Significance
Many of the important functions in the cell are performed by high-molecular-weight complexes that contain both protein and nucleic acid components. Although developments in solution NMR spectroscopy have facilitated the study of the protein portions of these complexes, similar quantitative studies of the nucleic acids have proven challenging. Here we describe an approach for labeling DNA molecules with methyl-group probes at specific sites, followed by the use of optimized NMR methods to obtain high-quality data in studies of these large molecular complexes. Studies of both the DNA and protein components of the nucleosome core particle are presented, including an investigation of how CpG methylation, an important epigenetic mark, affects the structural dynamics of the particle.
Keywords: high-molecular-weight protein–DNA complexes, one-pot synthesis of methyl-labeled DNA, structural dynamics of protein–DNA complexes, CpG methylation
Abstract
The development of methyl-transverse relaxation-optimized spectroscopy (methyl-TROSY)–based NMR methods, in concert with robust strategies for incorporation of methyl-group probes of structure and dynamics into the protein of interest, has facilitated quantitative studies of high-molecular-weight protein complexes. Here we develop a one-pot in vitro reaction for producing NMR quantities of methyl-labeled DNA at the C5 and N6 positions of cytosine (5mC) and adenine (6mA) nucleobases, respectively, enabling the study of high-molecular-weight DNA molecules using TROSY approaches originally developed for protein applications. Our biosynthetic strategy exploits the large number of naturally available methyltransferases to specifically methylate DNA at a desired number of sites that serve as probes of structure and dynamics. We illustrate the methodology with studies of the 153-base pair Widom DNA molecule that is simultaneously methyl-labeled at five sites, showing that high-quality 13C-1H spectra can be recorded on 100 μM samples in a few minutes. NMR spin relaxation studies of labeled methyl groups in both DNA and the H2B histone protein component of the 200-kDa nucleosome core particle (NCP) establish that methyl groups at 5mC and 6mA positions are, in general, more rigid than Ile, Leu, and Val methyl probes in protein side chains. Studies focusing on histone H2B of NCPs wrapped with either wild-type DNA or DNA methylated at all 26 CpG sites highlight the utility of NMR in investigating the structural dynamics of the NCP and how its histone core is affected through DNA methylation, an important regulator of transcription.
Solution NMR spectroscopy is a powerful technique for atomic resolution studies of the structure, dynamics, and function of biological molecules (1). Significant advances, including increased magnetic fields that have now reached 28 T, corresponding to a 1H resonance frequency of 1.2 GHz, cryogenically cooled probes that double sensitivity, facile isotope labeling schemes, and improved experiments that exploit the labeling so as to minimize sensitivity losses and optimize resolution, have transformed the field. This is particularly evident in studies of proteins where the introduction of transverse relaxation-optimized spectroscopy (TROSY)-based methods has significantly decreased the size limitations that have traditionally challenged NMR applications (2). Initial TROSY experiments for proteins focused on backbone amides where the interference between 1H-15N dipolar and 15N chemical shift anisotropy spin relaxation mechanisms was exploited to increase the lifetimes of NMR signals in high-molecular-weight protein systems (2) and later extended to aromatic side chains (3). Subsequently, a methyl-TROSY effect was described (4) that together with site-specific labeling of methyl groups as 13CH3 in an otherwise deuterated background (5) has enabled the study of protein systems with molecular weights as large as 1 MDa (6, 7). Applications to important molecular machines have been reported, emphasizing, in particular, the critical role that dynamics play in controlling function and malfunction in these systems (8, 9).
Impressive advances in studies of nucleic acids have also emerged (10–14), although NMR applications involving this important class of biomolecules have lagged behind protein studies. In part this reflects the much larger number of building blocks for proteins than for nucleic acids, 20 amino acids vs. 4 nucleobases, that often leads to poor chemical shift dispersion in nucleic acid spectra even at high magnetic fields, increased difficulties with sample preparation, and the fact that spectral quality often deteriorates more rapidly with molecular weight for nucleic acids than for proteins. As with proteins, isotopic labeling of DNA and RNA has proven critical for detailed NMR studies and strategies for sample production involving either chemical or enzymatic synthesis have emerged (15, 16). Most studies of nucleic acids by solution NMR have focused on relatively small molecules, typically on the order of 30 nucleotides or less, yet the success of this technology in structural studies is made clear by the fact that ∼40% of all Protein Data Bank (PDB) entries for this class of molecules are derived from NMR. Studies of larger nucleic acids have, however, been limited. Because enzymatic approaches for sample generation result in constructs where all nucleotides of at least one type are labeled, spectral resolution becomes limiting for large molecules. Some simplification can be achieved, however, by using partially deuterated precursors to generate samples (17), as has been convincingly demonstrated in studies of a number of RNA systems in excess of a hundred nucleotides (12, 18, 19). Problems with resolution can be further circumvented, in principle, via chemical synthesis where a labeled nucleotide can be added to a specific position along the sequence (20, 21), although the requirement for sufficient amounts of highly pure product limits this technology to lengths of ∼100 nucleotides.
Recognizing the importance of methyl labeling to NMR studies of large protein complexes, we asked whether a similar strategy might prove useful in applications involving nucleic acids, focusing presently on DNA. Although the thymine nucleobase in DNA contains a methyl group attached to C5, 13C labeling of all thymine methyl groups in DNA would be of limited utility, especially for large DNA molecules, due to peak overlap. Instead, we sought an approach whereby 13CH3 groups could be added to specific sites using a simple biosynthetic strategy that would then provide probes of structure and dynamics. The addition of methyl groups to nucleic acids is, of course, an idea that nature has exploited for specific functions. In prokaryotes, for example, DNA is methylated at N6, N4, and C5 positions of adenine (N6) and cytosine (N4 and C5) by DNA methyltransferases (MTases) that recognize specific DNA sequences to form N6-methyladenine (6mA), N4-methylcytosine (4mC), and C5-methylcytosine (5mC), respectively (22). The resulting modifications lead to a primitive immune system by providing resistance to enzymes that would otherwise cleave host DNA, while foreign DNA that is not methylated, derived from bacteriophages, for example, would be cleaved (23). In mammals the main methylation target is the C5 cytosine position in CpG dinucleotides (24), an important epigenetic mark in the regulation of gene expression, including X-chromosome inactivation (25) and gene silencing (26). DNA methylation patterns are preserved during replication by cellular MTases that recognize hemimethylated DNA (27). The importance of MTases in all kingdoms of life has led to a strong interest in understanding various facets of these enzymes and consequently many of these proteins are either commercially available or can be obtained as clones for recombinant expression so that they can be produced in sufficient amounts for the generation of methylated DNA for NMR experiments.
Herein, we exploit the array of DNA MTases that nature has provided to generate 13CH3-methyl–labeled DNA samples that are amenable to study via solution NMR. A simple biosynthetic strategy is presented based on a one-pot reaction in which 13CH3-methionine is added along with S-adenosylmethionine synthase (SAMS), adenosine 5′-triphosphate (ATP), the DNA of interest, and the required MTases. The efficacy of the approach is demonstrated first by labeling small DNA oligonucleotides using different MTases (one at a time). Subsequently, the thermodynamic and kinetic parameters for melting of a 12-base pair (bp) sequence is established using a pair of methyl probes that are attached to N6 positions of separate adenine bases, with relaxation dispersion experiments quantifying the rate of rotation about the adenine C6–N6 bond. Applications to the 153-bp Widom 601 nucleosome positioning DNA sequence (28) and a 200-kDa nucleosome core particle (NCP) in which two copies each of histones H2A, H2B, H3, and H4 are wrapped by the Widom DNA (29) are presented using methyl-TROSY–based approaches that highlight the utility of this labeling strategy for NMR studies of very-high-molecular-weight DNA and DNA–protein complexes.
Results and Discussion
Methylation of DNA via a One-Pot Reaction.
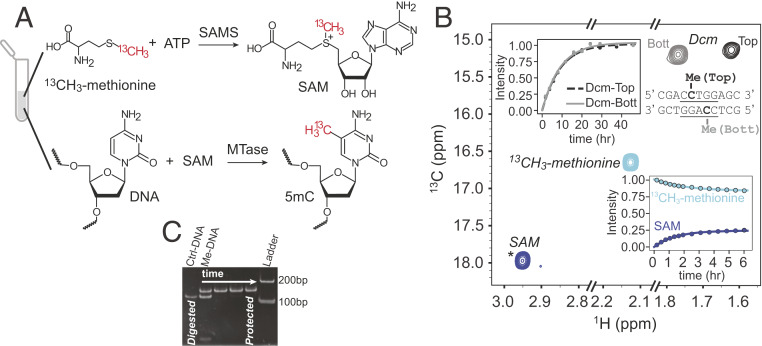
Fig. 1A illustrates the reactions used to generate 13CH3-labeled DNA, starting from the inexpensive 13CH3-methionine precursor. In addition to Met, the DNA to be labeled must be added (either 2H or 1H, discussed below), along with ATP, SAMS that catalyzes the conversion of Met to S-adenosyl methionine (SAM) (30), and the appropriate MTases that then facilitate the addition of 13CH3 moieties (red) to the DNA at the locations governed by the recognition sites of the MTases. Although a significant number of MTases are available commercially, their purchase becomes expensive in the amounts needed for the preparation of NMR quantities of labeled DNA and we have therefore produced all of the required enzymes, including SAMS, in-house using bacterial overexpression and purification protocols that are detailed in SI Appendix. Fig. 1B shows a 13C-1H heteronuclear multiple-quantum coherence (HMQC) spectrum at the completion of the labeling reaction in which 1 mM 13CH3-methionine, 2 mM ATP, 1.35 μM SAMS, 2 μM Escherichia coli Dcm MTase, and 0.2 mM double-stranded DNA containing the Dcm recognition sequence were mixed in an NMR tube and allowed to incubate at 25 °C. The Dcm MTase recognizes the sequence 5′-CC(A/T)GG-3′ (Fig. 1B, Inset) and catalyzes methylation of the C5 position of the second cytosine (i.e., closest to the 3′ end) on each strand of the DNA to 5mC. The buildup of cross-peaks derived from methyl groups of SAM and labeled DNA, as well as the concomitant decrease in 13CH3-methionine, are highlighted in separate panels. As expected, the reaction kinetics for the addition of methyl groups to the top (black) and bottom (gray) DNA strands are the same. We have observed that under our reaction conditions formation of SAM is much faster than the subsequent DNA labeling step and that the conversion of Met to SAM does not go to completion (Fig. 1 B, Lower), which reflects the strong product inhibition of SAMS (31). We have deliberately chosen to carry out the reaction at room temperature, enabling high-quality NMR data to be recorded so as to capture the complete time course of methylation; in practice, when preparative quantities are required, DNA labeling is performed at 37 °C where the reaction is significantly faster. As a final note, we have established that the Dcm MTase reaction can be made to go to completion by using the 153-bp Widom DNA sequence (discussed below) and monitoring protection against cleavage via the PspGI restriction enzyme that would normally cut at Dcm sites in the absence of methylation (32). The gel presented in Fig. 1C shows that protection occurs rapidly after addition of the MTase, with the DNA completely protected (i.e., methylated) within 1 h after initiation of the methylation reaction under the conditions of our experiment (Fig. 1C legend). We have verified that all of the MTase reactions on Widom DNA reported in this paper proceed to completion, as above, in particular as it is known that the S-adenosyl homocysteine by-product of the SAM reaction is a potent inhibitor of some of the MTases (33).
Fig. 1.
A one-pot DNA methylation reaction. (A) General scheme for the methylation of DNA in which SAM is produced by the reaction of ATP and 13CH3-methionine, catalyzed by SAMS (30). SAM serves as the methyl donor in the subsequent methylation of the DNA template by a specific MTase. (B) Methylation of an 11-bp oligonucleotide containing the Dcm recognition sequence (Inset), as monitored by the buildup of NMR peaks of the methyl groups of SAM and the labeled DNA 11-mer, and the decay of the Met peak (25 °C). The signal from SAM is aliased (*), from 26.0 ppm. (C) Verification that Dcm-based methylation of DNA can be driven to completion using a 153-bp DNA with two copies of the Dcm recognition sequence. Addition of the PspGI restriction enzyme leads to DNA cleavage at nonmethylated Dcm recognition sites (Ctrl-DNA lane). As two such sites are present in 601 DNA, three fragments are produced with 113, 28, and 2 bp, in addition to 5-base overhangs on each of the 5′ ends. Addition of PspGI at 0, 1, 2, and 3.5 h after initiation of the methylation reaction shows that complete protection is achieved after 1 h (37 °C) under the reaction conditions used to generate methyl-labeled DNA (SI Appendix). Shown is a 7% native polyacrylamide gel in 1× Tris–borate–(ethylenediamine)tetraacetic acid buffer, run for 1 h at 200 V.
Methylation of Oligonucleotides Using a Variety of Different MTases and Chemical Shift Assignments.
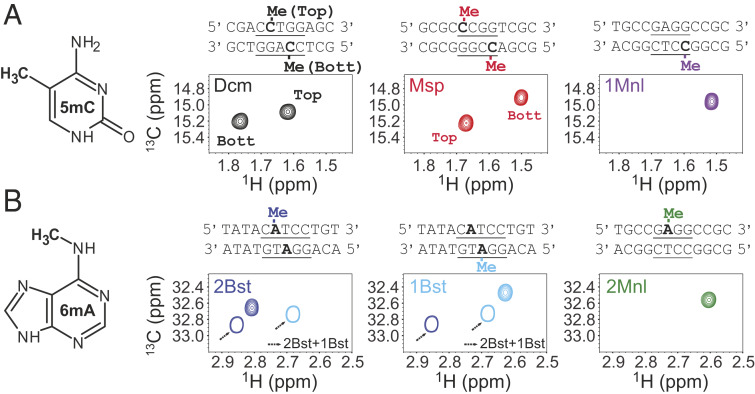
Having established the efficacy of the one-pot protocol of Fig. 1A when using the Dcm MTase we next wanted to confirm that other MTases would similarly methylate DNA in an efficient manner and to see if DNA methyl correlations are sufficiently well resolved in NMR spectra so that several sites could be methylated simultaneously in the context of large DNA molecules. Methylation reactions were therefore carried out on a collection of oligonucleotides, either 11 or 12 bp, each containing a sequence that is specifically recognized by a different MTase, leading to methylation at adenine N6 or cytosine C5 positions of the DNA. All reactions were performed using the in vitro procedure detailed above for Dcm (Fig. 1), except that the reactions were carried out overnight at 37 °C to ensure complete methylation by all of the different enzymes. The 13C-1H HMQC spectra for the resulting methyl labeled samples are illustrated in Fig. 2, along with the recognition sequences for each enzyme (underlined) and the bases that become methylated (bold). Some of the MTases act in an asymmetric manner, methylating a single strand of the double helix so that only a single correlation is present in spectra (M1.MnlI, M2.MnlI, M1.BstF5I, and M2.BstF5I, referred to in what follows as 1Mnl, 2Mnl, 1Bst, and 2Bst, respectively), while others place a methyl group on each of the strands, giving rise to a pair of cross-peaks in HMQC datasets (Dcm and M.MspI, the latter referred to in what follows as Msp). In the latter case we have obtained assignments of the methyl groups using a pair of oligonucleotides where positions immediately adjacent to the recognition site were modified. For example, consider the assignment of the Dcm spectrum of Fig. 2A (black). As the sequence of interest in this case is the duplex, 5′-ACCTGGA-3′ + 3′-TGGACCT-5′, where the methylated base is in bold and the recognition site is underlined, a pair of duplexes, 5′-TCCTGGA-3′ + 3′-AGGACCT-5′, as well as 5′-ACCTGGT-3′ + 3′-TGGACCA-5′, were prepared and methylated with Dcm. Comparing the resulting NMR spectra of these assignment variants with the spectrum of the desired sequence allowed the identification of which peak was derived from each of the strands of the DNA (SI Appendix, Fig. S1A). In a similar manner, peaks from methylated DNA generated with the Msp MTase were assigned (SI Appendix, Fig. S1B).
Fig. 2.
The 13C-1H HMQC spectra of methylated oligonucleotides (25 °C) by (A) C5 cytosine-specific and (B) N6 adenine-specific MTases. The sequences of the oligonucleotides are given at the top of each spectrum, with the recognition site underlined and the methylated base in bold. MTases can either methylate both strands of a recognized sequence (Msp, Dcm), resulting in pairs of peaks, or one strand (1Bst, 2Bst, 1Mnl, 2Mnl). 1Bst and 2Bst recognize the same sequence (as do 1Mnl and 2Mnl); however, each is able to place a methyl on only one of the strands (Top, 2Bst) or (Bottom, 1Bst); methylation with both 1Bst and 2Bst MTases leads to chemical shift changes relative to incorporation of a single methyl, as indicated (arrows). Spectra were referenced using sodium trimethylsilylpropanesulfanate (DSS).
The spectra of the different assignment mutants suggested a relation between the identity of the base at the i−2 position and the 1H chemical shift of the 5mC methyl group (i is the site of the methyl group and positive numbering extends to the 3′ DNA end). We explored this by generating spectra of oligonucleotides where the i−2 positions were one of the four nucleobases, with methylation using the Dcm enzyme (SI Appendix, Fig. S1C). A clear trend in 1H chemical shifts was noted, with A (most upfield) < G < C < T (most downfield), with a similar trend for 13C shifts although these were smaller (in hertz). We were also able to establish how the nucleobase at the i+1 position affects methyl 1H shifts, at least for i+1 = A or T (for which we have data; compare magenta peaks in SI Appendix, Fig. S1A). Notably, the methyl 1H shift for i+1 = T is upfield of that for A, with little change in the 13C shifts, so that the effect of purines vs. pyrimidines at the i+1 and i−2 positions on 5mC 1H chemical shifts is swapped. We have also investigated how the i−1 position influences methyl 1H and 13C shifts using separate oligonucleotides synthesized with each of the four bases at i−1 and labeled using the Msp enzyme. A large difference in 1H chemical shifts is noted for sequences with pyrimidines or purines at i−1, with significant downfield shifts (∼0.2 ppm) in the case of pyrimidines (SI Appendix, Fig. S1D). A similar trend regarding the influence of the i−1 position on 6mA 1H chemical shifts seems also to hold, as a comparison of methyl peak positions in an oligonucleotide labeled with either 1Bst or 2Bst, where the i−1 base is either G or C, respectively, shows an upfield shift for the purine (Fig. 2B).
Methyl Group Probes of the Thermodynamics and Kinetics of DNA Melting.
In what follows we provide an illustration of the utility of methyl groups as reporters of nucleic acid dynamics by considering a small 12-mer DNA that contains the recognition site for the E. coli Dam MTase (referred to in what follows as 12-merDam). Unlike all oligonucleotides illustrated in Fig. 2 that were generated by chemical synthesis and subsequently methylated with the appropriate MTase in vitro, the 12-mer considered here was generated via in vivo expression, as a by-product of the overexpression of the 153-bp 601 Widom DNA sequence that we will discuss subsequently. SI Appendix, Fig. S2 illustrates the high-copy-number plasmid that was used to generate 12-merDam, consisting of ∼30 repeats of the 601 sequence each separated by insertions of the 12-merDam that is used here. The small 12-mer oligonucleotides can be easily released from the large DNA fragment via restriction digestion and separated from the other, much larger, 153-bp DNA fragments as described in SI Appendix. Because Dam is endogenous to E. coli that was used for overexpression of the plasmid, production of the desired methyl-labeled sample was accomplished simply through the addition of 13CH3-methionine to the growth medium. As we required samples of highly deuterated 601 DNA for other studies, 12-merDam was, by default, also perdeuterated, although this is not essential for the applications considered here.
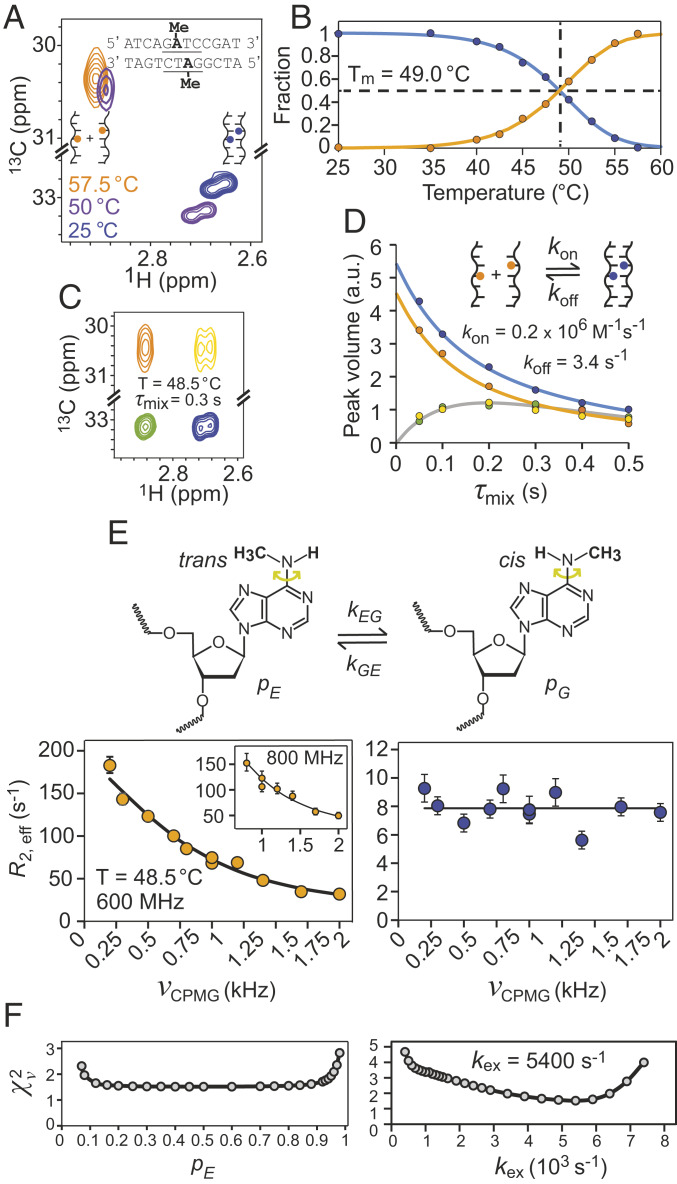
Fig. 3A shows a superposition of three 13C-1H HSQC spectra recorded of 12-merDam at different temperatures. The 12-bp sequence is shown, along with a pair of adenine N6 attached methyl groups. At temperatures far below the DNA melting temperature, TM, a pair of peaks is observed, one from each of the two methyl groups of the DNA duplex. As the temperature is raised an additional, well-resolved peak emerges from the separated strands, indicating that the methyl probes in the isolated strands are degenerate; this peak grows with temperature until the correlations from the duplex disappear. Peak volumes were quantified as a function of temperature and the fraction of strands in either the annealed (blue) or melted (orange) conformations plotted in Fig. 3B. The resulting curves were fit to SI Appendix, Eq. S17 to give TM = 49.0 °C, ΔH = −99 ± 3 kcal/mol and ΔS = (−2.9 ± 0.1) × 10−1 kcal/mol K for annealing. In order to quantify the kinetics of the duplexsingle-strand interconversion, we recorded a magnetization exchange experiment (34). Fig. 3C shows one plane from a series obtained with different mixing times, τmix, during which exchange occurs (48.5 °C). A pair of cross-peaks, green and yellow, are observed that derive from the conversion from duplex to single-strand (green) and vice versa (yellow), in addition to the diagonal peaks where magnetization both originates on and is detected from the same state (i.e., duplex or single strand). Peak volumes were quantified in Fig. 3D, from which kon and koff values of (0.2 ± 0.06) × 106 M−1s−1 and 3.4 ± 0.9 s−1, respectively, were obtained (SI Appendix).
Fig. 3.
Methyl probes of DNA melting, kinetics of strand annealing, and rotational dynamics. (A) Superposition of 13C-1H HSQC spectra of 12-merDam (6mA) recorded at different temperatures, from which the fraction of DNA strands in single- and double-stranded DNA as a function of temperature are calculated (B). (C) A series of magnetization exchange spectra (34) were recorded to quantify the kinetics of duplexsingle-strand interconversion; one plane at a mixing time, τmix, of 0.3 s is shown (48.5 °C) with the green and yellow peaks resulting from the interconversion process. (D) Peak volumes vs. τmix from which kon and koff values of (0.2 ± 0.06) × 106 M−1s−1 and 3.4 ± 0.9 s−1, respectively, were obtained (25 mM sodium phosphate and 100 mM sodium chloride). The fraction of double-stranded DNA obtained from analysis of magnetization exchange data at 48.5 °C, 0.55, is in excellent agreement with the value obtained in B, 0.54. (E) The 13C-1H multiple-quantum relaxation dispersion profiles (35) of 6mA from single- (orange) and double-stranded (blue) DNA, recorded at 48.5 °C and 600 MHz or 800 MHz (Inset). (F) Reduced χ2 values from fits of dispersion data as a function of pE (Left) or kex (Right). The population of the excited state, pE, corresponding to the trans conformation of the methyl group cannot be determined from the data, but kex = kEG + kGE = 5,400 ± 800 s−1 can be quantified. Errors for all magnetization exchange (relaxation dispersion) parameters were calculated via the covariance matrix (Monte Carlo, 200 simulations) method (83).
The 12-merDam spectrum clearly shows that the methyl peak assigned to the single strands (orange) is broader in the 13C dimension than the corresponding peaks from the duplex. This differential linewidth most likely arises from differences in rotation rates about the C6–N6 bond (Fig. 3 E, Top), with hydrogen bonding of the Watson–Crick base pairs in the context of the duplex eliminating rotation. To quantify these rates we have carried out 13C-1H multiple-quantum relaxation dispersion experiments (35) on the sample at 48.5 °C, where correlations from both single-strand and duplex DNA are present. Large dispersions were recorded for 12-merDam in the single-strand form (Fig. 3 E, Left), while flat lines indicating no exchange contributions to linewidths were obtained for the duplex (Fig. 3 E, Right). An exchange rate kex = kGE + kEG = 5,400 ± 800 s−1 was quantified from fits of the dispersion data for the single-strand conformation, sufficiently fast so that it was not possible to estimate the relative populations of the two conformers (Fig. 3F). It is known, however, that in the context of isolated 6mA bases there is a strong preference for conformation G in Fig. 3E that places the CH3 moiety in the cis position (relative to N1 of the base), with a fractional population, pG, on the order of 0.9 (36). In the context of a Watson–Crick base pair, however, the unfavorable trans conformation E is adopted that enables the required interbase hydrogen bonding. It is expected, therefore, that for single-stranded DNA pG ∼ 0.9.
Methyl-TROSY NMR of High-Molecular-Weight DNA and DNA–Protein Complexes.
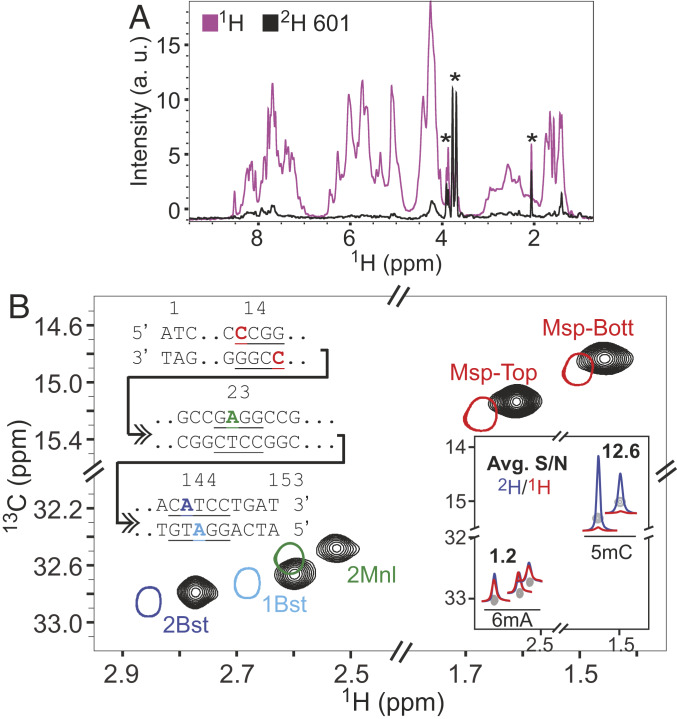
The NCP is the basic building block of chromatin, comprising four types of histones (H2A, H2B, H3, and H4, each in two copies) and ∼150 bp of DNA that wraps around the octameric protein core (37, 38). Methylation of DNA produces epigenetic marks that are maintained through cell division and that play a critical role in a plethora of different biological processes (39). The development of methodology for quantifying the effects of methylation on the structural dynamics of DNA, and on the associated protein components, in the context of large DNA–protein complexes is thus of considerable interest. As a first step we choose to work with Widom 601 DNA, a strong-positioning sequence designed for optimal wrapping of DNA in NCPs (28). As described above, NMR quantities of 601 DNA were produced using a high-copy-number plasmid (40), as shown schematically in SI Appendix, Fig. S2, with DNA generated in either rich growth medium or in a deuterated minimal medium using a protocol that is similar to that used for the expression of highly deuterated proteins (SI Appendix). Fig. 4A shows an overlay of one-dimensional proton NMR spectra of 1H- and 2H-601 DNA (not methylated), establishing that ∼95% of the protons are replaced with deuterons in the deuterated sample; peaks labeled with * derive from buffer or residual glycerol in the sample. Both 1H- and 2H-601 DNA samples were subsequently methylated in a one-pot mixture containing the reaction precursors shown in Fig. 1 and four MTases: Msp (5mC), 2 Bst (6mA), 1Bst (6mA), and 2Mnl (6mA). The advantage of an in vitro methylation procedure is that different methyl groups can be added simply by using different enzymes. In contrast, in vivo approaches would involve suppression of endogenous MTases if the labels that they produce are not of interest and adding vectors containing genes of the MTases of interest for each desired methyl-labeling pattern.
Fig. 4.
Deuteration is critical for recording high-sensitivity spectra of 5mC correlations. (A) One-dimenstional 1H spectral overlays of deuterated (black) and fully protonated (magenta) 601 DNA, both unmethylated (45 °C, 800 MHz). The asterisks indicate peaks arising from traces of glycerol in the samples, as well as from buffer components. (B) The 13C-1H HMQC spectrum of 2H-601 DNA (black) methylated simultaneously by 1Bst, 2Bst, 2Mnl, and Msp. The 601 DNA methyl assignments were obtained from the corresponding spectra of singly and doubly (1Bst and 2Bst) methylated oligonucleotides (single contours). One inset shows the positions of methylated bases highlighted on the sequence of the 153-bp 601 DNA. All spectra were recorded at 25 °C, 800 MHz, and referenced with DSS. Comparative 1H traces from 13C-1H HMQC spectra of 1H- (red) and 2H- (blue) 601 DNA, shown in a second inset (45 °C, 800 MHz); average relative signal-to-noise ratios (S/N of peaks from 2H vs. 1H samples) for 6mA and 5mC correlations are listed. Spectra have been processed identically, with either pure cosine-bell or exponential multiplication window functions in each dimension with similar relative S/N ratios obtained in both cases.
Fig. 4B shows the spectrum of 2H-601 DNA (25 °C) that is methylated at five sites by the enzymes listed above (black contours), overlaid with the corresponding peaks from oligonucleotides highlighted in Fig. 2, which were used for the assignment of the resonances in the 601 DNA (single contours). The primary 601 DNA sequence is also shown as an inset with the four recognition sites for the chosen MTases indicated along with the sites of methylation (five positions). Interestingly, slight, but consistent, chemical shift differences are observed between the methyl groups from 601 DNA and the oligonucleotides, despite the fact that they have the same base composition in the immediate vicinity of the recognition sites (with the exception of the Msp sequence where the i−2 positions on the top and bottom oligonucleotide strands have G and G replaced by T and C in 601 DNA, respectively). The discrepancy in peak positions is likely the result of end effects in the context of the short oligonucleotides. Indeed, chemical shift perturbations were noted in a comparison of spectra recorded on DNA fragments containing the Dcm recognition sequence, where the methylated base was either 10 bp or 3 bp from the DNA end.
The utility of deuteration in studies of high-molecular-weight proteins is well established (41, 42). We wondered whether similar advantages would be observed in NMR spectra recorded on highly deuterated DNA or whether the lower average proton density in nucleic acids (43) might obviate the need for 2H-labeled samples. Fig. 4 B, Inset shows comparative traces from 13C-1H HMQC spectra of 1H- and 2H-601 DNA (spectrum shown in gray). Average ratios of signal-to-noise (S/N) for 6mA or 5mC correlations from peak intensities measured for each of the DNA samples (S/N of peaks from 2H vs. 1H samples) are also indicated. The 13C-1H HMQC datasets were recorded with a 1H proton excitation pulse of 60° that improves sensitivity per measurement time over a 90° pulse. However, rigorous optimization via the Ernst-angle condition (44) is not possible as the 1H longitudinal relaxation rates of the 6mA and 5mC labels are different (discussed below). Notably, very large gains in S/N ratios are observed for 5mC methyls in the 2H-601 sample, while only modest gains are noted in the context of the 6mA label. In addition, within the 2H-601 sample the intensities of the 5mC correlations are two- to fourfold higher than for the 6mA peaks. In principle, this could result from differences in efficiencies of 5mC and 6mA MTases under the labeling conditions that we have used. However, assays that monitor the extent of methylation show that the MTase-based reactions have gone to completion at all sites (SI Appendix, Fig. S2).
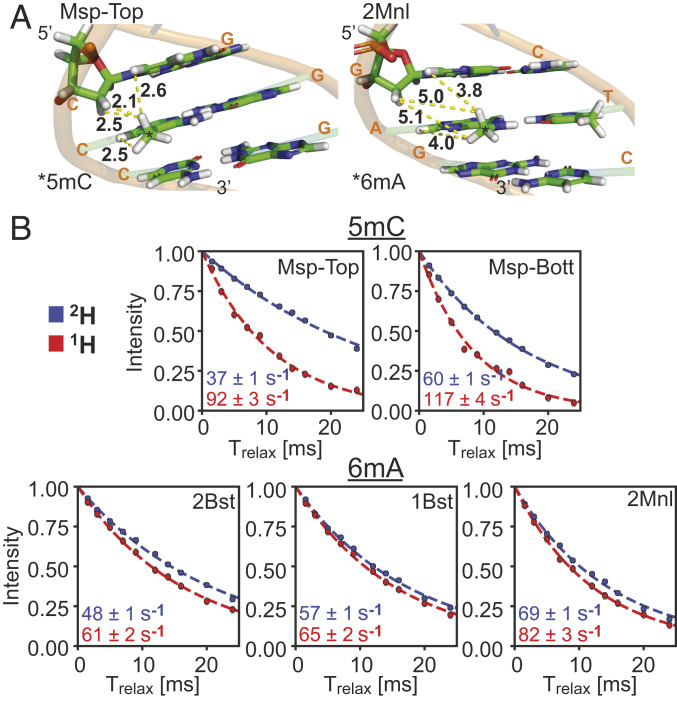
The differences in S/N enhancements for 5mC and 6mA peaks afforded by deuteration can be understood, at least semiquantitatively, by a comparative analysis of methyl 1H transverse (R2) and longitudinal (R1) relaxation rates. R2 rates are directly proportional to linewidths of cross-peaks in spectra, with spectral sensitivity and resolution improving as R2 decreases. In contrast, R1 rates quantify the recovery of magnetization to equilibrium after a perturbation via pulses, with a faster recovery (larger R1) allowing more rapid data acquisition and hence increased S/N per unit measurement time. Fig. 5A shows regions of ideal B-form DNA structures focused on the recognition sites for the Msp and 2Mnl MTases, with methyl groups added to the C5 (Msp) and N6 (2Mnl) positions, respectively. The four shortest distances between one of the methyl protons and neighboring protons are highlighted (yellow broken lines) in each case for a fully protonated molecule, establishing that the proton density is significantly reduced for 6mA relative to 5mC. Methyl 1H R2 rates have been measured using an experiment that selects for the slowly relaxing component of magnetization (45), producing the single exponential decay curves shown in Fig. 5B for fully protonated (red) and highly deuterated (blue) 601 DNA samples. For all of the five methyl groups considered the rates are attenuated with deuteration, but there are much larger changes in 1H R2 rates upon deuteration for the 5mC methyl groups (Msp) relative to the 6mA methyls (1Bst, 2Bst, and 2Mnl), consistent with differences in proton densities for the fully protonated samples in the two cases. Thus, the significant changes in relative S/N values of 5mC correlations in 2H- vs. 1H-601 DNA samples can be understood, in part, in terms of the large reduction in methyl 1H R2 values that accompany deuteration.
Fig. 5.
Effect of deuteration on methyl 1H R2 relaxation rates in 2H- and 1H-601 DNA. (A) Segments of ideal B-form 601 DNA structures with methyl groups added to the C5 (Msp, Left) and N6 (2Mnl, Right) positions of the central C and A bases. Yellow dashed lines connect methyl protons with the closest neighboring protons in the structure. (B) The 1H transverse relaxation rates (slowest component) measured at 45 °C, 800 MHz (circles) and single exponential fits (dashed lines) of data from the five 13CH3 labeled positions in the 2H- (blue) and 1H- (red) 601 DNA samples. Extracted R2 values are listed in the bottom left corner.
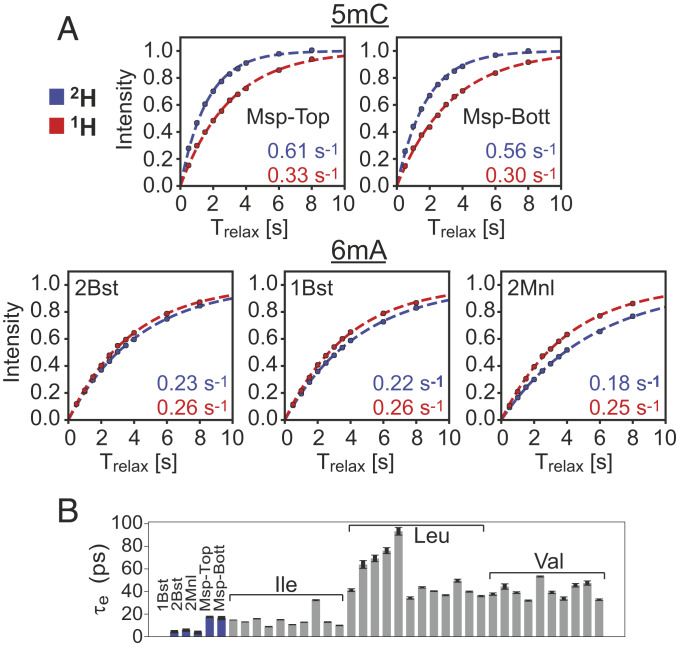
A second contributing factor to the differences in S/N enhancements between 2H- and 1H-601 DNA samples, at least in so far as the 5mC correlations are concerned, arises from R1 relaxation rates, which are approximately twofold larger for the deuterated sample (Fig. 6A; compare red and blue profiles for Msp). In protein NMR applications involving protonated samples it is widely appreciated that methyl groups serve as relaxation sinks because the presence of three protons increases the relaxation efficiency, with the three-fold methyl rotation an important determinant of the longitudinal relaxation (discussed below), such that methyl protons typically relax faster than neighboring nonmethyl proton spins (46, 47). This leads to efficient cross-relaxation between the spins, increasing the effective longitudinal relaxation rates of nonmethyl protons with a concomitant decrease in methyl 1H R1 rates. In contrast, in highly deuterated, methyl protonated samples these cross-relaxation pathways are eliminated, effectively enhancing methyl 1H longitudinal relaxation. This phenomenon is clearly observed in the Msp profiles in Fig. 6A. The reverse situation is noted for the 1H R1s of the 6mA methyl groups. Although the R1 differences are smaller between fully protonated and deuterated samples, methyl 1H longitudinal relaxation in the protonated sample is clearly more efficient for the three labeled 6mA sites considered. Moreover, the intrinsic 6mA R1 rates (in the 2H sample where there is little cross-relaxation) are ∼2.5-fold smaller than for 5mC. Thus, the slowly relaxing 6mA methyl protons are no longer effective relaxation sinks since they relax slowly; in fact, our data suggest that the nonmethyl protons in the protonated sample enhance relaxation of the 6mA protons via cross-relaxation (compare red and blue traces of 2Bst, 1Bst, and 2Mnl).
Fig. 6.
Effect of deuteration on methyl 1H R1 relaxation rates in 2H- and 1H-601 DNA. (A) The 1H magnetization recovery curves (starting from zero magnetization) of 5mC (Msp) and 6mA (2Bst, 1Bst, and 2Mnl) methyl groups in 2H- (blue) and 1H- (red) 601 DNA samples, 45 °C, 800 MHz. R1 rates, obtained from exponential fits (dashed lines) of the experimental data (circles), are shown in the bottom right-hand side of each panel (errors are in the third decimal point and are not included). (B) Bar plot of τe values (correlation times for the rapid rotation of methyl groups about their threefold axes) from fits of 13C R1 relaxation rates of methyls in free 601 DNA (blue) or of the methyl groups of H2B (gray) as part of the NCP complex.
One of the advantages of using methyl-group probes in studies of high-molecular-weight proteins is that, unlike the case for backbone amides buried in the protein core, 1H R1 rates increase very little with molecular weight. For an isolated 13CH3 group the dominant contributions to 1H longitudinal relaxation are given by one-bond intramethyl 1H-13C and 1H-1H dipolar interactions. In the macromolecular limit, ωcτc,eff >> 1, where ωc is the 13C Larmor frequency (radians per second) and τc,eff is the effective overall rotational correlation time for the methyl group in question, the 1H relaxation rate can be well approximated by (48)
| [1] |
independent of τc,eff. In Eq. 1 γj is the gyromagnetic ratio of spin j, h is Planck’s constant, rHC is the length of the 1H-13C bond, τe is the correlation time for the rapid rotation about the methyl threefold axis, is the square of an order parameter that quantifies the amplitude of the methyl axis motion, and the factors of 1/4 and 1/9 take into account the orientation of the methyl 1H-1H and 1H-13C bond vectors, respectively, with respect to the threefold rotation axis. In studies of a range of highly deuterated, methyl protonated proteins we have measured R1 values on the order of 0.8 s−1 – 1.5 s−1 (5, 49), with the variability largely reflecting differences in site-specific τe values.
The slow longitudinal relaxation rates of 6mA methyl groups in DNA, which fall well outside the typical range of measured R1 values in proteins, are consistent with very rapid rotation about the methyl threefold axis (i.e., small τe). In order to quantify methyl rotation rates we have measured methyl 13C R1 values (50) in the 2H-601 DNA sample described above. In the macromolecular limit the 13C relaxation rate can be well approximated by (48)
| [2] |
so that values of τe can be readily obtained from the measured values. The τe values for the five methyl labels in the 2H-601 DNA sample (free DNA) are indicated in Fig. 6B (blue). Also shown are the corresponding values for the Ile, Leu, and Val methyl groups of histone H2B in an NCP sample (gray; discussed below). Notably, τe ∼ 5 ps and ∼ 20 ps for the 6mA and 5mC methyl groups, respectively, accounting for the significant differences in the 1H R1 rates that were measured for each methyl type in DNA. In contrast, while the τe values for Ileδ1 and 5mC methyls are similar, those for Leu and Val are significantly larger, on average, so that higher 1H R1 rates are measured for these methyls in proteins.
Applications to the NCP.
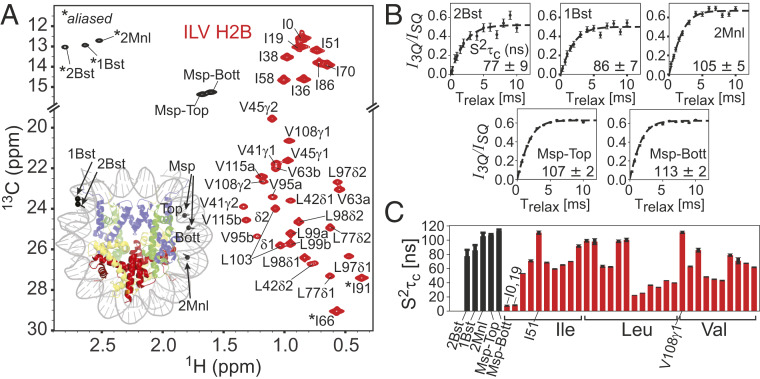
A major goal of the present work is to develop an NMR approach to study structural and motional properties of both the DNA and the protein components in high-molecular-weight nucleic acid–protein complexes. The NCP is one such example and to date NMR studies of this important complex have focused on the histone proteins exclusively, using largely 15N-based experiments to quantify the dynamics of the disordered protein tails (51, 52) and methyl-TROSY approaches to study the more rigid and structured elements of the histones and interactions between histones and other proteins that play important roles in regulating NCP function (53–58). Fig. 7A shows the 13C-1H HMQC spectrum of a complex in which highly deuterated 153-bp Widom DNA, 13CH3-methyl–labeled at the five sites discussed above, is wrapped around a deuterated octamer core in which the Ile, Leu, and Val methyl groups of deuterated H2B are labeled as 13CH3 (Ile δ1) and 13CH3,12CD3 [Leu, Val; labeling is not stereospecific (5)] and the remaining histones are perdeuterated. A comparison of spectra of methyl labels in the free and wrapped DNA is provided in SI Appendix, Fig. S3. In what follows deuterated H2B methyl-labeled in this manner will be referred to as ILV-H2B. The methyl-group peaks from DNA are well resolved in spectra from their protein-based counterparts, so that site-specific information can be obtained for each component of the complex simultaneously. Notably, high-quality datasets can be measured rapidly; in the present case the measurement time was 40 min and a sample concentration of 150 μM in NCP, 45 °C (800 MHz) was used. Shown also in Fig. 7A is a cartoon representation of the NCP (PBD ID code 6ESF; ref. 59) with each histone color-coded (H2A, yellow; H2B, red; H3, blue; and H4, green) and the positions of the methyl groups on the DNA indicated by black balls.
Fig. 7.
Methyl groups of 601 DNA in the nucleosome are well-ordered. (A) The 13C-1H HMQC spectrum of ILV-H2B, and 13CH3-5mC + 13CH3-6mA at the five positions indicated in Fig. 4B in an otherwise deuterated NCP particle. The protein resonances are contoured in red, with assignments (53) as indicated (correlations for I66 and I91 are aliased). In cases where stereospecific assignments are not available for Leu and Val methyl groups they are denoted by “a” (upfield in 1H) or “b.” Peaks derived from the methyl groups of the DNA are in black (assignments transferred from the free 601 DNA sample), with the positions of the 6mA methyls aliased (*). We have not observed the peak doubling of methyl resonances (one from each of the two H2B copies in the NCP) that might be expected for a nonpalindromic sequence like the Widom 601 DNA (53). This reflects the fact that the methyl groups in H2B are either not sufficiently close to DNA or localize to a local symmetry site of the 601 DNA (I36δ1; see main text). (Inset) The structure of the NCP with H2A, H2B, H3, and H4 indicated in yellow, red, blue, and green, respectively, with the DNA methyl sites highlighted by black spheres on the canonical structure of the NCP (PDB ID code 6ESF; ref. 59). (B) Plots of intensity ratios, I3Q/ISQ, of DNA methyl cross-peaks derived from 1H triple-quantum (I3Q) and single-quantum (ISQ) coherences as a function of an evolution time, Trelax, during which transverse relaxation occurs (60). These can be fit (dashed lines) to extract values (lower right-hand side of panels). (C) Bar plot of values for methyl groups from 601 DNA (black) and H2B (red) of the NCP complex. All experiments were recorded at 45 °C, 800 MHz.
The R1 relaxation experiments discussed above indicate significant differences in rotation rates about the methyl threefold axis for 5mC and 6mA labels, and also between DNA- and protein-based methyl groups. We were interested, therefore, in establishing whether these differences extend to other motional parameters as well. With this in mind we carried out methyl 1H triple-quantum–based experiments (60), Fig. 7B. The ratio of signal intensities derived from triple-quantum vs. single-quantum methyl 1H coherences (y axis) as a function of the time, Trelax, during which transverse 1H relaxation occurs (x axis) generates profiles which can be fit (SI Appendix) to quantify the product on a per-residue basis for methyl groups attached to both DNA and H2B in the NCP sample. Fig. 7C plots the obtained values for the different labels in DNA (black) and for different residues in H2B (red). As expected, a wide range of values is observed for the methyl groups in H2B, from very low for Ile0δ1 and Ile19δ1 that are localized to the H2B unstructured N-terminal tail to high for Ile51δ1 and Va108γ1 that are part of the folded H2B histone domain and that make important hydrophobic contacts that help stabilize the H2A–H2B dimer interface and the H2B fold, respectively. In contrast, values for the DNA probes are more uniform, reflecting their attachment to bases that are Watson–Crick-paired.
Extraction of motional parameters from methyl spin relaxation studies of high-molecular-weight systems is hampered by the fact that it is difficult to separate from τc,eff, as R1 measurements report on τe and R2 rates are sensitive to the product . In contrast, R1 and R2 relaxation rates of backbone spins, that can only be measured on relatively small complexes, are sensitive to and , respectively, so that the separation of and τc,eff becomes possible. In the present case we have used HYDROPRO (61) and the X-ray structure of a histone octamer from Xenopus laevis wrapped with Widom 601 DNA (PDB ID code 3LZ0; ref. 29) to estimate τc,eff, with a value of 114 ns obtained from the calculated diffusion tensor. We have also considered a more rigorous approach where the diffusion tensor at each of a series of molecular dynamics trajectory snapshots was averaged (the averaged tensor was kindly provided by Nikolai Skrynnikov, Purdue University, West Lafayette, IN) from which we have calculated τc,eff values for each DNA methyl group based on its orientation in the principal axis system of the diffusion tensor. Very little difference was obtained between the two methods, with values of 0.94 ± 0.02 (Msp-top), 0.99 ± 0.02 (Msp-bottom), 0.75 ± 0.06 (1Bst), 0.68 ± 0.08 (2Bst), and 0.93 ± 0.04 (2Mnl). The Msp methyl groups are the most rigid of those studied, while 2Bst and 1Bst have more mobility, likely due to their proximity to one of the DNA ends (6 and 7 bases away from the end). Of interest, the 1Bst and 2Bst methyl groups (or Msp methyls) that are proximal to each other, and hence report on the dynamics of the same DNA region, have similar order parameters, suggesting that very local, site-specific perturbations resulting from the placement of a given methyl group probe are likely to be small, at least in these cases.
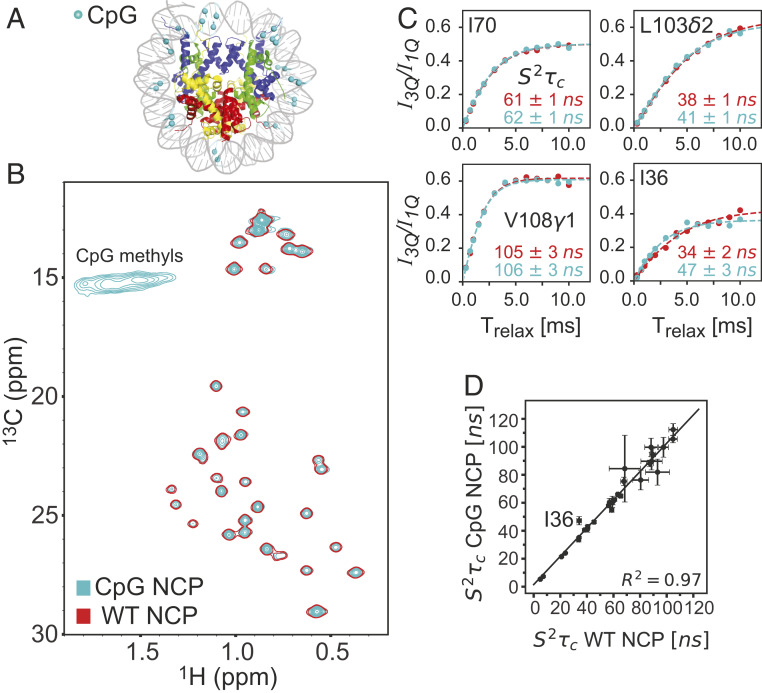
Previous studies have shown that in the context of small DNA oligonucleotides methylation at the cytosine C5 and adenosine N6 positions can lead to changes in stability, with 5mC resulting in either increases or decreases in stability in a manner that is sequence specific (62) and 6mA in decreases (63), an effect that has recently been confirmed in the context of RNA where 6mA labeling was shown to significantly slow the kinetics of duplex annealing with little change to the rate of strand dissociation (64). The DNA methyl labeling approaches that we have developed provide an avenue for addressing by NMR how DNA methylation might affect the structural and dynamical properties of the NCP. The main site of methylation in eukaryotes is the 5mC position (65) and in animals these are mostly localized to CpG dinucleotides (24). While CpGs throughout mammalian genomes are highly methylated, long repeats of unmethylated CpGs exist, so-called CpG islands, which are usually localized to promoter regions (66). Methylation of CpG islands leads to transcriptional repression, presumably via direct interference with the binding of transcription factors or indirectly, by recruitment of DNA binding proteins that recognize methylated CpG sites and block interactions with proteins required for transcription (67, 68). X-ray studies at the level of mononucleosomes establish that CpG methylation does not perturb the structure of the NCP (69). A number of investigations, however, indicate that a high degree of methylation of nucleosomal DNA does result in changes to nucleosome compaction and rigidity. Single-molecule fluorescence resonance energy transfer (FRET)-based studies report that NCPs become more tightly wrapped when the DNA is CpG-methylated, potentially leading to the transcriptional repression observed in highly methylated chromatin (70, 71). In contrast, subsequent studies by Jimenez-Useche et al. (72) present evidence that methylation leads to a decreased level of compaction of nucleosomal DNA ends. Yet another study, based on single-molecule FRET and optical tweezers, concludes that methylation both loosens packing of DNA ends, leading to faster unwrapping of DNA, and causes NCP mechanical destabilization (73). The authors speculate that loosening of the DNA ends of NCPs in the larger context of chromatin may permit histone tails of adjacent nucleosomes to interact, resulting in nucleosome condensation and repression of transcription. All of these studies have focused on the “DNA side” of the NCP and it is of interest to extend such investigations to include an analysis of whether the dynamics of the protein components are affected. Here we focus on H2B by preparing an NCP sample with ILV-H2B, perdeuterated histones H2A, H3, and H4, and fully protonated 5mCpG 601 DNA. The methylated DNA was generated using the in vitro approach illustrated in Fig. 1 by adding the M.SssI CpG MTase that specifically methylates the C5 positions of all cytosine bases in CpG dinucleotide sequences. As the added Met was 13C-labeled in the methyl position, all 5mCpG sequences in the DNA are 13C-labeled. The methyl-labeling reaction was shown to go to completion by establishing protection against the HhaI restriction enzyme that recognizes and cuts DNA at (unmethylated) 5′-GCGC-3′ sites (SI Appendix, Fig. S4). The 26 CpG sites in the 153-bp 601 Widom DNA used here are distributed across the sequence, as illustrated in Fig. 8A. Fig. 8B presents an overlay of 13C-1H HMQC spectra (45 °C, 800 MHz) of wild-type (WT; single contour, red) and CpG-methylated (light blue) NCPs, showing ILV-methyl correlations from H2B, as well as weak and very broad cross-peaks that arise from the 5mCpG-labeled DNA. As we were not interested in quantifying the DNA peaks in this application the broad 5mC correlations are not a concern, but their poor quality does emphasize the importance of deuteration in studies of high-molecular-weight complexes that focus on spectra of 5mC-labeled DNA. HMQC correlation maps of methyl-labeled proteins are, in general, exquisitely sensitive to the overall structure of the biomolecule under investigation, so that the superposition of the spectra shown here provides strong evidence that DNA methylation has no effect on the overall conformation of H2B, consistent with prior X-ray studies of 5mCpG NCPs (69). A comparative methyl 1H triple-quantum dynamics study was performed on NCP samples containing either 5mCpG 601 or WT 601 DNA, Fig. 8C, from which values were calculated and compared in Fig. 8D. Notably, a very strong correlation is obtained, indicating no differences in picosecond to nanosecond timescale dynamics for the ILV side chains of H2B in either of the two samples. Of the 26 residues compared, only in one case (Ile36) were the values different between the two samples and in this instance the difference may reflect the close proximity between Ile36δ1 and the and sugar protons of a deoxyguanosine in the DNA (<3.5 Å); short distances between methyl protons of interest and external protons can lead to errors in quantitation (74).
Fig. 8.
CpG methylation does not perturb the structure or picosecond to nanosecond timescale dynamics of histone H2B in the context of the nucleosome. (A) NCP model (PDB ID code 6ESF; ref. 59) highlighting the positions of the 26 CpG methyl groups in 601 DNA as cyan-colored balls. (B) Overlay of 13C-1H HMQC spectra of CpG-methylated (cyan, multiple contours) and unmethylated (WT, red, single contour) NCPs, prepared with ILV-H2B, deuterated H2A, H3, and H4, as well as protonated 601 DNA. The 5mC methyl groups of the 13 CpG sites per strand were 13CH3; correlations from these methyl groups were of low quality due to relaxation with adjacent protons (see main text). Significant differences in peak linewidths in 13C and 1H dimensions were not observed between WT and CpG NCP samples. (C) Superposition of plots of I3Q/ISQ intensity ratios of selected ILV peaks in CpG and WT NCPs as a function of Trelax and (D) extracted values from fits of the data, presented as a linear correlation plot. With the exception of I36, for which small differences in times are obtained between the two samples, all other values are the same to within experimental error. The line y = x is plotted to guide the eye. All experiments were recorded at 45 °C, 800 MHz.
Concluding Remarks
The development of facile labeling methods has been critical to the success of biomolecular NMR (75, 76). New labeling technologies invariably stimulate advances in experiments that are designed to exploit the label, leading, for example, to NMR studies of high-molecular-weight proteins (77, 78). Here we sought to develop similar strategies for focusing on DNA, so as to begin to address structure–dynamics–function relationships in the NCP. Labeling of DNA with NMR-active spins has been challenging as in vitro enzymatic synthesis requires a new template for each copy of DNA produced, unlike the case for the biosynthesis of RNA, which limits quantities that can be generated (16). In vivo-based labeling approaches have emerged in which multiple copies of the desired sequence are inserted into a plasmid that is then replicated (79); however a drawback with such enzymatic approaches is that all nucleotides of a given type become labeled, challenging spectral resolution in applications involving all but the smallest of DNA sequences. Chemical synthesis of DNA has become a powerful option as it is possible to add labeled phosphoramidite building blocks at various stages of the synthesis so as to position the NMR labels at the desired locations in the DNA sequence (20).
Here we describe a one-pot reaction in which the appropriate MTases are added to a reaction mixture containing 13CH3-methinone, ATP, S-adenosylmethionine synthase, and the desired DNA to produce 5mC- or 6mA-labeled DNA. Because the number of labeled sites can be controlled by either judicious choice of the DNA sequence or the MTases that are used, or both, spectral resolution is not limiting, even in studies of very high-molecular-weight DNAs, such as the 153-bp Widom sequence. The use of methyl labels as probes benefits from a wide range of experiments developed for protein applications exploiting a methyl-TROSY effect that provides high spectral resolution and sensitivity. Although replacement of DNA protons with deuterons is likely not necessary when 6mA probes exclusively are used and when the i−1 base is either C or G, as in the present applications, 1H distances to the methyl probe are smaller when the i−1 base is T and advantages with deuteration may occur (SI Appendix, Fig. S5). Replacement of DNA protons with deuterons is an absolute requirement for studies involving 5mC methyl groups due to higher proton densities in the regions surrounding these methyl protons.
A comparison of relaxation properties of 5mC and 6mA methyl groups in 2H-601 DNA establishes that the 6mA 1H T1 values are significantly larger than for 5mC, and over a factor of three larger, on average, than in proteins. This follows directly from very rapid rotation about the methyl symmetry axis, with τe values ranging from 3 to 6 ps for the 6mA methyls, based on 13C R1 relaxation measurements. These values are in excellent agreement with τe estimated from a solid-state 2H-NMR study of 2′-deoxythymidine, where a room-temperature value of τe = 4.2 ps was obtained (80). The long 1H T1 values of the 6mA labels result in correlations of decreased S/N in spectra in relation to peaks derived from 5mC. In principle, 1H relaxation can be enhanced through the use of paramagnetic relaxation agents (81), but we have not pursued that here. Notably, high-quality 13C-1H HMQC datasets can be recorded on samples of modest concentrations (100 μM in NCP) using measurement times of well under 1 h on spectrometers with state-of-the-art cryoprobes.
Methyl labeling of DNA is a common epigenetic mark and it is of interest to understand how 5 mC labeling, the most common mark in eukaryotes (82) and occurring predominantly at CpG sites in mammals, affects the structural dynamics of the nucleosome. Using an NCP sample prepared with 601-DNA containing 26 5mCpG sites, and ILV-H2B as a read-out, we observed no structural changes, consistent with previous X-ray studies (69). FRET experiments have shown, however, that there are changes in DNA compaction levels (71) and in the dynamics of the DNA ends (73). The effect of DNA methylation on histone dynamics has, to our knowledge, not been explored; our study establishes little relative change in H2B picosecond to nanosecond timescale dynamics in NCPs prepared with 5mCpG- DNA or with unlabeled DNA. Further studies will focus on larger constructs involving multiple copies of NCPs to establish whether interactions between adjacent core particles are affected by methylation.
The ability to generate 13CH3-labeled DNA in a facile manner, in concert with methyl-TROSY–based experiments, provides an avenue for solution NMR studies of high-molecular-weight DNA and protein–DNA complexes, as illustrated here in the context of the NCP. The combination of high-resolution electron cryomicroscopy and X-ray based structural studies and NMR applications focusing on dynamics over a wide range of timescales is expected to be a powerful and complementary strategy for elucidating structure–dynamics–function paradigms in large protein-DNA systems.
Materials and Methods
Details of DNA and protein expression and purification and NMR measurements, along with data fitting, are provided in SI Appendix.
Data Availability.
All relevant data are included in the paper and SI Appendix, and expression vectors for production of MTases and histones are available upon request from the authors.
Supplementary Material
Acknowledgments
We thank Professor Nikolai Skrynnikov (Purdue University) and Dr. Sevastayan Rabdano (St. Petersburg State University) for providing an average rotational diffusion tensor for the NCP from their molecular dynamics trajectory. This research was funded by Canadian Institutes of Health Research Grant MOP-133408. L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004317117/-/DCSupplemental.
References
- 1.Wüthrich K., NMR of Proteins and Nucleic Acids, (Wiley, 1986). [Google Scholar]
- 2.Pervushin K., Riek R., Wider G., Wüthrich K., Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pervushin K., Riek R., Wider G., Wüthrich K., Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 120, 6394–6400 (1998). [Google Scholar]
- 4.Tugarinov V., Hwang P. M., Ollerenshaw J. E., Kay L. E., Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Tugarinov V., Kay L. E., An isotope labeling strategy for methyl TROSY spectroscopy. J. Biomol. NMR 28, 165–172 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Rosenzweig R., Kay L. E., Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 83, 291–315 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y., Kalodimos C. G., NMR Studies of large proteins. J. Mol. Biol. 429, 2667–2676 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Schuetz A. K., Kay L. E., A dynamic molecular basis for malfunction in disease mutants of p97/VCP. eLife 5, e20143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Religa T. L., Sprangers R., Kay L. E., Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 328, 98–102 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Kimsey I. J., Petzold K., Sathyamoorthy B., Stein Z. W., Al-Hashimi H. M., Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 519, 315–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke J. E., Sashital D. G., Zuo X., Wang Y. X., Butcher S. E., Structure of the yeast U2/U6 snRNA complex. RNA 18, 673–683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keane S. C. et al., RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348, 917–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kocman V., Plavec J., Tetrahelical structural family adopted by AGCGA-rich regulatory DNA regions. Nat. Commun. 8, 15355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlomagno T., Present and future of NMR for RNA-protein complexes: A perspective of integrated structural biology. J. Magn. Reson. 241, 126–136 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Flamme M., McKenzie L. K., Sarac I., Hollenstein M., Chemical methods for the modification of RNA. Methods 161, 64–82 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Nelissen F. H. T., Tessari M., Wijmenga S. S., Heus H. A., Stable isotope labeling methods for DNA. Prog. Nucl. Magn. Reson. Spectrosc. 96, 89–108 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Lu K., Miyazaki Y., Summers M. F., Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR 46, 113–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai S., Kumar P., Hellen C. U. T., D’Souza V. M., Wagner G., An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 23, 859–864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki Y. et al., Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 404, 751–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nußbaumer F. et al., Synthesis and incorporation of 13C-labeled DNA building blocks to probe structural dynamics of DNA by NMR. Nucleic Acids Res. 45, 9178–9192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y. et al., Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature 522, 368–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casadesús J., ““Bacterial DNA methylation and methylomes”” in DNA Methyltransferases: Role and Function, Jeltsch A., Jurkowska R., Eds. (Advances in Experimental Medicine and Biology, Springer, New York, 2016), Vol. 945, pp. 35–61. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E. V., Makarova K. S., Wolf Y. I., Evolutionary genomics of defense systems in archaea and bacteria. Annu. Rev. Microbiol. 71, 233–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laird P. W., Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Riggs A. D., Pfeifer G. P., X-chromosome inactivation and cell memory. Trends Genet. 8, 169–174 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Li E., Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Jaenisch R., Bird A., Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Vasudevan D., Chua E. Y. D., Davey C. A., Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Markham G. D., Hafner E. W., Tabor C. W., Tabor H., S-Adenosylmethionine synthetase from Escherichia coli. J. Biol. Chem. 255, 9082–9092 (1980). [PubMed] [Google Scholar]
- 31.Park J., Tai J., Roessner C. A., Scott A. I., Overcoming product inhibition of S-Adenosyl-L-methionine (SAM) synthetase: Preparation of SAM on the 30 mM scale. Bioorg. Med. Chem. Lett. 5, 2203–2206 (1995). [Google Scholar]
- 32.Morgan R., Xiao J., Xu S., Characterization of an extremely thermostable restriction enzyme, PspGI, from a Pyrococcus strain and cloning of the PspGI restriction-modification system in Escherichia coli. Appl. Environ. Microbiol. 64, 3669–3673 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman D. R., Cornatzer W. E., Duerre J. A., Relationship between tissue levels of S-adenosylmethionine, S-adenylhomocysteine, and transmethylation reactions. Can. J. Biochem. 57, 56–65 (1979). [DOI] [PubMed] [Google Scholar]
- 34.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E., A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Korzhnev D. M., Kloiber K., Kanelis V., Tugarinov V., Kay L. E., Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: Application to a 723-residue enzyme. J. Am. Chem. Soc. 126, 3964–3973 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Engel J. D., von Hippel P. H., Effects of methylation on the stability of nucleic acid conformations: Studies at the monomer level. Biochemistry 13, 4143–4158 (1974). [DOI] [PubMed] [Google Scholar]
- 37.McGhee J. D., Felsenfeld G., Nucleosome structure. Annu. Rev. Biochem. 49, 1115–1156 (1980). [DOI] [PubMed] [Google Scholar]
- 38.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Bird A., DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Dyer P. N. et al., Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Farmer B. T., Venters R. A., ““NMR of perdeuterated large proteins”” in Modern Techniques in Protein NMR, Krishna N. R., Berliner L. J., Eds. (Kluwer Academic Publishers, 2006), pp. 75–120. [Google Scholar]
- 42.Gardner K. H., Kay L. E., The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 27, 357–406 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Barnwal R. P., Yang F., Varani G., Applications of NMR to structure determination of RNAs large and small. Arch. Biochem. Biophys. 628, 42–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst R. R., Anderson W. A., Application of fourier transform spectroscopy to magnetic resonance. Rev. Sci. Instrum. 37, 93–102 (1966). [Google Scholar]
- 45.Tugarinov V., Kay L. E., Relaxation rates of degenerate 1H transitions in methyl groups of proteins as reporters of side-chain dynamics. J. Am. Chem. Soc. 128, 7299–7308 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Sykes B. D., Hull W. E., Snyder G. H., Experimental evidence for the role of cross-relaxation in proton nuclear magnetic resonance spin lattice relaxation time measurements in proteins. Biophys. J. 21, 137–146 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalk A., Berendsen H. J. C., Proton magnetic relaxation and spin diffusion in proteins. J. Magn. Reson. 24, 343–366 (1976). [Google Scholar]
- 48.Rennella E., Huang R., Velyvis A., Kay L. E., (13)CHD2-CEST NMR spectroscopy provides an avenue for studies of conformational exchange in high molecular weight proteins. J. Biomol. NMR 63, 187–199 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Religa T. L., Kay L. E., Optimal methyl labeling for studies of supra-molecular systems. J. Biomol. NMR 47, 163–169 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Bouvignies G., Kay L. E., A 2D 13C-CEST experiment for studying slowly exchanging protein systems using methyl probes: An application to protein folding. J. Biomol. NMR 53, 303–310 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Zhou B. R. et al., Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. J. Mol. Biol. 421, 30–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato H., Gruschus J., Ghirlando R., Tjandra N., Bai Y., Characterization of the N-terminal tail domain of histone H3 in condensed nucleosome arrays by hydrogen exchange and NMR. J. Am. Chem. Soc. 131, 15104–15105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato H. et al., Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U.S.A. 108, 12283–12288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horn V. et al., Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat. Commun. 10, 1751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitevski-LeBlanc J. et al., The RNF168 paralog RNF169 defines a new class of ubiquitylated histone reader involved in the response to DNA damage. eLife 6, e23872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitevski-LeBlanc J. L. et al., Investigating the dynamics of destabilized nucleosomes using methyl-TROSY NMR. J. Am. Chem. Soc. 140, 4774–4777 (2018). [DOI] [PubMed] [Google Scholar]
- 57.van Emmerik C. L., van Ingen H., Unspinning chromatin: Revealing the dynamic nucleosome landscape by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 110, 1–19 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Sinha K. K., Gross J. D., Narlikar G. J., Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science 355, eaaa3761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bilokapic S., Strauss M., Halic M., Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 25, 101–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun H., Kay L. E., Tugarinov V., An optimized relaxation-based coherence transfer NMR experiment for the measurement of side-chain order in methyl-protonated, highly deuterated proteins. J. Phys. Chem. B 115, 14878–14884 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Ortega A., Amorós D., García de la Torre J., Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101, 892–898 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Severin P. M. D., Zou X., Gaub H. E., Schulten K., Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res. 39, 8740–8751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Engel J. D., von Hippel P. H., Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J. Biol. Chem. 253, 927–934 (1978). [PubMed] [Google Scholar]
- 64.Shi H. et al., NMR chemical exchange measurements reveal that N6-methyladenosine slows RNA annealing. J. Am. Chem. Soc. 141, 19988–19993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iyer L. M., Abhiman S., Aravind L., Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 101, 25–104 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Takai D., Jones P. A., Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U.S.A. 99, 3740–3745 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tate P. H., Bird A. P., Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 3, 226–231 (1993). [DOI] [PubMed] [Google Scholar]
- 68.Leonhardt H., Rahn H. P., Cardoso M. C., Functional links between nuclear structure, gene expression, DNA replication, and methylation. Crit. Rev. Eukaryot. Gene Expr. 9, 345–351 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Fujii Y., Wakamori M., Umehara T., Yokoyama S., Crystal structure of human nucleosome core particle containing enzymatically introduced CpG methylation. FEBS Open Bio 6, 498–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J. Y., Lee T. H., Effects of DNA methylation on the structure of nucleosomes. J. Am. Chem. Soc. 134, 173–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choy J. S. et al., DNA methylation increases nucleosome compaction and rigidity. J. Am. Chem. Soc. 132, 1782–1783 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jimenez-Useche I. et al., DNA methylation regulated nucleosome dynamics. Sci. Rep. 3, 2121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ngo T. T. M. et al., Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 7, 10813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tugarinov V., Sprangers R., Kay L. E., Probing side-chain dynamics in the proteasome by relaxation violated coherence transfer NMR spectroscopy. J. Am. Chem. Soc. 129, 1743–1750 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Zhang H., van Ingen H., Isotope-labeling strategies for solution NMR studies of macromolecular assemblies. Curr. Opin. Struct. Biol. 38, 75–82 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Kainosho M. et al., Optimal isotope labelling for NMR protein structure determinations. Nature 440, 52–57 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Sprangers R., Kay L. E., Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445, 618–622 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Fiaux J., Bertelsen E. B., Horwich A. L., Wüthrich K., NMR analysis of a 900K GroEL GroES complex. Nature 418, 207–211 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Louis J. M., Martin R. G., Clore G. M., Gronenborn A. M., Preparation of uniformly isotope-labeled DNA oligonucleotides for NMR spectroscopy. J. Biol. Chem. 273, 2374–2378 (1998). [DOI] [PubMed] [Google Scholar]
- 80.Kintanar A. et al., Solid-state 2H NMR investigation of internal motion in 2′-deoxythymidine. J. Am. Chem. Soc. 110, 6367–6372 (1988). [Google Scholar]
- 81.Eletsky A., Moreira O., Kovacs H., Pervushin K., A novel strategy for the assignment of side-chain resonances in completely deuterated large proteins using 13C spectroscopy. J. Biomol. NMR 26, 167–179 (2003). [DOI] [PubMed] [Google Scholar]
- 82.Luo C., Hajkova P., Ecker J. R., Dynamic DNA methylation: In the right place at the right time. Science 361, 1336–1340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Press W. H., Teukolsky S. A., Vetterling W. T., Flannery B. P., Numerical Recipes in C: The Art of Scientific Computing, (Cambridge University Press, ed. 2, 1988). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and SI Appendix, and expression vectors for production of MTases and histones are available upon request from the authors.