Abstract
Studies examining the associations between the interleukin‐6 (IL‐6) rs1800795 and rs1800796 gene polymorphisms and risk of coronary artery disease (CAD) remain controversial. Our aim was to evaluate the accurately determine role of these two polymorphisms in CAD risk. PubMed, Embase, VIP, Wan fang and China National Knowledge Infrastructure databases were searched. The odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. The trial sequential analysis (TSA) was conducted, and bioinformatics tools were employed. A total of thirty‐seven articles were obtained. For the IL‐6 rs1800795 polymorphism, 9411 CAD patients and 3161 controls were included, 4720 patients with CAD, and 5000 controls were included for the IL‐6 rs1800796 polymorphism. In the pooled analysis, significant associations were only observed for the rs1800796 polymorphism (allelic: OR [95%CI] = 1.28 [1.13, 1.44], dominant: OR [95%CI] = 1.35 [1.17, 1.57], recessive: OR [95%CI] = 1.35 [1.18, 1.55], heterozygote: OR [95%CI] = 1.26 [1.15, 1.37], homozygote: OR [95%CI] = 1.62 [1.23, 2.13]). Significant associations were detected in the Asian and Mongoloid populations and ‘more than 500’ subgroup for the rs1800795 polymorphism. TSA confirmed the true‐positive results for the rs1800796 polymorphism. The bioinformatics analysis showed that the two polymorphisms played important roles in the gene transcription. The IL‐6 rs1800796 polymorphism is associated with an increased susceptibility to CAD and is a risk factor for CAD. The IL‐6 rs1800795 polymorphism is associated with an increased risk of CAD in Asians, particularly in Chinese, and a decreased risk of CAD in an African population is remarkably observed.
Keywords: coronary artery disease, IL6 rs1800795, IL6 rs1800796, polymorphism
1. INTRODUCTION
Coronary artery disease (CAD) is the leading cause of death both in developed and developing countries. 1 , 2 The aetiology of CAD remains obscure. Environmental and genetic factors, as well as the interactions between them, play a crucial role in the pathophysiology of CAD. 3 , 4 The heritability of CAD was estimated to range from 40%‐60% based on family and twin studies. 5 Furthermore, the greatest genetic influence was observed on early‐onset CAD events, 6 which implies a more vital role for genetic factors in determining CAD risk. Genotyping common single nucleotide polymorphisms (SNPs) within a potential CAD–related gene is an essential and efficient method to detect genetic risk markers, and many significant SNPs associated with CAD risk have been reported, such as matrix metalloproteinase‐9, 7 interleukin‐27 8 and Toll‐like receptor 4. 9
Inflammation plays a key role in the pathophysiology of CAD by promoting the development of atherosclerosis. 10 As a pro‐inflammatory and immune‐regulatory cytokine, IL‐6 plays an important role in the genesis and maintenance of the inflammatory response in atherosclerosis. The IL‐6 gene is located on chromosome 7p21‐24 and comprises 5 introns and 6 exons. 11 Many SNPs in the IL‐6 gene related to CAD risk have been reported, including IL‐6‐174G/C, 12 IL‐6‐572C/G, 13 IL‐6‐597G/A, 14 IL‐6‐634C/G 15 and IL‐6+2954G/C 16 ; however, some of them were not associated with CAD risk (IL6‐597G/A and +2954G/C) or only one study reported the increased risk of CAD (IL‐6‐634C/G). Among them, two common polymorphisms (IL‐6 rs1800795 −174G/C and IL‐6 rs1800796 −572C/G) have been extensively investigated; however, the results were inconclusive. Several previous studies have been conducted in an attempt to draw significant conclusions, but the limitations in sample size and potential false‐positive results caused by systematic errors may bias the results. We therefore performed a study to more accurately determine associations between IL‐6 polymorphisms and CAD risk; in addition, the bioinformatics analysis was conducted to explore the potential molecular mechanism.
2. METHODS
2.1. Identification of the related studies
A comprehensive document retrieval procedure was conducted to identify for all relevant studies published prior to October 2019. PubMed, Embase, VIP, Wan fang and China National Knowledge Infrastructure databases were thoroughly searched by the first three investigators to identify potential studies examining the associations between polymorphisms in the interleukin‐6 (IL‐6) gene and coronary artery disease. The terms ‘coronary artery disease’, ‘coronary heart disease’, ‘myocardial infarction’, ‘CAD’, ‘heart disease’, ‘interleukin‐6’, ‘IL‐6’, ‘polymorphism’, ‘variant’ and ‘polymorphisms’ were used. The citations of review articles and all eligible studies were also browsed for additional potentially relevant study data. In addition, the language of the published studies was restricted to English.
2.2. Inclusion and exclusion criteria
For inclusion in our analysis, studies must have met the following inclusion criteria: (a) evaluation of the relationship between the IL‐6 polymorphisms and coronary artery disease; (b) coronary artery disease was defined as 50% stenosis in the left main coronary artery, or multiple significant (≥70% stenosis) in more than one coronary artery 17 ; (c) a case‐control or cohort design; (d) genotype distribution data were able to be acquired to calculate odds ratios (ORs) and 95% confidence intervals (CIs), particularly detailed data from the control group for testing Hardy‐Weinberg equilibrium. Exclusion criteria were as follows: (a) duplication of previous studies; (b) comments, reviews and editorials; (c) non‐English or non‐Chinese articles; and (d) studies lacking controls. Based on the inclusion and exclusion criteria, the first two authors independently reviewed the references and included the relevant studies. Any disagreement was solved by discussion with the third author (Wang).
2.3. Data extraction
For all included studies, the first two authors independently extracted the following data using a standardized form: first author's last name, year of publication, study country, study region, age and body mass index (BMI), source of the control population, genotyping method, sample size and genotype frequency of polymorphisms in the IL6 gene in patients and controls. Disagreement was settled by rechecking the data or discussion with a third author.
2.4. Quality assessment
The quality of the included studies was independently assessed by all the authors according to a set of criteria that were modified based on the Newcastle‐Ottawa quality assessment scale (Table S1).
2.5. Statistical analysis
Hardy‐Weinberg equilibrium (HWE) was tested in control groups from each study using the chi‐squared test, and P < .05 was considered a significant departure from HWE. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to evaluate the strength of the associations between IL‐6 gene polymorphisms and coronary artery disease risk. Pooled ORs were calculated for the allelic model (IL‐6 rs1800795: C versus G and IL‐6 rs1800796: G versus C), recessive model (IL‐6 rs1800795: CC versus CG + GG and IL‐6 rs1800796: GG versus GC+CC), dominant model (IL‐6 rs1800795: CC+CG versus GG and IL‐6 rs1800796: GG+GC versus CC), heterozygote model (IL‐6 rs1800795: CG versus GG and IL‐6 rs1800796: GC versus CC) and homozygote model (IL‐6 rs1800795: CC versus GG and IL‐6 rs1800796: GG versus CC), respectively. Heterogeneity was evaluated using the Q statistic (significance level of P < .1) and I2 statistic (greater than 50% as evidence of a significant inconsistency). Heterogeneity between studies was evaluated with the I2 test, and a higher I2 values indicated higher levels of heterogeneity (I2 > 90%: extreme heterogeneity; I2 = 70% to 90%: substantial heterogeneity; I2 = 50% to 70%: moderate heterogeneity; I2 < 50%: low heterogeneity). In the heterogeneity evaluation, the fixed‐effects model was used when I2 < 50%, a random‐effects model was used if I2 = 50% to 90%, and the studies were not pooled if I2 > 90%. A sensitivity analysis was performed to detect heterogeneity by sequentially omitting each study. Additionally, analyses were performed in subgroups stratified by accordance with HWE, region, ethnicity, source of controls and sample size. The potential for publication bias was assessed with Begg's funnel plot and Egger's test. We applied the Bonferroni method, 18 which controls for the false discovery rate (FDR), to adjust for multiple comparisons. All tests reported in this study were conducted with the REVMAN 5.3 software and the STATA software (version 12.0; State Corporation).
2.6. Trial sequential analysis
Systematic bias and random errors are inevitable when conducting a meta‐analysis because of the sparse data and repeated significance testing; moreover, trials with low methodological quality, publication bias and a small sample size may generate a false‐positive result. Trial sequential analysis (TSA) is an approach that provides the required amount of information (number of samples) and further reveals potentially false‐positive results in a meta‐analysis. 19 Therefore, TSA was employed to calculate the required amount information for obtaining reliable data of our study. 20 , 21 The TSA was performed by anticipating a 10% relative risk reduction, an overall 5% risk of type I error and a statistical test power of 80%.
2.7. Bioinformatics analysis
Ensembl is a genome browser for vertebrate genomes that supports research in comparative genomics, evolution, sequence variation and transcriptional regulation, and this database provides the genomic context, genes and regulatory elements, flanking sequence, population genetics, phenotype data, sample genotypes, linkage disequilibrium and phylogenetic context of a single nucleotide polymorphism (http://asia.ensembl.org/index.html). SNPinfo is an important bioinformatics analysis tool that predicts SNP function. The SNPinfo database can help researches specify genes or linkage regions and select SNPs based on GWAS results, calculate linkage disequilibrium (LD) and predict functional characteristics of both coding and non‐coding SNPs (https://snpinfo.niehs.nih.gov/). 22 In addition, the RNAfold web server is one of the core programmes of the Vienna RNA package that has been used to predict the minimum free energy of single sequences that influence the stability of the structure. 23 Therefore, we conducted bioinformatics analyses using the aforementioned databases and methods to identify the potential molecular mechanisms for further research.
3. RESULT
3.1. Characteristics of the included studies
The PRISMA flow diagram of our analysis was shown in Table S2. Two hundred and thirty articles were retrieved by searching the international and Chinese databases. After removing duplicates and screening title and abstracts, 54 articles were subjected to the full‐text assessment and 12 articles were excluded due to the lack of detailed genotype distribution data. Finally, 37 articles 12 , 13 , 14 , 16 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 were included in the qualitative and quantitative synthesis.
The characteristics of all included studies regarding the associations between IL6 gene polymorphism and coronary artery disease are presented in Table 1. For the IL‐6 rs1800795 polymorphism, 33 studies involving 9411 CAD patients and 3161 controls were included; 21 studies of 4720 patients with CAD and 5000 controls were included for the IL‐6 rs1800796 polymorphism. Based on the modified Newcastle‐Ottawa Quality Assessment Scale, the score of each included study was greater than 7, which implied a sufficient methodological quality for analysis.
TABLE 1.
Characteristics of included studies
| Study | Year | Country | Region | Age | BMI | Control | Genotyping | Sample | Case | Control | Quality | HWE* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAD | Control | CAD | Control | Source | Method | Size | XX | XY | YY | XX | XY | YY | Score | |||||
| GG | GC | CC | GG | GC | CC | |||||||||||||
| IL6 rs1800795 polymorphism | ||||||||||||||||||
| Nauck 24 | 2002 | Germany | Europe | 63.77 ± 9.89 | 58.30 ± 11.83 | 27.52 ± 4.04 | 27.44 ± 4.34 | HB | PCR‐RFLP | 3304 | 838 | 1238 | 499 | 230 | 355 | 144 | 8 | .739 |
| Georges 25 | 2003 | France | Europe | 62 ± 10 | 61 ± 7 | 26.8 ± 3.6 | 26.6 ± 5.0 | PB | PCR‐SSCP | 495 | 124 | 223 | 82 | 25 | 25 | 16 | 9 | .064 |
| Yang 26 | 2004 | China | Asian | 55 ± 14 | 52 ± 18 | 26.0 ± 3.3 | 23.1 ± 2.8 | HB | PCR‐RFLP | 295 | 110 | 2 | 0 | 179 | 4 | 0 | 8 | .881 |
| Sekuri 28 | 2007 | Turkey | Asian | 46.3 ± 7.8 | 44.3 ± 7.2 | 26.5 ± 2.8 | 24.3 ± 2.6 | PB | PCR‐RFLP | 220 | 61 | 49 | 5 | 57 | 41 | 7 | 8 | .919 |
| Sarecka 30 | 2008 | Poland | Europe | 43.0 ± 5.5 | 42.3 ± 6.5 | 26.7 ± 4.4 | 25.4 ± 3.5 | PB | PCR‐RFLP | 263 | 35 | 74 | 33 | 36 | 64 | 21 | 8 | .413 |
| Banerjee 31 | 2009 | India | Asian | 56.3 ± 12.1 | 56.0 ± 9.5 | NA | NA | HB | PCR‐RFLP | 442 | 159 | 43 | 8 | 171 | 57 | 4 | 9 | .763 |
| Rios1 47 | 2010 | Brazil | South America | 55.7 ± 7.9 | 51.8 ± 8.4 | NA | NA | HB | PCR‐TaqMan | 253 | 96 | 36 | 6 | 69 | 43 | 3 | 8 | .217 |
| Rios2 47 | 2010 | Brazil | South America | 55.7 ± 6.7 | 53.0 ± 7.7 | NA | NA | HB | PCR‐TaqMan | 414 | 158 | 90 | 28 | 82 | 46 | 10 | 9 | .323 |
| Coker 48 | 2011 | Turkey | Asian | 53.4 ± 9.5 | 53.9 ± 9.3 | 28.4 ± 3.7 | 28.1 ± 3.6 | PB | PCR‐RFLP | 402 | 102 | 56 | 9 | 141 | 81 | 13 | 9 | .761 |
| Ghazouani 33 | 2011 | Tunisia | Europe | 58.1 ± 12.0 | 56.7 ± 14.12 | 27.08 ± 4.20 | 25.22 ± 2.35 | HB | PCR‐RFLP | 824 | 298 | 110 | 10 | 297 | 102 | 7 | 9 | .602 |
| Vakili 49 | 2011 | Iran | Asian | NA | NA | NA | NA | PB | PCR‐TaqMan | 900 | 153 | 234 | 63 | 202 | 229 | 19 | 9 | .000 |
| Fan 56 | 2011 | China | Asian | 52.1 ± 6.8 | 52.3 ± 8.8 | NA | NA | HB | PCR‐RFLP | 214 | 84 | 0 | 0 | 129 | 1 | 0 | 8 | .965 |
| Liu 34 | 2011 | China | Asian | 60.6 ± 12.7 | 61.3 ± 13.7 | NA | NA | HB | PCR‐RFLP | 276 | 123 | 3 | 0 | 148 | 2 | 0 | 8 | .934 |
| Bhanushali 35 | 2013 | India | Asian | 48 ± 11 | 50 ± 11 | NA | NA | HB | PCR‐SNaPshot a | 250 | 77 | 20 | 3 | 121 | 25 | 4 | 8 | .068 |
| Phulukdaree1 36 | 2013 | South Africa | Africa | NA | NA | NA | NA | HB | PCR‐RFLP | 102 | 29 | 11 | 1 | 34 | 19 | 8 | 8 | .062 |
| Phulukdaree2 36 | 2013 | South Africa | Africa | NA | NA | NA | NA | HB | PCR‐RFLP | 120 | 38 | 16 | 5 | 34 | 19 | 8 | 8 | .062 |
| Satti 38 | 2013 | Pakistan | Asian | 46.4 ± 18.7 | 35.2 ± 17.4 | 25.9 ± 3.5 | 25.2 ± 3.5 | PB | PCR‐RFLP | 88 | 18 | 11 | 7 | 38 | 14 | 0 | 7 | .262 |
| Tong 50 | 2013 | China | Asian | 61.4 ± 8.7 | 60.6 ± 9.6 | 23.2 ± 3.1 | 22.7 ± 2.8 | HB | PCR‐TaqMan | 667 | 201 | 87 | 38 | 220 | 98 | 23 | 9 | .011 |
| Zhang 37 | 2013 | China | Asian | NA | NA | NA | NA | HB | PCR‐HRM | 506 | 221 | 10 | 0 | 264 | 11 | 0 | 9 | .735 |
| Elsaid 51 | 2014 | Egypt | Africa | 53.54 ± 9.1 | 45.3 ± 7.2 | NA | NA | PB | PCR‐TaqMan | 208 | 26 | 55 | 23 | 0 | 49 | 55 | 8 | .000 |
| Galimudi 39 | 2014 | India | Asian | 65 ± 5 | 64 ± 6 | NA | NA | PB | PCR‐RFLP | 400 | 72 | 102 | 26 | 113 | 69 | 18 | 9 | .123 |
| Hatzis1 40 | 2014 | Greece | Europe | NA | NA | NA | NA | HB | PCR‐RFLP | 361 | 109 | 76 | 12 | 64 | 72 | 28 | 9 | .733 |
| Hatzis2 40 | 2014 | Greece | Europe | NA | NA | NA | NA | HB | PCR‐RFLP | 285 | 36 | 71 | 43 | 67 | 57 | 11 | 8 | .817 |
| Sun 14 | 2014 | China | Asian | 61.2 ± 8.5 | 56.4 ± 11.6 | NA | NA | HB | PCR‐TaqMan | 623 | 191 | 61 | 44 | 236 | 63 | 28 | 9 | .000 |
| Celik 16 | 2015 | Turkey | Asian | 14.56 ± 1.73 | 13.91 ± 1.31 | 20.29 ± 3.59 | 19.78 ± 3.25 | HB | PCR‐RFLP | 82 | 24 | 12 | 0 | 29 | 16 | 1 | 7 | .476 |
| Li 41 | 2015 | China | Asian | NA | NA | NA | NA | HB | PCR‐RFLP | 730 | 213 | 113 | 39 | 245 | 105 | 15 | 9 | .382 |
| Wang 42 | 2015 | China | Asian | 65.4 ± 8.4 | 64.9 ± 8.2 | 22.8 ± 2.9 | 22.6 ± 2.6 | HB | PCR‐RFLP | 804 | 153 | 171 | 78 | 176 | 187 | 39 | 9 | .292 |
| Yang 43 | 2015 | China | Asian | NA | NA | NA | NA | HB | PCR‐RFLP | 820 | 198 | 163 | 49 | 239 | 146 | 25 | 9 | .669 |
| Hongmei 12 | 2016 | China | Asian | 62.64 ± 8.43 | 61.43 ± 7.85 | 26.41 ± 2.56 | 25.75 ± 2.54 | HB | PCR‐RFLP | 571 | 256 | 19 | 0 | 282 | 14 | 0 | 8 | .679 |
| Mao 52 | 2016 | China | Asian | 62.65 ± 9.72 | 56.82 ± 9.80 | 24.61 ± 4.16 | 21.57 ± 3.64 | HB | PCR‐RFLP | 584 | 142 | 45 | 37 | 267 | 63 | 30 | 7 | .000 |
| Jabir 53 | 2017 | Saudi Arabia | Asian | 60.6 ± 8.85 | 47.7 ± 5.06 | 28.69 ± 4.34 | 30.89 ± 2.90 | HB | PCR‐TaqMan | 179 | 62 | 25 | 3 | 63 | 23 | 3 | 8 | .620 |
| Mitrokhin 54 | 2017 | Russian | Europe | 70.37 ± 13.45 | 74.94 ± 7.43 | 30.71 ± 2.75 | 30.33 ± 6.09 | HB | PCR‐TaqMan | 314 | 62 | 100 | 36 | 32 | 58 | 26 | 9 | .977 |
| Chen 55 | 2018 | China | Asian | 61.00 ± 10.49 | 60.37 ± 10.38 | 25.13 ± 8.12 | 23.47 ± 8.72 | HB | Multiplex PCR | 779 | 155 | 218 | 56 | 190 | 133 | 27 | 9 | .581 |
| IL6 rs1800796 polymorphism | CC | CG | GG | CC | CG | GG | ||||||||||||
| Fu 45 | 2006 | China | Asian | 61.8 ± 12.4 | 59.89 ± 14.35 | NA | NA | HB | PCR‐RFLP | 505 | 128 | 101 | 16 | 166 | 90 | 4 | 7 | .034 |
| Wei 27 | 2006 | China | Asian | 61 ± 11 | 60 ± 10 | NA | NA | HB | PCR‐RFLP | 335 | 89 | 67 | 9 | 113 | 55 | 2 | 8 | .095 |
| Gao 29 | 2008 | China | Asian | 65.2 ± 9.8 | 62.5 ± 11.8 | NA | NA | HB | PCR‐RFLP | 234 | 65 | 51 | 10 | 72 | 32 | 4 | 8 | .850 |
| Jia 46 | 2010 | China | Asian | NA | NA | NA | NA | HB | PCR | 441 | 79 | 130 | 22 | 88 | 107 | 15 | 7 | .021 |
| Liang 32 | 2010 | China | Asian | 57.6 ± 7.4 | 56.4 ± 8.2 | 26.4 ± 3.1 | 24.2 ± 2.6 | HB | PCR‐RFLP | 851 | 259 | 161 | 14 | 283 | 126 | 8 | 8 | .156 |
| Fan 56 | 2011 | China | Asian | 52.1 ± 6.8 | 52.3 ± 8.8 | NA | NA | HB | PCR‐RFLP | 214 | 42 | 38 | 4 | 95 | 32 | 3 | 8 | .875 |
| Liu 34 | 2011 | China | Asian | 60.6 ± 12.7 | 61.3 ± 13.7 | NA | NA | HB | PCR‐RFLP | 276 | 63 | 52 | 11 | 92 | 55 | 3 | 9 | .107 |
| Coker 48 | 2011 | Turkey | Asian | 53.4 ± 9.5 | 53.9 ± 9.3 | 28.4 ± 3.7 | 28.1 ± 3.6 | PB | PCR‐RFLP | 402 | 126 | 30 | 11 | 169 | 45 | 21 | 7 | .000 |
| Zhang 37 | 2013 | China | Asian | NA | NA | NA | NA | HB | PCR‐HRM | 506 | 86 | 106 | 39 | 128 | 117 | 30 | 9 | .675 |
| Tong 50 | 2013 | China | Asian | 61.4 ± 8.8 | 60.6 ± 9.7 | 23.2 ± 3.2 | 22.7 ± 2.9 | HB | PCR‐TaqMan | 667 | 179 | 110 | 37 | 180 | 120 | 41 | 7 | .004 |
| Sun 14 | 2014 | China | Asian | 61.2 ± 8.5 | 56.4 ± 11.6 | NA | NA | HB | PCR‐TaqMan | 623 | 190 | 69 | 37 | 215 | 73 | 39 | 7 | .000 |
| Wang 42 | 2015 | China | Asian | 65.4 ± 8.4 | 64.9 ± 8.2 | 22.8 ± 2.9 | 22.6 ± 2.6 | HB | PCR‐RFLP | 804 | 176 | 187 | 39 | 192 | 181 | 29 | 9 | .119 |
| Li 41 | 2015 | China | Asian | NA | NA | NA | NA | HB | PCR‐RFLP | 729 | 132 | 165 | 68 | 166 | 155 | 43 | 9 | .462 |
| Fragoso 13 | 2015 | Mexico | South America | NA | NA | NA | NA | HB | PCR‐TaqMan | 244 | 7 | 39 | 32 | 11 | 77 | 78 | 8 | .163 |
| Celik 16 | 2015 | Turkey | Asian | 14.56 ± 1.73 | 13.91 ± 1.31 | 20.29 ± 3.59 | 19.78 ± 3.25 | HB | PCR‐RFLP | 82 | 25 | 10 | 1 | 42 | 3 | 1 | 7 | .013 |
| Mao 52 | 2016 | China | Asian | 62.65 ± 9.72 | 56.82 ± 9.80 | 24.61 ± 4.16 | 21.57 ± 3.64 | HB | PCR‐RFLP | 584 | 97 | 110 | 17 | 147 | 176 | 37 | 8 | .137 |
| Hongmei 12 | 2016 | China | Asian | 62.64 ± 8.43 | 61.43 ± 7.85 | 26.41 ± 2.56 | 25.75 ± 2.54 | HB | PCR‐RFLP | 572 | 87 | 134 | 55 | 135 | 129 | 32 | 8 | .886 |
| Chen 44 | 2016 | China | Asian | 63.22 ± 9.40 | 53.81 ± 8.45 | NA | NA | HB | PCR‐RFLP | 399 | 72 | 98 | 27 | 108 | 83 | 11 | 8 | .333 |
| Jabir 53 | 2017 | Saudi Arabia | Asian | 60.6 ± 8.85 | 47.7 ± 5.06 | 28.69 ± 4.34 | 30.89 ± 2.90 | HB | PCR‐TaqMan | 159 | 3 | 22 | 59 | 0 | 21 | 54 | 8 | .159 |
| Mitrokhin 54 | 2017 | Russian | Europe | 70.37 ± 13.45 | 74.94 ± 7.43 | 30.71 ± 2.75 | 30.33 ± 6.09 | HB | PCR‐TaqMan | 314 | 0 | 16 | 182 | 2 | 10 | 104 | 7 | .010 |
| Chen 55 | 2018 | China | Asian | 61.00 ± 10.49 | 60.37 ± 10.38 | 25.13 ± 8.12 | 23.47 ± 8.72 | HB | Multiplex PCR | 779 | 228 | 158 | 43 | 176 | 141 | 33 | 9 | .539 |
For rs1800795 polymorphism, XX, XY and YY represent GG, GC and CC, respectively; for rs1800796 polymorphism, XX, XY and YY represent CC, CG and GG, respectively.
Abbreviations: BMI, body mass index; CAD, coronary artery disease; HB, hospital based; NA, not available; PB, population based; PCR, polymorphism chain reaction‐restriction; PCR‐HRM, polymorphism chain reaction high‐resolution melting; PCR‐RFLP, polymorphism chain reaction‐restriction fragment length polymorphism; PCR‐SSCP, polymorphism chain reaction single‐strand conformation polymorphism; PCR‐TaqMan, polymorphism chain reaction‐restriction TaqMan polymorphism.
The polymorphism was determined by a variation of the allele termination assay reported by Bhanushali et al 35
P value for Hardy‐Weinberg equilibrium test in controls.
3.2. The pooled analysis of IL‐6 polymorphisms and CAD risk
The main results of our analysis and the heterogeneity test of the associations between IL‐6 gene polymorphisms and coronary artery disease risk are shown in Table 2. In the pooled analysis, no significant association was observed for the IL‐6 rs1800795 polymorphism; significant associations with heterogeneity were detected in all five genetic models for the IL‐6 rs1800796 polymorphism: allelic genetic model (OR [95% CI] = 1.28 [1.13, 1.44], P = 2*10−4), dominant genetic model (OR [95% CI] = 1.35 [1.17, 1.57], P = 2*10−4), heterogeneity genetic model (OR [95% CI] = 1.26 [1.15, 1.37], P = 2*10−4), recessive genetic model (OR [95% CI]=1.35 [1.18, 1.55], P = 2*10−4 (Figure 1)) and homozygote genetic model (OR [95% CI] = 1.62 [1.23, 2.13], P = .001).
TABLE 2.
Pooled and Subgroup analysis of the associations between IL‐6 polymorphisms and CAD risk
| Subgroup analysis | No. of the studies | Allelic genetic model | Dominant genetic model | Recessive genetic model | Heterozygote genetic model | Homozygote genetic model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR [95%CI] | P 1/Bon/FDR | P 2/I2/EM | OR [95%CI] | P 1/Bon/FDR | P 2/I2/EM | OR [95%CI] | P 1/Bon/FDR | P 2/I2/EM | OR [95%CI] | P 1/Bon/FDR | P 2/I2/EM | OR [95%CI] | P 1/Bon/FDR | P 2/I2/EM | ||
| IL6 rs1800795 polymorphism | ||||||||||||||||
| Pooled results | 33 | 1.40 [1.12, 1.75] | .003/.015/.015 | 2*10−4/93%/R | 1.21 [1.05, 1.40] | .01/.050/.0167 | 2*10−4/69%/R | 1.34 [1.02, 1.76] | .04/.200/.040 | 2*10−4/78%/R | 1.15 [1.01, 1.30] | .03/.150/.038 | 2*10−4/54%/R | 1.48 [1.10, 2.00] | .01/.050/.017 | 2*10−4/78%/R |
| Subgroup Results | ||||||||||||||||
| HWE | ||||||||||||||||
| In accordance with HWE | 28 | 1.31 [1.08, 1.59] | .007/.035/.035 | 2*10−4/88%/R | 1.18 [1.00, 1.39] | .04/.200/.060 | 2*10−4/69%/R | 1.30 [0.99, 1.72] | .06/.300/.060 | 2*10−4/69%/R | 1.15 [0.99, 1.32] | .06/.300/.060 | 2*10−4/55%/R | 1.40 [1.01, 1.94] | .04/.200/.060 | 2*10−4/75%/R |
| Departure from HWE | 5 | 1.97 [1.01, 3.85] | .05/.25/.20 | 2*10−4/97%/R | 1.34 [0.96, 1.88] | .09/.45/.20 | .005/73%/R | 1.49 [0.65, 3.40] | .35/1.00/.41 | 2*10−4/92%/R | 1.14 [0.83, 1.58] | .41/1.00/.41 | .03/63%/R | 1.79 [0.86, 3.74] | .12/.60/.20 | 2*10−4/84%/R |
| Region | ||||||||||||||||
| Asian | 21 | 1.84 [1.42, 2.39] | 2*10−4/2*10−4/2*10−4 | 2*10−4/91%/R | 1.40 [1.24, 1.58] | 2*10−4/2*10−4/2*10−4 | .05/36%/R | 1.99 [1.63, 2.42] | 2*10−4/2*10−4/2*10−4 | .20/22%/R | 1.26 [1.11, 1.43] | 2*10−4/.002/2*10−4 | .10/30%/R | 2.21 [1.78, 2.73] | 2*10−4/2*10−4/2*10−4 | .14/28%/R |
| Europe | 7 | 1.20 [0.87, 1.65] | .27/1/.800 | 2*10−4/90%/R | 1.13 [0.80, 1.59] | .48/1/.800 | 2*10−4/82%/R | 1.05 [0.65, 1.69] | .84/1/.840 | 2*10−4/81%/R | 1.11 [0.85, 1.46] | .43/1/.800 | .005/67%/R | 1.15 [0.62, 2.11] | .66/1/.825 | 2*10−4/86%/R |
| Africa | 3 | 0.45 [0.26, 0.77] | .004/.02/.01 | .06/64%/R | 0.33 [0.08, 1.33] | .12/.60/.15 | .009/79%/R | 0.29 [0.17, 0.50] | 2*10−4/.00/.00 | .37/1%/R | 0.41 [0.11, 1.58] | .20/1.00/.20 | .02/74%/R | 0.11 [0.01, 1.37] | .09/.45/.15 | 0.009/79%/R |
| South America | 2 | 0.97 [0.67, 1.41] | .88/1/.880 | .17/46%/R | 0.87 [0.53, 1.43] | .58/1/.725 | .13/56%/R | 1.50 [0.77, 2.91] | .23/1/.650 | .84/0%/R | 0.80 [0.48, 1.33] | .39/1/.650 | .14/53%/R | 1.45 [0.74, 2.85] | .28/1/.650 | .99/0%/R |
| Ethnicity | ||||||||||||||||
| Caucasian | 17 | 1.44 [1.10, 1.90] | .009/.045/.045 | 2*10−4/93%/R | 1.20 [0.98, 1.47] | .07/.350/.150 | 2*10−4/73%/R | 1.27 [0.89, 1.81] | .18/.900/.180 | 2*10−4/74%/R | 1.15 [0.98, 1.37] | .09/.450/.150 | .003/56%/R | 1.40 [0.92, 2.14] | .12/.600/.150 | 2*10−4/79%/R |
| Mongoloid | 12 | 1.97 [1.44, 2.70] | 2*10−4/2*10−4/2*10−4 | 2*10−4/90%/R | 1.47 [1.31, 1.65] | 2*10−4/2*10−4/2*10−4 | .40/4%/R | 2.07 [1.72, 2.50] | 2*10−4/2*10−4/2*10−4 | .94/0%/R | 1.28 [1.11, 1.49] | .001/.004/2*10−4 | .24/21%/R | 2.28 [1.87, 2.77] | 2*10−4/2*10−4/2*10−4 | .92/0%/R |
| African | 4 | 0.52 [0.32, 0.86] | .01/.05/.050 | .01/72%/R | 0.49 [0.23, 1.05] | .07/.35/.125 | .03/66%/R | 0.46 [0.18, 1.17] | .10/.50/.125 | .06/59%/R | 0.55 [0.28, 1.11] | .10/.50/.125 | .08/56%/R | 0.24 [0.03, 1.61] | .14/.70/.140 | .003/79%/R |
| Source of Controls | ||||||||||||||||
| Hospital based | 25 | 1.48 [1.14, 1.94] | .004/.020/.020 | 2*10−4/94%/R | 1.16 [0.99, 1.35] | .07/.350/.088 | 2*10−4/68%/R | 1.46 [1.10, 1.94] | .009/.045/.0225 | 2*10−4 /72%/R | 1.10 [0.96, 1.25] | .18/.900/.180 | .004/48%/R | 1.50 [1.08, 2.09] | .02/.100/.033 | 2*10−4/78%/R |
| Population based | 8 | 1.18 [0.78, 1.80] | .43/1.0/.5375 | 2*10−4/91%/R | 1.39 [0.98, 1.96] | .06/.3/.200 | .001/72%/R | 1.17 [0.57, 2.40] | .68/1.0/.6800 | 2*10−4/87%/R | 1.34 [0.97, 1.84] | .08/.4/.200 | .007/64%/R | 1.37 [0.64, 2.92] | .41/1.0/.538 | 2*10−4/81%/R |
| Sample size | ||||||||||||||||
| Less than 300 | 14 | 1.21 [0.68, 2.18] | .52/1.0/.97 | 2*10−4/93%/R | 1.05 [0.72, 1.54] | .80/1.0/.97 | 2*10−4/68%/R | 1.02 [0.47, 2.21] | .97/1.0/.97 | 2*10−4/79%/R | 1.04 [0.77, 1.41] | .80/1.0/.97 | .03/46%/R | 1.04 [0.77, 1.41] | .94/1.0/.97 | 2*10−4/78%/R |
| Between 300 and 500 | 7 | 1.11 [0.80, 1.53] | .54/1.0/.99 | 2*10−4/85%/R | 1.05 [0.73, 1.51] | .80/1.0/.99 | 2*10−4/79%/R | 0.92 [0.59, 1.43] | .71/1.0/.99 | .02/61%/R | 1.08 [0.76, 1.54] | .66/1.0/.99 | .001/75%/R | 1.08 [0.76, 1.54] | .99/1.0/.99 | .001/73%/R |
| More than 500 | 12 | 1.81 [1.33, 2.45] | 2*10−4/2*10−4/2*10−4 | 2*10−4/95%/R | 1.37 [1.17, 1.59] | 2*10−4/2*10−4/2*10−4 | .001/64%/R | 1.95 [1.42, 2.67] | 2*10−4/2*10−4/2*10−4 | 2*10−4/78%/R | 1.22 [1.06, 1.40] | .004/2*10−4/.004 | .03/48%/R | 1.22 [1.06, 1.40] | 2*10−4/2*10−4/2*10−4 | 2*10−4/80%/R |
| IL6 rs1800796 polymorphism | ||||||||||||||||
| Pooled results | 21 | 1.28 [1.13, 1.44] | 2*10−4/.001/2*10−4 | 2*10−4/68%/R | 1.35 [1.17, 1.57] | 2*10−4/.001/2*10−4 | 2*10−4/62%/R | 1.35 [1.18, 1.55] | 2*10−4/.001/2*10−4 | .01/46%/F | 1.26 [1.15, 1.37] | 2*10−4/2*10−4/2*10−4 | .001/47%/F | 1.62 [1.23, 2.13] | .001/.003/.001 | 2*10−4/60%/R |
| Subgroup results | ||||||||||||||||
| HWE | ||||||||||||||||
| In accordance with HWE | 14 | 1.32 [1.15, 1.53] | 2*10−4/2*10−4/2*10−4 | 2*10−4/70%/R | 1.42 [1.19, 1.69] | 2*10−4/2*10−4/2*10−4 | .001/62%/R | 1.48 [1.15, 1.91] | .002/2*10−4/.0020 | .02/50%/R | 1.33 [1.14, 1.55] | 2*10−4/2*10−4/.001 | .02/49%/R | 1.78 [1.28, 2.47] | .001/2*10−4/.001 | .002/60%/R |
| Departure from HWE | 7 | 1.18 [0.94, 1.47] | .15/.75/.283 | .01/63%/R | 1.22 [0.92, 1.62] | .16/.80/.283 | .02/60%/R | 1.16 [0.84, 1.61] | .37/1.00/.370 | .21/29%/R | 1.20 [0.92, 1.56] | .17/.85/.283 | .07/48%/R | 1.31 [0.82, 2.07] | .26/1.00/.325 | .06/51%/R |
| Region | ||||||||||||||||
| Asian | 19 | 1.30 [1.15, 1.47] | 2*10−4/.001/2*10−4 | 2*10−4/69%/R | 1.36 [1.17, 1.58] | 2*10−4/.001/2*10−4 | 2*10−4/63%/R | 1.44 [1.16, 1.78] | .001/.005/.001 | .02/45%/R | 1.29 [1.13, 1.48] | 2*10−4/.001/2*10−4 | .008/50%/R | 1.66 [1.26, 2.19] | 2*10−4/.002/2*10−4 | 2*10−4/61%/R |
| South America a | 1 | 0.83 [0.55, 1.24] | .36/1.0/.6375 | NA | 0.72 [0.27, 1.93] | .51/1.0/.638 | NA | 0.78 [0.46, 1.35] | .38/1.0/.638 | NA | 0.80 [0.29, 2.21] | .66/1.0/.660 | NA | 0.64 [0.23, 1.81] | .4/1.0/.638 | NA |
| Europe a | 1 | 1.53 [0.73, 3.19] | .26/1.0/.325 | NA | 8.67 [0.41, 182.13] | .16/.8/.325 | NA | 1.31 [0.60, 2.88] | .5/1.0/0.5 | NA | 7.86 [0.34, 180.34] | .2/1.0/.325 | NA | 8.73 [0.42, 183.62] | .16/.8/.325 | NA |
| Ethnicity | ||||||||||||||||
| Caucasian | 16 | 1.33 [1.17, 1.50] | 2*10−4/2*10−4/2*10−4 | 2*10−4/68%/R | 1.38 [1.19, 1.59] | 2*10−4/2*10−4/2*10−4 | .001/62%/R | 1.54 [1.23, 1.93] | 2*10−4/2*10−4/2*10−4 | .02/47%/R | 1.30 [1.14, 1.47] | 2*10−4/2*10−4/.0001 | .02/46%/R | 1.78 [1.34, 2.36] | 2*10−4/2*10−4/2*10−4 | 2*10−4/62%/R |
| Mongoloid | 5 | 1.02 [0.71, 1.48] | .90/1.0/.90 | .08/52%/R | 1.23 [0.50, 3.04] | .65/1.0/.813 | .04/61%/R | 0.88 [0.63, 1.23] | .47/1.0/.813 | .82/0%/R | 1.31 [0.52, 3.30] | .57/1.0/.813 | .05/58%/R | 0.73 [0.41, 1.31] | .30/1.0/.813 | .41/0%/R |
| Source of controls | ||||||||||||||||
| Hospital based | 20 | 1.30 [1.16, 1.47] | 2*10−4/2*10−4/2*10−4 | 2*10−4/67%/R | 1.39 [1.19, 1.61] | 2*10−4/2*10−4/2*10−4 | 2*10−4/61%/R | 1.42 [1.16, 1.74] | .001/2*10−4/.001 | .02/44%/R | 1.31 [1.14, 1.50] | 2*10−4/2*10−4/.002 | .01/47%/R | 1.70 [1.29, 2.24] | 2*10−4/2*10−4/2*10−4 | .001/58%/R |
| Population based a | 1 | 0.81 [0.56, 1.18] | .28/1.0/.5375 | NA | 0.83 [0.53, 1.31] | .43/1.0/.538 | NA | 0.72 [0.34, 1.53] | .39/1.0/.538 | NA | 0.89 [0.53, 1.50] | .67/1.0/.670 | NA | 0.70 [0.33, 1.51] | .37/1.0/.538 | NA |
| Sample size | ||||||||||||||||
| Less than 300 | 6 | 1.46 [0.99, 2.14] | .05/.25/.083 | .005/71%/R | 1.46 [0.99, 2.14] | .01/.05/.050 | .07/52%/R | 1.37 [0.78, 2.40] | .28/1.00/.280 | .12/42%/R | 1.71 [1.06, 2.77] | .03/ .15/.075 | .06/52%/R | 1.75 [0.71, 4.31] | .22/1.00/.275 | .08/50%/R |
| Between 300 and 500 | 5 | 1.35 [1.01, 1.80] | .04/.20/.067 | .02/66%/R | 1.35 [1.01, 1.80] | .04/.20/.067 | .04/60%/R | 1.56 [0.90, 2.69] | .11/.55/.110 | .07/54%/R | 1.40 [1.07, 1.83] | .01/.05/.050 | .24/27%/R | 2.17 [0.95, 4.93] | .06/.30/.075 | .01/68%/R |
| More than 500 | 10 | 1.21 [1.05, 1.39] | .008/.04/.010 | .001/70%/R | 1.21 [1.05, 1.39] | .01/.05/.013 | .006/61%/R | 1.36 [1.07, 1.74] | .01/.05/.013 | .03/50%/R | 1.18 [1.03, 1.35] | .02/.10/.020 | .10/38%/R | 1.48 [1.09, 2.01] | .01/.05/.013 | .002/65%/R |
Results with P < .05 even after the Bonferroni adjusted and a tolerable heterogeneity (I2 < 85%) were regarded as significant.
Abbreviations: BMI, body mass index; CI, confidence interval; EM, effect model; F, fixed effect model; OR, odds ratio; P1, P value for meta‐analysis; P2, P value for heterogeneity test; R, random effect model.
Only one study was included in the subgroup, and heterogeneity was not applicable.
Results with P < .05 even after the Bonferroni adjusted and a tolerable heterogeneity (I2 < 85%)
FIGURE 1.
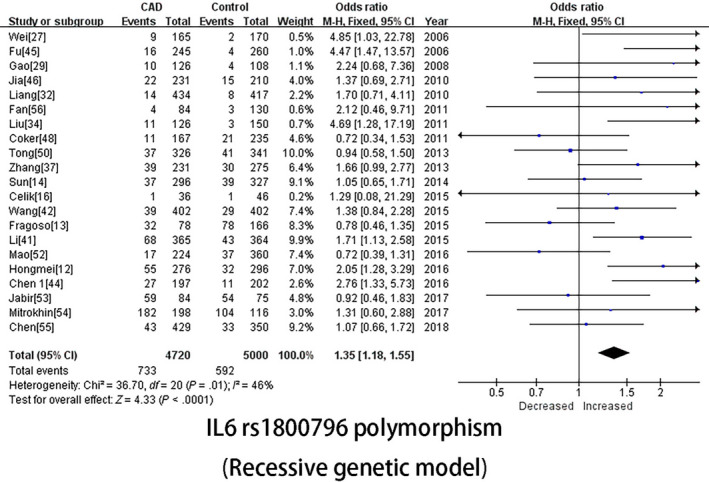
IL‐6 rs1800795 polymorphism (Recessive genetic model)
3.3. Subgroup analyses of the associations between IL‐6 polymorphisms and CAD risk
Subgroup analyses were introduced to identify the source of heterogeneity and further reveal additional information about the associations between IL‐6 polymorphisms and CAD risk. Table 2 summarizes the results of the subgroup analyses based on HWE, region, ethnicity, the source of controls and sample size.
For the subgroup in accordance with HWE, significant associations were only detected for the IL‐6 rs1800796 polymorphism, and all five genetic models indicated strong associations with an increased OR compared with the pooled OR: allelic (OR [95% CI] = 1.32 [1.15, 1.53], P = 2*10−4), dominant (OR [95% CI] = 1.42 [1.19, 1.69], P = 2*10−4), recessive (OR [95% CI] = 1.48 [1.15, 1.91], P = 2*10−4), heterozygote (OR [95% CI] = 1.33 [1.14, 1.55], P = 2*10−4) and homozygote (OR [95% CI] = 1.78 [1.28, 2.47], P = .001) genetic models.
In the analysis of the rs1800795 polymorphism stratified by region, significant associations with reduced heterogeneity in the Asian population were observed in the dominant (OR [95% CI] = 1.36 [1.17, 1.58], P = 2*10−4), recessive (OR [95% CI] = 1.44 [1.16, 1.78], P = .001) (Figure 2A), heterozygote (OR [95% CI] = 1.29 [1.13, 1.48], P = 2*10−4) and homozygote (OR [95% CI] = 1.66 [1.26, 2.19], P = 2*10−4) genetic models. In addition, decreased risks for the African population were remarkably identified in the allelic (OR [95% CI] = 0.45 [0.26, 0.77], P = .004) and recessive (OR [95% CI] = 0.29 [0.17, 0.50], P = 2*10−4) genetic models. Regarding the rs1800796 polymorphism, all five genetic models suggested a strong relationship with CAD risk in Asian volunteers (allelic: OR [95% CI] = 1.30 [1.15, 1.47], P = 2*10−4; dominant: OR [95% CI] = 1.36 [1.17, 1.58], P = 2*10−4; recessive: OR [95% CI] = 1.44 [1.16, 1.78], P = .001 (Figure 2B); heterozygote: OR [95% CI] = 1.29 [1.13, 1.48], P = 2*10−4; and homozygote: OR [95% CI] = 1.66 [1.26, 2.19], P = 2*10−4). Along with region as a geographic factor, ethnicity is also an important factor. In the Caucasian population, no association was observed for the rs1800795 polymorphism, but significant associations were widely observed with the rs1800796 polymorphism in all five genetic models (allelic: OR [95% CI] = 1.33 [1.17, 1.50], P = 2*10−4; dominant: OR [95% CI] = 1.38 [1.19, 1.59], P = 2*10−4; recessive: OR [95% CI] = 1.54 [1.23, 1.93], P = .001; heterozygote: OR [95% CI] = 1.30 [1.14, 1.47], P = 2*10−4; homozygote: OR [95% CI] = 1.78 [1.34, 2.36], P = 2*10−4). Regarding the Mongoloid population, significant associations were observed in the dominant (OR [95% CI] = 1.47 [1.31, 1.65], P = 2*10−4), recessive (OR [95% CI] = 2.07 [1.72, 2.50], P = 2*10−4), heterozygote (OR [95% CI] = 1.28 [1.11, 1.49], P = .001) and homozygote (OR [95% CI] = 2.28 [1.87, 2.77], P = 2*10−4) genetic models that implied a strong association between CAD risk and the IL6 rs1800795 polymorphism.
FIGURE 2.
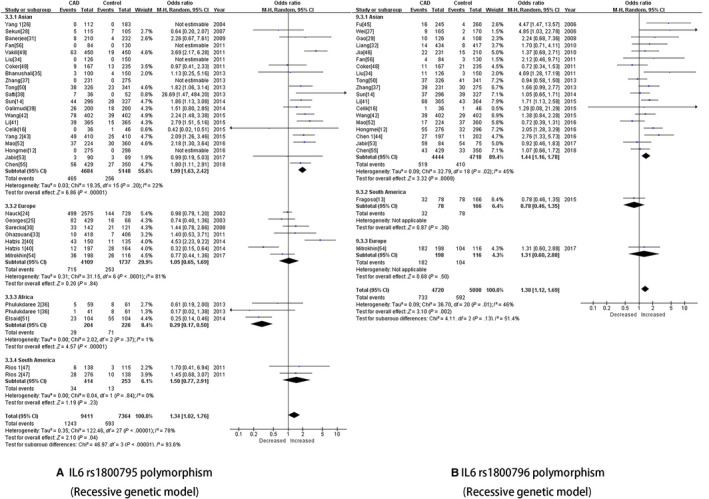
A, IL‐6 rs1800795 polymorphism (Recessive genetic model). B, IL‐6 rs1800796 polymorphism (Recessive genetic model)
In the subgroup analysis stratified by source of controls, significant associations were observed between CAD risk in the hospital‐based population and the rs1800795 polymorphism (recessive: OR [95% CI] = 1.46 [1.10, 1.94], P = .009) and the rs1800796 polymorphism (allelic: OR [95% CI] = 1.30 [1.16, 1.47], P = 2*10−4; dominant: OR [95% CI] = 1.39 [1.19, 1.61], P = 2*10−4; recessive: OR [95% CI] = 1.42 [1.16, 1.74], P = .001; heterozygote: OR [95% CI] = 1.31 [1.14, 1.50], P = 2*10−4; and homozygote: OR [95% CI] = 1.70 [1.29, 2.24], P = 2*10−4).
We stratified studies into three subgroups by sample size based on the modified quality scale score (less than 300, between 300 and 500, and greater than 500) to evaluate the effect of sample size on the associations between the two polymorphisms and CAD risk. In the greater than 500 subgroup, significant associations were observed between the rs1800795 polymorphism and CAD risk (dominant: OR [95% CI] = 1.37 [1.17, 1.59], P = 2*10−4; recessive: OR [95% CI] = 1.95 [1.42, 2.67], P = 2*10−4; heterozygote: OR [95% CI] = 1.22 [1.06, 1.40], P = .004; and homozygote: OR [95% CI] = 1.22 [1.06, 1.40], P = 2*10−4); for the rs1800796 polymorphism, a significant association was only observed in the allelic genetic model (OR [95% CI] = 1.21 [1.05, 1.39], P = .008). We discovered that a larger sample produced more significant associations with the two polymorphisms.
3.4. The sensitivity analysis of IL‐6 polymorphisms and CAD risk
A sensitivity analysis was conducted by sequentially omitting each individual study to detect the effect of each study on the results of the overall meta‐analysis. None of the studies changed the corresponding pooled ORs; thus, the results of our meta‐analysis were stable and reliable (Figure 3A‐B).
FIGURE 3.
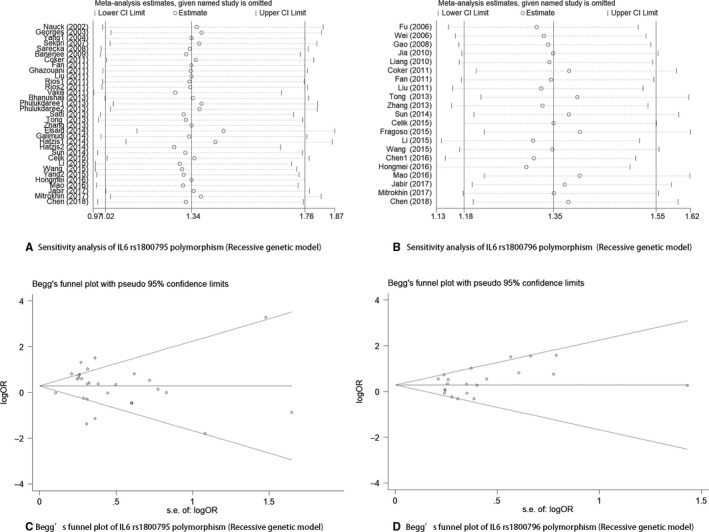
A, Sensitivity analysis of IL‐6 rs1800795 polymorphism (Recessive genetic model). B, Sensitivity analysis of IL‐6 rs1800796 polymorphism (Recessive genetic model). C, Begg's funnel plot of IL‐6 rs1800795 polymorphism (Recessive genetic model). D, Begg's funnel plot of IL‐6 rs1800796 polymorphism (Recessive genetic model)
3.5. Publication bias
The P values for the Egger's test of the IL‐6 rs1800795 and rs1800796 polymorphisms were .459 and .114, respectively; in addition, Begg's funnel plots of the two polymorphisms were symmetrical and all P values were greater than .05, indicating a lack of publication bias (Figure 3C‐D).
3.6. Trial sequential analysis
A previous meta‐analysis of the associations between IL‐6 polymorphisms and CAD risk reported negative results. For our pooled analysis of IL‐6 rs1800795 and rs1800796 polymorphisms, significant associations were only observed for the IL‐6 rs1800796 polymorphism. Hence, a trial sequential analysis was required to verify that our significant association was not a false‐positive result. Similar strength associations were discovered in five different genetic models. The allelic genetic model produced the best value and is a natural model of inheritance with a stronger genotype‐phenotype association, which also does not pre‐assume any interactions between the numbers of variant alleles. Therefore, we chose the allelic genetic model of the rs1800796 polymorphism to conduct the trial sequential analysis. The results of trial sequential analysis are shown in Figure 4. The x‐axis and y‐axis represent the number of patients and the cumulative Z score, respectively. Within the designed assumptions of confidence and effect size, the information size for the IL‐6 rs1800796 polymorphism is 24 788, and the Z curves not only cross the statistical significance line (Z = 1.96, P = .05), but also cross the O’ Brien Fleming boundaries, indicating that the significance level of our study was a true‐positive result and the previously reported negative association was due to a lower number of volunteers.
FIGURE 4.
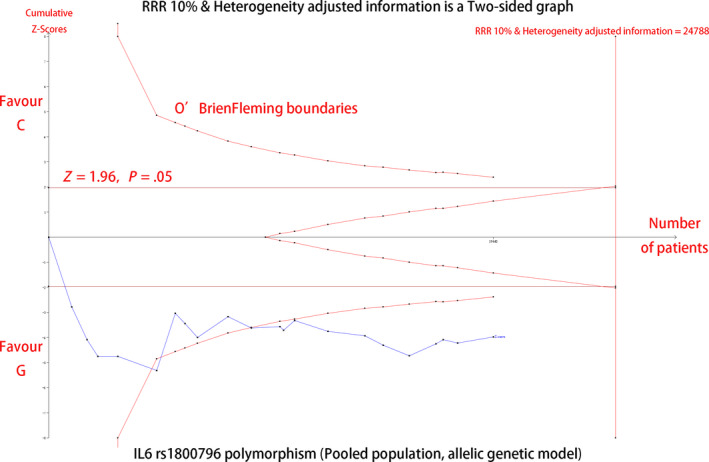
IL6 rs1800795 polymorphism (Pooled population, allelic genetic model)
3.7. Bioinformatics analysis
Based on the genomic context obtained from the Ensembl database, we constructed the summary genetic diagram for the rs1800795 and rs1800796 polymorphisms (Figure 5A). The two polymorphisms were both located in the promoter region near exon 2, implying that these sequences are potential transcription factor binding sites. Hence, we analysed the sequences of the two polymorphisms and the results from the SNPinfo database showed both polymorphisms are located in potential transcription factor binding sites (Figure 5B). In addition, the secondary structure of DNA at the rs1800795 and rs1800796 sequences was predicted using RNAfold. The minimum free energy (MFE) and the free energy of the thermodynamic ensemble (FETE) of the rs1800795 polymorphism were −142.50 kcal/mol and −170 kcal/mol for the wild G allele, and 141.60 kcal/mol and 169.58 kcal/mol for the mutant C allele, respectively. For the 1800796 polymorphism, the MFE and RFTE were −136.10 kcal/mol and −162.81 kcal/mol for the wild C allele, and −133.80 kcal/mol and −162.57 kcal/mol for the mutant G allele. Based on the predicted free energy of the two polymorphisms, the secondary structure of the two polymorphisms was determined. Compared with the wild allele, the mutant alleles of both polymorphisms caused a structure change, which were pointed with arrows in Figure 6.
FIGURE 5.
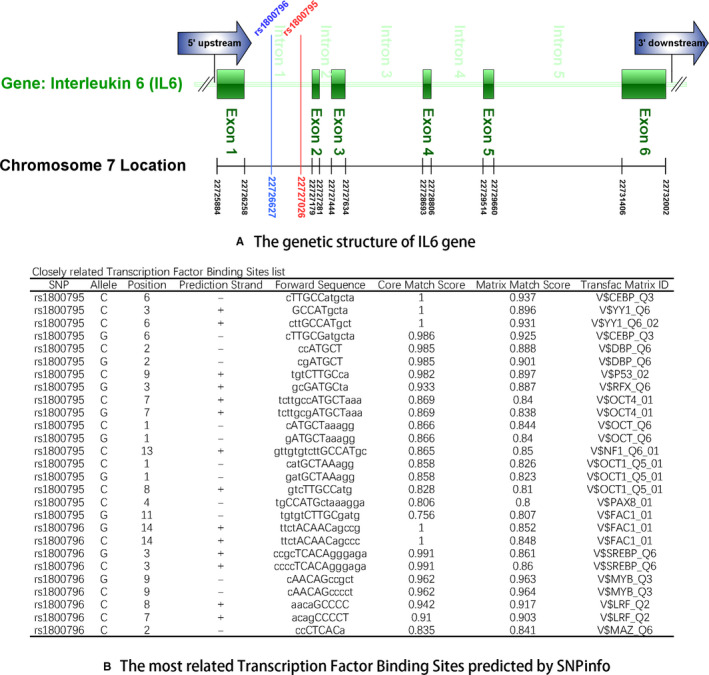
A, The genetic structure of IL‐6 gene. B, The most related Transcription Factor Binding Sites predicted by SNP ratio
FIGURE 6.
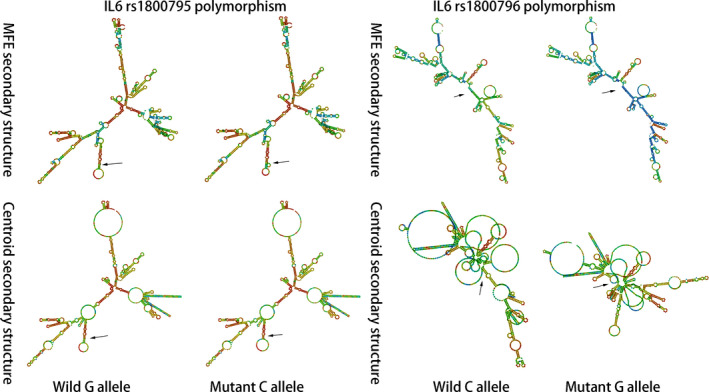
The RNAfold analysis of the IL‐6 polymorphisms
4. DISCUSSION
In our study, two polymorphisms (rs1800795 and rs1800796) in the IL‐6 gene were analysed for associations with CAD risk. The two common polymorphisms have been extensively studied in depth over the past few decades, providing sufficient enough data for a subgroup analysis designed to discover potential intriguing associations. Moreover, the two polymorphisms are located in the promoter region of the IL‐6 gene, and may influence the expression of the IL‐6 gene, and result in susceptibility to CAD. Several meta‐analyses have been conducted to explore the associations between the two polymorphisms and CAD risk, but the results were inconsistent. Significant associations between the IL‐6 rs1800795 polymorphism and CAD risk were reported in some meta‐analyses, 57 , 58 , 59 but not in the latest meta‐analysis reported by Liu et al 60 Interestingly, the opposite result was obtained for the IL‐6 rs1800796 polymorphism. The most recent studies by Song et al 61 and Hou et al 57 reported that this polymorphism may decrease the risk of CAD, which contradicts the conclusions of most previous meta‐analysis. 59 , 60 , 62 We comprehensively reviewed these two studies and found that the opposite results may due to the relatively small sample size. Additionally, the adjusted alpha was not used to adjust for multiple tests, and thus, that conclusion that the result was a true positive is questionable. The controversial results from previous meta‐analysis and case‐control studies prompted us to examine the associations between the IL6 rs1800795 and rs1800796 polymorphisms and CAD risk. Therefore, we chose these two common polymorphisms in the IL6 gene to analyse the potential CAD risk.
No association between the IL‐6 rs1800795 polymorphism and CAD risk with high heterogeneity was observed in the pooled results. Hence, we employed a detailed subgroup analysis to determine the potential sources of heterogeneity and associations. For the subgroup analysis stratified by region, significant associations with reduced heterogeneity were observed in the Asian population, which indicated an increased CAD risk for the rs1800795 polymorphism. Interesting results emerged in the analysis of the African subgroup and the decreased CAD risk for the mutant C allele and CC genotype were observed compared with the wild G allele and GC+CC genotypes, respectively. However, only three studies were included in the analysis of the African subgroup, and thus, we are sceptical about the conclusions and further studies are required. Similar findings were obtained for the Asian population. When stratified by ethnicity, the increased CAD risks for the rs1800795 polymorphism were observed in the Mongoloid population. The studies in the Mongoloid subgroup were performed in China, indicating that Chinese patients carrying the rs1800795 polymorphism would exhibit an increased risk of CAD. Moreover, high heterogeneity was significantly reduced when volunteers were stratified by region and ethnicity, indicating that these two factors are potential sources of high heterogeneity for the rs1800795 polymorphism. Sample size plays an important role in interpreting the conclusions of a case‐control study; thus, we conducted an analysis of subgroups stratified by sample size and discovered the increased CAD risk for the rs1800795 polymorphism in the larger sample size group. Therefore, if additional well‐designed studies are conducted, a lager sample size is required. The IL‐6 rs1800796 polymorphism is associated with an increased risk of CAD. Extensive associations with an increased risk of CAD were observed in the pooled analysis and subgroup analyses. In addition, the trial sequential analysis confirmed the true‐positive results for the rs1800796 polymorphism, indicating that carriers of the rs1800796 mutant G allele are likely predisposed to CAD.
Coronary atherosclerosis is the main pathophysiological process in coronary artery disease, and inflammation plays a predominant role in atherosclerosis. As an important pro‐inflammatory cytokine, IL‐6 has been proven to be an independent risk factor for coronary artery disease and is expressed at relatively high levels in human atherosclerotic plaques. 63 , 64 , 65 Considering the vital role of IL‐6 in atherosclerosis, the mechanism responsible for producing IL‐6 is a pivotal question. Researchers have not clearly determined whether polymorphisms in the IL‐6 gene may also be an answer to this question. We conducted a bioinformatics analysis to predict the potential molecular mechanism. The two polymorphisms are located in the promoter region of the IL‐6 gene, implying that the underlying mechanism occurs at the transcriptional level. The analysis of the SNPinfo database revealed potential transcription factor binding sites in the two polymorphisms, indicating the ability of these two polymorphisms to alter the expression of the IL‐6 gene. In addition, an analysis of the sequence and secondary structure was performed using the RNAfold web server. The minimum free energy (MFE) and the free energy of the thermodynamic ensemble of the mutant alleles of the rs1800795 and rs1800796 polymorphisms were reduced compared with the wild alleles. The principle of minimum energy states that for a closed system, with constant external parameters and entropy, the internal energy will decrease and approach a minimum value at equilibrium. 66 The mutant allele in a gene sequence may alter the free energy. The minimum free energy and the free energy of the thermodynamic ensemble are two thermodynamics parameters that have been used as a measure the required energy to reach the equilibrium for the stability of a sequence. 23 The reduction implies that less energy is needed to form the secondary structure of the sequence containing the mutant allele, indicating that the sequence of the mutant alleles of the rs1800795 and rs1800796 polymorphisms is easier to disperse from the DNA double helix structure to serve as the template strand during transcription. Hence, these structural changes may affect the expression of the IL‐6 gene. However, a bioinformatics prediction is not sufficient, and further fundamental research on the effect of the two polymorphisms on the transcription of the IL6 gene is needed.
Several limitations existed in our study. First, only English and Chinese articles were included as a language restriction, which may bias the results. Second, the number of included studies was relatively small in some subgroups, such as the African population in the subgroup analysis of the IL‐6 rs1800795 polymorphism, and thus, the results should be interpreted with caution. Third, only two common SNPs were evaluated in our study and other relevant SNPs in the IL‐6 gene that are unknown or understudied may also have potential associations with CAD risk. Forth, the distance between the two common polymorphisms is relatively close, and the potential interactions between the two polymorphisms or other unknown polymorphisms need to be studied. In addition, the potential influence of environmental factors on genotype‐CAD associations is worth considering.
In conclusion, the IL‐6 rs1800796 polymorphism is associated with an increased susceptibility to CAD and is a risk factor for CAD. In addition, the IL‐6 rs1800795 polymorphism is associated with an increased risk of CAD in Asian, particularly in Chinese volunteers. Remarkably, a decreased risk of CAD was observed in the African population.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTION
Shuai Lu, Ya Wang and Zhaohui Wang wrote the main manuscript text. Yijun Wang and Shuai Lu prepared Figures 1 and 2. Wenhan Ma and Jing Hu prepared Figures 3 and 4. Shuangye Liu, Di Wu and Xiaohui Zeng draw the Figures 5 and 6. Yijun Wang made the Table 1. Shuai Lu, Ya Wang and Yijun Wang forged the Table 2. Guo Yu performed the statistical analysis of evaluating the strengths of associations between these two polymorphisms and CAD risk. Zhaohui Wang hosted the meeting addressing the disagreements. All authors reviewed and revised the manuscript.
Supporting information
Table S1
Table S2
ACKNOWLEDGEMENT
This work is supported by grants from the National Natural Science Foundation of China (Nos. 81170205). The funder, Zhaohui Wang, was responsible for the article.
Lu S, Wang Y, Wang Y, et al. The IL-6 rs1800795 and rs1800796 polymorphisms are associated with coronary artery disease risk. J Cell Mol Med. 2020;24:6191–6207. 10.1111/jcmm.15246
Shuai Lu, Ya Wang and Yijun Wang contributed equally to this work.
DATA AVAILABILITY STATEMENT
All data included in this study are available upon request by contact with the corresponding author.
REFERENCES
- 1. Abdallah MH, Arnaout S, Karrowni W, Dakik HA. The management of acute myocardial infarction in developing countries. Int J Cardiol. 2006;111:189‐194. [DOI] [PubMed] [Google Scholar]
- 2. Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart disease and stroke statistics‐2017 update: a report from the American Heart Association. Circulation. 2017;135:e146‐e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosselman KE, Navas‐Acien A, Kaufman JD. Environmental factors in cardiovascular disease. Nat Rev Cardiol. 2015;12:627‐642. [DOI] [PubMed] [Google Scholar]
- 4. Thomas MR, Lip GY. Novel Risk Markers And Risk Assessments For Cardiovascular Disease. Circ Res. 2017;120:133‐149. [DOI] [PubMed] [Google Scholar]
- 5. Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM. Estimation and partition of heritability in human populations using whole‐genome analysis methods. Annu Rev Genet. 2013;47:75‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schunkert H, König IR, Kathiresan S, et al. Large‐scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El‐Aziz TA, Mohamed RH. Matrix metalloproteinase ‐9 polymorphism and outcome after acute myocardial infarction. Int J Cardiol. 2017;227:524‐528. [DOI] [PubMed] [Google Scholar]
- 8. Posadas‐Sanchez R, Perez‐Hernandez N, Rodriguez‐Perez JM, et al. Interleukin‐27 polymorphisms are associated with premature coronary artery disease and metabolic parameters in the Mexican population: the genetics of atherosclerotic disease (GEA) Mexican study. Oncotarget. 2017;8(38):64459‐64470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun D, Wu Y, Wang H, Yan H, Liu W, Yang J. Toll‐like receptor 4 rs11536889 is associated with angiographic extent and severity of coronary artery disease in a Chinese population. Oncotarget. 2017;8:2025‐2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Libby P, Ridker PM, Hansson GK. Leducq Transatlantic Network on A. Inflammation in atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol. 2009;54:2129‐2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benson MD, Lapedis CJ, Adler DS, Feinberg MW, Bhatt DL. Bulging at the root: an inflammatory tale. Circulation. 2016;133:1969‐1977. [DOI] [PubMed] [Google Scholar]
- 12. Hongmei Y, Yongping J, Jiyuan L. Interleukin‐6 polymorphisms and risk of coronary artery diseases in a Chinese population: a case‐control study. Pak J Med Sci. 2016;32:880‐885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fragoso JM, Zuniga‐Ramos J, Arellano‐Gonzalez M, et al. The T29C (rs1800470) polymorphism of the transforming growth factor‐beta1 (TGF‐beta1) gene is associated with restenosis after coronary stenting in Mexican patients. Exp Mol Pathol. 2015;98:13‐17. [DOI] [PubMed] [Google Scholar]
- 14. Sun GQ, Wu GD, Meng Y, Du B, Li YB. IL‐6 gene promoter polymorphisms and risk of coronary artery disease in a Chinese population. Genet Mol Res. 2014;13:7718‐7724. [DOI] [PubMed] [Google Scholar]
- 15. Murase Y, Yamada Y, Hirashiki A, et al. Genetic risk and gene‐environment interaction in coronary artery spasm in Japanese men and women. Eur Heart J. 2004;25:970‐977. [DOI] [PubMed] [Google Scholar]
- 16. Celik A, Ozcetin M, Ates O, et al. Analyses of C‐reactive protein, endothelial nitric oxide synthase and interleukin‐6 gene polymorphisms in adolescents with a family history of premature coronary artery disease: a pilot study. Balkan Med J. 2015;32:397‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farooq V, van Klaveren D, Steyerberg EW, et al. Anatomical and clinical characteristics to guide decision making between coronary artery bypass surgery and percutaneous coronary intervention for individual patients: development and validation of SYNTAX score II. Lancet (London, England). 2013;381:639‐650. [DOI] [PubMed] [Google Scholar]
- 18. Xin XY, Ding JQ, Chen SD. Apolipoprotein E promoter polymorphisms and risk of Alzheimer's disease: evidence from meta‐analysis. J Alzheimers Dis. 2010;19:1283‐1294. [DOI] [PubMed] [Google Scholar]
- 19. Wetterslev J, Thorlund K, Brok J, Gluud C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta‐analysis. J Clin Epidemiol. 2008;61:64‐75. [DOI] [PubMed] [Google Scholar]
- 20. Brok J, Thorlund K, Gluud C, Wetterslev J. Trial sequential analysis reveals insufficient information size and potentially false positive results in many meta‐analyses. J Clin Epidemiol. 2008;61:763‐769. [DOI] [PubMed] [Google Scholar]
- 21. Thorlund K, Anema A, Mills E. Interpreting meta‐analysis according to the adequacy of sample size. An example using isoniazid chemoprophylaxis for tuberculosis in purified protein derivative negative HIV‐infected individuals. Clin Epidemiol. 2010;2:57‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600‐W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70‐W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nauck M, Winkelmann BR, Hoffmann MM, Bohm BO, Wieland H, Marz W. The interleukin‐6 G(‐174)C promoter polymorphism in the LURIC cohort: no association with plasma interleukin‐6, coronary artery disease, and myocardial infarction. J Mol Med. 2002;80:507‐513. [DOI] [PubMed] [Google Scholar]
- 25. Georges JL, Rupprecht HJ, Blankenberg S, et al. Impact of pathogen burden in patients with coronary artery disease in relation to systemic inflammation and variation in genes encoding cytokines. Am J Cardiol. 2003;92:515‐521. [DOI] [PubMed] [Google Scholar]
- 26. Chao Y, Li Y, Pa Z, Jiang X, Huang C. The interleukin‐6 gene polymorphism in patients with coronary heart disease in Chinese population. Sichuan Med J. 2004;25:404‐406. [Google Scholar]
- 27. Yesheng W, Lan Y, Liu Y, Tang R, Lan J. Relationship between interleukin‐6 gene polymorphism and coronary heart disease and its effect on plasma lipid levels. Chin Crit Care Med. 2006;18(4):233‐236. [PubMed] [Google Scholar]
- 28. Sekuri C, Cam FS, Sagcan A, et al. No association of interleukin‐6 gene polymorphism (‐174 G/C) with premature coronary artery disease in a Turkish cohort. Coron Artery Dis. 2007;18:333‐337. [DOI] [PubMed] [Google Scholar]
- 29. Cuixiang G, Wang Y. Interleukin‐6 gene polymorphism in elderly patients with coronary heart disease. J Fourth Mil Med Uni. 2008;29:2183‐2185. [Google Scholar]
- 30. Sarecka B, Zak I, Krauze J. Synergistic effects of the polymorphisms in the PAI‐1 and IL‐6 genes with smoking in determining their associated risk with coronary artery disease. Clin Biochem. 2008;41:467‐473. [DOI] [PubMed] [Google Scholar]
- 31. Banerjee I, Pandey U, Hasan OM, Parihar R, Tripathi V, Ganesh S. Association between inflammatory gene polymorphisms and coronary artery disease in an Indian population. J Thromb Thrombolysis. 2009;27:88‐94. [DOI] [PubMed] [Google Scholar]
- 32. Liang Z, Zhang X, Sun Y, et al. Association of interleukin‐6 gene ‐572C/G single nucleotide polymorphism with coronary artery disease in Han population of North China. Prog Modern Biomed. 2010;4:4675‐4678. [Google Scholar]
- 33. Ghazouani L, Abboud N, Ben Hadj Khalifa S, et al. 174G>C interleukin‐6 gene polymorphism in Tunisian patients with coronary artery disease. Ann Saudi Med. 2011;31:40‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu YC, Duan ZM, Zhang MJ. Association between polymorphisms in interleukin‐6 gene promoter region and coronary artery disease. Clin Focus. 2011;26(14):1200‐1203. [Google Scholar]
- 35. Bhanushali AA, Das BR. Promoter variants in interleukin‐6 and tumor necrosis factor alpha and risk of coronary artery disease in a population from Western India. Indian J Hum Genet. 2013;19:430‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phulukdaree A, Khan S, Ramkaran P, Govender R, Moodley D, Chuturgoon AA. The interleukin‐6 ‐147 g/c polymorphism is associated with increased risk of coronary artery disease in young South African Indian men. Metab Syndr Relat Disorders. 2013;11:205‐209. [DOI] [PubMed] [Google Scholar]
- 37. Rui Z, Feng B, Guang‐li XU, Fei‐fei L, Xin‐ling G, Chong‐ge Y. High resolution melting genotyping for detection of single nucleotide polymorphisms of IL‐6 and its receptor genes in patients with coronary heart disease. Chin J Clin Lab Sci. 2013;31:415‐418. [Google Scholar]
- 38. Satti HS, Hussain S, Javed Q. Association of interleukin‐6 gene promoter polymorphism with coronary artery disease in Pakistani families. Sci World J. 2013;2013:538365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galimudi RK, Spurthi MK, Padala C, et al. Interleukin 6(‐174G/C) variant and its circulating levels in coronary artery disease patients and their first degree relatives. Inflammation. 2014;37:314‐321. [DOI] [PubMed] [Google Scholar]
- 40. Hatzis G, Tousoulis D, Papageorgiou N, et al. Combined effects of smoking and interleukin‐6 and C‐reactive protein genetic variants on endothelial function, inflammation, thrombosis and incidence of coronary artery disease. Int J Cardiol. 2014;176:254‐257. [DOI] [PubMed] [Google Scholar]
- 41. Li L, Li E, Zhang LH, Jian LG, Liu HP, Wang T. IL‐6‐174G/C and IL‐6‐572C/G polymorphisms are associated with increased risk of coronary artery disease. Genet Mol Res. 2015;14:8451‐8457. [DOI] [PubMed] [Google Scholar]
- 42. Wang K, Dong PS, Zhang HF, Li ZJ, Yang XM, Liu H. Role of interleukin‐6 gene polymorphisms in the risk of coronary artery disease. Genet Mol Res. 2015;14:3177‐3183. [DOI] [PubMed] [Google Scholar]
- 43. Yang HT, Wang SL, Yan LJ, Qian P, Duan HY. Association of interleukin gene polymorphisms with the risk of coronary artery disease. Genet Mol Res. 2015;14:12489‐12496. [DOI] [PubMed] [Google Scholar]
- 44. Dan C, Kui Z, Hong‐xia W, Lei L, Wei Q, Xin‐qin W. Association between interleukin‐6 gene‐572C/G polymorphism and coronary heart disease in Han population of Suqian city, Jiangsu province, China. Chin J Clin Lab Sci. 2016;34:366‐370. [Google Scholar]
- 45. Fu HX, Zhang JY, Li GS, Li Y, Xu JL, Zhao ZN. [Study on linkage between polymorphism of interleukin 6 gene ‐572C/G and susceptibility to myocardial infarction]. Chin J Med Genet. 2006;23:245‐249. [PubMed] [Google Scholar]
- 46. Jia X, Tian Y, Wang Y, et al. Association between the interleukin‐6 gene ‐572G/C and ‐597G/A polymorphisms and coronary heart disease in the Han Chinese. Med Sci Monit. 2010;16:CR103‐8. [PubMed] [Google Scholar]
- 47. Rios DL, Cerqueira CC, Bonfim‐Silva R, et al. Interleukin‐1 beta and interleukin‐6 gene polymorphism associations with angiographically assessed coronary artery disease in Brazilians. Cytokine. 2010;50:292‐296. [DOI] [PubMed] [Google Scholar]
- 48. Coker A, Arman A, Soylu O, Tezel T, Yildirim A. Lack of association between IL‐1 and IL‐6 gene polymorphisms and myocardial infarction in Turkish population. Int J Immunogenet. 2011;38:201‐208. [DOI] [PubMed] [Google Scholar]
- 49. Vakili H, Ghaderian SM, Akbarzadeh Najar R, Tabatabaei Panah AS, Azargashb E. Genetic polymorphism of interleukin‐6 gene and susceptibility to acute myocardial infarction. Coron Artery Dis. 2011;22:299‐305. [DOI] [PubMed] [Google Scholar]
- 50. Tong Z, Li Q, Zhang J, Wei Y, Miao G, Yang X. Association between interleukin 6 and interleukin 16 gene polymorphisms and coronary heart disease risk in a Chinese population. J Int Med Res. 2013;41:1049‐1056. [DOI] [PubMed] [Google Scholar]
- 51. Elsaid A, Abdel‐Aziz AF, Elmougy R, Elwaseef AM. Association of polymorphisms G(‐174)C in IL‐6 gene and G(‐1082)A in IL‐10 gene with traditional cardiovascular risk factors in patients with coronary artery disease. Indian J Biochem Biophys. 2014;51:282‐292. [PubMed] [Google Scholar]
- 52. Mao L, Geng GY, Han WJ, Zhao MH, Wu L, Liu HL. Interleukin‐6 (IL‐6) ‐174G/C genomic polymorphism contribution to the risk of coronary artery disease in a Chinese population. Genet Mol Res. 2016;15(2). 10.4238/gmr.15027803 [DOI] [PubMed] [Google Scholar]
- 53. Jabir NR, Firoz CK, Kamal MA, et al. Assessment of genetic diversity in IL‐6 and RANTES promoters and their level in Saudi coronary artery disease patients. J Clin Lab Anal. 2017;31(5):e22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitrokhin V, Nikitin A, Brovkina O, et al. Association between interleukin‐6/6R gene polymorphisms and coronary artery disease in Russian population: influence of interleukin‐6/6R gene polymorphisms on inflammatory markers. J Inflamm Res. 2017;10:151‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen H, Ding S, Liu XI, Wu Y, Wu X. Association of interleukin‐6 genetic polymorphisms and environment factors interactions with coronary artery disease in a chinese han population. Clin Exp Hypertension. 2018;40:514‐517. [DOI] [PubMed] [Google Scholar]
- 56. Fan WH, Liu DL, Xiao LM, Xie CJ, Sun SY, Zhang JC. Coronary heart disease and chronic periodontitis: is polymorphism of interleukin‐6 gene the common risk factor in a Chinese population? Oral Dis. 2011;17:270‐276. [DOI] [PubMed] [Google Scholar]
- 57. Hou H, Wang C, Sun F, Zhao L, Dun A, Sun Z. Association of interleukin‐6 gene polymorphism with coronary artery disease: an updated systematic review and cumulative meta‐analysis. Inflamm Res. 2015;64:707‐720. [DOI] [PubMed] [Google Scholar]
- 58. Yin YW, Li JC, Zhang M, et al. Influence of interleukin‐6 gene ‐174G>C polymorphism on development of atherosclerosis: a meta‐analysis of 50 studies involving 33,514 subjects. Gene. 2013;529:94‐103. [DOI] [PubMed] [Google Scholar]
- 59. Yang Y, Zhang F, Skrip L, et al. IL‐6 gene polymorphisms and CAD risk: a meta‐analysis. Mol Biol Rep. 2013;40:2589‐2598. [DOI] [PubMed] [Google Scholar]
- 60. Liu SL, Yin YW, Sun QQ, Hu AM, Zhang SJ. Genetic polymorphisms of interleukin‐6 gene and susceptibility to coronary artery disease in Chinese population: evidence based on 4582 subjects. Hum Immunol. 2015;76:505‐510. [DOI] [PubMed] [Google Scholar]
- 61. Song C, Liu B, Yang D, et al. Association between Interleukin‐6 gene ‐572G>C polymorphism and coronary heart disease. Cell Biochem Biophys. 2015;71:359‐365. [DOI] [PubMed] [Google Scholar]
- 62. Li YY, Zhou CW, Xu J, Qian Y, Wang XM. Interleukin‐6 C‐572G gene polymorphism and coronary artery disease in Asian: a meta‐analysis of 2511 subjects. Int J Clin Exp Med. 2015;8:8995‐9003. [PMC free article] [PubMed] [Google Scholar]
- 63. Yudkin JS, Kumari M, Humphries SE, Mohamed‐Ali V. Inflammation, obesity, stress and coronary heart disease: is interleukin‐6 the link? Atherosclerosis. 2000;148:209‐214. [DOI] [PubMed] [Google Scholar]
- 64. Luc G, Bard JM, Juhan‐Vague I, et al. C‐reactive protein, interleukin‐6, and fibrinogen as predictors of coronary heart disease: the PRIME Study. Arterioscler Thromb Vasc Biol. 2003;23:1255‐1261. [DOI] [PubMed] [Google Scholar]
- 65. Schieffer B, Schieffer E, Hilfiker‐Kleiner D, et al. Expression of angiotensin II and interleukin 6 in human coronary atherosclerotic plaques: potential implications for inflammation and plaque instability. Circulation. 2000;101:1372‐1378. [DOI] [PubMed] [Google Scholar]
- 66. Hovgaard W. The principle of minimum energy and the motion of fluids. Proc Natl Acad Sci U S A. 1923;9(11):363‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Data Availability Statement
All data included in this study are available upon request by contact with the corresponding author.


