown are representative of at least three independent experiments. IB, immunoblotting; IP, immunoprecipitation.
Validation of DOPr-Interacting Partners.
In order to validate our data, we compared the positive hits obtained from our LC-MS/MS analyses with other studies that used MS analyses, coimmunoprecipitation (coIP), and bioluminescence resonance energy transfer (BRET) to identify proteins interacting with DOPr. We confirmed 24 proteins that were previously reported by us, and others, to interact with DOPr, including β-arrestin-1 (18, 19), the cannabinoid receptor 1 (20, 21), heterotrimeric G proteins (22), the cyclin-dependent–like kinase 5 (23), calnexin (24), as well as subunits of the coatomer (25, 26) and the V-type proton ATPase complexes (27) (Table 1).
Table 1.
Validation of DOPr interactors identified by LC-MS/MS analysis performed on KI mice
| Gene symbol | Protein identity | N/2 | Fold intensity KI/KO | Unique peptide | Coverage (%) | Method | References |
| Proteins appearing in both experiments with an intensity ratio of at least 1.5-fold | |||||||
| ARF6 | ADP ribosylation factor 6 | 2 | 2.3–2.0 | 2–4 | 18.3–33.1 | CoIP | This study (Fig. 4E) |
| ARRB1 | β-Arrestin-1 | 2 | 3.0 – N/A | 2–2 | 3.3–10.0 | CoIP | Refs. 18 and 19 |
| ATP1A2 | Sodium/potassium transporting ATPase subunit α-2 | 2 | 1.6–1.7 | 18–18 | 35.7–36.1 | CoIP | 83 |
| COPA | Coatomer subunit α | 2 | 1.5–1.9 | 11–12 | 13.0–13.2 | MS | Ref. 25 |
| CNR1 | Cannabinoid receptor 1 | 2 | N/A – 1.9 | 2–1 | 6.3–3.3 | BRET/coIP | Refs. 20 and 21 |
| RAB10 | Ras-related protein Rab-10 | 2 | 51.0–1.5 | 2–2 | 17.0–23.0 | CoIP | This study (Fig. 5A) |
| Proteins appearing in KI group only | |||||||
| COPE | Coatomer subunit ε | 1 | N/A | 2 | 21.4 | MS | Ref. 25 |
| ITSN2 | Intersectin-2 | 1 | N/A | 1 | 11.6 | APEX-MS | Ref. 27 |
| RABEP1 | Rab GTPase-binding effector protein 1 | 1 | N/A | 1 | 6.6 | CoIP | This study (Fig. 4F) |
| TFRC | Transferrin receptor protein 1 | 1 | N/A | 1 | 1.7 | APEX-MS | Ref. 27 |
| VPS41 | Vacuolar protein sorting-associated protein 41 homolog | 1 | N/A | 1 | 1.8 | CoIP | This study (Fig. 4D) |
| Proteins appearing with an intensity ratio ≥1.5-fold | |||||||
| ARCN1 | Coatomer subunit δ | 1 | 1.9 | 3 | 6.7 | MS | Ref. 25 |
| ATP2A2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 1 | 1.7 | 30 | 31.0 | CoIP | Ref. 84 |
| ATP6V1A | V-type proton ATPase catalytic subunit A | 1 | 1.5 | 28 | 57.1 | APEX-MS | Ref. 27 |
| ATP6V1E1 | V-type proton ATPase catalytic subunit E1 | 1 | 1.8 | 5 | 35.4 | APEX-MS | Ref. 27 |
| CANX | Calnexin | 1 | 2.6 | 7 | 15.2 | CoIP | Ref. 24 |
| CCT7 | T-complex protein 1 subunit eta | 1 | 2.4 | 13 | 35.6 | CoIP | Ref. 56 |
| CDK5 | Cyclin-dependent-like kinase 5 | 1 | 1.7 | 9 | 30.8 | CoIP | Ref. 23 |
| GNAO1 | Guanine nucleotide-binding protein G(o) subunit α | 1 | 1.6 | 1 | 39.5 | CoIP | Ref. 22 |
| GNAQ | Guanine nucleotide-binding protein G(q) subunit α | 1 | 1.7 | 4 | 27.6 | CoIP | Ref. 22 |
| GNAZ | Guanine nucleotide-binding protein G(z) subunit α | 1 | 1.7 | 8 | 32.1 | CoIP | Ref. 22 |
| GPM6A | Neuronal membrane glycoprotein M6 | 1 | 1.7 | 2 | 6.7 | BRET | Ref. 85 |
| NAPG | γ-Soluble NSF attachment protein | 1 | 1.8 | 9 | 35.9 | APEX-MS | Ref. 27 |
| RPN1 | Ribophorin I | 1 | 3.1 | 5 | 11.4 | CoIP | Ref. 86 |
| RRAGC | Ras-related GTP-binding protein C | 1 | 2.6 | 2 | 5.5 | APEX-MS | Ref. 27 |
| SCAMP3 | Secretory carrier-associated membrane protein 3 | 1 | 2.4 | 2 | 13.0 | APEX-MS | Ref. 27 |
| SEC31A | Protein transport protein Sec31A | 1 | 2.3 | 3 | 3.5 | APEX-MS | Ref. 27 |
| VAPA | Vesicle-associated membrane protein-associated protein A | 1 | 1.5 | 2 | 20.5 | CoIP | Ref. 87 |
List of interactions between FLAG-DOPr immunoprecipitated from the forebrains of KI mice and identified proteins by LC-MS/MS analysis that have already been reported by us and others. A protein was considered as a positive hit when the ratio of peptide intensity KI/KO displayed at least a 1.5-fold increase. The number of experiments in which a protein was considered positive (N/2) is shown, and results from both experiments are separated by a hyphen when applicable. N/A indicates that peptides were detected only in KI condition. APEX-MS, engineered ascorbate peroxidase proximity labeling coupled with MS; BRET, bioluminescence resonance energy transfer; coIP, coimmunoprecipitation; MS, mass spectrometry.
As opposed to the other members of the opioid receptor family, and GPCRs in general, DOPr exhibits particular trafficking properties, contributing to our lack of knowledge regarding its regulation. First, although it has been shown to recycle after endocytosis, DOPr preferentially traffics to lysosomal compartments, resulting in its down-regulation (28). Interestingly, we have identified the vacuolar protein sorting-associated protein 41 homolog (VPS41), which is involved in the vesicle-mediated protein trafficking to lysosomal compartments (29), as a DOPr-interacting protein in our LC-MS/MS analyses, specifically in the KI condition (Fig. 4C and SI Appendix, Table S1). We confirmed the interaction between VPS41 and DOPr at the endogenous level in vivo by immunoprecipitation of FLAG-DOPr from brain lysates of KO and KI mice, followed by Western blot analysis using an anti-VPS41 antibody (Fig. 4D), thereby confirming our LC-MS/MS results. Further validation of our proteomic data was performed using coIP experiments of human orthologs of FLAG-DOPr and three newly identified interactors tagged with an HA epitope expressed in HEK293 cells. Fig. 4 E and F show that ADP ribosylation factor 6 (ARF6), which was previously reported to regulate the trafficking and signaling of MOPr (30), and Ras GTPase-binding effector protein 1 (also known as Rabaptin-5), involved in the regulation of endosome morphology and functions (31), coimmunoprecipitated with DOPr.
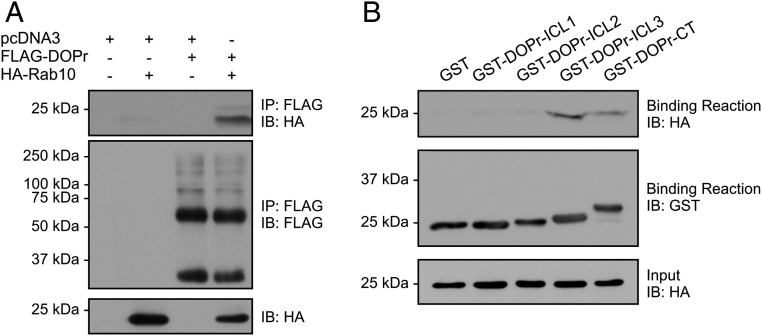
Although this remains controversial, another unique feature of the DOPr concerns its intracellular localization (extensively reviewed and discussed in ref. 32). Interestingly, we and others have shown that, under specific conditions, such as chronic morphine administrations or inflammatory pain, the cell surface targeting of DOPr is increased, which is also correlated with an augmentation in DOPr agonists-mediated analgesia (9–11). Therefore, among the newly identified interactors, Rab10 was of a particular interest since it was previously shown to regulate the cell surface delivery of several membrane proteins (33, 34), including the somatostatin receptor subtype 2 (SSTR2) (35), and because it displayed at least a 1.5-fold peptide intensity ratio KI/KO in both LC-MS/MS experiments (Fig. 4C and SI Appendix, Table S1). Noteworthy, a set of proteins related to the functional network of Rab10, such as Llgl1, EHBP1, and members of the exocyst complex (EXOC2, 3, 4, 5, and 8), were also identified in our proteomic analyses. The interaction between Rab10 and DOPr was confirmed in HEK293 cells (Fig. 5A). Moreover, we and others have previously shown that GPCRs can interact directly with Rab GTPases (36–39). In order to determine whether DOPr and Rab10 can interact directly, we generated and purified recombinant proteins of the three intracellular loops (ICL1, 2, 3) and the C-terminal tail (CT) of the human DOPr fused to GST, as well as an hexahistidine-tagged HA-Rab10 construct. As shown in Fig. 5B, GST-pulldown assays carried out using purified proteins indicated that Rab10 can bind directly to DOPr intracellular domains. The ICL3 and the CT of DOPr appeared to be the major molecular determinants for this interaction.
Fig. 5.
Identification of the ICL3 and CT of DOPr as the major molecular determinants for the interaction with Rab10. (A) Immunoprecipitation of FLAG-DOPr was performed in HEK293 cells transiently expressing FLAG-DOPr and HA-Rab10 using a M2 mouse monoclonal anti-FLAG antibody. (B) In vitro binding assays were carried out using purified GST or the DOPr intracellular loops (ICLs) or C terminus as GST-fusion proteins incubated with purified recombinant (His)6-HA-Rab10. Sequences of the GST-fused protein constructs are listed in SI Appendix, Table S6. Rab10 binding to the receptor was detected by immunoblotting using HA-specific HRP-conjugated antibody, and the GST fusion proteins present in the binding reaction were detected using an anti-GST polyclonal antibody.
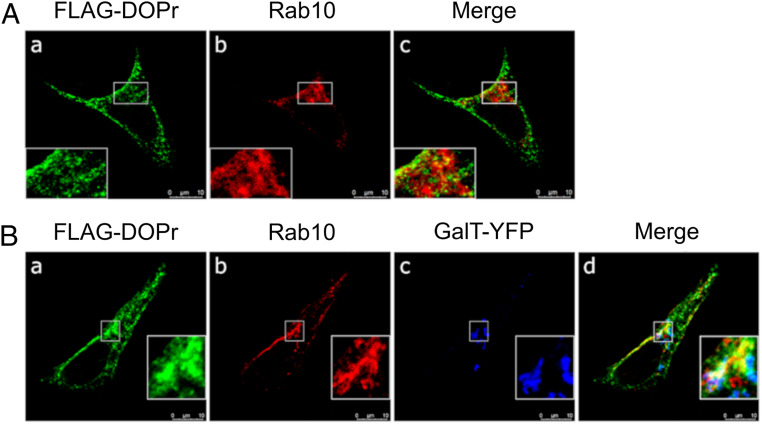
Colocalization between Rab10 and DOPr.
Our previous confocal microscopy studies showed that DOPr partially colocalized with the coatomer subunit β (β-COP) in Golgi-associated structures (25). Rab10 distribution was also described to overlap with β-COP in the perinuclear region (40). Confocal microscopy experiments performed in HEK293 cells revealed intracellular colocalization of FLAG-DOPr (green, Fig. 6A, a) and endogenous Rab10 (red, Fig. 6A, b) in the perinuclear region, as seen by the yellow pixels in Fig. 6A, c. Chen et al. (40) suggested that Rab10 may reside in the distal part of the Golgi apparatus. The possibility that DOPr and Rab10 colocalized in the trans-Golgi was therefore investigated using the galactosyltransferase enzyme (GalT) as a marker of the Golgi apparatus trans cisternae (41, 42). Using triple labeling, we observed that FLAG-DOPr (green, Fig. 6B, a), endogenous Rab10 (red, Fig. 6B, b), and GalT-YFP (blue, Fig. 6B, c) partly colocalized in HEK293 cells in a perinuclear structure resembling the Golgi (Fig. 6B, d, triple colocalization of FLAG-DOPr, Rab10, and GalT is evidenced by the white pixels). Interestingly, GalT-independent colocalization between Rab10 and FLAG-DOPr is also visible (yellow, Fig. 6B, d).
Fig. 6.
Rab10 colocalizes with DOPr in Golgi-associated compartments. HEK293 cells transiently transfected with FLAG-DOPr alone (A) or together with GalT-YFP (B) a marker of the Golgi apparatus trans cisternae were fixed and prepared as described in SI Appendix. FLAG-DOPr was labeled with a rabbit polyclonal anti-FLAG antibody and an Alexa Fluor 594 goat anti-rabbit IgG antibody (a, green). Endogenous Rab10 was labeled with a specific mouse monoclonal anti-Rab10 antibody and an Alexa Fluor 647 goat anti-mouse IgG antibody (b, red). GalT-YFP was labeled with a chicken anti-GFP antibody and an Alexa Fluor 488 goat anti-chicken IgY antibody (B, c, blue). In merge panels, colocalization between FLAG-DOPr and Rab10 appears in yellow (A, c, and B, d), and triple colocalization between FLAG-DOPr, Rab10, and GalT-YFP appears in white (B, d). The images shown are single confocal slices and are representative of ∼100 observed cells over three independent experiments. (Scale bars, 10 µm.)
Rab10 Regulates Cell Surface Expression and Agonist-Induced Trafficking of DOPr.
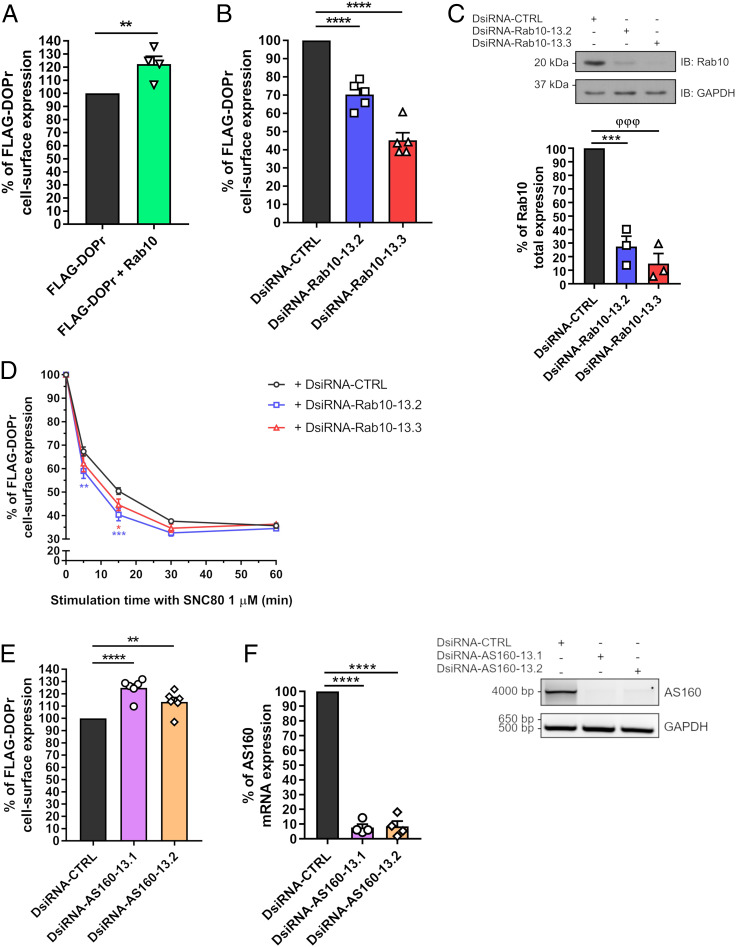
Since Rab10 was previously reported to be involved in the cell-surface delivery of membrane proteins such as the Toll-like receptor 4 (TLR4) and GLUT4 (33, 34), we investigated its role in DOPr expression at the cell surface. As shown in Fig. 7A, overexpression of Rab10 in HEK293 cells increased the cell surface expression of FLAG-DOPr by 22.3 ± 6.0%. Conversely, down-regulation of Rab10 expression by two distinct Dicer-substrate siRNAs (DsiRNAs) led to a significant reduction in DOPr cell surface expression by 30–55% in HEK293 cells stably expressing FLAG-DOPr (Fig. 7B). The effectiveness of Rab10 expression knockdown by both DsiRNAs is shown in Fig. 7C. Thereafter, HEK293 cells stably expressing FLAG-DOPr were transfected with control or Rab10 DsiRNAs, and DOPr cell surface expression was evaluated following stimulation with SNC80. As shown in Fig. 7D, after 15 min, the SNC80-induced decrease in DOPr cell surface expression was amplified by the down-regulation of Rab10, thereby supporting a role for Rab10 in the agonist-induced trafficking of DOPr. To get further mechanistic insights into the context of Rab10 involvement in DOPr trafficking, we investigated the role of AS160, a Rab10 GTPase-activating protein (GAP) also known as TBC1D4, in DOPr cell surface expression using DsiRNAs. As shown in Fig. 7E, a significant increase in the effect of Rab10 on DOPr cell surface expression was observed when AS160 DsiRNAs were transfected along with Rab10, suggesting that AS160 is involved in the mechanisms underlying DOPr membrane targeting by Rab10. The effectiveness of AS160 knockdown by both DsiRNAs is shown in Fig. 7F.
Fig. 7.
Rab10 and AS160 regulate DOPr trafficking. (A) HEK293 cells were transiently transfected with FLAG-DOPr alone or with HA-Rab10 for 48 h. Cell surface expression was measured by ELISA with FLAG-specific rabbit polyclonal and alkaline phosphatase-conjugated goat anti-rabbit antibodies. Results are shown as a percentage of cell surface FLAG-DOPr expression when HA-Rab10 is transfected compared with FLAG-DOPr alone set at 100% (n = 4, **P = 0.0100, two-tailed Student’s t test). (B–F) HEK293 cells stably expressing FLAG-DOPr were transfected with a control DsiRNA (DsiCTRL), (B–D) DsiRNAs targeting the exon 6 or 3 of the human Rab10 gene (DsiRNA-Rab10-13.2 or DsiRNA-Rab10-13.3, respectively), or (E and F) DsiRNAs targeting the exon 3 or 15 of the human AS160 gene (DsiRNA-AS160-13.1 or DsiRNA-AS160-13.2, respectively) for 72 h along with HA-Rab10 for 48 h. (B and E) Cell surface expression of FLAG-DOPr was measured as described above. Results are shown as a percentage of cell surface receptor expression when DsiRNAs targeting Rab10 or AS160 are transfected compared with the DsiCTRL condition set at 100% (B: n = 5, ****P < 0.0001; E: n = 6, **P = 0.0076 and ****P < 0.0001; one-way ANOVA with Dunnett’s multiple-comparisons test). (C) Total cell lysates were immunoblotted with a specific mouse monoclonal anti-Rab10 antibody to assess the effect of Rab10 DsiRNAs on total expression of Rab10. (F) Following RNA extraction, RT-PCRs were performed to assess the effect of AS160 DsiRNAs on total expression of AS160. Primers used are listed in SI Appendix, Table S5. Densitometry was performed using ImageJ software to quantify relative expression of Rab10 or AS160 normalized with GAPDH expression, and results were analyzed using one-way ANOVA with Dunnett’s multiple-comparisons test (C: n = 3, ***P = 0.0003 and φφφP = 0.0001; F: n = 4, ****P < 0.0001). IB, immunoblotting. (D) Cells were stimulated with SNC80 (1 µM) for up to 60 min, and cell surface expression of FLAG-DOPr was measured by ELISA as mentioned above (n = 4, *P = 0.0285, **P = 0.0017 and ***P = 0.0001; two-way ANOVA with Dunnett’s multiple-comparisons test).
Discussion
Despite their significant pharmacological importance, GPCRs remain among the most challenging targets to investigate. In addition to their transmembrane structure and diverse mechanisms of action, their localization and low endogenous expression levels contribute to the complexity of assessing this class of proteins. The current lack of highly potent and specific antibodies further limits the study of endogenous receptor protein–protein interactions and signaling, impeding our understanding of their biology in physiologically relevant conditions. In order to overcome this technical limitation, several groups relied on approaches based on epitope-tagged GPCRs expressed in heterologous systems. In vivo, only a few examples have thus far been reported. Indeed, GPCR KI mouse lines that have been generated and characterized include the HA-α2a adrenergic receptor (43), the nociceptin/orphanin FQ (NOP)-eGFP receptor (44), DOPr-eGFP (45), MOPr-mcherry (46), Rhodopsin-eGFP (47), and HA-DOPr (48). These studies investigated the expression, distribution, internalization, or functional responses by visualizing a specific GPCR using fluorescent proteins or, less frequently, antibodies. Here, using a KI mouse line, we report the proteomic analysis of a GPCR, namely the DOPr, expressed under endogenous control.
In the present study, we aimed to identify new protein partners involved in the regulation of DOPr in vivo. To this purpose, we first generated and characterized two mice models, namely the FLAG-DOPr-KO and FLAG-DOPr-KI mice, the latter being derived from the former following breeding rounds with Zp3-Cre mice. The analysis of the 125I-DLT binding patterns in brain slices of both KI and WT mice indicated that DOPr was predominantly expressed in the cortex, the olfactory bulb, the amygdala, and the striatum, which is consistent with the previously reported DOPr distribution (46, 49–51). Our binding experiments also revealed that the expression levels of FLAG-DOPr in brain of KI mice were similar to WT animals, while no 125I-DLT specific binding was observed in KO mice. Moreover, the functionality of FLAG-DOPr in KI mice was assessed by 35S-GTPγS binding, locomotor, and pain behavioral studies. In all cases, FLAG-DOPr-KI mice displayed nearly identical functional, locomotor, and antinociceptive properties to WT animals, suggesting that our newly developed FLAG-DOPr-KI mouse line is a physiologically relevant model to investigate the endogenous regulation of DOPr. Similarly, our FLAG-DOPr-KO mice resemble the already existing DOPr KO lines (52, 53). Most importantly, those newly generated mice models (i.e., FLAG-DOPr-KO and FLAG-DOPr-KI) are complementary to the existing DOPr-KO, DOPr-cKO, and DOPr-eGFP, and therefore add to our arsenal of genetically modified tools to elucidate the roles and functions of DOPr in the CNS (45, 52–54). In addition, our mice models are different from the recently reported HA-DOPr KI mouse (48), since it can be used to produce conditional KI within specific targeted regions using a tissue-specific Cre recombinase mouse line or viral approaches. This possibility was investigated by the injection of the recombinant adenoassociated virus rAAV2/9-CBA-Cre-GFP, which preferentially targets lumbar DRGs when injected intrathecally (16). In a CFA-induced chronic pain model, we observed that the antihyperalgesic effect of DLT II (specific DOPr agonist) was partially reinstated in KO mice intrathecally injected with the virus, thereby supporting a reexpression of DOPr in the spinal cord, presumably on central terminals of primary afferents. Such targeted expression of FLAG-DOPr will be an invaluable tool to decipher DOPr function in specific neuron populations and circuits.
Since DOPr expression is enriched in the forebrain, this region was used to immunoprecipitate FLAG-DOPr from KI and KO mice using anti–FLAG-M2 antibodies precoupled to magnetic beads. LC-MS/MS analysis of the immunoprecipitated complex revealed a total of 24 proteins that have already been shown by our group and others to interact with DOPr, thereby validating our approach to identify DOPr interaction partners at endogenous levels in KI mice. Moreover, numerous proteins that are part of molecular complexes with previously described DOPr-interacting partners were also observed in our analysis. For example, all of the subunits of the T-complex protein 1 (i.e., CCT-α, CCT-β, CCT-γ, CCT-δ, CCT-ε, CCT-ζ, CCT-η, and CCT-θ) were identified as putative interacting proteins with the DOPr. Given that the eight distinct CCT subunits are known to associate in order to form the chaperonin TCP-1 ring complex (TRiC) (55), and that we previously observed an interaction between CCT-η and DOPr (56), it is likely that DOPr interacts with the other CCT subunits of the TRiC, as suggested by our LC-MS/MS analyses. Interestingly, our LC-MS/MS analyses further revealed several hits previously identified by Lobingier et al. (27) with a proximity labeling strategy in HEK293 cells. These include the intersectin-2, transferrin receptor protein 1, γ-soluble NSF attachment protein, ras-related GTP-binding protein C, secretory carrier-associated membrane protein 3, protein transport protein Sec31A, and catalytic subunits of the V-type proton ATPase complex. Altogether, these results suggest that the FLAG-DOPr-KI mouse is a reliable and useful tool to guide further behavioral, cellular, and molecular studies.
Every step of GPCR trafficking, from biosynthesis, anterograde transport, exocytosis, endocytosis, recycling, and degradation involve a plethora of proteins in order to regulate physiological responses. In this regard, candidate proteins potentially relevant in the regulation of DOPr trafficking were classified according to their previously reported roles. As mentioned before, DOPr preferentially traffics to lysosomal compartments following internalization (28), a particularity that has also been observed and correlated with behavioral responses in vivo (57). Interestingly, our proteomic analyses identified VPS41, as well as other members (i.e., VPS16, VPS18, and VPS33A) of the HOPS complex, which mediates the fusion between endosomes and lysosomes, as potential DOPr-interacting proteins (29). Importantly, we further validated the DOPr-VPS41 endogenous interaction in vivo from brains of KI mice, confirming that our approach can be used to uncover novel molecular mechanisms underlying DOPr function. Moreover, among the identified proteins, several were reported to regulate the intracellular trafficking of other GPCRs. For example, ARAP1 was shown to promote the recycling of the angiotensin II type 1 receptor (58), while Rab21 seems to be involved in the early endosomal trafficking of the somatostatin receptor 3 (59). Chaperones and other proteins assisting folding or maturation of receptors are also essential for GPCR cell surface expression. Among the identified DOPr-interacting chaperones, CCT-η has already been reported to interact with MOPr and DOPr in addition to promote cell surface expression of the thromboxane A2 and the β2-adrenergic receptors (56). Whether or not CCT-η is involved in the trafficking of DOPr remains to be investigated.
Numerous studies have proposed heteromerization between DOPr and other GPCRs such as MOPr, KOPr, and members of the adrenergic receptors family (reviewed in ref. 32). Rozenfeld et al. (21) also described the formation of an interacting complex between DOPr, the cannabinoid type 1 receptor (CB1), and the adaptor proteins AP-2α and AP-3δ. Interestingly, we also observed CB1 as an endogenous DOPr-interacting partner, along with both AP-2 complex subunit α-1 and AP-3 complex subunit δ-1 proteins in our LC-MS/MS analyses. A potential interaction between DOPr and the GABAB receptor was also observed. To our knowledge, this has not been reported before, and could be of importance in DOPr function since several studies have linked both receptors through physiological and behavioral responses (60, 61). These observations are important because the entire concept of GPCR oligomerization is still controversial. However, if only little evidence of such a phenomenon exists in vivo (62, 63), our work supports the existence of endogenous GPCR heterocomplexes. Further studies will be needed to confirm and characterize the roles and impact of these interactions.
Several putative DOPr effectors linked to signal transduction were also identified in our analyses. The identification of the adenylate cyclase type 5 (AC5) as a potential DOPr-interacting protein is of particular interest since Kim et al. (64) have previously established AC5 as a primary effector for DOPr and MOPr signaling. However, a physical association between AC5 and the opioid receptors was not previously described. Gathering evidence suggests that GPCRs and ACs function as macromolecular complexes. Indeed, AC5, heteromers of adenosine A2A receptors and dopamine D2 receptors homodimers, and their cognate G proteins were proposed to exist as functional precoupled complexes (65). It has been reported that DOPr can couple to a broad range of heterotrimeric G proteins, generating a vast diversity of signaling pathways (reviewed in ref. 32). Our LC-MS/MS analyses revealed several subunits of heterotrimeric G proteins such as Gαo, Gα13, Gα11, Gα(olf), Gαq, and Gαz, as well as Gβ5, Gγ2, Gγ3, and Gγ4 to be potentially associated with DOPr. In addition to the traditional Gαi and Gαo, DOPr interaction with Gαq and Gαz has already been reported (22). The Gγ2 subunit was also shown to be involved in DOPr-mediated antinociception (66). To our knowledge, the other identified heterotrimeric G proteins have not yet been associated with DOPr signaling. It would thus be interesting to confirm these data and study their functional and physiological implications. Considering the observations of Navarro et al. (65) mentioned above, in which they reported the existence of functional precoupled complexes, it is also tempting to speculate that some of these heterotrimeric subunits could couple to other receptors oligomerizing with DOPr. For example, the identified Gα(olf) may not be coupled directly with DOPr, but rather with one of the olfactory receptors identified here as a potential DOPr-interacting partner. This assumption is further supported by the fact that Gαz, identified in our LC-MS/MS analyses, was shown to preferentially regulate the MOPr-DOPr heterodimer signal transduction (67). Finally, we also identified the phosphatidylethanolamine-binding protein 1, which was reported to regulate DOPr-mediated G protein signaling (68). Altogether, the identification of these classical GPCR interactors, in addition to β-arrestin-1, further supports the reliability of our model.
Among all of the identified proteins potentially implicated in the regulation of DOPr trafficking, 15 members of the Rab GTPase family of proteins were identified in our analysis. Rabs belong to the largest subfamily of Ras-related small GTPases with over 60 members (reviewed in ref. 69) and have been established as key regulators in nearly every stage of GPCR intracellular trafficking (70). Rab10 has been reported to increase cell surface targeting of membrane proteins such as GLUT4 (33) and TLR4 (34). Interestingly, in addition to Rab10, our LC-MS/MS analyses also revealed the presence of several proteins found in a Rab10-related functional network, including the Lethal (2) giant larvae protein homolog 1 (Llgl1), a protein previously reported to act as an activator of Rab10 in neurons by facilitating its dissociation from GDI (71), as well as the EH domain-binding protein 1 (EHBP1), a Rab10 effector (72). In HEK293 cells expressing human DOPr, we confirmed the interaction between DOPr and Rab10 by coIP assays. During the editorial process of this manuscript, a role for Rab10 in the trafficking of another GPCR, the somatostatin receptor subtype 2 (SSTR2), was reported for the first time, thereby supporting our observations (35). The existence of an interaction between Rab10 and a GPCR is further supported by a previous proteomic study identifying Rab10 as an interactor of the human melatonin receptor 1 (73). In addition, confocal microscopy experiments revealed intracellular colocalization between Rab10, DOPr, and GalT, indicating that the DOPr–Rab10 interaction is partially localized in trans-Golgi–associated compartments. This is consistent with the description of a Rab10 pool that colocalized with the TGN marker TGN38 in cultured hippocampal neurons (74). Given that Rab GTPases normally mediate the transport of cargo proteins between specific compartments, it is not surprising that the DOPr–Rab10 complex was found associated with, but not restricted to the TGN. Rab10 has also been described as a rather atypical Rab since its localization and functions were not as restrictive as for the other members of this small GTPase family (75).
Previous studies have shown that the phosphorylation of Akt following insulin receptor activation negatively regulates the Rab10 GAP AS160, which leads to increased levels of activated Rab10 resulting in the cell surface targeting of GLUT4 (33). Here, we observed that the overexpression or down-regulation of Rab10 respectively increased and reduced DOPr cell surface expression, whereas the down-regulation of AS160 significantly increased the Rab10-mediated effect on DOPr cell surface expression. Interestingly, stimulation of opioid receptors has also been reported to induce AS160 phosphorylation in a PI3K/Akt-dependent pathway, causing GLUT4 accumulation at the plasma membrane (76). Given that we and others have previously shown that chronic morphine treatments increased DOPr cell surface targeting (9, 10, 77), and that a relation between the inhibition of PI3K and the Golgi retention of DOPr was also reported by the Puthenveedu laboratory (78), these findings suggest that GLUT4 and DOPr are regulated through similar trafficking mechanisms.
Since the TGN represents the final compartment for receptor maturation prior to its membrane delivery, it is plausible that Rab10 could interact with DOPr in the TGN in order to regulate its cell surface targeting. Our results also suggest a role for Rab10 in the agonist-induced trafficking of DOPr. However, since this latter assay measures a dynamic process between internalization, recycling, and anterograde transport of the receptor, a decrease in DOPr cell surface expression following stimulation with SNC80 can be due to an augmentation in internalization, or to a decrease in either recycling or anterograde trafficking. Despite many reports of DOPr being targeted for degradation following internalization, this receptor was also shown to recycle (79, 80). More recently, Charfi et al. (81) proposed a mechanism in which Rab9 and TIP47 rescued DOPr from the degradation path by mediating its transport from late endosomes to the TGN, and therefore allowing its recycling back to the membrane. By being localized in the TGN, Rab10 may thus be implicated in the itinerary of those rescued receptors. Furthermore, given the recently reported role of Rab10 in the regulation of tubular endosome recycling (82), and the identification of several putative DOPr-interacting proteins involved in recycling pathways (e.g., EHD1, EHD3, SNX27), it is conceivable that Rab10 may indeed be implicated in various DOPr trafficking processes.
In summary, we have generated a transgenic FLAG-DOPr-KI mouse model and characterized the expression, distribution, and functional activation of the epitope-tagged DOPr. Using this unique mouse model, we further identified several potential endogenous DOPr-interacting partners, thus providing leads for the discovery of molecular and cellular mechanisms regulating DOPr signaling and trafficking in vivo. This work should also be pivotal in the study of many GPCRs, as it can be transposed to any other receptor and enables a myriad of possibilities, such as the analysis of their interactome in various tissues or following specific treatments in a physiologically relevant context.
Materials and Methods
Generation of FLAG-DOPr-KO (KO) mice was carried out by a classical homologous recombination-based approach. A mouse line expressing a N-terminal FLAG-tagged DOPr endogenously (FLAG-DOPr-KI or KI) was then generated by breeding KO with Zp3-Cre mice. For LC-MS/MS analyses, FLAG-DOPr was immunoprecipitated from the forebrain of five mice per condition (i.e., KO and KI) using M2 mouse monoclonal anti-FLAG antibodies immobilized on magnetic beads. A protein was considered as a potential DOPr-interacting partner when the ratio of peptide intensity displayed at least a 1.5-fold increase in the KI condition compared to KO.
Data Availability.
The authors declare that all data, protocols, and materials are detailed in the manuscript or in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to François-Michel Boisvert and Dominique Lévesque from the Plateforme de Protéomique at the Université de Sherbrooke for helpful advice on the LC-MS/MS data analysis. We also thank Dr. Michael Bruchas for constructive criticism of the manuscript, Dr. Brian Holleran for the preparation of the iodinated peptides, and Dr. Kenner C. Rice for providing SNC80 (National Institute on Drug Abuse, National Institutes of Health). This work was supported by a pilot project grant from the Quebec Pain Research Network to L.G. and C.M.C., and grants from the Canadian Institutes of Health Research to C.L.L., J.-L.P., and L.G. (MOP-136871 and PJT-162103). L.G. is the recipient of Chercheur-Boursier senior salary support from Fonds de la Recherche du Québec-Santé (FRQ-S), and J.-L.P. is the holder of the André-Lussier Research Chair. J.D. and S.G. received PhD scholarships from the FRQ-S. S.G. is also the recipient of a PhD scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). M.-P.L. received an undergraduate salary award from the NSERC.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917906117/-/DCSupplemental.
References
- 1.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong C., Filipeanu C. M., Duvernay M. T., Wu G., Regulation of G protein-coupled receptor export trafficking. Biochim. Biophys. Acta 1768, 853–870 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gavériaux-Ruff C., Kieffer B. L., Delta opioid receptor analgesia: Recent contributions from pharmacology and molecular approaches. Behav. Pharmacol. 22, 405–414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdallah K., Gendron L., The delta opioid receptor in pain control. Handb. Exp. Pharmacol. 247, 147–177 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Gallantine E. L., Meert T. F., A comparison of the antinociceptive and adverse effects of the mu-opioid agonist morphine and the delta-opioid agonist SNC80. Basic Clin. Pharmacol. Toxicol. 97, 39–51 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Codd E. E. et al., JNJ-20788560 [9-(8-azabicyclo[3.2.1]oct-3-ylidene)-9H-xanthene-3-carboxylic acid diethylamide], a selective delta opioid receptor agonist, is a potent and efficacious antihyperalgesic agent that does not produce respiratory depression, pharmacologic tolerance, or physical dependence. J. Pharmacol. Exp. Ther. 329, 241–251 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Feng P. et al., Effects of mu, kappa or delta opioids administered by pellet or pump on oral Salmonella infection and gastrointestinal transit. Eur. J. Pharmacol. 534, 250–257 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Gendron L., Mittal N., Beaudry H., Walwyn W., Recent advances on the δ opioid receptor: From trafficking to function. Br. J. Pharmacol. 172, 403–419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gendron L. et al., Morphine and pain-related stimuli enhance cell surface availability of somatic delta-opioid receptors in rat dorsal root ganglia. J. Neurosci. 26, 953–962 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahill C. M. et al., Prolonged morphine treatment targets delta opioid receptors to neuronal plasma membranes and enhances delta-mediated antinociception. J. Neurosci. 21, 7598–7607 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morinville A. et al., Morphine-induced changes in delta opioid receptor trafficking are linked to somatosensory processing in the rat spinal cord. J. Neurosci. 24, 5549–5559 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo M., Jung S. T., Engineering therapeutic antibodies targeting G-protein-coupled receptors. Exp. Mol. Med. 48, e207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel M. C., Wieland T., Tsujimoto G., How reliable are G-protein-coupled receptor antibodies? Naunyn Schmiedebergs Arch. Pharmacol. 379, 385–388 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Broom D. C. et al., Nonpeptidic δ-opioid receptor agonists reduce immobility in the forced swim assay in rats. Neuropsychopharmacology 26, 744–755 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Spina L., Longoni R., Mulas A., Chang K. J., Di Chiara G., Dopamine-dependent behavioural stimulation by non-peptide delta opioids BW373U86 and SNC 80: 1. Locomotion, rearing and stereotypies in intact rats. Behav. Pharmacol. 9, 1–8 (1998). [PubMed] [Google Scholar]
- 16.Abdallah K. et al., Adeno-associated virus 2/9 delivery of Cre recombinase in mouse primary afferents. Sci. Rep. 8, 7321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desroches J., Bouchard J. F., Gendron L., Beaulieu P., Involvement of cannabinoid receptors in peripheral and spinal morphine analgesia. Neuroscience 261, 23–42 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Cen B. et al., Direct binding of beta-arrestins to two distinct intracellular domains of the delta opioid receptor. J. Neurochem. 76, 1887–1894 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Zhang X. et al., β-arrestin1 and β-arrestin2 are differentially required for phosphorylation-dependent and -independent internalization of δ-opioid receptors. J. Neurochem. 95, 169–178 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Rios C., Gomes I., Devi L. A., mu opioid and CB1 cannabinoid receptor interactions: Reciprocal inhibition of receptor signaling and neuritogenesis. Br. J. Pharmacol. 148, 387–395 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozenfeld R. et al., Receptor heteromerization expands the repertoire of cannabinoid signaling in rodent neurons. PLoS One 7, e29239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law S. F., Reisine T., Changes in the association of G protein subunits with the cloned mouse delta opioid receptor on agonist stimulation. J. Pharmacol. Exp. Ther. 281, 1476–1486 (1997). [PubMed] [Google Scholar]
- 23.Xie W.-Y. et al., Disruption of Cdk5-associated phosphorylation of residue threonine-161 of the δ-opioid receptor: Impaired receptor function and attenuated morphine antinociceptive tolerance. J. Neurosci. 29, 3551–3564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leskelä T. T., Markkanen P. M. H., Pietilä E. M., Tuusa J. T., Petäjä-Repo U. E., Opioid receptor pharmacological chaperones act by binding and stabilizing newly synthesized receptors in the endoplasmic reticulum. J. Biol. Chem. 282, 23171–23183 (2007). [DOI] [PubMed] [Google Scholar]
- 25.St-Louis É. et al., Involvement of the coatomer protein complex I in the intracellular traffic of the delta opioid receptor. Mol. Cell. Neurosci. 79, 53–63 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Shiwarski D. J., Crilly S. E., Dates A., Puthenveedu M. A., Dual RXR motifs regulate nerve growth factor-mediated intracellular retention of the delta opioid receptor. Mol. Biol. Cell 30, 680–690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobingier B. T. et al., An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsao P. I., von Zastrow M., Type-specific sorting of G protein-coupled receptors after endocytosis. J. Biol. Chem. 275, 11130–11140 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Balderhaar H. J., Ungermann C., CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 126, 1307–1316 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Rankovic M. et al., ADP-ribosylation factor 6 regulates mu-opioid receptor trafficking and signaling via activation of phospholipase D2. Cell. Signal. 21, 1784–1793 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Kälin S., Hirschmann D. T., Buser D. P., Spiess M., Rabaptin5 is recruited to endosomes by Rab4 and Rabex5 to regulate endosome maturation. J. Cell Sci. 128, 4126–4137 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Gendron L., Cahill C. M., Von Zastrow M., Schiller P. W., Pineyro G., Molecular pharmacology of δ-opioid receptors. Pharmacol. Rev. 68, 631–700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sano H. et al., Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 5, 293–303 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Wang D. et al., Ras-related protein Rab10 facilitates TLR4 signaling by promoting replenishment of TLR4 onto the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 107, 13806–13811 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alshafie W. et al., Regulated resurfacing of a somatostatin receptor storage compartment fine-tunes pituitary secretion. J. Cell Biol. 219, e201904054 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binda C. et al., L-type prostaglandin D synthase regulates the trafficking of the PGD2 DP1 receptor by interacting with the GTPase Rab4. J. Biol. Chem. 294, 16865–16883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parent A., Hamelin E., Germain P., Parent J.-L., Rab11 regulates the recycling of the beta2-adrenergic receptor through a direct interaction. Biochem. J. 418, 163–172 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Hamelin E., Thériault C., Laroche G., Parent J. L., The intracellular trafficking of the G protein-coupled receptor TPbeta depends on a direct interaction with Rab11. J. Biol. Chem. 280, 36195–36205 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Esseltine J. L., Dale L. B., Ferguson S. S. G., Rab GTPases bind at a common site within the angiotensin II type I receptor carboxyl-terminal tail: Evidence that Rab4 regulates receptor phosphorylation, desensitization, and resensitization. Mol. Pharmacol. 79, 175–184 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Chen Y. T., Holcomb C., Moore H. P., Expression and localization of two low molecular weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc. Natl. Acad. Sci. U.S.A. 90, 6508–6512 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llopis J., McCaffery J. M., Miyawaki A., Farquhar M. G., Tsien R. Y., Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 6803–6808 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth J., Berger E. G., Immunocytochemical localization of galactosyltransferase in HeLa cells: Codistribution with thiamine pyrophosphatase in trans-Golgi cisternae. J. Cell Biol. 93, 223–229 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu R. et al., Epitope-tagged receptor knock-in mice reveal that differential desensitization of α2-adrenergic responses is because of ligand-selective internalization. J. Biol. Chem. 284, 13233–13243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozawa A. et al., Knock-in mice with NOP-eGFP receptors identify receptor cellular and regional localization. J. Neurosci. 35, 11682–11693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scherrer G. et al., Knockin mice expressing fluorescent delta-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 9691–9696 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erbs E., Faget L., Veinante P., Kieffer B. L., Massotte D., In vivo neuronal co-expression of mu and delta opioid receptors uncovers new therapeutic perspectives. Receptors Clin. Investig. 1, 1–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan F., Bradley A., Wensel T. G., Wilson J. H., Knock-in human rhodopsin-GFP fusions as mouse models for human disease and targets for gene therapy. Proc. Natl. Acad. Sci. U.S.A. 101, 9109–9114 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su D. et al., One-step generation of mice carrying a conditional allele together with an HA-tag insertion for the delta opioid receptor. Sci. Rep. 7, 44476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansour A., Thompson R. C., Akil H., Watson S. J., Delta opioid receptor mRNA distribution in the brain: Comparison to delta receptor binding and proenkephalin mRNA. J. Chem. Neuroanat. 6, 351–362 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Simonin F. et al., The human delta-opioid receptor: Genomic organization, cDNA cloning, functional expression, and distribution in human brain. Mol. Pharmacol. 46, 1015–1021 (1994). [PubMed] [Google Scholar]
- 51.Cahill C. M. et al., Immunohistochemical distribution of delta opioid receptors in the rat central nervous system: Evidence for somatodendritic labeling and antigen-specific cellular compartmentalization. J. Comp. Neurol. 440, 65–84 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Filliol D. et al., Mice deficient for δ- and μ-opioid receptors exhibit opposing alterations of emotional responses. Nat. Genet. 25, 195–200 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Zhu Y. et al., Retention of supraspinal delta-like analgesia and loss of morphine tolerance in delta opioid receptor knockout mice. Neuron 24, 243–252 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Gaveriaux-Ruff C. et al., Genetic ablation of delta opioid receptors in nociceptive sensory neurons increases chronic pain and abolishes opioid analgesia. Pain 152, 1238–1248 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Cong Y. et al., 4.0-Å resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. U.S.A. 107, 4967–4972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Génier S. et al., Regulation of GPCR expression through an interaction with CCT7, a subunit of the CCT/TRiC complex. Mol. Biol. Cell 27, 3800–3812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pradhan A. A. A. et al., In vivo delta opioid receptor internalization controls behavioral effects of agonists. PLoS One 4, e5425 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo D. F., Chenier I., Tardif V., Orlov S. N., Inagami T., Type 1 angiotensin II receptor-associated protein ARAP1 binds and recycles the receptor to the plasma membrane. Biochem. Biophys. Res. Commun. 310, 1254–1265 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Tower-Gilchrist C., Lee E., Sztul E., Endosomal trafficking of the G protein-coupled receptor somatostatin receptor 3. Biochem. Biophys. Res. Commun. 413, 555–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rady J. J., Fujimoto J. M., Spinal GABA receptors mediate brain delta opioid analgesia in Swiss Webster mice. Pharmacol. Biochem. Behav. 51, 655–659 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Watanabe Y., Aono Y., Komiya M., Waddington J. L., Saigusa T., Stimulation of accumbal GABAB receptors inhibits delta1- and delta2-opioid receptor-mediated dopamine efflux in the nucleus accumbens of freely moving rats. Eur. J. Pharmacol. 837, 88–95 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Gomes I. et al., G protein-coupled receptor heteromers. Annu. Rev. Pharmacol. Toxicol. 56, 403–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faron-Górecka A. et al., Understanding GPCR dimerization. Methods Cell Biol. 149, 155–178 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Kim K.-S. et al., Adenylyl cyclase type 5 (AC5) is an essential mediator of morphine action. Proc. Natl. Acad. Sci. U.S.A. 103, 3908–3913 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Navarro G. et al., Evidence for functional pre-coupled complexes of receptor heteromers and adenylyl cyclase. Nat. Commun. 9, 1242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hosohata K. et al., The role of the G protein gamma(2) subunit in opioid antinociception in mice. Eur. J. Pharmacol. 392, R9–R11 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Hasbi A. et al., Trafficking of preassembled opioid μ-δ heterooligomer-Gz signaling complexes to the plasma membrane: Coregulation by agonists. Biochemistry 46, 12997–13009 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Kroslak T., Koch T., Kahl E., Höllt V., Human phosphatidylethanolamine-binding protein facilitates heterotrimeric G protein-dependent signaling. J. Biol. Chem. 276, 39772–39778 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Zhen Y., Stenmark H., Cellular functions of Rab GTPases at a glance. J. Cell Sci. 128, 3171–3176 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Wang G., Wei Z., Wu G., Role of Rab GTPases in the export trafficking of G protein-coupled receptors. Small GTPases 9, 130–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang T. et al., Lgl1 activation of rab10 promotes axonal membrane trafficking underlying neuronal polarization. Dev. Cell 21, 431–444 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Wang P. et al., RAB-10 promotes EHBP-1 bridging of filamentous actin and tubular recycling endosomes. PLoS Genet. 12, e1006093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daulat A. M. et al., Purification and identification of G protein-coupled receptor protein complexes under native conditions. Mol. Cell. Proteomics 6, 835–844 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Liu Y. et al., Myosin Vb controls biogenesis of post-Golgi Rab10 carriers during axon development. Nat. Commun. 4, 2005 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Chua C. E. L., Tang B. L., Rab 10-a traffic controller in multiple cellular pathways and locations. J. Cell. Physiol. 233, 6483–6494 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Blaschke A., Heiss A., Eisinger D. A., Opioid-induced AS160 phosphorylation through TrkA transactivated PI3K/Akt signaling pathway implies neurite outgrowth. Pharmacologia 4, 428–440 (2013). [Google Scholar]
- 77.Lucido A. L., Morinville A., Gendron L., Stroh T., Beaudet A., Prolonged morphine treatment selectively increases membrane recruitment of δ-opioid receptors in mouse basal ganglia. J. Mol. Neurosci. 25, 207–214 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Shiwarski D. J., Darr M., Telmer C. A., Bruchez M. P., Puthenveedu M. A., PI3K class II α regulates δ-opioid receptor export from the trans-Golgi network. Mol. Biol. Cell 28, 2202–2219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapaidze N., Gomes I., Bansinath M., Devi L. A., Recycling and resensitization of delta opioid receptors. DNA Cell Biol. 19, 195–204 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hasbi A. et al., Internalization and recycling of delta-opioid receptor are dependent on a phosphorylation-dephosphorylation mechanism. J. Pharmacol. Exp. Ther. 293, 237–247 (2000). [PubMed] [Google Scholar]
- 81.Charfi I., Abdallah K., Gendron L., Pineyro G., Delta opioid receptors recycle to the membrane after sorting to the degradation path. Cell. Mol. Life Sci. 75, 2257–2271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Etoh K., Fukuda M., Rab10 regulates tubular endosome formation through KIF13A and KIF13B motors. J. Cell Sci. 132, jcs226977 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Deng H. et al., Interactions of Na+,K+-ATPase and co-expressed δ-opioid receptor. Neurosci. Res. 65, 222–227 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Tuusa J. T., Leskelä T. T., Petäjä-Repo U. E., Human δ opioid receptor biogenesis is regulated via interactions with SERCA2b and calnexin. FEBS J. 277, 2815–2829 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Wu D.-F. et al., Membrane glycoprotein M6a interacts with the micro-opioid receptor and facilitates receptor endocytosis and recycling. J. Biol. Chem. 282, 22239–22247 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Ge X., Loh H. H., Law P. Y., μ-Opioid receptor cell surface expression is regulated by its direct interaction with Ribophorin I. Mol. Pharmacol. 75, 1307–1316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petko J. et al., MOR is not enough: Identification of novel mu-opioid receptor interacting proteins using traditional and modified membrane yeast two-hybrid screens. PLoS One 8, e67608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data, protocols, and materials are detailed in the manuscript or in SI Appendix.