Significance
Understanding the mechanisms of retinal development and homeostasis is essential for the advancement of retinal regeneration strategies and conception of novel therapies for multifactorial retinal diseases. Forward genetic analysis allows for the unbiased identification of nonredundant genes involved in the generation and maintenance of retinal integrity. Here we combine a special forward genetics protocol, in which all mice in tested pedigrees have been pregenotyped at all mutant loci with a sensitive and reproducible retinal screening assay, optical coherence tomography. We show that this robust pipeline can detect subtle phenotype-genotype associations and characterize one of multiple novel associations. This approach will help discover new pathways and better define known pathways essential to retinal development and function.
Keywords: Sfxn3, forward genetics, ENU mutagenesis, CRISPR, retinal degeneration
Abstract
Retinal disease and loss of vision can result from any disruption of the complex pathways controlling retinal development and homeostasis. Forward genetics provides an excellent tool to find, in an unbiased manner, genes that are essential to these processes. Using N-ethyl-N-nitrosourea mutagenesis in mice in combination with a screening protocol using optical coherence tomography (OCT) and automated meiotic mapping, we identified 11 mutations presumably causative of retinal phenotypes in genes previously known to be essential for retinal integrity. In addition, we found multiple statistically significant gene-phenotype associations that have not been reported previously and decided to target one of these genes, Sfxn3 (encoding sideroflexin-3), using CRISPR/Cas9 technology. We demonstrate, using OCT, light microscopy, and electroretinography, that two Sfxn3−/− mouse lines developed progressive and severe outer retinal degeneration. Electron microscopy showed thinning of the retinal pigment epithelium and disruption of the external limiting membrane. Using single-cell RNA sequencing of retinal cells isolated from C57BL/6J mice, we demonstrate that Sfxn3 is expressed in several bipolar cell subtypes, retinal ganglion cells, and some amacrine cell subtypes but not significantly in Müller cells or photoreceptors. In situ hybridization confirmed these findings. Furthermore, pathway analysis suggests that Sfxn3 may be associated with synaptic homeostasis. Importantly, electron microscopy analysis showed disruption of synapses and synaptic ribbons in the outer plexiform layer of Sfxn3−/− mice. Our work describes a previously unknown requirement for Sfxn3 in retinal function.
The retina and the retinal pigment epithelium (RPE) are complex, specialized, highly organized tissues working in concert to transform light into neural messages ultimately manifested as vision. For this system to work, meticulously orchestrated development must be followed by an equally important set of homeostatic processes capable of resisting a variety of insults. Retinal disease and decreased vision can result from defects in these processes. Our goal is to increase our understanding of the mechanisms controlling retinal development and homeostasis.
Gene discovery using an unbiased forward genetic approach (1–6) is well suited to the identification of proteins with nonredundant function in retinal development and homeostasis. Important work in this area has been done by others using large screening efforts and forward genetics protocols (7–15). In this project, we made use of automated genetic mapping combined with screening by optical coherence tomography (OCT) to assemble a robust forward genetics pipeline targeting genes essential to the development and maintenance of retinal structure. Germ line mutations induced in C57BL/6J (B6J) mice using N-ethyl-N-nitrosourea (ENU) were bred to homozygosity on the same background and identified in G1 founders using whole-exome sequencing. The zygosity of each mutation in G2 dams and in all G3 mice of each pedigree was determined before phenotypic screening. As distinct from other mutagenesis/mapping protocols, our approach permits instantaneous identification of causative mutations as soon as screening is completed, allows assessment of multiple alleles at loci under consideration, identifies both noncausative and causative mutations, and tabulates saturation of the genome. Thus, we were able to find causative mutations in real time as described previously (1) and also to determine the fraction of genes severely damaged or destroyed (16). Mutations with strong statistical evidence of causation can then be retargeted in the germ line using CRISPR/Cas9 for verification. Causative alleles of genes not previously known to be required for retinal development or homeostasis, as well as alleles of genes with known retinal functions, were encountered in the course of this effort.
Here we describe in detail mutations in Sfxn3 encoding sideroflexin-3, a putative mitochondrial serine transporter recently reported to function in one-carbon metabolism (17). We demonstrate that Sfxn3 is necessary for retinal homeostasis, particularly for the maintenance of synapses.
Results
Selection of Reliable Retinal Phenotype Screening Tools.
Screening thousands of mice for phenotype-genotype associations on a continuous basis for a prolonged period requires efficient and reproducible assays/techniques. With this goal in mind, we tested the reproducibility of OCT measurements in SOD1/DJ-1/Parkin triple-knockout (TKO) mice, an established model for retinal degeneration (18, 19), compared with B6J control mice over several days. Mice were imaged and OCT measurements done in a masked manner. Three OCT measurements were taken from each eye over a 1-wk span (days 0, 3, and 6). We observed excellent day-to-day reproducibility in the OCT parameters; SI Appendix, Fig. S1A shows the ratio of Bruch’s membrane (BM) to internal limiting membrane (ILM) thickness. The average SD was ±0.9 μ, and the coefficient of variation (CV) was 0.42%. When the data were unmasked and separated by genotype (SI Appendix, Fig. S1B), we observed a significant difference between the two genotypes that was consistent from day to day. Finally, the same result was obtained when left eyes were analyzed instead of the right eyes (SI Appendix, Fig. S1B, last pair of bars).
In our previous work, similar experiments using electroretinography (ERG) revealed a much greater variability in the measurements (18). The average %CV for scotopic Ganzfeld ERG when analyzing three separate experiments using B6J mice was 34% for the a-wave and 22% for the b-wave when using the low-intensity flash (0.1 log cd.s.m−2) and 22% for the a-wave and 17% for the b-wave when using the high-intensity flash (3.1 log cd.s.m−2). Because the CV was 40- to 80-fold higher for ERG compared with OCT, we chose not to use ERG as a screening tool.
We also attempted to use several categorical parameters (discrete ordinal variables), including normal/abnormal retinal vasculature, presence/absence of chorioretinal scars, and presence/absence of retinal detachment. Screening using these parameters failed to reveal significant linkage with any genes.
Genes Identified by Gene-Phenotype Associations.
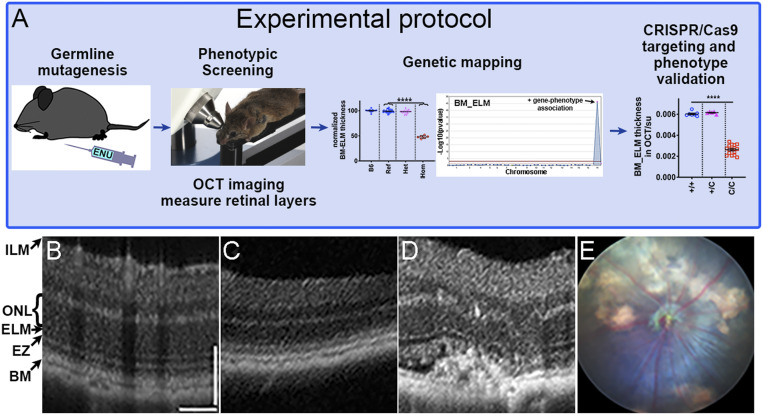
Having demonstrated that the OCT parameters can be used to easily, reliably, and reproducibly find small retinal differences between genotypes, we decided to focus on these to screen the ENU-mutagenized pedigrees for gene-phenotype associations using the protocol outlined in Fig. 1A. We examined 5,700 G3 mice carrying 11,305 coding/splicing mutations in 7,698 genes; we estimate that 4,326 of these genes carried likely damaging mutations (as judged by PolyPhen-2) (20) or likely null mutations (premature stop codons or critical splice site mutations). Genome saturation with mutations predicted to cause phenotypically detectable protein damage (16) and tested two times or more in the homozygous state was estimated at 4.9%. As a validation of our approach, we identified 11 genes (SI Appendix, Table S1) that were previously known to cause retinal degeneration when mutated [Slc6a6, Impdh1, Crx, Cngb1, Tmem135 (21), and Cacna2d4], to be important for retinal development or function (Epb41l2, Arntl, Plxnb1, and Llgl1), or to be associated with the development of subretinal neovascularization (Vldlr) (22). Examples of the OCT images obtained from a B6J control mouse (Fig. 1B), a mouse with a Cngb1 mutation (Fig. 1C), and a mouse with a Vldlr mutation (Fig. 1D) are shown for a side-by-side comparison. Fig. 1E presents a fundus photo of a mouse with the Vldlr mutation.
Fig. 1.
Genetic screening protocol and examples of OCT and fundoscopy images from mice with mutations identified in the forward genetic screen. (A) Schematic of the forward genetic retinal phenotype screen including ENU mutagenesis, screening of G3 mice, genetic mapping, and confirmation of gene-phenotype associations by CRISPR/Cas9 targeting. (C/C are mice homozygous for the CRISPR/Cas9 mutation.) (B–D) Representative OCT images for a B6J mouse showing normal retinal structure (B), a mouse with a Cngb1 homozygous mutation showing significant thinning of outer retina (C), and a mouse with a Vldlr homozygous mutation with neovascularization and distorted outer retina (D). (E) Corresponding fundus photo of the same Vldlr homozygous mutant mouse shown in D. BM, Bruch’s membrane; EZ, ellipsoid zone; ELM, external limiting membrane; ONL, outer nuclear layer; ILM, internal limiting membrane.
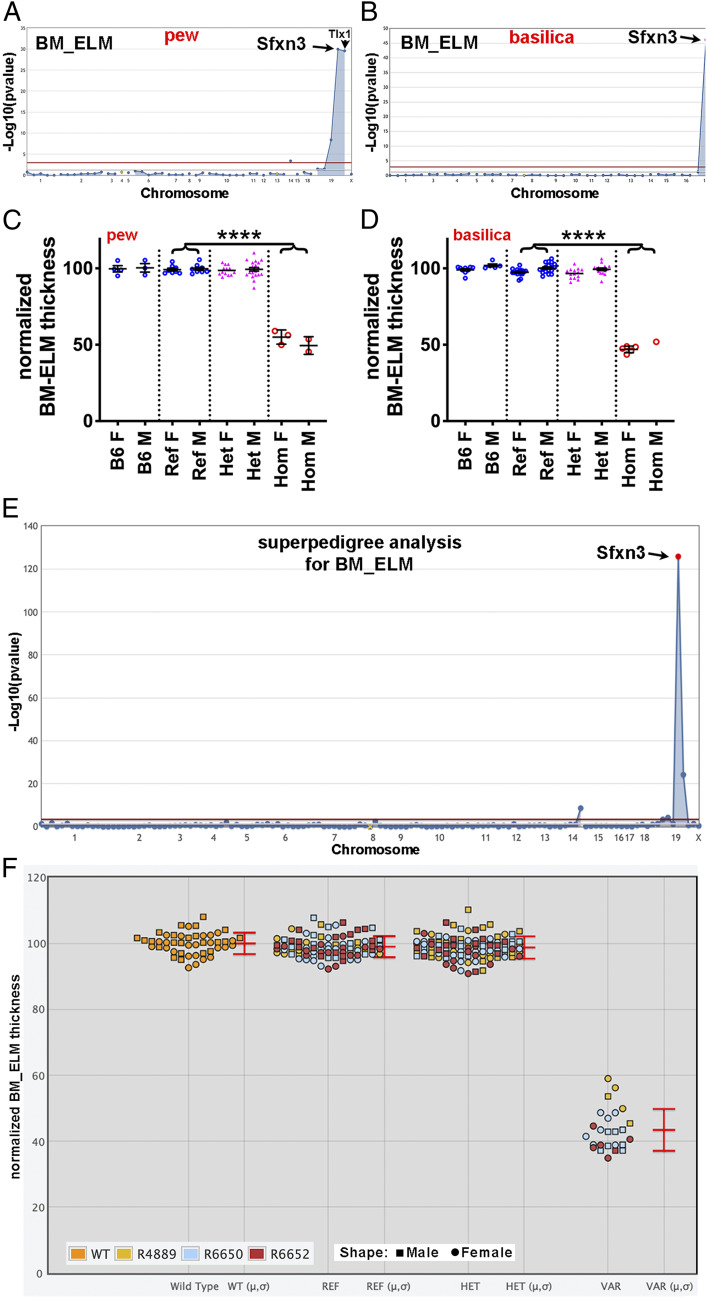
Interestingly, some genes were identified in pedigrees with a single homozygous mouse for the given mutation (e.g., SI Appendix, Table S1, rows 10 and 11). We also identified putative causative mutations in multiple candidate genes not previously associated with retinal diseases. Among these were two mutations in Sfxn3 associated with phenotypes termed pew and basilica. The basilica mutation is a G-to-A transition affecting the Sfxn3 exon 3 splice donor site (chromosome 19: 45,049,915 bp) and is likely a null mutation. The pew mutation is a T-to-C transition resulting in a phenylalanine-to-serine substitution at amino acid 78 (F78S) of the Sfxn3 protein (chromosome 19: 45,049,815 bp). At least three isoforms of Sfxn3 are produced by differential splicing, resulting in proteins of 321, 288, and 281 aa; all three isoforms are affected by the F78S mutation. Based on similarity with Sfxn1 (88% similarity), the longest Sfxn3 isoform is predicted to contain a 101-aa N-terminal tail that extends into the mitochondrial matrix, followed by five transmembrane domains inserted in the inner mitochondrial membrane and a 35-aa C-terminal tail in the intermembrane space. The F78S mutation affects the N-terminal tail and is immediately adjacent to the highly conserved His-Pro-Asp-Thr (HPDT) motif, which is present in all five mammalian Sfxn family proteins. Given the dissimilar properties of F vs. S residues (e.g., hydrophobicity, size, polarity), it is likely that the mutation alters local protein structure, including that of the HPDT motif, with ensuing functional impairment. Both the pew and basilica mutations led to highly significant gene-phenotype associations for all three OCT parameters (Fig. 2 A–D and SI Appendix, Fig. S2). Superpedigree analysis for the “Bruch’s membrane to external limiting membrane” (BM_ELM) parameter included roughly equal numbers of males and females (Fig. 2F) and showed a P value of 1.4 × 10−126 (Fig. 2E).
Fig. 2.
Decreased outer retinal thickness on in vivo OCT screening in mice from two different pedigrees with two different mutations in Sfxn3. (A and B) Manhattan plots showing P values calculated using a recessive transmission model are shown for two Sfxn3 mutations, pew (A) and basilica (B). The −log10 P values (y-axis) are plotted vs. the chromosomal positions of the mutations (x-axis) identified in the G1 founders of each pedigree. The horizontal red line represents a threshold of P = 0.05 with Bonferroni correction. Quantitative phenotypic data (continuous variable data) from OCT measurements of BM_ELM were used for linkage analysis and show highly significant associations with the two Sfxn3 mutations. The pedigree in which the pew mutation was identified (A) contained another mutation in linkage disequilibrium with Sfxn3 (Tlx1). However, in the second pedigree (B), the only mutation with a significant association with the OCT parameter was the basilica mutation. (C and D) Normalized measurements of outer retinal thickness (BM_ELM) on OCT images of mice from a pedigree carrying the pew mutation in Sfxn3 (C) show a 50% reduction in outer retinal thickness. A similar thinning of the outer retina is seen in mice carrying the basilica mutation in Sfxn3 (D). (E) A Manhattan plot of a superpedigree that includes three pedigrees (R4889, R6650, and R6652) containing the pew or basilica allele confirms a highly significant association (P = 1.4 × 10−126) of the Sfxn3 mutations and reduced outer retinal thickness. (F) The corresponding scatterplot shows a significant reduction in BM_ELM thickness in VAR (homozygous mutant) mice. Data points represent individual mice. ****P < 0.0001, Student’s t test. B6, wild-type C57BL/6J; Ref, G3 mice homozygous for the reference allele of Sfxn3; Het, G3 mice heterozygous for the mutant and reference alleles of Sfxn3; Hom, G3 mice homozygous for the mutant allele of Sfxn3.
Two CRISPR/Cas9-induced mutations were generated for Sfxn3, designated Sfxn3-1i (1-bp insertion) and Sfxn3-4d (4-bp deletion) (SI Appendix, Fig. S3), and separate mouse lines were bred. Because no phenotypic differences between the two alleles were observed, heterozygotes and homozygotes for either allele were analyzed together and denoted by the genotypes Sfxn3+/− and Sfxn3−/−, respectively. Fundus photographs revealed vascular attenuation and RPE mottling in Sfxn3−/− mice compared with Sfxn3+/+ littermates, which increased with age (SI Appendix, Fig. S4). We did not observe differences between female and male mutant mice (SI Appendix, Fig. S4G vs. Fig. S4D), or between the two CRISPR lines (SI Appendix, Fig. S4H vs. Fig. S4E).
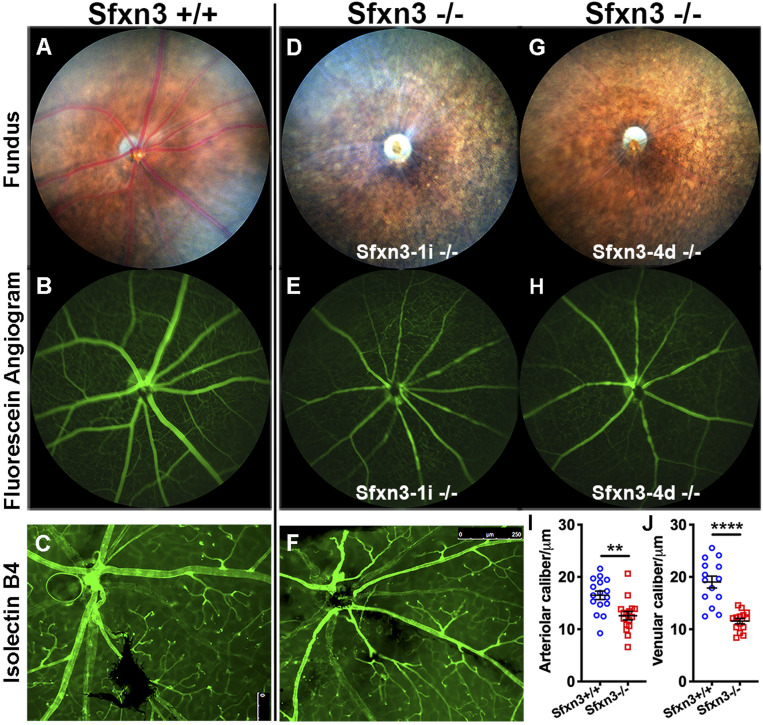
To further characterize these findings, we studied 10- to 12-mo-old mice (Fig. 3). By this age, the vascular attenuation and mottling of the RPE were striking in Sfxn3−/− mice (Fig. 3 D and G vs. Fig. 3A). Fluorescein angiography (Fig. 3 E and H vs. Fig. 3B) and isolectin B4-stained retinal flat mounts (Fig. 3F vs. Fig. 3C) confirmed the vascular attenuation. The difference in caliber was statistically significant both for arterioles (Fig. 3I) and venules (Fig. 3J). We did not observe any significant areas of nonperfusion or significant differences in the branching pattern of the vasculature of mutant vs. control mice. To determine whether the health of the RPE was affected, we prepared RPE flat mounts and stained them with an antibody to ZO-1, a component of the RPE tight junctions (SI Appendix, Fig. S5). Despite the absence of acute stimuli or injury, ZO-1 staining revealed a disruption of RPE morphology with loss of the normal hexagonal shape of RPE cells in Sfxn3−/− mice (SI Appendix, Fig. S5 C–F) compared with controls (SI Appendix, Fig. S5 A and B). This is a well-known sign of RPE pathology.
Fig. 3.
Sfxn3−/− mice show abnormal fundus and vasculature on fundoscopy, fluorescent angiography, and retinal flat mount staining. Sfxn3−/− mice (D–H) show a marked decrease in vessel caliber compared with controls (A–C). The difference is striking on fundus examination (D and G vs. A), in fluorescein angiography (E and H vs. B), and in isolectin B4-stained retinal flat mounts (F vs. C). The difference in caliber is statistically significant when comparing either arterioles (I) or venules (J) measured on the fluorescein angiography images. We did not observe any significant areas of nonperfusion in the Sfxn3−/− eyes. Mice were age 10 to 12 mo (n = 3 mice; 30 vessels/group). All retinal arterioles and venules were measured in three Sfxn3+/+ eyes and three Sfxn3−/− eyes. **P < 0.01, ****P < 0.0001, Student’s t test.
OCT Measurements Show Reduction of Retinal Thickness in Sfxn3−/− Mice.
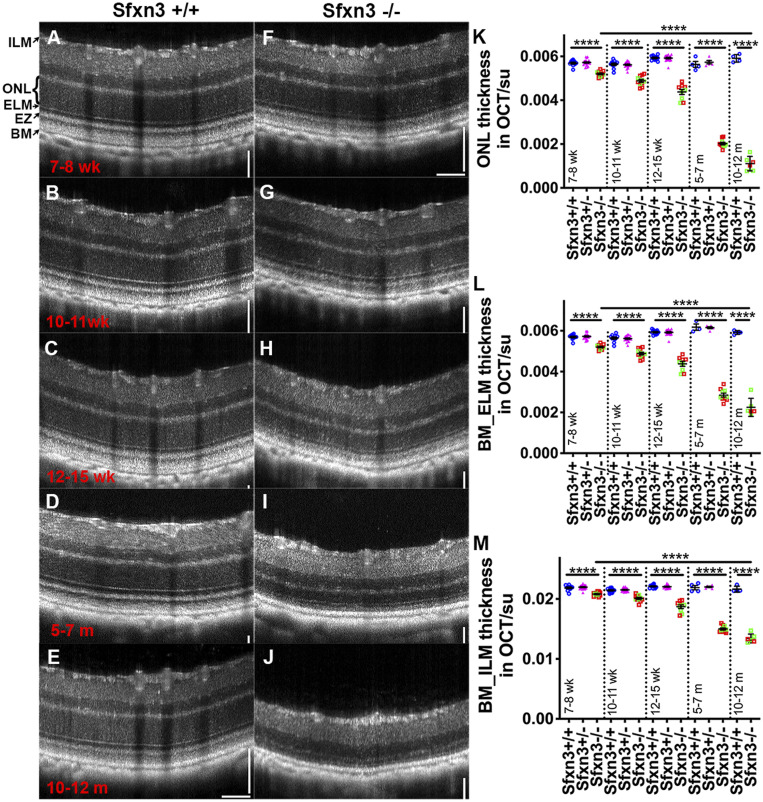
We analyzed retinal thickness in 7-wk to 12-mo-old Sfxn3−/−, Sfxn3+/−, and Sfxn3+/+ littermates using OCT (Fig. 4). We observed qualitative changes by 7 wk of age that worsened progressively with time. These changes included early hyperreflectivity of the photoreceptor outer segment band and hyporeflectivity of the interdigitation zone, accompanied by thinning of the outer retina (Fig. 4 A–J). These changes at this young age are similar to those that we previously described as the earliest OCT changes after light-induced retinal degeneration (23) and also in aging SOD1/DJ-1/Parkin TKO mice (18). By 7 wk of age, we found a small, but statistically significant reduction in all OCT measurements in the Sfxn3−/− mice compared with the Sfxn3+/+ and Sfxn3+/− mice (Fig. 4 K–M). These differences increased progressively as the mice aged and were highly significant. By 5 to 7 mo of age, the P values for the difference between Sfxn3+/+ and Sfxn3−/− were 1.7 × 10−12 (ONL [outer nuclear layer]), 2.1 × 10−9 (BM_ELM), and 1.3 × 10−10 (BM_ILM). We found no differences between Sfxn3+/+ and Sfxn3+/− eyes in OCT measurements. There was also no difference between the two CRISPR lines (Fig. 4 K–M; green squares, Sfxn3-1i−/−; red squares, Sfxn3-4d−/−). By age 10 to 12 mo, the retinas of Sfxn3−/− mice had degenerated further, having sustained a 57% reduction in BM_ELM thickness and a 79% reduction in ONL thickness.
Fig. 4.
OCT images reveal qualitatively and quantitatively abnormal retinal structures in Sfxn3−/− mice. (A–J) OCT images of Sfxn3+/+ and Sfxn3−/− mice were obtained at different ages (7 to 8 wk, 10 to 11 wk, 12 to 15 wk, 5 to 7 mo, and 10 to 12 mo; n = 5 to 11/group/age) to document the progression of retinal deterioration. Hyperreflectivity of the photoreceptor outer segment layer and hyporeflectivity of the interdigitation zone in Sfxn3−/− retinas started as early as 7 to 8 wk of age. There is also progressive thinning of the outer retina. (K–M) For quantitative retinal layer analyses, the following OCT parameters were measured using ImageJ (su, standard units): ONL thickness (K), BM_ELM (L), and BM_ILM (M). Compared with Sfxn3+/+ (blue circles) and Sfxn3+/− (pink triangles) mice, the Sfxn3−/− mice (1-bp insertion in green squares and 4-bp deletion in red squares) demonstrate significant thinning of the retina starting at 7 to 8 wk that worsens progressively with age. There was no difference between the two CRISPR lines. Data points represent individual mice.****P < 0.0001, Student’s t test.
Electrophysiology Analyses of Sfxn3−/− Mice Using Ganzfeld ERG.
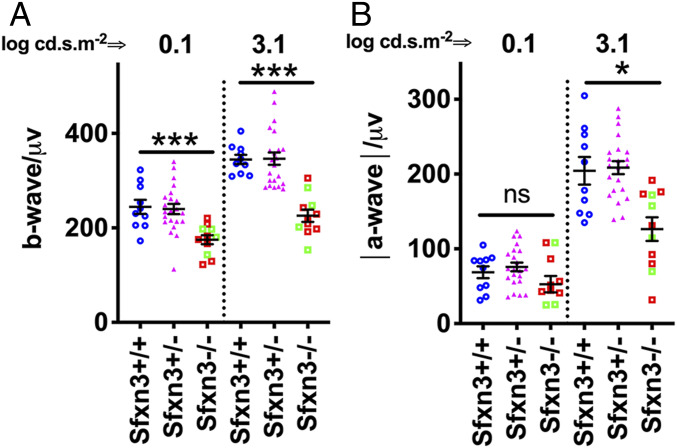
We then used full-field (Ganzfeld) scotopic ERG to assess whether the reduction in outer retinal thickness led to a loss in retinal function in Sfxn3−/− mice (Fig. 5 and SI Appendix, Fig. S6). A significant reduction in the b-wave signals of Sfxn3−/− retinas compared with Sfxn3+/+ controls was seen after stimulation with the 3.1 log cd.s.m−2 high-intensity light stimulus (P = 8 × 10−7) or with the 0.1 log cd.s.m−2 low-intensity stimulus (P = 0.0008) (Fig. 5A). A significant reduction was also observed in the a-wave signals of Sfxn3−/− retinas in response to the high stimulus flash (P = 0.004) (Fig. 5B). No differences were detected between the a- and b-wave ERG responses of Sfxn3+/+ and Sfxn3+/− retinas. We did not observe a difference between the two CRISPR lines (Fig. 5; green squares, Sfxn3-1i−/−; red squares, Sfxn3-4d−/−).
Fig. 5.
Sfxn3−/− mice have decreased retinal function compared with Sfxn3+/− and Sfxn3+/+ controls. Scotopic Ganzfeld ERG b-wave (A) and a-wave (B) amplitudes were measured in response to low (0.1 log cd.s.m−2) and high (3.1 log cd.s.m−2) flash intensities in 3-mo-old Sfxn3−/−, Sfxn3+/−, and Sfxn3+/+ control mice. Sfxn3−/− mice showed a significant reduction in both the a- and b-wave amplitudes at high flash intensities compared with both Sfxn3+/− and Sfxn3+/+ controls. A significant reduction was also seen at low flash intensity for the b-wave. There was no difference between the two CRISPR lines (1-bp insertion in green squares and 4-bp deletion in red squares). Data points represent individual mice (n per group: Sfxn3+/+ = 10, Sfxn3+/− = 22, Sfxn3−/− = 10). *P < 0.05, ***P < 0.001, Student’s t test.
Expression of Sfxn3 and Other Homologs in the Mouse Retina.
Five Sfxn family members exist in mice and display varying levels of amino acid sequence similarity. Given the paucity of data regarding the expression of the different Sfxn homologs in the retina, and the lack of homolog-specific antibodies, we applied single-cell RNA sequencing (scRNA-seq) to retinal cells isolated from four wild-type B6J mice to identify the Sfxn transcripts expressed in retinal cells. We first identified unique cell-type markers for the clusters generated by the Seurat R package (v3.0.0) (24) in an unbiased fashion. We correlated our cell-type marker findings with those described by others (25–28) and found a high degree of consistency, confirming the identity of our clusters (SI Appendix, Fig. S7). Violin plots showing the normalized expression level of each Sfxn homolog in different cell types are presented in SI Appendix, Fig. S8. For visualization purposes, since the vast majority of photoreceptor cells did not express Sfxn transcripts, we removed rods and cones from the plots and show the resulting simplified violin plots in Fig. 6. A more detailed summary of the expression data is provided in SI Appendix, Table S2.
Fig. 6.
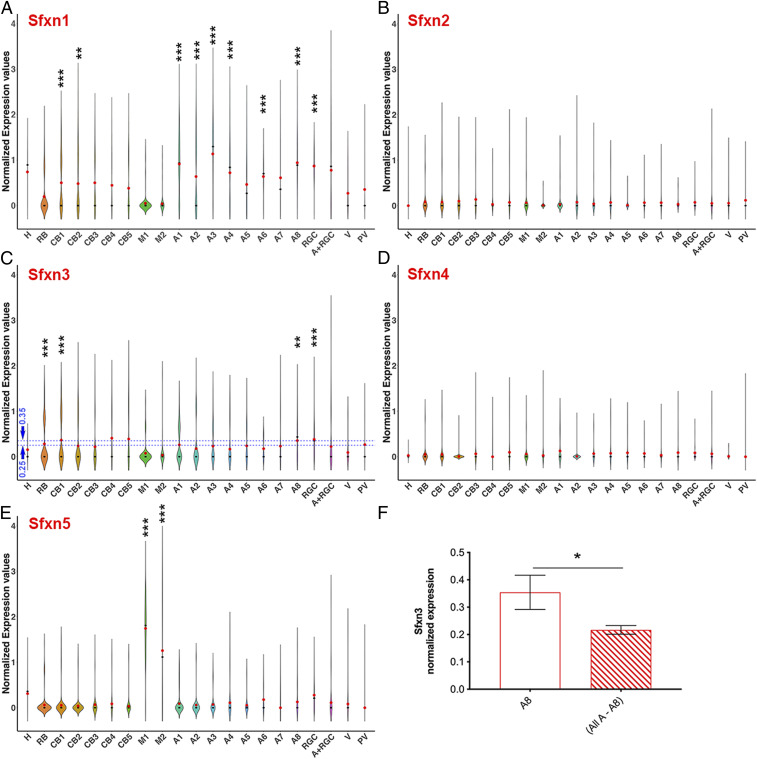
Analysis of the expression of Sfxn homologs in retinal cell types using scRNA-seq. Violin plots show normalized transcript expression values for Sfxn1 (A), Sfxn2 (B), Sfxn3 (C), Sfxn4 (D) and Sfxn5 (E) in different cell types. The average normalized expression of Sfxn3 in the amacrine-8 subtype is higher than in the combination of other amacrine cell subtypes (F). H, horizontal cells; RB, rod bipolar cells; CB, cone bipolar cells; A, amacrine cells; RGC, retinal ganglion cells; V, vascular cells; PV, perivascular cells; M, Müller cells. Differential gene expression analysis in Seurat was performed using the nonparametric Wilcoxon rank-sum test. *P < 0.05; **P < 0.01; ***P < 0.001.
A few general observations can be made. First, Sfxn2 and Sfxn4 transcripts were not detected at significant levels in the mouse retina (Fig. 6 B and D and SI Appendix, Table S2). Sfxn1 mRNA was expressed in multiple inner retina neuronal cell types, particularly retinal ganglion cells (RGCs), amacrine cells, cone bipolar cells, and horizontal cells, but not in photoreceptors or Müller cells (Fig. 6A and SI Appendix, Table S2). The expression of Sfxn3 mRNA varied; it was selectively expressed in several bipolar cell types, RGCs, and some amacrine cell types (Fig. 6C and SI Appendix, Table S2). Sfxn3 mRNA was not significantly expressed in Müller cells or photoreceptors (SI Appendix, Fig. S8 and Table S2). Sfxn5 mRNA was particularly highly expressed in Müller cells and RGCs (Fig. 6E and SI Appendix, Table S2). It should be noted that a small percentage of photoreceptor cells did express some level of these molecules; for Sfxn3, it was only 4% of rods and 10% of cones.
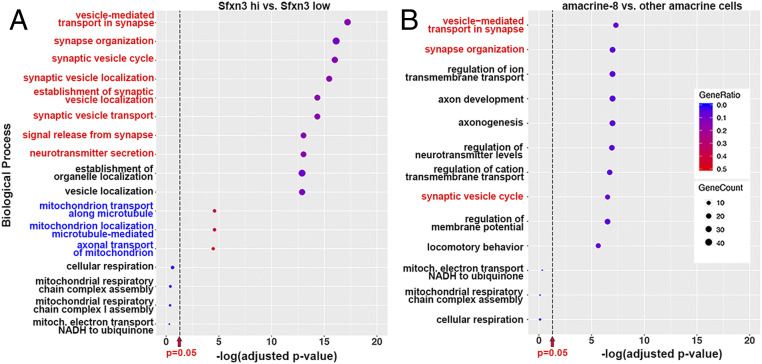
To determine which biological pathways may be affected by Sfxn3, we analyzed the scRNA-seq data in three ways. First, we used Gene Ontology (GO) analysis to identify enriched gene sets in all cells with a normalized expression of Sfxn3 >0.35 (“high” expression) vs. cells with a normalized expression <0.25 (“low” expression) (Fig. 7A). We chose these values based on the violin plots (Fig. 6C), because they allowed for a clear distinction between groups. We found that high–Sfxn3-expressing cells were enriched in the expression of genes involved in synaptic function, organization, and localization (top eight statistically significant pathways, red; Fig. 7A). Since Sfxn3 is thought to be a mitochondrial protein, we looked specifically for pathways related to mitochondria. We found that pathways related to mitochondrial transport, but not those related to mitochondrial respiration, were significantly associated with high Sfxn3 expression (Fig. 7A, blue).
Fig. 7.
GO pathway enrichment analysis of the scRNA-seq data identifies a relationship between Sfxn3 expression and synaptic pathways. (A) GO analysis comparing all cells expressing Sfxn3 at a normalized expression value >0.35 (“high” expression) vs. cells with a normalized expression <0.25 (“low” expression). The bubble plot shows the top 10 pathways with gene set enrichment in high-expressing cells vs. low-expressing cells [red, most related to synaptic processes, highly significant, −log(adjusted P value) > 10], followed by the top three pathways related to mitochondria [blue, significant, −log(adjusted P value) = 4], followed by four pathways related to mitochondrial respiration (black, not significant). (B) A similar analysis of the top 10 pathways with gene set enrichment in amacrine-8 cells vs. other amacrine cell subtypes (three related to synaptic processes, red), followed by three pathways related to mitochondrial respiration (not significant). GeneRatio, represented by the color scale, is the ratio of the number of genes in the dataset to the total number of genes in a pathway. GeneCount, represented by the bubble size, is the actual number of genes in the dataset found in a pathway.
We next performed a similar GO pathway enrichment analysis comparing amacrine-8 (A8) cells to the rest of the amacrine cells (Fig. 7B). Although A8 cells were not the highest-expressing cell type for Sfxn3, and although they might not be mainly responsible for the phenotypes we observed, we examined them in relation to other amacrine cells because they had higher Sfxn3 transcript expression than the other seven amacrine cell subtypes (Fig. 6F). We found that three of the top 10 pathways distinguishing A8 cells from the rest of the amacrine cells were related to synaptic function and organization (Fig. 7B, red).
We then applied Ingenuity Pathway Analysis software to our data, which indicated up-regulation of the synaptogenesis signaling pathway in high–Sfxn3-expressing cells relative to low-expressing cells (P = 1.6 × 10−18, z-score = 5.1). These data suggest that Sfxn3 may play a role in retinal synapse function, organization, and/or maintenance.
Finally, via comparisons with published analyses of gene expression in human organoids (28) and mouse retina (25–27), we classified our eight amacrine clusters into GABAergic (clusters A1, A3, and A8), glycinergic (clusters A2, A4, A5, and A7) or Starburst (cluster A6) amacrine subtypes (SI Appendix, Fig. S9). We also classified our cone bipolar clusters (SI Appendix, Fig. S10) into ON bipolars (clusters CB1 and CB3) and OFF bipolars (clusters CB2, CB4, and CB5). Of interest, GABAergic clusters (including the A8 cluster discussed above) tended to have a higher expression of the ribbon synapse marker Ctbp2 and also higher expression of Sfxn3 (SI Appendix, Fig. S9). In contrast, there was no segregation of Sfxn3-expressing cone bipolar cells into ON or OFF subtypes (SI Appendix, Fig. S10).
In Situ Hybridization Corroborates Location of Sfxn3 Expression in the Retina.
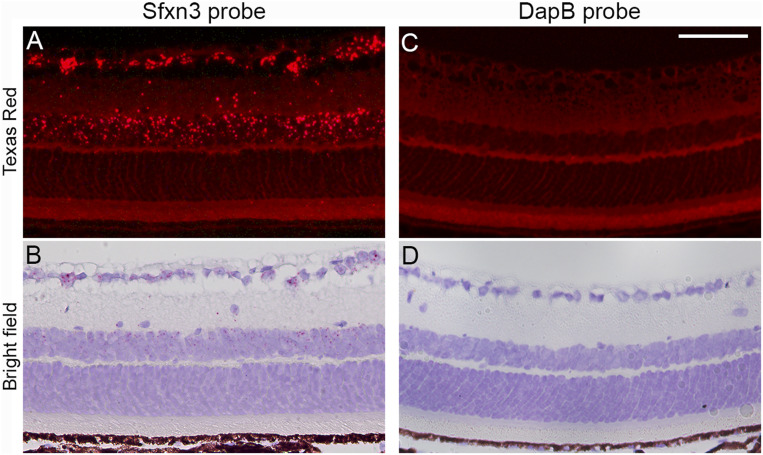
Since antibodies to Sfxn3 have a high degree of cross-reactivity with other Sfxn homologs that are also expressed in the retina, we decided to use in situ hybridization (ISH) to corroborate our scRNA-seq findings. A probe for RNAscope ISH (SI Appendix, Materials and Methods) targeting multiple short sequences within the Sfxn3 transcript was designed with the goal of specifically recognizing Sfxn3 transcripts (vs. other Sfxn homologs). Based on the scRNA-seq data, we expected to find Sfxn3 labeling mainly in the cell bodies of bipolar, amacrine, and RGCs. Our RNAscope ISH results showed exactly that pattern of labeling in the ganglion cells and inner nuclear layers (Fig. 8). Importantly, there were very few fluorescent spots in both the ONL and the RPE.
Fig. 8.
In situ hybridization of Sfxn3 in the retina of B6J mice. Representative images of retinal sections from B6J mice showing ISH of an Sfxn3 probe (A and B) or a DapB probe (C and D). DapB is a bacterial gene serving as a negative control. Strong signals for Sfxn3 are seen mainly in the RGC layer and inner nuclear layer. Few Sfxn3-positive signals are seen in the ONL or RPE. The assay labels with both fluorescent signals (Fast Red imaged with Texas Red filters) and also chromogenic red staining (bright field imaging). The bright field images show better the retinal layers and RPE. (Scale bar: 50 μm.)
Histological and Ultrastructural Analyses Show Degeneration of Outer Retinal Layers in Sfxn3−/− Mice.
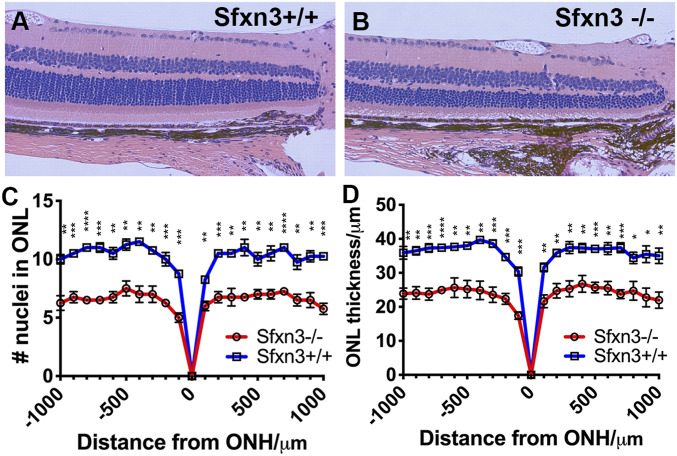
To further analyze the anatomic changes caused by Sfxn3 deficiency, we prepared histological sections from eyes collected from Sfxn3−/− and Sfxn3+/+ mice at age 3 to 4 mo. We found that both the number of nuclei in the ONL and the thickness of the ONL were significantly decreased in Sfxn3−/− mice (Fig. 9), and this was seen across the span of the retina (out to 1,000 µm on either side of the optic nerve head). Furthermore, thinning of the photoreceptor outer segments was also documented in Sfxn3−/− mice. The inner retinal structure appeared to be preserved. The general layered appearance of the retina was also preserved; there were no undulations or focal disruptions of the layers as are seen in some retinal dystrophies, such as that of the rd8 mutant mice (29).
Fig. 9.
Histology shows thinning of the outer retinal layers in Sfxn3−/− mice. (A and B) Representative H&E images taken at 20× magnification of retinas from Sfxn3+/+ (A) and Sfxn3−/− (B) mice are shown. (C and D) The number of cells in the ONL (C) and the thickness of the ONL (D) measured on histology sections at 100 μm intervals starting from the optic nerve head (ONH). Sfxn3−/− mice had a significantly thinner outer retina in comparison to Sfxn3+/+ controls based on both the nuclear count (C) and ONL thickness measurements (D). (n per group; Sfxn3+/+ = 10, Sfxn3+/− = 22, Sfxn3−/− = 10). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Student’s t test.
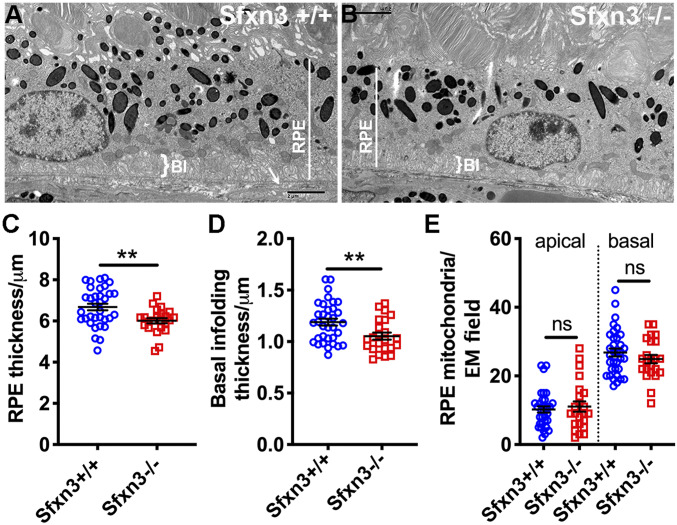
We used electron microscopy (EM) to examine ultrastructural changes in Sfxn3−/− mice (Figs. 10 and 11 and SI Appendix, Figs. S11 and S12). In particular, we focused on the RPE and outer retina and found several anomalies in the RPE of Sfxn3-deficient mice (Fig. 10B vs. Fig. 10A), including a decrease in average RPE thickness (Fig. 10C; P = 0.0039) and a decrease in average basal infolding thickness (Fig. 10D; P = 0.0077). Interestingly, we did not find a difference in the number of either basal or apical mitochondria in the RPE (Fig. 10E). Another ultrastructural anomaly that we found was a disruption in the ELM (SI Appendix, Fig. S11), as has been reported in other models of retinal dystrophy/degeneration (30, 31); loss or disorganization of the electron-dense adherens junctions was seen in Sfxn3−/− eyes.
Fig. 10.
Thinning of the RPE in Sfxn3−/− mice. (A and B) Representative transmission electron microscopy (TEM) images show the reduction of RPE (white line) and basal infolding (BI, white bracket) thickness in the Sfxn3−/− (B) compared to the Sfxn3+/+ mouse (A). (C and D) RPE thickness (C) and basal infolding thickness (D) on TEM micrographs measured using ImageJ. There is a significant thinning of these layers in Sfxn3−/− mice compared to Sfxn3+/+ mice. (E) Number of mitochondria present at either the apical or the basal aspects of the RPE cells (n per group; Sfxn3+/+ = 4 eyes [34 TEM fields], Sfxn3−/− = 3 eyes [23 TEM fields]). Data points represent individual EM fields. ns, not significant; *P < 0.05; **P < 0.01, Student’s t test.
Fig. 11.
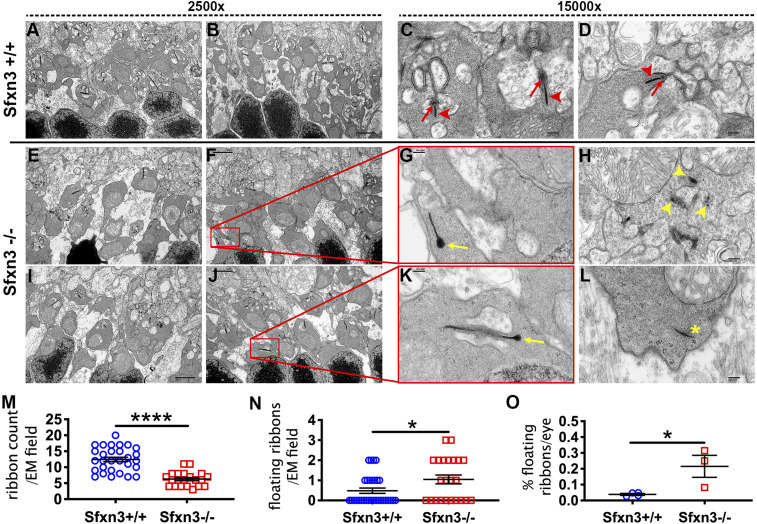
Electron microscopy analysis of ribbon synapses in the outer plexiform layer shows abnormal synapses in Sfxn3−/− eyes. (A–L) Representative TEM images. Eyes from Sfxn3+/+ mice (A and B) have a higher number of synaptic ribbons than Sfxn3−/− mice (E, F, I, and J). Higher magnification images show the normal appearance of synaptic ribbons (arrowheads) including the arciform density (arrows) in Sfxn3+/+ controls (C and D). In contrast, many synaptic ribbons in Sfxn3−/− eyes show abnormalities, including club-shaped ribbons (yellow arrows in G and K), degenerating ribbons (yellow arrowheads in H show pieces of a degenerating ribbon), and floating ribbons (yellow asterisk in L). (M–O) Standardized quantitative analysis of the 2,500× images show decreased numbers of normal synaptic ribbons (M) and increased numbers of abnormal synaptic ribbons (N) per EM field in Sfxn3−/− eyes. (O) There is also an increased percentage of abnormal floating ribbon synapses in Sfxn3−/− eyes. Data points represent individual EM fields (n per group; Sfxn3+/+ = 4 eyes [29 EM fields], Sfxn3−/− = 3 eyes [21 EM fields]). *P < 0.05; ****P < 0.0001, Student’s t test.
Given the synaptic gene set enrichment in high–Sfxn3-expressing retinal cells identified by GO analysis, we analyzed synapses in EM images of the outer plexiform layer of Sfxn3−/− mice. In particular, we examined the morphology of the ribbon synapses and quantified synaptic ribbons, which are essential to the functioning of the photoreceptor-to-inner nuclear layer cell synapses (32, 33). Interestingly, we found a reduction in synaptic ribbons per EM field in Sfxn3−/− eyes (Fig. 11 E, F, I, and J) compared with Sfxn3+/+ eyes (Fig. 11 A and B), which was highly significant (Fig. 11M). Most Sfxn3+/+ ribbon synapses had a normal morphology, including the “horseshoe” cap connection to the synapsis (red arrows, Fig. 11 C and D) and multiple horizontal cells and bipolar cells attaching directly or indirectly to the synapse (Fig. 11 C and D). In contrast, many Sfxn3−/− retinas had synaptic ribbons with morphological signs of damage as seen in other models of retinal degeneration (34–38), including club ribbons (yellow arrows, Fig. 11 G and K) and degenerating ribbons (yellow arrowhead, Fig. 11H). We also found a statistically significant increase in the number of floating ribbons per EM field (abnormal ribbons that are not forming a synapse; Fig. 11 L and N) in Sfxn3−/− eyes. The ratio of floating ribbons to total ribbons per eye (% floating ribbons) was significantly increased in Sfxn3−/− eyes relative to Sfxn3+/+ eyes (Fig. 11O). Finally, quantitative analysis of the EM images (39) demonstrated a statistically significant reduction in the number of cytoplasmic synaptic vesicles in the photoreceptor terminals of Sfxn3−/− mice (SI Appendix, Fig. S12). Taken together, these findings reveal a disruption of ribbon synapses in the outer retina of Sfxn3−/− mice.
Discussion
In this work, we identified two independent Sfxn3 loss-of-function mutations in a forward genetic screen for retinal defects observed using OCT. We demonstrated that Sfxn3 deficiency causes significant thinning of the retina, especially affecting the outer layers, as documented by both OCT and histology. Retinal function as measured by ERG was also decreased. Importantly, mouse and human Sfxn3 proteins are 94% identical in amino acid sequence, suggesting that findings from the mouse will be relevant to humans.
Sfxn gene expression has been detected in the brain (Sfxn1, 3, 4, and 5) and retina (Sfxn1 and 3) of Xenopus embryos by ISH (40). Sfxn3 and Sfxn5 are abundant in the mouse brain (41–43). Rivell et al. (41) reported that brain expression of Sfxn3 in rats is low at birth but increases early in life and remains at a high level in mature brains. The expression pattern of Sfxn3 in the mouse and human retina is not known, however. Using scRNA-seq (26, 27, 44, 45) of wild-type B6J retinal tissue, we detected moderate Sfxn3 mRNA expression in several neuronal retinal cell types, including rod bipolar cells, cone bipolar cells, RGCs, and amacrine cell subtypes. This pattern overlapped with Sfxn1 expression in some cone bipolar cells, RGCs, and amacrine cells. Sfxn2, Sfxn4, and Sfxn5 were minimally expressed or absent from all retinal cell types with Sfxn3 expression.
The overlapping tissue expression patterns of Sfxn homologs observed in Xenopus embryos (40) and the detection of coexpression of Sfxn3 and Sfxn1 transcripts in several retinal cell types are suggestive of functional redundancy among Sfxn family members. Indeed, Sfxn3 can partially compensate for the loss of mitochondrial serine transport in Jurkat cells caused by a deficiency of Sfxn1, with which Sfxn3 is 77% identical in mice (46). It is possible that Sfxn1 also compensates for Sfxn3 deficiency in the retina, which may explain why we did not see inner retinal degeneration given the significant level of Sfxn3 expression in RGCs. Whether Sfxn1 can functionally substitute for Sfxn3 in RGCs, bipolar cells, or amacrine cells remains to be tested. In light of the probable cross-reactivity of commercially available Sfxn3 antibodies with Sfxn1, mice with endogenous expression of HA-tagged Sfxn3 will be useful for better localizing the expression of Sfxn3 and exploring its molecular interactions. Further studies should also test the effect of light exposure on the progression of retinal degeneration in Sfxn3−/− mice.
An important issue concerns the molecular functions of Sfxn3 in the retina. Current understanding of Sfxn protein function is limited. The five known mammalian Sfxn proteins are reportedly localized to mitochondria (41–43, 46–48), consistent with our finding of mitochondrial transport gene set enrichment in high– Sfxn3-expressing retinal cells. Sfxn1 and Sfxn3 have been implicated in mitochondrial serine transport (46), and Amorim et al. (42) reported that “loss of SFXN3 did not disturb mitochondrial electron transport chain function in mouse synapses” and concluded that “its function in mitochondria is likely to be independent of canonical bioenergetic pathways.” No obvious ocular abnormalities were detected in Drosophila with transgenic overexpression of the fly Sfxn3 ortholog, despite abnormal neuromuscular synaptic development in those animals (42). Based on our scRNA-seq data, we hypothesize that Sfxn3 deficiency in the retina may compromise the transport of mitochondria or their function in the postsynaptic cell, resulting in abnormal synaptic formation.
Paradoxically, the retinal degenerative changes in Sfxn3−/− mice seemed to particularly affect photoreceptors, despite the absence of Sfxn3 expression in rods and cones. We did observe Sfxn3 expression, sometimes at high levels, in a small percentage of rod and cone cells (approximately 4% of rods and 10% of cones); however, the low proportion of cells showing this expression does not seem to match the diffuse damage observed. An alternative explanation may involve the impact of a dysfunctional synapse on photoreceptor health. Rapid photoreceptor degeneration has been documented in mice with abnormal photoreceptor synapses (49, 50). Considering previous reports of Sfxn3 synaptic localization in brain neurons (41) and neuromuscular synapses (42), synaptic gene set enrichment in high–Sfxn3-expressing retinal cells (Fig. 7A), and the increased occurrence of abnormal synapses in the outer plexiform layer of Sfxn3−/− mice (Fig. 10), we speculate that Sfxn3 in the postsynaptic terminal (inner nuclear cells, such as bipolar and horizontal cells) may be needed for normal synaptic formation/stability/function, without which photoreceptors may degenerate. The widespread degeneration of photoreceptors may in turn cause RPE pathology.
Finally, we have demonstrated the utility of our forward genetics pipeline in identifying novel gene-phenotype associations important to the retina. The identification of 11 genes known to be associated with retinal homeostasis or pathogenesis indicates that our screening tool (OCT) is effective. Others have documented that OCT is a clinically relevant method for monitoring retinal degeneration in mice (51). Moreover, by screening large pedigrees (50 mice on average), we have been able to identify mild phenotypes (e.g., a 12% reduction in BM_ELM thickness) and detect linkages to genes relevant to retinal physiology, such as Plxnb1 (52, 53). We intend to investigate other genes identified in our screen that have not previously been linked to retinal development or homeostasis. Improving our understanding of how the retina develops and functions will ultimately aid the development of retinal regeneration strategies and novel treatment strategies for a variety of multifactorial retinal diseases.
Materials and Methods
Animals.
We followed all international, national, and institutional guidelines for the care and use of animals, including the NIH’s Guide for the Care and Use of Laboratory Animals and the Association for Research in Vision and Ophthalmology’s Statement for the Use of Animals in Ophthalmic and Vision Research. The UT Southwestern Medical Center’s Institutional Animal Care and Use Committee approved all experiments. Sod1−/−Park7−/−Prkn−/− TKO mice on a B6J background, which have been described previously (18, 54), were used to validate the screening protocol. For the forward genetics studies, germ line mutations were randomly induced using ENU treatment of B6J male mice, followed by a special breeding protocol as detailed in SI Appendix, Materials and Methods.
Whole-Exome Sequencing and Determination of Candidate Genes.
Whole-exome sequencing and mapping were performed as described previously (1). In brief, exome-enriched DNA from G1 mice was sequenced using the Illumina HiSeq 2500 platform. G2 dams and G3 mice were genotyped across coding mutations specific for their pedigrees using Ion Torrent AmpliSeq custom primer panels. To correlate phenotype with the genotyping results, three OCT parameters (BM_ELM, BM_ILM, and ONL) were used as variables in linkage analysis. Linkage analysis was performed as detailed in SI Appendix, Materials and Methods.
Generation of Sfxn3 Null Mice Using CRISPR/Cas9 Targeting.
CRISPR/Cas9 technology was used to generate KO mice targeting the identified candidate gene, as detailed in SI Appendix, Materials and Methods; the resulting mice were tested for phenotypes as in the original screen.
Image-Guided OCT and Retinal Layer Analyses.
After anesthesia was achieved, the pupils were dilated and GenTeal liquid gel (Novartis) was applied to the corneal surface. OCT images were obtained with image-guided tomography (Micron IV-OCT2; Phoenix Research Laboratories). These images were analyzed to obtain measurements of the thickness of multiple retinal layers at the target location using the Freehand tool in ImageJ (https://imagej.nih.gov/ij/) as described previously (19). Three locations 200 µm apart and in the center of the image were measured for three parameters: BM_ELM, ONL, and BM_ILM. Finally, the three measurements for each parameter in an image were averaged for statistical analysis.
ERG.
ERG responses of the CRISPR/Cas9-mutated lines were recorded in 3-mo-old dark-adapted homozygous (Sfxn3−/−), heterozygous (Sfxn3+/−), and control (Sfxn3+/+) mice, using a scotopic Ganzfeld ERG system (Phoenix Research Labs) as described previously (19), following protocols outlined by the manufacturer and detailed in SI Appendix, Materials and Methods.
Histology of Retinal Sections.
Right eyes of Sfxn3−/− and Sfxn3+/+ mice (n = 4 eyes per group) were enucleated and processed as described previously (23, 24) and as detailed in SI Appendix, Materials and Methods.
EM.
Left eyes of Sfxn3−/− and Sfxn3+/+ mice were collected and immediately fixed in 2% glutaraldehyde and 2% paraformaldehyde in 0.2 M cacodylate buffer. Fixed eyes were removed and sectioned behind the limbus, and the resulting posterior eye cups were processed as described previously (55) and as detailed in SI Appendix, Materials and Methods.
Processing of Retinal Samples into Single-Cell Suspensions.
Retinal single-cell suspensions were prepared using the Papain Dissociation System (Worthington Biochemical; catalog no. LK003150) according to the manufacturer’s instructions with minor modifications (56–58) and as detailed in SI Appendix, Materials and Methods.
Droplet-Based scRNA-Seq of Dissociated Retinal Cells.
Dissociated single retinal cells were processed through the GemCode Single-Cell Platform using the GemCode Gel Bead, Chip, and Library Kits (10x Genomics) according to the manufacturer’s protocol and with the help of the UT Southwestern Next-Generation Sequencing Core Facility. In brief, single cells were sorted into 0.4% BSA in Dulbecco’s PBS. Approximately 14,000 single retinal cells were added to a channel. The cells were then partitioned into Gel Beads in emulsion in the 10x Genomics Chromium Controller, followed by cell lysis and barcoded oligo-deoxythymine priming and reverse transcription of polyadenylated RNA. Finally, amplified cDNAs were sheared for adapter and sample index attachment. Libraries were sequenced on an Illumina NextSeq 500.
Analyses of Single-Cell Transcriptomes.
scRNA-seq data were analyzed (59, 60) in collaboration with the director of the UT Southwestern McDermott Center of Bioinformatics (C.X.) and a computational biologist (A.K.), as detailed in SI Appendix, Materials and Methods.
Statistical Analysis.
SigmaPlot 11.0 (Systat Software) was used for conventional statistical analysis, as detailed in SI Appendix, Materials and Methods.
Data Deposition and Availability.
The scRNA-seq dataset has been deposited in the Gene Expression Omnibus database (accession no. GSE147573). All other data are contained in the main text and SI Appendix.
Supplementary Material
Acknowledgments
Support was provided by NIH grants 1R01EY022652, U19AI100627, and R01AI125581; and National Eye Institute Visual Science Core Grant EY020799. Additional support was provided by an unrestricted grant from Research to Prevent Blindness, the Patricia and Col. William Massad Retina Research Fund, and the David M. Crowley Foundation.
Footnotes
The authors declare no competing interest.
Data deposition: The scRNA-seq dataset has been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE147573).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921224117/-/DCSupplemental.
References
- 1.Wang T. et al., Real-time resolution of point mutations that cause phenovariance in mice. Proc. Natl. Acad. Sci. U.S.A. 112, E440–E449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull K. R. et al., Unlocking the bottleneck in forward genetics using whole-genome sequencing and identity by descent to isolate causative mutations. PLoS Genet. 9, e1003219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler B., Du X., Xia Y., Precis on forward genetics in mice. Nat. Immunol. 8, 659–664 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Wang Y. et al., Large-scale forward genetics screening identifies Trpa1 as a chemosensor for predator odor-evoked innate fear behaviors. Nat. Commun. 9, 2041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D. et al., Syndromic immune disorder caused by a viable hypomorphic allele of spliceosome component Snrnp40. Nat. Immunol. 20, 1322–1334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyoshi C. et al., Methodology and theoretical basis of forward genetic screening for sleep/wakefulness in mice. Proc. Natl. Acad. Sci. U.S.A. 116, 16062–16067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krebs M. P. et al., Mouse models of human ocular disease for translational research. PLoS One 12, e0183837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J. et al., An ENU mutagenesis screen in zebrafish for visual system mutants identifies a novel splice-acceptor site mutation in patched2 that results in Colobomas. Invest. Ophthalmol. Vis. Sci. 53, 8214–8221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanovice N. J., Daly C. M., Gross J. M., N-Ethylmaleimide-Sensitive factor b (nsfb) is required for normal pigmentation of the zebrafish retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 56, 7535–7544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vetrivel S. et al., Mutation in the mouse histone gene Hist2h3c1 leads to degeneration of the lens vesicle and severe microphthalmia. Exp. Eye Res. 188, 107632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jadeja S. et al., A CNS-specific hypomorphic Pdgfr-beta mutant model of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 54, 3569–3578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore B. A., et al. ; International Mouse Phenotyping Consortium , Identification of genes required for eye development by high-throughput screening of mouse knockouts. Commun. Biol. 1, 236 (2018). Correction in: Commun. Biol. 2, 97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charette J. R. et al., A mutagenesis-derived Lrp5 mouse mutant with abnormal retinal vasculature and low bone mineral density. Mol. Vis. 23, 140–148 (2017). [PMC free article] [PubMed] [Google Scholar]
- 14.Song D. et al., A murine RP1 missense mutation causes protein mislocalization and slowly progressive photoreceptor degeneration. Am. J. Pathol. 184, 2721–2729 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenwald S. H. et al., Mouse models of NMNAT1-leber congenital amaurosis (LCA9) recapitulate key features of the human disease. Am. J. Pathol. 186, 1925–1938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T. et al., Probability of phenotypically detectable protein damage by ENU-induced mutations in the Mutagenetix database. Nat. Commun. 9, 441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kory N. et al., SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 362, eaat9528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y. et al., Mice with a combined deficiency of superoxide dismutase 1 (Sod1), DJ-1 (Park7), and parkin (Prkn) develop spontaneous retinal degeneration with aging. Invest. Ophthalmol. Vis. Sci. 60, 3740–3751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B. et al., A mouse model of retinal recovery from photo-oxidative/photo-inflammatory injury: Nrf2, SOD1, DJ-1, and parkin are not essential to recovery. Invest. Ophthalmol. Vis. Sci. 60, 1165–1174 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Adzhubei I. A. et al., A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee W. H. et al., Mouse Tmem135 mutation reveals a mechanism involving mitochondrial dynamics that leads to age-dependent retinal pathologies. eLife 5, e19264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia C. H. et al., The role of Vldlr in intraretinal angiogenesis in mice. Invest. Ophthalmol. Vis. Sci. 52, 6572–6579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong X. et al., Fundus camera-delivered light-induced retinal degeneration in mice with the RPE65 Leu450Met variant is associated with oxidative stress and apoptosis. Invest. Ophthalmol. Vis. Sci. 57, 5558–5567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuart T. et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark B. S. et al., Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macosko E. Z. et al., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shekhar K. et al., Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowan C. S., et al. , Cell types of the human retina and its organoids at single-cell resolution: Developmental convergence, transcriptomic identity, and disease map. bioRxiv:10.1101/703348 (16 July 2019).
- 29.Aredo B. et al., Differences in the distribution, phenotype and gene expression of subretinal microglia/macrophages in C57BL/6N (Crb1 rd8/rd8) versus C57BL6/J (Crb1 wt/wt) mice. J. Neuroinflammation. 12, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehalow A. K. et al., CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum. Mol. Genet. 12, 2179–2189 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Stuck M. W., Conley S. M., Naash M. I., Defects in the outer limiting membrane are associated with rosette development in the Nrl−/− retina. PLoS One 7, e32484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitz F., The making of synaptic ribbons: How they are built and what they do. Neuroscientist 15, 611–624 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Schmitz F., Presynaptic [Ca(2+)] and GCAPs: Aspects on the structure and function of photoreceptor ribbon synapses. Front. Mol. Neurosci. 7, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J. et al., CNGA3 deficiency affects cone synaptic terminal structure and function and leads to secondary rod dysfunction and degeneration. Invest. Ophthalmol. Vis. Sci. 53, 1117–1129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller T. M. et al., A multiple piccolino-RIBEYE interaction supports plate-shaped synaptic ribbons in retinal neurons. J. Neurosci. 39, 2606–2619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J. et al., Early degeneration of photoreceptor synapse in Ccl2/Cx3cr1-deficient mice on Crb1(rd8) background. Synapse 67, 515–531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Won J. et al., NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum. Mol. Genet. 20, 482–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuchs M. et al., Rod photoreceptor ribbon synapses in DBA/2J mice show progressive age-related structural changes. PLoS One 7, e44645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaithianathan T., Henry D., Akmentin W., Matthews G., Functional roles of complexin in neurotransmitter release at ribbon synapses of mouse retinal bipolar neurons. J. Neurosci. 35, 4065–4070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X. et al., Developmental expression of sideroflexin family genes in Xenopus embryos. Dev. Dyn. 239, 2742–2747 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Rivell A., Petralia R. S., Wang Y. X., Mattson M. P., Yao P. J., Sideroflexin 3 is a mitochondrial protein enriched in neurons. Neuromolecular Med. 21, 314–321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amorim I. S. et al., Sideroflexin 3 is an α-synuclein-dependent mitochondrial protein that regulates synaptic morphology. J. Cell Sci. 130, 325–331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleming M. D., Campagna D. R., Haslett J. N., Trenor C. C. 3rd, Andrews N. C., A mutation in a mitochondrial transmembrane protein is responsible for the pleiotropic hematological and skeletal phenotype of flexed-tail (f/f) mice. Genes Dev. 15, 652–657 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng G. X. et al., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rheaume B. A., et al. , Single-cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun. 9, 2759 (2018). Correction in: Nat. Commun.9, 3203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshikumi Y. et al., Roles of CTPL/Sfxn3 and Sfxn family members in pancreatic islet. J. Cell. Biochem. 95, 1157–1168 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Hildick-Smith G. J. et al., Macrocytic anemia and mitochondriopathy resulting from a defect in sideroflexin 4. Am. J. Hum. Genet. 93, 906–914 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mon E. E. et al., Regulation of mitochondrial iron homeostasis by sideroflexin 2. J. Physiol. Sci. 69, 359–373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz F. et al., CSPalpha-deficiency causes massive and rapid photoreceptor degeneration. Proc. Natl. Acad. Sci. U.S.A. 103, 2926–2931 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grossman G. H., Pauer G. J., Narendra U., Peachey N. S., Hagstrom S. A., Early synaptic defects in tulp1−/− mice. Invest. Ophthalmol. Vis. Sci. 50, 3074–3083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Massengill M. T. et al., Clinically relevant outcome measures for the I307N rhodopsin mouse: A model of inducible autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 59, 5417–5430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swiercz J. M., Kuner R., Behrens J., Offermanns S., Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron 35, 51–63 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Bulloj A., Maminishkis A., Mizui M., Finnemann S. C., Semaphorin4D-PlexinB1 signaling attenuates photoreceptor outer segment phagocytosis by reducing Rac1 activity of RPE cells. Mol. Neurobiol. 55, 4320–4332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding Y., Aredo B., Zhong X., Zhao C. X., Ufret-Vincenty R. L., Increased susceptibility to fundus camera-delivered light-induced retinal degeneration in mice deficient in oxidative stress response proteins. Exp. Eye Res. 159, 58–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ufret-Vincenty R. L. et al., Transgenic mice expressing variants of complement factor H develop AMD-like retinal findings. Invest. Ophthalmol. Vis. Sci. 51, 5878–5887 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Barres B. A., Silverstein B. E., Corey D. P., Chun L. L., Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron 1, 791–803 (1988). [DOI] [PubMed] [Google Scholar]
- 57.Feodorova Y., Koch M., Bultman S., Michalakis S., Solovei I., Quick and reliable method for retina dissociation and separation of rod photoreceptor perikarya from adult mice. MethodsX 2, 39–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi H. J., Wang R., Jakobs T. C., Single-cell dissociation and characterization in the murine retina and optic nerve. Methods Mol. Biol. 1695, 311–334 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Chen Y. et al., ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 128, 1937–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z. et al., Dermal adipose tissue has high plasticity and undergoes reversible dedifferentiation in mice. J Clin. Invest. 129, 5327–5342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.