Significance
The bactericidal action delivered by rigid nanopillar arrays stems from the mechanical rupture of the bacterial cell membrane; however, the precise mechanism may be unique to the individual nanopillar geometries. In this study, we demonstrate that the bactericidal action of highly ordered, high-aspect-ratio nanopillar arrays may be associated with the relative flexibility of the individual nanopillars and the mechanical energy stored within the nanopillars. We propose that the lateral stretching of the cell membrane and interactions at the cell edge are induced by elastic pillar deformations that occur during bacterial adhesion. The results obtained in this study provide insight into a previously unknown category of mechano-bactericidal mechanism, highlighting another facet to the mechano-bactericidal action of nanostructured surfaces.
Keywords: mechano-bactericidal, nanostructured surface, antibacterial
Abstract
The mechano-bactericidal activity of nanostructured surfaces has become the focus of intensive research toward the development of a new generation of antibacterial surfaces, particularly in the current era of emerging antibiotic resistance. This work demonstrates the effects of an incremental increase of nanopillar height on nanostructure-induced bacterial cell death. We propose that the mechanical lysis of bacterial cells can be influenced by the degree of elasticity and clustering of highly ordered silicon nanopillar arrays. Herein, silicon nanopillar arrays with diameter 35 nm, periodicity 90 nm and increasing heights of 220, 360, and 420 nm were fabricated using deep UV immersion lithography. Nanoarrays of 360-nm-height pillars exhibited the highest degree of bactericidal activity toward both Gram stain-negative Pseudomonas aeruginosa and Gram stain-positive Staphylococcus aureus bacteria, inducing 95 ± 5% and 83 ± 12% cell death, respectively. At heights of 360 nm, increased nanopillar elasticity contributes to the onset of pillar deformation in response to bacterial adhesion to the surface. Theoretical analyses of pillar elasticity confirm that deflection, deformation force, and mechanical energies are more significant for the substrata possessing more flexible pillars. Increased storage and release of mechanical energy may explain the enhanced bactericidal action of these nanopillar arrays toward bacterial cells contacting the surface; however, with further increase of nanopillar height (420 nm), the forces (and tensions) can be partially compensated by irreversible interpillar adhesion that reduces their bactericidal effect. These findings can be used to inform the design of next-generation mechano-responsive surfaces with tuneable bactericidal characteristics for antimicrobial surface technologies.
The development of antimicrobial resistance to commonly used antibiotics heralds a serious worldwide health epidemic (1–5). Currently, bacterial resistance exists for all available antimicrobial chemotherapies and, as such, is considered one of the most urgent threats to public health. In particular, the emergence of methicillin-resistant Staphylococcus aureus (MRSA), the primary perpetrator of all hospital-acquired infections, generates significant cause for concern (6–8). Due to the increasingly widespread occurrence of antibiotic resistance in common, infection-causing bacteria, serious implications have arisen for the prevention of bacterial colonization of orthopedic implant surfaces. Microbial colonization of implants is frequently associated with implant failure in patients (1, 3, 9, 10). In response, considerable scientific and medical research has focused on the development of antibacterial surfaces (11). This includes surfaces that may resist the initial attachment of bacteria to the surface by repelling any cells that come into close contact (termed the “anti-biofouling effect”) (12–22) or that can kill cells that do contact the surface (“biocidal effect”) (23–43).
A well-established fabrication approach for preparing antibacterial surfaces includes the chemical functionalization of the surface using antibiotics (44), heavy metals (45), quaternary-ammonium compounds (46), fluorine (47), phenols (48, 49), or iodine (50) implanted into a durable coating. However, these surface coatings release their compounds gradually over time and do not kill bacteria by direct contact. Eventually, the surfaces become ineffective when the antimicrobial agent is exhausted (51).
Indeed, the long-term solution to preventing implant-associated infections altogether, and to circumvent the manifestation of bacterial resistance to conventional antimicrobial technologies [which arises due to the continued overprescription or misuse of antibacterial treatments (2, 52)], may rely upon the physical elimination of bacterial cells upon contact, which will restrict the subsequent possibility of an infection developing. One such promising technology relies upon engineering biomaterial surface to exhibit high-aspect-ratio nanostructures. The nanofeatures simultaneously prevent microbial colonization by providing unfavorable attachment points and mechanically lysing any bacterial cells that encounter the surface (9, 26, 32).
This technology was inspired by the naturally occurring nanopillar arrays found on the surface of insect wings that can efficiently kill bacteria by mechanical inactivation of the cells (11, 26, 27, 36, 40). Upon bacterial attachment to the nanopillar surface, the adhesion forces acting on the bacterial membrane induces stretching of the membrane between adjacent nanopillars, suspending the cell. This suspension exerts enough stress upon the bacterial membrane such that the membrane eventually ruptures, causing cell death (26, 29). This finding has stimulated the controlled fabrication of synthetic biomimetic nanotopographies that have emerged as a viable technology for the mitigation of biofilm formation on abiotic surfaces. It is noteworthy to add that the mechano-bactericidal activity of nanostructured surfaces is governed by surface geometry and is independent of surface chemistry. Therefore, these surfaces are a promising alternative strategy for providing a durable antibacterial substratum (9, 11–16, 23–32, 35, 36, 38, 40, 53).
The factors that determine the precise antibacterial activity of a nanostructured surface (whether antifouling or biocidal) are still largely unknown. Previous studies were limited to irregular surface patterns that exhibit large variation in the structural parameters of the individual nanopatterns (27, 31). The relative contributions of each of the surface parameters to the biocidal mechanism could not readily be determined. Therefore, the relationship between the spatial geometry of the nanopillar surface and the resultant antibacterial activity remains unclear. As a result, a considerable amount of uncertainty exists regarding the optimal surface geometry of nanopillar arrays required to maximize the degree of bactericidal activity exhibited by the surface (24, 28–30, 38, 54, 55). This debate stems from a lack of systematic studies (56).
Therefore, this work aimed to investigate the role of nanopillar height, and the increase in nanopillar flexibility, in delivering antibacterial activity toward two clinically relevant, pathogenic bacteria. Systematic alteration of a single surface parameter, while keeping all other structural parameters constant, will allow for determination of the specific mechanical actions that are directly responsible for bacterial cell death. This study will provide insights into the way that mechano-responsive, high-aspect-ratio nanopillared surfaces can impart sufficiently large mechanical forces to induce bacterial cell lysis. This information would advance the current understanding of mechano-bactericidal surfaces and will aid in the development of next-generation surfaces for the effective prevention of bacterial colonization.
Results
Characterization of the Silicon Nanopillared Surfaces.
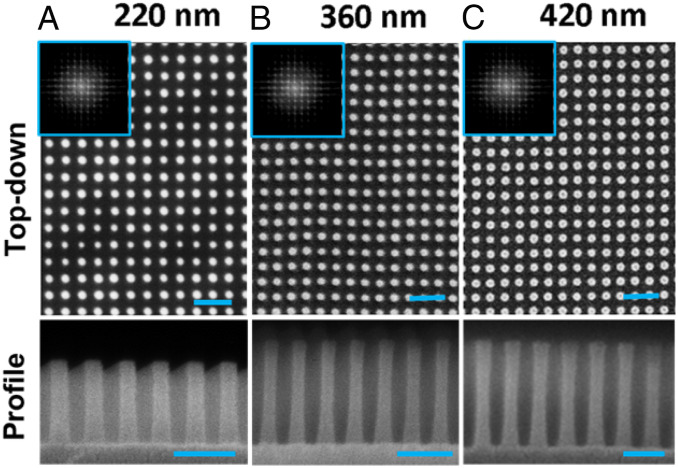
Surfaces possessing a highly ordered array of perpendicularly oriented silicon nanopillars of increasing height, equal diameter, and interpillar distance were fabricated using carefully controlled deep ultraviolet (UV) immersion lithography and plasma etching (57, 58). Nanopillar arrays of increasing height, L, of 224.7 ± 4.3 nm, 364.7 ± 2.4 nm, and 419.4 ± 5.4 (further referred to as 220-nm, 360-nm, and 420-nm pillars, respectively), were achieved by incrementally increasing the reactive ion etching time. Scanning electron microscopy (SEM) micrographs of the resulting nanopillar arrays (in air) are shown in Fig. 1. Three-dimensional (3D) surface reconstructions were computationally rendered to highlight the evolving nanoarchitectures (SI Appendix, Fig. S3). Side-view (90° tilted) SEM images and two-dimensional fast Fourier transforms (2D FFT) revealed differences in the spatial geometries as the nanopillar height was incrementally increased.
Fig. 1.
Nanopillar surface characterization. (Top) Top-down SEM micrographs of the native nanopillared surfaces fabricated with increasing heights L of (A) 220 nm, (B) 360 nm, and (C) 420 nm, as observed in air. (Bottom) Side profile SEM images (90° tilted) of each nanopillar array. Two-dimensional FFT are provided as insets to highlight the relative spatial periodicities and symmetry of the surface nanostructure for each image. (Scale bars, 200 nm.)
For the as-fabricated surfaces, SEM and 2D FFT analyses (Fig. 1, Insets) confirmed that the surfaces were composed of symmetric, free-standing nanopillar arrays with an average pillar diameter d ≈ 35.9 ± 2.2 nm and pillar-to-pillar spacing of a ≈ 92 nm. Therefore, all of the geometric parameters except for nanopillar height were held constant across samples.
The surface chemical composition of the nanopillar substrata was characterized using X-ray photoelectron spectroscopy (XPS) (SI Appendix, Fig. S4 and Table S1) (31–34, 59). XPS characterization indicated that each of the substrata were crystalline silicon with a significant presence of silicon dioxide. Surface chemistry does not play a significant role in the resultant antibacterial properties of these surfaces (11, 32–34, 58, 60), so it will not be considered further in this work.
The advantage of using highly ordered arrays of pillars of the same height L, diameter d, and interpillar distance a (SI Appendix, Fig. S1), is that the mechanical and elastic properties of the pillars are precisely known. Importantly, the inherent flexibility of the underlying nanopillars intrinsically increases as a function of pillar height, meaning that the elasticity of the surface features evolves across the series of substrata. Indeed, considering the material and shape of the pillars, the classical Euler–Bernoulli beam theory allows us to calculate the specific geometric parameters required for the pillar tips to touch each other under an applied external force (SI Appendix, Fig. S2).
Antibacterial Activity of the Silicon Nanopillar Arrays.
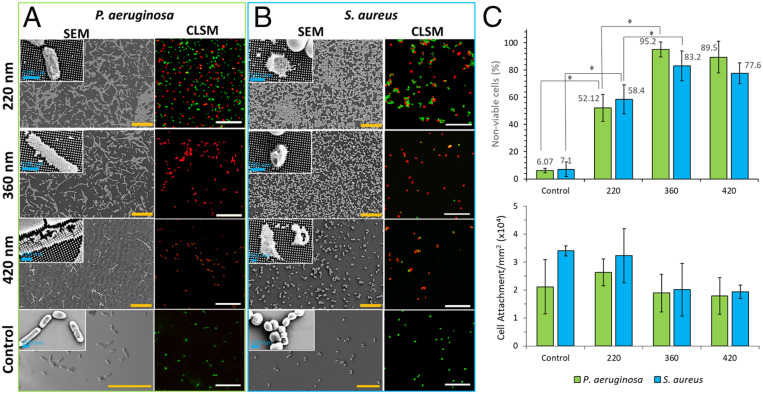
The antibacterial activity against Gram stain-negative Pseudomonas aeruginosa American Type Culture Collection (ATCC) 9721 and Gram stain-positive Staphylococcus aureus Culture Collection of the Institute Pasteur (CIP) 65.8T bacterial cells was assessed for each of the silicon nanopillar arrays. Confocal laser scanning microscopy (CLSM) was used to visualize and quantify the antibacterial efficacy of each surface. For the P. aeruginosa cells incubated on the surfaces possessing pillar lengths of 220, 360, and 420 nm, the average proportion of nonviable cells was found to be 53%, 95%, and 89%, respectively. For S. aureus cells incubated on the same substrata, the average proportion of dead cells was found to be 58%, 83%, and 77%, respectively. Therefore, the highest percentage of nonviable cells was achieved on the substrata possessing pillar heights of 360-nm, although no statistical significance was determined between the antibacterial activity of 360- and 420-nm-high pillars toward both bacterial species. This bactericidal efficacy is comparable to that recently observed on black silicon surfaces (85% for S. aureus and 89% for P. aeruginosa) (31). Analysis of the CLSM micrographs and quantification of the proportions of live (green) and dead (red) bacterial cells revealed that a high number of attached cells were dead across all of the investigated substrata (Fig. 2). Analysis of the attachment propensity on the nanopillared surfaces is included in SI Appendix.
Fig. 2.
Viability, morphology, and attachment of P. aeruginosa and S. aureus bacterial cells on the silicon nanopillar arrays. Representative SEM (Left) and CLSM (Right) images of (A) P. aeruginosa and (B) S. aureus bacterial cells incubated on substrates containing silicon nanopillars of differing heights (indicated in above images) and on planar silicon control surfaces. High-magnification images are shown as insets to the SEM images revealing the altered cell morphology. Fluorescent staining of samples with LIVE/DEAD BacLight display dead cells as red and viable cells as green in the CLSM micrographs. (Scale bars, 20 µm unless otherwise indicated.) (C) Graphs displaying the antibacterial activity of each nanopillar substratum as a percentage of total nonviable cells and the total numbers of attached cells per respective substratum. Error bars are one SD. SDs are calculated based on three independent trials containing duplicate samples. Statistical significance is denoted by ɸp < 0.05.
Inspection of the SEM images (Fig. 2) indicated that significant morphological changes had occurred to both bacterial cell types in each system. When compared to bacteria incubated on the flat silicon control surfaces, bacteria attached to the nanopillar surfaces appeared to be deflated or elongated, with the tips of the nanopillars emerging through the top side of the collapsed cells. This deformation was most evident for the P. aeruginosa cells but also observed to a lesser extent for the S. aureus cells. This result is in good agreement with previously published works (32, 25) and attributable to the differences in the cell outer wall rigidity of Gram stain-positive and Gram stain-negative bacteria (29).
As mentioned in the above sections, the length of the pillars is directly correlated to their elasticity. Analysis of SEM micrographs in Fig. 2 indicated bending of the nanopillars toward the attached bacterial cells. According to estimates made in the previous sections of this work (SI Appendix, Figs. S1 and S2), the nanopillars can contact each other at the tips with the application of sufficient external force, such as that of an adsorbing bacterial cell membrane. While the nanopillars may be bent at the external perimeter of the cells when the pillar heights are 220 and 360 nm, the pillars which are visible through the center of the cells remain perpendicular and independent. Therefore, minimal nanopillar bending took place beneath the central mass of the bacterial cells, and more considerable nanopillar bending occurred at the perimeter of the cells. However, the forces applied by the adsorbing membrane of the bacteria (either P. aeruginosa or S. aureus) was not sufficient to cause irreversible clustering of the nanopillars.
In contrast, the pillar–pillar interactions are more prominent for surfaces possessing the taller (420-nm) nanopillars. For this surface, pillar clusters are also observed to be present underneath the central mass of the cells, indicating our previous assumptions to be correct; pillars of larger heights require less external force (two orders of magnitude less) in order to bend and contact at the tips. This implies that we would observe a higher incidence of pillar clustering for longer pillars interacting with bacteria than for shorter pillars. This finding prompted detailed studies of the bacteria–nanopillar interface using focused ion-beam (FIB) milling, which are presented in the next section.
The Bacteria–Nanopillar Interface.
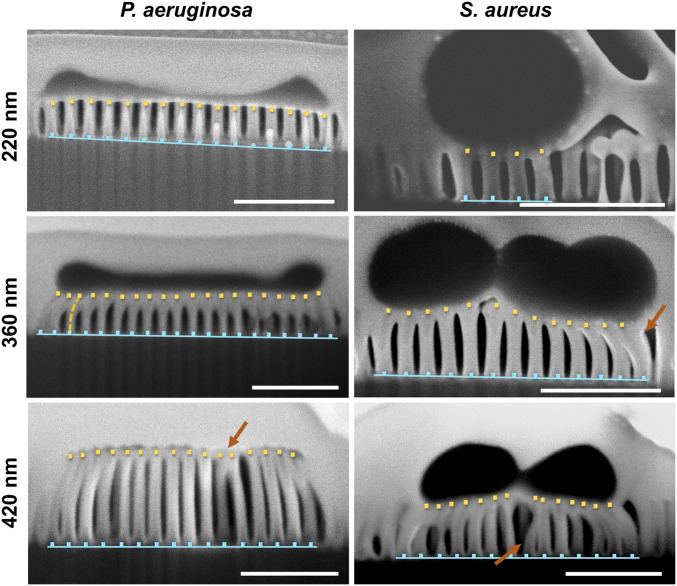
The biointerface formed between the substrata and the attached P. aeruginosa and S. aureus cells following a 30-min incubation period is shown in Fig. 3. During the FIB-SEM experiments, a dual-beam system was used to coat the cells with a protective Pt layer and precisely mill cross-sections for imaging, which provided an insight into the events occurring at the bacteria–nanopillar interface. A representative FIB-SEM image of the native substratum possessing 360-nm pillars following surface ion-beam milling can be seen in SI Appendix, Fig. S5, where the nanopillars appear to be unaffected by the Pt deposition, remaining aligned perpendicularly, unclustered, and identically arranged to those of the uncoated surfaces (SI Appendix, Fig. S5).
Fig. 3.
The biointerface between P. aeruginosa and S. aureus cells and the nanopillared substrata. FIB-SEM side profile images of the substrata with 220-nm, 360-nm, and 420-nm pillars exposed to P. aeruginosa and S. aureus cells. For all samples, FIB milling was completed after an incubation of 30 min in bacterial suspension to reveal the interaction of the bacterial cells with the underlying substratum. (Scale bars, 500 nm.) The arrows highlight areas where nanopillars have clustered.
Close inspection of the FIB-SEM images confirmed the direct contact of bacterial cells with the surfaces (60) and the altered cell morphology of the Gram stain-positive and Gram stain-negative cells that were attached to the surfaces, as well as the proportional bending of the underlying nanopillars. The FIB-SEM analysis further reinforces the findings of Linklater et al. (61), showing that the bacterial cells were in direct contact with the nanopillars on the surface. The nanopillars caused the stretching of the cell membrane but did not pierce the cell.
The FIB-SEM images also highlight the differences in the bacteria–substratum interface and reinforce what was observed for the SEM micrographs in Fig. 2. Relatively small nanopillar bending was observed for the 220-nm pillars interacting with bacterial cells, whereas the 360-nm pillars surrounding the bacterial cell edges were observed to be bending inward. For the substrata with 420-nm pillars the SEM data (Figs. 2 and 3) showed that the underlying surfaces contained nanopillars that were significantly more clustered, both at the cell periphery and underneath the central mass of the cells. On these surfaces, cells appeared to be elongated, substantially damaged, and “deflated,” bearing little morphological resemblance to that of the healthy bacterial cells (Fig. 2).
Estimate of Pillar-Induced Forces and Energies of Pillar Deformation.
With further analysis of the bacteria–nanopillar interface revealed by the FIB milling shown in Fig. 3, the pillar bending information from the cross-sections can be extracted. Ghibaudo et al. (62) followed an analogous idea for measuring traction forces of fibroblasts on microstructured surfaces. Based on the horizontal pillar deflections , between tip position and grafting point , the pillars' elastic energy is written as
| [1] |
In Eq. 1, k is the elastic constant that corresponds to k = (3EI/L3) in the case of the ideal beam deformation (Eq. 1). To avoid overestimation of energies and forces, however, it is assumed that substratum tilting at the pillar bases, in addition to shear contribute to the total deflection of the pillar tips, as described by Schoen et al. (63), leading to a reduced effective elastic constant k,
| [2] |
with d being the diameter of a pillar and
| [3] |
being proportional to the tilt angle of the pillar base due to the deformation of neighboring substrate as a function of Poisson’s ratio [ (64)]. We use the Young’s modulus of E = 66.5 GPa found for thin silicon sheets (65) by interpolating linearly between the given values of ∼53 GPa (12-nm thickness) and 68 GPa (38.5-nm thickness).
Furthermore, a softening of the elastic moduli with a decrease in the size of the nanoscale silicon structures has been reported (65, 66). In this work, we assumed a reduced silicon Young’s modulus of E = 66.5 GPa based on experimental results for silicon structures of comparable dimensions (65). The particular distribution of surface stresses is expected to influence the effective elastic properties of silicon nanostructures (67). The elastic moduli can be subject to deviations in the shape of ideal cylinders and the angle dependencies of elastic constants in crystals (68), surface textures (SI Appendix, Fig. S6B) that reduce the contributing cross-section, as well as the presence of silicon oxide layers (69, 70). Due to the strong dependency of the area moment on pillar diameter, it should be noted that an error of ±2.2 nm for the pillar diameter (SI Appendix, Table S1) leads to a relative variation between 20% and 30% of the elastic constant, k (Eq. 2). In view of the mentioned uncertainties of the elastic constant, this paper discusses our results based on the order of magnitudes, as well as a relative comparison between bacteria and pillar heights. For each system shown in the FIB-SEM images presented in Fig. 3, the deflection of each pillar in contact with bacteria was estimated in order to calculate mean absolute forces and mechanical energy of the pillar tips resulting from lateral deflection. For this purpose, we approximate the lateral grid coordinates of pillar base points (blue lines in Fig. 3) as well as the pillar tip positions (yellow points) to compute the deflection for pillars and refer to the known pillar spacing of a = 92 nm for deflection measurements. The average deflection value is subtracted from the result in order to compensate eventual force imbalances caused by Pt deposition (See, e.g., the apparent left shifts for P. aeruginosa, 220 nm, and S. aureus, 360 nm). The results are summarized in Table 1.
Table 1.
Estimated values for pillar deflection, δ, listing pillars from left to right in Fig. 3, the resulting absolute force |P| = k|δ|, elastic energy U, and perpendicular distance of the pillar tops from the next neighboring pillar atop ≈ (a + δnext) − δ − d, where δnext is the next pillar’s deflection
| 220, k(N/m): 1.25 | 360, k(N/m): 0.31 | 420, k(N/m): 0.21 | ||||||||||
| δ (nm) | |P| (nN) | U (fJ) | atop (nm) | δ (nm) | |P| (nN) | U (fJ) | atop (nm) | δ (nm) | |P| (nN) | U (fJ) | atop (nm) | |
| P. aeruginosa | ||||||||||||
| −5.3 | 6.6 | 0.017 | 61.6 | 99.0 | 30.5 | 1.511 | 30.6 | 109.5 | 22.4 | 1.229 | 23.2 | |
| 0.3 | 0.3 | 0.000 | 60.7 | 73.5 | 22.7 | 0.834 | 31.1 | 76.6 | 15.7 | 0.601 | 45.2 | |
| 4.9 | 6.1 | 0.015 | 55.2 | 48.6 | 15.0 | 0.364 | 43.8 | 65.7 | 13.5 | 0.443 | 43.6 | |
| 3.9 | 4.9 | 0.010 | 60.7 | 36.3 | 11.2 | 0.203 | 47.8 | 53.2 | 10.9 | 0.291 | 40.2 | |
| 8.5 | 10.7 | 0.046 | 57.9 | 28.0 | 8.6 | 0.121 | 57.3 | 37.4 | 7.7 | 0.143 | 34.2 | |
| 10.4 | 13.0 | 0.067 | 50.6 | 29.2 | 9.0 | 0.131 | 50.9 | 15.5 | 3.2 | 0.024 | 48.3 | |
| 4.9 | 6.1 | 0.015 | 55.2 | 24.0 | 7.4 | 0.089 | 44.0 | 7.6 | 1.6 | 0.006 | 53.7 | |
| 3.9 | 4.9 | 0.010 | 52.4 | 11.9 | 3.7 | 0.022 | 48.9 | 5.2 | 1.1 | 0.003 | 46.5 | |
| 0.3 | 0.3 | 0.000 | 58.9 | 4.8 | 1.5 | 0.003 | 42.6 | −4.3 | 0.9 | 0.002 | 47.8 | |
| 3.0 | 3.8 | 0.006 | 48.7 | −8.8 | 2.7 | 0.012 | 58.0 | −12.7 | 2.6 | 0.016 | 48.2 | |
| −4.3 | 5.4 | 0.012 | 57.9 | −6.8 | 2.1 | 0.007 | 54.4 | −20.6 | 4.2 | 0.043 | 21.8 | |
| −2.5 | 3.1 | 0.004 | 53.3 | −8.6 | 2.6 | 0.011 | 45.4 | −54.9 | 11.3 | 0.309 | 63.4 | |
| −5.3 | 6.6 | 0.017 | 57.9 | −19.3 | 6.0 | 0.058 | 48.9 | −47.6 | 9.8 | 0.232 | 47.0 | |
| −3.4 | 4.3 | 0.007 | 50.6 | −26.5 | 8.2 | 0.108 | 51.4 | −56.7 | 11.6 | 0.330 | 38.3 | |
| −8.9 | 11.2 | 0.050 | 54.8 | −31.2 | 9.6 | 0.150 | 51.2 | −74.5 | 15.3 | 0.569 | 31.2 | |
| −10.2 | 12.8 | 0.065 | −36.0 | 11.1 | 0.200 | 53.8 | −99.4 | 20.4 | 1.013 | |||
| −38.3 | 11.8 | 0.227 | 52.1 | |||||||||
| −42.4 | 13.1 | 0.277 | 39.1 | |||||||||
| −59.3 | 18.3 | 0.543 | 37.4 | |||||||||
| −78.0 | 24.1 | 0.939 | ||||||||||
| mean: | 6.2 | 0.021 | 55.8 | 11.0 | 0.290 | 46.8 | 9.5 | 0.328 | 42.2 | |||
| edge: | 9.7 | 0.041 | 58.2 | 27.3 | 1.225 | 34.0 | 21.4 | 1.121 | 27.2 | |||
| S. aureus | ||||||||||||
| 7.8 | 9.8 | 0.038 | 51.5 | 37.5 | 11.6 | 0.217 | 49.5 | 111.3 | 22.8 | 1.271 | 26.5 | |
| 3.2 | 4.0 | 0.006 | 50.6 | 30.9 | 9.5 | 0.147 | 43.5 | 81.7 | 16.8 | 0.685 | 29.3 | |
| −2.3 | 2.9 | 0.003 | 49.7 | 18.3 | 5.6 | 0.052 | 42.6 | 57.7 | 11.8 | 0.341 | 43.3 | |
| −8.7 | 10.9 | 0.048 | 4.8 | 1.5 | 0.003 | 62.2 | 46.5 | 9.5 | 0.222 | 35.6 | ||
| 10.9 | 3.4 | 0.018 | 81.7 | 27.1 | 5.5 | 0.075 | 41.6 | |||||
| 36.5 | 11.3 | 0.205 | 41.1 | 12.9 | 2.6 | 0.017 | 31.0 | |||||
| 21.5 | 6.6 | 0.071 | 48.9 | −12.1 | 2.5 | 0.015 | 121.1 | |||||
| 14.4 | 4.4 | 0.032 | 47.7 | 53.1 | 10.9 | 0.289 | −16.5 | |||||
| 5.9 | 1.8 | 0.005 | 31.8 | −19.2 | 3.9 | 0.038 | 55.8 | |||||
| −18.3 | 5.7 | 0.052 | 35.4 | −19.3 | 4.0 | 0.038 | 34.7 | |||||
| −39.0 | 12.0 | 0.234 | 42.4 | −40.5 | 8.3 | 0.168 | 23.3 | |||||
| −52.7 | 16.3 | 0.429 | 38.2 | −72.7 | 14.9 | 0.542 | 22.5 | |||||
| −70.6 | 21.8 | 0.769 | −104.4 | 21.4 | 1.117 | 33.7 | ||||||
| −122.1 | 25.0 | 1.528 | ||||||||||
| mean: | 6.9 | 0.024 | 50.6 | 8.6 | 0.172 | 47.1 | 11.4 | 0.453 | 37.1 | |||
| edge: | 10.3 | 0.043 | 50.6 | 16.7 | 0.493 | 43.8 | 23.9 | 1.399 | 30.1 | |||
Note that negative values for atop have not been excluded and can be the result of apparent pillar overlap in the view plane (S. aureus, 420 nm). For “edge” results we take the mean for the two outermost pillars, which typically coincide with maximum force and energy.
As seen in Tables 1 and 2, the mechanical energies (U/U0) increased by one order of magnitude between shorter pillars, L = 220 nm, and taller pillars, L = 360 and 420 nm. In contrast, the elastic forces (|P|/|P|0) were similar for all six samples in Table 1 (three pillar heights and two bacterial species), and only a slight increase can be observed for the taller pillars. The increase of mechanical energy and elastic forces for nanopillar arrays L = 360 and 420 nm could lead to additional stress being placed on the bacterial cell membrane. This is the most likely cause of the enhanced mortality observed for P. aeruginosa and S. aureus attached to 360- and 420-nm-height pillar arrays (Fig. 2C). The order of magnitude of ∼10 nN (Table 1) matches well with adhesion forces of ∼15 nN observed for S. aureus on stainless steel (71) and 8.53 ± 1.40 nN for P. aeruginosa adhesion measured for metal surfaces (72). It should be emphasized that in both studies by Alam and Balani and Harimawan et al., adhesion time scales were shorter than in our experiments (10 s and 60 s, respectively). A trend of increasing forces with increasing time was observed in both studies (71, 72) whereas a 30-min incubation was used for this work. For P. aeruginosa, attachment forces known from silicon AFM tips (73) were on the order of 0.5 nN, which is smaller than the observed forces in our study. The pillars fabricated in our study, however, are thicker in comparison (radius <10 nm). These structure differences and variability in experimental setups may contribute to the enhanced adhesion forces measured in this study.
Table 2.
Summary of the mean absolute forces and mechanical energy of the pillar tips labeled in Fig. 3 resulting from a lateral deflection
| P. aeruginosa | S. aureus | |||
| L, nm | |P|/|P|0 | U/U0 | |P|/|P|0 | U/U0 |
| 220 | 1.0 | 1.0 | 1.1 | 1.1 |
| 360 | 1.8 | 13.6 | 1.4 | 8.1 |
| 420 | 1.5 | 15.4 | 1.8 | 21.3 |
Both |P| and U are given relative to the result “0” for P. aeruginosa, L = 220 nm.
The calculated forces can be associated with the effective surface tensions imposed on bacterial cells scaling as Σ∼P/a, resulting on the order of 0.1 N/m. A more precise estimate integrating the contribution of many pillars within vertically aligned carbon nanotube arrays (61) revealed that values of the dynamic surface tension range between the orders of 0.1 and 1 N/m, following equivalent trends as the energies listed in Table 1, as a function of pillar height (L). It is, however, noteworthy to add that forces (and tensions) can be partially compensated by interpillar adhesion, which would reduce their bactericidal effect. This was particularly obvious for the outermost pillars of L = 420 nm, or in the case of induced clusters (arrows, Fig. 3) where relatively small interpillar spaces near the top region were observed (atop in Table 1). The corresponding pillars are subjected to the most significant elastic forces (20 to 30 nN), eventually exceeding the scale of bacterial adhesion forces (Table 1).
Discussion
Traditionally, many nanofabrication techniques used to fabricate synthetic nanostructured biocidal surfaces result in the inadvertent alteration of multiple surface parameters (31, 33, 37, 41, 49, 74) and, as a result, the precise relationship between the spatial geometry and the resultant antibacterial activity still remains poorly understood. Here, our fabrication technique allowed for the isolation of a single geometric parameter (nanopillar height) to study the change in antibacterial activity vs. the systematic increase of nanopillar height.
Specifically, the antibacterial activity of substrata with three nominal nanopillar heights of 220, 360, and 420 nm were investigated using Gram stain-positive S. aureus and Gram stain-negative P. aeruginosa bacteria. The nanoarrays of 360-nm pillars (Fig. 1) exhibited the highest degree of antibacterial activity, inducing 95 ± 5% and 83 ± 12% cell death for P. aeruginosa and S. aureus cells, respectively. Appraisal of similar experimental studies of highly performing naturally occurring (25–27, 40) and synthetic (31, 32, 34, 59) mechano-bactericidal surfaces revealed the presence of widely variable nanopillar diameters and interpillar spacing, making direct comparison of the systems difficult.
When compared with the regularly arrayed nanopillars of cicada wings, which exhibit similar surface topographical dimensions (i.e., height and spacing), the pillar heights of 220 nm fabricated in this study were observed to inactivate a large percentage (58%) of Gram stain-positive cells while the cicada wing nanopillars were not effective against S. aureus at all (25). By contrast, previous reports showed that the cicada wing nanopattern eliminated almost 100% of all Gram stain-negative cells, and herein only 50% of cells were killed. Nevertheless, a direct comparison is difficult to make between the two nanopatterns of pillars of similar heights as the overall dimensions, including width, pillar density, and general shape (tapered or blunt) of the pillars, are very different (SI Appendix, Fig. S6), which contribute significantly to the overall effectiveness of the nanopattern against each of the bacterial species.
While the substrata containing the 220- and 360-nm pillars were expected to be highly biocidal based on previous observations (25–27, 31, 32, 34, 40, 59), a distinct disparity in activity was observed (Fig. 2C). This strongly suggests that a separate surface parameter was responsible for the substantial increase in the observed bactericidal behavior when the nanopillar height was increased from 220 to 360 nm. For a selection of substrata possessing nanopillars, where only the geometric height of the pillars is varied, the antibacterial activity should be identical between systems because the bacteria are primarily interacting with the same geometric plane—a flat array of perpendicularly arranged nanopillars with a constant diameter (35 nm) and interpillar spacing of ∼90 nm.
An explanation for the enhanced killing effects when the nanopillar height is increased beyond 220 nm lies in the ability of the nanopillars to deflect in response to externally applied forces which are governed by the material and surface structure properties such as aspect ratio and bending stiffness (Fig. 3). The higher rates of bactericidal activity can be attributed to stress-induced deflection of the nanopillars upon bacterial membrane adsorption, which generates increased stretching of the membrane as the pillars deflect. This is in addition to the mechanical action developed for rigid nanopillars, that is, those found on cicada wing surfaces and pyramidal black silicon surfaces.
Therefore, based on theoretical and experimental observations in this work, at heights of 360 nm the pillars have sufficient elasticity to bend in response to bacterial membrane adsorption. The deflection of the pillars in this study is hypothesized to be responsible for the enhanced bactericidal action toward both Gram stain-negative and Gram stain-positive bacteria. It has been reported that biological samples prepared for electron microscopy by primary fixation using 3% glutaraldehyde and dried by critical point drying can lead to a cell boundary retraction of ∼60 nm in eukaryotic cells (75, 76). However, in our control images, the bacterial cells show no evidence of the wrinkling or flattening associated with drying/shrinkage artifacts (Fig. 2), and P. aeruginosa have maintained their flagellum, evidence of the maintenance of structural integrity. Furthermore, previous studies measuring the adhesion forces during bacterial attachment on nanopillar arrays have confirmed, using multiple independent experiments, that the deflection of nanowires can accurately probe the forces present during the surface interactions of bacterial cells (77).
We observed that the elastic forces acting between the nanostructures and bacteria are in the same order of magnitude irrespective of pillar height. In contrast, the mechanical energies stored within the pillars and corresponding deflections substantially increase by one order of magnitude when increasing the pillar height from 220 to 360 nm. It appears that similar adhesion forces, but higher mechanical energies, are accompanied by enhanced stress for the microorganism. This effect, however, would be limited when the pillar height is increased to a point where it becomes energetically cheap to assemble an effectively continuous surface comprising tightly packed pillar tips, that is, the pillars collapse, providing a surface topography that is closer to the class of flat surfaces for cell adhesion. Indeed, a subtle decrease in bactericidal behavior was observed when the nanopillar height was further increased to 420 nm. In cases of much larger aspect ratios, we expect that, upon contact with bacterial membranes, the surface nanoarchitecture would change insomuch that bacteria would no longer be interacting with the same perpendicularly arranged free-standing pillars across all samples. With the increased aspect ratio, the nanopillars would become more flexible and easily cluster during bacterial interactions, forming stable bunches of nanopillars with a greater surface area, thus changing the surface nanoarchitecture and resulting in lower levels of bactericidal activity. In contrast, when bacteria are adsorbed onto the substrate possessing pillars with an average aspect ratio, the pillars bend and impose additional mechanical stress on the cell, as confirmed by theoretical modeling.
Conclusions
High-aspect-ratio, nanostructured antibacterial surfaces have generated a considerable amount of scientific interest in recent years. This interest stems from the surfaces' capacity to mechanically rupture bacteria upon contact. While there is a consensus that the bactericidal behavior of such surfaces is physical in nature, the precise mechanism(s) of this behavior requires comprehensive in-depth investigation. In this study we have demonstrated that the antibacterial activity of symmetric nanopillar arrays, with increasing heights of 220, 360, and 420 nm, was associated with the relative flexibility of the individual nanopillars. We have shown that the mechanical energy stored in the pillars increases by one order of magnitude when increasing the pillar height from L = 220 nm to 360 and 420 nm. Corresponding elastic forces remain in the same order of magnitude throughout all three pillar lengths associated with two separate bacteria. Nevertheless, a moderate increase in the elastic forces (by a factor of ∼2) corresponds to the enhanced bactericidal effect observed for pillars of L ≥ 360 nm as compared to pillars with L = 220 nm. We have shown that the observed forces on the order of 10 nN per pillar are reasonable when compared to previous bacterial adhesion measurements published in the literature. Therefore, our results support the hypothesis that pillar deformation is induced during the cell adhesion processes. Furthermore, the similarity of force values throughout pillars of remarkably different elasticity suggests that lateral stretching of the cell membrane and interactions at the cell edge seem to be determined by a characteristic biophysical force. For the group of pillars with higher aspect ratio, similar forces are accompanied by a substantially more massive load of mechanical energy and lateral stretching that may expose the organism to additional stress as compared to relatively rigid substrates with smaller aspect ratio. Furthermore, bacteria-induced deflections on the order of 100 nm (Table 1) are large enough to shift the onset of pillar clustering toward smaller pillars.
This mechano-bactericidal action is, therefore, categorized separately from the mechano-bactericidal action delivered by rigid nanopillar arrays, as observed previously on cicada wing surfaces. The enhanced bactericidal activity was observed for the substratum containing 360-nm pillars against both Gram stain-negative (P. aeruginosa, 95% cell death) and Gram stain-positive (S. aureus, 83% cell death) bacteria. Given that all other surface parameters were held constant, the results obtained in this study provide insight into a previously unknown category of mechano-bactericidal mechanism, which is attributed to the elasticity of ordered nanopillars. The nanopatterns presented in this study can be replicated on other materials with controllable bactericidal behavior for biomedical and industrial applications.
Materials and Methods
Fabrication of Silicon Surfaces Containing Nanopillar Arrays.
Surfaces with nanopillar arrays were fabricated using a 300-mm silicon wafer subjected to deep UV immersion lithography and plasma etching using protocols previously described (57, 58). All studies were conducted under ambient atmospheric conditions.
SEM Analysis.
High-resolution electron micrographs of the native substratum surfaces were recorded using a field-emission scanning electron microscope (FE-SEM; ZEISS SUPRA 40 VP) at 3 kV using the method described in previously published studies (32, 40). The tilted (45° and 90°) sample images were obtained using a higher-resolution FE-SEM, FEI NOVA nanoSEM at 10 kV using the same method as described elsewhere (32, 40).
Bacterial Strains, Growth Conditions, and Sample Preparation.
Two strains of pathogenic bacteria, P. aeruginosa ATCC 9721 and S. aureus CIP 65.8T, were obtained from the ATCC and the CIP, respectively. Prior to each experiment, bacterial cultures were refreshed on nutrient agar from stocks (BD). New bacterial suspensions were grown overnight at 37 °C in 5 mL of nutrient broth (BD). Bacterial cells were collected and the density of bacterial suspensions was adjusted to an optical density of 0.1 using UV−visible spectrophotometry (Dynamica HALO RB-10) at a wavelength of 600 nm. For each experiment, Si nanopillar samples were submersed in duplicate in 1 mL of bacterial suspension in a UV-sterilized 24-well plate (In Vitro Technologies) under dark and static conditions at 25 °C for 18 h. Silicon wafer samples were used as control surfaces to monitor cell attachment, morphology, and viability.
Cell Viability Analysis.
CLSM was used to visualize the proportions of live and dead cells using a LIVE/DEAD BacLight Bacterial Viability Kit L7012 that contains a mixture of SYTO 9 and propidium iodide fluorescent dyes (Molecular Probes, Invitrogen). SYTO 9 permeated both intact and damaged membranes of the cells, binding to nucleic acids and fluorescing green when excited by a 488-nm-wavelength laser. Propidium iodide alone entered the cells with significant levels of membrane damage, which were then considered to be nonviable, and binds with higher affinity to nucleic acids than SYTO 9. Bacterial suspensions were stained according to the manufacturer’s protocol and as described in our previously published articles (26, 32, 78) and imaged using a Fluoview FV3000 inverted microscope (Olympus). Images were taken at 15 to 20 different fields of view to generate statistically reliable data. In addition, at least three independent technical replicates were performed to reconfirm the results. Cell attachment and viability were assessed using the CellC module of the MATLAB software. Statistical significance was determined using a t test.
Cell Morphology Analysis.
SEM (FE-SEM; Zeiss Supra 40 VP) was used to assess cell morphology postincubation on nanopillared Si substrata. Prior to imaging, the surfaces were gently rinsed twice with phosphate-buffered saline (pH 7.4) and fixed with 2.5% glutaraldehyde for 30 min. The surfaces were then dehydrated with a graded ethanol series and were then transferred to a critical point dryer (Polaron E3100; Quorum Technologies Ltd.). The samples were immersed in liquid CO2 via a series of 100% ethanol/liquid CO2 exchanges. The liquid CO2 was then heated to supercritical temperature and pressure (31.1 °C and 1,000 psi). The dried samples were sputtered with gold using a NeoCoater MP-19020NCTR prior to imaging.
FIB-SEM Measurements.
Prior to this process, the surfaces with bacteria were prepared according to the protocol above. To reveal the biointerface between nanopillars and bacterial surfaces, the FEI Nova NanoLab DualBeam 200 FIB system was utilized. Bacteria attached to the substratum surfaces were coated with a ∼1-µm-thick Pt protection layer using an e-beam Pt deposition process, followed by another 2-µm layer of ion-beam Pt deposition. A gallium ion beam was used to mill through the surface, and the biointerface was imaged at a tilt angle of 52°.
Data Availability.
The authors confirm that the data supporting the findings of this study are available within the paper and SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge the facilities as well as scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the RMIT University Microscopy & Microanalysis Facility. We thank the Bio21 Advanced Microscopy Facility (The University of Melbourne) for the use of their FEI Nova SEM. N.V. acknowledges the Fonds Voor Wetenschappelijk Onderzoek for a research grant. Funding from the Australian Research Council (ARC) Industrial Transformation Research Hubs Scheme (Project IH130100017) and ARC Industrial Transformation Training Centre (ITTC) scheme (Project IC180100005) to E.P.I. are gratefully acknowledged.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916680117/-/DCSupplemental.
References
- 1.Boucher H. W. et al., Bad bugs, no drugs: No ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1–12 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Spellberg B. et al.; Infectious Diseases Society of America , The epidemic of antibiotic-resistant infections: A call to action for the medical community from the infectious diseases society of America. Clin. Infect. Dis. 46, 155–164 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Levy S. B., Marshall B., Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 10 (12, suppl.), S122–S129 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Control CfD & Prevention , Antibiotic Resistance Threats in the United States, 2013, (Centres for Disease Control and Prevention, US Department of Health and Human Services, 2013). [Google Scholar]
- 5.Neu H. C., The crisis in antibiotic resistance. Science 257, 1064–1073 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Jacob J. T. et al.; Centers for Disease Control and Prevention (CDC) , Vital signs: Carbapenem-resistant enterobacteriaceae. MMWR Morb. Mortal. Wkly. Rep. 62, 165–170 (2013). [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta N., Limbago B. M., Patel J. B., Kallen A. J., Carbapenem-resistant enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 53, 60–67 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Rodvold K. A., McConeghy K. W., Methicillin-resistant staphylococcus aureus therapy: Past, Present, and Future. Clin. Infect. Dis. 58 (suppl. 1), S20–S27 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Hasan J., Crawford R. J., Ivanova E. P., Antibacterial surfaces: The quest for a new generation of biomaterials. Trends Biotechnol. 31, 295–304 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Nguyen S. H., Webb H. K., Crawford R. J., Ivanova E. P., “Natural antibacterial surfaces” in Antibacterial Surfaces, Ivanova E., Crawford R., Eds. (Springer International Publishing, Cham, 2015), pp. 9–26. [Google Scholar]
- 11.Elbourne A., Crawford R. J., Ivanova E. P., Nano-structured antimicrobial surfaces: From nature to synthetic analogues. J. Colloid Interface Sci. 508, 603–616 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Magin C. M., Cooper S. P., Brennan A. B., Non-toxic antifouling strategies. Mater. Today 13, 36–44 (2010). [Google Scholar]
- 13.Serrano C. et al., Nanostructured medical sutures with antibacterial properties. Biomaterials 52, 291–300 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Fadeeva E. et al., Bacterial retention on superhydrophobic titanium surfaces fabricated by femtosecond laser ablation. Langmuir 27, 3012–3019 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Ivanova E. P. et al., Impact of nanoscale roughness of titanium thin film surfaces on bacterial retention. Langmuir 26, 1973–1982 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Truong V. K. et al., Air-directed attachment of coccoid bacteria to the surface of superhydrophobic lotus-like titanium. Biofouling 28, 539–550 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Truong V. K. et al., The influence of nano-scale surface roughness on bacterial adhesion to ultrafine-grained titanium. Biomaterials 31, 3674–3683 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Truong V. K. et al., Effect of ultrafine-grained titanium surfaces on adhesion of bacteria. Appl. Microbiol. Biotechnol. 83, 925–937 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Variola F., Zalzal S. F., Leduc A., Barbeau J., Nanci A., Oxidative nanopatterning of titanium generates mesoporous surfaces with antimicrobial properties. Int. J. Nanomedicine 9, 2319–2325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng C. Y. et al., Enhanced in vitro biocompatibility of ultrafine-grained titanium with hierarchical porous surface. Appl. Surf. Sci. 257, 5634–5640 (2011). [Google Scholar]
- 21.Chung K. K. et al., Impact of engineered surface microtopography on biofilm formation of Staphylococcus aureus. Biointerphases 2, 89–94 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto A. et al., Antibacterial effects of protruding and recessed shark skin micropatterned surfaces of polyacrylate plate with a shallow groove. FEMS Microbiol. Lett. 361, 10–16 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Bhadra C. M. et al., Antibacterial titanium nano-patterned arrays inspired by dragonfly wings. Sci. Rep. 5, 16817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diu T. et al., Cicada-inspired cell-instructive nanopatterned arrays. Sci. Rep. 4, 7122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasan J. et al., Selective bactericidal activity of nanopatterned superhydrophobic cicada Psaltoda claripennis wing surfaces. Appl. Microbiol. Biotechnol. 97, 9257–9262 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Ivanova E. P. et al., Natural bactericidal surfaces: Mechanical rupture of Pseudomonas aeruginosa cells by cicada wings. Small 8, 2489–2494 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Kelleher S. M. et al., Cicada wing surface topography: An investigation into the bactericidal properties of nanostructural features. ACS Appl. Mater. Interfaces 8, 14966–14974 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Li X., Bactericidal mechanism of nanopatterned surfaces. Phys. Chem. Chem. Phys. 18, 1311–1316 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Pogodin S. et al., Biophysical model of bacterial cell interactions with nanopatterned cicada wing surfaces. Biophys. J. 104, 835–840 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue F., Liu J., Guo L., Zhang L., Li Q., Theoretical study on the bactericidal nature of nanopatterned surfaces. J. Theor. Biol. 385, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Linklater D. P., Nguyen H. K. D., Bhadra C. M., Juodkazis S., Ivanova E. P., Influence of nanoscale topology on bactericidal efficiency of black silicon surfaces. Nanotechnology 28, 245301 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Ivanova E. P. et al., Bactericidal activity of black silicon. Nat. Commun. 4, 2838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May P. W. et al., Diamond-coated “black silicon” as a promising material for high-surface-area electrochemical electrodes and antibacterial surfaces. J. Mater. Chem. B Mater. Biol. Med. 4, 5737–5746 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Nguyen D. H. K. et al., Adsorption of human plasma albumin and fibronectin onto nanostructured black silicon surfaces. Langmuir 32, 10744–10751 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hasan J. et al., Spatial variations and temporal metastability of the self-cleaning and superhydrophobic properties of damselfly wings. Langmuir 28, 17404–17409 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Truong V. K. et al., The susceptibility of Staphylococcus aureus CIP 65.8 and Pseudomonas aeruginosa ATCC 9721 cells to the bactericidal action of nanostructured Calopteryx haemorrhoidalis damselfly wing surfaces. Appl. Microbiol. Biotechnol. 101, 4683–4690 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Tsimbouri P. M. et al., Osteogenic and bactericidal surfaces from hydrothermal titania nanowires on titanium substrates. Sci. Rep. 6, 36857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandara C. D. et al., Bactericidal effects of natural nanotopography of dragonfly wing on Escherichia coli. ACS Appl. Mater. Interfaces 9, 6746–6760 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Ivanova E. P. et al., Molecular organization of the nanoscale surface structures of the dragonfly Hemianax papuensis wing epicuticle. PLoS One 8, e67893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mainwaring D. E. et al., The nature of inherent bactericidal activity: Insights from the nanotopology of three species of dragonfly. Nanoscale 8, 6527–6534 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Green D. W. et al., High quality bioreplication of intricate nanostructures from a fragile gecko skin surface with bactericidal properties. Sci. Rep. 7, 41023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X. et al., The nanotipped hairs of gecko skin and biotemplated replicas impair and/or kill pathogenic bacteria with high efficiency. Nanoscale 8, 18860–18869 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Watson G. S. et al., A gecko skin micro/nano structure - A low adhesion, superhydrophobic, anti-wetting, self-cleaning, biocompatible, antibacterial surface. Acta Biomater. 21, 109–122 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Cloutier M., Mantovani D., Rosei F., Antibacterial coatings: Challenges, perspectives, and opportunities. Trends Biotechnol. 33, 637–652 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Chernousova S., Epple M., Silver as antibacterial agent: Ion, nanoparticle, and metal. Angew. Chem. Int. Ed. Engl. 52, 1636–1653 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Asri L. A. T. W. et al., A shape-adaptive, antibacterial-coating of immobilized quaternary-ammonium compounds tethered on hyperbranched polyurea and its mechanism of action. Adv. Funct. Mater. 24, 346–355 (2013). [Google Scholar]
- 47.Lepoittevin B. et al., Antibacterial surfaces obtained through dopamine and fluorination functionalizations. Prog. Org. Coat. 82, 17–25 (2015). [Google Scholar]
- 48.Bound D. J., Murthy P. S., Srinivas P., Synthesis and antibacterial properties of 2,3-dideoxyglucosides of terpene alcohols and phenols. Food Chem. 185, 192–199 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Glinel K., Thebault P., Humblot V., Pradier C. M., Jouenne T., Antibacterial surfaces developed from bio-inspired approaches. Acta Biomater. 8, 1670–1684 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Kato T., Shirai T., Temporal attenuation of iodine content and its effect on the antibacterial activity of iodine-supported titanium implants. J. Microb. Biochem. Technol. 8, 285–289 (2016). [Google Scholar]
- 51.Tiller J. C., Liao C. J., Lewis K., Klibanov A. M., Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. U.S.A. 98, 5981–5985 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bush K. et al., Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivanova E. P. et al., Differential attraction and repulsion of Staphylococcus aureus and Pseudomonas aeruginosa on molecularly smooth titanium films. Sci. Rep. 1, 165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowlin K., Boseman A., Covell A., LaJeunesse D., Adhesion-dependent rupturing of Saccharomyces cerevisiae on biological antimicrobial nanostructured surfaces. J. R. Soc. Interface 12, 20140999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X., Chen T., Enhancement and suppression effects of a nanopatterned surface on bacterial adhesion. Phys. Rev. E 93, 52419 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Tuson H. H., Weibel D. B., Bacteria-surface interactions. Soft Matter 9, 4368–4380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vos I., Hellin D., Vertommen J., Demand M., Boullart W., Silicon nano-pillar test structures for quantitative evaluation of wafer drying induced pattern collapse. ECS Trans. 41, 189–196 (2011). [Google Scholar]
- 58.Xu X. et al., Capturing wetting states in nanopatterned silicon. ACS Nano 8, 885–893 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Wang X. et al., A bactericidal microfluidic device constructed using nano-textured black silicon. RSC Adv. 6, 26300–26306 (2016). [Google Scholar]
- 60.Linklater D. P., Juodkazis S., Rubanov S., Ivanova E. P., Comment on “bactericidal effects of natural nanotopography of dragonfly wing on Escherichia coli”. ACS Appl. Mater. Interfaces 9, 29387–29393 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Linklater D. P. et al., High aspect ratio nanostructures kill bacteria via storage and release of mechanical energy. ACS Nano 12, 6657–6667 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Ghibaudo M. et al., Traction forces and rigidity sensing regulate cell functions. Soft Matter 4, 1836 (2008). [Google Scholar]
- 63.Schoen I., Hu W., Klotzsch E., Vogel V., Probing cellular traction forces by micropillar arrays: Contribution of substrate warping to pillar deflection. Nano Lett. 10, 1823–1830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hess P., Laser diagnostics of mechanical and elastic properties of silicon and carbon films. Appl. Surf. Sci. 106, 429–437 (1996). [Google Scholar]
- 65.Li X., Ono T., Wang Y., Esashi M., Ultrathin single-crystalline-silicon cantilever resonators: Fabrication technology and significant specimen size effect on Young’s modulus. Appl. Phys. Lett. 83, 3081–3083 (2003). [Google Scholar]
- 66.Wang G., Li X., Predicting Young’s modulus of nanowires from first-principles calculations on their surface and bulk materials. J. Appl. Phys. 104, 113517 (2008). [Google Scholar]
- 67.Park H. S., Surface stress effects on the resonant properties of silicon nanowires. J. Appl. Phys. 103, 123504 (2008). [Google Scholar]
- 68.Wortman J. J., Evans R. A., Young’s modulus, shear modulus, and Poisson’s ratio in silicon and germanium. J. Appl. Phys. 36, 153–156 (1965). [Google Scholar]
- 69.Calahorra Y., Shtempluck O., Kotchetkov V., Yaish Y. E., Young’s modulus, residual stress, and crystal orientation of doubly clamped silicon nanowire beams. Nano Lett. 15, 2945–2950 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Kizuka T., Takatani Y., Asaka K., Yoshizaki R., Measurements of the atomistic mechanics of single crystalline silicon wires of nanometer width. Phys. Rev. B 72, 35333 (2005). [Google Scholar]
- 71.Alam F., Balani K., Adhesion force of staphylococcus aureus on various biomaterial surfaces. J. Mech. Behav. Biomed. Mater. 65, 872–880 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Harimawan A., Rajasekar A., Ting Y.-P., Bacteria attachment to surfaces–AFM force spectroscopy and physicochemical analyses. J. Colloid Interface Sci. 364, 213–218 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Atabek A., Camesano T. A., Atomic force microscopy study of the effect of lipopolysaccharides and extracellular polymers on adhesion of Pseudomonas aeruginosa. J. Bacteriol. 189, 8503–8509 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dickson M. N., Liang E. I., Rodriguez L. A., Vollereaux N., Yee A. F., Nanopatterned polymer surfaces with bactericidal properties. Biointerphases 10, 21010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Demignot S., Domurado D., Elimination of artifacts due to glutaraldehyde fixation in the histochemical detection of glucose oxidase with tetrazolium salts. Histochem. J. 20, 11–18 (1988). [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y. et al., Quantitating morphological changes in biological samples during scanning electron microscopy sample preparation with correlative super-resolution microscopy. PLoS One 12, e0176839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sahoo P. K. et al., Nanowire arrays as cell force sensors to investigate adhesin-enhanced holdfast of single cell bacteria and biofilm stability. Nano Lett. 16, 4656–4664 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Boulos L., Prévost M., Barbeau B., Coallier J., Desjardins R., LIVE/DEAD BacLight : Application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37, 77–86 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the paper and SI Appendix.