Abstract
Community‐acquired urinary tract infections (UTIs) are one of the most common bacterial infections worldwide. Escherichia coli is the most common cause of community‐acquired UTI. In general, UTI results from E. coli in the intestine that enters the bladder via the urethra. However, whether these E. coli strains that cause UTI represent members of the intestinal commensal E. coli or a distinct subgroup of pathogenic E. coli remains unestablished. Here, we analyzed E. coli isolates from fecal samples of healthy volunteers and urine samples of UTI patients obtained from a university‐affiliated health center. The E. coli isolates were genotyped by multilocus sequence typing (MLST). From May to October 2018, we analyzed 89 E. coli isolates from 76 (75%) rectal swabs from 113 unique healthy volunteers. We also analyzed 106 (27%) E. coli isolates from 398 unique urine samples collected between August and October 2018. Fecal and urine E. coli isolates each contained 31 distinct sequence types (STs). Nine STs were shared by fecal and urine E. coli isolates, which accounted for approximately 50% of urine isolates typed by MLST. Among the shared genotypes, ST10 and ST131 were significantly more frequently found in fecal samples, whereas ST95 and ST127 were significantly more frequently recovered from UTI samples. ST73 was found only among urine samples. These E. coli genotypes clustered and fluctuated over time. These observations suggest that E. coli genotypes found to cause UTI transiently colonize the intestine and that their primary reservoir may reside outside of the human intestine.
Keywords: commensal E. coli, Escherichia coli, molecular epidemiology, multilocus sequence typing, urinary tract infection, uropathogenic E. coli
We address an unresolved question regarding the source of Escherichia coli that causes community‐acquired urinary tract infection (CA‐UTI)—whether E. coli strains that cause UTI represent members of the intestinal commensal E. coli or a distinct subgroup of pathogenic E. coli. Our results based on multilocus sequence type (MLST) analysis of E. coli isolates from healthy volunteers versus urine isolates from patients with UTI indicate that uropathogenic E. coli has a source outside of the human intestine.

1. INTRODUCTION
Urinary tract infections (UTIs) are one of the most common bacterial infections that greatly challenge the quality of life of patients (Foxman, 2010). A recent study in southern California showed that between 2008 and 2017, UTI in virtual care (health care via telephone, video, and the Internet) settings increased by 21% in women and 29% in men each year (Bruxvoort et al., 2019). More than half of women will experience at least one episode in their lifetime. Foxman reported 27% of college women experienced at least one recurrence within 6 months following their initial infection (Foxman, 1990). Eighty percent of all community‐acquired UTIs are caused by Escherichia coli (Flores‐Mireles, Walker, Caparon, & Hultgren, 2015).
In general, UTI results from the dissemination of E. coli from the intestine into the bladder via the urethra (Hooton, 1990). Escherichia coli that causes UTI are variously labeled as uropathogenic E. coli (UPEC) or extraintestinal pathogenic E. coli (ExPEC). Previous studies showed that UPEC strains isolated from UTI patients are also found in fecal samples from the same patient, which would be expected since the immediate source of UPEC is the infected person's intestine (Chen et al., 2013; Nielsen, Dynesen, Larsen, & Frimodt‐Moller, 2014; Russo, Stapleton, Wenderoth, Hooton, & Stamm, 1995; Yamamoto et al., 1997). However, it is not known if UPECs are part of the commensal flora of the human intestine that causes UTI when they breach a sterile barrier, or if they represent a distinct pathogenic E. coli group that transiently colonizes the intestine. If they are distinct pathogens, their main reservoir should be outside of the human intestine.
Here, we collected E. coli isolates from feces of healthy individuals and the urine of patients with community‐acquired UTI (CA‐UTI) at a northern California university community and compared their genotypic distribution to determine whether ExPEC strains represent commensal E. coli or E. coli pathotype strains that transiently colonize the human intestine. Our observations suggest that UPECs comprise a group of pathogens distinct from the commensal E. coli.
2. MATERIALS AND METHODS
2.1. Study design
We cultured E. coli from fecal samples of healthy volunteers at a northern California university campus and from urine samples from patients diagnosed to have UTIs at an outpatient health clinic located at the same university. The fecal samples were collected between May and October 2018, while the urine samples were collected between August and October 2018. We examined fecal samples collected from 113 volunteers and urine samples from 398 UTI patients. We could not collect any samples in July due to the summer semester break period.
After obtaining approval from the Committee on Human Subjects, we recruited healthy volunteers that included participants between 18 and 45 years of age, comprised of students, staff, and faculty of the university community. We excluded subjects with any medical history of urinary tract corrective surgery or abnormality, bladder catheterization, or hospitalization in the 6 months before the fecal sample collection. As part of a separate study (Rubin, Mussio, Xu, Suh, & Riley, 2020), participants were given a pre‐addressed kit containing a rectal swab in Cary‐Blair transport media (Becton Dickinson BBLTM), two biosafety bags, detailed collection instructions, and a questionnaire asking about age, gender, recent antibiotic use, history of UTI, and lifestyle characteristics. Participants were instructed to send the rectal swab and the completed questionnaire back to the study laboratory via the United States Postal Service (USPS) immediately after collection. The rectal swabs were analyzed at the study laboratory within 48 hr of their delivery.
Urine samples were collected consecutively from patients diagnosed with UTI at the health center. All urine samples were first tested at the health center by dipstick, and those specimens found to test positive for leukocytes, nitrates, protein, blood, or glucose were collected for our study. No personal information was obtained from patients with UTI due to HIPAA restriction.
2.2. Isolation of E. coli from fecal samples
Fecal swab tips were placed in a 1.5 ml Eppendorf tube containing 1 ml of Luria–Bertani broth and vortexed for 60 s. A 10 µl aliquot of the vortexed samples was placed on separate MacConkey agar plates, each containing ampicillin (AMP) (32 μg/ml), trimethoprim–sulfamethoxazole (TMP‐SMZ) (4–76 μg/ml), gentamicin (GEN) (16 μg/ml), or colistin (2 μg/ml)) as well as on one MacConkey agar plate containing no drug. The plates were incubated overnight at 37°C and colonies that grew on the drug‐containing plates were subjected to multilocus sequence typing (MLST) tests.
2.3. 16S ribosomal RNA sequence and ERIC2‐PCR of fecal isolates
Five colonies were randomly selected from each plate. If less than five colonies were present, all colonies were selected for analysis. Basic procedures for DNA extraction by a freeze–thaw method were performed as previously described (Adams‐Sapper et al., 2012; Raphael, Wong, & Riley, 2011). Species identification was performed by 16S ribosomal RNA gene sequencing. We used the primer pairs (27f‐CM: 5′‐AGAGTTTGATCMTGGCTCAG, where M is A or C/1492r: 5′‐TACCTTGTTACGACTT) to generate 16S ribosomal RNA amplicons and performed bidirectional amplicon sequencing (Frank et al., 2008). Each sequence was compared with sequences in GenBank® by BLAST (National Center for Biotechnology Information). Species were determined by the criteria of >98% sequence identity and genus by >95% sequence identity. If isolates were identified as E. coli, we randomly selected one of the E. coli isolates from each plate.
We subtyped all the selected E. coli isolates by ERIC2‐PCR, as described previously (Wilson & Sharp, 2006). Isolates that had identical ERIC2 electrophoretic banding patterns by visual inspection were considered to belong to the same clonal group, and one of them was selected for further analysis by MLST.
2.4. Isolation of E. coli from urine samples
We defined a case of UTI as a symptomatic patient with a urine specimen that contained more than 102 CFU of E. coli per milliliter. We cultured a 10 µl aliquot of urine on a MacConkey agar plate. We presumptively identified lactose‐positive and indole‐positive colonies as E. coli and tested them for further analysis. One colony was randomly selected among the lactose‐positive and indole‐positive colonies from each plate. As outlined above, DNA extraction by a freeze–thaw method was performed.
2.5. Genotyping E. coli by MLST
Selected E. coli isolates from fecal and urine samples were genotyped by MLST based on the seven‐gene Achtman scheme described at the website https://pubmlst.org/ (Tartof, Solberg, Manges, & Riley, 2005). The allelic number and the corresponding genotype number were designated by the curator of the MLST website.
2.6. Antibiotic resistance gene identification in fecal E. coli isolates
We identified antibiotic resistance genes of ST69 and ST131 strains obtained from fecal samples of healthy volunteers. We selected these genotypes for further analysis because they are two of the most common multidrug‐resistant ExPEC lineages reported worldwide. Bacterial isolates that grew in the presence of AMP were examined for β‐lactamase gene families by multiplex PCR as described previously (Dallenne, Costa, Decré, Favier, & Arlet, 2010). Bacteria that grew on plates containing gentamicin or trimethoprim–sulfamethoxazole were examined for 5′ and 3′ conserved sequences flanking the class Ι integron gene cassettes. The entire cassette sequences harbored in the integron were analyzed. If present, these gene cassettes were sequenced to detect the presence of aad and dhfr gene types for GEN and TMP‐SMZ resistance, respectively.
2.7. Data analysis
Differences in proportions among collections were tested by Fisher's exact test. Statistical significance was defined as a p‐value of ≤.05. All analyses were carried out with the SAS version 6.12.
3. RESULTS
3.1. E. coli isolates from fecal samples
We collected rectal swab samples from 113 volunteers between May and October 2018. Eleven rectal swabs exhibited no growth on a MacConkey agar plate. Escherichia coli was isolated from 76 (75%) of the 102 remaining samples, yielding 119 E. coli isolates. Of the 76 volunteers, 44 (57%) were female, 35 (46%) were graduate students, 19 (25%) were undergraduates, and 22 (29%) were staff/faculty. In the 1 year before rectal swab collection, 48 (63%) had travel history abroad, 2 (3%) had been hospitalized, and 28 (37%) had used antibiotics. Ten (13%) had a history of UTI in the previous year. Of 119 E. coli isolates, 30 isolates showed ERIC‐PCR patterns similar to those of other isolates. Of the 89 E. coli isolates, 88 were genotyped by MLST and were found to belong to 31 unique STs (Figure 1a and Appendix). The other one isolate could not be assigned to a known ST type.
FIGURE 1.
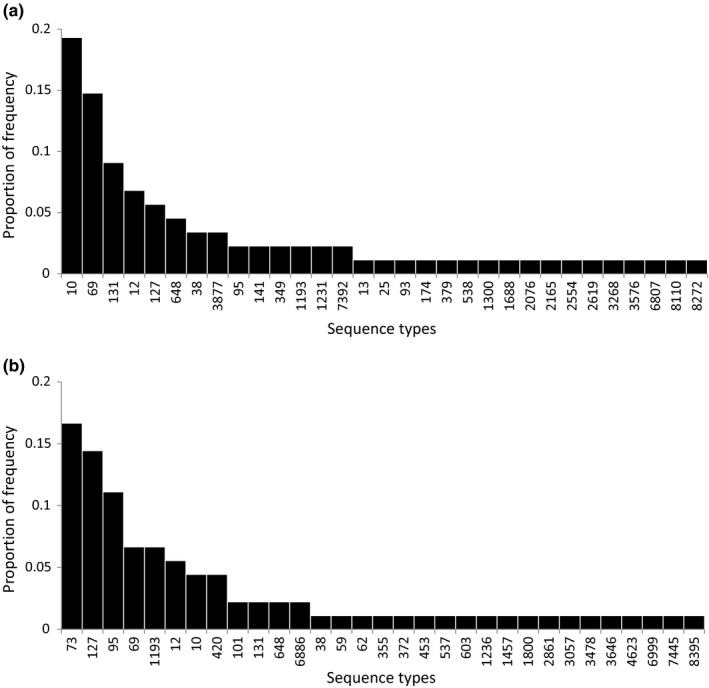
Distribution of multilocus sequence types of Escherichia coli isolates recovered from fecal samples of volunteers in a college community (a) and from urine samples of patients diagnosed to have urinary tract infection in the same community (b)
3.2. E. coli isolates from urine samples
We obtained 398 unique urine samples between August and October 2018. One hundred and six (27%) E. coli isolates were isolated from them. Of 106 E. coli isolates, 90 were genotyped and found to belong to 31 unique STs (Figure 1b and Appendix). The other 16 isolates could not be assigned an ST designation because we could not obtain complete sequencing data for all seven housekeeping genes.
3.3. Distribution of MLST genotypes of fecal and urine E. coli isolates
The most frequent sequence types among E. coli isolates from fecal samples were ST10 (19%), ST69 (15%), ST131 (9%), ST12 (7%), ST127 (6%), and ST648 (5%). These six ST isolates accounted for 60% of all E. coli isolates typed by MLST. The major common genotypes among E. coli isolates from urine samples were ST73 (17%), ST127 (14%), ST95 (11%), ST69 (7%), ST1193 (7%), and ST12 (6%) accounting for 61% of urine E. coli isolates genotyped by MLST (Figure 1). Nine genotypes, ST10, ST12, ST38, ST69, ST95, ST127, ST131, ST648, and ST1193, were shared by fecal and urine E. coli isolates (Figure 2). Of these genotypes, ST10, ST12, ST69, ST95, ST127, and ST1193 were among the most common lineages in urine E. coli isolates. Overall, the nine genotypes accounted for 54% of the urine isolates typed by MLST. Also, of 22 genotypes unique to fecal E. coli isolates, 4 (ST93, ST141, ST538, and ST2076) have been previously reported to be associated with human infections in the EnteroBase (https://enterobase.warwick.ac.uk/species/index/E.coli).
FIGURE 2.
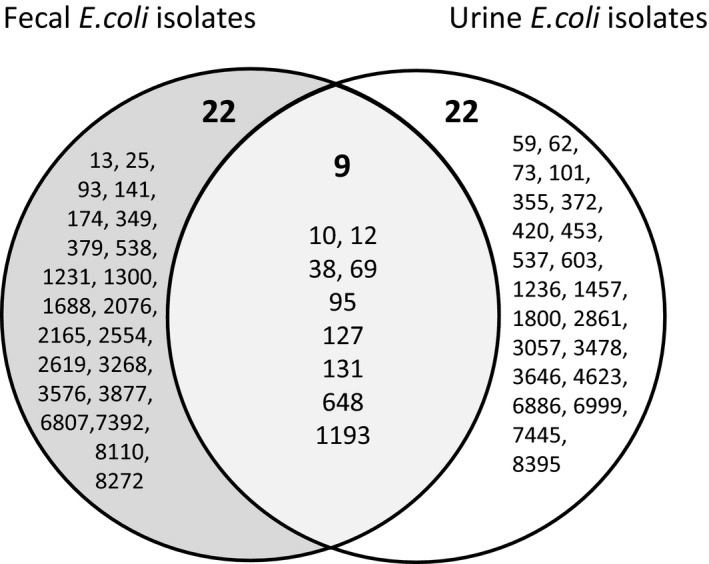
Venn diagram of the multilocus sequence types of Escherichia coli isolates recovered from fecal samples of volunteers in a college community (left circle) and from urine samples of patients diagnosed to have urinary tract infection at the same community (right circle)
3.4. Distribution of pandemic ExPEC lineages among fecal and urine E. coli isolates
ST69, ST73, ST95, and ST131 comprise members of so‐called pandemic lineages of ExPEC, and ST10 and ST127 are also increasingly reported from many regions of the world (Doumith et al., 2015; Gibreel et al., 2012; Olesen et al., 2012). Here, these ExPEC lineages together accounted for 56% of urine isolates and 51% of fecal isolates. Among them, 17 (19%) of 88 fecal isolates versus 4 (4%) of 90 urine isolates were ST10 (p = .002) (Table 1). Thirteen (15%) of 88 fecal isolates versus 6 (7%) of 90 urine isolates were ST69 (p = .08). Eight (9%) of 88 fecal isolates versus 2 (2%) of 90 urine isolates were ST131 (p = .05). On the other hand, 5 (6%) of 88 fecal isolates versus 13 (14%) of 90 urine isolates were ST127 (p = .07). Two (2%) of 88 fecal isolates versus 10 (11%) of 90 urine isolates were ST95 (p = .03). ST73 from urine samples was never found in fecal isolates (p < .001). In summary, ST10 and ST131 were significantly more frequently found in fecal samples, whereas ST95 and ST127 were significantly more frequently recovered from UTI samples. ST73 was found only among urine samples.
TABLE 1.
Escherichia coli isolates belonging to the shared 9 MLST genotypes and ST73 isolates from fecal samples of healthy volunteers and urine samples from patients diagnosed to have urinary tract infection
| Sequence type | No. (%) of isolates | p‐value a | |
|---|---|---|---|
| Fecal samples | Urine samples | ||
| ST10 | 17 (19) | 4 (4) | .002 |
| ST12 | 6 (7) | 5 (6) | .76 |
| ST38 | 3 (3) | 1 (1) | .36 |
| ST69 | 13 (15) | 6 (7) | .08 |
| ST73 | 0 (0) | 15 (17) | <.001 |
| ST95 | 2 (2) | 10 (11) | .03 |
| ST127 | 5 (6) | 13 (14) | .07 |
| ST131 | 8 (9) | 2 (2) | .05 |
| ST648 | 4 (5) | 2 (2) | .44 |
| ST1193 | 2 (2) | 6 (7) | .27 |
| Total no. of Isolates | 88 | 90 | |
p values based on Fisher's exact test.
3.5. Fecal and urine MLST genotype isolation by time
We plotted the month of isolation of the 88 fecal isolates by genotype over 6 months from May to October 2018 (Figure 3a) and 90 UTI isolates from August to October 2018 (Figure 3b). We found that each major genotype clustered in time and fluctuated month to month. Proportions of the genotypes also changed over time. The peak frequency of isolated fecal ST10 strains was in May, followed by its gradual decrease. Isolated fecal ST95 and ST127 strains were concentrated in 2 months (in August and September, and in September and October, respectively). No urine samples were obtained from May to July 2018 due to the summer semester break (Figure 3b). In contrast to the fecal genotype isolations, there was little change in month‐to‐month (August–October 2018) frequencies of E. coli genotypes isolated from urine samples (Figure 3b).
FIGURE 3.
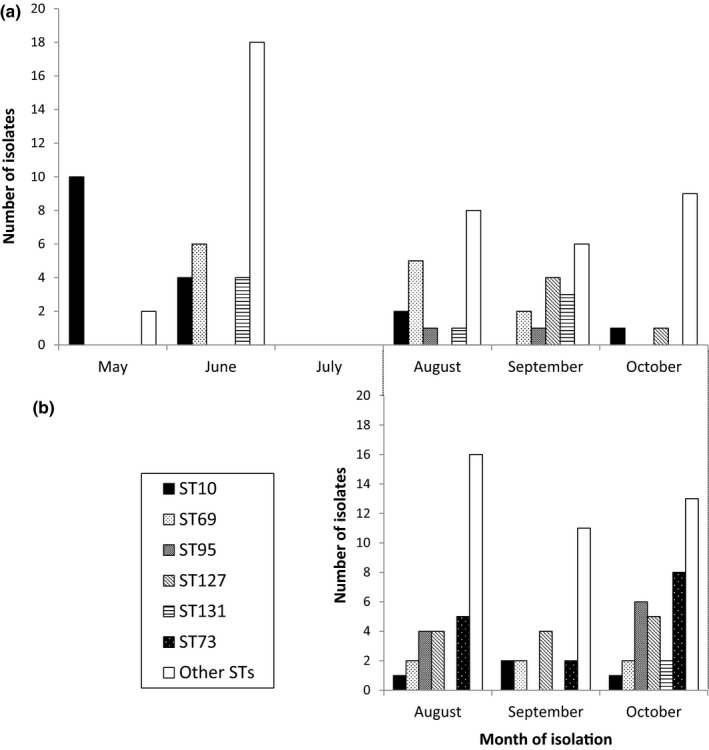
Isolation by month of Escherichia coli genotypes recovered from fecal samples of volunteers in a college community from May to October 2018 (a) and from urine samples of patients diagnosed to have urinary tract infection in the same community from August to October 2018 (b)
3.6. Antibiotics resistance genes among ST69 and ST131 isolated from fecal samples
Of 13 ST69 isolates, bla TEM‐type gene was detected in 12 strains that were resistant to AMP. One was AMP‐susceptible. Five of 12 isolates possessed other β‐lactamase genes (bla SHV type and bla CTX‐M group1). We also detected dhfr‐A17, dhfr‐A7, and aadA5 in the class Ι integron gene cassettes of seven isolates (Table 2). Similarly, of eight ST131 isolates, bla TEM‐type gene was detected in 6. One isolate had only bla CTX‐M group1 gene. Dhfr‐A17 and aadA5 were detected in 3 (Table 2).
TABLE 2.
Antibiotics resistance gene identified among ST69 and ST131 isolates from fecal samples of healthy volunteers
| Sequence type | Antibiotic resistance gene type (no. of isolates) | ||
|---|---|---|---|
| β‐lactamase gene (number of isolates) | Dihydrofolate reductase gene | Aminoglycoside resistance gene | |
| ST69 | bla TEM type (7) | dhfr‐A17 (5) | aadA5 (4) |
| bla TEM type + bla SHV type (3) | dhfr‐A7 (2) | NA (9) | |
| bla TEM type + bla CTX‐M group1 type (1) | NA (6) | ||
| bla TEM type + bla SHV type + bla CTX‐M group1 type (1) | |||
| NA (1) | |||
| ST131 | bla TEM type (3) | dhfr‐A17 (2) | aadA5 (2) |
| bla TEM type + bla SHV type (2) | NA (6) | NA (6) | |
| bla TEM type + bla CTX‐M group1 type (1) | |||
| bla CTX‐M group1 type (1) | |||
| NA (1) | |||
Abbreviation: NA, not applicable.
4. DISCUSSION
We compared the genotype distribution of E. coli isolates prospectively collected from a fecal swab of healthy volunteers and patients with UTI at a northern California university community. We found nine genotypes shared by both fecal E. coli isolates obtained from healthy volunteers and urine E. coli isolates obtained from UTI patients. These genotypes accounted for 68% of the fecal isolates and nearly half of the urine isolates. Strikingly, nearly all of the shared genotypes (ST10, ST12, ST38, ST69, ST95, ST127, ST131, ST648, and ST1193) belonged to pandemic ExPEC lineages or ExPEC lineages reported from multiple regions of the world (Manges et al., 2019).
While it is expected that the E. coli genotypes of the UTI isolates will overlap with those of E. coli isolates from fecal samples of the same subject, it was unexpected that the same ExPEC genotypes will be identified at such high frequencies in fecal swab samples of unrelated healthy subjects residing in the same community. Although pandemic ExPEC lineages dominated in both fecal and urine isolates, their frequency differed significantly in the two sources. No ST73 strains and only two ST95 strains were seen among fecal isolates, whereas ST73 was the most common and ST95 was the third most common genotype observed to cause UTI during this study period. The association of ST127 with UTI, the second most common cause of UTI, approached significance (p = .07). On the other hand, ST10 and ST131 were significantly more frequently isolated from fecal samples of healthy volunteers. ST69 was more commonly observed among fecal isolates (15%) than UTI isolates (7%), but the difference was borderline significant (p = .08).
Interestingly, we observed that the frequency of major genotypes of fecal E. coli isolates from this healthy population fluctuated month to month (Figure 3). Such an observation suggests external common‐source exposures or “outbreaks” of intestinal colonization. We have previously suggested that CA‐UTI may indeed occur as outbreaks (Manges, Natarajan, Solberg, Dietrich, & Riley, 2006; Smith, Manges, & Riley, 2008; Yamaji et al., 2018). Thus, cases of CA‐UTI may represent a subset of people intestinally colonized with ExPEC strains circulating in the community. Among similar numbers of isolates typed by MLST each month, ST69 peaked in August 2018 only to disappear completely by October 2018 (Figure 3). ST131 was observed in August and September but not in October. In May, 10 (83%) of all 12 fecal isolates belonged to ST10, but only three ST10 isolates were identified during 3 months from August through October. These observations suggest that intestinal colonization by ExPEC genotypes is common, but that UTI is consistently caused by a limited set of these genotypes, some of which were not found at all in the feces (e.g., ST73) or found in very low frequency (ST95). Commensal E. coli would be expected to persistently colonize the intestine. The changing monthly frequency of isolated fecal E. coli genotypes from healthy volunteers in this university community suggests that these genotypes are not likely to be members of the human commensal intestinal flora, but rather strains orally introduced into the residents of this community from external sources, such as contaminated food or environment.
A previous study in this same community has shown that 12 sequence types including ST10, ST69, and ST131 were shared between UTI and retail meat (pork, chicken, beef, and turkey) E. coli isolates (Yamaji et al., 2018). Other studies have shown ExPEC strain types shared by food E. coli isolates (Bergeron et al., 2012; Jakobsen et al., 2010, 2012; Leverstein‐van Hall et al., 2011; Maluta et al., 2014; Ramchandani et al., 2005; Vincent et al., 2010). Once introduced into the intestine, these strains may establish colonization for some time but then may be replaced later by other E. coli genotypes introduced into the intestine by food. A recent study has shown that ESBL‐producing ST131 strain was detected in the intestine of a 74‐year‐old woman for over 5 years, although repeated exposures to sources containing this genotype could not be ruled out (Forde et al., 2019).
Nielsen et al. (2017) examined fecal E. coli isolates from healthy volunteers with no previous history of UTI and compared their whole‐genome sequences to those of E. coli isolates from feces and urine of patients with UTI residing in the same community. They found that fecal and urine isolates from patients with UTI as well as fecal isolates from healthy volunteers were closely related and that they could be distinguished, if at all, only by their accessory genes in the genome (Nielsen et al., 2017). Our study analyzed fecal E. coli isolates from a college population at multiple time points, which enabled us to discover that human intestinal E. coli genotypes change over time. Thus, the intestinal microbiota may constitute a commensal population at the species level but may have a more dynamic population structure at the subspecies level.
We found ST1193 to be also common (7%) among the 90 urine E. coli isolates. ST1193 has not been previously found in this community based on past studies conducted in 1999–2000 or from a more recent study from 2016 to 2017 (Yamaji et al., 2018). Two ST1193 isolates were found among our fecal isolates. ST1193 is an emerging lineage of fluoroquinolone‐resistant E. coli belonging to phylogenetic group B2, reported in several countries, including Australia, China, South Korea, Norway, Germany, and the United States since 2012 (Jørgensen et al., 2017; Platell et al., 2012; Tchesnokova et al., 2019; Valenza et al., 2019; Wu, Lan, Lu, He, & Li, 2017). Again, the new appearance of this genotype of UTI‐causing E. coli in this community suggests that UPECs are not members of the human commensal E. coli.
ST69 and ST131 are internationally recognized as multiresistant ExPEC. A previous study conducted at the same college community between 2016 and 2017 found that 17 of 29 ST69 obtained from UTI samples contained at least one β‐lactamase gene. Similarly, nine of 14 ST131 harbored at least one β‐lactamase gene. In the present study, we found fecal isolates of ST69 and ST131 to frequently harbor the same β‐lactamase genes (Table 2).
One limitation of our study is that we did not obtain fecal samples from UTI patients. However, as others have found (Moreno et al., 2008; Nielsen et al., 2014, 2017), we would expect to find the same UTI genotypes from such patients fecal samples during the UTI episode or shortly afterward. Instead, we analyzed fecal E. coli isolates from healthy volunteers residing in the same community and found that while the overall genotypic distribution was similar, the frequency of genotypes of major ExPEC genotypes differed significantly between intestinal and UTI isolates.
Another limitation of this study is the relatively small sample size and short observation period. Sixteen UTI isolates could not be assigned an ST designation. Nevertheless, we were able to demonstrate statistically significant differences in nearly all (ST10, ST69, ST95, ST127, and ST131) of the major pandemic lineages of ExPEC. We did not find ST73 isolates from fecal samples. In another study based on the whole‐genome sequence of UPEC isolates from patients and fecal isolates from healthy controls, a few clonal complex 73 isolates were found in the fecal environment, but they were significantly more common among UTI isolates (Nielsen et al., 2017). These findings are consistent with our study.
In conclusion, this study showed that several UPEC isolates, including pandemic ExPEC strains, colonize the human intestine of healthy people and that they fluctuate over time. During the colonization period, some of the individuals may develop UTI. These observations, therefore, suggest a need to identify sources of ExPEC outside of the human intestine, including food and the environment. A more detailed understanding of the relationship between commensal E. coli and ExPEC can lead to novel and effective control strategies to prevent this ever‐increasing public health problem.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Yusuke Matsui: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); writing – original draft (equal); writing – review and editing (equal). Yuan Hu: Data curation (equal); investigation (equal). Julia Rubin: data curation (equal); investigation (equal). Reginara Souza de Assis: Data curation (equal); investigation (equal). Joy Suh: Data curation (equal); investigation (equal). Lee W. Riley: Conceptualization (equal); funding acquisition (equal); methodology (equal); supervision (equal); writing – original draft (equal).
ETHICS STATEMENT
The study protocol was approved by the Committee for Protection of Human Subjects of the University of California, Berkeley (protocol number 2017‐08‐10264), and all participants were informed about the study and signed the study consent forms.
ACKNOWLEDGMENTS
We thank staff members of the Tang Center of UC Berkeley‐affiliated health care service for their time and support of this project. We also thank the University instructors who were instrumental in recruiting healthy volunteers for this study. We thank all the members of the Riley laboratory for fruitful discussions and their contributions to this study. This study was supported by the Centers for Disease Control and Prevention's Combating Antibiotic Resistance BAA 200‐2016‐91939.
APPENDIX 1.
TABLE A1.
Number of Escherichia coli genotyped by MLST from healthy volunteers (fecal samples) and patients with UTIs (urine samples) at a northern California university community
| Genotype | Fecal samples | Urine samples |
|---|---|---|
| 10 | 17 | 4 |
| 12 | 6 | 5 |
| 13 | 1 | 0 |
| 25 | 1 | 0 |
| 38 | 3 | 1 |
| 59 | 0 | 1 |
| 62 | 0 | 1 |
| 69 | 13 | 6 |
| 73 | 0 | 15 |
| 93 | 1 | 0 |
| 95 | 2 | 10 |
| 101 | 0 | 2 |
| 127 | 5 | 13 |
| 131 | 8 | 2 |
| 141 | 2 | 0 |
| 174 | 1 | 0 |
| 349 | 2 | 0 |
| 355 | 0 | 1 |
| 372 | 0 | 1 |
| 379 | 1 | 0 |
| 420 | 0 | 4 |
| 453 | 0 | 1 |
| 537 | 0 | 1 |
| 538 | 1 | 0 |
| 603 | 0 | 1 |
| 648 | 4 | 2 |
| 1193 | 2 | 6 |
| 1231 | 2 | 0 |
| 1236 | 0 | 1 |
| 1300 | 1 | 0 |
| 1457 | 0 | 1 |
| 1688 | 1 | 0 |
| 1800 | 0 | 1 |
| 2076 | 1 | 0 |
| 2165 | 1 | 0 |
| 2554 | 1 | 0 |
| 2619 | 1 | 0 |
| 2861 | 0 | 1 |
| 3057 | 0 | 1 |
| 3268 | 1 | 0 |
| 3478 | 0 | 1 |
| 3576 | 1 | 0 |
| 3646 | 0 | 1 |
| 3877 | 3 | 0 |
| 4623 | 0 | 1 |
| 6807 | 1 | 0 |
| 6886 | 0 | 2 |
| 6999 | 0 | 1 |
| 7392 | 2 | 0 |
| 7445 | 0 | 1 |
| 8110 | 1 | 0 |
| 8272 | 1 | 0 |
| 8395 | 0 | 1 |
| Total | 88 | 90 |
Matsui Y, Hu Y, Rubin J, de Assis RS, Suh J, Riley LW. Multilocus sequence typing of Escherichia coli isolates from urinary tract infection patients and from fecal samples of healthy subjects in a college community. MicrobiologyOpen. 2020;9:e1032 10.1002/mbo3.1032
DATA AVAILABILITY STATEMENT
All data are provided in full in the results section of this paper.
REFERENCES
- Adams‐Sapper, S. , Sergeevna‐Selezneva, J. , Tartof, S. , Raphael, E. , Diep, B. A. , Perdreau‐Remington, F. , & Riley, L. W. (2012). Globally dispersed mobile drug‐resistance genes in gram‐negative bacterial isolates from patients with bloodstream infections in a US urban general hospital. Journal of Medical Microbiology, 61, 968–974. 10.1099/jmm.0.041970-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron, C. R. , Prussing, C. , Boerlin, P. , Daignault, D. , Dutil, L. , Reid‐Smith, R. J. , … Manges, A. R. (2012). Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerging Infectious Diseases, 18, 415–421. 10.3201/eid1803.111099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruxvoort, K. J. , Bider‐Canfield, Z. , Casey, J. A. , Qian, L. , Pressman, A. , Liang, A. S. , … Tartof, S. Y. (2019). Outpatient urinary tract infections in an era of virtual health care: Trends from 2008 to 2017. Clinical Infectious Diseases, ciz764 10.1093/cid/ciz764. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Wu, M. , Henderson, J. P. , Hooton, T. M. , Hibbing, M. E. , Hultgren, S. J. , & Gordon, J. I. (2013). Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Science Translational Medicine, 5, 184ra160 10.1126/scitranslmed.3005497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallenne, C. , Da Costa, A. , Decré, D. , Favier, C. , & Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β‐lactamases in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy, 65, 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Doumith, M. , Day, M. , Ciesielczuk, H. , Hope, R. , Underwood, A. , Reynolds, R. , … Woodford, N. (2015). Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. Journal of Clinical Microbiology, 53, 160–166. 10.1128/JCM.02562-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores‐Mireles, A. L. , Walker, J. N. , Caparon, M. , & Hultgren, S. J. (2015). Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nature Reviews Microbiology, 13, 269–284. 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde, B. M. , Roberts, L. W. , Phan, M. D. , Peters, K. M. , Fleming, B. A. , Russell, C. W. , … Beatson, S. A. (2019). Population dynamics of an Escherichia coli ST131 lineage during recurrent urinary tract infection. Nature Communications, 10, 3643 10.1038/s41467-019-11571-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, B. (1990). Recurring urinary tract infection: Incidence and risk factors. American Journal of Public Health, 80, 331–333. 10.2105/ajph.80.3.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, B. (2010). The epidemiology of urinary tract infection. Nature Reviews Urology, 7, 653–660. 10.1038/nrurol.2010.190 [DOI] [PubMed] [Google Scholar]
- Frank, J. A. , Reich, C. I. , Sharma, S. , Weisbaum, J. S. , Wilson, B. A. , & Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Applied and Environment Microbiology, 74, 2461–2470. 10.1128/AEM.02272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibreel, T. M. , Dodgson, A. R. , Cheesbrough, J. , Fox, A. J. , Bolton, F. J. , & Upton, M. (2012). Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. Journal of Antimicrobial Chemotherapy, 67, 346–356. 10.1093/jac/dkr451 [DOI] [PubMed] [Google Scholar]
- Hooton, T. (1990). Recurrent urinary tract infection in women. International Journal of Antimicrobial Agents, 17, 259–268. 10.1016/s0924-8579(00)00350-2 [DOI] [PubMed] [Google Scholar]
- Jakobsen, L. , Garneau, P. , Bruant, G. , Harel, J. , Olsen, S. S. , Porsbo, L. J. , … Frimodt‐Moller, N. (2012). Is Escherichia coli urinary tract infection a zoonosis? Proof of direct link with production animals and meat. European Journal of Clinical Microbiology and Infectious Diseases, 31, 1121–1129. 10.1007/s10096-011-1417-5 [DOI] [PubMed] [Google Scholar]
- Jakobsen, L. , Spangholm, D. J. , Pedersen, K. , Jensen, L. B. , Emborg, H. D. , Agerso, Y. , … Frimodt‐Moller, N. (2010). Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community‐dwelling humans and UTI patients. International Journal of Food Microbiology, 142, 264–272. 10.1016/j.ijfoodmicro.2010.06.025 [DOI] [PubMed] [Google Scholar]
- Jørgensen, S. , Sunde, M. , Fladberg, Ø. , Leegaard, T. , Berg, E. , & Steinbakk, M. (2017). Fluoroquinolone resistant Escherichia coli ST1193‐another global successful clone? 27th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), P0204. [Google Scholar]
- Leverstein‐van Hall, M. A. , Dierikx, C. M. , Cohen Stuart, J. , Voets, G. M. , van den Munckhof, M. P. , van Essen‐Zandbergen, A. , … National ESBL Surveillance Group (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clinical Microbiology & Infection, 17, 873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- Maluta, R. P. , Logue, C. M. , Casas, M. R. , Meng, T. , Guastalli, E. A. , Rojas, T. C. , … da Silveira, W. D. (2014). Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra‐intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE, 9, e105016 10.1371/journal.pone.0105016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges, A. R. , Geum, H. M. , Guo, A. , Edens, T. J. , Fibke, C. D. , & Pitout, J. D. D. (2019). Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clinical Microbiology Reviews, 32, e00135‐18 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges, A. R. , Natarajan, P. , Solberg, O. D. , Dietrich, P. S. , & Riley, L. W. (2006). The changing prevalence of drug‐resistant Escherichia coli clonal groups in a community: Evidence for community outbreaks of urinary tract infections. Epidemiology and Infection, 134, 425–431. 10.1017/S0950268805005005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, E. , Andreu, A. , Pigrau, C. , Kuskowski, M. A. , Johnson, J. R. , & Prats, G. (2008). Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. Journal of Clinical Microbiology, 46, 2529–2534. 10.1128/JCM.00813-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K. L. , Dynesen, P. , Larsen, P. , & Frimodt‐Moller, N. (2014). Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. Journal of Medical Microbiology, 63, 582–589. 10.1099/jmm.0.068783-0 [DOI] [PubMed] [Google Scholar]
- Nielsen, K. L. , Stegger, M. , Kiil, K. , Godfrey, P. A. , Feldgarden, M. , Lilje, B. , … Frimodt‐Moller, N. (2017). Whole‐genome comparison of urinary pathogenic Escherichia coli and faecal isolates of UTI patients and healthy controls. International Journal of Medical Microbiology, 307, 497–507. 10.1016/j.ijmm.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen, B. , Scheutz, F. , Andersen, R. L. , Menard, M. , Boisen, N. , Johnston, B. , … Johnson, J. R. (2012). Enteroaggregative Escherichia coli O78:H10, the cause of an outbreak of urinary tract infection. Journal of Clinical Microbiology, 50, 3703–3711. 10.1128/JCM.01909-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platell, J. L. , Trott, D. J. , Johnson, J. R. , Heisig, P. , Heisig, A. , Clabots, C. R. , … Cobbold, R. N. (2012). Prominence of an O75 clonal group (clonal complex 14) among non‐ST131 fluoroquinolone‐resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrobial Agents and Chemotherapy, 56, 3898–3904. 10.1128/AAC.06120-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani, M. , Manges, A. , DebRoy, C. , Smith, S. , Johnson, J. , & Riley, L. (2005). Possible animal origin of human‐associated, multidrug resistant, uropathogenic Escherichia coli . Clinical Infectious Diseases, 40, 251–257. 10.1086/426819 [DOI] [PubMed] [Google Scholar]
- Raphael, E. , Wong, L. K. , & Riley, L. W. (2011). Extended‐spectrum Bet alactamase gene sequences in gram‐negative saprophytes on retail organic and nonorganic spinach. Applied and Environment Microbiology, 77, 1601–1607. 10.1128/AEM.02506-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, J. , Mussio, K. , Xu, Y. , Suh, J. , & Riley, L. W. (2020). Prevalence of antimicrobial resistance genes and integrons in commensal gram‐negative bacteria in a college community. Microbial Drug Resistance. 10.1089/mdr.2019.0279. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Russo, T. , Stapleton, A. , Wenderoth, S. , Hooton, T. M. , & Stamm, W. E. (1995). Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. Journal of Infectious Diseases, 172, 440–445. 10.1093/infdis/172.2.440 [DOI] [PubMed] [Google Scholar]
- Smith, S. P. , Manges, A. R. , & Riley, L. W. (2008). Temporal changes in the prevalence of community‐acquired antimicrobial‐resistant urinary tract infection affected by Escherichia coli clonal group composition. Clinical Infectious Diseases, 46, 689–695. 10.1086/527386 [DOI] [PubMed] [Google Scholar]
- Tartof, S. Y. , Solberg, O. D. , Manges, A. R. , & Riley, L. W. (2005). Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. Journal of Clinical Microbiology, 43, 5860–5864. 10.1128/JCM.43.12.5860-5864.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokova, V. L. , Rechkina, E. , Larson, L. , Ferrier, K. , Weaver, J. L. , Schroeder, D. W. , … Sokurenko, E. V. (2019). Rapid and extensive expansion in the United States of a new multidrug‐resistant Escherichia coli clonal group, sequence type 1193. Clinical Infectious Diseases, 68, 334–337. 10.1093/cid/ciy525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza, G. , Werner, M. , Eisenberger, D. , Nickel, S. , Lehner‐Reindl, V. , Holler, C. , & Bogdan, C. (2019). First report of the new emerging global clone ST1193 among clinical isolates of extended‐spectrum bet alactamase (ESBL)‐producing Escherichia coli from Germany. Journal of Global Antimicrobial Resistance, 17, 305–308. 10.1016/j.jgar.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Vincent, C. , Boerlin, P. , Daignault, D. , Dozois, C. M. , Dutil, L. , Galanakis, C. , … Manges, A. R. (2010). Food reservoir for Escherichia coli causing urinary tract infections. Emerging Infectious Diseases, 16, 88–95. 10.3201/eid1601.091118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L. A. , & Sharp, P. M. (2006). Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: Evolution and implications for ERIC‐PCR. Molecular Biology and Evolution, 23, 1156–1168. 10.1093/molbev/msj125 [DOI] [PubMed] [Google Scholar]
- Wu, J. , Lan, F. , Lu, Y. , He, Q. , & Li, B. (2017). Molecular characteristics of ST1193 clone among phylogenetic group B2 non‐ST131 fluoroquinolone‐resistant Escherichia coli . Frontiers in Microbiology, 8, 2294 10.3389/fmicb.2017.02294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji, R. , Friedman, C. R. , Rubin, J. , Suh, J. , Thys, E. , McDermott, P. , … Riley, L. W. (2018). A population‐based surveillance study of shared genotypes of Escherichia coli isolates from retail meat and suspected cases of urinary tract infections. mSphere, 3, e00179‐18 10.1128/mSphere.00179-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S. , Tsukamoto, T. , Terai, A. , Kurazono, H. , Takeda, Y. , & Yoshida, O. (1997). Genetic evidence supporting the fecal perineal urethral hypothesis in cystitis caused by Escherichia coli . Journal of Urology, 157, 1127–1129. 10.1016/S0022-5347(01)65154-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in full in the results section of this paper.


