Significance
How organisms sense and respond to changes in nutrient dose is a basic unanswered question that is relevant to agriculture. Here, we demonstrate that genome-wide expression levels in the Arabidopsis root are nutrient dose-responsive. We find that such dose-responsive gene expression patterns are driven largely by Michaelis–Menten (MM) kinetics, indicating that genome-wide transcriptional responses to nutrient dose resemble a simple principle of enzyme kinetics. Transcription factors (TFs) can act as “catalysts” driving rates of transcript change in response to nutrient dose. Supporting this, we identified TGA1 as a TF that controls nitrogen-dose–dependent rates of transcriptional change and plant growth. Thus, our study of the molecular mechanisms that underlie N-dose–responsive transcriptome kinetics could lead to enhanced crop growth.
Keywords: nitrogen dose, Michaelis–Menten kinetics, transcriptome regulation
Abstract
An increase in nutrient dose leads to proportional increases in crop biomass and agricultural yield. However, the molecular underpinnings of this nutrient dose–response are largely unknown. To investigate, we assayed changes in the Arabidopsis root transcriptome to different doses of nitrogen (N)—a key plant nutrient—as a function of time. By these means, we found that rate changes of genome-wide transcript levels in response to N-dose could be explained by a simple kinetic principle: the Michaelis–Menten (MM) model. Fitting the MM model allowed us to estimate the maximum rate of transcript change (Vmax), as well as the N-dose at which one-half of Vmax was achieved (Km) for 1,153 N-dose–responsive genes. Since transcription factors (TFs) can act in part as the catalytic agents that determine the rates of transcript change, we investigated their role in regulating N-dose–responsive MM-modeled genes. We found that altering the abundance of TGA1, an early N-responsive TF, perturbed the maximum rates of N-dose transcriptomic responses (Vmax), Km, as well as the rate of N-dose–responsive plant growth. We experimentally validated that MM-modeled N-dose–responsive genes included both direct and indirect TGA1 targets, using a root cell TF assay to detect TF binding and/or TF regulation genome-wide. Taken together, our results support a molecular mechanism of transcriptional control that allows an increase in N-dose to lead to a proportional change in the rate of genome-wide expression and plant growth.
Due to their unique ability for indeterminate growth, plants can adapt their size and growth rate in response to the amount of nutrient available in their environment. Nitrogen (N) is a key nutrient that has such a dose-responsive effect on plant growth and development (1). For this reason, N-based fertilizers are routinely employed to enhance crop biomass and boost grain yields. However, such fertilizers are costly and harmful to the environment (2).
To help identify ways to improve plant N-use efficiency and reduce reliance on N fertilizers, the signaling mechanisms that allow plants to sense and respond to N availability are under intense scrutiny. Numerous transcriptome studies have detected an impressive catalog of genes that are differentially expressed in response to a change in N status, which participate in a wide range of biological processes related to growth and development (3–7). Together, these studies indicate that a large fraction of the plant genome, ∼10%, is transcriptionally responsive to N treatment.
Indeed, previous studies have shown that N nutrient dose sensing enables plants to reprogram both their gene expression patterns (8, 9) as well as their phenotype, including the rate of N uptake and plant growth (10, 11). However, what remains unknown are the molecular mechanisms that enable the dose of N to inform gene expression levels, and how such gene expression changes lead to dose-responsive changes in plant phenotype.
To address this question, we assayed transcriptome responses of Arabidopsis thaliana roots exposed to different N-doses over time. By modeling gene expression responses to N-dose as a function of time, we were able to reveal the in vivo kinetics that tailor gene expression responses to the dose of N available. By these means, we demonstrate that the dynamics of plant transcriptomic responses to N-dose follows simple kinetics, as described by the Michaelis–Menten (MM) model. While the MM model was designed to explain in vitro enzyme kinetics (12, 13), due to its simplicity it has enjoyed broad applicability (14, 15), including describing the rates of N uptake by plants and N-dose–dependent plant growth (10, 11). Using this model, we found that the rate of transcript accumulation is a function of N-dose, which reached a saturation point at the highest N-doses tested.
The MM model predicts the rate at which a catalyst can convert a substrate to its product. By analogy, transcription factors (TFs) act in part as the “catalyst” that drive rates of transcript change. To test this, we investigated whether altering TF abundance in planta could impact the maximum rate at which transcript abundance changed (Vmax) in responses to N-dose. We show that the overexpression of the TF TGA1 (TGACG SEQUENCE-SPECIFIC BINDING PROTEIN 1) leads to an increase in the maximum rate of transcript change in response to N-dose (Vmax), as predicted by the MM model. Furthermore, we validated TGA1’s direct gene targets genome-wide through RNA sequencing (RNA-seq), chromatin immunoprecipitation sequencing (ChIP-seq), and 4-thiouracil (4tU) labeling of nascent transcripts, revealing that TGA1 directly regulates both N-metabolic genes and their TF regulators. Importantly, we show that TGA1’s impact on the kinetics of the N-dose–response at the molecular level leads to an acceleration in N-dependent plant growth rates. In this way, our study of the basic mechanisms that underlie the transcriptome kinetics responding to changes in N-dose could potentially enhance plant growth and improve N-use efficiency.
Results
N-dose Informs Rates of Transcript Change Genome-Wide in Arabidopsis Roots.
To understand the transcriptional mechanisms underlying plant responses to N-dose, we assayed how the Arabidopsis transcriptome responds to N-dose as a function of time. To achieve this, we treated hydroponically grown, wild-type Arabidopsis seedlings to four increasing doses of N (0, 1, 10, and 60 mM N, provided as KNO3 + NH4NO3) over five time points (15, 30, 60, 120, and 240 min) (Fig. 1A). The maximum N-dose used reflected the amount of N present in standard Murashige and Skoog (MS) plant growth media (16). The time points were selected to capture the early transcriptional events that occur in response to N sensing (5). This experimental setup generated a factorial matrix holding 20 unique treatment conditions (Fig. 1A). For each condition, we harvested the root tissue of ∼30 plants and assayed their transcriptome by RNA-seq.
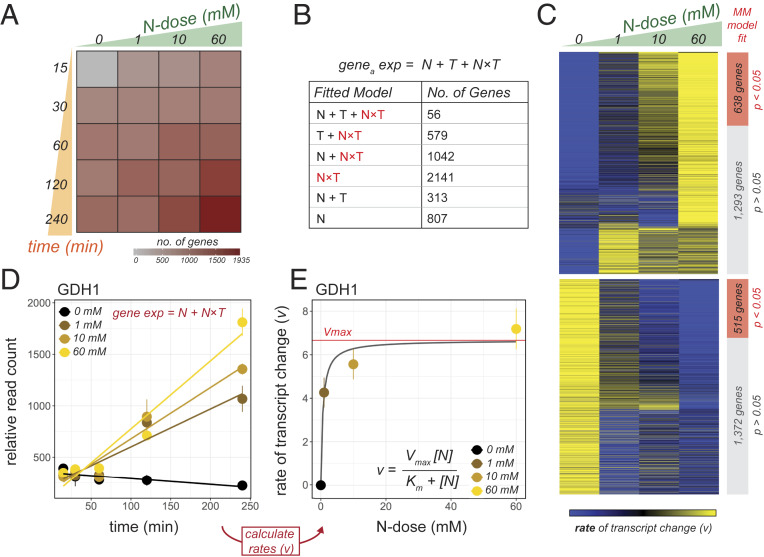
Fig. 1.
Michaelis–Menten (MM) kinetics underlie transcriptomic responses to N-dose in Arabidopsis roots. (A) A factorial treatment matrix systematically varied both the exposure time to N and the N-dose provided. Colors indicate how many genes were differentially expressed (fold change cutoff ± 1.5) in response to N-dose in each condition. (B) Differentially expressed genes were detected using a multivariate linear model. Seventy-seven percent of genes found by linear modeling (3,818 genes) were fit by a model holding an NT term. (C) Heatmap displaying rates of transcript change for each of the 3,818 genes under each N-dose. Genes whose rate change in N-dose–responsive expression significantly fit the MM model (1,153 genes) are indicated in red. (D) GDH1 is an example of a gene fit by model holding a “NT” term. (E) GDH1 is also an example of a gene whose rate of N-dose–responsive expression is significantly fit by the MM model.
To identify genes differentially expressed in response to N-dose as a function of time, we fit normalized gene expression patterns with a linear model. This model was designed to detect genes that were either responsive to the dose of N provided (N), time (T), or the interaction between the two (NT) (Fig. 1B). We employed model simplification to determine the best linear model that explained each gene’s expression. After excluding genes that were time-responsive only—and thus not relevant to our study—we found 4,938 genes that were differentially expressed as a function of N (adjusted P < 0.01) (Fig. 1B and SI Appendix, Table S1). We found that 77% of these N-dose–response genes (3,818 genes) were explained by a model holding an NT interaction term, indicating that the majority of genes that responded to N-dose did so as a function of time (Fig. 1B). Among these were well-known genes involved in N transport and assimilation such as the NITRATE TRANSPORTER 1.1 (NRT1.1/NPF6.3), NITRATE REDUCTASE 1 (NIA1), GLUTAMINE SYNTHETASE (GLN1), ASPARTATE AMINOTRANSFERASE (ASP3), and GLUTAMATE DEHYDROGENASE (GDH3) (SI Appendix, Table S1). Genes that were modeled by an NT interaction term included many known N-responsive genes found through previous transcriptomic analyses (SI Appendix, Fig. S1), suggesting that many previously reported N-responsive genes are N-dose responsive.
To further understand how the expression of these 3,818 “NT” genes was governed by both N-dose and time (T), we performed a fold-change analysis. To do this, we counted how many genes regulated at each time-point passed a ±1.5-fold change cutoff. This revealed that a higher N-dose led to more genes being differentially expressed at earlier time points (Fig. 1A). We next asked whether higher doses of N lead to higher rates of transcript change for each gene. To answer this, we calculated the change in each gene’s transcript abundance over time under each N-dose. Each rate was calculated by fitting a linear model to each N-dose tested (Materials and Methods). Comparing these rates revealed a genome-wide trend; we found that genes displayed rates of transcript change that either increased or decreased in proportion to the dose of N provided (Fig. 1C). An example of a gene that fits this trend is the N-assimilation gene GLUTAMATE DEHYDROGENASE 1 (GDH1) (Fig. 1D).
Next, we assessed whether such transcriptome responses to N-dose fit a general biological model that describes how reaction rates change. Specifically, we tested whether the rate changes in transcript levels could be explained by MM kinetics (12, 13). To do this, we fit each gene’s expression pattern to the MM model (Materials and Methods). A gene significantly fit by this MM model allowed us to estimate the maximum rate of transcript change (Vmax), as well as the dose of N at which one-half of Vmax was achieved (Km). We found that the MM kinetic model was able to explain the expression responses of 1,153 N-dose responsive genes (adjusted P < 0.05; SI Appendix, Table S2), including GDH1 (Fig. 1 D and E). Additionally, we found that the MM model was able to describe genes either activated or repressed by N-dose (Fig. 1C). The significant gene ontology (GO) terms enriched in up-regulated genes included “response to abiotic stimulus” and “response to inorganic substance” (SI Appendix, Table S3). Moreover, our MM modeling approach indicated that at a N-dose of 60 mM, many genes were being induced or repressed close to their estimated maximum rate (Vmax).
Transcription Factor TGA1 Regulates Transcriptional Response Rates to N-dose.
According to the MM model, changing enzyme abundance will impact the maximum rate of reaction possible (Vmax). TFs likely serve as the main catalyst driving rates of transcript change in response to N-dose. To assess the robustness of our MM models, as well as identify TFs that signal N-dose, we sought to test how altering TF abundance could perturb N-dose transcriptomic responses, with the goal of observing whether their Vmax estimates change.
We took two complementary approaches to identify candidate TFs responsible for mediating the rates of transcriptional change in response to N-dose. First, we investigated the expression patterns of TFs regulated by N-dose as a function of time within our experiment. We hypothesized that TFs that responded early might be involved in governing N-dose–responsive transcription. We thus ranked all expressed TFs within the Arabidopsis genome by their fold change in response to N-dose at the earliest time point tested (15 min) (Fig. 2A). Of these, the top three early N-dose–responsive TFs were LOB DOMAIN-CONTAINING PROTEIN 37 (LBD37), LBD38, and TGA1, with 32-, 20-, and 18-fold change in gene expression, respectively (Fig. 2A and SI Appendix, Fig. S2). All three TFs have been previously implicated in transcriptional regulation of N-responsive genes in planta (17, 18). Second, we searched for overrepresented cis-regulatory elements in the promoters of the 1,153 genes whose expression in response to N-dose could be explained by the MM model (Fig. 1C). We found that the TGA1 binding site was significantly overrepresented in the genes whose N-dose–responsive expression could be modeled by MM kinetics (Fig. 2B). Moreover, we found that genes whose N-dose–response is modeled by MM kinetics are preferentially expressed within inner cell layers of the root (pericycle and stele), where TGA1 itself is expressed (18, 19) (SI Appendix, Fig. S3).
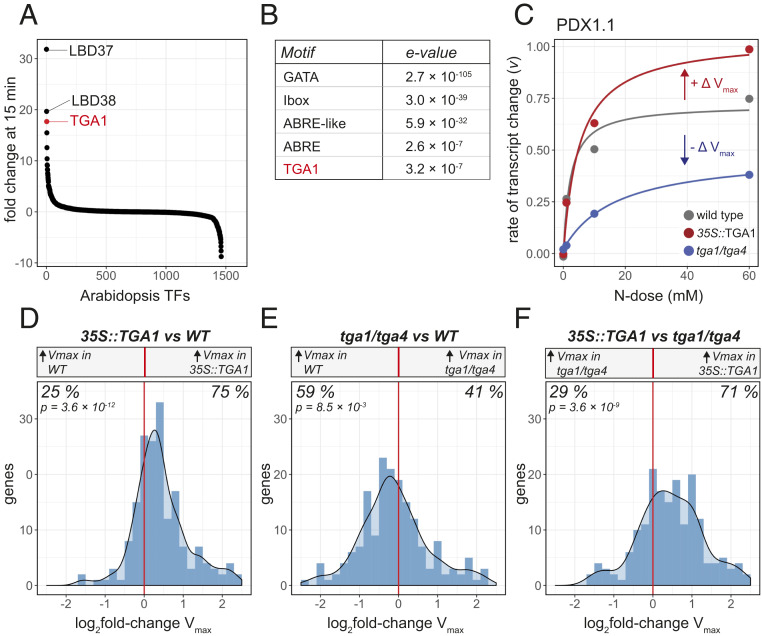
Fig. 2.
N-dose–response rates of transcripts mediated by changes in TGA1 levels in planta. (A) A ranking in expression fold change for mRNAs of all plant TFs after 15 min of exposure to N treatment. (B) cis-regulatory analysis identifies overrepresented motifs among the 1,153 N-dose–responsive genes captured by the MM model. (C) Changes in N-dose–dependent transcription rate of PDX1.1 is fit by the MM model in wild type, and Vmax is altered in the 35S::TGA1 and tga1/tga4 lines. (D–F) A histogram of log2 fold changes in Vmax shown for 192 MM-modeled genes up-regulated by N-dose across wild-type and TGA1 genotypes. D compares these changes in Vmax between 35S::TGA1 and wild type; E, between tga1/tga4 and wild type; and F, between 35S::TGA1 and tga1/tga4. A binomial test was used to assess bias toward an increase or decrease in Vmax.
Together, these findings suggested that TGA1 is a TF involved in regulating transcriptional responses to N-dose. To test this hypothesis, we repeated our factorial matrix experiment that varied N-dose over time (Fig. 1A), but now assessing the transcriptomic responses of a TGA1-overexpressing line (35S::TGA1) (20) and a tga1/tga4 double mutant (18). We note it was necessary to use the tga1/tga4 mutant line, as TGA4 is the closest gene homolog to TGA1, and as such these TFs have been shown to act redundantly (18). Our results indicated that overexpression of TGA1 in planta led to increases in rates of transcriptional change in response to N-dose (Fig. 2 D and F), while knocking out TGA1 and TGA4 led to decreased rates of transcriptional change (Fig. 2 E and F). This effect was captured by fitting the MM model to gene expression patterns in the 35S::TGA1 and tga1/tga4 lines to estimate Vmax, and comparing these Vmax estimates to those of wild type (Fig. 2). Of the 192 genes we were able to significantly model using MM kinetics in all three genotypes (wild type, 35S::TGA1, tga1/4) (adjusted P < 0.05), we found that 75% of these genes held higher Vmax values when TGA1 was overexpressed (35S::TGA1) (Fig. 2D and SI Appendix, Table S2). GO terms for genes with higher Vmax estimates in the TGA1-overexpressing lines included “translation,” “glucose metabolic process,” and “generation of precursor metabolites and energy” (SI Appendix, Table S2). In addition to these 192 MM-modeled genes, we also observed higher Vmax values for all N–up-regulated genes in the TGA1-overexpressing line, even if their resulting model P value did not pass the significance threshold (SI Appendix, Fig. S4). This suggested higher expression of TGA1 leads to increased rates of transcriptional change (i.e., increased Vmax) in response to changes in N-dose. This was further supported through conducting the same analysis in the tga1/tga4 knockout line, where Vmax estimates for these genes were found to be lower compared either to wild-type or the 35S::TGA1 line (Fig. 2 E and F and SI Appendix, Fig. S4). An example of a gene fitting this expression pattern is illustrated in Fig. 2C, where the MM model explains the rates of transcript change of PYRIDOXINE BIOSYNTHESIS 1.1 (PDX1.1), a glutamine amidotranferase (21), within each genotype. Here, the 35S::TGA1 line held a higher Vmax estimate compared to wild type, while the tga1/tga4 mutant line held a lower Vmax estimate. Changing levels of TGA1 in planta also impacted Vmax estimates for GDH1, in a similar fashion to PDX1.1 (SI Appendix, Fig. S5). Collectively, these data support the conclusion that TGA1 acts as transcriptional activator, increasing transcriptional response rates to N-dose. This is further supported by our finding that perturbing levels of TGA1 expression had a weaker effect on genes down-regulated by N (SI Appendix, Fig. S6).
Additionally, for MM-modeled genes that were up-regulated by N-dose genome-wide, we found a global increase in Km estimates within both the tga1/tga4 mutant and in the 35S::TGA1 lines, relative to wild type (SI Appendix, Fig. S7). Specifically, in the tga1/tga4 mutant background, not only were the majority of estimated maximum rates of transcriptional change (Vmax) lower relative to wild type (Fig. 2E), but the N-dose required to achieve half the maximum rate (Km) increased (SI Appendix, Fig. S7B). For the 35S::TGA1 line, the Km estimates also increased (SI Appendix, Fig. S7A). Since the estimated maximum rate of transcriptional change (Vmax) was higher in the 35S::TGA1 background (Fig. 2D), the N-dose required to reach this higher rate of transcriptional change is reflected in an increase in Km (SI Appendix, Fig. S7A). The response of gene ATDF2, encoding a ferredoxin-like superfamily protein, illustrates how Km values can increase when TGA1 is overexpressed (35S::TGA1), as well as when it is absent (tga1/tga4) (SI Appendix, Fig. S7D).
TF-Perturbation Assays Uncover TGA1 as a Transcriptional Activator in Root Cells.
Our transcriptomic analysis identified genes whose response to N-dose is mediated by TGA1 in planta; overexpression of TGA1 showed increased rates of transcript change relative to wild-type plants (Fig. 2 C–F). To experimentally determine whether this is a direct result of transcriptional activation by TGA1, we used the plant cell-based TARGET TF-perturbation assay, which can identify direct, actively transcribed targets of a TF (22). To perform this assay, we transiently expressed TGA1 in root protoplasts as a TGA1 glucocorticoid receptor fusion protein (TGA1-GR). To control TF import into the nucleus, transfected root cells expressing TGA1-GR were sequentially treated with 1) +/− N, 2) cycloheximide (+/− CHX), and 3) dexamethasone (+/− DEX) (Fig. 3A and Materials and Methods). DEX treatment induces nuclear import of the TGA1-GR fusion protein (23). Genes regulated by DEX-induced TF import are deemed direct “primary” targets of TGA1, since a +CHX pretreatment blocks translation of downstream regulators (22, 24). N treatment is included to induce any posttranslational modifications of TGA1 (25) or influence TGA1 partners by transcriptional or posttranscriptional mechanisms.
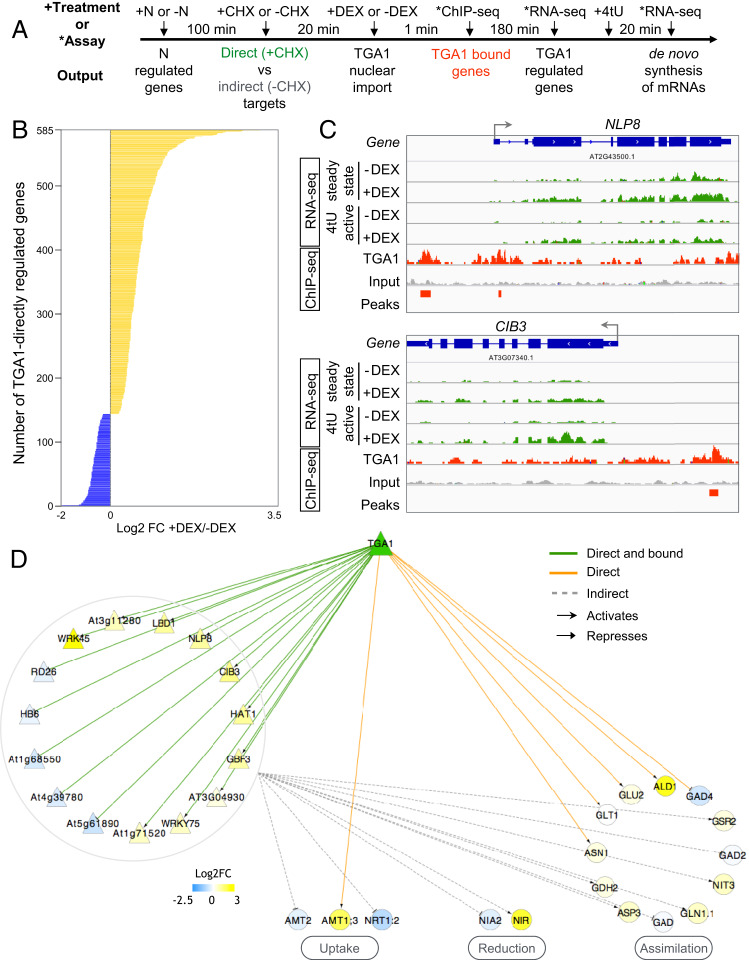
Fig. 3.
TGA1 is a transcriptional activator that regulates both TFs and N-uptake/assimilation genes in root cells. (A) Workflow of TARGET TF-perturbation assay that can detect the gene targets that TGA1 binds to and regulates in isolated root cells. (B) The TARGET TF-perturbation assay reveals TGA1 directly up-regulates (yellow, 77%) or down-regulates (blue, 23%) the expression of 584 genes in root cells (SI Appendix, Table S4). (C) NLP8 and CIB3 TFs are examples of direct targets transcriptionally activated by TGA1. The expression of NLP8 and CIB3 is affected by TGA1 nuclear import, as assayed by both RNA-seq (steady-state mRNA) and by 4tU affinity capture (de novo mRNA) (green bars). TGA1 binding at these loci was captured by ChIP-seq (red bars). (D) TGA1 transcriptional subnetwork in root cells, where nodes represent genes and edges represent regulatory interactions detected by our assay. TGA1 directly or indirectly—through intermediate TFs (triangles)—regulate the expression genes involved in N uptake/metabolism (circles).
Through analysis of mRNA-seq data, we identified 584 direct gene targets of TGA1 in root cells (adjusted P < 0.05), 77% of which were up-regulated by TGA1 nuclear import, a higher proportion than expected by chance (binomial test, P = 9 10−40) (Fig. 3B and SI Appendix, Table S4). This provided evidence that TGA1 serves largely as a transcriptional activator. Additionally, genes up-regulated by TGA1 held higher significance values and greater fold-change differences than down-regulated genes (SI Appendix, Fig. S8A). Among the direct regulated targets of TGA1 were N-related genes AMMONIUM TRANSPORTER 1.3 (AMT1.3), GLUTAMATE SYNTHASE 1 (GLT1), GLUTAMATE SYNTHASE 2 (GLU2), and GLUTAMATE DECARBOXYLASE (GAD4) (SI Appendix, Table S4). In line with this, GO terms enriched in TGA1 direct targets were related to metabolism including “regulation of cellular metabolic process” and “regulation of nitrogen compound metabolic process” (SI Appendix, Table S5).
To confirm that TGA1 directly initiates the transcription of its 584 direct targets as identified by mRNA-seq, we also tracked de novo synthesis of mRNAs made in response to TGA1 nuclear import. To do this, we repeated our TARGET TF-perturbation experiment, but spiked the isolated root cells with 4tU after DEX-induced nuclear import of TGA1, as described in ref. 26 (Fig. 3A). 4tU is a uracil analog that is incorporated into nascent mRNA transcripts, which can then be selectively isolated using a streptavidin mediated pull-down assay (27). Thus, we used 4tU to label, affinity purify, and sequence de novo transcripts that were actively transcribed in response to TGA1 nuclear localization (26). Using 4tU, we validated that TGA1 nuclear import resulted in de novo transcription of 83% of TGA1’s 584 direct targets (SI Appendix, Table S4). In addition, the magnitude of de novo transcript synthesis in response to TGA1 nuclear import correlated with the magnitude of steady-state mRNA levels (R = 0.82) (SI Appendix, Fig. S8B). This result indicates that active transcriptional regulation by TGA1 can largely explain changes in target gene transcript levels.
Next, we aimed to detect TGA1-DNA binding events in root cells using the TARGET assay. To do this, we employed ChIP-seq, using anti-GR antibodies against the TGA1-GR fusion protein (28). The aggregate binding profile of gene targets bound by TGA1 in root cells shows that TF binding occurs close to the transcription start site as well as the transcription termination site (SI Appendix, Fig. S9 and Table S4). We found a significant overlap between genes found bound by TGA1 and target genes directly regulated by TGA1 (SI Appendix, Fig. S8C). We also found a significant overlap between genes bound to TGA1 by ChIP-seq, and those bound to TGA1 as assayed by in vitro DNA affinity purification sequencing (DAP-seq) (29) (SI Appendix, Fig. S8C).
Combined, our mRNA-seq, 4tU, and ChIP-seq experiments allowed us to detect gene targets that TGA1 both binds to and actively regulates. For example, the NODULE INCEPTION PROTEIN 8 (NLP8) and CRY2-INTERACTING bHLH 3 (CIB3) TFs were among genes bound to and directly regulated by TGA1. NLP8 has been previously implicated in regulating N-promoted seed germination (30), while CIB3 has been previously implicated in light signaling (31) (Fig. 3B). The expression of both genes was induced by TGA1 nuclear import under +CHX treatments, indicating that NLP8 and CIB3 are primary TGA1 targets. ChIP-seq confirmed TGA1 binding in the promoter of NLP8 and CIB3, and the 4tU labeling showed that both genes were actively transcribed due to TGA1 binding (Fig. 3B).
From these datasets, we found that TGA1 directly regulates a large number of TFs (92 TFs out of 584 TGA1 direct targets, 16%). Indeed, the GO term “regulation of transcription” was significantly enriched among the 584 direct targets of TGA1 (P = 0.02) (SI Appendix, Table S5). The large amount of TFs directly controlled by TGA1 suggests that a main function of TGA1 is to induce a transcriptional cascade. To characterize the downstream effects of TGA1, we performed our TARGET TF-perturbation experiment without blocking protein translation (i.e., in the absence of CHX). This allowed us to also detect gene expression responses that were downstream of TGA1’s direct TF targets (Fig. 3A). We identified a total of 2,280 “indirect” targets of TGA1, defined as genes that are regulated by DEX-induced nuclear import of TGA1 under −CHX conditions, but not in +CHX conditions (SI Appendix, Table S4). GO term enrichment analysis of TGA1 indirect targets included “response to nutrient levels” and “growth.” To make sense of how the direct and indirect targets of TGA1 identified in root cells impact the expression of genes involved in N uptake and assimilation, we built a TF network (Fig. 3C). We used Cytoscape (32) to visualize the resulting network that included 32 genes in the N-uptake/assimilation pathway that are downstream targets of TGA1. This network revealed two modes of action of TGA1 on the N-uptake/assimilation pathway. First, TGA1 directly regulates the expression of AMMONIUM TRANSPORTER 1.3 (AMT1.3), GLUTAMATE SYNTHASE 1 (GLT1), GLUTAMATE SYNTHASE 2 (GLU2), GLUTAMATE DECARBOXYLASE (GAD4), and ASPARAGINE SYNTHASE 1 (ASN1). Second, through the regulation of secondary TFs, TGA1 indirectly influences the expression of additional N-uptake/assimilation genes, such as NITRATE TRANSPORTER 1.2 (NRT1.2), NITRATE REDUCTASE 2 (NIA2), NITRITE REDUCTASE (NIR), and ASPARTATE AMINOTRANSFERASE 3 (ASP3) (Fig. 3C).
Importantly, through repeating our TARGET TF-perturbation experiments in root cells isolated from the tga1/tga4 mutant, we found that the absence of TGA4 did not impact the regulation of TGA1 direct or indirect targets, as identified by regulated TGA1 nuclear import (SI Appendix, Fig. S10). This suggests that TGA1 and TGA4 are functionally redundant in N-signaling responses, in agreement with previous reports (18).
Last, we investigated the extent to which this TGA1 regulated gene network identified in root cells captured genes whose N-dose–responsive expression was explained by the MM model in planta (Fig. 1C). We found a significant overlap of both direct and indirect targets of TGA1 with genes whose N-dose–dependent expression rates fit the MM model in planta (SI Appendix, Fig. S8C). In line with TGA1 acting as an activator of gene expression, we found that the majority of TGA1 direct targets held higher Vmax estimates in the 35S::TGA1 line compared to wild type (SI Appendix, Fig. S8D). We found that the majority of TGA1 downstream indirect targets also held higher Vmax estimates, suggesting that TGA1 impacts rates of N-dose–dependent gene expression through secondary TFs.
TGA1 Levels Mediate Accelerated Arabidopsis Growth Rates in Response to N-dose.
Since we found evidence that TGA1 plays a role in regulating rates of transcription in response to N-dose, including the regulation of N-uptake/assimilation genes, we next investigated TGA1’s effect on plant phenotype under increasing N-dose treatments. To do this, we examined how wild-type plants, the 35S::TGA1-overexpressing line, and the tga1/tga4 double-knockout mutant line grew in response to four different N doses (0.1, 1, 10, and 60 mM N, provided as KNO3 + NH4NO3). By sampling whole-plant dry weight 6, 9, 12, and 15 d after sowing, we found that levels of TGA1 expression impacted plant growth rates in response to N-dose (Fig. 4 A–F and I).
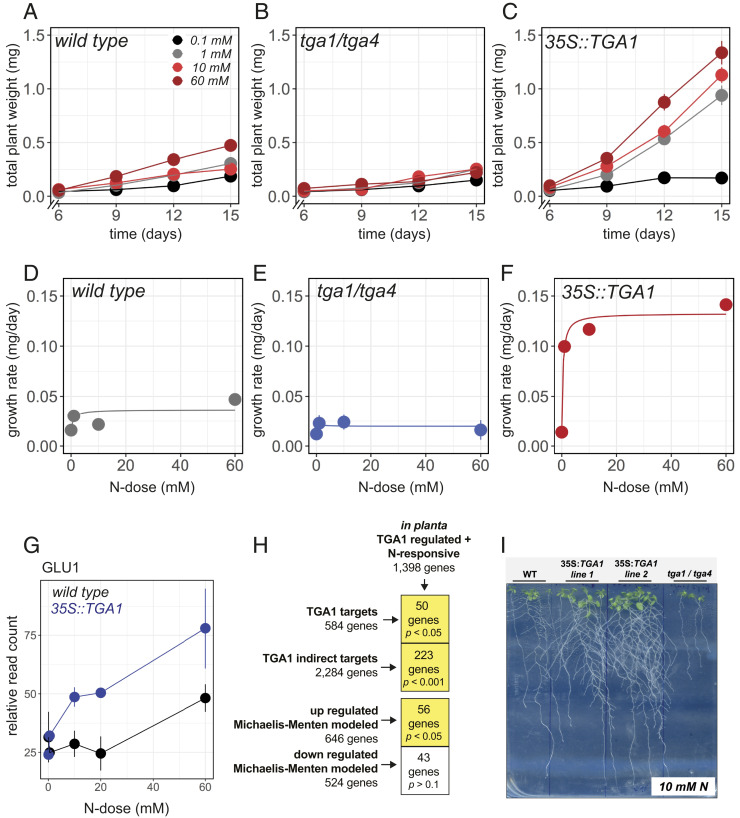
Fig. 4.
Changes in TGA1 levels impacts N-dose–responsive growth rates in planta. (A–C) Growth rates of (A) wild-type, (B) tga1/tga4, and (C) 35S::TGA1 seedling growth over time (days) under different N-doses. The impact of N-dose on growth rates differs significantly between wild-type and the 35S::TGA1 line (three-way ANCOVA, P = 4.1 10−5), and between wild-type and the tga1/tga4 line (P = 1.5 10−7). (D–F) Rates of plant growth of (D) wild-type, (E) tga1/tga4, and (F) 35S::TGA1 plants, fit with the MM model. (G) GLU1 gene expression assayed under these N-dose conditions in wild-type and 35S::TGA1. (H) Genes differentially expressed in response to N-dose in 35S::TGA1 plants significantly intersect with direct and indirect targets of TGA1, as well as MM-modeled genes (Monte Carlo test). (I) Phenotype of 15-d-old wild-type plants, a tga1/tga4 mutant, and two independent TGA1-overexpressing lines grown on plates containing 10 mM N.
Compared to wild type, overexpression of TGA1 led to significantly higher growth rates and increased biomass (three-way ANCOVA, P = 4.1 10−5). This effect was dependent on N dose; higher rates of growth were achieved with higher doses of N. At the highest N-dose of 60 mM, 35S::TGA1 plants grew three times faster than wild type, reaching a dry weight that was 2.8 times greater (Fig. 4 C and F and SI Appendix, Fig. S11). This result was confirmed using an independent transgenic line of 35S::TGA1 (SI Appendix, Fig. S11). Consistent with these findings, the tga1/tga4 mutant grew 2.9 times slower compared to wild type, with a final dry weight that was 2.1 times smaller (P = 1.5 10−7) (Fig. 4 B and E and SI Appendix, Fig. S11).
We also assayed the root transcriptomes of wild-type and 35S::TGA1 plants grown under the N-dose conditions described above. By these means, we found evidence that TGA1’s ability to accelerate plant growth was associated with changes in expression of TGA1 gene targets. Specifically, the N-dose–response of 1,398 genes were significantly perturbed in the 35S::TGA1 background, as identified by two-way ANCOVA analysis (SI Appendix, Fig. S11 and Table S6). Among these genes were known N-assimilation genes such as GLUTAMATE SYNTHASE (GLU1), which displayed a higher N-dose–response in the 35S::TGA1 line (Fig. 4G). We found that these 1,398 genes whose N-dose–response was perturbed in the 35S::TGA1 line significantly intersected with genes characterized as direct or indirect targets of TGA1, or whose N-dose expression was explained by MM kinetics (Fig. 4H). Collectively, these results suggest TGA1’s influence on transcription rates of target genes at the molecular level translates to increased rates of growth in response to N-dose.
Discussion
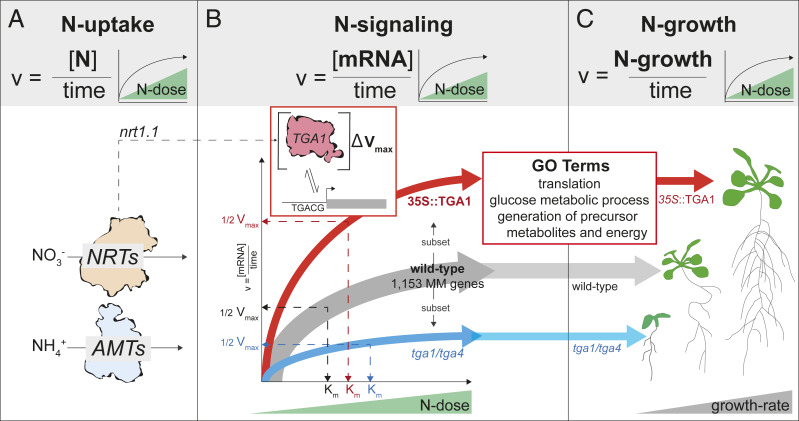
As key nutrient, N has a dose-responsive effect on plant growth and development. Indeed, N-dose–responsive traits, including the rate of N uptake as well as the rate of N-responsive plant growth, have been shown to follow MM kinetics (Fig. 5 A and C) (10, 11). However, the molecular mechanisms that align the rate of N uptake with the rate of N-mediated changes in plant growth have remained poorly understood. Results presented herein help bridge this gap. Specifically, we show that N-dose–responsive transcriptome changes driven by MM kinetics underlie changes in plant growth rate. Additionally, we identify TGA1 as a TF involved in mediating this N-dose– response (Fig. 5B).
Fig. 5.
The molecular basis for Michaelis–Menten (MM) kinetics that underlies N-dose–dependent regulation of plant growth. The MM model (upper gray panel) can explain the effect of N-dose on rates of N uptake (A), N signaling (B), and N growth (C). (A) The rate of N uptake by plant nitrate (NRTs) and ammonium (AMTs) transporters follows MM kinetics (11). The N-regulated expression of the NRT1.1 nitrate transceptor has also been shown to follow MM kinetics (53). (B) Results presented herein show that transcriptome responses to N-dose follows MM kinetics (1,153 genes) (Fig. 1). Moreover, a portion of this MM-mediated transcriptome response (192 genes) to N-dose is affected by TGA1, as shown via changes in Vmax and Km in tga1/4 and 35S::TGA1 lines (Figs. 2 and 4 and SI Appendix, Fig. S7). TGA1’s role in signaling N-dose at the molecular level impacts genes involved in translation, glucose metabolism, and energy metabolism. The N response of TGA1 mRNA levels is altered in the nrt1.1 transceptor mutant (SI Appendix, Fig. S12) (18). (C) Overexpression of TGA1 (35S::TGA1) leads to higher N-dose–dependent growth rates, while it is reduced in the tga1/4 mutant (Fig. 4 D–F). Thus, the role of TGA1 in mediating rate of transcript change in response to N-dose provides a molecular basis for how N-dose regulates rates of plant growth, a phenotype that can also be explained by MM kinetics (Fig. 4 D–F) (10).
Previous studies have shown that mRNA responses of individual genes to changes in endogenous or exogenous signals can be explained by the MM model (33, 34). Herein, we found the MM model could explain transcriptome-wide transcriptional changes in response to N-dose. Specifically, for 30% of all N-dose–responsive genes (1,153 genes), we found that the rates at which they respond transcriptionally to N-dose could be explained by MM kinetics (Fig. 1). Importantly, MM model revealed the relationship between N-dose and rates of transcriptional change was not linear—higher doses of N had diminishing effects on the rate at which genes were induced or repressed. This effect is similar to what occurs in in vitro enzymatic reactions the MM model describes; the rate of the reaction will peak at saturating levels of substrate (35). Likely, the expression of the remaining 70% of N-dose–responsive genes that were not explained by the MM model were driven by more complex kinetics. For example, such genes may be influenced by both activating and repressing TFs, resulting in more complex patterns of expression. This is supported by previous reports showing multiple TFs can target the same N-responsive gene (20, 36). Additionally, such transcripts may hold distinct mRNA degradation rates, or be impacted by posttranscriptional regulatory mechanisms (37, 38); parameters that are not included in the MM model.
Modeling transcriptome kinetics led us to identify TGA1 as a candidate TF involved in establishing rates of transcript change in response to N-dose (Fig. 2). This was based on TGA1’s early N-dose–responsive expression, and the enrichment of its cis-binding motif within the 1,153 genes whose N-dose–response fit the MM model. By modeling the kinetics of transcript change, we found that increasing levels of TGA1 in planta can increase the maximum rate of gene expression (Vmax). This agrees with the MM model, where an increase in the amount of catalyst—in our case, a TF—allows for higher rates of reaction (Vmax) to occur. By illustrating in vivo and genome-wide that the catalytic effects a TF can have on transcription rates follows this simple kinetic principle, we provide biological context to TF kinetic properties that are absent from in vitro assays of TF–target binding.
We note that within our MM modeling, the parameters of Vmax and Km not only depend on the transcriptional activity of TGA1, but are also informed by additional gene regulatory mechanisms, including other TFs. This is exemplified by N-dose–dependent expression of some MM-modeled genes occurring even in the absence of TGA1 (i.e., in the tga1/tga4 mutant background) (Fig. 2E). Moreover, according to the MM model, an increase in enzyme concentration in vitro leads to an increase in Vmax, without a change in Km (12). Thus, our observation of a change in Km values in both tga1/4 mutants and in 35S::TGA1 (SI Appendix, Fig. S7) suggests additional transcriptional or posttranscriptional mechanisms are at play in regulating rates of N-dose gene expression in vivo.
We show that the impact of TGA1 on the rate of transcriptional change in response to N-dose is linked to TGA1’s ability to accelerate plant growth (Fig. 4 and SI Appendix, Fig. S11). This is supported by our data; a significant portion of genes whose expression can be modeled by MM kinetics in response to N-dose are also differentially expressed in TGA1-overexpressing plants, plants that displayed accelerated growth rates (Fig. 4 and SI Appendix, Fig. S11). In this way, TGA1 likely plays a key role in aligning the rate of N uptake with rate of N growth (Fig. 5). For this reason, our findings provide context as to why overexpression of TFs can lead to improvements in plant growth responses. Such a mechanism may be at work in other studies in which overexpression of TFs involved in N signaling have led to improvements in plant N-use efficiency, such as DOF1 and NLP7 (39, 40). For our study, we speculate that the increase in growth rate caused by TGA1 overexpression is due to improvements in N-assimilation efficiency, rather than an increase in N uptake. This is suggested by our TGA1 network, which reveals that TGA1 directly—or indirectly through intermediate TFs—regulates 12 genes involved in N assimilation in root cells, compared to only three genes involved in N transport (Fig. 3C). This interpretation is also supported by previous studies showing that TGA1 regulates genes involved in N metabolism in roots (18, 20), and that N uptake is not perturbed in tga1/tga4 double mutant (18). However, we note that TGA1’s effect on N-dose–responses likely involves more than simply targeting N-assimilation genes. We found evidence that TGA1 regulates the expression of 92 other TFs—agreeing with a previous report showing TGA1 regulates the expression of many N-responsive TFs (20). Consequently, TGA1 does not act alone in regulating N-dose–responses.
Since TGA1’s expression itself is responsive to N-dose, it is likely that TGA1 sits downstream of the molecular mechanisms that sense N-dose. One such mechanism is NRT1.1, a nitrate transceptor that allows plants to sense a wide range of nitrate doses, and whose expression is also driven by MM kinetics (33). To test the hypothesis that TGA1 sits downstream of the NRT1.1 nitrate transceptor, we overlapped genes whose expression is dependent on NRT1.1 (41) with genes regulated by TGA1 (20), as well as genes whose expression is modeled by MM kinetics in our study (SI Appendix, Fig. S12). The significant overlap between these gene lists suggest that NRT1.1 and TGA1 are in the same pathway that control rates of transcriptional change in response to N-dose (Fig. 5 and SI Appendix, Fig. S12).
Our finding that increasing TGA1 mRNA levels results in proportional increases in mRNA of its target genes supports time-based methods that rely on TF mRNA levels to infer TF–target interactions (4, 5, 42, 43). In agreement with this, one recent study that employed time-series mRNA data to infer TF–target interactions used 33 TFs—including TGA1—to experimentally validate the resulting gene-regulatory network (20). In that study, TGA1 mRNA levels were able to predict target gene expression with one of the highest precision rates (75%) of all 33 TFs tested (20).
Finally, our study demonstrates that assaying for rates of transcript change in response to nutrient dose, rather than simply a change in gene expression at one time point, represents a fruitful approach to identify loci of interest. Indeed, this approach could be applied to identify TF regulators of other types of environmental cues in plants or other species. In our case, we identified TGA1, a TF that accelerates plant growth responses to N in part by impacting transcription rates of N-metabolism genes. As such, future work should investigate whether overexpression of TGA1 enhances crop growth in the field. More broadly, our study of the basic mechanisms that underlie the transcriptome kinetics that respond to changes in N-dose has the potential to enhance plant growth rates and improve N-use efficiency.
Materials and Methods
N-Dose-by-Time Factorial Experiment.
Approximately 100 seedlings of Arabidopsis Col-0, 35S::TGA1 (sourced from ref. 20) and tga1/tga4 (sourced from ref. 18) genotypes were grown for 13 d hydroponically on MS media (16), supplemented with 1 mM KNO3 and 1% sucrose. Light conditions were diurnal (16-h light and 8-h dark), with light intensity 120 μmol⋅m−2⋅s−1 at constant temperature of 22 °C. After 13 d, plants were starved for N for 1 d. After 24 h of N starvation, 2 h after subjective dawn, plants were provided with one of four N-doses, either no N, 0.3 mM KNO3 + 0.3 mM NH4NO3, 3.3 mM KNO3 + 3.3 mM NH4NO3 or 20 mM KNO3 + 20 mM NH4NO3. This created total-N doses of 0, 1, 10, or 60 mM. For each N-dose, N exposure time lasted for one of five time periods: 15, 30, 60, 120, or 240 min. At the respective time point, roots were cut and frozen immediately in liquid nitrogen. Each genotype was tested under each dose–time condition in duplicate.
RNA was extracted from root tissue using the RNeasy Mini Kit (Qiagen) with on-column DNase treatment. RNA quality was assessed using Agilent Tapestation using High Sensitivity RNA ScreenTape. One microgram of total RNA per sample was depleted of rRNA by Thermo Fisher Scientific mRNA Purification Kit. RNA-seq libraries were made using the NEBNext Ultra RNA Library Prep Kit and sequenced using Next Seq Illumina platform with 175-bp single read-end chemistry. Reads were then aligned to the Arabidopsis TAIR10 genome using Tophat (44), and gene counts called using HT-seq (45) with Araport11 annotation (46). Statistical analyses were then performed in R, as detailed below.
To control for the effect of potassium ions inducing differential expression within our assay, we compared Arabidopsis seedlings (Col-0) treated for 240 min with a dose of 20 mM KNO3 + 20 mM NH4NO3 with seedlings treated for 240 min with a mock dose of 20 mM KCl. We found 44 differentially expressed genes due to salt treatment using DESeq2 (47) (adjusted P < 0.01). These genes were removed from subsequent analyses.
N-Dose-by-Time Statistical Analysis.
Our factorial matrix experiment held two continuous variables—time and N-dose. To capture genes that were differentially expressed by either or both of these factors, we used the following linear model:
The full linear model was fit to the RNA-seq read counts of each gene (implemented in DESeq2 using design ∼ N + T + N × T), where N and T variables were logged to the base 2. We then performed model simplification as follows:
-
1)
Using the “LRT” command, an adjusted P value was computed for each of the factors within the model across all fit genes.
-
2)
If a gene were fit significantly by all three terms (adjusted P < 0.01), then this gene was deemed fit by the full model and removed from remaining model simplification steps.
-
3)
For all remaining genes, the factor with the least significance (highest adjusted P value) was removed, and the model was refit with the remaining terms. This allows for one of three variations of a simplified model to be fit for each gene.
-
4)
If a gene was fit significantly by two terms (both adjusted P values < 0.01), then this gene was deemed fit by a two-term model and removed from remaining model simplification steps.
-
5)
Steps 3 and 4 were repeated fitting one term models.
If a gene was not fit by any model form, then it was removed from further analysis. GO terms were called using VirtualPlant software (48).
To calculate rates of change in transcript abundance of genes at each N-dose, we fit each gene’s quantile-normalized expression values using the lm() function in R, using time as the dependent variable. These estimated rates of transcription under each N dose were then fit to the MM model using the drm() function in R. We note that rates were first normalized by subtracting the rate calculated for the 0 mM condition. After this subtraction, we then took the absolute values of each rate, which allowed us to model genes that had decreases in rates of transcription at higher N-doses (conferring to increases in gene repression over N-dose). We calculated the significance of the model by correlating model predicted values with input values (Pearson false-discovery rate-normalized P < 0.05). Additionally, we tested whether a simpler model could explain changes in transcription dynamics. We assessed whether any of the 3,818 genes followed first-order kinetics—a model in which the rate of mRNA change is linearly proportional to N-dose. We found no genes that significantly fit this model.
RNA-Seq, 4tU, and ChIP-Seq TARGET TF-Perturbation Experiments.
The cell-based TARGET assay for TF perturbation was performed as described in ref. 22 for steady-state mRNA and as in ref. 26 to capture de novo transcripts with 4tU. Briefly, Col-0 or tga1/tga4 seedlings were grown on vertical plates supplemented with MS media at 1 mM KNO3 and 1% sucrose under light conditions as described above. After 14 d, 2 h after subjective dawn, plant roots were harvested, finely cut, and placed in protoplast solution for 3 h. Root cell protoplasts were then washed and then PEG mediated transfected with pBOB11_C-term (available through https://gatewayvectors.vib.be/; National Center for Biotechnology Information accession number MN991175), a derivative of the pBeaconRFP_GR TARGET TF expression vector (22), which contains the TGA1-GR fusion protein and a 35S::RFP gene for selection of transfected cells by fluorescence-activated cell sorting (FACS) (22). Transfected root cells were incubated overnight to express the TF-GR fusion protein. The following day, the root cells were treated 2 h after subjective dawn with N-dose present in standard MS media (20 mM KNO3 + 20 mM NH4NO3), or control 20 mM KCl treatment. Cells were treated with CHX 100 min thereafter, or with a mock treatment of DMSO to enable the identification only of direct targets of the TF. To induce TF-GR localization to the nucleus, cells were treated with DEX 20 min thereafter, or with a mock ethanol treatment. Each treatment combination was tested in triplicate. Treated root cells were incubated for a further 2 h, then successfully transfected cells were FACS purified using RFP expression. To perform 4tU mRNA labeling, we repeated the above experimental treatment on transfected root protoplasts, treating three replicates of transfected root protoplasts with 1.5 mM 4tU in the presence of DEX and CHX, and three replicates in the presence of CHX only (26). All RNA-seq libraries were prepared through purifying polyadenylated transcripts as previously described above, and sequenced using the HiSeq Illumina platform with 150-bp single read-end chemistry or Next Seq Illumina platform with 175-bp single read-end chemistry.
For ChIP-seq analysis of TF binding, Col-0 seedlings were grown, and root cells were isolated and transfected as described above. Approximately 1 million root cells were treated for 100 min with 20 mM KNO3 + 20 mM NH4NO3, followed by a 20-min treatment with CHX, followed by a treatment of DEX lasting 1 min. Protoplasts were fixed with formaldehyde for 10 min, and then treated with 2 M glycine for 5 min, before being flash frozen in liquid nitrogen. ChIP was performed on samples as outlined in ref. 28, using an anti-GR antibody (GR P-20; Santa Cruz Biotechnology; 200 μg/mL). ChIP-seq libraries were made using NEBNext Ultra II DNA Library Prep Kit and sequenced using Next Seq Illumina platform with 175-bp single read-end chemistry.
TARGET RNA-Seq Statistical Analysis.
To find direct targets of TGA1, we implemented a three-way ANODEV model on our +CHX RNA-seq datasets in DESeq2 with design ∼ N + DEX + biological replicate. To find genes directly regulated by TGA1 in the presence of CHX, we took those genes that were significantly differentially expressed due to DEX treatment using the contrast function (adjusted P value < 0.01). To control for the effects of CHX on gene expression, we performed a two-way ANODEV model on our –CHX RNA-seq dataset, using design ∼ N + DEX. In this way, we found those genes that were significantly differentially expressed due to DEX treatment in the absence of CHX (adjusted P value < 0.01). We deemed a gene a direct target of TGA1 if it was differentially expressed due to DEX-induced TF nuclear import in both +CHX and –CHX conditions. A gene was deemed an indirect TARGET of TGA1 if it was found to be regulated in response to DEX-induced TF nuclear import only in the absence of CHX. GO terms were called using VirtualPlant software (48).
ChIP-Seq Analysis.
Reads obtained from the ChIP DNA and Input DNA were filtered and aligned to the Arabidopsis thaliana genome (TAIR10) using Bowtie2 (49), where clonal reads were removed. The ChIP alignment data were compared to its partner Input DNA and peaks were called using MACS2 (q = 0.05) (50). These regions were overlapped with the genome annotation to identify genes within 2 kbp downstream of the peak using Bedtools (SI Appendix, Table S5) (51). Genome browser images were made using the Integrative Genomics Viewer (52).
Plant Biomass Phenotyping.
Arabidopsis seedlings—Col-0, 35S::TGA1 (line 1), 35S:TGA1 (line 2), and tga1/tga4—were grown on vertical plates. Light and temperature conditions were identical to those described above. Plants were grown on MS media, with total N concentrations 0.1, 1, 10, and 60 (where N was supplied as KNO3 + NH4NO3). Total plant dry weight was measured 6, 9, 12, and 15 d after sowing.
Steady-State Transcriptomics.
Arabidopsis seedlings Col-0 and 35S::TGA1, were grown on vertical plates for 15 d on 0.1, 0.5, 10, 20, or 60 mM N KNO3 + NH4NO3. Root tissue was then flash frozen and sequenced as described previously. To discover differentially expressed genes, we fit an ANODEV model in DESeq2 (using design ∼ Genotype + log2Nitrogen). We used genes that were found differentially expressed in response to both log2Nitrogen and Genotype factors to intersect with other gene sets. The significance of intersects were assessed using Monte Carlo simulations (10,000 iterations), using the intersect of differentially expressed genes within each dataset as background.
Data Availability.
The data and R code that support the findings of this study are available from the corresponding author upon reasonable request. Raw sequencing data can be found at the National Center for Biotechnology Information Sequence Read Archive (accession number PRJNA522060).
Supplementary Material
Acknowledgments
This work was supported by a grant on Plant Genomics to G.M.C. from the Zegar Family Foundation (A16-0051), the Beachell-Borlaug International Scholarship to J.S., and grants to G.M.C. from the National Science Foundation (NSF) (NSF–Plant Genome Research Program Grants IOS-1339362 and NSF-DBI-0445666). We thank Joan Doidy for providing pBOB11_C-term vector, and Laurie Leonelli for critical reading of the manuscript.
Footnotes
The authors declare no competing interest.
Data deposition: The raw sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA522060). Vectors are available from National Center for Biotechnology Information (accession no. MN991175).
See QnAs on page 12508.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918619117/-/DCSupplemental.
References
- 1.Wang Y. Y., Hsu P. K., Tsay Y. F., Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Breitburg D., et al. , Declining oxygen in the global ocean and coastal waters. Science 359, eaam7240 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Canales J., Moyano T. C., Villarroel E., Gutiérrez R. A., Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci 5, 22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krouk G., Mirowski P., LeCun Y., Shasha D. E., Coruzzi G. M., Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11, R123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varala K., et al. , Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. U.S.A. 115, 6494–6499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson K., et al. , Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 33, 1486–1501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R., et al. , Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136, 2512–2522 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swift J., Adame M., Tranchina D., Henry A., Coruzzi G. M., Water impacts nutrient dose responses genome-wide to affect crop production. Nat. Commun. 10, 1374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X. S., et al. , Gene expression biomarkers provide sensitive indicators of in planta nitrogen status in maize. Plant Physiol. 157, 1841–1852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lana R., et al. , Application of Lineweaver–Burk data transformation to explain animal and plant performance as a function of nutrient supply. Livest. Prod. Sci. 98, 219–224 (2005). [Google Scholar]
- 11.McNickle G. G., Brown J. S., When Michaelis and Menten met Holling: Towards a mechanistic theory of plant nutrient foraging behaviour. AoB Plants 6, plu066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menten L., Michaelis M. I., Die kinetik der invertinwirkung. Biochem. Z. 49, 333–369 (1913). [Google Scholar]
- 13.Michaelis L., Menten M. L., Johnson K. A., Goody R. S., The original Michaelis constant: Translation of the 1913 Michaelis–Menten paper. Biochemistry 50, 8264–8269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Uden N., Transport-limited fermentation and growth of Saccharomyces cerevisiae and its competitive inhibition. Arch. Mikrobiol. 58, 155–168 (1967). [DOI] [PubMed] [Google Scholar]
- 15.López S., et al. , A generalized Michaelis–Menten equation for the analysis of growth. J. Anim. Sci. 78, 1816–1828 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Murashige T., Skoog F., A revised medium for rapid growth and Bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962). [Google Scholar]
- 17.Rubin G., Tohge T., Matsuda F., Saito K., Scheible W. R., Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez J. M., et al. , Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Gifford M. L., Dean A., Gutierrez R. A., Coruzzi G. M., Birnbaum K. D., Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U.S.A. 105, 803–808 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks M. D., et al. , Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 10, 1569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titiz O., et al. , PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 48, 933–946 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Bargmann B. O. R., et al. , TARGET: A transient transformation system for genome-wide transcription factor target discovery. Mol. Plant 6, 978–980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi N., Winter C. M., Wellmer F., Wagner D., Identification of direct targets of plant transcription factors using the GR fusion technique. Methods Mol. Biol. 1284, 123–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Para A., et al. , Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 10371–10376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindermayr C., Sell S., Müller B., Leister D., Durner J., Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doidy J., et al. , “Hit-and-Run” transcription: De novo transcription initiated by a transient bZIP1 “hit” persists after the “run.” BMC Genomics 17, 92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleary M. D., Meiering C. D., Jan E., Guymon R., Boothroyd J. C., Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23, 232–237 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Para A., Li Y., Coruzzi G. M., μChIP-Seq for genome-wide mapping of in vivo TF-DNA interactions in Arabidopsis root protoplasts. Methods Mol. Biol. 1761, 249–261 (2018). [DOI] [PubMed] [Google Scholar]
- 29.O’Malley R. C., et al. , Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan D., et al. , NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7, 13179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Li X., Li K., Liu H., Lin C., Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9, e1003861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho C. H., Lin S. H., Hu H. C., Tsay Y. F., CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Nachman I., Regev A., Friedman N., Inferring quantitative models of regulatory networks from expression data. Bioinformatics 20 (suppl. 1), i248–i256 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Segel I. H., Enzyme kinetics: Behavior and analysis of rapid equilibrium and steady state enzyme systems. FEBS Lett. 60, 102–103 (1975). [Google Scholar]
- 36.Gaudinier A., et al. , Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563, 259–264 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Vidal E. A., et al. , Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 4477–4482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honkela A., et al. , Genome-wide modeling of transcription kinetics reveals patterns of RNA production delays. Proc. Natl. Acad. Sci. U.S.A. 112, 13115–13120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu L. H., et al. , Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6, 27795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagisawa S., Akiyama A., Kisaka H., Uchimiya H., Miwa T., Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. U.S.A. 101, 7833–7838 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouguyon E., et al. , Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1, 15015 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Mirowski P., LeCu Y., “Dynamic factor graphs for time series modeling” in Machine Learning and Knowledge Discovery in Databases, Buntine W., Grobelnik M., Mladenic D., Shawe-Taylor J., Eds. (Springer, Berlin, 2009), vol. 2, pp. 128–143. [Google Scholar]
- 43.Krouk G., Lingeman J., Colon A. M., Coruzzi G., Shasha D., Gene regulatory networks in plants: Learning causality from time and perturbation. Genome Biol. 14, 123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S., Pyl P. T., Huber W., HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng C. Y., et al. , Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katari M. S., et al. , VirtualPlant: A software platform to support systems biology research. Plant Physiol. 152, 500–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., et al. , Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu H. C., Wang Y. Y., Tsay Y. F., AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57, 264–278 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and R code that support the findings of this study are available from the corresponding author upon reasonable request. Raw sequencing data can be found at the National Center for Biotechnology Information Sequence Read Archive (accession number PRJNA522060).