Significance
HIF2α protein up-regulation, caused by loss of VHL tumor suppressor, is a major oncogenic driver in clear cell renal cell carcinoma (ccRCC). It is well-known that HIF2α is targeted by VHL E3 ligase complex for degradation through the ubiquitin-proteasome system. However, whether HIF2α is regulated by deubiquitinase remains largely unknown. In this study, we identified USP37 as a HIF2α deubiquitinase that can bind with and promote HIF2α protein stability. As a result, loss of USP37 can decrease ccRCC cell proliferation as well as ccRCC tumor growth in an orthotopic xenograft model. Our results suggest that USP37 is a potential therapeutic target in ccRCC.
Keywords: HIF2α, USP37, deubiquitination, ccRCC
Abstract
Clear cell renal cell carcinoma (ccRCC) is characterized by loss of tumor suppressor Von Hippel Lindau (VHL) function, which leads to accumulation of hypoxia inducible factor α (including HIF1α and HIF2α). HIF2α was previously reported to be one of the major oncogenic drivers in ccRCC, however, its therapeutic targets remain challenging. Here we performed a deubiquitinase (DUB) complementary DNA (cDNA) library binding screen and discovered that ubiquitin-specific peptidase 37 (USP37) is a DUB that binds HIF2α and promotes HIF2α deubiquitination. As a result, USP37 promotes HIF2α protein stability in an enzymatically dependent manner, and depletion of USP37 leads to HIF2α down-regulation in ccRCC. Functionally, USP37 depletion causes decreased cell proliferation measured by MTS, two-dimensional (2D) colony formation as well as three-dimensional (3D) anchorage- independent growth. USP37 is also essential for maintaining kidney tumorigenesis in an orthotopic xenograft model and its depletion leads to both decreased primary kidney tumorigenesis and spontaneous lung metastasis. Our results suggest that USP37 is a potential therapeutic target in ccRCC.
Kidney cancer incidence has been increasing steadily for the past several decades although the reasons for this are unclear. Von Hippel Lindau (VHL) is the most important tumor suppressor gene in kidney cancer and is lost or mutated in over 70% of cases (1). Work from many laboratories shows that the pVHL-associated complex has E3 ubiquitin ligase activity, and VHL loss leads to accumulation of hypoxia inducible factor α (HIF-α, including HIF1α and HIF2α) as well as other potential substrates including ZHX2 and SFMBT1 (1–5). As a result of accumulation and translocation of HIF-α factors into the nucleus, dimerization occurs with a constitutively expressed HIFβ-subunit, which transactivates genes involved in angiogenesis (e.g., VEGF), glycolysis, and glucose transport (e.g., GLUT1), and erythropoiesis (e.g., EPO) (6). In kidney cancer, HIF2α is well established to be an oncoprotein that is sufficient and necessary to promote xenograft tumor growth (7, 8). HIF2α depletion decreased xenograft tumor growth in models using pVHL-deficient kidney cancer cell lines, and HIF2α overexpression can override the tumor suppressor effect of pVHL (7–9). Therefore, targeting HIF2α is an important therapeutic target in kidney cancer especially with pVHL loss. However, it is challenging to target HIF2α therapeutically. For example, recently developed HIF2α inhibitors could only inhibit the tumor growth in certain preclinical kidney cancer models but not others (10, 11). These inhibitors were designed to inhibit HIF2α transcriptional activity, rather than its protein stability. Therefore, it remains critical to identify the pathway regulating HIF2α protein stability, which potentially could be therapeutically targeted.
Substrate ubiquitination is regulated by a delicate balance between E3 ligase and deubiquitinases (DUBs). Importantly, inactivating the DUB for a particular substrate can tip the balance in favor of ubiquitination and lead to rapid substrate degradation. While HIF2α can undergo ubiquitination and degradation mediated by pVHL E3 ligase complex, it remains unclear whether HIF2α can also be regulated by deubiquitination pathways. Furthermore, it remains critical to identify the deubiquitinase that may regulate HIF2α protein stability such that development of deubiquitinase inhibitors may be beneficial for potential kidney cancer therapy. In our current study, we identify that ubiquitin-specific peptidase 37 (USP37) serves as a deubiquitinase for HIF2α. As a result, USP37 promotes HIF2α protein stability and kidney tumorigenesis. Our study therefore suggests that USP37 may be a therapeutic target in kidney cancer.
Results
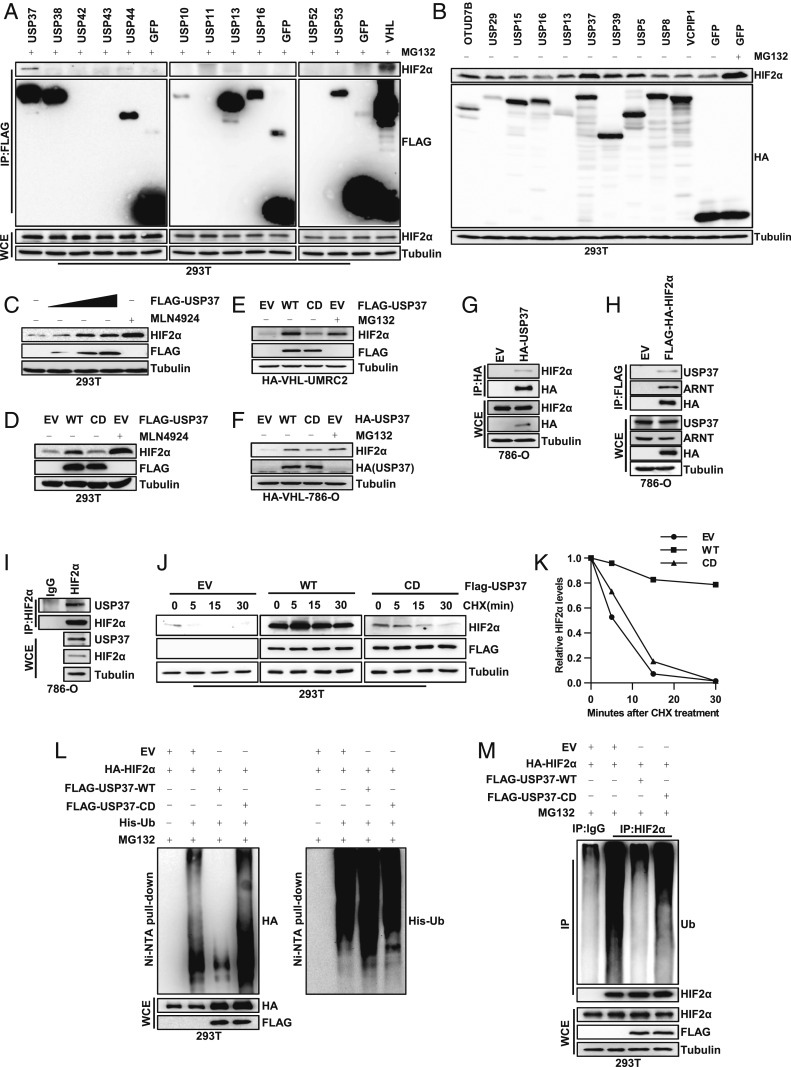
In order to identify the potential deubiquitinase(s) that regulates HIF2α, we obtained a FLAG (DYKDDDDK) and HA (human influenza hemagglutinin) double tagged deubiquitinase (DUB) complementary DNA (cDNA) library that contains 60 deubiquitinases, as described previously (12). First, we transfected 293T cells with these DUBs followed by the proteasomal inhibitor MG132 treatment and immunoprecipitation with FLAG antibody-conjugated beads. Among the interacting proteins, we found that some of DUBs displayed binding with endogenous HIF2α, including OTUD7B/Cezanne, USP29, USP39, and USP37 (SI Appendix, Fig. S1 A–D and Fig. 1A). Next, to examine whether these potential HIF2α binding DUBs may regulate HIF2α protein levels, we overexpressed this subset of DUBs in 293T cells followed by examination of HIF2α protein levels. Among these DUBs, USP37 displayed the most robust up-regulation of HIF2α, to a similar extent to these cells treated with the proteasome inhibitor MG132 or NAE (Nedd8 activating enzyme) inhibitor MLN4924 (Fig. 1 B and C). HIF2α also displayed a dose-dependent up-regulation upon USP37 overexpression (Fig. 1C). To examine whether this regulation of HIF2α is dependent on the enzymatic activity of USP37, we overexpressed wild-type (WT) USP37 or the USP37 C350S catalytic dead (CD) mutant in different cell lines, including 293T cells, UMRC2, or 786-O expressing exogenous HA-VHL. Consistently, WT, but not catalytic inactive USP37, up-regulated HIF2α protein levels (Fig. 1 D–F), suggesting that USP37 promotes HIF2α protein stability in an enzymatically dependent manner.
Fig. 1.
USP37 interacts with HIF2α and regulates HIF2α protein degradation. (A) Immunoblots of whole cell extracts (WCE) and immunoprecipitations (IP) from 293T cells transfected with Flag-tagged proteins as indicated and then treated with 10 μM MG132 under confluent condition. (B) Immunoblots of lysates from 293T cells transfected with HA-tagged proteins and then treated with 10 μM MG132 as indicated. Cells were treated under confluent condition to accumulate a high amount of HIF2α. (C) Immunoblots of lysates from 293T cells transfected with Flag-USP37 and then treated with 1 μM MLN4924 as indicated. (D) Immunoblots of lysates from 293T cells transfected with EV, WT, or CD form of USP37 and then treated with 1 μM MLN4924 as indicated. (E and F) Immunoblots of lysates from HA-VHL restored UMRC2 or 786-O cells infected with lentivirus encoding either EV, WT, or CD form of USP37 and then treated with 10 μM MG132 as indicated. (G–I) Immunoblots of WCE and IP from 786-O cells infected with lentivirus encoding either EV, HA-USP37, or Flag-HA-HIF2α as indicated. (J) Immunoblots of lysates from 293T cells transfected with EV, WT, or CD form of USP37 and then treated with 10 μg/mL CHX for indicated time. (K) Quantification of HIF2α expression level shown in J. (L and M) Immunoblots of WCE, nickel nitrilotriacetic acid (Ni-NTA) pull-down, or IP from 293T cells transfected with indicated plasmids and then treated with 10 μM MG132 as indicated.
Since HIF2α is a major oncogenic driver in kidney cancer, especially clear cell renal cell carcinoma (ccRCC), next we aimed to determine whether USP37 binds with HIF2α and acts as its deubiquitinase in ccRCC cells. First, we performed reciprocal immunoprecipitation with either exogenous USP37 or HIF2α and found that HIF2α and USP37 interact (Fig. 1 G and H). Next, we examined endogenous interaction between HIF2α and USP37 in 786-O cells and indeed confirmed the interaction (Fig. 1I). Motivated by our findings with the binding and overexpression data for USP37 and HIF2α, we hypothesized that USP37 may promote HIF2α deubiquitination and therefore affect HIF2α protein stability. To test this, first we used cycloheximide (CHX), a protein synthesis inhibitor, to treat 293T cells transfected with either empty vector (EV), WT USP37 (WT) or CD USP37 (CD) and then monitored the HIF2α protein half-life over time. Importantly, WT USP37 expression promoted HIF2α protein stability, while EV or CD USP37 displayed faster HIF2α protein turnover (Fig. 1 J and K). Furthermore, we also examined HIF2α protein ubiquitination in cells upon USP37 overexpression. For both exogenous or endogenous settings, our results consistently showed that WT, but not CD USP37, decreased HIF2α ubiquitination (Fig. 1 L and M). Taken together, our results suggest that USP37 acts as a previously uncharacterized HIF2α deubiquitinase in kidney cancer.
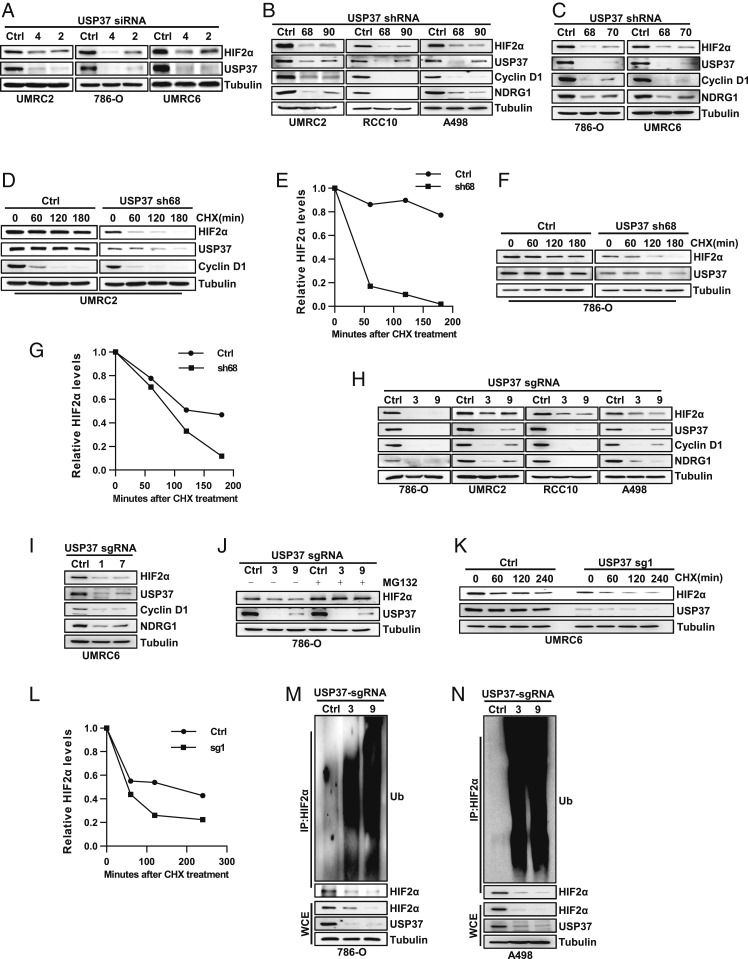
Next, to examine whether USP37 is required for HIF2α regulation in ccRCC cells, we tested multiple independent USP37 small interfering RNAs (siRNAs; #2, #4) in several ccRCC cell lines, including UMRC2, 786-O, and UMRC6 cells. USP37 depletion by these siRNAs led to decreased HIF2α protein levels in these cells (Fig. 2A). Here we also tested the effect of another two DUBs, USP39 and USP29, which also up-regulated HIF2α protein level in overexpression assay (Fig. 1B). Compared to USP37, knock down of USP39 or USP29 by siRNA had weak or no effect on HIF2α protein level (SI Appendix, Fig. S2 A and B). In addition, we also expressed multiple USP37 short hairpin RNAs (shRNAs) in UMRC2, RCC10, A498, 786-O, and UMRC6 cells. These independent shRNAs efficiently depleted endogenous USP37 protein levels and consistently led to decreased HIF2α protein levels in these cells (Fig. 2 B and C). In these settings, we also examined the protein level of some canonical HIF2α regulated genes, including CCND1 (encoding for Cyclin D1) and NDRG1, which displayed decreased protein levels upon USP37 depletion (Fig. 2 B and C). Next, we also wanted to examine whether USP37 depletion would affect HIF2α protein stability in these cells. To this end, we performed CHX pulse-chase experiments in UMRC2 and 786-O cells. In both cell lines, USP37 depletion led to decreased HIF2α protein stability (Fig. 2 D–G). To further strengthen our conclusion on the regulation of USP37 on HIF2α, we also designed multiple CRISPR-Cas9 guided single guide RNAs (sgRNAs) against USP37. These sgRNAs efficiently depleted USP37 protein levels in multiple ccRCC cell lines (786-O, UMRC2, RCC10, A498, and UMRC6), which corresponded with decreased protein levels for HIF2α as well as its downstream target protein expression, including Cyclin D1 and NDRG1 (Fig. 2 H and I). USP37-depletion mediated HIF2α down-regulation can be ameliorated by the proteasomal inhibitor MG132 treatment (Fig. 2J), suggesting that USP37 mainly affects HIF2α protein stability. Indeed, CHX pulse-chase experiments showed that USP37 depletion profoundly shortened the HIF2α protein half-life in UMRC6 cells (Fig. 2 K and L). Furthermore, we also examined HIF2α ubiquitination levels in 786-O and A498 cells. In both cell lines, depletion of USP37 by independent sgRNAs promoted HIF2α ubiquitination, which corresponded with decreased HIF2α protein levels in these cells (Fig. 2 M and N). In summary, our data suggest that USP37 depletion leads to decreased HIF2α by promoting HIF2α ubiquitination and degradation. Therefore, USP37 serves as a bona fide deubiquitinase for HIF2α in ccRCC.
Fig. 2.
Loss of USP37 destabilizes HIF2α by promoting HIF2α ubiquitination in kidney cancer cells. (A–C) Immunoblots of lysates from indicated cells transfected siRNA targeting USP37 or from cells infected with lentivirus encoding shRNA targeting USP37.(D–G) Immunoblots of lysates from UMRC2 or 786-O cells infected with either Ctrl shRNA or USP37 sh68 and then treated with 10 μg/mL CHX for indicated time. E and G are quantification of HIF2α expression level shown in D and F. (H and I) Immunoblots of lysates from indicated cells infected with either Ctrl sgRNA or sgRNA targeting USP37. (J–L) Immunoblots of lysates from indicated cells infected with either Ctrl sgRNA or sgRNA targeting USP37 and then treated with 10 μM MG132 or 10 μg/mL CHX. L is quantification of HIF2α expression level shown in K. (M and N) Immunoblots of whole cell extracts (WCE) and immunoprecipitations (IP) from 786-O and A498 cells infected with lentivirus encoding either Ctrl sgRNA or sgRNA targeting USP37.
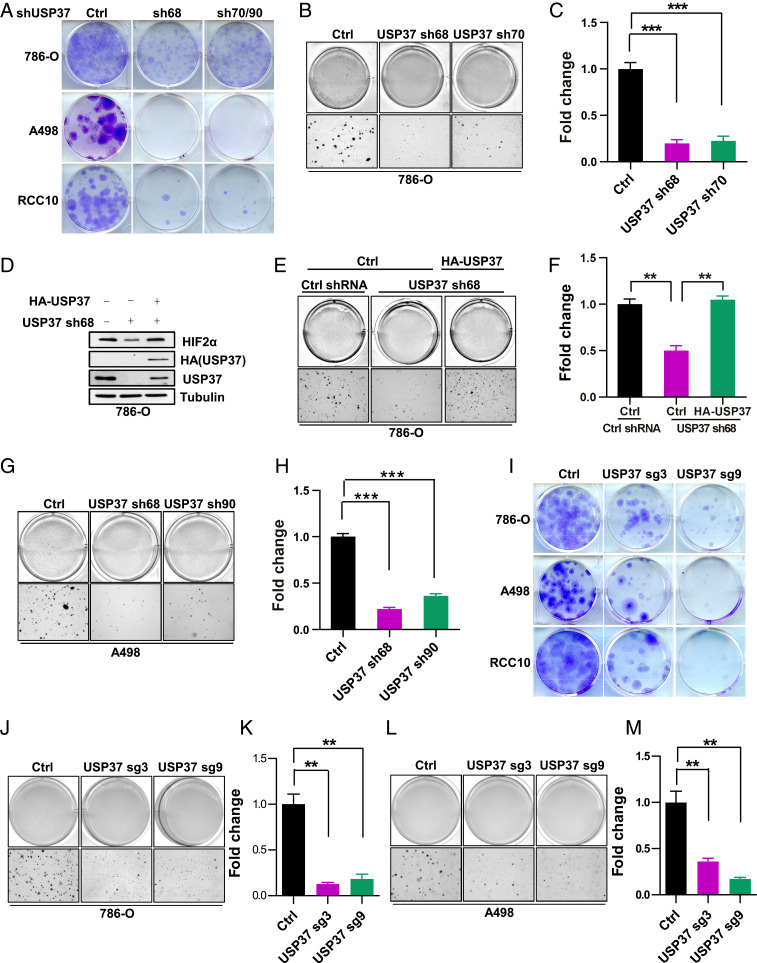
Given the important role of USP37 regulating HIF2α protein stability and that HIF2α is a critical oncoprotein in ccRCC, we aimed to examine the functional role of USP37 depletion in ccRCC. It is important to point out that only around 50% of ccRCC cell lines with VHL loss respond to HIF2α depletion or PT2385, a HIF2α specific inhibitor (10). Thus we selected a few cell lines that respond to HIF2α depletion to examine the functional role of USP37. First, we performed cell proliferation MTS assay and two-dimensional (2D) colony growth assays for ccRCC cells (786-O, A498, and RCC10). We found that USP37 depletion by multiple independent shRNAs led to decreased cell proliferation and colony formation (SI Appendix, Fig. S3 A–C and Fig. 3A). Next, we also examined three-dimensional (3D) soft agar growth with 786-O cells and consistently found that USP37 depletion led to decreased soft agar growth (Fig. 3 B and C). To determine whether the effect of the USP37 shRNA is on-target, we also infected 786-O cells with exogenous USP37 that is resistant to USP37 knockdown. Our soft agar growth assay showed that USP37 shRNA led to decreased HIF2α protein levels and soft agar growth, while both phenotypes were reversed by the USP37 shRNA-resistant cDNAs (Fig. 3 D–F), suggesting that the effect of USP37 shRNA on HIF2α and soft agar growth was on-target. We also used two independent USP37 shRNAs and examined their effect on soft agar growth in A498 cells. Consistently, USP37 depletion in these cells led to decreased soft agar growth (Fig. 3 G and H). To further confirm the phenotypes that we observed with USP37 depletion, we also examined colony growth on 2D or 3D in several different ccRCC cell lines with USP37 depletion induced by CRISPR-Cas9 sgRNAs. Concordantly, we observed that USP37 sgRNA-infected cell lines displayed decreased cell proliferation as measured by MTS assay, as well as decreased colony growth in 2D as well as in 3D (SI Appendix, Fig. S3 D–F and Fig. 3 I–M). It is worth mentioning that we also depleted USP37 in UMRC2 cells, the cell line that was shown to be resistance toward HIF2α depletion or inhibition and found that USP37 depletion also led to decreased cell proliferation in 2D and 3D for these cells (SI Appendix, Fig. S4 A–C). This may reflect the possibility that USP37 might regulate other substrates besides HIF2α that are important for UMRC2 cell proliferation. In summary, our data suggest that USP37 is necessary for cell proliferation of ccRCC cell lines.
Fig. 3.
Loss of USP37 suppresses ccRCC cell growth. (A) Representative crystal violet staining of indicated cells infected with either Ctrl shRNA or shRNA targeting USP37 (sh70 for 786-O, sh90 for A498 and RCC10). (B and C) Representative 3D soft agar growth pictures and quantification of colony numbers (triplicate wells) of 786-O cells infected with either Ctrl shRNA or shRNA targeting USP37. (D–F) Immunoblots of lysates (D), representative 3D soft agar growth pictures (E), and quantification of colony numbers (F, triplicate wells) of 786-O cells infected with lentivirus encoding indicated genes. (G and H), Representative 3D soft agar growth pictures and quantification of colony numbers (triplicate wells) of A498 cells infected with Ctrl shRNA or shRNA targeting USP37. (I) Representative crystal violet staining of indicated cells infected with either Ctrl sgRNA or sgRNA targeting USP37. (J–M) Representative 3D soft agar growth pictures and quantification of colony numbers (triplicate wells) of 786-O or A498 cells infected with either Ctrl sgRNA or sgRNA targeting USP37. Error bars represent SEM, **P < 0.01, ***P < 0.001.
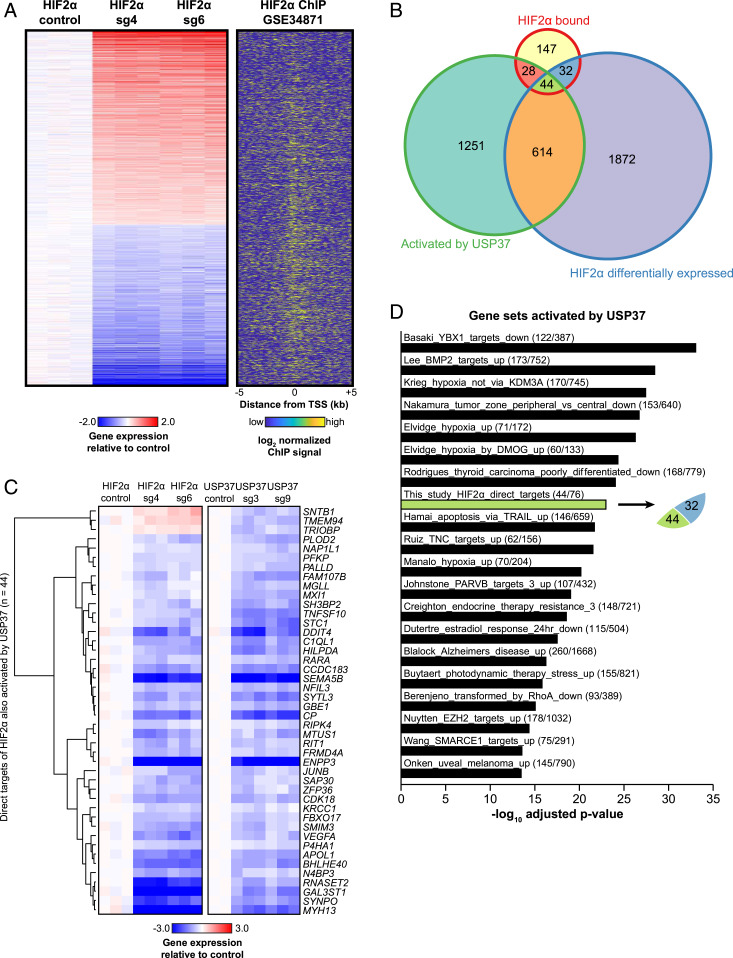
To determine the genome-wide level regulation induced by either USP37 or HIF2α, we performed RNA sequencing (RNA-seq) studies for 786-O cells with control (sgCtrl), USP37 sgRNAs (#3, #9), or HIF2α sgRNAs (#4, #6). Our Western blot showed that HIF2α and Cyclin D1 levels were efficiently down-regulated in these sgRNA-expressing ccRCC cells (SI Appendix, Fig. S5). In order to specifically examine HIF2α direct target genes in ccRCC, we integrated these data with previously published HIF2α chromatin immunoprecipitation sequencing (ChIP-seq). First, we generated a list of differentially expressed genes upon HIF2α depletion by both HIF2α sgRNAs (2562 total genes, adj.P < 0.05) (Fig. 4A). Next, we determined the overlap of these genes with HIF2α binding sites from the same cell line and derived 76 HIF2α direct target genes that displayed both HIF2α binding in the promoter (TSS ± 5 kb) as well as dysregulation upon HIF2α depletion by sgRNAs (Fig. 4B). Next, we found that 44 out of these 76 genes were differentially expressed following USP37 depletion in ccRCC (Fig. 4B). We found that the majority of these 44 genes shared very similar regulation by HIF2α and USP37 (Fig. 4C), including some of the canonical HIF2α direct target genes (e.g., DDIT4, VEGFA, and P4HA1). It is worth noting that CCND1 was not identified although Cyclin D1 was regulated by both USP37 and HIF2α at the protein level. This could be due to two possibilities. First, CCND1 was considered to be a HIF2α-regulated gene but did not meet our criteria for a direct target gene since HIF2α was reported not to directly bind the CCND1 proximal promoter but rather bind at a distal enhancer region (13). The second possibility could be the limitation of sequencing depth of previously published HIF2α ChIP-seq data (13). Nonetheless, analysis of over-enriched pathways among the genes activated by USP37 (down-regulated following USP37 sgRNA) did reveal that hypoxia is one of the most significant pathways enriched (Fig. 4D). Additionally, the overlap we observed between genes activated by USP37 and those we determined to be HIF2α direct targets was indeed statistically significant (adj.P = 1.1 × 10−23; Fig. 4D, green bar).
Fig. 4.
(A) Heatmap of genes differentially expressed following HIF2α depletion. Normalized expression is plotted such that values are centered around the mean of the control replicates. HIF2α ChIP-seq signal is plotted for the promoters (TSS ± 5 kb) of the genes shown, where yellow indicates higher enrichment. (B) Venn diagram showing overlapping gene numbers between HIF2α bound genes, genes differentially expressed following HIF2α depletion, and genes activated by USP37 (down-regulated following USP37 silencing). (C) Heatmap of the 44 genes determined to be direct targets of HIF2α. Normalized expression is plotted such that values are centered around the mean of the respective control replicates. (D) Top 20 significantly over-enriched pathways among genes activated by USP37, ranked by −log10 adjusted P value. The fractions given for each pathway represent the number of genes activated by USP37 relative to the total number of genes in the given pathway.
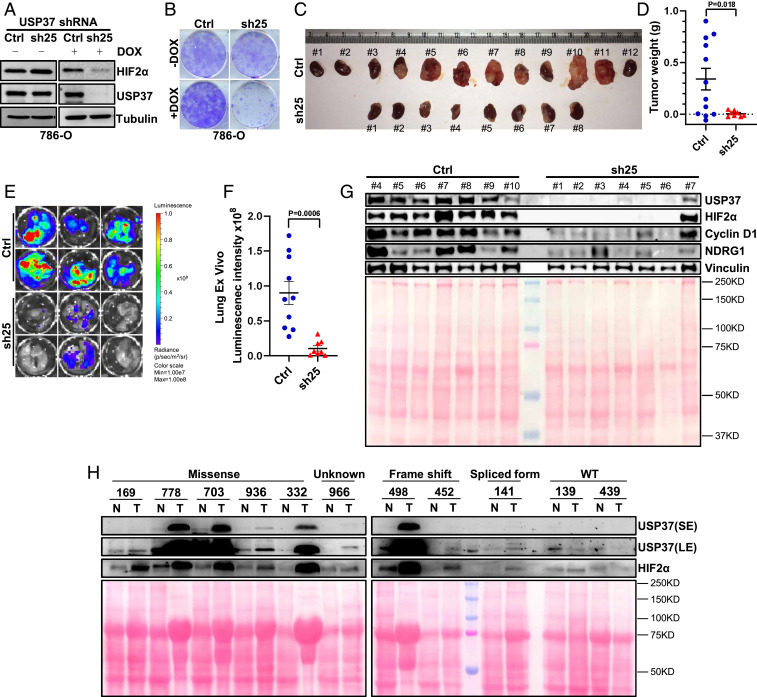
Next, to examine the ability of USP37 to maintain ccRCC tumor growth in vitro and in vivo, we generated 786-O cells infected with a doxycycline-inducible USP37 shRNA. Upon doxycycline addition, we can achieve efficient depletion of USP37 protein level in the cells, which corresponded with decreased colony formation (Fig. 5 A and B). Next, we expressed firefly luciferase in the doxycycline-inducible USP37 shRNA or control shRNA cells before injecting these cells orthotopically into the kidney capsules of NoD SCID Gamma (NSG)-deficient mice. We performed weekly bioluminescence imaging to ensure the successful implantation and growth of ccRCC tumor cells. Then, we fed these mice doxycycline chow and continued monitoring tumor growth over time. Upon necropsy, USP37 shRNA-infected ccRCC cells displayed reduced tumor burden retrieved from tumor-bearing mice compared to control mice (Fig. 5 C and D). We also measured the bioluminescence intensity ex vivo for spontaneous lung metastasis and found that USP37 depletion led to decreased lung metastasis (Fig. 5 E and F). In addition, tumor cell lysates from the USP37-depleted group displayed decreased HIF2α as well as HIF2α-regulated proteins, including NDRG1 and Cyclin D1 (Fig. 5G). To examine potential clinical relevance of USP37 in ccRCC, we generated cell lysates from ccRCC tumors as well as their corresponding normal tissues. Among 8 pairs of tissues that displayed VHL missense mutation, frame-shift mutation, or a splice variant, USP37 levels were up-regulated in tumor compared to normal tissue. In contrast, for the other two pairs of tissues that were VHL wild type, neither USP37 nor HIF2α showed distinctive up-regulation in ccRCC tumors compared to normal tissue (Fig. 5H). Taken together, our data strongly indicate that USP37 is a HIF2α deubiquitinase that contributes to VHL-deficient ccRCC tumorigenesis.
Fig. 5.
Loss of USP37 suppresses ccRCC tumor growth. (A and B) Immunoblots of lysates and representative crystal violet staining of 786-O cells infected with either inducible Ctrl shRNA or inducible shRNA targeting USP37 (sh25) and then treated with doxycycline as indicated. (C) Orthotopic kidney tumor formed by 786-O cells infected with either inducible Ctrl shRNA or inducible shRNA targeting USP37 (sh25). (D) Quantification of kidney tumor weight from Ctrl and sh25 group. (E and F) Representative pictures and quantification of lung ex vivo bioluminescence signal from Ctrl and sh25 group. (G) Immunoblots of lysates from representative tumors (C) containing either Ctrl shRNA or shRNA targeting USP37 (sh25). Total input protein amount is shown by Ponceau S staining. (H) Immunoblots of lysates from paired normal and kidney tumor tissues. VHL status is as labeled. Total input protein amount is shown by Ponceau S staining. SE, short exposure. LE, long exposure.
Discussion
In this study, by performing a screen for potential HIF2α DUBs, we identified USP37 as a critical DUB that regulates HIF2α ubiquitination and protein stability. In addition, we showed that USP37 depletion led to decreased cell proliferation, 2D colony growth, as well as 3D anchorage-independent growth in ccRCC. USP37 and HIF2α coregulated a subset of downstream target genes enriched for hypoxia signaling. Lastly, depletion of USP37 led to decreased ccRCC tumorigenesis in vivo using an orthotopic xenograft model. Therefore, our findings suggest that USP37 may be a therapeutic target in ccRCC. Since USP37 regulates HIF2α protein stability through its enzymatic activity, this study will motivate the development of USP37 inhibitors for potential treatment of ccRCC.
ccRCC is characterized by the lack of expression/function of the tumor suppressor VHL in most patients. VHL is the E3 ligase complex component that governs HIFα (including HIF1α and HIF2α) protein stability. Although HIF1α and HIF2α shared some common target genes since their response elements are largely identical, HIF1α preferentially regulates certain metabolic genes including BNIP3 and GLUT1 while HIF2α preferentially regulates NDRG1 and CCND1 (10). Previous results also showed that HIF1α serves as a tumor suppressor in kidney cancer (14) whereas HIF2α serves mainly as a potent oncoprotein (7). A recent discovery showed that the HIF2α-specific inhibitor PT-2385 disrupts the binding between HIF2α and HIF1β while not affecting HIF1α and HIF1β interaction (10, 11). PT-2385 elicited an inhibitory response in ∼50% of ccRCC cell lines by decreasing colony formation in 3D assays and also decreased the tumorigenesis/metastasis in some ccRCC tumors grown in immunodeficient mice or patient-derived xenografts grown in mice. Currently, this HIF2α inhibitor is in the recruitment phase of Phase 3 clinical trials. Although VHL regulates HIF2α protein ubiquitination and degradation, it was largely unclear how HIF2α may be regulated by deubiquitination. The DUB USP8 was previously suggested to regulate HIF2α since it serves as the deubiquitinase for HIF1α (15). However, we did not find robust interaction between HIF2α and USP8. In addition, USP8 overexpression did not affect HIF2α protein levels in cells, suggesting that USP37 is a HIF2α deubiquitinase. In addition, USP37 depletion in VHL null ccRCC cells also leads to decreased HIF2α protein levels, suggesting that there exists a VHL-independent regulation of HIF2α ubiquitination. Besides VHL-mediated HIF2α protein stability regulation, there exists multiple mechanisms that may regulate HIF2α protein levels in ccRCC. For example, Int6/eIF3e may target HIF2α for degradation in a VHL- and hypoxia-independent manner (16). Silencing Int6/eIF3e in subcutaneous (s.c.) tissues of mice promoted neoangiogenesis (17). Based on previous report, ubiquitinated HIF2α could be degraded by autophagy as well (18). From this perspective, the identification of USP37 could serve as a universal regulatory mechanism for HIF2α protein stability irrespective of VHL status.
USP37 has previously been reported to regulate ubiquitination of other targets, including Myc, Cyclin A, Wings Apart-Like (WAPL) and Cdt1. For example, previous results showed that USP37 promoted the deubiquitination of c-Myc in lung cancer (19), therefore promoting lung cancer cell proliferation. The potential effect of USP37 in lung tumorigenesis was not examined in xenograft or mouse models in that particular study. In another study, USP37 was shown to remove ubiquitin chains from Cyclin A (20). As a result, USP37 overexpression promoted Cyclin A accumulation and accelerated S phase entry, which suggests a potentially critical role for USP37 in regulating the cell cycle in a cancer setting. However, this study was not focused on any specific cancer type and did not examine the role of USP37 in tumorigenesis. Consistent with the potential role of USP37 in regulating cell cycle, previous research also showed that USP37 regulates WAPL, thereby affecting chromosomal cohesion and mitotic progression (21). Additionally, USP37 was also shown to deubiquitinate Cdt1, therefore regulating DNA replication (22). Our study examines the role of USP37 in kidney tumorigenesis, both using cell culture approaches and animal-based tumor models. The broader role of USP37 in regulating other substrates in cancers remains to be determined. However, identification of USP37 as a HIF2α deubiquitinase will hopefully lead to additional studies focused on USP37 as a therapeutic target in ccRCC. In addition, we observed that USP37 depletion also caused growth defects in ccRCC cell lines that were shown to be resistant to HIF2α depletion/inhibition, suggesting that USP37 may regulate the phenotype in these cells through HIF2α-independent mechanisms, such as by regulating other substrates. These findings will broaden the perspective of USP37 since its inhibition might be effective in both HIF2α-dependent as well as HIF2α-independent ccRCC.
Methods and Materials
Cell Culture.
The 786-O, A498, RCC10, UMRC2, and 293T cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS) plus 1% penicillin streptomycin. For lentivirus packaging and infection, following lentivirus infection, cells were maintained in the presence of blasticidin (10 μg/mL) or puromycin (2 μg/mL) depending on the vector. All cells were maintained at 37 °C with 5% CO2.
Immunoprecipitation and Western Blot.
Cells were lysed in EBC lysis buffer (50 mM Tris pH 8.0, 120 mM NaCl, 0.5% Nonidet P-40, 0.1 mM ethylenediaminetetraacetic acid and 10% glycerol) supplemented with complete protease inhibitors (Roche Applied Bioscience). The lysates were clarified by centrifugation and then mixed with primary antibodies, HA (Roche Applied Bioscience) or FLAG (Sigma-Aldrich) conjugated beads overnight. For primary antibody incubation overnight, cell lysates were incubated further with protein G sepharose beads (Roche Applied Bioscience) for 4 h. The bound protein complexes were washed with EBC buffer 3 times and then boiled in sodium dodecyl sulfate (SDS) loading buffer for 10 min. Bound proteins were resolved in SDS/polyacrylamide gel electrophoresis (PAGE) followed by Western blot analysis.
For direct Western blot assay, EBC buffer supplemented with complete protease inhibitor (Roche Applied Biosciences) was used to harvest whole cell lysates. Cell lysate concentrations were measured by Bradford assay. Equal amount of cell lysates was resolved by SDS/PAGE.
Antibodies and Reagents.
Rabbit anti-HA tag (3724), rabbit anti-FLAG tag (14793), mouse, anti–α-Tubulin (3873) were obtained from Cell Signaling Technology. Mouse anti-HIF2α (ab157249), mouse anti-NDRG1 (ab63989), rabbit anti-USP37 (ab72199), rabbit anti-USP37 (ab190184), and rabbit anti-Cyclin D1 (ab24249) were obtained from Abcam. Mouse anti-HIF2α (NB100-132) was obtained from Novus Biologicals. Mouse anti-ARNT (611078) was from BD Biosciences. Mouse anti-Ub (8017) was obtained from Santa Cruz. Peroxidase conjugated goat anti-mouse secondary antibody (31430) and peroxidase conjugated goat anti-rabbit secondary antibody (31460) were obtained from Thermo Scientific. MG132 (IZL-3175-v) was obtained from Peptide International, MLN4924 (SI9830) was obtained from LifeSensors, CHX (C7698) was obtained from Sigma-Aldrich.
Plasmids.
All DUB plasmids were purchased from Addgene. USP37 catalytic dead mutant (C350S) was generated by QuikChange XL Site-Directed Mutagenesis Kit (200516, Agilent Technologies). To generate lentivirus-based vector, USP37 coding sequence was cloned into pLenti6 vector by standard molecular cloning techniques.
Ubiquitination Assay.
Cells were collected and extracted in 100 μL of lysis buffer containing 1% SDS. Cell extracts were heat-denatured for 10 min and then diluted with 900 μL EBC lysis buffer supplemented with complete protease inhibitors (Roche Applied Bioscience) and 20 mM N-ethylmaleimide (E3876, Sigma). Diluted cell lysates were sonicated and clarified by centrifugation, followed by immunoprecipitation as described above. Immunoprecipitates were analyzed by Western Blot.
Lentiviral sgRNA, shRNA Vectors, and siRNA.
SgRNA or shRNA sequences targeting USP37 were cloned into pLenti CRISPR-V2 puro or pLKO puro vectors respectively. Doxycycline-inducible shUSP37 plasmids (V3IHSHEG_6429625, named sh25 in this study) and USP37/USP39/USP29 siRNA (siGENOME Human siRNA) were purchased from Horizon. The sh, sg target sequences are as follows: USP37 sh25: ATTTGGAGTTTGAGGTTCA; USP37 sh68: CCGGATTTGCAGAAGATGATA; USP37 sh70: GCTACCGAGTTAAGTCTTCAA; USP37 sh90: CGGAGTGGCTACATCTTCTTT; USP37 sg1: CTAGTAGTTCACTACAATAC; USP37 sg3: ATTGAATGAAGATTACCCTA; USP37 sg7: CTGTAATTCAGAGACCAAAA; USP37 sg9: TAGAATAGCATTCATATAGC; HIF2α sg4: AATCTCCTCATGGTCGCA; HIF2α sg6: TCATGAGGATGAAGTGCA.
Virus Production and Infection.
The 293T packaging cell line was used for lentivirus generation. Lentiviral infection was performed as previously described (23). Briefly, posttransfection with lipofectamine 3000, medium containing viruses was collected twice after 48 and 72 h. After passing through 0.45 μM filters, an appropriate amount of virus was used to infect target cells in the presence of 8 μg/mL polybrene (sc-134220, Santa Cruz). After incubation for additional 1 d, target cell lines were selected with the appropriate antibiotic.
Cell Proliferation Assays.
Cells were plated in 96-well plates (1,000 cells per well, triplicate) in 100 μL growth medium. At indicated time points, cells were replaced with 90 μL fresh growth medium supplemented with 10 μL MTS reagents (ab197010, Abcam) followed by incubation at 37 °C for 1 h. The absorbance optical density (OD) value was measured at 490 nm using a BioTek Synergy LX plate reader.
2D Colony Formation Assay.
To perform the 2D colony formation assay, cells infected with viruses were first selected for 4 to 6 d and then were plated in a 6-well plate (2,000 cells per well) in 1.5 mL growth medium. Medium was changed every 3 d for all wells. After growing for 2 to 3 wk, cells were incubated with fixation buffer (5% acetic acid, 5% methanol) for 30 min and then stained with 0.5% crystal violet solution for 30 min. After discarding crystal violet solution, pure water was added to wash away residual crystal violet.
Anchorage-Independent Growth Assay.
To perform the 3D soft agar growth assay, cells were plated at a density of 5,000 cells/mL for 786-O and 15,000 cells/mL for A498 in complete medium containing 0.4% agarose (BP165-25, Fisher Scientific), onto bottom layers composed of medium with 1% agarose. Afterward, cells were kept at 37 °C. For every 3 d, five drops of complete media were added onto the plate. After 4 wk for 786-O or 2 wk for UMRC2 and UMRC6 incubation, the liquid on the plate was aspirated and then colonies were stained by adding 500 μL medium containing 100 μg/mL iodonitrotetrazolium chloride (I8377, Sigma) into each well. After 1-d incubation, plates were scanned and colony numbers were counted by Image J.
Orthotopic Tumor Growth.
Six-week old female NOD SCID Gamma mice (NSG, Jackson Laboratory) were used for xenograft study. Approximately 5 × 105 viable 786-O cells were resuspended in 20 μL fresh growth medium and injected orthotopically into the left kidney of each mouse as described previously (24, 25). Bioluminescence imaging was performed as described previously (26). For inducible Teton-USP37 shRNA, after injection and following bioluminescence imaging over consecutive weeks to make sure tumors were successfully implanted in the kidney, mice were fed with purina rodent chow #5001 with doxycycline (Research Diets, Inc.); mice were euthanized 9 wk after treatment with doxycycline. All animal experiments were in compliance with NIH guidelines and were approved by the University of North Carolina at Chapel Hill Animal Care and Use Committee.
RNA-seq and ChIP-seq.
Total RNA from 786-O cells (triplicates) was extracted by using an RNeasy kit with on column DNase digestion (Qiagen). Libraries were prepared using TruSeq RNA Library Prep Kit v2 (Illumina) according to the manufacturer’s instructions. Samples were sequenced on an Illumina HiSeq2500 with paired-end 150 bp reads. Reads were then filtered for adapter contamination using cutadapt (27) and filtered such that at least 90% of bases of each read had a quality score >20. Reads were aligned to the reference genome (hg19) using STAR version 2.5.2b retaining only primary alignments (28). Reads overlapping blacklisted regions of the genome were then removed. Transcript abundance was then estimated using salmon (29), and differential expression was detected using DESeq2 (30). Variance-stabilizing transformed (VST) expression values were used for plotting. Pathway enrichment was performed using gProfiler (31), specifying a custom gene set database comprising MSigDB C7 gene sets and the 76 total direct target genes of HIF2α identified from this study.
ChIP-seq data for HIF2α were obtained from GSE34871, and reprocessed as above, except duplicated sequences were capped at a maximum of five occurrences. The filtered aligned reads were extended in silico to a fragment size of 250 bp, and regions of significant enrichment were identified using MACS2 (32).
Statistical Analysis.
Unless specifically indicated, the unpaired two-tail Student’s t test was used for experiments comparing two sets of data. Data represent mean ± SEM from three independent experiments. *, **, and *** denote P values < 0.05, 0.01, and 0.001, respectively. NS denotes not significant.
Data Availability.
RNA-seq and ChIP-seq reads have been deposited to Gene Expression Omnibus (GEO) under accession number GSE149005.
Supplementary Material
Acknowledgments
We thank the University of North Carolina (UNC) Lineberger Comprehensive Cancer Center Tissue Procurement Facility and UNC Animal Studies Core for excellent help. This work was supported in part by the National Cancer Institute (Q.Z., R01CA211732 and R21CA223675) and Cancer Prevention and Research Institute of Texas (CPRIT, RR190058 to Q.Z.). J.M.S. and T.S.P. were supported by The Eunice Kennedy Shriver National Institute of Child Health and Human Development (U54HD079124) and the National Institute of Neurological Disorders and Stroke (NINDS) (P30NS045892). Q.Z. is an American Cancer Society Research Scholar, CPRIT Scholar in Cancer Research, V Scholar, Kimmel Scholar, Susan G. Komen Career Catalyst awardee, and Mary Kay Foundation awardee. Q.Z. is also supported by the Kidney Cancer Research Alliance (KCCure).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq and ChIP-seq reads have been deposited to Gene Expression Omnibus (GEO) under accession number GSE149005.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002567117/-/DCSupplemental.
References
- 1.Kaelin W. G., Jr., Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2, 673–682 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Ivan M. et al., HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292, 464–468 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P. et al., Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Zhang J. et al., VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 361, 290–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X. et al., Genome-wide screening identifies SFMBT1 as an oncogenic driver in cancer with VHL loss. Mol. Cell 77, 1294–1306 e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., Kaelin W. G. Jr., Molecular pathogenesis of the von Hippel-Lindau hereditary cancer syndrome: Implications for oxygen sensing. Cell Growth Differ. 12, 447–455 (2001). [PubMed] [Google Scholar]
- 7.Kondo K., Klco J., Nakamura E., Lechpammer M., Kaelin W. G. Jr., Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1, 237–246 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Raval R. R. et al., Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25, 5675–5686 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo K., Kim W. Y., Lechpammer M., Kaelin W. G. Jr., Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, E83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho H. et al., On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W. et al., Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schodel J., et al. , Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat. Genet. 44, 420–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen C., Kaelin W. G. Jr., The VHL/HIF axis in clear cell renal carcinoma. Semin. Cancer Biol. 23, 18–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troilo A. et al., HIF1α deubiquitination by USP8 is essential for ciliogenesis in normoxia. EMBO Rep. 15, 77–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Uchida K., Endler A., Shibasaki F., Mammalian tumor suppressor Int6 specifically targets hypoxia inducible factor 2 alpha for degradation by hypoxia- and pVHL-independent regulation. J. Biol. Chem. 282, 12707–12716 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Chen L. et al., Int6/eIF3e silencing promotes functional blood vessel outgrowth and enhances wound healing by upregulating hypoxia-induced factor 2alpha expression. Circulation 122, 910–919 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Liu X. D. et al., Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene 34, 2450–2460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J. et al., USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene 34, 3957–3967 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Huang X. et al., Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol. Cell 42, 511–523 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Yeh C. et al., The deubiquitinase USP37 regulates chromosome cohesion and mitotic progression. Curr. Biol. 25, 2290–2299 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Hernández-Pérez S. et al., USP37 deubiquitinates Cdt1 and contributes to regulate DNA replication. Mol. Oncol. 10, 1196–1206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q. et al., Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell 16, 413–424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L. et al., SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell 24, 738–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu L. et al., TBK1 is a synthetic lethal target in cancer with VHL loss. Cancer Discov. 10, 460–475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J. et al., EglN2 associates with the NRF1-PGC1α complex and controls mitochondrial function in breast cancer. EMBO J. 34, 2953–2970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12 (2011). [Google Scholar]
- 28.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimand J. et al., Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y. et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq reads have been deposited to Gene Expression Omnibus (GEO) under accession number GSE149005.