Significance
Epstein–Barr virus (EBV) is a human herpesvirus linked to the development of several cancers. We demonstrate that XPB, a component of the TFIIH transcription factor complex, is required for EBV lytic gene expression and virus production. We show that EBV SM protein regulates lytic gene transcription by utilizing XPB as a cofactor to activate specific target genes. Spironolactone blocks transcriptional activation by SM and spironolactone’s antiviral activity against EBV is mediated by degrading XPB protein. XPB is thus particularly important for expression of an SM-dependent group of EBV genes. Because SPR destabilizes XPB protein, inhibiting SM transcription function without affecting cellular gene-transcription machinery, XPB may be a useful therapeutic target to control EBV and other human herpesviruses.
Keywords: Epstein–Barr virus, transcription, XPB, herpesvirus
Abstract
Epstein–Barr virus (EBV) is associated with epithelial and lymphoid malignancies, establishes latent infection in memory B cells, and intermittently produces infectious virions through lytic replication. Released virions play a key role in latent reservoir maintenance and transmission. Lytic EBV transcription differs from cellular transcription in requiring a virus-encoded preinitiation complex that binds to TATT motifs unique to EBV late lytic promoters. Expression of 15 late lytic genes that are important for virion production and infectivity is particularly dependent on the EBV SM protein, a nuclear protein expressed early during lytic reactivation that binds to viral RNAs and enhances RNA stability. We recently discovered that spironolactone blocks EBV virion production by inhibiting EBV SM function. Since spironolactone causes degradation of xeroderma pigmentosum group B-complementing protein (XPB), a component of human transcription factor TFIIH, in both B lymphocytes and epithelial cells, we hypothesized that SM utilizes XPB to specifically activate transcription of SM target promoters. While EBV SM has been thought to act posttranscriptionally, we provide evidence that SM also facilitates EBV gene transcription. We demonstrate that SM binds and recruits XPB to EBV promoters during lytic replication. Depletion of XPB protein, by spironolactone treatment or by siRNA transfection, inhibits SM-dependent late lytic gene transcription but not transcription of other EBV genes or cellular genes. These data indicate that SM acts as a transcriptional activator that has co-opted XPB to specifically target 15 EBV promoters that have uniquely evolved to require XPB for activity, providing an additional mechanism to differentially regulate EBV gene expression.
Epstein–Barr virus (EBV) is a human gammaherpesvirus associated with several lymphoid and epithelial malignancies (1). EBV infects the majority of people worldwide and establishes a life-long latent infection in B lymphocytes, from which it intermittently reactivates and produces infectious virion particles (2–4). EBV lytic proteins may also play an important role in oncogenesis and in maintaining the reservoir of latently infected B cells in vivo (5–7). EBV late lytic gene transcription varies in several important respects from cellular mRNA transcription, requiring ongoing lytic EBV DNA genome replication (8). Furthermore, most late genes utilize a virus-specific preinitiation complex composed of six EBV-encoded proteins, including a viral homolog of the TATA-binding protein (TBP), which binds to viral TATT motifs in late gene promoters rather than to conventional TATA boxes found in cellular genes and latent and early lytic EBV genes (9–11).
While EBV lytic cycle transcription is initiated by the immediate-early (IE) Z and R transactivators (12, 13), EBV late lytic gene expression and virion production also require the EBV SM protein (14–16). SM is a nuclear phosphoprotein expressed early during lytic reactivation that is essential for efficient expression of a specific subset of late lytic genes (14, 17–19). SM enhances accumulation of many lytic EBV mRNAs, but preferentially up-regulates 15 such genes, including EBV capsid, tegument, and glycoproteins that are essential for virion production and infectivity (14). The basis of this selective enhancement of EBV genes by SM has remained elusive. SM is a multifunctional protein that regulates gene expression by posttranscriptional mechanisms, including RNA splicing, RNA stability, mRNA export, and translation (20–28). SM binds directly to RNA and enhances target mRNA expression in reporter assays (15, 29). Differential GC content, the presence of specific RNA motifs in responsive transcripts, atypical codon usage, and differential transcriptional activation have all been proposed to explain specificity of action of SM and its homologs in other herpesviruses (14, 30–32). However, no clear relationship of SM activity to any of the above characteristics in the SM-dependent gene set could be established (14). Previous experiments also indicated that SM did not activate transcription of target genes (20, 33, 34). However, these early studies utilized nuclear run-on assays and reporter constructs that did not directly assess transcriptional effects of SM in EBV-infected cells (20).
Recently, using a cell-based, high-throughput screening assay, we found that a mineralocorticoid receptor blocker, spironolactone (SPR) (35), blocks EBV virion production by inhibiting SM function (36). SPR has been shown to inhibit nucleotide excision repair activity, and to lead to rapid degradation of a cellular transcription factor xeroderma pigmentosum group B-complementing protein (XPB) through a proteasome-dependent pathway (37). XPB is a component of the transcription factor II H (TFIIH) complex—composed of 10 subunits: XPB, XPD, P62, P52, P44, P34, P8, cdk7, cyclin H, and MAT1—that is involved in gene transcription and the DNA damage repair pathway (38–40). These findings raised the possibility that SM may specifically activate EBV genes transcriptionally in addition to its posttranscriptional effects on RNA processing and stability.
We used SPR as a tool to block SM function and investigate the mechanisms by which SM enhances gene-specific lytic gene expression. We also utilized bromouridine (BrU) to label nascent mRNA transcripts in EBV-infected cells undergoing lytic replication to investigate a role for SM in transcriptional activation. We show that SM acts as a transcriptional activator in addition to its known function as a posttranscriptional regulator. We show that SPR specifically inhibits SM-dependent EBV gene transcription without globally inhibiting EBV lytic gene transcription or cellular transcription. We provide mechanistic evidence that EBV SM co-opts XPB, a component of the TFIIH complex, and uses it as a cofactor for specific transcription of SM-dependent genes. Thus, SPR selectively blocks SM-mediated RNA transcription and virus production by degrading XPB. These findings demonstrate the importance of cellular XPB in EBV lytic gene transcription and SPR or its derivatives as potential antiviral drugs.
Results
SPR Inhibits EBV SM Function by Targeting a Cellular Transcription Factor XPB.
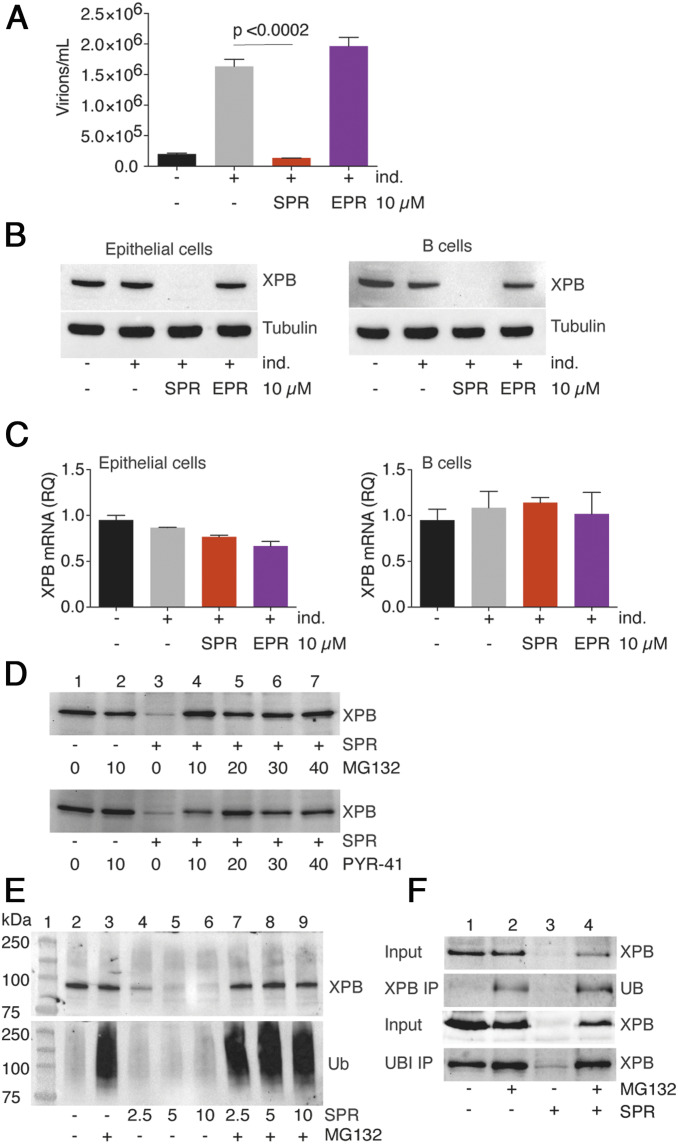
We have previously demonstrated that SPR exerts potent antiviral activity against EBV (36). Treatment of EBV-infected epithelial cells with SPR inhibited infectious virus production that occurs after induction of lytic EBV replication (Fig. 1A). EBV lytic replication was induced in the epithelial cell line AGSiZ, which contains a doxycycline-inducible transactivator of lytic EBV gene expression, Zta, and a recombinant GFP-expressing EBV bacmid. Concurrently, cells were either mock-treated or treated with SPR. Production of infectious viral particles in cell supernatants was measured by an assay in which uninfected 293T cells were incubated with supernatant and GFP transduction of 293T cells was quantitated by flow cytometry (41). SPR treatment led to an ∼88% reduction in the titer of infectious EBV in the cell supernatant. Treatment of AGSiZ cells with a chemically similar compound eplerenone (EPR), that also has potent mineralocorticoid blocking activity (36), resulted in no antiviral activity, confirming that mineralocorticoid blockade does not mediate antiviral activity (Fig. 1A). Recently, SPR was identified as an inhibitor of the nucleotide excision repair pathway and the mechanism was linked to rapid degradation of a cellular transcriptional factor XPB (37). Therefore, we investigated whether XPB loss in EBV-infected cells caused by SPR treatment was linked to SPR’s antiviral activity, possibly by affecting viral gene transcription. SPR treatment of EBV-infected epithelial and B lymphoma cells led to a complete loss of detectable XPB protein in both cell types (Fig. 1B). Again, EPR had no effect on XPB abundance, confirming that the antiviral activity of SPR against EBV is correlated to XPB loss (Fig. 1B).
Fig. 1.
Antiviral activity of SPR is associated with XPB protein degradation. (A) SPR but not EPR inhibits EBV production in epithelial cells. EBV lytic replication was induced (+ind.) in a GFP-EBV infected gastric carcinoma cell line (AGSiZ) treated with either SPR or EPR. Virion production was measured by incubating 293 cells with induced AGS cell supernatants containing infectious virions. GFP+ 293 cells representing infectious EBV particles were quantitated by flow cytometry. Error bars indicate the SEM from three biological replicates. (B) Effect of SPR on XPB degradation in epithelial cells and B lymphocytes. AGSiZ and P3HR1-ZHT cells were treated with SPR or EPR for 24 h after induction of lytic reactivation. Cell lysates were immunoblotted using anti-XPB antibody. Blots were stripped and reprobed with antitubulin antibody as a loading control. (C) SPR and EPR do not affect XPB RNA levels in epithelial cells and B lymphocytes. RNA was isolated from cells treated in B and XPB mRNA was quantitated by qRT-PCR. (D) SPR-induced XPB protein degradation is rescued by proteasome inhibitors or ubiquitin activation inhibitors. AGSiZ cells were pretreated for 2 h with various concentrations of proteasome inhibitor (MG132) or the ubiquitin activating enzyme inhibitor (PYR-41) before addition of SPR. Cell lysates were harvested 4 h after SPR treatment and immunoblotted with anti-XPB antibody. (E) Effect of proteasome inhibition on accumulation of ubiquitinated proteins. AGSiZ cells were treated with MG132 (+) or mock treated (−) and exposed to various concentrations of SPR as shown. Cell lysates were immunoblotted with anti-XPB antibody. Blot was stripped and reprobed with antiubiquitin antibody (Ub). (F) Effect of SPR on ubiquitination of XPB. P3HR1 cells were pretreated for 2 h with proteasome inhibitor MG132 before addition of SPR. Protein cell lysates were immunoprecipitated with either XPB or ubiquitin antibody and immunoblotted with antiubiquitin and anti-XPB antibody, respectively.
We next measured XPB RNA expression in SPR- or EPR-treated cells to determine whether XPB loss was mediated by effects on XPB mRNA. XPB RNA expression was not affected by treatment with SPR in both epithelial and B cells (Fig. 1C), indicating that SPR-mediated XPB protein degradation occurred posttranscriptionally. XPB mRNA levels were also not affected by lytic EBV replication. To further investigate the mechanism of XPB degradation by SPR, we pretreated AGS cells with either proteasome inhibitor (MG132) or the ubiquitin-activating enzyme inhibitor (PYR41) 2 h before SPR treatment, as previously described (37). Pretreatment of cells with various concentration of either MG132 or PYR41 completely rescued SPR-dependent XPB degradation (Fig. 1D, lanes 4 to 7), suggesting that SPR leads to ubiquitination and proteasome targeting of XPB by SPR. These finding were consistent with previously described XPB degradation mechanisms in HeLa, Jurkat T, and PAECs cells (37, 42, 43).
Because ubiquitinated proteins are degraded rapidly by the proteasome pathway, it was possible that higher molecular weight forms of ubiquitinated XPB might be detected when proteasome function was blocked in the presence of SPR. We therefore treated AGSiZ cells with various concentration of either SPR or SPR plus MG132 (Fig. 1E) and performed immunoblotting with antiubiquitin or anti-XPB antibodies. Increased accumulation of ubiquitinated proteins occurred with MG132 treatment, as expected; however, no higher molecular-weight forms of XPB were detected (Fig. 1E). These results suggest that either SPR leads to ubiquitination of other cellular targets that promote XPB degradation or that direct ubiquitination of XPB occurs but does not lead to an obvious increase in size or quantity of ubiquitinated XPB detectable by immunoblotting.
To further investigate whether ubiquitination of XPB is promoted by SPR, we performed immunoprecipitation (IP) with either XPB or ubiquitin antibody in B lymphoma cells, followed by immunoblotting with ubiquitin or XPB antibodies, respectively. This allows detection of ubiquitinated XPB using reciprocal IP and immunoblotting. As expected, ubiquitinated XPB was only detectable when cells were pretreated with proteasome inhibitors (Fig. 1F, lanes 2 and 4), but not when they were either untreated or SPR-treated. Importantly, the ratio of ubiquitinated XPB to total XPB increased by 3.29 when cells were treated with SPR in the presence of MG132 (Fig. 1F, lane 4) as opposed to 1.17 when treated with MG132 alone (Fig. 1F, lane 2). The intensities of the band on the Western blots were calculated using Bio-Rad Chemi Doc XRS+ with Image Lab Software. These results indicate that SPR may promote direct ubiquitination of XPB that rapidly targets XPB to the proteasome.
In order to verify the generalizability of these findings to nontumor-derived, EBV-infected cells, we examined the effect of SPR on normal B cells transformed and immortalized with EBV. SPR had similar inhibitory effects on virus production, XPB stability, and SM-dependent gene expression in these cells (SI Appendix, Fig. S1).
XPB Is Important for SM-Dependent EBV Gene Expression and EBV Virion Production.
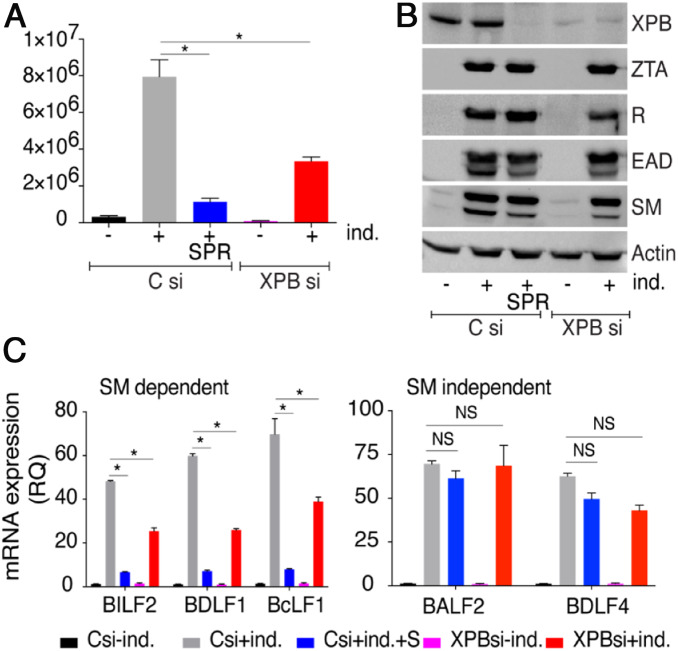
XPB protein is a component of the TFIIH complex and plays a complex role in cellular gene transcription (44). It is therefore likely that SPR’s antiviral activity operates by inhibiting a function of XPB in EBV lytic gene transcription. Alternatively, it is possible that SPR inhibits SM-dependent gene expression via other functions of XPB or even unrelated to XPB. To determine whether the SM-specific antiviral activity of SPR against EBV could directly be linked to XPB degradation, we examined the effects of XPB knockdown in EBV-infected cells. XPB was depleted in EBV+ AGS cells using small-interfering RNA (siRNA) and the effect on EBV production and EBV gene expression was measured. XPB depletion either by SPR treatment or by using siRNA had a clearly inhibitory effect on infectious EBV production as measured by a GFP transduction assay. SPR treatment led to an ∼85% reduction in infectious virion production (Fig. 2A). Knockdown (KD) of XPB with siRNA also inhibited virion production but only by ∼58%. Western blots of whole lysates demonstrated that SPR treatment or siRNA transfection both reduced XPB protein levels. However, SPR’s depletion of XPB was actually greater than that of XPB siRNA transfection, perhaps explaining its greater inhibitory activity (Fig. 2B). This conclusion is supported by a dose-related inhibition of virion production by SPR (SI Appendix, Fig. S2).
Fig. 2.
XPB is required for efficient EBV virion production and SM-dependent EBV gene expression. (A) Infectious EBV virion production in cells depleted of XPB. AGSiZ cells were transfected with control siRNA (C) or XPB siRNA. Virion production was measured by infecting 293 cells with induced cell supernatants as above. (B) Depletion of XPB does not affect EBV immediate-early and early protein expression. Forty-eight hours after transfection of control (C) or XPB siRNAs, cells were treated with doxycycline to induce EBV lytic replication (+ind.) and mock-treated or treated with SPR. Protein cell lysates were harvested at 48 h postinduction and immunoblotted using anti-XPB, -Z, -R, -EAD, and -SM antibodies. (C) Gene-specific effect of XPB depletion on EBV lytic RNA expression. RNA was isolated 48 h after EBV lytic induction in the absence or presence of XPB. qRT-PCR was performed to measure the effect of XPB depletion on expression of SM-dependent (BILF2, BDLF1, and BcLF1) and SM-independent RNAs (BALF2, BDLF4). The error bars indicate the SEM from three replicates. *P < 0.15; NS, P = 0.02 to 0.4.
Since the most SM-dependent and SPR-sensitive EBV genes are a subset of the late lytic genes, we examined the effect of XPB KD to see if it paralleled the effect of SPR or SM depletion. We first measured the effect of XPB KD on EBV immediate-early protein (ZTA or RTA) and early protein (EAD or SM) expression by immunoblotting. As expected, neither SPR treatment (Fig. 2B, lane 3) nor XPB KD (Fig. 2B, lane 5) had any effect on immediate-early or early protein expression (Fig. 2B, lane 2). Next, we examined the effect of XPB knockdown on both SM-dependent and SM-independent EBV mRNA expression during lytic replication. As shown in Fig. 2 C and D, depletion of XPB protein either by SPR or by XPB siRNA significantly inhibited SM-dependent gene expression (BILF2, BDLF1, and BcLF1) but not SM-independent (BALF2, BDLF4) gene expression as compared to control siRNA transfection. These data indicate that XPB plays a direct role in facilitating SM-dependent EBV lytic gene expression and virion production.
SPR Inhibits SM-Dependent EBV Gene Transcription but Does Not Globally Block Transcription.
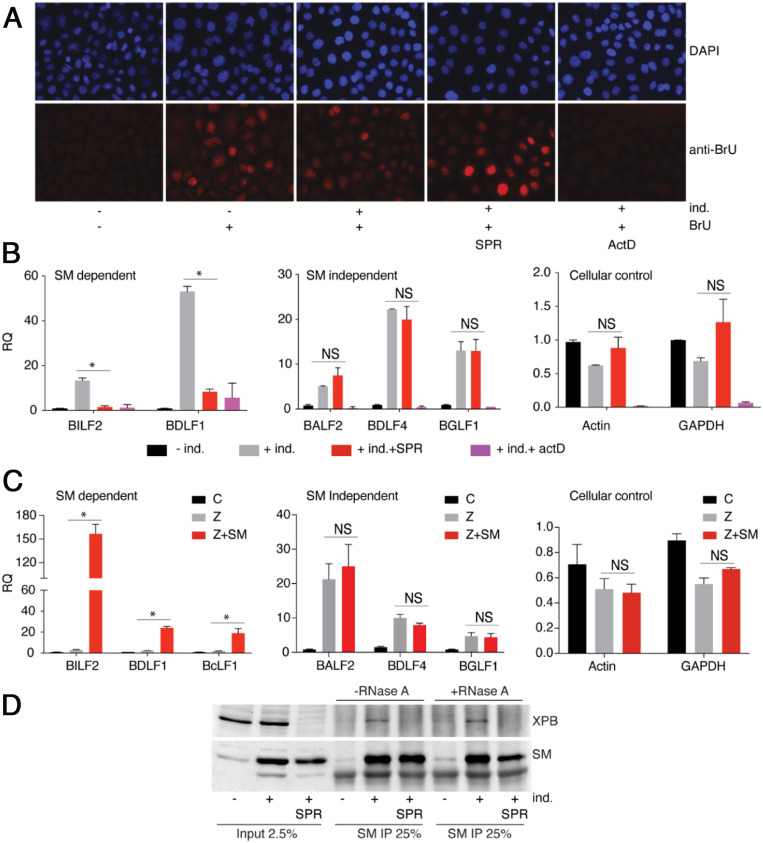
XPB is a subunit of the TFIIH core complex and has an ATP-dependent DNA helicase and translocase activity that is involved in transcriptional initiation and elongation (45). We hypothesized that XPB may be important for transcriptional initiation or elongation of EBV genes, particularly SM-dependent genes, leading to preferential inhibition of SM-dependent gene transcription upon SPR treatment. In order to test this hypothesis, we investigated the effect of SPR on EBV gene-transcription initiation. We employed a 2-BrU incorporation assay in which newly initiated transcripts are identified by transient inclusion of BrU in the growth medium, followed by IP with anti-BrU antibodies. The specificity of the assay was first validated by immunostaining BrU-pulsed cells with anti-BrU antibody. As shown in Fig. 3A, BrU incorporation was observed only in BrU-pulsed cells and was inhibited by simultaneous incubation with the transcription inhibitor actinomycin D (ActD), showing that BrU incorporation is transcription-specific. Importantly, global transcription initiation, as measured by BrU incorporation, was not affected by either induction of EBV lytic replication or by SPR treatment (Fig. 3A). To investigate the effect of SPR on EBV lytic gene-transcription initiation, we captured newly synthesized (BrU-labeled) RNA by IP with anti-BrU antibody and performed qRT-PCR for EBV lytic mRNAs. Robust increases in EBV lytic mRNA labeling with BrU occurred after induction of EBV lytic replication, demonstrating that BrU labels nascent EBV transcripts (Fig. 3B). However, SPR treatment during lytic replication completely abolished BrU incorporation of SM-dependent genes (BDLF1 and BILF2), but had no effect on SM-independent genes (BALF2, BDLF4, and BGLF1). Similarly, labeling of cellular gene transcripts (actin and GAPDH) was unaffected or was only slightly changed. As expected, ActD treatment completely blocked all transcription, confirming that BrU incorporation is transcription-specific. These data demonstrate that SPR preferentially blocks transcription and elongation of SM-dependent genes, suggesting that XPB plays an important role in SM-dependent EBV gene transcription.
Fig. 3.
SPR does not block general transcription but inhibits SM-dependent EBV gene transcription. (A) Effect of SPR on global transcription. EBV lytic replication was induced (+ind.) with doxycycline in EBV-infected AGSiZ cells and the effect of SPR on transcription was assessed by measuring 5- BrU incorporation into nascent RNA. Cells were treated with SPR or mock-treated at time of induction by doxycycline. Forty-eight hours postinduction, cells were pulsed with BrU for 30 min, then fixed and stained for BrU. Cells were also treated with ActD as a control to show that BrU incorporation is due to transcription initiation. Magnification was 40×. (B) Effect of SPR on EBV lytic gene transcription initiation. EBV lytic replication in AGSiZ cells was induced (+ind.) by addition of doxycycline and either mock-treated or treated with SPR. Forty-eight hours postinduction, cells were pulsed with BrU for 30 min. Newly synthesized RNA was immunoprecipitated with anti-BrU antibody and SM-dependent (BILF2 and BDLF1), SM-independent (BALF2, BDLF4, and BGLF1), and cellular (actin and GAPDH) transcripts were measured by qRT-PCR. (C) Effect of SM on EBV lytic transcription. EBV lytic replication in 293 SMKO EBV-infected cells was induced by transfecting with either empty vector (C), Z plasmid, or Z+SM. Forty-eight hours after lytic induction, cells were pulsed with BrU for 30 min. Newly synthesized RNA was immunoprecipitated with anti-BrU antibody. SM-dependent (BILF2, BDLF1, and BcLF1), SM-independent (BALF2, BDLF4, and BGLF1) and cellular (actin and GAPDH) RNAs were analyzed by qRT-PCR. The error bars indicate the SEM from three replicates. *P = 0.0001 to 0.018; NS, P = 0.072 to 0.96. (D) EBV SM interacts with XPB. EBV lytic replication was induced (+ind.) in AGSiZ EBV-infected cells in the absence or presence of SPR. Lysates of cells were immunoprecipitated with anti-SM serum and immunoblotting was performed with anti-XPB monoclonal antibody. IPs were mock-treated or treated with RNase A to determine whether the SM–XPB protein interaction was resistant to RNase. The blot was stripped and reprobed with anti-SM antibody.
EBV SM Acts as a Transcriptional Regulator.
All previous studies have shown that SM acts as a posttranscriptional regulator and enhances RNA accumulation by RNA stabilization and RNA export mechanisms (20, 21, 46–48). The data above suggest that SM may also activate SM-dependent lytic gene expression by enhancing transcription initiation or elongation. We hypothesized that SM possesses a transcriptional activation function, and that XPB is preferentially important for transcription of SM-dependent late genes. In order to better understand the role of EBV SM protein in EBV gene transcription, we directly measured the effect of SM on EBV transcription initiation using the BrU incorporation assay. We compared transcription initiation in cells infected with SM-knockout EBV (SMKO EBV) during lytic reactivation and in cells rescued by exogenous expression of SM. We captured newly synthesized (BrU-labeled) RNA by IP with BrU antibody and measured the abundance of EBV SM-dependent and -independent mRNAs by qRT-PCR (Fig. 3C). We observed a robust increase in nascent SM-dependent late lytic mRNAs (BILF2, BDLF1, and BcLF1) after the induction of EBV lytic replication only in the presence of SM (Z+SM), as compared to induction in the absence of SM (+Z). However, BrU incorporation for SM-independent genes (BALF2, BDLF4, and BGLF1) and cellular genes (actin and GAPDH) was unaffected by the presence of SM (Z+SM). These results clearly demonstrate that EBV SM protein also acts as a transcriptional regulator and that the antiviral activity of SPR against EBV observed here is due to inhibition of the SM transcriptional activation, implicating XPB as a cofactor important for SM-dependent gene transcription.
EBV SM Interacts with XPB but Does Not Bind Directly to EBV DNA.
The above experiments demonstrate that SM plays a direct role in transcription of specific EBV genes and that XPB is required for SM-dependent gene transcription. Since SM is an RNA binding protein and we have previously shown that SM does not bind directly to the EBV genome (14), we speculated that an interaction between SM and XPB, which does bind DNA, may be critical for SM activation of RNA transcription and elongation. Therefore, we tested for a possible SM and XPB protein–protein interaction in EBV-infected AGS cells during EBV lytic replication. SM produced during lytic replication was able to coimmunoprecipitate (co-IP) endogenous XPB (Fig. 3D). As expected, XPB co-IP did not occur in uninduced cells in which SM is not present or in induced cells after SPR treatment, where XPB is degraded. Because SM binds to RNA directly (28, 46, 47), we also tested whether the observed SM–XPB interaction was RNA-dependent. IP samples were mock-treated or treated with RNase A and analyzed by immunoblotting with XPB or SM antibody. As shown in Fig. 3D, there was no difference in XPB co-IP with SM with or without RNase treatment, indicating that the SM–XPB interaction is not mediated by an RNA bridge but is due to protein–protein interactions.
SPR Blocks SM-Dependent EBV Gene Transcription by Degrading XPB.
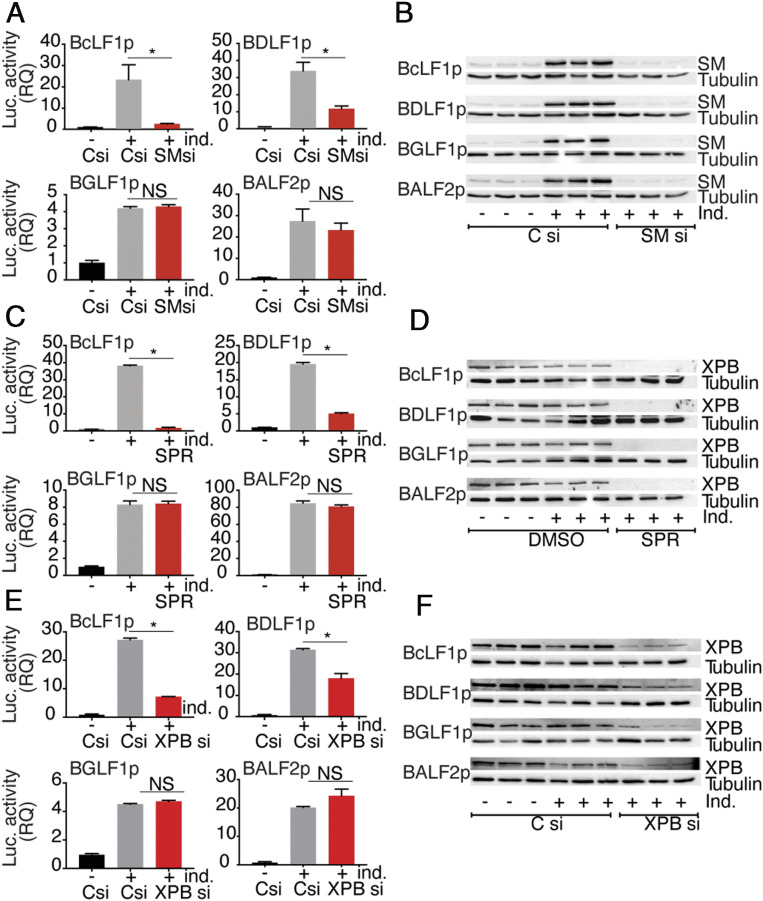
In order to better understand the potential SM transcriptional effect on EBV promoters and to validate the model whereby XPB is critical for transcription of SM-dependent promoters, we developed a reporter assay for SM-dependent transcription. We cloned a promoter from each EBV gene of interest (SM-dependent and -independent), comprised of 500 bp upstream of the initiator ATG codon and 100 bp downstream of ATG to include any downstream elements, in frame with the luciferase gene in the luciferase reporter vector pGL3. Since lytic replication in cis is required for efficient transcription of EBV late genes (8, 10, 11, 49), the origin of lytic EBV replication (oriLyt) was also cloned in each plasmid. Reporter plasmids were constructed containing promoters from two SM-dependent late (BcLF1 and BDLF1), an SM-independent late (BGLF1), and an SM-independent early (BALF2) gene. In order to compare the effects of SM on these promoters, we depleted or mock-depleted SM in AGSiZ Akata EBV cells by siRNA transfection followed by induction of EBV lytic replication. Twenty-four hours after lytic induction, reporter plasmids were transfected into cells and the effect of SM on each promoter was analyzed by luciferase assay. As shown in Fig. 4A, there was an increase in promoter activity with induction of lytic replication in the presence of SM. However, when we depleted SM, a significant decrease was observed in SM-dependent promoter activity (BcLF1 and BDLF1). Conversely, the SM-independent promoters (BGLF1 and BALF2) functioned equally well upon lytic replication regardless of whether SM was depleted. The efficiency of SM KD in this experiment was confirmed by performing a Western blot on protein cell lysates (Fig. 4B).
Fig. 4.
Gene-specific effect of SM on EBV promoters is mediated by XPB. (A) Effect of SM on SM-dependent and -independent promoters. AGSiZ cells were transfected with either control (Csi) or SM siRNA (SMsi) and EBV lytic replication was induced 3 h post-siRNA transfection. Twenty-four hours after lytic induction, SM-dependent or SM-independent promoter reporter constructs were transfected in parallel with reporter plasmids. Cells were lysed and SM-dependent late (BcLF1 and BDLF1), SM-independent late (BGLF1) and early (BALF2) promoter activities were measured by luciferase assay. Data are normalized to luciferase activity in uninduced control siRNA transfected cells and shown as the RQ. All transfections were done in biological triplicates and luciferase activity was measured in technical triplicates. Error bars represent the SEM. (B) Efficacy of SM KD. Western blots were performed using the above protein lysates and anti-SM serum. The blot was stripped and reprobed with antitubulin as a loading control. (C) Effect of SPR on SM-dependent and -independent promoters. AGSiZ cells were treated with SPR at time of lytic induction with doxycycline. Twenty-four hours after induction, cells were transfected with SM-dependent or -independent promoter constructs as stated previously. Cells were lysed and SM-dependent late (BcLF1 and BDLF1), SM-independent late (BGLF1), and early (BALF2) promoter activities were measured as in A. (D) Effect of SPR on XPB expression. Western blots were done using the above protein lysates and anti-XPB antibody. The blot was stripped and reprobed with antitubulin. (E) Effect of XPB depletion on SM-dependent and SM-independent promoters. AGSiZ cells were transfected with either control (Csi) or XPB siRNA (XPBsi) and EBV lytic replication was induced 3 h post-siRNA transfection. Twenty-four hours after lytic induction, SM-dependent or SM-independent promoter reporter constructs were transfected in parallel. Cells were lysed and SM-dependent late (BcLF1 and BDLF1), SM-independent late (BGLF1), and early (BALF2) promoter activities were measured by luciferase assay as in A. (F) Efficacy of XPB KD. Western blots were done using above protein lysates and anti-XPB antibody. The blot was stripped and reprobed with antitubulin as a loading control. The error bars indicate the SEM from three replicates. *P = 0.0001 to 0.042; NS, P = 0.12 to 0.56.
Having validated the reporter assay for SM transactivation, we asked if SPR specifically inhibited SM-dependent promoter activity through XPB degradation by testing the effect of SPR on the promoter constructs described above. SPR only inhibited SM-dependent promoter activity and had no effect on SM-independent promoters, exactly paralleling the SM dependence of each promoter (Fig. 4C). SPR-mediated XPB degradation in all SPR-treated cells was confirmed by immunoblotting of protein cell lysates (Fig. 4D). To further confirm that the SM-specific effect on EBV promoters was directly linked to XBP, we specifically knocked down XPB in EBV+ AGS cells using siRNA and measured the effect on SM-dependent and -independent EBV promoters. XPB depletion using siRNA transfection had a clear inhibitory effect on SM-dependent promoter activity only and had no effect on SM-independent promoters, indicating that XPB is critical for SM to exert gene-specific transcription function (Fig. 4E). XPB depletion by siRNA treatment was confirmed by immunoblotting of protein cell lysates (Fig. 4F).
EBV SM Enhances XPB Recruitment to EBV Promoters during Lytic Replication.
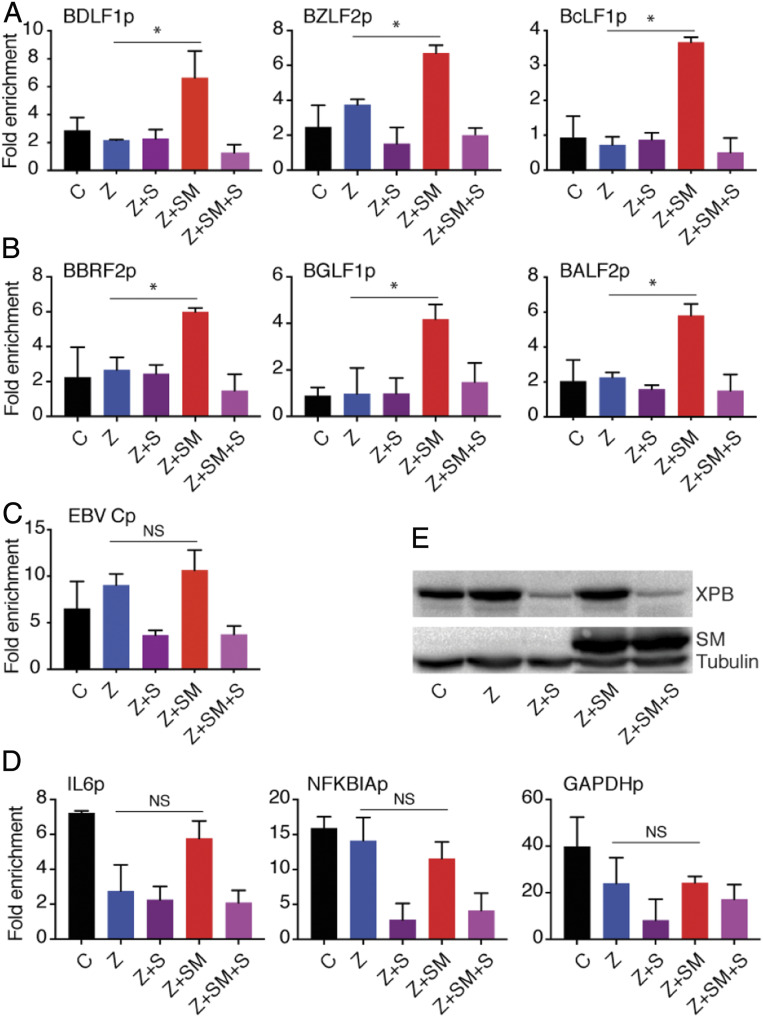
The linkage of SM-dependent EBV promoter activity to XPB could be explained by a model where XPB is recruited by SM to EBV promoters. To determine whether XPB occupancy at EBV promoters was enhanced by SM, we performed a chromatin IP (ChIP) assay in cells infected with SMKO EBV during lytic replication and compared the XPB signal at EBV promoters in the presence or absence of SM. Lytic replication in SMKO EBV-infected cells was induced by transfecting the cells with EBV Zta plasmid and SM expression was rescued by SM plasmid transfection. We also tested the effect of SPR on XPB recruitment to EBV promoters in the presence or absence of SM by treating cells with SPR at the time of lytic induction. Forty-eight hours after lytic induction, the cells were harvested, the proteins were cross-linked to DNA with formaldehyde, and the sheared chromatin was immunoprecipitated using an XPB polyclonal antibody. The precipitated DNA was purified, and qPCR was performed for both SM-dependent and -independent genes. As shown in Fig. 5, XPB recruitment to SM-dependent promoters (BDLF1p, BZLF2p, BcLF1p) was increased upon EBV lytic replication in the presence of SM (Z+SM) (Fig. 5A). Somewhat surprisingly, although BBRF2p, BGLF1p, and BALF2p are SM-independent promoters whose activity was not affected by SPR treatment or XPB KD, they nevertheless displayed increased XPB occupancy in the presence of SM (Fig. 5B). XPB occupancy of the EBV promoters was not increased when lytic induction was induced in the absence of SM. XPB occupancy was also decreased by SPR treatment (Z+SM+S), as expected. Importantly, SM did not increase XPB recruitment to the major EBV latent promoter Cp (Fig. 5C).
Fig. 5.
EBV SM increases XPB recruitment to EBV lytic promoters but not to cellular promoters or the major EBV latent promoter. ChIP assays to measure effect of SM on XPB binding to EBV lytic promoters during lytic replication. EBV lytic replication in 293 SMKO EBV-infected cells was induced by transfecting with Z plasmid, or with Z and SM plasmids to both induce replication and rescue SM expression. Cells were also transfected with empty vector as an uninduced control. Induced cells were also treated with SPR (+S) or mock-treated with vehicle. Forty-eight hours after lytic induction, proteins were cross-linked to DNA, sheared, and chromatin was immunoprecipitated using an XPB polyclonal antibody. DNA was extracted from IPs and qPCR was performed for several EBV SM-dependent lytic promoters (A), SM-independent lytic promoters (B), the major EBV latency C promoter (C), or cellular promoters (D), to quantitate XPB occupancy during lytic replication. The fold-enrichment over background was calculated using IgG antibody IP as the control in each sample. The error bars indicate the SEM from three different IPs. *P = 0.0004 to 0.03; NS, P = 0.16 to 0.9. (E) Efficacy of XPB depletion by SPR. Western blots was performed from above protein samples and blotted with anti-XPB and anti-SM antibody. The blot was stripped and reprobed with antitubulin as a loading control.
In order to ask whether SM increases recruitment of XPB to cellular promoters, we measured XPB occupancy on several cellular promoters (IL-6 and NFKBIA) previously shown to be bound by XPB (42), along with the GAPDH promoter. As shown in Fig. 5C, unlike EBV promoters, XPB recruitment to cellular promoters was not consistently increased by SM during lytic replication as compared to uninduced cells. In fact, there was a slight decrease of XPB occupancy of the cellular promoters tested during lytic EBV replication. These results, therefore, suggest that SM may enhance recruitment of XPB to EBV lytic promoters generally, but that only the SM-dependent lytic promoters require XPB for full activity. The linkage between XPB and SM activity is therefore not due to gene-specific recruitment of XPB but may involve postbinding effects unique to SM-dependent promoters.
Inhibition of XPB ATPase Activity Blocks EBV Lytic mRNA Synthesis.
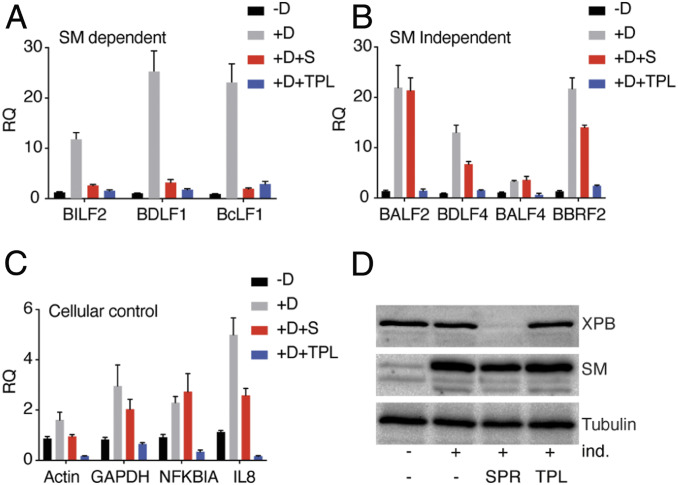
As shown above, XPB is required for transcription of SM-dependent genes and depletion of XPB by SPR specifically inhibits SM-dependent gene transcription but does not affect SM-independent gene transcription. XPB has an ATP-dependent DNA helicase and translocase activity that is involved in the transcriptional engagement of RNA pol II (45). It has been reported recently that triptolide (TPL), a small molecule that directly binds to XPB, selectively inhibits the XPB ATPase activity (50). Interestingly, TPL blocks cellular transcription, whereas loss of XPB altogether does not. This phenomenon has been explained as due to a blocking effect of XPB on the promoter when its ATPase function is inhibited, although it is not essential for transcription. In order to determine whether EBV promoters exhibit a similar behavior with regards to XPB function, we investigated the effect of TPL on EBV gene-transcription initiation using the BrU incorporation assay. We compared transcription initiation in EBV-infected cells during lytic reactivation and in cells treated with either SPR or TPL. We captured newly synthesized (BrU-labeled) RNA by BrU antibody and measured the abundance of SM-dependent, SM-independent, and cellular mRNAs by qRT-PCR. A robust increase in EBV lytic mRNA synthesis occurred after induction of EBV lytic replication, as expected, while TPL treatment for 1 h completely abolished synthesis of all viral and cellular transcripts tested (Fig. 6 A–C). However, SPR treatment during lytic replication only inhibited transcription of SM-dependent genes (BILF2, BDLF1, and BcLF1) but had no effect or a minimal effect on SM-independent genes (BALF2, BDLF4, BALF4, and BBRF2).
Fig. 6.
Inhibition of XPB ATPase activity blocks EBV lytic mRNA synthesis. (A) BrU incorporation assay to measure the effect of TPL on transcription initiation. EBV lytic replication in AGSiZ cells was induced (+ind.) by addition of doxycycline and either mock-treated or treated with SPR. Forty-eight hours postinduction, cells were treated with TPL for 1 h and then pulsed with BrU for 30 min. Newly synthesized RNA was immunoprecipitated with anti-BrU antibody and analyzed by qRT-PCR. (A) SM-dependent genes (BILF2, BDLF1, and BcLF1). (B) SM-independent genes (BALF2, BDLF4, BALF4, and BBRF2). (C) Cellular genes (actin GAPDH, NFKBIA, and IL-8). (D) Efficacy of XPB depletion by SPR. Western blots were performed with protein samples above and blotted with anti-XPB and anti-SM antibody. The blot was stripped and reprobed with antitubulin as a loading control. The error bars indicate the SEM from three replicates. All reductions in transcription initiation due to TPL were significant with a P value from 0.009 to 0.02.
Similarly, transcription initiation of cellular gene transcripts (actin and GAPDH) were unaffected or only slightly changed after SPR treatment. We also measured transcription initiation of cellular genes IL-8 and NFKBIA, which are reported as XPB-dependent and -independent genes, respectively (42). Consistent with this finding, we also observed that SPR suppressed IL-8 mRNA synthesis but did not affect NFKBIA mRNA synthesis (Fig. 6C). In summary, TPL treatment completely abolished transcription of all mRNAs tested here, while XPB depletion affected only SM-dependent gene transcription but had no effect or a slight effect on SM-independent gene transcription. Thus, global transcription takes place in the absence of XPB, indicating that XPB is not absolutely required for TFIIH function but is critical for SM-dependent EBV gene transcription. However, blocking XPB-ATPase activity in the presence of XPB inhibits both viral and cellular gene transcription.
XPB Is Required for Early Events in Transcription of SM-Dependent Genes.
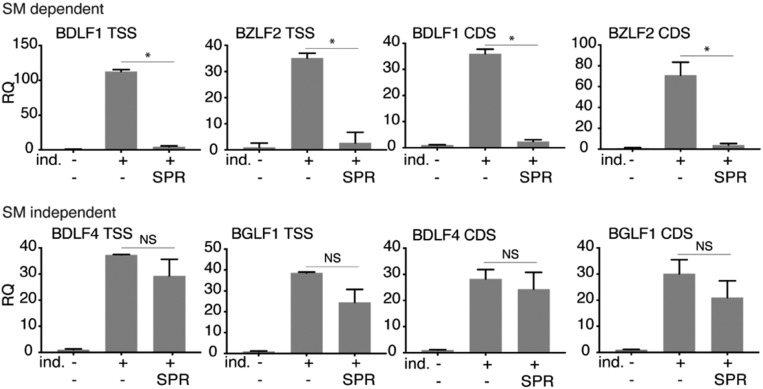
The particular requirement of SM-dependent promoters for XPB could be due to an XPB requirement for promoter opening or later events, such as promoter escape and elongation. To ask whether transcriptional inhibition by SPR would still allow production of short nascent transcripts, we assessed the presence of short transcripts produced prior to an elongation block in the absence of XPB. We performed qPCR using primers very close to the previously mapped EBV transcriptional start sites (11) to detect such transcripts in BrU immunoprecipitates from EBV-infected cells, either SPR- or mock-treated. qPCR was also performed with primers in the gene body to serve as an internal control for RNA abundance of each transcript. As shown in Fig. 7, in contrast to SM-independent genes, SM-dependent promoters did not produce any detectable transcripts when treated with SPR, even when primers capable of detecting transcripts shorter than 42 nt were used. This suggests that XPB is required for promoter opening or very early events in transcription from SM-dependent promoters.
Fig. 7.
XPB is essential for early events in transcription of SM-dependent EBV genes. AGSiZ cells were induced (+ind.) to permit lytic EBV replication by the addition of doxycycline and were either mock-treated or treated with SPR. Forty-eight hours postinduction, cells were pulsed with BrU for 30 min. Newly synthesized RNA was immunoprecipitated with anti-BrU antibody. SM-dependent (BDLF1 and BZLF2) and SM-independent (BDLF4 and BGLF1) gene transcripts were measured by qRT-PCR at transcriptional start site (TSS) and gene body (CDS). The error bars indicate the SEM from three different replicates. *P < 0.0001, NS, P = 0.08–0.65.
Discussion
The lytic phase of EBV replication, during which lytic proteins are expressed and infectious virions are produced, plays an important role in maintaining the human viral reservoir during persistent infection and may promote oncogenesis (8, 51, 52). The EBV SM protein is essential for progression of lytic replication and specifically enhances expression of 15 late EBV genes (14). While SM has been shown to facilitate EBV mRNA export, regulate splicing, and enhance EBV mRNA stability (15, 21, 28, 34, 53), it has not previously been implicated in EBV transcription. In this study, we have shown that SM protein enhances transcription of the SM-dependent subset of EBV late genes. Based on our finding that SPR, which preferentially inhibits SM-dependent gene expression, also leads to efficient proteasomal degradation of XPB, a cellular transcription factor, we investigated the possible role of SM as a gene-specific transcriptional activator. Using BrU to label nascent transcripts, we demonstrated that SM enhanced transcription of SM-dependent EBV genes and that SPR treatment abolished transcriptional activation by SM. Furthermore, XPB depletion had similar effects to SPR treatment, specifically inhibiting SM-dependent gene expression. Taken as a whole, these data indicate that SM co-opts XPB to preferentially activate transcription of a subset of late EBV genes.
One simple model to explain the specific requirement of XPB for SM-dependent promoter activity is that SM recruits XPB only to promoters that are SM-dependent. However, ChIP assays demonstrated that while SM does increase XPB occupancy at EBV promoters, it does so at both SM-dependent and SM-independent EBV promoters. These findings make it highly unlikely that XPB dependence of SM-activated promoters is due to selective enrichment of XPB at these promoters by SM.
Conventional models of RNA pol II-mediated transcription involve TFIIH opening the promoter after assembly of a preinitiation complex (PIC) consisting of pol II, TBP, and basal transcription factors (54). The ATPase-dependent helicase function of XPB was thought to play a role in unwinding the DNA duplex with the kinase functions of TFIIH phosphorylating the C-terminal domain (55). However, this model has recently been challenged by the finding that XPB helicase mutants are functional (56). Interestingly, inhibition of XPB ATPase blocks pol II transcription efficiently, whereas depletion of XPB has virtually no effect (56). These seemingly contradictory observations may be reconciled by a model where XPB exerts its action by acting as a molecular wrench, rotating the DNA downstream of the transcriptional initiation site, combined with a translocase function, whereby it pumps DNA into the active cleft of RNA pol II, partially opening the double-stranded DNA at the initiation site (57). The findings that blocking the ATPase function inhibits transcription, whereas XPB depletion does not, may be explained if the XPB ATPase function is required to relieve a block imposed by the presence of XPB itself, but pol II transcription tolerates the absence of XPB. Thus, removal of XPB altogether allows transcription to proceed, but inhibiting the ATPase function imposes a barrier to transcription/elongation.
The finding that TPL, which inhibits the XPB ATPase, globally inhibits EBV transcription suggests that XPB plays a similar role at EBV promoters and cellular promoters, with the exception of SM-dependent promoters that always require XPB. Understanding the involvement of TFIIH in beta- and gammaherpesvirus transcription is complicated by the fact that late gene transcription is carried out by a virally encoded PIC of six proteins, including a viral TBP, rather than the host cell PIC (8–10, 58). While it is possible that due to these differences, EBV late gene promoters have a particular requirement for TFIIH, and specifically XPB, this is unlikely since depletion of XPB with SPR or by siRNA only affected SM-dependent late promoters. These findings therefore suggest that SM-dependent promoters may uniquely require the function of XPB to allow promoter opening and escape. A model based on the local structure of the viral chromatin at SM-dependent promoters can be envisioned to explain the unique requirement of SM-dependent promoters for XPB, invoking a role for either the helicase or translocase function of XPB. We postulate that SM-dependent promoters possess a structure that is more resistant to melting or opening than other EBV late promoters, although they are both bound by the viral PIC. Such refractoriness to opening in the absence of XPB is unlikely to be due simply to local sequence variation, as the DNA sequences in the vicinity of SM-dependent promoters are not obviously different from SM-independent promoters in terms of GC content (14). The requirement for TFIIH in pol II transcription can, however, be affected by the degree of template supercoiling (59). Local supercoiling may be variable along the latent EBV genome, which is chromatinized and bound by chromatin conformation-modifying proteins, such as CTCF and cohesin (60). Further investigation directed at elucidating the requirements for open complex formation at SM and XPB-dependent EBV promoters will be informative in this regard.
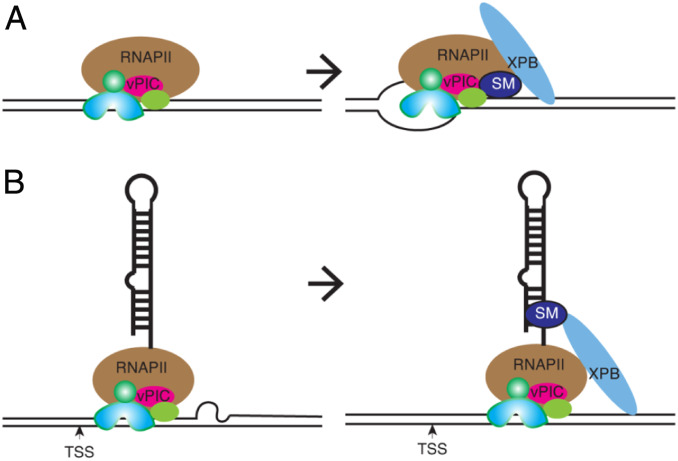
The involvement of SM in transcriptional activation was surprising since we have been unable to detect any direct interaction of SM with the EBV genome in EBV-infected cells by ChIP assay (14). SM was, however, shown to bind to a highly repetitive site on chromosome 11 of the human genome in the same cells, suggesting that SM, while capable of binding to certain DNA sequences and RNA, does not directly interact with EBV DNA with high avidity. Nevertheless, SM could be bound by XPB and the presence of SM in a multiprotein complex with XPB, and the viral PIC, could increase affinity of the complex for EBV promoters (Fig. 8A). Increased XPB recruitment would then promote open-complex formation and transcription initiation as shown.
Fig. 8.
Models for the role of XPB in SM-dependent transcription. (A) Assembly of viral preinitiation complex at an SM-dependent late EBV promoter without open-complex formation (Left). XPB and SM cooperate in forming an active structure that enhances open-complex formation necessary for transcriptional initiation (Right). (B) Transcription initiation occurs but is blocked prior to transition to elongation (Left). SM binds to nascent RNA and recruits XPB to facilitate transition to early elongation (Right). XPB is shown in isolation for simplicity but may be recruited to promoters individually or as a component of TFIIH.
Alternatively, based on the known ability of SM to interact broadly with EBV mRNAs, SM may bind to the 5′ terminus of nascent RNAs and enhance XPB recruitment to the promoter (Fig. 8B). XPB, in addition to facilitating transcriptional initiation, is also involved in permitting promoter escape of pol II (61). Thus, SM, by recruiting XPB to sites where transcription has been initiated, would promote RNA pol II escape, which also requires the ATPase-dependent functions of XPB (56). In this model, EBV mRNA binding by SM is required, although the interaction with XPB itself is not RNA-dependent, consistent with our IP experiments. XPB in this scenario, enhances early steps in the transition to elongation. We consider this alternative scenario less likely, since XPB depletion led to an inability to detect even short nascent SM-dependent gene transcripts. However, a very early elongation block (<40 nt) remains possible.
In summary, we find that EBV has co-opted the cellular transcription factor XPB to facilitate transcription of specific lytic genes that are essential for late steps in virion assembly and for infectivity. The EBV SM protein has evolved to bind XPB and recruit it to EBV lytic promoters. Although general pol II transcription can proceed in the absence of XPB, SM-dependent promoters cannot, and require XPB activity. EBV may therefore utilize XPB to overcome transcriptional hurdles unique to a subset of its promoters, allowing EBV SM to activate those promoters specifically, adding another layer of transcriptional regulation with which to fine-tune late gene expression.
Materials and Methods
Cell Cultures.
Gastric carcinoma cell line AGS was infected with GFP-expressing EBV Akata BX1 virus (62). AGSiZ was derived from AGS-BX1 by stably transducing with a lentivirus expressing doxycycline-inducible EBV lytic transactivator protein Zta that induces lytic replication and infectious virion production (36). AGSiZ cells were cultured in Ham’s F-12 medium with 10% fetal bovine serum (FBS) (tetracycline system-approved FBS, Clontech #631106), 1% GlutaMAX (Life Technologies), 0.5 mg/mL neomycin, and 0.5 μg/mL puromycin. HEK293 cells carrying an SM-null EBV 2089 bacmid with the SM gene deleted by insertion of a kanamycin-resistance gene (EBV BMLF1 KO) (19), referred to as SMKO EBV, were cultured in DMEM with 10% FBS, 1% GlutaMAX, and 100 μg/mL hygromycin. Transfection of 293 SMKO cells was performed with TransIT293 reagent (Mirus Bio) and 1 μg of total DNA in a six-well plate according to the manufacturer’s protocol. P3HR1-ZHT and 3BLCL-ZHT are an EBV+ Burkitt lymphoma and an EBV-transformed lymphoblastoid B cell line, respectively. P3HR1-ZHT contains the EBV BZLF1 gene fused to the hormone-binding domain of the 4-hydroxytamoxifen (4HT) receptor that allows robust induction of EBV replication upon addition of 4HT to the growth medium (63), and the 3BLCL-ZHT cell line was generated similarly (64). Raji is an EBV+ human Burkitt’s lymphoma cell line (65). All B cell lines were grown in RPMI medium 1640 supplemented with 10% FBS and 1% GlutaMAX.
Depletion of Cellular XPB and EBV SM by siRNA Transfection.
XPB KD was performed with cellular XPB SMARTpool: ON-TARGETplus XPBsi (L-011028-00, Dharmacon). EBV SM siRNA sequences used in this study were designed using an RNAi design tool (IDT) and synthesized by IDT. Sequences of a pool of three siRNAs used for EBV SM KD were as follows. The sense strand of each siRNA is shown followed by the antisense strand. The last two nucleotides of each sense strand were DNA. 5′CACUACAUCAAGAAUUACAACCCTG3′; 5′CAGGGUUGUAAUUCUUGAUGUAGUGGC3′; 5′GAAGCAACUCUUCUACAUCACCUGT3′; 5′ACAGGUGAUGUAGAAGAGUUGCUUCAC3′; 5′CUACGUGAGUUUUUCACCAAGUCAA3′; 5′UUGACUUGGUGAAAAACUCACGUAGUG3′.
KD in AGSiZ cells was performed by transfecting specific siRNAs targeting XPB, SM, or negative control siRNA (D-001210-03-05, Dharmacon) with Lipofectamine RNAiMAX transfection reagent (Invitrogen,13778150) according to the manufacturer’s protocols. For XPB depletion, lytic reactivation was induced 48 h after siRNA transfection.
Quantification of Infectious Virus Production.
For quantification of infectious virus production, 250,000 AGSiZ cells were plated in six-well plates 1 d before induction of replication. Cells were treated with doxycycline (0.5 μg/mL) to induce lytic replication and cells were either mock-treated or treated with SPR or EPR at a concentration of 10 μM. Five days after induction, cell supernatants were collected, passed through 0.8-μm cellulose acetate filters, and serial dilutions of supernatants were used to infect 293 cells. The infected 293 cells turn green 2 d postinfection, representing infectious viral particles, which were visualized under fluorescent microscopy and quantitated by flow cytometry (8, 15, 41).
For virus production in LCL-ZHT, 2 million cells were induced with the addition of 100 nM 4HT, 20 ng/mL TPA (12-O-tetradecanoylphorbol-13-acetate), and 1 mM sodium butyrate. Cells were either mock-treated or treated with SPR at the time of induction. The next day, growth medium was replaced with medium containing 10 μM SPR or vehicle for 5 d. Cell supernatants were collected and stored at 4 °C. The cell pellets were lysed by freezing and thawing on dry ice three times, mixed with cell supernatant, and filtered through 0.8-μm cellulose acetate filters. Next, 250 μL of filtered supernatant was used to infect Raji cells. Virus titer was measured and quantitated in infected cells by flow cytometry (66).
Induction Reagents and Compounds.
Doxycycline (D9891), 4HT (H6278), TPA (P8139), SPR (S3378), EPR (E6657), and TPL (T3652) were purchased from Sigma.
Western Blotting and IP.
For Western blotting and IP, 250,000 AGSiZ cells were plated on six-well plate 1 d before induction. Cells were treated with doxycycline to induce EBV lytic induction and either mock-treated or treated with SPR, as indicated. Whole-cell lysates were prepared 48 h after EBV lytic induction and analyzed by Western blotting with anti-Zta (Argene,11-007), anti-Rta (Argene,11-008), anti-SM, anti-EAD (Capricorn,18-48180), anti-Ubiquitin (PA1-187), anti-Tubulin (Sigma, SAB3501072), and anti-XPB (Millipore, MABE1123) antibodies. The signal was visualized by incubation with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody using Bio-Rad chemiluminescence detection reagent.
EBV SM and XPB IP were performed in AGSiZ cells. EBV lytic reactivation in AGSiZ cells was induced by doxycycline treatment and cells were either mock-treated or treated with SPR at the time of induction. Forty-eight hours after lytic induction, cells were lysed and clarified by centrifugation as described previously (53). Supernatant was precleared by incubation with 5 μg of rabbit IgG (Bethyl, P120-101) for 1 h followed by incubation with Protein A agarose beads. IP was performed at 4 °C overnight by incubating with either control antibody (rabbit IgG) or SM-specific serum. Immunocomplexes were captured by incubating with Protein A agarose beads (Invitrogen,15918014) for 2 h at 4 °C. Beads were washed two times with wash buffer (1× TBS, 1% Nonidet P-40) and RNase treatment was performed by incubating with 100 μg/mL of RNase A for 30 min at 37 °C. Beads were washed twice in wash buffer and eluted in sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) buffer by boiling at 95 °C for 5 min.
XPB or ubiquitin IP were performed in P3HR1-ZHT cells. Cells were treated with 10 μM MG132 for 2 h before SPR treatment. Cells were harvested 4 h after SPR treatment, lysed as described above, and immunoprecipitated overnight at 4 °C with either XPB or ubiquitin polyclonal antibodies. Immunocomplexes were captured by incubating with Protein A agarose beads for 2 h at 4 °C. Beads were washed twice in 1× TBS buffer and the complexes were eluted in SDS/PAGE buffer.
Immunofluorescence Microscopy.
For immunofluorescence analysis, AGSiZ cells were plated on coverslips in six-well plates. Lytic replication was induced with doxycycline and cells were either mock-treated or treated with SPR the following day. Forty-eight hours after lytic induction, cells were incubated with 2 mM 5-BrU for 30 min at 37 °C. Pulsed cells were washed three times with phosphate-buffered saline (PBS) to remove unincorporated BrU. Washed cells were fixed in 4% paraformaldehyde and stored at 4 °C. Cells were permeabilized with 0.1% Triton X-100 and incubated with 20% goat serum and anti-BrU antibody (BD Pharmingen, 555627). Cells were washed three times with PBS and incubated with Alexa Fluor 594 conjugated goat anti-mouse IgG antibody for 1 h. After three washes, nuclei were stained by incubating with ProLong Gold antifade reagent with DAPI (Molecular Probes, P36935). Mounted slides were imaged using a Zeiss Axio Imager M2 microscope.
RNA Isolation and qRT-PCR.
Cells were harvested 48 h after lytic induction and RNA was isolated using a miRNeasy kit followed by on column DNase treatment (Qiagen). qRT-PCR was performed with RNA-to-Ct SYBR green PCR mix (Applied Biosystems, 4389986) using a StepOne Plus real-time PCR thermocycler (ABI). Next, 50 ng of RNA was used in each reaction with gene-specific primers and relative quantity (RQ) was calculated using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as an endogenous control, as reported previously (14). Each PCR was performed in triplicate and RQs were calculated from three biological replicates. The gene-specific primers used in this study are provided in SI Appendix, Table S1.
Measuring Transcription Using 5-BrU Incorporation and IP.
AGSiZ cells were plated on a 100-mm cell culture dish followed by lytic reactivation with doxycycline the following day. Cells were also either mock-treated or treated with SPR. Forty-eight hours after lytic induction, cells were incubated with 2 mM 5-BrU (Alfa Aesar, AAA1850701) for 30 min at 37 °C. As indicated, AGSiZ cells were treated with 5 μg/mL actinomycin. After BrU labeling, cells were washed three times with PBS to remove unincorporated BrU and lysed in 700 μL Qiazol reagent. RNA was isolated using a Qiagen miRNeasy kit with on column DNase digestion and the BrU labeled RNAs were immunoprecipitated using Dynabeads Protein G (Invitrogen,10004D) and anti-BrdU antibody (BD Pharmingen, 555627), as previously reported (67). Immunoprecipitated EBV and cellular gene transcripts were measured by qRT-PCR using gene-specific primers, as described above.
Transcription assays in SMKO 293 cells were performed as described above, except induction of lytic replication in SMKO cells was performed by transfection with Z plasmid DNA. Mock induction was performed by transfection of empty vector and SM rescue by transfection of EBV SM plasmid in addition to Z plasmid. For TPL treatment, AGSiZ cells were plated on a 100-mm cell culture dish, lytic replication was induced with doxycycline the following day, and cells were either mock-treated or treated with SPR. For TPL experiments, cells were treated with 125 nM TPL for 1 h 48 h after lytic induction and pulsed with 2 mM 5-BrU for 30 min at 37 °C. Cells were harvested and transcription was measured by a BrU incorporation and qRT-PCR assay, as described above.
Luciferase Reporter Assay.
AGSiZ Akata EBV cells were seeded at 250,000 cells in six-well plates and grown overnight. The following day, KD was performed by transfection with siRNA targeting SM or with negative control siRNA in triplicate with Lipofectamine RNAiMAX. Three hours later, the control siRNA and the SM siRNA transfected cells were treated with doxycycline to induce lytic replication or mock-treated. Twenty-four hours later, each promoter luciferase construct was transfected using Mirus Transit-293 reagent and cells were harvested 24 h after transfection and were lysed in reporter lysis buffer (Promega). Effects of SPR on promoter function were measured similarly except that cells were treated at the time of lytic induction with either 10 μM SPR or vehicle. Twenty-four hours after drug treatment and lytic induction, cells were transfected with each promoter luciferase construct as above. Luciferase activity was measured in biological triplicate transfections with technical triplicates using firefly luciferase reagent (Promega) and a Turner Biosystems 20/20 luminometer. Data were normalized to luciferase activity in uninduced control siRNA transfected cells and shown as the RQ. Error bars represent the SEM. Aliquots of each transection were harvested in protein loading buffer in order to check for efficient knockdown by Western blot.
Plasmids.
DNA fragments extending 500 bp upstream and 100 bp downstream of the initiator ATG of two SM-dependent late (BcLF1 and BDLF1), one SM-independent late (BGLF1), and one SM-independent early (BALF2) gene were generated by PCR amplification. PCR products were fused to the luciferase gene by ligation between BglII to NcoI sites in frame with the start site in the luciferase gene in pGL3-basic plasmid (Promega). The oriLyt sequence (nucleotide 53449 to 54441 in the EBV genome GenBank: V01555.2) was amplified from B95-8 EBV and cloned in cis to the promoter–luciferase construct. Plasmid constructs were verified by restriction analysis and sequencing.
ChIP Assay.
The ChIP assay was performed as described previously with slight modifications (41). Briefly, 5 million SMKO cells were harvested and cross-linking of proteins to DNA was performed in 1% methanol-free formaldehyde (Thermo Fisher, 28906) for 10 min at room temperature. Cross-linking was stopped by addition of 128 mM glycine. The fixed cells were washed three times with cold PBS with protease inhibitors (Sigma, P8340) and cell pellets were resuspended in 500 μL of ice-cold swelling buffer (5 mM Pipes pH 8.0, 85 mM KCl, 0.5% Nonidet P-40) for 10 min on ice. Cell nuclei were pelleted and resuspended in 100 μL SDS lysis buffer (10 mM EDTA, 50 mM Tris⋅HCl pH 8.0, and 1% SDS) with protease inhibitors for 15 min on ice. Lysed nuclei were diluted 1:10 in ChIP dilution buffer (16.7 mM Tris⋅HCl, pH 8, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, and 1% Triton X-100). Chromatin shearing was performed using a Branson Sonifier 450 to produce DNA fragments ∼400 bp in size. Cleared supernatants were incubated with 5 μg of rabbit polyclonal IgG (Bethyl, P120-101) and 50 μL of 50% Protein-A agarose beads slurry to preclear supernatants for 2 h at 4 °C. The beads were pelleted by quick centrifugation, and 10% of cleared supernatants was reserved to measure input amounts. The remaining supernatant was divided into two tubes and either 5 μg of XPB polyclonal antibody (Novus, NB100-61060) or control antibody was added. 50 μL of 50% Dynabeads Protein G (Invitrogen, 10004D) were added and the slurry was washed three times with PBS containing 0.1% BSA, and incubated at 4 °C overnight on a rocker. Beads were magnetically separated for 5 min and were washed three times with ice-cold low salt buffer (20 mM Tris⋅HCl pH 8.1, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100) and once with high salt buffer (20 mM Tris⋅HCl pH 8.1, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100) and once with TE. Protein–antibody complexes were eluted twice at 65 °C for 15 min on a Thermomixer R (Eppendorf) with 250 μL of freshly prepared elution buffer (0.84% NaHCO3, 1% SDS). Sodium chloride was added to 500 μL of eluted samples and to input samples to a final concentration of 200 mM NaCl and cross-linking was reversed by incubating samples at 65 °C for 4 h. All samples were sequentially treated with RNase A and Proteinase K, and DNA was purified using a PCR purification kit (Qiagen, 28106). qPCR was performed with diluted DNA-to-SYBR green PCR mix (Applied Biosystems, 4309155) using a StepOne Plus real-time PCR thermocycler (ABI) using primers provided in SI Appendix, Table S2. Each PCR was performed in technical triplicates using purified immunoprecipitated DNA and fold-enrichment was calculated relative to IgG control IPs. RQs were calculated from three biological replicates.
Statistical Analysis.
Data for qPCR and ChIP assays were analyzed using GraphPad Prism 8 software. The error bars indicate the SEM from three different biological replicates. P values were calculated using unpaired t tests and are shown in each figure.
Materials and Data Availability.
Relevant data are provided in the main text and SI Appendix.
Supplementary Material
Acknowledgments
This work was funded by Public Health Service Grant R01 CA081133 (to S.S.) from the National Cancer Institute. D.V. is also supported by a Department of Internal Medicine Academic Seed Grant, University of Utah. Flow cytometry and DNA sequencing were performed at Health Sciences Core Facilities, University of Utah.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000625117/-/DCSupplemental.
References
- 1.Khan G., Hashim M. J., Global burden of deaths from Epstein-Barr virus attributable malignancies 1990-2010. Infect. Agent. Cancer 9, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi I. et al., Prevalence of Epstein-Barr virus in oral squamous cell carcinoma. J. Pathol. 189, 34–39 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Murata T., Tsurumi T., Switching of EBV cycles between latent and lytic states. Rev. Med. Virol. 24, 142–153 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Rickinson A., Kieff E., ““Epstein Barr virus”” in Fields Virology, Knipe D., Howley P., Eds. (Lippincott Williams, Philadelphia, 2001), Vol. 2, pp. 2575–2627. [Google Scholar]
- 5.Hong G. K. et al., Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 79, 13993–14003 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiorini S., Ooka T., Secretion of Epstein-Barr virus-encoded BARF1 oncoprotein from latently infected B cells. Virol. J. 5, 70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang C. Y. et al., Recurrent chemical reactivations of EBV promotes genome instability and enhances tumor progression of nasopharyngeal carcinoma cells. Int. J. Cancer 124, 2016–2025 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Li D., Fu W., Swaminathan S., Continuous DNA replication is required for late gene transcription and maintenance of replication compartments in gammaherpesviruses. PLoS Pathog. 14, e1007070 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruffat H., Kadjouf F., Mariamé B., Manet E., The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late gene expression. J. Virol. 86, 6023–6032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aubry V. et al., Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J. Virol. 88, 12825–12838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djavadian R., Chiu Y. F., Johannsen E., An Epstein-Barr virus-encoded protein complex requires an origin of lytic replication in cis to mediate late gene transcription. PLoS Pathog. 12, e1005718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenney S. et al., The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J. Virol. 63, 1729–1736 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragoczy T., Heston L., Miller G., The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72, 7978–7984 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J., Verma D., Li D., Mosbruger T., Swaminathan S., Identification and characterization of the physiological gene targets of the essential lytic replicative epstein-barr virus SM protein. J. Virol. 90, 1206–1221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Z. et al., Multiple roles of Epstein-Barr virus SM protein in lytic replication. J. Virol. 81, 4058–4069 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batisse J., Manet E., Middeldorp J., Sergeant A., Gruffat H., Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J. Virol. 79, 14102–14111 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaminathan S., “Post-transcriptional gene regulation by EBV SM protein” in Epstein-Barr Virus, Robertson E., Ed. (Caister Press, 2005), chap. 29, pp. 631–650. [Google Scholar]
- 18.Swaminathan S., Kenney S., “The Epstein-Barr virus lytic lifecycle” in DNA Tumor Viruses, Damania B., Pipas J., Eds. (Springer, 2009), chap. 13. [Google Scholar]
- 19.Gruffat H. et al., Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76, 9635–9644 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruvolo V., Wang E., Boyle S., Swaminathan S., The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. U.S.A. 95, 8852–8857 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiriart E. et al., A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278, 335–342 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Farjot G. et al., Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74, 6068–6076 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiriart E. et al., Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 280, 36935–36945 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Juillard F. et al., Epstein-Barr virus protein EB2 stimulates cytoplasmic mRNA accumulation by counteracting the deleterious effects of SRp20 on viral mRNAs. Nucleic Acids Res. 40, 6834–6849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juillard F. et al., Epstein-Barr virus protein EB2 contains an N-terminal transferable nuclear export signal that promotes nucleocytoplasmic export by directly binding TAP/NXF1. J. Virol. 83, 12759–12768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle S. M., Ruvolo V., Gupta A. K., Swaminathan S., Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J. Virol. 73, 6872–6881 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D. J., Verma D., Swaminathan S., Binding of cellular export factor REF/Aly by Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF57 protein is not required for efficient KSHV lytic replication. J. Virol. 86, 9866–9874 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma D., Swaminathan S., Epstein-Barr virus SM protein functions as an alternative splicing factor. J. Virol. 82, 7180–7188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semmes O. J. et al., Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72, 9526–9534 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt C. et al., ORF57 overcomes the detrimental sequence bias of Kaposi’s sarcoma-associated herpesvirus lytic genes. J. Virol. 89, 5097–5109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilello J., Morgan J., Desrosiers R., Extreme dependence of gH and gL expression on ORF57 and association with highly unusual codon usage in rhesus monkey rhadinovirus. J. Virol. 82, 7231–7237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massimelli M. et al., Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5’ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 7, 1145–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenney S. et al., The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J. Virol. 63, 3870–3877 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicewonger J., Suck G., Bloch D., Swaminathan S., Epstein-Barr virus (EBV) SM protein induces and recruits cellular Sp110b to stabilize mRNAs and enhance EBV lytic gene expression. J. Virol. 78, 9412–9422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrario C. M., Schiffrin E. L., Role of mineralocorticoid receptor antagonists in cardiovascular disease. Circ. Res. 116, 206–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma D., Thompson J., Swaminathan S., Spironolactone blocks Epstein-Barr virus production by inhibiting EBV SM protein function. Proc. Natl. Acad. Sci. U.S.A. 113, 3609–3614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alekseev S. et al., A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chem. Biol. 21, 398–407 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Ranish J. A. et al., Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat. Genet. 36, 707–713 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Giglia-Mari G. et al., A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36, 714–719 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Egly J. M., The 14th Datta Lecture. TFIIH: From transcription to clinic. FEBS Lett. 498, 124–128 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Li D. J., Verma D., Mosbruger T., Swaminathan S., CTCF and Rad21 act as host cell restriction factors for Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. + Pathog. 10, e1003880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elinoff J. M. et al., Spironolactone-induced degradation of the TFIIH core complex XPB subunit suppresses NF-κB and AP-1 signalling. Cardiovasc. Res. 114, 65–76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacombe B., Morel M., Margottin-Goguet F., Ramirez B. C., Specific inhibition of HIV infection by the action of spironolactone in T cells. J. Virol. 90, 10972–10980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuda A., Nogi Y., Hisatake K., The regulatory role for the ERCC3 helicase of general transcription factor TFIIH during promoter escape in transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 99, 1206–1211 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreland R. J. et al., A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J. Biol. Chem. 274, 22127–22130 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Ruvolo V., Gupta A. K., Swaminathan S., Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 75, 6033–6041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Z., Verma D., Hilscher C., Dittmer D. P., Swaminathan S., General and target-specific RNA binding properties of Epstein-Barr virus SM posttranscriptional regulatory protein. J. Virol. 83, 11635–11644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiriart E. et al., A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278, 37790–37798 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Amon W. et al., Lytic cycle gene regulation of Epstein-Barr virus. J. Virol. 78, 13460–13469 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Titov D. V. et al., XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutt-Fletcher L. M., The long and complicated relationship between Epstein-Barr virus and epithelial cells. J. Virol. 91, e01677-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H. et al., Epstein-Barr virus lytic reactivation regulation and its pathogenic role in carcinogenesis. Int. J. Biol. Sci. 12, 1309–1318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma D., Bais S., Gaillard M., Swaminathan S., Epstein-Barr virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J. Virol. 84, 11781–11789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kandiah E., Trowitzsch S., Gupta K., Haffke M., Berger I., More pieces to the puzzle: Recent structural insights into class II transcription initiation. Curr. Opin. Struct. Biol. 24, 91–97 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Ohkuma Y., Roeder R. G., Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature 368, 160–163 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Alekseev S. et al., Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol. Cell 65, 504–514.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Kim T. K., Ebright R. H., Reinberg D., Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Isomura H. et al., The human cytomegalovirus gene products essential for late viral gene expression assemble into prereplication complexes before viral DNA replication. J. Virol. 85, 6629–6644 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parvin J. D., Sharp P. A., DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 73, 533–540 (1993). [DOI] [PubMed] [Google Scholar]
- 60.Holdorf M. M., Cooper S. B., Yamamoto K. R., Miranda J. J., Occupancy of chromatin organizers in the Epstein-Barr virus genome. Virology 415, 1–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spangler L., Wang X., Conaway J. W., Conaway R. C., Dvir A., TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 5544–5549 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molesworth S. J., Lake C. M., Borza C. M., Turk S. M., Hutt-Fletcher L. M., Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74, 6324–6332 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verma D., Ling C., Johannsen E., Nagaraja T., Swaminathan S., Negative autoregulation of Epstein-Barr virus (EBV) replicative gene expression by EBV SM protein. J. Virol. 83, 8041–8050 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johannsen E. et al., Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 101, 16286–16291 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pulvertaft J. V., A study of malignant tumours in Nigeria by short-term tissue culture. J. Clin. Pathol. 18, 261–273 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Church T. M., Verma D., Thompson J., Swaminathan S., Efficient translation of Epstein-Barr virus (EBV) DNA polymerase contributes to the enhanced lytic replication phenotype of M81 EBV. J. Virol. 92, e01794-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paulsen M. T. et al., Use of Bru-seq and BruChase-seq for genome-wide assessment of the synthesis and stability of RNA. Methods 67, 45–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.