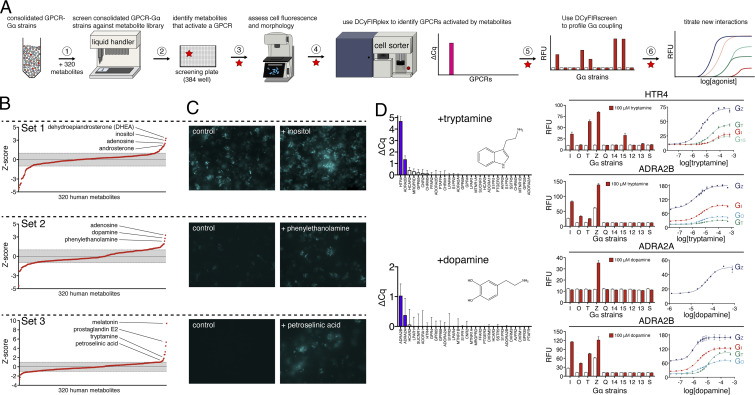
Significance
G protein-coupled receptors (GPCRs) are the largest class of membrane receptors in humans. As such, GPCR signaling is central to human biology and medicine. While more than 30% of approved drugs target roughly 150 GPCRs, most receptors lack well-defined endogenous ligands and are currently not druggable. To address this challenge, we created DCyFIR, a GPCR screening platform for ligand and drug discovery. This technology enables the cost-effective profiling of ligands and drug compounds against mixtures of hundreds of GPCR-barcoded cell strains in a single experiment. Because ligands are tested against many receptors simultaneously, DCyFIR profiling will eventually enable the physical screening of huge numbers of compounds, bringing wet-laboratory experimental throughput a step closer to what is done in silico.
Keywords: G protein-coupled receptors, DCyFIR, multiplex, pharmacologically dark GPCR, yeast
Abstract
More than 800 G protein-coupled receptors (GPCRs) comprise the largest class of membrane receptors in humans. While there is ample biological understanding and many approved drugs for prototypic GPCRs, most GPCRs still lack well-defined biological ligands and drugs. Here, we report our efforts to tap the potential of understudied GPCRs by developing yeast-based technologies for high-throughput clustered regularly interspaced short palindromic repeats (CRISPR) engineering and GPCR ligand discovery. We refer to these technologies collectively as Dynamic Cyan Induction by Functional Integrated Receptors, or DCyFIR. A major advantage of DCyFIR is that GPCRs and other assay components are CRISPR-integrated directly into the yeast genome, making it possible to decode ligand specificity by profiling mixtures of GPCR-barcoded yeast strains in a single tube. To demonstrate the capabilities of DCyFIR, we engineered a yeast strain library of 30 human GPCRs and their 300 possible GPCR–Gα coupling combinations. Profiling of these 300 strains, using parallel (DCyFIRscreen) and multiplex (DCyFIRplex) DCyFIR modes, recapitulated known GPCR agonism with 100% accuracy, and identified unexpected interactions for the receptors ADRA2B, HCAR3, MTNR1A, S1PR1, and S1PR2. To demonstrate DCyFIR scalability, we profiled a library of 320 human metabolites and discovered several GPCR–metabolite interactions. Remarkably, many of these findings pertained to understudied pharmacologically dark receptors GPR4, GPR65, GPR68, and HCAR3. Experiments on select receptors in mammalian cells confirmed our yeast-based observations, including our discovery that kynurenic acid activates HCAR3 in addition to GPR35, its known receptor. Taken together, these findings demonstrate the power of DCyFIR for identifying ligand interactions with prototypic and understudied GPCRs.
G protein-coupled receptors (GPCRs) mediate cellular decision-making and physiological processes by detecting a wide variety of chemical signals, such as small molecule regulators, peptides, and proteins. GPCRs transduce these extracellular signals across the plasma membrane to activate intracellular G proteins that amplify the receptor response through a variety of downstream second messengers (cAMP, IP3, DAG, and Ca2+). While more than 360 nonolfactory GPCRs comprise the largest and most therapeutically targeted class of membrane receptors in humans, only 30% to 40% have well-defined biological ligands and are currently druggable (1). The remaining 60% to 70%, more than a hundred of which are classified as pharmacologically dark (2), represent an enormous potential for developing therapeutics and advancing our understanding of GPCR biology. While computational methods for screening GPCRs have evolved to accommodate millions of ligands (3), experimental methods for validating virtual screening predictions and performing large-scale experimental GPCR ligand screens cannot match this throughput in the current paradigm of one receptor versus one ligand. To overcome this limitation, we have designed a yeast-based experimental screening platform that enables many human GPCRs to be profiled against many ligands simultaneously.
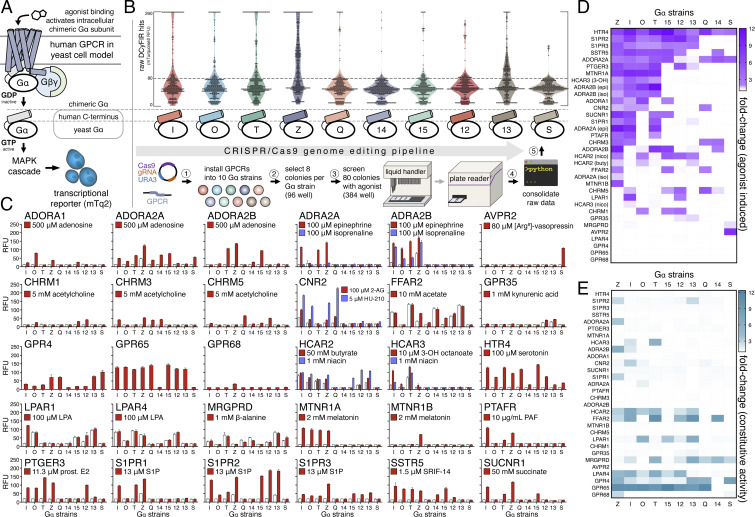
The yeast Saccharomyces cerevisiae is a useful model for studying human GPCRs (4, 5). Haploid yeast cells have only one insulated GPCR pathway, known as the pheromone pathway (Fig. 1A) (6, 7), that can be adapted to human GPCRs (4, 5). In this system, a human GPCR is coupled to the yeast Gα subunit using C-terminal Gα chimeras in which the last five residues of the yeast Gα are replaced with the last five residues of a human Gα (Fig. 1A and SI Appendix, Fig. S1). Activation of a GPCR–Gα chimera pair will stimulate a downstream mitogen-activated protein kinase cascade that drives the expression of pheromone-responsive genes. Replacing a dispensable pheromone-responsive gene with a reporter, such as a fluorescent protein, provides a GPCR signaling readout compatible with high-throughput screening formats.
Fig. 1.
High-throughput CRISPR engineering and DCyFIRscreen validation of the DCyFIR yeast strain library. (A) Simplified schematic of our yeast pheromone pathway model for studying human GPCRs (see SI Appendix, Fig. S1 for further details). (B) Workflow of our high-throughput CRISPR/Cas9 genome editing pipeline showing primary screening data (more than 5,000 mTq2 fluorescence measurements) for our exploratory panel of 30 human GPCRs. All fluorescence values are reported as RFUs. (C) DCyFIRscreen profiles for 300 GPCR–Gα strains against their known agonists, with error bars representing the SD of n = 4 experimental replicates. Untreated/treated conditions are represented by white/colored bars. All RFU measurements were quantified using the same gain setting. (D) Heat map of agonist-induced signaling in the 300 GPCR–Gα strains. (E) Heat map of constitutive activity in the 300 GPCR–Gα strains.
Yeast is also an excellent system for high-throughput CRISPR engineering, as genome editing with accuracy, speed, and scale is far easier than in mammalian cells (8–10). This difference in editing efficiency stems from the contrasting mechanisms by which yeast and mammalian cells repair CRISPR/Cas9 targeted double-stranded (DS) DNA breaks. Unlike mammalian cells, which repair DS breaks primarily by nonhomologous end joining, yeast repair DS breaks by homologous recombination, readily integrating DNA payloads having sequence homology to CRISPR/Cas9 cut sites. As such, it is possible to build yeast-based reporter systems that contain genomically stable assay components, preserve available yeast auxotrophies, and obviate heterologous protein overexpression from episomal plasmids.
Here, we report our efforts to build, validate, and apply a multiplexed GPCR profiling platform that exploits the experimental advantages of the yeast pheromone pathway and the power of yeast CRISPR. We demonstrate that this discovery platform, which we refer to as Dynamic Cyan Induction by Functional Integrated Receptors (DCyFIR), is highly scalable and capable of simultaneously profiling ligands against hundreds of GPCR–Gα combinations with single-cell and single-receptor resolution.
Results
High-Throughput CRISPR Engineering of the DCyFIR Yeast Strain Library.
To create the DCyFIR yeast strain library, we first built a panel of 10 Gα reporter strains that covered all possible GPCR–Gα subunit coupling combinations. Each reporter strain contained a pheromone-responsive fluorescent transcriptional reporter mTq2 (11); a CRISPR-addressable expression cassette in a safe harbor locus on chromosome X, known as X-2 (10); and a unique C-terminal Gα chimera. In addition, the endogenous GPCR Ste2, GTPase-activating protein Sst2, and cell cycle arrest factor Far1 were deleted from each strain. These deletions were necessary to avoid potential interference from the native yeast GPCR (ste2Δ), sensitize the pheromone pathway (sst2Δ), and prevent cell cycle arrest on pathway activation (far1Δ). The collection and characterization of the CRISPR deletions, knock-ins, and edits used to build the 10 Gα reporter strains are summarized in SI Appendix, Fig. S1.
As shown in Fig. 1B, our high-throughput CRISPR/Cas9 genome-editing pipeline enabled us to install a set of 30 human GPCRs into all 10 Gα reporter strains, generating all 300 possible GPCR–Gα coupling combinations. We installed each GPCR into the CRISPR-addressable X-2 expression cassette. As a result, each GPCR–Gα strain was barcoded with a genome-integrated GPCR sequence. Although we typically achieved genome-integration efficiencies >80%, we accounted for occasional CRISPR failures by screening 80 candidate colonies for each receptor (8 colonies for each GPCR–Gα combination) against one or more known agonists, or no agonist, as in the case of constitutively active receptors such as GPR4, GPR65, and GPR68. As shown in raw CRISPR screening data in Fig. 1B, the collection of GPCR–Gα strains exhibited a continuum of strong (>200 mTq2 relative fluorescence units [RFUs]), moderate (>80 mTq2 RFUs), and weak (30 to 80 mTq2 RFUs) signaling.
DCyFIRscreen Validation of the DCyFIR Yeast Strain Library.
Remarkably, we recapitulated known GPCR agonism in the library of 300 GPCR–Gα yeast strains with 100% accuracy. Our validation process involved two steps. First, we tested each of the 30 sets of 80 candidate GPCR–Gα strains to verify GPCR genome integration by PCR. A single PCR-confirmed hit was selected for each GPCR–Gα reporter strain to establish 10 Gα reporter strains per receptor. For constitutively active and agonist-inducible GPCR–Gα strains, we selected PCR-verified colonies that exhibited mTq2 fluorescence. We observed minimal variation in raw mTq2 fluorescence for successfully edited CRISPR colonies (SI Appendix, Fig. S2). In the second validation step, we used a parallel DCyFIR application, called DCyFIRscreen, to analyze the 300 GPCR–Gα strains in a 384-well plate format for known agonism and constitutive activity (Fig. 1C). This procedure showed that most GPCRs coupled to multiple Gα chimeras (Fig. 1 C–E), 24 (80%) of 30 GPCRs coupled to the Gαz chimera (Fig. 1 D and E), and 15 (50%) of 30 GPCRs exhibited some degree of constitutive activity (Fig. 1E). In addition, we demonstrated that DCyFIR can be used to study antagonists (SI Appendix, Fig. S3). As shown in SI Appendix, Fig. S4, Z-score (12) and strictly standardized mean difference (13) analyses confirmed the robustness of the signal-to-noise level of DCyFIRscreen for identifying known agonists.
Developing and Validating DCyFIRplex for GPCR–Ligand Discovery.
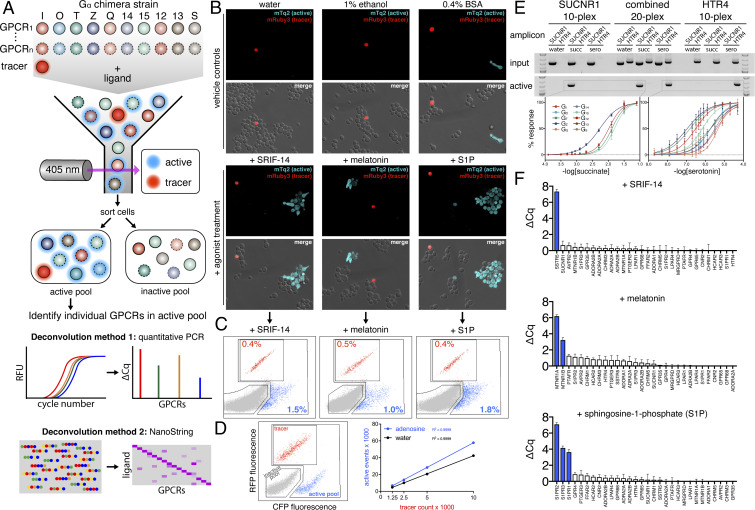
A major innovation of our DCyFIR strain library is that each strain is barcoded with genome-integrated sequences of a single human GPCR and Gα chimera. This barcoding feature enabled us to develop a second DCyFIR application, called DCyFIRplex (Fig. 2A), that we used to profile ligand binding specificity against hundreds of GPCR–Gα strains in a single tube. In the DCyFIRplex method, we monitor agonist-induced mTq2 fluorescence in consolidated mixtures of GPCR–Gα strains, using fluorescence-activated cell sorting (FACS). Using FACS, we can collect pools of active GPCR–Gα strains (Fig. 2 A and B) and identify the GPCR barcode or barcodes that are present, using automated quantitative PCR and NanoString deconvolution. As shown in Fig. 2A, in a DCyFIRplex experiment, equal parts of growth-normalized GPCR–Gα strains are combined with a mRuby3 tracer strain (14) necessary for normalizing different DCyFIRplex runs (Fig. 2C) and to empirically determine the optimal duration for FACS (Fig. 2D).
Fig. 2.
Developing and validating DCyFIRplex. (A) Schematic of the DCyFIRplex workflow showing strain consolidation to build the multiplex, FACS to collect active receptor strain pools, and our two primary multiplex deconvolution techniques. (B) Confocal microscopy images of treated and untreated (vehicle) samples of the GPCR–Gα 300-plex with the added mRuby3 tracer strain (maximum intensity projections, 63X magnification). (C) FACS analysis of the inactive (gray), active (cyan), and tracer (red) pools for the 300-plex shown in B. (D) FACS analysis of negative (gray), positive (cyan), and tracer (red) controls (see Methods for details); also, a representative standard curve of tracer event counts versus active pool event counts for our reference conditions of ±adenosine used to calibrate the FACS sorting procedure. Tracer event counts between 3,000 and 5,000 gave the most consistent deconvolution results. (E) PCR deconvolution of SUCNR1 and HTR4 10-plexes and combined 20-plex visualized by gel electrophoresis; also, the family of normalized titration curves that corresponded to the active GPCR–Gα strains in each 10- and 20-plex (errors bars represent the SD of n = 4 experimental replicates). (F) DCyFIRplex profiles deconvoluted via qPCR for the agonist-treated 300-plexes characterized in B and C. Expected hits are colored blue. ΔCq values correspond to the Cq difference between treated and untreated conditions, with error bars representing the SEM of n = 6 repeats derived from 3 independent 300-plex consolidations deconvoluted in technical duplicate. ΔCq values correspond to a log2 scale.
The probabilistic character of DCyFIRplex is illustrated in Fig. 2 B and C. Adding the mRuby3 tracer strain to a 300-plex of GPCR–Gα strains results in a 1 in 301 chance of a tracer sorting event. However, the chance of a sorting event for a given GPCR depends on its number of active GPCR–Gα strains in the multiplex. For example, the melatonin receptor MTNR1A, which signaled in 4 GPCR–Gα strains (Fig. 1C), has an expected sorting probability of 4 in 301. As expected, confocal images of untreated samples captured solitary tracer cells surrounded by reporter cells comprising the GPCR–Gα strain pool (Fig. 2B). However, we occasionally observed constitutively active GPCR–Gα cells (see Fig. 2B, lower right corner of BSA vehicle control). Confocal images taken after agonist treatment showed solitary tracer cells now surrounded by an increased number of active, cyan fluorescent, GPCR–Gα strains (Fig. 2B). These active strains tended to form filamentous arrays, a natural process triggered by activation of the pheromone pathway (15). In liquid culture, these cell clusters did not interfere with FACS.
As shown in Fig. 2 C and D, we designed our FACS gates to discern tracer, active, and inactive cell pools. To test this gating strategy, we studied the response of the 300-plex to representative peptide (SRIF-14), small-molecule (melatonin), and lipid (sphingosine-1-phosphate [S1P]) agonists that activate one (SSTR5), two (MTNR1A and MTNR1B), and three (S1PR1, S1PR2, and S1PR3) receptors, respectively (Fig. 2C). Furthermore, we extensively tested the robustness of our gating strategy with panels of negative (the 10 base Gα reporter strains lacking GPCRs) and positive (two strains that constitutively expressed the mRuby3 tracer or mTq2 reporter) controls (Fig. 2D). We typically used 3,000 to 5,000 tracer counts to collect an active pool of 20,000 to 25,000 sorting events (Fig. 2D). As shown in Fig. 2C, only a small fraction of sorting events corresponded to tracer (0.4%, 0.5%, and 0.4%) and active (1.5%, 1.0%, and 1.8%) cell pools after agonist treatments.
To further validate the DCyFIRplex method, we next identified GPCR genes sorted into active pools using a 20-plex of SUCNR1 and HTR4 GPCR–Gα strains (Fig. 2E). As expected, treatment of the 20-plex with succinate and serotonin resulted in active pools that exclusively contained barcoded gene sequences for the receptors SUCNR1 or HTR4, respectively. Following on this success, we expanded our experiments to include all 300 GPCR–Gα strains. For these DCyFIRplex runs, we pooled individual strains via automation, using liquid-handling robotics. This procedure was performed in triplicate to mix multiple independent experiments that were then treated with ligand, incubated, and sorted. FACS samples for each plex were then deconvoluted via a second round of automated liquid-handling in which qPCR reactions were mixed in technical duplicate for each of the three independent DCyFIRplex runs. Representative results for the 300-plex treated with agonists SRIF-14 (SSTR5), melatonin (MTNR1A and MTNR1B), and S1P (S1PR1, S1PR2, and S1PR3) are shown in Fig. 2F. As anticipated, each agonist treatment resulted in active pools exclusively containing the expected receptor genes (Fig. 2F). Having demonstrated our ability to sort activated GPCR–Gα strains, we were poised to advance the speed, scope, and scale of GPCR profiling using DCyFIRplex.
DCyFIRplex Profiling Identifies Interactions for Known Agonists.
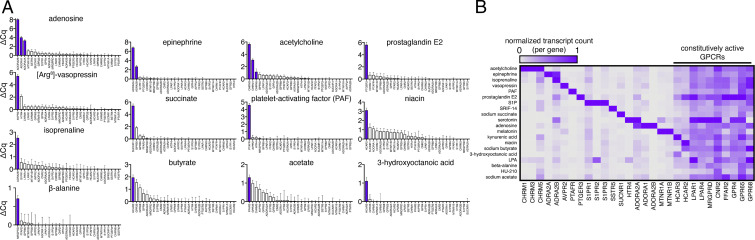
To demonstrate the ability of DCyFIRplex to discover GPCR–ligand interactions, we first challenged the full 300-plex with all 21 of the known agonists used in our initial development. Using the DCyFIRplex strategy, we again recapitulated known GPCR agonism with 100% accuracy (Fig. 3 A and B). More surprisingly, the DCyFIRplex experimental format also led us to discover interactions for known agonists (Fig. 4A), including lysophosphatidic acid (LPA) with S1PR2, serotonin with MTNR1A, and the metabolite kynurenic acid (KYNA) with both ADRA2B and the dark receptor HCAR3. As shown in Fig. 4B, we used DCyFIRscreen to confirm each interaction. In only one case (KYNA) did we observe a reduction in signaling upon ligand treatment (ADRA2B; Fig. 4A), suggesting that KYNA was an inverse agonist or negative allosteric modulator of ADRA2B. To investigate these possibilities and validate our discoveries, we performed detailed titrations that quantified micromolar interactions of serotonin with MTNR1A and ADRA2B (Fig. 4C), KYNA with HCAR3 and ADRA2B (Fig. 4D), and LPA with S1PR2 (SI Appendix, Fig. S5). Additional HCAR3 and ADRA2B titrations with their known ligands (3-hydroxyoctanoic acid and epinephrine) in the presence of fixed subsaturating concentrations of KYNA strongly suggest that KYNA is an orthosteric agonist of HCAR3 (SI Appendix, Fig. S5B) and a negative allosteric modulator of ADRA2B (Fig. 4D). Last, head-to-head KYNA titrations of GPR35 and HCAR3 revealed that KYNA has a nearly 20-fold lower EC50 for HCAR3 (EC50 41 μM) compared with its known target, GPR35 (EC50 795 μM; Fig. 4D).
Fig. 3.
Using DCyFIRplex profiling to recapitulate known agonist interactions. (A) DCyFIRplex profiles for known GPCR agonists in our 30-receptor panel deconvoluted via qPCR. (B) Same samples as in A deconvoluted using NanoString. (A and B) ΔCq values correspond to the Cq difference between treated and untreated conditions, with error bars representing the SEM of n = 6 repeats derived from 3 independent 300-plex consolidations deconvoluted in technical duplicate. ΔCq values correspond to a log2 scale. NanoString transcript counts were collected in technical duplicate, averaged, and normalized to the maximal RNA transcript count for each GPCR gene.
Fig. 4.
Using DCyFIRplex to discover interactions for known GPCR agonists. (A) DCyFIRplex profiles identifying GPCR–ligand interactions (pink bars) discovered in the process of screening known agonists (purple bars) within our panel of 30 exploratory receptors. (B) DCyFIRscreen profiles confirming the DCyFIRplex discoveries in A, with error bars representing the SD of n = 4 experimental replicates. (C and D) Select titrations confirming the DCyFIRplex discoveries in A, with error bars representing the SD of n = 4 experimental replicates. Dashed lines and boxes indicate datasets showing that KYNA activates HCAR3 with greater potency than GPR35 and is also an endogenous negative allosteric modulator of ADRA2B. Full titration datasets are available in SI Appendix, Fig. S5.
DCyFIR Profiling of 320 Metabolites Identifies GPCR–Metabolite Interactions.
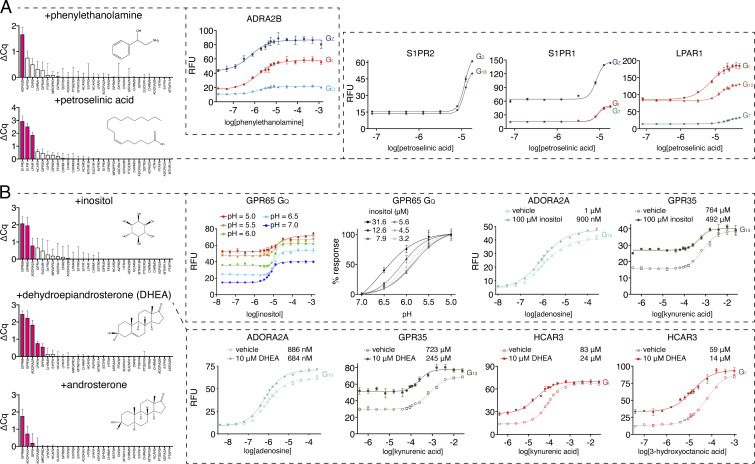
To demonstrate scalability of the DCyFIR platform, we next profiled the 300-plex of GPCR–Gα strains against a library of 320 endogenous human metabolites, using a 6-step procedure (Fig. 5A). As shown in Fig. 5B, we divided the 300-plex into three sets. The GPCR–Gα strains comprising sets 2 (100-plex) and 3 (90-plex) exhibited lower constitutive activity than those in set 1 (110-plex). We classified potential metabolite hits as having a Z-score >1 (Fig. 5B) and fluorescence microscopy images with marked increases in mTq2 fluorescence, filamentous arrays, and shmooing (Fig. 5C). Using these criteria, we recovered known agonists for GPCRs in our exploratory panel, such as adenosine, melatonin, and prostaglandin E2 (Fig. 5B). However, most metabolite hits corresponded with known interactions, including tryptamine and dopamine agonism of adrenergic receptors (Fig. 5D), phenylethanolamine agonism of ADRA2B (Fig. 6A and SI Appendix, Fig. S6), and petroselinic acid agonism of S1PR1, S1PR2, and LPAR1 (Fig. 6A and SI Appendix, Fig. S6).
Fig. 5.
DCyFIR profiling of a human metabolite library. (A) Step-by-step workflow used to screen a library of 320 endogenous human metabolites. (B) Z-score profiles for metabolite screens of receptor set 1 (ADORA1, ADORA2A, FFAR2, GPR4, GPR65, GPR68, HCAR2, HCAR3, LPAR1, LPAR4, MRGPRD), set 2 (ADORA2B, ADRA2A, ADRA2B, AVPR2, CHRM1, CHRM3, CHRM5, CNR2, GPR35), and set 3 (HTR4, MTNR1A, MTNR1B, PTAFR, PTGER3, S1PR1, S1PR2, S1PR3, SSTR5, SUCNR1). Gray bands indicate Z-scores between ± 1. (C) Representative fluorescence microscopy images for Z-score hits in receptor subsets 1 (110-plex), 2 (90-plex), and 3 (100-plex). (D) Discovery workflow illustrating tryptamine agonism of HTR4 and ADRA2B and dopamine agonism of ADRA2A and ADRA2B. Once tryptamine and dopamine were identified as hits (steps 1 to 3 in A), we used DCyFIRplex profiling to identify their GPCR target or targets, DCyFIRscreen profiling to identify their Gα coupling pattern or patterns, and titrations to quantify their EC50 values (steps 4 to 6 in A).
Fig. 6.
Identification and validation of GPCR–metabolite interactions. (A) DCyFIRplex profiles and titrations for metabolite agonists and (B) positive allosteric modulators. For B, full titration datasets for all GPCR–Gα coupling combinations are available in SI Appendix, Fig. S7. DCyFIRplex error bars represent the SEM of n = 6 repeats derived from 3 independent 300-plex consolidations deconvoluted in technical duplicate, and titration error bars represent the SD of n = 4 experimental replicates.
The sensitivity of DCyFIR profiling also enabled us to discover metabolites that appear to function as allosteric modulators in the yeast model. Remarkably, most of these findings involved the understudied, pharmacologically dark receptors GPR4, GPR65, GPR68, and HCAR3. As shown in Fig. 6B and SI Appendix, Fig. S7, inositol and the steroid metabolites dehydroepiandrosterone (DHEA) and androsterone interacted with multiple prototypic and understudied GPCRs in the DCyFIRplex experiment. To rigorously assess ligand specificity, we DCyFIRscreen profiled more promiscuous ligands such as inositol, DHEA, and androsterone against many GPCRs (>150 control experiments are available in SI Appendix, Figs. S6–S9). This process indicated that inositol modulated 5 receptors (ADORA2A, CNR2, GPR35, GPR65, GPR68), and that the structurally similar steroid metabolites DHEA and androsterone modulated 8 receptors (ADORA2A, GPR35, GPR4, GPR68, HCAR2, HCAR3, LPAR1, LPAR4). Interestingly, all three metabolites modulated a trio of GPCRs (ADORA2A, GPR35, and GPR68). On the basis of these observations, we speculated that inositol, DHEA, and androsterone may act as broad-spectrum allosteric modulators. As shown in Fig. 6B, we tested this idea by titrating select receptors with inositol (GPR65, GPR68) or orthosteric agonists in the presence of inositol and DHEA (ADORA2A, GPR35, HCAR3). These experiments suggested that inositol and DHEA are weak positive allosteric modulators of ADORA2A, and DHEA is a relatively stronger positive allosteric modulator of HCAR3 and GPR35. In addition, inositol titrations of the proton-sensing receptors GPR65 (Fig. 6B) and GPR68 (SI Appendix, Fig. S7A) at several pH values suggested that inositol is a positive allosteric modulator of GPR65 and GPR68 pH sensing, with higher inositol concentrations causing an increase in the pH50 values of both receptors. Taken together, these results demonstrate the ability of DCyFIR profiling to identify even the weakest allosteric modulators, an attribute that will likely serve to benefit lead discovery efforts.
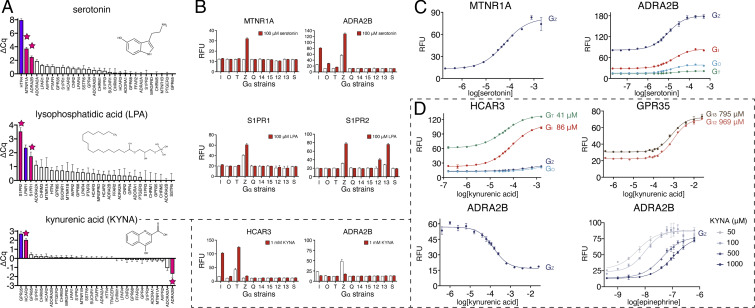
Validating Select GPCR-Ligand Interactions in Mammalian Cells.
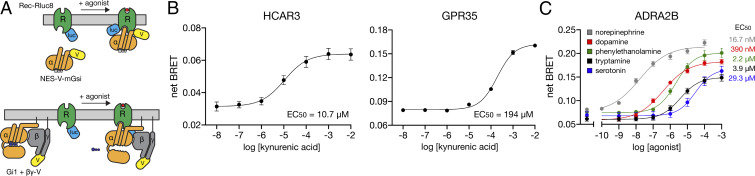
In addition to validating known GPCR agonism with DCyFIRscreen and DCyFIRplex (Figs. 1C, 2F, and 3), we used Bioluminescence Resonance Energy Transfer (BRET) assays to confirm a subset of our discovered GPCR–ligand interactions in HEK 293 cells. We monitored the recruitment of mini G (mG) proteins (Fig. 7 A, Top) or full heterotrimers (Fig. 7 A, Bottom) to luciferase-tagged receptors. Using these BRET assays, we confirmed our yeast-based discovery that KYNA is an orthosteric agonist of HCAR3, and reproduced the known KYNA agonism of GPR35 (Fig. 7B). Remarkably, as in yeast, we found that KYNA was ∼20-fold more potent for HCAR3 than its known receptor, GPR35. In addition, we confirmed several of our yeast-based findings, using the well-studied adrenergic receptor, ADRA2B (Fig. 7C). In all cases, the pEC50 values measured for ADRA2B agonists in yeast and HEK cells were in excellent agreement: norepinephrine 7.8 in HEK and epinephrine 7.8 in yeast, tryptamine 5.4 in HEK and 5.3 to 5.8 in yeast, dopamine 6.4 in HEK and 4.9 to 6.0 in yeast, and phenylethanolamine 5.7 in HEK and 5.9 to 6.2 in yeast. In contrast, we could not reproduce the robust inverse agonism of ADRA2B by KYNA we observed in yeast (Fig. 4D) in HEK cells. This result suggests that KYNA’s inverse agonism of ADRA2B may be limited to conditions specific to the yeast system.
Fig. 7.
Validation of DCyFIR discoveries in mammalian cells. GPCR signaling as measured by BRET between a GPCR fused to the Renilla luciferase (Rec-Rluc8) and Venus fluorescent protein (V) fused to either a mini G protein (A, Top) or split between the βγ subunits (βγ-V) of a full G protein heterotrimer (A, Bottom). (B) Net BRET for KYNA titrations of HCAR3 (using the mini G protein V-mGsi containing an N-terminal nuclear export signal, NES-V-mGsi) and GPR35 (using full-length Gα12 with βγ-V). Error bars represent SEM of at least n = 6 independent experiments. (C) Net BRET for titrations of ADRA2B (using full-length Gαi1 with βγ-V) with a known agonist (norepinephrine) and discovered agonists (dopamine, phenylethanolamine, tryptamine, and serotonin). Error bars represent SEM of at least n = 4 independent experiments.
Discussion
The objective of this study was to demonstrate the feasibility and potential of our innovative DCyFIR platform. Using this technology, we have identified a relatively large number of GPCR–ligand interactions by profiling a relatively small number of compounds and receptors. These discoveries are solely attributable to the multiplex format of the DCyFIR experiment, which enables simultaneous ligand profiling of many GPCRs at once. In the discussion that follows, we characterize a select number of our findings in the context of their potential biological relevance.
Interactions between GPCRs and Amino Acid Metabolites.
In this work we showed that the DCyFIR platform can be used to identify interactions between GPCRs and small molecule regulators. For example, we identified several GPCR–ligand interactions involving three amino acid metabolites that belong to the tryptophan pathway: KYNA, serotonin, and tryptamine. Of these findings, our discovery that KYNA is a more potent agonist of HCAR3 than GPR35 is particularly notable. KYNA is known to play important roles in neuroprotection, depression, schizophrenia, obesity, diabetes, and cancer (16–18). Before this study, the only GPCR known to respond to KYNA was GPR35 (19). However, we have shown that KYNA activates HCAR3 with a nearly 20-fold greater potency than GPR35. Interestingly, we also found that similarities between HCAR3 and GPR35 extended beyond KYNA agonism, as our findings suggest that both receptors exhibit positive allosteric modulator (PAM) interactions with the steroid metabolite DHEA. In the case of HCAR3, DHEA PAM interactions appear to extend to both new (KYNA) and known (3-hydroxyoctanoic acid) orthosteric agonism, similar to the dual regulation imposed by KYNA and DHEA on the N-methyl-D-aspartate receptor (20). Based on these observations, we believe a comprehensive reassessment of HCAR3, GPR35, and KYNA agonism in relevant cell types and physiological systems is warranted.
Cross-Activation of Lipid-Binding GPCRs.
Our set of 30 GPCRs included members of two evolutionarily related lipid receptor families, LPAR and S1PR. Both receptor families are involved in inflammatory responses, fibrosis, and a variety of other disorders (21, 22). They also share similar spatially conserved residues that determine their respective specificities for LPA and S1P metabolites (23). Here we confirmed the known cross-activation of LPA for S1PR1 (24, 25) and report LPA cross-activation of S1PR2. Consistent with previous LPA studies, we found that LPA activated both S1PR1 and S1PR2 with higher effective concentration 50 (EC50) values than LPAR1, indicating that S1PR1 and S1PR2 are low-affinity LPA receptors. Although it has been suggested that the micromolar LPA concentrations needed to activate S1PR1 and S1PR2 receptors may not be biologically relevant (24), local (paracrine and autocrine) and systemic (endocrine) metabolite concentrations can regularly occur at micromolar levels in response to biological cues. For example, the succinate receptor SUCNR1 has a low affinity for succinate; however, the accumulation of succinate under stress conditions, such as ischemia, activates SUCNR1 (26, 27). Interestingly, we did not observe LPA cross-activation of S1PR3.
Allosteric Modulators of Understudied GPCRs.
The sensitivity of the DCyFIR platform was demonstrated by our discovery of several metabolites that appear to be allosteric regulators of prototypic and understudied GPCRs. Although most of these interactions had EC50 values in the micromolar range, they were repeatedly detectable in both DCyFIRscreen and DCyFIRplex experimental modes. Remarkably, this sensitivity even extended to GPCR–ligand interactions having near-millimolar EC50 values (e.g., the EC50 of KYNA for GPR35). Given that allosteric modulators are often attractive drug candidates, we believe that this unique feature of DCyFIR profiling will enable the identification of chemical leads over a wide range of affinities, greatly expediting development of pharmacological tools and drugs.
Notably, all the apparent allosteric modulators we discovered in our screen interacted with pharmacologically dark GPCRs. Inositol, a structural isoform of glucose, appears to be a PAM of GPR65 and GPR68, while DHEA, the most abundant circulating steroid in humans (28), appears to be a PAM of GPR4, GPR68, and HCAR3. To the best of our knowledge, these are the potential endogenous PAMs for these understudied receptors. Furthermore, there has been a long-standing interest in discovering mediators of steroid responses outside the nucleus (29). Our finding that DHEA and androsterone are apparent PAMs of prototypic and dark GPCRs suggests there are areas of steroid metabolism waiting to be explored. Last, our steroid-related discoveries showcase one of the major advantages of our yeast-based platform: Because yeast lack cholesterol and have primitive steroid pathways (30), there is little to no steroid interference from the model system or its genetic background in the discovery process. Based on our DHEA and androsterone findings, we are currently taking advantage of this feature of the yeast system to comprehensively examine the unexplored frontier of GPCR–steroid pharmacology.
Translating Yeast-Based Discoveries to Mammalian Cell Models.
Our findings show that yeast-based studies of human GPCRs translate well to mammalian cell models. We recapitulated known mammalian cell responses for more than 30 GPCR-agonist interactions, and confirmed several of our discoveries in HEK cell assays, suggesting that human GPCRs expressed in yeast typically do not have altered functional properties. Our survey of available mammalian and yeast Gα coupling data (Dataset S5), as well as the comparable EC50 values we measured in both models, further support this notion. We believe these results suggest that translational failures between yeast and mammalian cell models will be uncommon. However, our inability to confirm KYNA inhibition of ADRA2B in mammalian cells emphasizes the importance of confirming yeast-based discoveries in more biologically relevant cell models (31–35). Other caveats should also be considered. For example, DCyFIR relies on the coupling of C-terminal Gα chimeras to human GPCRs. Although large-scale experiments in mammalian cells have shown that such chimeras generally show appropriate Gα coupling (31), in some cases, structural features other than the Gα C terminus will be necessary for efficient coupling (34). Similarly, functional expression of some receptors at the cell surface may pose additional challenges in yeast, as is sometimes the case in mammalian cells. In the present study, we truncated the third intracellular loop (iL3) of three muscarinic receptors (CHRM1, CHRM3, and CHRM5), rendering these receptors compatible with yeast (36). Similar engineering interventions, including fusions to signal peptides that expedite receptor trafficking, may be necessary to maximize the number of receptors that can be ported to the yeast system. Having said this, our work here demonstrates that expressing unaltered GPCR sequences in yeast is often straightforward, which we believe is a remarkable observation, given the large evolutionary distance between yeast and humans.
The Advantage and Potential of the DCyFIR Platform.
The primary advantage of the DCyFIR platform, relative to other GPCR assays, is that many receptors are simultaneously profiled against a single ligand. By combining the scalability of a yeast-strain receptor plex with the low requirement for ligand material, it is highly practical to expand our many-GPCR versus many-ligand format to entire compound libraries, as we have demonstrated for human metabolites, effectively screening hundreds of GPCR–Gα combinations versus hundreds (or thousands) of ligands at once. More traditional GPCR assays that use readouts such as second messengers, β-arrestin recruitment, and BRET have tremendous value, but are much more resource intensive (and likely cost prohibitive), both in terms of the amounts of transfected cells and ligands required. The DCyFIR platform serves to lower these barriers.
In conclusion, we have demonstrated the utility of the DCyFIR platform for simultaneously profiling many GPCRs against individual ligands to efficiently explore chemical space. We have established that this innovative approach facilitates the rapid, inexpensive, and comprehensive assessment of ligand specificity and receptor promiscuity. We believe that combining the DCyFIR platform with other GPCR screening approaches has the potential to open avenues in GPCR research. By quickly and openly sharing the results of such studies, the longer-term process of validating the biological and clinical relevance of GPCR–ligand interactions, as well as developing lead compounds into future drugs, can be expedited within the community. As our library of GPCR–Gα strains continues to grow, so too will the scale of our DCyFIR profiling capabilities. Ultimately, we envision substantially larger receptor plexes that enable the physical screening of huge numbers of compounds, bringing wet-laboratory throughput a step closer to what can be done in silico.
Methods
Data Availability.
All relevant data, protocols, and results of analyses are included in the main text and within the supplementary materials and datasets. Code and yeast strains are available upon request.
Media.
The different media types used in this study are detailed in the SI Appendix, Table S1.
Plasmids.
All plasmid vector sequences are provided in Dataset S3.
Strains.
The 323 yeast strains created in this work are listed in Dataset S1.
Engineering CRISPR-Optimized Yeast Strains for Human GPCR Studies.
The collection of 323 yeast strains created and used in this study is detailed in Dataset S1. The major steps we used to build the 10 base GPCR–Gα strains are summarized here. We used CRISPR to make every gene deletion, replacement, edit, and knock-in in this study (see CRISPR transformation reaction supplementary method for further detail). See Dataset S3 for a complete list of CRISPR plasmids.
Deletion of signaling components to sensitize the pheromone pathway.
In the first steps of our strain engineering efforts, we created our DI2Δ strain by sequentially deleting the pheromone pathway components FAR1 and SST2. The factor arrest protein (FAR1) was deleted to prevent cell-cycle arrest on pathway activation, and the GTPase-activating protein (SST2) was deleted to sensitize the pheromone pathway by prolonging Gα activation. Our CRISPR gene deletion procedure employed two CRISPR vectors, pML107 and pT040, each having their own selectable markers, LEU and URA. Vector pT040 contained a guide RNA sequence that targeted the N-terminal/C-terminal region of the gene to be deleted. These vectors were cotransformed with DNA payload comprising homology arms generally having 60 to 100 bp of sequence immediately upstream and downstream of the targeted ORF.
Installation of the mTq2 transcriptional reporter.
After the creation of the DI2Δ strain, we replaced the pheromone-responsive gene FIG1 ORF with the cyan fluorescence protein mTq2. As with our CRISPR gene deletion procedure, we replaced the FIG1 ORF with the mTq2 gene, using two CRISPR vectors, pML107 and pT040, each having their own selectable markers, LEU and URA. Vectors pML107 and pT040 contained a guide RNA sequence that targeted the N-terminal/C-terminal region of the FIG1 gene. These vectors were cotransformed with DNA payload comprising homology arms having 60 bp of sequence immediately upstream and downstream of FIG1 ORF. The resultant genotype of this strain, which we refer to as DI2Δ fig1Δ::mTq2, was BY4741 far1Δ sst2Δ fig1Δ::mTq2.
Deleting the endogenous yeast GPCR STE2 gene.
After the creation of the DI2Δ fig1Δ::mTq2 strain, we deleted the native yeast GPCR gene (STE2), using the same plasmids and procedure described in Deletion of signaling components to sensitize the pheromone pathway. This new strain, DI3Δ fig1Δ::mTq2, had the genotype BY4741 far1Δ sst2Δ ste2Δ fig1Δ::mTq2.
Installation of the X-2 landing pad.
After the creation of the DI3Δ fig1Δ::mTq2 strain, we installed our CRISPR-addressable expression cassette (see Design of the X-2 CRISPR-addressable expression cassette in the supplementary methods for details) into the X-2 locus of chromosome X. To install the X-2 landing pad into the X-2 safe harbor locus, we PCR amplified the landing pad sequence from the pMARQ vector and cotransformed the resultant DNA payload with the CRISPR vector pML104 X2 The resultant genotype of this strain, which we refer to as DI3Δ fig1Δ::mTq2 P1, was BY4741 far1Δ sst2Δ ste2Δ fig1Δ::mTq2 X-2:PTEF1a-UnTS-TCYC1b.
Genome editing to create humanized yeast C-terminal Gα chimeras.
To build our panel of 10 GPCR–Gα base reporter strains, we created 10 different versions of our DI3Δ fig1Δ::mTq2 P1 strain, each having its own unique Gα C-terminal yeast/human chimera. In each Gα chimera, the last five yeast residues of the yeast Gα subunit, Gpa1, were replaced by the last five residues of a human Gα subunit (see SI Appendix, Fig. S1 and Table S1 for sequence details). As a result of C-terminal degeneracies, all 16 human Gα genes could be represented by 10 Gα C-terminal chimeras. A codon-optimized DNA payload for each Gα chimeric sequence was designed as a gBlock gene fragment (Integrated DNA Technologies) comprising the 15 bp sequence of a human Gα C-termini flanked by 123 bp homology arms that targeted the C terminus of the yeast Gα subunit sequence. These synthetic DNA payloads were cotransformed with the CRISPR vectors pML107 and pT040 GPA1:1373, each having their own selectable markers, LEU and URA. The resultant genotypes of these strains, which we refer to as DI DCyFIR P1 I, DI DCyFIR P1 O, DI DCyFIR P1 T, DI DCyFIR P1 Z, DI DCyFIR P1 Q, DI DCyFIR P1 14, DI DCyFIR P1 15, DI DCyFIR P1 12, DI DCyFIR P1 13, and DI DCyFIR P1 S, are described in Dataset S1.
Installation of human GPCRs into X-2 CRISPR-addressable expression cassette.
All human GPCR DNA sequences were sourced from the Presto-TANGO plasmid library (32), using primers to PCR amplify only the GPCR ORF, avoiding the additional N- and C-terminal DNA sequence elements in the Presto-TANGO plasmid constructs. Using two rounds of PCR amplification, we extended each GPCR sequence with homology arms corresponding to sequences within the TEF promoter and CYC1b terminator of the X-2 landing pad. For the first round of PCR, we used receptor-specific primers having ∼45-bp homology overhangs. In a second round of PCR, we used universal primers to extend both homology arms to a final length of 60 bp. All primer and GPCR sequences are listed in Datasets S2 and S4. With the exception of the muscarinic receptors CHRM1, CHRM3, and CHRM5, native GPCR sequences were used (i.e., no affinity tags or localization sequences were added). However, residues corresponding to iL3 of CHRM1, CHRM3, and CHRM5 were deleted to reproduce iL3 deletion results that were previously published in a similar yeast system (36). As with these original studies, we found that full-length CHRM1, CHRM3, and CHRM5 did not functionally express in our system. The CHRM1, CHRM3, and CHRM5 sequences in SI Appendix, Table S1 correspond to the iL3 loop deletion variants. To install human GPCRs into the X-2 landing pad, the amplified GPCR PCR product with 60 bp homology arms was cotransformed with the CRISPR vector pML104 X2 UnTS, using the approach described in the supplementary methods section CRISPR transformation reactions. Because we installed each human GPCR into all 10 base GPCR–Gα reporter strains, we produced a library of 300 GPCR–Gα strains barcoded with a human GPCR.
DCyFIRscreen Protocol.
Individual Gα reporter strains were grown in SCD LoFo at pH 7.0 at 30 °C to an OD of 1.0 in a 2.0-mL 96-well DeepWell block (Greiner; 780271-FD). Cells were normalized to an OD of 0.1 in SCD LoFo at pH 7.0, using a Biomek NXp liquid-handling robot. Next, 10X ligand/vehicle stocks were prepared (see Key Resource Table) and 4 μL were distributed to each well of a 384-well plate (Greiner; 781096) in quadruplicate, using a Biomek NXp. Then, 36 μL normalized cells was distributed to each well containing the appropriate 10X ligand/vehicle. Plates were sealed with a breathable cover (Diversified Biotech; BERM-2000) and incubated at 30 °C. Fluorescence readings were collected after 18 h, using a plate reader (ClarioStar, BMG LabTech, Offenburg, Germany; bottom read, 10 flashes/well, excitation filter: 430–10 nm; dichroic filter: LP 458 nm; emission filter: 482–16 nm; gain = 1,300 [1,500 for Fig. 1B]). Absorbance readings were also collected after 18 h, using the same instrument (22 flashes/well, excitation filter: 600 nm).
DCyFIRplex Protocol.
Control/tracer strains (DI P1 mTq2, individual Gα reporter strains lacking an integrated receptor, DI P1 mRuby3) and the 300 GPCR–Gα strains were grown in SCD LoFo at pH 5.0 to saturation in individual wells of a 2.0-mL 96-well DeepWell block (Greiner; 780271-FD). The 10 GPCR–Gα strains for a single receptor were then consolidated in growth-normalized amounts into single wells of a DeepWell block, using a Biomek NXp (each well is comprised of one unique receptor in all 10 Gα reporter strains). Each receptor-consolidated well and control well was grown to midlog phase in SCD LoFo at pH 7.0 and then further consolidated into a single tube (300-plex; 30 receptors, 300 Gα strains) in growth-normalized amounts (this consolidation was performed three times for each sorting procedure [n = 3]). Next, 10X ligand/vehicle stocks were added to individual wells of a DeepWell block. The consolidated 300-plex or control strains were then added to each well and grown in the presence of ligand/vehicle overnight, so that each culture would reach an OD of 4.0 before DCyFIRplex profiling. Samples were washed with sterile ddH2O and normalized to an OD of 2.0 in SCD LoFo at pH 7.0. Tracer cells were added to each sample at a 1:301 ratio. The final mixture was then transferred into a glass sample tube (USA Scientific; 1485-2810) and used for cell sorting. A BD FACSAria-II cell sorter was used for all DCyFIRplex experiments to assess mTq2 (405 nm excitation, 450/50 nm emission) and mRuby3 fluorescence (535 nm excitation, 610/20 nm emission). A gating strategy was set using the three control samples (DI P1 mTq2, individual Gα reporter strains lacking an integrated receptor, and DI P1 mRuby3), such that tracer cells and any cell expressing mTq2 was sorted into a 14-mL collection tube (USA Scientific; 1485-2810) containing 500 μL YPD. False-positive and false-negative sorting events are shown in SI Appendix, Fig. S10 for ligand activation of the native yeast receptor, Ste2, and the 10 pooled Gα reporter strains of the serotonin receptor, HTR4, in SI Appendix, Fig. S10. Samples treated with water or 500 μM adenosine (well-characterized using DCyFIRscreen, inexpensive, and water-soluble) were used to build a standard curve measuring total events in the mRuby3 and mTq2 positive gates. The standard curve from a water-treated 300-plex was used to determine the number of tracer events that would correspond to 15,000 events in the mTq2 gate for a water-treated 300-plex. Each sample was sorted until the standardized tracer count was reached. Sorted cells were enriched by outgrowth in 5 mL YPD at 30 °C with shaking (200 rpm) for 18 h. Cells were harvested by centrifugation at 3,000 × g for 5 min and resuspended in 1 mL ddH2O. Cells were either processed immediately for qPCR deconvolution or frozen in 100 μL aliquots for storage at −20 °C. The set of samples comparatively deconvoluted by qPCR and NanoString methods were derived from aliquots of the same DCyFIRplex experiments (see SI Appendix, Methods for details). The consistency of the 300-plex cell composition for n = 100 deconvolution experiments is shown in SI Appendix, Fig. S11.
Experimental Replicates.
In all DCyFIRscreen, DCyFIRplex, and follow-up characterization experiments, we conducted multiple (n = 3 or n = 4) independent experiments. DCyFIRscreen and follow-up titration experiments were performed in 4 independent replicates, each treated with ligand and vehicle. As shown in SI Appendix, Fig. S12, titrations performed using 4 colonies versus 1 colony in 4 independent experiments gave the same results. The average of these n = 4 replicates were included in each plot, with error representing the SD of n = 4 independent experiments. All dose–response curve fit parameters and error/confidence intervals are provided in SI Appendix, Table S2. All DCyFIRplex experiments were performed in triplicate by three independent plex consolidations on our liquid handling robot, ligand treatments, sorts, and post processing. This process produced three independent sorting results that were independently deconvoluted in technical duplicate, giving us a total of n = 6 observations for three independent DCyFIRplex runs.
Supplementary Material
Acknowledgments
We thank Dr. Antonio Barrientos, Dr. Grace Zhai, and Dr. Vladlen Slepak for their critical review of this manuscript. This work was supported by the NIH through the National Institute of General Medical Sciences (R35GM119518) and NIH Common Fund for Illuminating the Druggable Genome (R03TR002908 to D.G.I.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000430117/-/DCSupplemental.
References
- 1.Sriram K., Insel P. A., G protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 93, 251–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodgers G. et al., Glimmers in illuminating the druggable genome. Nat. Rev. Drug Discov. 17, 301–302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyu J. et al., Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowell S. J., Brown A. J., Yeast assays for G-protein-coupled receptors. Receptors Channels 8, 343–352 (2002). [PubMed] [Google Scholar]
- 5.Dowell S. J., Brown A. J., Yeast assays for G protein-coupled receptors. Methods Mol. Biol. 552, 213–229 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Bardwell L., A walk-through of the yeast mating pheromone response pathway. Peptides 26, 339–350 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Dohlman H. G., Pheromone signaling mechanisms in yeast: A prototypical sex machine. Science 306, 1508–1509 (2004). [DOI] [PubMed] [Google Scholar]
- 8.DiCarlo J. E. et al., Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laughery M. F. et al., New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32, 711–720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronda C. et al., CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 14, 97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goedhart J. et al., Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3, 751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J. H., Chung T. D., Oldenburg K. R., A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Zhang X. D., Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screen. 16, 775–785 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Bajar B. T. et al., Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erdman S., Snyder M., A filamentous growth response mediated by the yeast mating pathway. Genetics 159, 919–928 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cervenka I., Agudelo L. Z., Ruas J. L., Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 357, eaaf9794 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Contreras M. et al., Metabolomics study of the effects of inflammation, hypoxia, and high glucose on isolated human pancreatic islets. J. Proteome Res. 16, 2294–2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urenjak J., Obrenovitch T. P., Neuroprotective potency of kynurenic acid against excitotoxicity. Neuroreport 11, 1341–1344 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Wang J. et al., Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 281, 22021–22028 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Mok M. H., Fricker A. C., Weil A., Kew J. N., Electrophysiological characterisation of the actions of kynurenic acid at ligand-gated ion channels. Neuropharmacology 57, 242–249 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Blaho V. A., Hla T., An update on the biology of sphingosine 1-phosphate receptors. J. Lipid Res. 55, 1596–1608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yung Y. C., Stoddard N. C., Chun J., LPA receptor signaling: Pharmacology, physiology, and pathophysiology. J. Lipid Res. 55, 1192–1214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D. A. et al., A single amino acid determines lysophospholipid specificity of the S1P1 (EDG1) and LPA1 (EDG2) phospholipid growth factor receptors. J. Biol. Chem. 276, 49213–49220 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Blaho V. A., Hla T., Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem. Rev. 111, 6299–6320 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M. J., Thangada S., Liu C. H., Thompson B. D., Hla T., Lysophosphatidic acid stimulates the G-protein-coupled receptor EDG-1 as a low affinity agonist. J. Biol. Chem. 273, 22105–22112 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Correa P. R. et al., Succinate is a paracrine signal for liver damage. J. Hepatol. 47, 262–269 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husted A. S., Trauelsen M., Rudenko O., Hjorth S. A., Schwartz T. W., GPCR-mediated signaling of metabolites. Cell Metab. 25, 777–796 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Rutkowski K., Sowa P., Rutkowska-Talipska J., Kuryliszyn-Moskal A., Rutkowski R., Dehydroepiandrosterone (DHEA): Hypes and hopes. Drugs 74, 1195–1207 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Lösel R., Wehling M., Nongenomic actions of steroid hormones. Nat. Rev. Mol. Cell Biol. 4, 46–56 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Parks L. W., Casey W. M., Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49, 95–116 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Inoue A. et al., Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947.e25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroeze W. K. et al., PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuho I. et al., Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal. 8, ra123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okashah N. et al., Variable G protein determinants of GPCR coupling selectivity. Proc. Natl. Acad. Sci. U.S.A. 116, 12054–12059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan Q. et al., Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J. Biol. Chem. 293, 7466–7473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erlenbach I. et al., Functional expression of M(1), M(3) and M(5) muscarinic acetylcholine receptors in yeast. J. Neurochem. 77, 1327–1337 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data, protocols, and results of analyses are included in the main text and within the supplementary materials and datasets. Code and yeast strains are available upon request.