Significance
A hallmark of numerous inflammatory diseases is the overexpression of chemokines giving rise to inappropriate leukocyte recruitment. The inhibition of chemokines is therefore recognized as an attractive strategy for the development of antiinflammatories. Blood-feeding ticks produce sulfated proteins called “evasins” in their saliva that modulate the host inflammatory response by binding to host chemokines. In this work we employ a semisynthetic approach to generate variants of the ACA-01 evasin from the Amblyomma cajennense tick bearing homogeneous sulfation patterns. We reveal that sulfation significantly enhances chemokine binding as well as chemokine receptor inhibition. Our work lays the foundation for the development of engineered sulfopeptides and sulfoproteins to target a range of inflammatory diseases associated with dysregulated chemokine-receptor signaling.
Keywords: chemokines, sulfation, ticks, evasins, antiinflammatory
Abstract
Blood-feeding arthropods produce antiinflammatory salivary proteins called evasins that function through inhibition of chemokine-receptor signaling in the host. Herein, we show that the evasin ACA-01 from the Amblyomma cajennense tick can be posttranslationally sulfated at two tyrosine residues, albeit as a mixture of sulfated variants. Homogenously sulfated variants of the proteins were efficiently assembled via a semisynthetic native chemical ligation strategy. Sulfation significantly improved the binding affinity of ACA-01 for a range of proinflammatory chemokines and enhanced the ability of ACA-01 to inhibit chemokine signaling through cognate receptors. Comparisons of evasin sequences and structural data suggest that tyrosine sulfation serves as a receptor mimetic strategy for recognizing and suppressing the proinflammatory activity of a wide variety of mammalian chemokines. As such, the incorporation of this posttranslational modification (PTM) or mimics thereof into evasins may provide a strategy to optimize tick salivary proteins for antiinflammatory applications.
A defining feature of inflammation is the accumulation of leukocytes in the affected tissues. Leukocyte recruitment is orchestrated by the interactions of chemokine receptors, localized in the leukocyte cell membrane, with chemokines, small proteins that are up-regulated in tissues as a response to injury or infection (1, 2). Dysregulation of the chemokine–chemokine receptor axis underpins a number of immune and inflammatory diseases, for example allergic asthma (3) and atherosclerosis (4). As such, blockade of chemokine signaling is recognized as a viable avenue for the development of therapeutics for diseases characterized by excessive recruitment of leukocytes. The importance of chemokine-receptor signaling for the control of inflammatory responses is further highlighted by the mechanisms that pathogens have evolved to evade chemokine-mediated processes (5, 6). For example, a number of mammalian viruses interfere with chemokine-receptor signaling through the expression of chemokine mimics, chemokine receptor mimics, or chemokine-binding proteins (CKBPs) that are structurally distinct from chemokines and their receptors (7–11).
The salivary glands of hematophagous arthropods possess a mixture of bioactive proteins that facilitate essential blood-feeding activities. In particular numerous species of ticks express two distinct families of CKBPs (called the “evasins”) that bind and inhibit mammalian chemokines, resulting in suppression of host inflammation to enable prolonged feeding without detection (12, 13). The archetypal evasins, evasin-1, evasin-3, and evasin-4 from Rhipicephalus sanguineus (14, 15), have demonstrated efficacy in several murine models of inflammation, thus highlighting the potential of the evasins as antiinflammatory therapeutic candidates (16–19). We and others have recently exploited bioinformatics and yeast display approaches to discover hundreds of putative evasins from tick species spanning the Rhipicephalus, Amblyomma, and Ixodes genera (20, 21). Among these evasin candidates several have been validated to exhibit CKBP activity and possess different spectra of target chemokine selectivity, suggesting that these proteins could be repurposed or engineered for the development of chemokine-targeted therapeutics.
Our initial sequence analyses of the evasins revealed that the N-terminal regions of a large family of evasins (homologous to R. sanguineus evasin-1 and evasin-4) contain one or more tyrosine residue(s) flanked by acidic amino acids—a consensus motif for tyrosine sulfation (22, 23). Remarkably, despite chemokine receptors having unrelated protein structures to the evasins, their N-terminal regions also possess conserved tyrosine residues and posttranslational sulfation of these is known to enhance receptor binding and signaling (24). Interestingly, in a structure of evasin-1 in complex with the chemokine CCL3, the evasin N-terminal region interacts with the same electropositive crevice of the chemokine that binds to the sulfated N termini of chemokine receptors (25). Based on this information, we hypothesized that the tyrosine residues within the evasins may mimic those found on the chemokine receptors and may therefore be posttranslationally sulfated, thus serving to increase the affinity and inhibitory activity for chemokines. To test this hypothesis, we investigated tyrosine sulfation of the recently discovered ACA-01 evasin [also called P974_AMBCA or EVA-P974 (13, 20, 21)] from the tick Amblyomma cajennense. Nonsulfated ACA-01, expressed in Escherichia coli, binds to several proinflammatory chemokines that signal via chemokine receptors CCR2 or CCR3 (20). Herein, we show that ACA-01 secreted by eukaryotic cells is indeed sulfated at two tyrosine residues on the N terminus of the protein, albeit as an inseparable mixture of sulfated variants. To enable functional evaluation of sulfation at each site, we implemented a semisynthetic approach to prepare homogeneous samples of the four possible sulfated variants of ACA-01, namely unsulfated (1), monosulfated on Tyr10 (2), monosulfated on Tyr12 (3), and doubly sulfated (both Tyr10 and Tyr12, 4) (Fig. 1). We demonstrate that tyrosine sulfation at both sites significantly enhances the chemokine-binding affinity and inhibitory activity of ACA-01. These results suggest that tyrosine sulfation is a receptor mimetic posttranslational modification of evasins that ticks have evolved to enhance the effectiveness of their antiinflammatory protein arsenal.
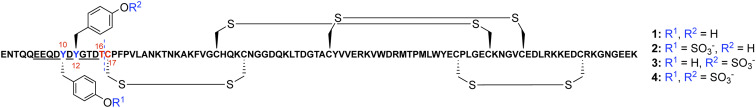
Fig. 1.
Schematic of the ACA-01 evasin sequence. Two predicted Tyr sulfation sites (Y10 and Y12) are shown in blue flanked by a highly acidic stretch of amino acids from E6–D15 (underlined). T16–C17 (shown in red) was selected as the site for semisynthetic assembly of the ACA-01 sulfoproteins by native chemical ligation. The disulfide bond connectivity is predicted based on conservation of cysteine positioning with evasin-1 where disulfide bond connectivity has been confirmed through X-ray crystallography (25).
Results
ACA-01 Is Sulfated at Tyr10 and Tyr12.
Tyrosine sulfation of proteins occurs during secretion and is catalyzed by the enzymes tyrosylprotein sulfotransferase-1 and -2 (TPST-1 and -2) that are localized in the trans-Golgi network (26, 27). The N terminus of ACA-01 (1ENTQQEEQDY10DY12GTDT16) contains two tyrosine residues (Tyr10 and Tyr12) within a highly acidic sequence that we proposed could be posttranslationally sulfated by TPSTs. To investigate whether ACA-01 is sulfated during secretion from eukaryotic cells, a codon-optimized sequence of ACA-01, containing an N-terminal mouse immunoglobulin Kappa leader sequence and C-terminal Myc-His6 tag, was expressed in HEK293 cells. Protein secreted into the cell culture medium was purified via nickel-affinity chromatography followed by size-exclusion chromatography. The purified protein was first analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (Fig. 2A). SDS-PAGE revealed that the protein had a molecular weight significantly higher than predicted from the amino acid sequence, which is consistent with the presence of a potential site for N-glycosylation as has been reported for other evasins (20, 28). Treatment of the purified protein with the glycosidase PNGase F led to a substantial reduction of the observed molecular weight, confirming that ACA-01 was indeed N-glycosylated. Moreover, the protein could be readily detected by Western blotting using a pan-specific antisulfotyrosine monoclonal antibody (29), clearly showing the presence of tyrosine sulfation.
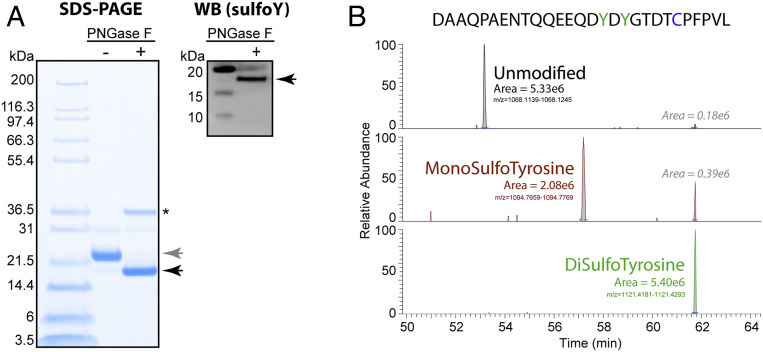
Fig. 2.
Tyrosine sulfation of evasin ACA-01 secreted by eukaryotic cells. (A) SDS-PAGE and Western blot analysis of purified ACA-01 secreted by HEK293 cells, before (−) and after (+) treatment with PNGase F to remove N-glycans. Gray and black arrows indicate N-glycosylated ACA-01-Myc-His6 and deglycosylated ACA-01-Myc-His6, respectively. PNGase F is indicated by an asterisk. (Left) Total protein was detected by Coomassie staining. (Right) Proteins containing sulfotyrosine were detected with immunoblotting using a pan-specific antisulfotyrosine antibody (see SI Appendix, Fig. S29 for full Western blot including nonsulfated ACA-01 as a negative control). (B) LC-MS/MS analysis of chymotryptic peptides derived from a digest of ACA-01 expressed in HEK293 cells. Extracted ion chromatograms were generated for the peptide sequence shown above that was modified at the tyrosine residues (green in sequence) with either both sulfated (green), only one sulfated (brown), or neither being modified (black). The peak area is shown for each ion type. The disulfotyrosine ion showed some artifactual desulfation during the process of ESI and ion transfer and the area of these peaks is shown.
To both gauge the degree of protein sulfation and pinpoint the sulfation site(s) of ACA-01 secreted by HEK293, we used a bottom-up proteomic nano LC-MS/MS (liquid chromatography–tandem mass spectrometry) approach, whereby the protein was digested to peptides using chymotrypsin, with subsequent neutral-pH peptide cleanup to preserve labile sulfotyrosine modifications. Analysis of these peptides with high-resolution MS revealed the presence of doubly sulfated ACA-01 (at Tyr10 and Tyr12), monosulfated ACA-01 (at either Tyr10 or Tyr12), and unmodified forms of the protein (Fig. 2B) (30). While the integrated peak area for the doubly sulfated and unmodified forms was similar, this has not been adjusted for possible different ionization efficiencies induced by the sulfate modifications. Previous studies have shown that introduction of sulfation onto tyrosine residues of peptides greatly suppresses their ionization (31), so it is likely that the modified forms are substantially more abundant than the unmodified form.
Semisynthesis of Homogeneous, Differentially Sulfated Forms of ACA-01.
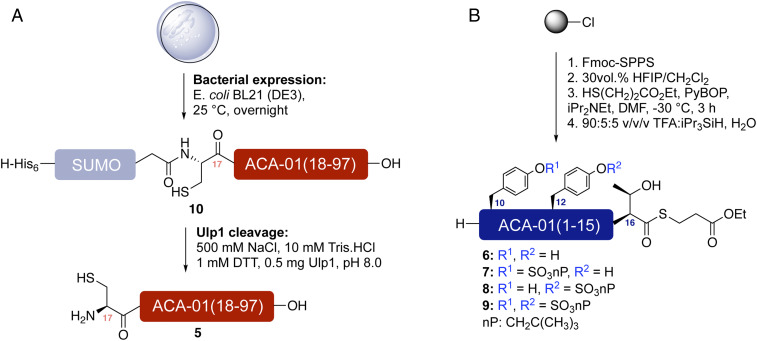
Due to the heterogeneity of sulfation in the expressed material (discussed above) and the size of ACA-01, it was not possible to obtain homogeneously sulfated forms of this evasin by either protein expression or direct chemical synthesis. However, we envisaged that the pure sulfoforms of ACA-01 (1–4) could be prepared via a convergent, semisynthetic strategy. In this approach, the large C-terminal fragment of ACA-01(17–97) 5, bearing an N-terminal cysteine residue, would be obtained by recombinant expression, while chemical synthesis would be used to generate four differentially sulfated N-terminal ACA-01(1–16) peptides 6–9, functionalized at the C terminus as thioesters. Subsequently, the expressed protein fragment 5 would be fused to the synthetic (sulfo)peptide thioesters 6–9 by native chemical ligation (32), followed by folding and purification to produce the target ACA-01 sulfoproteins. The C-terminal fragment 5 was successfully generated by expression of a N-terminal hexahistidine-tagged SUMO-ACA-01(17–97) fusion (10) in BL21 (DE3) E. coli cells (Fig. 3A) and purified using a HiTrap TALON Crude column. Cleavage of the hexahistidine-SUMO tag was carried out during dialysis with Ulp1 protease (33) and the protein was purified on a HiTrap TALON column followed by reverse-phase high-performance liquid chromatography (HPLC) to afford 3 to 4 mg of purified ACA-01(17-97) 5 per liter of culture (SI Appendix, Fig. S1). Synthesis of the (sulfo)peptide thioesters 6–9 was carried out on 2-chlorotrityl chloride resin via Fmoc-based solid-phase peptide synthesis (SPPS) (Fig. 3B and see SI Appendix for full synthetic details). Each of the sulfotyrosine residues were incorporated as neopentyl (nP) protected sulfate esters (34, 35) to prevent the loss of the labile aryl sulfate ester functionalities in the final acidic cleavage step (see SI Appendix for full synthetic details). Following elongation of the target sequence, side chain protected (sulfo)peptides were cleaved from the solid support with hexafluoroisopropanol and the free C-terminal carboxylate thioesterified. Side-chain deprotection using a trifluoroacetic acid-based acidic mixture and purification by reverse-phase HPLC provided 6–9 in 4 to 25% yield over the 33 synthetic steps (SI Appendix, Figs. S2–S5).
Fig. 3.
Expression and synthesis of ACA-01 fragments. (A) Strategy for the generation of ACA-01(17–97) (5) through the expression of a His6-SUMO-ACA-01(17–97) (10) fusion followed by proteolysis with Ulp1. (B) Synthesis of ACA-01(1–15) (sulfo)peptide thioesters fragments (6–9) via Fmoc-strategy SPPS.
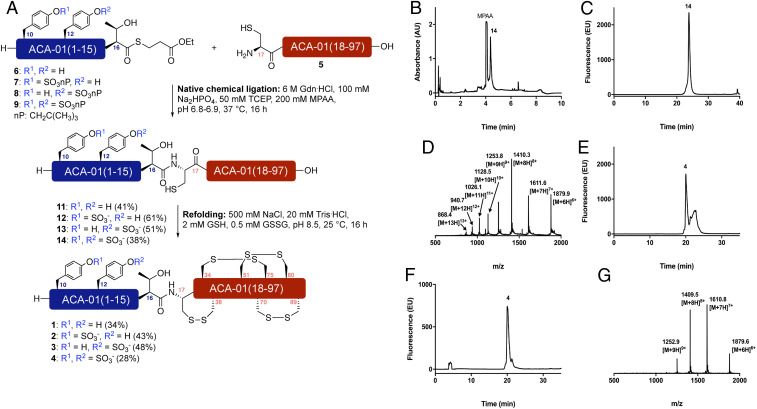
With expressed C-terminal protein fragment 5 and synthetic (sulfo)peptide thioesters 6–9 in hand we next undertook the assembly of the ACA-01 (sulfo)proteins by native chemical ligation (Fig. 4A). Toward this end, bacterially expressed ACA-01(17–97) 5 (1 equivalent [equiv], 2.5 mM) was reacted with synthetic ACA-01(1-16) thioesters 6, 7, 8, or 9 (2 equiv, 5 mM) in 6 M guanidine hydrochloride, 200 mM disodium phosphate (Na2HPO4) buffer in the presence of 50 mM Tris-(2-carboxyethyl)phosphine (TCEP) and 200 mM 4-mercaptophenylacetic acid (MPAA) at pH 6.8 to 6.9. The reactions were monitored by ultraperformance liquid chromatography (UPLC)-MS analysis and reached completion in 16 h to generate proteins 11–14. Each of the reactions proceeded with concomitant neopentyl sulfate ester deprotection to generate the native sulfotyrosine residues in the full-length ACA-01 proteins. The (sulfo)proteins were purified by reverse-phase HPLC using 10 mM ammonia in H2O and 10 mM ammonia in MeCN as eluents to prevent acidolysis of the sulfate ester, which can occur using traditionally employed acidic solvents, for example with trifluoroacetic acid. Following lyophilization, the library of ACA-01 (sulfo)proteins 11–14 were isolated in 38 to 61% yields (see Fig. 4 B–D for representative data for doubly sulfated ACA-01 14 and SI Appendix, Figs. S7–S10; see SI Appendix for details).
Fig. 4.
Assembly of differentially sulfated ACA-01 via native chemical ligation. (A) Semisynthesis of ACA-01 evasin (sulfo)proteins 1–4 via native chemical ligation and oxidative folding. Exemplar data for doubly sulfated ACA-01 4. NB: nP = CH2C(CH3)3; (B) UPLC trace of crude reaction of 5 with doubly sulfated peptide thioester 9; (C) analytical HPLC chromatogram of purified ligation product 14; (D) ESI mass spectrum of purified ligation product 14; (E) analytical HPLC trace of crude folding reaction of 14 to afford 4; (F) analytical HPLC trace of purified folded doubly sulfated ACA-01 protein 4; (G) ESI mass spectrum of purified folded doubly sulfated ACA-01 protein 4.
Having successfully generated the ACA-01 sulfoforms 11–14, we next sought to oxidatively fold the proteins. Gratifyingly, rapid dissolution of lyophilized 11–14 into an optimized refolding buffer, comprising 500 mM NaCl, 20 mM Tris⋅HCl, 2 mM reduced l-glutathione , 0.5 mM oxidized glutathione at pH 8.5 at 0.1 mg⋅mL−1 at 25 °C overnight led to conversion to a folded form with four disulfide bridges. Each of the folding reactions was monitored by UPLC-MS and analytical HPLC and led to a distinct shift in the retention time and mass envelope (−8 Da) in the electrospray ionization (ESI) spectrum. Following complete refolding each of the ACA-01 (sulfo)proteins was purified via reverse-phase HPLC, using 0.1vol % ammonia in the H2O and MeCN eluents as above to prevent acidolysis of the sulfate ester (see Fig. 4 E–G for exemplar data for doubly sulfated ACA-01 4 and SI Appendix, Figs. S11–S18). Following purification, the homogeneously sulfated ACA-01 proteins 1–4 were isolated in 28 to 48% yield. To determine the disulfide bond connectivity of the proteins we analyzed the proteins by digestion and subsequent LC-MS/MS analysis of the peptides without prior reduction of disulfides. This allowed us to observe the disulfide cross-linked peptides directly and confirm the presence of three of the four disulfide bonds known to be present in the folded form of evasins (SI Appendix, Fig. S19). To verify this disulfide bond arrangement in the ACA-01 expressed in HEK293 cells, we also examined this protein using the same method and observed the same disulfide bond structure (SI Appendix, Fig. S20).
Tyrosine Sulfation Enhances the Chemokine-Binding Affinity of ACA-01.
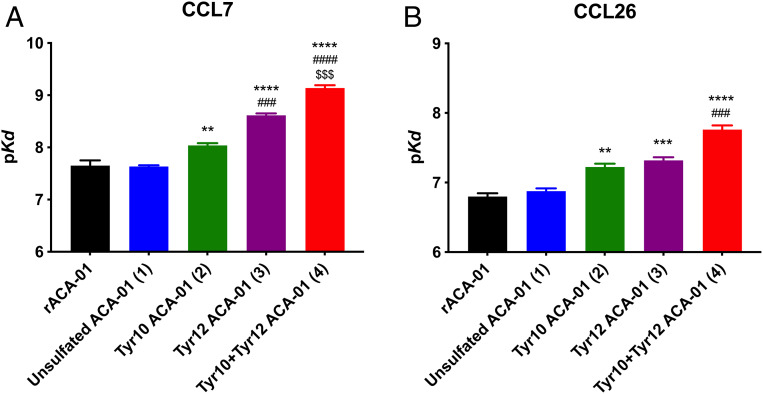
We have previously shown that the unsulfated form of ACA-01 binds to chemokine ligands of CCR2 (a major chemokine receptor on monocytes) and CCR3 (a major chemokine receptor on eosinophils). These include CCR2 ligands CCL2 (monocyte chemoattractant protein-1, MCP-1), CCL7 (MCP-3), and CCL8 (MCP-2) and CCR3 ligands CCL11 (eotaxin-1), CCL24 (eotaxin-2), and CCL26 (eotaxin-3) (20). To test the hypothesis that posttranslational sulfation of ACA-01 modulates its binding affinity for these chemokines, we used a competitive fluorescence anisotropy assay (36), which monitored the ability of evasins 1–4 to displace fluorescein-labeled, receptor-derived sulfopeptides (36, 37) from the target chemokines (SI Appendix, Figs. S21 and S22). Due to the concentration of chemokine (100 nM) required for this assay, the concentration of evasin needed to displace 50% of the fluorescent peptide was higher than the equilibrium dissociation constant (Kd) (20). Nevertheless, Kd values could be readily determined by fitting the displacement data to a competitive binding model (Kd; see Fig. 5 and Table 1).
Fig. 5.
Binding of semisynthetic ACA-01 (sulfo)proteins 1–4 to CC chemokines. Affinities (pKd) derived from the competitive fluorescence anisotropy data for binding of all ACA-01 isoforms (1–4) and recombinantly expressed ACA-01 (rACA-01). Each panel shows the binding affinity of all isoforms for one chemokine: CCL7 (A) and CCL26 (B). Data represent the average pKd ± SEM of values from three independent experiments, each recorded in duplicate. *, #, and $ indicate significant differences from unsulfated ACA-01, Tyr10 ACA-01, and Tyr12 ACA-01, respectively. Significance (one-way ANOVA) is shown as ** P < 0.01; ***, ###, $$$ P < 0.001; ****, ####, $$$$ P < 0.0001. See SI Appendix, Figs. S21 and S22 for binding data.
Table 1.
Binding affinities between ACA-01 (sulfo)proteins 1–4 and CC chemokines*
| ACA-01 | Chemokine | |||||
| CCL11 | CCL24 | CCL26 | CCL2 | CCL8 | CCL7 | |
| rACA-01† | 47.5 (7.32 ± 0.08) | 302.7 (6.52 ± 0.05) | 159.0 (6.80 ± 0.05) | 55.3 (7.26 ± 0.06) | 6.9 (8.12 ± 0.09) | 25.8 (7.59 ± 0.08) |
| Unsulfated (1) | 135.0 (6.87 ± 0.05) | 391.9 (6.41 ± 0.05) | 133.2 (6.88 ± 0.04) | 65.1 (7.20 ± 0.04) | 10.9 (7.97 ± 0.12) | 23.2 (7.64 ± 0.02) |
| Tyr10 (2) | 82.1 (7.09 ± 0.03) | 183.0 (6.74 ± 0.04) | 59.7 (7.23 ± 0.05) | 23.1 (7.65 ± 0.05) | 10.6 (7.98 ± 0.12) | 9.1 (8.04 ± 0.04) |
| Tyr12 (3) | 40.0 (7.40 ± 0.03) | 51.1 (7.30 ± 0.06) | 48 (7.32 ± 0.04) | 5.3 (8.29 ± 0.09) | 3.0 (8.52 ± 0.10) | 2.4 (8.62 ± 0.04) |
| Tyr10+12 (4) | 14.9 (7.83 ± 0.03) | 31.4 (7.51 ± 0.06) | 17.3 (7.77 ± 0.06) | 1.9 (8.73 ± 0.24) | 1.5 (8.84 ± 0.15) | 0.7 (9.14 ± 0.05) |
Affinities are reported as Kd values, in nanomolar. The corresponding pKd values (−log10 of the Kd, in molar) ± SEM are in parentheses.
rACA-01 = recombinant ACA-01 expressed in E. coli.
Sulfation of Tyr10 and/or Tyr12 of ACA-01 led to significant enhancement of chemokine-binding affinity (Fig. 5 and Table 1). Specifically, sulfation of Tyr10 (2 compared to 1) increased binding affinity to most chemokines by approximately twofold, whereas sulfation of Tyr12 (3 compared to 1) had a greater impact, increasing the binding affinity by 3- to 10-fold for all chemokines tested. Sulfation of both tyrosine residues (4) led to the strongest influence on binding to all chemokines. Thus, in comparison with the unsulfated form, the affinity of doubly sulfated ACA-01 is ∼10-fold higher for the eotaxins (CCL11, CCL24, and CCL26; Kd = 14.9 to 31.4 nM) and ∼30-fold higher for the MCPs (CCL2, CCL7, and CCL8; Kd = 0.7 to 1.5 nM) chemokines.
Tyrosine Sulfation of ACA-01 Enhances Chemokine Inhibition.
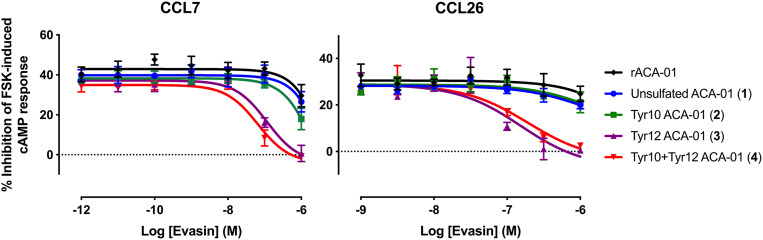
Having demonstrated the effect of ACA-01 sulfation on binding to chemokines, we next assessed the semisynthetic evasin (sulfo)proteins 1–4 for their ability to inhibit chemokine activity. Chemokines activate their cognate receptors to elicit both G protein-dependent (e.g., Gαi-dependent inhibition of adenosine 3′,5′-cyclic monophosphate [cAMP] production) and G protein-independent signaling pathways. Here, we detected CCR2 or CCR3 activation by monitoring chemokine-stimulated inhibition of cAMP production in HEK293 cells stably expressing the chemokine receptor and transiently transfected with a cAMP biosensor (38). Concentration-response curves (SI Appendix, Fig. S25) yielded the concentration of each chemokine (EC80) required to elicit 80% of the maximal signal in this assay. To address the hypothesis that binding of ACA-01 (sulfo)proteins 1–4 would abrogate the chemokine-dependent cAMP response, we stimulated receptor-expressing cells with chemokine agonists (at EC80 concentration) either alone or preincubated with an ACA-01 (sulfo)protein. A preliminary screen, using each ACA-01 (sulfo)protein at a single concentration (100 nM) (SI Appendix, Fig. S26), indicated that the chemokines CCL2, CCL8, CCL7, CCL11, and CCL26 were all inhibited by one or more of the ACA-01 (sulfo)proteins. We subsequently confirmed and quantified the concentration-dependent inhibition of these chemokines (Fig. 6, Table 2, and SI Appendix, Fig. S27).
Fig. 6.
Inhibition of chemokine activity by ACA-01 proteins 1–4. Shown are the inhibition profiles of chemokine activity by ACA-01 (sulfo)proteins (1–4) and recombinantly expressed ACA-01 (rACA-01) at concentrations ranging from 1 pM to 1 µM or 1 nM to 1 µM for CCL26 (80 nM) or CCL7 (30 nM), respectively, acting at receptor CCR2 (CCL7) or CCR3 (CCL26) in FlpIn TREx HEK293 cells. Chemokine activity was detected as the capacity of the chemokine to inhibit forskolin-induced production of cAMP, as detected via the BRET sensor, CAMYEL; differentially sulfated ACA-01 proteins (1–4) inhibit the cAMP-inhibitory activity of chemokines. Data points represent the average ± SEM of three independent experiments, each conducted in duplicate.
Table 2.
Inhibition of chemokines CCL2, CCL7, and CCL8 signaling at CCR2 and CCL11 and CCL26 signaling at CCR3 by ACA-01 evasin (sulfo)proteins 1–4*
| ACA-01 | Chemokine† | ||||
| CCL11 | CCL26 | CCL2 | CCL8 | CCL7 | |
| rACA-01‡ | —§ | — | — | 29 (7.54 ± 0.36) | — |
| Unsulfated (1) | — | — | — | 30 (7.52 ± 0.26) | — |
| Tyr10 (2) | 475 (6.32 ± 0.10) | — | — | 37 (7.43 ± 0.28) | — |
| Tyr12 (3) | 82 (7.08 ± 0.12) | 155 (6.81 ± 0.30) | 600 (6.22 ± 0.46) | 43 (7.37 ± 0.28) | 116 (6.94 ± 0.18) |
| Tyr10+12 (4) | 99 (7.00 ± 0.10) | 193 (6.72 ± 0.22) | 109 (6.96 ± 0.25) | 37 (7.43 ± 0.34) | 65 (7.19 ± 0.15) |
Inhibition constants are reported as IC50 values, in nanomolar. The corresponding pIC50 (−log of the IC50 in molar) ± SEM values are in parentheses.
EC80 values are 10 nM (CCL2), 20 nM (CCL8 and CCL7), and 80 nM (CCL11 and CCL26). Chemokines were acting at receptor CCR2 (CCL2, CCL7, and CCL8) or CCR3 (CCL11 and CCL26).
rACA-01 = recombinant ACA-01 expressed in E. coli.
Dash indicates no inhibition detected or IC50 > 1 µM.
The ability of ACA-01 variants 1–4 to inhibit chemokine signaling (Table 2) was largely consistent with the enhanced binding affinities observed for chemokines upon sulfation (Table 1). Specifically, for the chemokines CCL2 and CCL7 (signaling through CCR2) and the chemokines CCL11 and CCL26 (signaling through CCR3), the Tyr12-sulfated ACA-01 variant 3 and doubly sulfated ACA-01 (4) suppressed chemokine activity with 50% inhibitory concentrations (IC50) of 65 to 600 nM, whereas E. coli-expressed or semisynthetic unsulfated ACA-01 1 and Tyr10-sulfated ACA-01 2 showed relatively weak or no measurable inhibition. In the case of CCL8 (signaling through CCR2), the differentially sulfated ACA-01 proteins (1–4) did not show significant differences in inhibitory activity, with all IC50 values in the range ∼30 to 40 nM. This is because there is a lower limit of ∼30 nM (the concentration of CCL8 used) for IC50 values measured in this assay and all sulfoforms of ACA-01 bind to CCL8 with Kd values below this detection limit (Table 1). In summary, the effects of sulfation on the inhibitory potency of ACA-01 depend on the inhibitory potency of the unsulfated form as well as the concentration of the target chemokine.
Discussion
Due to the critical function of blood feeding for survival, ticks secrete an arsenal of biologically active salivary proteins into the bite sites on their hosts. Among these, various analgesic agents, anticoagulants, and vasodilators have been investigated as potential biologics for a range of human diseases (14–19). In an additional strategy to regulate the host response to tick infestation, the two recently discovered families of evasins are capable of suppressing the host inflammatory response by targeting host chemokines. Given this privileged bioactivity, evasins could potentially be repurposed to suppress chemokine-driven inflammation in human disease. However, the development of such therapeutic candidates would require overcoming the usual challenges of protein therapeutics (e.g., stability, delivery, bioavailability, and immunogenicity) as well as improving our understanding of the factors influencing the chemokine affinity and selectivity of evasin proteins.
The two families of evasins have distinct structures and chemokine selectivity (39). Homologs of evasin-1 and evasin-4 (class A evasins), such as ACA-01, exclusively target CC chemokines, which are characterized by a pair of adjacent cysteine residues near the protein N terminus (20, 21). Within this family, different evasins exhibit distinct spectra of chemokine-binding affinity (20, 21). However, the features of evasins that control chemokine-binding affinity have not been characterized in detail. Based on bioinformatics and sequence analyses, we made the prediction that ACA-01 would be posttranslationally sulfated on two specific tyrosine residues and that sulfation would affect chemokine-binding affinity and potentially selectivity. The data presented here unequivocally demonstrate that the ACA-01 is sulfated at two Tyr residues (Tyr10 and Tyr12) when secreted from eukaryotic cells, albeit heterogeneously. Through the exploitation of a semisynthetic strategy we were able to efficiently assemble and fold the three homogeneously sulfated forms of ACA-01, and the nonsulfated form, and show that sulfation enhances the chemokine-binding and inhibitory activities of the protein for a range of human CC chemokines.
Our conclusion that ticks utilize tyrosine sulfation to modulate the activity of evasins is consistent with previous evidence that the activity of other tick salivary proteins can be modulated upon tyrosine sulfation. In particular, tyrosine sulfation of two thrombin inhibitors from the tick Haemaphysalis longicornis substantially enhances their inhibitory potency (35). Although relatively little is known about the mechanism of tyrosine sulfation by ticks, database searches reveal that A. cajennense encodes predicted proteins with 65% and 60% identity to human TPST-1 and TPST-2, respectively, and other tick species encode proteins with similar identities to human TPSTs. Moreover, similar target sequences, rich in acidic amino acids, are known to be sulfated in organisms as diverse as humans and leeches (23, 40, 41). Therefore, it is reasonable to expect that tyrosine sulfation of evasins by ticks occurs, as in many other organisms, via the activity of trans-Golgi–localized TPST enzymes selective for tyrosine residues within acidic sequences. Such sequences are present in the N-terminal regions of many putative evasins (20), so it appears that tyrosine sulfation may be a general mechanism for modulating the activity of these proteins.
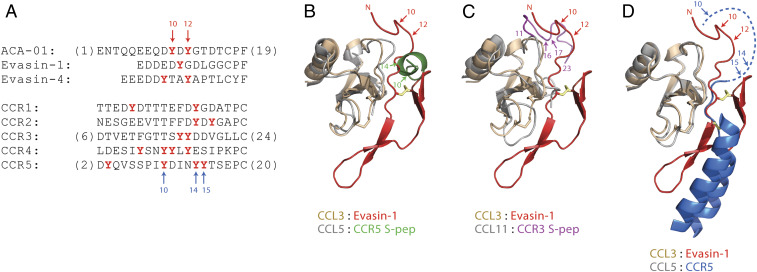
Our results show that tyrosine sulfation not only affects chemokine-binding affinity but also significantly modifies the selectivity of ACA-01 between two pairs of chemokines tested (SI Appendix, Fig. S24 and Table S1). Specifically, unsulfated ACA-01 binds to CCL7 and CCL11 with Kd values of 23 and 135 nM, respectively, an approximately sixfold difference in affinity, whereas doubly sulfated ACA-01 binds with Kd values of 0.7 and 15 nM, a ∼21-fold affinity difference. Similar affinity ratios were observed for CCL7 versus CCL26. The consequences of modified selectivity must be considered with the knowledge that sulfation of ACA-01 in tick saliva is likely to be heterogeneous, as observed here for ACA-01 expressed in mammalian cells, and also typically observed in other biochemical analyses of expressed proteins bearing tyrosine sulfation modifications (35, 42). Moreover, many tick species encode numerous (up to a dozen) evasin proteins, which substantially differ from each other in their target chemokine selectivity (20, 21). Therefore, it appears that ticks produce a complex mixture of evasins with different patterns of tyrosine sulfation (and potentially other posttranslational modifications [PTMs]) as an evolutionary strategy to target a broad range of chemokines in host species. It is tempting to speculate that this strategy may not only suppress the ability of a typical host to detect infesting ticks but also support the ability of ticks to migrate to new hosts.
To what extent is tyrosine sulfation of evasins a receptor mimetic strategy? Almost all chemokine receptors contain potential tyrosine sulfation sites in their N-terminal regions (24). For several receptors (and for N-terminal peptides derived from several receptors), sulfation of these sites enhances chemokine affinity and modulates selectivity among cognate chemokines (23, 24, 36). In these respects, sulfation of evasins is likely to have similar consequences to sulfation of chemokine receptors. To probe whether tyrosine sulfation could be a widespread modification of the evasins, we have analyzed all 21 validated chemokine-binding C8 “class A evasins” for predicted tyrosine sulfation using the online Sulfinator tool (43) (see SI Appendix for details). Of the 21 evasins assessed, 12 (57%) are predicted to have sulfated tyrosine sites—all except one in the first ∼20 residues (SI Appendix, Figs. S30–S32). Upon closer inspection, this placement of the sulfated tyrosine residues, as well as the sequences of the tyrosine sulfation sites of evasins, is strikingly similar to those in chemokine receptors (Fig. 7A). In particular, both ACA-01 and chemokine receptor CCR2 contain the pentapeptide sequence DYDYG in this region. A doubly sulfated variant of DYDYG (see SI Appendix, Fig. S6 for synthetic details) does not possess significant chemokine affinity (SI Appendix, Fig. S23), but sulfation of these tyrosine residues is known to affect chemokine recognition in larger N-terminal fragments of chemokine receptors (44, 45). Insights into the structural roles of the sulfated tyrosine residues in evasins and chemokine receptors can be obtained by comparing the structure of evasin-1 bound to CCL3 with those of a chemokine receptor, or receptor N-terminal fragments, bound to CC chemokines (Fig. 7 B–D) (25, 46–48). The N-terminal region of evasin-1 binds to the “N-loop” and β3 strand regions of CCL3 (25). Sulfopeptides derived from the N-terminal regions of CCR3 and CCR5 interact with the same regions of their cognate chemokines (Fig. 7 B and C) (47, 48), although the orientations of the sulfopeptides in these complexes may not be truly reflective of the orientation with which the actual receptor N terminus binds to the chemokines (39). Nevertheless, based on the structure of CCR5 bound to chemokine variant 5P7-CCL5 (46), it appears likely that the orientation of the receptor N terminus is similar to that of the evasin-1 N terminus in complex with CCL3 (Fig. 7D). Thus, the chemokine-binding and inhibition data, interpreted within the context of sequence similarities and structural comparisons, strongly suggest that tick evasins and mammalian chemokine receptors have independently evolved the ability to be posttranslationally sulfated to optimize chemokine recognition. Our data prompt an extensive and systematic characterization of the role of PTMs on the activity and specificity of other recently discovered evasins using a combination of semisynthetic and detailed biochemical studies. We anticipate that a detailed understanding of how tyrosine sulfation underpins chemokine binding, inhibition, and selectivity for the evasins will provide the foundation from which to develop engineered peptides and proteins to target inflammatory diseases associated with dysregulated chemokine–chemokine receptor signaling.
Fig. 7.
Sequence and structural comparisons of tyrosine sulfation sites in evasins and chemokine receptors. (A) Aligned partial sequences of the N-terminal regions of selected evasins (Top) and human CC chemokine receptors (Bottom). Alignment is based on the structural overlay of chemokine ligands in structures of evasin-1 bound to CCL3 and CCR5 bound to 5P7-CCL5 (D). Potentially sulfated tyrosine residues are in red bold type. (B–D) Partial structure of evasin-1 (residues 1–39 shown as red ribbons, with the first disulfide in yellow) bound to CCL3 (wheat ribbons, disulfides as sticks) (25) overlayed with (B) the structure of a CCR5-derived doubly sulfated peptide (CCR5 S-pep, green) bound to CCL5 (gray) (48), (C) the structure of a CCR3-derived doubly sulfated peptide (CCR3 S-pep, magenta) bound to CCL11 (gray) (47), and (D) the structure of CCR5 (transmembrane helices TM1 and TM7 shown as blue ribbons, with the connecting disulfide in yellow) bound to 5P7-CCL5 (gray) (25, 46). The dashed blue line indicates the N-terminal region of CCR5, which is not defined in the crystal structure. Numbered arrows indicate the known or likely positions of Tyr sulfation. The structural overlays are based on chemokine sequence alignments extending from the CC motif to end of the C-terminal α-helix. Disordered regions of chemokines and peptides are omitted for clarity.
Materials and Methods
Full details for peptide synthesis, bacterial and mammalian protein expression, ligation and refolding reactions, proteomics, and chemokine-binding and chemokine-inhibition assays of ACA-01 variants can be found in SI Appendix.
Data Availability.
HPLC chromatograms and ESI and matrix-assisted laser desorption/ionization time-of-flight mass spectra for synthetic (sulfo)peptides 6–9, expressed ACA-01 protein fragment 5 and semisynthetic ACA-01 (sulfo)proteins 11–14 (unfolded) and 1–4 (folded) can be found in SI Appendix. One- and two-dimensional NMR characterization data for (sulfo)peptides 6–9 are also included in SI Appendix. RAW MS data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD016778. RAW data were analyzed using Byonic (Protein Metrics) and the search output has also been uploaded to the ProteomeXchange Consortium under the same identifier. All chemokine-binding and chemokine inhibition data can be found in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge funding from an Australian Research Council Discovery Project (DP190101526 to R.J.P. and M.J.S.) and a National Health and Medical Research Council Project Grant APP1140867 to M.J.S. and R.J.P. and an Investigator Grant APP1174941 to R.J.P. We thank the John A. Lamberton Research Scholarship (to C.F. and J.J.-L.) and a Research Training Scholarship (to J.J.-L.). M.L. is a Cancer Institute New South Wales Future Research Leader Fellow. SydneyMS, The University of Sydney, is acknowledged for providing the mass spectrometry instrumentation used in this study.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.L. is a guest editor invited by the Editorial Board.
Data deposition: RAW MS data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD016778. RAW data were analyzed using Byonic (Protein Metrics) and the search output has also been uploaded to the ProteomeXchange Consortium under the same identifier.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000605117/-/DCSupplemental.
References
- 1.Bonecchi R. et al., Chemokines and chemokine receptors: An overview. Front. Biosci. 14, 540–551 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Rollins B. J., Chemokines. Blood 90, 909–928 (1997). [PubMed] [Google Scholar]
- 3.Lloyd C., Chemokines in allergic lung inflammation. Immunology 105, 144–154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charo I. F., Peters W., Chemokine receptor 2 (CCR2) in atherosclerosis, infectious diseases, and regulation of T-cell polarization. Microcirculation 10, 259–264 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Epperson M. L., Lee C. A., Fremont D. H., Subversion of cytokine networks by virally encoded decoy receptors. Immunol. Rev. 250, 199–215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felix J., Savvides S. N., Mechanisms of immunomodulation by mammalian and viral decoy receptors: Insights from structures. Nat. Rev. Immunol. 17, 112–129 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Alejo A. et al., A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. U.S.A. 103, 5995–6000 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant N. A., Davis-Poynter N., Vanderplasschen A., Alcami A., Glycoprotein G isoforms from some alphaherpesviruses function as broad-spectrum chemokine binding proteins. EMBO J. 22, 833–846 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parry C. M. et al., A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191, 573–578 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González-Motos V., Kropp K. A., Viejo-Borbolla A., Chemokine binding proteins: An immunomodulatory strategy going viral. Cytokine Growth Factor Rev. 30, 71–80 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Seet B. T., McFadden G., Viral chemokine-binding proteins. J. Leukoc. Biol. 72, 24–34 (2002). [PubMed] [Google Scholar]
- 12.Bonvin P., Power C. A., Proudfoot A. E., Evasins: Therapeutic potential of a new family of chemokine-binding proteins from ticks. Front. Immunol. 7, 208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhusal R. P. et al., Evasins: Tick salivary proteins that inhibit mammalian chemokines. Trends Biochem. Sci. 45, 108–122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Déruaz M. et al., Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. J. Exp. Med. 205, 2019–2031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frauenschuh A. et al., Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus. J. Biol. Chem. 282, 27250–27258 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Montecucco F. et al., Treatment with Evasin-3 abrogates neutrophil-mediated inflammation in mouse acute pancreatitis. Eur. J. Clin. Invest. 44, 940–950 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Vieira A. T. et al., Treatment with a novel chemokine-binding protein or eosinophil lineage-ablation protects mice from experimental colitis. Am. J. Pathol. 175, 2382–2391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copin J.-C. et al., Treatment with Evasin-3 reduces atherosclerotic vulnerability for ischemic stroke, but not brain injury in mice. J. Cereb. Blood Flow Metab. 33, 490–498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo R. C. et al., Therapeutic effects of evasin-1, a chemokine binding protein, in bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 45, 72–80 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Hayward J. et al., Ticks from diverse genera encode chemokine-inhibitory evasin proteins. J. Biol. Chem. 292, 15670–15680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh K. et al., Yeast surface display identifies a family of evasins from ticks with novel polyvalent CC chemokine-binding activities. Sci. Rep. 7, 4267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenquist G. L., Nicholas H. B. J. Jr., Analysis of sequence requirements for protein tyrosine sulfation. Protein Sci. 2, 215–222 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone M. J., Payne R. J., Homogeneous sulfopeptides and sulfoproteins: Synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Acc. Chem. Res. 48, 2251–2261 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Ludeman J. P., Stone M. J., The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 171, 1167–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dias J. M. et al., Structural basis of chemokine sequestration by a tick chemokine binding protein: The crystal structure of the complex between Evasin-1 and CCL3. PLoS One 4, e8514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kehoe J. W., Bertozzi C. R., Tyrosine sulfation: A modulator of extracellular protein-protein interactions. Chem. Biol. 7, R57–R61 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Nicholas H. B. Jr., Chan S. S., Rosenquist G. L., Reevaluation of the determinants of tyrosine sulfation. Endocrine 11, 285–292 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Eaton J. R. O. et al., The N-terminal domain of a tick evasin is critical for chemokine binding and neutralization and confers specific binding activity to other evasins. J. Biol. Chem. 293, 6134–6146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J. et al., Tyrosylprotein sulfotransferase-1 and tyrosine sulfation of chemokine receptor 4 are induced by Epstein-Barr virus encoded latent membrane protein 1 and associated with the metastatic potential of human nasopharyngeal carcinoma. PLoS One 8, e56114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franck C., et al. , Semi-synthesis of an evasin from tick saliva reveals a critical role of tyrosine sulfation for chemokine binding and inhibition. ProteomeXchange. https://www.ebi.ac.uk/pride/archive/projects/PXD016778. Deposited 17 December 2019. [DOI] [PMC free article] [PubMed]
- 31.Chen G., Zhang Y., Trinidad J. C., Dann C. 3rd, Distinguishing sulfotyrosine containing peptides from their phosphotyrosine counterparts using mass spectrometry. J. Am. Soc. Mass Spectrom. 29, 455–462 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. H., Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Guerrero F., Ciragan A., Iwaï H., Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif. 116, 42–49 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Simpson L. S., Zhu J. Z., Widlanski T. S., Stone M. J., Regulation of chemokine recognition by site-specific tyrosine sulfation of receptor peptides. Chem. Biol. 16, 153–161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson R. E. et al., Tyrosine sulfation modulates activity of tick-derived thrombin inhibitors. Nat. Chem. 9, 909–917 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Ludeman J. P. et al., Phosphate modulates receptor sulfotyrosine recognition by the chemokine monocyte chemoattractant protein-1 (MCP-1/CCL2). Org. Biomol. Chem. 13, 2162–2169 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Sanchez J., Stone M. J., Payne R. J., Sulfation of the human cytomegalovirus protein UL22A enhances binding to the chemokine RANTES. Angew. Chem. Int. Ed. Engl. 56, 8490–8494 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Jiang L. I. et al., Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J. Biol. Chem. 282, 10576–10584 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhusal R. P., Foster S. R., Stone M. J., Structural basis of chemokine and receptor interactions: Key regulators of leukocyte recruitment in inflammatory responses. Protein Sci. 29, 420–432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh Y. S. Y., Wijeyewickrema L. C., Wilkinson B. L., Pike R. N., Payne R. J., Total synthesis of homogeneous variants of hirudin P6: A post-translationally modified anti-thrombotic leech-derived protein. Angew. Chem. Int. Ed. Engl. 53, 3947–3951 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Stone M. J., Chuang S., Hou X., Shoham M., Zhu J. Z., Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. N. Biotechnol. 25, 299–317 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Watson E. E. et al., Mosquito-derived anophelin sulfoproteins are potent antithrombotics. ACS Cent. Sci. 4, 468–476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monigatti F., Gasteiger E., Bairoch A., Jung E., The sulfinator: Predicting tyrosine sulfation sites in protein sequences. Bioinformatics 18, 769–770 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Tan J. H. et al., Tyrosine sulfation of chemokine receptor CCR2 enhances interactions with both monomeric and dimeric forms of the chemokine monocyte chemoattractant protein-1 (MCP-1). J. Biol. Chem. 288, 10024–10034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preobrazhensky A. A. et al., Monocyte chemotactic protein-1 receptor CCR2B is a glycoprotein that has tyrosine sulfation in a conserved extracellular N-terminal region. J. Immunol. 165, 5295–5303 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y. et al., Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity 46, 1005–1017.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Millard C. J. et al., Structural basis of receptor sulfotyrosine recognition by a CC chemokine: The N-terminal region of CCR3 bound to CCL11/eotaxin-1. Structure 22, 1571–1581 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Abayev M. et al., The solution structure of monomeric CCL5 in complex with a doubly sulfated N-terminal segment of CCR5. FEBS J. 285, 1988–2003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HPLC chromatograms and ESI and matrix-assisted laser desorption/ionization time-of-flight mass spectra for synthetic (sulfo)peptides 6–9, expressed ACA-01 protein fragment 5 and semisynthetic ACA-01 (sulfo)proteins 11–14 (unfolded) and 1–4 (folded) can be found in SI Appendix. One- and two-dimensional NMR characterization data for (sulfo)peptides 6–9 are also included in SI Appendix. RAW MS data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD016778. RAW data were analyzed using Byonic (Protein Metrics) and the search output has also been uploaded to the ProteomeXchange Consortium under the same identifier. All chemokine-binding and chemokine inhibition data can be found in SI Appendix.