Abstract
An understanding of how memory is acquired and how it can be modified in fear-related anxiety disorders, with the enhancement of failing memories on one side and a reduction or elimination of traumatic memories on the other, is a key unmet challenge in the fields of neuroscience and neuropsychiatry. The latter process depends on an important form of learning called fear extinction, where a previously acquired fear-related memory is decoupled from its ability to control behaviour through repeated non-reinforced exposure to the original fear-inducing cue. Although simple in description, fear extinction relies on a complex pattern of brain region and cell-type specific processes, some of which are unique to this form of learning and, for better or worse, contribute to the inherent instability of fear extinction memory. Here we explore an emerging layer of biology that may compliment and enrich the synapse-centric perspective of fear extinction. As opposed to the more classically defined role of protein synthesis in the formation of fear extinction memory, a neuroepigenetic view of the experience-dependent gene expression involves an appreciation of dynamic changes in the state of the entire cell: from a transient change in plasticity at the level of the synapse, to potentially more persistent long-term effects within the nucleus. A deeper understanding of neuroepigenetic mechanisms and how they influence the formation and maintenance of fear extinction memory has the potential to enable the development of more effective treatment approaches for fear-related neuropsychiatric conditions.
Keywords: neuroepigenetics, extinction, learning, epitranscriptomics, memory, epigenetics, DNA modification, DNA structure, Histone modification, RNA modification, RNA editing
Introduction
Theories and models of fear-related memory exist in many forms and, historically, they have been primarily focused on the idea that memories are created by mechanisms that translate an initially labile experience to a persistent state (Muller & Pilzecker 1900; Lechner & Squire 1999; McGaugh 2000) which can then return to a labile state upon reactivation (“reconsolidation”, Misanin et al. 1968; Nader et al. 2000; although see Dudai & Eisenberg 2004). The concept of dynamic memory states is most apparent in fear extinction where it has been observed, since the time of Pavlov, that fear responses can be reduced following repeated exposure to cues related to the original fear evoking experience, but then return with the passage of time (Pavlov 1927; Baum 1988), by a return to original context in which fear was acquired (Bouton & Bolles 1979a; Bouton & Bolles 1979b), and by exposure to an unrelated but stress evoking cue (Rescorla & Heth 1975). However, what has never truly been established following Pavlov’s (1927) original observations of extinction is an understanding of whether the reduction in behavioural responding results in the erasure of the original trace, the creation of a new trace, or something else, and a complete picture of what may be happening at the molecular level in the brain has yet to be achieved. In this review, we compare and contrast transient extinction learning-induced electrical signalling events at the level of the synapse with potentially more persistent experience-dependent changes in the capacity for gene expression, which occur against the backdrop of the nuclear environment. We highlight newly emerging neuroepigenetic mechanisms surrounding DNA, RNA and the local chromatin environment, and describe how they contribute to this important form of learning and memory when extinction is considered as a dynamic equilibrium between erasure and inhibition of the original fear memory trace.
Theoretical perspectives on fear extinction and the role of LTP
It is notable that since the early days of investigation into fear extinction as a unique inhibitory learning process, the question of whether fear memories are ‘erased’ or inhibited following extinction has been strongly debated. Pavlov (1927) rejected any form of erasure as a mechanism for fear extinction learning and proclaimed that only a new inhibitory trace following extinction training was possible due to the observation of spontaneous recovery. But, he also stated that “being a definite circumscribed material system, [a reflex] can only continue to exist so long as it is in continuous equilibrium with the forces external to it: so soon as this equilibrium is seriously disturbed the organism will cease to exist as the entity it was. Reflexes are the elemental units in the mechanism of perpetual equilibrium,” suggesting, at the very least, an appreciation of the importance of an equilibrium between different memory states. This idea, as opposed to a dichotomy between fear and extinction, is theoretically attractive because both models, ‘erasure of fear’ and ‘inhibitory control over fear’, have repeatedly been observed over the course of 50 years of fear extinction research (Quirk & Mueller 2008; Flavell et al. 2013).
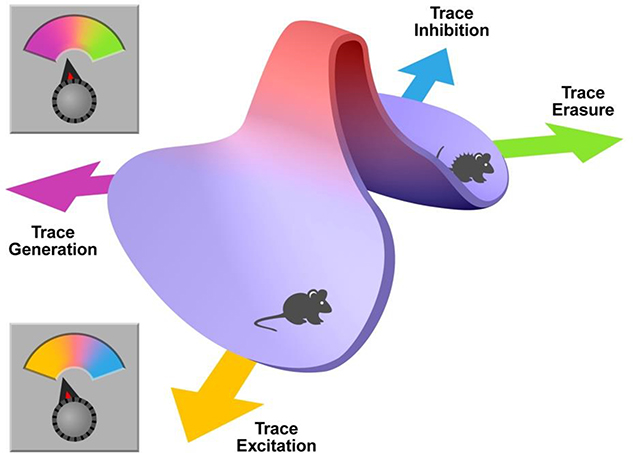
If one were to view extinction learning as a process that exists as an equilibrium between erasure and inhibition, it may therefore be posited that any behavioural manipulation which influences fear expression would also have impact on a variety of underlying continuous biological variables, such as intrinsic excitability, which could either promote a return to baseline in the original cells resulting in functional erasure of the original trace, or have permissive effects on new ‘inhibitory control’ cells leading to the inhibition of the original trace. Behavioural expression also exists on a continuous scale, from absolute fear expression to no expression, and this could be overlaid as a combined model in which sufficient activation of either of these two mechanisms enables the transition from one behavioural state to another, such as from freezing to exploration (Figure 1). The emphasis would then be placed on the subthreshold variables that summate in both the original fear-related cells and the cells encoding the new inhibitory trace.
Figure 1.
Visual representation of behavioural tuning where modulation of both fear cells and inhibitory control cells can occur along a continuum. When erasure of original fear (green) and/or fear trace inhibition (blue) occurs, animals may transition from fear behaviours such as freezing and escape behaviours, to normal grooming and exploratory behaviour. However, when original fear traces are activated (purple) and/or engagement of inhibitory control is low (yellow) animals will fail to overcome the threshold to re-engage normal behaviours and remain freezing or performing escape behaviours. Thus, the dynamic equilibrium of behavioural expression relies on mechanisms which modulate either of these aspects.
To describe how these two processes may operate and functionally interact, the most well supported and utilized model of memory storage and retrieval, the electrochemical model of plasticity, where synapse-related mechanisms play a dominant role, has often been invoked (Poo et al. 2016). The electrochemical model posits that signals in the nervous system originate via membrane depolarization, which then propagate via spreading activation along axons, and transmit their signal to surrounding neurons via chemical messengers. With respect to fear-related learning and memory, this particular model aligns with Hebb’s postulate, whereby a collection of neurons representing a memory trace is formed and maintained through modulating mechanisms which allow firing of one neuron to increase the probability of triggering firing in the network of temporally coincident neurons distributed across the brain. This idea has been validated at the cellular level, in part, with the discovery of long-term potentiation (LTP; Bliss & Lomo 1973) and long-term depression (LTD; Ito 1989). Because of its potential link with memory formation, this correlate for experience-dependent plasticity has thus taken hold of our conception of cellular memory, and supporting evidence can be found throughout the literature, supporting a relationship between LTP/LTD like processes and fear-related learning and memory. For example, a synapsin-driven channelrhodopsin called oChIEF, was used to demonstrate that modulation of LTP and LTD in the medial geniculate nucleus and auditory cortex, can either induce or impair fear memory respectively (Saucier & Cain 1995; although see Nabavi et al. 2014 for evidence that LTP is not necessary for the formation of other forms of memory).
Electrophysiological recordings have also been used to define anxiety, fear, and extinction cells in associated brain areas, such as the hippocampus and prefrontal cortex, which have made it possible to examine LTP in neurons selectively activated by fear and extinction learning (Milad & Quirk 2002; Herry et al. 2010; Tovote et al. 2015; Jimenez et al. 2018). It has even been possible to demonstrate that manipulations which appear to completely erase cued fear to either a 7kHz or 2kHz tone which share overlapping cells, can be rescued if just the cells of one representation are activated with oChIEF (Abdou et al. 2018). From this, it is clear that electrophysiological changes must occur in particular regions and cells of the brain for extinction to occur; however, it is still debated as to whether these changes theoretically constitute erasure or inhibition. In a recent extension of this idea, An et al. (2017) demonstrated that the training conditions are critical and the mechanisms of erasure or inhibition can be activated depending on the behavioural protocol employed. Specifically, they found that a single training session resulted in firing in the prelimbic PFC and basal lateral amygdala, which is known to be associated with inhibition of fear. Whereas repeated training sessions, failed to activate these areas and instead resulted in the activation of ‘erasure’ mechanisms including depotentiation of local circuits in the lateral amygdala.
As indicated, it has been shown that the neuronal circuits responsible for fear memory can be electrophysiologically de-potentiated and therefore relate to ‘erasure’, or, when alternative inhibitory traces are created, the two processes can compete (Clem & Schiller 2016). Although this conceptual framing of extinction in relation to equilibrium is a step in the right direction, this premise remains incomplete as it fails to integrate molecular mechanisms, which may serve as primary components of the extinction engram (Lashley 1950; Marshall & Bredy 2016). This is important because there are many instances where LTP-related changes in synaptic efficacy and synaptic structure following fear extinction learning do not persist even when the memory is maintained over long periods of time. For example, following a significant reduction in LTP associated with fear and its extinction, there is typically a return of fear (Nabavi et al. 2014). Mechanisms known to promote LTP such as spine formation (Hayashi-Takagi et al. 2015) have also been shown to be impermanent (Chen et al. 2014; Attardo et al. 2015; Lai et al. 2018). Furthermore, performance, memory and electrophysiological recordings can be decoupled; although recent findings suggest that this may be in part due to not clearly differentiating which outputs, among many, are involved in memory during the manipulation of local synaptic plasticity (Saucier & Cain 1995; Herry et al. 2008; Hong & Kim 2017; Marek et al. 2018). Fortunately, in recent years an entirely new model of molecular memory has emerged, which may advance the understanding of experience-dependent plasticity and address the problem of lingering fear memory following fear extinction.
Neuroepigenetics: a new frontier in the molecular underpinnings of fear extinction
An emerging perspective that may overcome current limitations in fully understanding the process of fear extinction involves recent discoveries in the burgeoning field of cognitive neuroepigenetics. Broadly defined, neuroepigenetic mechanisms represent a variety of molecular processes traditionally associated with cell lineage specification during early development, which have been co-opted in the adult brain to facilitate experience-dependent gene expression (Day & Sweatt 2011; Marshall & Bredy 2016). These mechanisms are rapid, long lasting and are well suited to govern the fine tuning of experience-dependent gene expression and memory in ways that are potentially independent, yet complementary, to protein synthesis-dependent changes in synaptic plasticity and behavioural performance at the time of learning.
DNA Modification
The first neuroepigenetic mechanism that satisfies these criteria is DNA methylation, which is known to be directly involved in memory formation (Ashapkin et al. 1982; Miller & Sweatt 2007; Stafford & Lattal 2011; Baker-Andresen et al. 2013). Importantly, this molecular process does not simply turn on and off genes necessary for fear learning or for its extinction. The accumulation of 5-methylcytosine (5mC) on DNA can both activate and inhibit gene expression associated with fear extinction depending on its position within the genome (Figure 2). In gene promoters, 5mC often silences gene expression and when it accumulates in gene bodies, this epigenetic mark tends to be permissive (Day & Sweatt 2010; Vanyushin & Ashapkin 2017). Other recently identified DNA modifications, including 5-hydroxymethylcytosine (5hmC) and N6-methyldeoxyadenosine (m6da), appear to be biased towards gene activation in fear extinction (Li et al. 2014; Li et al. 2018).
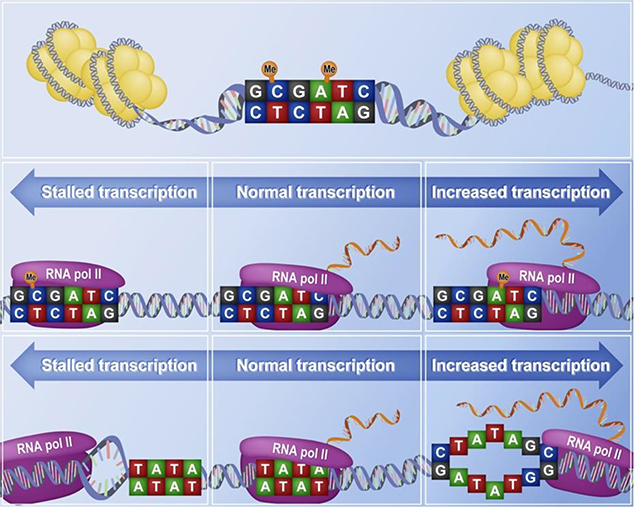
Figure 2.
A proposed mechanism for controlling behavioural equilibrium during fear extinction is genomic tuning. This is a process in which both DNA modifications and DNA structure can modulate transcription rate along a continuum. In the case of DNA modifications, while 5mC may entirely inhibit transcription, m6dA enhances transcription rate above baseline. Similarly DNA structure can completely stall RNA polymerase II (RNA pol II) and block transcription, or facilitate RNA pol II by opening DNA and thus enhance transcription rate above baseline. Combination of either of these mechanisms may thus dial transcription rate up or down.
Beyond on-off states, DNA modification can also influence the rate of transcription (Roundtree & Selker 1997). If one were to consider fear extinction as a process that involves a dynamic equilibrium between erasure and inhibition, perhaps in this context, changes in DNA methylation and the regulation of transcription rates would be important. In the case of functional erasure, this might involve transcriptional repression by DNA methylation or, similar to depotentiation, active DNA demethylation may occur as a function of fear extinction learning, which may then functionally erase transcriptional blocks that occur during fear learning (Li et al. 2013; Rudenko et al. 2013; Li et al. 2014). For inhibitory control, transcription rate may be controlled along an analogue scale with many DNA modifications required to govern gene expression associated with the new trace. DNA modifications could therefore serve to tune gene expression and behavioural outcomes in a manner akin to a dimmer switch, dialling down the original fear, or dialling up inhibitory control, with the behavioural impact depending on both sequence context, and surrounding DNA modifications (Figure 1 and Figure 2).
However, much in the way that LTP is only observed after repeated stimulation, there are many cases when the effect of DNA modification on gene expression and behavioural regulation is only evident at the time of reactivation (Baker-Andresen et al. 2013; Liu et al. 2017). This has led to the intriguing hypothesis that DNA modification may serve as a form of genomic meta-plasticity where it can effectively tune the genome in response to experience, which could then dynamically modify the strength of fear extinction memory and performance depending on the context and future experience (Baker-Andresen et al. 2013). For example, it is possible that one reason why only repeated training leads to fear erasure is because fear cells must first be epigenetically primed. Future studies may explore the conditions under which this may occur in order to facilitate more permanent changes.
DNA Structure State
DNA structure states represent another level of DNA that may follow this concept of dynamic tuning. Initially thought to be static, based on the original double-helix structure proposed by Watson and Crick, there is now a long history of observation of alternate and dynamic DNA structures, which may functionally regulate transcription and, possibly, memory traces (Watson & Crick 1953; Felsenfeld et al. 1957). Specifically, particular structures like particular DNA modifications, have been observed to both impair or promote transcription rate, suggesting a non-binary structure-switch potentially playing a role in tuning fear memory (Figure 2 and Figure 1; Naylor & Clark 1990; Siddiqui-Jain et al. 2002). In addition, like DNA modification, it has been proposed that these structures once induced can modify the probability of future structure-switch induction in this genomic context thus providing further support for the idea of genomic priming and tuning (Pohl 1987). The idea of a structure switch has been demonstrated in the context of regulation of the immediate early gene expression thought to be important for fear conditioning. Specifically, Top2B activation is required to cut and repair DNA to promote the expression of immediate early genes (Madabhushi et al. 2015). In unpublished experiments, we have also found that DNA breaks produced by Top2B generate changes in DNA methylation, which must be actively removed to produce multiple waves of transcription, which are then critical for the formation of fear memory (Li et al. 2018). Further work will be required to determine the extent of structure-function relationships, and their interaction with other layers such as DNA modification in fear extinction regulation.
Histone Modification
Histone modifications represent another layer in which epigenetic modification appears to play a role in the formation of extinction memory. Schmitt & Matthies (1979) first observed a relationship between histones and learning, and since then histone modifications have become some of the best established epigenetic mediators of behaviour ranging from acetylation to methylation of histone proteins (Swank & Sweatt 2001; Gupta-Agarwal et al. 2012; Damez-Werno et al. 2016). Furthermore, histone acetylation and methylation are also specifically engaged during reconsolidation (Gupta-Agarwal et al. 2014; Webb et al. 2017; Jarome et al. 2018), and extinction (Itzhak et al. 2012; Stafford et al. 2012; Li et al. 2014). As a result of these marks transcriptional activity is enhanced or inhibited by modifying chromatin accessibility, specifically methylation tends to inhibit transcription by closing the chromatin, while acetylation tends to do the opposite (Rice & Allis 2001). Thus, blocking proteins which promote acetylation such as PCAF impairs extinction (Wei et al. 2012). Similarly, facilitating acetylation by blocking enzymes which remove acetylation either indirectly with histone deacetylase inhibitor drugs (Bredy et al. 2007; Bredy & Barad 2008), or directly by knocking down specific histone deacetylases (HDACs) such as HDAC1 (Bahari-Javan et al. 2012) HDAC2 (Morris et al. 2013) or HDA3 (Malvaez et al. 2013) facilitates extinction.
More interestingly, like DNA modifications, histone modifications have also been shown to be involved in epigenetic priming. Specifically, HDAC inhibitors when given alone appear to have no effect on destabilizing or stabilizing memory, but when given just before or after behavioural experience, or in conjunction with other memory modulators, they can drastically alter memory memory updating (Gräff & Tsai 2013; Gräff et al. 2014). Furthermore, early studies demonstrated that massed vs. spaced extinction training in the presence of an HDAC inhibitor biased the expression of memory from extinction to the enhanced reconsolidation of the original fear (Bredy & Barad 2008). Thus, these reversible modifications appear also to interact with the behavioural conditions, and through these interactions modulate both the original trace and the fear extinction trace by modifying transcription.
Histone Variant Exchange
In addition to posttranslational modification of histone proteins, a variety of histone variants including: H3.3, H2A.Lap1, H2Az and H2BE have been shown to regulate experience dependent gene expression within the context of learning and memory. While H2BE, has been shown to be mainly for learning in the olfactory bulb (Santoro & Dulac 2012) H3.3 (Maze et al. 2015) H2A.Lap1 (Anuar 2018), and H2A.Z (Zovkic et al. 2014) appear to be critical for fear learning. Additionally, like RNA and DNA there appear to be 100’s of unexplored variants that may also participate in extinction learning, each with a potentially specialized role (Draizen et al. 2016). For example, H2A.Z appear to regulate the equilibrium of chromatin architecture such that it biases regions to remain in a transcriptionally poised state (Subramanian et al. 2015).
Furthermore, recent data has made clear that variant presence or absence is not all that governs their function. The turnover rate of these histone variants is also critical to their function such that impairing histone turnover impairs: expression of genes critical for fear expression, cell to cell signalling, and fear expression itself (Maze et al. 2015). Additionally, as animals age the distribution of these variants also changes (Stefanelli et al. 2018). Both of these data can be interpreted within the proposed framework that these variants specify regions of transcriptional activity or silencing (Hake & Allis 2006). Thus one shift that may be observed following repeated behavioural training is local variants composition, such that genomic regions containing transcriptional repressors are activated in fear cells, and disinhibited in inhibitory control cells following extinction training. Together this suggests this epigenetic mechanism can modify transcriptional rate through bidirectionally modifying the histone code, and tune the activity of both the original fear and inhibitory control cells involved in extinction learning.
RNA Modification
A bidirectional and graded regulatory signal driving fear extinction may also extend to RNA-mediated regulatory processes (Figure 3 and Figure 1). For example, RNA methylation in the form of N6-methyldeoxyadenosine (m6A) has been implicated in fear learning (Widagdo et al. 2016; Walters et al. 2017) and that this occurs in part as a consequence of RNA degradation. But with over 140 RNA modifications identified to date (Machnicka et al. 2013), “epitranscriptomic” mechanisms may expand beyond just acting as transcription termination signals by directly modifying the rate of RNA degradation (Machnicka et al. 2013; Nainar et al. 2016; Widagdo et al. 2016) and the efficiency of translation (Wang et al. 2015). RNA modifications may also serve modify the localization of the RNA itself (Wang et al. 2015; Ohtan Wang) or other yet to be characterized interactions in the context of fear extinction learning (Nainar et al. 2016). It is therefore likely that we will see the emergence of novel RNA modifications that influence both the rate of translation and RNA degradation during fear extinction learning, which could then facilitate or inhibit the extinction processes via altering the molecular pathways underlying the memory trace. Theoretically speaking, this might serve to enhance the formation of extinction trace substrates in new inhibitory cells, while enhancing RNA degradation in the cells supporting the original fear trace, which could lead to memory erasure, suggesting yet another opportunity for the continuous tuning of gene expression to control behaviour (Figure 1) which may arise independent of local changes in synaptic activity.
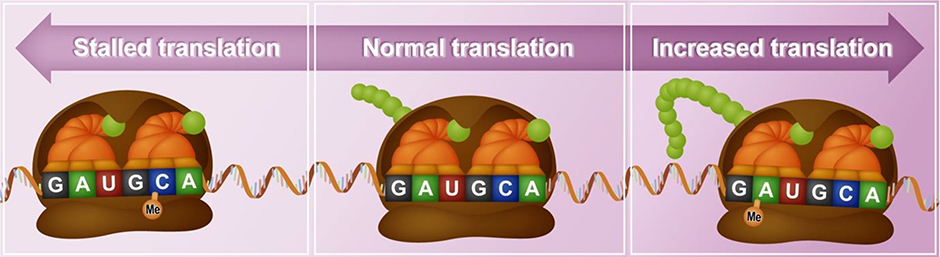
Figure 3.
A second proposed mechanism for controlling behavioural equilibrium during fear extinction is translational tuning in which RNA modifications can modulate translation rate along a continuum. RNA modifications are shown altering physical interactions with the ribosome machinery to modify its translation rate. Here, 5mC methylation in RNA may stall translation, while m6A methylation may enhance translation rate. Thus RNA modifications may dial translation rate up or down depending on the presence of absence of these marks.
RNA Editing
Interestingly, the potential impact of RNA modification on fear extinction may not end here. Another mechanism of RNA metabolism that may play a critical role in fear extinction is RNA editing. This is a biochemical process that enzymatically converts one nucleotide to another to produce diverse types of transcripts without altering the genetic code. For example, ADAR-mediated-editing of the AMPA receptor subunit GluR2, in which conversion of adenosine to inosine changes a glutamine to arginine, can dramatically modify channel permeability and, therefore, fear extinction learning (Clem & Huganir 2010; Wright & Vissel 2012). In addition, the RNA editing-related molecule ADAR3 has recently been shown to be critical for the formation of contextual fear memory (Mladenova et al. 2018). Furthermore, in a series of recent studies, we have found a functional role for another RNA editing enzyme, activation-induced cytidine deaminase (AID), in the regulation of gene expression (Ratnu et al, 2014) and in the formation of fear extinction memory (Marshall, unpublished observations).
These observations are not surprising given the fact that RNA editing has increased throughout evolution in organisms with increased cognitive complexity (Mattick & Mehler 2008). RNA editing may therefore be playing a direct role in fear and extinction memory by regulating translational efficiency such that the presence or absence of critical proteins for inhibiting and facilitating extinction are being bidirectionally ‘tuned’. In this case, the biological variable impacting fear extinction could be described as a ratio of edited to non-edited RNA, such that the complete absence of RNA editing facilitates plasticity and behaviour whereas a ratio above a certain threshold may lead to a reduction in plasticity and behavioural inhibition. This process could have its most relevant impact in the context of a new trace model for extinction by facilitating the expression of functionally distinct proteins, which could impact the stability of the original fear memory and/or the instability of the fear extinction trace.
Limitations of a cognitive neuroepigenetic perspective
Epigenetic modifications may contribute to our understanding of how the formation of fear extinction memory could proceed by either ‘erasure’ of the original trace, the formation of inhibitory memory traces, or the tuning of both in real-time during behaviour. However, much akin to the fact that at the psychological level one cannot differentiate between a lack of retrieval due to permanent erasure or a lack of appropriate retrieval cues, a limitation of this neuroepigenetic view is the inability to determine whether the degradation of the substrates of epigenetic modifications are complete and can therefore be deemed ‘erasure’, or whether the processes have been tuned temporarily to zero. We predict that similar to the electrophysiological perspective, there exists an equilibrium in epigenetic states that can be pushed in one direction or another at multiple levels of molecular control within the cell; likely leading to both erasure and new inhibitory traces depending on: the context, time since acquisition, length of training, or time between sessions (Cain et al. 2003; Myers 2006; Auber et al. 2013; Flavell et al. 2013; Clem & Schiller 2016).
This leads to the second limitation around what to call this process, as acknowledging that retrieval can activate both original fear cells and new inhibitory cells blurs the line between classical definitions of extinction and reconsolidation (Hemstedt et al. 2017). This said, there is little debate that retrieval activates a variety of epigenetic mechanisms that seem to be critical for storage of the fear memory including enzymes which promote DNA and histone modifications (Zhao et al. 2014; Liu et al. 2017). Thus, to overcome this limitation in future, as opposed to labelling manipulations as solely effecting reconsolidation or extinction, it may prove useful to find drugs which can be combined without interfering with each other to maximally potentiate changes in both populations of cells.
Predicting behavioural strength from neuroepigenetic changes within the nucleus
The most significant problem with fear extinction is the persistence or re-emergence of fear after extinction learning and, therefore, clinical preference is given to treatments that aim to achieve total erasure. One way in which total erasure may be achieved is to utilize manipulations which engage genomic priming in conjunction with both behavioural and electrophysiological manipulations, such that they prime the epigenome for a more permanent change. For example, a mechanism which initially only partially modifies the transcription rate, may poise the same locus to be disengaged more robustly thereby leading to a complete cessation of transcription, and resultant electrophysiological activity, which we may present itself as cellular erasure. However, this strategy presents an issue, such that global manipulations which would favourably prime gene activation in extinction cells might also reactive the fear cells, or treatments that prime fear cells for erasure may also erase inhibitory cells. Thus, to achieve the desired behavioural balance with minimal side-effects one must selectively target the cells in which these behavioural relevant changes occur. More specifically, in addition to electrophysiologically defined “fear”, “extinction”, and “anxiety” cells (Milad & Quirk 2002; Herry et al. 2010; Tovote et al. 2015; Jimenez et al. 2018), the precise molecular marks within a cell that allow it to compete more effectively for resources leading to aforementioned behavioural tuning must be identified and linked to neuroepigenetic mechanisms.
One attractive explanation that has been described among others is that higher levels of CREB are critical for inclusion into a fear memory engram, specifically higher levels of CREB predict the cells where the largest changes in intrinsic membrane excitability occur (Silva et al. 1998; Kida et al. 2002; Kim et al. 2013). These observations are likely to represent the physiological correlate of the dominant trace hypothesis that proposes there are mechanisms, which bias a memory trace towards one that commands resources to update or modify a memory, hence guiding which cells are tuned and when (Eisenberg et al. 2003). Recent observations from our laboratory suggest this may be the case for neuroepigenetic states. Specifically, higher levels of Activity-regulated cytoskeleton-associated protein, which occurs in cells with high CREB expression, tend to co-occur in neurons that exhibit higher rates of RNA editing and the accumulation of DNA modifications such as m6dA (Li et al, 2018). Other marks which can differentiate between reconsolidation and consolidation of new extinction traces such as zif268 and BDNF, may also co-occur with neuroepigenetic changes, but this remains to be investigated (Lee et al. 2004). Together, the data suggest the intriguing possibility that if these markers are experimentally specified and utilized, certain neuroepigenetic mechanisms may be selectively engaged in these two discrete population of cells and lead to more permanent behavioural changes.
Implications, impact and future directions
Based on a neuroepigenetic perspective, promising new directions to answer long held questions about the stability of fear extinction memory, and how to target it, are emerging. For example, if epigenetic priming is required for long-term erasure, as has been suggested, re-consolidation protocols with epigenetic modifiers as adjuncts to therapy could be implemented in the clinic (Gräff et al. 2014). In addition, if one accepts that memory is not primarily contained within the synapse, a cognitive neuroepigenetic view leaves room for Lamarkian-like phenomena where information can be acquired in one generation and passed on to the next (Franklin et al. 2010; Fischer 2014). Although the evidence is inconclusive, it has been shown that paternal sperm of defeated mice, potentially through a miRNA-mediated signal effecting DNA methylation, can bias the next generations towards a similar fearful phenotype (Dietz et al. 2011; Rodgers et al. 2015). In this case adolescent behaviour associated with mental health issues may therefore be viewed as epigenetic tendencies, which can be predicted and thus tuned behaviourally in order to minimise the future development of anxiety disorders.
But with so many levels and interactions, where might development of new therapies begin testing? Because of the large amount of data on HDAC effects in animal models and limited side effects (Gräff & Tsai 2013), these have been some of the first epigenetic targets to be tested clinically (Whittle & Singewald 2014). But there still remains a variety of other actionable epigenetic targets which can enhance the tuning of fear memory traces at every level. For example, with respect to RNA, it has been shown that increasing either miR-128b in the prefrontal cortex or miR144–3p in the amygdala can enhance extinction learning (Lin et al. 2011; Murphy et al. 2017). With respect to DNA modification, it has been demonstrated that activity of Tet3, which leads to the accumulation of 5hmC, can also enhance the formation of extinction memory (Li et al. 2014). The problem remains that unlike globally manipulating HDACs global manipulation of these targets can have detrimental side effects. Thus in order to alter these and other levels mentioned it will require the development of more targetted manipulations ranging from specific drugs to gene editing tools like CRISPR (Adli 2018). As well as advancing delivery systems such as self-deleting viral vectors (Russ et al. 1996) or nanoparticles (Gao 2016) to deliver these directly to the affected areas. Treatments of the future may also take this one step further by developing tools which apply knowledge of which molecular markers occur in fear and extinction cells and use this to differentially target these areas following central infusion.
In conclusion, a neuroepigenetic view of fear extinction suggests that, although the formation and stability of fear extinction memory occurs at more levels within the cell than currently appreciated, a better understanding of this tunable equilibrium and how to target it may be another key step to addressing the issue of lingering fear.
Acknowledgements
The authors gratefully acknowledge grant support from the NIH (5R01MH105398-TWB) and the Australian Research Council (SR120300015-TWB). The authors would also like to thank Ms. Rowan Tweedale for helpful editing of the manuscript.
Footnotes
Conflicts of interest: None to declare
References
- Abdou K, Shehata M, Choko K, Nishizono H, Matsuo M, Muramatsu S, Inokuchi K. 2018. Synapse-specific representation of the identity of overlapping memory engrams. Science (80). 360:1227–1231. [DOI] [PubMed] [Google Scholar]
- Adli M 2018. The CRISPR tool kit for genome editing and beyond. Nat Commun. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B, Kim J, Park K, Lee S, Song S, Choi S. 2017. Amount of fear extinction changes its underlying mechanisms. Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anuar ND. 2018. Using TALENs to Knockout H2A.Lap1 Function in Mice. Aust Natl Univ. PhD thesis:1–274. [Google Scholar]
- Ashapkin VV, Romanov GA, Tushmalova NA, Vanyushin BF. 1982. Selective DNA Synthesis in the Rate Brain Induced by Learning. 48:355–362. [Google Scholar]
- Attardo A, Fitzgerald JE, Schnitzer MJ. 2015. Impermanence of dendritic spines in live adult CA1 hippocampus. Nature. 523:592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auber A, Tedesco V, Jones CE, Monfils MH, Chiamulera C. 2013. Post-retrieval extinction as reconsolidation interference: Methodological issues or boundary conditions? Psychopharmacology (Berl). 226:631–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bahr M, Burkhardt S, Delalle I, Kugler S, Fischer A, Sananbenesi F. 2012. HDAC1 Regulates Fear Extinction in Mice. J Neurosci. 32:5062–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Andresen D, Ratnu VS, Bredy TW. 2013. Dynamic DNA methylation: A prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 36:3–13. [DOI] [PubMed] [Google Scholar]
- Baum M 1988. Spontaneous recovery from the effects of flooding (exposure) in animals. Behav Res Ther. 26:185–186. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Lomo T. 1973. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol. 232:331–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton ME, Bolles RC. 1979a. Contextual control of the extinction of conditioned fear. Learn Motiv. 10:445–466. [Google Scholar]
- Bouton ME, Bolles RC. 1979b. Role of conditioned contextual stimuli in reinstatement of extinguished fear. J Exp Psychol Anim Behav Process. 5:368–378. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Barad M. 2008. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 15:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. 2007. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 14:268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain CK, Blouin AM, Barad M. 2003. Temporally Massed CS Presentations Generate More Fear Extinction than Spaced Presentations. J Exp Psychol Anim Behav Process. 29:323–333. [DOI] [PubMed] [Google Scholar]
- Chen S, Cai D, Pearce K, Sun PY, Roberts AC, Glanzman DL. 2014. Reinstatement of long-term memory following erasure of its behavioral and synaptic expression in Aplysia. :1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RL, Huganir RL. 2010. Calcium-permeable AMPA receptor dynamics mediate fear memory erasure. Science (80). 330:1108–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RL, Schiller D. 2016. New Learning and Unlearning: Strangers or Accomplices in Threat Memory Attenuation? Trends Neurosci. 39:340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damez-Werno DM, Sun H, Scobie KN, Shao N, Rabkin J, Dias C, Calipari ES, Maze I, Pena CJ, Walker DM, et al. 2016. Histone arginine methylation in cocaine action in the nucleus accumbens. Proc Natl Acad Sci. 113:9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. 2010. DNA methylation and memory formation. Nat Neurosci. 13:1319–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. 2011. Cognitive neuroepigenetics: A role for epigenetic mechanisms in learning and memory. Neurobiol Learn Mem. 96:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz DM, LaPlant Q, Watts EL, Hodes GE, Russo SJ, Feng J, Oosting RS, Vialou V, Nestler EJ. 2011. Paternal Transmission of Stressed-Induced Pathologies. Biol Psychiatry. 70:408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draizen EJ, Shaytan AK, Mariño-Ramírez L, Talbert PB, Landsman D, Panchenko AR. 2016. HistoneDB 2.0: A histone database with variants - An integrated resource to explore histones and their variants. Database. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y, Eisenberg M. 2004. Rites of passage of the engram: Reconsolidation and the lingering consolidation hypothesis. Neuron. 44:93–100. [DOI] [PubMed] [Google Scholar]
- Eisenberg M, Kobilo T, Berman DE, Dudai Y. 2003. Stability of retrieved memory: Inverse correlation with trace dominance. Science (80). 301:1102–1104. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Davies DR, Rich A. 1957. Formation of a Three-Stranded Polynucleotide Molecule. J Am Chem Soc. 79:2023–2024. [Google Scholar]
- Fischer A 2014. Epigenetic memory: The Lamarckian brain. EMBO J. 33:945–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell CR, Lambert E a, Winters BD, Bredy TW. 2013. Mechanisms governing the reactivation-dependent destabilization of memories and their role in extinction. Front Behav Neurosci. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Grff J, Linder N, Michalon A, Vizi S, Mansuy IM. 2010. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 68:408–415. [DOI] [PubMed] [Google Scholar]
- Gao H 2016. Progress and perspectives on targeting nanoparticles for brain drug delivery. Acta Pharm Sin B. 6:268–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, et al. 2014. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 156:261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. 2013. Histone acetylation: Molecular mnemonics on the chromatin. Nat Rev Neurosci. 14:97–111. [DOI] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin A V., DeRamus T, Wheelock M, Davis RL, McMahon LL, Lubin FD. 2012. G9a/GLP Histone Lysine Dimethyltransferase Complex Activity in the Hippocampus and the Entorhinal Cortex Is Required for Gene Activation and Silencing during Memory Consolidation. J Neurosci. 32:5440–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Jarome TJ, Fernandez J, Lubin FD. 2014. NMDA receptor- and ERK-dependent histone methylation changes in the lateral amygdala bidirectionally regulate fear memory formation. Learn Mem. 21:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD. 2006. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis.” Proc Natl Acad Sci. 103:6428–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, Loshbaugh AL, Kuhlman B, Hahn KM, Kasai H. 2015. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 525:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemstedt TJ, Lattal KM, Wood MA. 2017. Reconsolidation and extinction: Using epigenetic signatures to challenge conventional wisdom. Neurobiol Learn Mem. 142:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herry C, Ciocchi S, Senn V, Demmou L, Müller C, Lüthi A. 2008. Switching on and off fear by distinct neuronal circuits. Nature. 454:600–606. [DOI] [PubMed] [Google Scholar]
- Herry C, Ferraguti F, Singewald N, Letzkus JJ, Ehrlich I, Lüthi A. 2010. Neuronal circuits of fear extinction. Eur J Neurosci. 31:599–612. [DOI] [PubMed] [Google Scholar]
- Hong J, Kim D. 2017. Freezing response-independent facilitation of fear extinction memory in the prefrontal cortex. Sci Rep. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M 1989. Long-Term Depression. Annu Rev Neurosci. 12:85–102. [DOI] [PubMed] [Google Scholar]
- Itzhak Y, Anderson KL, Kelley JB, Petkov M. 2012. Histone acetylation rescues contextual fear conditioning in nNOS KO mice and accelerates extinction of cued fear conditioning in wild type mice. Neurobiol Learn Mem. 97:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Perez GA, Hauser RM, Hatch KM, Lubin FD. 2018. EZH2 Methyltransferase Activity Controls Pten Expression and mTOR Signaling During Fear Memory Reconsolidation. J Neurosci:0538–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JC, Su K, Goldberg AR, Luna VM, Biane JS, Ordek G, Zhou P, Ong SK, Wright MA, Zweifel L, et al. 2018. Anxiety Cells in a Hippocampal-Hypothalamic Circuit. Neuron. 97:670–683.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, De Ortiz SP, Kogan JH, Chevere I, Masushige S, Silva AJ. 2002. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 5:348–355. [DOI] [PubMed] [Google Scholar]
- Kim J, Kwon J-T, Kim H-S, Han J-H. 2013. CREB and neuronal selection for memory trace. Front Neural Circuits. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CSW, Adler A, Gan W-B. 2018. Fear extinction reverses dendritic spine formation induced by fear conditioning in the mouse auditory cortex. Proc Natl Acad Sci:201801504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashley K 1950. In search of the engram. Exp Biol Symp No 4 Physiol Mech Anim Behav:454–482. [Google Scholar]
- Lechner HA, Squire LR. 1999. 100 Years of Consolidation— Remembering Müller and Pilzecker. Learn Mem (Cold Spring Harb NY). [PubMed] [Google Scholar]
- Lee JLC, Everitt BJ, Thomas KL. 2004. Independent Cellular Processes for Hippocampal Memory Consolidation and Reconsolidation. Science (80- ). 304:839–843. [DOI] [PubMed] [Google Scholar]
- Li X, Marshall PR, Leighton LJ, Zajaczkowski EL. 2018. The DNA repair associated protein Gadd45g regulates the temporal coding of immediate early gene expression and is required for the consolidation of associative fear memory. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wei W, Lin Q, Magnan C, Emami M, Wearick-Silva LE, Viola T, Marshall P, Grassi-Oliveira R, Nainar S, et al. 2016. The formation of extinction memory requires the accumulation of N6-methyl-2-deoxyadenosine in DNA. BioRxiv. [Google Scholar]
- Li X, Wei W, Ratnu VS, Bredy TW. 2013. On the potential role of active DNA demethylation in establishing epigenetic states associated with neural plasticity and memory. Neurobiol Learn Mem. 105:125–132. [DOI] [PubMed] [Google Scholar]
- Li X, Wei W, Zhao Q-Y, Widagdo J, Baker-Andresen D, Flavell CR, D’Alessio A, Zhang Y, Bredy TW. 2014. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci. 111:7120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, Bredy TW. 2011. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 14:1115–1117. [DOI] [PubMed] [Google Scholar]
- Liu C, Sun X, Wang Z, Le Q, Liu P, Jiang C, Wang F, Ma L. 2017. Retrieval-Induced Upregulation of Tet3 in Pyramidal Neurons of the Dorsal Hippocampus Mediates Cocaine-Associated Memory Reconsolidation. Int J Neuropsychopharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: a database of RNA modification pathways−−2013 update. Nucleic Acids Res. 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao P-C, et al. 2015. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 161:1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. 2013. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci. 110:2647–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek R, Jin J, Goode TD, Giustino TF, Wang Q, Acca GM, Holehonnur R, Ploski JE, Fitzgerald PJ, Lynagh T, et al. 2018. Hippocampus-driven feed-forward inhibition of the prefrontal cortex mediates relapse of extinguished fear. Nat Neurosci. 21:384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall P, Bredy TW. 2016. Cognitive neuroepigenetics: the next evolution in our understanding of the molecular mechanisms underlying learning and memory? NPJ Sci Learn. 1:16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Mehler MF. 2008. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 31:227–233. [DOI] [PubMed] [Google Scholar]
- Maze I, Wenderski W, Noh KM, Bagot RC, Tzavaras N, Purushothaman I, Elsässer SJ, Guo Y, Ionete C, Hurd YL, et al. 2015. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron. 87:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. 2000. Memory--a century of consolidation. Science. 287:248–251. [DOI] [PubMed] [Google Scholar]
- Milad MRR, Quirk GJJ. 2002. Neurons in medial prefrontal cortex signal memory for fear extinction. Nature. 420:70–74. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. 2007. Covalent Modification of DNA Regulates Memory Formation. Neuron. 53:857–869. [DOI] [PubMed] [Google Scholar]
- Misanin JR, Miller RR, Lewis DJ. 1968. Retrograde Amnesia Produced by Electroconvulsive Shock after Reactivation of a Consolidated Memory Trace. Science (80- ). 160:554–555. [DOI] [PubMed] [Google Scholar]
- Mladenova D, Barry G, Konen LM, Pineda SS, Guennewig B, Avesson L, Zinn R, Schonrock N, Bitar M, Jonkhout N, et al. 2018. Adar3 Is Involved in Learning and Memory in Mice. Front Neurosci. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Mahgoub M, Na ES, Pranav H, Monteggia LM. 2013. Loss of Histone Deacetylase 2 Improves Working Memory and Accelerates Extinction Learning. J Neurosci. 33:6401–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CP, Li X, Maurer V, Oberhauser M, Gstir R, Wearick-Silva LE, Viola TW, Schafferer S, Grassi-Oliveira R, Whittle N, et al. 2017. MicroRNA-Mediated Rescue of Fear Extinction Memory by miR-144–3p in Extinction-Impaired Mice. Biol Psychiatry. 81:979–989. [DOI] [PubMed] [Google Scholar]
- Myers KM. 2006. Different mechanisms of fear extinction dependent on length of time since fear acquisition. Learn Mem. 13:216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S, Fox R, Proulx CD, Lin JY, Tsien RY, Malinow R. 2014. Engineering a memory with LTD and LTP. Nature. 511:348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader K, Schafe GE, Le Doux JE. 2000. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 406:722–726. [DOI] [PubMed] [Google Scholar]
- Nainar S, Marshall PR, Tyler CR, Spitale RC, Bredy TW. 2016. Evolving insights into RNA modifications and their functional diversity in the brain. Nat Neurosci. 19:1292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor LH, Clark EM. 1990. D(TG)n·d(CA)nsequences upstream of the rat prolactin gene form z-DNA and inhibit gene transcription. Nucleic Acids Res. 18:1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov IP. 1927. Conditioned reflexes: an investigation of the physiological activity of the cerebral cortex. Oxford Univ Press:xv–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl FM. 1987. Hysteretic behaviour of a Z-DNA-antibody complex. Biophys Chem. 26:385–390. [DOI] [PubMed] [Google Scholar]
- Poo M ming, Pignatelli M, Ryan TJ, Tonegawa S, Bonhoeffer T, Martin KC, Rudenko A, Tsai LH, Tsien RW, Fishell G, et al. 2016. What is memory? The present state of the engram. BMC Biol. 14:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk GJ, Mueller D. 2008. Neural mechanisms of extinction learning and retrieval. Neuropsychopharmacology. 33:56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescorla R a, Heth CD. 1975. Reinstatement of fear to an extinguished conditioned stimulus. Journal of Experimental Psychology: Anim Behav Process. 104:88–96. [PubMed] [Google Scholar]
- Rice JC, Allis CD. 2001. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol. 13:263–273. [DOI] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Leu NA, Bale TL. 2015. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci. 112:13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Selker EU. 1997. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 11:2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty M, Seo J, Cheng A, Meng J, Le T, Faull K, Jaenisch R, Tsai LH. 2013. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 79:1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ a P, Friedel C, Grez M, von Melchner H. 1996. Self-deleting retrovirus vectors for gene therapy. J Virol. 70:4927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. 2012. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. Elife. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucier D, Cain DP. 1995. Spatial learning without NMDA receptor-dependent long-term potentiation. Nature. 378:186–189. [DOI] [PubMed] [Google Scholar]
- Schmitt M, Matthies H. 1979. [Biochemical studies on histones of the central nervous system. III. Incorporation of [14C]-acetate into the histones of different rat brain regions during a learning experiment]. Acta Biol Med Ger. 38:683–689. [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci. 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. 1998. Creb and Memory. Annu Rev Neurosci. 21:127–148. [DOI] [PubMed] [Google Scholar]
- Stafford JM, Lattal KM. 2011. Is an epigenetic switch the key to persistent extinction? Neurobiol Learn Mem. 96:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM. 2012. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol Psychiatry. 72:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanelli G, Azam AB, Walters BJ, Brimble MA, Gettens CP, Bouchard-Cannon P, Cheng HYM, Davidoff AM, Narkaj K, Day JJ, et al. 2018. Learning and Age-Related Changes in Genome-wide H2A.Z Binding in the Mouse Hippocampus. Cell Rep. 22:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Fields PA, Boyer LA. 2015. H2A.Z: a molecular rheostat for transcriptional control. F1000Prime Rep. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD. 2001. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci. 21:3383–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovote P, Fadok JP, Lüthi A. 2015. Neuronal circuits for fear and anxiety. Nat Rev Neurosci. 16:317–331. [DOI] [PubMed] [Google Scholar]
- Vanyushin BF, Ashapkin V V. 2017. History and Modern View on DNA Modifications in the Brain. In: DNA Modif Brain. [place unknown]; p. 1–25. [Google Scholar]
- Walters BJ, Mercaldo V, Gillon CJ, Yip M, Neve RL, Boyce FM, Frankland PW, Josselyn SA. 2017. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology. 42:1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FHC. 1953. Molecular structure of nucleic acids. Nature. 171:737–738. [DOI] [PubMed] [Google Scholar]
- Webb WM, Sanchez RG, Perez G, Butler AA, Hauser RM, Rich MC, O’Bierne AL, Jarome TJ, Lubin FD. 2017. Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol Learn Mem. 142:66–78. [DOI] [PubMed] [Google Scholar]
- Wei W, Coelho CM, Li X, Marek R, Yan S, Anderson S, Meyers D, Mukherjee C, Sbardella G, Castellano S, et al. 2012. p300/CBP-associated factor selectively regulates the extinction of conditioned fear. J Neurosci. 32:11930–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle N, Singewald N. 2014. HDAC inhibitors as cognitive enhancers in fear, anxiety and trauma therapy: where do we stand? Biochem Soc Trans. 42:569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Zhao Q-Y, Kempten M-J, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V, Bredy TW. 2016. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J Neurosci.:In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo XJ, Zhao XQ, Kempen XM, Tan XMC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono XV, Bredy XTW. 2016. Experience-Dependent Accumulation of N 6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J Neurosci. 36:6771–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Vissel B. 2012. The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, Park J, Ji SJ. 2018. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46:1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W-N, Malinin N, Yang F-C, Staknis D, Gekakis N, Maier B, Reischl S, Kramer A, Weitz CJ, Sun M, et al. 2014. Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. PLoS One. 9:690–699. [Google Scholar]
- Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD. 2014. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature. 515:582–586. [DOI] [PMC free article] [PubMed] [Google Scholar]