Significance
Solar UV generates abundant carcinogenic DNA lesions, and consequently, cells need to cope with these deleterious lesions to survive UV damage. This study used lesion-specific photolyases to isolate the biologically relevant effects of each major type of UV-induced DNA lesions (cyclobutane pyrimidine dimers [CPDs] and 6-4 photoproducts [6-4PPs]) on ATR activation, a crucial DNA damage response. A newly developed, flow cytometric single-cell analysis of UV-induced DNA lesions, thymidine analog incorporation, and DNA damage response enabled unprecedented determination of cellular events following UV irradiation. The striking finding is that only 6-4PPs, the shorter-lived and less abundant lesion type, cause replication blockage and activation of the ATR DNA damage response.
Keywords: CPD, 6-4PP, DNA replication, DNA damage response, Chk1
Abstract
The most prevalent human carcinogen is sunlight-associated ultraviolet (UV), a physiologic dose of which generates thousands of DNA lesions per cell, mostly of two types: cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs). It has not been possible, in living cells, to precisely characterize the respective contributions of these two lesion types to the signals that regulate cell cycle progression, DNA replication, and cell survival. Here we coupled multiparameter flow cytometry with lesion-specific photolyases that eliminate either CPDs or 6-4PPs and determined their respective contributions to DNA damage responses. Strikingly, only 6-4PP lesions activated the ATR-Chk1 DNA damage response pathway. Mechanistically, 6-4PPs, but not CPDs, impeded DNA replication across the genome as revealed by microfluidic-assisted replication track analysis. Furthermore, single-stranded DNA accumulated preferentially at 6-4PPs during DNA replication, indicating selective and prolonged replication blockage at 6-4PPs. These findings suggest that 6-4PPs, although eightfold fewer in number than CPDs, are the trigger for UV-induced DNA damage responses.
Ultraviolet (UV) irradiation is the most prevalent carcinogen in humans, leading to diverse skin malignancies that outnumber all other cancers combined (1). UV irradiation mostly damages DNA by forming dimers at dipyrimidine sites, and ∼100,000 to 200,000 DNA lesions are generated per diploid cell in human skin by a moderate dose of UV (1 h of sunlight; equivalent to ∼30 mJ/cm2 UVB [280 to 320 nm]) (2, 3). UV-induced DNA lesions are critical in the pathogenesis of UV-induced skin cancer (4), and these DNA lesions are typically removed through the nucleotide excision repair (NER) pathway (5). NER is defective in xeroderma pigmentosum (XP), a genetic disorder characterized by UV hypersensitivity and predisposition to UV-induced skin cancer (6). UV generates two major types of DNA lesions: cyclobutane pyrimidine dimer (CPD) and pyrimidine-pyrimidone (6-4) photoproduct (6-4PP). These two lesions are structurally distinct: 6-4PP is more DNA distorting (44° bend of DNA helix) than CPD (9° helix bend) (7). Compared to CPDs, 6-4PPs are eightfold less frequently generated by the cancer-relevant UVB spectrum (8) and are much more efficiently repaired by NER (2 h half-life for 6-4PP versus 33 h for CPD) (9). If unrepaired, a single 6-4PP lesion is severalfold more likely to induce apoptosis than a CPD lesion (10, 11).
In response to UV damage, cells activate DNA damage response pathways that elicit cell cycle checkpoints and DNA repair processes to maintain genomic integrity (3). During S phase, UV-damaged DNA causes replication fork stalling (12) that triggers activation of the ATR (ataxia telangiectasia and Rad3-related) kinase (13). Specifically, UV irradiation causes helicase-polymerase uncoupling: the replicative DNA polymerase stalls at a DNA lesion while the MCM (minichromosome maintenance) replicative helicase continues unwinding the DNA duplex (13). This uncoupling leads to generation of long stretches of single-stranded DNA (ssDNA) (14) that are then bound by replication protein A (RPA) (15). The RPA-coated ssDNA recruits not only ATR kinase but also more than 10 mediator proteins to sites of DNA damage for full activation of the ATR pathway (16–18). ATR activates downstream effectors by phosphorylating numerous targets including Chk1 and p53 (3, 19). ATR-mediated phosphorylation of the Chk1 kinase at Ser345 is critical for Chk1 activation (20) and preventing mitotic entry (21, 22). ATR is also required to inhibit new origin firing (23), thereby ensuring a sufficient quantity of available RPA to bind ssDNA and stabilizing stalled replication forks (24). Inhibition of ATR in cells with DNA damage results in premature chromatin condensation, a hallmark of premature entry into mitosis before completion of DNA replication (25). Thus, the ATR pathway plays an important role in ensuring DNA replication after genotoxic stress. However, it remains unclear whether CPD and 6-4PP have distinct effects on activation of the ATR-Chk1 pathway that is a central regulator of cellular responses to UV irradiation.
In the present study, we revealed the respective roles of CPD and 6-4PP in ATR activation by generating cells with only one major type of UV lesion. Surprisingly, we found that less abundant 6-4PPs (but not CPDs) potently activate the ATR-Chk1 pathway. We also determined that only 6-4PP markedly blocks replication progression, a plausible mechanism for robust ATR activation mediated by 6-4PP. Taken together, these findings provide mechanistic insight into how the two major types of DNA lesions that occur at dipyrimidine sites have remarkably different effects on cellular DNA damage responses.
Results
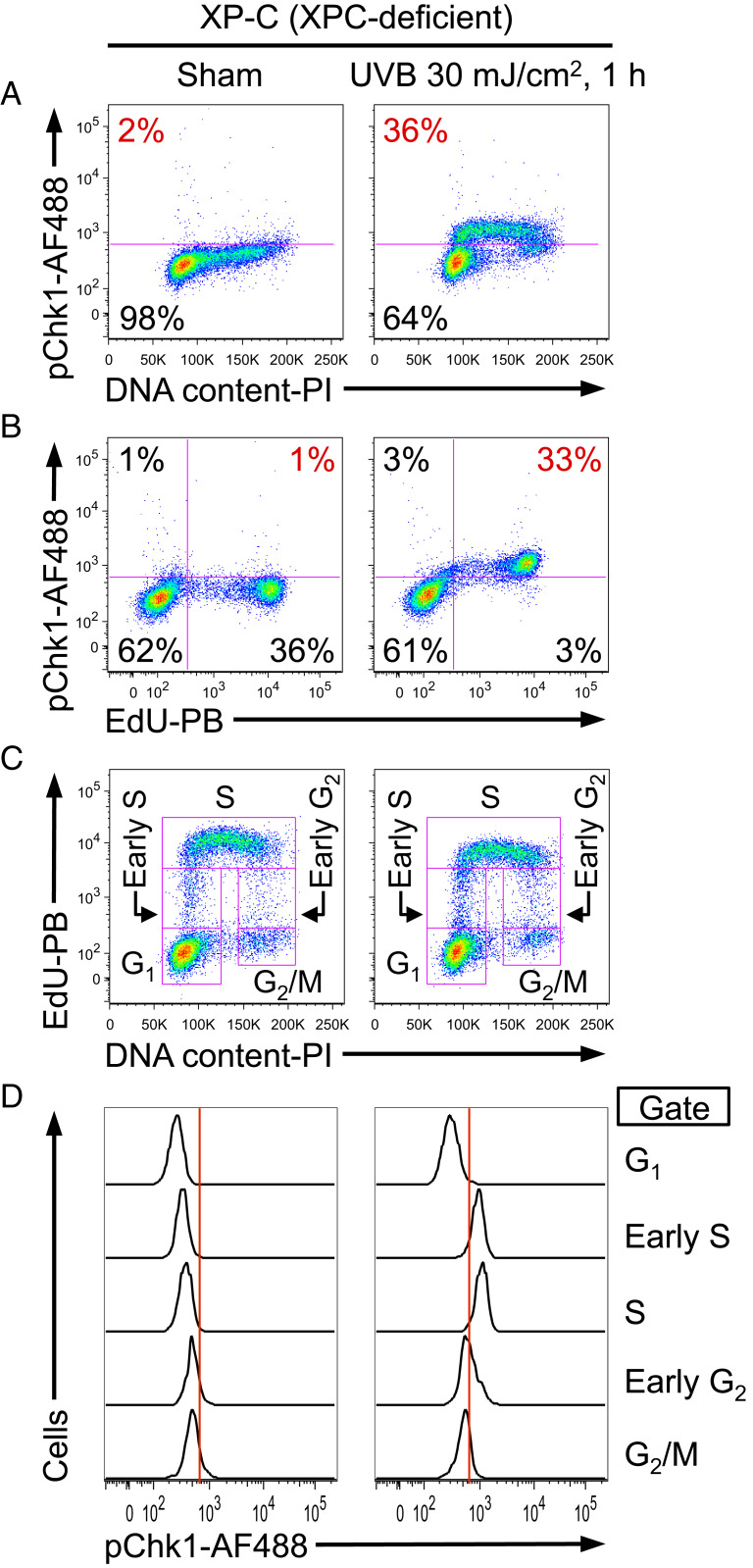
UV Activates the ATR-Chk1 Pathway Exclusively in S Phase.
Phosphorylation of Chk1 at Ser345 by the ATR kinase is the hallmark indicator of ATR activation following UV irradiation as validated by using kinase-dead ATR, ATR-Seckel mutation, ATR knockdown, and an ATR-selective small-molecule inhibitor (19, 21, 26–28). This particular phosphorylation event following UV irradiation depends on ATR, but not the closely related kinase ATM (26). To characterize the effect of UV lesions on the ATR pathway, we investigated the cell cycle-specific induction of Chk1 phosphorylation in cells that could not efficiently remove UV-induced DNA lesions by NER. In NER-deficient cells (XP-C: immortalized fibroblasts derived from xeroderma pigmentosum complementation group C patient with no expression of XPC protein) (29), UV-induced phosphorylation of Chk1 was detected in an S-phase pattern, as assessed by multiparameter flow cytometry (Fig. 1A). For a more detailed assessment of the S-phase specificity of Chk1 phosphorylation, the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) was used to label cells undergoing DNA replication. Indeed, Chk1 was phosphorylated exclusively in cells that were synthesizing DNA (EdU incorporating), but not in non-S phase (EdU negative) cells after UV irradiation (Fig. 1B). Cell cycle status was more precisely determined by using a combination of EdU incorporation and DNA content (Fig. 1C). Among five subpopulations based on cell cycle status (G1, early S, S, early G2, and G2/M), UV-induced Chk1 phosphorylation was mostly restricted to early S phase and S phase (Fig. 1D). Importantly, phosphorylated Chk1 was entirely absent in G1 phase (Fig. 1D).
Fig. 1.
UV-induced phosphorylation of Chk1 is strictly limited to S phase as revealed by flow cytometry. (A–D) XP-C cells (GM15983) were pulse labeled with EdU for 1 h, followed by sham (Left) or UVB irradiation (Right), and harvested 1 h after irradiation. (A) Phosphorylation of Chk1 at Ser345 (pChk1) was evaluated as a function of DNA content. The percentage of pChk1(+) cells is shown in red. (B) UV-induced pChk1 strongly correlates with EdU incorporation. The percentage of EdU(+)pChk1(+) cells is shown in red. (C) EdU incorporation and DNA content were used to identify five cell cycle subpopulations (G1, early S, S, early G2, and G2/M phases; pink boxes identify gates used). (D) pChk1 was evaluated for the cell cycle subpopulations defined in C. Chk1 phosphorylation was essentially restricted to UV-irradiated cells that were in early S or S phase. Data from one representative experiment of three independent experiments are shown.
We also investigated the cell cycle-specific induction of Chk1 phosphorylation following UV irradiation in NER-proficient cells. HCT116 cells (human colon carcinoma cell line with wild-type p53) (30, 31) and fully corrected XP-C cells (XP-C cells stably transfected with wild-type XPC cDNA, regaining XPC expression and full NER functional capacity) (29) both exhibited strong correlation of Chk1 phosphorylation with S phase (SI Appendix, Fig. S1 A–C). Although primary normal human fibroblasts proliferate very slowly, Chk1 was also phosphorylated exclusively in EdU-incorporating cells (SI Appendix, Fig. S1D). Thus, the specificity of Chk1 phosphorylation for S phase is not affected by the ability of cells to repair UV-induced DNA damage.
To determine the kinetics of Chk1 phosphorylation after UV irradiation, NER-proficient (primary normal human fibroblasts and HCT116) and NER-deficient (XP-C) cells were UV irradiated and harvested at different time points up to 10 h post-UV. Phosphorylation of Chk1 was rapidly induced by UV (within 30 min) in S phase in both cell types (SI Appendix, Fig. S2). After peak induction, UV-induced phosphorylation of Chk1 declined in primary normal human fibroblasts and HCT116 cells but was sustained in XP-C cells (SI Appendix, Fig. S2). Taken together, phosphorylation of Chk1 is induced rapidly and exclusively in S phase in both NER-proficient and NER-deficient cells. However, persistence of phosphorylated Chk1 following UV may be associated with the continued presence of UV lesions in NER-deficient cells.
Development of a Flow Cytometry Assay That Allows Detection of both Phosphoproteins and UV Lesions.
To dissect the role of each type of UV lesion in phosphorylation-mediated ATR signaling, an assay that allows detection of both phosphoproteins and UV-induced DNA lesions was needed. For UV lesion detection, antibodies that specifically recognize CPD or 6-4PP have been widely used (32). Because these epitopes (CPD and 6-4PP) are likely obscured in double-stranded DNA (dsDNA), detection of UV lesions by these antibodies may require DNA denaturation. Hydrochloric acid (HCl) has been previously used to denature DNA prior to detection of UV lesions by flow cytometry (31), and we also observed that HCl allowed detection of the UV lesion signal (SI Appendix, Fig. S3A). However, this strong acid treatment eliminated our ability to detect phosphorylated Chk1 (SI Appendix, Fig. S3B). Because DNase I is an endonuclease that introduces random single-stranded nicks and can facilitate formation of ssDNA (33), we tested whether DNase I could be used to expose UV lesions to antibodies without destroying phosphoproteins. Indeed, DNase treatment enabled UV lesions to be efficiently detected while also preserving the ability to detect Chk1 phosphorylation (SI Appendix, Fig. S3). With DNase treatment, rapid repair of 6-4PP lesions in NER-proficient cells was detected by flow cytometry (SI Appendix, Fig. S4). Therefore, this DNase-based flow cytometry assay represents a useful tool to investigate interactions between UV lesions and phosphorylation-associated signaling pathways.
Identification of Cells Expressing Photolyase by Flow Cytometry.
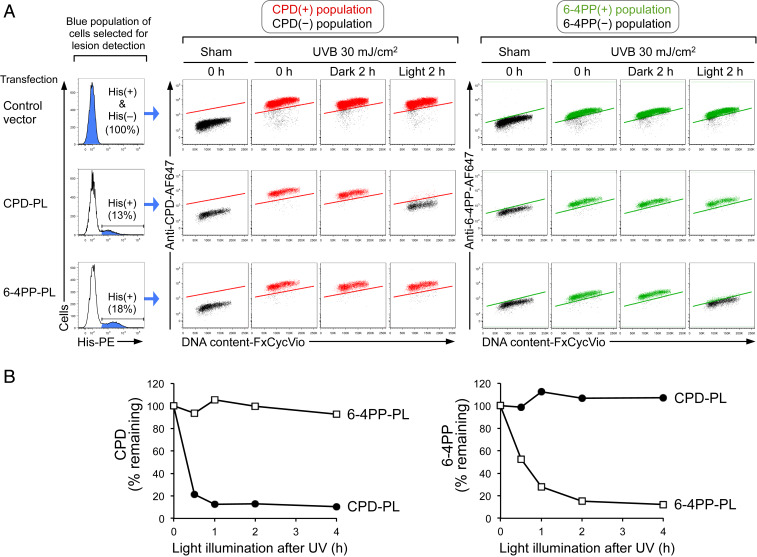
To determine the individual effect of CPD and 6-4PP lesions on ATR activation, it was necessary to generate cells that had a single type of UV lesion. First, to retain both types of UV lesions for investigation, a global genome-NER (GG-NER)-deficient XP-C cell line (GM15983) (29) was selected for this study. In normal cells, 6-4PP lesions are rapidly repaired by GG-NER (34), making it difficult to assess the effect of 6-4PP on ATR activation. Second, we made use of lesion-specific photolyases, which are not present in placental mammals, to selectively repair a single type of lesion (either CPD or 6-4PP) using the energy of visible light (35, 36).
In XP-C cells transfected with control vector (empty vector without photolyase), both CPD and 6-4PP persisted 2 h after UV irradiation in the presence or absence of visible light (Fig. 2A, first row). This validates the NER deficiency of this cell type and ensures that illuminating cells with visible light does not affect the levels of CPD and 6-4PP. After transfection of the relevant photolyase vector, cells expressing polyhistidine (His)-tagged CPD-photolyase (CPD-PL) or 6-4PP-photolyase (6-4PP-PL) were identified by flow cytometry for subsequent analysis (Fig. 2 A, Left). As expected, repair of each lesion type was restricted to cells that expressed the corresponding photolyase and were also exposed to visible light (Fig. 2A, second and third rows). This light-dependent, lesion-specific repair (photorepair) occurred with the same efficiency regardless of cell cycle phase (DNA content) (Fig. 2A). Notably, the difference in 6-4PP signals between sham and UV was smaller than that of CPD, likely because 6-4PP lesions are generated less frequently than CPD by the same dose of UV irradiation (eightfold less by UVB) (8).
Fig. 2.
Selection of cells with a single type of UV lesion using photolyase and flow cytometry. (A) Flow cytometry identifies photolyase-expressing cells that exhibit light-dependent, lesion-specific repair (photorepair) in all cell cycle phases. XP-C cells (GM15983) transfected with control vector, polyhistidine (His)-tagged CPD-photolyase (CPD-PL), or His-tagged 6-4PP-photolyase (6-4PP-PL) were sham or UVB irradiated and were harvested immediately (0 h) or after 2 h with (light) or without (dark) visible light illumination. Control vector-transfected cells [whole population including His(+) and His(–)] or photolyase-expressing cells [His(+)] were selected for lesion detection (as indicated in blue in histogram with percentage). Middle and Right show the levels of CPD or 6-4PP as a function of DNA content (FxCycle Violet). CPD(+), 6-4PP(+), and lesion-negative populations are indicated in red, green, and black, respectively. (B) Time course of CPD and 6-4PP photorepair by lesion-specific photolyase. XP-C cells were transfected with CPD-PL (closed circles) or 6-4PP-PL (open squares). Cells were sham or UVB irradiated (30 mJ/cm2) and subsequently illuminated with visible light until harvest at the indicated time points. Remaining lesions in photolyase-expressing cells (His tag-positive) were assessed by flow cytometry. Background signal (based on sham-treated cells) was subtracted, and lesion signal at 0 h (immediately after UV) was set to 100%. Data from one representative experiment of three independent experiments are shown.
To determine the duration of visible light illumination required for sufficient photorepair, the kinetics of photorepair were investigated. Near-complete photorepair of CPD or 6-4PP lesions required 1- to 2-h exposure of visible light (Fig. 2B). By combining flow cytometry with lesion-specific photolyase transfection in NER-deficient cells as well as sufficient visible light illumination, we were able to select cells carrying only a single major type of UV lesion (CPD or 6-4PP).
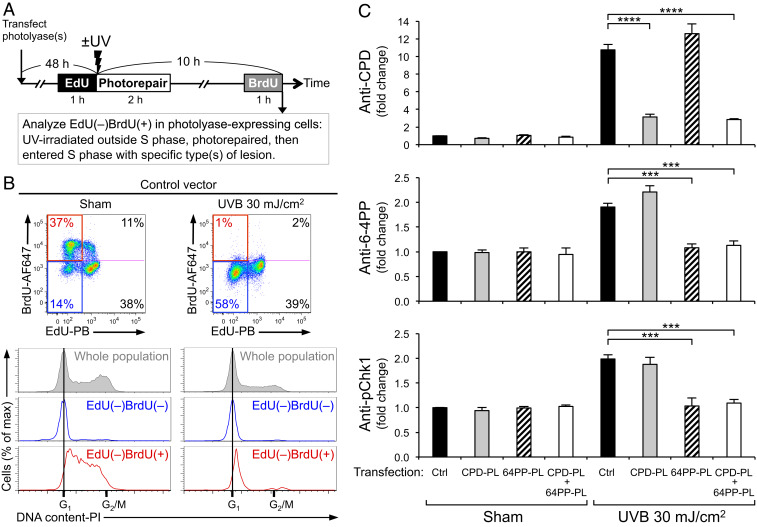
The 6-4PP, but Not CPD, Potently Activates the ATR-Chk1 Pathway.
Although photolyases can selectively eliminate either CPD or 6-4PP, the specific role of each lesion type in DNA damage responses still cannot be determined if these responses had already been induced before lesion-specific photorepair was complete. To circumvent this problem, we designed a system in which CPD or 6-4PP lesions were eliminated in cells that had not yet entered S phase, hence had not yet activated the ATR-Chk1 pathway. In this experimental setting (Fig. 3A), 5-ethynyl-2′-deoxyuridine (EdU)-negative, 5-bromo-2′-deoxyuridine (BrdU)-positive cells [EdU(–)BrdU(+)] should represent the desired population: UV-irradiated outside S phase, underwent photorepair, and subsequently entered S phase with only the desired type(s) of UV lesion present. The cell cycle status of nonreplicating cells [EdU(–)BrdU(–)] showed DNA content equivalent to G1 phase (Fig. 3 B, Right, blue line), suggesting that this double negative population was likely in G1 at the time of irradiation and remained in G1 at harvest. In contrast, the EdU(–)BrdU(+) population had a cell cycle profile that was right-shifted toward higher DNA content (Fig. 3 B, Right, red line) compared to that of EdU(–)BrdU(–), indicating that, as expected, EdU(–)BrdU(+) cells had newly entered S phase after UV. Photorepair of CPD and 6-4PP facilitated S phase entry (SI Appendix, Fig. S5).
Fig. 3.
The 6-4PP, but not CPD, potently induces phosphorylation of Chk1. (A) Experimental design for analyzing cells that enter S phase with a specific type of lesion. XP-C cells (GM15983) transfected with control vector (Ctrl), CPD-photolyase (CPD-PL), and/or 6-4PP-photolyase (64PP-PL) were labeled with EdU prior to UV. Following UVB 30 mJ/cm2 irradiation, cells were illuminated with visible light for photorepair. Cells were labeled with BrdU prior to harvest. Photolyase-expressing cells (polyhistidine tag-positive) were analyzed for levels of CPD, 6-4PP, and Chk1 phosphorylation at Ser345 (pChk1) within the EdU(–)BrdU(+) population. (B) The EdU(–)BrdU(+) population of UV-irradiated cells is in early S phase. Cells were stained with propidium iodide (PI) for DNA content. Cell cycle profiles of whole population (gray) and its subpopulations of EdU(–)BrdU(–) (blue) and EdU(–)BrdU(+) (red) are shown. Data from one representative experiment of four independent experiments are shown. (C) Phosphorylation of Chk1 is potently induced in cells with 6-4PP, but not in cells with CPD, upon S-phase entry. The levels of CPD, 6-4PP, and pChk1 (Ser345) in cells that newly entered S phase [EdU(–)BrdU(+) population] were evaluated using antibody-based flow cytometry assay. The mean ± SEM (four independent experiments) of fluorescence signals (fold change compared with sham-irradiated control vector cells) are shown. Statistical significance was determined by one-way ANOVA and Dunnett’s test, comparing each UV-irradiated, photolyase-transfected group with a single control (UV-irradiated control group). ***P < 0.001; ****P < 0.0001.
The relative impact of CPD or 6-4PP on ATR-Chk1 activation can thus be determined in cells that newly enter S phase with the desired lesion type (Fig. 3C). Phosphorylation of Chk1 was robustly induced upon S-phase entry in UV-irradiated cells that carried both CPD and 6-4PP (control) and in cells in which CPD was repaired but 6-4PP remained (CPD-PL). In contrast, in cells that had newly entered S phase with only CPD remaining (6-4PP-PL), phosphorylation of Chk1 was not induced and stayed at the basal level equivalent to sham-irradiated control cells. In cells that had neither CPD nor 6-4PP (CPD-PL + 6-4PP-PL), phosphorylation of Chk1 also remained at the basal level. These results demonstrate that 6-4PP, but not CPD, has a potent ability to induce phosphorylation of Chk1. The same results were obtained from a separate XP-C cell line (GM16093) that is derived from a different XP-C patient (37) (SI Appendix, Fig. S6), suggesting that the requirement for 6-4PP in inducing phosphorylation of Chk1 was not limited to a particular cell line. To exclude the possibility that lack of ATR activation by CPD may be due to impaired recognition and repair of UV-induced DNA lesions in XP-C cell lines, we used primary normal human fibroblasts that are NER proficient. Strikingly, in this more physiologically relevant setting, 6-4PP is again the lesion responsible for UV-induced ATR activation, and CPD does not have a role in activating the ATR-Chk1 pathway in S phase (SI Appendix, Fig. S7).
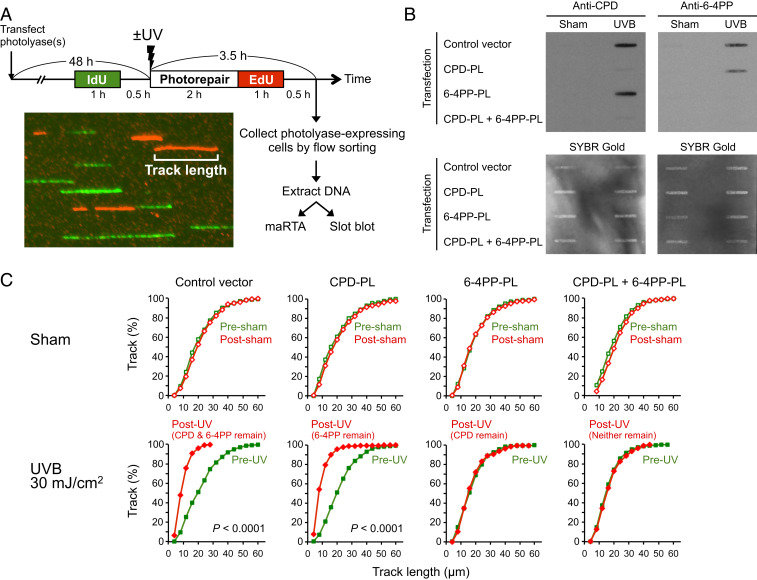
The 6-4PP Lesion, but Not CPD, Impedes DNA Replication Progression.
To assess replication progression in cells with or without a specific type of lesion, photolyase-transfected cells were pulse labeled with two thymidine analogs (5-iodo-2′-deoxyuridine [IdU] and EdU) separately for 1 h each (Fig. 4A). After harvest, cells expressing His-tagged photolyase were collected by flow sorting, and DNA was extracted from these sorted cells and subjected to immuno-slot blot analysis for photorepair validation and microfluidic-assisted replication track analysis (maRTA) (38).
Fig. 4.
DNA replication is impeded by 6-4PP, but not CPD lesions. (A) Experimental design to quantitate DNA replication progression in cells with specific type(s) of lesion(s). XP-C cells (GM15983) transfected with the indicated photolyase (PL) were sham or UVB irradiated and pulse labeled with IdU and EdU separately for 1 h each. Photolyase-expressing cells (polyhistidine tag-positive) were collected by flow sorting. Genomic DNA extracted from sorted (photolyase-expressing cells) or unsorted (for control vector) cells was subjected to immuno-slot blot and microfluidic-assisted replication track analysis (maRTA; a representative image of segments of labeled DNA “tracks” is shown). (B) Slot-blot validation of lesion-specific photorepair of DNA used for maRTA. Extracted genomic DNA was heat denatured and then spotted onto membranes. Membranes were probed with anti-CPD or anti-6-4PP antibodies and subsequently stained with SYBR Gold for total DNA detection. Data from one representative experiment of two independent experiments are shown. (C) DNA replication is impeded in the presence of 6-4PP, but not CPD. Genomic DNA was aligned through microchannels and stained for IdU and EdU (maRTA). Replication track lengths of first (IdU; presham/UV) and second (EdU; postsham/UV) labels were measured in two independent experiments, and combined data are shown as cumulative distributions (n = 500 tracks for each label). The vertical axis indicates the cumulative fraction of tracks that are equal to or shorter than the corresponding track length indicated on the horizontal axis. The remaining lesion type(s) (validated in B) is indicated in parentheses above each UV graph. Statistical significance was determined by Mann–Whitney U test. Two conditions showed P < 0.0001 as indicated.
Immuno-slot blot analysis using extracted DNA showed that each lesion type was repaired, consistent with the expression of the relevant photolyase, validating lesion-specific photorepair in cells collected by flow sorting (Fig. 4B). To assess replication progression in the presence of a specific type of lesion, replication track lengths of the first (pre-UV) and second (post-UV and photorepair) labels were compared (Fig. 4C). The lengths of replication tracks were not altered after sham irradiation and exposure to visible light, indicating that these treatments had no effect on replication. In UV-irradiated cells with both CPD and 6-4PP remaining (control), track lengths of post-UV replication (second label, red) were shorter than those of pre-UV replication (first label, green), indicating that replication is impeded by UV lesions. Similarly, in UV-irradiated cells that had only 6-4PP remaining (CPD-PL), track lengths were shorter in post-UV replication than in pre-UV replication. In contrast, in UV-irradiated cells that had only CPD remaining (6-4PP-PL), track lengths of pre-UV and post-UV replication were essentially identical. Consistently, in cells that had neither CPD nor 6-4PP (CPD-PL + 6-4PP-PL), no difference of track lengths was observed between pre-UV and post-UV replication. This finding strongly suggests that, under these experimental conditions, no other types of UV-induced DNA lesions contribute significantly to replication blockage and that the interactions of CPD- and 6-4PP-photolyases with their cognate lesions per se do not block DNA replication. Taken together, a combination of photolyase, flow sorting, and maRTA enabled the analysis of replication progression in cells with a single major type of lesion. Strikingly, we found that 6-4PP, but not CPD, is the critical type of lesion that effectively impedes replication progression.
The 6-4PP Lesion Preferentially Becomes Surrounded by ssDNA during Post-UV Replication.
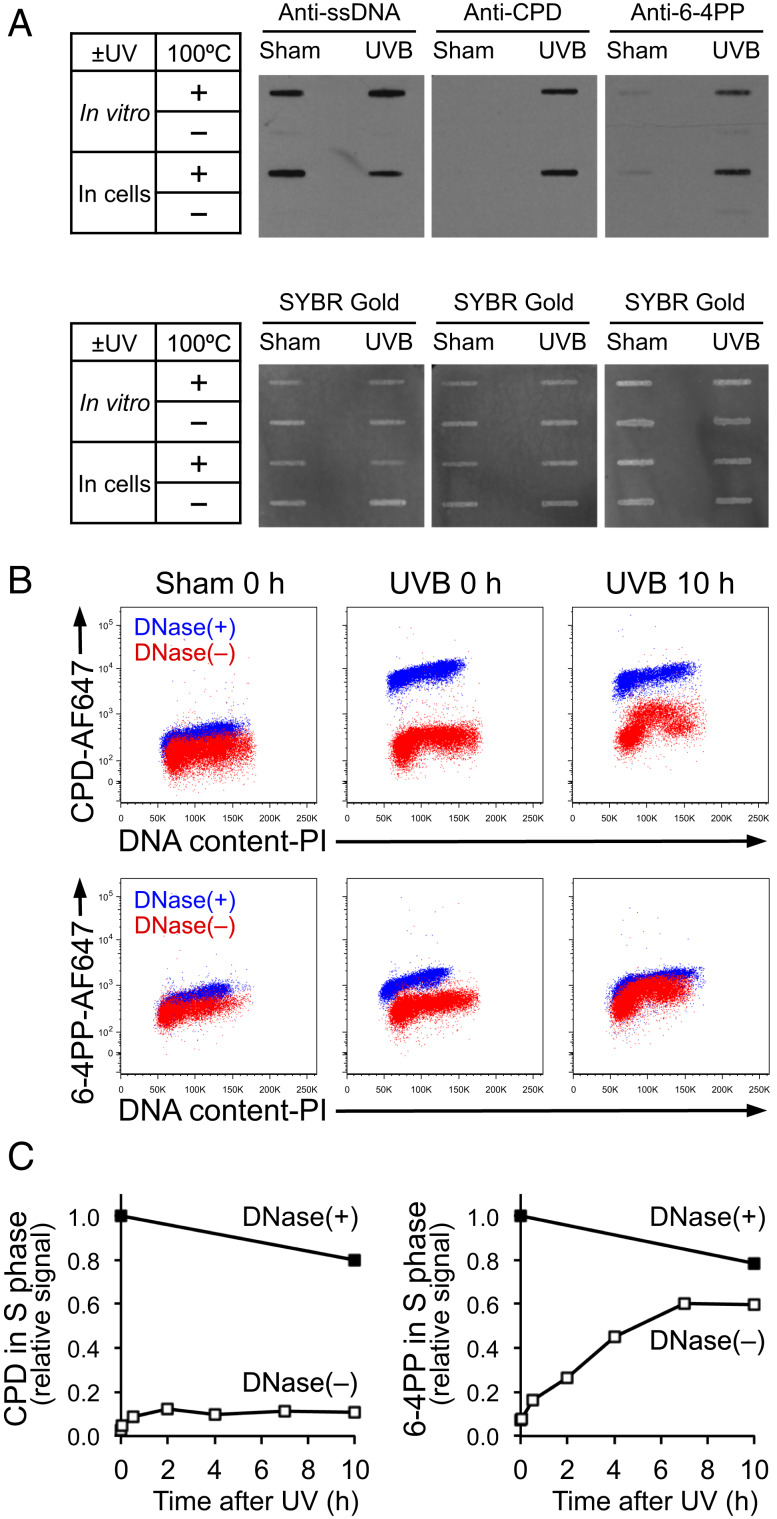
Upon DNA replication blockage, ssDNA is typically generated adjacent to replication-blocking lesions after replicative helicases are uncoupled from stalled DNA polymerases (13, 14). If the antibodies used for detection of either CPD or 6-4PP are specific to lesions on ssDNA, replication-blocking lesions would thus be preferentially detected. To validate the specificity of the available antibodies (anti-CPD [TDM-2] and anti-6-4PP [64M-2]) (32) for their respective epitopes on ssDNA (but not on dsDNA), we compared lesion detection using DNA that was heat denatured or left unheated. Immuno-slot blot analysis using an anti-ssDNA antibody showed strong signals in heat-denatured DNA receiving sham or UV irradiation (Fig. 5 A, Left). This finding indicates that 10 min of 100 °C heat treatment is sufficient to generate ssDNA for antibody recognition. In these heat-denatured samples, CPD and 6-4PP were detected in UV-irradiated DNAs using the relevant antibody (Fig. 5 A, Middle and Right). In contrast, ssDNA was not detected in unheated samples, indicating DNA in these samples remained double stranded (Fig. 5 A, Left). In these unheated samples that persisted as dsDNA, essentially no signals of CPD or 6-4PP were detected using lesion-specific antibodies even when DNAs were UV irradiated (Fig. 5 A, Middle and Right). These results demonstrate that lesion-specific antibodies preferentially recognize CPD or 6-4PP lesions that are on ssDNA, but not on dsDNA.
Fig. 5.
The 6-4PP lesion preferentially becomes surrounded by ssDNA. (A) Lesion-specific antibodies recognize CPD or 6-4PP on ssDNA, but not on dsDNA. For “in vitro” UV irradiation, genomic DNA was extracted from unirradiated cells and subsequently irradiated with UVB 30 mJ/cm2 in vitro. For “in cells” irradiation, genomic DNA was extracted from cells that were irradiated with UVB 30 mJ/cm2. DNA was left untreated or heat denatured at 100 °C for 10 min and spotted onto membranes for immuno-slot blot using antibodies specific for ssDNA, CPD, or 6-4PP. The same membranes were stained with SYBR Gold for total DNA detection. (B) UV lesions on ssDNA under non-DNase condition become detectable in S phase long after UV, but not immediately after UV. XP-C cells (GM15983) were harvested immediately (0 h) or 10 h after UVB 30 mJ/cm2. Fixed cells were left untreated or treated with DNase. CPD and 6-4PP were detected using lesion-specific antibodies validated in A to recognize the lesions only when on ssDNA. DNA content was assessed using propidium iodide (PI). Flow cytometry data from two samples [DNase(+) and DNase(–)] were overlaid on each plot. (C) In S-phase cells, 6-4PP lesions are increasingly surrounded by ssDNA. Signal intensities of CPD and 6-4PP in S phase at various time points following UV were measured using the same flow cytometry assay as in B. Relative signal intensities of CPD or 6-4PP in S phase were calculated relative to 0 (sham irradiation at 0 h) and 1 (UVB 30 mJ/cm2 at 0 h with DNase treatment). Data from one representative experiment of three independent experiments are shown.
Using these validated lesion-specific antibodies under nondenaturing conditions, we detected replication-blocking lesions that are left on ssDNA (Fig. 5B). As a positive control, we used DNase to detect UV lesions regardless of whether or not lesions were surrounded by ssDNA. In cells treated with DNase, CPD and 6-4PP were detectable immediately after irradiation, and these signals were strong even 10 h following irradiation (XP-C cells could not repair lesions) (Fig. 5B, blue population). In contrast, in UV-irradiated cells without subsequent DNase treatment, both types of lesion were barely detectable immediately after UV irradiation (Fig. 5B, red population, UVB 0 h). This indicates that UV lesions were not surrounded by ssDNA immediately after irradiation and therefore not accessible to the antibodies, likely because replication across UV lesions had not yet occurred. However, 10 h after UV irradiation, CPD and 6-4PP were partially detectable in S-phase cells in the absence of DNase treatment (Fig. 5B, red population, UVB 10 h). Because lesion detection under nondenaturing conditions was limited to S phase, it is likely that this signal resulted from replication blockage that generated ssDNA at these lesions.
We further characterized the kinetics of replication blockage at CPD and 6-4PP under nondenaturing conditions. In S phase, CPD became detectable after UV irradiation in the absence of DNase treatment, but the CPD signal remained at a low level for the duration of the experiment [Fig. 5 C, Left, DNase(–)], suggesting that ssDNA-surrounded CPD lesions were not accumulating. In contrast, the 6-4PP signal markedly and continuously increased [Fig. 5 C, Right, DNase(–)], indicating that 6-4PP preferentially became surrounded by ssDNA. This striking time-dependent increase in the signal of ssDNA-surrounded 6-4PP suggests that DNA synthesis across 6-4PP is impeded in a prolonged manner.
Discussion
UV irradiation provokes diverse cellular responses to cope with DNA damage, most prominently via the ATR-Chk1 pathway (3). However, no prior studies have revealed the precise contributions of each of the two UV-induced DNA lesions (CPD or 6-4PP) in activating this pathway. Carrying out such a study has proven elusive because thousands of both types of lesion are instantaneously generated in UV-irradiated cells, and activation of the ATR-Chk1 pathway is rapidly initiated after UV (within 3 min) in S-phase cells. Although photolyases are capable of selectively removing a specific type of UV-induced DNA lesion, photolyase-mediated repair requires more than an hour of visible light illumination for near-complete repair. We overcame these issues by combining photolyases, cell cycle tracking, and multiparameter flow cytometry optimized to detect UV lesions, thymidine analogs, and phosphoproteins. Flow cytometry-based single-cell analysis to select only photolyase-expressing cells and the desired cell population was essential to precisely determine the contributions of CPD and 6-4PP to UV-induced replication blockage and ATR activation. Cell synchronization to increase the number of cells entering S phase was not suitable for this study because it is known that cell synchronization itself increases replication stress (39).
The present study identified 6-4PP as the critical lesion that causes replication blockage and activation of the ATR-Chk1 pathway in S phase. This raises the question of how DNA replication proceeds essentially unimpeded in the presence of CPD, whereas 6-4PP lesions are strongly inhibitory to replication progression. Prior studies suggest that there are two major mechanisms by which DNA replication can continue following replicative polymerase blockage: direct bypass across a lesion via translesion synthesis (TLS) (40, 41), or replication restart downstream of a lesion via repriming (42). Repriming leaves an ssDNA gap that could span up to 400 nucleotides (14). Of note, these ssDNA gaps are too short to be reliably detected in the maRTA assay we used (detection limit is 3 µm, equivalent to 12 kb). However, ssDNA gaps of this size are more than sufficient to be identified by lesion-specific antibodies that can recognize either CPD or 6-4PP contained within as little as 8 bases of ssDNA (32). After verifying the ssDNA specificity of these antibodies, we found that the signal of 6-4PP lesions surrounded by ssDNA continuously increased in S phase over several hours to nearly 10-fold above the level detected shortly after UV (Fig. 5C). In contrast, the signal of CPD lesions surrounded by ssDNA was also observed exclusively in S phase but remained at a low level. Therefore, a plausible model for our findings is that DNA replication promptly resumes downstream of a CPD lesion through repriming, initially generating an ssDNA gap (transiently detectable by the antibody) that is soon filled by TLS. In this model, an ssDNA gap at a CPD lesion is transient, and new short-lived gaps are created as DNA replication progresses without significant slowing. In contrast, 6-4PP lesions persistently block DNA replication, perhaps because TLS and/or replication restart is persistently impeded at 6-4PP lesions. This prolonged replication blockage corresponds to the ability of 6-4PP to activate the ATR-Chk1 pathway, presumably by recruiting RPA to regions of persistent ssDNA surrounding 6-4PP lesions. The inability of CPD lesions to activate the ATR-Chk1 pathway is likely due to its efficient bypass in cells that have intact TLS polymerases. Indeed, in cells that are unable to bypass CPD lesions (XP-variant cells with defective DNA polymerase η), ATR pathway activation is markedly augmented and prolonged (43, 44). This suggests that when they cannot be bypassed, CPD lesions can activate the ATR pathway similarly to 6-4PP lesions, which are difficult to bypass even in normal cells (40).
To precisely determine the effects of CPD and 6-4PP on DNA replication blockage, we carefully selected appropriate experimental conditions for maRTA (Fig. 4). We used 30 mJ/cm2 UVB irradiation that generates ∼180,000 CPDs and 21,600 6-4PPs in the human genome (3 billion bp) (8). Mammalian replicons are heterogeneous in size but most fall into the range of 30 to 450 kbp (45). Thus, a typical replicon (∼140 kbp) will have eight CPDs and one 6-4PP following 30 mJ/cm2 UVB irradiation. Furthermore, ∼140 kbp corresponds to 35 µm of track length in maRTA (1 µm = 4 kb) (38), and 35 µm is within the range of track lengths generated by 1 h of pulse labeling with thymidine analogs (Fig. 4C). Thus, 30 mJ/cm2 UVB irradiation generates approximately eight CPDs and one 6-4PP per typical track and is thus appropriate to determine the effects of CPD and 6-4PP on DNA replication progression. For photolyase expression, we initially used adenoviruses carrying CPD- or 6-4PP-photolyases. However, we found that ATR activation was deeply attenuated in response to UV in adenovirus-infected cells (SI Appendix, Fig. S8, blue population). This finding is consistent with a previous study showing that adenovirus inhibits ATR activation by promoting degradation of TopBP1, a required cofactor for ATR activation (46). In turn, ATR is important for CPD bypass because the activity of DNA polymerase η (the enzyme that mediates CPD bypass) is augmented by ATR-mediated phosphorylation (47). Thus, in adenovirus-infected cells, DNA replication may be impeded at CPD lesions as observed by Quinet et al. (48), potentially due to diminished ATR activation and attenuated DNA polymerase η function. Another contributing factor that may underlie the different findings regarding the role of CPD lesions in Quinet et al. versus the present study may be the timing of thymidine analog labeling relative to photorepair. Since photorepair takes minutes to hours to complete, if photorepair and pulse labeling are performed simultaneously, it is likely that DNA replication forks predominantly encounter CPD lesions (due to their abundance and their long half-life) rather than 6-4PP lesions that are fewer in number. Thus, it is critical to label cells after lesion-specific photorepair is complete in order to accurately assess DNA replication progression in the presence of only one type of UV lesion. Taken together, the use of nonadenoviral vectors for photolyase expression and thymidine analog labeling after photorepair, is likely what enabled the present study to identify the highly selective replication-blocking effect of 6-4PP.
Activation of the ATR-Chk1 pathway by replication-blocking lesions promotes survival of DNA-damaged cells; however, this would also increase mutation incorporation. In this manner, 6-4PP-mediated ATR activation would likely promote “mutagenic survival” because cells with DNA damage/mutations would persist. Therefore, targeting ATR-activated cells should lead to eliminating 6-4PP-containing cells and inhibiting UV mutagenesis and carcinogenesis. Indeed, previous mouse studies showed that genetic or pharmacological inhibition of ATR suppresses UV carcinogenesis (49, 50). In parallel, multiple human epidemiological studies have shown that intake of caffeine (a nonspecific ATR inhibitor) is associated with decreased risk of developing multiple types of UV-induced skin cancers (51–53). The cancer-preventive effect of ATR inhibition is likely due in part to increased UV-induced apoptosis (27), which eliminates damaged skin cells that are prone to incorporating DNA mutations and are more likely to undergo malignant transformation. Furthermore, unlike XP, patients with trichothiodystrophy (TTD) have deficient repair of CPD but proficient repair of 6-4PP, and TTD patients do not have an increased frequency of skin cancer (54, 55). This suggests that removal of 6-4PP is crucial for preventing skin cancer development. Intriguingly, during unperturbed DNA replication, a minority of ATR molecules are activated to limit origin firing at sites of ongoing replication, and ATR and Chk1 kinase inhibitors induce unscheduled dormant origin firing throughout S phase (56, 57). Therefore, preserving some degree of ATR function is critical for cell survival and genome integrity (58, 59), whereas modest inhibition of ATR, for example via caffeine, may be well tolerated and beneficial for preventing UV-induced skin cancer development.
ATR inhibition may also directly prevent error-prone TLS that ultimately contributes to skin cancer development. This notion is supported by a prior study showing that caffeine abolishes TLS that is mediated by Rev3 (60), a catalytic subunit of DNA polymerase ζ that promotes error-prone bypass across 6-4PP. Furthermore, XP-V (polymerase η-deficient) cells exhibit reduced levels of recovery of replicative DNA synthesis following UV irradiation in the presence of caffeine, implying the existence of TLS that is not mediated by polymerase η (potentially mediated by polymerase ζ) and is sensitive to caffeine (61). Because TLS across 6-4PP lesions is a highly mutagenic process (40, 62), it will be relevant to determine whether ATR is required for this process. In summary, the present study identifying the respective roles of the two major types of UV lesions provides a molecular basis for their distinct effects on DNA damage responses and insight into the mechanisms of UV carcinogenesis.
Materials and Methods
Cell Lines and Culture Conditions.
SV40-transformed, human XP-C (XPC-deficient) skin fibroblast cell lines derived from two different patients [GM15983 (line: XP4PA-SV-EB) (29, 63) and GM16093 (line: XP14BRneo17) (37, 64)] and primary normal human skin fibroblasts (AG13145; untransformed) were purchased from Coriell Institute for Medical Research and were grown in Dulbecco’s modified Eagle medium (DMEM) (11995-040, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (10438-026, Thermo Fisher Scientific) and 1% penicillin–streptomycin (15140-122, Thermo Fisher Scientific). Fully corrected XP-C cells [GM16248 (line: XP4PA-SE2); derived from GM15983 (XP4PA-SV-EB) by stable transfection with wild-type XPC cDNA, regaining XPC expression and full NER functional capacity (29)] were also purchased from Coriell Institute for Medical Research and grown in DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, and 0.2 mg/mL hygromycin B (10687-010, Thermo Fisher Scientific). HCT116 (human colon carcinoma) p53+/+ cell line was a kind gift from Bert Vogelstein, Johns Hopkins University, Baltimore, MD. U2OS (human osteosarcoma) cell line (HTB-96) was purchased from American Type Culture Collection (ATCC). HCT116 and U2OS cells were grown in McCoy’s 5A medium (16600-082, Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin–streptomycin. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. Cells were harvested by trypsinization using 0.05% trypsin-ethylenediaminetetraacetic acid (EDTA) (25300, Thermo Fisher Scientific) for passage and for experiments.
Construction and Transfection of Lesion-Specific Photolyase Plasmids.
cDNAs of marsupial Potorous tridactylus CPD-PL (GenBank accession No. D26020) and the plant Arabidopsis thaliana 6-4PP-PL (GenBank accession No. NM_112432) were kind gifts from Carlos F. M. Menck, University of São Paulo, São Paulo, Brazil (35, 65). Photolyase cDNA was subcloned into pcDNA6/myc-His B mammalian expression vector with human cytomegalovirus promoter (V221-20, Thermo Fisher Scientific) to fuse with a polyhistidine tag (6× His) at the C terminus of photolyase. Specifically, for constructing a plasmid encoding His-tagged CPD-PL, the XhoI site at 5′ and SacII site at 3′ were used to insert cDNA of CPD-PL into the expression vector. To construct a plasmid encoding His-tagged 6-4PP-PL, cDNA of 6-4PP-PL was first amplified by PCR using the following primers to add restriction sites: forward 5′-ACTCGAGACCATGCAACGATTCTGCGTCTG-3′ (XhoI site); reverse 5′-ATATCCGCGGTTTGAGTTTTGGTCGTTGG-3′ (SacII site). The amplified product was then subcloned into pcDNA6/myc-His B mammalian expression vector using the XhoI site at 5′ and SacII site at 3′. pcDNA6/myc-His B mammalian expression vector without insert was used as a control vector.
Transfection experiments for transformed cell lines were performed using FuGENE HD transfection reagent (E231, Promega; F500, Switchgear Genomics) according to the manufacturer’s instructions. For transfection of human XP-C fibroblast cell lines, 3 × 105 cells were plated on a 35-mm dish and allowed to grow at 37 °C in a humidified atmosphere of 5% CO2 for 16 h. Prior to transfection, culture medium was removed, and fresh medium was added to cells. Transfection mixture in a final volume of 100 µL per 35-mm dish was prepared by diluting 2 µg plasmid DNA and 7 µL FuGENE HD in Opti-MEM I reduced serum medium (11058-021, Thermo Fisher Scientific) that was prewarmed to room temperature. For double transfection of CPD-PL and 6-4PP-PL, 1 µg plasmid DNA of each photolyase was mixed. The transfection mixture was incubated at room temperature for 15 min and then added to cells in a dropwise manner. Cells were incubated at 37 °C in a humidified atmosphere of 5% CO2 for 48 h for subsequent experiments (UV irradiation followed by flow cytometry or replication track analysis).
Primary normal human fibroblasts were transfected with photolyase plasmids by using the Amaxa Human Dermal Fibroblast Nucleofector Kit (VPD-1001, Lonza) and Nucleofector Program U-20.
Adenovirus Encoding Photolyase-Enhanced Green Fluorescent Protein.
Photolyase cDNA (CPD-PL or 6-4PP-PL) was subcloned into pEGFP-N1 using the XhoI site at 5′ and SacII site at 3′ to fuse with enhanced green fluorescent protein (EGFP) at the C terminus of photolyase. CPD-PL-EGFP or 6-4PP-PL-EGFP was then subcloned into pAdeno-X adenoviral vector (Clontech). Recombinant adenovirus was produced as previously described (65). Primary Xpc−/− mouse keratinocytes were isolated as previously described (49) and transduced with adenovirus encoding CPD-PL-EGFP or 6-4PP-PL-EGFP at a multiplicity of infection of 850.
UV Irradiation and Photorepair.
UVB irradiation was performed using a panel of four UVB bulbs (RPR-3000, Southern New England Ultraviolet) emitting radiation centered around 311 nm. UVB bulbs were covered by a Kodacel filter (K6808, Eastman Kodak) to eliminate any UVC irradiation (<290 nm). UVB dosage was monitored using a Photolight IL400A radiometer equipped with a SEL240/UVB detector (International Light Technologies), and 30 mJ/cm2 UVB irradiation typically required 40 s. Prior to UVB irradiation, cells were rinsed once with warm Hank’s balanced salt solution without phenol red (HBSS; 14025-092, Thermo Fisher Scientific). Immediately after HBSS was removed, cells on a cell culture dish were placed under UVB bulbs without a dish lid and irradiated with UVB. Sham-irradiated cells were treated exactly the same way, except that cells were not exposed to UV irradiation. For experiments without photorepair, 2 mL of fresh medium was added to a 35-mm dish immediately after UV irradiation, and cells were incubated at 37 °C in the dark. For photorepair by visible light illumination, 2 mL of warm HBSS without phenol red was added to a 35-mm dish immediately following UV irradiation. The dish of cell culture was then illuminated by visible light from the bottom by placing a dish on a 3-mm thick glass surface sitting 6 mm above one fluorescent lamp (F13T5/WW, warm white, 766 lm [mean], Sylvania) at room temperature for 2 h for photorepair (denoted as “light”). For mock photorepair control (“dark”), dishes were wrapped with aluminum foil and placed on the glass above the same fluorescent lamp used for visible light-illuminated samples.
Thymidine Analog Preparation.
For stock solution, BrdU (B5002, Sigma-Aldrich; 15 mM), IdU (I7125, Sigma-Aldrich; 2 mM), and EdU (A10044, Thermo Fisher Scientific; 15 mM) were dissolved in Dulbecco’s phosphate-buffered saline (PBS) (14190-250, Thermo Fisher Scientific) at the indicated concentrations and stored at −20 °C. Stock solutions were diluted with cell culture medium to have final optimized concentrations: 15 µM EdU and 20 µM BrdU for identifying replicating cells (flow cytometry analysis); 50 µM IdU and 10 µM EdU for labeling DNA (replication track analysis).
Flow Cytometry Analyses.
For experiments determining the effect of each lesion type in activating the ATR-Chk1 pathway (Fig. 3), XP-C cells (GM15983) were transfected with photolyase plasmids (or control vector) 48 h prior to UV irradiation. Cell culture medium was replaced with 15 µM EdU-containing medium 1 h prior to UV irradiation (47 h after transfection), and cells were incubated at 37 °C for 1 h. UVB irradiation (48 h after transfection) and subsequent photorepair were performed as described above. After 2 h of visible light illumination for photorepair, HBSS was replaced with warm cell culture medium, and cells were incubated at 37 °C for 7 h without light illumination. Cell culture medium was then replaced with 20 µM BrdU-containing medium for a subsequent 1-h incubation at 37 °C. After the 1-h incubation with BrdU (total 10 h post-UV), cells on a 35-mm dish were harvested by trypsinization. Cell suspension was centrifuged at 200 × g for 5 min at 4 °C, and cells were resuspended in 1 mL of cold PBS. To fix cells with 2% formaldehyde, 143 µL of 16% paraformaldehyde (formaldehyde) aqueous solution (15710, Electron Microscopy Sciences) was added into 1 mL of cell suspension, followed by incubation of cells in a 37 °C water bath for 10 min. Fixed cells were centrifuged at 700 × g for 5 min at 4 °C and were washed with 1 mL of cold PBS. Cells were pelleted by centrifugation at 700 × g for 10 min at 4 °C and then resuspended in ice-cold 90% methanol and incubated at −20 °C overnight for permeabilization. To detect UV lesions regardless of whether or not lesions were surrounded by ssDNA, cells were centrifuged at 700 × g for 5 min at 4 °C (this centrifugation speed and time was used hereafter), washed with 500 µL PBS, and then incubated with 125 µL of DNase reaction buffer containing 12.5 units of RQ1 RNase-free DNase (M6101, Promega) at 37 °C for 1 h. DNase-treated cells were centrifuged and washed with 1 mL of 1% bovine serum albumin (BSA) (A9647, Sigma-Aldrich) in PBS, and cells were incubated in 200 µL of 1% BSA in PBS at room temperature for 10 min for blocking. Each sample was split into three aliquots for different staining patterns: 1) Pacific Blue (PB) for EdU, phycoerythrin (PE) for His tag, and Alexa Fluor 647 for CPD; 2) PB for EdU, PE for His tag, and Alexa Fluor 647 for 6-4PP; and 3) PB for EdU, PE for His tag, Alexa Fluor 647 for BrdU, and Alexa Fluor 488 for phosphorylation of Chk1 at Ser345. Each split sample (equivalent to half of the cells from a 35-mm dish) was stained with 100 µL of antibody dilution buffer (0.25% Tween-20-containing 1% BSA in PBS) containing the following primary antibodies at the indicated dilutions: anti-CPD mouse monoclonal IgG2aκ (1:1,000, clone TDM-2, CAC-NM-DND-001, Cosmo Bio), anti-6-4PP mouse monoclonal IgG2aκ (1:1,000, clone 64M-2, CAC-NM-DND-002, Cosmo Bio), anti-BrdU mouse monoclonal (1:25, clone MoBU-1, B35141, Thermo Fisher Scientific), or anti-phospho-Chk1 (Ser345) (133D3) rabbit monoclonal (1:100, 2348, Cell Signaling Technology) antibodies. After overnight incubation with primary antibodies at 4 °C, cells were centrifuged and washed twice with 1 mL wash buffer (0.05% Tween-20-containing 1% BSA in PBS). For EdU detection, Pacific Blue azide was conjugated to EdU via CuSO4-mediated click chemistry reaction for 30 min at room temperature using the Click-iT EdU Pacific Blue Flow Cytometry Assay Kit (C10418, Thermo Fisher Scientific; 250-µL reaction volume for one sample). After washing cells twice with 1 mL wash buffer, cells were resuspended in 100 µL of antibody dilution buffer containing the following secondary antibodies at the indicated dilutions and incubated for 30 min at room temperature in the dark: Alexa Fluor 647-conjugated donkey anti-mouse IgG (H+L) (1:800, A31571, Thermo Fisher Scientific) for staining patterns 1 and 2; or Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) (1:200, A11034, Thermo Fisher Scientific) and Alexa Fluor 647-conjugated donkey anti-mouse IgG (H+L) (1:200, A31571, Thermo Fisher Scientific) for staining pattern 3. Cells were washed twice with 1 mL wash buffer, and the expression of His-tagged photolyase was detected by incubating cells in 100 µL of antibody dilution buffer containing anti-His-tag mouse monoclonal antibody (1:2,000, clone OGHis, D291-3, IgG2aκ, 1 µg/µL, MBL) that was conjugated with R-phycoerythrin (R-PE) using the Zenon R-PE Mouse IgG2a Labeling Kit (Z25155, Thermo Fisher Scientific) at an antigen-binding fragment (Fab):antibody molar ratio of 3:1. Following 1-h incubation at room temperature in the dark, cells were washed twice and resuspended in 200 µL PBS. Cells were analyzed on a FACSCanto II Flow Cytometer (BD Biosciences), and the acquired data were analyzed using FlowJo version 9 (Tree Star).
Detailed methods for flow cytometry experiments shown in Figs. 1, 2, 3B, and 5 B and C, and SI Appendix, Figs. S1–S8 are provided in SI Appendix, SI Materials and Methods.
Replication Track Analysis Combined with Flow Sorting.
For experiments quantitating DNA replication progression in cells before and after UV irradiation and photorepair (Fig. 4), XP-C cells (GM15983) were transfected with photolyase plasmids (or control vector) 48 h prior to UV irradiation. Thirty-four hours after transfection, cell culture medium was replaced with fresh medium to increase viability of transfected cells. Cell culture medium was replaced with 50 µM IdU-containing medium 1.5 h prior to UV irradiation (46.5 h after transfection) and incubated at 37 °C for 1 h. Cells were then rinsed with fresh medium twice and incubated in fresh medium without thymidine analogs for 30 min at 37 °C. UVB irradiation and subsequent photorepair were performed as mentioned above. Following 2 h of visible light illumination, HBSS was replaced with 10 µM EdU-containing medium. After 1-h incubation with EdU at 37 °C, cells were rinsed with fresh medium twice and incubated in fresh medium without thymidine analogs for 30 min at 37 °C. Cells were then harvested by trypsinization (3.5 h after UV irradiation). In this experimental design, 30-min incubation in fresh medium without thymidine analogs prior to sham or UV irradiation was critical to eliminate a potential discrepancy in lengths of labeled DNA between UV and sham samples in the first labeling, because incorporation of thymidine analogs that were already in cells may be inhibited immediately after UV irradiation but not after sham irradiation. In addition, to have a consistent labeling schedule between the first (presham/UV) and second (postsham/UV) labeling periods, another 30-min incubation without thymidine analogs was included immediately before harvest. After harvest, cells were fixed with 2% formaldehyde and permeabilized with 90% methanol at −20 °C overnight as described above. After blocking with 1% BSA in PBS at room temperature for 10 min, the expression of His-tagged photolyase was detected by incubating cells (from a 35-mm dish) in 200 µL of antibody dilution buffer containing anti-His-tag mouse monoclonal antibody (1:2,000, clone OGHis, D291-3, IgG2aκ, 1 µg/µL, MBL) that was conjugated with R-PE using the Zenon R-PE Mouse IgG2a Labeling Kit (Z25155, Thermo Fisher Scientific) at a Fab:antibody molar ratio of 3:1. Following 1-h incubation at room temperature in the dark, cells were washed and resuspended in 100 µL of 1% BSA in PBS and sorted using a FACSAria Cell Sorter (BD Biosciences). Approximately 1 × 105 single cells that showed high signals of His tag (photolyase-expressing cells) were collected via flow sorting. Cells transfected with control vector (no photolyase) were collected without flow sorting. Flow-sorted (photolyase-expressing) and unsorted (control vector-transfected) cells were centrifuged at 700 × g for 5 min and washed with 500 µL agarose insert buffer (10 mM Tris pH 7.5, 20 mM NaCl, 50 mM EDTA solution in water). Cells were centrifuged again and resuspended in 30 µL agarose insert buffer. To embed cells in agarose, an equal amount of melted 2% low-gelling-temperature (LGT) DNase-free agarose was added to the cell suspension, and the mixed solution was immediately transferred into an agarose insert mold and incubated at 4 °C for 1 h. After solidification, the agarose gel insert was stored in agarose insert buffer in a 2-mL tube at 4 °C until use. To release genomic DNA from embedded cells, the agarose gel insert was incubated in 500 µL lysis buffer (100 mM EDTA pH 8.0, 0.2% sodium deoxycholate, 2% sodium lauryl sarcosine, 1 mg/mL proteinase K in water) at 55 °C overnight. After 55 °C incubation, the agarose gel insert was kept at room temperature for 5 min to harden the gel. The agarose gel insert was washed four times at room temperature in 1 mL 1× TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) for 30 min each on a rocking platform. To release DNA from the agarose gel insert, 300 µL 1× β-Agarase I reaction buffer (B0392S, New England Biolabs) was added to the agarose gel insert, which was then melted by heating at 71 °C for 10 min. Melted agarose was digested by adding 3 µL of β-Agarase I (1,000 units/mL, M0392L, New England Biolabs) and incubating at 42 °C for 20 h. After digestion of agarose, 30 µL of 10× TE buffer (pH 8.0) was added to the agarose-digested DNA solution to neutralize the β-Agarase buffer (pH 6.5). The final volume of DNA solution was ∼393 µL (30 µL agarose insert buffer, 30 µL 2% agarose, 300 µL 1× β-Agarase I reaction buffer, 3 µL β-Agarase I, 30 µL 10× TE buffer), and this DNA solution was used for maRTA and immuno-slot blot. To align DNA fibers on glass coverslips using microfluidics, maRTA was performed as previously described (38), with modifications. Detailed methods for maRTA are provided in SI Appendix, SI Materials and Methods.
Immuno-Slot Blot Assay.
For experiments validating the antibody specificity for UV lesions on ssDNA (Fig. 5A), UV-irradiated DNA was prepared using two different methods. For “in cells” UV irradiation samples, XP-C cells (GM15983) were irradiated with UVB 30 mJ/cm2 as described above and then harvested by scraping immediately after irradiation. Genomic DNA was extracted from these UV-irradiated cells using the QIAamp DNA Blood Mini Kit (51104, Qiagen) and RNase A (19101, Qiagen), and DNA concentrations were measured using the NanoDrop 1000 UV-Vis spectrophotometer (Thermo Fisher Scientific). Next, DNA solutions were prepared in 100 µL of PBS per sample to have 120 ng (for ssDNA detection), 200 ng (for CPD detection), or 800 ng (for 6-4PP detection) of genomic DNA. For “in vitro” UV irradiation samples, genomic DNA was extracted from unirradiated XP-C cells (GM15983) using the QIAamp DNA Blood Mini Kit and RNase A, and 100 µL of PBS containing the above-mentioned amount of extracted DNA was placed on a dish lid and irradiated with UVB 30 mJ/cm2. For heat denaturation, DNA solutions of in vitro and in cells UV irradiation samples in 1.5-mL tubes were boiled in 100 °C water for 10 min and rapidly chilled on ice for 15 min. Heat-denatured or nondenatured DNA solutions were spotted onto Hybond-N+ positively charged nylon membrane (RPN1210B, GE Healthcare) using the Minifold II Slot-Blot System (SRC 072/0, Schleicher & Schuell BioScience) under vacuum, and 100 µL of Milli-Q water was used to rinse each slot after loading DNA. Membranes were kept in the slot-blot system under vacuum for 10 min and then baked at 80 °C using a gel dryer (Savant SGD4050) for 2 h for DNA immobilization. After baking, membranes were incubated in blocking buffer (5% dry milk [170-6404, Bio-Rad], 0.1% sodium azide [S2002, Sigma-Aldrich], 0.1% Tween-20 in PBS) on a shaker at room temperature for 30 min. Each membrane was then incubated in 3 mL of buffer (5% dry milk, 0.1% Tween-20 in PBS) containing mouse monoclonal antibodies against ssDNA (1:150, clone F7-26, IgM, MAB3299, Millipore), CPD (1:2,000, TDM-2, IgG2aκ, Cosmo Bio), or 6-4PP (1:500, 64M-2, IgG2aκ, Cosmo Bio) on a shaker at 4 °C overnight. After washing three times (1 min each) using wash buffer (0.1% Tween-20 in PBS), membranes were incubated in 15 mL of buffer (1% dry milk, 0.1% Tween-20 in PBS) containing horseradish peroxidase (HRP)-linked goat anti-mouse IgM antibody (1:1,500, sc-2973, Santa Cruz Biotechnology) or HRP-linked whole antibody against mouse IgG (from sheep) (1:2,500, NA931, GE Healthcare) at room temperature for 30 min. Membranes were washed three times (15 min each), and chemiluminescence was assessed using 2 mL of Amersham ECL Western Blotting Detection Reagent (RPN2209, GE Healthcare) or 1 mL of Luminata Forte Western HRP Substrate (WBLUF0100, Millipore). Membranes were then exposed to Kodak BioMax XAR Film (165-1454, Carestream Health) for chemiluminescence detection. For total DNA (including dsDNA and ssDNA) detection as loading control, membranes were subsequently washed three times with wash buffer for 1 min each, rinsed with PBS, and incubated with 1× SYBR Gold Nucleic Acid Gel Stain (S11494, Thermo Fisher Scientific) that was diluted in PBS on a shaker at room temperature for 10 min. Membranes were then washed three times with wash buffer for 1 min each, and SYBR Gold signals were detected by FluorChem Q Imaging System (ProteinSimple) with 475-nm excitation channel and 537-nm emission filter.
For validation of lesion-specific photorepair in flow-sorted cells (Fig. 4B), DNA prepared for maRTA was used for immuno-slot blot assay. DNA concentrations were determined by interpolating from logarithmic trendline of band intensities of standard DNA in agarose gel electrophoresis. DNA isolated from sorted cells was diluted in 100 µL of PBS to have 30 ng (for CPD detection) or 120 ng (for 6-4PP detection) of DNA. DNA solutions were boiled, rapidly chilled, and spotted onto Hybond-N+ positively charged nylon membranes. Subsequent steps of immuno-slot blot assay and SYBR Gold staining were performed as described above.
Data Availability.
All data are presented within this paper and in the accompanying SI Appendix. Source data for Fig. 3C and SI Appendix, Fig. S7B have been provided in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
The authors thank Dr. Allan H. Conney for critical advice and support of this study, and Dr. Carlos F. M. Menck (University of São Paulo) for the gift of cDNAs of CPD-photolyase and 6-4PP-photolyase. This work was supported by the NIH (R01-AR049832 and R01-AR067722 to P.N. and R21-ES019485 to J.M.S.), a Dermatology Foundation Research Career Development Award (to M.K.), University of Washington Royalty Research Fund (to J.M.S.), and the Michael Piepkorn Endowment for Dermatology Research at University of Washington. Parts of this article were adapted from K.-F.H.’s doctoral dissertation (University of Washington).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917196117/-/DCSupplemental.
References
- 1.Rogers H. W., Weinstock M. A., Feldman S. R., Coldiron B. M., Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the U.S. population, 2012. JAMA Dermatol. 151, 1081–1086 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Mouret S. et al., Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. U.S.A. 103, 13765–13770 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciccia A., Elledge S. J., The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jans J. et al., Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 15, 105–115 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Sancar A., DNA excision repair. Annu. Rev. Biochem. 65, 43–81 (1996). [DOI] [PubMed] [Google Scholar]
- 6.DiGiovanna J. J., Kraemer K. H., Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 132, 785–796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J. K., Patel D., Choi B. S., Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: Implication in mutagenesis and repair activity. Photochem. Photobiol. 62, 44–50 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Perdiz D. et al., Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J. Biol. Chem. 275, 26732–26742 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Young A. R. et al., The in situ repair kinetics of epidermal thymine dimers and 6-4 photoproducts in human skin types I and II. J. Invest. Dermatol. 106, 1307–1313 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Nakajima S. et al., UV light-induced DNA damage and tolerance for the survival of nucleotide excision repair-deficient human cells. J. Biol. Chem. 279, 46674–46677 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Lo H. L. et al., Differential biologic effects of CPD and 6-4PP UV-induced DNA damage on the induction of apoptosis and cell-cycle arrest. BMC Cancer 5, 135 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudolph C. J., Upton A. L., Lloyd R. G., Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 21, 668–681 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A., Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes M., Foiani M., Sogo J. M., Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21, 15–27 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Wold M. S., Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Zou L., Elledge S. J., Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Dart D. A., Adams K. E., Akerman I., Lakin N. D., Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J. Biol. Chem. 279, 16433–16440 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Kemp M. G. et al., Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J. Biol. Chem. 285, 16562–16571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokes M. P. et al., Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H., Piwnica-Worms H., ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21, 4129–4139 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q. et al., Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000). [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Girona A. et al., Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 98, 11289–11294 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santocanale C., Diffley J. F., A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395, 615–618 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Toledo L. I. et al., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Nghiem P., Park P. K., Kim Y., Vaziri C., Schreiber S. L., ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. U.S.A. 98, 9092–9097 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helt C. E., Cliby W. A., Keng P. C., Bambara R. A., O’Reilly M. A., Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J. Biol. Chem. 280, 1186–1192 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Heffernan T. P. et al., ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: Potential basis for the UV protective effects of caffeine. J. Invest. Dermatol. 129, 1805–1815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp M. G., Sancar A., ATR kinase inhibition protects non-cycling cells from the lethal effects of DNA damage and transcription stress. J. Biol. Chem. 291, 9330–9342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmert S., Kobayashi N., Khan S. G., Kraemer K. H., The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc. Natl. Acad. Sci. U.S.A. 97, 2151–2156 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adimoolam S., Lin C. X., Ford J. M., The p53-regulated cyclin-dependent kinase inhibitor, p21 (cip1, waf1, sdi1), is not required for global genomic and transcription-coupled nucleotide excision repair of UV-induced DNA photoproducts. J. Biol. Chem. 276, 25813–25822 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Rouget R., Auclair Y., Loignon M., Affar B., Drobetsky E. A., A sensitive flow cytometry-based nucleotide excision repair assay unexpectedly reveals that mitogen-activated protein kinase signaling does not regulate the removal of UV-induced DNA damage in human cells. J. Biol. Chem. 283, 5533–5541 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Mori T. et al., Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 54, 225–232 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Takagi S., McFadden M. L., Humphreys R. E., Woda B. A., Sairenji T., Detection of 5-bromo-2-deoxyuridine (BrdUrd) incorporation with monoclonal anti-BrdUrd antibody after deoxyribonuclease treatment. Cytometry 14, 640–648 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Hu J., Adar S., Selby C. P., Lieb J. D., Sancar A., Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Lima-Bessa K. M. et al., CPDs and 6-4PPs play different roles in UV-induced cell death in normal and NER-deficient human cells. DNA Repair (Amst.) 7, 303–312 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Sancar A., Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Arlett C. F. et al., Clinical and cellular ionizing radiation sensitivity in a patient with xeroderma pigmentosum. Br. J. Radiol. 79, 510–517 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Sidorova J. M., Li N., Schwartz D. C., Folch A., Monnat R. J. Jr., Microfluidic-assisted analysis of replicating DNA molecules. Nat. Protoc. 4, 849–861 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darzynkiewicz Z., Halicka H. D., Zhao H., Podhorecka M., Cell synchronization by inhibitors of DNA replication induces replication stress and DNA damage response: Analysis by flow cytometry. Methods Mol. Biol. 761, 85–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shachar S. et al., Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 28, 383–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prakash S., Johnson R. E., Prakash L., Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 74, 317–353 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Elvers I., Johansson F., Groth P., Erixon K., Helleday T., UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 39, 7049–7057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bomgarden R. D. et al., Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 25, 2605–2614 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Despras E., Daboussi F., Hyrien O., Marheineke K., Kannouche P. L., ATR/Chk1 pathway is essential for resumption of DNA synthesis and cell survival in UV-irradiated XP variant cells. Hum. Mol. Genet. 19, 1690–1701 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Berezney R., Dubey D. D., Huberman J. A., Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108, 471–484 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Blackford A. N. et al., Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. U.S.A. 107, 12251–12256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y. W. et al., Human DNA polymerase eta activity and translocation is regulated by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 16578–16583 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinet A. et al., Translesion synthesis mechanisms depend on the nature of DNA damage in UV-irradiated human cells. Nucleic Acids Res. 44, 5717–5731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawasumi M. et al., Protection from UV-induced skin carcinogenesis by genetic inhibition of the ataxia telangiectasia and Rad3-related (ATR) kinase. Proc. Natl. Acad. Sci. U.S.A. 108, 13716–13721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conney A. H., Lu Y. P., Lou Y. R., Kawasumi M., Nghiem P., Mechanisms of caffeine-induced inhibition of UVB carcinogenesis. Front. Oncol. 3, 144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abel E. L. et al., Daily coffee consumption and prevalence of nonmelanoma skin cancer in Caucasian women. Eur. J. Cancer Prev. 16, 446–452 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Song F., Qureshi A. A., Han J., Increased caffeine intake is associated with reduced risk of basal cell carcinoma of the skin. Cancer Res. 72, 3282–3289 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Loftfield E. et al., Coffee drinking and cutaneous melanoma risk in the NIH-AARP diet and health study. J. Natl. Cancer Inst. 107, dju421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marionnet C., Armier J., Sarasin A., Stary A., Cyclobutane pyrimidine dimers are the main mutagenic DNA photoproducts in DNA repair-deficient trichothiodystrophy cells. Cancer Res. 58, 102–108 (1998). [PubMed] [Google Scholar]
- 55.Kraemer K. H. et al., Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience 145, 1388–1396 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moiseeva T. N. et al., An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 116, 13374–13383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moiseeva T. N., Bakkenist C. J., Dormant origin signaling during unperturbed replication. DNA Repair (Amst.) 81, 102655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Klein A. et al., Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 10, 479–482 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Brown E. J., Baltimore D., Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17, 615–628 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takezawa J., Aiba N., Kajiwara K., Yamada K., Caffeine abolishes the ultraviolet-induced REV3 translesion replication pathway in mouse cells. Int. J. Mol. Sci. 12, 8513–8529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh T. et al., Xeroderma pigmentosum variant heterozygotes show reduced levels of recovery of replicative DNA synthesis in the presence of caffeine after ultraviolet irradiation. J. Invest. Dermatol. 115, 981–985 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Jansen J. G. et al., Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol. Cell. Biol. 29, 3113–3123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Legerski R., Peterson C., Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature 359, 70–73 (1992). [DOI] [PubMed] [Google Scholar]
- 64.Abbaszadeh F. et al., A novel splice variant of the DNA-PKcs gene is associated with clinical and cellular radiosensitivity in a patient with xeroderma pigmentosum. J. Med. Genet. 47, 176–181 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Chiganças V., Sarasin A., Menck C. F., CPD-photolyase adenovirus-mediated gene transfer in normal and DNA-repair-deficient human cells. J. Cell Sci. 117, 3579–3592 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented within this paper and in the accompanying SI Appendix. Source data for Fig. 3C and SI Appendix, Fig. S7B have been provided in SI Appendix, Table S1.