Significance
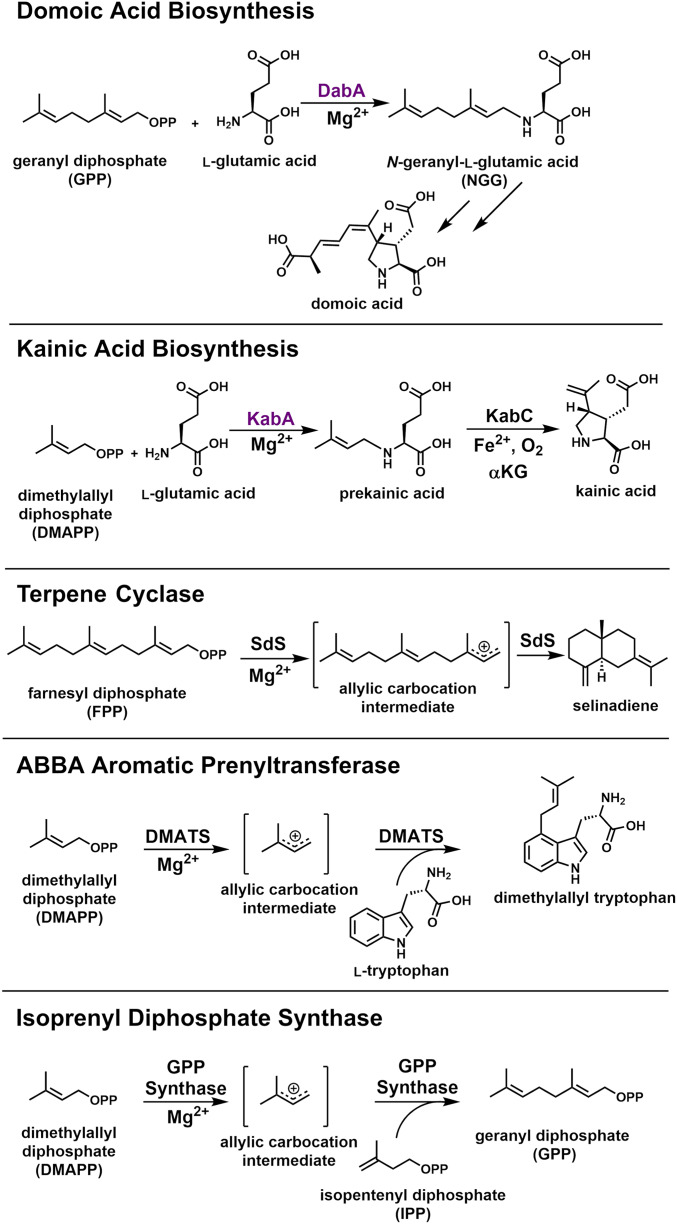
Domoic acid is a neurotoxin produced by marine algae that readily bioaccumulates in shellfish with significant impacts for both human and animal lives. The first committed step of domoic acid biosynthesis is the N-prenylation of l-glutamic acid catalyzed by DabA. By solving the crystal structure of DabA, we demonstrate that DabA repurposed the phylogenetically ubiquitous terpene cyclase fold to catalyze a rarer reaction, N-prenylation of a primary amine. These structural insights enabled the rational engineering of two N-prenyltransferases to accept alternative prenyl donors. These results expand our understanding of the reaction scope catalyzed by terpene cyclase folds and will inform future domoic acid environmental monitoring efforts.
Keywords: prenyltransferase, domoic acid, natural products, X-ray crystallography, terpene cyclase
Abstract
Prenylation is a common biological reaction in all domains of life wherein prenyl diphosphate donors transfer prenyl groups onto small molecules as well as large proteins. The enzymes that catalyze these reactions are structurally distinct from ubiquitous terpene cyclases that, instead, assemble terpenes via intramolecular rearrangements of a single substrate. Herein, we report the structure and molecular details of a new family of prenyltransferases from marine algae that repurposes the terpene cyclase structural fold for the N-prenylation of glutamic acid during the biosynthesis of the potent neurochemicals domoic acid and kainic acid. We solved the X-ray crystal structure of the prenyltransferase found in domoic acid biosynthesis, DabA, and show distinct active site binding modifications that remodel the canonical magnesium (Mg2+)-binding motif found in terpene cyclases. We then applied our structural knowledge of DabA and a homologous enzyme from the kainic acid biosynthetic pathway, KabA, to reengineer their isoprene donor specificities (geranyl diphosphate [GPP] versus dimethylallyl diphosphate [DMAPP]) with a single amino acid change. While diatom DabA and seaweed KabA enzymes share a common evolutionary lineage, they are distinct from all other terpene cyclases, suggesting a very distant ancestor to the larger terpene synthase family.
Marine and freshwater algae produce neurotoxins that encompass a range of structural classes and biological targets (1, 2). During harmful algal blooms, these neurotoxins accumulate in the environment leading to a number of damaging effects including fish and bird mortality, beach and fishery closures, and human toxicity (2, 3). For example, the ionotropic glutamate receptor agonist domoic acid is produced by marine diatoms of the genus Pseudo-nitzschia (Scheme 1). Blooms of these diatoms regularly occur globally, and, as a consequence, human consumption of seafood contaminated with domoic acid leads to amnesic shellfish poisoning, which is characterized by memory loss, seizures, and, in severe cases, death (4, 5).
Scheme 1.
Examples of glutamate N-prenyltransferases, terpene cyclases, and prenyltransferases. Selinadiene synthase is abbreviated as SdS and dimethylallyl tryptophan synthase is abbreviated as DMATS.
Domoic acid is a member of the kainoid family of natural products along with the red macroalgal compound kainic acid (Scheme 1). While kainic acid shares a conserved cyclic core with domoic acid, the prenyl moiety in kainic acid is shorter, resulting in different bioactive properties. Even though it also targets ionotropic glutamate receptors, kainic acid is less potent (6) and has instead been used as both an anthelminthic drug (7, 8) and a reagent to study neurological disorders (9).
Recent work on the biosyntheses of domoic acid and kainic acid established the origin of the prenyl groups (10, 11) (Scheme 1). In the case of domoic acid, DabA adds a ten-carbon geranyl group to l-glutamate to generate N-geranyl-l-glutamate (NGG), while KabA uses the five-carbon DMAPP prenyl donor during kainic acid biosynthesis to produce the intermediate prekainic acid (Scheme 1).
Even though terpene biochemistry is shared across all domains of life, the DabA and KabA N-prenylating enzymes are unusual for several reasons. First, the reaction catalyzed by DabA and KabA is uncommon in nature. Numerous examples exist of prenylation of hydroxyl groups or nucleophilic carbons, but N-prenylation, especially of primary amines, is comparatively rare. The few known reactions include prenylation of primary amines found in adenines (12, 13) or in the cyanobactin class of natural products (14, 15).
In addition to unusual biochemistry, DabA and KabA possess distinct amino acid sequences. Querying DabA by a BLASTp search revealed that the closest homologs outside of those found in kainic biosynthesis have E values of 10−4. A more detailed analysis based on hidden Markov models (HMMs), suggested that both DabA and KabA are distantly related to terpene synthases and more specifically terpene cyclases (16). This initial classification was further supported by structural predictions using both Phyre2 (17) and I-TASSER (18). The terpene cyclase assignment by both HMM and structural prediction was unexpected as terpene cyclases are not known to catalyze intermolecular prenylation reactions. Instead, the larger terpene synthase family generally catalyzes intramolecular cyclization reactions or loss of the diphosphate moiety of the substrate to yield a linear product (Scheme 1). While this reaction may appear different from the one catalyzed by KabA and DabA, the key allylic carbocation intermediate is likely conserved. Instead, the feature that differentiates KabA and DabA from typical terpene cyclases is that the catalyzed reaction is no longer intramolecular and now two substrates need to be positioned in the active site.
Even though terpene cyclases are not known to catalyze intermolecular prenylation reactions, Nature has evolved separate structural classes of enzymes that catalyze such reactions. One, called ABBA prenyltransferases, is regularly found in secondary metabolism (Scheme 1) (19). These enzymes utilize prenyl diphosphate substrates and catalyze the transfer of the prenyl group onto a variety of electron-rich moieties. ABBA prenyltransferases are found in numerous natural product biosynthetic pathways. They are defined by an αββα secondary structural motif (20, 21). This arrangement contrasts with the exclusively α-helical fold of terpene cyclases and the one predicted to comprise DabA/KabA. Despite this, the chemical reaction catalyzed by ABBA prenyltransferases is similar in that it also proceeds through an allylic carbocation that ultimately serves as an electrophile.
A second class of prenyltransferases, called isoprenyl diphosphate synthases or sometimes simply referred to as prenyltransferases, are primary metabolic enzymes in all three domains of life (Scheme 1). They are responsible for the biosynthesis of the different sized prenyl diphosphate donors, such as GPP (10 carbons), farnesyl diphosphate (FPP) (15 carbons), and geranylgeranyl diphosphate (20 carbons) (16, 22). They also catalyze an intermolecular prenyl transfer reaction wherein two isoprenoid diphosphates typically serve as substrates, such as DMAPP and isopentenyl diphosphate (IPP). Enzymes in this class are structurally related to terpene cyclases despite very low sequence similarities and distinct active site residues.
To better understand the basis for DabA’s unusual activity as a putative member of the terpene cyclase protein family, we solved high-resolution crystal structures of DabA in complex with both substrate analogs and products. Validation of our structural observations by mutagenesis revealed how the active site differs from canonical terpene synthases to enable intermolecular prenylation reactions. We further employed this structural foundation to uncover and functionally demonstrate a key residue that dictates prenyl group chain length specificity, and thus, catalytically differentiates DabA and KabA.
Results and Discussion
Overall Structure.
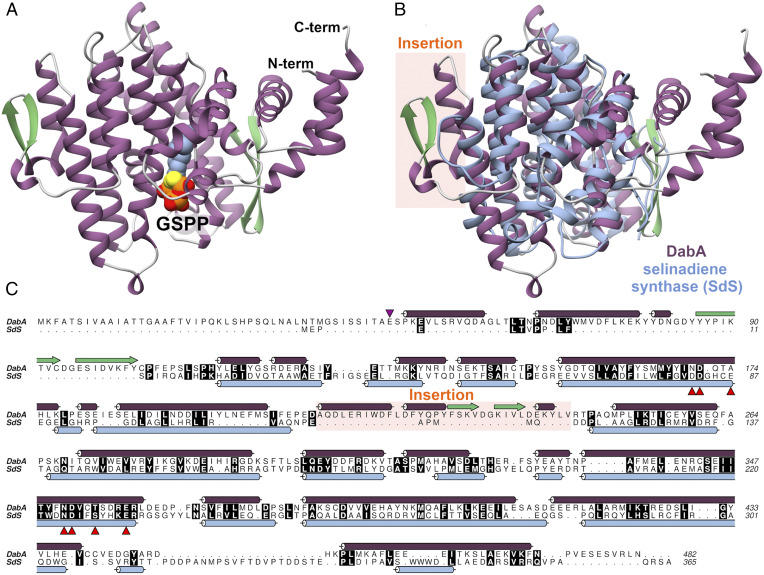
In order to determine the structure of DabA and the basis for its activity, we obtained crystals of DabA. While crystals could be formed using a DabA construct that lacked the predicted N-terminal signal sequence for plastid import (10), efforts to optimize these crystals to diffraction quality were unsuccessful. We next generated a DabA variant that lacked an additional N-terminal region suggested to be unstructured based on modeling and secondary structure predictions using Phyre2 (17) (Fig. 1C). After confirming this variant retained N-prenylation activity (SI Appendix, Fig. S1), we successfully crystallized, solved, and refined the structure of this latter N-terminally truncated DabA construct in complex with geranyl S-thiolodiphosphate (GSPP) and Mg2+ to 2.1 Å resolution.
Fig. 1.
(A) Secondary structure of DabA with GSPP also shown. (B) Structural alignment of DabA (purple) and SdS (PDB code 4OKZ) (blue). (C) Sequence alignment of DabA and SdS. Secondary structure is overlaid next to the sequence and colored according to B. The terpene cyclase motifs are indicated by red arrows. The purple arrow indicates the start of the DabA construct used for crystallography.
DabA is a monomer in the asymmetric unit, and gel filtration analysis suggests that this is the physiological assembly (SI Appendix, Fig. S2). Structurally, DabA is composed primarily of an α-helical core that is commonly observed in terpene cyclase α-domains, consistent with previous structural prediction (Fig. 1A) (10). Analysis by the Dali server (23) indicates structural homology to bacterial terpene cyclases, and DabA is most similar to selinadiene synthase from the bacterium Streptomyces pristinaespiralis (2.5 Å rmsd over 298 aligned Cα's) (Fig. 1B and Scheme 1) (24). This high level of structural conservation is observed despite low levels of amino acid sequence identity (<15% identity) (Fig. 1C). While the terpene cyclase core is conserved, DabA possesses two additional features not observed in typical terpene cyclases. First, DabA contains N- and C-terminal extensions that together form a small α-helical bundle (Fig. 1 B and C). This region connects to the α-helical core via a β-hairpin. A second insertion is found within the terpene cyclase core and forms a short helical β-hairpin containing extension.
Active Site.
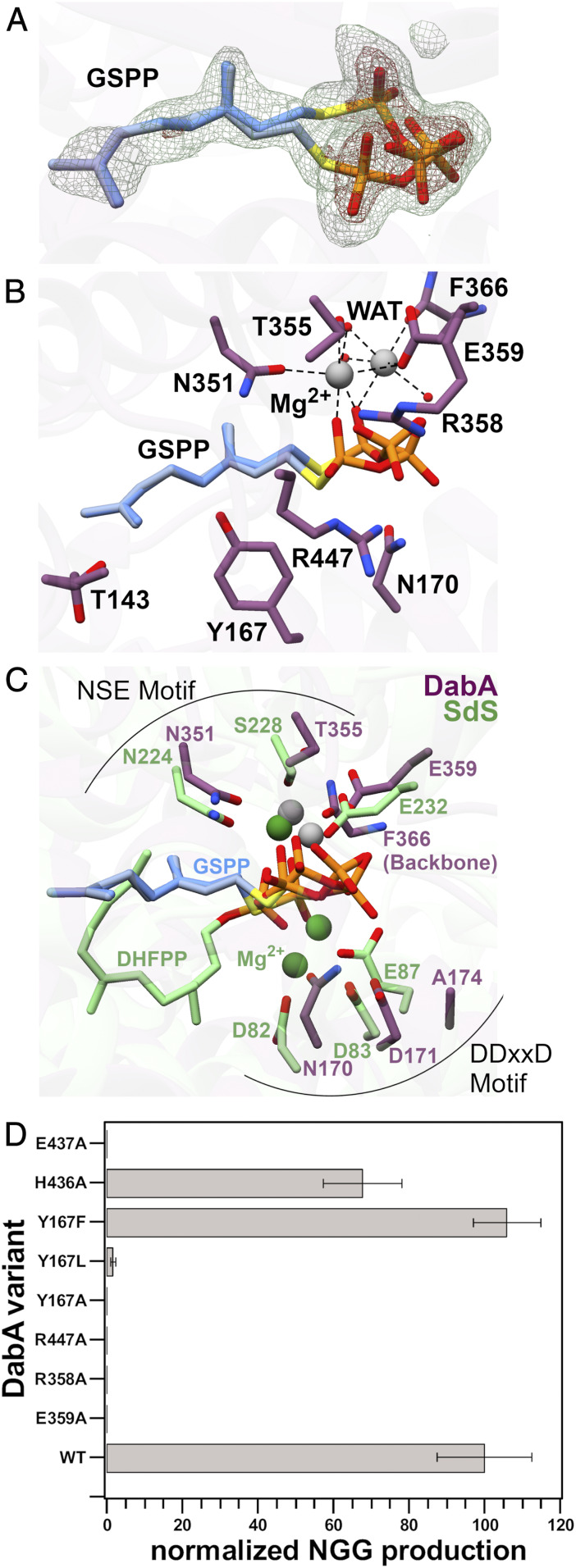
While the overall structure and location of the active site are conserved between DabA and terpene cyclases, many of the molecular details are very different. Examination of the DabA active site reveals the less-hydrolyzable GPP mimic GSPP bound in at least two distinct conformations (Fig. 2A). In both cases, the 10-carbon geranyl group of GSPP extends into a narrow hydrophobic pocket while the positions of the α-phosphate differ between the two conformations. Two Mg2+ ions are coordinated in the active site. DabA crystals soaked with the more electron dense MnCl2 confirm the location of one of the bound divalent cations crystallographically (SI Appendix, Fig. S3). The first Mg2+ coordinates to the α-phosphate in each of the two GSPP conformations along with the side chains of Asn351, Glu359, Thr355, and a water molecule. The second Mg2+ sits adjacent to the first and interacts with one of the α-phosphate conformations, three water molecules, the side chain of Glu359, and the backbone carbonyl of Phe366. This arrangement contrasts with canonical class I terpene cyclases which contain two key Mg2+-binding motifs that typically bind three Mg2+ ions. The first motif, NSE, is typically observed as a NDxxSxxxE sequence. This NSE motif is found in DabA encompassing Asn351, Thr355, and Glu359 (Figs. 1C and 2B). Superimposition of the DabA active site with selinadiene synthase reveals clear alignment of both residues and Mg2+ (Fig. 2C). Notably, DabA also appears to coordinate an additional Mg2+ ion near the NSE motif not observed in the structure of other terpene cyclases.
Fig. 2.
(A) Active site of DabA in complex with GSPP, which is shown in two orientations. Fo-Fc maps were generated by removing GSPP and completing refinement without a ligand. Meshes are contoured to 3σ (green) and 8σ (red). (B) Active site of DabA with important residues highlighted. (C) Alignment of DabA (purple) and SdS (green) active sites. DHFPP is 2,3-dihydroFPP. (D) Activity assays showing the effect of different mutations on the production of NGG by DabA.
The second Mg2+-binding motif found in terpene cyclases is defined by an aspartate rich DDxxD/E motif. Sequence alignments of DabA with selinadiene synthase show that this motif is different, NDxxA, and its absence may have contributed to the difficulty in initially identifying DabA as a member of the terpene cyclase family (Fig. 1C) (10). Examination of the crystal structure shows that the canonical DDxxD motif has not been replaced by other Mg2+-binding residues; no Mg2+ is present in this region of the active site (Fig. 2C). The active site arrangement of DabA is also distinct from the recently described class IB terpene synthases that are responsible for the biosynthesis of large terpene products (C25/C30/C35) (25). Class IB terpene synthases lack the canonical NSE and DDxxD motifs. Instead, they have new aspartate rich motifs that occupy the same 3D location within the active site, even though they do not align in primary sequence comparisons (25). This contrasts with DabA which retains the NSE motif but completely lacks the DDxxD motif or any substitute.
Other features prevalent in terpene cyclase substrate binding, such as cationic Arg or Lys residues interacting with the diphosphate moiety, are also well conserved. Specifically, Arg358 and Arg447 are both less than 3.2 Å away from the α-phosphate and likely engage in important electrostatic hydrogen bonding interactions (Fig. 2B).
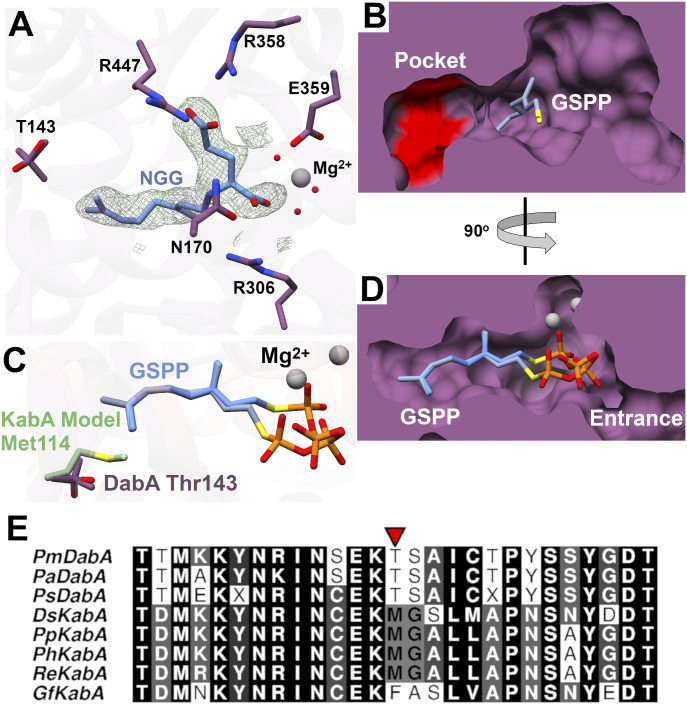
Glutamic acid was present in the crystallization buffer, but it was not observed in the refined structure. In an effort to determine the location of the glutamate-binding pocket, we crystallized DabA in the presence of Mg2+ and the NGG reaction product and solved the structure to a resolution of 2.1 Å (Fig. 3A). Examination of the active site revealed density for NGG with the geranyl group again extended into the hydrophobic tunnel (Fig. 3). Surprisingly, the glutamic acid derived portion of NGG was bound in the same location as the diphosphate end of GSPP. This would suggest a mechanism wherein DabA must first lose diphosphate from the active site before glutamic acid binds. While possible, we do not favor this mechanism because it requires prolonged stabilization of the carbocation intermediate and protection from hydration in a solvent exposed pocket. Instead, we propose that a small side pocket in the active site may serve as the binding site for glutamic acid (Fig. 3B). In both the NGG and the GSPP cocrystal structures, the pocket contains five ordered water molecules and is located near the carbocation intermediate that is predicted to form during the course of the reaction. We evaluated this putative glutamate-binding pocket by mutating two residues that occupy this area, Glu437 and His436. Activity assays were completed with both DabA variants (Fig. 2D). The Glu437Ala DabA variant was completely inactive, while the His436Ala variant had reduced activity. We further evaluated the His436Ala mutation by obtaining Michaelis–Menten kinetic parameters. Compared to wild-type (WT) DabA, the Km app for glutamic acid increased threefold, suggesting that this residue may be involved in glutamate binding (Table 1 and SI Appendix, Fig. S4).
Fig. 3.
(A) Active site of DabA in complex with NGG. Fo-Fc maps were generated by removing NGG and completing refinement without a ligand. Mesh was contoured to 3σ (green). (B) Surface model of DabA indicates a side pocket is present that may accommodate l-Glu binding. His436 and Glu437 are shown in red. (C) Overlay of the DabA active site and a model of KabA. (D) Surface modeling of the active site shows a narrow tunnel that accommodates GSPP. (E) Sequence alignment of all DabA/KabA homologs. The residue of interest is indicated by a red arrow.
Table 1.
Michaelis–Menten kinetic constants for DabA, KabA, and amino acid variants
| GPP | DMAPP | l-Glu | |||||||
| Enzyme | kcat app (min−1) | Km (μM) | kcat app/Km (M−1min−1) × 106 | kcat app (min−1) | Km (μM) | kcat app/Km (M−1min−1) × 106 | kcat app (min−1) | Km (mM) | kcat app/Km (M−1min−1) × 103 |
| DabA | 4.3 ± 0.1 | 0.33 ± 0.03 | 12.9 | Non MM | Non MM | 4.8 ± 0.3 | 21 ± 4 | 0.23 | |
| DabA T143M | 0.65 ± 0.04 | 13 ± 2 | 0.052 | 1.2 ± 0.1 | 130 ± 20 | 0.0092 | |||
| DabA H436A | 6.9 ± 0.6 | 68 ± 16 | 0.10 | ||||||
| KabA | 0.29 ± 0.01 | 89 ± 10 | 0.0033 | 1.3 ± 0.1 | 1.1 ± 0.2 | 1.2 | |||
| KabA M114T | 0.51 ± 0.02 | 0.51 ± 0.06 | 1 | 0.28 ± 0.01 | 2.2 ± 0.3 | 0.13 | |||
Mutagenesis Supports Active Site Residues.
Based on the cocrystal structures, we designed a series of mutations and tested the resulting DabA variants for their ability to catalyze production of NGG (Fig. 2D). DabA contains several charged residues in the active site to coordinate both the positively charged Mg2+ ions and the negatively charged phosphates. Consistent with this hypothesis, mutation of the Mg2+ coordinating Glu359 residue to Ala completely abolishes activity. Similarly, mutation of either Arg358 or Arg447 to Ala results in an inactive DabA variant. We next explored the significance of Tyr167 by creating a series of mutations. This residue is located directly below GSPP in the cocrystal structure and could be responsible for different roles in catalysis including stabilizing of the carbocation, maintaining the shape of the hydrophobic tunnel, or deprotonating the α-amine of the glutamic acid cosubstrate. Mutation of Tyr167 to Phe resulted in no change in activity, indicating that Tyr167 does not serve as a catalytic base. Instead, the DabA Tyr167Leu and Tyr167Ala variants exhibited dramatically reduced or no activity, respectively. This indicates that Tyr167 is important in shaping the tunnel as the much smaller Tyr167Ala residue appeared completely inactive under the reaction conditions, while the slightly smaller Tyr167Leu retained some activity. Furthermore, the observation that only phenylalanine can successfully substitute for tyrosine is consistent with a carbocation stabilizing activity for Tyr167. Terpene cyclases often employ aromatic residues in the active site, and both crystal structures and modeling studies have suggested that they are responsible for stabilizing the carbocation of the mechanistic intermediates through the use of cation-π interactions (26–28).
DabA Kinetic Analysis.
Based on the unusual activity of DabA as a structural member of the terpene cyclase family, we sought to determine the kinetic parameters of DabA and compare them to both canonical terpene cyclases and prenyltransferases (SI Appendix, Fig. S4). WT DabA used the native GPP substrate with a Km app of 0.33 μM and a kcat app of 4.3 min−1. These constants are similar to those found in other characterized prenylating or cyclizing enzymes. Specifically, the low Km app for the prenyl donor is similar to the submicromolar values observed in structurally similar terpene cyclases, such as geosmin synthase (29), pentalenene synthase (30), and aristolochene synthase (31). Moreover, DabA has turnover numbers similar to slower members of the terpene cyclase family (29, 31) as well as ABBA aromatic prenyltransferases, such as FtmPT2 (32).
In addition to GPP, kinetics parameters were also determined for l-glutamic acid, and a kcat app of 4.8 min−1 was observed, consistent with the value obtained for GPP. The Km app for l-glutamic acid was 21 mM, much higher than the Km app of GPP. Enzymes typically possess Km app values within 10-fold of the physiological concentration of substrates (33, 34). DabA is expected to be localized to the diatom plastid (10), and glutamic acid is among the most concentrated metabolites in that organelle with values of 14 (35) and 74 mM (36) reported from different plants. Therefore, even though the Km app value for glutamic acid is high, it is well aligned with the expected in vivo concentrations.
Basis for Prenyl Group Selectivity.
The major structural difference between the two marine kainoid natural products, domoic acid and kainic acid (Scheme 1), is the biosynthetic origin of the prenyl donor, either GPP or DMAPP, respectively (10, 11). The cocrystal structure of DabA with GSPP indicated that the carbon chain is positioned within a hydrophobic tunnel (Fig. 3D). We hypothesized that alterations to the length of the tunnel would be major factors modulating the selectivity of DabA for GPP and KabA for DMAPP. Therefore, we created a model of the Digenea simplex KabA structure using the I-TASSER server (18) and DabA as a template. Overlay of the DabA structure and KabA model shows that the active site is largely conserved (SI Appendix, Fig. S5). Examination of the hydrophobic tunnel revealed a threonine to methionine substitution in KabA (Fig. 3C). Moreover, sequence alignments of the known DabA and KabA enzymes suggest that the identity of this residue can predict the prenylation activity (Fig. 3E). The DabA homologs found in diatoms all contain a threonine, while the red algal kainic acid biosynthetic KabA proteins have a larger residue, either methionine or phenylalanine. It should be noted that other amino acid substitutions are found near Thr143, such as Ser144 to Gly115 and Tyr150 to Asn121. However, based on the structural modeling, those residues are not expected to directly interact with the GPP or DMAPP substrates (SI Appendix, Fig. S5).
We evaluated the significance of DabA Thr143 by completing the Michaelis–Menten kinetics of DabA, KabA, and variants wherein this tunnel residue was switched (SI Appendix, Fig. S4). Exchange of the native threonine or methionine tunnel residue in both DabA and KabA dramatically decreased catalytic efficiency for the native substrate and, instead, promoted the use of the nonnative substrate (Table 1). Specifically, the DabA Thr143Met mutation decreases catalytic efficiency for GPP by ∼250-fold. WT DabA did not display canonical Michaelis–Menten kinetics with DMAPP but had a maximum turnover of 0.6 min−1 at a DMAPP concentration of 500 μM. Conversely, the DabA Thr143Met variant does display typical steady state kinetics with a kcat app of 1.2 min−1 and a Km app of 130 μM.
A parallel analysis was completed for KabA to evaluate the effect of Met114 on substrate specificity. Similar to DabA, the KabA Met114Thr mutation decreases catalytic efficiency for DMAPP by 10-fold and increases catalytic efficiency for GPP by ∼300-fold. In both DabA and KabA, activities with the native substrates decrease, while activities with the nonnative substrates improve substantially.
This “molecular ruler” basis for substrate selectivity has been observed in other prenylating enzymes. For example, FPP synthase is an iterative enzyme that first produces GPP from DMAPP and IPP. It then uses GPP and IPP to generate FPP. Constriction of the hydrophobic pocket with bulky residues biases formation of GPP over FPP (37, 38), while expansion of the pocket promotes prenylation of FPP to generate products of 20 carbons and longer (39). In another case, the tyrosine prenyltransferase PagF natively utilizes DMAPP, but a single point mutation can expand the hydrophobic-binding site to generate a PagF variant that, instead, favors GPP as a substrate (40).
Alternative Substrate Selectivity.
The kinetic and mutagenic analyses revealed that both DabA and KabA are selective for their respective prenyl donors and exhibit greatly reduced catalytic efficiencies when nonnative prenyl donors are used. To further explore substrate selectivities, we tested the ability of both DabA and KabA to utilize alternative substrates. We began by synthesizing a suite of alkynyl and alkenyl diphosphate derivatives: propargyl, 2-butynyl, allyl, crotyl, and farnesyl. Each of these was evaluated as a potential substrate for DabA and KabA and monitored by liquid chromatography coupled with mass spectrometry (SI Appendix, Fig. S6). Both DabA and KabA exhibited high levels of selectivity with both enzymes unable to utilize the alkyne-containing carbon chain donors and the allyl diphosphate substate. From a mechanistic standpoint, all of these substrates could form resonance-stabilized carbocations. However, these alternative substrates lack a tertiary carbon in the resonance system, which is the most stabilizing. Crotyl diphosphate, however, could employ two secondary carbons to stabilize the allyl carbocation. KabA alone used this substrate to form the crotyl-l-glutamate product. Finally, we tested the 15-carbon FPP substate and found that only DabA catalyzed formation of the farnesyl-l-glutamate product.
DabA was also tested for its ability to utilize alternative divalent cations as cofactors (SI Appendix, Fig. S7). DabA activity was highest when Mg2+ was used as the cofactor, and no product was observed when a divalent cation was omitted from the reaction conditions. Mn2+ could substitute for Mg2+ but led to ∼28% lower activity. Other metals were poor replacements for Mg2+ with Ni2+ and Zn2+ reducing DabA activity by 69% and 96%, respectively, compared to Mg2+. Finally, Ca2+ was unable to serve as a cofactor.
Gene Phylogeny.
While the structure of DabA shows high similarity to bacterial terpene cyclases, BLASTp analysis of sequences in the National Center for Biotechnology Information (NCBI) protein database reveals only distant homologs outside of kainic acid biosynthetic genes with the closest homolog having a BLASTp E value greater than 10−4. We next probed the evolutionary history of these glutamate N-prenyltransferases by constructing a maximum likelihood phylogenetic tree. Due to the low sequence similarity of DabA/KabA to any publicly available protein sequence, we generated a global view of the entire terpene cyclase family (InterPro families IPR034741 and IPR034686). We seeded the analysis with characterized red algae terpene cyclases (41, 42) along with the closest DabA sequence homologs identified from PSI- and DELTA-BLAST (SI Appendix, Table S3). Additionally, 50 representative isoprenyl diphosphate synthases, such as FPP synthase, were added for comparison.
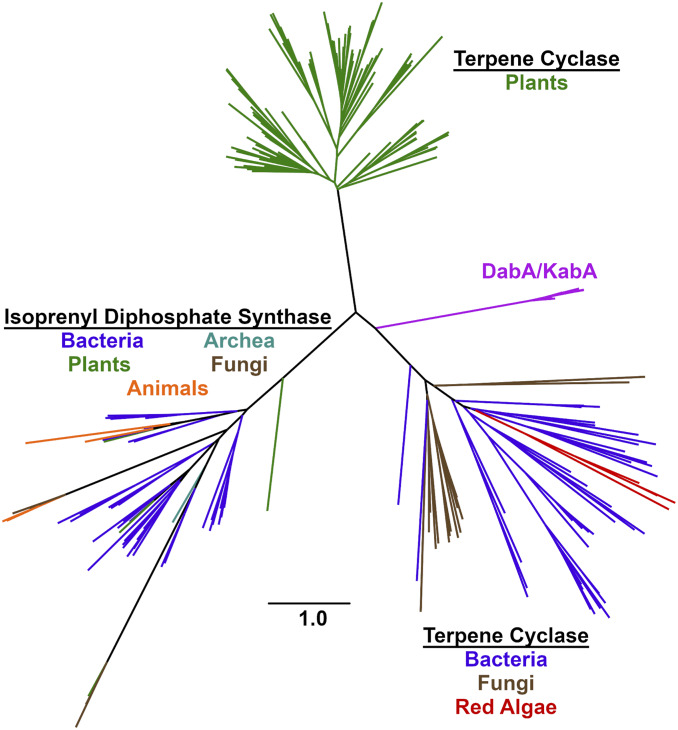
The tree forms three distinct sections composed of: 1) plant terpene cyclases, 2) a mixture of bacterial/algal/fungal terpene cyclases, and 3) the isoprenyl diphosphate synthases (Fig. 4). The terpene cyclase portion of the tree corresponds with α/β-domain organization of many plant terpene cyclases and the lone α-domain of the enzymes from the remaining organisms. The DabA proteins from Pseudo-nitzschia spp. diatoms cluster together and are closely related to the red algal KabA proteins. This N-prenyltransferase protein region is most related to the bacterial, red algal, and fungal enzymes. In contrast, the four characterized red algal terpene cyclases form a distinct region within the bacterial branch of terpene cyclases, consistent with previous phylogenetic analysis (41). Even though both KabA and red algae terpene cyclase sequences are associated with the microbial clade, the distance between these proteins suggests that the kainoid biosynthetic genes did not simply arise from algal terpene cyclases through a recent gene duplication event. Instead, a more distant evolutionary relationship is likely. Furthermore, the phylogeny of the diatom DabA and the red algal KabA proteins does not match the evolution of the organisms themselves. Diatoms are not closely related to red algae and are thought to have originated from a secondary endosymbiotic event from an ancestral red alga (43). Instead, red algae are more closely related to green plants (44). This inconsistency again suggests a curious evolutionary history that may employ mechanisms such as horizontal gene transfer.
Fig. 4.
A representative selection of all terpene cyclase (149 sequences) and isoprenyl diphosphate synthase (50 sequences) family members were used to generate a maximum likelihood phylogenetic tree.
Conclusions and Discussion
The biosynthesis of the toxic algal natural products domoic acid and kainic acid begins with an uncommon N-prenylation reaction of a primary amine. Our work demonstrated that DabA and KabA catalyze these reactions by repurposing the ubiquitous terpene cyclase fold. However, DabA and KabA catalyze an intermolecular prenylation reaction instead of the more common intramolecular cyclization reaction. Our structural work revealed that DabA uses the same active site location and some of the conserved ligand-binding features of canonical terpene cyclases. In contrast, DabA lacks the canonical DDxxD motif and substantially narrows the active site to create an elongated hydrophobic tunnel to sequester an extended prenyl moiety incapable of intramolecular cyclization upon ionization. However, the structural differences that enable binding of the l-glutamic acid cosubstrate remain to be elucidated. Curiously, DabA is not the first instance of an N-prenyltransferase reusing a prenyl diphosphate synthase fold. For example, Rv3378c, an adenosine N-prenyltransferase, uses a conserved fold typically found in the decaprenyl diphosphate synthase-like superfamily (IPR036424) (45).
Our results also uncover the structural differences between DabA and KabA that dictate substrate specificity. By creating single point mutations that change the length of the prenyl donor-binding tunnels in DabA and KabA, we successfully altered the specificity of this family of N-prenyltransferases. Moreover, this selectivity filter can serve as an indicator for the physiological prenyl substrate of functionally uncharacterized homologs. Finally, phylogenetic analysis reinforces the unusual evolutionary history of these enzymes first suggested by amino acid sequence alignments. Instead of being related to plant or red algal enzymes, they form their own distinct branch. This affiliation is despite the distant relationship between red algae and diatoms. Examination of hypothetical homologs from the acromelic acid producing mushroom may enable better insight into how these enzymes evolved (46). In addition, the detailed understanding of DabA described here may help inform the creation of PCR based assays that can detect DabA expression in the environment to serve as early warning systems before domoic acid accumulates to toxic levels. This approach is analogous to the successful methods used in freshwater neurotoxin monitoring (47). Finally, our results reinforce the observation that understudied organisms create opportunities to discover new types of enzyme biocatalysts (48).
Materials and Methods
The SI Appendix includes methods for expression, purification, and mutagenesis of DabA and KabA. Methodologies for crystallization, activity assays, kinetics, and phylogenetics are also detailed. Finally, synthetic procedures and small molecule characterization are available in the SI Appendix. Crystallographic data are available in the Protein Data Bank (see Data Deposition), while the remaining data and methods are present in the SI Appendix. The NCBI accession nos. for proteins discussed in this work are: AYD91073.1 (https://www.ncbi.nlm.nih.gov/protein/AYD91073.1/) (DabA) and QCC62379.1 (https://www.ncbi.nlm.nih.gov/protein/QCC62379.1/) (KabA).
Supplementary Material
Acknowledgments
We thank G. Louie (Salk Institute) and the staff of the Advanced Light Source (ALS) at beamlines 8.2.1 and 8.2.2 (Berkeley, CA) for assistance with data collection, G. Rouse (Scripps Institution of Oceanography) for helpful discussions, and D. Nguyen for assistance with synthesis. This research was supported by the National Oceanic and Atmospheric Administration under Award NA19NOS4780181 to B.S.M. and the Life Science Research Foundation through a Simons Foundation Fellowship to J.R.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors and coordinates have been deposited in the Protein Data Bank, www.wwpdb.org: DabA in complex with Mg2+ and GSPP (PDB ID code 6VKZ), DabA in complex with Mn2+ and GSPP (PDB ID code 6VL0), and DabA in complex with Mg2+ and NGG (PDB ID code 6VL1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001325117/-/DCSupplemental.
References
- 1.Mello F. D. et al., Mechanisms and effects posed by neurotoxic products of cyanobacteria/microbial eukaryotes/dinoflagellates in algae blooms: A review. Neurotox. Res. 33, 153–167 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Grattan L. M., Holobaugh S., Morris J. G., Harmful algal blooms and public health. Harmful Algae 57, 2–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson D. M., Cembella A. D., Hallegraeff G. M., Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 4, 143–176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefebvre K. A., Robertson A., Domoic acid and human exposure risks: A review. Toxicon 56, 218–230 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Wright J. L. C. et al., Identification of domoic acid, a neuroexcitatory amino acid, in toxic mussels from eastern Prince Edward Island. Can. J. Chem. 67, 481–490 (1989). [Google Scholar]
- 6.Alt A. et al., Pharmacological characterization of glutamatergic agonists and antagonists at recombinant human homomeric and heteromeric kainate receptors in vitro. Neuropharmacology 46, 793–806 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Lee S. H., Kang S. C., Ahn J. H., Lee J. W., Rim H. J., Santonin-kainic acid complex as a mass chemotherapeutic of Ascaris lumbricoides control in Korea. Korean J. Parasitol. 10, 79–85 (1972). [DOI] [PubMed] [Google Scholar]
- 8.Komiya Y., Kobayashi A., Techniques applied in Japan for the control of Ascaris and hookworm infections-A review. Jpn. J. Med. Sci. Biol. 18, 1–17 (1965). [PubMed] [Google Scholar]
- 9.Lévesque M., Avoli M., The kainic acid model of temporal lobe epilepsy. Neurosci. Biobehav. Rev. 37, 2887–2899 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunson J. K. et al., Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science 361, 1356–1358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chekan J. R. et al., Scalable biosynthesis of the seaweed neurochemical, kainic acid. Angew. Chem. Int. Ed. Engl. 58, 8454–8457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugawara H. et al., Structural insight into the reaction mechanism and evolution of cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 2734–2739 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum N., Gefter M. L., Δ 2 -isopentenylpyrophosphate: Transfer ribonucleic acid 2 -isopentenyltransferase from Escherichia coli. Purification and properties of the enzyme. J. Biol. Chem. 247, 5675–5680 (1972). [PubMed] [Google Scholar]
- 14.Sardar D. et al., Enzymatic N- and C-protection in cyanobactin RiPP natural products. J. Am. Chem. Soc. 139, 2884–2887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattila A. et al., Biosynthesis of the bis-prenylated alkaloids muscoride A and B. ACS Chem. Biol. 14, 2683–2690 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Christianson D. W., Structural and chemical biology of terpenoid cyclases. Chem. Rev. 117, 11570–11648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tello M., Kuzuyama T., Heide L., Noel J. P., Richard S. B., The ABBA family of aromatic prenyltransferases: Broadening natural product diversity. Cell. Mol. Life Sci. 65, 1459–1463 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuzuyama T., Noel J. P., Richard S. B., Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435, 983–987 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzger U. et al., The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 14309–14314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang P. H., Ko T. P., Wang A. H. J. H.-J., Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269, 3339–3354 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Holm L., Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Baer P. et al., Induced-fit mechanism in class I terpene cyclases. Angew. Chem. Int. Ed. Engl. 53, 7652–7656 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Fujihashi M. et al., Crystal structure and functional analysis of large-terpene synthases belonging to a newly found subclass. Chem. Sci. 9, 3754–3758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aaron J. A., Lin X., Cane D. E., Christianson D. W., Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry 49, 1787–1797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wendt K. U., Schulz G. E., Corey E. J., Liu D. R., Enzyme mechanisms for polycyclic triterpene formation. Angew. Chem. Int. Ed. 31, 2813–2833 (2010). [PubMed] [Google Scholar]
- 28.Schrepfer P. et al., Identification of amino acid networks governing catalysis in the closed complex of class I terpene synthases. Proc. Natl. Acad. Sci. U.S.A. 113, E958–E967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J., He X., Cane D. E., Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J. Am. Chem. Soc. 128, 8128–8129 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Cane D. E. et al., Pentalenene synthase. Purification, molecular cloning, sequencing, and high-level expression in Escherichia coli of a terpenoid cyclase from Streptomyces UC5319. Biochemistry 33, 5846–5857 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Felicetti B., Cane D. E., Aristolochene synthase: Mechanistic analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 126, 7212–7221 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Grundmann A., Kuznetsova T., Afiyatullov S. S., Li S.-M., FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. ChemBioChem 9, 2059–2063 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Copeland R. A., Reversible Modes of Inhibitor Interactions with Enzymes, (Wiley Interscience, ed. 2, 2013). [Google Scholar]
- 34.Bennett B. D. et al., Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter H., Robinson D. G., Heldt H. W., Subcellular volumes and metabolite concentrations in spinach leaves. Planta 193, 530–535 (1994). [Google Scholar]
- 36.Winter H., Robinson D. G., Heldt H. W., Subcellular volumes and metabolite concentrations in barley leaves. Planta 191, 180–190 (1993). [Google Scholar]
- 37.Narita K., Ohnuma S., Nishino T., Protein design of geranyl diphosphate synthase. Structural features that define the product specificities of prenyltransferases. J. Biochem. 126, 566–571 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Stanley Fernandez S. M., Kellogg B. A., Poulter C. D., Farnesyl diphosphate synthase. Altering the catalytic site to select for geranyl diphosphate activity. Biochemistry 39, 15316–15321 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Tarshis L. C., Proteau P. J., Kellogg B. A., Sacchettini J. C., Poulter C. D., Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. U.S.A. 93, 15018–15023 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrada P., Morita M., Hao Y., Schmidt E. W., Nair S. K., A single amino acid switch alters the isoprene donor specificity in ribosomally synthesized and post-translationally modified peptide prenyltransferases. J. Am. Chem. Soc. 140, 8124–8127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei G. et al., Terpene biosynthesis in red algae is catalyzed by microbial type but not typical plant terpene synthases. Plant Physiol. 179, 382–390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kersten R. D. et al., A red algal bourbonane sesquiterpene synthase defined by microgram-scale NMR-coupled crystalline sponge X-ray diffraction analysis. J. Am. Chem. Soc. 139, 16838–16844 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Keeling P. J., The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burki F., Roger A. J., Brown M. W., Simpson A. G. B., The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Layre E. et al., Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc. Natl. Acad. Sci. U.S.A. 111, 2978–2983 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konno K., Shirahama H., Matsumoto T., Isolation and structure of acromelic acid A and B. New kainoids of Clitocybe acromelalga. Tetrahedron Lett. 24, 939–942 (1983). [Google Scholar]
- 47.Al-Tebrineh J., Pearson L. A., Yasar S. A., Neilan B. A., A multiplex qPCR targeting hepato- and neurotoxigenic cyanobacteria of global significance. Harmful Algae 15, 19–25 (2012). [Google Scholar]
- 48.Cragg G. M., Newman D. J., Biodiversity: A continuing source of novel drug leads. Pure Appl. Chem. 77, 7–24 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.