Significance
Metabolism in a biological system is ultimately determined by the organisms’ ability to self-organize metabolism for maintaining cellular homeostasis. Quantification of the limits of metabolic robustness is essential for understanding cellular behavior. Using physiological, metabolomics, and proteomics data coupled to thermodynamic modeling, we show that metabolic oscillations in acetogens are controlled at the thermodynamic level. This work can contribute to advancing the understanding of an industrially relevant cell factory used in gas fermentation for sustainable production of fuels and chemicals from waste feedstocks.
Keywords: metabolic robustness, oscillations, acetogen, gas fermentation, Wood-Ljungdahl pathway
Abstract
Living biological systems display a fascinating ability to self-organize their metabolism. This ability ultimately determines the metabolic robustness that is fundamental to controlling cellular behavior. However, fluctuations in metabolism can affect cellular homeostasis through transient oscillations. For example, yeast cultures exhibit rhythmic oscillatory behavior in high cell-density continuous cultures. Oscillatory behavior provides a unique opportunity for quantitating the robustness of metabolism, as cells respond to changes by inherently compromising metabolic efficiency. Here, we quantify the limits of metabolic robustness in self-oscillating autotrophic continuous cultures of the gas-fermenting acetogen Clostridium autoethanogenum. Online gas analysis and high-resolution temporal metabolomics showed oscillations in gas uptake rates and extracellular byproducts synchronized with biomass levels. The data show initial growth on CO, followed by growth on CO and H2. Growth on CO and H2 results in an accelerated growth phase, after which a downcycle is observed in synchrony with a loss in H2 uptake. Intriguingly, oscillations are not linked to translational control, as no differences were observed in protein expression during oscillations. Intracellular metabolomics analysis revealed decreasing levels of redox ratios in synchrony with the cycles. We then developed a thermodynamic metabolic flux analysis model to investigate whether regulation in acetogens is controlled at the thermodynamic level. We used endo- and exo-metabolomics data to show that the thermodynamic driving force of critical reactions collapsed as H2 uptake is lost. The oscillations are coordinated with redox. The data indicate that metabolic oscillations in acetogen gas fermentation are controlled at the thermodynamic level.
Cells are capable of self-organizing their metabolism, using chemical reactions that constantly break and build molecules to extract energy. As energy is dissipated into the environment, intrinsic characteristic and dynamic chemical patterns emerge (1). Metabolism has evolved not merely to optimize a biological objective but, equally, to maintain metabolic robustness (i.e., capacity to maintain metabolic homeostasis) (2, 3). Metabolism is hardwired to anticipate and rapidly respond to abrupt internal and external perturbations through adjusting metabolic flux distributions to ensure cellular homeostasis.
Metabolic responses are generally fast, making it difficult to analyze their dynamics and understand the specific mechanisms underlying self-organization in a given species. However, some biological systems will spontaneously develop stable oscillations in continuous culture at high cell density (4–11). These oscillating cultures offer an opportunity to understand fundamental principles of metabolic regulation and what limits metabolism.
Oscillation in continuous yeast cultures has been studied extensively, and several theories have been developed. One possibility is that metabolite concentrations regulate gene expression and trigger natural oscillators such as the cell cycle and circadian oscillators. For example, it was observed that acetyl-coenzyme A (CoA) induces transcription of the G1 cyclin CLN3 in yeast (12). Another theory is that oscillations are an inevitable side effect of a trade-off between robustness and efficiency (13). Chandra et al. demonstrated, using a simple glycolysis two-state model, that in an autocatalytic system where 1 ATP is invested in the first step in return for 2 ATP in the second step, the feed-forward regulated system would tend to oscillate when pushed to its limit unless a vast excess of enzyme capacity was available for the first step (13). This hypothesis is aligned with the more general observation that the sophisticated regulatory behavior observed in living systems is a compromise between minimizing levels of metabolite pools and the ability to efficiently utilize enzymes (14).
Spontaneous oscillations have been studied extensively in heterotrophs like yeast and Escherichia coli with highly evolved, complex metabolisms. However, it is unclear whether the observations made for regulation of these systems translate to the regulation of primitive chemoautotrophic organisms. We recently reported oscillations in continuous cultures of the acetogen Clostridium autoethanogenum (15), where metabolism collapses, then recovers, and repeats without external interference (e.g., adding antibiotics, environmental shock). This self-oscillating system provides an excellent opportunity for deciphering the limits of metabolic robustness. Notably, acetogen metabolism operates at the limit of thermodynamic feasibility (16, 17) while utilizing CO, CO2, and H2 as carbon and energy sources, using the Wood-Ljungdahl pathway (WLP) (18, 19). Potential insights into the factors behind oscillations come from studies showing that maintenance of ATP homeostasis controls autotrophic carbon distribution (15), and that changes in metabolic flux rates are regulated at the posttranslational level (20, 21).
In this study, we replicated the oscillations to explore robustness of acetogen metabolism. We performed high-resolution, temporal sampling throughout the oscillatory cycles for proteomics and metabolomics analyses. Online gas analysis showed that the downcycle in biomass concentration coincided with a loss of H2 uptake, which was only restored after a prolonged period with CO as the sole energy and carbon source until cells recovered H2 uptake to replenish energy and accelerate growth. Thermodynamic metabolic flux analysis (tMFA) using endo- and exo-metabolomics data revealed that the thermodynamic driving force of several critical reactions collapsed at the time of the loss of H2 uptake. The results show that oscillations in an acetogen growing on syngas is a result of the highly efficient redox and energy metabolism combined with suppression of H2 metabolism by CO. Our data suggest that oscillations are potentially a mean of energy conservation, as previously suggested by others (22).
Results
Oscillatory Behavior of C. autoethanogenum Continuous Cultures.
We first triggered oscillations in continuous cultures of C. autoethanogenum (15) and performed extensive sampling throughout the cycles. Two biological replicate chemostat cultures at a dilution rate of 1 d−1 were grown on syngas (CO, H2, and CO2) as the carbon and energy sources. Oscillations were triggered at 800 rpm, at which point cultivation parameters remained constant. We reproducibly observed oscillations for ∼330 h (Fig. 1A), until the supply of the gas feed from cylinders was exhausted. Cultures were sampled around every 8 h for biomass, extracellular and intracellular metabolomics, and proteomics covering a total of four cycles. Gas uptake and production rates were obtained continuously from the online mass spectrometer attached to the bioreactors.
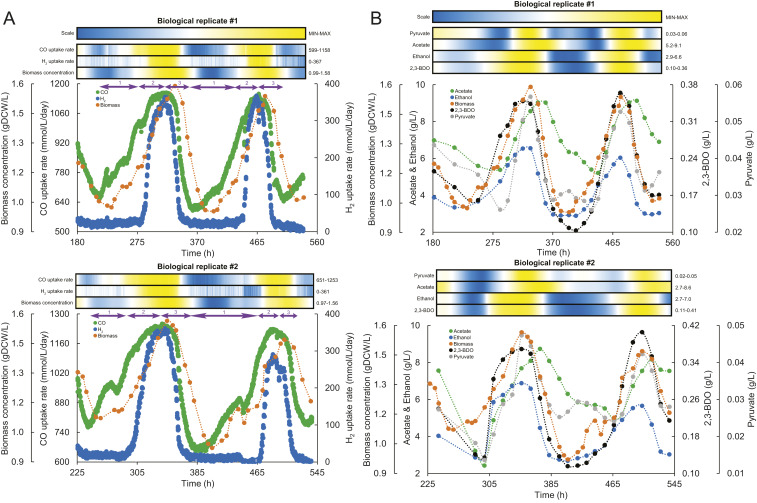
Fig. 1.
Oscillatory behavior of Clostridium autoethanogenum continuous cultures. (A) Oscillations in gas uptake rates and biomass concentration for two biological replicate cultures (#1 and #2). Numbers 1, 2, and 3 in purple font together with arrows above the graphs denote three phases (1, growth on CO; 2, growth on CO and H2; 3, crash). (B) Oscillations in extracellular metabolite levels and biomass concentration for two biological replicate cultures (#1 and #2). Heat maps show the relative change of the respective parameter between its minimum and maximum value (denoted on the right of heat map) with color scale shown on the top. Heat maps were generated with values derived from using spline (third-order polynomial cubic spline with point count 1) on the measured values to achieve higher resolution. time, fermentation duration from culture inoculation. 2,3-BDO, 2,3-butanediol.
Our first observation from the gas data showed that CO uptake rate (mmol/L/d) exhibited a nearly perfect synchronized oscillatory behavior with the biomass concentration (Fig. 1A), suggesting that continuous cultures were CO limited. The oscillation cycles had a period of ∼6 d, whereas the amplitudes for biomass concentration and CO uptake rate were ∼0.6 g dry cell weight (gDCW)/L (minimum, ∼1; maximum, ∼1.6) and ∼580 mmol/L/d (minimum, ∼600; maximum, ∼1,200), respectively. While it took the cultures ∼100 h to recover to maximum values, the decrease to minimum values took half the time (∼50 h). Strikingly, the H2 uptake rate was well synchronized with the previous characteristics, with an amplitude of ∼360 mmol/L/d (Fig. 1A). However, H2 uptake was lost entirely, in contrast to CO, showing that oscillations are composed of sequential growth phases: growth on CO, growth on CO and H2, and crash (slow growth) on CO as energy and carbon source (purple arrows in Fig. 1A).
The online gas analysis also revealed that cells immediately counterbalanced lowered supply of reduced ferredoxin (Fdred) from the loss of H2 uptake (see Fig. 3) by dissipating ∼1/3 more CO as CO2 (SI Appendix, Fig. S1). Interestingly, it took ∼24 h less for H2 uptake to recover compared wth the recovery of biomass concentration and CO uptake rate (Fig. 1A). Recovery of H2 uptake was potentially triggered by the culture becoming CO-limited again (SI Appendix, Fig. S2), as prerecovery excess levels of CO inhibited cellular hydrogenases, and thus H2 uptake (23–25).
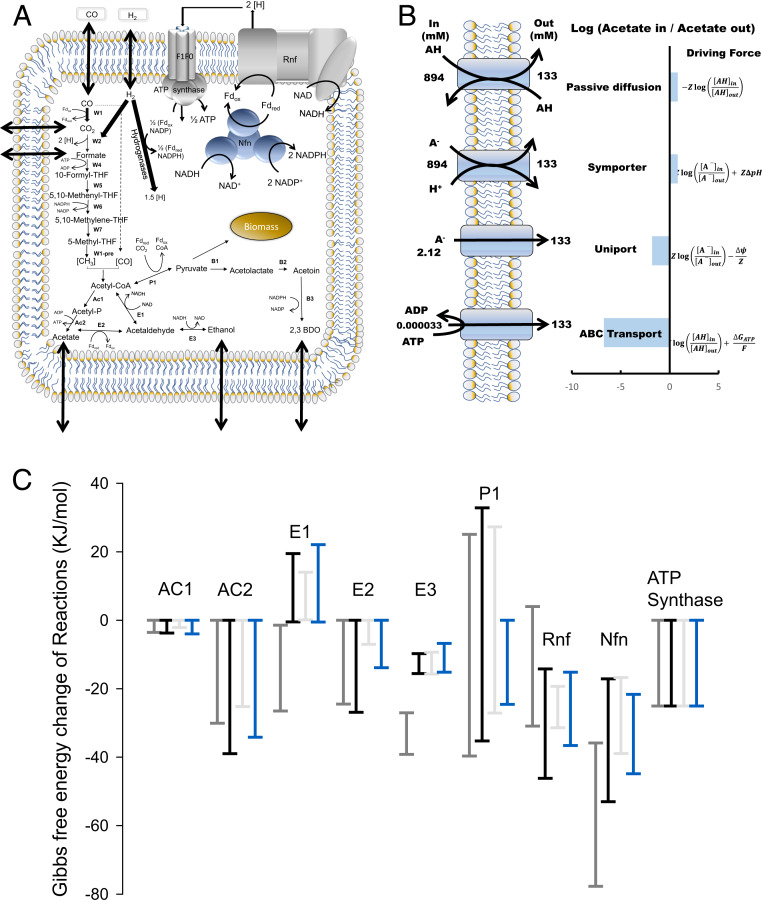
Fig. 3.
tMFA of C. autoethanogenum. (A) Schematic representation of our tMFA model of C. autoethanogenum. Arrows across the cell membrane denote product transport. (B) Modeling four mechanisms for acetate transport which were then used in our tMFA model. See Results and Dataset S1 . 2,3-BDO, 2,3-butanediol; THF, tetrahydrofolate; Fd, ferredoxin; A−, acetate; AH, acetic acid; H+, proton. (C) The plot shows the maximum allowable range of Gibbs free energy for respective reactions at given conditions calculated using thermodynamic variability analysis. Gray and black show the three phases from this work, and blue shows data from the steady state. See Results for details; Each bar denotes mean and error bars SD of that allowable range. *Positive reactions are thermodynamically not feasible at given conditions (have positive Gibbs free energy change).
No Changes in Protein Expression Levels Were Observed during Oscillations.
To establish whether changes in cellular protein expression levels were responsible for the oscillatory behavior, we conducted a thorough proteomics investigation, hypothesizing first that translational changes control oscillations (26). We performed quantitative proteomics using data-independent acquisition mass spectrometry (27) to confidently quantitate expression of 596 proteins on average, with at least two peptides per protein with high reproducibility (SI Appendix, Fig. S3A). We compared protein expression between samples with lowest (i.e., recovery) and highest (i.e., crash) biomass concentration (Figs. 1 and 2) within each biological replicate culture. Intriguingly, however, we detected no proteins being significantly differentially expressed [fold-change, >1.5; q-value, <0.05 after false discovery rate correction (28)] (SI Appendix, Fig. S3B). This result is consistent with previous observations that show that changes in protein expression do not regulate flux or product distribution during autotrophic growth of acetogens (20, 21).
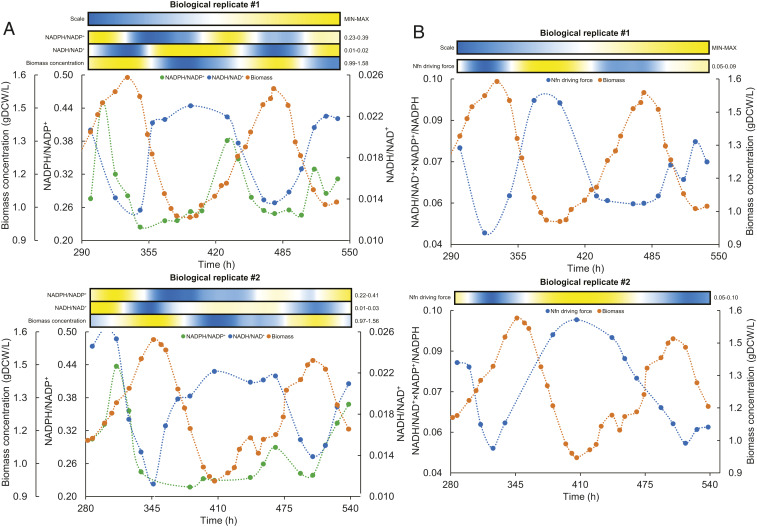
Fig. 2.
Oscillatory behavior of intracellular redox in C. autoethanogenum continuous cultures. (A) Oscillations in intracellular redox ratios and biomass concentration for two biological replicate cultures (#1 and #2). (B) Oscillations in Nfn driving force and biomass concentration for two biological replicate cultures (#1 and #2). Heat maps show the relative change of the respective parameter between its minimum and maximum value (denoted on the right of heat map) with color scale shown on the top. Heat maps were generated with values derived from using spline (third order polynomial cubic spline with point count 1) on the measured values to achieve higher resolution. time, fermentation duration from culture inoculation; Nfn driving force, NADH/NAD+×NADP+/NADPH.
The lack of changes in protein expression levels points toward posttranslational regulation of metabolism during oscillations. Given the energy-limited nature of acetogen metabolism (16, 17), regulation of metabolic fluxes through the energetically expensive processes of protein posttranslational modifications is unlikely. Hence, as previously suggested by others, regulation by thermodynamics through metabolite levels seemed feasible. It has been suggested that acetogen metabolism operates at the thermodynamic edge of feasibility (16, 17); that is, transformed Gibbs free energy values of key reactions are close to zero. We therefore next investigated the levels of metabolite concentrations throughout the oscillation cycles.
Oscillations Are Coordinated by Redox.
High-resolution temporal sampling throughout the oscillation cycles revealed oscillations in extracellular levels of acetate, ethanol, 2,3-butanediol, and pyruvate (Fig. 1B). Interestingly, while changes in ethanol, 2,3-butanediol, pyruvate, and biomass levels were synchronized, acetate levels peaked ∼24 h after the rest.
Extensive sampling and liquid chromatography–mass spectrometry (LC-MS) analysis failed to show clear oscillations in individual intracellular metabolite concentrations (Dataset S1). However, the intracellular redox ratio of nicotinamide adenine dinucleotide forms (NADH/NAD+) was synchronized with biomass level, but with an opposite trend; for example, biomass started to decline when NADH/NAD+ dropped to ∼0.012 and recovered once a ratio of ∼0.022 was reached (Fig. 2A). The ratio of nicotinamide adenine dinucleotide phosphate forms (NADPH/NADP+) showed a peak just before onset of hydrogen metabolism, which led to a rapid drop, and a possible second peak in the decline phase (Fig. 2B). These trends potentially indicated the importance of the Nfn transhydrogenase complex (CAETHG_07665) behind oscillations, as it is a central control valve of redox in acetogens (Fig. 3A). Indeed, the correlation between Nfn’s thermodynamic driving force (i.e., NADH/NAD+×NADP+/NADPH) and biomass levels suggests that Nfn might play a central role also behind metabolic oscillations in C. autoethanogenum (Fig. 2B).
tMFA.
tMFA (29) was next used to determine the thermodynamic driving force across central carbon and energy metabolism in acetogens. The model comprises the WLP (18, 19), energy and redox metabolism, acetate, ethanol, and 2,3-butanediol production pathways for a total of 32 reactions and 32 metabolites (Fig. 3A). We used the component contribution method to calculate standard Gibbs free energy of reactions (30) together with metabolomics data to determine reaction directionalities using thermodynamic variability analysis (see Materials and Methods for details).
Mechanism for acetate transport.
Since the efficiency of product transport across the cell membrane influences the thermodynamic equilibrium of reactions, especially for charged products (31, 32), and because acetogens often divert a significant fraction of substrate carbon into acetate production, we considered four candidate mechanisms for acetate transport that each requires different concentration gradients (Fig. 3B): transport of the undissociated acid (i.e., passive diffusion), symport of the anion with a proton, transport of anion via uniport, and ATP-consuming transport (i.e., ABC type). We calculated the required acetate concentration gradients (Dataset S1) and used them to constrain tMFA to check for thermodynamic feasibility.
Passive diffusion and symport of the anion with a proton are thermodynamically equivalent and require almost molar level intracellular acetate concentration to export against 8 g/L (∼0.134 M) of extracellular acetate (Fig. 3B). tMFA shows that there is no feasible flux distribution with such high internal acetate concentration, and hence both these mechanisms can be ruled out. While the ABC transporter is thermodynamically feasible at very low intracellular concentrations, this mechanism is rejected because acetate production generates less than 1 ATP per acetate formed. In contrast, thermodynamically and energetically feasible solutions were observed for the acetate uniport, and we will use the uniport in our tMFA, rather than the proton symporter commonly assumed in bacterial models (33). The distinction is important because the uniport is associated with indirect energy cost, since a proton must be transported independently to ensure charge balance. This proton can be transported by the membrane-bound, multisubunit Fd-NAD+ oxido-reductase Rnf complex that generates the proton motive force required to drive ATP synthesis through ATPase in C. autoethanogenum (34, 35) or, alternatively, by the ATPase operating in proton efflux mode; either option is associated with an energy cost of a fraction of an ATP.
Pivotal role of the acetaldehyde:ferredoxin oxidoreductase in ethanol production.
Together with acetate, ethanol is the main byproduct of acetogen growth. Generally, acetogens, including C. autoethanogenum (36), have two routes for ethanol biosynthesis: the conventional direct route from acetyl-CoA, using the bifunctional aldehyde/alcohol dehydrogenase (AdhE), or the ATP-coupled route through acetate, using the acetaldehyde:ferredoxin oxidoreductase (AOR) (Fig. 3A). Previous acetogen -omics studies (15, 20, 21, 37, 38), as well as the study of AOR knockouts (39), have shown that AOR plays a significant role in ethanol production. Consistent with the latter, tMFA shows that AdhE can only be active during the initial phase of CO-only growth (E1 in Fig. 3C). Under CO+H2 growth and during the crash, the reaction is thermodynamically infeasible in the ethanol production direction (Gibbs free energy range >0), and AOR (E2) is solely responsible (Gibbs free energy range <0) for the production of ethanol, assuming free diffusion of ethanol across the membrane (40).
Interestingly, applying tMFA to our previous study of nonoscillating (performed at lower gas transfer) CO-limited syngas chemostat culture of C. autoethanogenum (15) shows that ethanol production through AdhE is also infeasible under true steady state syngas conditions (Fig. 3C). This is consistent with the conclusion reached in a recent study of a kinetic ensemble model of C. autoethanogenum (41) that most kinetic parameters are favorable for AdhE to operate toward acetyl-CoA. While they used the same data set (15), they performed enzyme-by-enzyme rather than network-based analysis and did not use measured metabolite concentrations for calculating the reaction Gibbs free energies, which leads to loose bounds (30) and may explain why some models allowed flux through E1 in the ethanol direction.
Loss of AOR Driving Force May Trigger Crash.
A high flux through AOR is essential for simultaneous growth on CO and H2. AOR is important for energy generation, reconstitution of oxidized ferredoxin needed in the WLP, and regulation of intracellular acetate levels (Fig. 3A). tMFA reveals that the AOR driving force is very low in the crash; that is, a few hours after peak cell density (Fig. 3C). It is much lower than the thermodynamic driving force in the two growth phases and only half the level seen in the previous steady state at lower cell density. Operating near equilibrium and with no changes in enzyme concentration, the flux is expected to be proportional to the driving force. Thus, it is likely that the crash (i.e., slowing of growth to below dilution) is caused by a loss in AOR driving force as the external ethanol concentrations increases.
Discussion
Homeostatic regulation is a fundamental biological process, underlying cellular metabolism and physiology. Ideal homeostasis robustly rejects internal and external disturbances, which makes it very difficult to elucidate the detailed mechanism of regulation. Observed in many systems, oscillations indicate an imbalance between metabolic requirements affecting robustness (13) and provides a unique window into homeostatic regulation. Recently, oscillations without external interference were reported in acetogen continuous cultures (15). Here, we explored the underlying mechanisms using proteomics, metabolomics, and thermodynamic metabolic flux analysis.
The oscillations were relatively slow, lasting ∼150 h or 6 residence times at D = 1 d−1 (Fig. 1). While this might have suggested a transcriptional component to regulation, the absence of changes in the proteome between high and low cell density indicates that the mechanism is posttranscriptional.
The oscillations are not inherent to high-density cultivations. We have previously achieved steady state cultures at high cell density, using pure CO (20) (SI Appendix, Fig. S6). Rather, the oscillations are characterized by two key events around cometabolism of H2 and CO (Fig. 1). Onset of H2 uptake coincides with increased growth rate and increased ethanol production, which last until briefly before the onset of the crash (Fig. 1 and SI Appendix, Figs. S1 and S5).
Cellular hydrogenases and H2 uptake are inhibited by excess CO (23–25); hence, the onset of cometabolism only occurs when CO is almost exhausted. Prior to the onset of cometabolism, cells are effectively unaware of the H2 and are converging on a (lower) steady state cell concentration reflecting CO transfer only. Sudden access to H2 fuels rapid growth and ethanol production until the AOR driving force (Fig. 3C) is exhausted and cell growth slows. The crash is dramatically amplified by the increase of CO leading to loss of H2 cometabolism. When this happens, the cells are effectively located at a point with too high biomass, ethanol, and acetate relative to the steady state point defined by CO transfer only, and hence biomass quickly falls until the cycle can repeat.
Here we show that redox imbalances, caused by cometabolizing H2 and CO, trigger these metabolic oscillations in acetogens. Acetogen metabolism operates at the thermodynamic edge of life (16, 17). Our work describes in detail the sequence of events that eventually trigger metabolic oscillations as follows.
During the CO-only growth phase, CO inhibited cellular hydrogenases, and thus H2 uptake (SI Appendix, Fig. S2). As the biomass levels increase, H2 uptake eventually resumes, resulting in a seemingly unsustainable acetate to ethanol ratio (SI Appendix, Fig. S4). The increased fraction of carbon excreted as ethanol (refs. 15, 20 and SI Appendix, Figs. S4 and S5) and the higher gas uptake rate inherently demands more redox in the form of NADH and/or NADPH (Fig. 4). That redox imbalance potentially serves as the initial starting point of the events leading to oscillations. Regeneration of both redox types needs the Rnf activity that consumes Fdred. This, however, causes direct competition for Fdred between the Rnf complex and the AOR (Fig. 4), which is responsible for ethanol production (Fig. 3A). The competition is evident from the rapidly decreasing thermodynamic driving force of the Nfn transhydrogenase complex, with increasing gas uptake (Figs. 2B and 3C). At the same time, elevated throughput of the Rnf complex is required to meet higher energy demands for acetate export at increasing extracellular acetate levels (Fig. 1B), as the Rnf complex also generates the proton motive force to drive the ATPase in C. autoethanogenum (34, 35).
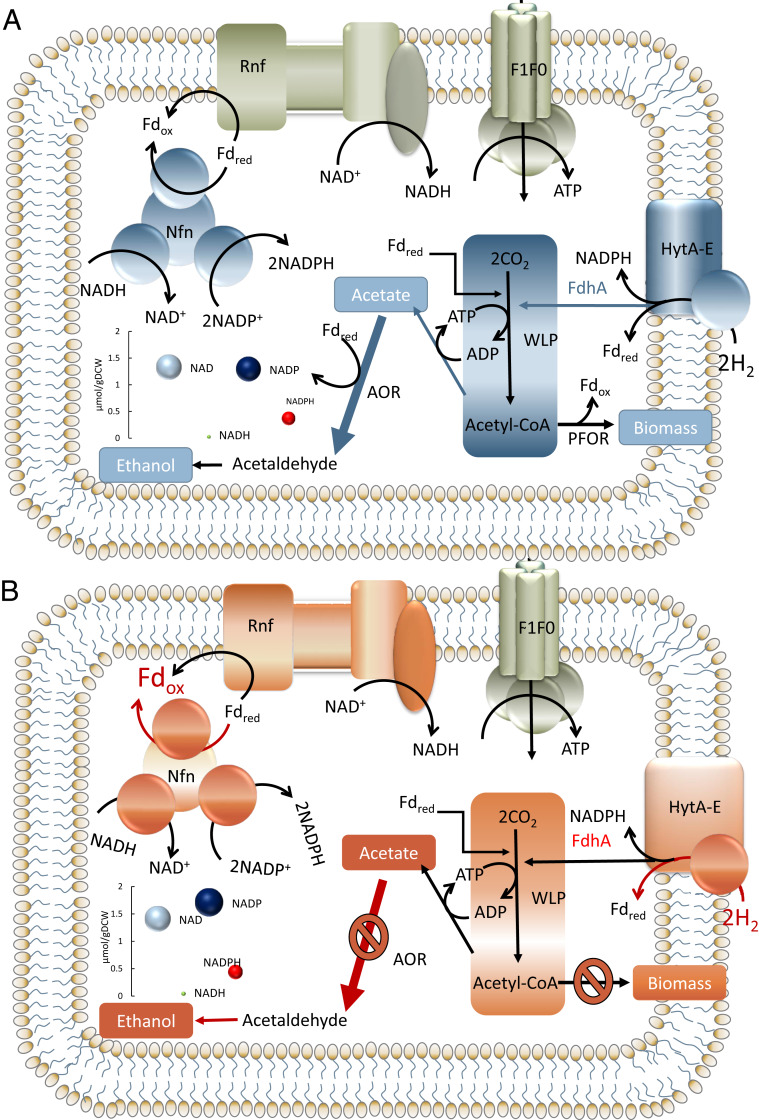
Fig. 4.
Two extremes of Clostridium autoethanogenum metabolism during oscillatory behavior in continuous cultures. (A) Metabolism enabling growth/recovery of the oscillating culture. (B) Metabolism precrash triggering oscillations of the culture. (Left Bottom Inset) Intracellular concentrations (measured; area of circle scales with concentration) of respective redox species for biological replicate culture #1 at lowest (i.e., recovery) or highest (i.e., crash) biomass concentration levels (see Figs. 1 and 2).
As indicated by the model, AOR is a near-equilibrium reaction (see Results); thus, its directionality is sensitive to changes in substrate/product concentrations. We hypothesize that the increasing demand for Fdred by the Rnf complex coupled with increase in ethanol concentration due to CO and H2 metabolism, leads the AOR to loose its thermodynamic driving force and the arrest of ethanol production, thus reducing the availability of oxidized ferredoxin (Fig. 4B). This is evidenced by the delay between the peaks of ethanol and acetate observed during oscillations (Fig. 1B). The arrest of the AOR results in an increase in the intracellular acetate concentration that further enhances the demand for Fdred by the Rnf complex to maintain the proton motive force. As a result, the WLP stops operation, since the required reduced-to-oxidized Fd ratio for the oxidation of CO cannot be maintained. This also rapidly shuts off H2 uptake due to its close link to CO uptake for recycling oxidized ferredoxin (Fig. 3A). Since the cells cannot uptake H2 for redox, metabolism shifts to growth on CO only. Since the AOR is unable to produce ethanol, cells mainly produce acetate, and this is evident from the increasing acetate concentrations even after ethanol levels drop.
Concomitantly, Nfn driving force starts to recover (Fig. 2B) while ethanol and acetate levels decrease (Fig. 2A). Eventually, AOR can again operate in the direction toward ethanol synthesis, and thus CO uptake is recovered. H2 uptake is resumed, resulting in an increased growth rate and a redox surplus which eventually results in the crash (SI Appendix, Fig. S2).
Metabolite levels have been shown to regulate oscillatory behavior in yeast (4, 8–10, 12). Also, keeping the ratios of oxidized/reduced redox species in check is generally fundamental to maintain cellular homeostasis. Similar to glycolysis, the pathway for carbon fixation in acetogens (the WLP) is tightly linked to ratios of redox pairs (Fig. 4) and is aiming at producing ATP for growth. Our results indicate that the imbalance of redox through the driving force of the Nfn ultimately limits acetogen metabolism, at least under oscillatory conditions. We thus conclude that the Nfn transhydrogenase complex plays a pivotal role in acetogen metabolism by operating as a valve. This is consistent with the observation that an Nfn deletion strain of C. autoethanogenum shows nearly no growth during growth on syngas and CO2+H2 (37).
Maximizing energy conservation when resources are scarce is a fundamental principle of biological systems that adapt to perturbations rapidly. As acetogens live at the edge of thermodynamic feasibility, where energy conservation is essential, acetogens offer an ideal platform to understand and explore the hard limits of metabolic robustness. Glycolytic oscillations in yeast have been studied extensively for over half a century. Despite the enormous number of studies looking at glycolytic oscillations, it is surprising that the biological function of oscillations remains a mystery. Similar to the WLP of acetogens, glycolysis is a central energy-producing pathway with feed-forward regulation. In the WLP, the first step requires redox as in glycolysis, where ATP is required in the early steps. It has been suggested that glycolytic oscillations are “a necessary consequence of autocatalysis and hard trade-offs between robustness and efficiency (or fragility and overhead)” (13). We show here that acetogen metabolism is controlled at the thermodynamic level through metabolite concentrations, and propose that an overshoot in the regulatory response potentially triggers oscillations. This is in accordance with a previous study which shows that cellular Gibbs energy dissipation rate limits metabolism (42). However, it remains unclear whether these oscillations can be a mechanism of energy conservation, as previously suggested (22), or are just a limitation of the system.
Our study is important, as our comprehensive experimental and computational analyses of oscillations in the gas-fermenting acetogen C. autoethanogenum suggest factors potentially limiting robustness of cellular metabolism. We provide a fundamental understanding between carbon, redox, and energy metabolism and product formation for a system relying on what was potentially the first biochemical pathway on Earth (43–46). Our work can also contribute toward expanding the product spectrum of acetogens and increasing the efficiency of the acetogen gas fermentation bioprocess through providing insights into potential limits in acetogen metabolism.
Materials and Methods
tMFA.
To calculate Gibbs free energy ranges (∆rG’min, ∆rG’max) for each metabolic reaction, it is necessary to calculate the standard Gibbs free energy values (∆rG°) and the corresponding metabolite concentrations (29). We used the component contribution method (30) to calculate the standard transformed Gibbs free energies of metabolites and the corresponding SDs to constrain reaction Gibbs free energies in the tMFA model, using an implementation in Python. Reaction directionalities were determined using thermodynamic variability analysis. Detailed methodology is available in SI Appendix.
Bacterial Strain, Growth Medium, and Continuous Chemostat Culture Conditions.
As we hereby replicated the previously observed oscillatory behavior, full details of continuous cultures are reported in another work (15). Shortly, C. autoethanogenum strain DSM 19630 was grown on syngas (50% CO, 20% H2, 20% CO2, and 10% Ar; BOC Australia) in chemically defined medium (without yeast extract) in continuous chemostat cultures (dilution rate of 1.0 ± 0.03 d−1) under strictly anaerobic conditions at 37 °C and at pH of 5. The gas-liquid transfer rate was increased by changing the agitation rate until oscillations were triggered at ∼800 rpm, after which the culture parameters remained constant.
Biomass Concentration Analysis.
To estimate the culture biomass concentration (gDCW/L), we measured its optical density (OD) at 600 nm and used a correlation coefficient of 0.21 determined in ref. 15 between sample OD and dry cell weight.
Bioreactor Off-Gas Analysis.
Bioreactor off-gas analysis for quantification of gas uptake and production rates was performed by an online Hiden HPR-20-QIC mass spectrometer as described in ref. 15 using the Faraday Cup detector.
Extracellular and Intracellular Metabolomics.
Samples collected for extracellular metabolomics were filtered and stored at −20 °C until analysis using high-performance liquid chromatography (HPLC), as described before (47). Sampling, sample storage, and sample processing (extraction) for intracellular metabolomics were performed as described before (15, 37, 48). Targeted LC-MS/MS analysis was performed using a Shimadzu UPLC coupled to a Shimadzu 8060 triple quadrupole mass spectrometer operated in negative ion mode while chromatographic separation was achieved on a Shim-pack Velox SP-C18 UHPLC column (227-32001-04). Full details of LC-MS/MS analysis are in SI Appendix.
Proteomics.
Quantitative proteome analysis was carried out using a data-independent acquisition mass spectrometry approach (27), with full details in SI Appendix. Briefly, sampling, sample storage, and sample preparation were performed as described earlier (20), with slight modifications specified in SI Appendix. Mass spectrometry using LC-MS/MS was performed using a Thermo Fisher Scientific UHPLC system coupled to a Q-Exactive HF-X mass spectrometer. Analysis of data-independent acquisition data were performed using Skyline (49), and differential protein expression analyzed using MSstats (50) as described before (20), with modifications specified in SI Appendix.
Data Availability.
Proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD016381. Intracellular metabolite concentration data of oscillating chemostat cultures are available as Dataset S1.
Supplementary Material
Acknowledgments
This work was funded by the Australian Research Council (ARC LP140100213) in collaboration with LanzaTech. We thank the following investors in LanzaTech’s technology: Sir Stephen Tindall, Khosla Ventures, Qiming Venture Partners, Softbank China, the Malaysian Life Sciences Capital Fund, Mitsui, Primetals, China International Capital Corporation Limited (CICC) Growth Capital Fund I, L.P. and the New Zealand Superannuation Fund. The research utilised equipment and support provided by the Queensland node of Metabolomics Australia, an initiative of the Australian Government being conducted as part of the National Collaborative Research Infrastructure Strategy (NCRIS) National Research Infrastructure for Australia. There was no funding support from the European Union for the experimental part of the study. However, K.V. acknowledges support also from the European Union’s Horizon 2020 research and innovation programme under grant agreement N810755.
Footnotes
Competing interest statement: LanzaTech has interest in commercial gas fermentation with Clostridium autoethanogenum. R.T., M.K., and S.D.S. are employees of LanzaTech.
This article is a PNAS Direct Submission.
Data deposition: Proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (dataset identifier PXD016381).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919531117/-/DCSupplemental.
References
- 1.Muchowska K. B. et al., Metals promote sequences of the reverse Krebs cycle. Nat. Ecol. Evol. 1, 1716–1721 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuetz R., Zamboni N., Zampieri M., Heinemann M., Sauer U., Multidimensional optimality of microbial metabolism. Science 336, 601–604 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Ng C. Y., Wang L., Chowdhury A., Maranas C. D., Pareto optimality explanation of the glycolytic alternatives in nature. Sci. Rep. 9, 2633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray D. B., Beckmann M., Kitano H., Regulation of yeast oscillatory dynamics. Proc. Natl. Acad. Sci. U.S.A. 104, 2241–2246 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber J., Kayser A., Rinas U., Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. II. Dynamic response to famine and feast, activation of the methylglyoxal pathway and oscillatory behaviour. Microbiology 151, 707–716 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Robert M. et al., “Extracellular metabolite dynamics and temporal organization of metabolic function in E. coli” in Proceedings of 2012 ICME International Conference on Complex Medical Engineering (CME), (IEEE, 2012), pp. 197–202. [Google Scholar]
- 7.Ofiţeru I. D., Ferdeş M., Knapp C. W., Graham D. W., Lavric V., Conditional confined oscillatory dynamics of Escherichia coli strain K12-MG1655 in chemostat systems. Appl. Microbiol. Biotechnol. 94, 185–192 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Tu B. P., McKnight S. L., The yeast metabolic cycle: Insights into the life of a eukaryotic cell. Cold Spring Harb. Symp. Quant. Biol. 72, 339–343 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Xu Z., Tsurugi K., A potential mechanism of energy-metabolism oscillation in an aerobic chemostat culture of the yeast Saccharomyces cerevisiae. FEBS J. 273, 1696–1709 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Satroutdinov A. D., Kuriyama H., Kobayashi H., Oscillatory metabolism of Saccharomyces cerevisiae in continuous culture. FEMS Microbiol. Lett. 77, 261–267 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Kaspar von Meyenburg H., Energetics of the budding cycle of Saccharomyces cerevisiae during glucose limited aerobic growth. Arch. Mikrobiol. 66, 289–303 (1969). [DOI] [PubMed] [Google Scholar]
- 12.Shi L., Tu B. P., Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 110, 7318–7323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra F. A., Buzi G., Doyle J. C., Glycolytic oscillations and limits on robust efficiency. Science 333, 187–192 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Tepper N. et al., Steady-state metabolite concentrations reflect a balance between maximizing enzyme efficiency and minimizing total metabolite load. PLoS One 8, e75370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valgepea K. et al., Maintenance of ATP homeostasis triggers metabolic shifts in gas-fermenting acetogens. Cell Syst. 4, 505–515.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Schuchmann K., Müller V., Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Molitor B., Marcellin E., Angenent L. T., Overcoming the energetic limitations of syngas fermentation. Curr. Opin. Chem. Biol. 41, 84–92 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Ragsdale S. W., Pierce E., Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta 1784, 1873–1898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood H. G., Life with CO or CO2 and H2 as a source of carbon and energy. FASEB J. 5, 156–163 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Valgepea K. et al., H2 drives metabolic rearrangements in gas-fermenting Clostridium autoethanogenum. Biotechnol. Biofuels 11, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter H. et al., Ethanol production in syngas-fermenting Clostridium ljungdahlii is controlled by thermodynamics rather than by enzyme expression. Energy Environ. Sci. 9, 2392–2399 (2016). [Google Scholar]
- 22.Richter P. H., Ross J., Concentration oscillations and efficiency: glycolysis. Science 211, 715–717 (1981). [DOI] [PubMed] [Google Scholar]
- 23.Shima S., Thauer R. K., A third type of hydrogenase catalyzing H2 activation. Chem. Rec. 7, 37–46 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Adams M. W. W., The structure and mechanism of iron-hydrogenases. BBA Bioenerg. 1020, 115–145 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Wang S. et al., NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valgepea K., Marcellin E., Data from "Redox controls metabolic robustness in the gas-fermenting acetogen Clostridium autoethanogenum." PRIDE. http://proteomecentral.proteomexchange.org/cgi/PXD016381. Deposited 21 November 2019. [DOI] [PMC free article] [PubMed]
- 27.Gillet L. C. et al., Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 29.Henry C. S., Broadbelt L. J., Hatzimanikatis V., Thermodynamics-based metabolic flux analysis. Biophys. J. 92, 1792–1805 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noor E., Haraldsdóttir H. S., Milo R., Fleming R. M. T., Consistent estimation of Gibbs energy using component contributions. PLOS Comput. Biol. 9, e1003098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Maris A. J., Konings W. N., van Dijken J. P., Pronk J. T., Microbial export of lactic and 3-hydroxypropanoic acid: Implications for industrial fermentation processes. Metab. Eng. 6, 245–255 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Jol S. J., Kümmel A., Hatzimanikatis V., Beard D. A., Heinemann M., Thermodynamic calculations for biochemical transport and reaction processes in metabolic networks. Biophys. J. 99, 3139–3144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beauprez J. J. et al., Influence of C4-dicarboxylic acid transporters on succinate production. Green Chem. 13, 2179–2186 (2011). [Google Scholar]
- 34.Hess V. et al., Occurrence of ferredoxin:NAD(+) oxidoreductase activity and its ion specificity in several Gram-positive and Gram-negative bacteria. PeerJ 4, e1515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tremblay P. L., Zhang T., Dar S. A., Leang C., Lovley D. R., The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. MBio 4, e00406–e00412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bengelsdorf F. R. et al., Industrial acetogenic biocatalysts: A comparative metabolic and genomic analysis. Front. Microbiol. 7, 1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcellin E. et al., Low carbon fuels and commodity chemicals from waste gases – Systematic approach to understand energy metabolism in a model acetogen. Green Chem. 18, 3020–3028 (2016). [Google Scholar]
- 38.Nagarajan H. et al., Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Fact. 12, 118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liew F. et al., Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab. Eng. 40, 104–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cueto-Rojas H. F., van Maris A. J. A., Wahl S. A., Heijnen J. J., Thermodynamics-based design of microbial cell factories for anaerobic product formation. Trends Biotechnol. 33, 534–546 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Greene J., Daniell J., Köpke M., Broadbelt L., Tyo K. E. J., Kinetic ensemble model of gas fermenting Clostridium autoethanogenum for improved ethanol production. Biochem. Eng. J. 148, 46–56 (2019). [Google Scholar]
- 42.Niebel B., Leupold S., Heinemann M., An upper limit on Gibbs energy dissipation governs cellular metabolism. Nat. Metab. 1, 125–132 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Varma S. J., Muchowska K. B., Chatelain P., Moran J., Native iron reduces CO2 to intermediates and end-products of the acetyl-CoA pathway. Nat. Ecol. Evol. 2, 1019–1024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss M. C. et al., The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Fuchs G., Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Russell M. J., Martin W., The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 29, 358–363 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Valgepea K. et al., Arginine deiminase pathway provides ATP and boosts growth of the gas-fermenting acetogen Clostridium autoethanogenum. Metab. Eng. 41, 202–211 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Marcellin E., Nielsen L. K., Abeydeera P., Krömer J. O., Quantitative analysis of intracellular sugar phosphates and sugar nucleotides in encapsulated streptococci using HPAEC-PAD. Biotechnol. J. 4, 58–63 (2009). [DOI] [PubMed] [Google Scholar]
- 49.MacLean B. et al., Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi M. et al., MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD016381. Intracellular metabolite concentration data of oscillating chemostat cultures are available as Dataset S1.