Significance
Adoptive T cell therapy (ACT) is an experimental cancer immunotherapy with partial clinical responses (∼20%). A mechanistic understanding of T cell receptors (TCR) isolated from tumors and defining the biochemical attributes associated with tumor elimination may improve the ACT success rate. Here, we studied a successful case of ACT targeting HLA-C–restricted KRAS-G12D neoantigens. Through biochemical, structural, and cellular approaches, we define how these TCRs recognized their neoantigens. Key findings were that these TCRs were of higher affinity than most tumor-specific TCRs identified to date and these TCRs were specific for two distinct neoantigens. Whether these are general features of successful ACT remains to be seen, but this approach may serve as a basis for future rational improvement of ACT.
Keywords: TCR–HLA-C complex structure, tumor-infiltrating TCR–antigenic affinity, adoptive T cell transfer immunotherapy, KRAS-G12D neoantigen, cancer therapy
Abstract
Complete cancer regression occurs in a subset of patients following adoptive T cell therapy (ACT) of ex vivo expanded tumor-infiltrating lymphocytes (TILs). However, the low success rate presents a great challenge to broader clinical application. To provide insight into TIL-based immunotherapy, we studied a successful case of ACT where regression was observed against tumors carrying the hotspot mutation G12D in the KRAS oncogene. Four T cell receptors (TCRs) made up the TIL infusion and recognized two KRAS-G12D neoantigens, a nonamer and a decamer, all restricted by human leukocyte antigen (HLA) C*08:02. Three of them (TCR9a, 9b, and 9c) were nonamer-specific, while one was decamer-specific (TCR10). We show that only mutant G12D but not the wild-type peptides stabilized HLA-C*08:02 due to the formation of a critical anchor salt bridge to HLA-C. Therapeutic TCRs exhibited high affinities, ranging from nanomolar to low micromolar. Intriguingly, TCR binding affinities to HLA-C inversely correlated with their persistence in vivo, suggesting the importance of antigenic affinity in the function of therapeutic T cells. Crystal structures of TCR–HLA-C complexes revealed that TCR9a to 9c recognized G12D nonamer with multiple conserved contacts through shared CDR2β and CDR3α. This allowed CDR3β variation to confer different affinities via a variable HLA-C contact, generating an oligoclonal response. TCR10 recognized an induced and distinct G12D decamer conformation. Thus, this successful case of ACT included oligoclonal TCRs of high affinity recognizing distinct conformations of neoantigens. Our study revealed the potential of a structural approach to inform clinical efforts in targeting KRAS-G12D tumors by immunotherapy and has general implications for T cell-based immunotherapies.
In cancer, cytotoxic CD8+ T cells can eliminate tumor cells through recognition of peptide epitopes presented on major histocompatibility complex class I (MHC-I) molecules by the alpha-beta T cell receptor (αβTCR). Each TCR chain contains three complementarity-determining regions (CDRs) generated by V(D)J recombination, which form six flexible loops that contact MHC-I (1–4). In general, the germline-encoded CDR1 and CDR2 contact the MHC-I heavy chain, while the hypervariable CDR3 binds the MHC-I–bound peptide (1, 2, 5). T cells can recognize peptides derived from tumor-associated antigens, cancer-testis antigens, viral antigens (in the case of virally derived tumors), and neoantigens (6). Neoantigens are peptides derived from mutated “self” proteins that the immune system detects as “nonself.” Many neoantigens are “private” to individual tumors, and immunity to these antigens can be exploited with immunotherapies such as checkpoint blockade or personalized vaccines (7, 8). Some neoantigens are derived from common or “hotspot” mutations such as those arising in the RAS proteins and p53. The RAS family (H, N, and KRAS) of small GTPases are among the most commonly mutated oncogenes in cancer (27%) (9). Among them, the G12D mutation in KRAS occurs most frequently and is found in ∼45% of pancreatic, 13% of colorectal, and 4% of lung cancers (10, 11). The high frequency of this mutation makes it an ideal drug target but attempts to chemically target mutant RAS have proven challenging (10, 11).
Adoptive T cell therapy (ACT) with expanded tumor-infiltrating lymphocytes (TILs) has demonstrated clinical efficacy; however, the complete response rate is only 20 to 25% (12, 13). In antibody-based immunotherapy, clinical efficacy is known to depend on antigenic specificity, affinity, and antibody half-life (14). In contrast, the desirable traits of TCRs for favorable ACT clinical outcomes remain unclear and, with the expansion of TIL-based immunotherapy (15), there is a need to evaluate and predict the potential clinical efficacy of TIL TCRs. To provide insight into tumor recognition by TILs and to facilitate the development of effective ACT, we combined biochemical, structural, and cellular approaches to elucidate 1) cell-based tumor antigenic presentation by class I human leukocyte antigen (HLA-I), 2) the specificity and affinity of TIL-TCR recognition of tumor antigen in solution and on reconstituted T cells, and 3) structural recognition of tumor antigen by TIL TCRs (Fig. 1A). In particular, we studied five KRAS-G12D–specific tumor-infiltrating TCRs obtained from two patients (16, 17). One patient (no. 4095) with metastatic colorectal cancer was treated with an infusion of ex vivo expanded TILs carrying four TCRs (TCR9a to 9c and TCR10), which resulted in successful tumor regression. Another KRAS-G12D–specific TCR (TCR9d) was identified from the TILs of patient no. 3995 but was a minority of the TIL infusion used to treat this patient (16). All five TCRs are HLA-C*08:02–restricted and recognize either a nonamer (10GADGVGKSA) or decamer (10GADGVGKSAL) KRAS peptide, in which the wild-type (WT) Gly is mutated to Asp (D) at position 12 (Table 1). Nonamer-specific TCRs (9a to 9d) shared the same V genes but varied in their CDR3β sequence. The decamer-specific TCR10 used a different V gene and CDR3 sequences.
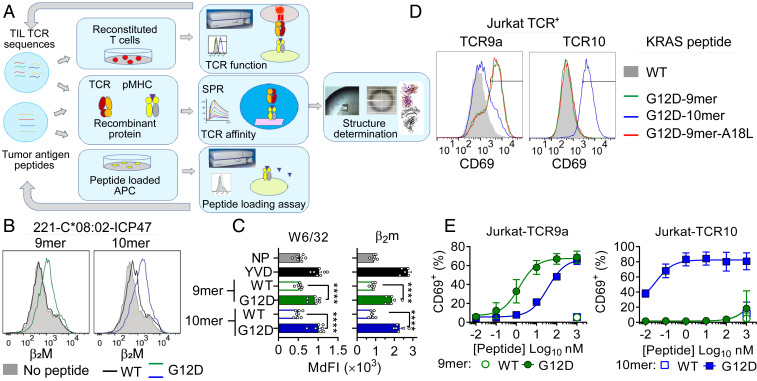
Fig. 1.
G12D mutation is critical for presentation and T cell recognition of KRAS-G12D neoantigens. (A) Strategy to study TIL TCRs. (B and C) HLA-I stabilization on TAP-deficient 221-C*08:02-ICP47 cells incubated overnight at 26 °C with 100 μM WT and G12D KRAS 9- and 10-mer and control peptide YVD (YVDEHGTRL). NP, no peptide. A representative experiment is shown in B and data from five or six independent experiments are summarized in C. Statistical significance was assessed by one-way ANOVA with Tukey’s multiple comparison test (****P < 0.0001). (D and E) Frequency of TCR+ Jurkat T cells expressing CD69 after incubation with 221-C*08:02-ICP47 cells loaded with WT and G12D KRAS 9- and 10-mer, as measured by flow cytometry. Peptides were tested from 1,000 to 0.01 nM. A representative experiment is the data shown in D with 10 nM peptide and four to six independent experiments are summarized in E with mean and SEM values. MdFI = median fluorescence intensity.
Table 1.
KRAS-G12D–specific T cell receptors
| Patient no. | TCR name | V alpha | V beta | CDR3α (length) | CDR3β (length) | TCR frequency (infusion), % | TCR frequency (post transfer), % | KRASG12D specificity |
| 4095 | 9a | TRAV4*01 | TRBV5-6*01 | CLVGDMDQAGTALIF (13) | CASSLGEGRVDGYTF (13) | 50 | 0 | 10GADGVGKSA |
| 4095 | 9b | TRAV4*01 | TRBV5-6*01 | CLVGDMDQAGTALIF (13) | CASSLGRASNQPQHF (13) | 7 | 5 | 10GADGVGKSA |
| 4095 | 9c | TRAV4*01 | TRBV5-6*01 | CLVGDRDQAGTALIF (13) | CASSFGQSSTYGYTF (13) | 0.04 | 0.05 | 10GADGVGKSA |
| 4095 | 10 | TRAV12-2*01 | TRBV10-2*01 | CAAAMDSSYKLIF (11) | CASSDPGTEAFF (10) | 20 | 10 | 10GADGVGKSAL |
| 3995 | 9d | TRAV4*01 | TRBV5-6*01 | CLVGDMDQAGTALIF (13) | CASSLGQTNYGYTF (12) | 0.002 | 0 | 10GADGVGKSA |
We show that only G12D but not WT KRAS peptides are presented by HLA-C*08:02, explaining the lack of recognition of WT KRAS peptides by G12D-specific TCRs. All five KRAS-G12D–specific TCRs displayed nanomolar to low micromolar peptide–HLA-C (pHLA-C) binding affinities, which inversely correlated with their in vivo persistence, suggesting antigenic affinity impacts the T cell’s in vivo persistence. Multiple TCR–HLA-C complex structures revealed that TCR9a to 9d share a common recognition mode, dependent on their CDR2β and CDR3α interactions with peptide and HLA-C. The lack of peptide-contacting CDR3β residues allowed its sequence variation among TCR9a to 9d, while maintaining neoantigen specificity. The variable CDR3β of the four TCR9s produced different contacts with HLA-C that modulated their binding affinities. The structure of TCR10 in complex with decamer–HLA-C showed the receptor recognized a distinct G12D peptide conformation not present in TCR9–HLA-C structures. This suggests both TCR9 and 10 are required to target a distinct conformation of KRAS-G12D neoantigens. Our study provides a molecular understanding of TCRs from a successful case of ACT and suggests that oligoclonal, tumor-specific TCRs with high affinities can deliver effective tumor clearance. We anticipate rules for selecting TILs for better immunotherapy will emerge as more clinical cases are explored using the current approach. It is our hope that these studies will eventually yield predicative power to facilitate the design of ACT with effective tumor clearance, better antigenic coverage, and optimal therapeutic persistence.
Results
HLA-C Presents the G12D Neoantigens but Not Wild-Type KRAS for T Cell Recognition.
Tumor specificity is essential to a T cell-based antitumor therapy. While the four transferred TIL-expanded T cells to patient 4095 successfully cleared metastatic tumors through targeting KRAS-G12D in the context of HLA-C*08:02, the source of their tumor specificity remained unknown (17). The therapy’s specificity against neoantigen but not wild-type KRAS was assumed the property of the four TCRs (18). Namely, these TCRs possess higher affinity to HLA-C presenting the G12D mutant than WT KRAS. To assess antitumor TCR preference for neoantigen, we utilized a cell-based functional peptide-binding assay using transporter associated with antigen presentation (TAP)-deficient cells (221-C*08:02-ICP47). The HLA-I–negative cell line 221 was transfected with HLA-C*08:02 and the TAP inhibitor ICP47 (19). Peptide binding was assessed as an increase in HLA-C expression by flow cytometry after overnight incubation with peptide at 26 °C. Both the KRAS-G12D mutant nonamer and decamer peptides stabilized HLA-C*08:02 expression, measured by staining for class I HLA (W6/32) or beta-2 microglobulin (β2m) (Fig. 1 B and C). This increase in HLA-C stabilization was similar to a control peptide YVD (YVDEHGTRL), previously shown to bind HLA-C*08:02 (Fig. 1C) (19). However, the WT KRAS peptides failed to stabilize HLA-C expression on the TAP-deficient cells (Fig. 1 B and C), suggesting the TCR tumor specificity is derived from preferential tumor antigen presentation by HLA-C rather than differential TCR recognition of G12D over WT KRAS.
To evaluate the specificity of the KRAS-G12D–specific TCRs, we generated Jurkat T cells transfected with TCR9a (Vα4Vβ5) and TCR10 (Vα12Vβ10), specific for the nonamer and decamer, respectively. TCR-transfected Jurkat cells were specifically activated by their G12D but not WT KRAS epitopes loaded on 221-C*08:02-ICP47 cells, measured by CD69 expression (Fig. 1 D and E). TCR10-transfected Jurkat cells recognized the G12D decamer exclusively with a half maximal effective concentration (EC50) of 0.03 nM, while TCR9a-transfected Jurkat cells recognized both the nonamer and decamer G12D with an EC50 of 1 and 35 nM, respectively.
The G12D Mutation and Peptide C Terminus Form Critical Anchors for Binding HLA-C*08:02.
The role of the G12D mutation in peptide binding was illustrated in the crystal structures of HLA-C*08:02 in complex with the KRAS-G12D nonamer (10GADGVGKSA) and decamer (10GADGVGKSAL) peptides (Fig. 2 A and B and SI Appendix, Table S1). Both structures were refined to 1.9-Å resolution and were nearly identical to each other. Comparing the Cα residues 2 to 272 of the HLA-C heavy chains gave an rmsd of 1.06 Å. The two peptides were anchored at the N and C termini by similar interactions with HLA-C*08:02 but showed distinct conformations in the middle between the p5 and p8 positions (Fig. 2 B and C and SI Appendix, Fig. S1 A and B). The decamer bulged out of the peptide-binding groove peaking at the p7 position Lys, similar to previous studies of longer peptides (20, 21). The most contacts between the HLA-C*08:02 peptide-binding groove and peptide side chains were at peptide p3, where the negatively charged Asp formed a salt bridge with the positively charged Arg-156 on the α2-helix (Fig. 2C and SI Appendix, Fig. S1C). Critically, p3 Asp is the result of the KRAS-G12D mutation and the salt bridge cannot be formed with the WT Gly, suggesting G12D but not WT KRAS is selectively presented by HLA-C*08:02. This is consistent with a previous study of the immunopeptidome of HLA-C*08:02 showing the p3 position contained exclusively negatively charged Asp or Glu residues (22) (Fig. 2D). Consistent with the crucial role of p3D in antigen presentation, other KRAS mutations such as G12V, G12R, and G12S were not recognized by the G12D-specific TCRs (SI Appendix, Fig. S1D).
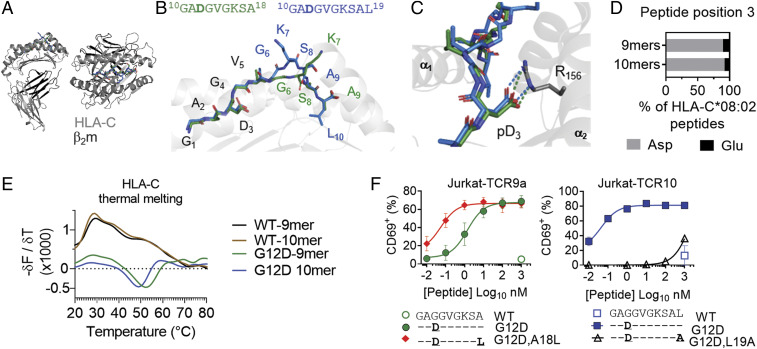
Fig. 2.
KRAS-G12D mutation and peptide C terminus form critical anchors for binding HLA-C*08:02. (A and B) Structures of HLA-C*08:02 (gray) bound to the KRAS-G12D-9-mer (green) and 10-mer (blue); β2m, dark gray. (C) Salt bridge between peptide position 3 Asp and HLA-C*08:02 Arg-156. (D) Amino acid frequency at position 3 of 9- and 10-mer peptides eluted from HLA-C*08:02. Sequences are from ref. 22. (E) Thermal melting profiles of HLA-C*08:02 refolded with WT or G12D KRAS peptides. HLA-C molecules were heated from 10 to 90 °C in the presence of SYPRO orange dye. The negative of the change in fluorescence over the change in temperature (−δF/δT, first derivative) is shown at each temperature. (F) Frequency of TCR+ Jurkat T cells expressing CD69 after incubation with 221-C*08:02-ICP47 cells loaded with WT and G12D KRAS 9- and 10-mer peptides and C-terminally modified G12D peptides. Amino acids identical to the KRAS sequence are indicated with “–.” Peptides were tested from 1,000 to 0.01 nM. Data shown are a mean and SEM of three independent experiments.
To further assess the role of the G12D mutation in peptide binding to HLA-C*08:02, we refolded HLA-C*08:02 with WT and G12D KRAS peptides. By size-exclusion chromatography, HLA-C refolded with WT peptides appeared largely misfolded, with no major peak at the expected elution volume of 82 mL (SI Appendix, Fig. S2A). In contrast, HLA-C refolded with G12D peptides eluted as expected. Furthermore, we determined the melting temperature of these proteins by differential scanning fluorimetry with a fluorescent dye (SYPRO orange) that binds hydrophobic regions of proteins (23). HLA-C refolded with WT peptides displayed strong binding to the dye at low temperatures with no discernable melting point, indicative of misfolded protein (Fig. 2E). In contrast, HLA-C*08:02 refolded with either the KRAS-G12D nonamer or decamer exhibited well-behaved thermal melting curves with melting points of 51 ± 1.3 and 45 ± 1.8 °C, respectively, similar to other peptide–HLA-C molecules (24). Thus, only the G12D mutation but not WT KRAS peptides stabilized the peptide–HLA-C complex for antigen presentation.
Understanding the structural mechanism of peptide presentation by HLA molecules can lead to rational peptide substitutions for the development of peptide vaccines (25–27). The C-terminal peptide residue often anchors the bound peptide to the MHC, such as Leu-19 in the G12D decamer (SI Appendix, Fig. S2B). However, the C-terminal anchor in the G12D nonamer is the less hydrophobic Ala, which has 44.5 Å2 less buried surface area than Leu. While Leu is the most common C-terminal residue, Ala at the C terminus is absent from nonamer or decamer peptides previously eluted from HLA-C*08:02 (22) (SI Appendix, Fig. S2C). We hypothesized that substituting the C-terminal Ala with Leu (KRAS-G12D-A18L) would improve HLA-C stabilization and confer better recognition by nonamer-specific TCRs. Indeed, this substitution improved the stabilization of HLA-C*08:02, and enhanced the EC50 of TCR9a by 20-fold (Fig. 2F and SI Appendix, Fig. S2D). Conversely, substitution of Leu for Ala in the decamer C terminus abolished HLA-C stabilization and recognition by TCR10 (Fig. 2F). Thus, Ala is a suboptimal G12D nonamer C-terminal anchor for HLA-C*08:02, and a Leu-substituted G12D nonamer may lead to more sensitive T cell recognition. Together, we conclude that the G12D mutation is critical for neoantigen presentation by HLA-C*08:02 (Fig. 2), which is essential for T cell recognition.
Therapeutic TCRs Display High Affinities That Inversely Correlate with In Vivo Persistence.
The molecular properties of TIL TCRs are poorly understood. For most TIL-based therapy, the antigenic affinities of therapeutic T cells are not known, nor are considered as a selection criterion. Patient 4095 was treated as part of a clinical trial using ex vivo expanded TILs (17). As part of this trial, TIL cultures were determined to be neoantigen-reactive regardless of neoantigen identity or the number of TCR clonotypes. This TIL infusion contained four KRAS-G12D–specific TCRs, TCR9a to 9c and TCR10. After adoptive transfer of this TIL infusion product, TCR9a to 9c and TCR10 exhibited differences in their in vivo persistence, despite similar in vitro functional avidities, and expression of cell-surface markers (17). TCR9a to 9d also exhibit CDR3β variation, the impact of which is unknown but could impact TCR specificity and affinity.
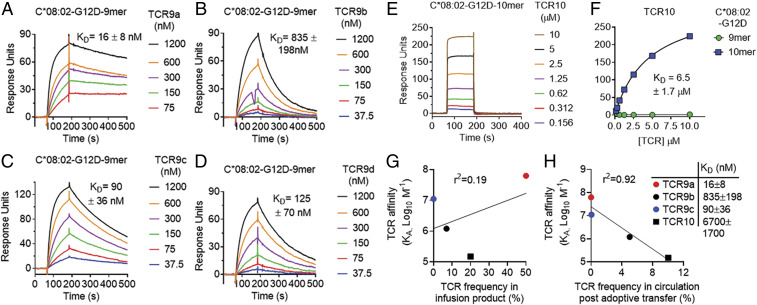
To investigate the impact of TCR–pHLA binding affinity on the success of KRAS-G12D–specific immunotherapy, we measured the solution binding affinities of KRAS-G12D–specific TCRs to their cognate HLA-C by surface plasmon resonance (SPR) using BIAcore (Fig. 3). Recombinant HLA-C was captured by an immobilized class I HLA monoclonal antibody (mAb) on CM5 sensorchips and recombinant TCRs were used as analytes. TCR9a to 9d exhibited affinities in the range of 16 to 835 nM for the G12D nonamer-complexed HLA-C*08:02 (Fig. 3 A–D). The best binder was TCR9a, with a KD of 16 (±8) nM, the highest to our knowledge for an unmodified tumor-specific TCR (28–30). TCR10 bound to the decamer presenting HLA-C*08:02 with a KD of 6.7 (±1.7) μM. No detectable binding was observed between TCR10 and the G12D nonamer-complexed HLA-C*08:02 at these TCR concentrations (Fig. 3 E and F). Compared with typical 10 to 100 μM TCR–MHC affinities (28–30), these therapeutic TCRs exhibited higher affinities. Critically, these data uncovered previously unappreciated differences between the nonamer-specific TCRs, with dissociation constants ranging from 16 nM (TCR9a) to 835 nM (TCR9b). These differences in binding affinity were not due to differences in thermal stability between the TCRs (SI Appendix, Table S2). While limited to four data points, we observed an intriguing inverse correlation between TCR affinity and TCR frequency 40 d post adoptive transfer (Fig. 3H). Notably, TCR9a (KD 16 nM) made up 50% of the infusion product but was undetectable in circulation 40 d post transfer (Fig. 3 G and H). Conversely, TCR10 (KD 6.7 μM) made up 20% of the infusion product but was maintained as 10% of circulating T cells at 9 mo post transfer (Fig. 3 G and H) (17).
Fig. 3.
KRAS-G12D–specific TCRs display high affinities that inversely correlate with in vivo persistence. (A–D) Binding of TCR9a (A), 9b (B), 9c (C), and 9d (D) to captured HLA-C*08:02–KRAS-G12D-9-mer at the indicated nanomolar concentrations determined by surface plasmon resonance. Dissociation constants were determined by kinetic curve fitting. Data are representative of two independent experiments. (E) Binding of TCR10 to captured HLA-C*08:02–KRAS-G12D-10-mer at the indicated micromolar concentrations determined by SPR. Data are representative of three independent experiments. (F) Equilibrium binding and affinity (steady state) of TCR10 to HLA-C*08:02–KRAS-G12D-10-mer and KRAS-G12D-9-mer. Data are representative of three independent experiments. (G and H) Correlation of TCR affinity (KA) with TCR frequency in the infusion product used to treat patient 4095 (G) and in the periphery of patient 4095, 9 mo after T cell transfer (H). TCR frequencies are from ref. 17.
Contrasting TCR Binding Modes for Recognition of KRAS-G12D Neoepitopes.
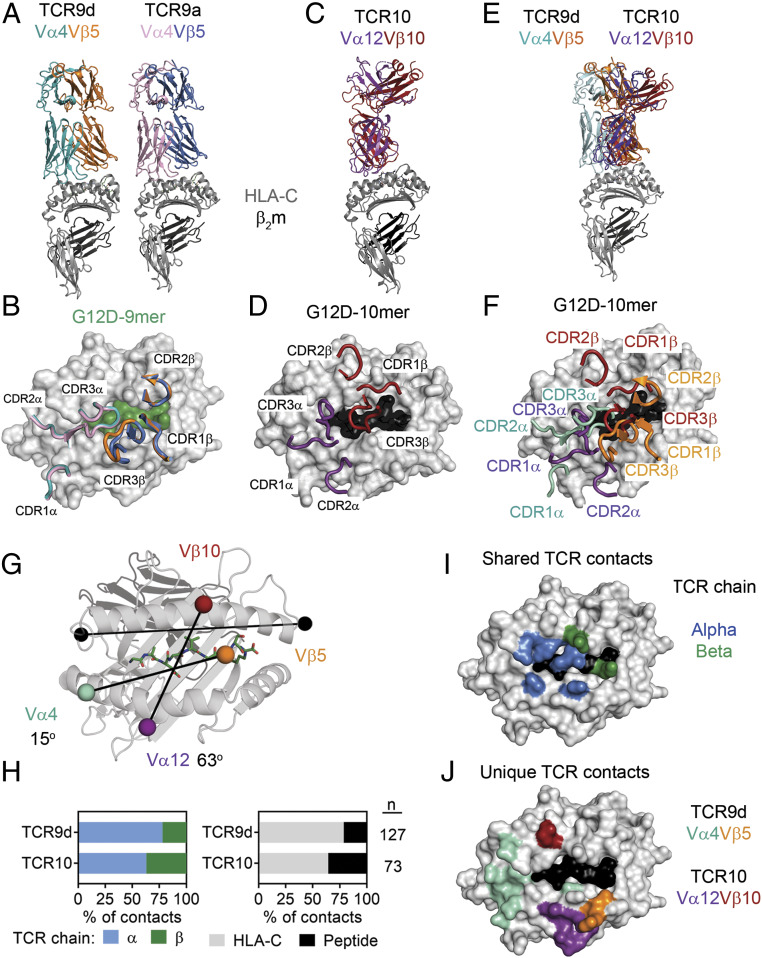
To understand how these therapeutic TCRs exhibit non–cross-reactive binding to the two similar G12D neoantigens, we solved the X-ray crystal structures of three TCRs (9a, 9d, 10) in complex with HLA-C*08:02 (SI Appendix, Table S1). TCR9a and 9d recognized the HLA-C*08:02–presenting G12D nonamer and these structures were refined to 3.2- and 2.0-Å resolution, respectively (Fig. 4 A and B and SI Appendix, Table S1). TCR9a and 9d (both Vα4Vβ5) bind HLA-C*08:02 in an identical orientation with almost complete overlap of their six CDR loops (Fig. 4B). The third structure was a complex of TCR10 with HLA-C*08:02–decamer, which was refined to a resolution of 3.5 Å (Fig. 4 C and D). Given the similarity of the TCR9a and 9d complex structures, comparisons to the TCR10 complex were made only with the TCR9d complex. Unlike the conserved structural recognition of HLA-C between TCR9a and 9d, TCR9d and TCR10 have limited overlap when aligned onto HLA-C*08:02 (Fig. 4 E and F). The two TCRs have docking geometries that differ by 48° (Fig. 4G). TCR9d docks diagonally (relative to the HLA-C α1-helix vector) with a crossing angle of 15°, while TCR10 docks more vertically with a crossing angle of 63°. Overall, TCR9d makes more contacts with the peptide–HLA-C complex than TCR10, consistent with its higher antigenic affinity (Figs. 3 and 4H and SI Appendix, Tables S3 and S4). Proportionally, the α-chain of both TCRs made more contacts to pHLA-C than their β-chain, a difference that was more pronounced for TCR9d (Fig. 4H). Importantly, TCR9d and TCR10 exhibited unique contacts to HLA-C to accommodate the nonamer and decamer antigens that are similar in sequence but divergent in structure (Fig. 4 H–J and SI Appendix, Tables S3 and S4).
Fig. 4.
Contrasting TCR binding modes for recognition of KRAS-G12D neoepitopes. (A) Structures of TCR9d and TCR9a with HLA-C*08:02 and the KRAS-G12D-9-mer. TCR9d α-chain, turquoise; β-chain, orange; TCR9a α-chain, pink; β-chain, blue; HLA-C*08:02, gray; β2m, dark gray; KRAS-G12D-9-mer, green. (B) Placement of TCR9d and TCR9a CDR loops over HLA-C*08:02–KRAS-G12D-9-mer. (C) Structure of TCR10 with HLA-C*08:02 and the KRAS-G12D-10-mer. TCR10 α-chain, purple; β-chain, red; HLA-C*08:02, gray; β2m, dark gray; KRAS-G12D-10-mer, black. (D) Placement of TCR10 CDR loops over HLA-C*08:02–KRAS-G12D-10-mer. (E) Overlay of TCR9d and TCR10 with HLA-C*08:02. TCR9d and the HLA-C*08:02–9-mer complex was aligned with HLA-C*08:02 of the TCR10–HLA-C*08:02 complex. (F) Placement of TCR9d and TCR10 CDR loops over HLA-C*08:02–KRAS-G12D-10-mer. TCR9d α-chain, turquoise; β-chain, orange; TCR10 α-chain, purple; β-chain, red. (G) Crossing angles of TCR9d and TCR10 are shown in reference to the HLA-C α1-helix. Vectors are shown drawn between the centers of mass for each TCR Vα- and Vβ-chain, colored as above. Crossing angles were determined as where the TCR vector crosses the HLA-C α1-helix vector (black), drawn between positions 50 and 86 as described (2). (H) Percentage of TCR contacts with HLA-C*08:02 or peptide (Right) and percentage of TCR contacts derived from the alpha or beta chain (Left). Data were analyzed from the complex of TCR9d–HLA-C*08:02–KRAS-G12D-9-mer and TCR10–HLA-C*08:02–KRAS-G12D-10-mer. (I) Location of HLA-C*08:02 residues that form contacts in both the TCR9d and TCR10 complexes. HLA-C*08:02 residues are color-coded depending on whether the contact was with the alpha (blue) or beta (green) TCR chain. Other HLA-C residues, gray; KRAS-G12D-10-mer, black. (J) Location of HLA-C*08:02 residues that form unique contacts with TCR9d or TCR10. Color depicts HLA-C residues in contact with the TCR9d α-chain, turquoise; β-chain, orange; TCR10 α-chain, purple; β-chain, red. Other HLA-C residues, gray; KRAS-G12D-10-mer, black.
T Cell Recognition of the KRAS-G12D Nonamer.
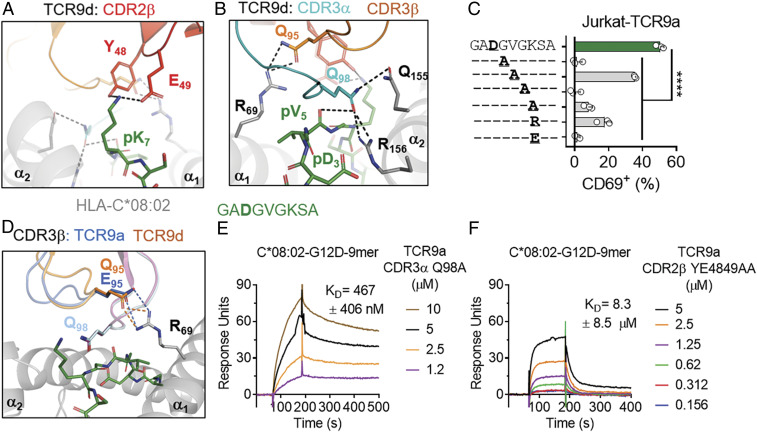
TCR9a and 9d were identified from TILs of two individuals and share almost identical binding modes (Fig. 4). Specifically, the nonamer TCRs share almost identical TCR sequences, differing significantly only in their CDR3β. The common recognition mode was dominated by shared CDR3α and CDR2β interactions with pHLA-C (Fig. 5 A and B and SI Appendix, Fig. S3A). Lys at peptide p7 is the most prominent exposed residue of the peptide (Figs. 1D and 5A). It forms a hydrogen bond (h-bond) with Tyr-48 and a salt bridge with Glu-49 of CDR2β. T cell activation was substantially reduced when p7 Lys was replaced with Ala and abolished when Lys was replaced with Glu, likely due to charge repulsion with Glu-49 of CDR2β (Fig. 5C and SI Appendix, Fig. S3D). Arg replacement at p7 did not fully restore the T cell activation, suggesting that Lys is preferred at p7 (Fig. 5C). In addition, Gln-98 of CDR3α forms h-bonds with Gln-155 and Arg-156 of HLA-C*08:02, and with the carbonyl group of the peptide p5 (Val) (Fig. 5B). Ala replacement of Gln-98 resulted in a 30-fold loss in TCR9a binding affinity to the nonamer–HLA-C ligand, whereas Ala substitutions of Tyr-48 and Glu-49 in CDR2β decreased the binding by 500-fold, suggesting the CDR2β interactions contribute more than CDR3α to the high affinity of TCR9a (Fig. 5 E and F). Ala substitution at p4 or p6 Gly, but not p5 Val, abolished T cell recognition, suggesting conformational flexibility was necessary for T cell recognition (Fig. 5C and SI Appendix, Fig. S3D).
Fig. 5.
T cell recognition of the KRAS-G12D nonamer via CDR2β. (A) TCR9d CDR2β interactions with p7 Lys of the KRAS-G12D-9-mer. CDR2β, red; HLA-C, gray; KRAS-G12D-9-mer, green. (B) TCR9d CDR3α interactions with HLA-C*08:02 and the KRAS-G12D-9-mer. CDR3α, turquoise; CDR3β, orange; HLA-C, gray; KRAS-G12D-9-mer, green.(C) Frequency of TCR+ Jurkat T cells expressing CD69 after incubation with 221-C*08:02-ICP47 cells loaded with KRAS-G12D-9-mer peptides with the indicated amino acid substitutions. Amino acids identical to the KRAS sequence are indicated with “–.” Peptides were tested from 1,000 to 1 nM, shown here at 10 nM; data are a mean of three independent experiments. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison test (****P < 0.0001). (D) TCR9a/d CDR3β interactions with HLA-C*08:02 Arg-69. TCR9a-CDR3β, blue; TCR9d-CDR3β, orange; HLA-C, gray; KRAS-G12D-9-mer, green. (E and F) Binding of TCR9a-CDR3α Q98A (E) and TCR9a-CDR2β YE48,49AA (F) to captured HLA-C*08:02–KRAS-G12D-9-mer at the indicated nanomolar concentrations determined by SPR. Dissociation constants were determined by kinetic curve fitting. Data are representative of two independent experiments.
TCR9a to 9d differ substantially in their CDR3β sequence and exhibit a range of affinities (Fig. 3). Consistently, the conformation of the CDR3β loop was the main difference between the structures of the TCR9a and 9d complexes with HLA-C (Figs. 4B and 5D and SI Appendix, Fig. S3 B and C). CDR3α extends underneath CDR3β, obscuring its contact with the peptide (Fig. 5 B and D). Consequently, both TCR9a and 9d CDR3βs make no contacts with the peptide and instead form either a salt bridge (from TCR9a Glu-95) or h-bond (TCR9d Gln-95) to Arg-69 of HLA-C*08:02, respectively (Fig. 5D). Variation at CDR3β position 95 among the four nonamer-specific TCRs is consistent with the variable HLA-C binding affinities observed among the nonamer-specific TCRs (Fig. 5G). Glu-95 of TCR9a forms a salt bridge with Arg-69 of HLA-C, leading to the highest ligand binding affinity of 16 nM (Fig. 5G). Gln-95 of TCR9c and 9d forms an h-bond with Arg-69, resulting in similar binding affinities of 90 and 125 nM, respectively. In contrast, Arg-95 of TCR9b would not be able to interact with Arg-69, and thus TCR9b displays the lowest pHLA-C binding affinity of 825 nM (Fig. 5G). Consistently, substitution of Glu-95 with Arg in the CDR3β of TCR9a reduced its affinity from 16 to 377 nM, similar to that of TCR9b (SI Appendix, Fig. S3E). Together, our data show that the G12D nonamer-specific TCRs share an identical HLA binding mode that relies on their CDR3α and CDR2β for peptide recognition. CDR3β did not contact peptide but made a variable HLA-C contact. These structures provide a molecular explanation for how multiple TCRs can maintain neoantigen specificity with variable affinities by adopting a shared binding mode.
TCR10 Recognizes an Altered KRAS-G12D Decamer Peptide Conformation.
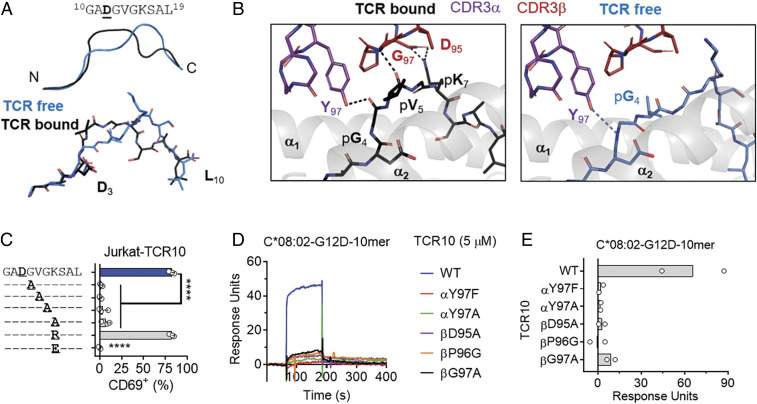
Notably, in the crystal structure of TCR10 (Vα12Vβ10) complexed with HLA-C*08:02-G12D, the decamer exhibited yet a different conformation compared with that of the “TCR-free” (HLA-C–alone) structure (Figs. 2B and 6A and SI Appendix, Fig. S4 A–C). While the decamer bulges out of the HLA-C peptide-binding groove between the p6 and p8 positions in the TCR-free HLA-C structure (Figs. 2B and 6A), the bulge is shifted to the p4 to p6 positions in the TCR-bound HLA-C (Fig. 6A). Specifically, the positions of Val-Gly-Lys-Ser (p5 to p8) of the peptide shifted 3.5 to 4.8 Å upon TCR complex, breaking a van der Waals contact and two h-bonds between the peptide and HLA-C observed in the TCR-free structure (SI Appendix, Fig. S4D). The two h-bonds were between Tyr-97 of CDR3α and the carbonyl of Gly (p4) and between the amide of CDR3β Gly-97 and the carbonyl of Val (p5). The salt bridge was between CDR3β Asp-95 and Lys (p7) (Fig. 6B). Substitution of the p7 Lys with Arg preserved activation of TCR10-transfected T cells, while Ala or Glu substitutions abolished activation, demonstrating the importance of the salt bridge between Asp-95 of CDR3β and Lys at p7 of the peptide (Fig. 6D and SI Appendix, Fig. S4D). None of these CDR3β contacts could be made with the TCR-free peptide by modeling (Fig. 6 A–C). Ala substitution of p4 to p6 (GVG) abolished recognition, further supporting the importance of a peptide conformational change (Fig. 6D and SI Appendix, Fig. S4D). By SPR, substitutions in CDR3α (Y97F, Y97A) and CDR3β (D95A, P96G, G97A) each resulted in loss of the TCR binding to the decamer–HLA-C (Fig. 6 E and F). We conclude that the G12D-decamer conformation with a bulge at p5 Val is necessary for TCR10 activation.
Fig. 6.
T cell recognition of the KRAS-G12D decamer via an altered peptide conformation. (A) Cartoon (Top) and stick (Bottom) models of the KRAS-G12D-10-mer in two conformations. TCR-free conformation, blue; the conformation in complex with TCR10, black. (B) Interactions of TCR10 with the KRAS-G12D-10-mer in TCR-bound (Left) conformation. H bonds are between Tyr-97 of CDR3α and the carbonyl of Gly (p4) and between the amide of CDR3β Gly-97 and the carbonyl of Val (p5). The salt bridge was between CDR3β Asp-95 and Lys (p7). CDR3α, purple; CDR3β, red; HLA-C, gray; KRAS-G12D-10-mer, black. (Right) Modeling of the TCR10 interaction with the KRAS-G12D-10-mer in the TCR-free conformation. KRAS-G12D-10-mer, blue. (C) Frequency of TCR+ Jurkat T cells expressing CD69 after incubation with 221-C*08:02-ICP47 cells loaded with KRAS-G12D-10-mer peptides with the indicated amino acid substitutions. Amino acids identical to the KRAS sequence are indicated with “–.” Peptides were tested from 1,000 to 1 nM, shown here at 10 nM; data are a mean of three independent experiments. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison test (****P < 0.0001). (D and E) Binding of WT and mutant TCR10 with indicated amino acid substitutions to captured HLA-C*08:02–KRAS-G12D-10-mer at 5 μM, determined by SPR. Representative of two independent experiments (D) and summary (E).
Modeling TCR9 onto the decamer–HLA-C structure revealed the importance of TCR10 in recognition of the “TCR10-bound” decamer conformation. TCR9d could partially accommodate the decamer peptide in its TCR-free conformation, as evidenced by the cross-reactive recognition of nonamer-specific TCR9a to the G12D decamer (Fig. 1E and SI Appendix, Fig. S5). However, significant steric hindrance between the TCR10-bound decamer conformation and TCR9 CDR3α would prevent the receptor binding to this conformation. The distinct antigenic recognition is further supported by peptide substitutions, where Ala substitutions of p4 to p7 eliminated both TCR9a and TCR10 activation (SI Appendix, Fig. S5D). In contrast, Arg at p7 reduced TCR9a but not TCR10 activation. Our functional and structural studies suggest the decamer exists in two conformational states, one that is weakly recognized by TCR9 and one that is effectively recognized by TCR10. It is likely that KRAS-G12D+ tumors present both the nonamer and decamer peptides and therefore the existence of TCR10 in the TIL infusion ensured efficient targeting of both structurally distinct neoantigens.
Discussion
ACT with ex vivo expanded TILs is a promising cancer immunotherapy, but predicting clinical success remains challenging. This is in part due to the lack of understanding of the structural and sequence requirements of TCRs for effective ACT. Here, we studied multiple TCRs from a successful case of ACT against the clinically important, common oncogenic mutation KRAS-G12D. Our study demonstrates that effective ACT, in this case, is associated with oligoclonal TCRs that exhibit high binding affinities for two structurally distinct neoantigens. The nonamer-specific TCRs differed in their binding affinities due to a CDR3β-dependent variable contact with HLA-C. However, neoantigen specificity by TCR9 was maintained via a shared binding mode dependent on their CDR3α and CDR2β. Effective recognition of the G12D decamer required a peptide conformation only recognized by TCR10, not TCR9. Tumor specificity was in large part due to the selective binding of mutant KRAS-G12D to HLA-C*08:02, sparing healthy cells that express WT KRAS. Together, this successful case of ACT was associated with the infusion of multiple, high-affinity TCRs which exhibit oligoclonal-like recognition of the same mutation.
Antigen-specific TCRs generally consist of diverse populations of sequences (31–33) and CDR3β sequences often play a dominant role in determining peptide specificity (1, 2). For the nonamer-specific TCRs this was not the case, as CDR3β variation generated oligoclonality leading to variable binding affinities but neoantigen specificity was maintained through a shared binding mode. Crystal structures of TCR9a and 9d complexed with HLA-C*08:02–KRAS-G12D nonamer revealed that TCR9 recognition was dependent on the germline-encoded CDR2β of TRBV5-6*01 and the CDR3α chain. The shared binding mode is indicative of “public” TCRs that could be exploited by vaccine strategies (34). Explanations for public TCRs include convergent recombination, where many V(D)J recombination events converge on the same sequence (34). By mutational analysis, germline-encoded CDR2β TRBV5-6*01 Tyr-48 and Glu-49 appeared largely responsible for nonamer binding affinity and thus multiple TCRβ recombination events, with variable CDR3β sequences, could derive nonamer specificity. This likely explains why nonamer-specific TCRs were found in two patients (16, 17). As these TCRs derive from only two individuals, it is yet to be seen whether these TCRs are truly public. Nonetheless, it would be interesting to examine the frequency of TCR9-like TCRs from HLA-C*08:02+ patients who carry KRAS-G12D+ tumors.
The presence of both TCR9 and TCR10 ensured a more complete antigenic coverage than the use of either alone. The nonamer and decamer KRAS-G12D neoantigens contact HLA-C*08:02 in similar ways but form structurally distinct T cell epitopes. While the nonamer-specific TCRs may recognize the TCR-free conformation of the decamer, TCR10 more effectively recognized a structurally distinct conformation of the decamer that is not cross-reactive to the nonamer. This different binding mode was associated with fewer pHLA-C contacts and a weaker affinity than the nonamer-specific TCRs, but was still in the higher range for natural TCRs (28–30).
Patient 4095 was treated with an infusion of expanded TILs containing four KRAS-G12D–specific TCRs (17). These TCRs displayed similar functional responses in vitro; however, post adoptive transfer they displayed variable in vivo persistence (17). Most dramatic was the disappearance of TCR9a and the substantial engraftment of TCR10 at 10% of peripheral T cells 9 mo post transfer (17). Prior to our study, no molecular feature could explain the differences in in vivo TCR persistence. Limited of course by the number of TCRs studied from this case, we nonetheless observed that TCR frequency post adoptive transfer was inversely correlated with TCR affinity. Specifically, those TCRs with lower affinity (TCR9b and TCR10) had the highest frequencies. Somewhat consistent with this, previous studies suggested that intermediate-affinity TCRs are optimal for ACT and there is an optimal affinity window for TCR signaling with a KD near 1 μM (35–40). The highest-affinity TCRs (9a and 9c) were almost undetectable 9 mo post transfer (17). For TCR9c, this could be explained by its low frequency in the TIL infusion (0.04%) but not for TCR9a, which made up 50% of the infusion. While the high-affinity TCRs (9a and 9c) displayed no reactivity to WT KRAS, in vivo these TCRs may have experienced chronic stimulation due to their higher affinity for HLA-C*08:02 or cross-reactivity to other peptide antigens. Indeed, it would be interesting, and important for the clinical development of these TCRs, to study the peptide cross-reactivity of TCR9a to 9d to examine the impact of the variable HLA-C contact on neoantigen specificity. Previous studies of high-affinity TCR–MHC-I interactions suggest they can induce an anergic-like response, perhaps due to increased levels of SHP-1 (41, 42). Supporting the idea that TCRa+ cells exhibit poor in vivo functionality, a recent single-cell RNA-sequencing analysis revealed nonpersistent TCR9a+ T cells exhibited a differential gene expression pattern compared with the persistent TCR9b+ and TCR10+ T cells (43). Specifically, TCR9a+ T cells expressed lower transcripts for IL-7R and higher levels of the EOMES transcription factor, hallmarks of exhausted T cells (44). Monoclonal TIL-expanded or TCR-transduced T cells have been successfully used in immunotherapy (45, 46); however, it is not clear whether in the case studied here the inclusion of multiple TCRs with a range of affinities contributed to its clinical success. It would be interesting to examine in an in vivo setting whether the inclusion of high-affinity TCRs, in addition to intermediate-affinity TCRs, may confer an advantage compared with the intermediate-affinity TCRs alone. In addition, high-affinity TCRs such as TCR9a, 9c, and 9d could be of advantage for ACT with TCR-transduced T cells as they are likely to be CD8-independent and can be functionally expressed in CD4+ T cells (47, 48).
The KRAS-G12D neoantigens exhibit features of “high-quality” neoantigens, which may also have contributed to the success of this case (49–51). The KRAS-G12D neoantigens have no similarity to self as WT KRAS peptides lack the critical Asp residue to stabilize HLA-C*08:02 (Fig. 1). This suggests G12D-specific T cells would exhibit minimal off-target effects, which have been reported in previous ACT trials (52–55). A recent analysis of immunogenic neoantigens revealed that for anchor residue mutations, the relative binding comparing WT with mutant was a strong indicator of immunogenicity (56). In contrast to G12D mutants, the WT KRAS peptides were very poor ligands for HLA-C*08:02, conferring no HLA-C stabilization of cell-surface or recombinant HLA-C, suggesting the relative binding affinity was very high for these epitopes. For the design of peptide vaccines, we showed that a C-terminally modified nonamer G12D peptide (G12D-A18L) would be a better candidate as it improved the sensitivity of TCR9a+ Jurkat cells by ∼20-fold (Fig. 2C). Future work should explore the impact of this C-terminal substitution on T cell recognition as anchor modifications can have subtle effects on peptide side-chain orientation and T cell recognition (27).
There is considerable interest in determining which tumor mutations lead to immunogenic neoantigens (56, 57). A deeper understanding of how TIL TCRs recognize immunogenic neoantigens could lead to improvements in predicting targets for neoantigen vaccines and the development of other TCR-based therapeutics (58, 59). Here we evaluated how the immune system detects a common cancer mutation of great clinical interest. A recent analysis of colorectal adenocarcinoma patients identified 18.4% (687/3,734) carried HLA-C*08:02 and 2.3% (85/3,734) carried HLA-C*08:02 in combination with tumors bearing KRAS-G12D (60). It was estimated that over 130,000 new cases of colorectal cancer would be diagnosed in 2016 (61), suggesting thousands of patients would be eligible for KRAS-G12D–specific immunotherapy in this one cancer type alone. Our study revealed a set of favorable TCR attributes associated with a successful TIL-based immunotherapy in addition to deepening our understanding of T cell recognition and informing peptide vaccine design.
Materials and Methods
Cell Culture and TCR Transfection.
Jurkat T cells and 221-C*08:02-ICP47 cells were cultured in Iscove’s modified Dulbecco’s medium (Gibco) and 10% fetal calf serum. DNA encoding TCR9a and TCR10 was synthesized and cloned into pCDNA3.1 (GenScript). The constructs contained a P2A site separating the alpha and beta chains, as described (62). Jurkat cells were transfected by nucleofection using Amaxa (Lonza) with the program X-001 in Cell Line Nucleofector Solution V. After G418 selection, cells were cloned by limiting dilution and screened for functional responses to cognate peptide loaded on 221-C*08:02-ICP47 cells.
Peptide Loading Assay.
Peptide loading assays were as previously described (19). Peptides were synthesized at >95% purity (GenScript). Peptide was incubated overnight at 100 μM with 105 221-C*08:02-ICP47 cells at 26 °C. The following day, cells were washed once with phosphate-buffered saline (PBS) and stained with anti–HLA-I mAb (APC [allophycocyanin], W6/32; BioLegend; 311410). Expression of HLA-C was determined by flow cytometry. Each peptide was tested at least twice in independent experiments.
T Cell Activation Assay.
The day prior to the assay, 105 221-C*08:02-ICP47 target cells were incubated overnight at 26 °C with peptide ranging from 1 μM to 0.01 nM. The following day, target cells were mixed with TCR+ Jurkat T cells for 6 h at 37 °C. Cells were then washed twice in PBS and stained with mAbs to CD69 (APC, FN50; BD Biosciences; 555533) and CD3 (APC-Cy7, UCHT1; BioLegend; 300426). Expression of CD69 was measured on CD3+ cells by flow cytometry. All peptides were tested at the indicated concentrations at least twice in independent experiments.
Flow Cytometry.
Flow cytometry was performed on an LSR II or Fortessa X-20 (BD Biosciences) and data were analyzed using FlowJo software (Treestar; v10). Cytometer setup and tracking beads were run daily and single-mAb–stained beads were used to determine compensation settings for multicolor experiments
Protein Expression, Purification, and Crystallization.
DNA encoding residues 1 to 278 of HLA-C*08:02, 1 to 99 of β2m, and the TCR extracellular portions were synthesized and cloned into the bacterial expression vector pET30a via NdeI and XhoI (GenScript). All proteins were expressed as inclusion bodies in BL21 (DE3) cells (Invitrogen). The HLA-Cβ2m–peptide complex and TCR heterodimers were refolded by rapid dilution as previously described (63–65). Proteins were purified first via ion exchange followed by size-exclusion chromatography with a Superdex 200 column (GE Healthcare). Proteins were concentrated to 10 mg/mL and screened for optimal crystallization conditions using commercial and in-house screens using a Crystal Gryphon (ARI). HLA-C*08:02–nonamer and HLA-C*08:02–decamer crystals formed in 0.1 M Bis-Tris (pH 6.5), 0.05 M CaCl2 dihydrate, 30% poly(ethylene) glycol monomethyl ether (PEG MME) 500. TCR9a and TCR9d, complexed with HLA-C*08:02–nonamer, formed crystals in 22% PEG 3350, 0.1 M Mops (pH 7.1), and 0.25 M MgSO4. TCR10 complexed with HLA-C*08:02–decamer formed crystals in 12.5% PEG 4000, 0.1 M Na cacodylate (pH 5.8), 0.2 M (NH4)2SO4, and 10% glycerol.
Data Collection, Structure Determination, and Refinement.
Crystals were immersed in cryoprotectant (crystallization conditions plus 20% glycerol) before flash cooling in liquid nitrogen. Single datasets were collected for all structures except the TCR10–HLA-C*08:02–decamer complex, where a complete and partial dataset from the same crystal were merged. All data were collected on the SER-CAT 22 ID or BM beamlines (Argonne National Laboratory) and processed and merged using HKL2000 (66). All structures were solved by the molecular replacement method with Phaser in the CCP4 package and models were built and refined with Coot and Phenix (67–70). The structure of HLA-C*08:01 (71) (Protein Data Bank [PDB] ID code 4NT6) with the peptide omitted was used as the search model for the HLA-C*08:02–alone structures. Our HLA-C*08:02 structure with the peptide omitted and a Vα4 TCR (72) (PDB ID code 6AVG) with CDR loops omitted were used as search models for the TCR9 complexes. For the TCR10–HLA-C*08:02–decamer complex, a Vα12 TCR with the CDR loops omitted was used as the search model (PDB ID code 4ZDH). HLA-C*08:02–alone structures contained one molecule per asymmetric unit and belonged to the C2 space group. The TCR9a/d–HLA-C*08:02–nonamer complexes contained one complex per asymmetric unit and belonged to the P21 space group. The TCR10–HLA-C*08:02–decamer complex contained two complexes per asymmetric unit and belonged to the P21 space group. Peptide and CDR loops were added manually using 2Fo − Fc electron density maps. Graphical figures were generated in PyMOL.
Surface Plasmon Resonance.
SPR was performed with a BIAcore 3000 instrument and analyzed with BIAevaluation software v4.1 (GE Healthcare). The pan HLA-I–specific mAb W6/32 (BioLegend) was immobilized to CM5 chips (GE Healthcare) at 5,000 to 7,000 response units (RUs) by primary amine coupling with a 2 μL/min flow rate in 10 mM sodium acetate (pH 5.5). HLA-C was captured by W6/32 at 400 to 700 RUs in PBS. The analytes were TCR heterodimers in 10 mM Hepes (pH 7.5) and 0.15 M NaCl with a flow rate of 50 μL/min. TCRs were injected for 2 min followed by a dissociation of 10 min. Binding was measured with serial dilutions of TCR from 10 to 0.15 μM for TCR10 and 1,200 to 37.5 nM for TCR9a to 9d. Dissociation constants were obtained by modeling steady-state kinetics for TCR10 and kinetic curve fitting for TCR9a to 9d with BIAevaluation software.
Differential Scanning Fluorimetry.
Thermal denaturation assays were performed by differential scanning fluorimetry largely as described (23). Proteins were diluted to 20 and 10 μM in duplicate on ice and then mixed with 10 and 5× (final concentration) SYPRO orange dye (Thermo Fisher Scientific). Assays were diluted in 10 mM Hepes (pH 7.5) and 0.15 M NaCl and proteins were heated from 10 to 90 °C at 1 °C/min. Fluorescence was measured every minute on a CFX Connect Real-Time PCR Detection System on the fluorescence resonance energy transfer setting (Bio-Rad).
Statistical Analysis.
All statistical analyses were carried out in GraphPad Prism (version 7).
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases, and National Cancer Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The X-ray crystallographic data and coordinates reported in this paper have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes are 6ULI, 6ULK, 6ULN, 6ULR, and 6UON).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921964117/-/DCSupplemental.
References
- 1.Rossjohn J. et al., T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Rudolph M. G., Stanfield R. L., Wilson I. A., How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Garcia K. C. et al., An alphabeta T cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex. Science 274, 209–219 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Garboczi D. N. et al., Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Garcia K. C., Adams J. J., Feng D., Ely L. K., The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat. Immunol. 10, 143–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilyas S., Yang J. C., Landscape of tumor antigens in T cell immunotherapy. J. Immunol. 195, 5117–5122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havel J. J., Chowell D., Chan T. A., The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z., Ott P. A., Wu C. J., Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 18, 168–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobbs G. A., Der C. J., Rossman K. L., RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephen A. G., Esposito D., Bagni R. K., McCormick F., Dragging Ras back in the ring. Cancer Cell 25, 272–281 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg S. A. et al., Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff S. L. et al., Randomized, prospective evaluation comparing intensity of lymphodepletion before adoptive transfer of tumor-infiltrating lymphocytes for patients with metastatic melanoma. J. Clin. Oncol. 34, 2389–2397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott A. M., Wolchok J. D., Old L. J., Antibody therapy of cancer. Nat. Rev. Cancer 12, 278–287 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto T. N., Kishton R. J., Restifo N. P., Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat. Med. 25, 1488–1499 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Tran E. et al., Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran E. et al., T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.June C. H., Drugging the undruggable Ras—Immunotherapy to the rescue? N. Engl. J. Med. 375, 2286–2289 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Sim M. J. et al., Canonical and cross-reactive binding of NK cell inhibitory receptors to HLA-C allotypes is dictated by peptides bound to HLA-C. Front. Immunol. 8, 193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josephs T. M., Grant E. J., Gras S., Molecular challenges imposed by MHC-I restricted long epitopes on T cell immunity. Biol. Chem. 398, 1027–1036 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Guo H. C. et al., Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360, 364–366 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Di Marco M. et al., Unveiling the peptide motifs of HLA-C and HLA-G from naturally presented peptides and generation of binding prediction matrices. J. Immunol. 199, 2639–2651 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Hellman L. M. et al., Differential scanning fluorimetry based assessments of the thermal and kinetic stability of peptide-MHC complexes. J. Immunol. Methods 432, 95–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur G. et al., Structural and regulatory diversity shape HLA-C protein expression levels. Nat. Commun. 8, 15924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borbulevych O. Y., Baxter T. K., Yu Z., Restifo N. P., Baker B. M., Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: Implications for vaccine design. J. Immunol. 174, 4812–4820 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buhrman J. D., Slansky J. E., Improving T cell responses to modified peptides in tumor vaccines. Immunol. Res. 55, 34–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J. L. et al., Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 201, 1243–1255 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leem J., de Oliveira S. H. P., Krawczyk K., Deane C. M., STCRDab: The Structural T-Cell Receptor Database. Nucleic Acids Res. 46, D406–D412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borrman T. et al., ATLAS: A database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes. Proteins 85, 908–916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aleksic M. et al., Different affinity windows for virus and cancer-specific T-cell receptors: Implications for therapeutic strategies. Eur. J. Immunol. 42, 3174–3179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G. et al., Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens. Cell Rep. 19, 569–583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song I. et al., Broad TCR repertoire and diverse structural solutions for recognition of an immunodominant CD8+ T cell epitope. Nat. Struct. Mol. Biol. 24, 395–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su L. F., Kidd B. A., Han A., Kotzin J. J., Davis M. M., Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 38, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venturi V., Price D. A., Douek D. C., Davenport M. P., The molecular basis for public T-cell responses? Nat. Rev. Immunol. 8, 231–238 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Corse E., Gottschalk R. A., Krogsgaard M., Allison J. P., Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol. 8, e1000481 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMahan R. H. et al., Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J. Clin. Invest. 116, 2543–2551 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Presotto D. et al., Fine-tuning of optimal TCR signaling in tumor-redirected CD8 T cells by distinct TCR affinity-mediated mechanisms. Front. Immunol. 8, 1564 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong S. et al., T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 110, 6973–6978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slansky J. E., Jordan K. R., The Goldilocks model for TCR—Too much attraction might not be best for vaccine design. PLoS Biol. 8, e1000482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid D. A. et al., Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J. Immunol. 184, 4936–4946 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Hebeisen M. et al., SHP-1 phosphatase activity counteracts increased T cell receptor affinity. J. Clin. Invest. 123, 1044–1056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith T. R., Verdeil G., Marquardt K., Sherman L. A., Contribution of TCR signaling strength to CD8+ T cell peripheral tolerance mechanisms. J. Immunol. 193, 3409–3416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y. C. et al., Single-cell transcriptome analysis reveals gene signatures associated with T-cell persistence following adoptive cell therapy. Cancer Immunol. Res. 7, 1824–1836 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paley M. A. et al., Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robbins P. F. et al., A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: Long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran E. et al., Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone J. D., Kranz D. M., Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front. Immunol. 4, 244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerry S. E. et al., Interplay between TCR affinity and necessity of coreceptor ligation: High-affinity peptide-MHC/TCR interaction overcomes lack of CD8 engagement. J. Immunol. 171, 4493–4503 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGranahan N., Swanton C., Neoantigen quality, not quantity. Sci. Transl. Med. 11, eaax7918 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Łuksza M. et al., A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balachandran V. P. et al.; Australian Pancreatic Cancer Genome Initiative , Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linette G. P. et al., Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 122, 863–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkhurst M. R. et al., T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 19, 620–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson L. A. et al., Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmer D. C. et al., Effective tumor treatment targeting a melanoma/melanocyte-associated antigen triggers severe ocular autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 105, 8061–8066 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capietto A. H. et al., Mutation position is an important determinant for predicting cancer neoantigens. J. Exp. Med. 217, e20190179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Garijo A., Fajardo C. A., Gros A., Determinants for neoantigen identification. Front. Immunol. 10, 1392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riley T. P. et al., Structure based prediction of neoantigen immunogenicity. Front. Immunol. 10, 2047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowe K. L. et al., Novel TCR-based biologics: Mobilising T cells to warm “cold” tumours. Cancer Treat. Rev. 77, 35–43 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Maoz A., Rennert G., Gruber S. B., T-cell transfer therapy targeting mutant KRAS. N. Engl. J. Med. 376, e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marley A. R., Nan H., Epidemiology of colorectal cancer. Int. J. Mol. Epidemiol. Genet. 7, 105–114 (2016). [PMC free article] [PubMed] [Google Scholar]
- 62.Yang S. et al., Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Ther. 15, 1411–1423 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garboczi D. N., Hung D. T., Wiley D. C., HLA-A2-peptide complexes: Refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. U.S.A. 89, 3429–3433 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tikhonova A. N. et al., αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity 36, 79–91 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clements C. S. et al., The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr. D Biol. Crystallogr. 58, 2131–2134 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 67.McCoy A. J. et al., Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Adams P. D. et al., PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Collaborative Computational Project, Number 4 , The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 71.Choo J. A., Liu J., Toh X., Grotenbreg G. M., Ren E. C., The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans. J. Virol. 88, 10613–10623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan K. F. et al., Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide. Nat. Commun. 9, 1026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.