Abstract
The cytokine content in tissue microenvironments shapes the functional capacity of a T cell. This capacity depends on the integration of extracellular signaling through multiple receptors, including the T-cell receptor (TCR), co-receptors, and cytokine receptors. Transforming growth factor β (TGF-β) signals through its cognate receptor, TGFβR, to SMAD family member proteins and contributes to the generation of a transcriptional program that promotes regulatory T-cell differentiation. In addition to transcription, here we identified specific signaling networks that are regulated by TGFβR. Using an array of biochemical approaches, including immunoblotting, kinase assays, immunoprecipitation, and flow cytometry, we found that TGFβR signaling promotes the formation of a SMAD3/4-protein kinase A (PKA) complex that activates C-terminal Src kinase (CSK) and thereby down-regulates kinases involved in proximal TCR activation. Additionally, TGFβR signaling potentiated CSK phosphorylation of the P85 subunit in the P85–P110 phosphoinositide 3-kinase (PI3K) heterodimer, which reduced PI3K activity and down-regulated the activation of proteins that require phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3) for their activation. Moreover, TGFβR-mediated disruption of the P85–P110 interaction enabled P85 binding to a lipid phosphatase, phosphatase and tensin homolog (PTEN), aiding in the maintenance of PTEN abundance and thereby promoting elevated PtdIns(4,5)P2 levels in response to TGFβR signaling. Taken together, these results highlight that TGF-β influences the trajectory of early T-cell activation by altering PI3K activity and PtdIns levels.
Keywords: T-cell receptor (TCR), phosphatidylinositide 3-kinase (PI 3-kinase), computer modeling, phosphatidylinositol, phosphatidylinositol kinase (PI kinase), T cell, T-cell biology, phosphatase and tensin homolog (PTEN), protein kinase A (PKA), phosphatidylinositol signaling, C-terminal Src kinase (CSK), immunity, transforming growth factor β (TGF-β), cytokine, phosphoinositide 3-kinase (PI3K), phospholipid signaling
Introduction
The cytokine content in the microenvironment shapes the functional capacity of a T cell. An exquisite example of the synergy between T cell receptor (TCR) and cytokine receptor signaling is T-cell differentiation. Polarization of a naïve CD4+ T cell into distinct subsets requires unique combinations of cytokines in addition to TCR signaling (1, 2). For example, signaling through the TCR and TGF-β receptor (TGFβR) drives the induction of regulatory T cells (3). Canonical TCR signaling triggers CD45 to dephosphorylate an inhibitory phosphorylation site on LCK, which allows for autophosphorylation and activation of LCK kinase activity (4, 5). LCK then phosphorylates multiple substrates including the ζ chain of the TCR, which recruits and leads to the activation of the ZAP70 kinase (6). Additionally, TCR activation engages PI3K to generate the PtdIns(3,4,5)P3 lipid second messenger to activate multiple kinases including PDK1 and AKT (7). Together, these early signaling events drive a downstream signaling cascade leading to nuclear translocation of transcription factors including NFAT that result in T-cell activation (8).
In canonical TGFβR signaling in T cells, TGF-β binding drives the formation of a heterotetrameric receptor complex (2TGFβRI–2TGFβRII), which stimulates the kinase activity of the TGFβRI subunit to phosphorylate SMAD3, which promotes the formation of a SMAD3/SMAD4 complex (9, 10). Formation of SMAD3/4 complexes results in nuclear translocation, where the SMAD complexes directly regulate transcriptional programs. Although TGFβR signaling is required for the induction of regulatory CD4+ T cells (11), TGFβR signaling reduces Th1 CD4+ T-cell differentiation (12). In addition to its role in T-cell differentiation, TGFβR signaling reduces T-cell proliferation and limits CD8+ T cell–mediated cytotoxicity (13).
Although both TCR and TGF-β signaling are required for cytokine-driven Treg induction, little is known about how these pathways cooperate during the early phase of T-cell activation. Previous work demonstrated that TGFβR signaling during T-cell activation reduced the activation of the TEC kinase family member ITK (14), which is downstream of ZAP70. Additionally, TGF-β down-modulated ERK activation and calcium mobilization (14). In other cell types, TGFβR via SMAD3/4 can regulate kinase activity including PKA (15), which raises the possibility that other kinases and signaling proteins in T cells might be regulated by TGFβR signaling. Herein, we established a circuit in murine CD4+ T cells where TGFβR signaling globally regulates tyrosine kinase–mediated signaling, activates CSK to down-modulate proximal TCR signaling, and suppresses PI3K activity to modulate AKT activation. This work in part defines the biochemical basis by which CD4+ T cells sense the TGF-β cytokine, how TGF-β suppresses early signaling events and identifies pathways in addition to transcriptional networks that are regulated by TGFβR signaling.
Results
TGF-β receptor signaling regulates phospho-tyrosine signaling networks during early T-cell activation
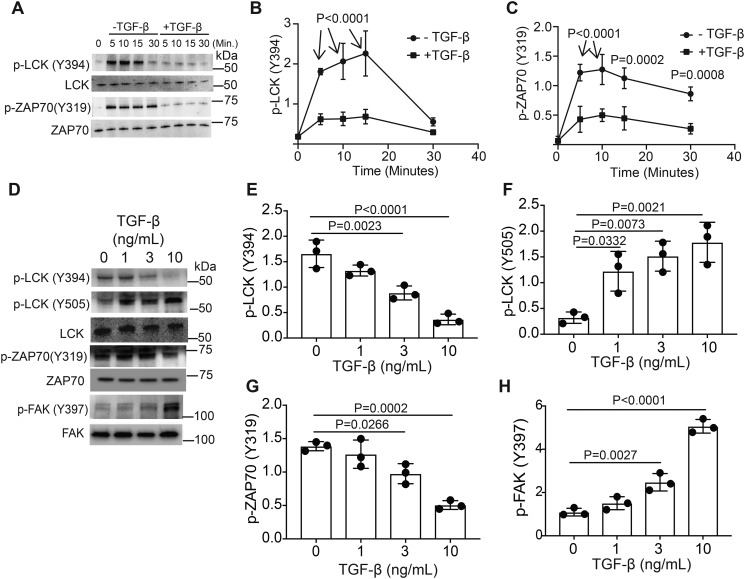
TCR signaling engages tyrosine kinases to activate downstream signaling pathways. LCK is one of the first kinases activated in response to TCR signaling. In the resting state, CSK phosphorylates LCK on Tyr-505, maintaining inhibition of LCK enzymatic activity (16). Upon T-cell activation, CD45 dephosphorylates Tyr-505, allowing for LCK activation (4, 5). LCK autophosphorylation occurs on Tyr-394 (17). We took an immunoblotting approach to monitor the phosphorylation of LCK and ZAP70, which is another TCR proximal kinase downstream of LCK, in murine CD4+ T cells activated through TCR, CD28 with and without TGF-β (Fig. 1A). During activation, signaling through the TGF-β receptor resulted in decreased p-LCK (Tyr-394) (Fig. 1B) and p-ZAP70 (Tyr-319) (Fig. 1C). To establish the threshold required for TGF-β receptor signaling to modulate LCK and ZAP70 phosphorylation, murine T cells were activated through the TCR and CD28 receptors in the presence of varying doses of TGF-β (Fig. 1D). Activation of TGFβR reduces Tyr-394 phosphorylation and increases Tyr-505 phosphorylation on LCK (Fig. 1, E and F). Phosphorylation of Tyr-319 on ZAP70 occurs downstream of LCK activation (18). T cells activated in the presence of TGF-β have reduced p-ZAP70 (Tyr-319) levels (Fig. 1G). FAK recruits CSK to the plasma membrane to inhibit LCK (19). TGF-β addition increases autophosphorylation of Tyr-397 on FAK (Fig. 1H). This demonstrates that although TGF-β suppresses the activation of some kinases, TGF-β can also potentiate the phosphorylation and activation of other kinases including FAK.
Figure 1.
TGF-β signaling alters LCK and ZAP70 activation in murine CD4+ T cells. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for various time points with or without 10 ng/ml TGF-β. A, immunoblotting was performed on cell lysates for the phosphorylated and total protein LCK and ZAP-70 abundance. B and C, densitometry was performed across three independent experiments to quantitate the abundance of tyrosine phosphorylation for (B) LCK (Tyr-394) and (C) ZAP70 (Tyr-319). The abundance of phosphorylated protein was normalized to total protein. Shown are mean ± S.D.; p values were calculated by two-way ANOVA. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min with various doses of TGF-β. D, immunoblotting was performed on cell lysates for the phosphorylated and total protein abundance for LCK, ZAP-70, and FAK. E–H, densitometry was performed across three independent experiments to quantitate the abundance of tyrosine phosphorylation for (E) LCK (Tyr-394), (F) LCK (Tyr-505), (G) ZAP70 (Tyr-319), and (H) FAK (Tyr-397). The abundance of phosphorylated protein was normalized to total protein. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test.
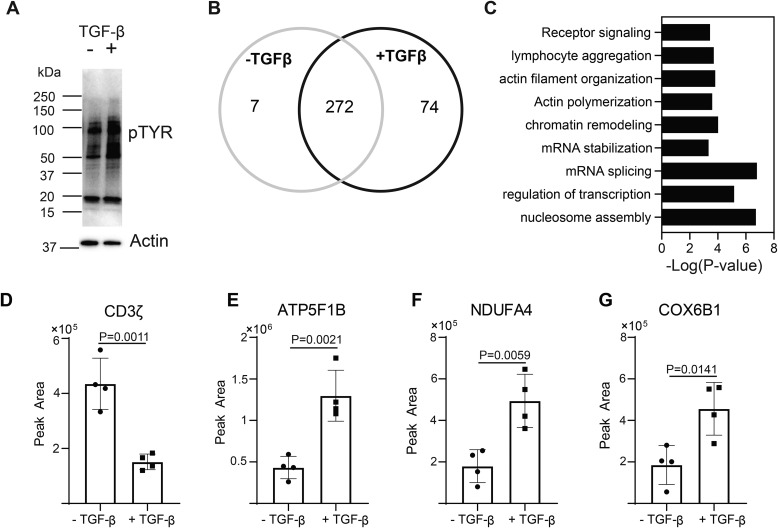
To more globally profile the impact TGFβR signaling had on the phospho-tyrosine proteome, immunoblotting was performed with an antibody that binds to proteins containing a phospho-tyrosine residue. This analysis demonstrated that TGFβR signaling globally regulated phospho-tyrosine signaling networks during T-cell activation (Fig. 2A). To identify specific proteins that were tyrosine phosphorylated in response to TGFβR signaling, T cells were activated through the TCR and CD28 receptors in the presence or absence of TGF-β. Proteins containing a phospho-tyrosine residue were immunoprecipitated with the phospho-tyrosine specific antibody, and the resulting proteins were subjected to a quantitative mass spectrometric analysis (Table S1). 7 unique proteins in the phospho-tyrosine immunoprecipitate from CD4+ T cells activated without TGF-β, 74 unique proteins in CD4+ T cells activated in the presence of TGF-β, and 272 proteins shared (Fig. 2B). TGFβR signaling combined with TCR and CD28 stimulation resulted in the preferential tyrosine phosphorylation of proteins involved in regulating diverse processes including receptor signaling, actin cytoskeleton, mRNA processes, and transcription (Fig. 2C). In this dataset, TGFβR signaling reduced the amount of CD3ζ in the phospho-tyrosine immunoprecipitate (Fig. 2D and S1), consistent with the observed reduction of LCK activation (Fig. 1, A, B, D, and E). The relative quantitation from the mass spectrometric data is depicted for ATP5F1B, NDUFA1, and COX6B1 (Fig. 2, E–G). Taken together, these data demonstrated that TGF-β alters global phospho-tyrosine signaling during T-cell activation.
Figure 2.
TGF-β signaling alters phospho-tyrosine signaling networks in CD4+ T cells. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 stimulation for 10 min ± 10 ng/ml TGF-β. A, immunoblotting was performed on cell lysates using the anti–phospho-tyrosine antibody. One representative blot from three independent experiments is shown. Tyrosine phosphorylated proteins were immunoprecipitated from T cells activated for 10 min ± 10 ng/ml TGF-β. Label-free quantitative MS was used to identify proteins in the phospho-tyrosine IPs. B, a Venn diagram was constructed using a 2-fold cutoff based on the label-free quantitative data to illustrate the number of tyrosine-phosphorylated proteins enriched in T cells activated in the presence or absence of TGF-β from four or five independent experiments. C, the distribution of tyrosine-phosphorylated proteins specific to T cells activated through TCR-CD28 plus TGF-β were compared with the mouse proteome with a binomial test for gene ontology and biological pathway in the PANTHER software package. The probability (p value) that the number of proteins in each group occurred by chance was calculated. D–G, representative examples of the label-free quantitation of the mass spectrometric data are illustrated for (D) CD3ζ, (E) ATP5F1B, (F) NDUFA4, and (G) COX5B1. Shown are mean ± S.D.; p values were calculated with a two-tailed Student's t test.
TGFβR signaling promotes phosphorylation of P85 and disruption of the PI3K heterodimer in CD4+ T cells
The phospho-tyrosine proteomic screen identified that T-cell activation with or without TGF-β promoted the phosphorylation of the lipid kinase PIP4K2α (Fig. 3A and S2), which converts PtdIns(4)P to PtdIns(4,5)P2. The P85 subunit of PI3K was in the phospho-tyrosine immunoprecipitate from T cells activated with TGF-β (Fig. 3B and S3). PI3K is a critical enzyme that phosphorylates PtdIns(4,5)P2 to yield PtdIns(3,4,5)P3 and activates kinases including AKT (7). Phosphorylation of P85 on Tyr-458 was observed in mass spectrometric datasets of multiple cell types (20, 21), which raised the possibility that TGF-β receptor signaling could promote P85 phosphorylation on Tyr-458. Using an immunoblotting approach with an antibody specific for the phosphorylated Tyr-458 proteoform of P85, we observed that murine CD4+ T cells activated in the presence of TGF-β had significantly more phosphorylated Tyr-458 P85 compared with CD4+ T cells activated without addition of TGF-β (Fig. 3, C and D). To establish the threshold required for TGF-β receptor signaling to modulate P85 phosphorylation on Tyr-458, murine T cells were activated through the TCR and CD28 receptors in the presence of varying doses of TGF-β. Immunoblotting revealed that TGF-β receptor signaling antagonized the phosphorylation of Tyr-458 in a dose-dependent manner (Fig. 3, E and F).
Figure 3.
TGF-β signaling induces the phosphorylation of P85 (Y458) and disruption of the PI3K heterodimer. Murine CD4+ T cells were isolated by negative selection. Tyrosine phosphorylated proteins were immunoprecipitated from T cells activated for 10 min ± 10 ng/ml TGF-β. Label-free quantitative MS was used to identify proteins in the phospho-tyrosine IPs. A and B, label-free quantitation of the mass spectrometric data are illustrated for (A) PIP4K2α and (B) P85α. Shown are mean ± S.D.; p values were calculated with a two-tailed Student's t test. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for various time points with or without 10 ng/ml TGF-β. C, immunoblotting was performed on cell lysates for the phosphorylated and total protein abundance for P85. D, densitometry was performed across three independent experiments to quantitate the abundance of tyrosine phosphorylation for P85 (Tyr-458). The abundance of phosphorylated protein was normalized to total protein. Shown are mean ± S.D.; p values were calculated by two-way ANOVA. CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 mins with various doses of TGF-β. E) immunoblotting was performed for p-P85 (Tyr-458) and total P85 on cell lysates. One representative blot from three independent experiments is shown. F, densitometry was performed across three independent experiments to quantitate the abundance of tyrosine P85 (Tyr-458) phosphorylation normalized to total P85 abundance as a function of TGF-β concentration. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test. G, the Thr-458 phosphorylation site on P85 was mapped onto the P85–P110 heterodimer crystal structure (PDB ID, 2RD0). CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min in the absence or presence of 10 ng/ml TGF-β. P85 was immunoprecipitated from total cell lysates and immunoblotting was performed for P110 and P85. H, one representative blot from three independent experiments is shown. I, densitometry was performed across three independent experiments to quantitate the amount of P110 that coimmunoprecipitated with P85, which was normalized to the abundance of P85 immunoprecipitated. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test. J, mass ELISA assays were utilized to measure PtdIns(3,4,5)P3 in murine CD4+ T cells activated for 10 min ± 10 ng/ml TGF-β. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test.
PI3K is a lipid kinase comprised of a P85 regulatory subunit and a P110 catalytic subunit. Structural modeling revealed that Tyr-458 of P85 is at the P85–P110 heterodimer interface (Fig. 3G). Thus, phosphorylation of Tyr-458 could impact the P85–P110 interaction. To determine whether TGFβR signaling regulated the assembly of the P85–P110 heterodimer, P85 was immunoprecipitated from cell lysates derived from T cells activated through the TCR and CD28 receptor with and without TGF-β. Immunoblotting demonstrated that less P110 coprecipitated with P85 from cell lysates derived from T cells activated in the presence of TGF-β (Fig. 3, H and I).
P85 contains two SH2 domains and an SH3 domain. Following T-cell activation, the SH2 and SH3 domains of P85 bind to phosphorylated tyrosine residues on intracellular domains of receptors, including CD28 (22, 23), which recruits the PI3K complex to the cell membrane and promotes PtdIns(3,4,5)P3 synthesis. Because PtdIns(4,5)P2, the substrate for PI3K, localizes to the plasma membrane, we hypothesized that the disruption of the P85–P110 dimer by TGFβR signaling would reduce PtdIns(3,4,5)P3 levels during T-cell activation, which was confirmed by biochemical measurement (Fig. 3J). Together, these data are consistent with a model where TGFβR signaling down-modulated PI3K activity in T cells by disrupting the P85–P110 dimer.
Inhibition of SMAD3 phosphorylation alleviates PI3K suppression by TGF-β
We wished to determine the molecular mechanism by which TGFβR signaling regulated PI3K. An interesting mediator of TGFβR signaling is SMAD3, which can bind to multiple targets and regulate their activity. TGF-β binding stimulates TGFβR phosphorylation of SMAD3 and the dissociation of SMAD3 from the TGFβR (9, 10). In turn, phosphorylated SMAD3 binds to SMAD4, leading to nuclear translocation of the SMAD3/4 complex to bind DNA or other transcriptional complexes and regulate gene transcription (24). SMAD complexes can also function in other subcellular compartments besides the nucleus. TGF-β receptor signaling promotes SMAD3/4 localization to the mitochondria and regulation of metabolism (25). In other cell types, SMAD3/4 binds to and activates kinases including PKA (15). We therefore hypothesize that TGFβR signaling suppresses PI3K activity in CD4+ T cells via SMAD3.
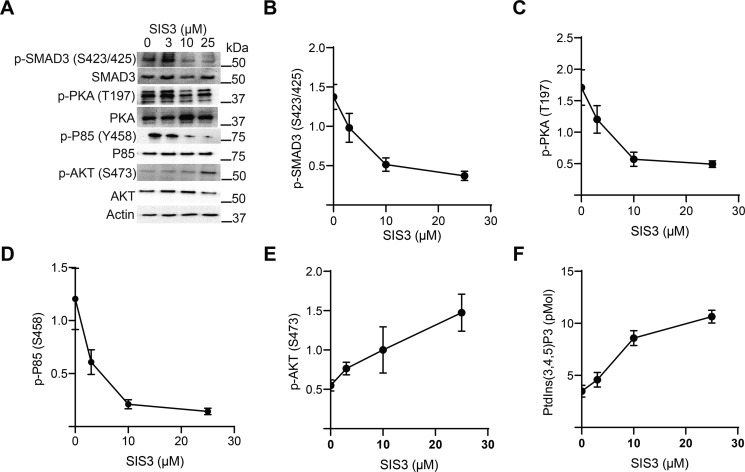
Murine CD4+ T cells were pretreated with multiple doses of the SIS3 inhibitor, which specifically inhibits SMAD3 phosphorylation (26), and then stimulated through TCR and CD28 in the presence of TGF-β. As expected, SMAD3 phosphorylation decreased with increased SIS3 dose (Fig. 4, A and B). SIS3 treatment also reduced the autophosphorylation of PKA (Fig. 4, A and C) and decreased phosphorylation of P85 (Fig. 4, A and D). The prediction was that inhibition of TGFβR signaling and SMAD3 phosphorylation would maintain the P85–P110 complex and restore PI3K activity. High levels of PtdIns(3,4,5)P3 activate mTORC2 to phosphorylate Ser-473 on AKT, and therefore Ser-473 phosphorylation serves as a proxy for PtdIns(3,4,5)P3 generation (27). As predicted, SIS3 treatment resulted in increased AKT Ser-473 phosphorylation (Fig. 4, A and E). Additionally, direct biochemical measurement revealed that PtdIns(3,4,5)P3 levels increased with SIS3 dose (Fig. 4F). Taken together, these data supported a model where TGFβR phosphorylation of SMAD3 was required to down-modulate PI3K activity in activated CD4+ T cells.
Figure 4.
SMAD3 inhibition restores PI3K activity. A, murine CD4+ T cells were pretreated with various doses of SIS3 inhibitor for 2 h and activated through the TCR, CD28, and with 10 ng/ml TGF-β for 10 min. Immunoblotting was performed for various phosphoproteins. One representative blot from two independent experiments is shown. B–E, immunoblots from panel A were quantitated by densitometry and normalized to total protein abundance for (B) p-SMAD3 (Ser-423/425), (C) p-PKA (Thr-197), (D) p-P85 (Ser-458), and (E) p-AKT (Ser-473). Shown are mean ± S.D. from two independent experiments per data point. F, lipids were extracted from activated T cells and mass ELISA were utilized to measure the levels of PtdIns(3,4,5)P3 as a function of SIS3 dose (n = 2 per data point).
TGF-β receptor signaling activates PKA via SMAD3/4
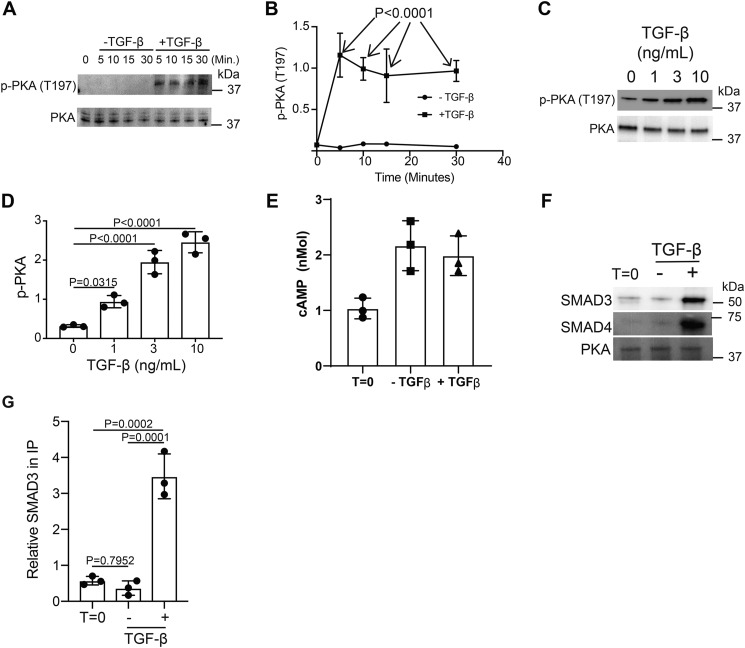
We sought to determine the biochemical pathway by which TGFβR signaling down-modulated PI3K (Fig. 3J). One possible mediator is PKA, which is a known suppressor of TCR signaling (28). PKA is an attractive target because its activity is enhanced by TGFβR signaling (15). Therefore, we hypothesize that TGFβR signaling activates PKA in T cells. Thr-197 is an autophosphorylation site on PKA (29) and therefore serves as a proxy to monitor PKA activity. Using an immunoblotting approach, we observe that murine CD4+ T cells activated in the presence of TGF-β had significantly more phosphorylated Thr-197 PKA than CD4+ T cells activated without addition of TGF-β (Fig. 5, A and B). To establish the threshold required for TGF-β receptor signaling to modulate PKA phosphorylation on Thr-197, murine T cells were activated through the TCR and CD28 receptors in the presence of varying doses of TGF-β. Immunoblotting revealed that TGF-β receptor signaling antagonized the phosphorylation of Thr-197 in a dose-dependent manner and showed a significant difference starting at 3 ng/ml of TGF-β (Fig. 5, C and D).
Figure 5.
TGF-β signaling activates PKA via SMAD3/4 binding. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for various time points with or without 10 ng/ml TGF-β. A, immunoblotting was performed on cell lysates for the phosphorylated and total protein abundance for PKA. B, densitometry was performed across three independent experiments to quantitate the abundance of tyrosine phosphorylation for PKA (Thr-197). The abundance of phosphorylated protein was normalized to total protein. Shown are mean ± S.D.; p values were calculated by two-way ANOVA. T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min with various amounts of TGF-β. C, immunoblotting was performed for p-PKA (Thr-197) and total PKA. D, densitometry across three independent immunoblots was performed for p-PKA (Thr-197) normalized to total PKA. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test. E, cAMP levels were measured in resting (T = 0) or T cells activated for 10 min in the presence or absence of 10 ng/ml TGF-β (n = 3). CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min ± 10 ng/ml TGF-β. F, PKA was immunoprecipitated and immunoblotted for SMAD3, SMAD4, and PKA. One representative blot from three independent experiments is shown. G, densitometry across three independent immunoblots was performed for SMAD3 and normalized to total PKA in the IP. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test.
There are at least two known mechanisms that can activate PKA. Elevated cAMP generation activates PKA (30). Alternatively, the SMAD3/4 heterotrimer generated by TGFβR signaling binds to and activates PKA (15). We first biochemically measured cAMP levels in murine CD4+ T cells activated in the presence or absence of TGF-β. This analysis reveals that cAMP levels are similar between T cells activated in the presence or absence of TGF-β (Fig. 5E), suggesting that TGFβR signaling uses a cAMP independent mechanism to activate PKA. To determine whether TGFβR signaling promotes SMAD3/4 binding to PKA, CD4+ T cells were activated with and without TGF-β and PKA was immunoprecipitated. In T cells activated in the presence of TGF-β, both SMAD3 and SMAD4 coprecipitated with PKA (Fig. 5, F and G). Together, these data support a model where TGFβR signaling in murine CD4+ T cells promotes PKA activation via SMAD3/4 binding.
TGF-β signaling activates CSK via PKA to disrupt the PI3K P85–P110 heterodimer
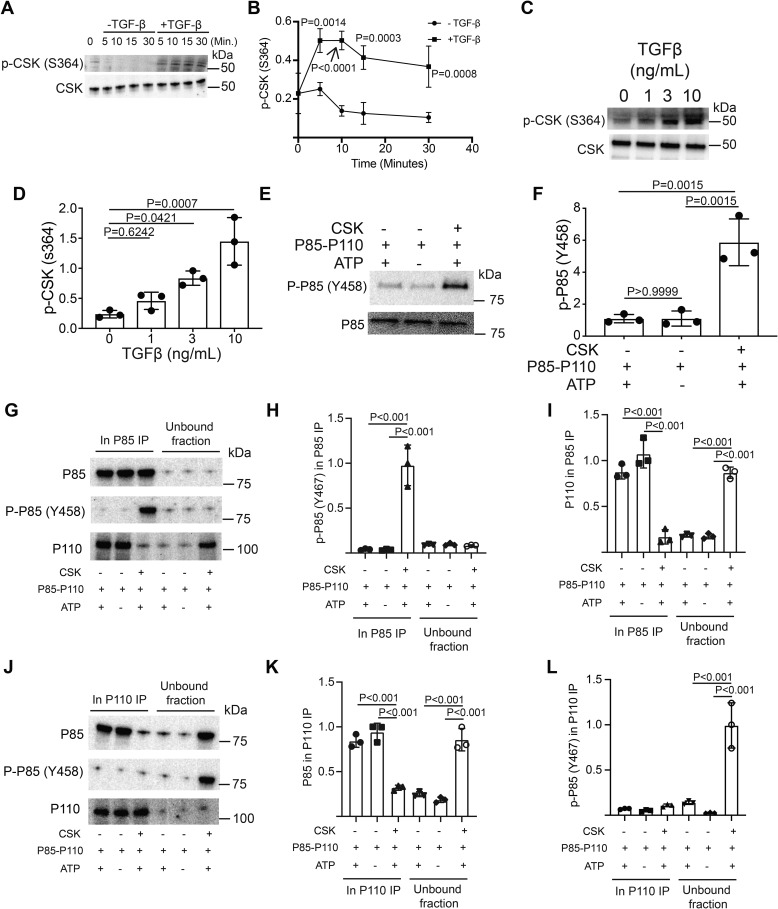
The kinase that phosphorylates P85 on Tyr-458 in T cells was not defined. The ScanSite algorithm predicted that CSK was a putative kinase for P85–Tyr-458 (31). The CSK substrate, Tyr-505 on LCK, was hyperphosphorylated in response to TGFβR signaling (Fig. 1, D and F), which suggested that CSK was activated by TGFβR signaling. Previous work demonstrated that PKA phosphorylated CSK on Ser-364, which stimulated CSK activity (32). Thus, TGFβR signaling could activate CSK via PKA. Immunoblotting for p-CSK (Ser-364) revealed that phosphorylation of Ser-364 increased in T cells activated in the presence of TGF-β (Fig. 6, A and B). To establish the threshold required for TGF-β receptor signaling to promote Ser-364 phosphorylation of CSK, murine T cells were activated through the TCR and CD28 receptors in the presence of varying doses of TGF-β. Immunoblotting revealed that Ser-364 phosphorylation increased as a function of TGF-β in the culture (Fig. 6, C and D). Together, these data demonstrated that TGFβR signaling activated CSK.
Figure 6.
TGF-β signaling activates CSK via PKA to disrupt the PI3K P85–P110 heterodimer. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for various time points with or without 10 ng/ml TGF-β. A, immunoblotting was performed on cell lysates for the phosphorylated and total protein abundance for CSK. B, densitometry was performed across three independent experiments to quantitate the abundance of tyrosine phosphorylation for CSK (Ser-364). The abundance of phosphorylated protein was normalized to total protein. Shown are mean ± S.D.; p values were calculated by two-way ANOVA. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min with various doses of TGF-β. C, immunoblotting was performed for p-CSK (Ser-364) and total CSK on cell lysates. One representative blot from three independent experiments is shown. D, densitometry was performed across three independent experiments to quantitate the abundance of phosphorylated CSK (Ser-364) as a function of TGF-β dose. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test. E, in vitro kinase reactions with recombinant CSK and recombinant PI3K heterodimer were performed. Immunoblotting was performed on the in vitro reactions for p-P85 (Tyr-458) and total P85. One representative blot from three independent experiments is shown. F, densitometry was performed across three independent experiments to quantitate abundance of phosphorylated P85 (Tyr-458) generated in the CSK-PI3K in vitro reactions. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test. Recombinant PI3K was reacted with ATP or CSK+ATP for 1 h. P85 was immunoprecipitated. G, immunoblotting was performed for total P85, p-P85 (Tyr-458), or P110 in the immunoprecipitated or unbound fractions. H and I, densitometry was performed across three independent experiments to quantitate abundance of p-P85 (Tyr-458) or P110 in the P85 immunoprecipitate. Additionally, recombinant PI3K was reacted with ATP or CSK+ATP and P110 was immunoprecipitated. J, immunoblotting was performed for total P85, p-P85 (Tyr-458) or P110 in the immunoprecipitated or unbound fractions. K and L, densitometry was performed across three independent experiments to quantitate abundance of P85 or p-P85 (Tyr-458) in the P110 immunoprecipitate. p values were calculated by one-way ANOVA with a Tukey multiple comparison test for panels H, I, K, and L.
To determine whether CSK directly phosphorylated P85, in vitro kinase assays were performed with recombinant CSK and PI3K complex. Immunoblotting of the in vitro kinase reactions revealed that CSK could phosphorylate Tyr-458 on P85 (Fig. 6, E and F). TGFβR signaling in combination with stimulation through TCR and CD28 receptors resulted in the disruption of the P85–P110 heterodimer (Fig. 3, H and I). CSK phosphorylated P85 on Tyr-458, which is positioned at the P85–P110 dimer interface (Fig. 3G). Therefore, we hypothesized that CSK catalyzed phosphorylation of P85 would disrupt the P85–P110 dimer. Recombinant PI3K complex was incubated with ATP or with both ATP and CSK. P85 was immunoprecipitated. Immunoblotting was performed on the IP and unbound fractions for P85, p-85 (Tyr-458), and P110. As expected, P85 was strongly detected in the IP (Fig. 6G). p-P85 was only detected in the IP where both CSK and ATP were added to the reaction (Fig. 6, G and H). P110 coprecipitated with P85 in the ATP only and control conditions (Fig. 6, G and I). However, CSK-catalyzed phosphorylation of P85 resulted in a significant reduction of P110 coprecipitating with P85 and significantly more P110 in the unbound fraction (Fig. 6, G and I).
To confirm the results of the P85 IP, Recombinant PI3K complex was incubated with ATP or with both ATP and CSK. P110 was immunoprecipitated. P110 was strongly detected in all IPs with little in the unbound fractions (Fig. 6J). P85 coprecipitated with P110 in reactions were ATP was incubated with PI3K or buffer control and little P85 was detected in the unbound fraction (Fig. 6, J and K). However, CSK-mediated phosphorylation of P85 resulted in less P85 coprecipitating with P110 and increased the levels of P85 and p-P85 (Tyr-458) in the unbound fraction (Fig. 6, J–L). Taken together, these data support a model where TGFβR signaling disrupts the PI3K P85–P110 dimer via CSK.
TGF-β receptor signaling regulates phosphatidylinositol metabolism
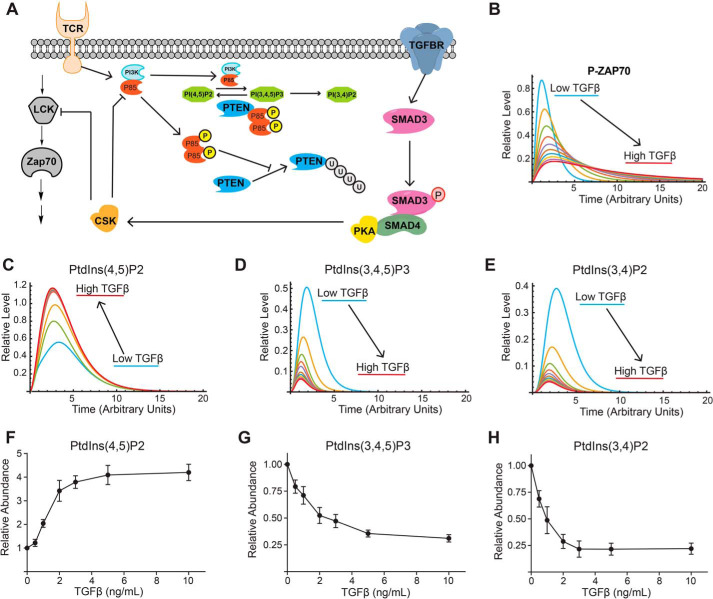
A clear prediction from our signaling data are that TGFβR signaling would regulate phosphatidylinositol metabolism and downstream pathways sensitive to PI3K signaling. To systematically test this hypothesis, we built a computational model based on our observed experimental and published data to better conceptualize how TGFβR signaling regulates early activation events in T cells (Fig. 7A). In this model, signaling through the TCR results in LCK activation (measured by increased phosphorylation on LCK Tyr-394) and downstream activation of ZAP70 (measured by phosphorylation of Tyr-319). TCR signaling also activates PI3K, which would convert PtdIns(4,5)P2 to PtdIns(3,4,5)P3. We experimentally observe that PIP4KA, which synthesizes PtdIns(4,5)P2 from PtdIns(4)P, is phosphorylated in T cells activated with or without TGF-β and could stimulate its enzymatic activity (Fig. 3A). We therefore assume in the model that TCR signaling stimulated the synthesis PtdIns(4,5)P2. Signaling through the TGF-β receptor results in phosphorylation of SMAD3 and formation of the SMAD3/4 complex (15), which binds and activates PKA (Fig. 5, F and G). In turn, PKA phosphorylates and activates CSK (Fig. 6, A–D), which down-modulates the activity of both LCK (Fig. 1, D and F) and PI3K by disrupting the P85–P110 dimer (Figs. 3, H and I, and 6, E–L). The PTEN lipid phosphatase, which converts PtdIns(3,4,5)P3 to PtdIns(4,5)P2, is also included in the model. Our previous work demonstrates that strong TCR signals drive PTEN degradation via the ubiquitin and caspase pathways whereas weak TCR signals maintained PTEN levels (33). The TCR strength used in all experiments falls into the “strong” regime. One mechanism that could protect PTEN from degradation is binding to P85. In other cell types, P85 can dimerize and bind to PTEN, which sterically inhibits ubiquitination and proteasomal degradation of PTEN (34). Additionally, P85 binding can enhance the phosphatase activity of PTEN (35). Because TGF-β receptor signaling disrupted the P85–P110 heterodimer (Figs. 3, H and I, and 6, E–L), we reasoned more P85 would be free to bind to and prevent PTEN degradation, which was included in the computational model.
Figure 7.
TGF-β receptor signaling regulates phosphatidylinositol metabolism. A, a model for the crosstalk between TCR and TGFβR signaling was constructed. Simulations were performed using a constant amount of TCR signal and varying amounts of TGFβR signal. B–E, the trajectories for (B) p-ZAP70, (C) PtdIns(4,5)P2, (D) PtdIns(3,4,5)P3, and (E) PtdIns(3,4)P2 are presented. CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min with various doses of TGF-β. F–H, imaging flow cytometry was utilized to measure the relative abundance of (F) PtdIns(4,5)P2, (G) PtdIns(3,4,5)P3, and (H) PtdIns(3,4)P2 normalized to the zero TGF-β data point. Shown are mean ± S.D. for three independent experiments.
Simulations were performed where the strength of the TCR signal remained constant and the level of TGFβR signaling was modulated. Increased TGFβR signal reduced phosphorylation and activation of both LCK and ZAP70 (Fig. 7B). This was consistent with our experimental data where the activating phosphorylation sites on both p-LCK (Tyr-394) and p-ZAP70 (Tyr-319) decreased as a function of TGF-β dose (Fig. 1, D, E, and G). Focusing on PI3K signaling, the simulations predicted that increased TGFβR signaling would result in increased PtdIns(4,5)P2 (Fig. 7C), while decreasing both PtdIns(3,4,5)P3 (Fig. 7D) and PtdIns(3,4)P2 (Fig. 7E). To test the model prediction that TGFβR signaling regulated phosphatidylinositol metabolism, the abundance of PtdIns(4,5)P2 (Fig. 7F), PtdIns(3,4,5)P3 (Fig. 7G), and PtdIns(3,4)P2 (Fig. 7H) were measured using imaging flow cytometry in murine CD4+ T cells activated in the presence of varying doses of TGF-β. These data supported the modeling prediction that TGF-β receptor signaling increased PtdIns(4,5)P2 and decreased both PtdIns(3,4,5)P3 and PtdIns(3,4)P2.
TGF-β receptor signaling promotes a P85–PTEN complex that protects PTEN from degradation and dampens AKT activation
The dose of anti-CD3 antibody used in our assays and in standard in vitro Treg polarization assays would result in a strong TCR signal leading to PTEN degradation via proteasomal and caspase pathways (33). Yet, PTEN is required for Treg induction and maintenance (33, 36). Based on previously published work in other cell types (34), we invoked in our computational model that the dissociation of the P85–P110 PI3K heterodimer would allow P85 to bind to and prevent PTEN degradation (Fig. 7A). In simulations where there is no TGFβR signaling, PTEN is rapidly degraded (Fig. 8A). Increasing the strength of TGFβR signal in the simulations stabilized PTEN abundance via P85 binding (Fig. 8A). Experimentally, addition of TGF-β during T-cell activation increased PTEN abundance (Fig. 8, B and C). To determine whether TGFβR signaling promoted P85 binding to PTEN, P85 was immunoprecipitated from T cells activated in the presence or absence of TGF-β. Immunoblotting the P85 immunoprecipitates demonstrated that more PTEN coprecipitated with P85 in T cells activated in the presence of TGF-β (Fig. 8, D and E). Taken together, these data supported a model where TGFβR signaling maintained PTEN levels via P85 binding.
Figure 8.
TGF-β receptor signaling promotes P85 stabilization of PTEN and AKT activation. Simulations were performed using a constant amount of TCR signal and varying amounts of TGFβR signal. A, trajectories for total PTEN abundance is presented. B, CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min with various doses of TGF-β. Immunoblotting was performed for total PTEN and actin. C, densitometry was performed from three independent experiments where PTEN abundance was normalized to actin. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test. D, CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for 10 min with ± 10 ng/ml TGF-β. P85 was immunoprecipitated from cell lysates. Immunoblotting was performed for PTEN and P85. One representative blot from three independent experiments is shown. E, densitometry across three independent immunoblots was performed for PTEN normalized to total P85 in the IP. Shown are mean ± S.D.; p values were calculated by a Student's t test. Simulations were performed using a constant amount of TCR signal and varying amounts of TGFβR signal. F, trajectories for p-AKT (Ser-473) abundance are presented. Murine CD4+ T cells isolated by negative selection were in vitro activated through TCR and CD28 costimulation for various time points with or without 10 ng/ml TGF-β. G, immunoblotting was performed on cell lysates for the phosphorylated and total protein abundance for phosphorylated AKT (Ser-473). H, densitometry was performed across three independent experiments to quantitate the abundance of phosphorylation for AKT (Ser-473). The abundance of phosphorylated protein was normalized to total protein. Shown are mean ± S.D.; p values were calculated by two-way ANOVA. I, CD4+ T cells were activated in the presence of different doses of TGF-β and immunoblotting was performed for p-AKT (Thr-308), p-AKT (Ser-473), and total AKT. J and K, densitometry across three independent immunoblots was performed for (J) p-AKT (Thr-308) normalized to total AKT and (K) p-AKT (Ser-473) normalized to total AKT. Shown are mean ± S.D.; p values were calculated by one-way ANOVA with a Tukey multiple comparison test.
One kinase regulated by the PtdIns(4,5)P2:PtdIns(3,4,5)P3 ratio is AKT. PKD1 is activated by low levels of PtdIns(3,4,5)P3 and phosphorylates Thr-308 on AKT. mTORC2 requires high levels of PtdIns(3,4,5)P3 for activation and phosphorylation of Ser-473 on AKT (27). In simulations, TGFβR signaling reduced Ser-473 phosphorylation (Fig. 8F), driven by reduced PtdIns(3,4,5)P3 (Fig. 7, D and G) and PTEN abundance (Fig. 8, A–C). To confirm this prediction, we took an immunoblotting approach to monitor the phosphorylation of Ser-473 on AKT in murine CD4+ T cells activated through TCR, CD28 with and without TGF-β (Fig. 8G). We observed that murine CD4+ T cells activated in the presence of TGF-β had significantly reduced p-AKT (Ser-473) than CD4+ T cells activated without TGF-β (Fig. 8H). To establish the threshold required for TGF-β receptor signaling to down-modulate AKT Ser-473 phosphorylation, murine T cells were activated through the TCR and CD28 receptors with varying doses of TGF-β. Immunoblotting was performed for p-AKT (Thr-308) and p-AKT (Ser-473) (Fig. 8I). p-AKT (Thr-308) abundance was not significantly impacted by TGF-β (Fig. 8J). Our previous work demonstrated that even weak TCR signals generate enough PtdIns(3,4,5)P3 to activate PDK1 (27), so it was not surprising that TGF-β did not impact p-AKT (Thr-308). However, TGFβR signaling did reduce p-AKT(Ser-473) in a dose-dependent manner (Fig. 8K), consistent with mTORC2 activation requiring high levels of PtdIns(3,4,5)P3. Taken together, these data supported a model where TGFβR signaling regulated the activation of AKT by altering the PtdIns(4,5)P2: PtdIns(3,4,5)P3 ratio.
Discussion
Herein, we defined at the biochemical level how TGFβR signaling regulated early events during T-cell activation. The conventional view is that TGFβR regulates T cells at the level of transcription by driving SMAD3/4 nuclear translocation and transcriptional regulation. Our data argue that the mechanism of action of TGF-β extends beyond transcriptional regulation. We find that TGFβR signaling globally regulates kinase signaling networks that could regulate multiple biological processes including the actin cytoskeleton, metabolism, chromatin organization and mRNA splicing (Fig. 2C). A key kinase that is regulated by TGF-β is CSK, which is a negative regulator of TCR signaling. TGFβR signaling activated CSK via SMAD3/4 activation of PKA (Fig. 5, F and G), resulting in LCK inhibition via Tyr-505 phosphorylation (Fig. 1, D and F). Additionally, TGFβR signaling promoted the phosphorylation of the P85 subunit of PI3K (Fig. 3, B–F), which disrupted the P85–P110 heterodimer to reduce PI3K activity at the plasma membrane (Fig. 3, H–J). In turn, TGFβR signaling altered the PtdIns(4,5)P2 to PtdIns(3,4,5)P3 ratio to favor increased PtdIns(4,5)P2 (Fig. 7, F and G). Reduced PtdIns(3,4,5)P3 down-modulated p-AKT(Ser-473) (Fig. 8, G–I and K), which requires high levels of PtdIns(3,4,5)P3 to activate the mTORC2 kinase (27). Increased PtdIns(4,5)P2 correlated with FAK activation (Fig. 1, D and H), which is a kinase activated by PtdIns(4,5)P2 (37). These data demonstrate TGFβR signaling controlled the activation of kinases by regulating the generation of phosphatidylinositol species.
TGF-β is a regulator of T-cell physiology and differentiation (38–40). Mice lacking either TGF-β1 or specific deletion of TGFβR generated T cells that were hyperproliferative and more sensitive to activation. We found that TGF-β signaling reduced PI3K activity which resulted in reduced PtdIns(3,4,5)P3 and increased PtdIns(4,5)P2 levels (Fig. 7, F and G). The activation of many kinases and pathways are controlled by PtdIns(3,4,5)P3 generation. For example, PtdIns(3,4,5)P3 generation recruits ITK to the plasma membrane through its pleckstrin homology domain, which ultimately leads to ITK activation and downstream signaling (41–43). Previous work demonstrates that TGF-β blunts ITK activation (14). Our work provides a biochemical basis for how TGF-β signaling suppresses kinases like ITK that are activated by PtdIns(3,4,5)P3. TGF-β suppresses the generation of PtdIns(3,4,5)P3 in T cells (Fig. 7G). By reducing PtdIns(3,4,5)P3 generated by T-cell activation, TGF-β signaling selects kinases that are activated during T-cell activation. There is a range of affinities for proteins that bind to PtdIns(3,4,5)P3. A direct example relevant to T cells is AKT. PDK1 phosphorylates Thr-308 on AKT and mTORC2 phosphorylates Ser-473. Both PDK1 and mTORC2 kinase activity are activated by PtdIns(3,4,5)P3. PDK1 binds to PtdIns(3,4,5,)P3 with a 10-fold greater affinity than mTORC2 (44). We have previously seen that TCR signal strength regulates PtdIns(3,4,5)P3 generation whereas weak TCR signals generate low PtdIns(3,4,5)P3 levels and strong TCR signals generate high levels (27). Low levels of PtdIns(3,4,5)P3 are sufficient to activate PDK1 but not mTORC2, which requires high PtdIns(3,4,5)P3 levels (27). Additionally, our work demonstrates that AKT activation is altered by TGF-β that blunts high PtdIns(3,4,5)P3 generation and phosphorylation of Ser-473 phosphorylation on AKT (Fig. 8, F–K). The AKT/mTORC1 axis supports cellular growth and proliferation. Therefore, we propose that TGF-β suppresses T-cell proliferation in part by diminishing PtdIns(3,4,5)P3 generation by directly regulating PI3K and down-modulating signaling through the AKT/mTOR axis.
Our work highlights that the SMAD3/4 heterotrimer generated by TGFβR activation binds to and activates PKA (Fig. 5, F and G). These data suggest that SMAD3/4 directly regulates cytoplasmic kinases during T-cell activation, which to our knowledge is novel in the context of T-cell biology. Other work demonstrates that TGF-β signaling can regulate mitochondrial processes. TGFβR signaling promotes SMAD mitochondrial localization and inhibition of ATP synthase activity (25). Consistent with TGFβR signaling regulating metabolic processes that occur in the mitochondria, our proteomic data identified that the ATP5F1B subunit of ATP synthase (Fig. 2E) was hyperphosphorylated in CD4+ T cells activated in the presence of TGF-β. Additionally, components of the electron transport chain including the NDUFA4 subunit of complex I (Fig. 2F) and the COX6B1 subunit of complex I (Fig. 2G) were also hyperphosphorylated in CD4+ T cells activated in the presence of TGFβR. Together with other studies (45), these results demonstrate that differential signaling in T cells can regulate metabolic processes and that TGF-β impacts kinase signaling pathways.
The inhibitory mechanism used by TGFβR to suppress TCR signaling closely resembles that of prostaglandin E2 (46), which signals through EP2 and EP4 GPCR prostanoid receptors. Signaling through EP2 or EP4 activates adenylate cyclase to generate cAMP, which stimulates PKA to phosphorylate and activate CSK. Activation of CSK results in to phosphorylation of Tyr-505 on LCK, which down-modulates LCK activity and TCR signaling (28, 30, 46). cAMP levels are similar between T cells activated with and without TGF-β in our experiments (Fig. 5E), suggesting that TGFβR does not activate PKA via cAMP generation. However, TGFβR signaling utilizes an alternate mechanism to activate PKA. Signaling through TGFβR results in the phosphorylation of SMAD3, which in turn binds SMAD4. Previous work (15) and our current work demonstrate that SMAD3/4 binds to and activates PKA (Fig. 5, F and G). As with prostaglandin E2, TGFβR signaling activates CSK via PKA to inhibit LCK activation via Tyr-505 phosphorylation (Fig. 1, D and E). These data demonstrate that there are multiple pathways in T cells that can signal through PKA and CSK to down-modulate T-cell signaling.
In addition to regulating proximal TCR signaling via LCK (Fig. 1, A–C), we found that TGFβR signaling also regulated PI3K by disrupting the P85–P110 heterodimer (Fig. 3, H and I). In T cells activated without TGF-β, high levels of PtdIns(3,4,5)P3 were generated. However, TGFβR signaling suppressed PtdIns(3,4,5)P3 generation and favored elevated PtdIns(4,5)P2 in part by P85 binding to and preventing PTEN degradation (Fig. 8, B–E). Generation of quantitatively different amounts of phosphatidylinositols could serve as a switch to activate specific signaling pathways (27). In the context of TGFβR signaling, FAK activation correlated with the generation of PtdIns(4,5)P2 (Figs. 1, A and E, and 7F), which is known to activate FAK through an integrin-independent mechanism (37, 47). Generation of PtdIns(3,4,5)P3 in T cells activated without TGF-β resulted in mTORC2 catalyzed phosphorylation of Ser-473 on AKT (Fig. 8, G–I and K). In both B cells and T cells, differential phosphorylation on Ser-473 controlled AKT substrate specificity (48, 49). In our previous work, we found that strong TCR signals drove PtdIns(3,4,5)P3 generation which favored phosphorylation on both Thr-308 and Ser-473 on AKT and weak TCR signals only generated phosphorylation on Thr-308 (48). Differential activation of AKT by TCR signal strength regulated mRNA splicing of key signaling proteins including CD247 (48). Together, these data demonstrate that T cells utilize phosphatidylinositol metabolism to interpret multiple extracellular stimuli including the measurement of TCR signal strength and gauge the level TGF-β cytokine in the microenvironment. Although we have documented that differential phosphatidylinositol generation controls the activation of FAK and AKT in T cells, likely other proteins are regulated by differential phosphatidylinositol generation, which will be the focus of our future work.
Recent work highlights the importance of utilizing proteomic approaches to profile signaling in T cells. Growing evidence demonstrates that T cells remodel their proteomes in response to different stimulation (50). Additionally, the phospho-tyrosine proteome generated by naïve versus memory T cells was markedly different (51), demonstrating that T-cell subsets are poised to respond differently to stimulation through the TCR. Although these and other studies highlight the importance of global proteomic changes in interpreting different extracellular stimuli and that signaling differences exist between T-cell subsets, more work using proteomic profiling technologies is needed to characterize signaling networks in T cells activated with different stimuli to understand more fully the diversity of biochemical signaling modalities used by T cells to promote adaptive immune responses and maintain tolerance.
Experimental procedures
Murine CD4+ T-cell isolation and activation
Spleens from C57BL/6 mice were a generous gift from the laboratory of Dr. Mark Shlomchik at the University of Pittsburgh. Mice were housed at the University of Pittsburgh in a pathogen-free facility and handled under Institutional Animal Care and Use Committee–approved guidelines. Negative selection (Miltenyi Biotech) was utilized to isolate CD4+ T cells from C57BL/6 spleens. T cells were rested for 1 h at 37 ° Celsius. T cells were activated using plates that were coated with 3 μg/ml of anti-CD3 mAb (clone 17A2, BioLegend) and 3 μg/ml soluble anti-CD28 mAb (clone 37.51, BioLegend), and IL-2 (5 ng/ml). For some experiments, variable doses of recombinant TGF-β1 (BioLegend) noted in the text were added. For experiments using the small molecule SIS3 inhibitor, CD4+ T cells were incubated with various does of SIS3 (Sigma-Aldrich) noted in the text for 2 h prior to activation.
Western blotting
PAGE was performed using Bio-Rad precast Protein TGX gels. Proteins were transferred to PVDF membranes using a Bio-Rad Trans-Blot Turbo transfer system with the preprogramed mixed molecular weight setting. Antibodies used for Western blotting purchased from Cell Signaling Technology included: p-LCK (Tyr-505) (2751), LCK (Asp-88), p-tyrosine (P-Tyr-1000), p-P85 (Tyr-458), (E3U1H), P85 (19H8), P110 (D1Q7R), SMAD3 (C67H9), p-PKA (Thr-197) (D45D3), PKA (D38C6), CSK (C74C1), p-ZAP70 (Tyr-319) (65E4), ZAP70 (D1C10E), AKT (C67E7), p-AKT (Ser-473) (D9E), actin (13E5), FAK (D2R2E), and p-FAK (Tyr-397) (D20B1). Antibodies used for Western blotting purchased from Thermo Fisher included p-CSK (Ser-364) (PA5–40214) and p-LCK (Tyr-394) (PA5–37628). Antibodies used for Western blotting purchased from Abcam included p-SMAD3 (Ser-423 + Ser-425) (EP823Y). All primary antibodies utilized were rabbit. An anti-rabbit IgG-HRP antibody (Cell Signaling Technology, 7074) was used with the SuperSignal West Femto chemiluminescent substrate for detection on a protein simple FluorChem M system. Densitometry quantitation was performed using the ImageJ software package.
Immunoprecipitation of tyrosine phosphorylated proteins and label-free LC-MS analysis
Murine CD4+ T cells were activated for 10 min in the presence or absence of 10 ng/ml TGF-β. Four biological replicates were included per activation condition. Cells were lysed in a buffer containing 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 5% glycerol, cOmplete C protease (Roche) and PhosSTOP phosphatase inhibitors (Roche). The resulting lysates were sonicated and centrifuged. The remaining lysate was incubated with an anti-tyrosine antibody conjugated to magnetic beads (Cell Signaling Technology, clone P-Tyr-100, catalogue no. 8095) for 12 h at 4 °C. The beads were washed twice with lysis buffer. The remaining proteins were eluted from the beads with a buffer containing 8 m urea (U5128, Sigma) and 0.1 m Tris-HCl, pH 8.5. Trypsin was utilized to generate tryptic peptides and processed with the filter-aided sample preparation method (52), desalted utilizing C18 spin columns, and dried in a SpeedVac. Peptides were reconstituted in 0.5% formic acid in 96:4 water:acetonitrile and resolved with LC tandem MS using a system composed of a Waters nanoACQUITY UPLC in-line with a Q Exactive mass spectrometer (Thermo Fisher). Solvent A (0.1% formic acid in water, Burdick & Jackson) and solvent B (0.1% formic acid in acetonitrile, Burdick & Jackson) were used as the mobile phase. Peptides were then eluted from a capillary column (100 μm inner diameter × 100 mm long; ACQUITY UPLC M-Class Peptide BEH C18 Column, 1.7-μm particle size, 300 Å Waters), and resolved using a 100-min gradient at a flow rate of 0.9 μl/min (4–33% B for 90 min, 33–80% B for 2 min, constant at 80% B for 6 min, and then 80–0% B for 2 min to equilibrate the column). Data were collected in positive ionization mode.
PEAKS Studio 10.0 build 20190129 was used to sequence and identify peptides. The UniProt-SwissProt Mus musculus (house mouse) sequence database was used, which contained 17,013 entries. One missed cleavage by trypsin was permitted in the database search. Carbamidomethylation was treated as a fixed modification. Acetylation (Protein N-term), oxidation (M) and phosphorylation (Ser, Thr, or Tyr) were set as variable modifications. The mass tolerance for precursor ions was set to 20 ppm and mass tolerance for fragment ions set to 0.8 Da. The FDR for peptide/spectrum matches is reported as 9.5%, calculated by decoy fusion. Label-free quantitation was performed using the quantitative module in the PEAKSX software. Individual values for all quantitative mass spectrometric measurements are depicted and the mean peak area is depicted as a bar graph. Error bars represent ± the S.D. p values were calculated with a two-tailed Student's t test to assess statistical significance.
Biological pathway analysis
The following metrics were used to identify proteins that were differentially tyrosine phosphorylated in CD4+ T cells activated in the presence or absence of TGF-β: 1) peptide(s) corresponding to a protein had to be identified by the mass spectrometric analysis, and 2) if a protein was identified in multiple groups, its abundance had to be more than 2-fold greater based on the mass spectrometric label-free quantitation to be assigned as specific to a group. The PANTHER software was used to perform a statistical overrepresentation test to identify pathways differentially targeted by tyrosine phosphorylation between CD4+ T cells activated in the presence or absence of TGF-β.
Immunoprecipitation of P85 or PKA
Murine CD4+ T cells were activated for 10 min in the presence or absence of 10 ng/ml TGF-β. Cells were lysed in a buffer containing 1% Nonidet P-40, 50 mm Tris, pH 8.0, 150 mm NaCl and cOmplete C phosphatase inhibitor mixture. Lysates were incubated at room temperature for 2 h with antibodies specific for either P85 (Cell Signaling Technology, 19H8) or PKA (Cell Signaling Technology, D38C6). Magnetic protein A beads (Cell Signaling Technology) were utilized to capture the immune complexes, which were analyzed by Western blotting as described above.
PIP3 mass ELISA
Murine CD4+ T cells were activated in the presence or absence of TGF-β. Pellets from 2 million cells were washed with ice-cold 0.5 m TCA and treated with 750 μl of MeOH:CHCl3:12N HCl (80:40:1), vortexing for 30 min and centrifuging for 10 min at 3000 rpm to remove neutral lipids. The supernatant was treated with 250 μl of CHCl3 and 450 μl of 0.1 N HCl and centrifuged. The organic phase was dried under a stream of nitrogen gas and reconstituted in PBS. Mass ELISA kits from Echelon Biosciences were used to measure PIP3 using the manufacturer's protocol using a Molecular Devices SpectraMax i3 plate reader.
In vitro CSK enzyme assay and immunoprecipitation of P85 and P110
0.1 μg of recombinant CSK (Abcam, ab42617) was incubated for 1 h at 37 ° Celsius with 1.0 μg of recombinant PI3K heterodimer (Abcam, ab125633) in a reaction buffer containing 50 mm HEPES, pH 7.4, 3 mm MgCl2, 1 mm DTT and 0.2 m ATP. Reactions were quenched by adding 2× Laemmli Sample Buffer (Bio-Rad, 1610737) and heated at 95 °C for 10 mins. Reactions were resolved by PAGE, and Western blotting was performed with antibodies specific for P85 (19H8, Cell Signaling Technology) and p-P85 (Tyr-458, Cell Signaling Technology).
Following in vitro enzyme reactions, P85 or P110 was immunoprecipitated and was incubated at room temperature for 2 h with antibodies specific for either P85 (Cell Signaling Technology, 19H8) or P110 containing complexes (Cell Signaling Technology, D1Q7R). Magnetic protein A beads (Cell Signaling Technology) were utilized to capture the immune complexes, which were analyzed by Western blotting as described above.
Measurement of cAMP levels in T cells
Five million activated murine CD4+ T cells collected per data point. A cAMP colorimetric assay kit was utilized (Abcam, catalogue no. ab65355). Cells were lysed in a 0.1 m HCL buffer. The lysates were processed per the manufacturer's instructions. Absorbance readings at 450 nm were recorded on a Molecular Devices SpectraMax i3 plate reader. A standard curve with cAMP standards was fit by linear regression in the Prism GraphPad8 software package and used to determine the amount of cAMP in the experimental samples.
Computational modeling
Computational models of T cell signaling were constructed in the SystemModeler 12.0 software package. Simulations were performed in the Mathmatica 12.0 software package using the DSSL solver.
Imaging flow cytometric measurement of phosphatidylinositol abundance
Murine CD4+ T cells activated in the presence or absence of TGF-β were incubated in 1 volume of 2× Cytofix/Perm/Wash Buffer (3% PFA + BD Perm/Wash Buffer (catalogue no. 554723)) at room temperature for 15 min and on ice for 30 min. Cells were washed with Perm/Wash buffer and resuspended in Perm/Wash buffer with FC blocking antibody (2.4G2). Antibodies against cells were stained with antibodies against TCR-APC (BD Biosciences, clone H57–597) and CD4-PerCP-5.5 (BD Biosciences, clone RM4–5). Additionally, cells were stained with antibodies against Pi(4,5)P2-PE (Echelon Biosciences, Z-B045), PIP3-PE (Echelon Biosciences, Z-B3345B) or Pi(3,4)P2-PE (Echelon Biosciences, Z-B034). Cells were washed with Perm/Wash buffer and resuspended in PBS containing 3% FBS, 2 mm EDTA, 0.02% sodium azide. Samples were analyzed on an AMNIS Image Stream MarkII imaging flow cytometer. Cells were gated on the TCR+CD4+ population and 2000 cells per sample were collected. The IDEAS software package was utilized to compensate and analyze the imaging flow cytometry data.
Data availability
Data will be shared upon request. Please contact William Hawse, Dept. of Immunology at the University of Pittsburgh, whawse@pitt.edu. The raw mass spectrometric data were deposited to the PRIDE database under the accession number PXD017871.
Supplementary Material
Acknowledgments
We greatly thank Dr. Mark Shlomchik and members of the lab for useful discussions. We thank Dewayne Falkner and Aarika MacIntyre for superb technical assistance with flow cytometry and imaging flow cytometric analysis. This work benefitted from an IMAGESTREAMX MARKII funded by the National Institutes of Health 1S10OD019942[hyhen]01, for which we thank Lisa Borghesi.
This article contains supporting information.
Author contributions—R. T. C. and W. F. H. conceptualization; R. T. C. and W. F. H. data curation; R. T. C. and W. F. H. formal analysis; R. T. C., M. L., W. C. B., and W. F. H. investigation; R. T. C., M. L., W. C. B., and W. F. H. methodology; M. L. and W. C. B. resources; W. F. H. supervision; W. F. H. funding acquisition; W. F. H. validation; W. F. H. writing-original draft; W. F. H. project administration; W. F. H. writing-review and editing.
Funding and additional information—This work was supported by Competitive Medical Research Fund Grant from the University of Pittsburgh Medical Center (to W. F. H.), startup funds generously provided by the University of Pittsburgh Department of Immunology (to W. F. H.) and by the National Institutes of Health Grant 1R21AI144558-01A1 (to W. F. H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TCR
- T-cell receptor
- TGF
- transforming growth factor
- CSK
- C-terminal Src kinase
- PI3K
- phosphoinositide 3-kinase
- PtdIns(3,4,5)P3
- phosphatidylinositol (3,4,5)-trisphosphate
- PTEN
- phosphatase and tensin homolog
- IP
- immunoprecipitation
- ANOVA
- analysis of variance.
References
- 1. O'Shea J. J., and Paul W. E. (2010) Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu J., and Paul W. E. (2010) Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 238, 247–262 10.1111/j.1600-065X.2010.00951.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Horwitz D. A., Zheng S. G., and Gray J. D. (2003) The role of the combination of IL-2 and TGF-β or IL-10 in the generation and function of CD4+ CD25+ and CD8+ regulatory T cell subsets. J. Leukoc. Biol. 74, 471–478 10.1189/jlb.0503228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cahir McFarland E. D., Hurley T. R., Pingel J. T., Sefton B. M., Shaw A., and Thomas M. L. (1993) Correlation between Src family member regulation by the protein-tyrosine-phosphatase CD45 and transmembrane signaling through the T-cell receptor. Proc. Natl. Acad. Sci. U.S.A. 90, 1402–1406 10.1073/pnas.90.4.1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sieh M., Bolen J. B., and Weiss A. (1993) CD45 specifically modulates binding of Lck to a phosphopeptide encompassing the negative regulatory tyrosine of Lck. EMBO J. 12, 315–321 10.1002/j.1460-2075.1993.tb05659.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iwashima M., Irving B. A., van Oers N. S., Chan A. C., and Weiss A. (1994) Sequential interactions of the TCR with two distinct cytoplasmic tyrosine kinases. Science 263, 1136–1139 10.1126/science.7509083 [DOI] [PubMed] [Google Scholar]
- 7. Costello P. S., Gallagher M., and Cantrell D. A. (2002) Sustained and dynamic inositol lipid metabolism inside and outside the immunological synapse. Nat. Immunol. 3, 1082–1089 10.1038/ni848 [DOI] [PubMed] [Google Scholar]
- 8. Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., and Crabtree G. R. (1988) Identification of a putative regulator of early T cell activation genes. Science 241, 202–205 10.1126/science.3260404 [DOI] [PubMed] [Google Scholar]
- 9. Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., and Wrana J. L. (1997) TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272, 27678–27685 10.1074/jbc.272.44.27678 [DOI] [PubMed] [Google Scholar]
- 10. Souchelnytskyi S., Tamaki K., Engström U., Wernstedt C., ten Dijke P., and Heldin C. H. (1997) Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J. Biol. Chem. 272, 28107–28115 10.1074/jbc.272.44.28107 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., Zhang P., Li J., Kulkarni A. B., Perruche S., and Chen W. (2008) A critical function for TGF-β signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9, 632–640 10.1038/ni.1607 [DOI] [PubMed] [Google Scholar]
- 12. Li M. O., Wan Y. Y., and Flavell R. A. (2007) T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity 26, 579–591 10.1016/j.immuni.2007.03.014 [DOI] [PubMed] [Google Scholar]
- 13. Chen M. L., Pittet M. J., Gorelik L., Flavell R. A., Weissleder R., von Boehmer H., and Khazaie K. (2005) Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-β signals in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 419–424 10.1073/pnas.0408197102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C. H., Seguin-Devaux C., Burke N. A., Oriss T. B., Watkins S. C., Clipstone N., and Ray A. (2003) Transforming growth factor β blocks Tec kinase phosphorylation, Ca2+ influx, and NFATc translocation causing inhibition of T cell differentiation. J. Exp. Med. 197, 1689–1699 10.1084/jem.20021170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L., Duan C. J., Binkley C., Li G., Uhler M. D., Logsdon C. D., and Simeone D. M. (2004) A transforming growth factor β-induced Smad3/Smad4 complex directly activates protein kinase A. Mol. Cell Biol. 24, 2169–2180 10.1128/MCB.24.5.2169-2180.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bergman M., Mustelin T., Oetken C., Partanen J., Flint N. A., Amrein K. E., Autero M., Burn P., and Alitalo K. (1992) The human p50csk tyrosine kinase phosphorylates p56lck at Tyr-505 and down-regulates its catalytic activity. EMBO J. 11, 2919–2924 10.1002/j.1460-2075.1992.tb05361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hardwick J. S., and Sefton B. M. (1995) Activation of the Lck tyrosine protein kinase by hydrogen peroxide requires the phosphorylation of Tyr-394. Proc. Natl. Acad. Sci. U.S.A. 92, 4527–4531 10.1073/pnas.92.10.4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neumeister E. N., Zhu Y., Richard S., Terhorst C., Chan A. C., and Shaw A. S. (1995) Binding of ZAP-70 to phosphorylated T-cell receptor zeta and eta enhances its autophosphorylation and generates specific binding sites for SH2 domain-containing proteins. Mol. Cell Biol. 15, 3171–3178 10.1128/MCB.15.6.3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chapman N. M., Connolly S. F., Reinl E. L., and Houtman J. C. (2013) Focal adhesion kinase negatively regulates Lck function downstream of the T cell antigen receptor. J. Immunol. 191, 6208–6221 10.4049/jimmunol.1301587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., et al. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oudart J. B., Doué M., Vautrin A., Brassart B., Sellier C., Dupont-Deshorgue A., Monboisse J. C., Maquart F. X., Brassart-Pasco S., and Ramont L. (2016) The anti-tumor NC1 domain of collagen XIX inhibits the FAK/ PI3K/Akt/mTOR signaling pathway through αvβ3 integrin interaction. Oncotarget 7, 1516–1528 10.18632/oncotarget.6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cai Y. C., Cefai D., Schneider H., Raab M., Nabavi N., and Rudd C. E. (1995) Selective CD28pYMNM mutations implicate phosphatidylinositol 3-kinase in CD86-CD28-mediated costimulation. Immunity 3, 417–426 10.1016/1074-7613(95)90171-X [DOI] [PubMed] [Google Scholar]
- 23. Pagès F., Ragueneau M., Klasen S., Battifora M., Couez D., Sweet R., Truneh A., Ward S. G., and Olive D. (1996) Two distinct intracytoplasmic regions of the T-cell adhesion molecule CD28 participate in phosphatidylinositol 3-kinase association. J. Biol. Chem. 271, 9403–9409 10.1074/jbc.271.16.9403 [DOI] [PubMed] [Google Scholar]
- 24. Heldin C. H., Miyazono K., and ten Dijke P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 10.1038/37284 [DOI] [PubMed] [Google Scholar]
- 25. Dimeloe S., Gubser P., Loeliger J., Frick C., Develioglu L., Fischer M., Marquardsen F., Bantug G. R., Thommen D., Lecoultre Y., Zippelius A., Langenkamp A., and Hess C. (2019) Tumor-derived TGF-β inhibits mitochondrial respiration to suppress IFN-γ production by human CD4+ T cells. Sci. Signal 12, eaav3334 10.1126/scisignal.aav3334 [DOI] [PubMed] [Google Scholar]
- 26. Jinnin M., Ihn H., and Tamaki K. (2006) Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β-induced extracellular matrix expression. Mol. Pharmacol. 69, 597–607 10.1124/mol.105.017483 [DOI] [PubMed] [Google Scholar]
- 27. Hawse W. F., and Cattley R. T. (2019) T cells transduce T-cell receptor signal strength by generating different phosphatidylinositols. J. Biol. Chem. 294, 4793–4805 10.1074/jbc.RA118.006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wehbi V. L., and Taskén K. (2016) Molecular mechanisms for cAMP-mediated immunoregulation in T cells—role of anchored protein kinase A signaling units. Front. Immunol. 7, 222 10.3389/fimmu.2016.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Girod A., Kinzel V., and Bossemeyer D. (1996) In vivo activation of recombinant cAPK catalytic subunit active site mutants by coexpression of the wild-type enzyme, evidence for intermolecular cotranslational phosphorylation. FEBS Lett. 391, 121–125 10.1016/0014-5793(96)00717-X [DOI] [PubMed] [Google Scholar]
- 30. Vang T., Torgersen K. M., Sundvold V., Saxena M., Levy F. O., Skålhegg B. S., Hansson V., Mustelin T., and Taskén K. (2001) Activation of the COOH-terminal Src kinase (Csk) by cAMP-dependent protein kinase inhibits signaling through the T cell receptor. J. Exp. Med. 193, 497–507 10.1084/jem.193.4.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Obenauer J. C., Cantley L. C., and Yaffe M. B. (2003) Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 10.1093/nar/gkg584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yaqub S., Abrahamsen H., Zimmerman B., Kholod N., Torgersen K. M., Mustelin T., Herberg F. W., Taskén K., and Vang T. (2003) Activation of C-terminal Src kinase (Csk) by phosphorylation at serine-364 depends on the Csk-Src homology 3 domain. Biochem. J. 372, 271–278 10.1042/bj20030021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hawse W. F., Sheehan R. P., Miskov-Zivanov N., Menk A. V., Kane L. P., Faeder J. R., and Morel P. A. (2015) Cutting edge: Differential regulation of PTEN by TCR, Akt, and FoxO1 controls CD4+ T cell fate decisions. J. Immunol. 194, 4615–4619 10.4049/jimmunol.1402554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheung L. W., Walkiewicz K. W., Besong T. M., Guo H., Hawke D. H., Arold S. T., and Mills G. B. (2015) Regulation of the PI3K pathway through a p85α monomer-homodimer equilibrium. Elife 4, e06866 10.7554/eLife.06866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chagpar R. B., Links P. H., Pastor M. C., Furber L. A., Hawrysh A. D., Chamberlain M. D., and Anderson D. H. (2010) Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 107, 5471–5476 10.1073/pnas.0908899107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huynh A., DuPage M., Priyadharshini B., Sage P. T., Quiros J., Borges C. M., Townamchai N., Gerriets V. A., Rathmell J. C., Sharpe A. H., Bluestone J. A., and Turka L. A. (2015) Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16, 188–196 10.1038/ni.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goñi G. M., Epifano C., Boskovic J., Camacho-Artacho M., Zhou J., Bronowska A., Martín M. T., Eck M. J., Kremer L., Gräter F., Gervasio F. L., Perez-Moreno M., and Lietha D. (2014) Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc. Natl. Acad. Sci. U.S.A. 111, E3177–E3186 10.1073/pnas.1317022111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gorelik L., Constant S., and Flavell R. A. (2002) Mechanism of transforming growth factor β-induced inhibition of T helper type 1 differentiation. J. Exp. Med. 195, 1499–1505 10.1084/jem.20012076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gorelik L., and Flavell R. A. (2002) Transforming growth factor-β in T-cell biology. Nat. Rev. Immunol. 2, 46–53 10.1038/nri704 [DOI] [PubMed] [Google Scholar]
- 40. Lucas P. J., Kim S. J., Melby S. J., and Gress R. E. (2000) Disruption of T cell homeostasis in mice expressing a T cell-specific dominant negative transforming growth factor β II receptor. J. Exp. Med. 191, 1187–1196 10.1084/jem.191.7.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. August A., Sadra A., Dupont B., and Hanafusa H. (1997) Src-induced activation of inducible T cell kinase (ITK) requires phosphatidylinositol 3-kinase activity and the Pleckstrin homology domain of inducible T cell kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 11227–11232 10.1073/pnas.94.21.11227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ching K. A., Grasis J. A., Tailor P., Kawakami Y., Kawakami T., and Tsoukas C. D. (2000) TCR/CD3-Induced activation and binding of Emt/Itk to linker of activated T cell complexes: Requirement for the Src homology 2 domain. J. Immunol. 165, 256–262 10.4049/jimmunol.165.1.256 [DOI] [PubMed] [Google Scholar]
- 43. Yang W. C., Ching K. A., Tsoukas C. D., and Berg L. J. (2001) Tec kinase signaling in T cells is regulated by phosphatidylinositol 3-kinase and the Tec pleckstrin homology domain. J. Immunol. 166, 387–395 10.4049/jimmunol.166.1.387 [DOI] [PubMed] [Google Scholar]
- 44. Gokhale N. A., Zaremba A., Janoshazi A. K., Weaver J. D., and Shears S. B. (2013) PPIP5K1 modulates ligand competition between diphosphoinositol polyphosphates and PtdIns(3,4,5)P3 for polyphosphoinositide-binding domains. Biochem. J. 453, 413–426 10.1042/BJ20121528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hawse W. F., Cattley R. T., and Wendell S. G. (2019) Cutting edge: TCR signal strength regulates acetyl-CoA metabolism via AKT. J. Immunol. 203, 2771–2775 10.4049/jimmunol.1900749 [DOI] [PubMed] [Google Scholar]
- 46. Chemnitz J. M., Driesen J., Classen S., Riley J. L., Debey S., Beyer M., Popov A., Zander T., and Schultze J. L. (2006) Prostaglandin E2 impairs CD4+ T cell activation by inhibition of Lck: Implications in Hodgkin's lymphoma. Cancer Res. 66, 1114–1122 10.1158/0008-5472.CAN-05-3252 [DOI] [PubMed] [Google Scholar]
- 47. Zhou J., Bronowska A., Le Coq J., Lietha D., and Gräter F. (2015) Allosteric regulation of focal adhesion kinase by PIP(2) and ATP. Biophys. J. 108, 698–705 10.1016/j.bpj.2014.11.3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hawse W. F., Boggess W. C., and Morel P. A. (2017) TCR signal strength regulates Akt substrate specificity to induce alternate murine Th and T regulatory cell differentiation programs. J. Immunol. 199, 589–597 10.4049/jimmunol.1700369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luo W., Hawse W., Conter L., Trivedi N., Weisel F., Wikenheiser D., Cattley R. T., and Shlomchik M. J. (2019) The AKT kinase signaling network is rewired by PTEN to control proximal BCR signaling in germinal center B cells. Nat. Immunol. 20, 736–746 10.1038/s41590-019-0376-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Howden A. J. M., Hukelmann J. L., Brenes A., Spinelli L., Sinclair L. V., Lamond A. I., and Cantrell D. A. (2019) Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol. 20, 1542–1554 10.1038/s41590-019-0495-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Piccirillo A. R., Cattley R. T., D'Cruz L. M., and Hawse W. F. (2019) Histone acetyltransferase CBP is critical for conventional effector and memory T-cell differentiation in mice. J. Biol. Chem. 294, 2397–2406 10.1074/jbc.RA118.006977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be shared upon request. Please contact William Hawse, Dept. of Immunology at the University of Pittsburgh, whawse@pitt.edu. The raw mass spectrometric data were deposited to the PRIDE database under the accession number PXD017871.