Significance
CD8+ T lymphocytes protect from intracellular pathogens by recognizing pathogen's antigenic peptides presented on MHC-I molecules. Many viruses evade T cell control by using genes that subvert MHC-I presentation on infected cells. Here we shift this paradigm and show that viral proteins that block the effector branch of T cells are more efficient for T cell evasion than MHC-I downregulation. The block of signaling in infected target cells at the level of caspase-8 as a crucial convergence point of death-receptor and perforine/granzyme pathways in the viral evasion of cytotoxic T cells. We show that these principles apply in the clinically relevant HCMV infection and in the biologically relevant in vivo context, using the MCMV experimental model.
Keywords: immune evasion, cytomegalovirus, apoptosis inhibition, apoptosis, CD8 T cells
Abstract
Viral immune evasion is currently understood to focus on deflecting CD8 T cell recognition of infected cells by disrupting antigen presentation pathways. We evaluated viral interference with the ultimate step in cytotoxic T cell function, the death of infected cells. The viral inhibitor of caspase-8 activation (vICA) conserved in human cytomegalovirus (HCMV) and murine CMV (MCMV) prevents the activation of caspase-8 and proapoptotic signaling. We demonstrate the key role of vICA from either virus, in deflecting antigen-specific CD8 T cell-killing of infected cells. vICA-deficient mutants, lacking either UL36 or M36, exhibit greater susceptibility to CD8 T cell control than mutants lacking the set of immunoevasins known to disrupt antigen presentation via MHC class I. This difference is evident during infection in the natural mouse host infected with MCMV, in settings where virus-specific CD8 T cells are adoptively transferred. Finally, we identify the molecular mechanism through which vICA acts, demonstrating the central contribution of caspase-8 signaling at a point of convergence of death receptor-induced apoptosis and perforin/granzyme-dependent cytotoxicity.
Human cytomegalovirus (HCMV) is a highly prevalent virus that belongs to the subfamily of strictly species-specific β-herpesviruses that coevolved with their natural hosts (1). Mouse CMV (MCMV) displays striking parallels with HCMV and is commonly used as an infection model to study pathogenesis and immune control of this herpesvirus subgroup. Primary HCMV or MCMV infection in the immunocompetent host is typically not associated with disease, but is characterized by lifelong latency and sporadic shedding with natural transmission reaching a majority of the population. CD8 T cells play a prominent role in controlling acute infection as well as maintaining latency despite the elaboration of viral immunoevasins that undermine MHC class I antigen recognition (2). In the immunocompromised host, reactivation of HCMV or MCMV leads to significant disease, which gives this virus its hallmark characteristics as an opportunist in tissue and organ transplant recipients as well as hosts with genetic or acquired immunodeficiency.
Cytotoxic CD8 T cells are central to host immune control over CMV infection and reactivation (3, 4). The antiviral mechanisms of CD8 T cells include production of cytokines, death receptor ligands, and cytotoxic granules that kill target cells. The relative contribution of perforin-mediated and death receptor-mediated killing by CD8 T cells varies, with both crosstalk between pathways and overlapping mechanisms that are context-dependent (5, 6). Either pathway induces caspase-dependent apoptosis, via granzymes or initiating caspases such as caspase-8 (Casp8), either directly (7, 8) or indirectly (9). CMV expresses multiple immunoevasins, collectively known as viral regulators of antigen presentation (vRAP), that disrupt antigen presentation by downregulating MHC class I (MHC-I). HCMV genes US2, US3, US6, and US11 interfere with MHC-I expression (2). In vitro experiments show that MHC class I is progressively diminished from infected cells. Similarly, MCMV-encoded m06 and m152 disrupt antigen presentation via MHC-I, while m04 is a positive regulator of MHC-I expression (10). Overall, the impact of virus-encoded suppressors of antigen presentation is to reduce recognition of virus-infected cells. However, in vivo studies showed that MCMVs lacking vRAP genes display only modest growth defects (11–13). Thus, despite clear in vitro results, the in vivo functional significance of immunoevasins remains to be determined.
CMVs encode several suppressors of programmed cell death (14). Apoptosis is a cell-autonomous defense mechanism against virus infections that may be triggered by cell stress (intrinsic pathway) or by cytokines (extrinsic pathway) upon engagement of membrane-associated death receptors (DR). FasL and TNFα are well-studied DR ligands that engage Fas (also known as CD95) and TNFR1, respectively (15, 16). DR activation results in the recruitment of adaptor molecules like Fas-associated protein with death domain (FADD), which recruits Casp8 via its death effector domain (DED) (17). Casp8 activation in the death-inducing signaling complex (DISC) results in caspase-dependent apoptosis, either by directly activating caspase-3, or indirectly via the mitochondrial (intrinsic) pathway (18). Casp8-induced apoptosis is inhibited by the HCMV protein vICA (viral inhibitor of caspase-8 activation), a product of the viral UL36 gene (19), or by the MCMV homolog, M36 (20). Both of these gene products inhibit Casp8 activation and are interchangeable across species (21). MCMV lacking the M36 is attenuated in vivo (22, 23) because of a failure to inhibit FADD signaling to Casp8 (22). M36 is required for virus growth in the presence of macrophages and soluble factors, such as TNF, released upon activation (24). However, the contribution of M36 to evasion of cytotoxic immune effector cells has not been explored.
Here we show that CMV proteins UL36 and M36 protect infected cells from CD8 T cell control by inhibiting Casp8-dependent apoptosis. We developed a set of methods to in vitro measure the antiviral capacity of CD8 T cells and show that UL36/M36 protect the virus from T cell cytotoxicity more effectively than the combined effect of the immune evasins that target MHC class I antigen presentation. M36 increased viral fitness in animals where CD8 T cells control infection, preventing CD8 T cell-induced apoptosis triggered by DR or perforin–granzyme-dependent signaling at a point of their convergence.
Results
CMV Antiapoptotic Proteins Evade CD8 T Cells’ Control More Effectively than vRAPs.
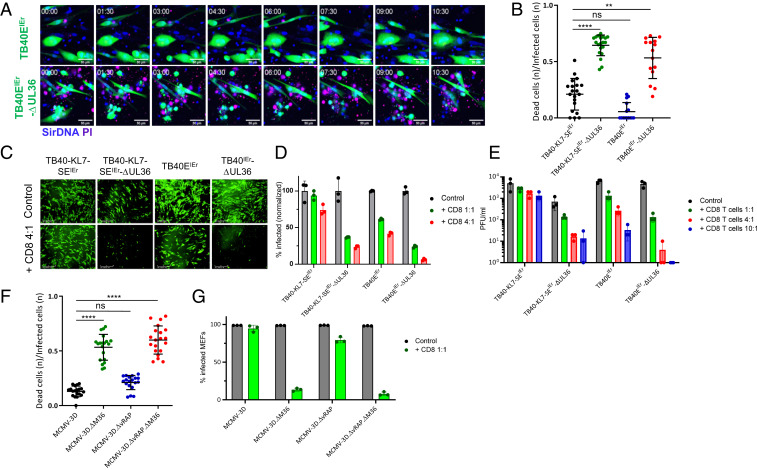
We generated reporter HCMVs on the background of the clinical isolate TB40-E (TB40EIEr), lacking the vRAPs US2, US3, and US6, or the antiapoptotic gene UL36, or both and compared control of these viruses by CD8 T cells to wild-type (WT) virus expressing these immunoevasins (25). CD8 T cells specific for the HLA-A02–restricted pp65 epitope (NLVPMVATV) killed MRC-5 cells infected with HCMV lacking the UL36 gene much more efficiently than TB40EIEr-KL7-SE or the TB40EIEr variant lacking US2, US3, and US6 (Fig. 1 A and B). In order to exclude an effect of allogenic response on the killing of the virus-infected cells, we cocultured HCMV-infected fibroblasts with autologous HLA-B35–restricted pp65 epitope (IPSINVHHY) specific CD8 T cells (26), which controlled the growth of ΔUL36 mutants substantially better than that of the UL36 sufficient viruses (Fig. 1 C–E). The absence of vRAP genes made HCMV more susceptible to CD8 T cell control, while control over HCMV lacking both vRAPs and UL36 was the most substantial (Fig. 1 D and E). Furthermore, time-lapse imaging of reporter HCMV in autologous coculture showed that the onset of virus control occurs roughly 20–24 h post CD8 T cell coculture (SI Appendix, Fig. S1 and Movie S1).
Fig. 1.
CMVs lacking antiapoptotic proteins UL36/M36 are susceptible to CD8 T cells' control. (A and B) MRC-5 cells were infected with indicated HCMV variants at a multiplicity of infection (MOI) of 0.1 and cocultured with HLA-A2–restricted pp65-specific CD8 T cells at 3:1 (E:T). (A) Still frames from live-cell imaging of coculture. Reporter viruses expressed mNeonGreen (green). Propidium iodide (PI, purple) was used to label dead cells. Time 00:00 is 24 h after T cell addition. (Scale bar, 50 μm.) (B) Time-lapse imaging was used to quantify fraction of dead cells in the infected population for each frame by counting the dead cells within the infected population. The cumulative n of such events for 48 h post T cells’ coculture is plotted. Each symbol represents one time-lapse imaging field. Data are compiled from at least three independent experiments. (C–E) BFF2 skin fibroblast cells were infected with HCMV at an MOI of 0.1 and cocultured with autologous HLA-B35–restricted pp65-specific CD8 T cells at 4:1 (E:T). (C) Representative images from the isogeneic coculture show HCMV-infected BFF2 cells 6 d post CD8 T cell coculture (control = without T cells). (Scale bar, 400 μm.) (D) HCMV-infected BFF2 cells were analyzed 6 d post CD8 T cell coculture with flow cytometric analysis. (E) Supernatants were collected from infected BFF2 cocultured with autologous CD8 T cells 6 d post infection (dpi), and titrated with standard plaque assay on BFF2 cells. (F and G) BL/6 MEF cells were infected with indicated viruses and cocultured at 4:1 (E:T) with syngeneic CD8 T cells (CD44+CD62L−) sorted from spleen of BL/6 animals latently infected with MCMV. (F) MEFs were infected at an MOI of 0.2, 24 h prior to CD8 T cell coculture. Fraction of dead cells in the infected population for initial 48 h post T cells’ coculture is plotted. Each symbol represents one time-lapse imaging field. Data are compiled from at least four independent experiments. (G) MEFs were infected at an MOI of 0.1 and cocultured with CD8 T cells. Cells were analyzed 6 d post CD8 T cell coculture with flow cytometric analysis to quantify mCherry positive (infected) cells. Panels represent typical data from at least two independent experiments. Data with error bars depict mean ± SD. B and F show statistical significance calculated with Dunn’s post test following Kruskal-Wallis test. **P < 0.01, ****P < 0.0001, P > 0.05 not significant (n.s.).
To determine whether this extends to the evasion of MHC class I antigen presentation in the mouse model, we cocultured broad MCMV-specific CD8 T cells with MCMV-infected mouse embryonic fibroblast (MEF) cells. We used different variants of MCMV-3D (27) that lack M36 (MCMV-3D.ΔM36), vRAPs m06 and m152 (MCMV-3D.∆vRAP), or all of the above (MCMV-3D.∆vRAP.∆M36), to infect MEF cells. MCMV-3D was derived from MCMV smith strain pSM3fr (28) by inserting mCherry in place of m157 gene, and it expresses the Kb-restricted SIINFEKL epitope in the m164 gene (27). CD8 T cells controlled MCMV-3D.ΔM36 and MCMV-3D.∆vRAP.∆M36 growth substantially better than that of WT virus (Fig. 1G) and lysed the infected cells much more efficiently in absence of M36 (Fig. 1F and Movie S2). Killing of the vRAP mutant, on the other hand, was modestly increased as compared to MCMV-3D. Similarly, ΔM36 mutant was also controlled in endothelial cells (SI Appendix, Fig. S2). CD8 T cells’ control over virus growth was dose-dependent. While ΔvRAP mutant was controlled at higher effector-to-target ratio (E:T), control of M36 mutant virus occurred at lower E:T ratios (SI Appendix, Fig. S3A). Thus, suppression of Casp8-dependent signaling provided more efficient evasion of CD8 T cells' control than MHC-I down-regulation by HCMV and MCMV.
CD8 T Cells with Different MCMV Epitope Specificity Show Distinct Pattern of Virus Control.
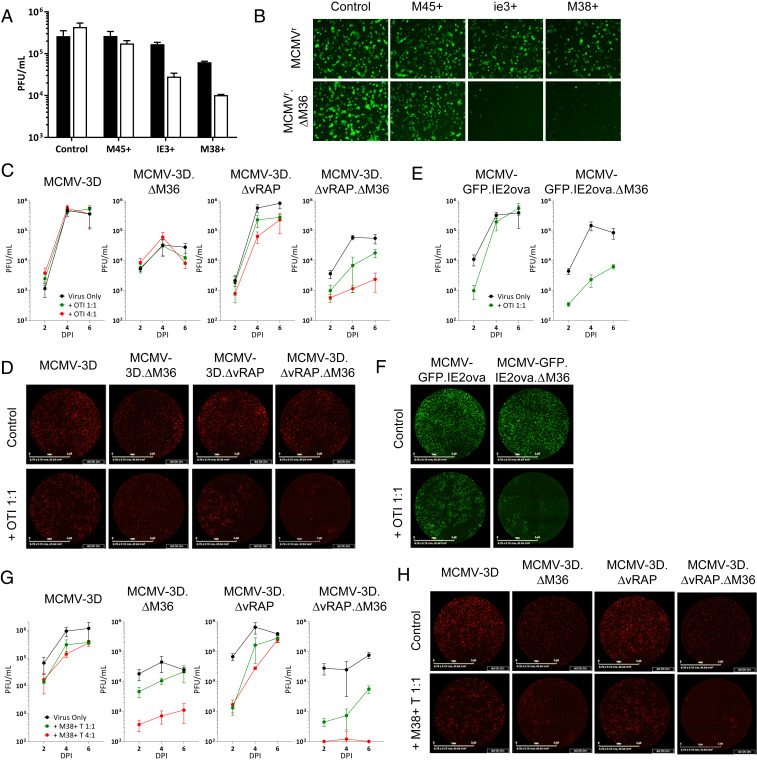
To identify the epitope-specific CD8 T cells capable of controlling MCMV growth, we cocultured tetramer-sorted CD8 T cells with MEF cells infected with reporter MCMVr, which expresses YFP and tdTomato under the control of the MIEP (major immediate-early promoter) as proxies of ie1 and ie2, respectively (29). In the presence of vRAPs, CD8 T cells specific for the immediate-early epitopes IE3 and M38 reduced MCMVr.∆M36 levels more efficiently than they acted against M36-expressing MCMVr, but CD8 T cells recognizing the early M45 epitope failed to reduce levels of either MCMVr or MCMVr.∆M36 (Fig. 2 A and B). This difference could have been explained by the kinetics of epitope expression, but also by differences in epitope processing, where the M45 epitope requires immunoproteasomal processing and is thus less efficiently processed in MEF cells (30). To compare the M36 and vRAP immune evasion of antigens expressed with early (E) or immediate-early (IE) kinetics, we compared CD8 T cells recognizing the SIINFEKL epitope expressed with E kinetics within the m164 protein (31) to M38-specific CD8 T cells that target this IE epitope. Both of these epitopes induce inflationary responses and therefore can be processed independently of the immunoproteasome (32). OT-I cells were unable to reduce the levels of MCMV-3D virus and only moderately impaired the growth of mutants lacking M36 or the vRAP genes. However, the combined deletion of both vRAPs and M36 resulted in a severe growth impairment in the presence of OT-I cells (Fig. 2 C and D), suggesting that the combined activity of the two immune evasion mechanisms provides superior evasion against CD8 T cells that target an E epitope.
Fig. 2.
Control of MCMV.∆M36 by different epitope-specific CD8 T cells. MEFs were infected with indicated viruses at an MOI of 0.1 and cocultured with epitope-specific CD8 T cells from latently infected BL/6 mice. (A) MEFs were infected with MCMVr (filled bars) or MCMVr.∆M36 (empty bars) and cocultured with CD8 T cells at 1:1 (E:T) or in their absence (control). (B) Representative fluorescent microscopy images of coculture wells from A at 6 dpi. (C–H) MEFs were infected with indicated viruses at an MOI of 0.1 and cocultured with either OT-I cells (C–F) or M38-specific CD8 T cells (G and H). Both OT-I and M38-specific CD8 T cells were isolated from BL/6 animals latently infected with an MCMV-inducing inflationary responses against either epitope. D, F, and H show representative images at 4 dpi from C, E, and F, respectively. (Scale bar, 3.4 mm.) Panels represent typical data from at least two independent experiments. Data with error bars depict mean from biological triplicates ± SD.
Importantly, M38-specific CD8 T cells, which showed some antiviral activity against WT-MCMV, exerted a highly efficient control over ∆M36 virus, despite the retention of vRAP function (Fig. 2 G and H and SI Appendix, Fig. S3B). In contrast, the reduction in ∆vRAP virus levels by M38-specific CD8 T cells was less dramatic in the presence of M36. Immune control mediated by M38 CD8 T cells was most efficient in the combined absence of Casp8 suppression and MHC class I down-regulation. In order to ascertain that observed hierarchy of control manifested by CD8 T cell targeting epitopes expressed with different kinetics is not due to difference in their cytotoxic activity, we generated MCMV that expresses the SIINFEKL epitope with IE kinetics within IE2 gene (MCMV-GFP.IE2ova). OT-I cells isolated from latently infected host controlled the MCMV-GFP.IE2ova.ΔM36 mutant in the presence of vRAP proteins (Fig. 2 E and F). Similarly, OT-I cells isolated from animals with acute infection also showed similar pattern of control of viruses expressing SIINFEKL epitope with IE and E kinetics (SI Appendix, Fig. S4). These results clearly indicated that only M36 allows the virus to efficiently evade control by CD8 T cells recognizing the immediate-early antigen. This point will be revisited in more detail in the discussion.
M36 Protects MCMV from CD8 T Cell Control In Vivo.
MCMV.∆M36 replication is not normalized (rescued) in the absence of host cellular immunity (RAG2−/−γc−/− mice) because vICA function protects virus from proapoptotic cytokines released from macrophages (SI Appendix, Fig. S5 and ref. 24). Hence, to study the role of CD8 T cells in MCMV.∆M36 in vivo control, we opted for a gain-of-function approach. We used MCMV-3D variants to infect RAG2−/−γc−/− mice and adoptively transferred antigen-specific OT-I cells to provide T cell-mediated immune control (Fig. 3A). MCMV-3D.∆M36 and MCMV-3D.∆vRAP.∆M36 were slightly attenuated in control mice lacking OT-I (Fig. 3B), likely due to macrophage presence (24). Upon OT-I transfer, all groups showed a reduction in virus titer, but the growth of MCMV-3D.∆vRAP.∆M36 was significantly impaired (Fig. 3B). These data matched the in vitro coculture data (Fig. 2C), where only the virus lacking both vRAPs and M36, was efficiently controlled by CD8 T cells. Since the virus control by macrophages deters ∆M36 virus growth and spread in RAG2−/−γc−/− knockout mice (24), and poly(I:C) stimulation could have also affected the macrophage function, we considered that this protocol may mask effects that are driven by CD8 T cell-mediated control. In order to minimize the macrophage-mediated interference with ∆M36 MCMV spread, we resorted to measuring the ability of T cells to influence the infection of spleen mesenchymal (CD45-Ter119-CD71- CD31-) and endothelial (CD45-Ter119-CD71- CD31+) cells at 24 hpi, in absence of poly(I:C) stimulation. MCMV-3D and mutant viruses lacking M36 or vRAPs showed a similar percentage of infected cells in the absence of OT-I cells, but OT-I transfer controlled mutant viruses (Fig. 3 C–F). However, MCMV-3D.∆M36 control increased to statistically significant levels, while the control of the virus lacking vRAPs did not. This pattern was observed in endothelial splenic cells (Fig. 3 C and D) and in the mesenchymal ones (Fig. 3 E and F).
Fig. 3.
In vivo control of ∆M36 mutant by CD8 T cells. (A and B) OT-I cells (104) were adoptively transferred to RAG2−/−γc−/− host and activated with SIINFEKL peptide plus Poly(I:C). Animals were infected with MCMV via i.p. route and sacrificed 3 dpi. (A) Prior to sacrification, animals were bled to ensure activation and proliferation of OT-I cells. (B) Virus replication in spleen and liver was assayed with plaque assay. Each symbol represents one mouse, and the horizontal line represents the median value and error bars show SD. Data pooled from two experiments. (C–F) OT-I cells were adoptively transferring to BL/6 host and activated by infection with MCMV-IE2ova. Then, spleens from donors were isolated 7 dpi, and OT-I cells were sorted. After sorting, 106 primed OT-I cells were transferred to RAG2−/−γc−/− host 24 h prior to infection with 106 pfu of MCMV via i.v. route. Animals were sacrificed 24 h post infection (hpi), and the spleen was isolated. Flow cytometric analysis was performed to quantify percent infected cells among spleen endothelial cells (C and D) and spleen mesenchymal cells (E and F). Data with error bars depict mean ± SD. Statistical significance was calculated with Kruskal-Wallis test followed by Dunn’s postanalysis. *P < 0.05, **P < 0.01, P > 0.05 not significant (n.s.).
CD8 T Cells Induce Casp8-Dependent Apoptosis in MCMV.∆M36-Infected Cells.
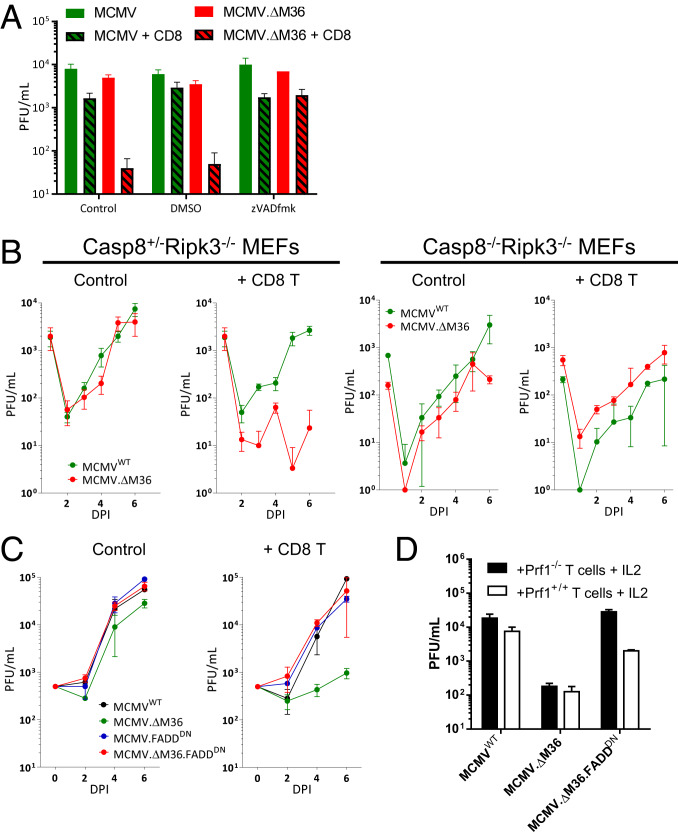
Time-lapse imaging of cocultures showed that CD8 T cells rapidly kill MCMV.∆M36-infected cells (Movies S2 and S3). To demonstrate that CD8 T cell-induced apoptosis controls MCMV.∆M36 growth, we measured virus titers in the presence of CD8 T cells and of the pan-caspase inhibitor zVADfmk. While CD8 T cells reduced MCMV.∆M36 titers, MCMV.∆M36 growth was rescued by zVADfmk, strongly arguing that CD8 cells controlled MCMV.∆M36 by inducing apoptosis (Fig. 4A).
Fig. 4.
CD8 T cells control the MCMV lacking M36 by inducing apoptosis via caspase-8. MEFs were infected at an MOI of 0.1 and cocultured with CD8 T cells at 4:1 (E:T). (A) Supernatant collected 6 dpi from wells treated with either vehicle or zVADfmk. (B) MEFs from Casp8+/−Ripk3−/− or Casp8−/−Ripk3−/− mice were infected with indicated viruses and cocultured with CD8 T cells. (C) MEFs were infected with indicated MCMV-expressing FADDDN and cocultured with CD8 T cells. (D) Supernatants were collected 6 dpi from coculture of infected MEF and either Prf1−/− or Prf1+/+ CD8 T cells stimulated with IL2. All of the experiments were performed at least twice, and typical data are shown. Error bars represent mean from biological triplicates ± SD.
The M36 protein binds to Casp8 (20), and the in vivo growth defect of MCMV.∆M36 is rescued in Casp8−/−Ripk3−/− animals (33). Thus, we cocultured CD8 T cells with virus-infected MEFs lacking either RIPK3 or both RIPK3 and Casp8 and measured virus titers over time. MCMV.∆M36 levels were reduced by coculture of CD8 T cells with Casp8+/−Ripk3−/− MEFs, but not with Casp8−/−Ripk3−/− MEF cells (Fig. 4B). Therefore, CD8 T cells controlled MCMV.∆M36 growth via a Casp8-dependent mechanism, most likely apoptosis, in the infected cells.
M36 Prevents CD8 T Cell Control of MCMV Growth by Inhibiting DR and Perforin/Granzyme Signaling.
CD8 T cells use multiple effector mechanisms to control virus infection. The crosstalk between death pathways in T cell cytotoxicity has not been fully elucidated. Known effectors include DR ligands like FasL and TRAIL and cytokines like IFNγ and TNFα functioning in addition to the perforin/granzyme pathway. We quantified the IFNγ release in coculture assay with epitope-specific cells and observed varying levels of IFNγ release when cultured with virus-infected cells (SI Appendix, Fig. S6). Transfer of these supernatants inhibited MCMV growth, but both MCMVWT and MCMV.∆M36 were inhibited to a similar extent. Furthermore, MCMV growth was restored by an antibody-neutralizing IFNγ, but not by a TNFα-specific antibody, arguing that the effect was IFNγ-dependent (SI Appendix, Fig. S6). Thus, cytokines released by CD8 T cells restrict MCMV replication, but this is not impacted by viral suppression of Casp8.
We have previously shown that a dominant-negative variant of FADD (FADDDN) neutralizes DR apoptosis and rescues MCMV growth in absence of M36. We used the FADDDN-expressing ∆M36 (MCMV.∆M36.FADDDN) to determine whether CD8 T cells relied on FADD signaling in the MCMV.∆M36-infected cells. The growth of MCMV.∆M36.FADDDN in the presence of CD8 T cells was completely rescued (Fig. 4C). Therefore, M36 protected the virus from CD8 T cells by a mechanism that requires FADD-Casp8 interaction within virus-infected cells. To assess the role of the perforin/granzyme pathway in MCMV control and the role of M36 in its evasion, we considered that granzyme B cleaves Casp8 directly, without FADD involvement (7), and that IL2 enhances the perforin/granzyme pathway (34). Hence, we tested MCMV.∆M36 and MCMV.∆M36.FADDDN growth in cells cocultured with Prf1+/+ or Prf1−/− CD8 T cells in the presence of IL2, which reduced the infectious titers of all tested viruses, especially in coculture with Prf1+/+ CD8 T cells (SI Appendix, Fig. S7 A and B). IL-2–treated CD8 T cells controlled MCMV.∆M36 very efficiently in IL-2–treated cocultures of WT MEFs and Prf1+/+ and Prf1−/− CD8 T cells (Fig. 4D), and this control was caspase-8–dependent because it did not occur in Casp8-deficient MEFs (SI Appendix, Fig. S7C). FADDDN expression entirely rescued the growth of MCMV.∆M36 in coculture with Prf1−/− CD8 T cells. However, FADDDN could not replace M36 in the presence of Prf1+/+ CD8 T cells, where MCMV.∆M36.FADDDN growth was only partly rescued (Fig. 4D). Therefore, FADDDN completely substituted M36, but only in absence of perforin/granzyme. Taken together, our data argue that the interaction of vICA with Casp8 protects infected cells from redundant DR and perforin/granzyme signaling that crosstalk at the Casp8 level.
Discussion
Cytomegalovirus evades immune effector mechanisms by an array of fine-tuned approaches that span from antigen presentation interference to the manipulation of cytokine responses (35–37). Similarly, CMV uses multiple proteins to counter the cell-intrinsic control mechanisms, including apoptosis (14, 33). The vICA protein from HCMV is a determinant of virus fitness in cultured macrophages (38). The MCMV homolog M36 confers virus growth fitness in vivo (21–23), where the macrophages have been shown to be the major cell type that limits the virus spread and growth (24). Here, we show that UL36 and M36 proteins also protect the virus-infected cells from CD8 T cell control by inhibiting cell-extrinsic apoptosis at the Casp8 level.
It has been shown that the UL36 gene can protect cells from lysis by CD95L and CAR-T cells, when expressed in isolation (39). However, its function in the context of infection has not been studied. We deleted the UL36 from the TB40-E genome, either alone, or in combination with MHC-I downregulating genes. Surprisingly, UL36 was much more efficient at protecting virus-infected cells from CD8 T cell killing and promoting virus replication than the vRAPs US3 and US6. While we did not study the effects of another HCMV vRAP (US11), we observed the same hierarchy of T cell evasion in MCMV mutants lacking the homologous genes. Therefore, our data argue that inhibition of Casp8 autoactivation and consequent apoptosis is a hitherto unappreciated mechanism of CD8 T cell evasion and a critical determinant of CMV fitness in the presence of CD8 T cells. While our study did not address putative M36/UL36 effects on T cell responses, our data argue that T cell control of M36/UL36-deficient viruses was independent of T cell activation because the same T cells controlled the mutant viruses more efficiently than the parental ones.
CD8 T cells recognizing inflationary epitopes expressed within IE genes (e.g., IE3 or M38) were very efficient in controlling MCMV.∆M36, but not MCMVWT, arguing for a critical contribution of the M36 function in protecting MCMV from CD8 cells recognizing IE epitopes. On the other hand, cells targeting epitopes expressed with E kinetics (e.g., M45 and m164) were less efficient in controlling MCMV, both in the presence or absence of M36. This was explained by redundant immune evasion by vRAPs. The vRAP genes m06 and m152 are expressed with early kinetics in MCMV infection (37, 40) and thus can block MHC-I trafficking to the cell surface in the early and late phase of the virus cycle, but not in the IE phase. Therefore, the SIINFEKL peptide expressed within the early m164 gene is not presented on MCMV-infected cells in the presence of vRAPs (41). Consequently, OT-I cells did not efficiently control MCMV mutants lacking only M36, or only vRAPs, but were able to robustly control the MCMV mutant that lacked both the antiapoptotic gene and the MHC-I repressor genes, and this phenomenon was observed in vivo as well, although the in vivo data have to be considered critically, as M36 protects the virus not only from CD8 T cells, but also from macrophages, and hence a slight in vivo growth deficit of M36-deficient viruses was observed even in the absence of T cells. Nevertheless, the expression of SIINFEKL peptide with IE kinetics led to ΔM36 mutant control even in the presence of vRAPs. These unique MCMV variants expressing the SIINFEKL epitope with IE or E kinetics allowed us to demonstrate the hierarchy of immune evasion mechanism, where vRAPs and M36 have to cooperate to evade control from CD8 T cells targeting an E epitope, but Casp8 suppression clearly protects from CD8 T cells that recognize an IE epitope.
The initial description of MCMV immune evasion repressing CD8 T cell-mediated cytotoxic effects was published 30 y ago (42). The data presented here argue that the initial observation was likely due to the impact of M36-encoded vICA, rather than MHC repression by vRAPs. In their paper, Del Val et al. showed that CD8 T cells recognizing an IE epitope are prevented from killing virus-infected cells once genes from the early phase were expressed (42). However, subsequent work demonstrated that vRAPs block MHC-I trafficking to the surface (43, 44) and thus do not down-regulate efficiently the MHC-I molecules that have already reached the surface (41, 43, 44). Our observations resolve this dilemma because vICA efficiently protected the virus against CD8 T cells recognizing IE epitopes, whereas the same cells controlled virus growth in the presence of m06 and m152 vRAPs. This also implies that Casp8 inhibition might be a more efficient mechanism of immune evasion of CD8 T cells than the block of peptide presentation on MHC-I, particularly for antigenic epitopes expressed in the IE phase.
CD8 T cells controlled the growth of MCMV.ΔM36 by inducing apoptosis. Consequently, CD8 T cells could not control this virus in the presence of a caspase inhibitor, akin to the situation in MCMV.ΔM36-infected macrophages (22). Furthermore, we have shown that overexpression of FADDDN and the consequent block of DR signaling completely rescue the growth of MCMV.ΔM36 in the presence of CD8 T cells, unless they are stimulated with IL-2, which suggests that IL-2–unstimulated CD8 T cells control MCMV.ΔM36 exclusively through DR signaling. IL2 stimulation, known to up-regulate the granzymes, improved that antiviral activity of CD8 T cells, and FADDDN expression was not sufficient to compensate for the loss of M36 in presence of IL2-stimulated cells. We demonstrated that this was due to the perforin/granzyme pathway by using perforin deficient CD8 T cells, indicating that M36 binding to Casp8 protects the infected cells from both granzyme and DR signaling. This was additionally confirmed when we observed a lack of MCMV.ΔM36 control by IL-2–activated CD8 T cells in Casp8-deficient target cells. Our results align with previous reports that direct cleavage of Casp8 by granzyme B (7) can short-circuit the canonical requirement for Casp8 recruitment to the DISC during the initiation of extrinsic apoptosis (45). Taken together our data implicate M36 not only in suppression of apoptosis induced via DR but also in suppression of caspase-dependent apoptosis induced by granzymes. Hence, we showed that CMV targeting of Casp8 by highly conserved viral genes M36 and UL36 manipulates and neutralizes multiple CD8 T cell-induced signaling pathways at a critical juncture in the infected cell. More broadly, this provides, to our knowledge, firm evidence of apoptosis inhibition as a mechanism of T cell evasion.
Materials and Methods
In Vivo Work.
C57BL/6JRj mice were purchased from Janvier. RAG2−/−γc−/−BL/6 and Thy1.1 OT-I breeding pairs were purchased from Jackson Laboratory. Prf1−/−BL/6 mice were bred at the central animal facility of the Medical Faculty, University of Rijeka. Casp8-/−Ripk3−/− mice were defined previously (33, 46) and were bred at the Emory University Division of Animal Resources. Animal experiments were approved by the local responsible office, namely, Animal Welfare Committee of University of Rijeka, Emory University Institutional Animal Care and Use Committee of Emory University, and Lower Saxony State Office of Consumer Protection and Food Safety (under permit nos. 33.19-42502-04-12/0838, 33.19-42502-04-17/2679, and 33.19-42502-05-18A293). Typically, 8- to 12-wk-old animals were used for infection. For isolating CD8 T cells, mice were intraperitoneally infected with 106 plaque-forming units (pfu) of MCMVWT and housed in SPF conditions for at least 3 months before spleen isolation.
OT-I cells were isolated from spleen of Thy1.1 OT-I mice using Naive CD8a+ T Cell Isolation Kit (Miltenyi Biotec) according to manufacturers’ guidelines. Naive OT-I cells (104) were adoptively transferred into 8- to 12-wk-old RAG2−/−γc−/− animals (i.v.) and activated the next day by administering 10 µg SIINFEKL peptide and 50 µg poly(I:C) (i.p.). Control mice were not subject to poly(I:C) treatment. Mice were infected intraperitoneally 3 d post OT-I transfer with 2 × 106 pfu of MCMV-3D variants and sacrificed 3 dpi.
Alternatively, 105 OT-I cells were transferred to BL/6 mice and primed by infecting animals with MCMV-IE2ova. After 7 d, spleens were isolated and the OT-I cells were enriched using CD90.1 microbeads (Miltenyi Biotec) and further purified with FACS. After priming, 106 OT-I were transferred to RAG2−/−γc−/−mice 24 h prior to infection with 106 pfu of MCMV-3D variants via i.v. route. Animals were sacrificed 24 hpi, and the spleen was isolated to analyze the infected cells. Spleens were digested in R5 media (Roswell Park Memorial Institute [RPMI] 1640 medium with 5% fetal calf serum (FCS), 1 mM sodium pyruvate, 10 mM Hepes, 100 IU/mL penicillin, and 100 g/mL streptomycin) supplemented with Dnase I (50 U/mL), dispase (2 mg/mL), and collagenase P (0.4 mg/mL). Single-cell suspensions were filtered through a 100-µm cell strainer followed by depletion of CD45+ and Ter119+ cells using magnetic-activated cell sorting. The resulting stromal-cell–enriched fractions were labeled with antibodies and analyzed by flow cytometry. Virus infection rates were measured by determining the percentage of mCherry-positive cells within 7AAD-CD45-Ter119-CD71- stromal fractions distinguished into CD31+ endothelial and CD31− mesenchymal compartments.
Cell Lines.
M2-10B4 (CRL-1972) and MRC-5 (CCL-171) cells were purchased from American Type Culture Collection. C57BL/6 primary MEF cells were prepared in-house from C57BL/6JRj, Casp8+/−Ripk3−/−, or Casp8−/−Ripk3−/− mice. BFF2 are primary human cutaneous fibroblast cells (HLA A0201, A0301, B3501, B4001, C0304, and C1502) isolated from donor no. 4 (47). BFF2, M2-10B4, and MEFs were maintained in DMEM supplemented with 10% FCS, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin. MRC-5 cells were cultured in Eagle's minimum essential medium with 10% FCS, 1 mM sodium pyruvate, 100 IU/mL penicillin, and 100 μg/mL streptomycin.
Viruses and Viral Mutagenesis.
MCMVWT refers to pSM3fr-MCK-2fl clone 3.3 (48). MCMV-3D and MCMV-3D.∆vRAP were kindly provided by Martin Messerle (27). MCMVr (29) and MCMV.FADDDN (22) have been described before. Virus mutants were generated with en passant mutagenesis as described before (49, 50). MCMV-GFP.ie2ova was generated by fusing the SIINFEKL epitope at the C terminus of ie2 protein sequence, and P2A-linked GFP was inserted before the second exon of ie1/ie3 (GAGA before the ATG was left intact, and two nucleotides were added to keep P2A in frame with ie1/3). The ∆M36 mutants were generated by replacing the start codon and methionine codon in exon 1 of M36 ORF, at position 49,270 and 49,090, respectively, with stop codons as described previously (21). Similarly, UL36 gene ORF was disrupted from HCMV genome by replacing the methionine in first exon at position 49,087 with stop codon. Generation of the reporter TB40EIEr has been described in detail previously (51). Briefly, mNeonGreen gene linked to the P2A peptide was inserted before the start codon of UL122/123 exon 2. TB40-KL7-SEIEr and TB40-KL7-SEIEr-ΔUL36 viruses were generated using the repaired TB40-KL7-SE bacterial artificial chromosome (BAC) (25). Sequence of the recombinant viruses was confirmed by Sanger sequencing. Furthermore, off-target deleterious mutations were ruled out by whole-genome sequencing of the recombinant virus BACs and virus growth kinetics in the fibroblast cell lines. See SI Appendix for virus reconstitution from BAC and virus stock production.
CD8 T Cell Coculture.
MEFs were cocultured with CD8 T cells in an isogeneic fashion, where both cells were isolated from C57BL/6JRj mice. MEFs were seeded 1 d prior to infection with MCMV at an MOI of 0.1. Cells were cocultured in RPMI medium supplemented with 10% FBS, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin. CD8 T cell were sorted from splenocytes by labeling them with florescent-labeled antibodies against CD3 (17A2), CD8a (53-6.7), CD44 (IM7), and CD62L (MEL-14). In some assays, antigen-specific cells were used that were sorted with tetramer staining. CD8 T cells were stimulated with 10 ng/mL recombinant murine IL2 (PeproTech, Inc.).
For HCMV-infected cell coculture, MRC-5 cells were infected with HCMV at defined MOIs with centrifugal enhancement protocol. However, the virus stocks were also titrated in a similar manner. Virus suspension was centrifuged at 2,000 rpm for 10 min at room temperature essentially as described before (51). Infected cells were cocultured with CD8 T cells 48 hpi. HCMV-infected MRC-5 were cocultured with ex vivo sorted pp65-specific CD8 T cells in CTS OpTmizer T Cell Expansion SFM medium, supplemented with 5% Serum Replacement CTS Immune Cell SR and 50 U/mL human IL-2. CD8 T cells against pp65 NLV epitope were isolated from peripheral blood mononuclear cells (PBMC) using anti-pp65 streptamer labeling.
Autologous coculture was performed by seeding BFF2 cells 1 d prior to HCMV infection and CD8 T cell coculture. HLA-B35-restricted IPS (IPSINVHHY) specific CD8 T cell clone was generated by transducing PBMC from autologous donor (donor no. 4) with IPS-specific TCR sequence as described earlier (26). CD8 T cells were cocultured with autologous pp65-expressing mLCL cells in the presence of 100 U/mL of IL2 for T cell proliferation. The CD8 T cell coculture with BFF2 cells was performed in RPMI medium supplemented with 10% FBS, 50 U/mL human IL-2, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin.
Microscopy and Image Analysis.
Time-lapse imaging was performed with either BioStation IM-Q live-cell screening system (Nikon Instruments, Inc.), Leica TCS SP5 confocal microscope (Leica Microsystems), or ZEISS LSM 980 confocal laser scanning microscope (Carl Zeiss Microscopy) for 72–96 h post T cell transfer. Usually, cells were infected 24 h prior to CD8 T cell coculture (human autologous coculture was imaged with infection and CD8 T cell addition on the same day). All of the images and time series image stacks were analyzed with Fiji (ImageJ). The fraction of infected cells within the infected population was quantified manually or by using an automated in-house Fiji macro (51) to track death events in the virus-infected population (reporter signal+). In some experiments, CellEvent Caspase-3/7 Green was used at 5 µM concentration during imaging for labeling apoptotic cells. Cell death was defined either by CellEvent Caspase-3/7 or propidium iodide (PI) labeling along with loss of florescent signal from the infected cell and appearance of apoptotic bodies. The dead and the infected cells were counted for each frame by counting the dead and the alive infected cells, and the fraction of dead cells in the infected population within the initial 48 h post T cell coculture was plotted using GraphPad Prism.
Statistical Analyses.
Kruskal-Wallis test followed by Dunn’s postanalysis was performed with Graphpad prism 7.0 to quantify the statistical significance. P values < 0.05 were considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001); P > 0.05 not significant (n.s.). Error bars denote ± SD.
Materials and Data Availability.
The data are made available in the manuscript. Further information and requests for resources, reagents, and primary data should be directed to and will be fulfilled by the lead contact, Luka Cicin-Sain (Luka.Cicin-Sain@helmholtz-hzi.de).
Supplementary Material
Acknowledgments
We thank Inge Hollatz-Rangosch, Ayse Barut, and Linda Roback for excellent technical support. This work was supported by the Impulse and Networking Fund of the Helmholtz Association Grant VH-VI-424, DZIF through the research projects TTU 07.804 and TTU 07.817, and the DFG (Deutsche Forschungsgemeinschaft) Excellence Cluster RESIST, Research Area B2.1.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914667117/-/DCSupplemental.
References
- 1.McGeoch D. J., Cook S., Dolan A., Jamieson F. E., Telford E. A., Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247, 443–458 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Lin A., Xu H., Yan W., Modulation of HLA expression in human cytomegalovirus immune evasion. Cell. Mol. Immunol. 4, 91–98 (2007). [PubMed] [Google Scholar]
- 3.Quinnan G. V. Jr. et al., Cytotoxic t cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N. Engl. J. Med. 307, 7–13 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Jonjić S., Mutter W., Weiland F., Reddehase M. J., Koszinowski U. H., Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J. Exp. Med. 169, 1199–1212 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassin D., Garber O. G., Meiraz A., Schiffenbauer Y. S., Berke G., Cytotoxic T lymphocyte perforin and Fas ligand working in concert even when Fas ligand lytic action is still not detectable. Immunology 133, 190–196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meiraz A., Garber O. G., Harari S., Hassin D., Berke G., Switch from perforin-expressing to perforin-deficient CD8(+) T cells accounts for two distinct types of effector cytotoxic T lymphocytes in vivo. Immunology 128, 69–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medema J. P. et al., Cleavage of FLICE (caspase-8) by granzyme B during cytotoxic T lymphocyte-induced apoptosis. Eur. J. Immunol. 27, 3492–3498 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Adrain C., Murphy B. M., Martin S. J., Molecular ordering of the caspase activation cascade initiated by the cytotoxic T lymphocyte/natural killer (CTL/NK) protease granzyme B. J. Biol. Chem. 280, 4663–4673 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Sutton V. R. et al., Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J. Exp. Med. 192, 1403–1414 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtappels R. et al., Cytomegalovirus encodes a positive regulator of antigen presentation. J. Virol. 80, 7613–7624 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Böhm V. et al., The immune evasion paradox: Immunoevasins of murine cytomegalovirus enhance priming of CD8 T cells by preventing negative feedback regulation. J. Virol. 82, 11637–11650 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold M. C. et al., Murine cytomegalovirus interference with antigen presentation has little effect on the size or the effector memory phenotype of the CD8 T cell response. J. Immunol. 172, 6944–6953 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Lu X., Pinto A. K., Kelly A. M., Cho K. S., Hill A. B., Murine cytomegalovirus interference with antigen presentation contributes to the inability of CD8 T cells to control virus in the salivary gland. J. Virol. 80, 4200–4202 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brune W., Andoniou C. E., Die another day: Inhibition of cell death pathways by cytomegalovirus. Viruses 9, 130–146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strand S. et al., Lymphocyte apoptosis induced by CD95 (APO-1/Fas) ligand-expressing tumor cells–A mechanism of immune evasion? Nat. Med. 2, 1361–1366 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Smith C. A., Farrah T., Goodwin R. G., The TNF receptor superfamily of cellular and viral proteins: Activation, costimulation, and death. Cell 76, 959–962 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Chinnaiyan A. M., O’Rourke K., Tewari M., Dixit V. M., FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81, 505–512 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Scaffidi C. et al., Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17, 1675–1687 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skaletskaya A. et al., A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U.S.A. 98, 7829–7834 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ménard C. et al., Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77, 5557–5570 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhry M. Z. et al., UL36 rescues apoptosis inhibition and In vivo replication of a chimeric MCMV lacking the M36 gene. Front. Cell. Infect. Microbiol. 7, 312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cicin-Sain L. et al., Dominant-negative FADD rescues the in vivo fitness of a cytomegalovirus lacking an antiapoptotic viral gene. J. Virol. 82, 2056–2064 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cicin-Sain L., Podlech J., Messerle M., Reddehase M. J., Koszinowski U. H., Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J. Virol. 79, 9492–9502 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebermann L. et al., Block of death-receptor apoptosis protects mouse cytomegalovirus from macrophages and is a determinant of virulence in immunodeficient hosts. PLoS Pathog. 8, e1003062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampaio K. L., Weyell A., Subramanian N., Wu Z., Sinzger C., A TB40/E-derived human cytomegalovirus genome with an intact US-gene region and a self-excisable BAC cassette for immunological research. Biotechniques 63, 205–214 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Schub A., Schuster I. G., Hammerschmidt W., Moosmann A., CMV-specific TCR-transgenic T cells for immunotherapy. J. Immunol. 183, 6819–6830 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Marquardt A. et al., Single cell detection of latent cytomegalovirus reactivation in host tissue. J. Gen. Virol. 92, 1279–1291 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Wagner M., Jonjic S., Koszinowski U. H., Messerle M., Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73, 7056–7060 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dag F. et al., A new reporter mouse cytomegalovirus reveals maintained immediate-early gene expression but poor virus replication in cycling liver sinusoidal endothelial cells. Virol. J. 10, 197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dekhtiarenko I., Jarvis M. A., Ruzsics Z., Čičin-Šain L., The context of gene expression defines the immunodominance hierarchy of cytomegalovirus antigens. J. Immunol. 190, 3399–3409 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Holtappels R. et al., Processing and presentation of murine cytomegalovirus pORFm164-derived peptide in fibroblasts in the face of all viral immunosubversive early gene functions. J. Virol. 76, 6044–6053 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchinson S. et al., A dominant role for the immunoproteasome in CD8+ T cell responses to murine cytomegalovirus. PLoS One 6, e14646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daley-Bauer L. P. et al., Mouse cytomegalovirus M36 and M45 death suppressors cooperate to prevent inflammation resulting from antiviral programmed cell death pathways. Proc. Natl. Acad. Sci. U.S.A. 114, E2786–E2795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janas M. L., Groves P., Kienzle N., Kelso A., IL-2 regulates perforin and granzyme gene expression in CD8+ T cells independently of its effects on survival and proliferation. J. Immunol. 175, 8003–8010 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Hengel H., Brune W., Koszinowski U. H., Immune evasion by cytomegalovirus–Survival strategies of a highly adapted opportunist. Trends Microbiol. 6, 190–197 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Hengel H. et al., Macrophages escape inhibition of major histocompatibility complex class I-dependent antigen presentation by cytomegalovirus. J. Virol. 74, 7861–7868 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krmpotic A. et al., The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J. Exp. Med. 190, 1285–1296 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCormick A. L., Skaletskaya A., Barry P. A., Mocarski E. S., Goldmacher V. S., Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology 316, 221–233 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Proff J. et al., Cytomegalovirus-infected cells resist T cell mediated killing in an HLA-recognition independent manner. Front. Microbiol. 7, 844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto A. K., Munks M. W., Koszinowski U. H., Hill A. B., Coordinated function of murine cytomegalovirus genes completely inhibits CTL lysis. J. Immunol. 177, 3225–3234 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Lemmermann N. A. et al., Immune evasion proteins of murine cytomegalovirus preferentially affect cell surface display of recently generated peptide presentation complexes. J. Virol. 84, 1221–1236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Val M., Münch K., Reddehase M. J., Koszinowski U. H., Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell 58, 305–315 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Ziegler H. et al., A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-golgi compartments. Immunity 6, 57–66 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Reusch U. et al., A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18, 1081–1091 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barry M. et al., Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 20, 3781–3794 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser W. J. et al., RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 471, 368–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ameres S., Besold K., Plachter B., Moosmann A., CD8 T cell-evasive functions of human cytomegalovirus display pervasive MHC allele specificity, complementarity, and cooperativity. J. Immunol. 192, 5894–5905 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Jordan S. et al., Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary Glands due to a fixed mutation of MCK-2. J. Virol. 85, 10346–10353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tischer B. K., Smith G. A., Osterrieder N., En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 634, 421–430 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Dekhtiarenko I., Cičin-Šain L., Messerle M., Use of recombinant approaches to construct human cytomegalovirus mutants. Methods Mol. Biol. 1119, 59–79 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Kasmapour B. et al., Myeloid dendritic cells repress human cytomegalovirus gene expression and spread by releasing interferon-unrelated soluble antiviral factors. J. Virol. 92, e01138-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.