Abstract
Obstructive sleep apnea (OSA) has been linked to dysregulated metabolic states, and treatment of sleep apnea may improve these conditions. Subcutaneous adipose tissue is a readily samplable fat depot that plays an important role in regulating metabolism. However, neither the pathophysiologic consequences of OSA nor the effects of continuous positive airway pressure (CPAP) in altering this compartment’s molecular pathways are understood. This study aimed to systematically identify subcutaneous adipose tissue transcriptional programs modulated in OSA and in response to its effective treatment with CPAP. Two subject groups were investigated: Study Group 1 was comprised of 10 OSA and 8 controls; Study Group 2 included 24 individuals with OSA studied at baseline and following CPAP. For each subject, genome-wide gene expression measurement of subcutaneous fat was performed. Differentially activated pathways elicited by OSA (Group 1) and in response to its treatment (Group 2) were determined using network and Gene Set Enrichment Analysis (GSEA). In Group 2, treatment of OSA with CPAP improved apnea-hypopnea index, daytime sleepiness, and blood pressure, but not anthropometric measures. In Group 1, GSEA revealed many up-regulated gene sets in OSA subjects, most of which were involved in immuno-inflammatory (e.g. interferon-γ signaling), transcription, and metabolic processes such as adipogenesis. Unexpectedly, CPAP therapy in Group 2 subjects was also associated with up-regulation of several immune pathways as well as cholesterol biosynthesis. Collectively, our findings demonstrate that OSA alters distinct inflammatory and metabolic programs in subcutaneous fat, but these transcriptional signatures are not reversed with short-term effective therapy.
Keywords: sleep apnea, subcutaneous adipose, gene expression, CPAP, microarray, pathway
Statement of Significance.
Obstructive sleep apnea is linked to metabolic abnormalities and subcutaneous adipose tissue is an important regulator of metabolism. In this study, we investigated how the presence of sleep apnea changes gene expression in subcutaneous fat and whether CPAP therapy influences this signal. Identifying the genes and pathways altered in adipose tissue due to obstructive sleep apnea and in response to its effective treatment can help elucidate molecular mechanisms driving metabolic and cardiovascular morbidities associated with this complex disorder.
Introduction
Obstructive sleep apnea (OSA) is commonly associated with insulin resistance and type 2 diabetes independent of obesity [1, 2], and emerging data suggest that OSA may be an independent risk factor for incident type 2 diabetes [3, 4] The mechanisms by which OSA may impact diabetes risk are not completely clear, but effects on adipose tissue represent one potential pathway. Growing evidence suggests adipose tissue hypoxia through an outstripping of blood supply plays a key role in mediating the development of inflammation and insulin resistance in the setting of obesity [5, 6], and clearly, the recurrent breathing stoppages seen in OSA may further exacerbate adipose tissue hypoxia. Given the known importance of adipose tissue in regulating inflammation and insulin resistance [7], it is highly likely that many of the systemic effects of OSA are due to changes in adipocyte biology. Adipose tissue is a major site of production of pro-inflammatory mediators and modulators of insulin resistance such as adiponectin [8, 9]. In addition, the regulatory role of adipose tissue on free fatty acid levels can importantly affect insulin resistance [10, 11]. We have previously demonstrated that OSA is associated with an inflammatory and insulin-resistant expression phenotype in visceral adipose tissue [12]. However, reversibility of this phenotype cannot be easily demonstrated given the difficulties in accessing visceral adipose tissue in a longitudinal fashion. In contrast, subcutaneous fat can be repeatedly sampled through percutaneous biopsies. In this study, we assessed the cross-sectional association of OSA with the transcriptome in subcutaneous adipose tissue as well as the impact of OSA therapy using continuous positive airway pressure (CPAP) in order to better define the causal effects of OSA on adipose tissue function.
Methods
Protocol for Study Group 1
Adult patients with a body mass index (BMI) ≥ 20 kg/m2 and on no OSA treatment scheduled to undergo ventral hernia repair surgery were recruited from a general surgery clinic to undergo sleep apnea screening and abdominal subcutaneous fat biopsy under general anesthesia at the time of hernia repair [12]. Study procedures were approved by the University Hospitals Case Medical Center institutional review board and informed consent was obtained from each subject. OSA severity was assessed using the ARES Unicorder (Watermark Medical, Boca Raton, FL) a previously validated portable sleep monitor worn two consecutive nights prior to surgery [13, 14]. Respiratory events were defined as a ≥50% decrease in airflow for ≥ 10 seconds as assessed by nasal pressure associated with a ≥3% decrease in oxygen saturation. The respiratory disturbance index (RDI) was calculated as the total number of respiratory events divided by total recording time in hours averaged over both nights of recording. Subcutaneous fat biopsies were obtained at the initiation of surgery using the incisions made for the procedure. Biopsy tissue was rinsed in PBS to remove excess blood, minced, and immediately frozen in liquid nitrogen.
Protocol for Study Group 2
Adult patients with a BMI ≥ 25 kg/m2 diagnosed with severe OSA as evidenced by an apnea-hypopnea index (AHI) ≥30 event/hour on overnight polysomnography and ≥2% of total sleep time spent with oxyhemoglobin saturation <90%, planning treatment with CPAP and not taking medications (e.g. thiazolidinediones, glucocorticoids) that alter adipose tissue function were recruited from a sleep disorders clinic [15]. Study procedures were approved by the University Hospitals Case Medical Center institutional review board and informed consent was obtained from each subject. All participants underwent in-laboratory overnight polysomnography (Compumedics, Abbottsford AU) with lights off at 10:30 pm and lights on at 06:30 am using oronasal thermocouple and nasal pressure to assess airflow and inductive plethysmography to assess respiratory effort. Apneas were defined as complete cessation of airflow, and hypopneas were defined as a ≥ 30% decrease in airflow ≥ 10 seconds associated with a ≥3% decrease in oxygen saturation. The AHI was computed by dividing the total number of apneas and hypopneas by total sleep time.
A percutaneous biopsy of the subcutaneous adipose tissue was performed at 07:00 am in the peri-umbilical region using a 13 g needle. The tissue was immediately rinsed with PBS to remove excess blood and then frozen in liquid nitrogen and stored at −80°C.
Following the biopsy, patients were initiated on CPAP therapy. After at least 2 weeks of self-reported CPAP usage greater than 4 hours per night, a follow-up overnight visit was performed identical to the initial visit except the subject used CPAP during the overnight PSG. The follow-up biopsy was performed on the opposite side of the abdomen from the initial site. CPAP adherence data were downloaded from the subject’s machine and nightly usage was averaged over the 2 weeks prior to the follow-up visit.
RNA isolation and microarray experiments
Total RNA was isolated from subcutaneous adipose tissue in both experiments using RNeasy Lipid Tissue Mini Kit with DNase treatment (Qiagen, Valencia, California) according to the manufacturer’s protocol. The integrity of purified total RNA samples was assessed qualitatively on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and verified by the presence of two discrete electropherogram peaks corresponding to the 28S and 18S rRNA at a ratio approaching 2:1. Of note, in Study Group 2, RNA extraction was only performed from samples of patients who had maintained an average of > 4 hours of CPAP use per night over the 2 weeks prior to the second biopsy.
RNA samples were processed using sense target GeneChip Human Gene 1.0 ST arrays (Affymetrix, Inc., Santa Clara, CA), which analyze the expression levels of 36 079 transcripts from 21 014 annotated genes. For each sample, 100 ng of total RNA was labeled using Affymetrix GeneChip Whole Transcript (WT) Sense Target Labeling Assay, which included cRNA synthesis and generation of amplified and biotinylated sense-strand DNA targets, covering the entire expression genome. After hybridization and scanning, image acquisition was performed using the GeneChip Operating System (GCOS). Gene expression levels from probe intensities were estimated using a Robust Multiarray Analysis (RMA) method with quantile normalization and background correction [16]. Detailed information meeting Minimum Information About a Microarray Experiment (MIAME) requirements has been deposited for both experiments at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/, GSE135917).
Data analysis
Multidimensional scaling.
To assess whether the presence of OSA influenced global transcriptional responses, we applied both unsupervised [17] (correspondence analysis) and supervised [18] (partial least squares discriminant analysis, PLSDA) multidimensional scaling on subcutaneous fat transcriptome in Study Group 1 subjects (n = 10 OSA, n = 8 no OSA).
Gene-based statistical testing.
We applied empirical Bayes moderated t-statistics algorithm as implemented in the R package “Limma” [19] to test for differential gene expression between OSA vs. no OSA (Study 1) and paired analysis for pre vs. post-CPAP in OSA (Study 2). Probe intensities were log-transformed and a false discovery rate (FDR) < 0.05 cutoff was used to designate statistical significance.
Gene set enrichment analysis.
For each study group (OSA vs. no OSA; OSA: pre- vs. post-CPAP) we performed identical Gene Set Enrichment Analysis (GSEA) separately, using 1379 curated canonical biological pathways (e.g. Hallmark, Reactome, KEGG, Biocarta, etc.) [20]. We applied an FDR threshold < 0.05 to identify significantly enriched gene sets. We performed leading edge analysis in GSEA to identify the genes with the largest contribution to pathway enrichment [21]. Network-based visualization of GSEA results was achieved using Enrichment Map [22], a plug-in application within the bioinformatics software platform, Cytoscape [23].
Interaction network analysis.
We implemented gene product interaction analysis using Ingenuity Pathway Analysis software [24] (Qiagen Bioinformatics, Redwood City, CA) by combining leading edge gene members of “Interferon-γ” associated gene sets enriched in subcutaneous fat of OSA subjects relative to those without OSA (Study 1). To enhance biological relevance, we limited nodal relationships to those based on high confidence, experimentally verified direct interactions.
Results
Study subject demographics and CPAP adherence
There were 18 subjects in Study Group 1 (n = 10 OSA and n = 8 no OSA) as has been previously described [12]. As presented in Table 1, this cohort was middle-aged, primarily female and obese with mean (SD) BMI 35.7 (7.7) kg/m2 with no significant differences in age, sex, or BMI between groups. Of note, three of the control subjects and two of the OSA subjects were morbidly obese with BMI > 40 kg/m2. As expected, those found to have OSA on screening had greater mean RDI (19.2 vs. 0.6 events/hour, p = 0.05) and lower minimum O2 saturation (79.7% vs. 87.8%, p = 0.001).
Table 1.
Subject characteristics at baseline
| Study Group 1 | Study Group 2 | ||
|---|---|---|---|
| Control (n = 8) | OSA (n = 10) | OSA (n = 24) | |
| Age (years) | 54.5 ± 11.6 | 56.1 ± 10.8 | 49.4 ± 10.9 |
| Male | 1 (13%) | 3 (30%) | 13 (54%) |
| Body mass index (kg/m2) | 35.2 ± 5.8 | 36.1 ± 9.3 | 42.6 ± 9.4 |
| RDI or AHI (events/hour) | 0.6 ± 0.5 | 19.2 ± 25.9 | 41.5 ± 23.8 |
| Diabetes | 0 (0%) | 3 (30%) | 5 (21%) |
| Hypertension | 3 (38%) | 3 (30%) | 9 (38%) |
| Heart disease | 0 (0%) | 0 (0%) | 2 (8%) |
Study Group 1 consisted of individuals undergoing ventral hernia repair surgery. Study Group 2 consisted of individuals diagnosed with severe obstructive sleep apnea (OSA) getting initiated on continuous positive airway pressure (CPAP) therapy. Data reported as mean ± standard deviation or N (%). Respiratory disturbance index (RDI) reported for Study Group 1 and apnea hypopnea index (AHI) reported for Study Group 2.
There were 31 subjects initially recruited to Study Group 2. Of these, seven subjects had suboptimal CPAP adherence so follow-up fat biopsy was either not performed or the tissue was not analyzed. The 24 subjects included in this analysis were middle-aged, 45.8% female, and had severe OSA (Table 1). Mean BMI did not change between baseline and follow-up biopsies (42.6 vs. 42.3 kg/m2, p = 0.23). The follow-up biopsy occurred a mean of 44 (26) days after initiating CPAP therapy and mean adherence for the 2 weeks prior to biopsy was 6.6 (1.1) hours/night. Subject-specific demographics for both Study Groups are provided in Supplementary Table S1.
OSA elicits immuno-inflammatory and metabolic signatures in subcutaneous fat
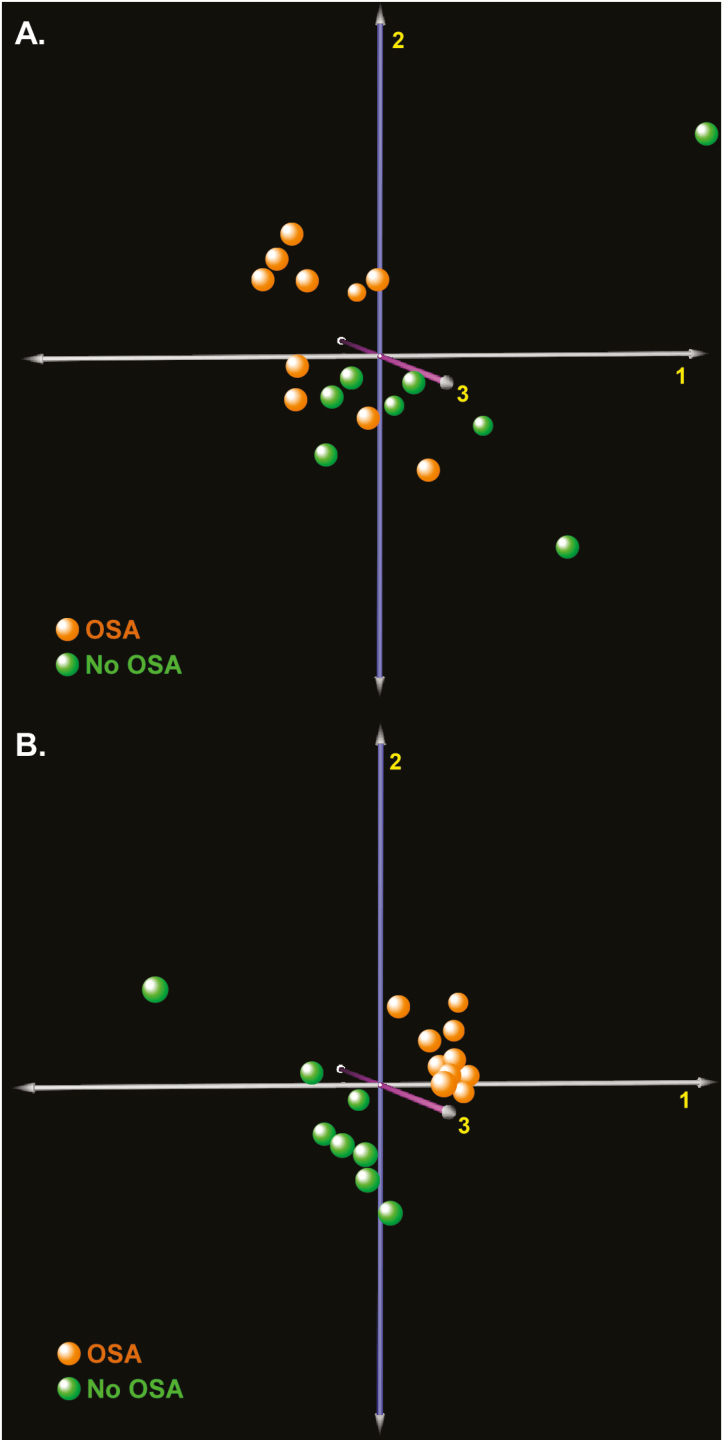
To evaluate the global consequences of OSA in subcutaneous adipose tissue transcriptome, we applied unsupervised and supervised multidimensional scaling to Study Group 1 (n = 10 OSA, n = 8 no OSA). As depicted in Figure 1, we found segregation between the subject groups, indicating that the presence of sleep apnea is associated with widespread changes in gene expression. Since the supervised algorithm (PLSDA) incorporates a priori information about the two subgroups, it outperforms the unsupervised (correspondence analysis) method in discriminating between subjects but may be susceptible to overfitting. Nevertheless, both classification approaches confirmed that OSA influences subcutaneous fat transcriptome.
Figure 1.
Multidimensional scaling of subcutaneous fat gene expression in subjects with OSA (n = 10) and those without OSA (n = 8) assesses whether sleep apnea alters global transcriptional signals. (A) Unsupervised correspondence analysis (COA) of subcutaneous fat transcriptome. (B) Supervised partial least squares discriminant analysis (PLSDA) of subcutaneous fat transcriptome improves separation between subjects with and without OSA.
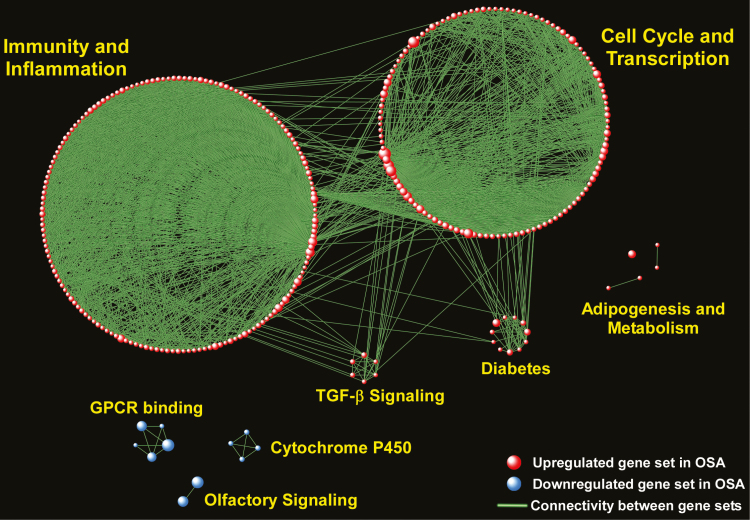
We next performed gene-based testing using the “Limma” statistical algorithm but did not find any differentially expressed genes after adjustment for multiple hypothesis testing, indicating modest changes in any single gene’s expression. However, most biological processes and complex diseases are the result of coordinated effects of multiple interacting genes [25, 26]. To map the landscape of pathways altered in OSA, we applied GSEA using the entire subcutaneous fat tissue transcriptome. We limited the enrichment analysis to well-curated canonical pathways (n = 1379) and identified 361 up-regulated and 22 down-regulated gene sets in OSA patients relative to subjects without OSA (complete list in Supplementary Tables S2 and S3). To visualize this large information, we performed network enrichment analysis by grouping gene sets that shared at least 25% of their member genes into larger biological modules with common functional themes (Figure 2). “Immunity and Inflammation” and “Cell Cycle and Transcription” were the largest up-regulated biological modules in OSA, but metabolism-associated processes such as “Adipogenesis” and “Diabetes” were also prominent, as was “TGF-β signaling.” Down-regulated modules included “Olfactory Signaling,” “Cytochrome P450,” and “G-protein Coupled Receptors.” Taken together, these results indicate that the presence of sleep apnea affects diverse pathways in subcutaneous fat tissue, with prominent up-regulation of immuno-inflammatory, cell cycle/transcription, and metabolic signals.
Figure 2.
Network-based visual depiction of gene set enrichment analysis (GSEA) of subcutaneous fat tissue in OSA subjects vs. those without OSA. Red spheres correspond to gene sets up-regulated and blue spheres to gene sets down-regulated in OSA. Connectivity between the gene sets is based on 25% or greater overlap among their member genes. Note that the topology of the network is characterized by the emergence of biological modules comprises of highly interconnected gene sets with similar functional themes. Notable modules up-regulated in OSA include “Immunity and Inflammation,” “Cell Cycle and Transcription,” “Adipogenesis,” “Diabetes,” TGF-β Signaling,” whereas down-regulated modules include “Olfactory Signaling,” “GPCR Binding,” and “Cytochrome P450.” Complete list of enriched gene sets in included in Supplementary Tables S2 and S3.
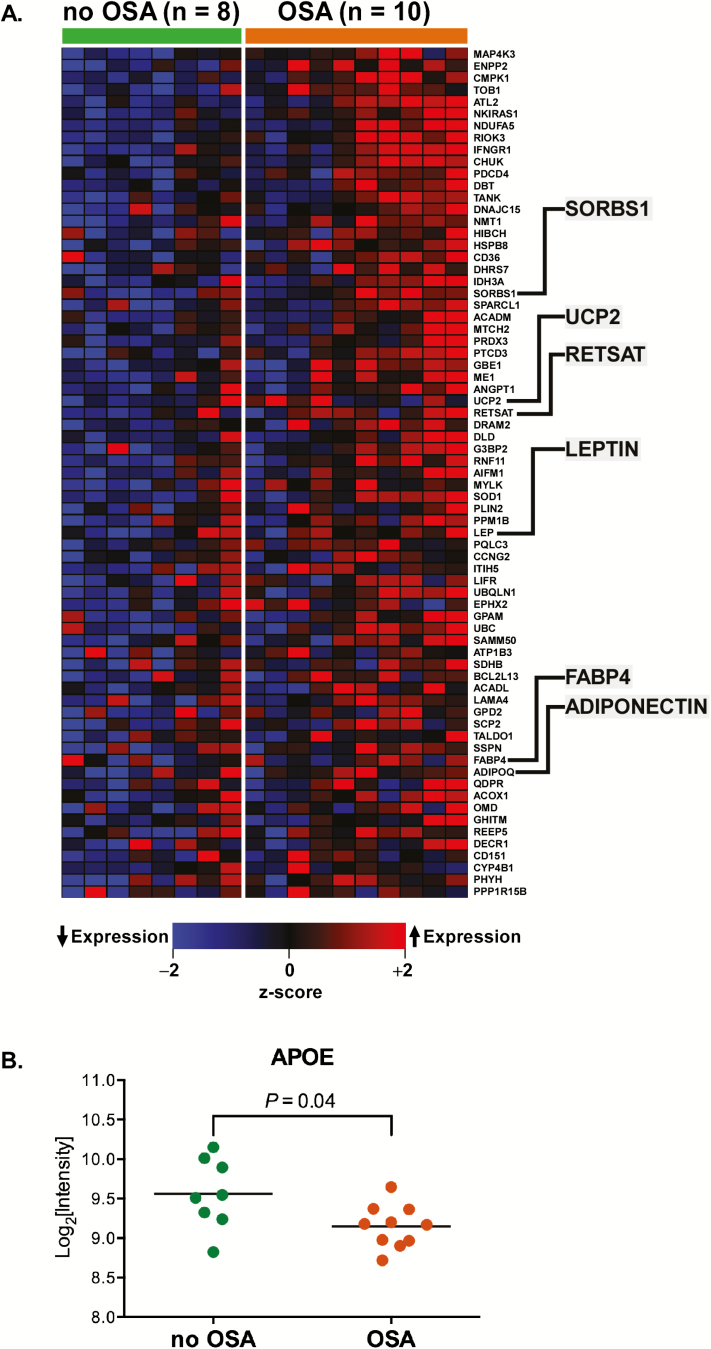
To gain more insight into specific altered processes, we performed “leading edge” analysis on the “Adipogenesis” gene set to identify the genes that were significant contributors to its enrichment. Figure 3A summarizes this analysis using a heatmap to display the expression patterns of adipogenesis-associated leading edge genes in subjects with and without OSA. These genes include several well-known regulators of adipogenesis such as leptin, adiponectin, retinol saturase, and fatty acid-binding protein 4 (Supplementary Table S4). However, not every member of the “Adipogenesis” gene set was up-regulated in OSA; for example, the expression of apolipoprotein E (APOE)—a key carrier of fat molecules—was down-regulated (Figure 3B).
Figure 3.
(A) Heatmap of the leading edge members of “Adipogenesis,” an up-regulated gene set in OSA subjects. Several representative genes have been highlighted (details provided in Supplementary Table S4). (B) However, some members of the “Adipogenesis” pathway, such as APOE, were down-regulated in OSA.
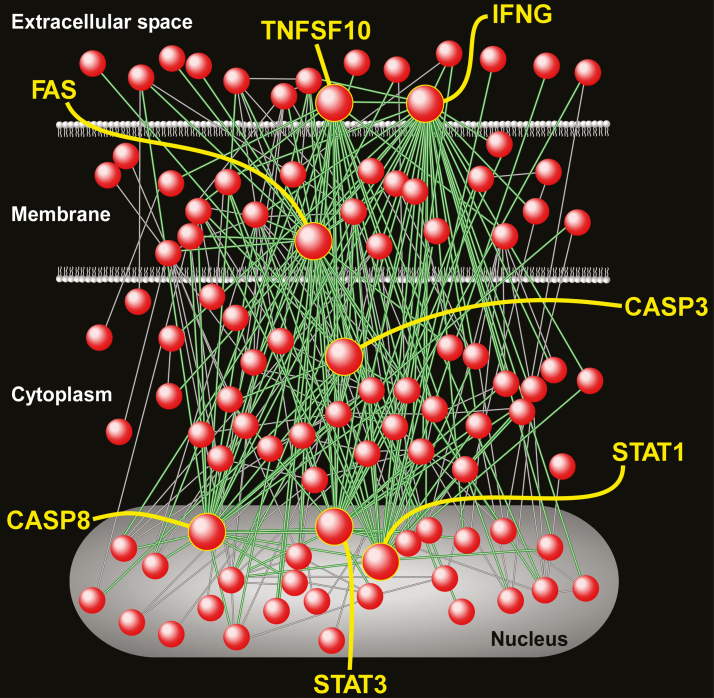
The “Immune and Inflammation” module represented the largest grouping of up-regulated gene sets and included many canonical processes such as “FAS pathway,” “T-cell receptor,” “Cytokine signaling in immune system,” “TNF pathway,” “Toll receptor cascade,” as well as several gene sets involved in interferon-γ signaling. To better elucidate the components of interferon-γ signaling cascade that are activated in the subcutaneous fat of OSA patients, we performed gene product interaction network analysis on the “leading edge” members of this gene set. The resultant relational network (Figure 4) demonstrated complex molecular interactions among its gene members, and was characterized by several highly connected nodes. These hubs may represent critical regulators of interferon-γ signaling in subcutaneous adipose tissue of individuals with OSA and included IFNG, FAS, STAT1, STAT3, TNFSF10, CASP3, and CASP8 (Supplementary Table S5).
Figure 4.
Gene product interaction network of “Interferon-γ” gene set—an up-regulated pathway in subcutaneous fat of OSA subjects relative to those without OSA. A number of the most highly connected nodes (known as hubs) have been labeled, including interferon-γ (IFNG), FAS, STAT1, STAT3, CASP3, CASP8, and TNFSF10. Note that the interactions among these densely connected nodes (shown as green lines) capture the majority of the network’s overall connectivity (green and gray lines). These hubs represent key drivers of OSA-induced activation of interferon-γ signaling in subcutaneous adipose tissue. Full list of network nodes is provided in Supplementary Table S5.
Subcutaneous and visceral fat share similar pathway enrichment patterns in sleep apnea
We previously reported on the transcriptional effects of OSA in visceral adipose tissue in the same subject cohort (Study Group 1) [12]. To compare those findings with the present data generated from subcutaneous fat, we re-analyzed the visceral fat gene expression data using identical pathway enrichment program (GSEA) and curated gene set databases. We identified 285 up-regulated gene sets (FDR < 0.05) in visceral adipose tissue of OSA patients, of which 228 (80%) were also up-regulated in subcutaneous fat of the same subjects, including many of the same immuno-inflammatory pathways such as interferon-γ signaling. Similarly, we found 32 down-regulated gene sets (FDR < 0.05) in visceral fat of OSA subjects, of which 15 (47%) were identical to pathways down-regulated in subcutaneous adipose tissue (including the top four most significant gene sets). The complete lists are provided in Supplementary Tables S6 and S7. Taken together, our results indicate that the transcriptional responses induced by OSA are broadly similar in adipose and subcutaneous fat tissue, although differences do exist. For example, the peroxisome proliferator-activated receptor (PPAR) pathway—a key regulator of metabolism—was down-regulated in visceral but not subcutaneous fat.
CPAP therapy alters distinct transcriptional programs in subcutaneous fat of OSA patients
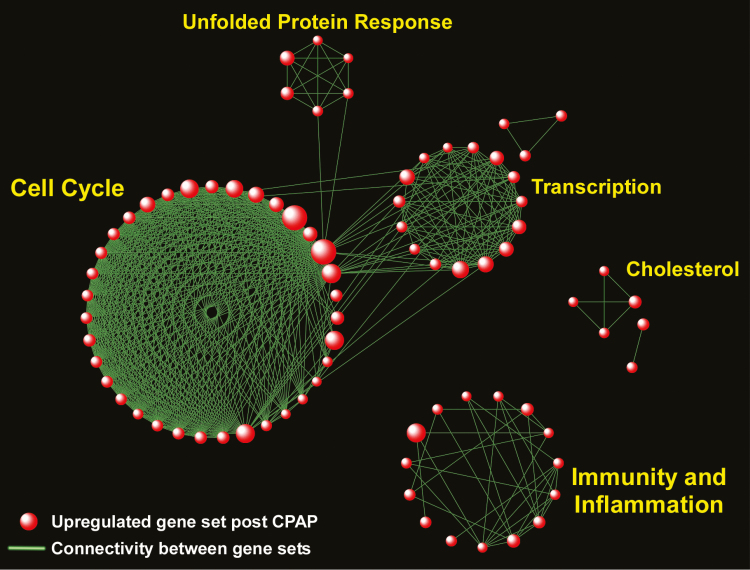
We evaluated the transcriptional consequences of effective sleep apnea treatment on subcutaneous adipose tissue of 24 subjects with OSA at baseline (pre-CPAP) and after CPAP (Study Group 2). Using a paired gene-specific statistical analysis (“Limma”), we did not observe any differentially expressed genes in pre- vs. post-CPAP comparison after adjustment for multiple hypothesis testing. We next applied GSEA to identify pathways enriched in response to CPAP therapy. We found 88 gene sets up-regulated in subcutaneous fat of OSA patients after therapeutic CPAP (FDR < 0.05), but no significantly down-regulated gene sets (Supplementary Table S8). Using the same network-based approach used for Study Group 1, we grouped enriched gene sets following CPAP use into larger functional aggregates representing biological modules (Figure 5). Surprisingly, several of the modules previously found to be up-regulated in the OSA vs. no OSA cohort (Study Group 1) were also enriched after CPAP treatment of OSA in Study Group 2, including “Cell Cycle,” “Transcription,” and “Immunity and Inflammation.” This observation suggests that immuno-inflammatory signals in subcutaneous fat are not moderated within the first few months of effective OSA therapy, but rather enhanced in response to CPAP.
Figure 5.
Network-based visualization of gene set enrichment analysis of subcutaneous fat tissue in OSA subjects following treatment with CPAP. Red spheres correspond to pathways up-regulated following CPAP therapy. No pathway was significantly down-regulated post-CPAP. Note the emergence of several biological modules in response to CPAP, including “Immunity and Inflammation,” “Cell Cycle,” “Transcription,” “Unfolded Protein Response,” and “Cholesterol.” Complete list of enriched gene sets in included in Supplementary Table S8.
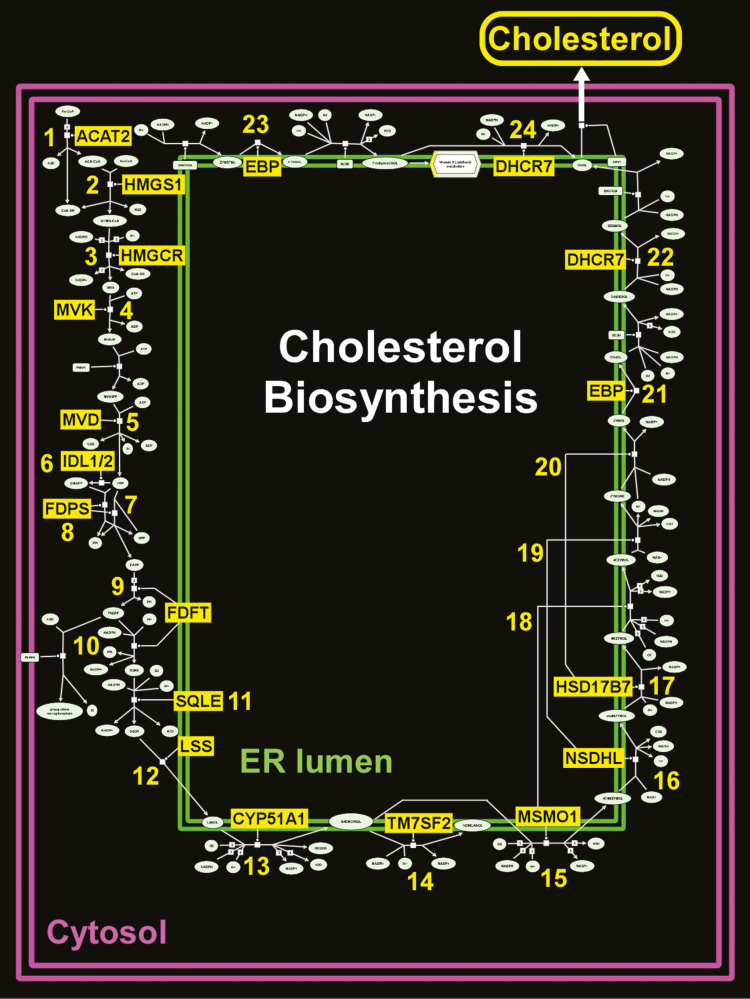
Several gene sets involved in cholesterol biosynthesis were also up-regulated after CPAP therapy in Study Group 2 (Figure 5). We performed “leading edge” analysis to identify the main drivers of this network and integrated our results within the canonical cholesterol biosynthesis pathway, as depicted in Figure 6. We found that up-regulated “leading edge” genes mapped to 24 key steps of the multi-component processes involved in steroid synthesis (Supplementary Table S9).
Figure 6.
Schematic diagram of cholesterol biosynthesis—a pathway that was up-regulated in OSA subjects treated with CPAP. Leading edge members of the “cholesterol biosynthesis” gene set have been highlighted in yellow background and populate 24 critical steps for steroid synthesis and are detailed in Supplementary Table S9. Adapted from Reactome database.
Discussion
To our knowledge, this study is the first comprehensive evaluation of the transcriptional profile of subcutaneous adipose tissue in OSA and its response to CPAP therapy. Leveraging two different subject cohorts, we found that while gene-specific expression changes were modest, pathway-based analyses revealed a diverse repertoire of differentially activated processes in both untreated OSA and following its short-term effective treatment.
Adipose tissue is a key regulator of metabolic state: increasing evidence implicates OSA as a disruptor of this homeostasis, with an important role in insulin resistance, diabetes, and obesity [27]. While much of the research efforts to date have focused on the visceral fat compartment, subcutaneous adipose tissue is also an important contributor to metabolism [28, 29]. Furthermore, repeated sampling of subcutaneous fat in human subjects is much more feasible than invasive biopsies of visceral adipose tissue and allowed us to assess the molecular consequences of pre- vs. post-CPAP therapy in OSA patients.
We initially compared subjects with versus without OSA and found global alterations in subcutaneous fat transcriptome due to the presence of sleep apnea. When applying gene-specific statistical models with multiple comparisons adjustments, we did not find significant differential gene expression, an observation that has been previously reported in genome-wide transcriptional profiling of adipose tissue [30]. However, gene set enrichment analysis revealed up-regulation of inflammatory and immune pathways in OSA, which is consistent with other reports of elevated circulating pro-inflammatory markers in sleep apnea and our previous study in visceral fat tissue [12, 31–33]. Activation of immuno-inflammatory signals in fat tissue has been well-characterized in obesity, insulin resistance and diabetes. Our study extends these findings and demonstrates that among subjects with similar BMI, those with OSA have profound enrichment of immune-related programs in their subcutaneous fat. As an example, interferon-γ-associated gene sets were highly enriched in our OSA cohort, and gene interaction network analysis revealed several putative drivers of interferon-γ signaling, such as STAT1/STAT3, in subcutaneous fat tissue of OSA subjects. Interestingly, interferon-γ activation in adipose has been recently implicated as a contributor to metabolic abnormalities in human obesity [34], in vitro studies have shown that interferon-γ suppresses insulin signaling and lipid storage in human adipocytes via STAT pathway [35], and animal experiments have established a critical role for interferon-γ in regulating fat inflammation [36].
Other up-regulated gene sets in OSA included those involved in transforming growth factor-β (TGF-β) signaling, which plays a multifaceted role in fat cells and obesity [37], as well as gene sets mapping to metabolic processes such as diabetes and adipogenesis. A number of well-known regulators of adipocyte biology and metabolism such as leptin [38], adiponectin [9, 39], and fatty acid-binding protein 4 [40], were up-regulated in OSA, indicating that the effects of sleep apnea in subcutaneous fat depots may have system-wide metabolic consequences.
We also found that a number of biological modules were down-regulated in OSA patients, with the topmost significant gene sets being involved in “Olfactory Signaling.” While the sensory role of smell has been shown to influence metabolic state [41], there is emerging evidence that olfactory receptors are also present in adipose tissue and may be directly involved in regulating adipogenesis, energy metabolism and obesity [42, 43]. Our findings, therefore, suggest another potential signaling mechanism by which OSA can alter fat tissue homeostasis.
We compared pathway enrichment between subcutaneous and visceral adipose tissue in Study Group 1 subjects (OSA vs. control), and observed generally similar up- and down-regulated patterns between the two fat depots. This finding implies that the more readily accessible subcutaneous fat may be a reasonable surrogate for sampling visceral adipose tissue in sleep apnea.
In a second set of experiments, we applied a similar transcriptomics analysis to subcutaneous fat tissue biopsies of 24 patients with OSA at baseline (pre-CPAP) and after effective therapy (post-CPAP). Unexpectedly, we observed that pathway enrichment after treatment with CPAP was characterized by up-regulation of immuno-inflammatory programs, as well as processes involved in transcription, cell cycle, and cholesterol synthesis. Activation of immune-associated gene sets such as “TNF signaling via NF-κB,” “AP1 pathway,” and several cytokine signaling processes following CPAP therapy indicates that the inflammatory signal associated with OSA in subcutaneous fat tissue is not attenuated with treatment, but maybe even enhanced. There may a number of explanations for this finding. First, the duration of therapy was relatively short, and the transcriptional effects of long-term CPAP use were not captured in our study. In this context, obtaining additional time points during CPAP therapy will be important in future studies. It is also possible that chronic disease exposure moderated the OSA-induced inflammatory state of adipose tissue and short-term treatment with CPAP elicited a paradoxical re-setting of pro-inflammatory programs. There is conflicting evidence on whether CPAP reduces inflammatory markers in sleep apnea [44–46]; for example, we did not find down-regulation of pro-inflammatory transcriptional signals in circulating leukocytes of OSA patients after effective CPAP therapy [15], and a more recent re-analysis of our original study using different statistical methods also did not find alteration of immune or inflammatory processes following CPAP therapy [47]. Furthermore, our study, as well as several other reports, have found that treatment of sleep apnea with CPAP does not result in weight loss [48, 49], and does not lead to reduced volume of visceral or subcutaneous fat content [50]. Since adipose tissue inflammation is a prominent characteristic of obesity, it is possible that the pro-inflammatory state of fat depots persists despite CPAP therapy. Interestingly, recent reports suggest that even with weight loss, the inflammatory profile of subcutaneous fat in humans may not become attenuated [51, 52].
Another prominent up-regulated module after CPAP therapy in OSA was “Cholesterol Biosynthesis.” When we mapped the “leading edge” genes that drive the enrichment of these cholesterol-associated pathways, we found that almost every step of steroid synthesis included one such gene member, highlighting the widespread effects of OSA treatment in modulating cholesterol production.
Our study has a number of limitations. The sample size for each Study Group was relatively small, and our findings need to be validated in larger cohorts before they can be generalized. Three of the OSA subjects in Study Group 1 had diabetes, whereas none of the controls were diabetic. It is possible that the presence of diabetes in the OSA subjects may have influenced the transcriptional profile of subcutaneous adipose tissue and confounded our results. Subcutaneous fat is a complex mixture of several cell types including adipocytes, macrophages, and connective tissue—each of which likely contributed to the overall gene expression that was measured in our study. Obtaining cell-specific transcriptional profiles of this fat compartment can provide more granular information and deeper insight into the role of subcutaneous adipose tissue constituents in OSA. Differences between the two Study Groups may also have impacted findings. Although both groups were obese, the CPAP intervention group was conducted in a population with morbid obesity (mean BMI 42.6 kg/m2). It is possible that this severity of obesity led to severe tissue hypoxia independent of OSA such that the impact of CPAP therapy was minimal. The duration of CPAP therapy in Study Group 2 was brief and only one time point after initiation of treatment was captured, precluding assessment of long-term consequences. Therefore, temporal changes in gene expression and pathway enrichment, and chronic molecular effects of CPAP therapy on subcutaneous fat were not investigated. We also did not have a “control” group of OSA subjects treated with sham CPAP in Study Group 2 to compare with subjects undergoing effective therapy. Finally, it is possible that the level of CPAP adherence achieved (mean 6.6 hours/night) was insufficient to fully eliminate the ongoing exposure to intermittent hypoxia and other OSA-associated physiologic insults. Nevertheless, this level of CPAP adherence does reflect what is typically observed in clinical populations initiated on CPAP and therefore reflects the impact of current approaches to OSA therapy.
Despite these limitations, our study represents the largest effort to date to investigate the transcriptional consequences of OSA and its treatment in subcutaneous fat tissue. We found that compared to subjects without sleep apnea, OSA patients have a pro-inflammatory and dysregulated metabolic transcriptional state in this important fat depot. However, the immuno-inflammatory signal associated with sleep apnea was not reversed with effective but short-term CPAP therapy, highlighting a complex relationship between the treatment of OSA and its molecular sequelae in adipose tissue.
Supplementary Material
Acknowledgments
We would like to thank all subject participants in this study.
Conflict of interest statement. This was not an industry-supported study. S.R.P. has received grant funding through his institution from Bayer Pharmaceuticals and Philips Respironics. S.A.G. has consulted for Revalesio Corporation. R.M.’s institution has received investigator-initiated grants and research support from Philips Respironics, Resmed, GE Healthcare, and Natus. She serves on the American Board of Internal Medicine examination committee and has received royalties from Up to Date, Inc. None of the other authors have any financial disclosures.
Funding
This work was funded by the American Thoracic Society (S.R.P.), NIH HL081385 (S.R.P.), HL127307 (S.R.P.), HL079114 (R.M.), HL125177 (R.M.), and AI137111 (S.A.G.).
References
- 1. Ip MS, et al. . Obstructive sleep apnea is independently associated with insulin resistance. Am J Respir Crit Care Med. 2002;165(5):670–676. [DOI] [PubMed] [Google Scholar]
- 2. Foster GD, et al. ; Sleep AHEAD Research Group. Obstructive sleep apnea among obese patients with type 2 diabetes. Diabetes Care. 2009;32(6):1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kendzerska T, et al. . Obstructive sleep apnea and incident diabetes. A historical cohort study. Am J Respir Crit Care Med. 2014;190(2):218–225. [DOI] [PubMed] [Google Scholar]
- 4. Xu PH, et al. . Incident type 2 diabetes in OSA and effect of CPAP treatment: a retrospective clinic cohort study. Chest. 2019;156(4):743–753. [DOI] [PubMed] [Google Scholar]
- 5. Hosogai N, et al. . Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes. 2007;56(4):901–911. [DOI] [PubMed] [Google Scholar]
- 6. Trayhurn P. Hypoxia and adipose tissue function and dysfunction in obesity. Physiol Rev. 2013;93(1):1–21. [DOI] [PubMed] [Google Scholar]
- 7. Tilg H, et al. . Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol. 2006;6(10):772–783. [DOI] [PubMed] [Google Scholar]
- 8. Hotamisligil GS, et al. . Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. [DOI] [PubMed] [Google Scholar]
- 9. Hu E, et al. . AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem. 1996;271(18):10697–10703. [DOI] [PubMed] [Google Scholar]
- 10. Randle PJ, et al. . The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1(7285):785–789. [DOI] [PubMed] [Google Scholar]
- 11. Ferrannini E, et al. . Effect of fatty acids on glucose production and utilization in man. J Clin Invest. 1983;72(5):1737–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gharib SA, et al. . A pathway-based analysis on the effects of obstructive sleep apnea in modulating visceral fat transcriptome. Sleep. 2013;36(1):23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westbrook PR, et al. . Description and validation of the apnea risk evaluation system: a novel method to diagnose sleep apnea-hypopnea in the home. Chest. 2005;128(4): 2166–2175. [DOI] [PubMed] [Google Scholar]
- 14. Ayappa I, et al. . Validation of a self-applied unattended monitor for sleep disordered breathing. J Clin Sleep Med. 2008;4(1):26–37. [PMC free article] [PubMed] [Google Scholar]
- 15. Gharib SA, et al. . Treatment of obstructive sleep apnea alters cancer-associated transcriptional signatures in circulating leukocytes. Sleep. 2014;37(4):709–714, 714A–714T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bolstad BM, et al. . A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. [DOI] [PubMed] [Google Scholar]
- 17. Fellenberg K, et al. . Correspondence analysis applied to microarray data. Proc Natl Acad Sci U S A. 2001;98(19):10781–10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chong J, et al. . MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ritchie ME, et al. . limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramanian A, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Subramanian A, et al. . GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23(23):3251–3253. [DOI] [PubMed] [Google Scholar]
- 22. Isserlin R, et al. . Enrichment map - a cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. 2014;3:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cline MS, et al. . Integration of biological networks and gene expression data using cytoscape. Nat Protoc. 2007;2(10):2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calvano SE, et al. ; Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–1037. [DOI] [PubMed] [Google Scholar]
- 25. Hartwell LH, et al. . From molecular to modular cell biology. Nature. 1999;402(6761 Suppl):C47–C52. [DOI] [PubMed] [Google Scholar]
- 26. Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461(7261):218–223. [DOI] [PubMed] [Google Scholar]
- 27. Ryan S, et al. . Adipose tissue as a key player in obstructive sleep apnoea. Eur Respir Rev. 2019;28(152):190006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ebbert JO, et al. . Fat depots, free fatty acids, and dyslipidemia. Nutrients. 2013;5(2):498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jialal I, et al. . Subcutaneous adipose tissue biology in metabolic syndrome. Horm Mol Biol Clin Investig. 2018;33(1):498–508. [DOI] [PubMed] [Google Scholar]
- 30. Arner P, et al. . Weight gain and impaired glucose metabolism in women are predicted by inefficient subcutaneous fat cell lipolysis. Cell Metab. 2018;28(1):45–54.e3. [DOI] [PubMed] [Google Scholar]
- 31. Murphy AM, et al. . Intermittent hypoxia in obstructive sleep apnoea mediates insulin resistance through adipose tissue inflammation. Eur Respir J. 2017;49(4):1601731. [DOI] [PubMed] [Google Scholar]
- 32. Shamsuzzaman AS, et al. . Elevated C-reactive protein in patients with obstructive sleep apnea. Circulation. 2002;105(21):2462–2464. [DOI] [PubMed] [Google Scholar]
- 33. Ryan S, et al. . Selective activation of inflammatory pathways by intermittent hypoxia in obstructive sleep apnea syndrome. Circulation. 2005;112(17):2660–2667. [DOI] [PubMed] [Google Scholar]
- 34. Wentworth JM, et al. . Interferon-gamma released from omental adipose tissue of insulin-resistant humans alters adipocyte phenotype and impairs response to insulin and adiponectin release. Int J Obes (Lond). 2017;41(12):1782–1789. [DOI] [PubMed] [Google Scholar]
- 35. McGillicuddy FC, et al. . Interferon gamma attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J Biol Chem. 2009;284(46):31936–31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rocha VZ, et al. . Interferon-gamma, a Th1 cytokine, regulates fat inflammation: a role for adaptive immunity in obesity. Circ Res. 2008;103(5):467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee MJ. Transforming growth factor beta superfamily regulation of adipose tissue biology in obesity. Biochim Biophys Acta Mol Basis Dis. 2018;1864(4 Pt A):1160–1171. [DOI] [PubMed] [Google Scholar]
- 38. Cui H, et al. . The cellular and molecular bases of leptin and ghrelin resistance in obesity. Nat Rev Endocrinol. 2017;13(6):338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kadowaki T, et al. . Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116(7):1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garin-Shkolnik T, et al. . FABP4 attenuates PPARγ and adipogenesis and is inversely correlated with PPARγ in adipose tissues. Diabetes. 2014;63(3):900–911. [DOI] [PubMed] [Google Scholar]
- 41. Riera CE, et al. . The sense of smell impacts metabolic health and obesity. Cell Metab. 2017;26(1):198–211.e5. [DOI] [PubMed] [Google Scholar]
- 42. Wu C, et al. . Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J Clin Invest. 2017;127(11):4118–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tong T, et al. . Regulation of adipogenesis and thermogenesis through mouse olfactory receptor 23 stimulated by alpha-cedrene in 3T3-L1 Cells. Nutrients. 2018;10(11):E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gottlieb DJ, et al. . CPAP versus oxygen in obstructive sleep apnea. N Engl J Med. 2014;370(24):2276–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stradling JR, et al. . Markers of inflammation: data from the MOSAIC randomised trial of CPAP for minimally symptomatic OSA. Thorax. 2015;70(2):181–182. [DOI] [PubMed] [Google Scholar]
- 46. Thunstrom E, et al. . CPAP does not reduce inflammatory biomarkers in patients with coronary artery disease and nonsleepy obstructive sleep apnea: a randomized controlled trial. Sleep. 2017;40(11). doi:10.1093/sleep/zsx157. [DOI] [PubMed] [Google Scholar]
- 47. Peng J, et al. . Effects of CPAP on the transcriptional signatures in patients with obstructive sleep apnea via coexpression network analysis. J Cell Biochem. 2019;120(6):9277–9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Drager LF, et al. . Effects of CPAP on body weight in patients with obstructive sleep apnoea: a meta-analysis of randomised trials. Thorax. 2015;70(3):258–264. [DOI] [PubMed] [Google Scholar]
- 49. Ou Q, et al. ; SAVE investigators. The effects of long-term CPAP on weight change in patients with comorbid OSA and cardiovascular disease: data from the SAVE trial. Chest. 2019;155(4):720–729. [DOI] [PubMed] [Google Scholar]
- 50. Hoyos CM, et al. . Cardiometabolic changes after continuous positive airway pressure for obstructive sleep apnoea: a randomised sham-controlled study. Thorax. 2012;67(12):1081–1089. [DOI] [PubMed] [Google Scholar]
- 51. Alemán JO, et al. . Effects of rapid weight loss on systemic and adipose tissue inflammation and metabolism in obese postmenopausal women. J Endocr Soc. 2017;1(6): 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Magkos F, et al. . Effects of moderate and subsequent progressive weight loss on metabolic function and adipose tissue biology in humans with obesity. Cell Metab. 2016;23(4):591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.