Abstract
The Ser/Thr protein kinase MELK (maternal embryonic leucine zipper kinase) has been considered an attractive therapeutic target for managing cancer since 2005. Studies using expression analysis have indicated that MELK expression is higher in numerous cancer cells and tissues than in their normal, nonneoplastic counterparts. Further, RNAi-mediated MELK depletion impairs proliferation of multiple cancers, including triple-negative breast cancer (TNBC), and these growth defects can be rescued with exogenous WT MELK, but not kinase-dead MELK complementation. Pharmacological MELK inhibition with OTS167 (alternatively called OTSSP167) and NVS-MELK8a, among other small molecules, also impairs cancer cell growth. These collective results led to MELK being classified as essential for cancer proliferation. More recently, in 2017, the proliferation of TNBC and other cancer cell lines was reported to be unaffected by genetic CRISPR/Cas9-mediated MELK deletion, calling into question the essentiality of this kinase in cancer. To date, the requirement of MELK in cancer remains controversial, and mechanisms underlying the disparate growth effects observed with RNAi, pharmacological inhibition, and CRISPR remain unclear. Our objective with this review is to highlight the evidence on both sides of this controversy, to provide commentary on the purported requirement of MELK in cancer, and to emphasize the need for continued elucidation of the functions of MELK.
Keywords: RNA interference (RNAi), inhibitor, CRISPR/Cas, cancer, cell proliferation, serine/threonine protein kinase, drug target, maternal embryonic leucine zipper kinase (MELK), OTS167, target specificity
A brief overview of MELK: From discovery to controversy
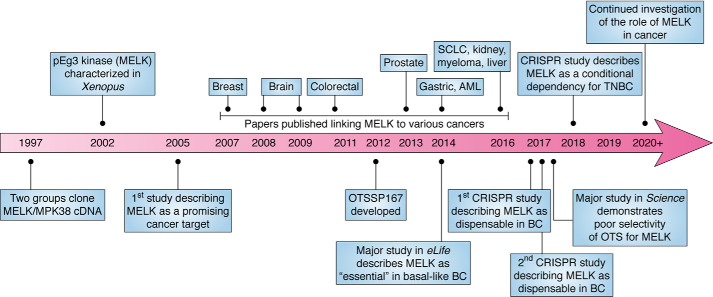
We begin this review with a brief chronological history of MELK, to provide context before analyzing the controversy concerning the requirement of this kinase in cancer (see Fig. 1 for an accompanying timeline). In 1997, MELK cDNA was cloned for the first time, in two studies published in rapid succession. The first group sought to identify genes that were important in embryonic genome activation. They ultimately cloned the cDNA of a gene containing a kinase domain sharing significant sequence identity with the Snf1/AMPK family kinases. The cloned kinase also contained a leucine zipper motif, and transcription of this gene was increased in mouse preimplantation embryos. Accordingly, the kinase was named MELK (maternal embryonic leucine zipper kinase) (1). Shortly after this, another group cloned the same gene from mice, naming it MPK38 (murine protein serine/threonine kinase 38) (2). Five years later, the MELK homolog was characterized in Xenopus and named pEg3 (no acronym defined) (3), not to be confused with PEG3 (paternally expressed gene 3) (4). In this study, pEg3 was demonstrated to be phosphorylated in a cell cycle–dependent manner to regulate its activity, which is maximal in mitosis (3). Naming conventions converged on MELK within a few years, although the name MPK38 is still used sporadically in the literature.
Figure 1.
Timeline of major MELK milestones in cancer. The cancer types shown do not represent a comprehensive list of all cancers for which MELK has been described to play a role in proliferation. AML, acute myeloid leukemia; SCLC, small-cell lung cancer; BC, breast cancer.
In 2005, the first study characterizing MELK as a promising cancer target was published. Here, it was noted that MELK RNA levels were elevated in a panel of 20 cancer tissues, relative to matched normal samples, and that siRNA-mediated MELK knockdown decreased proliferation of cervical, breast, colorectal, and pancreatic cancer cell lines (5). This publication spawned dozens of subsequent studies over the next 15 years, demonstrating increased MELK expression in numerous cancers, including glioblastoma, breast, prostate, and gastric, and slowed proliferation of these and other cancers as a result of RNAi-mediated MELK depletion (6–9). As the body of literature implicating MELK as a promising therapeutic target grew, the first MELK inhibitor (OTSSP167 (OTS), also called OTS167) was developed in 2012 (10). OTS effectively impairs growth and induces apoptosis of numerous cancer types, including breast cancer, acute myeloid leukemia, and small-cell lung cancer, and is currently in four clinical trials (10–12). Because of its status as the leading MELK inhibitor, OTS has been used in most functional studies of MELK since its development. It has recently been definitively demonstrated that OTS has extremely poor selectivity for MELK (13–16), yet unfortunately, it is still regularly used as a tool compound to investigate MELK function.
The start of the controversy currently surrounding the requirement of MELK in cancer can be traced back to a comprehensive 2014 study, in which MELK was demonstrated to be essential for the proliferation of basal-like breast cancer (BLBC) cells, one of the most aggressive subtypes of breast cancer. This study utilized RNAi-mediated MELK knockdown to demonstrate slowed proliferation and essentiality and further showed that growth effects in cells and tumors could be rescued with exogenous WT MELK expression, but not with kinase-dead MELK (17). Three years later, in 2017, another group used CRISPR/Cas9 genomic knockout to show that MELK is not required in triple-negative breast cancer (TNBC) and other cancer types (18). This publication was quickly followed by a study from the group that originally showed essentiality, now demonstrating that MELK is not required for TNBC cell proliferation, based upon genomic MELK knockout and other experimental techniques (16). To further complicate the situation, members of this same group published another study in 2018, asserting that TNBC cells actually have a conditional dependence on MELK for proliferation (19). No additional studies have been completed to specifically address this controversy. The essentiality of MELK for cancer cell proliferation is thus currently contested.
Examining the requirement for MELK in cancer
The sole purpose of this review is to discuss the present controversy concerning the requirement of MELK in cancer. Thus, we will not chronicle what is known about MELK regulation, substrates, and functions in specific cancer processes. For more information on these topics, we refer the reader to an excellent review by Pitner et al. (20).
Importantly, while we will raise concerns about the interpretation of specific results and technical considerations, we will not ultimately take a stance on which side of the controversy we believe is correct. We feel that it would be impossible to justifiably do so at this point in time. Rather, we aim to describe the relevant findings on both sides of the controversy and identify key areas requiring further investigation, while also highlighting some important principles broadly applicable to the scientific community.
Evidence supporting the requirement for MELK in cancer
Perhaps the most robust evidence implicating MELK as an important mediator of cancer progression comes in the form of expression analyses between cancerous and normal tissues and cells. Numerous studies have used microarray and TCGA analysis or immunoblotting methods to demonstrate that expression of MELK RNA or protein is significantly increased in neoplastic cells. This effect seems to be a characteristic that broadly defines many cancer types, as it has been described in breast (17, 21, 22), brain (glioma (6, 23), astrocytoma (24), and neuroblastoma (25)), liver (26), prostate (8), bladder (27), and endometrial cancers (28), among others (5). Further, high levels of MELK expression correlate with high-grade tumors, increased aggressiveness, poor patient outcomes, and radioresistance (17, 21, 22, 25–29). Increased MELK expression has been linked to concurrent up-regulation of genes important or essential for cell cycle progression, including CDK1, CCNB1/2, TOP2A, AURKB, PLK1, and BUB1 (8, 24, 30–33), suggesting that MELK likely also plays a role in this process.
The effects of RNAi-mediated MELK knockdown have resolutely indicated that MELK expression is vital to the proliferation and survival of cancer cells. In cancers, including basal-like breast (17), endometrial (28), glioma (6), acute myeloid leukemia (11), high-risk neuroblastoma (25), and hepatocellular carcinoma (26), MELK depletion has been shown to slow or halt proliferation of cancer cells and tumors. Some studies have additionally demonstrated that knockdown of MELK sensitizes cells to radiation (17, 22, 34, 35) and impairs migration (8, 9, 28, 36). Crucially, a number of studies have rescued the antiproliferative effects of RNAi-mediated MELK knockdown with ectopic MELK expression, indicating that these effects are specifically due to loss of MELK (6, 17, 23, 26, 37, 38). These results were extended by two studies that demonstrated that complementation of knockdown with kinase-dead MELK-D150A or -T167A does not restore normal growth, indicating that catalytic activity is required for rescue (17, 38). Additionally, the oncogenic potential of MELK has been shown using rodent fibroblasts that expressed a dominant negative form of p53 (Rat1-p53DD), which necessitate only one oncogenic event for neoplastic transformation. When WT, but not kinase-dead, MELK was overexpressed, these cells gained the ability to grow in an anchorage-independent manner and form tumors in vivo, evidence of transformation indicating that MELK may function as a driver oncogene (17).
It was recently demonstrated that CRISPR/Cas9-mediated MELK knockout impaired proliferation and induced apoptosis in chronic lymphocytic leukemia cells (36). It should be noted that this is the only study that has shown a growth effect resulting from MELK knockout. Other studies that demonstrate no phenotype in MELK knockout cells will be discussed in the next section.
Multiple MELK inhibitors have been developed as potential cancer therapeutics (reviewed previously by Pitner et al. (20)). The molecules OTS (10), NVS-MELK8a (14, 39), HTH-01-091 (16), MELK-T1 (40), IN17 (41), and C1 (23) have all demonstrated preclinical efficacy in slowing cancer cell proliferation, with OTS exhibiting such potent, broad anti-neoplastic effects that it has advanced to phase I/II clinical trials (11, 12, 25, 36, 42–44). Certainly, all small-molecule inhibitors exhibit some degree of polypharmacology, so the antiproliferative effects of these compounds cannot be solely attributed to MELK inhibition. For the inhibitor NVS-MELK8a, the viability of TNBC cells has been shown to be reduced at concentrations at which NVS-MELK8a is highly selective for MELK, suggesting that inhibition of MELK is a major contributor to its antiproliferative effects (14).
Evidence against the requirement for MELK in cancer
On the other side of the controversy are a series of studies that used CRISPR/Cas9 to genetically knock out MELK, revealing no growth phenotype in MELK-null cell lines. The first of these studies used both single and double guide RNA (gRNA) strategies to genetically delete MELK from a panel of TNBC cell lines, with some experiments additionally completed in other cancers. For the single gRNA strategy, seven independent gRNAs were used to generate pooled MELK-null A375, Cal51, and MDA-MB-231 cell lines, and these lines were compared with Rosa26 (a nonessential, noncoding gene) knockout control lines in growth assays. Both cell proliferation and anchorage-independent growth were unaffected by MELK knockout. The double-gRNA strategy was used to induce DNA cutting in two places, excising a portion of the MELK gene that encodes for residues essential for ATP binding in two TNBC cell lines. Long-term growth assays were repeated with these MELK knockout cell lines, and again they were found to proliferate normally, indicating that MELK is not required for TNBC proliferation (18).
Reasoning that signaling pathway adaptations may have occurred during the extensive time required for clonal selection, which could restore normal cell proliferation, the authors employed a GFP dropout assay to assess cell proliferation more proximal to MELK deletion. Briefly, seven Cas9-expressing TNBC cell lines were transduced with seven independent gRNAs targeting MELK or three gRNAs targeting Rosa26, RPA, or PCNA. Transductions were done at a low multiplicity of infection to ensure that some portion of cells remained untransduced. gRNA plasmids additionally expressed GFP, which allowed for the proportion of GFP+ to GFP− cells to be monitored over five passages to assess the relative fitness of cells that had been transduced with MELK or control gRNA relative to their untransduced counterparts. Less than 2-fold dropout of MELK and Rosa26 knockout cells was observed, compared with 5–100-fold dropout of the positive control RPA or PCNA knockout cells, again suggesting that MELK is not a requirement for TNBC proliferation. Crucially, the methods for this study indicate that the baseline GFP+/GFP− measurement was not taken until 3 days post-transduction, with the first reading assessing GFP dropout measured 3–4 days later. Whereas the authors state that this GFP dropout assay was used in an effort to test the effects of MELK knockout immediately following MELK loss, the 3 days that passed between gRNA transduction and baseline readings could certainly still allow ample time for cellular reprogramming that restores normal growth (18).
The next CRISPR study replicated many of the results described above, while also using additional techniques to test MELK dependence (16). Again, MELK-null MDA-MB-468 cells were observed to have no growth phenotype relative to control cells. A chemical-induced protein degradation approach was additionally employed to show that MELK loss had no immediate or prolonged effects on BLBC proliferation. The authors state that MELK expression was maintained throughout the process of creating these cell lines, by first stably expressing an FKBP12-MELK fusion that could be selectively degraded with an engineered degrader molecule and then deleting endogenous MELK with CRISPR. Importantly, it was never demonstrated that this MELK fusion is similarly functional to endogenous MELK (i.e. interactions and substrate phosphorylations are unperturbed by FKBP12 fusion). It therefore cannot be ruled out that this cell line acted as another MELK knockout line, which has had ample time during clonal selection for reprogramming of signaling pathways, that additionally expresses a nonfunctional MELK fusion.
This study also demonstrated quite convincingly that MELK knockdown with CRISPR interference (CRISPRi) has no effect on BLBC cell proliferation. MDA-MB-468 cells were transduced with KRAB-dCas9, a catalytically dead version of Cas9 that allows for transcriptional repression of the gRNA-targeted gene. Multiple gRNAs targeting the MELK promoter were shown to efficiently decrease MELK transcript and protein levels, and five of these gRNAs were cloned into a doxycycline-inducible vector and stable cell lines were generated. Using this system, MELK knockdown by doxycycline treatment was not found to significantly decrease cell proliferation in any of the five cell lines. It should be noted that, while not statistically significant, all cell lines did exhibit slightly decreased growth (∼5–20%) when treated with doxycycline, compared with untreated controls (16).
A follow-on study to the first MELK CRISPR paper was completed by the same group roughly a year later, in which they expanded upon their previous results (45). In stark contrast to previous results by Wang et al. (17), MELK overexpression was found to be insufficient for neoplastic transformation of immortalized cell lines as measured by anchorage-independent growth assays (45). Multiple cell models were used to test transformation potential, including the Rat1-p53DD system used in the study by Wang et al. (17). Similarly, MELK knockout TNBC, melanoma, and colorectal cell lines did not exhibit impaired anchorage-independent growth. This study additionally found that genetic deletion of MELK did not impair proliferation of cancer cells plated at varying densities in crystal violet assays and had no effect on cancer cell sensitivity to five common chemotherapeutic agents or to metabolic stresses, including hypoxia, glucose limitation, or exposure to reactive oxygen species. CRISPR-mediated MELK knockout also had no effect on the proliferation of tumor xenografts in mice (45).
Commentary on the MELK requirement controversy
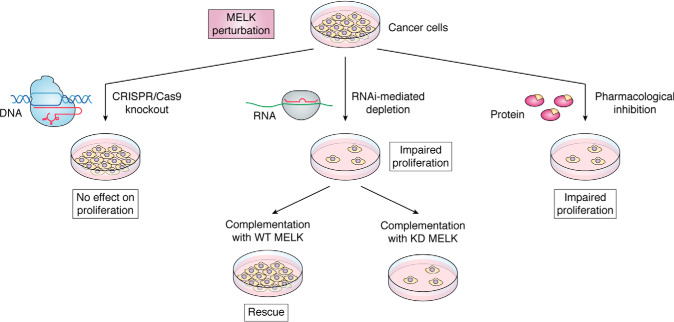
The disparate proliferative effects of perturbation of MELK expression or activity with RNAi, pharmacological inhibition, and CRISPR are summarized in Fig. 2. In this commentary, we will provide insights into these results, identify gaps in knowledge that should be addressed to improve the field's understanding of the importance of MELK in cancer, and make recommendations for future studies.
Figure 2.
Summary of the effects of different methods of MELK perturbation on cancer cell proliferation. Decreased MELK expression or activity has differing effects on cancer cell proliferation, depending upon the technique used (CRISPR/Cas9, RNAi, or pharmacological inhibition). CRISPR/Cas9 genetic knockout of MELK has no effect on proliferation of TNBC and other cancer cells. RNAi-mediated MELK depletion impairs proliferation of TNBC and other cancer cells. These anti-proliferative effects can be rescued with exogenous WT MELK, but not with kinase-dead (KD) MELK complementation. Pharmacological inhibition of MELK with OTS, NVS-MELK8a, and other inhibitors impairs proliferation of TNBC and other cancer cells.
Some of the results suggesting that MELK is a cancer requirement have been recently refuted or can now be viewed with added perspective. Specifically, neoplastic transformation of immortalized cells was demonstrated with MELK overexpression in one model (17), but these results were later convincingly refuted by another group using the same and additional models (45). Results from dozens of studies have demonstrated that MELK expression is up-regulated in cancer and that higher levels of MELK correlate with tumor grade and poor prognosis. However, these results are purely correlative in nature. It has been suggested that MELK may be up-regulated as part of a cell cycle/mitotic cluster of genes in cancer cells simply because these cells proliferate more rapidly than their nonneoplastic counterparts (45). Additionally, it must be recognized that cancer cells may up-regulate expression of certain proteins for reasons besides the regulation of cell proliferation. In the case of MELK, increased expression may affect DNA damage response to influence genetic instability of cancer cells, or it may be important for maintenance of stemness in the cancer stem cell niche. MELK has been linked to both processes previously (27, 34, 40, 46, 47). Finally, multiple MELK inhibitors have demonstrated antiproliferative effects, but it is not possible to definitively conclude that the observed effects are due solely to MELK inhibition. The possibility of off-target inhibition contributing to observed phenotypes cannot be wholly disproved. None of these results provide definitive evidence that MELK is a cancer dependency.
To expand upon the MELK inhibitor landscape, OTSSP167 is considered the leading MELK inhibitor, and it has been used nearly universally in functional studies of MELK. In the original publication that described the development and characterization of OTS, it was lauded as a MELK-specific inhibitor. However, the authors never showed any data to prove this claim—not a single selectivity experiment was presented in this or subsequent studies (10). It has since been revealed that OTS has remarkably poor selectivity for MELK. In a landmark study by Klaeger et al. (13), researchers used an established MS-based selectivity profiling method (kinobeads) to measure the selectivity of 243 Food and Drug Administration–approved and clinical kinase inhibitors. OTS was specifically labeled as a “very broad multikinase inhibitor,” with poorer selectivity than 75% of all molecules that were tested. While a potent inhibitor of MELK (Kdapp = 29 nm), OTS was classified as having 107 other protein kinase targets at this concentration. Further, the binding affinity of OTS for MELK is lower than its binding affinity for 15 other protein kinases (see supporting material of the Klaeger et al. study (13) for quantifications). Using a similar MS-based selectivity-profiling method (MIB/MS), our group recently confirmed the low specificity of OTS (14). We showed that 12 protein kinases were inhibited more than MELK by 1 μm OTS, and 52 kinases (of 235 that were quantified) were inhibited ≥75% at this concentration. Strikingly, 8 of the 12 kinases that we found to be higher-affinity OTS binders than MELK overlapped with the 15 high-affinity binders identified by Klaeger et al. (13). These kinases (CSNK2A2, RIPK2, CSNK2A1/3, GAK, STK10, DYRK1A, LATS1, and TAOK3) should thus be considered bona fide high-affinity targets of OTS. Finally, another study profiled the selectivity of this compound using an in vitro platform (DiscoverX), revealing that 100 nm OTS inhibited 189 of 403 kinases >65%, and 69 of 403 kinases >99% (48).
For certain, there is a lesson to be learned here: one should not simply take authors at their word without analyzing the data underlying a statement or conclusion. However, numerous groups have used, and continue to use, OTS for studies of MELK function, simultaneously parroting the original claim that it is a selective MELK inhibitor and failing to present any data to back up that claim. This has created a self-reinforcing cycle, whereby each group assumes that others have done the diligence of validating that OTS is truly MELK-specific.
Undoubtedly, the widespread use of OTS as the primary inhibitor for functional MELK studies has facilitated the enigmatic nature of this kinase. It is impossible to draw conclusions about the functions of a kinase using a small-molecule tool compound that inhibits over a dozen kinases with similar potency as that for MELK. We also would like to explicitly dispel the notion that OTS may have improved selectivity at low concentrations. This is a wholeheartedly counterintuitive statement, and one that has been disproven by the aforementioned work of Klaeger et al. (13). One would expect the selectivity profile of OTS to remain unchanged at lower concentrations, such that MELK and off-target kinases are all inhibited to a lesser extent, but the proportionality of inhibition is maintained. It would be quite surprising to find that, at lower concentrations, MELK remains highly inhibited, while off-target inhibition simply falls away. Unless, and until, data are presented proving that OTS has improved selectivity at low concentrations, it must be considered a poorly selective inhibitor regardless of the concentration at which it is used. Accordingly, we recommend that OTS not be used for any future functional studies of MELK. As an alternative, NVS-MELK8a should be used, as it has been shown to exhibit excellent selectivity in both enzyme- and cell-based assays (14, 39). Further characterization of other seemingly selective inhibitors, such as MELK-T1 (40, 49), would also be beneficial to the field.
Compelling evidence of MELK's importance in cancer is provided by some results. Numerous studies have demonstrated that RNAi-mediated MELK knockdown slows proliferation, induces apoptosis, and decreases migration and radioresistance of cancer cells. Whereas it has been documented that RNAi approaches often have off-target silencing effects (50), the use of multiple shRNAs or siRNAs in a single study hedges against this possibility. When it is observed that they all cause similar phenotypes, it suggests that depletion of the common target of the sh/siRNA (i.e. MELK) was responsible for the observed phenotypes. When this is expanded to a large number of studies (∼30, compiled in a commentary by Settleman et al. (51)) completed by separate groups, all showing evidence of similar antiproliferative phenotypes in varied cancers, it provides quite strong evidence that the phenotypes observed are due to MELK loss as opposed to off-target effects. Even stronger evidence that the observed antiproliferative effects are due specifically to MELK loss were provided by the studies showing successful rescue experiments. Six studies, all completed by different groups, have rescued the effects of MELK knockdown with exogenous MELK expression (6, 17, 23, 26, 37, 38), including at least two that demonstrated a failure to rescue with kinase-dead MELK (17, 38).
The only partially contrasting results come from the study that used CRISPRi to show that depletion of MELK transcript had no significant growth effect in TNBC cells (16). This has only been demonstrated in one study, compared with a large body of work demonstrating that MELK depletion with RNAi has antiproliferative effects. It is thus imperative to further test and expand upon these CRISPRi results. Further, it is important to note that CRISPRi is an orthogonal approach to RNAi, not an identical one. Differences in observed effects may therefore be due to technical differences between the approaches. This CRISPRi result also does not account for or explain the results observed with rescue experiments. The only effort ever made to rebut rescue experiment results appeared in the discussion of the paper that showed no growth phenotype following CRISPRi-mediated MELK depletion (16). The authors stated that, “Since the previous study was able to rescue the antiproliferative activity observed for shMELK-2 using an shRNA-resistant MELK (Wang et al., 2014), we postulated that the potential off-target of shMELK-2 might only manifest its effect in the presence of MELK knockdown, a so-called 'synthetic lethal' interaction.” Absent any data to support this claim, we find this to be an extremely unlikely scenario to have occurred in one study, and a near-impossibility when considering that six independent groups, each using different RNAi knockdown and rescue reagents, have successfully rescued growth phenotypes with exogenous MELK.
Still on the side of the controversy rebutting a MELK requirement, there are three studies that have demonstrated that CRISPR-mediated MELK knockout has no effect on proliferation of TNBC and other cancer cells (16, 18, 45). Recently, contrasting results have been presented, indicating that MELK knockout induces apoptosis and slows proliferation of chronic lymphocytic leukemia cells (36), although the former three studies were more thorough in their analyses of MELK deletion and subsequent growth assays. It is possible that technical or cell line–dependent differences account for the conflicting results between these studies. Importantly, in the studies that showed no growth phenotype as a result of MELK knockout, the methods used do not preclude the possibility that cellular reprogramming of signaling pathways occurred to circumvent MELK loss and restore normal growth. There are numerous examples in the literature of cellular reprogramming in response to pharmacological inhibition and RNAi. Duncan et al. (52) used a chemical proteomics technique known as MIB/MS to show that, in response to MEK inhibition with AZD6244 (selumetinib) or depletion with RNAi, kinome reprogramming occurs to increase signaling through PDGFRβ and other receptor tyrosine kinase pathways. Notably, this cellular reprogramming was observed as soon as 24 h after treatment and ultimately acted to restore normal proliferation (52). We direct the reader to a review by Graves et al. (53) for more examples and an in-depth discussion of the phenomenon of kinome reprogramming. To our knowledge, cellular reprogramming in response to CRISPR/Cas9 genome editing has not been similarly investigated. We recommend that MS-based approaches be employed to begin to interrogate signaling pathway reprogramming that may occur as a result of MELK knockout in cancer cells.
Whereas the CRISPR studies showed compelling evidence that MELK is not a cancer cell dependency, efforts should be made to more comprehensively characterize MELK knockout cell lines. There is at least one documented instance of purported knockout cell lines actually still expressing splice variants of the targeted protein. Bub1 was thought to be essential for proper functioning of the spindle assembly checkpoint (SAC), until CRISPR-mediated Bub1 knockout cells were shown to have a functional SAC. Recently, however, RT-PCR and MS-based approaches were used to show that these Bub1 “knockout” cells still expressed low levels of alternatively spliced Bub1 or Bub1 protein with a small deletion. Subsequent experiments demonstrated that Bub1 is indeed essential to the SAC and that very low levels of Bub1 were sufficient to restore normal SAC functioning (54–56). This level of evasion of CRISPR-mediated gene deletion is almost certainly more of an exception than a rule. Nonetheless, given the unexpected disparate findings in the MELK field, we advocate for further characterization of MELK knockout cell lines using MS-based and RNA-Seq approaches, to explore the possibility that these cells express alternatively spliced MELK or other gene- or protein-level adaptations that evade complete MELK knockout.
Overall, assuming no technical shortcomings, CRISPR-based results indicate that MELK is not a strict requirement for cancer cell proliferation. The abundant RNAi studies, however, indicate that MELK does play an important yet poorly defined role in cancer. How does the field rectify this apparent paradox? One possible explanation is that MELK is dispensable under some conditions, but required, or at least highly important, under others. In this vein, members of the group that initially labeled MELK essential for BLBC cell proliferation (17), and later showed it was not necessary for proliferation of these cells (16), recently published a study indicating that TNBC cells have a conditional dependence on MELK for growth (19). In this study, TNBC cells were transduced with a gRNA (plus Cas9) that caused efficient reduction of MELK expression and infected cells were selected with antibiotic. As with the approach utilized in the first MELK CRISPR study (selection of GFP+ MELK-null cells) (18), a heterogeneous population of cells was used as opposed to clonal selection. MELK knockout was shown to have no effect on the proliferation of TNBC cells in short-term growth assays, in which cells were seeded at medium to high density (MDA-MB-231 cells, 50,000 cells/well seeded on a 12-well plate, 3-day assay). However, when MELK was depleted and cells were seeded at low density (MDA-MB-231 cells, 500 cells/well seeded on a 12-well plate, 10-day assay), they exhibited markedly slower proliferation relative to control cells. These experiments were also completed with gRNAs targeting the essential mitotic genes AURKB and PLK1 and the oncogenes KRAS and MYC. CRISPR-mediated depletion of AURKB and PLK1 slowed proliferation of cells seeded at high or low densities, indicating that the effects of knockout of truly essential genes occur independently of assay conditions. Depletion of KRAS and MYC caused moderate decreases in proliferation (50%) when cells were seeded at high densities and more markedly impaired proliferation (>90%) when seeded at lower densities (19). Collectively, these results suggest that MELK is likely not universally essential in cancer cells, but under specific growth conditions, MELK expression seems to be important. MELK may conditionally function as an oncogene without being a strict essentiality for cancer cells.
Authors of some of the CRISPR studies have suggested that MELK does not play an important role, or may not even play any role, in mammalian biology and the cell cycle progression of cancer cells (18, 45). However, the fact that MELK expression is cell cycle–regulated (17, 28, 57, 58) and that the up-regulation of MELK in cancer correlates with up-regulation of many other important or essential mitotic genes, including CDK1, CCNB1/2, TOP2A, AURKB, PLK1, and BUB1 (8, 24, 30–33), suggests that MELK plays a role in cancer cell proliferation despite negative results from CRISPR studies. Why would MELK be cell cycle–regulated and consistently up-regulated in concert with many essential mitotic proteins if it did not contribute to these processes? There are a few possible explanations for the observations that MELK expression is tightly correlated with the cell cycle, yet MELK knockout does not perturb growth. In line with the conditional dependency hypothesis, it is possible that MELK acts as a functional redundancy for a specific cell cycle pathway, such that MELK is nonessential during normal cell cycling but becomes required under certain conditions. Conversely, there may be a functional redundancy for MELK, such that total loss of MELK (i.e. CRISPR knockout) allows for compensatory reprogramming of signaling networks, whereas RNAi-mediated partial MELK depletion does not trigger the reprogramming necessary to shift to this redundant pathway.
The MELK controversy has underscored a few important scientific principles. First, it is imperative not to overinterpret negative results, as there are a wide variety of potential explanations for the lack of any observed phenotype, ranging from functional or biological to purely technical or methodological, as the Bub1 CRISPR story illustrates. Second, technical considerations, while often only given cursory attention in publications, are vitally important. The observation that CRISPR-mediated MELK depletion has differing effects in TNBC cells depending upon cell density serves as an illustration of this. Third, when orthogonal approaches do not give identical results, it does not necessarily mean that one result is correct, whereas the other is incorrect. Rather, a more complex and interesting biological explanation may be underlying the seemingly discordant results.
To expand upon the third principle in the context of the MELK controversy, authors of the CRISPR studies have attempted to conclude that MELK is not important for cancer cell proliferation and that MELK is not a viable therapeutic target. Neither of these assertions are definitively supported by their results demonstrating no growth phenotype following CRISPR-mediated MELK deletion. Whereas these studies have provided strong evidence that MELK is not essential to cancer cells, they have done nothing to rebut the studies that coupled RNAi depletion with rescue, indicating that MELK is important for proliferation, and the therapeutic viability of inhibiting MELK should not yet be fully discounted, particularly in combination with other chemotherapeutic agents. It is vitally important to recognize that CRISPR-mediated genetic knockout, RNAi-mediated transcript depletion, and pharmacological inhibition are three orthogonal but fundamentally different approaches. Whereas some view CRISPR technologies as a universal improvement over RNAi-based approaches, this view is not entirely accurate. Off-target effects are less prevalent with CRISPR than RNAi, but both techniques still have their advantages and limitations (59–62). Results obtained with RNAi knockdown and subsequent rescue, long considered the gold standard for functional studies, still hold tremendous value, as this approach mitigates the off-target effects of RNAi. Further, RNAi offers greater temporal control of target depletion than CRISPR, which may be important especially when there is a possibility of signaling pathway reprogramming. Whereas CRISPR technologies are undeniably powerful, they are also still relatively new, with CRISPR first being used for genome editing in 2013 (63). Some aspects of CRISPR/Cas9 gene editing are still not entirely understood, including central parts of the technology, such as prediction of the on- and off-target DNA-cutting efficiency of specific gRNAs (59, 60). The scientific community is still uncovering some of the intricacies of this powerful yet complex approach. As such, negative results obtained with CRISPR should be continually re-evaluated as new information becomes available. Finally, the advent and widespread implementation of CRISPR for functional studies does not render RNAi-based approaches obsolete, particularly when coupled with exogenous complementation.
The field should continue to investigate the controversy concerning the requirement of MELK in cancer. The role of MELK in cancer, and in specific cellular processes, is poorly understood (reviewed in 2017 by Pitner et al. (20)). Future efforts to better understand the functions of MELK in cancer will likely shed light on the mechanism underlying discordant results observed with CRISPR and RNAi, which could be broadly applicable to functional studies of other proteins using these techniques. Some studies have begun to investigate the specific conditions under which MELK expression seems to be required, but the conditions tested thus far are certainly not comprehensive (19, 45). Efforts to elucidate the specific conditions under which MELK is important for cancer cell proliferation will be a crucial step toward a more complete functional understanding of MELK. There are seemingly more questions than answers in the MELK field at the present moment, underscoring the importance of continued efforts to investigate the functions of this enigmatic kinase.
Acknowledgments
We thank Dr. Mike Emanuele for insights into the effects of Bub1 knockout using CRISPR.
Funding and additional information—This work was supported by National Institutes of Health Grant R01 CA199064 (to L. M. G.). This work was also supported, in part, by a UNC dissertation completion fellowship (to I. M. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MELK
- maternal embryonic leucine zipper kinase
- MPK38
- murine protein serine/threonine kinase 38
- OTS
- OTSSP167
- BLBC
- basal-like breast cancer
- TNBC
- triple-negative breast cancer
- gRNA
- guide RNA
- CRISPRi
- CRISPR interference
- SAC
- spindle assembly checkpoint
- PCNA
- proliferating nuclear cell antigen
- RPA
- replication protein A
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
References
- 1. Heyer B. S., Warsowe J., Solter D., Knowles B. B., and Ackerman S. L. (1997) New member of the Snf1/AMPK kinase family, Melk, is expressed in the mouse egg and preimplantation embryo. Mol. Reprod. Dev. 47, 148–156 [DOI] [PubMed] [Google Scholar]
- 2. Gil M., Yang Y., Lee Y., Choi I., and Ha H. (1997) Cloning and expression of a cDNA encoding a novel protein serine/threonine kinase predominantly expressed in hematopoietic cells. Gene 195, 295–301 10.1016/S0378-1119(97)00181-9 [DOI] [PubMed] [Google Scholar]
- 3. Blot J., Chartrain I., Roghi C., Philippe M., and Tassan J. P. (2002) Cell cycle regulation of pEg3, a new Xenopus protein kinase of the KIN1/PAR-1/MARK family. Dev. Biol. 241, 327–338 10.1006/dbio.2001.0525 [DOI] [PubMed] [Google Scholar]
- 4. Li L., Keverne E. B., Aparicio S. A., Ishino F., Barton S. C., and Surani M. A. (1999) Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284, 330–333 10.1126/science.284.5412.330 [DOI] [PubMed] [Google Scholar]
- 5. Gray D., Jubb A. M., Hogue D., Dowd P., Kljavin N., Yi S., Bai W., Frantz G., Zhang Z., Koeppen H., de Sauvage F. J., and Davis D. P. (2005) Maternal embryonic leucine zipper kinase/murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 65, 9751–9761 10.1158/0008-5472.CAN-04-4531 [DOI] [PubMed] [Google Scholar]
- 6. Nakano I., Masterman-Smith M., Saigusa K., Paucar A. A., Horvath S., Shoemaker L., Watanabe M., Negro A., Bajpai R., Howes A., Lelievre V., Waschek J. A., Lazareff J. A., Freije W. A., Liau L. M., et al. (2008) Maternal embryonic leucine zipper kinase is a key regulator of the proliferation of malignant brain tumors, including brain tumor stem cells. J. Neurosci. Res. 86, 48–60 10.1002/jnr.21471 [DOI] [PubMed] [Google Scholar]
- 7. Lin M.-L., Park J.-H., Nishidate T., Nakamura Y., and Katagiri T. (2007) Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res. 9, R17 10.1186/bcr1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuner R., Fälth M., Chui Pressinotti N. C., Brase J. C., Puig S. B., Metzger J., Gade S., Schäfer G., Bartsch G., Steiner E., Klocker H., and Sültmann H. (2013) The maternal embryonic leucine zipper kinase (MELK) is upregulated in high-grade prostate cancer. J. Mol. Med. 91, 237–248 10.1007/s00109-012-0949-1 [DOI] [PubMed] [Google Scholar]
- 9. Du T., Qu Y., Li J., Li H., Su L., Zhou Q., Yan M., Li C., Zhu Z., and Liu B. (2014) Maternal embryonic leucine zipper kinase enhances gastric cancer progression via the FAK/Paxillin pathway. Mol. Cancer 13, 100 10.1186/1476-4598-13-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung S., Suzuki H., Miyamoto T., Takamatsu N., Tatsuguchi A., Ueda K., Kijima K., Nakamura Y., and Matsuo Y. (2012) Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget 3, 1629–1640 10.18632/oncotarget.790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alachkar H., Mutonga M. B., Metzeler K. H., Fulton N., Malnassy G., Herold T., Spiekermann K., Bohlander S. K., Hiddemann W., Matsuo Y., Stock W., and Nakamura Y. (2014) Preclinical efficacy of maternal embryonic leucine-zipper kinase (MELK) inhibition in acute myeloid leukemia. Oncotarget 5, 12371–12382 10.18632/oncotarget.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inoue H., Kato T., Olugbile S., Tamura K., Chung S., Miyamoto T., Matsuo Y., Salgia R., Nakamura Y., and Park J.-H. (2016) Effective growth-suppressive activity of maternal embryonic leucine-zipper kinase (MELK) inhibitor against small cell lung cancer. Oncotarget 7, 13621–13633 10.18632/oncotarget.7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.-A., Reinecke M., Ruprecht B., Petzoldt S., Meng C., Zecha J., Reiter K., Qiao H., Helm D., Koch H., et al. (2017) The target landscape of clinical kinase drugs. Science 358, eaan4368 10.1126/science.aan4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McDonald I. M., Grant G. D., East M. P., Gilbert T. S. K., Wilkerson E. M., Goldfarb D., Beri J., Herring L. E., Vaziri C., Cook J. G., Emanuele M. J., and Graves L. M. (2020) Mass spectrometry-based selectivity profiling identifies a highly selective inhibitor of the kinase MELK that delays mitotic entry in cancer cells. J. Biol. Chem. 295, 2359–2374 10.1074/jbc.RA119.011083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji W., Arnst C., Tipton A. R., Bekier M. E. 2nd, Taylor W. R., Yen T. J., and Liu S.-T. (2016) OTSSP167 abrogates mitotic checkpoint through inhibiting multiple mitotic kinases. PLoS ONE 11, e0153518 10.1371/journal.pone.0153518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang H.-T., Seo H.-S., Zhang T., Wang Y., Jiang B., Li Q., Buckley D. L., Nabet B., Roberts J. M., Paulk J., Dastjerdi S., Winter G. E., McLauchlan H., Moran J., Bradner J. E., et al. (2017) MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife 6, e26693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Lee Y.-M., Baitsch L., Huang A., Xiang Y., Tong H., Lako A., Von T., Choi C., Lim E., Min J., Li L., Stegmeier F., Schlegel R., Eck M. J., et al. (2014) MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells. Elife 3, e01763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin A., Giuliano C. J., Sayles N. M., and Sheltzer J. M. (2017) CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. Elife 6, e24179 10.7554/eLife.24179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Li B. B., Li J., Roberts T. M., and Zhao J. J. (2018) A conditional dependency on MELK for the proliferation of triple-negative breast cancer cells. iScience 9, 149–160 10.1016/j.isci.2018.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pitner M. K., Taliaferro J. M., Dalby K. N., and Bartholomeusz C. (2017) MELK: a potential novel therapeutic target for TNBC and other aggressive malignancies. Expert Opin. Ther. Targets 21, 849–859 10.1080/14728222.2017.1363183 [DOI] [PubMed] [Google Scholar]
- 21. Pickard M. R., Green A. R., Ellis I. O., Caldas C., Hedge V. L., Mourtada-Maarabouni M., and Williams G. T. (2009) Dysregulated expression of Fau and MELK is associated with poor prognosis in breast cancer. Breast Cancer Res. 11, R60 10.1186/bcr2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Speers C., Zhao S. G., Kothari V., Santola A., Liu M., Wilder-Romans K., Evans J., Batra N., Bartelink H., Hayes D. F., Lawrence T. S., Brown P. H., Pierce L. J., and Feng F. Y. (2016) Maternal embryonic leucine zipper kinase (MELK) as a novel mediator and biomarker of radioresistance in human breast cancer. Clin. Cancer Res. 22, 5864–5875 10.1158/1078-0432.CCR-15-2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minata M., Gu C., Joshi K., Nakano-Okuno M., Hong C., Nguyen C.-H., Kornblum H. I., Molla A., and Nakano I. (2014) Multi-kinase inhibitor C1 triggers mitotic catastrophe of glioma stem cells mainly through MELK kinase inhibition. PLoS ONE 9, e92546 10.1371/journal.pone.0092546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marie S. K. N., Okamoto O. K., Uno M., Hasegawa A. P. G., Oba-Shinjo S. M., Cohen T., Camargo A. A., Kosoy A., Carlotti C. G. Jr., Toledo S., Moreira-Filho C. A., Zago M. A., Simpson A. J., and Caballero O. L. (2008) Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int. J. Cancer 122, 807–815 10.1002/ijc.23189 [DOI] [PubMed] [Google Scholar]
- 25. Guan S., Lu J., Zhao Y., Yu Y., Li H., Chen Z., Shi Z., Liang H., Wang M., Guo K., Chen X., Sun W., Bieerkehazhi S., Xu X., Sun S., et al. (2018) MELK is a novel therapeutic target in high-risk neuroblastoma. Oncotarget 9, 2591–2602 10.18632/oncotarget.23515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia H., Kong S. N., Chen J., Shi M., Sekar K., Seshachalam V. P., Rajasekaran M., Goh B. K. P., Ooi L. L., and Hui K. M. (2016) MELK is an oncogenic kinase essential for early hepatocellular carcinoma recurrence. Cancer Lett. 383, 85–93 10.1016/j.canlet.2016.09.017 [DOI] [PubMed] [Google Scholar]
- 27. Chen S., Zhou Q., Guo Z., Wang Y., Wang L., Liu X., Lu M., Ju L., Xiao Y., and Wang X. (2020) Inhibition of MELK produces potential anti-tumour effects in bladder cancer by inducing G1/S cell cycle arrest via the ATM/CHK2/p53 pathway. J. Cell Mol. Med. 24, 1804–1821 10.1111/jcmm.14878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Q., Ge Q., Zhou Y., Yang B., Yang Q., Jiang S., Jiang R., Ai Z., Zhang Z., and Teng Y. (2020) MELK promotes endometrial carcinoma progression via activating mTOR signaling pathway. EBioMedicine 51, 102609 10.1016/j.ebiom.2019.102609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chlenski A., Park C., Dobratic M., Salwen H. R., Budke B., Park J.-H., Miller R., Applebaum M. A., Wilkinson E., Nakamura Y., Connell P. P., and Cohn S. L. (2019) Maternal embryonic leucine zipper kinase (MELK), a potential therapeutic target for neuroblastoma. Mol. Cancer Ther. 18, 507–516 10.1158/1535-7163.MCT-18-0819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Agarwal R., Narayan J., Bhattacharyya A., Saraswat M., and Tomar A. K. (2017) Gene expression profiling, pathway analysis and subtype classification reveal molecular heterogeneity in hepatocellular carcinoma and suggest subtype specific therapeutic targets. Cancer Genet. 216, 37–51 10.1016/j.cancergen.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 31. Sun C., Cheng X., Wang C., Wang X., Xia B., and Zhang Y. (2019) Gene expression profiles analysis identifies a novel two-gene signature to predict overall survival in diffuse large B-cell lymphoma. Biosci. Rep. 39, BSR20181293 10.1042/BSR20181293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng J. L., Xu Y. H., and Wang G. (2019) Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front. Genet. 10, 695 10.3389/fgene.2019.00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zou Y. F., Meng L. B., He Z. K., Hu C. H., Shan M. J., Wang D. Y., and Yu X. (2019) Screening and authentication of molecular markers in malignant glioblastoma based on gene expression profiles. Oncol. Lett. 18, 4593–4604 10.3892/ol.2019.10804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim S. H., Joshi K., Ezhilarasan R., Myers T. R., Siu J., Gu C., Nakano-Okuno M., Taylor D., Minata M., Sulman E. P., Lee J., Bhat K. P. L., Salcini A. E., and Nakano I. (2015) EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Rep. 4, 226–238 10.1016/j.stemcr.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi S., and Ku J. L. (2011) Resistance of colorectal cancer cells to radiation and 5-FU is associated with MELK expression. Biochem. Biophys. Res. Commun. 412, 207–213 10.1016/j.bbrc.2011.07.060 [DOI] [PubMed] [Google Scholar]
- 36. Zhang Y., Zhou X., Li Y., Xu Y., Lu K., Li P., and Wang X. (2018) Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia. Oncogene 37, 5520–5533 10.1038/s41388-018-0333-x [DOI] [PubMed] [Google Scholar]
- 37. Hebbard L. W., Maurer J., Miller A., Lesperance J., Hassell J., Oshima R. G., and Terskikh A. V. (2010) Maternal embryonic leucine zipper kinase is upregulated and required in mammary tumor-initiating cells in vivo. Cancer Res. 70, 8863–8873 10.1158/0008-5472.CAN-10-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Janostiak R., Rauniyar N., Lam T. T., Ou J., Zhu L. J., Green M. R., and Wajapeyee N. (2017) MELK promotes melanoma growth by stimulating the NF-κB pathway. Cell Rep. 21, 2829–2841 10.1016/j.celrep.2017.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Touré B. B., Giraldes J., Smith T., Sprague E. R., Wang Y., Mathieu S., Chen Z., Mishina Y., Feng Y., Yan-Neale Y., Shakya S., Chen D., Meyer M., Puleo D., Brazell J. T., et al. (2016) Toward the validation of maternal embryonic leucine zipper kinase: Discovery, optimization of highly potent and selective inhibitors, and preliminary biology insight. J. Med. Chem. 59, 4711–4723 10.1021/acs.jmedchem.6b00052 [DOI] [PubMed] [Google Scholar]
- 40. Beke L., Kig C., Linders J. T. M., Boens S., Boeckx A., van Heerde E., Parade M., De Bondt A., Van den Wyngaert I., Bashir T., Ogata S., Meerpoel L., Van Eynde A., Johnson C. N., Beullens M., et al. (2015) MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Biosci. Rep. 35, e00267 10.1042/BSR20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edupuganti R., Taliaferro J. M., Wang Q., Xie X., Cho E. J., Vidhu F., Ren P., Anslyn E. V., Bartholomeusz C., and Dalby K. N. (2017) Discovery of a potent inhibitor of MELK that inhibits expression of the anti-apoptotic protein Mcl-1 and TNBC cell growth. Bioorg. Med. Chem. 25, 2609–2616 10.1016/j.bmc.2017.03.018 [DOI] [PubMed] [Google Scholar]
- 42. Li S., Li Z., Guo T., Xing X.-F., Cheng X., Du H., Wen X.-Z., and Ji J.-F. (2016) Maternal embryonic leucine zipper kinase serves as a poor prognosis marker and therapeutic target in gastric cancer. Oncotarget 7, 6266–6280 10.18632/oncotarget.6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stefka A. T., Park J.-H., Matsuo Y., Chung S., Nakamura Y., Jakubowiak A. J., and Rosebeck S. (2016) Anti-myeloma activity of MELK inhibitor OTS167: effects on drug-resistant myeloma cells and putative myeloma stem cell replenishment of malignant plasma cells. Blood Cancer J. 6, e460 10.1038/bcj.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bolomsky A., Heusschen R., Schlangen K., Stangelberger K., Muller J., Schreiner W., Zojer N., Caers J., and Ludwig H. (2018) Maternal embryonic leucine zipper kinase is a novel target for proliferation-associated high-risk myeloma. Haematologica 103, 325–335 10.3324/haematol.2017.172973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giuliano C. J., Lin A., Smith J. C., Palladino A. C., and Sheltzer J. M. (2018) MELK expression correlates with tumor mitotic activity but is not required for cancer growth. Elife 7, e32838 10.7554/eLife.32838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gu C., Banasavadi-Siddegowda Y. K., Joshi K., Nakamura Y., Kurt H., Gupta S., and Nakano I. (2013) Tumor-specific activation of the C-JUN/MELK pathway regulates glioma stem cell growth in a p53-dependent manner. Stem Cells 31, 870–881 10.1002/stem.1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Joshi K., Banasavadi-Siddegowda Y., Mo X., Kim S. H., Mao P., Kig C., Nardini D., Sobol R. W., Chow L. M. L., Kornblum H. I., Waclaw R., Beullens M., and Nakano I. (2013) MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells 31, 1051–1063 10.1002/stem.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Allegretti P. A., Horton T. M., Abdolazimi Y., Moeller H. P., Yeh B., Caffet M., Michel G., Smith M., and Annes J. P. (2020) Generation of highly potent DYRK1A-dependent inducers of human β-cell replication via multi-dimensional compound optimization. Bioorg. Med. Chem. 28, 115193 10.1016/j.bmc.2019.115193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson C. N., Berdini V., Beke L., Bonnet P., Brehmer D., Coyle J. E., Day P. J., Frederickson M., Freyne E. J. E., Gilissen R. A. H. J., Hamlett C. C. F., Howard S., Meerpoel L., McMenamin R., Patel S., Rees D. C., Sharff A., Sommen F., Wu T., and Linders J. T. M. (2015) Fragment-based discovery of type I inhibitors of maternal embryonic leucine zipper kinase. ACS Med. Chem. Lett. 6, 25–30 10.1021/ml5001245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., and Linsley P. S. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 10.1038/nbt831 [DOI] [PubMed] [Google Scholar]
- 51. Settleman J., Sawyers C. L., and Hunter T. (2018) Challenges in validating candidate therapeutic targets in cancer. Elife 7, e32402 10.7554/eLife.32402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duncan J. S., Whittle M. C., Nakamura K., Abell A. N., Midland A. A., Zawistowski J. S., Johnson N. L., Granger D. A., Jordan N. V., Darr D. B., Usary J., Kuan P.-F., Smalley D. M., Major B., He X., et al. (2012) Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 10.1016/j.cell.2012.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Graves L. M., Duncan J. S., Whittle M. C., and Johnson G. L. (2013) The dynamic nature of the kinome. Biochem. J. 450, 1–8 10.1042/BJ20121456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rodriguez-Rodriguez J. A., Lewis C., McKinley K. L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I. M., and Jallepalli P. V. (2018) Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol. 28, 3422–3429.e5 10.1016/j.cub.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang G., Kruse T., Guasch Boldú C., Garvanska D. H., Coscia F., Mann M., Barisic M., and Nilsson J. (2019) Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 38, e100977 10.15252/embj.2018100977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meraldi P. (2019) Bub1—the zombie protein that CRISPR cannot kill. EMBO J. 38, e101912 10.15252/embj.2019101912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Badouel C., Chartrain I., Blot J., and Tassan J.-P. (2010) Maternal embryonic leucine zipper kinase is stabilized in mitosis by phosphorylation and is partially degraded upon mitotic exit. Exp. Cell Res. 316, 2166–2173 10.1016/j.yexcr.2010.04.019 [DOI] [PubMed] [Google Scholar]
- 58. Grant G. D., Brooks L. 3rd, Zhang X., Mahoney J. M., Martyanov V., Wood T. A., Sherlock G., Cheng C., and Whitfield M. L. (2013) Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell 24, 3634–3650 10.1091/mbc.e13-05-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Komor A. C., Badran A. H., and Liu D. R. (2017) CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 10.1016/j.cell.2016.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsai S. Q., and Joung J. K. (2016) Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 17, 300–312 10.1038/nrg.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seok H., Lee H., Jang E. S., and Chi S. W. (2018) Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol. Life Sci. 75, 797–814 10.1007/s00018-017-2656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wittrup A., and Lieberman J. (2015) Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543–552 10.1038/nrg3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]