Significance
CRC patients with KRAS mutations are confronted with limited targeted therapeutic options. In this study, we have shown that the median survival time for KRAS-mutation mCRC patients with diabetes on metformin is 37.8 mo longer than those treated with other hypoglycemic drugs in combination with standard systemic therapy. Metformin is preferentially accumulated in KRAS-mutation CRC cells in both primary cell cultures and patient-derived xenografts. The promising therapeutic activity of metformin has a negative correlation with MATE1 expression, which is proven to eliminate metformin from CRC cells. These findings indicate that KRAS-mutation mCRC patients could benefit from metformin treatment, and somatic KRAS status or MATE1 expression should be recommended to predict the therapeutic response of metformin in CRC.
Keywords: metastatic colorectal cancer, KRAS mutation, metformin, multidrug and toxic compound extrusion 1 (MATE1), DNA methyltransferase 1 (DNMT1)
Abstract
Metastatic colorectal cancer (mCRC) patients have poor overall survival despite using irinotecan- or oxaliplatin-based chemotherapy combined with anti-EGFR (epidermal growth factor receptor) drugs, especially those with the oncogene mutation of KRAS. Metformin has been reported as a potentially novel antitumor agent in many experiments, but its therapeutic activity is discrepant and controversial so far. Inspiringly, the median survival time for KRAS-mutation mCRC patients with diabetes on metformin is 37.8 mo longer than those treated with other hypoglycemic drugs in combination with standard systemic therapy. In contrast, metformin could not improve the survival of mCRC patients with wild-type KRAS. Interestingly, metformin is preferentially accumulated in KRAS-mutation mCRC cells, but not wild-type ones, in both primary cell cultures and patient-derived xenografts, which is in agreement with its tremendous effect in KRAS-mutation mCRC. Mechanistically, the mutated KRAS oncoprotein hypermethylates and silences the expression of multidrug and toxic compound extrusion 1 (MATE1), a specific pump that expels metformin from the tumor cells by up-regulating DNA methyltransferase 1 (DNMT1). Our findings provide evidence that KRAS-mutation mCRC patients benefit from metformin treatment and targeting MATE1 may provide a strategy to improve the anticancer response of metformin.
Colorectal cancer (CRC) is the third most common cancer and the leading cause of cancer death worldwide, and there is an increasing incidence of tumor metastasis before diagnosis, especially among young patients (1). For patients with metastatic colorectal cancer (mCRC), irinotecan- or oxaliplatin-based first-line chemotherapy combined with epidermal growth factor receptor (EGFR)-targeted drugs were found to have improved median overall survival to more than 2 y recently (2). However, CRC was perceived to be a genomic heterogeneous disease, in which genetic alterations of APC, KRAS, TP53, BRAF, PIK3CA, and so forth promote tumorigenesis, tumor metastasis, and drug resistance (3). To date, ∼30 to 50% of CRC patients are diagnosed with somatic KRAS mutation in exon 2, which is an established predictive biomarker of resistance to anti-EGFR drugs (4, 5). In addition, farnesyltransferase inhibitors or geranylgeranyltransferase inhibitors, which disrupt RAS posttranslational modification, also have had limited clinical efficacy in CRC clinical trials (6, 7). Thus, CRC patients with KRAS mutations are excluded from anti-EGFR therapy and confronted with limited novel targeted therapeutic options.
The biguanide metformin is one of the front-line hypoglycemic agents in the management of type 2 diabetes mellitus (T2DM) through lowering glucose levels and improving insulin sensitivity. Recent clinical (8, 9) and preclinical (10) studies have demonstrated that metformin has direct anticancer effects, which underlie pleiotropic mechanisms including AMP-activated protein kinase activation, the mammalian target of rapamycin (mTOR) inactivation, mitogen-activated protein kinase kinase 1 (MEK)/extracellular signal-regulated kinase (ERK), and the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway inhibition (11). However, there is also growing evidence indicating that metformin cannot improve the outcome of CRC patients (12, 13). Hence, the therapeutic activity of metformin is still controversial. In particular, clinical evidence or an underlying mechanism of the therapeutic activity of metformin use in KRAS-mutation mCRC is still absent. Of note, the alteration of cellular function is metformin concentration-relevant (14). Chowdhury et al. (15) have demonstrated that organic cation transporter 1 (OCT1) and multidrug and toxic compound extrusion 2 (MATE2), which mediate metformin absorption and elimination, respectively, are associated with the response to metformin in certain cancer cells in vitro.
Here, we aimed to identify the subgroup of mCRC patients who are responsive to metformin and to investigate the mechanisms determining metformin sensitivity.
Results
KRAS Mutation Determines Metformin Sensitivity in mCRC Patients.
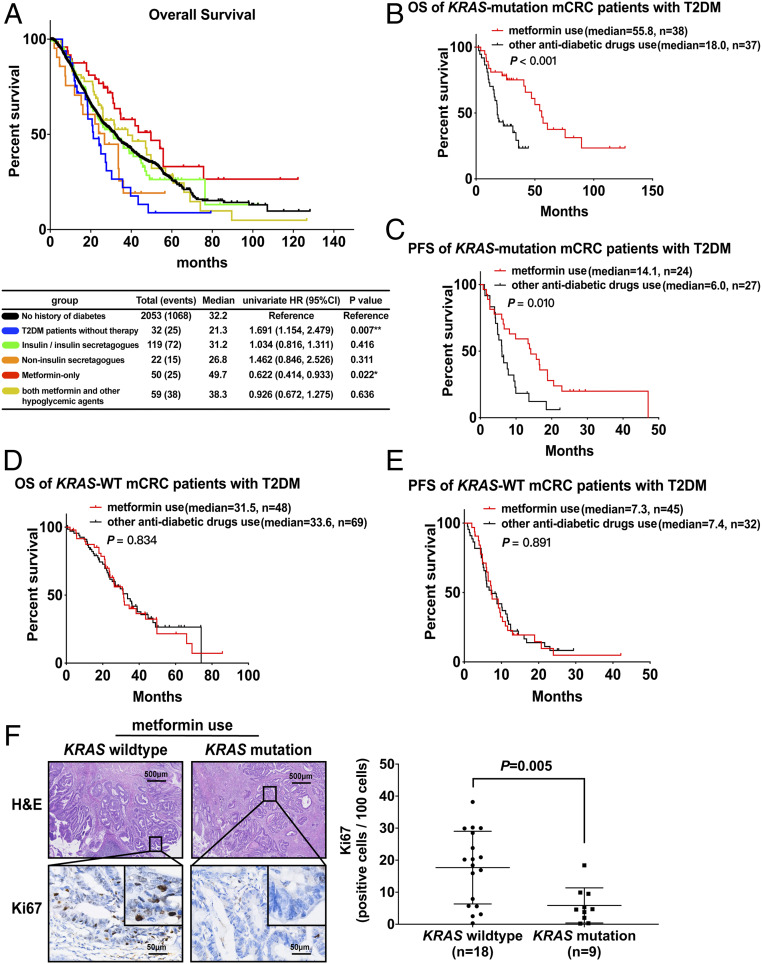
Most current studies have focused on the anticancer effect of metformin, but information is still limited as to the association between anticancer activity and personal characteristics. Here, we retrospectively studied the medical records of 282 T2DM patients in a 2,335-mCRC patient population, who were divided into a nonuser group and user group according to hypoglycemic agent use. The distribution of sex, age, body mass index (BMI), primary tumor location, metastatic sites, pathological grading, and KRAS genotype was nearly identical in these two groups (SI Appendix, Table S1). Further, Kaplan–Meier analysis in Fig. 1A shows that diabetic mCRC patients without hypoglycemic treatment had poor outcomes compared with mCRC patients without diabetes in overall survival (OS) (hazard ratio [HR], 1.691; 95% CI, 1.154 to 2.479) and hypoglycemic therapy could improve OS in diabetic mCRC patients. Unexpectedly, the OS of the metformin-only use group was even 17.5 mo longer than that of mCRC patients without diabetes (HR, 0.622; 95% CI, 0.414 to 0.933). Further, metformin users had ameliorative OS (metformin-only: HR, 0.356; 95% CI, 0.183 to 0.693; both metformin and other hypoglycemic agents: HR, 0.491; 95% CI, 0.269 to 0.894) after being adjusted for personal characteristics (SI Appendix, Table S2). Moreover, we found that nonmetformin users, including insulin, sulfonylureas, nonsulfonylureas, α-glucosidase inhibitors, and thiazolidinediones, had no improved outcome in either Kaplan–Meier analysis compared with mCRC patients without diabetes or Cox analysis compared with diabetic mCRC patients without hypoglycemic treatment. Considering results that indicated that the anticancer effect of metformin on mCRC and diabetes might not be attributable to its hypoglycemic effect, the underlying mechanism is worth studying.
Fig. 1.
KRAS mutation enhances the antitumor activity of metformin in CRC patients. (A) The overall survival analysis was conducted by the Kaplan–Meier method after dividing the mCRC patients into non-DM patients, T2DM metformin-use group, and other antidiabetic drug-use group. (B and C) The overall survival (B) and progression-free survival of first-line chemotherapy (C) analysis were conducted by the Kaplan–Meier method after dividing the KRAS-mutation mCRC and T2DM patients into a metformin-use group and other antidiabetic drug-use group. (D and E) The overall survival (D) and progression-free survival of first-line chemotherapy (E) analysis of the KRAS wild-type subgroup were conducted with T2DM and mCRC patients in a metformin-use group and other antidiabetic drug-use group. (F) Representative images of hematoxylin and eosin (H&E) and Ki67 immunohistochemistry on cross-sections from patients with mCRC with T2DM and metformin use (Left) and the proportion of Ki67-positive cells per 100 cells (Right) are shown. Data are shown as mean ± SEM. P = 0.005 was compared with the KRAS wild-type group. P values were determined by unpaired two-tailed Student’s t test.
Therefore, we established nonmetformin users as a critical control for metformin users, and further explored the key factors which impact the anticancer effect of metformin using hierarchical Cox proportional hazard (PH) analysis (SI Appendix, Fig. S1A). The distribution of clinical characteristics (e.g., sex, age, BMI, primary tumor location, metastatic sites, pathological grading, and KRAS genotype) was nearly identical in these two groups, and the listed characteristics satisfied the PH assumption (SI Appendix, Table S3; P > 0.05). However, as shown in Table 1, we found metformin use was not associated with OS (HR, 0.746; 95% CI, 0.496 to 1.121) or progression-free survival (PFS) of first chemotherapy (HR, 0.737; 95% CI, 0.501 to 1.086) compared with nonmetformin use. Hence, we hypothesized that the therapeutic activity of metformin might be individually different. When stratifying by KRAS genotype, we observed that the association between metformin use and OS was limited to individuals with the KRAS mutation (HR, 0.272; 95% CI, 0.120 to 0.617; P interaction < 0.001) as well as PFS (HR, 0.405; 95% CI, 0.212 to 0.774; P interaction = 0.02). Moreover, no evidence of interaction between metformin use and other individual characteristics was related to OS or PFS. Then, we also calculated median survival using Kaplan–Meier analysis. The results showed that the OS of KRAS-mutation patients with metformin use was 37.8 mo longer than those treated with other hypoglycemic drugs (P < 0.001), and the median PFS was 8.1 mo longer (P = 0.01) (Fig. 1 B and C). However, metformin use could not improve OS and PFS in KRAS wild-type (WT) patients (Fig. 1 D and E). In addition, we detected proliferating Ki67(+) cells in histological sections from the foci of mCRC patients with metformin use. The immunohistochemistry results showed that the proportion of Ki67(+) cells in KRAS-mutation colorectal cancer was significantly lower than in KRASWT, with a significant statistical difference (P = 0.005), as shown in Fig. 1F. These data suggested that metformin use was more effective for those mCRC patients with the KRAS mutation. One possible reason for the controversial therapeutic activity of metformin in colorectal cancer is that the KRAS mutation is not classified. Thus, our study to some extent explains the inconsistencies in the existing research.
Table 1.
Association between metformin use and OS and PFS, according to the characteristics of mCRC patients with T2DM, SYUCC 2004 to 2016
| Characteristics* | OS | Pinteraction | PFS | Pinteraction | ||
| No. of events† | HR (95% CI) | No. of events‡ | HR (95% CI) | |||
| Metformin use vs. nonmetformin use | 51:68 | 0.746 (0.496, 1.121) | 55:60 | 0.737 (0.501, 1.086) | ||
| Metformin use vs. nonmetformin use after stratifying with | ||||||
| Sex | 0.321 | 0.493 | ||||
| Male | 37:44 | 0.673 (0.401, 1.131) | 42:43 | 0.732 (0.468, 1.146) | ||
| Female | 14:15 | 0.944 (0.385, 2.318) | 13:17 | 0.697 (0.318, 1.529) | ||
| Age at diagnosis, y | 0.697 | 0.054 | ||||
| <60 | 21:19 | 0.869 (0.411, 1.715) | 23:22 | 1.074 (0.580, 1.990) | ||
| ≥60 | 30:40 | 0.596 (0.348, 0.988) | 32:38 | 0.562 (0.339, 0.930) | ||
| BMI, kg/m2 | 0.867 | 0.430 | ||||
| Normal (18.5∼23.0) | 33:33 | 0.582 (0.334, 1.013) | 37:33 | 0.884 (0.542, 1.442) | ||
| Pre & obese (>23.0) | 18:26 | 0.702 (0.357, 1.382) | 18:27 | 0.555 (0.287, 1.073) | ||
| Primary site | 0.444 | 0.426 | ||||
| Right colon | 10:17 | 0.729 (0.294, 1.807) | 8:15 | 0.622 (0.255, 1.517) | ||
| Left colon | 21:24 | 0.982 (0.527, 1.830) | 22:29 | 0.823 (0.451, 1.500) | ||
| Rectum | 20:18 | 0.475 (0.215, 1.051) | 25:16 | 0.544 (0.282, 1.048) | ||
| Metastatic site | — | — | ||||
| Liver | 23:25 | 1.151 (0.563, 2.354) | 23:29 | 0.949 (0.481, 1.871) | ||
| Other organs | 9:4 | — | 10:4 | — | ||
| Distant lymph nodes | 7:5 | — | 4:2 | — | ||
| Multiple sites | 10:12 | 1.008 (0.346, 2.939) | 14:16 | 0.818 (0.329, 2.031) | ||
| Peritoneum | 2:13 | — | 4:9 | — | ||
| Pathological grading | — | — | ||||
| Well-differentiated | 1:0 | — | 0:2 | — | ||
| Moderately differentiated | 33:38 | 0.676 (0.407, 1.125) | 32:35 | 0.644 (0.384, 1.080) | ||
| Poorly & undifferentiated | 17:21 | 0.629 (0.245, 1.616) | 23:23 | 0.811 (0.445, 1.477) | ||
| KRAS genotype | <0.001 | 0.020 | ||||
| Wild type | 32:37 | 1.487 (0.844, 2.620) | 32:39 | 1.194 (0.720, 1.979) | ||
| Mutation | 19:22 | 0.272 (0.120, 0.617) | 23:21 | 0.405 (0.212, 0.774) | ||
Pinteraction, P value for the interaction test. “—” indicates the variable is excluded because of fewer than five observations in this category.
Each characteristic was stratified by age at diagnosis and adjusted for the other variables listed above.
The number of cases of death after diagnosis as mCRC.
The number of cases dead or getting worse during first-line chemotherapy.
KRAS Mutation Determines Metformin Sensitivity in a CRC PDX Animal Model and CRC Cells.
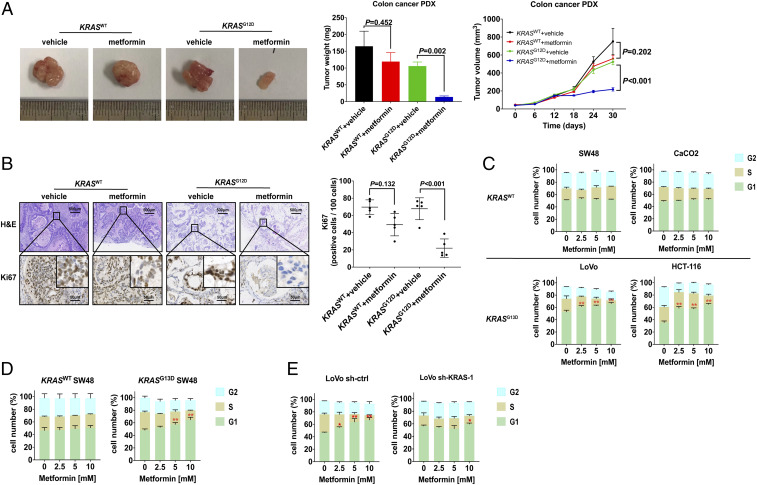
To further validate whether pharmacological metformin is sufficient to mediate antitumor effects in a preclinical model, we generated patient-derived xenografts (PDXs) from a KRASG12D CRC patient and one with KRAS wild type to assess the therapeutic activity of metformin at a pharmacological dose (200 mg/kg mouse weight, equal to 1,000 mg/60 kg human weight per day, orally). While treatment with metformin in KRASWT PDX models had only minor antitumor effects compared with the vehicle control, metformin treatment inhibited KRASG12D tumor growth significantly (Fig. 2A). Moreover, histology of the KRASG12D xenografts showed metformin could inhibit cancer cell proliferation, an effect not seen in KRASWT xenografts (Fig. 2B).
Fig. 2.
KRAS mutation enhances the antitumor activity of metformin in the CRC PDX animal model and CRC cells. (A and B) The representative morphology (A, Left), tumor weight (A, Middle), mean tumor growth rate (A, Right), and representative images of Ki67 immunohistochemistry and their corresponding H&E staining (B, Left) and statistical graph (B, Right) are shown as a result of 30-d treatment with metformin in 374469 KRASWT colon adenocarcinoma and 386650 KRASG12D colon mucinous adenocarcinoma patient-derived xenograft models. Data are shown as mean ± SEM, and differences between metformin and vehicle were analyzed by two-way ANOVA. (C–E) The distribution of G1, S, and G2 phases in KRASWT CRC cell lines SW48 and CaCO2, KRASG13D CRC cell lines LoVo and HCT-116 (C), KRASG13D SW48 established by the CRISPR-Cas9 system (D), and LoVo infected by shRNA lentivirus (E) were detected after treatment with 0, 2.5, 5, and 10 mM metformin for 24 h (n = 3). Data are shown as mean ± SEM. *P < 0.05, **P < 0.01 was compared with 0 mM metformin. All P values were determined by two-way ANOVA.
Next, we determined whether metformin had superior antitumor activity in KRAS-mutated CRC cell lines to those with KRAS wild type through a cell-viability test. As shown in SI Appendix, Fig. S2A, metformin inhibited the cell viability of CRC cells with the KRASG13D mutation (HCT-116 and LoVo) and KRASG12V mutation (SW480 and SW620) in a dose-dependent manner (P < 0.01) but not KRASWT CRC cells (SW48 and CaCO2) (P > 0.05). Moreover, we overexpressed KRASG12D, KRASG12V, and KRASG13D in KRASWT SW48 cells, and the results indicated that both codon 12 and 13 mutations could up-regulate the sensitivity to the antiproliferation therapy of metformin (SI Appendix, Fig. S2B). In addition, this result can be duplicated in the KRASG13D SW48 cell model (P < 0.01; SI Appendix, Fig. S2C), which was established by the CRISPR-Cas9 system. On the contrary, short hairpin RNA (shRNA)-mediated KRAS knockdown impaired the sensitivity to metformin in LoVo cells (SI Appendix, Fig. S2D).
To further determine whether metformin impairs cell viability by apoptosis induction or proliferation inhibition, CRC cells treated with an incremental concentration of metformin were analyzed by flow cytometry following Annexin V-fluorescein isothiocyanate and propidium iodide (PI) dual labeling. However, there was no significant alteration in the number of apoptotic cells (Annexin V+/PI− and Annexin V+/PI+) observed in each CRC cell line (SI Appendix, Fig. S3), while colony-forming inhibition and proliferating (Edu+) cell reduction were observed in KRAS-mutated CRC cell lines (SI Appendix, Fig. S4 A and B). We subsequently detected the cell-cycle distribution by PI monostained (Fig. 2 C–E and SI Appendix, Fig. S5), and it showed that metformin dose-dependently increased the proportions of KRAS-mutation CRC cell lines LoVo and HCT-116 in the sub-G1 phase of PI-stained cells, and the phenomenon did not occur in KRASWT SW48 and CaCO2 (Fig. 2C). Moreover, a G0/G1 arrest was also found in KRASG13D SW48 (Fig. 2D), while the antiproliferation activity was impaired after knocking down the KRAS expression in LoVo cells (Fig. 2E).
These in vivo and in vitro data suggested that the sensitivity to metformin in CRC was associated with the KRAS mutation. However, why the antiproliferation effect of metformin does not occur in KRASWT CRC cells is still to be determined.
Intracellular Concentration Determines the Anticancer Activity of Metformin In Vitro and In Vivo.
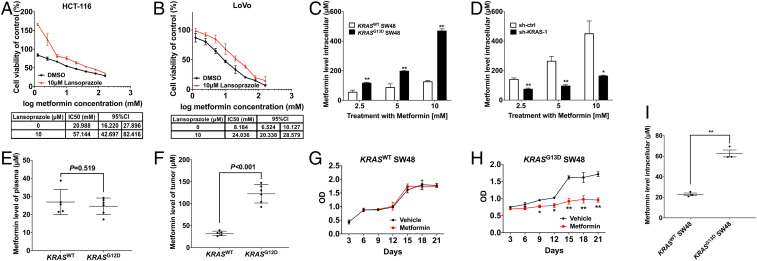
One study demonstrated that significant associations between metformin exposure and colorectal cancer-specific mortality were observed only for high-intensity metformin use (>1.1 g/d), not low-dose use (<1.1 g/d), in the diabetic cohort (16). It suggests that the antiproliferation effect of metformin in cancer cells may be directly associated with its intracellular concentration. Moreover, as metformin-mediated antiproliferation performance is related to the cellular accumulation of this drug, which is mainly taken up by the membrane transporters OCT1 to 3, we used the proton pump inhibitor (PPI) lansoprazole, an OCT1 inhibitor as described in other research (17), to inhibit the activity of OCT1. Our results showed that lansoprazole increased the half-maximum inhibitory concentration (IC50) of metformin by 1.7 and 2.5 times in KRAS-mutation CRC cell lines HCT-116 and LoVo (Fig. 3 A and B). These data indicated that the high metformin concentration remaining in the cancer cells caused a direct effect on cellular proliferation.
Fig. 3.
KRAS mutation up-regulates the intracellular accumulation of metformin in vitro and in vivo. (A and B) Forty-eight-hour growth of KRASG13D CRC cell lines HCT-116 (A) and LoVo (B) cultured with or without lansoprazole was determined after treatment with metformin (Top; n = 5), and the IC50 and 95% CI were calculated by SPSS 21.0 software (Bottom). (C and D) Levels of metformin in KRASWT SW48, KRASG13D SW48 (C), sh-KRAS LoVo, and its control cell strain sh-ctrl LoVo (D) were detected by the LC-MS method after treatment with 2.5, 5, and 10 mM metformin for 24 h (n = 3 at each time point). Data are shown as mean ± SEM. The significance of KRASG13D SW48 vs. KRASWT SW48 (C) and sh-KRAS LoVo vs. sh-ctrl LoVo (D) was determined as *P < 0.05, **P < 0.01. (E and F) Levels of metformin in the plasma (E) and tumor tissues (F) of PDX models were determined after treatment with 1 mg/mL metformin in drinking water for 30 d; n = 5 for KRASWT colon adenocarcinoma PDX analysis; n = 6 for KRASG12D colon mucinous adenocarcinoma PDX analysis. (G–I) Growth curves of KRASWT SW48 cells (G) and KRASG13D SW48 (H) treated with 40 μM metformin for the indicated time points. Data are shown as mean ± SEM. The difference between metformin and vehicle at each time point was determined as *P < 0.05, **P < 0.01. (I) Levels of metformin in KRASWT SW48 and KRASG13D SW48 were detected by the LC-MS method after 40 μM metformin treatment at 21 d. Data are shown as mean ± SEM. The difference between KRASG13D and KRASWT SW48 was determined as **P < 0.01. All P values were determined by unpaired two-tailed Student’s t test.
However, evidence is lacking as to how much metformin accumulating in CRC cells is sufficient to mediate the antiproliferation effect. We therefore quantified intracellular metformin levels through a liquid chromatography-mass spectrometry (LC-MS) method. As shown in Fig. 3C, the metformin levels in KRASG13D SW48 after 24 h were higher than KRASWT SW48. Likewise, in Fig. 3D, the drug accumulation in LoVo was reduced after shRNA-mediated KRAS knockdown compared with its control group. Our analysis of intracellular metformin levels indicated the discrepancy of the therapeutic activity of metformin between KRAS-mutation and KRASWT CRC cells was intracellular concentration-associated.
Further, we quantified the metformin levels in PDX mouse plasma and tumor tissue after treatment with metformin in drinking water for 30 d. As shown in Fig. 3 E and F, consistent with in vitro results, the metformin levels in KRASG12D xenografts reached 122.5 ± 8.6 μM, exhibiting considerably higher levels than that in plasma (24.6 ± 1.9 μM), while the levels of metformin in the tumors of KRASWT PDX models (32.5 ± 2.3 μM) paralleled those observed in plasma (26.9 ± 3.1 μM).
It has been suggested that the plasma concentration of metformin is ∼10 to 40 μM after a standard oral dose in humans and animals. In addition, in vitro, we treated KRASWT SW48 and KRASG13D SW48 with 40 μM metformin for 21 d. The growth curves in Fig. 3 G and H show that continuous metformin use at low concentrations could also exhibit antiproliferation activity in KRAS-mutation cells. In Fig. 3I, the intracellular level of metformin reached 62 μM in KRASG13D SW48, while remaining at 22 μM in KRASWT SW48, reflecting metformin can be accumulated in KRAS-mutation cells and plays a role in the anticancer effect. The results shown in Fig. 3 G–I are parallel to our clinical retrospective study that a longer duration of metformin use was associated with more favorable OS and PFS in the KRAS-mutation population (P trend < 0.05; SI Appendix, Table S4). Therefore, along with the in vitro and in vivo results, we concluded that the sufficient accumulation of metformin for inhibiting CRC cell proliferation is associated with KRAS mutation.
KRAS Mutation-Mediated MATE1 Down-Regulation Sensitizes CRC Cells to Metformin.
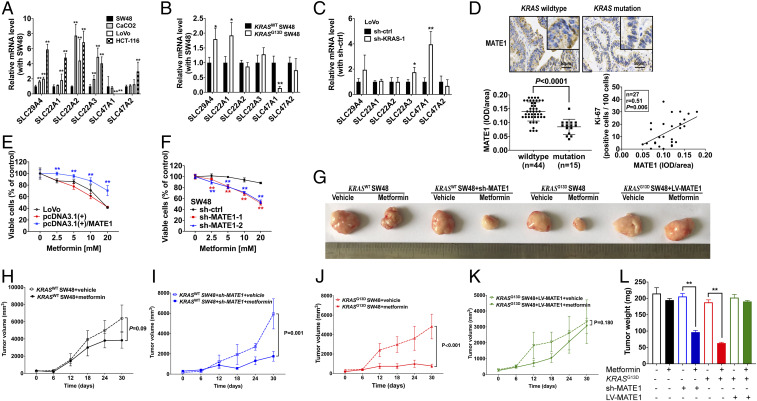
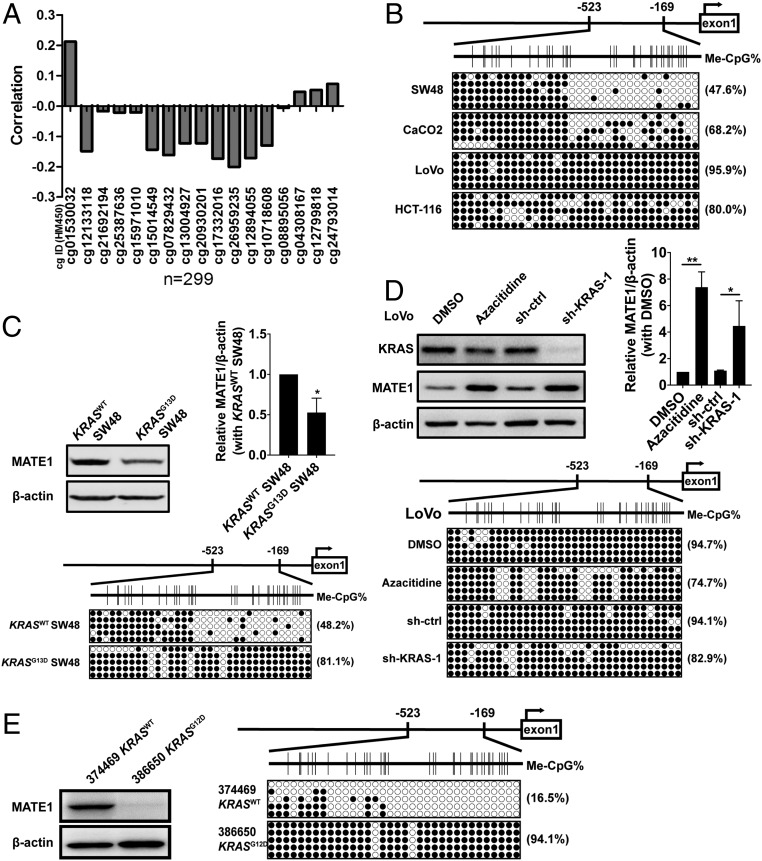
As our in vitro and in vivo studies define that the intracellular accumulation of metformin matters for its antitumor property, the underlying mechanism that KRAS mediates metformin absorption or elimination should be further demonstrated. We subsequently detected the transcriptional level of six recognized channel proteins by qRT-PCR, including SLC294A (encoding PMAT), SLC22A1 (encoding OCT1), SLC22A2 (encoding OCT2), SLC22A3 (encoding OCT3), SLC47A1 (encoding MATE1), and SLC47A2 (encoding MATE2K). Of note, SLC47A1, which encodes MATE1 for metformin elimination, is dramatically down-regulated in both LoVo and HCT-116 with KRAS mutation compared with SW48 and CaCO2 (Fig. 4A). Consistently, the SLC47A1 transcriptional level was decreased in KRASG13D SW48 and was rescued after knocking down KRAS in LoVo (Fig. 4 B and C).
Fig. 4.
Down-regulation of MATE1 under KRAS mutation is associated with the sensitivity of CRC cells to metformin. (A–C) Transcriptional levels of SLC29A4 (PMAT), SLC22A1 (OCT1), SLC22A2 (OCT2), SLC22A3 (OCT3), SLC47A1 (MATE1), and SLC47A2 (MATE2k) in KRASWT CRC cell lines SW48 and CaCO2, KRASG13D CRC cell lines HCT-116 and LoVo (A), KRASG13D SW48 and its counterpart SW48 (B), and sh-KRAS LoVo and sh-ctrl LoVo (C) were determined by qRT-PCR (n = 3). Data are shown as mean ± SEM. P values were determined with the black bar as a control in A–C as *P < 0.05, **P < 0.01. Each analysis was replicated three times. (D) Representative images of MATE1 immunohistochemistry on cross-sections from patients with mCRC with T2DM and metformin use, and the different integral optical density (IOD) of MATE1 between the KRAS wild type and mutation group is shown. The correlation between MATE1 expression and Ki67 level of 27 mCRC patients with T2DM and metformin use was determined by Pearson’s correlation analysis. (E and F) Forty-eight-hour growth of the KRASG13D CRC cell line LoVo with or without MATE1 overexpression (E) and KRASWT CRC cell line SW48 with or without MATE1 knockdown by small interfering RNA (F) was detected after treatment with 2.5, 5, 10, and 20 mM metformin (n = 5). Data are shown as mean ± SEM. The difference compared with the control group at each concentration was determined as **P < 0.01. (G–L) The representative morphology (G), tumor growth rate, and tumor weight (L) are shown as a result of 30-d treatment with metformin in SW48 xenograft (H), SW48 with sh-MATE1 xenograft (I), KRASG13D SW48 (J), and KRASG13D SW48 with LV-MATE1 xenograft (K) models. Data are shown as mean ± SEM. All P values were determined by two-way ANOVA. **P < 0.01.
To further determine whether MATE1 expression is associated with sensitivity to metformin and mediated by KRAS status, we first detected MATE1 of histological sections from the foci of mCRC patients and detected Ki67 expression of those using metformin. As shown in Fig. 4D, the expression of MATE1 was lower in KRAS-mutated CRC histological sections than KRASWT, and MATE1 had a positive correlation with Ki67 expression (r = 0.51, P = 0.006). Next, in vitro, we respectively overexpressed MATE1 by recombinant plasmid transfection in LoVo and knocked down MATE1 expression by small interfering RNA (siRNA) in SW48. We found that MATE1 elevation impaired the antitumor activity of metformin in LoVo (Fig. 4E), while MATE1 reduction rescued the sensitivity to metformin of SW48 (Fig. 4F).
Moreover, to address the key role of MATE1 in the metformin effect of CRC, we generated a cell line-derived xenograft model using SW48 cells, SW48 with sh-MATE1, KRASG13D SW48, and KRASG13D SW48 with MATE1 overexpression by lentiviral vector (LV-MATE1). Four groups of animals were treated with metformin at a pharmacological dose (200 mg/kg mouse weight, equal to 1,000 mg/60 kg human weight per day, orally). It showed that treatment with metformin in SW48 xenografts had no significant antitumor effects compared with their vehicle control, and metformin treatment inhibited the SW48 + sh-MATE1 tumor growth significantly. On the contrary, metformin was ineffective in KRASG13D SW48-overexpressing MATE1 xenografts (Fig. 4 G–L).
Above all, it was concluded that low expression of MATE1 is necessary for the sensitivity to metformin of KRAS-mutated CRC cells.
KRAS Mutation Silences MATE1 Transcription by Inducing CpG Island Hypermethylation at Its Promoter.
A previous study has shown that Sp1 was identified as the transcription factor binding to the MATE1 promoter and positively regulating MATE1 expression (18). However, we found no relationship between Sp1 protein level and MATE1 expression among CRC cell lines (SI Appendix, Fig. S6). Sp1 is known to bind to the GC box, and two GC-rich sites can be observed in the MATE1 promoter. We therefore analyzed the methyArray 450k dataset from The Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD) data collection and found that MATE1 expression had a negative correlation with the hypermethylation of −484 to +970 bp from the MATE1 transcription start site (Fig. 5A). Next, we detected the methylation site in a downstream promoter −523 to −169 bp of MATE1 in CRC cell lines by bisulfite DNA sequencing PCR (BSP), and the methylation degree was higher in LoVo and HCT-116 compared with SW48 and CaCO2 (Fig. 5B). Likewise, data also showed that the MATE1 promoter was hypermethylated with protein decreasing after KRASG13D editing (Fig. 5C), while the CpG island methylation level was reduced, along with the protein expression induction of MATE1 in LoVo treated with azacitidine (causing hypomethylation of DNA by inhibiting the DNA methyltransferase) (19) or KRAS knockdown (Fig. 5D). In addition, MATE1 hypermethylation was also observed in human CRC tissues, which were used for the PDX model generation (Fig. 5E). These data together suggested that the MATE1 level was transcriptionally regulated by KRAS-mediated hypermethylation in the promoter.
Fig. 5.
KRAS mutation down-regulates MATE1 through mediating the hypermethylation status on the CpG island of the MATE1 promoter. (A) The correlation between MATE1 transcriptional levels and methylation levels of CpG sites on the promoter of MATE1. Data from TCGA-COAD RNA-seq-HTseq-FPKM-521 (workflow type HTSeq-FPKM, normalized from RNA-seq of 521 samples) and methyArray 450k were conducted by Pearson’s correlation analysis. (B) Bisulfite sequencing PCR analysis showing the methylation status of CpG sites on the MATE1 promoter in CRC cell lines. (C) Immunoblot analysis of MATE1 levels and BSP analysis of the MATE1 promoter are shown between KRASG13D SW48 and its counterpart SW48. (D) Immunoblot analysis monitoring MATE1 levels and CpG-site methylation on the MATE1 promoter in KRAS shRNA-LoVo cells treated with dimethyl sulfoxide (DMSO) as a negative control or azacitidine as a positive control. (E) The MATE1 expression in 374469 KRASWT colon adenocarcinoma and 386650 KRASG12D colon mucinous adenocarcinoma was analyzed by immunoblot, and the methylation status of CpG sites on the MATE1 promoter was determined by BSP. Data are shown as mean ± SEM. All P values were determined by two-way ANOVA. *P < 0.05, **P < 0.01.
KRAS Mutation Induces MATE1 Promoter Methylation by Up-Regulating DNMT1.
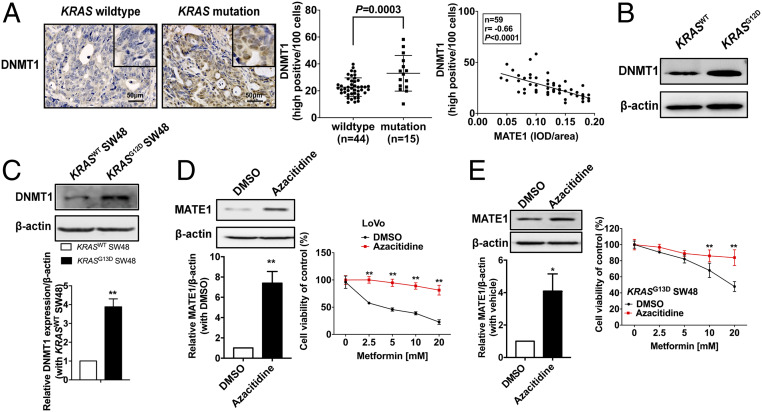
As DNA methylation is mainly regulated by the synergy of DNA methyltransferases and demethylases, we detected the transcriptional changes of the methylation-status maintenance enzyme DNA methyltransferase 1 (DNMT1) and DNA demethylase TET1/2 (ten-eleven translocation family proteins) (20, 21). DNMT1 and TET1 expression was first detected in the histological sections of CRC patients. We found that the proportion of high DNMT1-positive cells was larger in KRAS-mutation CRC than KRASWT and had a negative correlation with MATE1 expression (Fig. 6A), while the changes of TET1 between KRAS-mutation and KRASWT CRC were not significant (SI Appendix, Fig. S7). In addition, we also found that the level of DNMT1 was higher in KRASG12D CRC tissue than KRASWT of the two PDX model donors (Fig. 6B).
Fig. 6.
Transcriptional silencing of MATE1 by hypermethylation in KRAS-mutation CRC cells is associated with the up-regulation of DNMT1. (A) Representative images of DNMT1 immunohistochemistry on cross-sections from mCRC patients with T2DM were shown (Left); cells with high positive DNMT1 expression were counted by ImageJ software, and the proportion was presented by two-way ANOVA (Middle); the association between the IOD of MATE1 (shown in Fig. 3 D and E) and cell proportion with high positive DNMT1 expression was determined by Pearson’s correlation analysis (Right). (B and C) Expression levels of DNMT1 in 374469 KRASWT colon adenocarcinoma and 386650 KRASG12D colon mucinous adenocarcinoma (B) and in KRASG13D SW48 (C) were analyzed by immunoblot. (D and E) Immunoblot analysis of MATE1 level (Left) and 48-h cell viability (Right) in LoVo (D) or KRASG13D SW48 (E) cultured in 10 μM azacitidine for more than three generations (n = 3). Data are shown as mean ± SEM. *P < 0.05, **P < 0.01 compared with the DMSO group. All P values were determined by two-way ANOVA.
Next, we examined the expression of DNMT1 in CRC cells. In contrast with SW48, KRASG13D editing up-regulated DNMT1 expression (Fig. 6C), while it showed that DNMT1 was decreased in shKRAS LoVo cells (SI Appendix, Fig. S8). We further verified the effect of DNMT1 on MATE1 expression and subsequent sensitivity to metformin in cancer cells. DNMT1 function was inhibited by azacitidine, and subsequently the antitumor activity of metformin was significantly impaired with MATE1 up-regulation in LoVo cells (Fig. 6D) and KRASG13D SW48 cells (Fig. 6E). These data indicated that MATE1 was transcriptionally regulated by DNMT1 in CRC.
Metformin Inhibited KRAS-Mutation Cell Proliferation through Inactivating Both RAS/ERK and AKT/mTOR Signaling.
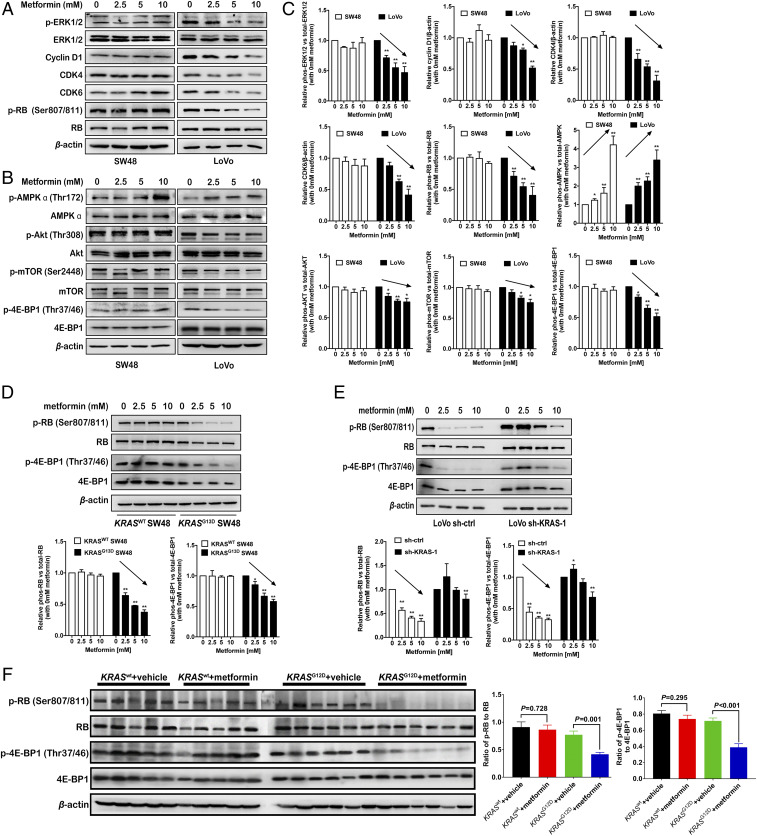
Next, we wanted to clarify the intracellular mechanism in which metformin inhibits KRAS-mutation cell proliferation. As is known, in the G1/S phase of the cell-cycle checkpoint, the cyclin D1–CDK4/6–RB complex promotes retinoblastoma (RB) protein phosphorylation, thus releasing the transcription factor E2F, which increases the expression of protein regulating the cell proliferation. Cyclin D1 is regulated by the RAS/RAF proto-oncogene serine/threonine-protein kinase (RAF)/MEK/ERK signaling pathway, and the PI3K-Akt-mTOR pathway also regulates cell proliferation, which has not been reported in CRC cells. Thus, we simultaneously detected the alterations in both ERK and Akt-mTOR signaling pathway under metformin treatment. Western blot showed that the level of phosphorylated ERK1/2 (p-ERK1/2), cyclin D1, CDK4/6, and phosphorylated RB protein (p-RB) was decreased in a dose-dependent manner in KRAS-mutation LoVo cells (Fig. 7 A–C). However, the AKT/mTOR pathway can usually be activated to compensate for the proliferative signal after inhibiting RAS/ERK transduction (22). Therefore, we also detected the level of phosphorylated AKT, mTOR, and its downstream eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), which is inactivated by phosphorylation and subsequently releases the transcription factor eIF4E (23). The results indicated that the levels of phosphorylated AKT at threonine 308 (Thr308) and mTOR at serine 2448 (Ser2448) were significantly down-regulated, followed by 4E-BP1 phosphorylation in LoVo cells (Fig. 7 B and C). Likewise, the changes of phosphorylated RB and 4E-BP1 were also found in KRASG13D SW48 and LoVo cells with KRAS knockdown (Fig. 7 D and E). Moreover, Western blot analysis of the KRASG12D xenografts showed metformin could impede both RB and 4E-BP1 signaling, an effect not seen in KRASWT xenografts (Fig. 7F). Our findings indicated that dual inhibition of RB (downstream of MEK/ERK) and 4E-BP1 (downstream of AKT/mTOR) by metformin had a promising antiproliferation effect on KRAS-mutation CRC cells.
Fig. 7.
Metformin inhibits both RB and 4E-BP1 activity in cell proliferation. (A and B) Immunoblot analysis of the MEK/ERK pathway, phosphorylated ERK1/2, total ERK1/2, Cyclin D1, CDK4/6, phosphorylated RB protein, and total RB protein (A), and the AKT/mTOR pathway, phosphorylated AKT (Thr308), total AKT, phosphorylated mTOR (Ser2448), total mTOR, phosphorylated 4E-BP1, and total 4E-BP1 (B) was conducted in SW48 and LoVo cells after treatment with 2.5, 5, and 10 mM metformin for 24 h. (C) Quantification of proteins in A and B by densitometry from three independent experiments, normalized by β-actin levels (mean ± SEM). *P < 0.05, **P < 0.01. (D–F) Phosphorylated RB protein, total RB protein, phosphorylated 4E-BP1, and total 4E-BP1 were detected by immunoblot analysis in KRASG13D SW48 and its control SW48 cells (D), sh-KRAS LoVo, and its control sh-ctrl LoVo cells (E) after treatment with 2.5, 5, and 10 mM metformin for 24 h and detected in 374469 KRASWT colon adenocarcinoma and 386650 KRASG12D colon mucinous adenocarcinoma patient-derived xenograft models after treatment with 1 mg/mL metformin in drinking water for 30 d (F). All P values were determined by two-way ANOVA. *P < 0.05, **P < 0.01.
Briefly, as the expression of the DNA methyltransferase was up-regulated in KRAS-mutation CRC cells, MATE1 was transcriptionally silenced by hypermethylation, followed by intracellular sufficient metformin accumulation for its direct antitumor activity (SI Appendix, Fig. S9).
Discussion
Aberrant KRAS signaling plays a crucial role in cancer cell proliferation and is commonly associated with poor prognosis and resistance to anti-EGFR therapy. Without exception, metastatic colorectal cancer patients with the KRAS mutation have a poor prognosis without effective drugs. Long-term efforts to target the cancer-associated KRAS mutation have been unsuccessful. Therefore, it has become imperative to look for new drugs targeting KRAS-mutation cancer. Recently, the preventive and antitumor activity of metformin has been extensively evaluated in various cancers (11, 24). Surprisingly, in our study, we found that metformin selectively inhibited metastatic colorectal cancer with the KRAS mutation and the median survival time in this group was prolonged for 37.8 mo. This dramatic effect was further proved in PDX and mutation cell models. Our investigation has put forward the possibility of a therapeutic strategy for KRAS-mutation mCRC patients by the use of metformin, an inexpensive and safe drug.
Although a clinical trial has failed to show a protective association between metformin and survival in CRC patients with T2DM (13), this finding is in line with several other retrospective investigations (25, 26). To date, except for our study, clinical retrospective studies or clinical trials in identifying the association between KRAS-mutation status and metformin use are still limited. A previous study reported an absence of effect of metformin treatment on KRAS-mutated stage III colorectal cancer (27). However, it is unknown whether metformin was used before diagnosis or during adjuvant therapy and follow-up in this study. In addition, it is quite different between stage III and stage IV in terms of clinical therapy. For instance, most stage III CRC patients will receive a recommendation of surgery to completely remove the tumor, while palliative operation and FOLFOX (fluorouracil, leucovorin, and oxaliplatin) or FOLFIRI (fluorouracil, leucovorin, and irinotecan) chemotherapy combinations are more recommended for stage IV (metastatic or recurrent) CRC patients. This difference may be a reason that metformin improves the survival of stage IV CRC patients (mCRC) but not stage III CRC patients. Our study has provided an explanation for the conflict between the different investigations. In particular, the median overall survival time of the metformin group was 17.5 mo longer than that of mCRC patients without diabetes (Fig. 1A), and metformin also exhibited superior improvement in a KRAS-mutation CRC PDX model without diabetes. These findings suggested that metformin could be applied in KRAS-mutation mCRC patients beyond specificity for diabetes.
The KRAS mutation is generally associated with poor outcomes; different mutated KRAS alleles predict different overall survival in a given cancer. For example, it has been reported that G12D and G12V mutations in advanced CRC are associated with worse overall survival than the G13D mutation, possibly because codon 12 mutations are associated with poor differentiation and cancer metastasis (28). After administration with metformin in this study, our retrospective data showed that G12D (n = 16), G12V (n = 4), G13D (n = 8), and another rare codon 12 mutation (n = 9) had respectively superior responses to metformin rather than CRC patients with KRAS wild type (n = 47), and there was no significant difference in whether metformin improved prognosis among different KRAS-mutation types (SI Appendix, Table S5). In addition, we have verified that codon 12-mutation KRAS oncoprotein also reinforces the response to metformin in vitro (Fig. 2 C and D), and the results indicated that both codon 12 and 13 mutations could up-regulate the sensitivity to the antiproliferation therapy of metformin. Herein, our investigation put forward the possibility of a precision therapeutic strategy that KRAS-mutation mCRC patients, regardless of mutation type, may benefit from metformin treatment.
As the KRAS mutation mediates aberrant RAS/RAF/MEK/ERK signaling transduction, agents targeting this pathway have been developed. However, RAS-GTPase inactivators (e.g., tipifarnib and lonafarnib) or the MEK inhibitor selumetinib failed to improve progression-free survival, as reported in several clinical trials (6, 7, 29, 30). The reason is mainly ascribed to the feedback activation of PI3K/AKT signaling after inhibiting RAS/RAF/MEK/ERK transduction (31, 32). Therefore, a combination of MEK and AKT inhibitors may be an important therapeutic strategy in KRAS-driven human malignancies. Our findings indicated that dual inhibition of RB (downstream of MEK/ERK) and 4E-BP1 (downstream of AKT/mTOR) by metformin has a promising antiproliferation effect on KRAS-mutation CRC cells.
Metformin is largely absorbed through the small intestine, mainly accumulates in hepatocytes, circulates without catabolism, and finally is excreted in urine, processes in which several transporters determine the concentration of metformin in target tissues. However, it remains unclear which transporter predominantly regulates the concentration of metformin in large intestinal epithelial cells, let alone in CRC cells. In this study, we identified that MATE1 was not only expressed in KRASWT CRC SW48 and CaCO2 cells (Fig. 4A) but also in normal colon tissues from TCGA (SI Appendix, Fig. S10A), while expression was significantly lower in KRAS-mutation CRC cells. The results were in accordance with immunohistochemical staining in primary cancer specimens (Fig. 4). Meanwhile, the RNA level of MATE1 was lower in tumor sections than in adjacent sections from TCGA-COAD database (SI Appendix, Fig. S10B) and GSE12398 dataset (33) (SI Appendix, Fig. S10C). These results indicated that MATE1 was the key operator for the metformin effect in CRC cells.
What is more, our study identifies the hypermethylation of CpG islands in the MATE1 promoter as a predictor for the response to metformin in KRAS-mutation CRC cells (Fig. 5). As KRAS and BRAF mutations are usually accompanied by the CpG island methylator phenotype, which is defined by poor prognosis (34), we found that BRAFV600E CRC cell line HT-29 was accompanied by a high-methylation phenotype in the CpG island of the MATE1 promoter and was also susceptible to metformin (SI Appendix, Fig. S11). Furthermore, we identified the enzymes that mediated the hypermethylation of CpG on the MATE1 promoter; those are proven to be regulated by aberrant KRAS signaling (35–37). It suggested DNMT1 is the key enzyme mediating the hypermethylation of MATE1, and methylation inhibitors (e.g., decitabine, azacitidine) or metformin absorption inhibitors (PPIs, e.g., lansoprazole, cimetidine) may be inappropriate for those CRC patients treated with metformin.
KRAS is the most frequently mutated oncogene also in pancreatic cancer (70 to 90%) (38), lung cancer (20 to 30%) (39), breast cancer (<5%) (40), and endometrial cancer of the endometrioid type (about 18%) and serious type (3%) (41). It is reasonable to speculate that metformin may benefit other KRAS-mutation cancers as well. However, clinical phase II trials suggest that metformin does not improve outcome in patients with advanced pancreatic cancer (42, 43) and non–small-cell lung cancer (44) treated with standard systemic therapy, while metformin improved the worse prognosis in breast cancer (45) and decreased proliferation markers in tumors of patients with endometrial cancer (46). Obviously, the KRAS mutation is not a common determinant for the metformin effect in all cancers; if so, it is hard to understand the invalidation of metformin in pancreatic cancer with high KRAS-mutation frequency and the effect in breast cancer with low mutation frequency. Of note, we think the key point is metformin cellular accumulation and the corresponding metformin transport channel changes rather than simple KRAS mutation in certain cancers. The failure in clinical trials in pancreatic cancer and lung cancer might be ascribed to the inability of the KRAS mutation to mediate metformin accumulation in cells. It is possible that MATE1 is not mediated by the KRAS mutation in other cancers and the expression pattern of metformin transport channels or DNMT is not identical to CRC. All of the assumptions need to be verified in retrospective studies and clinical trials to authenticate the clinical significance of metformin therapy in KRAS-mutation pancreatic and lung cancers, and to measure the concentration of metformin and expression changes of the transport channels in these cancer cells. So far, only the KRAS mutation in CRC could be considered as a marker for metformin treatment.
There are limitations to this study. Due to the inadequate medical records in this study, a more convincing subgroup analysis of KRAS mutations in codons 12 and 13 should be carried out in multiple cancer centers. We were not able to execute retrospective studies to authenticate the clinical significance of metformin therapy in pancreatic or lung cancers with a high frequency of the KRAS mutation. Although we were not able to present prospective clinical trial data in CRC patients for metformin use in the current study, we have already launched trials (metformin combined with FOLFIRI therapy [fluorouracil, leucovorin, and irinotecan] for CRC patients without diabetes) and may obtain more convincing data in the future to see whether metformin can be therapeutically employed in CRC treatment.
In conclusion, precision medicine is an emerging approach that directs the development of a more effective therapeutic strategy for patients according to individual variability. Our research demonstrates that metformin has a promising antitumor effect associated with its intracellular accumulation, while the sufficient metformin level is determined by MATE1, which is transcriptionally silenced by KRAS-directed DNA hypermethylation in CRC. This study suggests that KRAS-mutation status and the hypermethylation of MATE1 could be potential biomarkers for using metformin in CRC patients. Further clinical trials are necessary to verify the therapeutic benefit from metformin in CRC patients with or without T2DM.
Materials and Methods
Medical Records and Tissue Collection.
Patients (4,751) were diagnosed with mCRC at Sun Yat-sen University Cancer Center (SYUCC) between 2004 and 2016, and 2,378 patients with a complete past medical history were enrolled. Among them, 325 patients were diagnosed with mCRC with type 2 mellitus diabetes (SI Appendix, Fig. S1). Specimens were obtained from patients who had undergone CRC resection and with tumor grade categorized as well-differentiated, moderately differentiated, poorly differentiated, or undifferentiated by three pathologists. All patients’ informed consent was obtained before surgery, and the use of medical records and histological sections has also been approved by the ethics committee at SYUCC.
Establishment of Xenograft Models and Animal Studies.
For the patient-derived xenograft model, colorectal cancer specimens were obtained from patients undergoing resection of primary disease at SYUCC. All patients’ informed consent was obtained before surgery, and the samples used for PDX establishment were procured with the approval of the ethics committee at SYUCC. The cell line-derived xenografts mice were randomly divided into a metformin treatment group and its control group for each cell line model. All procedures related to animal feeding, treatment, and welfare were conducted in accordance with the Institutional Animal Care and Use Committee of Sun Yat-sen University.
Cell Culture and Gene Editing.
The human CRC cell lines (SW48, CaCO2, LoVo, HCT-116, HT-29) were obtained from the American Type Culture Collection. Cell lines were authenticated by Cellcook Biotech. KRASG13D SW48 was established by an improved CRISPR-Cas9–mediated precise genetic modification by using 1 μM nonhomologous end-joining inhibitor Scr7 (Selleck; S7742) (47).
Detection of Metformin Concentration.
The concentration of metformin in plasma, intracellular cell lines, and xenografts was determined by liquid chromatography-mass spectrometry.
Bisulfite Sequencing PCR.
Tumor or cell DNA was extracted using a Genomic DNA Extraction Kit (TaKaRa; T9765) and then modified by a DNA Bisulfite Conversion Kit (TIANGEN; DP215). The bisulfite-treated DNA was amplified using an EpiTaq HS PCR assay (TaKaRa; R110A) according to the manufacturer’s protocol.
Statistical Analysis.
For the clinical retrospective study, proportional hazard assumptions were examined using the Kolmogorov–Smirnov test and the Cramer–von Mises test, and P > 0.05 represents the interaction between metformin use and event (death or progression) is not time-dependent. Moreover, then a hierarchical PH regression analysis was performed to estimate the adjusted HR and 95% CI for the association of metformin use with overall survival and progression-free survival, after stratifying by each characteristic. The other experimental data are expressed as mean ± SEM, while statistical significance was determined by one-way ANOVA, unpaired two-tailed Student’s t test, or Pearson’s χ2 test using GraphPad Prism 7 or IBM SPSS Statistics 21.0 software, according to the data type. P value <0.05 indicated a statistically significant difference.
A detailed materials and methods section is provided in SI Appendix.
Data Availability.
All data generated or analyzed during this study are included in the published article (and its SI Appendix). No applicable resources were generated or analyzed during the current study.
Supplementary Material
Acknowledgments
We thank BEIJING IDMO Co., Ltd for their technical assistance with the PDX model, Dr. Yanxin Luo and Xiaoling Wang, PhD (Affiliated Sixth Hospital, Sun Yat-sen University) for technical assistance for the methylation test, and Prof. Yihan Lu (Fudan University) for statistical discussions. We also thank Dr. Yan-hua Zhang for help facilitating collaboration for this work. This study was supported by the National Nature Science Foundation of China (Grants 81770808, 81872165, 81701414, and 81871211); National Key R&D Program of China (Grant 2018YFA0800403); Guangdong Provincial Key R&D Program (Grants 2018B030337001 and 2019B020227003); Key Project of Nature Science Foundation of Guangdong Province, China (Grant 2019B1515120077); Guangdong Natural Science Fund (Grant 2019A1515011810); Guangdong Science Technology Project (Grant 2017A020215075); Key Sci-Tech Research Project of Guangzhou Municipality, China (Grants 201803010017, 201807010069, and 202002020022); 2017 and 2019 Milstein Medical Asian American Partnership Foundation Research Project Award in Translational Medicine; and China Postdoctoral Science Foundation (Grant 2019M 662991).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918845117/-/DCSupplemental.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Cremolini C. et al., FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: Updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 16, 1306–1315 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Yaeger R. et al., Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 33, 125–136.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allegra C. J. et al., Extended RAS gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: American Society of Clinical Oncology Provisional Clinical Opinion update 2015. J. Clin. Oncol. 34, 179–185 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Douillard J. Y. et al., Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 369, 1023–1034 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Sharma S. et al., A phase II trial of farnesyl protein transferase inhibitor SCH 66336, given by twice-daily oral administration, in patients with metastatic colorectal cancer refractory to 5-fluorouracil and irinotecan. Ann. Oncol. 13, 1067–1071 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Rao S. et al., Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J. Clin. Oncol. 22, 3950–3957 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z. J. et al., Reduced risk of colorectal cancer with metformin therapy in patients with type 2 diabetes: A meta-analysis. Diabetes Care 34, 2323–2328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higurashi T. et al., Metformin for chemoprevention of metachronous colorectal adenoma or polyps in post-polypectomy patients without diabetes: A multicentre double-blind, placebo-controlled, randomised phase 3 trial. Lancet Oncol. 17, 475–483 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Mohamed Suhaimi N. A. et al., Metformin inhibits cellular proliferation and bioenergetics in colorectal cancer patient-derived xenografts. Mol. Cancer Ther. 16, 2035–2044 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Pernicova I., Korbonits M., Metformin—Mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 10, 143–156 (2014). [DOI] [PubMed] [Google Scholar]
- 12.O’Donoghue D., Adebayo G., McHugh C. M., Is the use of a combination of insulin and metformin as safe as insulin alone in gestational diabetes. Ir. J. Med. Sci. 183, S459 (2014). [Google Scholar]
- 13.Mc Menamin U. C., Murray L. J., Hughes C. M., Cardwell C. R., Metformin use and survival after colorectal cancer: A population-based cohort study. Int. J. Cancer 138, 369–379 (2016). [DOI] [PubMed] [Google Scholar]
- 14.He L., Wondisford F. E., Metformin action: Concentrations matter. Cell Metab. 21, 159–162 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury S. et al., MATE2 expression is associated with cancer cell response to metformin. PLoS One 11, e0165214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spillane S., Bennett K., Sharp L., Barron T. I., A cohort study of metformin exposure and survival in patients with stage I-III colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 22, 1364–1373 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Ding Y. et al., The effect of lansoprazole, an OCT inhibitor, on metformin pharmacokinetics in healthy subjects. Eur. J. Clin. Pharmacol. 70, 141–146 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Kajiwara M. et al., Critical roles of Sp1 in gene expression of human and rat H+/organic cation antiporter MATE1. Am. J. Physiol. Renal Physiol. 293, F1564–F1570 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Christman J. K., 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Liao J. et al., Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 47, 469–478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pastor W. A., Aravind L., Rao A., TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman J. A. et al., Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 14, 1351–1356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay N., The Akt-mTOR tango and its relevance to cancer. Cancer Cell 8, 179–183 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Morales D. R., Morris A. D., Metformin in cancer treatment and prevention. Annu. Rev. Med. 66, 17–29 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Cossor F. I. et al., Diabetes, metformin use, and colorectal cancer survival in postmenopausal women. Cancer Epidemiol. 37, 742–749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanders M. M. J. et al., Are metformin, statin and aspirin use still associated with overall mortality among colorectal cancer patients with diabetes if adjusted for one another? Br. J. Cancer 113, 403–410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh P. P. et al., Relationship between metformin use and recurrence and survival in patients with resected stage III colon cancer receiving adjuvant chemotherapy: Results from North Central Cancer Treatment Group N0147 (Alliance). Oncologist 21, 1509–1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W. et al., Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer 15, 340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jänne P. A. et al., Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: The SELECT-1 randomized clinical trial. JAMA 317, 1844–1853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blumenschein G. R. Jr. et al., A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. 26, 894–901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wee S. et al., PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 69, 4286–4293 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Sun C. et al., Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7, 86–93 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Monticone M. et al., Gene expression deregulation by KRAS G12D and G12V in a BRAF V600E context. Mol. Cancer 7, 92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyota M. et al., CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 96, 8681–8686 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serra R. W., Fang M., Park S. M., Hutchinson L., Green M. R., A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. eLife 3, e02313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu B. K., Brenner C., Suppression of TET1-dependent DNA demethylation is essential for KRAS-mediated transformation. Cell Rep. 9, 1827–1840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kottakis F. et al., LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539, 390–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas M. et al., Extended RAS analysis and correlation with overall survival in advanced pancreatic cancer. Br. J. Cancer 116, 1462–1469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Arcangelo M., Cappuzzo F., K-ras mutations in non-small-cell lung cancer: Prognostic and predictive value. ISRN Mol. Biol. 2012, 837306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sánchez-Muñoz A. et al., Lack of evidence for KRAS oncogenic mutations in triple-negative breast cancer. BMC Cancer 10, 136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Hara A. J., Bell D. W., The genomics and genetics of endometrial cancer. Adv. Genomics Genet. 2012, 33–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kordes S. et al., Metformin in patients with advanced pancreatic cancer: A double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 16, 839–847 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Reni M. et al., (Ir)Relevance of metformin treatment in patients with metastatic pancreatic cancer: An open-label, randomized phase II trial. Clin. Cancer Res. 22, 1076–1085 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Parikh A. B., Kozuch P., Rohs N., Becker D. J., Levy B. P., Metformin as a repurposed therapy in advanced non-small cell lung cancer (NSCLC): Results of a phase II trial. Invest. New Drugs 35, 813–819 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Sonnenblick A. et al., Impact of diabetes, insulin, and metformin use on the outcome of patients with human epidermal growth factor receptor 2-positive primary breast cancer: Analysis from the ALTTO phase III randomized trial. J. Clin. Oncol. 35, 1421–1429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laskov I. et al., Anti-diabetic doses of metformin decrease proliferation markers in tumors of patients with endometrial cancer. Gynecol. Oncol. 134, 607–614 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Chu V. T. et al., Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in the published article (and its SI Appendix). No applicable resources were generated or analyzed during the current study.