Abstract
Peripheral nerves provide a supportive growth environment for developing and regenerating axons and are essential for maintenance and repair of many non-neural tissues. This capacity has largely been ascribed to paracrine factors secreted by nerve-resident Schwann cells. Here, we used single-cell transcriptional profiling to identify ligands made by different injured rodent nerve cell types and have combined this with cell-surface mass spectrometry to computationally model potential paracrine interactions with peripheral neurons. These analyses show that peripheral nerves make many ligands predicted to act on peripheral and CNS neurons, including known and previously uncharacterized ligands. While Schwann cells are an important ligand source within injured nerves, more than half of the predicted ligands are made by nerve-resident mesenchymal cells, including the endoneurial cells most closely associated with peripheral axons. At least three of these mesenchymal ligands, ANGPT1, CCL11, and VEGFC, promote growth when locally applied on sympathetic axons. These data therefore identify an unexpected paracrine role for nerve mesenchymal cells and suggest that multiple cell types contribute to creating a highly pro-growth environment for peripheral axons.
Keywords: growth factor, nerve, paracrine interactions, regeneration, scRNA-seq, peripheral neurons, neuronal growth, Schwann cell, mesenchymal cell
Significance Statement
This work expands our understanding of the cellular sources of ligands in the injured peripheral nerve that are potentially important for promoting axon growth. Here, we used single-cell RNA sequencing (scRNA-seq) to reveal that Schwann cells and, surprisingly, nerve mesenchymal cells are primary sources of ligands in the injured nerve. We then combined injured nerve scRNA-seq data with proteomic and transcriptomic data from sensory and sympathetic neurons and used a systems-biology/modeling approach to predict novel mesenchymal cell-derived factors that may promote peripheral axon growth. We tested some of these predictions and found three factors, ANGPT1, CCL11, and VEGFC, that promoted outgrowth of cultured sympathetic axons, supporting a potential role for mesenchymal-derived factors in axon growth.
Introduction
Following injury, mammalian peripheral neurons can regenerate and reinnervate their target tissues. Their ability to do so is thought to be a consequence of a peripheral nerve environment that is highly supportive of axonal growth. Support for this idea comes from classic studies with CNS neurons, which normally fail to regenerate following brain or spinal cord injury but will regrow their axons when peripheral nerve segments are transplanted into the damaged region (David and Aguayo, 1981; for review, see Benowitz et al., 2017). Intriguingly, peripheral nerves are also important for maintenance, repair and regeneration of the non-neural tissues that they innervate. For example, normal peripheral innervation is essential for mammalian hair follicle and hematopoietic stem cells (Brownell et al., 2011; Yamazaki et al., 2011), for cardiac and dermal repair (Mahmoud et al., 2015; Johnston et al., 2013, 2016) and for amphibian limb (for review, see Kumar and Brockes, 2012) and murine digit tip regeneration (Johnston et al., 2016).
The supportive peripheral nerve environment has largely been ascribed to growth factors made by nerve cells (for review, see Terenghi, 1999; Fledrich et al., 2019). These nerve-derived ligands have been particularly well studied with regard to axonal development and regeneration (Chen et al., 2007; Fledrich et al., 2019), although several studies have shown that they are also important for limb and digit tip regeneration (Kumar et al., 2007; Johnston et al., 2016). These growth factors are thought to be Schwann cell derived, since transplantation of Schwann cells alone is enough to promote CNS axon regeneration (for review, see Bunge, 2016) and murine digit tip regeneration (Johnston et al., 2016). In addition to growth factors, the peripheral nerve provides an extracellular matrix environment that is highly conducive to axonal growth, particularly by contrast to the CNS, where known axon growth inhibitors prevail (Chen et al., 2007; Benowitz et al., 2017). This supportive substrate is also thought to derive in part from Schwann cells, which generate a basal lamina and synthesize ECM proteins and cell adhesion molecules (Muir, 2010; Gardiner, 2011).
These studies all indicate that Schwann cells play an important role in establishing a nerve environment that is supportive of axonal growth. However, the nerve is a structurally-complex tissue containing many different cell types, including vasculature-associated cells, immune cells such as tissue-resident macrophages, and mesenchymal cells of both mesodermal and neural crest origin. In this regard, one recent study identified four transcriptionally and spatially-distinct populations of Pdgfra-positive mesenchymal cells within the injured peripheral nerve, including endoneurial mesenchymal cells that are tightly associated with Schwann cells and axons (Carr et al., 2019). These nerve mesenchymal cells were shown to directly contribute to the repair and regeneration of mesenchymal target tissues including the digit tip, bone, and dermis. Nerve mesenchymal cells have also been shown to play an essential role in forming bridges over gaps in injured nerves (for review, see Cattin and Lloyd, 2016). Together, these findings raise the possibility that mesenchymal cells might also be important for axonal growth in the peripheral nerve.
Here, we provide support for this concept, using an unbiased systems biology approach to define the sciatic nerve ligand environment. We show, using single-cell profiling, that under both homeostatic and injury conditions, mesenchymal cells and Schwann cells are the predominant sources of peripheral nerve ligands, including known and uncharacterized ligands, and that there is induction of ligand expression in both these cell types following injury. Moreover, using mass spectrometry, transcriptional profiling, and computational modeling, we show that peripheral neurons and CNS retinal ganglion neurons express receptors for many of these ligands. Finally, we validate three of these ligands, ANGPT1, CCL11, and VEGFC, as being synthesized and secreted by Pdgfra-positive nerve mesenchymal cells and show that they can promote growth when applied to axons of peripheral sympathetic neurons. Thus, our data support a model where nerve mesenchymal cells and Schwann cells collaborate to establish a generally supportive growth environment in the peripheral nerve.
Materials and Methods
Animals
All animal procedures were performed in accordance with Canadian Council on Animal Care regulations as approved by the Hospital for Sick Children animal care committee. Sprague Dawley rats (purchased from Charles River) used in this study ranged from embryonic day (E)15 to young adult (six weeks old) and CD1 mice (purchased from Charles River) ranged in age from eight to twelve weeks old. All rats and mice were healthy throughout the duration of the study and had free access to chow and water in a 12/12 h light/dark cycle room. In most cases, rats and mice of both sexes were used with the exception of six-week-old male rats for sciatic nerve injury microarray experiments. PdgfraEGFP/+ (B6.129S4-Pdgfrαtm11(EGFP)Sor/J; JAX stock #007669; Hamilton et al., 2003) mice were obtained from The Jackson Laboratory and were bred and genotyped as recommended by The Jackson Laboratory. Animals that underwent sciatic nerve injury surgeries were housed individually for recovery purposes.
Sciatic nerve resection surgeries
Sciatic nerve resections were performed on young adult male Sprague Dawley rats (microarray analysis), adult CD1 mice (scRNA-seq analysis) or adult PdgfraEGFP/+ mice [fluorescence in situ hybridization (FISH) and immunostaining]. Before surgery, animals were anesthetized with 2% isoflurane gas and the surgical site was shaved. Animals were kept under anesthesia for the duration of the surgery. To resect the sciatic nerve, an incision was made along the lateral aspect of the mid-thigh of the right hindlimb, the sciatic nerve was then raised, an ∼5- to 10-mm segment was removed, and the distal nerve ending was carefully tucked away (distally) from the injury site to prevent regeneration. The wound was then closed with 4–0 Polysorb sutures (Covidien). Animals were treated subcutaneously with ketoprofen or meloxicam (∼2–5 mg/kg) as well as buprenorphine (0.05 mg/kg) before surgery, along with a postoperative treatment of ketoprofen or meloxicam 24 h after surgery. Mice and rats were housed separately following surgery and remained healthy throughout the postoperative period and were monitored twice daily for 3 d following surgery.
Single-cell isolation and myelin removal for Drop-seq analysis
For preparation of the 3 d postinjury (DPI) nerve scRNA-seq dataset, young adult CD1 mice underwent unilateral surgical resections as described above, and injured distal sciatic nerve segments were collected 3 d following surgery. For the uninjured nerve and neonatal nerve analyses, bilateral sciatic nerve segments were collected from adult and postnatal day (P)2–P4 CD1 mice, respectively. Freshly dissected nerves were digested in a mixture of collagenase Type XI (1 mg/ml, Sigma) and 0.05% Trypsin-EDTA (Thermo Fisher Scientific) for 30 min at 37°C. Enzymatic digestion was halted by diluting the cell suspension with HBSS (Thermo Fisher Scientific). Following centrifugation (1200 rpm for 5 min) and removal of the supernatant, the cell pellet was resuspended in PBS containing 0.5% BSA and passed through a 70-μm cell strainer (BD Biosciences). For datasets purified with myelin removal beads (3 DPI, neonatal and uninjured nerve; as shown in Figs. 1C,E, 2C,E; referred to as set 2 for the neonatal analyses, where cells were prepared in two ways), myelin debris was removed from the single-cell suspension using Myelin Removal Beads II and a MidiMACS magnetic separator with LS columns (Miltenyi Biotec), according to the manufacturer’s instructions. Following myelin removal, the cell suspension was centrifuged (1200 rpm for 5 min), and the supernatant was removed before resuspending the pellet in 0.22-mm sterile-filtered PBS containing 0.01% BSA. For the second neonatal nerve dataset that was purified using fluorescence-activated cell sorting (FACS), a single-cell suspension of dissociated injured nerve cells was prepared as described above. After passing the cells through a 70-μm cell strainer and resuspending them in PBS containing 0.25% BSA, Hoechst 33258 was added to distinguish nucleated cells from myelin debris, in addition to propidium iodide (PI) to exclude dead cells. The Hoechsthigh and PI-negative cell fractions were FACS purified using a MoFlo XDP cell sorter (Beckmann Coulter) before proceeding with scRNA-seq analysis. In all cases, cells were then resuspended in PBS containing 0.01% BSA, counted with a hemocytometer, and the solution was adjusted to a final concentration of 140,000 cells/ml and run through the Drop-seq apparatus at the Princess Margaret Genomics Facility. Drop-seq, cDNA amplification, library preparation, sequencing, processing of FASTQ sequencing reads, and read alignment steps were all conducted including minor modifications according to previously published protocols (Macosko et al., 2015). For the 3 DPI nerve scRNA-seq analysis (as shown in Fig. 1C), a raw digital gene expression (DGE) matrix was generated from 2500 cell barcodes as described in the Drop-seq Alignment Cookbook (version 1.2, January 2016; http://mccarrolllab.com/dropseq/). Similarly, for the uninjured nerve scRNA-seq analysis (as shown in Fig. 2C), a raw DGE matrix was generated from 2000 cell barcodes and used for all further analyses. In the case of the two neonatal nerve datasets (FACS sorted and bead treated), 2500 and 6200 cell barcodes were used to generate the DGE matrices as described above. DGE matrices described here were used for all subsequent analyses. The previously published DGE matrices for the 9-d injured nerve datasets (both FACS and myelin bead treated; GEO:GSE120678) were described in Carr et al. (2019).
Figure 1.
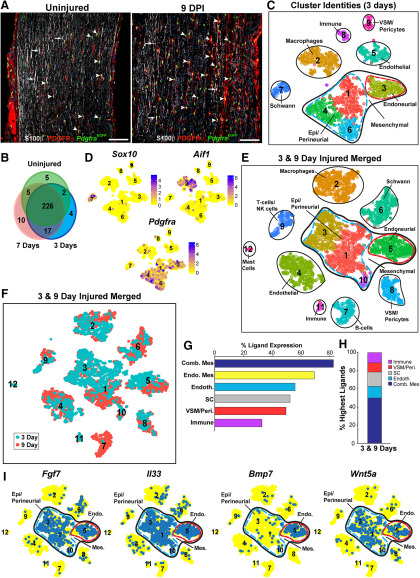
Characterization of ligand expression in the injured sciatic nerve (see also Extended Data Fig. 1-1). A, Images of longitudinal sections of an uninjured adult nerve and a 9 DPI distal sciatic nerve from PdgfraEgfp/+ mice analyzed for EGFP (green) and immunoreactivity for PDGFRα (red) and S100β (white). Arrowheads denote endoneurial cells positive for both PDGFRα protein and nuclear Pdgfra-EGFP and arrows indicate S100β immunoreactive Schwann cells. Scale bars = 100 μm. B, Venn diagram showing the number of ligands expressed in the uninjured versus 3 and 7 DPI distal sciatic nerves, based on microarray analysis. Ligand mRNAs were defined as expressed if their levels were ≥Ntf3. C–I, Characterization of ligand expression in injured distal sciatic nerve scRNA-seq datasets. C, t-SNE cluster visualization of 3 DPI sciatic nerve cell transcriptomes analyzed via the computational pipeline, with clusters annotated for cell types as identified by marker gene expression. D, t-SNE gene expression overlays on the dataset in C for the Schwann cell marker Sox10, the macrophage marker Aif1, and the mesenchymal cell marker Pdgfra. Relative transcript expression levels are color coded as per the adjacent color keys. E, t-SNE cluster visualization of the combined 3 and 9 DPI distal sciatic nerve cell transcriptomes with clusters annotated for cell types as identified by marker gene expression. F, t-SNE visualization of the dataset in E with cells color coded for their dataset of origin. Numbers correspond to cluster numbers in E. G, Bar graph showing the percentage of the 143 injured nerve ligand mRNAs detectably expressed in the combined 3 and 9 DPI sciatic nerve cell types (shown and annotated in E), including Pdgfra-positive mesenchymal cells (Comb. Mes), Pdgfra-positive endoneurial mesenchymal cells (Endo. Mes), endothelial cells (Endoth.), Schwann cells (SC), VSM/pericyte cells (VSM/Peri.), and immune cells. Ligand mRNAs were considered to be expressed if they were detected in 2% or more of the cell type of interest. H, Stacked bar graph showing the relative percentage of ligand mRNAs expressed at the highest levels in the different injured peripheral nerve cell types shown in E. I, t-SNE gene expression overlays of the combined 3 and 9 DPI sciatic nerve dataset (shown in E) for Fgf7, Il33, Bmp7, and Wnt5a. Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters.
Figure 2.
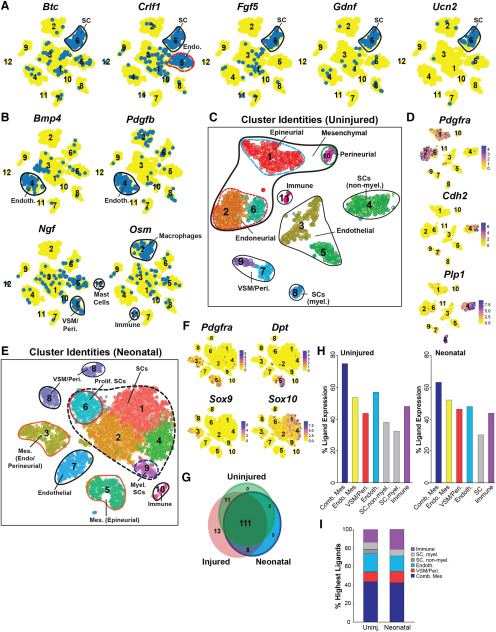
Ligand expression in the injured, uninjured and neonatal sciatic nerves (see also Extended Data Fig. 2-1). A, B, t-SNE gene expression overlays of the combined 3 and 9 DPI sciatic nerve dataset (shown in Fig. 1E) for Btc, Crlf1, Fgf5, Gdnf, and Ucn2 (A) and Bmp4, Pdgfb, Ngf, and Osm (B). Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Specific cell types with the highest ligand expression are circled and annotated, including Schwann cells (SC), endoneurial mesenchymal cells (Endo.), endothelial cells (Endoth.), VSM/pericytes (VSM/Peri.), and various types of immune cells. C, t-SNE cluster visualization of uninjured sciatic nerve single-cell transcriptomes annotated for cell types as identified by marker gene expression. D, t-SNE gene expression overlays of the dataset in C for the mesenchymal cell gene Pdgfra, and for the Schwann cell genes Cdh2 and Plp1. Relative transcript expression levels are color coded as per the adjacent color keys. E, t-SNE cluster visualization of neonatal sciatic nerve single-cell transcriptomes annotated for cell types identified by marker gene expression. Mes. = mesenchymal cells. F, t-SNE gene expression overlays of the dataset in E for the mesenchymal cell genes Pdgfra, Dpt, and Sox9, and the Schwann cell gene Sox 10. Relative transcript expression levels are color coded as per the adjacent color key. G, Venn diagram showing overlapping expression of the 143 injured nerve ligand mRNAs in the uninjured, neonatal, and injured nerve scRNA-seq datasets. Ligand mRNAs were considered to be expressed if they were detectable in 2% or more cells in any defined cell type cluster. H, Bar graphs showing the percentage of the 143 injured nerve ligand mRNAs detectably expressed in the uninjured or neonatal sciatic nerve cell types (shown and annotated in C, E), including Pdgfra-positive mesenchymal cells (Comb. Mes), Pdgfra-positive endoneurial mesenchymal cells (Endo. Mes), endothelial cells (Endoth.), Schwann cells (SC; for the uninjured, also designated as myelinating vs non-myelinating), VSM/pericyte cells (VSM/Peri.), or immune cells. Ligand mRNAs were considered to be expressed if they were detected in 2% or more cells of that particular cell type. I, Stacked bar graphs showing the relative percentages of ligand mRNAs expressed at the highest levels in the different uninjured and neonatal peripheral nerve cell types shown in C, E, respectively.
Characterization of the 3- and 9-d injured sciatic nerve scRNA-seq datasets. A, t-SNE gene expression overlays on the 3 DPI total cell dataset (shown in Fig. 1C and in the adjacent legend) for the endothelial cell marker Pecam1, the immune cell marker Trbc2, the VSM/pericyte cell marker Rgs5, and the Pdgfra-positive mesenchymal epineurial and endoneurial cell markers Dpp4 and Wif1. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. B, t-SNE gene expression overlays on the combined 3 and 9 DPI total cell datasets (shown in Figure 1E and in the adjacent legend) for the endothelial cell marker Pecam1, the immune cell marker Aif1, the VSM/pericyte cell marker Rgs5, the mesenchymal marker Pdgfra, the Schwann cell marker Sox10, and the B cell marker Cd19. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. C, t-SNE gene expression overlays on the combined 3 and 9 DPI total cell dataset (shown in Fig. 1E and in the adjacent legend) for markers for the different types of Pdgfra-positive mesenchymal cells, including Etv1-positive endoneurial cells, Pcolce2-positive epineurial cells, Msln-positive perineurial cells, and Dlk1-positive differentiating mesenchymal cells. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. D, t-SNE gene expression overlays of the combined 3 and 9 DPI total cell dataset for Fgf10, Adm, Pthlh, and Ntn1. Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Specific cell types are circled and annotated, including mesenchymal (Mes.), endoneurial (Endo.), and epineurial/perineurial (Epi/Perineurial) cells. Download Figure 1-1, TIF file (2.5MB, tif)
Characterization of the uninjured and neonatal sciatic nerve scRNA-seq datasets. A, t-SNE gene expression overlays of the combined 3 and 9 DPI total cell dataset (shown in Fig. 1E and the adjacent legend) for Bdnf, Dhh, and Shh. Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Schwann cells are circled and annotated (SC). B, t-SNE gene expression overlays on the uninjured sciatic nerve total cell dataset (shown in Fig. 2C and in the adjacent legend) for the VSM/pericyte cell marker Acta2, the endothelial cell marker Pecam1, the immune cell marker Cd52, and the proliferating cell marker Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. C, t-SNE cluster visualization of neonatal sciatic nerve single-cell transcriptomes (as in Fig. 2E and the adjacent legend) showing dataset of origin. Set 1 (red) refers to the neonatal nerve cells isolated by FACS and Set 2 (blue) to the neonatal nerve cells isolated by treatment with the myelin removal beads. The right t-SNE cluster visualization indicates the datasets of origin following Harmony data integration batch correction of the combined datasets. D, t-SNE gene expression overlays on the neonatal sciatic nerve total cell dataset (shown in Fig. 2E and in the adjacent legend) for Osr2, which marks endoneurial mesenchymal cells, Casq2, which is expressed in perineurial cells, the endothelial cell marker Pecam1, the VSM/pericyte cell marker Acta2, and the proliferating cell marker Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. Download Figure 2-1, TIF file (3.1MB, tif) .
Computational analysis of scRNA-seq data
Drop-seq data (DGE matrices) were analyzed used a previously described custom computational pipeline (Yuzwa et al., 2017; Carr et al., 2019; Storer et al., 2020; described in detail in Innes and Bader, 2019). Briefly, data were filtered to remove cells with low unique molecular identifier counts, cell doublets, contaminant red blood cells, and cells that contained high mitochondrial gene content. Genes detected in less than three cells were removed. Cell transcriptomes were then normalized as previously described (Lun et al., 2016) using an algorithm in the scran package in R that corrects for differences in sequencing depth by the use of scaling factors within each cell by pooling random subsets of cells, summing their library sizes, and comparing them with average library size across all cells in the group. This is iteratively performed, and the cell-wise scaling factors can be deconvolved from the set of pool-wise scaling factors. Following normalization, DGE matrices were imported into Seurat (v. 1.4.0.16) in R. Principal component (PC) analysis was then undertaken using highly variable genes, and clustering analysis was performed using top PCs. This analysis was conducted using the shared nearest neighbor (SNN) modularity optimization-based clustering algorithm implemented in Seurat (FindClusters function). Clustering was iteratively performed at increasing resolution until a lower limit of ∼10–20 differentially expressed genes [calculated by the Seurat FindMarkers function, p < 0.01 family-wise error rate (FWER), Holm’s method] was reached between the most similar clusters. For conservative analysis of all datasets analyzed, clusters were assigned at the lowest resolution that still distinguished distinct cell types, as defined by established marker genes. As a result, clusters were assigned at a resolution of 0.4 for analysis of all datasets.
For the 3 DPI dataset (2075 total cells), nine clusters were identified with 210 differentially expressed genes between most similar clusters (p < 0.01, FWER). Cells were sequenced to an average depth of >70,000 reads/cell. The average number of genes detected per cell was 1027 ± 588, and the average number of transcripts was 2257 ± 2855. For the 3 and 9 DPI nerve combined dataset, cell transcriptomes from the 3 DPI dataset and the myelin bead removal-treated 9 DPI dataset (from Carr et al., 2019; GEO:GSE120678) were merged using the unique cell identifier barcodes from all cells present in all clusters of the two datasets following pipeline processing. The constructed raw DGE matrices of the combined datasets were then re-run through the pipeline for re-clustering, resulting in 5395 total cells. Twelve clusters were identified with 200 differentially expressed genes between most similar clusters (p < 0.01, FWER). The uninjured nerve dataset was previously analyzed for Pdgfra-positive mesenchymal cells (Carr et al., 2019), but not for other cell types. Reanalysis of this dataset (1841 total cells) identified 11 clusters with 105 differentially expressed genes between the most similar clusters (p < 0.01, FWER). For the combined neonatal dataset, cell transcriptomes from both the FAC-sorted (set 1; Extended Data Fig. 2-1C) and myelin removal bead-treated (set 2; Extended Data Fig. 2-1C) samples were merged using the unique cell identifier barcodes from all cells present in all clusters of the two datasets following pipeline processing. The constructed raw DGE matrices of the combined datasets were then re-run through the pipeline for re-clustering, resulting in 6885 total cells. Ten clusters were identified with 540 differentially expressed genes between most similar clusters (p < 0.01, FWER). For the FAC-sorted sample, cells were sequenced to an average depth of >90,000 reads/cell. The average number of genes detected per cell was 1005 ± 728 and the average number of transcripts was 2044 ± 1985. For the bead treated sample, cells were sequenced to an average depth of >43,000 reads/cell. The average number of genes detected per cell was 732 ± 485, and the average number of transcripts was 1231 ± 1095.
For the combined Schwann cell dataset (Fig. 3A,B), the unique cell identifier barcodes from all cells present in Sox10-positive clusters in each of the six described datasets (Figs. 1E, 2C,E; FAC-sorted cells in Carr et al., 2019) were merged. For the combined mesenchymal cell dataset (Fig. 4A,B), the unique cell identifier barcodes from all cells present in Pdgfra-positive clusters in each of the same six datasets were merged. The constructed raw DGE matrices of the combined datasets were then re-run through the pipeline, resulting in 5331 total Schwann cells and 5416 total mesenchymal cells. Batch correction of these combined datasets as well as data shown in Extended Data Figure 2-1C was performed using the Harmony batch-effect-correction method (Korsunsky et al., 2019) with Seurat V2. Briefly, gene expression data from the combined datasets were transferred to Seurat, where highly variable genes were then used to carry out principal component analysis. The Harmony iterative algorithm was used to integrate datasets and adjust for dataset specific effects based on the top 20 principal components. Iterative clustering was performed using the FindClusters function in Seurat V2, with clusters being assigned at a resolution of 0.4. This resulted in seven and nine clusters in the combined Schwann cell and mesenchymal cell datasets, respectively. t-Distributed stochastic neighbor embedding (t-SNE) visualizations of batch-corrected data were generated using the FeaturePlot function in Seurat. t-SNE gene expression overlays displayed in the figures were generated using the FeaturePlot function in Seurat and binary expression overlays were performed using the SubsetCells and TSNEPlot functions in Seurat. These tSNE overlays were further edited using Adobe Illustrator (Adobe Systems Incorporated) as necessary to highlight features of the t-SNE visualization.
Figure 3.
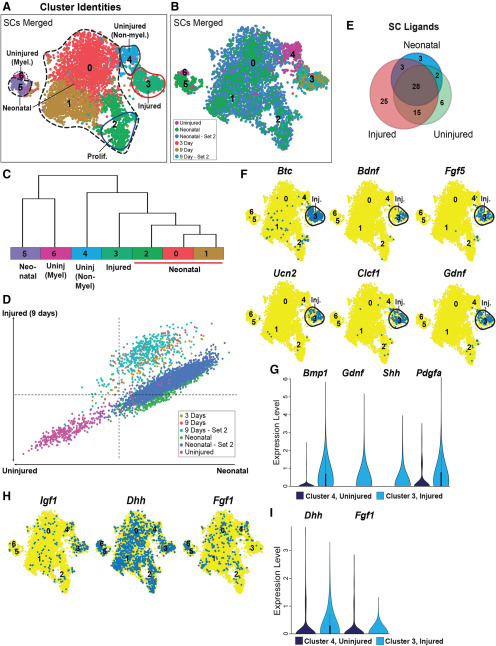
Characterization of ligand expression in sciatic nerve Schwann cells (see also Extended Data Fig. 3-1). Schwann cell transcriptomes from the injured, uninjured and neonatal nerve datasets (in Figs. 1E, 2C,E; also see Carr et al., 2019) were extracted, combined together, analyzed and batch-corrected using Harmony data integration and clustered in Seurat based on principal components. A, B, t-SNE visualizations of the combined Schwann cells (SCs), showing clusters (A) and datasets of origin (B). The clusters in A are also annotated based on marker gene expression (Extended Data Fig. 3-1A) and datasets of origin. Neonatal and 9 d refer to the FAC-sorted preparations at these timepoints while neonatal – set 2 and 9 d – set 2 refer to the cells prepared with myelin removal beads. C, A dendrogram showing hierarchical analysis of cell clusters from the combined Schwann cell dataset, with cluster identity numbers, annotations and colors as in A. D, Scatterplot showing differential correlation of single-cell transcriptomes from the combined Schwann cell dataset in A (individual colors represent different datasets) with bulk transcriptomes from uninjured adult versus neonatal Schwann cells on the x-axis and uninjured adult versus 9 DPI on the y-axis. E, Venn diagram showing the overlap of ligand mRNAs expressed in neonatal, injured and uninjured Schwann cell clusters from the combined Schwann cell dataset in A. Ligand mRNAs were considered to be expressed if they were detectable in 2% or more cells in any defined cell type cluster. F, t-SNE gene expression overlays of the combined Schwann cell data in A for Btc, Bdnf, Fgf5, Ucn2, Clcf1, and Gdnf. Cells that detectably express the ligand are colored blue, and the numbers correspond to the clusters. Injured Schwann cell cluster 3 (Inj.) is circled. G, Violin plots showing the relative expression of Bmp1, Gdnf, Shh, and Pdgfa in injured Schwann cluster 3 versus uninjured Schwann cell cluster 4 from the dataset in A. H, t-SNE gene expression overlays of the combined Schwann cell data in A for Igf1, Dhh, and Fgf1. Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. I, Violin plots showing the relative expression of Dhh and Fgf1 in injured Schwann cell cluster 3 versus uninjured cluster 4 from the dataset in A.
Figure 4.
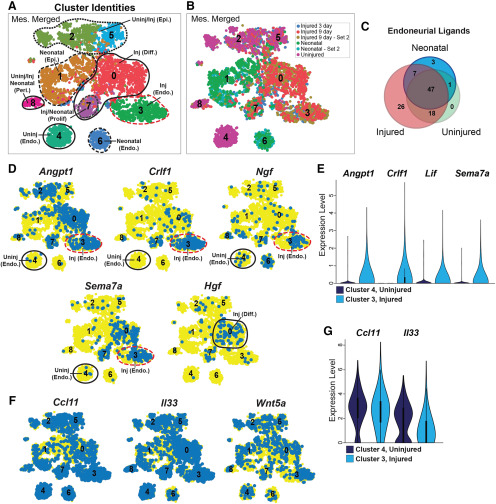
Characterization of ligand expression in sciatic nerve mesenchymal cells (see also Extended Data Fig. 4-1). Pdgfra-positive mesenchymal cell transcriptomes from the injured, uninjured and neonatal nerve datasets (in Figs. 1E, 2C,E; also see Carr et al., 2019) were extracted, combined together, analyzed and batch-corrected using Harmony data integration and clustered in Seurat based on principal components. A, B, t-SNE visualizations of the combined mesenchymal cells showing clusters (A) and datasets of origin (B). The clusters in A are also annotated based on marker gene expression (Extended Data Fig. 4-1) and datasets of origin. Neonatal and 9 d refer to the FAC-sorted preparations at these timepoints, while neonatal – set 2 and 9 d – set 2 refer to the cells prepared with myelin removal beads. C, Venn diagram showing the overlap of ligand mRNAs expressed in neonatal (cluster 6), injured (cluster 3), and uninjured (cluster 4) nerve endoneurial mesenchymal cells from the combined mesenchymal nerve dataset in A. Ligand mRNAs were considered to be expressed if they were detectable in 2% or more cells in the relevant cluster. D, t-SNE gene expression overlays of the combined mesenchymal cell data in A for Angpt1, Crlf1, Ngf, Sema7a, and Hgf. Cells that detectably express the ligand are colored blue, and the numbers correspond to the clusters. Relevant clusters are circled and annotated, including uninjured endoneurial (Uninj Endo.), injured endoneurial (Inj Endo.), and injured differentiating (Inj Diff.) mesenchymal cells. E, Violin plots showing the relative expression of Angpt1, Crlf1, Lif, and Sema7a in injured endoneurial mesenchymal cell cluster 3 versus uninjured endoneurial cell cluster 4 from the dataset in A. F, t-SNE gene expression overlays of the combined mesenchymal cell data in A for Ccl11, Il33, and Wnt5a. Cells that detectably express the ligand are colored blue, and the numbers correspond to the clusters. G, Violin plots showing the relative expression of Ccl11 and Il33 in injured endoneurial mesenchymal cell cluster 3 versus uninjured endoneurial cell cluster 4 from the dataset in A.
Characterization of the combined Schwann cell sciatic nerve scRNA-seq dataset. A, t-SNE gene expression overlays on the combined and batch-corrected neonatal, injured adult and uninjured adult Schwann cell data (shown in Fig. 3A and the adjacent legend) for Sox10, the myelination gene Mag, the pre-myelinating Schwann cell marker Pou3f1, the non-myelinating Schwann cell gene Emp1, and the proliferation marker Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. B, Plots show correlation analyses of average transcript expression in the in the combined injured nerve dataset (Fig. 1E) showing Schwann cell cluster 6 compared to endoneurial cell cluster 5 (left plot) and to epineurial cell cluster 3 (right plot). Outlier transcripts expressed in the mesenchymal cell clusters are highlighted red and labelled. C, D, t-SNE gene expression overlays of the combined Schwann cell data (shown in Fig. 3A and the adjacent legend) for Ccl3, Crlf1, Lif, Shh, and Tgfb1 (C) and Bmp1, Fgf7, Mdk, and Pdgfa (D). Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Injured Schwann cell cluster 3 is circled and annotated (C, Inj.). Download Figure 3-1, TIF file (2.7MB, tif) .
Characterization of the combined Pdgfra-positive mesenchymal cell sciatic nerve scRNA-seq dataset. t-SNE gene expression overlays on the combined and batch-corrected neonatal, injured adult and uninjured adult Pdgfra-positive mesenchymal cell data (shown in Fig. 4A and the adjacent legend) for Pdgfra, the epineurial markers Pcolce2 and Dpp4, the perineurial gene Slc2a1, the endoneurial gene Meox1, the differentiating injured cell genes Mest and Dlk1, and the proliferation gene Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. Download Figure 4-1, TIF file (1.5MB, tif) .
Cell types (clusters) were defined based on the expression of the following established marker genes: endothelial cells: Pecam1/Cd31, Plvap, and Esam; Schwann lineage cells: Ngfr/p75NTR and Sox10; non-myelinating Schwann cells: Ngfr/p75NTR, Cdh2, L1cam, Ednrb, Emp1, and Sema3e; premyelinating Schwann cells: Pou3f1 and Egr2; myelinating Schwann cells: Mag, Mbp, Pmp22, Mpz, and Plp1; macrophages/monocytes: Aif1/Iba1; lymphoid immune cells including B cells, T cells, and NK cells: Ptprcap, Trbc2, and Cd52; B cells: Cd19; vasculature-associated smooth muscle (VSM) and pericyte cells: Desmin, Mylk, Acta2, and Rgs5; mesenchymal cells: Pdgfra; epineurial mesenchymal cells: Pcolce2, Dpp4, Dpt, Ly6c1, and Comp; endoneurial mesenchymal cells: Etv1, Wif1, Sox9, Osr2, and Meox1; perineurial mesenchymal cells: Slc2a1, Casq2, and Msln; differentiating nerve bridge mesenchymal cells: Dlk1 and Mest; and proliferating cells: Mki67 and Top2a.
In combined datasets, dataset identities were distinguished by using the gg color hue and hcl functions in R. Correlation analysis comparing gene expression between different clusters was performed by averaging the expression of each gene across all cells in the individual clusters to be compared, then Pearson correlation analysis was performed using the Cor function and plotted in R. Genes of interest were then highlighted using Adobe Illustrator (Adobe Systems Incorporated; as in Extended Data Fig. 3-1B). Differential correlation of single-cell transcriptomes as shown in Figure 3D was performed as described previously (Gerber et al., 2018). Briefly, mock bulk transcriptomes were generated for the 9 DPI Schwann cells (both bead and FAC sorted), the uninjured non-myelinating Schwann cells and the neonatal Schwann cells (both bead and FAC sorted) by determining the mean expression of each gene in the total combined cells in each dataset. Each single-cell transcriptome was then correlated with each of the mock bulk transcriptomes. We then determined the differential correlation of each single cell with the bulk uninjured nerve versus the bulk 9 DPI transcriptomes (y-axis) and the differential correlation with the bulk uninjured nerve versus the bulk neonatal nerve transcriptomes (x-axis). Violin plots were generated using the VioPlot package in R. Hierarchical clustering of the batch corrected combined Schwann cell data in Figure 3C was performed based on the top 20 Principal Component (PC) using the BuildClusterTree and PlotClusterTree functions in Seurat. Node numbers were removed from the plot and cluster descriptions and colors were added for clarity using Adobe Illustrator (Adobe Systems Incorporated). The single cell heatmaps were generated (with scaled expression values) using the DoHeatMap function in Seurat at resolution 0.4.
Ligand mRNA expression and Venn diagram analysis
The expression of ligand mRNAs (Table 1) was characterized from the whole nerve microarray analysis using a custom curated ligand-receptor database (modified from Yuzwa et al., 2016). Extracellular matrix proteins and potential ligands without well-defined, receptor-mediated paracrine roles were excluded. The VennDiagram package in R was used to determine overlapping ligands in the uninjured, 3-d injured, and 7-d injured nerve datasets and was modified to show proportional representation of data using the eulerAPE tool (Micallef and Rodgers, 2014).
Table 1.
Ligand mRNAs expressed in uninjured, 3 DPI, and 7 DPI sciatic nerves using global transcriptomic analysis
| Uninjured (238) | 3 DPI (249) | 7 DPI (258) | Uninjured,3 DPI, and 7 DPIintersect (226) | |||
|---|---|---|---|---|---|---|
| Adipoq | Adipoq | Adipoq | Adipoq | |||
| Adm | Adm | Adm | Adm | |||
| Agt | Agt | Agt | Agt | |||
| Angpt1 | Angpt1 | Angpt1 | Angpt1 | |||
| Angpt2 | Angpt2 | Angpt2 | Angpt2 | |||
| Angpt4 | Angpt4 | Angpt4 | Angpt4 | |||
| Apln | Apln | Apln | Apln | |||
| Artn | Areg++ | Areg++ | Artn | |||
| Avp | Artn | Artn | Avp | |||
| Bdnf | Avp | Avp | Bdnf | |||
| Bmp1 | Bdnf | Bdnf | Bmp1 | |||
| Bmp2 | Bmp1 | Bmp1 | Bmp2 | |||
| Bmp4 | Bmp2 | Bmp2 | Bmp4 | |||
| Bmp5 | Bmp4 | Bmp4 | Bmp5 | |||
| Bmp6 | Bmp5 | Bmp5 | Bmp6 | |||
| Bmp7 | Bmp6 | Bmp6 | Bmp7 | |||
| Btc | Bmp7 | Bmp7 | Btc | |||
| Cck | Btc | Btc | Cck | |||
| Ccl11 | Cck | Btla+ | Ccl11 | |||
| Ccl19 | Ccl11 | Calca+ | Ccl19 | |||
| Ccl2 | Ccl17* * | Cck | Ccl2 | |||
| Ccl21 | Ccl19 | Ccl11 | Ccl21 | |||
| Ccl22 | Ccl2 | Ccl19 | Ccl22 | |||
| Ccl24 | Ccl20++ | Ccl2 | Ccl24 | |||
| Ccl25 | Ccl21 | Ccl20++ | Ccl25 | |||
| Ccl27 | Ccl22 | Ccl21 | Ccl27 | |||
| Ccl3 | Ccl24 | Ccl22 | Ccl3 | |||
| Ccl5 | Ccl25 | Ccl24 | Ccl5 | |||
| Ccl7 | Ccl27 | Ccl25 | Ccl7 | |||
| Clcf1 | Ccl3 | Ccl27 | Clcf1 | |||
| Clu | Ccl5 | Ccl3 | Clu | |||
| Cmtm8 | Ccl7 | Ccl5 | Cmtm8 | |||
| Cntf | Cga++ | Ccl7 | Cntf | |||
| Cntn1 | Clcf1 | Cga++ | Cntn1 | |||
| Cntn2 | Clu | Clcf1 | Cntn2 | |||
| Copa | Cmtm8 | Clu | Copa | |||
| Crlf1 | Cntf | Cmtm8 | Crlf1 | |||
| Csf1 | Cntn1 | Cntf | Csf1 | |||
| Cst3 | Cntn2 | Cntn1 | Cst3 | |||
| Ctf1 | Copa | Cntn2 | Ctf1 | |||
| Ctgf | Crlf1 | Copa | Ctgf | |||
| Cx3cl1 | Csf1 | Crlf1 | Cx3cl1 | |||
| Cxcl1 | Cst3 | Csf1 | Cxcl1 | |||
| Cxcl10 | Ctf1 | Cst3 | Cxcl10 | |||
| Cxcl12 | Ctgf | Ctf1 | Cxcl12 | |||
| Cxcl13 | Cx3cl1 | Ctgf | Cxcl13 | |||
| Cxcl16 | Cxcl1 | Cx3cl1 | Cxcl16 | |||
| Cxcl2 | Cxcl10 | Cxcl1 | Cxcl2 | |||
| Cxcl9 | Cxcl12 | Cxcl10 | Cxcl9 | |||
| Dhh | Cxcl13 | Cxcl11+ | Dhh | |||
| Dll1 | Cxcl16 | Cxcl12 | Dll1 | |||
| Dll4 | Cxcl2 | Cxcl13 | Dll4 | |||
| Ebi3 | Cxcl9 | Cxcl16 | Ebi3 | |||
| Eda | Dhh | Cxcl2 | Eda | |||
| Edn3 | Dll1 | Cxcl9 | Edn3 | |||
| Efna1 | Dll4 | Dhh | Efna1 | |||
| Efna2 | Ebi3 | Dll1 | Efna2 | |||
| Efna4 | Eda | Dll4 | Efna4 | |||
| Efna5 | Edn3 | Ebi3 | Efna5 | |||
| Efnb1 | Efna1 | Eda | Efnb1 | |||
| Efnb2 | Efna2 | Edn3 | Efnb2 | |||
| Efnb3 | Efna4 | Efna1 | Efnb3 | |||
| Epo | Efna5 | Efna2 | Epo | |||
| Esm1 | Efnb1 | Efna4 | Esm1 | |||
| Fgf1 | Efnb2 | Efna5 | Fgf1 | |||
| Fgf10 | Efnb3 | Efnb1 | Fgf17 | |||
| Fgf17 | Epo | Efnb2 | Fgf18 | |||
| Fgf18 | Esm1 | Efnb3 | Fgf19 | |||
| Fgf19 | Fgf1 | Epo | Fgf2 | |||
| Fgf2 | Fgf17 | Esm1 | Fgf4 | |||
| Fgf4 | Fgf18 | Fgf1 | Fgf5 | |||
| Fgf5 | Fgf19 | Fgf10 | Fgf7 | |||
| Fgf7 | Fgf2 | Fgf17 | Figf | |||
| Figf | Fgf4 | Fgf18 | Fjx1 | |||
| Fjx1 | Fgf5 | Fgf19 | Flt3lg | |||
| Flt3lg | Fgf7 | Fgf2 | Fstl1 | |||
| Fstl1 | Figf | Fgf23+ | Gal | |||
| Gal | Fjx1 | Fgf4 | Gap43 | |||
| Gap43 | Flt3lg | Fgf5 | Gas6 | |||
| Gas6 | Fstl1 | Fgf7 | Gdf10 | |||
| Gdf10 | Gal | Figf | Gdf11 | |||
| Gdf11 | Gap43 | Fjx1 | Gdf9 | |||
| Gdf9 | Gas6 | Flt3lg | Gdnf | |||
| Gdnf | Gdf10 | Fstl1 | Ghrh | |||
| Ghrh | Gdf11 | Gal | Gip | |||
| Gip | Gdf5++ | Gap43 | Gmfb | |||
| Gmfb | Gdf6* * | Gas6 | Gmfg | |||
| Gmfg | Gdf9 | Gdf10 | Gnrh1 | |||
| Gnrh1 | Gdnf | Gdf11 | Gpi | |||
| Gpi | Gh1* * | Gdf5++ | Grp | |||
| Grp | Ghrh | Gdf9 | Habp2 | |||
| Guca2a | Gip | Gdnf | Hbegf | |||
| Habp2 | Gmfb | Ghrh | Hcrt | |||
| Hbegf | Gmfg | Gip | Hdgf | |||
| Hcrt | Gnrh1 | Gmfb | Hdgfrp3 | |||
| Hdgf | Gpi | Gmfg | Hgf | |||
| Hdgfrp3 | Grp | Gnrh1 | Hmgb1 | |||
| Hgf | Habp2 | Gpi | Ifna4 | |||
| Hmgb1 | Hbegf | Grp | Igf1 | |||
| Ifna1 | Hcrt | Guca2a | Igf2 | |||
| Ifna4 | Hdgf | Habp2 | Igfbpl1 | |||
| Igf1 | Hdgfrp3 | Hbegf | Ihh | |||
| Igf2 | Hgf | Hcrt | Il13 | |||
| Igfbpl1 | Hmgb1 | Hdgf | Il15 | |||
| Ihh | Ifna4 | Hdgfrp3 | Il16 | |||
| Il13 | Igf1 | Hgf | Il17b | |||
| Il15 | Igf2 | Hmgb1 | Il18 | |||
| Il16 | Igfbpl1 | Ifna1 | Il19 | |||
| Il17b | Ihh | Ifna4 | Il23a | |||
| Il18 | Il12a++ | Igf1 | Il25 | |||
| Il19 | Il13 | Igf2 | Il33 | |||
| Il21* | Il15 | Igfbpl1 | Inha | |||
| Il23a | Il16 | Ihh | Inhba | |||
| Il25 | Il17b | Il12a++ | Inhbb | |||
| Il33 | Il18 | Il13 | Insl3 | |||
| Inha | Il19 | Il15 | Jag1 | |||
| Inhba | Il23a | Il16 | Jag2 | |||
| Inhbb | Il25 | Il17b | Kiss1 | |||
| Insl3 | Il27* * | Il18 | Kitlg | |||
| Jag1 | Il33 | Il19 | Lgals3 | |||
| Jag2 | Il6++ | Il1b+ | Lif | |||
| Kiss1 | Inha | Il23a | Lrsam1 | |||
| Kitlg | Inhba | Il25 | Ltb | |||
| Lgals3 | Inhbb | Il33 | Mdk | |||
| Lgi1 | Inhbe++ | Il6++ | Metrn | |||
| Lif | Insl3 | Inha | Mif | |||
| Lrrc4 | Jag1 | Inhba | Mln | |||
| Lrsam1 | Jag2 | Inhbb | Nampt | |||
| Ltb | Kiss1 | Inhbe++ | Nenf | |||
| Mdk | Kitlg | Insl3 | Ngf | |||
| Metrn | Lgals3 | Jag1 | Nodal | |||
| Mif | Lgi1 | Jag2 | Nov | |||
| Mln | Lif | Kiss1 | Npb | |||
| Mst1* | Lrsam1 | Kitlg | Npff | |||
| Nampt | Ltb | Lgals3 | Nppb | |||
| Nenf | Mdk | Lif | Nppc | |||
| Ngf | Metrn | Lrrc4 | Nrtn | |||
| Nodal | Mif | Lrsam1 | Ntf3 | |||
| Nov | Mln | Ltb | Ntf4 | |||
| Npb | Mmp12++ | Mdk | Ntn1 | |||
| Npff | Mmp9++ | Metrn | Osm | |||
| Nppb | Nampt | Mif | Oxt | |||
| Nppc | Nell2++ | Mln | Pcsk1n | |||
| Nrtn | Nenf | Mmp12++ | Pdap1 | |||
| Ntf3 | Ngf | Mmp9++ | Pdgfa | |||
| Ntf4 | Nodal | Nampt | Pdgfb | |||
| Ntn1 | Nov | Nell2++ | Pdgfc | |||
| Osm | Npb | Nenf | Pf4 | |||
| Oxt | Npff | Ngf | Pgf | |||
| Pcsk1n | Nppb | Nodal | Plg | |||
| Pdap1 | Nppc | Nov | Pnoc | |||
| Pdgfa | Nrtn | Npb | Prdx2 | |||
| Pdgfb | Ntf3 | Npff | Prdx6 | |||
| Pdgfc | Ntf4 | Nppb | Prlh | |||
| Pf4 | Ntn1 | Nppc | Proc | |||
| Pgf | Osm | Nrg1+ | Prok1 | |||
| Plg | Oxt | Nrtn | Prok2 | |||
| Pnoc | Pcsk1n | Ntf3 | Psip1 | |||
| Prdx2 | Pdap1 | Ntf4 | Pspn | |||
| Prdx6 | Pdgfa | Ntn1 | Pth2 | |||
| Prlh | Pdgfb | Osm | Pthlh | |||
| Proc | Pdgfc | Oxt | Ptn | |||
| Prok1 | Pf4 | Pcsk1n | Rabep1 | |||
| Prok2 | Pgf | Pdap1 | Rln3 | |||
| Psip1 | Plg | Pdgfa | Rspo1 | |||
| Pspn | Pnoc | Pdgfb | Rtn1 | |||
| Pth2 | Ppy++ | Pdgfc | Rtn4 | |||
| Pthlh | Prdx2 | Pf4 | S100b | |||
| Ptn | Prdx6 | Pgf | Scgb3a1 | |||
| Rabep1 | Prlh | Plg | Scrn1 | |||
| Rln3 | Proc | Pnoc | Sct | |||
| Rspo1 | Prok1 | Pomc+ | Sema3a | |||
| Rspo3 | Prok2 | Ppbp+ | Sema3b | |||
| Rspo4* | Psip1 | Ppy++ | Sema3c | |||
| Rtn1 | Pspn | Prdx2 | Sema3d | |||
| Rtn4 | Pth2 | Prdx6 | Sema3e | |||
| S100b | Pthlh | Prlh | Sema3f | |||
| Scg3* | Ptn | Proc | Sema3g | |||
| Scgb3a1 | Rabep1 | Prok1 | Sema4a | |||
| Scrn1 | Rln3 | Prok2 | Sema4b | |||
| Sct | Rspo1 | Psip1 | Sema4c | |||
| Sema3a | Rtn1 | Pspn | Sema4d | |||
| Sema3b | Rtn4 | Pth2 | Sema4g | |||
| Sema3c | S100b | Pthlh | Sema5a | |||
| Sema3d | Scgb3a1 | Ptn | Sema5b | |||
| Sema3e | Scrn1 | Rabep1 | Sema6a | |||
| Sema3f | Sct | Rln3 | Sema6b | |||
| Sema3g | Sema3a | Rspo1 | Sema6c | |||
| Sema4a | Sema3b | Rspo3 | Sema6d | |||
| Sema4b | Sema3c | Rtn1 | Sema7a | |||
| Sema4c | Sema3d | Rtn4 | Serpinh1 | |||
| Sema4d | Sema3e | S100b | Sfrp1 | |||
| Sema4g | Sema3f | Scgb3a1 | Sfrp2 | |||
| Sema5a | Sema3g | Scrn1 | Sfrp4 | |||
| Sema5b | Sema4a | Sct | Sfrp5 | |||
| Sema6a | Sema4b | Sema3a | Shh | |||
| Sema6b | Sema4c | Sema3b | Smoc1 | |||
| Sema6c | Sema4d | Sema3c | Sost | |||
| Sema6d | Sema4f++ | Sema3d | Sparc | |||
| Sema7a | Sema4g | Sema3e | Sparcl1 | |||
| Serpinh1 | Sema5a | Sema3f | Spp1 | |||
| Sez6 | Sema5b | Sema3g | Sst | |||
| Sfrp1 | Sema6a | Sema4a | Tgfa | |||
| Sfrp2 | Sema6b | Sema4b | Tgfb1 | |||
| Sfrp4 | Sema6c | Sema4c | Tgfb2 | |||
| Sfrp5 | Sema6d | Sema4d | Tgfb3 | |||
| Shh | Sema7a | Sema4f++ | Thpo | |||
| Smoc1 | Serpinh1 | Sema4g | Timp2 | |||
| Sost | Sez6 | Sema5a | Tnf | |||
| Sparc | Sfrp1 | Sema5b | Tnfrsf11b | |||
| Sparcl1 | Sfrp2 | Sema6a | Tnfsf10 | |||
| Spp1 | Sfrp4 | Sema6b | Tnfsf11 | |||
| Sst | Sfrp5 | Sema6c | Tnfsf12 | |||
| Tgfa | Shh | Sema6d | Tnfsf13 | |||
| Tgfb1 | Smoc1 | Sema7a | Tnfsf14 | |||
| Tgfb2 | Sost | Serpinh1 | Tnfsf15 | |||
| Tgfb3 | Sparc | Sfrp1 | Tymp | |||
| Thpo | Sparcl1 | Sfrp2 | Ucn2 | |||
| Timp2 | Spp1 | Sfrp4 | Ucn3 | |||
| Tnf | Sst | Sfrp5 | Vegfa | |||
| Tnfrsf11a* | Tgfa | Shh | Vegfb | |||
| Tnfrsf11b | Tgfb1 | Smoc1 | Vegfc | |||
| Tnfsf10 | Tgfb2 | Sost | Wnt2 | |||
| Tnfsf11 | Tgfb3 | Sparc | Wnt5a | |||
| Tnfsf12 | Thpo | Sparcl1 | Wnt11 | |||
| Tnfsf13 | Timp2 | Spp1 | Xcl1 | |||
| Tnfsf14 | Tnf | Sst | ||||
| Tnfsf15 | Tnfrsf11b | Tdgf1+ | ||||
| Tymp | Tnfsf10 | Tgfa | ||||
| Ucn2 | Tnfsf11 | Tgfb1 | ||||
| Ucn3 | Tnfsf12 | Tgfb2 | ||||
| Vegfa | Tnfsf13 | Tgfb3 | ||||
| Vegfb | Tnfsf13b++ | Thpo | ||||
| Vegfc | Tnfsf14 | Timp2 | ||||
| Wnt2 | Tnfsf15 | Tnf | ||||
| Wnt5a | Tnfsf8++ | Tnfrsf11b | ||||
| Wnt11 | Tnfsf9++ | Tnfsf10 | ||||
| Xcl1 | Tslp++ | Tnfsf11 | ||||
| Tymp | Tnfsf12 | |||||
| Ucn2 | Tnfsf13 | |||||
| Ucn3 | Tnfsf13b++ | |||||
| Vegfa | Tnfsf14 | |||||
| Vegfb | Tnfsf15 | |||||
| Vegfc | Tnfsf8++ | |||||
| Wnt2 | Tnfsf9++ | |||||
| Wnt5a | Tslp++ | |||||
| Wnt7a++ | Tymp | |||||
| Wnt11 | Ucn2 | |||||
| Xcl1 | Ucn3 | |||||
| Vegfa | ||||||
| Vegfb | ||||||
| Vegfc | ||||||
| Wnt1+ | ||||||
| Wnt2 | ||||||
| Wnt5a | ||||||
| Wnt7a++ | ||||||
| Wnt11 | ||||||
| Xcl1 |
Ligands identified in 3 DPI, 7 DPI, and uninjured rat sciatic nerves based on microarray analysis and our curated ligand-receptor database (modified from Yuzwa et al., 2016). Ligands were considered expressed if their expression levels exceeded that of Ntf3 mRNA. Ligands expressed only in the uninjured nerve are indicated by one asterisk, only in the 3 DPI nerve by two asterisks, and only in the 7 DPI nerve by one plus sign. Ligands expressed in the 3 and 7 DPI nerves but not in the uninjured nerve are indicated by two plus signs. Also shown in a separate column are ligands expressed in all populations (intersect).
* uninjured only.
** 3 DPI only.
+ 7 DPI only.
++ 3 DPI and 7 DPI intersect.
The combined 3 and 9 DPI nerve scRNA-seq dataset (Fig. 1E) was analyzed to identify the percentage of cells in each cell type expressing the ligand mRNAs identified by the microarray analysis. For this analysis, Pdgfra-positive mesenchymal cells were separated into endoneurial cells (cluster 5; Fig. 1E) and all other mesenchymal cells. Ligand mRNAs were considered further only if they were detectable in 2% or more cells of at least one cell type. The 143 resultant injured nerve ligands (Table 2) were further analyzed in the other scRNA-seq datasets. Venn diagrams comparing expression of the 143 injured nerve ligands in the various datasets (Figs. 2G, 3E, 4C) were prepared using the VennDiagram package, modified to show proportional representation with the eulerAPE tool (Micallef and Rodgers, 2014).
Table 2.
Gene abundance analysis of ligand mRNAs in the combined 3 and 9 DPI nerve scRNA-seq dataset
| Gene abundance (%) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Epineurial/perineurial | Endoneurial | Combinedmesenchymal | VSM/pericytes | Endothelialcells | Schwanncells | Immune |
| Adm* * | 11.6 | 34.2 | 17.4 | 2.8 | BT | BT | BT |
| Agt* | 2.2 | 10.4 | 4.2 | 13.2 | BT | BT | BT |
| Angpt1* * | 17.3 | 23.7 | 18.9 | 21.1 | 2.1 | BT | BT |
| Angpt2* | 3.2 | BT | 2.6 | 29.0 | 18.5 | BT | BT |
| Angpt4** | 2.6 | BT | 2.0 | BT | BT | BT | BT |
| Apln* | 2.0 | 6.0 | 3.0 | BT | 11.2 | BT | BT |
| Artn | BT | BT | BT | BT | BT | 3.3 | BT |
| Bdnf* | 2.6 | BT | 2.3 | 2.2 | BT | 9.5 | BT |
| Bmp1** | 53.1 | 35.7 | 48.7 | 14.8 | 10.8 | 50.6 | 2.7 |
| Bmp2* | BT | 3.0 | 2.1 | 24.6 | 3.3 | BT | BT |
| Bmp4* | 2.8 | BT | 2.3 | BT | 16.6 | BT | BT |
| Bmp5* | BT | 3.8 | BT | 11.7 | BT | BT | BT |
| Bmp7** | 2.2 | 31.1 | 9.5 | BT | BT | BT | BT |
| Btc* | BT | 2.0 | 2.0 | BT | BT | 60.4 | BT |
| Cck | BT | BT | BT | 5.0 | BT | BT | BT |
| Ccl11** | 29.4 | 87.0 | 44.0 | 34.1 | 7.5 | 6.2 | 4.7 |
| Ccl19 | BT | BT | BT | 2.5 | BT | BT | BT |
| Ccl2** | 27.0 | 81.5 | 40.9 | 21.1 | 12.9 | 13.8 | 15.2 |
| Ccl24 | BT | BT | BT | BT | BT | BT | 2.4 |
| Ccl25 | BT | BT | BT | BT | BT | 2.2 | BT |
| Ccl3* | 3.0 | 4.2 | 3.3 | 3.2 | 2.3 | 3.1 | 13.7 |
| Ccl5* | 3.0 | 2.8 | 2.9 | BT | BT | BT | 6.4 |
| Ccl7** | 21.8 | 78.0 | 36.1 | 10.4 | 7.7 | 6.6 | 7.7 |
| Ccl9** | 11.8 | 75.0 | 27.9 | 4.1 | 6.1 | 5.9 | 22.5 |
| Clcf1* | 3.7 | 4.5 | 3.9 | BT | 4.3 | 16.0 | BT |
| Crlf1* | 3.2 | 25.9 | 9.0 | BT | BT | 35.2 | BT |
| Csf1** | 32.3 | 31.2 | 32.0 | 13.9 | 11.7 | 11.6 | 3.3 |
| Ctgf** | 25.7 | 36.9 | 28.5 | 25.6 | 33.3 | 3.5 | 2.2 |
| Cx3cl1** | 4.0 | 14.4 | 6.7 | BT | 5.3 | BT | BT |
| Cxcl1** | 27.4 | 59.3 | 35.5 | 38.2 | 29.0 | 11.7 | 9.5 |
| Cxcl10** | 2.6 | 9.8 | 4.4 | 3.2 | 4.1 | 6.6 | 2.0 |
| Cxcl12** | 46.6 | 52.6 | 48.1 | 33.8 | 44.0 | 5.9 | 5.0 |
| Cxcl13** | 6.7 | BT | 5.1 | BT | BT | BT | BT |
| Cxcl16* | 6.3 | 4.2 | 5.7 | 2.8 | 3.7 | BT | 15.7 |
| Cxcl2* | 12.3 | 22.9 | 15.0 | 7.3 | 10.4 | 7.2 | 22.9 |
| Cxcl9** | 5.3 | 2.7 | 4.6 | BT | 4.6 | BT | BT |
| Dhh | BT | BT | BT | BT | 6.3 | 30.1 | BT |
| Dll1 | BT | BT | BT | BT | 6.4 | BT | BT |
| Dll4 | BT | BT | BT | BT | 10.0 | BT | BT |
| Ebi3 | BT | BT | BT | BT | BT | BT | 2.5 |
| Eda** | 6.0 | 9.7 | 7.0 | BT | BT | 3.7 | BT |
| Edn3** | 3.1 | BT | 2.3 | BT | BT | BT | BT |
| Efna1* | 3.4 | 8.0 | 4.5 | BT | 24.9 | BT | BT |
| Efna2** | 3.3 | 3.8 | 3.4 | 2.5 | BT | 3.5 | BT |
| Efna4** | 4.8 | 6.2 | 5.2 | 3.2 | BT | 4.0 | BT |
| Efna5** | 5.1 | 6.7 | 5.5 | BT | BT | BT | BT |
| Efnb1** | 22.5 | 39.2 | 26.8 | 9.5 | 11.4 | 11.4 | 2.3 |
| Efnb2** | 13.0 | 42.4 | 20.4 | 4.1 | 16.4 | 4.0 | BT |
| Fgf1* | 7.1 | 4.8 | 6.5 | 9.8 | BT | 2.2 | BT |
| Fgf10** | 7.5 | BT | 5.8 | BT | BT | BT | BT |
| Fgf18** | 8.5 | BT | 6.8 | BT | BT | BT | BT |
| Fgf5 | BT | BT | BT | BT | BT | 19.4 | BT |
| Fgf7** | 26.1 | 37.7 | 29.1 | 3.2 | BT | 12.5 | BT |
| Figf** | 15.8 | 11.2 | 14.6 | BT | BT | BT | BT |
| Fstl1** | 96.8 | 96.5 | 96.7 | 85.8 | 56.5 | 33.4 | 19.6 |
| Gas6** | 44.3 | 2.3 | 35.9 | 13.6 | 22.5 | 2.2 | 6.2 |
| Gdf10** | 24.5 | 4.7 | 19.5 | BT | 3.3 | 2.0 | BT |
| Gdf11** | 4.3 | 10.4 | 5.9 | 10.1 | 3.4 | 3.9 | BT |
| Gdnf | BT | BT | BT | BT | BT | 21.8 | BT |
| Gmfb** | 23.4 | 34.6 | 26.3 | 10.7 | 17.5 | 27.9 | 11.6 |
| Gmfg | BT | BT | BT | BT | 5.7 | BT | 17.9 |
| Gnrh1** | BT | 2.3 | BT | BT | BT | BT | BT |
| Grp | BT | BT | BT | BT | 3.4 | BT | BT |
| Hbegf* | 6.5 | 5.2 | 6.2 | 10.7 | 19.2 | 43.9 | BT |
| Hgf** | 4.0 | BT | 3.1 | BT | BT | BT | BT |
| Igf1** | 76.6 | 84.3 | 78.6 | 15.8 | 31.7 | 16.7 | 13.8 |
| Igf2* | 15.5 | 8.2 | 13.6 | 16.4 | 6.1 | 2.4 | BT |
| Il15** | 2.2 | 9.0 | 3.9 | BT | 7.4 | BT | BT |
| Il16* | BT | 3.0 | BT | BT | 2.3 | BT | 7.9 |
| Il18** | 6.4 | 5.3 | 6.1 | BT | BT | BT | 2.5 |
| Il1b* | 5.6 | 7.3 | 6.1 | 5.0 | 5.7 | 5.0 | 18.6 |
| Il33** | 39.1 | 63.3 | 45.3 | 2.5 | 3.6 | 6.4 | 2.2 |
| Il6* | 7.2 | 7.0 | 7.1 | 12.6 | 15.2 | BT | BT |
| Inha** | BT | 2.0 | BT | BT | BT | BT | BT |
| Inhba** | 12.6 | 11.9 | 12.4 | 9.8 | BT | BT | 1.9 |
| Inhbb** | BT | 12.7 | 4.5 | BT | 9.2 | BT | BT |
| Jag1* | 20.3 | 17.2 | 19.5 | 33.1 | 17.8 | 8.4 | 2.8 |
| Jag2 | BT | BT | BT | BT | 8.4 | BT | BT |
| Lif** | 2.6 | 13.2 | 5.3 | BT | BT | 5.9 | BT |
| Ltb | BT | BT | BT | BT | BT | BT | 12.2 |
| Mdk** | 39.0 | 51.1 | 42.1 | 9.1 | 4.0 | 25.1 | BT |
| Metrn* | 7.1 | 14.5 | 9.0 | 8.5 | 3.0 | 57.8 | 2.6 |
| Mif** | 32.7 | 40.2 | 34.6 | 26.5 | 31.4 | 29.5 | 26.0 |
| Nenf** | 54.7 | 59.1 | 55.8 | 42.6 | 29.4 | 50.8 | 8.4 |
| Ngf* | 2.4 | 6.7 | 3.5 | 10.4 | BT | BT | BT |
| Nov** | 25.0 | 11.4 | 21.5 | 6.0 | 5.4 | 5.0 | BT |
| Nppc** | 2.2 | 5.3 | 3.0 | BT | BT | BT | BT |
| Ntf3* | 3.4 | BT | 2.9 | 4.4 | 4.8 | BT | BT |
| Ntn1** | 19.2 | 25.5 | 20.8 | BT | 6.0 | 4.6 | BT |
| Osm | BT | BT | BT | BT | BT | BT | 8.3 |
| Pdgfa* | 12.1 | 7.8 | 11.0 | 49.5 | 6.8 | 50.6 | 5.7 |
| Pdgfb | BT | BT | BT | 2.5 | 22.3 | BT | 2.4 |
| Pdgfc** | 7.2 | BT | 5.8 | 6.0 | 4.3 | BT | BT |
| Pf4* | 2.3 | 4.3 | 2.8 | BT | BT | BT | 10.0 |
| Pgf** | 2.0 | 12.2 | 4.6 | 7.6 | BT | BT | BT |
| Pomc** | BT | 3.3 | BT | BT | 3.3 | 2.2 | BT |
| Pthlh** | 10.4 | 13.7 | 11.2 | BT | BT | 2.4 | BT |
| Ptn* | 35.9 | 10.4 | 29.4 | 2.2 | 19.9 | 52.5 | BT |
| Rspo1** | 2.2 | BT | BT | BT | BT | BT | BT |
| Rspo3** | 6.0 | BT | 4.7 | BT | 2.4 | BT | BT |
| Rtn4* | 57.9 | 68.3 | 60.5 | 51.7 | 47.1 | 73.6 | 28.3 |
| Sema3b* | 7.7 | 12.4 | 8.9 | BT | BT | 53.8 | BT |
| Sema3c** | 35.4 | 9.0 | 28.7 | BT | BT | 25.1 | BT |
| Sema3d** | 9.6 | 2.0 | 7.6 | BT | BT | BT | BT |
| Sema3e* | 2.0 | BT | BT | BT | BT | 31.0 | BT |
| Sema3f* | 2.3 | 4.5 | 2.9 | 2.8 | 14.5 | 2.9 | BT |
| Sema3g* | BT | 3.0 | BT | BT | 11.8 | 9.0 | BT |
| Sema4a* | BT | 3.7 | 2.0 | BT | BT | BT | 6.9 |
| Sema4b* | 2.0 | 3.2 | 2.3 | 3.5 | BT | 7.7 | BT |
| Sema4c* | 8.4 | 12.0 | 9.3 | 6.3 | 10.2 | 16.0 | BT |
| Sema4d | BT | BT | BT | BT | BT | BT | 14.5 |
| Sema4f | BT | BT | BT | BT | BT | 18.3 | BT |
| Sema5a* | 4.3 | 3.5 | 4.1 | 13.2 | BT | BT | BT |
| Sema5b | BT | BT | BT | 10.4 | BT | BT | BT |
| Sema6a* | 4.0 | 14.5 | 6.7 | BT | 35.6 | 9.4 | BT |
| Sema6b* | 3.1 | 7.0 | 4.1 | BT | 18.1 | BT | BT |
| Sema6c** | 7.2 | 4.0 | 6.4 | BT | BT | 2.0 | BT |
| Sema6d* | 5.9 | 11.7 | 7.3 | 18.9 | 9.5 | 16.7 | BT |
| Sema7a* | 7.5 | 19.5 | 10.5 | BT | 17.4 | 20.0 | BT |
| Sfrp1** | 42.3 | 31.9 | 39.6 | 4.1 | 3.4 | 8.8 | BT |
| Sfrp2** | 29.6 | 6.5 | 23.8 | 2.2 | 3.3 | BT | 2.4 |
| Sfrp4** | 61.5 | 51.9 | 59.1 | 8.5 | 9.4 | 11.0 | 6.3 |
| Sfrp5** | 8.8 | 3.2 | 7.4 | BT | 2.1 | 8.1 | BT |
| Shh | BT | BT | BT | BT | BT | 12.8 | BT |
| Tgfa | BT | BT | BT | BT | 2.6 | BT | BT |
| Tgfb1* | 6.5 | 12.0 | 7.9 | 6.0 | 13.4 | 10.3 | 12.4 |
| Tgfb2* | 13.3 | 2.7 | 10.6 | 16.4 | 5.1 | 6.2 | BT |
| Tgfb3* | 26.3 | 8.7 | 21.8 | 27.4 | 2.3 | 4.6 | BT |
| Tnf* | BT | 2.2 | BT | BT | BT | BT | 7.7 |
| Tnfrsf11b** | 6.6 | BT | 5.0 | BT | BT | BT | BT |
| Tnfsf10 | BT | BT | BT | BT | 22.8 | 1.1 | BT |
| Tnfsf12** | 15.0 | 16.7 | 15.4 | 11.4 | 9.1 | 6.8 | 7.3 |
| Tnfsf14 | BT | BT | BT | BT | BT | BT | 2.3 |
| Tnfsf8** | 3.8 | 2.0 | 3.3 | BT | BT | BT | BT |
| Tnfsf9** | 2.8 | 5.8 | 3.6 | 5.4 | BT | BT | BT |
| Tslp** | 2.5 | 4.8 | 3.1 | 2.8 | 2.4 | BT | BT |
| Ucn2 | BT | BT | BT | BT | BT | 15.0 | BT |
| Vegfa** | 28.2 | 18.7 | 25.8 | 3.8 | 4.1 | 2.6 | 7.0 |
| Vegfb* | 7.1 | 7.0 | 7.0 | 7.3 | 6.0 | 8.4 | 3.3 |
| Vegfc* | 2.0 | BT | BT | BT | 5.5 | BT | BT |
| Wnt11** | 2.7 | BT | 2.2 | BT | BT | BT | BT |
| Wnt2** | 6.5 | BT | 5.3 | BT | BT | BT | BT |
| Wnt5a** | 15.9 | 14.4 | 15.5 | 4.4 | BT | BT | BT |
The combined 3 and 9 DPI scRNA-seq dataset (Fig. 1E) was analyzed for the percentage of cells within the defined nerve cell types that expressed ligand mRNAs as identified by microarray analysis (Table 1). Ligand mRNAs were only considered to be detected if they were expressed in at least 2% of cells within at least one cell type. BT = below threshold and indicates that <2% of cells detectably expressed the ligand mRNA. Ligands annotated with one asterisk in the leftmost column were expressed in ≥2% Pdgfra-positive mesenchymal cells and two asterisks indicate ligands with the highest expression in either the epineurial/perineurial or endoneurial Pdgfra-positive mesenchymal cells.
* >2% expression in Pdgfra-positive cells.
** >2% expression and highest expression in Pdgfra-positive cells.
RNA isolation and microarray analysis
Total RNA was extracted from E15 rat dorsal root ganglion (DRG) sensory neurons and neonatal rat superior cervical ganglion (SCG) sympathetic neurons using the RNeasy Micro kit (QIAGEN) according to manufacturer’s instructions. RNA was isolated from the distal rat sciatic nerve following injury (3 and 7 d after resection) and contralateral uninjured sciatic nerve as follows. After harvesting, nerves were flash frozen in liquid nitrogen and stored at −80°C until RNA isolation was performed. Nerve tissue was lysed using a Dounce homogenizer with cooled TRIzol (1 ml/50–100 mg of tissue) followed by passing the lysate through a 23.5-gauge needle. The homogenate was then spun at 12,000 × g at 4°C for 10 min, with the clear supernatant transferred to a new tube and allowed to incubate at room temperature for 5 min, 0.2 ml of chloroform was then added per milliliter of homogenate, shaken vigorously for 15 s, incubated 2–3 min at room temperature, and spun at 12,000 × g for 15 min at 4°C. The resulting aqueous phase was removed and added to a new tube with equal volume of 70% ethanol before RNA isolation with the RNeasy Micro kit (QIAGEN) according to manufacturer’s instructions. Microarray analysis was then performed at the Center for Applied Genomics at the Hospital for Sick Children (Toronto, ON). A total of 250 ng of total RNA was processed using the Affymetrix WT Plus kit to generate cDNA following Bioanalyzer analysis to confirm the quality of the RNA. 5.5 μg of labeled cDNA was hybridized onto Rat Gene 2.0 ST arrays using the Affymetrix FS450_0002 hybridization protocol and scanned using the Affymetrix GeneChip Scanner 3000.
Normalization and differential gene expression analysis of microarray data
Raw probe intensity values were background corrected and then normalized with quantile normalization. Values were transformed into the log2 scale and summarized into probesets using the robust multichip analysis (RMA) algorithm in the oligo Bioconductor package in R. For all of the rat microarray datasets, gene annotation was performed using the “ragene20sttranscriptcluster.db” library in R. Motor neuron microarray data (Kaplan et al., 2014) was obtained from mouse P7 lumbar motor neurons from the GEO database under the accession number GSE52118. For these data, raw probe intensity values were normalized and summarized into probesets as described above, except for the Affymetrix Mouse Genome 430 2.0 Array (n = 3 replicates), and gene annotation was performed using the “mouse4302.db” library in R. The Limma Bioconductor package was used to calculate differential gene expression between the sensory neurons (DRGs, n = 6 replicates) and sympathetic neurons (SCGs, n = 4 replicates). Bayesian statistics were calculated and annotated receptor genes were considered to be differentially expressed if they were ≥2-fold different with p < 0.05 false discovery rate (FDR; Benjamini and Hochberg correction).
Cell-surface capture mass spectrometry
Cell-surface mass spectrometry was conducted based on modified published protocols (McDonald et al., 2009; Schiess et al., 2009; Yuzwa et al., 2016). Briefly, 6-cm dishes containing sensory and sympathetic neurons were washed with coupling buffer [1× PBS (pH 6.5) and 0.1% fetal bovine serum (FBS)] following removal of the medium. Cultures were treated with 5 mm NaIO4 in coupling buffer for 30 min at room temperature in the dark and lysed in buffer containing 20 mm Tris-HCl, 150 mm NaCl, 0.0002% NaN3, 1% NP-40 (pH 7.5), and one complete mini-protease inhibitor tablet (Roche) per 10 ml of lysis buffer. Lysates were passed through 23-gauge needles, protein concentrations determined using the Pierce BCA assay kit (catalog #23227, Thermo Fisher Scientific), and equal amounts of total protein (between 0.9 and 1.5 mg for sensory neurons and <1 mg for sympathetic neurons) were added to 200 μl of Ultralink Hydrazide resin (Pierce) pre-equilibrated with lysis buffer and rotated overnight at room temperature. The unbound protein was removed via centrifugation the following day, washed twice with 8 m urea and three times with 50 mm ammonium bicarbonate (pH 8; ABC). The resin was treated with 50 mm dithiothreitol (DTT; American Bioanalytical) in ABC at 37°C for 60 min, washed once with ABC, and incubated with 65 mm iodacetamide in ABC at room temperature in the dark for 30 min. The resin was washed once with ABC, once with 1.5 m NaCl and three times with ABC and incubated with 40 ng/μl of Trypsin (Worthington) in ABC overnight at 37°C. The following day, the resin was washed three times with 1.5 m NaCl, followed by 80% acetonitrile, methanol, water, and ABC and then incubated with 1300 U/ml of PNGaseF (New England Biolabs) at 37°C overnight in ABC. The following day, the eluted peptides were collected from the resin and the resin washed once with ABC and combined with the eluted peptides. The eluates were lyophilized overnight and prepared for mass spectrometry using C18 reverse-phase ZipTips (EMD Millipore). Peptides were lyophilized and resuspended in 11 μl of 0.1% formic acid following elution from ZipTips. Samples were then analyzed on an Orbitrap analyzer (Q-Exactive, Thermo Fisher) outfitted with a nano-spray source and EASY-nLC nano-LC system (Thermo Fisher); 5 μl of the resuspended peptide mixtures were loaded onto a 75 μm × 50 cm PepMax RSLC EASY-Spray column filled with 2 μm C18 beads (Thermo Fisher) at a pressure of 800 Bar. Peptides were eluted over 60 min at a rate of 250 nl/min using a 0–35% acetonitrile gradient in 0.1% formic acid. Peptides were introduced by nano-electrospray into the Q-Exactive mass spectrometer (Thermo Fisher). The instrument method consisted of one MS full scan (400–1500 m/z) in the Orbitrap mass analyzer with an automatic gain control (AGC) target of 1e6, maximum ion injection time of 120 ms, and a resolution of 70,000 followed by 10 data-dependent MS/MS scans with a resolution of 17,500, an AGC target of 1e6, maximum ion time of 120 ms, and one microscan. The intensity threshold to trigger a MS/MS scan was set to 1.7e4. Fragmentation occurred in the HCD trap with normalized collision energy set to 26. The dynamic exclusion was applied using a setting of 8 s. Peptide and protein identification was performed using PEAKS version 8 software (Bioinformatics Solutions Inc.). Peptides and proteins were identified at the 0–0.1% FDR level in the sensory neuron dataset and at the 0.8–1% FDR level in the sympathetic neuron dataset. With both sensory and sympathetic neuron culture datasets, data were pooled across the three samples, and we included all proteins where at least a single peptide was detected in at least one of the samples.
Identification of receptors based on the microarray and mass spectroscopy data
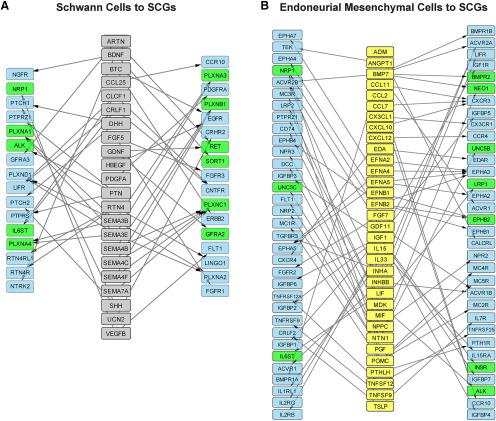
Receptors were identified in both the microarray and mass spectrometry data using the ligand-receptor database described in Yuzwa et al. (2016), with modifications. In addition, proteins that were classified as receptors using the PANTHER (http://pantherdb.org) protein classification system, but not necessarily present in the ligand-receptor database, were included. Manual curation of receptors with known signaling functions (such as the Plxn receptors) as well as other genes identified as receptors by Gene Ontology (GO) terms were also included as an update to the previously published version of the ligand-receptor database. Receptor classifications (Fig. 5B) were based on GO terms and descriptions provided by websites including GeneCards (http://genecards.org) and UniProt (http://uniprot.org).
Figure 5.
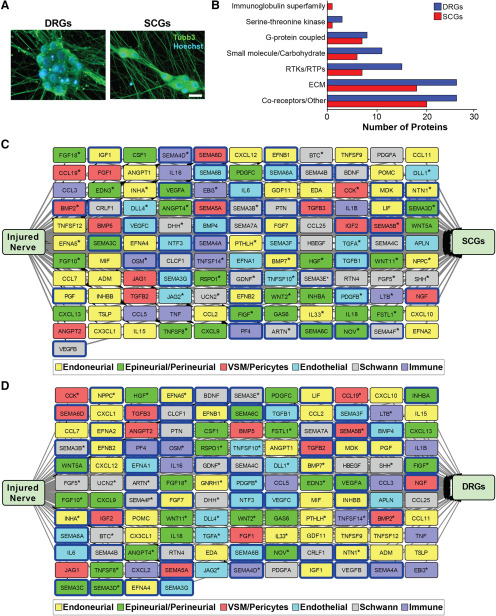
Sensory and sympathetic neuron receptor expression and modeling of their predicted interactions with injured nerve-derived ligands (see also Extended Data Fig. 5-1). A, Images of E15 rat DRG sensory neurons cultured for 9 d (left panel) and neonatal rat SCG sympathetic neurons cultured for 6 d (right panel), immunostained for βIII-tubulin (Tubb3; green). Cells were counterstained with Hoechst 33258 to show cell nuclei (blue). Scale bars = 20 μm. B, Bar graphs showing types of receptor proteins detected on the surface of sensory and sympathetic neurons following cell-surface capture mass spectrometry. Classification of receptors was based on GO terms identified via the UniProt database (http://uniprot.org) as well as manual curation of all detected cell-surface proteins. RTKs/RTPs = receptor tyrosine kinases and receptor tyrosine phosphatases; ECM = extracellular matrix. C, D, Models showing predicted unidirectional paracrine interaction networks between the 143 injured nerve ligands and receptors expressed by sympathetic (C, SCGs) and sensory (D, DRGs) neurons, as defined at the transcriptional and proteomics levels. Interactions were predicted by computational modeling using the ligand-receptor database, and then manually curated for well-validated interactions. Nodes represent ligands that are color coded to identify the injured nerve cell type with the highest expression of the ligand mRNA based on the scRNA-seq analysis. Predicted interactions where the receptors were detected at the protein level are indicated by a blue box around the corresponding ligand node. Asterisks indicate ligands that were expressed at 4-fold higher levels in the indicated cell type as compared with all other injured nerve cell types. Arrows indicate directionality of interactions.
Identification of cell-surface proteins on sensory and sympathetic neurons. A, Bar graphs showing the percentage of cells expressing the neuronal protein βIII-tubulin (Tubb3), the Schwann cell protein S100β, or the fibroblast protein Fibronectin in cultures of DRG sensory neurons or SCG sympathetic neurons as shown in Figure 5A. The total number of cells in the cultures was determined by counterstaining with Hoechst 33258. Values: mean ± SEM, n = 6 for DRGs, n = 4 for SCGs except for cultures immunostained for Fibronectin where n = 2. B, Venn diagram showing the overlap of cell-surface proteins detected by mass spectrometry in sensory neurons and sympathetic neurons. All proteins included were annotated by the terms “cell membrane” and/or “secreted” by the UniProtKB database (http://uniprot.org). C, Bar graphs showing classification of the proteins detected by cell-surface capture mass spectrometry on sensory neurons (DRGs, blue) and sympathetic neurons (SCGs, red). Proteins were classified as receptors based on the ligand-receptor database, GO terms in the PANTHER classification system, as well as by manual curation, and were further classified into receptor types as shown in Figure 5B. The remainder of the graph includes proteins classified using PANTHER (http://pantherdb.org). D, Graphs showing the distribution of proteins detected by cell-surface capture mass-spectrometry relative to their transcript expression levels (based on microarray analyses described in the text) in sensory neurons (DRGs, left) and sympathetic neurons (SCGs, right). The cutoffs used to define receptor expression in the microarray data were based on the receptors detected by mass spectrometry analysis that had the lowest levels of mRNA expression. This was Itgam for sensory neurons (DRGs) and Sorcs3 for sympathetic neurons (SCGs, shown in red). Download Figure 5-1, TIF file (573.7KB, tif) .
Computational modeling and pre-processing of data
To define receptors for generation of ligand-receptor models, gene expression values from the injured nerve and from the motor, sensory, and sympathetic neuron microarrays were first averaged across replicates, and in the case of multiple probes corresponding to the same receptor gene, the highest expressed probe was used (this was following averaging). For rat retinal ganglion cells (RGCs), we analyzed RNA-seq data from Blanco-Suarez et al. (2018) as obtained from the GEO database (accession GEO:GSE108484). Data from control RGCs were used (n = 3) and processed as described above for the microarray data. Expression values with fragments per kilobase of transcript per million mapped reads (FPKM) >1 were considered expressed and included for analysis. For the modeling, receptor mRNAs that had expression values exceeding the thresholds as described in the results were included. These receptors and the 143 injured nerve ligands (Table 2) were then analyzed using a custom Python script (“Cellcellinteractnet,” Python version 2.7.6) and custom ligand-receptor database (database modified from Yuzwa et al., 2016) to predict ligand-receptor interactions. Models were all constructed from this information using Cytoscape (3.7.0). The Venn diagram comparing predicted interactions in Figure 7C was prepared using the VennDiagram package, modified to show proportional representation with the eulerAPE tool (Micallef and Rodgers, 2014).
Figure 7.
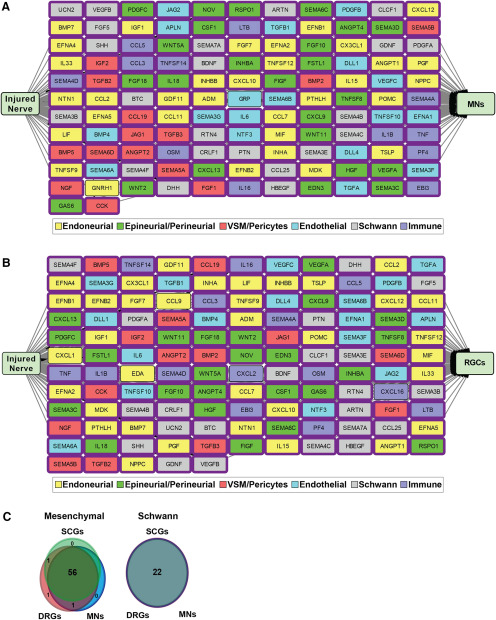
Modeling the potential paracrine interactions between ligands of the injured sciatic nerve and receptors on motor neurons and RGCs. A, B, Models showing predicted unidirectional paracrine interaction networks between the 143 injured nerve ligands and receptors expressed by motor neurons (A, MNs) and RGCs (B), as defined at the transcriptional level. Interactions were predicted by computational modeling using the ligand-receptor database, and then manually curated for well-validated interactions. Nodes represent ligands that are color coded to identify the injured nerve cell type with the highest expression of the ligand mRNA based on the scRNA-seq analysis. Arrows indicate directionality of interactions. Predicted interactions that were shared among all four injured nerve-neuron models are indicated by a purple box around the corresponding ligand nodes. C, Venn diagrams showing the overlap of predicted ligand-receptor interactions between injured nerve mesenchymal cells (left) or Schwann cells (right) and sympathetic neurons (SCGs), sensory neurons (DRGs), and motor neurons (MNs).
Sympathetic neuron cultures
SCGs of newborn (P1–P2) Sprague Dawley rats were dissected, dissociated and cells were plated at eight ganglia per 3.5-cm tissue culture-treated dish for microarrays, ∼1 × 106 cells per 6-cm dish for mass spectrometry, or ∼1.4 × 105 cells per 13-mm glass coverslip for immunostaining. Neurons were plated on dishes and coverslips coated with poly-d-lysine and laminin (1 μg/ml laminin; VWR). Neurons were cultured in growth medium composed of UltraCULTURE medium (Lonza), 2 mm l-glutamine (Lonza), 100 U/ml penicillin (Wisent) with 100 μg/ml streptomycin (pen/strep; Wisent), and 50 ng/ml NGF (Cedarlane). Neurons were cultured in growth medium containing 3% heat-inactivated FBS for 3 d with inclusion of 7.2 μm cytosine arabinoside (CA) for the first day, and 3.6 μm CA for the second and third days. Cultures were switched to growth medium alone for an additional 3 d, as described previously (Park et al., 2010; Feinberg et al., 2017).
Sensory neuron cultures
Sensory neurons from E15 rat DRGs were cultured as described (Feinberg et al., 2010, 2017). For immunostaining, neurons were plated at a density of 2 × 105 cells per 13-mm glass coverslip precoated with laminin (VWR), and poly-d-lysine (Sigma-Aldrich). Neurons were plated at a density of 1 × 106 cells per 6-cm dish for proteomics. For microarrays, neurons were plated under both conditions. Initially, cells were plated in a medium #1 containing Neurobasal medium (Invitrogen), GlutaMAX (Invitrogen), 50 ng/ml NGF (Cedarlane), B27 supplement (Invitrogen), and pen/strep. The day after plating, cultures were treated for 2 d in medium #2 composed of Eagles basal medium (Invitrogen), ITS supplement (Sigma), 0.2% BSA (Sigma), 4 mg/ml d-glucose (Sigma), GlutaMAX (Invitrogen), 50 ng/ml NGF (Cedarlane), and pen/strep with 0.8 μm CA to eliminate mitotic non-neuronal cells. Cells were then treated with another cycle of medium #1 for 2 d followed by a final 2-d cycle of medium #2. After the second CA treatment, cultures were grown in medium #1 for two additional days before cell harvest.
Culturing and sorting mesenchymal cells from the sciatic nerve for ELISA analysis
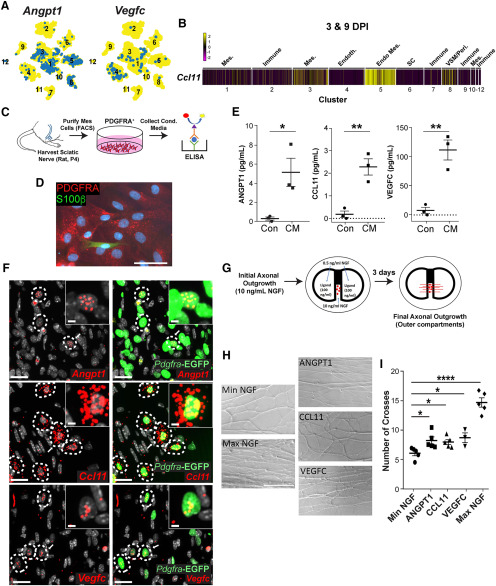
Sciatic nerves were harvested from P4 Sprague Dawley rat pups. Each biological sample included four litters of pups (two litters harvested per day over 2 d). Nerve cells from two litters were dissociated and plated on 6-cm dishes in medium consisting of low-glucose DMEM/F12 (3:1, Invitrogen) and 1% pen/strep (referred to as basal medium) with 10% FBS. In order to isolate PDGFRα-positive mesenchymal cells, 1–2 d after plating, cells were treated with an antibody solution containing basal medium with goat anti-PDGFRα antibody (1:250, R&D Systems, catalog #AF1062) and donkey anti-goat Alexa Fluor 488 secondary (1:500, Thermo Fisher Scientific, catalog #A11055) that was preincubated for 30 min at room temperature. Cells were treated for 1 h at 37°C with the antibody solution before being returned to basal medium with 10% FBS overnight. PDGFRα-positive cells were then isolated using a Mo-Flo XDP sorter (Beckman Coulter). Following the sort, between 4–9 × 105 cells were re-plated on 6-cm dishes with basal medium and 10% FBS. Approximately 72 h later, medium containing FBS was removed, cells were washed with HBSS, and basal medium without serum was added for conditioning. Cells were treated with cycles of basal medium for conditioning (24–96 h) and basal medium with FBS for a period of 2.5–4 weeks total. Collected conditioned medium was spun down at 1300 rpm for 5 min to removed cell debris and frozen at −80°C until ELISAs were performed. ELISA assays for rat ANGPT1 (catalog #LS-F10827, LSBio), CCL11 (catalog #LS-F11046, LSBio), and VEGFC (catalog #LS-F5482, LS Bio) were performed and results were analyzed according to manufacturer’s instructions.
Sympathetic neuron compartment cultures
Campenot cultures were established on 35-mm collagen-coated dishes as previously reported (Campenot et al., 1991); 20 × 20 mm Teflon chambers (Tyler Research) were grease-sealed and assembled on plates where the substratum was corrugated with 20 parallel tracks 200 μm wide using a pin rake (CAMP-PR, Tyler Research), forming lanes for outgrowth. Neonatal SCG sympathetic neurons were enzymatically and mechanically dissociated and plated in the central chamber at a density of three to four ganglia per compartment in 10 ng/ml NGF (Cedarlane, catalog #CLMCNET-001.1), methylcellulose Ultraculture media (Lonza) supplemented with 2 mm L-glutamine (Lonza), and antibiotics [100 U/ml penicillin (Wisent) and 100 μg/ml streptomycin (Wisent)]; 10 μm CA with 3% FBS was added to the cell bodies for 2–3 d as the axons were permitted to extend into the adjacent compartments containing 10 ng/ml NGF. After this period, central and side chambers were washed with fresh medium, medium in the cell body chamber was replaced with 10 ng/ml NGF, and medium on the axonal chambers was replaced with 100 ng/ml candidate ligand and varying amounts of NGF (0.5 ng/ml for experimental compartments, 50 ng/ml for maximum NGF control). Ligands included human recombinant ANGPT1 (Peprotech, catalog #130-06-5UG), murine recombinant Eotaxin (CCL11, Peprotech, catalog #250-01-5UG), and human recombinant VEGFC (Peprotech, catalog #100-20CD). Images of axon outgrowth were obtained at 3d post-ligand addition using a Zeiss AxioObserver Z1 microscope and outgrowth was analyzed using ImageJ software (NIH). To quantify the density of axonal growth in these compartments, a line was drawn perpendicular to the axis of the outgrowth within the furthest 1 mm of outgrowth where axons were maximally defasciculated and the number of axons crossing the line were quantified.
Immunostaining, imaging, and analysis of cultures
Immunostaining of cultured cells was performed on glass coverslips and tissue culture dishes. Cells were fixed for ∼10 min in 4% paraformaldehyde in PBS solution and washed three times (10 min per wash) in PBS before treatment with the primary antibody solution, which contained the antibody, 10% normal donkey or normal goat blocking serum (Jackson ImmunoResearch), and 0.3% Triton X-100 (Fisher) in PBS. After 1–2 h of incubation at room temperature, cells were washed three times (10 min per wash) with PBS. Cells were then incubated in a secondary antibody solution containing antibodies diluted 1:500 in 0.3% Triton X-100 (Fisher) in PBS. After 1 h, cells were then washed three times (10 min per wash) in PBS with Hoechst 33258 (Sigma) added in one of the washes to label nuclei. For glass slides, coverslips were mounted with PermaFluor mounting media (Thermo Fisher Scientific). For tissue culture dishes, mounting medium was added directly on to the cells, and coverslips were placed on top of this. Cells were imaged using either a Zeiss AxioImager M2 microscope with an X-Cite 120 LED light source and a C11440 Hamamatsu camera using Zen acquisition software in the case of glass coverslips, or with a Zeiss AxioObserver Z1 microscope with Zen software in the case of tissue culture dishes. To quantify cells, multiple regions per glass coverslip or dish were imaged and cells were counted. For purified sensory and sympathetic neuron culture quantifications, S100β-positive and βIII-tubulin-positive cells were counted in four regions per coverslip across two coverslips per biological replicate and summed before calculating percent positive immunoreactive cells. Fibronectin-positive cells were counted in three regions over two sympathetic neuron replicates. Total cell counts per biological replicate ranged from ∼100 to ∼600. In cases where cells in dishes were counted, between three and eight regions per dish were used. Images were then processed using Photoshop software (Adobe Systems Incorporated), where brightness and contrast were edited as appropriate.
Immunostaining of sections
Adult PdgfraEGFP/+ mice underwent unilateral sciatic nerve injury and injured distal sciatic nerve and uninjured contralateral sciatic nerve tissue was harvested at 9 DPI. Nerves were fixed overnight in 4% paraformaldehyde at 4°C, then cryoprotected overnight in 30% sucrose at 4°C. Nerves were then cryosectioned longitudinally at 12 μm and immunostained for PDGFRα, S100β and counterstained with Hoechst 33258 (Sigma). Images were acquired with a 20× objective lens on a Zeiss AxioImager M2 microscope with a light source, camera, and software setup as described above.
Antibodies
The following primary antibodies were used in this study at the indicated dilutions: rabbit anti-Fibronectin (1:500; Sigma; catalog #F3648), goat anti-PDGFRα (1:250 for cultures and 1:500 for sections; R&D Systems, catalog #AF1062), rabbit anti-S100β (1:500; Dako; catalog #Z0311), and mouse anti-βIII-tubulin (1:1000; BioLegend; catalog #801202). The following secondary antibodies were used in this study (all from Thermo Fisher Scientific and used at a dilution of 1:500, or two drops/ml in the case of the ReadyProbe): donkey anti-goat IgG Alexa Fluor 647 (catalog #A21447), donkey anti-mouse IgG Alexa Fluor 488 (catalog #A21202), donkey anti-mouse IgG ReadyProbes Alexa Fluor 488 (catalog #R37114), donkey anti-rabbit IgG Alexa Fluor 488 (catalog #A21206), and donkey anti-rabbit IgG Alexa Fluor 555 (catalog #A31572).
FISH
Single molecule FISH was performed on 9 DPI adult PdgfraEGFP/+ nerve sections with probes targeting Vegfc (catalog #492701), Ccl11 (catalog #464031), and Angpt1 (catalog #449271) mRNAs using the RNAscope kit (Advanced Cell Diagnostics), according to the manufacturer’s instructions. Briefly, freshly dissected nerves were fixed overnight in 4% PFA at 4°C and cryopreserved in 30% sucrose overnight (at 4°C) under RNase-free conditions using RNase-free reagents. Nerves were cryosectioned sagittally at a thickness of 14 μm. Sections were washed with ethanol, followed by tissue pretreatment (1:10 dilution) for 20 min, probe hybridization, and signal amplification. Positive signal was identified as red punctate dots. Digital images were acquired with a Quorum Spinning-Disk confocal microscope system using Volocity acquisition software (PerkinElmer). Z-stack confocal images were taken with an optical slick thickness of 0.3 μm, and projected Z-stacked images are shown. The scrambled probe provided with the RNAscope kit was used as a negative control.
Statistical analyses
With the exception of analyses related to the microarray and scRNA-seq data, all statistics were performed using GraphPad Prism 8 (GraphPad Software). Standard deviation values are given for the genes and transcripts per cell values for the scRNA-seq data. Scatter plots show standard error of the mean. Statistical significance was considered reached at p < 0.05. All statistical tests and conditions are described in the text. Graphs were generated in both GraphPad Prism 8 and Excel (Microsoft).
Data/software accessibility
Raw scRNA-seq and microarray datasets have been deposited in the GEO database under the ID codes GEO:GSE147285 and GSE146958. The Cellcellinteractnet code, the updated ligand-receptor database, and the raw proteomics data are available on request.
Results
Defining growth factors made within the injured and uninjured peripheral nerve
We hypothesized that peripheral nerves promote axon growth in part due to ligand secretion by resident nerve Pdgfra-positive mesenchymal cells. To test this idea, we analyzed the sciatic nerve, which contains axons from sensory, sympathetic and motor neurons. Initially, we confirmed that Pdgfrα-positive mesenchymal cells are found within the nerve endoneurium where axons are located. To do this, we analyzed control and injured sciatic nerve sections from mice carrying a transgene where EGFP is driven by Pdgfra regulatory sequences, and which thus tags Pdgfra-positive mesenchymal cells (PdgfraEgfp/+ mice; Hamilton et al., 2003). We resected sciatic nerves from these mice, and at 9 DPI immunostained for the PDGFRα protein product and for the Schwann cell marker S100β. For comparison, we performed a similar analysis of the contralateral uninjured sciatic nerve. This analysis identified many Pdgfra-EGFP-positive, PDGFRα protein-positive cells within the endoneurium of both the control and injured nerves (Fig. 1A), consistent with a similar recent analysis in Carr et al. (2019).
Having confirmed the presence of many mesenchymal cells in the endoneurium, we obtained an overview of ligands expressed in the sciatic nerve by performing global transcriptomic analysis of rat sciatic nerves that were either uninjured or that were injured by resection 3 or 7 d earlier. In all cases, we isolated total RNA from four independent biological replicates of the distal nerve segment and analyzed samples on Affymetrix GeneChip Rat Gene 2.0 ST Arrays. To define ligand mRNAs, we used a curated database of secreted ligands and their cognate receptors (Yuzwa et al., 2016), establishing an expression cutoff by considering only mRNAs expressed at similar or higher levels than neurotrophin 3 (Ntf3) mRNA, which is expressed in and important for peripheral nerves. Ntf3 mRNA was expressed in the top 67.6%, 70.3%, and 71.2% of mRNAs in control, 3-d injured, and 7-d injured distal nerves, respectively. This analysis identified 238, 249, and 258 ligand mRNAs in the uninjured, 3-d injured, and 7-d injured nerves, respectively (Fig. 1B; Table 1). Most (226) were detected in all three conditions, but 31 ligand mRNAs were found only in the injured nerves, including Areg, Ccl20, Il6, and Wnt7a and five ligands were detected only in the control nerve, including Il21 (Table 1).
Single-cell profiling defines similarities between peripheral nerves at 3 and 9 DPI
The microarray analysis defined the global nerve ligand environment. To ask which nerve cells express these ligands, we performed single-cell RNA sequencing (scRNA-seq), initially analyzing the distal sciatic nerve 3 d following a nerve transection. To do this, we dissociated the distal transected sciatic nerve, removed myelin debris with myelin-removal beads, and sequenced cells using high-throughput, droplet-based scRNA-seq (for details of all sequencing runs, see Materials and Methods). To analyze these individual transcriptomes, we then used a pipeline that incorporates extensive low-level data quality analysis with visualization and clustering methods that use evidence-based parameter selection (Innes and Bader, 2019), as previously described (Yuzwa et al., 2017; Carr et al., 2019; Storer et al., 2020). Genes with high variance were then used to compute principal components as inputs for projecting cells in two dimensions using t-SNE followed by graph-based clustering (Butler et al., 2018) with a range of resolution parameters.
This analysis defined nine distinct clusters containing 2075 cells in the 3-d injured sciatic nerve dataset (Fig. 1C). To assign cell types to these different clusters, we used well-characterized marker genes (for all cell type-specific marker genes used, see Materials and Methods; Fig. 1D; Extended Data Fig. 1-1A). This approach identified Pecam1/Cd31-positive endothelial cells, Sox10-positive Schwann cells, Aif1/Iba1-positive macrophages, Trbc2-positive immune cells, and Rgs5-positive VSM and pericyte cells. As previously seen in the 9-d injured sciatic nerve (Carr et al., 2019), we also identified distinct populations of Pdgfra-positive nerve mesenchymal cells, including Dpp4-positive epineurial cells (clusters 1, 4, and 6) and Wif1-positive endoneurial cells (cluster 3).
We next asked whether distal sciatic nerve cells were similar at 3 and 9 d following a transection, taking advantage of a recently published 9 DPI scRNA-seq analysis where nerve cells were also isolated using myelin beads (Carr et al., 2019). To do this, we extracted the relevant raw transcriptomes from the previously published 9 DPI dataset, combined them with the 3 DPI raw transcriptomes we had generated, and put this combined 5395 cell dataset through our computational pipeline, which corrects for any differences in sequencing depth and library size. As seen with the 3 DPI dataset alone, marker gene analysis (Fig. 1E; Extended Data Fig. 1-1B,C) identified clusters containing endothelial cells, VSM/pericyte cells, Schwann cells, macrophages, immune cells, and Pdgfra-positive mesenchymal cells. These mesenchymal cells included Etv1-positive endoneurial cells (cluster 5), Pcolce2-positive epineurial cells (cluster 3), Msln-positive perineurial cells (cluster 10), and differentiating nerve bridge cells enriched for Dlk1 (cluster 1; Fig. 1E; Extended Data Fig. 1-1B,C). Analysis of the dataset of origin (Fig. 1F) showed that all cell type clusters were comprised of intermingled 3 and 9 DPI cells except for cluster 7, which included Cd19-positive B cells that came almost exclusively (99.4%) from the 9 DPI time point (Extended Data Fig. 1-1B). Similar results were obtained following batch correction. Thus, 3- and 9-d distal transected sciatic nerve cells are transcriptionally similar, although there is an increased proportion of B cells at 9 d.
Schwann cells, mesenchymal cells, vasculature, and immune cells make distinct contributions to the peripheral nerve ligand environment
To understand how the different cell types contributed to the injured nerve environment, we analyzed the combined 3- and 9-d scRNA-seq dataset for expression of ligands identified in the microarray analysis (Fig. 1B; Table 1), excluding extracellular matrix proteins and ligands without well-defined, receptor-mediated paracrine roles. This analysis identified 143 ligands that were detectably expressed in ≥ 2% of at least one nerve cell type (Table 2), including many well-known nerve ligands such as the neurotrophins NGF, BDNF, and NT3, the Ret family ligands GDNF, Artemin and Neurturin, DHH, and various Semaphorin and FGF family members. Notably, Pdgfra-positive mesenchymal cells detectably expressed more ligand mRNAs than any other nerve cell type (118/143, or 82.5%; Fig. 1G; Table 2). Of these, 71 were expressed, proportionately, in more mesenchymal cells than any other cell type (Fig. 1H; Table 2), and some were highly mesenchymally enriched, including Adm, Bmp7, Cxcl13, Fgf7, Fgf10, Gdf10, Hgf, Il33, Ntn1, Pthlh, and Wnt5a (Fig. 1I; Extended Data Fig. 1-1D). A total of 39 of these mesenchymal ligands were most highly expressed in the endoneurial mesenchymal cells that are closely apposed to Schwann cells and axons (Fig. 1G; Table 2).
Schwann cells were a second major source of injured nerve ligands, detectably expressing 74 ligand mRNAs (Fig. 1G,H; Table 2). A total of 23 of these were expressed in more Schwann cells than any other cell type, including Artn, Bdnf, Btc, Clcf1, Crlf1, Dhh, Fgf5, Gdnf, Hbegf, Sema3e, Sema4f, Shh, and Ucn2 (Fig. 2A; Extended Data Fig. 2-1A; Table 2). Of the other cell types, endothelial and VSM/pericyte cells expressed 18 and 15 ligand mRNAs, respectively, at the highest levels (Fig. 1G,H; Table 2). These included Bmp4 and Pdgfb mRNAs in endothelial cells and Bmp2 and Ngf in VSM/pericytes (Fig. 2B). The immune cells expressed 16 ligand mRNAs in the highest proportions including well-characterized immune ligands like Osm and Tnf (Figs. 1G,H, 2B; Table 2). Thus, ligands known to be important for axon growth and tissue regeneration are expressed by diverse nerve cell populations after injury.
Single-cell transcriptional profiling of uninjured and developing nerves
We asked whether this cellular profile of ligand expression was exclusive to the injured nerve by analyzing single-cell transcriptional datasets of the uninjured and developing sciatic nerves. For the uninjured nerve, we used a dataset that was previously analyzed for Pdgfra-positive mesenchymal cells (Carr et al., 2019), but not for other cell types. Analysis of the entire 1841 uninjured nerve cell dataset (Fig. 2C,D; Extended Data Fig. 2-1B) identified clusters comprised of epineurial, perineurial, and endoneurial mesenchymal cells, as well as VSM/pericyte cells, endothelial cells, and a small population (1.9%) of immune cells. There were also two Sox10-positive Schwann cell populations; non-myelinating cells expressing Ngfr/p75NTR, Cdh2, and L1cam, and myelinating Schwann cells expressing high levels of Mbp, Pmp22, and Plp. This latter population was likely relatively reduced in numbers because myelin removal beads were used when isolating the nerve cells. There were very few proliferating cells, as indicated by expression of cell cycle genes like Top2a and Ki67 (Extended Data Fig. 2-1B).
For the developing neonatal nerve, we generated a new dataset, sequencing P2–P4 sciatic nerve cells after isolating them using either flow cytometry or myelin removal beads in two separate preparations. We combined raw transcriptomes from both preparations and analyzed them together. This analysis, which included 6885 total cells, identified 10 clusters containing intermingled cells from both preparations (Fig. 2E; Extended Data Fig. 2-1C,D). This intermingling was unaffected by batch correction (Extended Data Fig. 2-1C). Cell type-specific marker genes identified endothelial cells, VSM/pericyte cells, macrophages, and epineurial, endoneurial, and perineurial mesenchymal cells (Fig. 2E,F; Extended Data Fig. 2-1D). At this age, there were five Sox10-positive Schwann cell clusters, with one cluster (6) containing proliferative Schwann cells and another (9) myelinating Schwann cells expressing high levels of Mag, Mbp, Mpz, and Pmp22 (Fig. 2E,F).
Analysis of ligand mRNA expression in these datasets showed that cells in the uninjured and developing sciatic nerves expressed many but not all injured nerve ligands. Specifically, of 143 total ligands, 122 and 119 were detectably expressed in uninjured and neonatal nerves, respectively, and 111 were shared in all three conditions (Fig. 2G; Tables 3, 4). These ligand mRNAs were contributed by all of the different cell types in the uninjured and neonatal nerves (Fig. 2H,I). However, the pattern of ligand expression differed from the injured nerve (compare Figs. 2I, 1H), with endoneurial mesenchymal cells and Schwann cells contributing relatively fewer ligands in the developing and uninjured nerves (endoneurial cells, 11% and 27% in the uninjured vs injured nerves; Schwann cells, 5% and 16% in the uninjured vs injured nerves). Moreover, 13 injured nerve ligands were not detectably expressed in the neonatal or uninjured nerves (Artn, Btc, Ccl25, Crlf1, Fgf5, Gdnf, Gnrh1, Hgf, Rspo3, Sema4f, Shh, Tnfsf8, and Ucn2; Tables 3, 4). Notably, all of these were injured nerve Schwann cell or mesenchymal cell ligands (Table 2). Thus, multiple cell types contribute to the sciatic nerve ligand environment in all conditions, but mesenchymal and Schwann cells become relatively more important following injury.
Table 3.
Gene abundance of injured nerve ligand mRNAs in the uninjured nerve scRNA-seq dataset
| Gene abundance (%) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Epineurial/perineurial | Endoneurial | VSM/pericytes | Endothelial | Schwann(non-myelinating) | Schwann(myelinating) | Immune |
| Adm** | 14.4 | 8.4 | BT | 3.6 | BT | BT | 2.9 |
| Agt** | 2.9 | BT | BT | BT | BT | BT | 2.9 |
| Angpt1* | 12.2 | BT | 12.4 | BT | BT | BT | BT |
| Angpt2* | 2.2 | BT | 21.1 | 13.6 | BT | 6.0 | BT |
| Angpt4** | 6.2 | BT | BT | BT | BT | BT | 2.9 |
| Apln | BT | BT | BT | 7.1 | BT | BT | BT |
| Bdnf | BT | BT | 3.1 | BT | BT | BT | BT |
| Bmp1** | 42.0 | 16.1 | 6.8 | 6.2 | 4.6 | 4.8 | BT |
| Bmp2* | 3.5 | BT | 15.5 | BT | BT | BT | 11.4 |
| Bmp4** | 18.6 | 6.5 | BT | 17.6 | BT | BT | BT |
| Bmp5* | 2.4 | 6.8 | 8.1 | BT | BT | BT | BT |
| Bmp7** | 3.1 | 11.9 | BT | BT | BT | BT | BT |
| Cck | BT | BT | 3.1 | BT | BT | BT | BT |
| Ccl11** | 35.8 | 88.3 | 22.4 | 9.0 | 5.3 | 3.6 | 2.9 |
| Ccl19** | 6.2 | BT | 5.0 | BT | BT | BT | BT |
| Ccl2* | 2.2 | 16.1 | 4.3 | BT | BT | BT | 28.6 |
| Ccl3 | BT | BT | BT | BT | BT | BT | 11.4 |
| Ccl5 | BT | BT | BT | BT | BT | BT | 11.4 |
| Ccl7** | 6.0 | 35.5 | 2.5 | 5.0 | 2.7 | 3.6 | 8.6 |
| Ccl9** | BT | 16.6 | BT | BT | BT | 2.4 | 14.3 |
| Clcf1 | BT | BT | BT | 4.0 | BT | BT | 2.9 |
| Csf1** | 31.9 | 10.0 | 4.3 | 15.2 | 8.8 | 2.4 | 8.6 |
| Ctgf* | 16.6 | 2.1 | 10.6 | 28.8 | BT | BT | BT |
| Cx3cl1 | BT | BT | BT | 11.2 | BT | 2.4 | BT |
| Cxcl1** | 17.5 | 52.3 | 18.6 | 15.0 | 11.5 | 6.0 | 8.6 |
| Cxcl10 | BT | BT | BT | BT | BT | BT | 5.7 |
| Cxcl12* | 34.3 | 60.0 | 39.8 | 78.1 | 13.4 | 3.6 | 2.9 |
| Cxcl13** | 16.2 | BT | BT | BT | BT | BT | BT |
| Cxcl16** | 10.0 | BT | BT | 3.3 | BT | BT | 2.9 |
| Cxcl2 | BT | BT | BT | BT | BT | BT | 8.6 |
| Cxcl9 | BT | BT | BT | 3.8 | BT | BT | BT |
| Dhh | BT | BT | BT | 3.8 | 13.4 | 28.9 | BT |
| Dll1 | BT | BT | BT | 10.0 | BT | BT | BT |
| Dll4 | BT | BT | BT | 18.1 | BT | BT | BT |
| Eda** | 5.1 | 7.7 | BT | BT | 3.4 | 2.4 | 2.9 |
| Edn3** | 8.4 | BT | BT | BT | BT | BT | BT |
| Efna1* | 4.2 | 4.0 | 3.7 | 30.2 | BT | BT | BT |
| Efna2 | BT | BT | BT | BT | BT | BT | 2.9 |
| Efna5** | 4.4 | BT | BT | BT | BT | BT | BT |
| Efnb1** | 9.5 | 14.3 | 2.5 | 10.5 | 6.1 | 3.6 | 2.9 |
| Efnb2* | 3.5 | 11.0 | 4.3 | 15.7 | BT | BT | BT |
| Fgf1* | 4.9 | BT | 10.6 | BT | 10.3 | 26.5 | 2.9 |
| Fgf10** | 9.1 | BT | BT | BT | BT | BT | BT |
| Fgf18** | 8.4 | BT | BT | BT | BT | BT | BT |
| Fgf7** | 28.8 | 6.3 | 2.5 | 2.4 | BT | 8.4 | BT |
| Figf** | 12.8 | BT | BT | 3.1 | BT | BT | BT |
| Fstl1** | 90.9 | 75.5 | 47.2 | 34.5 | 29.8 | 10.8 | 8.6 |
| Gas6** | 61.5 | 9.3 | 18.0 | 49.8 | 3.8 | 6.0 | 2.9 |
| Gdf10** | 39.4 | 4.0 | BT | 3.1 | BT | 2.4 | 2.9 |
| Gdf11* | BT | 4.2 | 4.3 | BT | BT | BT | 2.9 |
| Gmfb* | 14.2 | 8.6 | 5.6 | 10.5 | 7.6 | 15.7 | 5.7 |
| Gmfg | BT | BT | BT | 3.1 | BT | BT | 28.6 |
| Hbegf* | BT | 2.3 | 12.4 | 17.4 | 25.2 | 8.4 | 5.7 |
| Igf1** | 56.9 | 43.9 | 8.1 | 17.6 | 2.7 | BT | BT |
| Igf2* | 3.5 | 8.6 | 7.5 | 14.3 | BT | BT | BT |
| Il15* | BT | 4.7 | BT | 5.0 | BT | BT | BT |
| Il16 | BT | BT | BT | 3.1 | BT | 8.4 | 25.7 |
| Il18** | 7.1 | 3.7 | BT | BT | BT | BT | BT |
| Il1b | BT | BT | BT | BT | BT | BT | 8.6 |
| Il33** | 29.4 | 71.5 | BT | 5.0 | 3.4 | 2.4 | 2.9 |
| Il6* | 2.4 | 2.8 | 4.3 | 5.2 | BT | BT | BT |
| Inha** | 2.0 | BT | BT | BT | BT | BT | BT |
| Inhba | BT | BT | 8.1 | BT | BT | BT | BT |
| Jag1* | 12.4 | 4.2 | 20.5 | 19.3 | BT | BT | BT |
| Jag2 | BT | BT | BT | 9.5 | BT | BT | BT |
| Lif** | BT | 3.0 | BT | BT | BT | BT | 2.9 |
| Ltb | BT | BT | BT | BT | BT | BT | 11.4 |
| Mdk** | 8.2 | 16.4 | BT | BT | 3.4 | BT | 2.9 |
| Metrn* | 2.7 | BT | 2.5 | BT | 14.5 | 12.0 | 2.9 |
| Mif* | 14.8 | 14.7 | 19.3 | 31.0 | 13.7 | 18.1 | 31.4 |
| Nenf** | 54.2 | 48.6 | 28.0 | 31.0 | 26.7 | 19.3 | 17.1 |
| Ngf* | BT | 2.6 | 11.2 | BT | BT | BT | BT |
| Nov** | 19.2 | 7.9 | 6.8 | 5.0 | 7.3 | 15.7 | BT |
| Nppc** | BT | 2.6 | BT | BT | BT | BT | BT |
| Ntf3* | 4.0 | BT | 11.8 | 5.7 | BT | BT | BT |
| Ntn1** | 31.2 | 12.6 | BT | 6.7 | BT | BT | BT |
| Osm | BT | BT | BT | BT | BT | BT | 8.6 |
| Pdgfa* | 7.3 | BT | 32.9 | 9.8 | 14.1 | 16.9 | 5.7 |
| Pdgfb | BT | BT | 3.1 | 28.1 | BT | BT | BT |
| Pdgfc** | 3.3 | BT | BT | BT | BT | BT | 2.9 |
| Pgf* | 2.4 | 4.7 | 7.5 | BT | BT | BT | BT |
| Pomc | BT | BT | BT | 5.0 | BT | BT | 2.9 |
| Pthlh** | 12.6 | 7.2 | BT | BT | BT | BT | BT |
| Ptn* | 7.7 | 4.0 | BT | 45.7 | 56.5 | BT | 5.7 |
| Rspo1** | 8.4 | BT | BT | BT | BT | BT | BT |
| Rtn4** | 37.6 | 33.4 | 21.7 | 33.8 | 24.0 | 21.7 | 11.4 |
| Sema3b* | 15.5 | 32.7 | 3.1 | 3.1 | 44.3 | 56.6 | BT |
| Sema3c** | 38.1 | 9.1 | 2.5 | 3.6 | 9.2 | 2.4 | BT |
| Sema3d** | 13.7 | BT | BT | BT | BT | BT | BT |
| Sema3e* | 3.5 | BT | BT | BT | 14.5 | BT | BT |
| Sema3f | BT | BT | 2.5 | 12.6 | BT | BT | BT |
| Sema3g* | BT | 3.0 | BT | 18.8 | 2.7 | 2.4 | BT |
| Sema4a | BT | BT | BT | BT | BT | BT | 5.7 |
| Sema4b** | 2.0 | 2.1 | BT | BT | BT | BT | BT |
| Sema4c* | 6.6 | 6.3 | BT | 11.2 | 5.7 | 22.9 | BT |
| Sema4d | BT | BT | BT | BT | 2.3 | BT | 8.6 |
| Sema5a* | 2.7 | 3.5 | 6.8 | BT | BT | 9.6 | BT |
| Sema6a* | 2.7 | 18.7 | BT | 26.2 | 2.7 | 2.4 | 2.9 |
| Sema6b* | 2.0 | 2.8 | BT | 12.4 | BT | BT | 2.9 |
| Sema6c* | BT | 3.3 | BT | BT | BT | 7.2 | BT |
| Sema6d* | 3.3 | 3.5 | 8.1 | 9.5 | 25.2 | 13.3 | BT |
| Sema7a | BT | BT | BT | 28.8 | 8.4 | BT | 2.9 |
| Sfrp1** | 45.4 | 15.0 | 2.5 | 4.0 | 6.9 | BT | BT |
| Sfrp2** | 45.1 | 5.1 | BT | 5.2 | 3.8 | 6.0 | 5.7 |
| Sfrp4** | 61.7 | 8.6 | 3.1 | 9.8 | 5.3 | 6.0 | 8.6 |
| Sfrp5* | 18.8 | 5.8 | 3.7 | 4.5 | 26.0 | 31.3 | 5.7 |
| Tgfa | BT | BT | BT | 4.0 | BT | BT | BT |
| Tgfb1* | 2.0 | 3.3 | BT | 7.9 | BT | BT | 17.1 |
| Tgfb2** | 8.2 | 2.8 | 5.6 | 6.0 | BT | BT | BT |
| Tgfb3** | 16.8 | 2.3 | 16.1 | 3.1 | BT | BT | BT |
| Tnf | BT | BT | BT | BT | BT | BT | 11.4 |
| Tnfsf10 | BT | BT | BT | 18.6 | BT | BT | BT |
| Tnfsf12** | 13.3 | 17.8 | 6.2 | 12.6 | 9.9 | 9.6 | 5.7 |
| Tnfsf14 | BT | BT | BT | BT | BT | BT | 8.6 |
| Tnfsf9* | BT | 4.0 | BT | BT | 5.7 | BT | BT |
| Tslp** | 3.5 | BT | BT | BT | BT | BT | 2.9 |
| Vegfa** | 14.2 | 7.9 | 8.1 | 6.2 | BT | 2.4 | 2.9 |
| Vegfb* | 5.5 | 4.7 | 6.2 | 5.7 | 3.4 | 7.2 | BT |
| Vegfc* | 3.3 | BT | BT | 6.2 | BT | BT | BT |
| Wnt11** | 6.6 | BT | BT | BT | BT | BT | 2.9 |
| Wnt2** | 6.9 | BT | BT | BT | BT | BT | BT |
| Wnt5a** | 17.5 | 10.3 | BT | BT | BT | BT | BT |
The uninjured nerve scRNA-seq dataset (Fig. 2C) was analyzed to determine the percentage of cells within a given cell type that detectably expressed (≥2%) the 143 injured nerve ligand mRNAs (Table 2). Cell populations were defined as for the injured nerve analysis except that Schwann cells were divided into myelinating (cluster 8) and non-myelinating (cluster 4) cells. BT = below threshold and indicates that <2% of cells detectably expressed the ligand mRNA. Ligands annotated with one asterisk in the leftmost column were expressed in ≥2% Pdgfra-positive mesenchymal cells and two asterisks indicate ligands with the highest expression in either the epineurial/perineurial or endoneurial Pdgfra-positive mesenchymal cells.
* >2% expression in Pdgfra-positive cells.
** >2% expression and highest expression in Pdgfra-positive cells.
Table 4.
Gene abundance of injured nerve ligand mRNAs in the neonatal nerve scRNA-seq dataset
| Gene abundance (%) | ||||||
|---|---|---|---|---|---|---|
| Gene | Epineurial | Endoneurial/perineurial | VSM/pericytes | Endothelial | Schwann cells | Immune cells |
| Adm** | 5.1 | 4.7 | BT | 3.0 | BT | BT |
| Agt* | 2.5 | BT | 12.2 | BT | BT | BT |
| Angpt1* | 5.1 | BT | 11.1 | BT | BT | BT |
| Angpt2* | 5.5 | BT | 18.1 | 6.9 | 2.0 | BT |
| Apln | BT | BT | BT | 29.0 | BT | BT |
| Bdnf | BT | BT | 3.6 | BT | BT | BT |
| Bmp1** | 34.2 | 21.5 | 10.9 | 11.0 | 10.8 | BT |
| Bmp2 | BT | BT | 6.7 | 3.4 | BT | 8.7 |
| Bmp4* | 3.4 | BT | BT | 6.9 | BT | BT |
| Bmp5 | BT | BT | 13.4 | BT | BT | BT |
| Bmp7** | BT | 3.9 | BT | BT | BT | BT |
| Ccl11** | 9.4 | 49.8 | 20.0 | BT | BT | 2.6 |
| Ccl2* | BT | 10.0 | 9.9 | BT | BT | 20.0 |
| Ccl24 | BT | BT | BT | BT | BT | 27.0 |
| Ccl3 | BT | BT | BT | BT | BT | 16.5 |
| Ccl5 | BT | BT | BT | BT | BT | 2.6 |
| Ccl7** | 3.4 | 18.5 | 2.7 | BT | BT | 14.8 |
| Ccl9 | BT | BT | BT | BT | BT | 28.7 |
| Clcf1 | BT | BT | BT | 2.3 | BT | BT |
| Csf1** | 15.2 | 4.3 | 4.8 | 4.1 | 2.4 | BT |
| Ctgf* | 7.1 | 4.0 | 7.4 | 7.4 | BT | BT |
| Cxcl1* | BT | 12.4 | 20.6 | 2.5 | BT | 2.6 |
| Cxcl10 | BT | BT | BT | BT | 2.6 | BT |
| Cxcl12** | 18.9 | 37.2 | 23.1 | 33.8 | BT | BT |
| Cxcl16 | BT | BT | BT | 4.2 | BT | 11.3 |
| Cxcl2 | BT | BT | BT | BT | BT | 4.3 |
| Cxcl9** | BT | 2.1 | BT | BT | BT | BT |
| Dhh | BT | BT | 2.3 | 7.1 | 25.6 | 3.5 |
| Dll1 | BT | BT | BT | 5.3 | BT | BT |
| Dll4 | BT | BT | BT | 15.2 | BT | BT |
| Ebi3 | BT | BT | BT | BT | BT | 13.0 |
| Eda** | 7.1 | 9.0 | BT | BT | 4.4 | BT |
| Edn3** | 4.1 | 3.5 | BT | BT | BT | BT |
| Efna1* | 3.1 | 4.3 | BT | 34.0 | BT | BT |
| Efna2** | 2.9 | 3.3 | BT | BT | 2.9 | BT |
| Efna4** | 2.6 | 3.6 | BT | BT | BT | BT |
| Efna5** | 6.0 | 4.0 | BT | BT | 2.3 | BT |
| Efnb1** | 18.3 | 30.6 | 6.3 | 9.7 | 9.1 | 2.6 |
| Efnb2** | 5.4 | 15.1 | 4.8 | 16.3 | BT | BT |
| Fgf1** | 5.1 | 5.6 | 3.6 | BT | 5.4 | 3.5 |
| Fgf10** | 5.7 | BT | BT | BT | BT | BT |
| Fgf18** | 7.8 | BT | BT | BT | BT | BT |
| Fgf7** | 21.0 | 10.3 | BT | BT | BT | BT |
| Figf** | 3.8 | 4.8 | BT | BT | BT | BT |
| Fstl1** | 96.0 | 92.2 | 72.9 | 59.3 | 43.0 | 11.3 |
| Gas6* | 22.4 | 14.3 | 7.4 | 24.6 | BT | 45.2 |
| Gdf10** | 8.4 | BT | BT | BT | BT | BT |
| Gdf11** | BT | 6.6 | 3.8 | 2.7 | BT | BT |
| Gmfb** | 9.5 | 17.6 | 6.1 | 13.8 | 14.1 | 9.6 |
| Gmfg | BT | BT | BT | 6.4 | BT | 36.5 |
| Grp | BT | BT | BT | 2.1 | BT | BT |
| Hbegf* | BT | 2.5 | 14.3 | 15.0 | 19.2 | 3.5 |
| Igf1** | 56.0 | 52.0 | 4.6 | 16.6 | 2.0 | 40.0 |
| Igf2** | 92.3 | 89.5 | 54.0 | 69.7 | 11.1 | 25.2 |
| Il15 | BT | BT | BT | BT | BT | 4.3 |
| Il16 | BT | BT | BT | 4.6 | BT | 12.2 |
| Il18 | BT | BT | BT | BT | BT | 9.6 |
| Il1b | BT | BT | BT | BT | BT | 3.5 |
| Il33** | 13.7 | 16.1 | BT | BT | BT | BT |
| Inha** | 2.6 | BT | BT | BT | BT | BT |
| Inhba** | 4.1 | BT | BT | BT | BT | BT |
| Inhbb | BT | BT | 8.0 | BT | BT | BT |
| Jag1** | 18.4 | 19.1 | 15.5 | 11.0 | BT | BT |
| Jag2 | BT | BT | BT | 7.1 | BT | BT |
| Ltb | BT | BT | BT | BT | BT | 2.6 |
| Mdk** | 52.3 | 68.1 | 20.2 | 8.8 | 20.9 | 8.7 |
| Metrn* | 3.4 | 8.7 | 10.5 | 5.0 | 43.2 | 10.4 |
| Mif* | 17.5 | 26.4 | 28.8 | 26.0 | 32.8 | 26.1 |
| Nenf** | 54.3 | 54.6 | 39.7 | 28.8 | 24.5 | BT |
| Ngf** | BT | 8.7 | 8.0 | BT | BT | 0.0 |
| Nov** | 23.8 | 4.3 | 3.8 | 3.4 | 2.8 | 3.5 |
| Nppc** | BT | 6.1 | BT | BT | BT | BT |
| Ntf3 | BT | BT | 2.3 | BT | BT | BT |
| Ntn1** | 22.2 | 10.0 | BT | BT | BT | BT |
| Osm | BT | BT | BT | BT | BT | 3.5 |
| Pdgfa* | 3.4 | 3.6 | 30.5 | 3.0 | 17.8 | BT |
| Pdgfb | BT | BT | 2.9 | 21.9 | BT | BT |
| Pdgfc | BT | BT | 3.2 | BT | 2.2 | 2.6 |
| Pf4 | BT | BT | BT | BT | BT | 61.7 |
| Pgf | BT | BT | 3.8 | BT | BT | BT |
| Pthlh** | 7.1 | 4.0 | BT | BT | BT | BT |
| Ptn* | 29.3 | 13.9 | 6.5 | 15.9 | 43.4 | 4.3 |
| Rspo1** | 7.8 | 6.5 | BT | BT | BT | BT |
| Rtn4* | 31.0 | 38.2 | 35.7 | 40.4 | 26.8 | 38.3 |
| Sema3b* | 5.4 | 6.5 | 2.3 | 2.8 | 34.7 | 7.8 |
| Sema3c** | 51.7 | 19.2 | BT | BT | 7.1 | 3.5 |
| Sema3d** | 10.4 | 15.9 | BT | BT | BT | BT |
| Sema3f* | 2.6 | BT | BT | 2.8 | BT | BT |
| Sema3g | BT | BT | BT | 9.6 | 7.5 | BT |
| Sema4a | BT | BT | BT | BT | BT | 10.4 |
| Sema4b | BT | BT | BT | BT | BT | 3.5 |
| Sema4c* | 5.1 | 5.7 | 2.7 | 8.1 | 8.5 | 5.2 |
| Sema4d | BT | BT | BT | BT | BT | 7.8 |
| Sema5a* | 5.4 | 5.5 | 12.4 | BT | BT | BT |
| Sema5b* | 4.4 | 8.6 | 14.1 | BT | 2.6 | BT |
| Sema6a* | 4.4 | 7.9 | BT | 22.8 | 4.5 | BT |
| Sema6b | BT | BT | BT | 2.5 | BT | BT |
| Sema6c** | 4.9 | 3.3 | 2.5 | BT | BT | BT |
| Sema6d* | 2.0 | 3.0 | 4.4 | 6.4 | 16.7 | 8.7 |
| Sema7a | BT | BT | 2.1 | 17.3 | 2.0 | BT |
| Sfrp1** | 50.5 | 48.5 | 2.9 | 4.1 | 28.0 | 3.5 |
| Sfrp2** | 17.2 | BT | BT | BT | BT | BT |
| Sfrp4** | 43.7 | 5.5 | BT | BT | BT | 2.6 |
| Sfrp5** | 5.4 | 30.7 | BT | BT | 8.1 | 2.6 |
| Tgfb1 | BT | BT | BT | 10.3 | BT | 20.9 |
| Tgfb2* | 12.1 | 2.7 | 18.3 | 9.9 | BT | BT |
| Tgfb3** | 21.5 | 8.3 | 10.9 | BT | BT | BT |
| Tnf | BT | BT | BT | BT | BT | 6.1 |
| Tnfrsf11b* | 2.9 | BT | BT | 3.9 | BT | BT |
| Tnfsf10 | BT | BT | BT | 6.4 | BT | BT |
| Tnfsf12** | 8.0 | 10.8 | 5.5 | 6.5 | 3.2 | 4.3 |
| Tnfsf9 | BT | BT | BT | BT | BT | 4.3 |
| Tslp | BT | BT | 2.7 | 2.8 | BT | 3.5 |
| Vegfa** | 16.0 | 3.6 | 3.8 | 3.2 | BT | BT |
| Vegfb** | 4.0 | 7.7 | 6.3 | 5.5 | 4.3 | 6.1 |
| Vegfc | BT | BT | BT | 8.5 | BT | BT |
| Wnt11 | BT | BT | 2.7 | BT | BT | BT |
| Wnt2** | 7.5 | BT | BT | BT | BT | BT |
| Wnt5a** | 17.0 | 6.8 | BT | BT | BT | BT |
The neonatal nerve scRNA-seq dataset (Fig. 2E) was analyzed to determine the percentage of cells within a given cell type that detectably expressed (≥2%) the 143 injured nerve ligand mRNAs (Table 2). Cell populations were defined as for the injured nerve analysis. BT = below threshold and indicates that <2% of cells detectably expressed the ligand mRNA. Ligands annotated with one asterisk in the leftmost column were expressed in ≥2% Pdgfra-positive mesenchymal cells and two asterisks indicate ligands with the highest expression in either the epineurial/perineurial or endoneurial Pdgfra-positive mesenchymal cells.
* >2% expression in Pdgfra-positive cells.
** >2% expression and highest expression in Pdgfra-positive cells.
Injured Schwann cells acquire a unique transcriptional phenotype following injury including upregulation of many growth factor genes
To ask about the apparent injury-associated increase in ligand expression, we analyzed the Schwann cells and Pdgfra-positive mesenchymal cells in more detail. We first combined transcriptomes from all Schwann cell clusters in the neonatal, uninjured, 3 DPI, and 9 DPI nerve datasets [cluster 6 (Fig. 1E), clusters 4 and 8 (Fig. 2C), clusters 1, 2, 4, 6, and 9 (Fig. 2E)]. We augmented this combined dataset by including Schwann cells from the FAC-sorted 9 DPI dataset from Carr et al. (2019). Once this combined dataset was put through the pipeline, we used the Harmony batch correction method (Korsunsky et al., 2019) to correct for any technical variation. Analysis of this combined dataset indicated that injured nerve Schwann cells were distinct from both developing and adult uninjured Schwann cells. Specifically, the combined dataset included 5331 Schwann cells in seven clusters (Fig. 3A,B). The differentiating neonatal Schwann cells were present in clusters 0, 1, and 2 with proliferating cells in cluster 2 (Fig. 3A,B; Extended Data Fig. 3-1A). Adult and neonatal myelinating Schwann cells were present in clusters 5 and 6, while the adult uninjured non-myelinating Schwann cells were in cluster 4. By contrast, almost all injured nerve Schwann cells were present in cluster 3.
To better understand these clusters, we performed hierarchical and correlation analyses of average gene expression (Fig. 3C). These analyses confirmed that the injured Schwann cells were distinct from the other populations, and indicated that they were more similar to the differentiating neonatal cells (r = 0.88 for the comparison between clusters 3 and 0) than to the adult non-myelinating Schwann cells (r = 0.76 for the comparison between clusters 3 and 4). To understand these similarities and differences at an individual cell level, we performed single-cell correlation analysis. As comparators for the analysis, we determined average gene expression for uninjured non-myelinating versus neonatal non-myelinating Schwann cells and for uninjured non-myelinating versus 9 DPI Schwann cells. We then compared each single-cell transcriptome with the averaged bulk transcriptomes using Pearson’s correlation and used the resultant correlation coefficients to assign a two-dimensional coordinate to each cell. This analysis (Fig. 3D) showed that (1) virtually all 3 and 9 DPI Schwann cells were more similar to the neonatal Schwann cells than to the uninjured non-myelinating cells, (2) most neonatal Schwann cells were more similar to the injured cells than to the uninjured non-myelinating cells, and (3) despite these similarities, there was very little direct overlap between the injured and neonatal cells.
These data indicate that following nerve injury Schwann cells become more like neonatal Schwann cells, but that they are nonetheless distinct. In this regard, it has been reported that this unique injury state might involve acquisition of mesenchymal-like characteristics (Arthur-Farraj et al., 2017; Clements et al., 2017). To explore this idea further, we compared injured nerve Schwann cells and Pdgfra-positive mesenchymal cells from the combined 3 and 9 DPI nerve dataset (Fig. 1E). Correlation analysis showed that the injured Schwann cells were very distinct from both endoneurial and epineurial cells in the injured nerve (Extended Data Fig. 3-1B; r = 0.74–0.78). Thus, after injury, Schwann cells acquire a unique transcriptional profile that is similar but not identical to neonatal Schwann cells.
We asked about ligand gene expression in this combined dataset. This analysis showed that nerve injury led to upregulation of a subset of ligand mRNAs in Schwann cells. Specifically, in the combined Schwann cell dataset, 82 of the 143 injured nerve ligands were detectably expressed, but only 28 of these were common to the neonatal, uninjured, and injured Schwann cells (Fig. 3E; Table 5). Notably, 36 ligand mRNAs were expressed in ≥3-fold more injured versus uninjured non-myelinating cells (Table 5), with some almost exclusive to the injured cells, including Artn, Bdnf, Btc, Ccl2, Ccl3, Clcf1, Crlf1, Cxcl2, Fgf5, Gdnf, Lif, Sema4f, Shh, Tgfb1, and Ucn2 mRNAs (Fig. 3F,G; Extended Data Fig. 3-1C). Other ligand mRNAs were also upregulated following injury but were still expressed by other Schwann cell populations, such as Bmp1, Fgf7, Igf1, and Pdgfa (Fig. 3G,H; Extended Data Fig. 3-1D). By contrast, some ligand mRNAs were expressed to some degree in all or most Schwann cell populations, including, for example, Dhh, Mdk, and Fgf1 (Fig. 3H,I; Extended Data Fig. 3-1D). Thus, injured nerve Schwann cells acquire a unique, development-like transcriptional state that includes upregulation of growth factors implicated in nerve development, nerve regeneration, and tissue repair, including, for example, GDNF (Trupp et al., 1995; Naveilhan et al., 1997), BDNF (Lindsay, 1988; Leibrock et al., 1989), and PDGFα (Johnston et al., 2016).
Table 5.
Gene abundance of injured nerve ligand mRNAs in the combined injured, uninjured and neonatal Schwann cell scRNA-seq dataset
| Gene abundance (%) | Fold change | ||||
|---|---|---|---|---|---|
| Gene | Neonatal | Uninjured (myelinating) | Uninjured (non-myelinating) | Injured | Injured:uninjured (non-myel.) |
| Angpt2 | 2.0 | 6.4 | BT (0.4) | BT (1.7) | 4.1 |
| Artn | BT (0.0) | BT (0.0) | BT (0.0) | 3.1* | >3.1 |
| Bdnf | BT (0.0) | BT (0.0) | BT (0.0) | 9.3* | >9.3 |
| Bmp1 | 10.9 | 3.8 | 4.9 | 46.1* | 9.3 |
| Btc | BT (0.8) | BT (0.0) | BT (0.0) | 58.8* | >58.8 |
| Ccl11 | BT (0.8) | 3.8 | 4.5 | 5.6* | 1.2 |
| Ccl2 | BT (0.7) | BT (0.0) | BT (1.6) | 12.7* | 7.7 |
| Ccl3 | BT (0.0) | BT (0.0) | BT (0.0) | 3.9* | >3.9 |
| Ccl7 | BT (0.2) | 2.6 | 2.9 | 6.1* | 2.1 |
| Ccl9 | BT (0.0) | 2.6 | BT (0.4) | 6.6* | 16.1 |
| Clcf1 | BT (0.2) | BT (0.0) | BT (0.0) | 18.6* | >18.6 |
| Crlf1 | BT (1.3) | BT (0.0) | 2.5 | 30.5* | 12.4 |
| Csf1 | 2.4 | BT (1.3) | 9.1 | 9.7* | 1.1 |
| Ctgf | BT (0.1) | BT (0.0) | BT (0.0) | 3.4* | >3.4 |
| Cx3cl1 | BT (0.0) | 2.6 | BT (0.0) | BT (0.2) | - |
| Cxcl1 | BT (1.4) | 6.4 | 11.9 | 10.7 | 0.9 |
| Cxcl10 | 2.6 | BT (0.0) | BT (0.4) | 5.9* | 14.4 |
| Cxcl12 | BT (1.4) | 2.6 | 12.3 | 5.4 | 0.4 |
| Cxcl2 | BT (0.1) | BT (0.0) | BT (0.0) | 7.1* | >7.1 |
| Dhh | 25.3 | 28.2 | 11.9 | 26.8 | 2.2 |
| Eda | 4.4 | 2.6 | 3.7 | 2.9 | 0.8 |
| Efna2 | 2.8 | BT (1.3) | BT (0.8) | 3.2* | 3.9 |
| Efna4 | BT (1.0) | BT (1.3) | BT (0.0) | 4.9* | >4.9 |
| Efna5 | 2.2 | BT (0.0) | BT (0.0) | BT (0.3) | - |
| Efnb1 | 8.9 | 2.6 | 7.0 | 10.0* | 1.4 |
| Efnb2 | BT (0.6) | BT (0.0) | BT (0.4) | 3.9* | 9.5 |
| Fgf1 | 5.4 | 28.2 | 10.7 | BT (1.5) | 0.1 |
| Fgf5 | BT (0.0) | BT (0.0) | BT (0.0) | 18.3* | >18.3 |
| Fgf7 | BT (1.3) | 9.0 | BT (0.8) | 12.9* | 15.7 |
| Fstl1 | 42.6 | 9.0 | 30.9 | 29.0 | 0.9 |
| Gas6 | BT (0.6) | 6.4 | 2.9 | 2.5 | 0.9 |
| Gdf11 | BT (1.2) | BT (0.0) | BT (0.4) | 3.1* | 7.4 |
| Gdnf | BT (0.0) | BT (0.0) | BT (0.0) | 20.5* | >20.5 |
| Gmfb | 14.2 | 16.7 | 7.0 | 21.9* | 3.1 |
| Hbegf | 19.0 | 9.0 | 26.7 | 39.0* | 1.5 |
| Igf1 | 2.1 | BT (1.3) | 3.3 | 15.4* | 4.7 |
| Igf2 | 11.2 | BT (1.3) | BT (1.2) | 3.6 | 2.9 |
| Il16 | BT (0.7) | 7.7 | BT (0.4) | BT (0.3) | 0.8 |
| Il18 | BT (0.3) | BT (0.0) | 2.1 | BT (0.5) | 0.2 |
| Il1b | BT (0.0) | BT (0.0) | BT (0.0) | 3.7* | >3.7 |
| Il33 | BT (0.2) | 2.6 | 2.9 | 4.2* | 1.5 |
| Jag1 | BT (0.9) | BT (1.3) | BT (1.2) | 6.9* | 5.6 |
| Lif | BT (0.0) | BT (0.0) | BT (0.0) | 4.9* | >4.9 |
| Mdk | 20.6 | BT (1.3) | 3.3 | 25.3* | 7.7 |
| Metrn | 42.8 | 11.5 | 14.0 | 53.6* | 3.8 |
| Mif | 32.5 | 17.9 | 11.9 | 26.1 | 2.2 |
| Nenf | 24.3 | 19.2 | 27.2 | 47.3* | 1.7 |
| Nov | 2.7 | 14.1 | 7.8 | 4.2 | 0.5 |
| Ntn1 | BT (0.6) | BT (0.0) | BT (0.8) | 3.2* | 3.9 |
| Pdgfa | 17.5 | 15.4 | 15.6 | 47.5* | 3.0 |
| Pdgfb | BT (0.2) | BT (0.0) | BT (0.0) | 2.2* | >2.2 |
| Pdgfc | 2.2 | BT (1.3) | BT (1.2) | BT (1.0) | 0.8 |
| Ptn | 42.9 | BT (0.0) | 57.2 | 47.3 | 0.8 |
| Rtn4 | 26.7 | 21.8 | 24.3 | 70.7* | 2.9 |
| Sema3b | 34.4 | 56.4 | 43.2 | 51.4 | 1.2 |
| Sema3c | 7.1 | BT (1.3) | 9.5 | 19.5* | 2.1 |
| Sema3e | BT (1.0) | BT (0.0) | 14.4 | 29.2* | 2.0 |
| Sema3f | BT (0.4) | BT (0.0) | 2.1 | 2.9* | 1.4 |
| Sema3g | 7.5 | 2.6 | BT (1.6) | 8.1* | 4.9 |
| Sema4b | BT (0.4) | BT (0.0) | BT (0.8) | 7.1* | 8.6 |
| Sema4c | 8.5 | 21.8 | 5.3 | 15.4 | 2.9 |
| Sema4d | BT (0.9) | BT (0.0) | 2.1 | BT (1.0) | 0.5 |
| Sema4f | BT (0.0) | BT (0.0) | BT (0.0) | 17.1* | >17.1 |
| Sema5a | BT (0.9) | 10.3 | BT (0.0) | BT (1.9) | >1.9 |
| Sema5b | 2.6 | BT (0.0) | BT (0.0) | BT (0.2) | - |
| Sema6a | 4.5 | 2.6 | 2.9 | 8.0* | 2.8 |
| Sema6c | BT (1.3) | 7.7 | BT (1.2) | 2.0 | 1.6 |
| Sema6d | 16.5 | 11.5 | 25.5 | 14.1 | 0.6 |
| Sema7a | 2.1 | BT (0.0) | 9.5 | 17.1* | 1.8 |
| Sfrp1 | 27.6 | BT (0.0) | 7.0 | 8.0 | 1.1 |
| Sfrp2 | BT (0.4) | 5.1 | 3.7 | 2.0 | 0.5 |
| Sfrp4 | BT (1.3) | 6.4 | 4.9 | 8.8* | 1.8 |
| Sfrp5 | 8.1 | 30.8 | 28.0 | 5.1 | 0.2 |
| Shh | BT (0.0) | BT (0.0) | BT (0.0) | 12.0* | >12.0 |
| Tgfb1 | BT (0.9) | BT (1.3) | BT (0.8) | 9.8* | 11.9 |
| Tgfb2 | BT (0.6) | BT (0.0) | BT (0.8) | 6.1* | 7.4 |
| Tgfb3 | BT (1.8) | BT (1.3) | BT (1.6) | 4.2* | 2.6 |
| Tnfsf12 | 3.2 | 9.0 | 9.5 | 6.3 | 0.7 |
| Tnfsf9 | BT (0.6) | BT (1.3) | 5.3 | BT (0.7) | 0.1 |
| Ucn2 | BT (0.1) | BT (0.0) | BT (0.0) | 14.1* | >14.1 |
| Vegfa | BT (1.1) | 2.6 | BT (0.8) | 2.4 | 2.9 |
| Vegfb | 4.2 | 7.7 | 3.3 | 7.5 | 2.3 |
The combined Schwann cell scRNA-seq dataset (Fig. 3A,B) was analyzed to determine the percentage of cells within the different Schwann cell clusters that detectably expressed (≥2%) the 143 injured nerve ligand mRNAs (Table 2). Also shown is the difference, expressed as fold change, in the percentage of positive cells in the injured versus uninjured, non-myelinating Schwann cell clusters. BT = below threshold and indicates that <2% of cells detectably expressed the ligand mRNA. Also shown are the absolute values, since these were used to determine the fold changes. Neonatal includes cells in clusters 0, 1, 2, and 5, uninjured myelinating includes cluster 6 cells, uninjured non-myelinating includes cluster 4 cells, and injured includes cluster 3 cells. Asterisks indicate ligand mRNAs with the highest expression in the injured Schwann cell cluster.
*Injured, highest expression.
Upregulation of ligands in endoneurial mesenchymal cells following nerve injury
We performed a similar analysis of nerve mesenchymal cells, combining transcriptomes from the neonatal, uninjured and injured nerve Pdgfra-positive clusters [clusters 1, 3, 5, and 10 (Fig. 1E), clusters 1, 2, 6, and 10 (Fig. 2C), clusters 3 and 5 (Fig. 2E)], as well as the Pdgfra-positive mesenchymal transcriptomes of the FAC-sorted 9 DPI cells from Carr et al. (2019). Once this combined dataset was put through the pipeline, we used Harmony batch correction (Korsunsky et al., 2019) to correct for any technical variation. Analysis of this combined dataset showed that, as published previously (Carr et al., 2019), some mesenchymal populations were transcriptionally altered by injury, while others were largely unaffected. Specifically, the combined dataset included 5416 cells in nine Pdgfra-positive clusters (Fig. 4A,B). The injured and uninjured epineurial cells were coclustered, as were the injured, uninjured, and neonatal perineurial cells (Fig. 4A,B; Extended Data Fig. 4-1). By contrast, the uninjured, injured, and neonatal endoneurial cells were all segregated from each other. The other segregated clusters included neonatal epineurial cells (cluster 1) and the injured nerve differentiating bridge cells (cluster 0).
We used this combined dataset to ask about injury-induced ligand induction in mesenchymal cells. This analysis indicated that the endoneurial mesenchymal cells were largely responsible for this induction. Specifically, 102 of the 143 injured nerve ligands were detectably expressed in endoneurial mesenchymal cells (Fig. 4C), and, of these, 49 were expressed in at least 3-fold more injured versus uninjured cells, with 26 detectably expressed only in the injured cells (Table 6). These upregulated ligand mRNAs included Angpt1, Ccl9, Crlf1, Cxcl2, Inhbb, Lif, Sema7a, and Ngf (Fig. 4D,E; Table 6). In addition to this endoneurial cell response, some ligands were highest in the injured bridge cells, such as Bdnf, Cxcl9, and Hgf (Fig. 4D; Table 6). By contrast, many ligand mRNAs were expressed to a greater or lesser degree in all nerve mesenchymal cell populations regardless of nerve injury, including for example Adm, Bmp1, Ccl11, Cxcl12, Il33, Pthlh, Fgf18, Pdgfa, Tgfb3, Vegfa, and Wnt5a (Fig. 4F,G). Thus, injury induces expression of many ligand mRNAs in endoneurial mesenchymal cells, but many ligands are also expressed under homeostatic conditions in uninjured nerve mesenchymal cells.
Table 6.
Gene abundance of injured nerve ligand mRNAs in the combined injured, uninjured and neonatal mesenchymal cell scRNA-seq dataset
| Gene abundance (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Neonatal | Inj/uninjured | Injured/uninjured/neonatal | Uninjured | Injured | Neonatal/injured | Fold change | |||
| Gene | Epineurial | Endoneurial | Epineurial | Perineurial | Endoneurial | Endoneurial | Differentiating | Proliferating | Injured:uninjuredendoneurial |
| Adm | 5.2 | 4.8 | 17.3 | 17.0 | 9.0 | 35.1 | 7.6 | 10.1 | 3.9 |
| Agt | BT (1.7) | BT (0.5) | 3.3 | BT (0.0) | BT (1.9) | 10.7 | BT (1.7) | 3.9 | 5.6 |
| Angpt1 | 3.7 | BT (0.5) | 11.6 | 5.2 | BT (0.7) | 23.9 | 24.6 | 20.8 | 33.6 |
| Angpt2 | 3.4 | BT (0.0) | 2.0 | BT (1.3) | BT (0.5) | BT (1.0) | 5.1 | 2.9 | 2.0 |
| Angpt4 | BT (0.5) | BT (0.0) | 4.9 | BT (1.3) | BT (0.2) | BT (0.4) | 2.0 | 2.6 | 1.7 |
| Apln | BT (0.7) | 2.1 | BT (0.5) | BT (0.7) | BT (0.7) | 5.3 | BT (1.9) | 6.5 | 7.5 |
| Bdnf | BT (0.6) | BT (1.1) | BT (0.2) | BT (0.7) | BT (0.0) | BT (1.9) | 5.2 | BT (1.6) | >1.9 |
| Bmp1 | 34.3 | 12.7 | 51.4 | 52.9 | 16.4 | 37.2 | 52.3 | 47.6 | 2.3 |
| Bmp2 | BT (0.2) | BT (0.0) | 2.9 | BT (0.7) | BT (0.9) | 3.1 | BT (0.7) | 3.9 | 3.3 |
| Bmp4 | 2.6 | 2.6 | 11.7 | BT (0.7) | 6.6 | BT (1.0) | BT (0.3) | BT (1.0) | 0.1 |
| Bmp5 | BT (0.4) | BT (1.6) | BT (1.8) | BT (1.3) | 6.6 | 4.0 | BT (1.1) | BT (1.6) | 0.6 |
| Bmp7 | BT (1.7) | 6.3 | BT (1.5) | 8.5 | 12.1 | 30.1 | BT (1.2) | 5.9 | 2.5 |
| Btc | BT (0.4) | BT (0.0) | BT (1.1) | BT (0.0) | BT (0.2) | BT (1.6) | BT (1.5) | 4.6 | 6.9 |
| Ccl11 | 18.0 | 70.9 | 42.6 | 19.0 | 89.3 | 85.9 | 21.4 | 40.4 | 1.0 |
| Ccl19 | BT (0.1) | BT (0.0) | 4.9 | BT (0.0) | BT (0.0) | BT (0.0) | BT (0.5) | BT (0.0) | - |
| Ccl2 | BT (0.7) | 19.8 | 14.4 | 10.5 | 16.1 | 81.6 | 25.6 | 49.5 | 5.1 |
| Ccl3 | BT (0.1) | BT (0.0) | 2.0 | 2.0 | BT (0.0) | 5.2 | 5.8 | 3.6 | >5.2 |
| Ccl5 | BT (0.2) | BT (0.0) | 2.2 | BT (1.3) | BT (0.2) | 3.0 | 2.5 | 3.3 | 12.7 |
| Ccl7 | 6.0 | 29.1 | 12.7 | 10.5 | 36.0 | 77.3 | 23.8 | 44.6 | 2.1 |
| Ccl9 | BT (0.2) | 2.9 | 5.2 | 2.6 | 16.8 | 72.1 | 12.0 | 26.1 | 4.3 |
| Clcf1 | BT (0.8) | BT (0.5) | BT (1.6) | BT (1.3) | BT (0.9) | 4.4 | 5.0 | 5.2 | 4.6 |
| Crlf1 | BT (0.2) | BT (0.0) | BT (1.1) | 5.2 | BT (0.0) | 26.1 | 3.4 | 11.1 | >26.1 |
| Csf1 | 12.2 | 3.4 | 40.9 | 11.8 | 10.9 | 31.0 | 24.0 | 30.9 | 2.8 |
| Ctgf | 7.3 | BT (1.9) | 18.9 | 19.0 | 2.4 | 37.2 | 29.2 | 25.4 | 15.7 |
| Cx3cl1 | BT (0.0) | BT (0.8) | BT (0.9) | BT (0.7) | BT (1.7) | 14.3 | 8.6 | 4.2 | 8.6 |
| Cxcl1 | 1.9 | 23.3 | 20.1 | 17.6 | 53.1 | 60.7 | 35.9 | 37.8 | 1.1 |
| Cxcl10 | BT (0.4) | BT (1.3) | BT (0.9) | BT (0.7) | BT (1.7) | 9.6 | 3.5 | 6.5 | 5.8 |
| Cxcl12 | 17.3 | 63.2 | 42.8 | 10.5 | 61.1 | 52.9 | 50.2 | 48.5 | 0.9 |
| Cxcl13 | BT (0.0) | BT (0.3) | 17.6 | BT (0.0) | BT (1.2) | BT (0.4) | BT (1.0) | BT (0.7) | 0.3 |
| Cxcl16 | BT (0.8) | BT (1.3) | 10.3 | 3.9 | BT (1.7) | 5.1 | 8.2 | 4.6 | 3.0 |
| Cxcl2 | BT (0.1) | BT (1.3) | 7.1 | 6.5 | BT (0.5) | 24.7 | 12.8 | 20.2 | 52.2 |
| Cxcl9 | BT (1.0) | BT (1.9) | BT (1.4) | BT (0.0) | BT (0.5) | 2.2 | 7.6 | 7.5 | 4.6 |
| Dhh | BT (1.3) | 2.4 | BT (0.0) | BT (0.7) | BT (0.2) | BT (0.5) | BT (0.8) | 2.3 | 2.3 |
| Eda | 8.2 | 10.6 | 4.0 | 19.6 | 7.8 | 9.4 | 7.7 | 7.5 | 1.2 |
| Edn3 | 5.3 | BT (0.5) | 6.6 | 6.5 | BT (1.7) | BT (0.3) | BT (1.6) | BT (1.0) | 0.2 |
| Efna1 | 4.1 | 3.7 | 3.2 | 5.9 | 4.0 | 7.8 | 3.9 | 4.2 | 1.9 |
| Efna2 | 3.6 | 3.7 | 2.4 | BT (1.3) | BT (1.2) | 4.4 | 4.9 | 5.2 | 3.7 |
| Efna4 | 4.2 | BT (1.9) | 2.5 | 6.5 | BT (0.7) | 6.1 | 9.0 | 3.9 | 8.6 |
| Efna5 | 6.1 | 4.2 | 4.7 | 11.1 | BT (1.2) | 5.2 | 2.0 | 4.9 | 4.4 |
| Efnb1 | 27.0 | 25.9 | 9.5 | 39.2 | 14.2 | 37.8 | 31.7 | 32.6 | 2.7 |
| Efnb2 | 9.5 | 17.2 | 3.4 | 23.5 | 10.9 | 41.9 | 19.7 | 20.5 | 3.8 |
| Fgf1 | 7.0 | 3.7 | 5.7 | 15.7 | BT (0.9) | 4.6 | 9.9 | 7.2 | 4.9 |
| Fgf10 | 4.0 | BT (0.0) | 13.6 | BT (0.0) | BT (0.0) | BT (0.7) | 3.9 | BT (1.6) | - |
| Fgf18 | 6.6 | BT (0.0) | 11.7 | 4.6 | BT (1.9) | BT (1.5) | 5.9 | 6.2 | 0.8 |
| Fgf5 | BT (1.9) | BT (0.0) | BT (0.5) | BT (0.0) | BT (0.2) | BT (0.1) | 2.1 | 2.0 | 0.6 |
| Fgf7 | 21.3 | 4.0 | 27.8 | 23.5 | 6.6 | 35.9 | 24.2 | 25.1 | 5.4 |
| Figf | 6.1 | BT (0.8) | 21.2 | 5.9 | 2.1 | 9.7 | 10.2 | 15.6 | 4.5 |
| Fstl1 | 96.2 | 92.3 | 96.9 | 86.3 | 76.1 | 94.7 | 96.1 | 90.6 | 1.2 |
| Gas6 | 26.6 | BT (0.5) | 51.4 | 88.2 | 9.7 | 11.1 | 38.7 | 20.5 | 1.1 |
| Gdf10 | 5.0 | BT (0.3) | 42.0 | 3.3 | 4.3 | 4.5 | 17.8 | 12.1 | 1.1 |
| Gdf11 | 4.0 | 7.1 | BT (1.6) | 3.3 | 4.3 | 9.8 | 5.3 | 4.6 | 2.3 |
| Gmfb | 13.7 | 18.3 | 15.2 | 28.1 | 9.2 | 34.4 | 25.9 | 30.6 | 3.7 |
| Gnrh1 | BT (0.8) | BT (0.8) | BT (1.9) | 3.3 | BT (0.5) | 2.2 | BT (1.5) | BT (1.6) | 4.6 |
| Hbegf | 2.3 | 2.1 | BT (1.4) | BT (1.3) | 2.4 | 4.9 | 9.3 | 13.4 | 2.1 |
| Hgf | BT (0.4) | BT (0.0) | BT (1.2) | BT (0.7) | BT (0.2) | BT (0.8) | 6.0 | 6.2 | 3.5 |
| Igf1 | 44.2 | 82.8 | 75.8 | 17.6 | 45.0 | 85.1 | 84.1 | 63.8 | 1.9 |
| Igf2 | 91.9 | 92.3 | 4.7 | 20.9 | 8.8 | 9.0 | 26.1 | 25.1 | 1.0 |
| Il15 | BT (0.6) | BT (1.3) | BT (1.4) | 9.8 | 4.7 | 9.3 | 4.2 | 2.6 | 2.0 |
| Il16 | BT (0.4) | BT (0.5) | 2.2 | BT (0.7) | BT (1.2) | 3.1 | BT (1.3) | BT (1.0) | 2.7 |
| Il18 | BT (0.3) | BT (0.3) | 9.4 | BT (0.7) | 4.0 | 5.5 | 5.2 | 5.2 | 1.4 |
| Il1b | BT (0.0) | BT (0.0) | 2.3 | BT (0.7) | BT (0.0) | 7.4 | 5.9 | 7.2 | >7.4 |
| Il33 | 17.2 | 13.8 | 32.7 | 56.2 | 72.7 | 63.7 | 40.8 | 44.6 | 0.9 |
| Il6 | BT (0.2) | 2.1 | 3.7 | BT (0.0) | 2.8 | 8.1 | 10.0 | 8.8 | 2.8 |
| Inha | 2.0 | BT (1.3) | BT (1.9) | 2.0 | BT (1.2) | 2.3 | 2.3 | 2.3 | 2.0 |
| Inhba | 3.4 | BT (0.5) | 2.1 | 6.5 | BT (1.4) | 12.8 | 15.8 | 23.5 | 9.0 |
| Inhbb | BT (0.0) | BT (0.0) | BT (1.3) | 2.6 | BT (0.0) | 12.8 | BT (1.6) | 2.3 | >12.8 |
| Jag1 | 22.3 | 13.8 | 13.4 | 25.5 | 4.5 | 16.7 | 23.4 | 23.1 | 3.7 |
| Lif | BT (1.3) | BT (1.1) | 2.4 | BT (0.0) | 3.1 | 11.6 | 2.1 | 6.2 | 3.8 |
| Mdk | 58.8 | 70.9 | 16.1 | 30.1 | 16.4 | 54.6 | 65.9 | 45.6 | 3.3 |
| Metrn | 5.2 | 10.3 | 3.1 | 5.9 | BT (1.2) | 13.3 | 5.1 | 23.5 | 11.2 |
| Mif | 21.1 | 30.4 | 18.6 | 15.0 | 14.9 | 42.2 | 42.5 | 54.7 | 2.8 |
| Nenf | 54.7 | 62.7 | 55.7 | 49.0 | 49.8 | 59.7 | 60.5 | 59.6 | 1.2 |
| Ngf | 2.3 | 13.5 | BT (1.2) | 8.5 | 2.6 | 7.7 | BT (1.7) | 6.8 | 2.9 |
| Nov | 16.7 | 5.6 | 30.0 | BT (0.7) | 8.1 | 9.7 | 17.6 | 22.5 | 1.2 |
| Nppc | 2.1 | 7.7 | BT (0.3) | BT (0.0) | 2.6 | 6.3 | 8.3 | 4.2 | 2.4 |
| Ntf3 | BT (1.3) | BT (0.5) | 4.6 | BT (0.0) | BT (0.7) | BT (1.4) | 5.3 | 3.6 | 1.9 |
| Ntn1 | 21.7 | BT (0.5) | 24.8 | 61.4 | 12.8 | 25.0 | 11.1 | 18.6 | 2.0 |
| Pdgfa | 4.0 | 2.6 | 6.5 | 9.8 | BT (1.7) | 7.9 | 11.0 | 25.1 | 4.8 |
| Pdgfc | BT (0.7) | BT (0.3) | 6.0 | BT (0.0) | BT (0.2) | BT (1.4) | 7.2 | 8.1 | 5.8 |
| Pf4 | BT (0.8) | BT (1.1) | 2.0 | BT (1.3) | BT (0.0) | 4.0 | 3.7 | 4.9 | >4.0 |
| Pgf | BT (1.4) | BT (1.1) | 2.0 | 3.3 | 5.0 | 12.7 | 2.5 | 3.9 | 2.6 |
| Pomc | BT (0.3) | BT (1.1) | BT (0.9) | BT (0.7) | BT (1.4) | 3.1 | BT (1.2) | BT (1.3) | 2.2 |
| Pthlh | 5.7 | 6.3 | 9.8 | 20.3 | 7.6 | 13.8 | 13.4 | 11.1 | 1.8 |
| Ptn | 21.1 | 21.7 | 15.5 | 2.6 | 4.0 | 11.1 | 56.8 | 45.6 | 2.7 |
| Rspo1 | 11.5 | BT (0.0) | 3.2 | 20.9 | BT (1.2) | BT (0.5) | BT (0.5) | BT (0.3) | 0.5 |
| Rspo3 | BT (0.4) | BT (0.0) | 3.1 | BT (1.3) | BT (0.0) | BT (0.7) | 11.6 | 6.2 | - |
| Rtn4 | 35.1 | 36.2 | 47.9 | 43.8 | 33.9 | 68.6 | 60.5 | 72.3 | 2.0 |
| Sema3b | 5.2 | 9.3 | 14.0 | 11.1 | 33.4 | 11.3 | 5.0 | 6.5 | 0.3 |
| Sema3c | 43.0 | 14.8 | 50.5 | 18.3 | 9.2 | 8.6 | 23.0 | 27.7 | 0.9 |
| Sema3d | 18.0 | 3.7 | 11.4 | 45.1 | BT (0.7) | BT (1.8) | 6.3 | 7.8 | 2.5 |
| Sema3e | BT (0.7) | BT (0.3) | 3.7 | BT (0.0) | BT (0.2) | BT (0.1) | BT (1.0) | 2.9 | 0.6 |
| Sema3f | 2.2 | BT (0.5) | BT (1.1) | 2.6 | BT (0.7) | 4.5 | 2.0 | 3.3 | 6.3 |
| Sema3g | BT (0.3) | BT (1.3) | BT (0.6) | BT (0.0) | 3.1 | 2.7 | BT (1.0) | 2.0 | 0.9 |
| Sema4a | BT (0.8) | BT (1.9) | BT (0.6) | 4.6 | BT (1.7) | 3.3 | BT (1.3) | BT (1.6) | 2.0 |
| Sema4b | BT (1.7) | BT (0.3) | BT (1.5) | 2.0 | 2.1 | 2.9 | 2.6 | 2.3 | 1.3 |
| Sema4c | 5.8 | 5.8 | 5.9 | 13.1 | 6.4 | 11.7 | 9.3 | 10.1 | 1.8 |
| Sema5a | 4.7 | 7.4 | BT (1.8) | 4.6 | 3.6 | 3.6 | 6.1 | 5.9 | 1.0 |
| Sema5b | 5.6 | 9.5 | BT (0.2) | BT (0.0) | BT (1.2) | BT (1.1) | BT (1.6) | 5.2 | 0.9 |
| Sema6a | 6.2 | 9.8 | 3.1 | 7.8 | 19.0 | 14.6 | 3.2 | 7.5 | 0.8 |
| Sema6b | BT (0.1) | BT (0.3) | 2.5 | BT (0.0) | 2.8 | 7.1 | BT (1.3) | 7.8 | 2.5 |
| Sema6c | 4.3 | 4.8 | 5.4 | BT (1.3) | 3.3 | 3.4 | 8.0 | 6.2 | 1.0 |
| Sema6d | 2.2 | 3.2 | 3.7 | 7.8 | 3.6 | 11.2 | 3.7 | 9.4 | 3.2 |
| Sema7a | BT (0.3) | BT (0.5) | BT (1.3) | BT (0.7) | BT (0.9) | 19.4 | 11.0 | 21.5 | 20.5 |
| Sfrp1 | 60.0 | 29.1 | 40.0 | 54.2 | 15.2 | 32.9 | 50.0 | 32.6 | 2.2 |
| Sfrp2 | 11.4 | BT (0.3) | 47.8 | 3.9 | 5.5 | 7.1 | 21.7 | 18.6 | 1.3 |
| Sfrp4 | 33.6 | BT (1.9) | 72.4 | 32.7 | 9.0 | 50.7 | 56.8 | 39.1 | 5.6 |
| Sfrp5 | 26.7 | 2.6 | 2.6 | 94.8 | 5.9 | 2.6 | 6.3 | 10.4 | 0.4 |
| Tgfb1 | BT (0.6) | BT (0.3) | 2.4 | 3.3 | 3.3 | 11.1 | 10.0 | 15.3 | 3.3 |
| Tgfb2 | 10.3 | BT (1.1) | 10.5 | 3.3 | 2.8 | 3.7 | 17.0 | 12.1 | 1.3 |
| Tgfb3 | 20.6 | 3.4 | 16.8 | 19.0 | 2.4 | 9.0 | 36.1 | 19.2 | 3.8 |
| Tnf | BT (0.0) | BT (0.3) | BT (1.7) | BT (0.7) | BT (0.0) | 2.5 | BT (0.7) | 2.6 | >2.5 |
| Tnfrsf11b | BT (1.9) | BT (0.0) | 4.6 | 8.5 | BT (0.5) | BT (0.7) | 4.4 | 10.7 | 1.4 |
| Tnfsf10 | BT (0.0) | BT (0.0) | BT (0.4) | 2.0 | BT (1.7) | BT (0.7) | BT (0.2) | BT (1.0) | 0.4 |
| Tnfsf12 | 9.6 | 11.6 | 15.7 | 15.0 | 17.8 | 17.3 | 14.6 | 14.7 | 1.0 |
| Tnfsf8 | BT (0.3) | BT (0.0) | BT (0.8) | 10.5 | BT (0.0) | 2.2 | 5.7 | 3.3 | >2.2 |
| Tnfsf9 | BT (0.5) | BT (1.6) | BT (1.4) | BT (1.3) | 4.0 | 5.9 | 4.5 | 7.2 | 1.5 |
| Tslp | BT (1.2) | 2.4 | 4.0 | 2.6 | BT (0.2) | 4.8 | BT (1.2) | 2.9 | 20.2 |
| Vegfa | 11.0 | 4.8 | 25.0 | 3.9 | 8.3 | 18.2 | 29.6 | 27.0 | 2.2 |
| Vegfb | 5.2 | 9.0 | 5.4 | 9.8 | 4.7 | 7.1 | 8.3 | 11.4 | 1.5 |
| Vegfc | BT (0.7) | BT (0.0) | 2.2 | BT (0.7) | BT (0.2) | BT (0.7) | 2.2 | 5.5 | 2.9 |
| Wnt11 | BT (1.2) | BT (0.0) | 6.2 | BT (0.0) | BT (0.5) | BT (0.5) | BT (0.9) | 2.9 | 1.2 |
| Wnt2 | 6.0 | BT (0.3) | 9.1 | BT (0.0) | BT (0.2) | BT (1.5) | 4.6 | 4.9 | 6.3 |
| Wnt5a | 15.5 | 4.0 | 19.1 | 7.8 | 10.4 | 13.9 | 13.0 | 14.0 | 1.3 |
The combined mesenchymal cell dataset (Fig. 4A,B) was analyzed to determine the percentage of cells within the different mesenchymal cell clusters that detectably expressed (≥2%) the 143 injured nerve ligand mRNAs (Table 2). Also shown is the difference, expressed as fold change, in the percentage of positive cells in the injured versus uninjured endoneurial mesenchymal cells. BT = below threshold and indicates that <2% of cells detectably expressed the ligand mRNA. Also shown are the absolute values, since these were used to determine the fold changes. Neonatal epineurial includes cluster 1 cells, neonatal endoneurial includes cluster 6 cells, inj/uninjured epineurial includes clusters 2 and 5 cells, neonatal/uninjured/injured perineurial includes cluster 8 cells, uninjured endoneurial includes cluster 4 cells, injured endoneurial includes cluster 3 cells, injured differentiating includes cluster 0 cells, and neonatal/injured proliferating includes cluster 7 cells.
Identification of growth factor receptors on peripheral sympathetic and sensory neurons
To determine which of these ligands are likely to be important for axonal growth, we characterized growth factor receptors on sensory and sympathetic neurons which project their axons via the sciatic nerve. To do this, we coupled cell-surface proteomics and transcriptome profiling on purified neuronal cultures. For sensory neurons, we cultured E15 rat DRG neurons for 9 d in medium containing NGF. Immunostaining showed that these cultures were comprised of relatively pure βIII-tubulin-positive neurons with 2–3% contaminating S100β-positive Schwann cells (Fig. 5A; Extended Data Fig. 5-1A). For sympathetic neurons, we isolated cells from the neonatal rat SCG and cultured them for 6 d in NGF. These cultures contained βIII-tubulin-positive neurons and low percentages of Fibronectin-positive fibroblasts and S100β-positive Schwann cells (Fig. 5A; Extended Data Fig. 5-1A).
Initially, we characterized the neuronal cell-surface proteomes, taking advantage of the fact that many cell-surface proteins are glycosylated. Specifically, we performed periodate oxidation of cell-surface glycans, bound modified proteins on a hydrazide resin after cell lysis, digested the bound proteins with trypsin and PNGase F, and identified peptides by mass spectrometry. For each sample, we analyzed three independent biological replicates. This analysis identified 608 and 271 unique proteins on sensory and sympathetic neurons, respectively, with 219 of these common to both populations (Extended Data Fig. 5-1B; Table 7). The lower number of unique proteins on sympathetic neurons is likely due to decreased protein that was isolated (samples averaged ≈1100 vs 300 μg/ml for sensory vs sympathetic neurons). PANTHER classification identified most of these proteins as receptors, transporters and hydrolases, indicating appropriate enrichment for cell-surface proteins (Extended Data Fig. 5-1C). We then used the ligand-receptor database and manual curation to identify 102 proteins as receptors of various types, including G-protein-coupled receptors, receptor tyrosine kinases and phosphatases, cytokine receptors, and ligand-gated ion channels (Fig. 5B; Table 8). Among these were well-characterized receptors found on both types of neurons such as TrkA (encoded by Ntrk1), BMP receptor 2, RET, gp130 (encoded by Il6st), and IGF2 receptor, receptors identified only on sensory neurons such as GFRα3 and receptors identified only on sympathetic neurons such as ALK and GFRα2.
Table 7.
Proteins identified in sensory (DRG) and sympathetic (SCG) neurons using mass spectrometry
| Sensory neurons(608) | Sympatheticneurons (271) | Sensory and sympatheticintersect (219) | ||
|---|---|---|---|---|
| Abca1 | 11/3R | Abca1 | ||
| Abca5 | Abca1 | Ache | ||
| Abca7 | AceIII | Acp2 | ||
| Ache | Ache | Adam22 | ||
| Acp2 | Acp2 | Adam23 | ||
| Actb | Adam22 | Adgre5 | ||
| Actg1 | Adam23 | Adgrl1 | ||
| Acvr2a | Adgre1 | Adgrl2 | ||
| Adam10 | Adgre5 | Ahsg | ||
| Adam11 | Adgrl1 | Alcam | ||
| Adam19 | Adgrl2 | Ano6 | ||
| Adam22 | Ahsg | Aplp1 | ||
| Adam23 | Alcam | Apmap | ||
| Adam9 | Alk | Asah1 | ||
| Adcy6 | Angpt2 | Atp1b1 | ||
| Adcy9 | Ano6 | Atp6ap1 | ||
| Adgrb3 | Anpep | B3glct | ||
| Adgre5 | Aplp1 | Bcam | ||
| Adgrl1 | Apmap | Bgn | ||
| Adgrl2 | Asah1 | Bmpr2 | ||
| Adgrl3 | Aspm | Bsg | ||
| Agrn | Atp1b1 | Bst2 | ||
| Ahsg | Atp6ap1 | Cacna2d1 | ||
| Alcam | B3glct | Cadm1 | ||
| Alpl | Bcam | Cadm2 | ||
| Alpl1 | Bgn | Cadm4 | ||
| Ano6 | Bmpr2 | CatL | ||
| Aplp1 | Bsg | Cd151 | ||
| Aplp2 | Bst2 | Cd200 | ||
| Apmap | Cacna2d1 | Cd276 | ||
| Arse | Cadm1 | Cd320 | ||
| Asah1 | Cadm2 | Cd47 | ||
| Asph | Cadm4 | Cd59 | ||
| Astn2 | CatL | Cd63 | ||
| Atg9a | Cd151 | Cdh2 | ||
| Atp1a1 | Cd200 | Celsr3 | ||
| Atp1b1 | Cd276 | Chl1 | ||
| Atp1b3 | Cd320 | Clmp | ||
| Atp5a1 | Cd47 | Clu | ||
| Atp6ap1 | Cd59 | Cntn1 | ||
| Atraid | Cd63 | Cntn2 | ||
| Atrn | Cdh17 | Cntnap1 | ||
| Atrnl1 | Cdh2 | Col1a1 | ||
| Avil | Cdig2 | Col5a2 | ||
| B3gat3 | Cdk5r2 | Colgalt1 | ||
| B3glct | Celsr3 | Cpd | ||
| Bace1 | Chl1 | Cpe | ||
| Bcam | Clmp | Ctsa | ||
| Bgn | Clu | Ctsc | ||
| Bmper | Cnnm2 | Ctsd | ||
| Bmpr2 | Cntn1 | Ctsl | ||
| Brinp1 | Cntn2 | Ctsz | ||
| Brinp2 | Cntnap1 | Dpp10 | ||
| Bscl2 | Col1a1 | Dpp6 | ||
| Bsg | Col2a1 | Ece1 | ||
| Bst2 | Col5a2 | Efna5 | ||
| Btd | Colgalt1 | Efnb2 | ||
| C11orf24 | Cp | Emb | ||
| Cacna1b | Cpd | Enpp4 | ||
| Cacna1c | Cpe | Entpd2 | ||
| Cacna2d1 | Cst3 | Ephb2 | ||
| Cacna2d2 | Ctsa | Ero1a | ||
| Cacng8 | Ctsc | Fam234b | ||
| Cadm1 | Ctsd | Fn1 | ||
| Cadm2 | Ctsl | Gaa | ||
| Cadm3 | Ctsz | Gabbr1 | ||
| Cadm4 | Cyp4f17 | Gba | ||
| Calm1 | Cyp4f40 | Gdpd5 | ||
| Calm2 | Dbh | Ggt7 | ||
| Calu | Dio1 | Glg1 | ||
| Cant1 | Dkk3 | Gns | ||
| Car11 | Dopey1 | Gpc1 | ||
| Casc4 | Dpp10 | Grik3 | ||
| Casd1 | Dpp6 | Grm7 | ||
| CatL | Ece1 | Hexa | ||
| Cd151 | Ecel1 | Hs2st1 | ||
| Cd164 | Efna5 | Hsp90b1 | ||
| Cd200 | Efnb2 | Hyou1 | ||
| Cd24 | Emb | Icam1 | ||
| Cd276 | Enpp4 | Igf2r | ||
| Cd320 | Entpd2 | Iglon5 | ||
| Cd44 | Ephb2 | Igsf3 | ||
| Cd47 | Ero1a | Il6st | ||
| Cd55 | F2r | Impad1 | ||
| Cd59 | Fam234b | Insr | ||
| Cd63 | Fcrl2 | Islr2 | ||
| Cd81 | Fn1 | Itga1 | ||
| Cdh18 | Folr2 | Itga3 | ||
| Cdh2 | Gaa | Itga5 | ||
| Cdh4 | Gabbr1 | Itga6 | ||
| Celsr2 | Gba | Itgam | ||
| Celsr3 | Gdpd5 | Itgav | ||
| Cemip | Gfra2 | Itgb1 | ||
| Chl1 | Ggt7 | L1cam | ||
| Chpf2 | Glg1 | Lamb1 | ||
| Chst3 | Gnas | Lamc1 | ||
| Clcn5 | Gns | Lamp1 | ||
| Clcn6 | Gpc1 | Ldlr | ||
| Clmp | Grik3 | Lgals3bp | ||
| Clptm1 | Grm7 | Lnpep | ||
| Clu | H2-Q10 | LOC100912445 | ||
| Cntfr | H2-Q7 | LOC679087 | ||
| Cntn1 | Hexa | Lrp1 | ||
| Cntn2 | Hs2st1 | Lrp11 | ||
| Cntn3 | Hsp90b1 | Lrrc8b | ||
| Cntn4 | Hyou1 | Lsamp | ||
| Cntn6 | Icam1 | Ly6h | ||
| Cntnap1 | Igf2r | Man2a2 | ||
| Cntnap4 | Iglon5 | Mcam | ||
| Col12a1 | Igsf3 | Mcoln1 | ||
| Col18a1 | Il6st | Mdga1 | ||
| Col1a1 | Impad1 | Megf8 | ||
| Col5a1 | Insr | Megf9 | ||
| Col5a2 | Islr2 | Mmp15 | ||
| Colgalt1 | Itga1 | Ncam1 | ||
| Colgalt2 | Itga3 | Ncam2 | ||
| Cpd | Itga5 | Ncstn | ||
| Cpe | Itga6 | Negr1 | ||
| Cpm | Itga8 | Nell1 | ||
| Cr1l | Itgam | Neo1 | ||
| Creld1 | Itgav | Nfasc | ||
| Crtac1 | Itgb1 | Npc1 | ||
| Csmd1 | L1cam | Nptn | ||
| Csmd2 | Lama1 | Nrcam | ||
| Cspg5 | Lamb1 | Nrp1 | ||
| Ctsa | Lamc1 | Nrxn1 | ||
| Ctsc | Lamp1 | Nrxn3 | ||
| Ctsd | Ldlr | Ntrk1 | ||
| Ctsl | Lgals3bp | Olfm1 | ||
| Ctsz | Lnpep | Ostm1 | ||
| Cxadr | LOC100912445 | P2rx4 | ||
| Daf1 | LOC286987 | P4htm | ||
| Dchs1 | LOC679087 | Panx1 | ||
| Dgcr2 | Lrp1 | Pcdh1 | ||
| Disp2 | Lrp11 | Pcdh17 | ||
| Dnase2 | Lrrc8b | Pcdh9 | ||
| Dpp10 | Lsamp | Pcdhgc3 | ||
| Dpp6 | Ly6h | Pcyox1 | ||
| Dpp7 | Man2a2 | pE4_antigen | ||
| Dpysl2 | Mcam | Plbd2 | ||
| Dpysl3 | Mcoln1 | Pld3 | ||
| Ece1 | Mdga1 | Plod1 | ||
| Ece2 | Megf8 | Plod3 | ||
| Edem3 | Megf9 | Plxna1 | ||
| Edil3 | Mlnr | Plxna3 | ||
| Eef1a1 | Mmp15 | Plxna4 | ||
| Efna1 | Mrc1 | Plxnb1 | ||
| Efna3 | Mtor | Plxnb2 | ||
| Efna5 | Ncam1 | Plxnc1 | ||
| Efnb1 | Ncam2 | Ppt1 | ||
| Efnb2 | Ncstn | Prnp | ||
| Efnb3 | Negr1 | Ptk7 | ||
| Elfn1 | Nell1 | Ptprg | ||
| Elfn2 | Neo1 | Pttg1ip | ||
| Emb | Nfasc | PVR | ||
| Enpp4 | Nkain3 | Pvrl1 | ||
| Enpp5 | Npc1 | Pvrl2 | ||
| Entpd2 | Nptn | Rbm12b | ||
| Epdr1 | Nrcam | Ret | ||
| Epha2 | Nrp1 | rt1-E | ||
| Ephb2 | Nrxn1 | Scarb2 | ||
| Ero1a | Nrxn3 | Scn2b | ||
| Extl3 | Nt5e | Scn3a | ||
| F11r | Ntng1 | Scn9a | ||
| F2rl2 | Ntrk1 | Sdk2 | ||
| Fam189b | Olfm1 | Sel1l | ||
| Fam234b | Ostm1 | Sema4c | ||
| Fat1 | P2rx4 | Sema4d | ||
| Fat3 | P4htm | Sez6l2 | ||
| Fat4 | Panx1 | Sgce | ||
| Fbn2 | Pcdh1 | Slc12a7 | ||
| Fkbp10 | Pcdh17 | Slc2a1 | ||
| Fkbp9 | Pcdh9 | Slc2a13 | ||
| Flrt1 | Pcdhgc3 | Slc2a3 | ||
| Fn1 | Pcyox1 | Slc39a6 | ||
| Foxred2 | pE4_antigen | Slc44a2 | ||
| Fras1 | Plbd2 | Slc6a15 | ||
| Fstl1 | Pld3 | Slco3a1 | ||
| Gaa | Plod1 | Slit1 | ||
| Gabbr1 | Plod3 | Slit2 | ||
| Gabbr2 | Plxna1 | Sorcs2 | ||
| Gabra2 | Plxna3 | Sort1 | ||
| Gabrb3 | Plxna4 | Spock2 | ||
| Galnt9 | Plxnb1 | Ssr2 | ||
| Gapdh | Plxnb2 | Stt3a | ||
| Gapdh-ps2 | Plxnc1 | Stt3b | ||
| Gba | Pon1 | Suco | ||
| Gdpd5 | Ppt1 | Sulf2 | ||
| Gfra3 | Prnp | Sv2a | ||
| Ggh | Ptk7 | Sv2b | ||
| Ggt5 | Ptprg | Sv2c | ||
| Ggt7 | Ptprm | Tage4 | ||
| Gla | Pttg1ip | Tenm3 | ||
| Glg1 | PVR | Tenm4 | ||
| Gnao1 | Pvrl1 | Tfrc | ||
| Gnptab | Pvrl2 | Thbs1 | ||
| Gns | Rbm12b | Thsd7a | ||
| Gpc1 | Ret | Thy1 | ||
| Gpm6b | RGD1560108 | Timp1 | ||
| Gpr158 | RT1-A2b | Tm9sf3 | ||
| Gria2 | RT1-Ak | Tmed4 | ||
| Grik3 | RT1-Aw2 | Tmed7 | ||
| Grin1 | rt1-E | Tmed9 | ||
| Grm7 | RT1.A1 | Tmeff1 | ||
| Grn | Rt1.L | Tmem106b | ||
| Hexa | Scarb2 | Tmem132a | ||
| Hist1h2ba | Scn2b | Tmem63b | ||
| Hist1h2bd | Scn3a | Tmem63c | ||
| Hist1h2bh | Scn9a | Tmem87a | ||
| Hist1h2bk | Scube1 | Tmem87b | ||
| Hist1h2bl | Sdk2 | Tpp1 | ||
| Hist1h2bo | Sel1l | Trpv2 | ||
| Hist1h2bq | Sema4c | Tspan3 | ||
| Hist2h2be | Sema4d | Tspan6 | ||
| Hist3h2ba | Sez6l2 | Tspan8 | ||
| Hist3h2bb | Sgce | Ttyh3 | ||
| Hnrnpa1 | Slc12a7 | Unc5b | ||
| Hs2st1 | Slc2a1 | Unc5c | ||
| Hs6st1 | Slc2a13 | Vwa7 | ||
| Hsp70 | Slc2a3 | |||
| Hsp90ab1 | Slc39a6 | |||
| Hsp90b1 | Slc44a2 | |||
| Hspa13 | Slc6a15 | |||
| Hspa2 | Slc6a2 | |||
| Hspa8 | Slco3a1 | |||
| Hyou1 | Slit1 | |||
| Icam1 | Slit2 | |||
| Icam5 | Sorcs1 | |||
| Ids | Sorcs2 | |||
| Idua | Sorcs3 | |||
| Ifnar1 | Sort1 | |||
| Igf1r | Spock2 | |||
| Igf2r | Ssr2 | |||
| Igfbpl1 | Stab1 | |||
| Iglon5 | Stt3a | |||
| Igsf3 | Stt3b | |||
| Ikbip | Suco | |||
| Il1rapl1 | Sulf2 | |||
| Il6st | Sv2a | |||
| Impad1 | Sv2b | |||
| Insr | Sv2c | |||
| Islr2 | Tage4 | |||
| Itfg1 | Tenm3 | |||
| Itga1 | Tenm4 | |||
| Itga3 | Tfrc | |||
| Itga4 | Thbs1 | |||
| Itga5 | Thsd7a | |||
| Itga6 | Thy1 | |||
| Itga7 | Timp1 | |||
| Itga9 | Tll2 | |||
| Itgal | Tm9sf3 | |||
| Itgam | Tmed4 | |||
| Itgav | Tmed7 | |||
| Itgb1 | Tmed9 | |||
| Itgb8 | Tmeff1 | |||
| Jag1 | Tmem106b | |||
| Jkamp | Tmem132a | |||
| Kcnc4 | Tmem63b | |||
| Kdelc2 | Tmem63c | |||
| Kiaa0319 | Tmem87a | |||
| Kirrel3 | Tmem87b | |||
| L1cam | Tpp1 | |||
| Lama4 | Trpv2 | |||
| Lama5 | TSLC1 | |||
| Lamb1 | Tspan3 | |||
| Lamc1 | Tspan6 | |||
| Lamp1 | Tspan8 | |||
| Lamp2 | Ttyh3 | |||
| Ldlr | Unc5b | |||
| Lgals3bp | Unc5c | |||
| Lgi2 | Vwa7 | |||
| Lifr | ||||
| Lingo1 | ||||
| Lingo2 | ||||
| Lman2l | ||||
| Lnpep | ||||
| LOC100294508 | ||||
| LOC100359563 | ||||
| LOC100360413 | ||||
| LOC100360548 | ||||
| LOC100364116 | ||||
| LOC100909441 | ||||
| LOC100909911 | ||||
| LOC100911252 | ||||
| LOC100912445 | ||||
| LOC100912446 | ||||
| LOC102549061 | ||||
| LOC102549957 | ||||
| LOC314016 | ||||
| LOC679087 | ||||
| LOC685186 | ||||
| Lphn3 | ||||
| Lppr1 | ||||
| Lrfn1 | ||||
| Lrfn4 | ||||
| Lrfn5 | ||||
| Lrig2 | ||||
| Lrp1 | ||||
| Lrp11 | ||||
| Lrp3 | ||||
| Lrp8 | ||||
| Lrrc8a | ||||
| Lrrc8b | ||||
| Lrrn1 | ||||
| Lsamp | ||||
| Ly6h | ||||
| M6pr | ||||
| Man2a2 | ||||
| Man2b1 | ||||
| Mcam | ||||
| Mcoln1 | ||||
| Mdga1 | ||||
| Mdga2 | ||||
| Megf8 | ||||
| Sensoryneurons(608) | ||||
| Megf9 | ||||
| Mfge8 | ||||
| Mme | ||||
| Mmp15 | ||||
| Mpz | ||||
| Mpzl1 | ||||
| Mrc2 | ||||
| Mst1r | ||||
| Myh7b | ||||
| Naglu | ||||
| Ncam1 | ||||
| Ncam2 | ||||
| Ncan | ||||
| Ncln | ||||
| Ncstn | ||||
| Negr1 | ||||
| Nell1 | ||||
| Neo1 | ||||
| Neu1 | ||||
| Nfasc | ||||
| Nid2 | ||||
| NKCC1 | ||||
| Nomo1 | ||||
| Npc1 | ||||
| Npr3 | ||||
| Nptn | ||||
| Nptx1 | ||||
| Nptx2 | ||||
| Nptxr | ||||
| Nrcam | ||||
| Nrp1 | ||||
| Nrxn1 | ||||
| Nrxn2 | ||||
| Nrxn3 | ||||
| Ntm | ||||
| Ntng2 | ||||
| Ntrk1 | ||||
| Ntrk2 | ||||
| Olfm1 | ||||
| Ostm1 | ||||
| P2rx3 | ||||
| P2rx4 | ||||
| P3h1 | ||||
| P3h4 | ||||
| P4ha1 | ||||
| P4htm | ||||
| Panx1 | ||||
| Pcdh1 | ||||
| Pcdh17 | ||||
| Pcdh19 | ||||
| Pcdh7 | ||||
| Pcdh9 | ||||
| Pcdha6 | ||||
| Pcdha8 | ||||
| Pcdhb2 | ||||
| Pcdhb3 | ||||
| Pcdhga11 | ||||
| Pcdhga3 | ||||
| Pcdhga5 | ||||
| Pcdhga7 | ||||
| Pcdhgb4 | ||||
| Pcdhgb5 | ||||
| Pcdhgb6 | ||||
| Pcdhgb7 | ||||
| Pcdhgb8 | ||||
| Pcdhgc3 | ||||
| Pcsk2 | ||||
| Pcyox1 | ||||
| pE4_antigen | ||||
| Pgap1 | ||||
| Piezo2 | ||||
| Pigs | ||||
| Pigt | ||||
| Pla2g15 | ||||
| Plaur | ||||
| Plbd2 | ||||
| Pld3 | ||||
| Plod1 | ||||
| Plod2 | ||||
| Plod3 | ||||
| Plxdc2 | ||||
| Plxna1 | ||||
| Plxna2 | ||||
| Plxna3 | ||||
| Plxna4 | ||||
| Plxnb1 | ||||
| Plxnb2 | ||||
| Plxnc1 | ||||
| Plxnd1 | ||||
| Pm20d1 | ||||
| Podxl | ||||
| Podxl2 | ||||
| Pofut2 | ||||
| Pol | ||||
| Pomgnt2 | ||||
| Postn | ||||
| Ppia | ||||
| Ppib | ||||
| Ppil1 | ||||
| Ppt1 | ||||
| Prdx2 | ||||
| Prnp | ||||
| Prph | ||||
| Psap | ||||
| Ptger2 | ||||
| SensoryNeurons(608) | ||||
| Ptgfrn | ||||
| Ptk7 | ||||
| Ptprf | ||||
| Ptprg | ||||
| Ptprn | ||||
| Ptpro2 | ||||
| Ptprs | ||||
| Pttg1ip | ||||
| PVR | ||||
| Pvrl1 | ||||
| Pvrl2 | ||||
| Pxdn | ||||
| Qpct | ||||
| ratASCT1 | ||||
| Rbm12b | ||||
| Rcn1 | ||||
| Ret | ||||
| RGD1562725 | ||||
| RGD1563124 | ||||
| RGD1563349 | ||||
| RGD1565368 | ||||
| Rgma | ||||
| Ror2 | ||||
| Rps16 | ||||
| Rps20 | ||||
| Rps27a | ||||
| Rps27a-ps6 | ||||
| rt1-E | ||||
| Rtn4r | ||||
| Rtn4rl1 | ||||
| Scarb2 | ||||
| Scg3 | ||||
| Scn10a | ||||
| Scn2b | ||||
| Scn3a | ||||
| Scn7a | ||||
| Scn9a | ||||
| Sdk2 | ||||
| Sel1l | ||||
| Sema3c | ||||
| Sema3f | ||||
| Sema3g | ||||
| Sema4b | ||||
| Sema4c | ||||
| Sema4d | ||||
| Sema4f | ||||
| Sema6a | ||||
| Sema6d | ||||
| Sema7a | ||||
| Serpinh1 | ||||
| Sez6l2 | ||||
| Sgcb | ||||
| Sgce | ||||
| Shisa7 | ||||
| Siae | ||||
| Sil1 | ||||
| Sirpa | ||||
| Slc12a2 | ||||
| Slc12a4 | ||||
| Slc12a7 | ||||
| Slc12a9 | ||||
| Slc1a2 | ||||
| Slc1a4 | ||||
| Slc22a23 | ||||
| Slc24a2 | ||||
| Slc24a3 | ||||
| Slc25a31 | ||||
| Slc25a4 | ||||
| Slc25a5 | ||||
| Slc2a1 | ||||
| Slc2a13 | ||||
| Slc2a3 | ||||
| Slc35a5 | ||||
| Slc38a2 | ||||
| Slc39a10 | ||||
| Slc39a14 | ||||
| Slc39a6 | ||||
| Slc39a8 | ||||
| Slc3a2 | ||||
| Slc44a1 | ||||
| Slc44a2 | ||||
| Slc46a1 | ||||
| Slc4a1 | ||||
| Slc52a2 | ||||
| Slc6a15 | ||||
| Slc6a17 | ||||
| Slc6a8 | ||||
| Slc7a1 | ||||
| Slc8a1 | ||||
| Slco3a1 | ||||
| Slit1 | ||||
| Slit2 | ||||
| Slitrk2 | ||||
| Slitrk3 | ||||
| Sorcs2 | ||||
| Sort1 | ||||
| Spock2 | ||||
| Spock3 | ||||
| Sppl2a | ||||
| Sppl2b | ||||
| Ssr2 | ||||
| St8sia1 | ||||
| St8sia3 | ||||
| Stt3a | ||||
| Stt3b | ||||
| Sensoryneurons(608) | ||||
| Suco | ||||
| Sulf2 | ||||
| Sun1 | ||||
| Sun2 | ||||
| Sv2a | ||||
| Sv2b | ||||
| Sv2c | ||||
| Syp | ||||
| Sypl1 | ||||
| Tage4 | ||||
| Tapbp | ||||
| Tctn1 | ||||
| Tctn2 | ||||
| Tenm2 | ||||
| Tenm3 | ||||
| Tenm4 | ||||
| Tfrc | ||||
| Tgfb2 | ||||
| Tgfbr3 | ||||
| Thbs1 | ||||
| Thsd7a | ||||
| Thsd7b | ||||
| Thy1 | ||||
| Timp1 | ||||
| Tm2d1 | ||||
| Tm9sf3 | ||||
| Tmed4 | ||||
| Tmed7 | ||||
| Tmed9 | ||||
| Tmeff1 | ||||
| Tmem106b | ||||
| Tmem132a | ||||
| Tmem132c | ||||
| Tmem132e | ||||
| Tmem158 | ||||
| Tmem181 | ||||
| Tmem2 | ||||
| Tmem200c | ||||
| Tmem231 | ||||
| Tmem255a | ||||
| Tmem63b | ||||
| Tmem63c | ||||
| Tmem87a | ||||
| Tmem87b | ||||
| Tmem9b | ||||
| Tmtc4 | ||||
| Tmx3 | ||||
| Tor1aip2 | ||||
| Tor2a | ||||
| Tpbg | ||||
| Tpcn1 | ||||
| Tpp1 | ||||
| Trhde | ||||
| Trpv2 | ||||
| Tspan13 | ||||
| Tspan3 | ||||
| Tspan6 | ||||
| Tspan7 | ||||
| Tspan8 | ||||
| Ttyh3 | ||||
| Tuba1a | ||||
| Tuba1b | ||||
| Tuba1c | ||||
| Tuba3a | ||||
| Tubb2a | ||||
| Tubb2b | ||||
| Tubb3 | ||||
| Tubb4b | ||||
| Tubb5 | ||||
| Txndc15 | ||||
| Uba52 | ||||
| Ubb | ||||
| Ubc | ||||
| Uggt1 | ||||
| Unc5b | ||||
| Unc5c | ||||
| Ust | ||||
| Vstm2a | ||||
| Vstm5 | ||||
| Vwa7 | ||||
| Wbscr17 | ||||
| Ywhag | ||||
| Ywhah | ||||
| Z043_117466 |
Gene symbols of proteins identified using cell-surface capture mass spectrometry on sensory neurons (column 1), sympathetic neurons (column 2), and both neuron types (column 3; intersect). Total numbers of proteins are indicated. Proteins included in this list were annotated by the terms “cell membrane” and/or “secreted” by the UniProtKB database (http://uniprot.org) and were verified by manual curation.
Table 8.
Receptors identified on sensory (DRG) and sympathetic (SCG) neurons using mass spectrometry and microarrays
| Mass spectrometry | ||
| DRGs (42) | SCGs (13) | DRGs and SCGs (47) |
| Acvr2a | Adgre1 | Adgre5 |
| Adgrb3 | Alk | Adgrl1 |
| Adgrl3 | F2r | Adgrl2 |
| Cd44 | Fcrl2 | Bcam |
| Celsr2 | Folr2 | Bmpr2 |
| Cntfr | Gfra2 | Cd320 |
| Epha3 | Itga8 | Cd63 |
| F2rl2 | Mrc1 | Celsr3 |
| Gabra2 | Mlnr | Ephb2 |
| Gabrb3 | Ntng1 | Gabbr1 |
| Gfra3 | Ptprm | Grik3 |
| Gpr158 | Sorcs1 | Grm7 |
| Gria2 | Sorcs3 | Icam1 |
| Grin1 | Igf2r | |
| Ifnar1 | Il6st | |
| Igf1r | Insr | |
| Itfg1 | Itga1 | |
| Itga4 | Itga3 | |
| Itga7 | Itga5 | |
| Itga9 | Itga6 | |
| Itgal | Itgam | |
| Itgb8 | Itgav | |
| Lifr | Itgb1 | |
| Lingo1 | Ldlr | |
| Mrc2 | Lrp1 | |
| Mst1r | Mcam | |
| Npr3 | Neo1 | |
| Ntng2 | Nptn | |
| Ntrk2 | Nrp1 | |
| P2rx3 | Ntrk1 | |
| Plaur | P2rx4 | |
| Plxna2 | Plxna1 | |
| Plxnd1 | Plxna3 | |
| Ptger2 | Plxna4 | |
| Ptprf | Plxnb1 | |
| Ptprn | Plxnb2 | |
| Ptprs | Plxnc1 | |
| Ror2 | Ptprg | |
| Rtn4r | PVR | |
| Rtn4rl1 | Pvrl1 | |
| Sirpa | Pvrl2 | |
| Tgfbr3 | Ret | |
| Sorcs2 | ||
| Sort1 | ||
| Spock2 | ||
| Unc5b | ||
| Unc5c | ||
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Acvr1 | Acvr1 | |
| Acvr1b | Acvr1b | |
| Acvr1c | Acvr2a | |
| Acvr2a | Acvr2b | |
| Acvr2b | Acvrl1 | |
| Acvrl1 | Adcyap1r1 | |
| Adcyap1r1 | Adipor1 | |
| Adipor1 | Adipor2 | |
| Adipor2 | Adora2a | |
| Adora2a | Adra1b | |
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Adra1a | Adrb2 | |
| Adra1b | Ager | |
| Adrb2 | Alk | |
| Ager | Amfr | |
| Alk | Aplnr | |
| Amfr | Avpr1a | |
| Amhr2 | Avpr2 | |
| Aplnr | Axl | |
| Ar | Bdkrb2 | |
| Avpr1a | Bmpr1a | |
| Avpr2 | Bmpr1b | |
| Axl | Bmpr2 | |
| Bdkrb2 | C3ar1 | |
| Bmpr1a | C5ar1 | |
| Bmpr1b | Calcrl | |
| Bmpr2 | Cckar | |
| Btn1a1 | Cckbr | |
| C3ar1 | Ccr10 | |
| C5ar1 | Ccr4 | |
| Calcr | Ccr7 | |
| Calcrl | Ccr8 | |
| Cckar | Cd14 | |
| Cckbr | Cd27 | |
| Ccr10 | Cd33 | |
| Ccr4 | Cd4 | |
| Ccr7 | Cd44 | |
| Ccr8 | Cd5l | |
| Cd14 | Cd7 | |
| Cd27 | Cd74 | |
| Cd33 | Cntfr | |
| Cd4 | Crhr1 | |
| Cd40 | Crhr2 | |
| Cd44 | Crlf1 | |
| Cd5l | Crlf2 | |
| Cd7 | Csf1r | |
| Cd74 | Csf2ra | |
| Cntfr | Csf2rb | |
| Crhr1 | Csf3r | |
| Crhr2 | Ctf1 | |
| Crlf1 | Cx3cr1 | |
| Crlf2 | Cxcr1 | |
| Csf1r | Cxcr3 | |
| Csf2ra | Cxcr4 | |
| Csf2rb | Dcc | |
| Csf3r | Ddr1 | |
| Ctf1 | Derl1 | |
| Cx3cr1 | Dip2a | |
| Cxcr1 | Edar | |
| Cxcr2 | Ednra | |
| Cxcr3 | Ednrb | |
| Cxcr4 | Egfr | |
| Cxcr5 | Eng | |
| Dcc | Epha1 | |
| Ddr1 | Epha2 | |
| Derl1 | Epha3 | |
| Dip2a | Epha4 | |
| Edar | Epha5 | |
| Ednra | Epha7 | |
| Ednrb | Ephb1 | |
| Egfr | Ephb2 | |
| Eng | Ephb3 | |
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Epha1 | Ephb4 | |
| Epha2 | Epor | |
| Epha3 | Eps15l1 | |
| Epha4 | Erbb2 | |
| Epha5 | Erbb3 | |
| Epha7 | Esr2 | |
| Ephb1 | F2r | |
| Ephb2 | F2rl1 | |
| Ephb3 | F2rl2 | |
| Ephb4 | F2rl3 | |
| Epor | Fas | |
| Eps15l1 | Fgfr1 | |
| Erbb2 | Fgfr2 | |
| Erbb3 | Fgfr3 | |
| Erbb4 | Fgfr4 | |
| Esr2 | Fgfrl1 | |
| F2r | Flt1 | |
| F2rl1 | Flt3 | |
| F2rl2 | Flt4 | |
| F2rl3 | Folr1 | |
| Fas | Fshr | |
| Fgfr1 | Fzd1 | |
| Fgfr2 | Fzd2 | |
| Fgfr3 | Fzd4 | |
| Fgfr4 | Fzd5 | |
| Fgfrl1 | Fzd9 | |
| Flt1 | Gabbr1 | |
| Flt3 | Galr1 | |
| Flt4 | Galr2 | |
| Folr1 | Gcgr | |
| Fshr | Gfra2 | |
| Fzd1 | Gfra3 | |
| Fzd2 | Gfra4 | |
| Fzd4 | Ghr | |
| Fzd5 | Ghrhr | |
| Fzd9 | Ghsr | |
| Gabbr1 | Gipr | |
| Galr1 | Glp1r | |
| Galr2 | Gosr1 | |
| Gcgr | Grik5 | |
| Gfra2 | Grin2a | |
| Gfra3 | Grin2b | |
| Gfra4 | Grin2c | |
| Ghr | Grin2d | |
| Ghrhr | Gucy2c | |
| Ghsr | Hcrtr1 | |
| Gipr | Hcrtr2 | |
| Glp1r | Hnf4a | |
| Glp2r | Hpn | |
| Gnrhr | Ifnar1 | |
| Gosr1 | Ifnar2 | |
| Grik5 | Ifngr1 | |
| Grin2a | Ifngr2 | |
| Grin2b | Igf1r | |
| Grin2c | Igf2r | |
| Grin2d | Igfbp1 | |
| Gucy2c | Igfbp2 | |
| Hcrtr1 | Igfbp3 | |
| Hcrtr2 | Igfbp4 | |
| Hnf4a | Igfbp5 | |
| Hpn | Igfbp6 | |
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Ifnar1 | Igfbp7 | |
| Ifnar2 | Il10ra | |
| Ifngr1 | Il10rb | |
| Ifngr2 | Il12rb1 | |
| Igf1r | Il15ra | |
| Igf2r | Il17ra | |
| Igfbp1 | Il17rc | |
| Igfbp2 | Il18r1 | |
| Igfbp3 | Il18rap | |
| Igfbp4 | Il1r1 | |
| Igfbp5 | Il1r2 | |
| Igfbp6 | Il1rap | |
| Igfbp7 | Il1rl1 | |
| Il10ra | Il1rl2 | |
| Il10rb | Il20rb | |
| Il12rb1 | Il21r | |
| Il12rb2 | Il22ra1 | |
| Il13ra2 | Il22ra2 | |
| Il15ra | Il27ra | |
| Il17ra | Il2ra | |
| Il17rb | Il2rb | |
| Il17rc | Il2rg | |
| Il18r1 | Il3ra | |
| Il18rap | Il4r | |
| Il1r1 | Il6r | |
| Il1r2 | Il6st | |
| Il1rap | Il7r | |
| Il1rl1 | Insr | |
| Il1rl2 | Irs1 | |
| Il20rb | Itga2 | |
| Il21r | Itga2b | |
| Il22ra1 | Itga5 | |
| Il22ra2 | Itga9 | |
| Il27ra | Itgal | |
| Il2ra | Itgav | |
| Il2rb | Itgb1 | |
| Il2rg | Itgb2 | |
| Il3ra | Itgb3 | |
| Il4r | Itgb5 | |
| Il6r | Itgb6 | |
| Il6st | Itgb8 | |
| Il7r | Itpr3 | |
| Il9r | Kit | |
| Insr | Ldlr | |
| Irs1 | Lgals3bp | |
| Itga2 | Lgr5 | |
| Itga2b | Lifr | |
| Itga5 | Lingo1 | |
| Itga9 | Loxl2 | |
| Itgal | Lrp1 | |
| Itgav | Lrp2 | |
| Itgb1 | Lrp5 | |
| Itgb2 | Lrp6 | |
| Itgb3 | Lsr | |
| Itgb5 | Ltbr | |
| Itgb6 | Marco | |
| Itgb8 | Mc1r | |
| Itpr3 | Mc2r | |
| Kdr | Mc3r | |
| Kit | Mc4r | |
| Ldlr | Mc5r | |
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Lgals3bp | Mchr1 | |
| Lgr5 | Met | |
| Lifr | Mpl | |
| Lingo1 | Mst1r | |
| Loxl2 | Ncoa3 | |
| Lrp1 | Ncor1 | |
| Lrp2 | Neo1 | |
| Lrp5 | Ngfr | |
| Lrp6 | Notch1 | |
| Lsr | Notch2 | |
| Ltbr | Notch3 | |
| Marco | Npffr1 | |
| Mc1r | Npffr2 | |
| Mc2r | Npr1 | |
| Mc3r | Npr2 | |
| Mc4r | Npr3 | |
| Mc5r | Npy1r | |
| Mchr1 | Npy2r | |
| Met | Npy5r | |
| Mpl | Nr3c1 | |
| Mrgprx2 | Nrp1 | |
| Mst1r | Nrp2 | |
| Mtnr1b | Ntng1 | |
| Ncoa3 | Ntng2 | |
| Ncor1 | Ntrk1 | |
| Neo1 | Ntrk2 | |
| Ngfr | Ntrk3 | |
| Notch1 | Ntsr1 | |
| Notch2 | Oprl1 | |
| Notch3 | Osmr | |
| Npffr1 | Oxtr | |
| Npffr2 | Pdgfa | |
| Npr1 | Pdgfra | |
| Npr2 | Pdgfrb | |
| Npr3 | Pgr | |
| Npy1r | Plaur | |
| Npy2r | Plgrkt | |
| Npy5r | Plxna1 | |
| Nr3c1 | Plxna2 | |
| Nrp1 | Plxna3 | |
| Nrp2 | Plxna4 | |
| Ntng1 | Plxnb1 | |
| Ntng2 | Plxnc1 | |
| Ntrk1 | Plxnd1 | |
| Ntrk2 | Procr | |
| Ntrk3 | Prokr1 | |
| Ntsr1 | Prokr2 | |
| Oprl1 | Ptch1 | |
| Osmr | Ptch2 | |
| Oxtr | Pth1r | |
| Pdgfa | Pth2r | |
| Pdgfra | Ptprk | |
| Pdgfrb | Ptprs | |
| Pgr | Ptprz1 | |
| Plaur | Ret | |
| Plgrkt | Robo3 | |
| Plxna1 | Ror1 | |
| Plxna2 | Ror2 | |
| Plxna3 | Rorb | |
| Plxna4 | Rtn4r | |
| Plxnb1 | Rtn4rl1 | |
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Plxnc1 | Rxrg | |
| Plxnd1 | Ryr1 | |
| Prlhr | Ryr2 | |
| Prlr | Sctr | |
| Procr | Sdc4 | |
| Prokr1 | Sfrp1 | |
| Prokr2 | Sfrp2 | |
| Ptch1 | Slc1a5 | |
| Ptch2 | Sorcs3 | |
| Pth1r | Sort1 | |
| Pth2r | Sstr1 | |
| Ptprh | Sstr2 | |
| Ptprk | Sstr3 | |
| Ptprs | Sstr4 | |
| Ptprz1 | Sstr5 | |
| Ret | Tek | |
| Robo3 | Tgfbr2 | |
| Ror1 | Tgfbr3 | |
| Ror2 | Thbd | |
| Rorb | Thra | |
| Rtn4r | Thrap3 | |
| Rtn4rl1 | Tnfrsf10b | |
| Rxfp1 | Tnfrsf11a | |
| Rxrg | Tnfrsf11b | |
| Ryr1 | Tnfrsf12a | |
| Ryr2 | Tnfrsf13c | |
| Sctr | Tnfrsf14 | |
| Sdc4 | Tnfrsf17 | |
| Sfrp1 | Tnfrsf18 | |
| Sfrp2 | Tnfrsf1a | |
| Slc1a5 | Tnfrsf1b | |
| Sorcs3 | Tnfrsf25 | |
| Sort1 | Tnfrsf4 | |
| Sstr1 | Tnfrsf8 | |
| Sstr2 | Tnfrsf9 | |
| Sstr3 | Tshr | |
| Sstr4 | Unc5b | |
| Sstr5 | Unc5c | |
| Tek | Uts2r | |
| Tgfbr2 | Vipr1 | |
| Tgfbr3 | Vldlr | |
| Thbd | Vtn | |
| Thra | Xcr1 | |
| Thrap3 | ||
| Tnfrsf10b | ||
| Tnfrsf11a | ||
| Tnfrsf11b | ||
| Tnfrsf12a | ||
| Tnfrsf13c | ||
| Tnfrsf14 | ||
| Tnfrsf17 | ||
| Tnfrsf18 | ||
| Tnfrsf1a | ||
| Tnfrsf1b | ||
| Tnfrsf25 | ||
| Tnfrsf4 | ||
| Tnfrsf8 | ||
| Tnfrsf9 | ||
| Tshr | ||
| Unc5b | ||
| Unc5c | ||
| Microarrays | ||
| DRGs (321) | SCGs (297) | |
| Uts2r | ||
| Vipr1 | ||
| Vipr2 | ||
| Vldlr | ||
| Vtn | ||
| Xcr1 | ||
Gene symbols of proteins identified by mass spectrometry exclusively on sensory neurons (DRGs), exclusively on sympathetic neurons (SCGs), or on both neuron types, which were identified from the protein lists shown in Table 7. Also shown are receptor mRNAs identified by microarrays as expressed by DRG and SCG neurons, defined using the updated ligand-receptor database (modified from Yuzwa et al., 2016). Only receptor mRNAs that had expression exceeding the cutoffs for each neuron type (DRGs; Itgam, 87% and SCGs; Sorcs3, 81%) are included. The total numbers of receptors in each column are indicated.
Cell-surface proteomics is relatively insensitive, and it ha5s previously been shown that more sensitive transcriptomic profiling can also be used to identify biologically relevant paracrine interactions (Johnston et al., 2016; Yuzwa et al., 2016; Voronova et al., 2017). We therefore complemented the proteomics by analyzing six and four independent biological replicates of cultured DRG and SCG RNA, respectively, on Affymetrix GeneChip Rat Gene 2.0 ST Arrays. To analyze these data, we defined an expression cutoff based on the proteomics data. Specifically, we identified the cell-surface receptor proteins with the lowest mRNA expression on the microarray for each neuron type and used those as the cutoffs. For sensory and sympathetic neurons, these were Itgam and Sorcs3 mRNAs, respectively (expressed at 87% and 81% of total mRNAs; Extended Data Fig. 5-1D). When these thresholds were applied to the microarray data, there were 321 and 297 receptor mRNAs in sensory and sympathetic neurons, respectively (Table 8). Importantly, there was good correspondence between the proteomics and microarray data; TrkA/Ntrk1, Bmpr2, Ret, and Igf2r mRNAs were similarly expressed in both populations of neurons; Rtn4r, Gfra3, and Acvr2a mRNAs were enriched in sensory neurons (2.8-, 10-, and 3.1-fold enriched, respectively); and Alk mRNA was 9-fold enriched in sympathetic neurons (p < 0.05 FDR for differences).
Computational modeling predicts that ligands deriving from multiple types of nerve cells, including mesenchymal cells, act on peripheral neurons
We performed computational modeling with the 143 injured nerve ligands and the sensory and sympathetic neuron receptors we had defined to predict how the injured nerve environment might regulate peripheral axon biology. This modeling predicted 122 and 125 potential unidirectional paracrine interactions between the injured nerve and sympathetic and sensory neurons, respectively (Fig. 5C,D; Table 9). Of these, cell-surface receptor protein expression was detected for 49 and 60 sympathetic and sensory neuron predicted interactions (Fig. 5C,D, blue boxes). Many predicted interactions involved known peripheral nerve ligands such as the neurotrophin and GDNF families. Notably, all but three ligands (GNRH1, CXCL1, and CXCL2) were predicted to act on both sympathetic and sensory neurons. The receptors for these predicted interactions were also largely the same, except for several sensory neuron receptors; the Erbb4 receptor for EGF/neuregulin family ligands, KDR for the VEGF family, ACVR1C for the activin/BMP family, and the CXCR5 chemokine receptor (Table 9). Thus, the injured nerve is predicted to produce ligands that act on both sympathetic and sensory neurons, largely through the same receptors.
Table 9.
Ligand-receptor modeling between injured nerve ligands and sympathetic neurons (SCGs), sensory neurons (DRGs), motor neurons (MNs), and retinal ganglion cells (RGCs)
| SCGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | ADM | SCGs | CALCRL |
| Injured nerve | ANGPT1 | SCGs | TEK |
| Injured nerve | ANGPT2 | SCGs | TEK |
| Injured nerve | ANGPT4 | SCGs | TEK |
| Injured nerve | APLN | SCGs | APLNR |
| Injured nerve | ARTN | SCGs | GFRA3 |
| Injured nerve | ARTN | SCGs | RET |
| Injured nerve | BDNF | SCGs | NTRK2 |
| Injured nerve | BDNF | SCGs | SORT1 |
| Injured nerve | BDNF | SCGs | NGFR |
| Injured nerve | BMP2 | SCGs | BMPR1A |
| Injured nerve | BMP2 | SCGs | BMPR1B |
| Injured nerve | BMP2 | SCGs | BMPR2 |
| Injured nerve | BMP2 | SCGs | ENG |
| Injured nerve | BMP2 | SCGs | NEO1 |
| Injured nerve | BMP4 | SCGs | BMPR1A |
| Injured nerve | BMP4 | SCGs | BMPR1B |
| Injured nerve | BMP4 | SCGs | BMPR2 |
| Injured nerve | BMP4 | SCGs | NEO1 |
| Injured nerve | BMP5 | SCGs | BMPR1A |
| Injured nerve | BMP7 | SCGs | ACVR1 |
| Injured nerve | BMP7 | SCGs | ACVR2A |
| Injured nerve | BMP7 | SCGs | ACVR2B |
| Injured nerve | BMP7 | SCGs | BMPR1A |
| Injured nerve | BMP7 | SCGs | BMPR1B |
| Injured nerve | BMP7 | SCGs | BMPR2 |
| Injured nerve | BMP7 | SCGs | NEO1 |
| Injured nerve | BTC | SCGs | EGFR |
| Injured nerve | BTC | SCGs | ERBB2 |
| Injured nerve | CCK | SCGs | CCKAR |
| Injured nerve | CCK | SCGs | CCKBR |
| Injured nerve | CCL11 | SCGs | CXCR3 |
| Injured nerve | CCL19 | SCGs | CCR10 |
| Injured nerve | CCL19 | SCGs | CCR7 |
| Injured nerve | CCL2 | SCGs | CCR10 |
| Injured nerve | CCL25 | SCGs | CCR10 |
| Injured nerve | CCL3 | SCGs | CCR4 |
| Injured nerve | CCL5 | SCGs | CCR4 |
| Injured nerve | CCL5 | SCGs | CXCR3 |
| Injured nerve | CCL5 | SCGs | SDC4 |
| Injured nerve | CCL7 | SCGs | CCR10 |
| Injured nerve | CCL7 | SCGs | CXCR3 |
| Injured nerve | CLCF1 | SCGs | CNTFR |
| Injured nerve | CLCF1 | SCGs | IL6ST |
| Injured nerve | CLCF1 | SCGs | LIFR |
| Injured nerve | CRLF1 | SCGs | CNTFR |
| Injured nerve | CRLF1 | SCGs | IL6ST |
| Injured nerve | CRLF1 | SCGs | LIFR |
| Injured nerve | CSF1 | SCGs | CSF1R |
| Injured nerve | CX3CL1 | SCGs | CX3CR1 |
| Injured nerve | CXCL10 | SCGs | CXCR3 |
| Injured nerve | CXCL12 | SCGs | CCR4 |
| Injured nerve | CXCL12 | SCGs | CXCR4 |
| Injured nerve | CXCL13 | SCGs | CCR10 |
| Injured nerve | CXCL13 | SCGs | CXCR3 |
| Injured nerve | CXCL9 | SCGs | CXCR3 |
| Injured nerve | DHH | SCGs | PTCH1 |
| Injured nerve | DHH | SCGs | PTCH2 |
| Injured nerve | DLL1 | SCGs | NOTCH1 |
| Injured nerve | DLL1 | SCGs | NOTCH2 |
| SCGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | DLL1 | SCGs | NOTCH3 |
| Injured nerve | DLL4 | SCGs | NOTCH1 |
| Injured nerve | EBI3 | SCGs | IL27RA |
| Injured nerve | EDA | SCGs | EDAR |
| Injured nerve | EDN3 | SCGs | EDNRA |
| Injured nerve | EDN3 | SCGs | EDNRB |
| Injured nerve | EFNA1 | SCGs | EPHA1 |
| Injured nerve | EFNA1 | SCGs | EPHA3 |
| Injured nerve | EFNA2 | SCGs | EPHA3 |
| Injured nerve | EFNA4 | SCGs | EPHA3 |
| Injured nerve | EFNA4 | SCGs | EPHA5 |
| Injured nerve | EFNA5 | SCGs | EPHA3 |
| Injured nerve | EFNA5 | SCGs | EPHA2 |
| Injured nerve | EFNA5 | SCGs | EPHA7 |
| Injured nerve | EFNA5 | SCGs | EPHA5 |
| Injured nerve | EFNB1 | SCGs | EPHB2 |
| Injured nerve | EFNB2 | SCGs | EPHB1 |
| Injured nerve | EFNB2 | SCGs | EPHA4 |
| Injured nerve | EFNB2 | SCGs | EPHA3 |
| Injured nerve | EFNB2 | SCGs | EPHB4 |
| Injured nerve | EFNB2 | SCGs | EPHB2 |
| Injured nerve | FGF1 | SCGs | FGFR1 |
| Injured nerve | FGF1 | SCGs | FGFR2 |
| Injured nerve | FGF1 | SCGs | FGFR3 |
| Injured nerve | FGF1 | SCGs | FGFR4 |
| Injured nerve | FGF10 | SCGs | FGFR2 |
| Injured nerve | FGF18 | SCGs | FGFR4 |
| Injured nerve | FGF5 | SCGs | FGFR1 |
| Injured nerve | FGF5 | SCGs | FGFR3 |
| Injured nerve | FGF7 | SCGs | FGFR2 |
| Injured nerve | FGF7 | SCGs | NRP1 |
| Injured nerve | FIGF | SCGs | FLT4 |
| Injured nerve | FIGF | SCGs | NRP1 |
| Injured nerve | FIGF | SCGs | NRP2 |
| Injured nerve | FSTL1 | SCGs | CD14 |
| Injured nerve | FSTL1 | SCGs | DIP2A |
| Injured nerve | GAS6 | SCGs | AXL |
| Injured nerve | GDF11 | SCGs | ACVR1B |
| Injured nerve | GDF11 | SCGs | ACVR2B |
| Injured nerve | GDNF | SCGs | GFRA2 |
| Injured nerve | GDNF | SCGs | RET |
| Injured nerve | HBEGF | SCGs | EGFR |
| Injured nerve | HGF | SCGs | MET |
| Injured nerve | IGF1 | SCGs | IGF1R |
| Injured nerve | IGF1 | SCGs | IGFBP1 |
| Injured nerve | IGF1 | SCGs | IGFBP2 |
| Injured nerve | IGF1 | SCGs | IGFBP3 |
| Injured nerve | IGF1 | SCGs | IGFBP4 |
| Injured nerve | IGF1 | SCGs | IGFBP5 |
| Injured nerve | IGF1 | SCGs | IGFBP6 |
| Injured nerve | IGF1 | SCGs | IGFBP7 |
| Injured nerve | IGF1 | SCGs | INSR |
| Injured nerve | IGF2 | SCGs | IGF1R |
| Injured nerve | IGF2 | SCGs | IGF2R |
| Injured nerve | IGF2 | SCGs | INSR |
| Injured nerve | IL15 | SCGs | IL15RA |
| Injured nerve | IL15 | SCGs | IL2RB |
| Injured nerve | IL15 | SCGs | IL2RG |
| Injured nerve | IL16 | SCGs | CD4 |
| Injured nerve | IL16 | SCGs | GRIN2A |
| Injured nerve | IL16 | SCGs | GRIN2B |
| SCGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | IL16 | SCGs | GRIN2C |
| Injured nerve | IL16 | SCGs | GRIN2D |
| Injured nerve | IL18 | SCGs | IL18R1 |
| Injured nerve | IL18 | SCGs | IL18RAP |
| Injured nerve | IL1B | SCGs | IL1R1 |
| Injured nerve | IL1B | SCGs | IL1R2 |
| Injured nerve | IL1B | SCGs | IL1RAP |
| Injured nerve | IL33 | SCGs | IL1RL1 |
| Injured nerve | IL6 | SCGs | IL6R |
| Injured nerve | IL6 | SCGs | IL6ST |
| Injured nerve | INHA | SCGs | ACVR2A |
| Injured nerve | INHA | SCGs | TGFBR3 |
| Injured nerve | INHBA | SCGs | ACVR1B |
| Injured nerve | INHBA | SCGs | ACVR2A |
| Injured nerve | INHBA | SCGs | ACVR2B |
| Injured nerve | INHBB | SCGs | ACVR1 |
| Injured nerve | INHBB | SCGs | ACVR1B |
| Injured nerve | INHBB | SCGs | ACVR2A |
| Injured nerve | INHBB | SCGs | ACVR2B |
| Injured nerve | JAG1 | SCGs | NOTCH1 |
| Injured nerve | JAG1 | SCGs | NOTCH2 |
| Injured nerve | JAG1 | SCGs | NOTCH3 |
| Injured nerve | JAG2 | SCGs | NOTCH1 |
| Injured nerve | JAG2 | SCGs | NOTCH2 |
| Injured nerve | JAG2 | SCGs | NOTCH3 |
| Injured nerve | LIF | SCGs | IL6ST |
| Injured nerve | LIF | SCGs | LIFR |
| Injured nerve | LTB | SCGs | LTBR |
| Injured nerve | MDK | SCGs | ALK |
| Injured nerve | MDK | SCGs | LRP1 |
| Injured nerve | MDK | SCGs | LRP2 |
| Injured nerve | MDK | SCGs | PTPRZ1 |
| Injured nerve | MIF | SCGs | CD74 |
| Injured nerve | NGF | SCGs | NGFR |
| Injured nerve | NGF | SCGs | NTRK1 |
| Injured nerve | NGF | SCGs | SORCS3 |
| Injured nerve | NGF | SCGs | SORT1 |
| Injured nerve | NOV | SCGs | NOTCH1 |
| Injured nerve | NPPC | SCGs | NPR2 |
| Injured nerve | NPPC | SCGs | NPR3 |
| Injured nerve | NTF3 | SCGs | NGFR |
| Injured nerve | NTF3 | SCGs | NTRK1 |
| Injured nerve | NTF3 | SCGs | NTRK2 |
| Injured nerve | NTF3 | SCGs | NTRK3 |
| Injured nerve | NTN1 | SCGs | DCC |
| Injured nerve | NTN1 | SCGs | NEO1 |
| Injured nerve | NTN1 | SCGs | UNC5B |
| Injured nerve | NTN1 | SCGs | UNC5C |
| Injured nerve | OSM | SCGs | IL6ST |
| Injured nerve | OSM | SCGs | LIFR |
| Injured nerve | OSM | SCGs | OSMR |
| Injured nerve | PDGFA | SCGs | PDGFRA |
| Injured nerve | PDGFB | SCGs | PDGFRA |
| Injured nerve | PDGFB | SCGs | PDGFRB |
| Injured nerve | PDGFC | SCGs | PDGFRA |
| Injured nerve | PF4 | SCGs | CXCR3 |
| Injured nerve | PF4 | SCGs | LDLR |
| Injured nerve | PF4 | SCGs | THBD |
| Injured nerve | PGF | SCGs | FLT1 |
| Injured nerve | PGF | SCGs | NRP1 |
| Injured nerve | PGF | SCGs | NRP2 |
| SCGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | POMC | SCGs | MC1R |
| Injured nerve | POMC | SCGs | MC2R |
| Injured nerve | POMC | SCGs | MC3R |
| Injured nerve | POMC | SCGs | MC4R |
| Injured nerve | POMC | SCGs | MC5R |
| Injured nerve | PTHLH | SCGs | PTH1R |
| Injured nerve | PTN | SCGs | ALK |
| Injured nerve | PTN | SCGs | PTPRS |
| Injured nerve | PTN | SCGs | PTPRZ1 |
| Injured nerve | RSPO1 | SCGs | LGR5 |
| Injured nerve | RTN4 | SCGs | LINGO1 |
| Injured nerve | RTN4 | SCGs | RTN4R |
| Injured nerve | RTN4 | SCGs | RTN4RL1 |
| Injured nerve | SEMA3B | SCGs | PLXNA1 |
| Injured nerve | SEMA3B | SCGs | PLXNA2 |
| Injured nerve | SEMA3B | SCGs | PLXNA3 |
| Injured nerve | SEMA3B | SCGs | PLXNA4 |
| Injured nerve | SEMA3B | SCGs | PLXND1 |
| Injured nerve | SEMA3C | SCGs | PLXNA1 |
| Injured nerve | SEMA3C | SCGs | PLXNA2 |
| Injured nerve | SEMA3C | SCGs | PLXNA3 |
| Injured nerve | SEMA3C | SCGs | PLXNA4 |
| Injured nerve | SEMA3C | SCGs | PLXND1 |
| Injured nerve | SEMA3D | SCGs | PLXNA1 |
| Injured nerve | SEMA3D | SCGs | PLXNA2 |
| Injured nerve | SEMA3D | SCGs | PLXNA3 |
| Injured nerve | SEMA3D | SCGs | PLXNA4 |
| Injured nerve | SEMA3D | SCGs | PLXND1 |
| Injured nerve | SEMA3E | SCGs | PLXNA1 |
| Injured nerve | SEMA3E | SCGs | PLXNA2 |
| Injured nerve | SEMA3E | SCGs | PLXNA3 |
| Injured nerve | SEMA3E | SCGs | PLXNA4 |
| Injured nerve | SEMA3E | SCGs | PLXND1 |
| Injured nerve | SEMA3F | SCGs | PLXNA1 |
| Injured nerve | SEMA3F | SCGs | PLXNA2 |
| Injured nerve | SEMA3F | SCGs | PLXNA3 |
| Injured nerve | SEMA3F | SCGs | PLXNA4 |
| Injured nerve | SEMA3F | SCGs | PLXND1 |
| Injured nerve | SEMA3G | SCGs | PLXNA1 |
| Injured nerve | SEMA3G | SCGs | PLXNA2 |
| Injured nerve | SEMA3G | SCGs | PLXNA3 |
| Injured nerve | SEMA3G | SCGs | PLXNA4 |
| Injured nerve | SEMA3G | SCGs | PLXND1 |
| Injured nerve | SEMA4A | SCGs | PLXNB1 |
| Injured nerve | SEMA4A | SCGs | PLXNC1 |
| Injured nerve | SEMA4A | SCGs | PLXND1 |
| Injured nerve | SEMA4B | SCGs | PLXNB1 |
| Injured nerve | SEMA4B | SCGs | PLXNC1 |
| Injured nerve | SEMA4C | SCGs | PLXNB1 |
| Injured nerve | SEMA4C | SCGs | PLXNC1 |
| Injured nerve | SEMA4D | SCGs | PLXNB1 |
| Injured nerve | SEMA4D | SCGs | PLXNC1 |
| Injured nerve | SEMA4F | SCGs | PLXNB1 |
| Injured nerve | SEMA4F | SCGs | PLXNC1 |
| Injured nerve | SEMA5A | SCGs | PLXNA3 |
| Injured nerve | SEMA5A | SCGs | PLXNA4 |
| Injured nerve | SEMA5A | SCGs | PLXNC1 |
| Injured nerve | SEMA5B | SCGs | PLXNA3 |
| Injured nerve | SEMA5B | SCGs | PLXNA4 |
| Injured nerve | SEMA5B | SCGs | PLXNC1 |
| Injured nerve | SEMA6A | SCGs | PLXNA1 |
| SCGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | SEMA6A | SCGs | PLXNA2 |
| Injured nerve | SEMA6B | SCGs | PLXNA1 |
| Injured nerve | SEMA6C | SCGs | PLXNA1 |
| Injured nerve | SEMA6D | SCGs | PLXNA1 |
| Injured nerve | SEMA7A | SCGs | PLXNC1 |
| Injured nerve | SHH | SCGs | PTCH1 |
| Injured nerve | SHH | SCGs | PTCH2 |
| Injured nerve | TGFA | SCGs | EGFR |
| Injured nerve | TGFA | SCGs | ERBB2 |
| Injured nerve | TGFB1 | SCGs | ACVRL1 |
| Injured nerve | TGFB1 | SCGs | ENG |
| Injured nerve | TGFB1 | SCGs | TGFBR2 |
| Injured nerve | TGFB1 | SCGs | TGFBR3 |
| Injured nerve | TGFB2 | SCGs | TGFBR2 |
| Injured nerve | TGFB2 | SCGs | TGFBR3 |
| Injured nerve | TGFB3 | SCGs | ACVRL1 |
| Injured nerve | TGFB3 | SCGs | TGFBR2 |
| Injured nerve | TNF | SCGs | TNFRSF1A |
| Injured nerve | TNF | SCGs | TNFRSF1B |
| Injured nerve | TNFSF10 | SCGs | TNFRSF10B |
| Injured nerve | TNFSF12 | SCGs | TNFRSF12A |
| Injured nerve | TNFSF12 | SCGs | TNFRSF25 |
| Injured nerve | TNFSF14 | SCGs | LTBR |
| Injured nerve | TNFSF14 | SCGs | TNFRSF14 |
| Injured nerve | TNFSF8 | SCGs | TNFRSF8 |
| Injured nerve | TNFSF9 | SCGs | TNFRSF9 |
| Injured nerve | TSLP | SCGs | CRLF2 |
| Injured nerve | TSLP | SCGs | IL7R |
| Injured nerve | UCN2 | SCGs | CRHR2 |
| Injured nerve | VEGFA | SCGs | FLT1 |
| Injured nerve | VEGFA | SCGs | NRP1 |
| Injured nerve | VEGFA | SCGs | NRP2 |
| Injured nerve | VEGFB | SCGs | FLT1 |
| Injured nerve | VEGFB | SCGs | NRP1 |
| Injured nerve | VEGFC | SCGs | FLT4 |
| Injured nerve | VEGFC | SCGs | NRP1 |
| Injured nerve | VEGFC | SCGs | NRP2 |
| Injured nerve | WNT11 | SCGs | FZD4 |
| Injured nerve | WNT2 | SCGs | FZD1 |
| Injured nerve | WNT2 | SCGs | FZD9 |
| Injured nerve | WNT5A | SCGs | FZD2 |
| Injured nerve | WNT5A | SCGs | FZD5 |
| Injured nerve Injured Nerve |
WNT5A WNT5A |
SCGs SCGs |
ROR1 ROR2 |
| DRGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | ADM | DRGs | CALCRL |
| Injured nerve | ANGPT1 | DRGs | TEK |
| Injured nerve | ANGPT2 | DRGs | TEK |
| Injured nerve | ANGPT4 | DRGs | TEK |
| Injured nerve | APLN | DRGs | APLNR |
| Injured nerve | ARTN | DRGs | GFRA3 |
| Injured nerve | ARTN | DRGs | RET |
| Injured nerve | BDNF | DRGs | NGFR |
| Injured nerve | BDNF | DRGs | NTRK2 |
| Injured nerve | BDNF | DRGs | SORT1 |
| Injured nerve | BMP2 | DRGs | BMPR1A |
| Injured nerve | BMP2 | DRGs | BMPR1B |
| Injured nerve | BMP2 | DRGs | BMPR2 |
| Injured nerve | BMP2 | DRGs | ENG |
| Injured nerve | BMP2 | DRGs | NEO1 |
| DRGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | BMP4 | DRGs | BMPR1A |
| Injured nerve | BMP4 | DRGs | BMPR1B |
| Injured nerve | BMP4 | DRGs | BMPR2 |
| Injured nerve | BMP4 | DRGs | NEO1 |
| Injured nerve | BMP5 | DRGs | BMPR1A |
| Injured nerve | BMP7 | DRGs | ACVR1 |
| Injured nerve | BMP7 | DRGs | ACVR2A |
| Injured nerve | BMP7 | DRGs | ACVR2B |
| Injured nerve | BMP7 | DRGs | BMPR1A |
| Injured nerve | BMP7 | DRGs | BMPR1B |
| Injured nerve | BMP7 | DRGs | BMPR2 |
| Injured nerve | BMP7 | DRGs | NEO1 |
| Injured nerve | BTC | DRGs | EGFR |
| Injured nerve | BTC | DRGs | ERBB2 |
| Injured nerve | BTC | DRGs | ERBB4 |
| Injured nerve | CCK | DRGs | CCKAR |
| Injured nerve | CCK | DRGs | CCKBR |
| Injured nerve | CCL11 | DRGs | CXCR3 |
| Injured nerve | CCL19 | DRGs | CCR10 |
| Injured nerve | CCL19 | DRGs | CCR7 |
| Injured nerve | CCL2 | DRGs | CCR10 |
| Injured nerve | CCL25 | DRGs | CCR10 |
| Injured nerve | CCL3 | DRGs | CCR4 |
| Injured nerve | CCL5 | DRGs | CCR4 |
| Injured nerve | CCL5 | DRGs | CXCR3 |
| Injured nerve | CCL5 | DRGs | SDC4 |
| Injured nerve | CCL7 | DRGs | CCR10 |
| Injured nerve | CCL7 | DRGs | CXCR3 |
| Injured nerve | CLCF1 | DRGs | CNTFR |
| Injured nerve | CLCF1 | DRGs | IL6ST |
| Injured nerve | CLCF1 | DRGs | LIFR |
| Injured nerve | CRLF1 | DRGs | CNTFR |
| Injured nerve | CRLF1 | DRGs | IL6ST |
| Injured nerve | CRLF1 | DRGs | LIFR |
| Injured nerve | CSF1 | DRGs | CSF1R |
| Injured nerve | CX3CL1 | DRGs | CX3CR1 |
| Injured nerve | CXCL1 | DRGs | CXCR2 |
| Injured nerve | CXCL10 | DRGs | CXCR3 |
| Injured nerve | CXCL12 | DRGs | CCR4 |
| Injured nerve | CXCL12 | DRGs | CXCR4 |
| Injured nerve | CXCL13 | DRGs | CCR10 |
| Injured nerve | CXCL13 | DRGs | CXCR3 |
| Injured nerve | CXCL13 | DRGs | CXCR5 |
| Injured nerve | CXCL2 | DRGs | CXCR2 |
| Injured nerve | CXCL9 | DRGs | CXCR3 |
| Injured nerve | DHH | DRGs | PTCH1 |
| Injured nerve | DHH | DRGs | PTCH2 |
| Injured nerve | DLL1 | DRGs | NOTCH1 |
| Injured nerve | DLL1 | DRGs | NOTCH2 |
| Injured nerve | DLL1 | DRGs | NOTCH3 |
| Injured nerve | DLL4 | DRGs | NOTCH1 |
| Injured nerve | EBI3 | DRGs | IL27RA |
| Injured nerve | EDA | DRGs | EDAR |
| Injured nerve | EDN3 | DRGs | EDNRA |
| Injured nerve | EDN3 | DRGs | EDNRB |
| Injured nerve | EFNA1 | DRGs | EPHA1 |
| Injured nerve | EFNA1 | DRGs | EPHA3 |
| Injured nerve | EFNA2 | DRGs | EPHA3 |
| Injured nerve | EFNA4 | DRGs | EPHA3 |
| Injured nerve | EFNA4 | DRGs | EPHA5 |
| Injured nerve | EFNA5 | DRGs | EPHA3 |
| DRGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | EFNA5 | DRGs | EPHA2 |
| Injured nerve | EFNA5 | DRGs | EPHA7 |
| Injured nerve | EFNA5 | DRGs | EPHA5 |
| Injured nerve | EFNB1 | DRGs | EPHB2 |
| Injured nerve | EFNB2 | DRGs | EPHB1 |
| Injured nerve | EFNB2 | DRGs | EPHA4 |
| Injured nerve | EFNB2 | DRGs | EPHA3 |
| Injured nerve | EFNB2 | DRGs | EPHB4 |
| Injured nerve | EFNB2 | DRGs | EPHB2 |
| Injured nerve | FGF1 | DRGs | FGFR1 |
| Injured nerve | FGF1 | DRGs | FGFR2 |
| Injured nerve | FGF1 | DRGs | FGFR3 |
| Injured nerve | FGF1 | DRGs | FGFR4 |
| Injured nerve | FGF10 | DRGs | FGFR2 |
| Injured nerve | FGF18 | DRGs | FGFR4 |
| Injured nerve | FGF5 | DRGs | FGFR1 |
| Injured nerve | FGF5 | DRGs | FGFR3 |
| Injured nerve | FGF7 | DRGs | FGFR2 |
| Injured nerve | FGF7 | DRGs | NRP1 |
| Injured nerve | FIGF | DRGs | FLT4 |
| Injured nerve | FIGF | DRGs | KDR |
| Injured nerve | FIGF | DRGs | NRP1 |
| Injured nerve | FIGF | DRGs | NRP2 |
| Injured nerve | FSTL1 | DRGs | CD14 |
| Injured nerve | FSTL1 | DRGs | DIP2A |
| Injured nerve | GAS6 | DRGs | AXL |
| Injured nerve | GDF11 | DRGs | ACVR1B |
| Injured nerve | GDF11 | DRGs | ACVR1C |
| Injured nerve | GDF11 | DRGs | ACVR2B |
| Injured nerve | GDNF | DRGs | GFRA2 |
| Injured nerve | GDNF | DRGs | RET |
| Injured nerve | GNRH1 | DRGs | GNRHR |
| Injured nerve | HBEGF | DRGs | EGFR |
| Injured nerve | HBEGF | DRGs | ERBB4 |
| Injured nerve | HGF | DRGs | MET |
| Injured nerve | IGF1 | DRGs | IGF1R |
| Injured nerve | IGF1 | DRGs | IGFBP1 |
| Injured nerve | IGF1 | DRGs | IGFBP2 |
| Injured nerve | IGF1 | DRGs | IGFBP3 |
| Injured nerve | IGF1 | DRGs | IGFBP4 |
| Injured nerve | IGF1 | DRGs | IGFBP5 |
| Injured nerve | IGF1 | DRGs | IGFBP6 |
| Injured nerve | IGF1 | DRGs | IGFBP7 |
| Injured nerve | IGF1 | DRGs | INSR |
| Injured nerve | IGF2 | DRGs | IGF1R |
| Injured nerve | IGF2 | DRGs | IGF2R |
| Injured nerve | IGF2 | DRGs | INSR |
| Injured nerve | IL15 | DRGs | IL15RA |
| Injured nerve | IL15 | DRGs | IL2RB |
| Injured nerve | IL15 | DRGs | IL2RG |
| Injured nerve | IL16 | DRGs | CD4 |
| Injured nerve | IL16 | DRGs | GRIN2A |
| Injured nerve | IL16 | DRGs | GRIN2B |
| Injured nerve | IL16 | DRGs | GRIN2C |
| Injured nerve | IL16 | DRGs | GRIN2D |
| Injured nerve | IL18 | DRGs | IL18R1 |
| Injured nerve | IL18 | DRGs | IL18RAP |
| Injured nerve | IL1B | DRGs | IL1R1 |
| Injured nerve | IL1B | DRGs | IL1R2 |
| Injured nerve | IL1B | DRGs | IL1RAP |
| Injured nerve | IL33 | DRGs | IL1RL1 |
| DRGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | IL6 | DRGs | IL6R |
| Injured nerve | IL6 | DRGs | IL6ST |
| Injured nerve | INHA | DRGs | ACVR2A |
| Injured nerve | INHA | DRGs | TGFBR3 |
| Injured nerve | INHBA | DRGs | ACVR1B |
| Injured nerve | INHBA | DRGs | ACVR2A |
| Injured nerve | INHBA | DRGs | ACVR2B |
| Injured nerve | INHBB | DRGs | ACVR1 |
| Injured nerve | INHBB | DRGs | ACVR1B |
| Injured nerve | INHBB | DRGs | ACVR1C |
| Injured nerve | INHBB | DRGs | ACVR2A |
| Injured nerve | INHBB | DRGs | ACVR2B |
| Injured nerve | JAG1 | DRGs | NOTCH1 |
| Injured nerve | JAG1 | DRGs | NOTCH2 |
| Injured nerve | JAG1 | DRGs | NOTCH3 |
| Injured nerve | JAG2 | DRGs | NOTCH1 |
| Injured nerve | JAG2 | DRGs | NOTCH2 |
| Injured nerve | JAG2 | DRGs | NOTCH3 |
| Injured nerve | LIF | DRGs | IL6ST |
| Injured nerve | LIF | DRGs | LIFR |
| Injured nerve | LTB | DRGs | LTBR |
| Injured nerve | MDK | DRGs | ALK |
| Injured nerve | MDK | DRGs | LRP1 |
| Injured nerve | MDK | DRGs | LRP2 |
| Injured nerve | MDK | DRGs | PTPRZ1 |
| Injured nerve | MIF | DRGs | CD74 |
| Injured nerve | NGF | DRGs | NGFR |
| Injured nerve | NGF | DRGs | NTRK1 |
| Injured nerve | NGF | DRGs | SORCS3 |
| Injured nerve | NGF | DRGs | SORT1 |
| Injured nerve | NOV | DRGs | NOTCH1 |
| Injured nerve | NPPC | DRGs | NPR2 |
| Injured nerve | NPPC | DRGs | NPR3 |
| Injured nerve | NTF3 | DRGs | NGFR |
| Injured nerve | NTF3 | DRGs | NTRK1 |
| Injured nerve | NTF3 | DRGs | NTRK2 |
| Injured nerve | NTF3 | DRGs | NTRK3 |
| Injured nerve | NTN1 | DRGs | DCC |
| Injured nerve | NTN1 | DRGs | NEO1 |
| Injured nerve | NTN1 | DRGs | UNC5B |
| Injured nerve | NTN1 | DRGs | UNC5C |
| Injured nerve | OSM | DRGs | IL6ST |
| Injured nerve | OSM | DRGs | LIFR |
| Injured nerve | OSM | DRGs | OSMR |
| Injured nerve | PDGFA | DRGs | PDGFRA |
| Injured nerve | PDGFB | DRGs | PDGFRA |
| Injured nerve | PDGFB | DRGs | PDGFRB |
| Injured nerve | PDGFC | DRGs | PDGFRA |
| Injured nerve | PF4 | DRGs | CXCR3 |
| Injured nerve | PF4 | DRGs | LDLR |
| Injured nerve | PF4 | DRGs | THBD |
| Injured nerve | PGF | DRGs | FLT1 |
| Injured nerve | PGF | DRGs | NRP1 |
| Injured nerve | PGF | DRGs | NRP2 |
| Injured nerve | POMC | DRGs | MC1R |
| Injured nerve | POMC | DRGs | MC2R |
| Injured nerve | POMC | DRGs | MC3R |
| Injured nerve | POMC | DRGs | MC4R |
| Injured nerve | POMC | DRGs | MC5R |
| Injured nerve | PTHLH | DRGs | PTH1R |
| Injured nerve | PTN | DRGs | ALK |
| DRGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | PTN | DRGs | PTPRS |
| Injured nerve | PTN | DRGs | PTPRZ1 |
| Injured nerve | RSPO1 | DRGs | LGR5 |
| Injured nerve | RTN4 | DRGs | LINGO1 |
| Injured nerve | RTN4 | DRGs | RTN4R |
| Injured nerve | RTN4 | DRGs | RTN4RL1 |
| Injured nerve | SEMA3B | DRGs | PLXNA1 |
| Injured nerve | SEMA3B | DRGs | PLXNA2 |
| Injured nerve | SEMA3B | DRGs | PLXNA3 |
| Injured nerve | SEMA3B | DRGs | PLXNA4 |
| Injured nerve | SEMA3B | DRGs | PLXND1 |
| Injured nerve | SEMA3C | DRGs | PLXNA1 |
| Injured nerve | SEMA3C | DRGs | PLXNA2 |
| Injured nerve | SEMA3C | DRGs | PLXNA3 |
| Injured nerve | SEMA3C | DRGs | PLXNA4 |
| Injured nerve | SEMA3C | DRGs | PLXND1 |
| Injured nerve | SEMA3D | DRGs | PLXNA1 |
| Injured nerve | SEMA3D | DRGs | PLXNA2 |
| Injured nerve | SEMA3D | DRGs | PLXNA3 |
| Injured nerve | SEMA3D | DRGs | PLXNA4 |
| Injured nerve | SEMA3D | DRGs | PLXND1 |
| Injured nerve | SEMA3E | DRGs | PLXNA1 |
| Injured nerve | SEMA3E | DRGs | PLXNA2 |
| Injured nerve | SEMA3E | DRGs | PLXNA3 |
| Injured nerve | SEMA3E | DRGs | PLXNA4 |
| Injured nerve | SEMA3E | DRGs | PLXND1 |
| Injured nerve | SEMA3F | DRGs | PLXNA1 |
| Injured nerve | SEMA3F | DRGs | PLXNA2 |
| Injured nerve | SEMA3F | DRGs | PLXNA3 |
| Injured nerve | SEMA3F | DRGs | PLXNA4 |
| Injured nerve | SEMA3F | DRGs | PLXND1 |
| Injured nerve | SEMA3G | DRGs | PLXNA1 |
| Injured nerve | SEMA3G | DRGs | PLXNA2 |
| Injured nerve | SEMA3G | DRGs | PLXNA3 |
| Injured nerve | SEMA3G | DRGs | PLXNA4 |
| Injured nerve | SEMA3G | DRGs | PLXND1 |
| Injured nerve | SEMA4A | DRGs | PLXNB1 |
| Injured nerve | SEMA4A | DRGs | PLXNC1 |
| Injured nerve | SEMA4A | DRGs | PLXND1 |
| Injured nerve | SEMA4B | DRGs | PLXNB1 |
| Injured nerve | SEMA4B | DRGs | PLXNC1 |
| Injured nerve | SEMA4C | DRGs | PLXNB1 |
| Injured nerve | SEMA4C | DRGs | PLXNC1 |
| Injured nerve | SEMA4D | DRGs | PLXNB1 |
| Injured nerve | SEMA4D | DRGs | PLXNC1 |
| Injured nerve | SEMA4F | DRGs | PLXNB1 |
| Injured nerve | SEMA4F | DRGs | PLXNC1 |
| Injured nerve | SEMA5A | DRGs | PLXNA3 |
| Injured nerve | SEMA5A | DRGs | PLXNA4 |
| Injured nerve | SEMA5A | DRGs | PLXNC1 |
| Injured nerve | SEMA5B | DRGs | PLXNA3 |
| Injured nerve | SEMA5B | DRGs | PLXNA4 |
| Injured nerve | SEMA5B | DRGs | PLXNC1 |
| Injured nerve | SEMA6A | DRGs | PLXNA1 |
| Injured nerve | SEMA6A | DRGs | PLXNA2 |
| Injured nerve | SEMA6B | DRGs | PLXNA1 |
| Injured nerve | SEMA6C | DRGs | PLXNA1 |
| Injured nerve | SEMA6D | DRGs | PLXNA1 |
| Injured nerve | SEMA7A | DRGs | PLXNC1 |
| Injured nerve | SHH | DRGs | PTCH1 |
| Injured nerve | SHH | DRGs | PTCH2 |
| DRGs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | TGFA | DRGs | EGFR |
| Injured nerve | TGFA | DRGs | ERBB2 |
| Injured nerve | TGFB1 | DRGs | ACVRL1 |
| Injured nerve | TGFB1 | DRGs | ENG |
| Injured nerve | TGFB1 | DRGs | TGFBR2 |
| Injured nerve | TGFB1 | DRGs | TGFBR3 |
| Injured nerve | TGFB2 | DRGs | TGFBR2 |
| Injured nerve | TGFB2 | DRGs | TGFBR3 |
| Injured nerve | TGFB3 | DRGs | ACVRL1 |
| Injured nerve | TGFB3 | DRGs | TGFBR2 |
| Injured nerve | TNF | DRGs | TNFRSF1A |
| Injured nerve | TNF | DRGs | TNFRSF1B |
| Injured nerve | TNFSF10 | DRGs | TNFRSF10B |
| Injured nerve | TNFSF12 | DRGs | TNFRSF12A |
| Injured nerve | TNFSF12 | DRGs | TNFRSF25 |
| Injured nerve | TNFSF14 | DRGs | LTBR |
| Injured nerve | TNFSF14 | DRGs | TNFRSF14 |
| Injured nerve | TNFSF8 | DRGs | TNFRSF8 |
| Injured nerve | TNFSF9 | DRGs | TNFRSF9 |
| Injured nerve | TSLP | DRGs | CRLF2 |
| Injured nerve | TSLP | DRGs | IL7R |
| Injured nerve | UCN2 | DRGs | CRHR2 |
| Injured nerve | VEGFA | DRGs | FLT1 |
| Injured nerve | VEGFA | DRGs | KDR |
| Injured nerve | VEGFA | DRGs | NRP1 |
| Injured nerve | VEGFA | DRGs | NRP2 |
| Injured nerve | VEGFB | DRGs | FLT1 |
| Injured nerve | VEGFB | DRGs | NRP1 |
| Injured nerve | VEGFC | DRGs | FLT4 |
| Injured nerve | VEGFC | DRGs | KDR |
| Injured nerve | VEGFC | DRGs | NRP1 |
| Injured nerve | VEGFC | DRGs | NRP2 |
| Injured nerve | WNT11 | DRGs | FZD4 |
| Injured nerve | WNT2 | DRGs | FZD1 |
| Injured nerve | WNT2 | DRGs | FZD9 |
| Injured nerve | WNT5A | DRGs | FZD2 |
| Injured nerve | WNT5A | DRGs | FZD5 |
| Injured nerve | WNT5A | DRGs | ROR1 |
| Injured nerve | WNT5A | DRGs | ROR2 |
| MNs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | ADM | MNs | CALCRL |
| Injured nerve | ANGPT1 | MNs | TEK |
| Injured nerve | ANGPT2 | MNs | TEK |
| Injured nerve | ANGPT4 | MNs | TEK |
| Injured nerve | APLN | MNs | APLNR |
| Injured nerve | ARTN | MNs | GFRA3 |
| Injured nerve | ARTN | MNs | RET |
| Injured nerve | BDNF | MNs | NGFR |
| Injured nerve | BDNF | MNs | NTRK2 |
| Injured nerve | BDNF | MNs | SORT1 |
| Injured nerve | BMP2 | MNs | BMPR1A |
| Injured nerve | BMP2 | MNs | BMPR1B |
| Injured nerve | BMP2 | MNs | BMPR2 |
| Injured nerve | BMP2 | MNs | ENG |
| Injured nerve | BMP2 | MNs | NEO1 |
| Injured nerve | BMP4 | MNs | BMPR1A |
| Injured nerve | BMP4 | MNs | BMPR1B |
| Injured nerve | BMP4 | MNs | BMPR2 |
| Injured nerve | BMP4 | MNs | NEO1 |
| Injured nerve | BMP5 | MNs | BMPR1A |
| MNs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | BMP7 | MNs | ACVR1 |
| Injured nerve | BMP7 | MNs | ACVR2A |
| Injured nerve | BMP7 | MNs | ACVR2B |
| Injured nerve | BMP7 | MNs | BMPR1A |
| Injured nerve | BMP7 | MNs | BMPR1B |
| Injured nerve | BMP7 | MNs | BMPR2 |
| Injured nerve | BMP7 | MNs | NEO1 |
| Injured nerve | BTC | MNs | EGFR |
| Injured nerve | BTC | MNs | ERBB2 |
| Injured nerve | BTC | MNs | ERBB4 |
| Injured nerve | CCK | MNs | CCKAR |
| Injured nerve | CCK | MNs | CCKBR |
| Injured nerve | CCL11 | MNs | CCR5 |
| Injured nerve | CCL11 | MNs | CXCR3 |
| Injured nerve | CCL19 | MNs | CCR10 |
| Injured nerve | CCL2 | MNs | CCR10 |
| Injured nerve | CCL2 | MNs | CCR2 |
| Injured nerve | CCL25 | MNs | CCR10 |
| Injured nerve | CCL25 | MNs | CCR9 |
| Injured nerve | CCL3 | MNs | CCR5 |
| Injured nerve | CCL5 | MNs | CCR5 |
| Injured nerve | CCL5 | MNs | CXCR3 |
| Injured nerve | CCL5 | MNs | SDC4 |
| Injured nerve | CCL7 | MNs | CCR10 |
| Injured nerve | CCL7 | MNs | CCR2 |
| Injured nerve | CCL7 | MNs | CCR5 |
| Injured nerve | CCL7 | MNs | CXCR3 |
| Injured nerve | CLCF1 | MNs | CNTFR |
| Injured nerve | CLCF1 | MNs | IL6ST |
| Injured nerve | CLCF1 | MNs | LIFR |
| Injured nerve | CRLF1 | MNs | CNTFR |
| Injured nerve | CRLF1 | MNs | IL6ST |
| Injured nerve | CRLF1 | MNs | LIFR |
| Injured nerve | CSF1 | MNs | CSF1R |
| Injured nerve | CX3CL1 | MNs | CX3CR1 |
| Injured nerve | CXCL10 | MNs | CXCR3 |
| Injured nerve | CXCL12 | MNs | CXCR4 |
| Injured nerve | CXCL13 | MNs | CCR10 |
| Injured nerve | CXCL13 | MNs | CXCR3 |
| Injured nerve | CXCL13 | MNs | CXCR5 |
| Injured nerve | CXCL9 | MNs | CXCR3 |
| Injured nerve | DHH | MNs | PTCH1 |
| Injured nerve | DHH | MNs | PTCH2 |
| Injured nerve | DLL1 | MNs | NOTCH1 |
| Injured nerve | DLL1 | MNs | NOTCH2 |
| Injured nerve | DLL1 | MNs | NOTCH3 |
| Injured nerve | DLL4 | MNs | NOTCH1 |
| Injured nerve | EBI3 | MNs | IL27RA |
| Injured nerve | EDN3 | MNs | EDNRA |
| Injured nerve | EDN3 | MNs | EDNRB |
| Injured nerve | EFNA1 | MNs | EPHA1 |
| Injured nerve | EFNA1 | MNs | EPHA3 |
| Injured nerve | EFNA2 | MNs | EPHA3 |
| Injured nerve | EFNA4 | MNs | EPHA3 |
| Injured nerve | EFNA4 | MNs | EPHA5 |
| Injured nerve | EFNA5 | MNs | EPHA3 |
| Injured nerve | EFNA5 | MNs | EPHA2 |
| Injured nerve | EFNA5 | MNs | EPHA7 |
| Injured nerve | EFNA5 | MNs | EPHA5 |
| Injured nerve | EFNB1 | MNs | EPHB2 |
| Injured nerve | EFNB2 | MNs | EPHB1 |
| MNs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | EFNB2 | MNs | EPHA4 |
| Injured nerve | EFNB2 | MNs | EPHA3 |
| Injured nerve | EFNB2 | MNs | EPHB4 |
| Injured nerve | EFNB2 | MNs | EPHB2 |
| Injured nerve | FGF1 | MNs | FGFR1 |
| Injured nerve | FGF1 | MNs | FGFR2 |
| Injured nerve | FGF1 | MNs | FGFR3 |
| Injured nerve | FGF1 | MNs | FGFR4 |
| Injured nerve | FGF10 | MNs | FGFR2 |
| Injured nerve | FGF18 | MNs | FGFR4 |
| Injured nerve | FGF5 | MNs | FGFR1 |
| Injured nerve | FGF5 | MNs | FGFR3 |
| Injured nerve | FGF7 | MNs | FGFR2 |
| Injured nerve | FGF7 | MNs | NRP1 |
| Injured nerve | FIGF | MNs | FLT4 |
| Injured nerve | FIGF | MNs | KDR |
| Injured nerve | FIGF | MNs | NRP1 |
| Injured nerve | FIGF | MNs | NRP2 |
| Injured nerve | FSTL1 | MNs | CD14 |
| Injured nerve | FSTL1 | MNs | DIP2A |
| Injured nerve | GAS6 | MNs | AXL |
| Injured nerve | GDF11 | MNs | ACVR1B |
| Injured nerve | GDF11 | MNs | ACVR1C |
| Injured nerve | GDF11 | MNs | ACVR2B |
| Injured nerve | GDNF | MNs | GFRA1 |
| Injured nerve | GDNF | MNs | GFRA2 |
| Injured nerve | GDNF | MNs | RET |
| Injured nerve | GNRH1 | MNs | GNRHR |
| Injured nerve | GRP | MNs | GRPR |
| Injured nerve | HBEGF | MNs | EGFR |
| Injured nerve | HBEGF | MNs | ERBB4 |
| Injured nerve | HGF | MNs | MET |
| Injured nerve | IGF1 | MNs | IGF1R |
| Injured nerve | IGF1 | MNs | IGFBP1 |
| Injured nerve | IGF1 | MNs | IGFBP2 |
| Injured nerve | IGF1 | MNs | IGFBP3 |
| Injured nerve | IGF1 | MNs | IGFBP4 |
| Injured nerve | IGF1 | MNs | IGFBP5 |
| Injured nerve | IGF1 | MNs | IGFBP6 |
| Injured nerve | IGF1 | MNs | IGFBP7 |
| Injured nerve | IGF1 | MNs | INSR |
| Injured nerve | IGF2 | MNs | IGF1R |
| Injured nerve | IGF2 | MNs | IGF2R |
| Injured nerve | IGF2 | MNs | INSR |
| Injured nerve | IL15 | MNs | IL15RA |
| Injured nerve | IL15 | MNs | IL2RB |
| Injured nerve | IL15 | MNs | IL2RG |
| Injured nerve | IL16 | MNs | CD4 |
| Injured nerve | IL16 | MNs | GRIN2A |
| Injured nerve | IL16 | MNs | GRIN2B |
| Injured nerve | IL16 | MNs | GRIN2C |
| Injured nerve | IL16 | MNs | GRIN2D |
| Injured nerve | IL18 | MNs | IL18R1 |
| Injured nerve | IL18 | MNs | IL18RAP |
| Injured nerve | IL1B | MNs | IL1R1 |
| Injured nerve | IL1B | MNs | IL1R2 |
| Injured nerve | IL1B | MNs | IL1RAP |
| Injured nerve | IL33 | MNs | IL1RL1 |
| Injured nerve | IL6 | MNs | IL6ST |
| Injured nerve | INHA | MNs | ACVR2A |
| Injured nerve | INHA | MNs | TGFBR3 |
| MNs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | INHBA | MNs | ACVR1B |
| Injured nerve | INHBA | MNs | ACVR2A |
| Injured nerve | INHBA | MNs | ACVR2B |
| Injured nerve | INHBB | MNs | ACVR1 |
| Injured nerve | INHBB | MNs | ACVR1B |
| Injured nerve | INHBB | MNs | ACVR1C |
| Injured nerve | INHBB | MNs | ACVR2A |
| Injured nerve | INHBB | MNs | ACVR2B |
| Injured nerve | JAG1 | MNs | NOTCH1 |
| Injured nerve | JAG1 | MNs | NOTCH2 |
| Injured nerve | JAG1 | MNs | NOTCH3 |
| Injured nerve | JAG2 | MNs | NOTCH1 |
| Injured nerve | JAG2 | MNs | NOTCH2 |
| Injured nerve | JAG2 | MNs | NOTCH3 |
| Injured nerve | LIF | MNs | IL6ST |
| Injured nerve | LIF | MNs | LIFR |
| Injured nerve | LTB | MNs | LTBR |
| Injured nerve | MDK | MNs | ALK |
| Injured nerve | MDK | MNs | LRP1 |
| Injured nerve | MDK | MNs | PTPRZ1 |
| Injured nerve | MIF | MNs | CD74 |
| Injured nerve | NGF | MNs | NGFR |
| Injured nerve | NGF | MNs | NTRK1 |
| Injured nerve | NGF | MNs | SORCS3 |
| Injured nerve | NGF | MNs | SORT1 |
| Injured nerve | NOV | MNs | NOTCH1 |
| Injured nerve | NPPC | MNs | NPR2 |
| Injured nerve | NPPC | MNs | NPR3 |
| Injured nerve | NTF3 | MNs | NGFR |
| Injured nerve | NTF3 | MNs | NTRK1 |
| Injured nerve | NTF3 | MNs | NTRK2 |
| Injured nerve | NTF3 | MNs | NTRK3 |
| Injured nerve | NTN1 | MNs | DCC |
| Injured nerve | NTN1 | MNs | NEO1 |
| Injured nerve | NTN1 | MNs | UNC5B |
| Injured nerve | NTN1 | MNs | UNC5C |
| Injured nerve | OSM | MNs | IL6ST |
| Injured nerve | OSM | MNs | LIFR |
| Injured nerve | OSM | MNs | OSMR |
| Injured nerve | PDGFA | MNs | PDGFRA |
| Injured nerve | PDGFB | MNs | PDGFRA |
| Injured nerve | PDGFB | MNs | PDGFRB |
| Injured nerve | PDGFC | MNs | PDGFRA |
| Injured nerve | PF4 | MNs | CXCR3 |
| Injured nerve | PF4 | MNs | LDLR |
| Injured nerve | PF4 | MNs | THBD |
| Injured nerve | PGF | MNs | FLT1 |
| Injured nerve | PGF | MNs | NRP1 |
| Injured nerve | PGF | MNs | NRP2 |
| Injured nerve | POMC | MNs | MC1R |
| Injured nerve | POMC | MNs | MC2R |
| Injured nerve | POMC | MNs | MC3R |
| Injured nerve | POMC | MNs | MC5R |
| Injured nerve | PTHLH | MNs | PTH1R |
| Injured nerve | PTN | MNs | ALK |
| Injured nerve | PTN | MNs | PTPRS |
| Injured nerve | PTN | MNs | PTPRZ1 |
| Injured nerve | RSPO1 | MNs | LGR5 |
| Injured nerve | RTN4 | MNs | LINGO1 |
| Injured nerve | RTN4 | MNs | RTN4R |
| Injured nerve | RTN4 | MNs | RTN4RL1 |
| MNs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | SEMA3B | MNs | PLXNA1 |
| Injured nerve | SEMA3B | MNs | PLXNA2 |
| Injured nerve | SEMA3B | MNs | PLXNA3 |
| Injured nerve | SEMA3B | MNs | PLXNA4 |
| Injured nerve | SEMA3B | MNs | PLXNB2 |
| Injured nerve | SEMA3B | MNs | PLXND1 |
| Injured nerve | SEMA3C | MNs | PLXNA1 |
| Injured nerve | SEMA3C | MNs | PLXNA2 |
| Injured nerve | SEMA3C | MNs | PLXNA3 |
| Injured nerve | SEMA3C | MNs | PLXNA4 |
| Injured nerve | SEMA3C | MNs | PLXNB2 |
| Injured nerve | SEMA3C | MNs | PLXND1 |
| Injured nerve | SEMA3D | MNs | PLXNA1 |
| Injured nerve | SEMA3D | MNs | PLXNA2 |
| Injured nerve | SEMA3D | MNs | PLXNA3 |
| Injured nerve | SEMA3D | MNs | PLXNA4 |
| Injured nerve | SEMA3D | MNs | PLXNB2 |
| Injured nerve | SEMA3D | MNs | PLXND1 |
| Injured nerve | SEMA3E | MNs | PLXNA1 |
| Injured nerve | SEMA3E | MNs | PLXNA2 |
| Injured nerve | SEMA3E | MNs | PLXNA3 |
| Injured nerve | SEMA3E | MNs | PLXNA4 |
| Injured nerve | SEMA3E | MNs | PLXNB2 |
| Injured nerve | SEMA3E | MNs | PLXND1 |
| Injured nerve | SEMA3F | MNs | PLXNA1 |
| Injured nerve | SEMA3F | MNs | PLXNA2 |
| Injured nerve | SEMA3F | MNs | PLXNA3 |
| Injured nerve | SEMA3F | MNs | PLXNA4 |
| Injured nerve | SEMA3F | MNs | PLXNB2 |
| Injured nerve | SEMA3F | MNs | PLXND1 |
| Injured nerve | SEMA3G | MNs | PLXNA1 |
| Injured nerve | SEMA3G | MNs | PLXNA2 |
| Injured nerve | SEMA3G | MNs | PLXNA3 |
| Injured nerve | SEMA3G | MNs | PLXNA4 |
| Injured nerve | SEMA3G | MNs | PLXNB2 |
| Injured nerve | SEMA3G | MNs | PLXND1 |
| Injured nerve | SEMA4A | MNs | PLXNB1 |
| Injured nerve | SEMA4A | MNs | PLXNB2 |
| Injured nerve | SEMA4A | MNs | PLXNC1 |
| Injured nerve | SEMA4A | MNs | PLXND1 |
| Injured nerve | SEMA4B | MNs | PLXNB1 |
| Injured nerve | SEMA4B | MNs | PLXNB2 |
| Injured nerve | SEMA4B | MNs | PLXNC1 |
| Injured nerve | SEMA4C | MNs | PLXNB1 |
| Injured nerve | SEMA4C | MNs | PLXNB2 |
| Injured nerve | SEMA4C | MNs | PLXNC1 |
| Injured nerve | SEMA4D | MNs | PLXNB1 |
| Injured nerve | SEMA4D | MNs | PLXNB2 |
| Injured nerve | SEMA4D | MNs | PLXNC1 |
| Injured nerve | SEMA4F | MNs | PLXNB1 |
| Injured nerve | SEMA4F | MNs | PLXNB2 |
| Injured nerve | SEMA4F | MNs | PLXNC1 |
| Injured nerve | SEMA5A | MNs | PLXNA3 |
| Injured nerve | SEMA5A | MNs | PLXNA4 |
| Injured nerve | SEMA5A | MNs | PLXNC1 |
| Injured nerve | SEMA5B | MNs | PLXNA3 |
| Injured nerve | SEMA5B | MNs | PLXNA4 |
| Injured nerve | SEMA5B | MNs | PLXNC1 |
| Injured nerve | SEMA6A | MNs | PLXNA1 |
| Injured nerve | SEMA6A | MNs | PLXNA2 |
| Injured nerve | SEMA6B | MNs | PLXNA1 |
| MNs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | SEMA6C | MNs | PLXNA1 |
| Injured nerve | SEMA6D | MNs | PLXNA1 |
| Injured nerve | SEMA7A | MNs | PLXNC1 |
| Injured nerve | SHH | MNs | PTCH1 |
| Injured nerve | SHH | MNs | PTCH2 |
| Injured nerve | TGFA | MNs | EGFR |
| Injured nerve | TGFA | MNs | ERBB2 |
| Injured nerve | TGFB1 | MNs | ACVRL1 |
| Injured nerve | TGFB1 | MNs | ENG |
| Injured nerve | TGFB1 | MNs | TGFBR1 |
| Injured nerve | TGFB1 | MNs | TGFBR2 |
| Injured nerve | TGFB1 | MNs | TGFBR3 |
| Injured nerve | TGFB2 | MNs | TGFBR1 |
| Injured nerve | TGFB2 | MNs | TGFBR2 |
| Injured nerve | TGFB2 | MNs | TGFBR3 |
| Injured nerve | TGFB3 | MNs | ACVRL1 |
| Injured nerve | TGFB3 | MNs | TGFBR1 |
| Injured nerve | TGFB3 | MNs | TGFBR2 |
| Injured nerve | TNF | MNs | TNFRSF1A |
| Injured nerve | TNF | MNs | TNFRSF1B |
| Injured nerve | TNFSF10 | MNs | TNFRSF10B |
| Injured nerve | TNFSF12 | MNs | TNFRSF12A |
| Injured nerve | TNFSF12 | MNs | TNFRSF25 |
| Injured nerve | TNFSF14 | MNs | LTBR |
| Injured nerve | TNFSF14 | MNs | TNFRSF14 |
| Injured nerve | TNFSF8 | MNs | TNFRSF8 |
| Injured nerve | TNFSF9 | MNs | TNFRSF9 |
| Injured nerve | TSLP | MNs | CRLF2 |
| Injured nerve | TSLP | MNs | IL7R |
| Injured nerve | UCN2 | MNs | CRHR2 |
| Injured nerve | VEGFA | MNs | FLT1 |
| Injured nerve | VEGFA | MNs | KDR |
| Injured nerve | VEGFA | MNs | NRP1 |
| Injured nerve | VEGFA | MNs | NRP2 |
| Injured nerve | VEGFB | MNs | FLT1 |
| Injured nerve | VEGFB | MNs | NRP1 |
| Injured nerve | VEGFC | MNs | FLT4 |
| Injured nerve | VEGFC | MNs | KDR |
| Injured nerve | VEGFC | MNs | NRP1 |
| Injured nerve | VEGFC | MNs | NRP2 |
| Injured nerve | WNT11 | MNs | FZD4 |
| Injured nerve | WNT2 | MNs | FZD1 |
| Injured nerve | WNT2 | MNs | FZD9 |
| Injured nerve | WNT5A | MNs | FZD2 |
| Injured nerve | WNT5A | MNs | FZD5 |
| Injured nerve | WNT5A | MNs | ROR1 |
| Injured nerve | WNT5A | MNs | ROR2 |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | ADM | RGCs | CALCRL |
| Injured nerve | ANGPT1 | RGCs | TEK |
| Injured nerve | ANGPT2 | RGCs | TEK |
| Injured nerve | ANGPT4 | RGCs | TEK |
| Injured nerve | APLN | RGCs | APLNR |
| Injured nerve | ARTN | RGCs | GFRA3 |
| Injured nerve | ARTN | RGCs | RET |
| Injured nerve | BDNF | RGCs | NGFR |
| Injured nerve | BDNF | RGCs | NTRK2 |
| Injured nerve | BDNF | RGCs | SORT1 |
| Injured nerve | BMP2 | RGCs | BMPR1A |
| Injured nerve | BMP2 | RGCs | BMPR1B |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | BMP2 | RGCs | BMPR2 |
| Injured nerve | BMP2 | RGCs | ENG |
| Injured nerve | BMP2 | RGCs | NEO1 |
| Injured nerve | BMP4 | RGCs | BMPR1A |
| Injured nerve | BMP4 | RGCs | BMPR1B |
| Injured nerve | BMP4 | RGCs | BMPR2 |
| Injured nerve | BMP4 | RGCs | NEO1 |
| Injured nerve | BMP5 | RGCs | BMPR1A |
| Injured nerve | BMP7 | RGCs | ACVR1 |
| Injured nerve | BMP7 | RGCs | ACVR2A |
| Injured nerve | BMP7 | RGCs | ACVR2B |
| Injured nerve | BMP7 | RGCs | BMPR1A |
| Injured nerve | BMP7 | RGCs | BMPR1B |
| Injured nerve | BMP7 | RGCs | BMPR2 |
| Injured nerve | BMP7 | RGCs | NEO1 |
| Injured nerve | BTC | RGCs | EGFR |
| Injured nerve | BTC | RGCs | ERBB2 |
| Injured nerve | BTC | RGCs | ERBB4 |
| Injured nerve | CCK | RGCs | CCKAR |
| Injured nerve | CCK | RGCs | CCKBR |
| Injured nerve | CCL11 | RGCs | CCR5 |
| Injured nerve | CCL11 | RGCs | CXCR3 |
| Injured nerve | CCL19 | RGCs | CCR10 |
| Injured nerve | CCL2 | RGCs | CCR1 |
| Injured nerve | CCL2 | RGCs | CCR10 |
| Injured nerve | CCL2 | RGCs | CCR2 |
| Injured nerve | CCL2 | RGCs | DARC |
| Injured nerve | CCL25 | RGCs | CCR10 |
| Injured nerve | CCL25 | RGCs | CCR9 |
| Injured nerve | CCL3 | RGCs | CCR1 |
| Injured nerve | CCL3 | RGCs | CCR4 |
| Injured nerve | CCL3 | RGCs | CCR5 |
| Injured nerve | CCL5 | RGCs | CCR1 |
| Injured nerve | CCL5 | RGCs | CCR4 |
| Injured nerve | CCL5 | RGCs | CCR5 |
| Injured nerve | CCL5 | RGCs | CXCR3 |
| Injured nerve | CCL5 | RGCs | DARC |
| Injured nerve | CCL5 | RGCs | SDC4 |
| Injured nerve | CCL7 | RGCs | CCR1 |
| Injured nerve | CCL7 | RGCs | CCR10 |
| Injured nerve | CCL7 | RGCs | CCR2 |
| Injured nerve | CCL7 | RGCs | CCR5 |
| Injured nerve | CCL7 | RGCs | CXCR3 |
| Injured nerve | CCL7 | RGCs | DARC |
| Injured nerve | CCL9 | RGCs | CCR1 |
| Injured nerve | CLCF1 | RGCs | CNTFR |
| Injured nerve | CLCF1 | RGCs | IL6ST |
| Injured nerve | CLCF1 | RGCs | LIFR |
| Injured nerve | CRLF1 | RGCs | CNTFR |
| Injured nerve | CRLF1 | RGCs | IL6ST |
| Injured nerve | CRLF1 | RGCs | LIFR |
| Injured nerve | CSF1 | RGCs | CSF1R |
| Injured nerve | CX3CL1 | RGCs | CX3CR1 |
| Injured nerve | CXCL1 | RGCs | CXCR2 |
| Injured nerve | CXCL1 | RGCs | DARC |
| Injured nerve | CXCL10 | RGCs | CXCR3 |
| Injured nerve | CXCL12 | RGCs | CCR4 |
| Injured nerve | CXCL12 | RGCs | CXCR4 |
| Injured nerve | CXCL13 | RGCs | CCR10 |
| Injured nerve | CXCL13 | RGCs | CXCR3 |
| Injured nerve | CXCL13 | RGCs | CXCR5 |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | CXCL16 | RGCs | CXCR6 |
| Injured nerve | CXCL2 | RGCs | CXCR2 |
| Injured nerve | CXCL9 | RGCs | CXCR3 |
| Injured nerve | DHH | RGCs | PTCH1 |
| Injured nerve | DHH | RGCs | PTCH2 |
| Injured nerve | DLL1 | RGCs | NOTCH1 |
| Injured nerve | DLL1 | RGCs | NOTCH2 |
| Injured nerve | DLL1 | RGCs | NOTCH3 |
| Injured nerve | DLL4 | RGCs | NOTCH1 |
| Injured nerve | EBI3 | RGCs | IL27RA |
| Injured nerve | EDA | RGCs | EDAR |
| Injured nerve | EDN3 | RGCs | EDNRA |
| Injured nerve | EDN3 | RGCs | EDNRB |
| Injured nerve | EFNA1 | RGCs | EPHA1 |
| Injured nerve | EFNA1 | RGCs | EPHA3 |
| Injured nerve | EFNA2 | RGCs | EPHA3 |
| Injured nerve | EFNA4 | RGCs | EPHA3 |
| Injured nerve | EFNA4 | RGCs | EPHA5 |
| Injured nerve | EFNA5 | RGCs | EPHA3 |
| Injured nerve | EFNA5 | RGCs | EPHA2 |
| Injured nerve | EFNA5 | RGCs | EPHA7 |
| Injured nerve | EFNA5 | RGCs | EPHA5 |
| Injured nerve | EFNB1 | RGCs | EPHB2 |
| Injured nerve | EFNB2 | RGCs | EPHB1 |
| Injured nerve | EFNB2 | RGCs | EPHA4 |
| Injured nerve | EFNB2 | RGCs | EPHA3 |
| Injured nerve | EFNB2 | RGCs | EPHB4 |
| Injured nerve | EFNB2 | RGCs | EPHB2 |
| Injured nerve | FGF1 | RGCs | FGFR1 |
| Injured nerve | FGF1 | RGCs | FGFR2 |
| Injured nerve | FGF1 | RGCs | FGFR3 |
| Injured nerve | FGF1 | RGCs | FGFR4 |
| Injured nerve | FGF10 | RGCs | FGFR2 |
| Injured nerve | FGF18 | RGCs | FGFR4 |
| Injured nerve | FGF5 | RGCs | FGFR1 |
| Injured nerve | FGF5 | RGCs | FGFR3 |
| Injured nerve | FGF7 | RGCs | FGFR2 |
| Injured nerve | FGF7 | RGCs | NRP1 |
| Injured nerve | FIGF | RGCs | FLT4 |
| Injured nerve | FIGF | RGCs | KDR |
| Injured nerve | FIGF | RGCs | NRP1 |
| Injured nerve | FIGF | RGCs | NRP2 |
| Injured nerve | FSTL1 | RGCs | CD14 |
| Injured nerve | FSTL1 | RGCs | DIP2A |
| Injured nerve | GAS6 | RGCs | AXL |
| Injured nerve | GDF11 | RGCs | ACVR1B |
| Injured nerve | GDF11 | RGCs | ACVR1C |
| Injured nerve | GDF11 | RGCs | ACVR2B |
| Injured nerve | GDNF | RGCs | GFRA1 |
| Injured nerve | GDNF | RGCs | GFRA2 |
| Injured nerve | GDNF | RGCs | RET |
| Injured nerve | HBEGF | RGCs | EGFR |
| Injured nerve | HBEGF | RGCs | ERBB4 |
| Injured nerve | HGF | RGCs | MET |
| Injured nerve | IGF1 | RGCs | IGF1R |
| Injured nerve | IGF1 | RGCs | IGFBP1 |
| Injured nerve | IGF1 | RGCs | IGFBP2 |
| Injured nerve | IGF1 | RGCs | IGFBP3 |
| Injured nerve | IGF1 | RGCs | IGFBP4 |
| Injured nerve | IGF1 | RGCs | IGFBP5 |
| Injured nerve | IGF1 | RGCs | IGFBP6 |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | IGF1 | RGCs | IGFBP7 |
| Injured nerve | IGF1 | RGCs | INSR |
| Injured nerve | IGF2 | RGCs | IGF1R |
| Injured nerve | IGF2 | RGCs | IGF2R |
| Injured nerve | IGF2 | RGCs | INSR |
| Injured nerve | IL15 | RGCs | IL15RA |
| Injured nerve | IL15 | RGCs | IL2RB |
| Injured nerve | IL15 | RGCs | IL2RG |
| Injured nerve | IL16 | RGCs | CD4 |
| Injured nerve | IL16 | RGCs | GRIN2A |
| Injured nerve | IL16 | RGCs | GRIN2B |
| Injured nerve | IL16 | RGCs | GRIN2C |
| Injured nerve | IL16 | RGCs | GRIN2D |
| Injured nerve | IL18 | RGCs | IL18RAP |
| Injured nerve | IL1B | RGCs | IL1R1 |
| Injured nerve | IL1B | RGCs | IL1RAP |
| Injured nerve | IL33 | RGCs | IL1RL1 |
| Injured nerve | IL6 | RGCs | IL6ST |
| Injured nerve | INHA | RGCs | ACVR2A |
| Injured nerve | INHA | RGCs | TGFBR3 |
| Injured nerve | INHBA | RGCs | ACVR1B |
| Injured nerve | INHBA | RGCs | ACVR2A |
| Injured nerve | INHBA | RGCs | ACVR2B |
| Injured nerve | INHBB | RGCs | ACVR1 |
| Injured nerve | INHBB | RGCs | ACVR1B |
| Injured nerve | INHBB | RGCs | ACVR1C |
| Injured nerve | INHBB | RGCs | ACVR2A |
| Injured nerve | INHBB | RGCs | ACVR2B |
| Injured nerve | JAG1 | RGCs | NOTCH1 |
| Injured nerve | JAG1 | RGCs | NOTCH2 |
| Injured nerve | JAG1 | RGCs | NOTCH3 |
| Injured nerve | JAG2 | RGCs | NOTCH1 |
| Injured nerve | JAG2 | RGCs | NOTCH2 |
| Injured nerve | JAG2 | RGCs | NOTCH3 |
| Injured nerve | LIF | RGCs | IL6ST |
| Injured nerve | LIF | RGCs | LIFR |
| Injured nerve | LTB | RGCs | LTBR |
| Injured nerve | MDK | RGCs | ALK |
| Injured nerve | MDK | RGCs | LRP1 |
| Injured nerve | MDK | RGCs | LRP2 |
| Injured nerve | MDK | RGCs | PTPRZ1 |
| Injured nerve | MIF | RGCs | CD74 |
| Injured nerve | NGF | RGCs | NGFR |
| Injured nerve | NGF | RGCs | NTRK1 |
| Injured nerve | NGF | RGCs | SORCS3 |
| Injured nerve | NGF | RGCs | SORT1 |
| Injured nerve | NOV | RGCs | NOTCH1 |
| Injured nerve | NPPC | RGCs | NPR2 |
| Injured nerve | NPPC | RGCs | NPR3 |
| Injured nerve | NTF3 | RGCs | NGFR |
| Injured nerve | NTF3 | RGCs | NTRK1 |
| Injured nerve | NTF3 | RGCs | NTRK2 |
| Injured nerve | NTF3 | RGCs | NTRK3 |
| Injured nerve | NTN1 | RGCs | DCC |
| Injured nerve | NTN1 | RGCs | NEO1 |
| Injured nerve | NTN1 | RGCs | UNC5B |
| Injured nerve | NTN1 | RGCs | UNC5C |
| Injured nerve | OSM | RGCs | IL6ST |
| Injured nerve | OSM | RGCs | LIFR |
| Injured nerve | OSM | RGCs | OSMR |
| Injured nerve | PDGFA | RGCs | PDGFRA |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | PDGFB | RGCs | PDGFRA |
| Injured nerve | PDGFB | RGCs | PDGFRB |
| Injured nerve | PDGFC | RGCs | PDGFRA |
| Injured nerve | PF4 | RGCs | CXCR3 |
| Injured nerve | PF4 | RGCs | DARC |
| Injured nerve | PF4 | RGCs | LDLR |
| Injured nerve | PF4 | RGCs | THBD |
| Injured nerve | PGF | RGCs | FLT1 |
| Injured nerve | PGF | RGCs | NRP1 |
| Injured nerve | PGF | RGCs | NRP2 |
| Injured nerve | POMC | RGCs | MC1R |
| Injured nerve | POMC | RGCs | MC3R |
| Injured nerve | POMC | RGCs | MC4R |
| Injured nerve | POMC | RGCs | MC5R |
| Injured nerve | PTHLH | RGCs | PTH1R |
| Injured nerve | PTN | RGCs | ALK |
| Injured nerve | PTN | RGCs | PTPRS |
| Injured nerve | PTN | RGCs | PTPRZ1 |
| Injured nerve | RSPO1 | RGCs | LGR5 |
| Injured nerve | RTN4 | RGCs | LINGO1 |
| Injured nerve | RTN4 | RGCs | RTN4R |
| Injured nerve | RTN4 | RGCs | RTN4RL1 |
| Injured nerve | SEMA3B | RGCs | PLXNA1 |
| Injured nerve | SEMA3B | RGCs | PLXNA2 |
| Injured nerve | SEMA3B | RGCs | PLXNA3 |
| Injured nerve | SEMA3B | RGCs | PLXNA4 |
| Injured nerve | SEMA3B | RGCs | PLXNB2 |
| Injured nerve | SEMA3B | RGCs | PLXND1 |
| Injured nerve | SEMA3C | RGCs | PLXNA1 |
| Injured nerve | SEMA3C | RGCs | PLXNA2 |
| Injured nerve | SEMA3C | RGCs | PLXNA3 |
| Injured nerve | SEMA3C | RGCs | PLXNA4 |
| Injured nerve | SEMA3C | RGCs | PLXNB2 |
| Injured nerve | SEMA3C | RGCs | PLXND1 |
| Injured nerve | SEMA3D | RGCs | PLXNA1 |
| Injured nerve | SEMA3D | RGCs | PLXNA2 |
| Injured nerve | SEMA3D | RGCs | PLXNA3 |
| Injured nerve | SEMA3D | RGCs | PLXNA4 |
| Injured nerve | SEMA3D | RGCs | PLXNB2 |
| Injured nerve | SEMA3D | RGCs | PLXND1 |
| Injured nerve | SEMA3E | RGCs | PLXNA1 |
| Injured nerve | SEMA3E | RGCs | PLXNA2 |
| Injured nerve | SEMA3E | RGCs | PLXNA3 |
| Injured nerve | SEMA3E | RGCs | PLXNA4 |
| Injured nerve | SEMA3E | RGCs | PLXNB2 |
| Injured nerve | SEMA3E | RGCs | PLXND1 |
| Injured nerve | SEMA3F | RGCs | PLXNA1 |
| Injured nerve | SEMA3F | RGCs | PLXNA2 |
| Injured nerve | SEMA3F | RGCs | PLXNA3 |
| Injured nerve | SEMA3F | RGCs | PLXNA4 |
| Injured nerve | SEMA3F | RGCs | PLXNB2 |
| Injured nerve | SEMA3F | RGCs | PLXND1 |
| Injured nerve | SEMA3G | RGCs | PLXNA1 |
| Injured nerve | SEMA3G | RGCs | PLXNA2 |
| Injured nerve | SEMA3G | RGCs | PLXNA3 |
| Injured nerve | SEMA3G | RGCs | PLXNA4 |
| Injured nerve | SEMA3G | RGCs | PLXNB2 |
| Injured nerve | SEMA3G | RGCs | PLXND1 |
| Injured nerve | SEMA4A | RGCs | PLXNB1 |
| Injured nerve | SEMA4A | RGCs | PLXNB2 |
| Injured nerve | SEMA4A | RGCs | PLXNC1 |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | SEMA4A | RGCs | PLXND1 |
| Injured nerve | SEMA4B | RGCs | PLXNB1 |
| Injured nerve | SEMA4B | RGCs | PLXNB2 |
| Injured nerve | SEMA4B | RGCs | PLXNC1 |
| Injured nerve | SEMA4C | RGCs | PLXNB1 |
| Injured nerve | SEMA4C | RGCs | PLXNB2 |
| Injured nerve | SEMA4C | RGCs | PLXNC1 |
| Injured nerve | SEMA4D | RGCs | PLXNB1 |
| Injured nerve | SEMA4D | RGCs | PLXNB2 |
| Injured nerve | SEMA4D | RGCs | PLXNC1 |
| Injured nerve | SEMA4F | RGCs | PLXNB1 |
| Injured nerve | SEMA4F | RGCs | PLXNB2 |
| Injured nerve | SEMA4F | RGCs | PLXNC1 |
| Injured nerve | SEMA5A | RGCs | PLXNA3 |
| Injured nerve | SEMA5A | RGCs | PLXNA4 |
| Injured nerve | SEMA5A | RGCs | PLXNC1 |
| Injured nerve | SEMA5B | RGCs | PLXNA3 |
| Injured nerve | SEMA5B | RGCs | PLXNA4 |
| Injured nerve | SEMA5B | RGCs | PLXNC1 |
| Injured nerve | SEMA6A | RGCs | PLXNA1 |
| Injured nerve | SEMA6A | RGCs | PLXNA2 |
| Injured nerve | SEMA6B | RGCs | PLXNA1 |
| Injured nerve | SEMA6C | RGCs | PLXNA1 |
| Injured nerve | SEMA6D | RGCs | PLXNA1 |
| Injured nerve | SEMA7A | RGCs | PLXNC1 |
| Injured nerve | SHH | RGCs | PTCH1 |
| Injured nerve | SHH | RGCs | PTCH2 |
| Injured nerve | TGFA | RGCs | EGFR |
| Injured nerve | TGFA | RGCs | ERBB2 |
| Injured nerve | TGFB1 | RGCs | ACVRL1 |
| Injured nerve | TGFB1 | RGCs | ENG |
| Injured nerve | TGFB1 | RGCs | TGFBR1 |
| Injured nerve | TGFB1 | RGCs | TGFBR2 |
| Injured nerve | TGFB1 | RGCs | TGFBR3 |
| Injured nerve | TGFB2 | RGCs | TGFBR1 |
| Injured nerve | TGFB2 | RGCs | TGFBR2 |
| Injured nerve | TGFB2 | RGCs | TGFBR3 |
| Injured nerve | TGFB3 | RGCs | ACVRL1 |
| Injured nerve | TGFB3 | RGCs | TGFBR1 |
| Injured nerve | TGFB3 | RGCs | TGFBR2 |
| Injured nerve | TNF | RGCs | TNFRSF1A |
| Injured nerve | TNF | RGCs | TNFRSF1B |
| Injured nerve | TNFSF10 | RGCs | TNFRSF10B |
| Injured nerve | TNFSF12 | RGCs | TNFRSF12A |
| Injured nerve | TNFSF12 | RGCs | TNFRSF25 |
| Injured nerve | TNFSF14 | RGCs | LTBR |
| Injured nerve | TNFSF14 | RGCs | TNFRSF14 |
| Injured nerve | TNFSF8 | RGCs | TNFRSF8 |
| Injured nerve | TNFSF9 | RGCs | TNFRSF9 |
| Injured nerve | TSLP | RGCs | CRLF2 |
| Injured nerve | TSLP | RGCs | IL7R |
| Injured nerve | UCN2 | RGCs | CRHR2 |
| Injured nerve | VEGFA | RGCs | FLT1 |
| Injured nerve | VEGFA | RGCs | KDR |
| Injured nerve | VEGFA | RGCs | NRP1 |
| Injured nerve | VEGFA | RGCs | NRP2 |
| Injured nerve | VEGFB | RGCs | FLT1 |
| Injured nerve | VEGFB | RGCs | NRP1 |
| Injured nerve | VEGFC | RGCs | FLT4 |
| Injured nerve | VEGFC | RGCs | KDR |
| Injured nerve | VEGFC | RGCs | NRP1 |
| RGCs | |||
| Source cell | Ligand | Target cell | Receptor |
| Injured nerve | VEGFC | RGCs | NRP2 |
| Injured nerve | WNT11 | RGCs | FZD4 |
| Injured nerve | WNT2 | RGCs | FZD1 |
| Injured nerve | WNT2 | RGCs | FZD9 |
| Injured nerve | WNT5A | RGCs | FZD2 |
| Injured nerve | WNT5A | RGCs | FZD5 |
| Injured nerve | WNT5A | RGCs | ROR1 |
| Injured nerve | WNT5A | RGCs | ROR2 |
Predicted unidirectional ligand-receptor interactions between the injured sciatic nerve ligands and receptors on sympathetic (SCGs) neurons, sensory (DRGs) neurons, motor neurons (MNs), and retinal ganglion cells (RGCs). Directionality of predicted paracrine interactions are indicated by the source cell (ligand) and target cell (receptor) column designations. Some ligands are predicted to act on more than one receptor. The interactions shown here accompany the models presented in Figures 5-7 and Extended Data Figure 6-1.
Predicted unidirectional ligand-receptor interactions between injured sciatic nerve Schwann cells or endoneurial mesenchymal cells and sensory neurons. Models showing predicted unidirectional interactions between the ligands most highly expressed by injured nerve Schwann cells (A) or endoneurial mesenchymal cells (B) and their receptors on cultured sensory neurons (DRGs). Ligands are shown in the central columns in A, B and are color coded as in Figure 5 (Schwann cell ligands in grey and endoneurial mesenchymal cell ligands in yellow). Receptors are shown on either side of the ligand column and also include coreceptors that are well-characterized components of receptor complexes. Receptors that were observed at both the transcriptomic and proteomic levels are colored green while those defined only at the transcriptomic level are colored blue. Arrows indicate directionality of interactions. Note that many ligands interact with multiple receptors and, conversely, that multiple ligands are sometimes predicted to share receptors. Download Figure 6-1, TIF file (1.4MB, tif) .
We then used the scRNA-seq data to define the nerve cell types that made the ligands involved in the predicted interactions (Figs. 5C,D, 6A,B; Extended Data Fig. 6-1A,B). Almost half (57) of 122 shared predicted sympathetic and sensory neuron interactions involved ligands that were expressed in the highest proportions in Pdgfra-positive mesenchymal cells including FGF10, FGF18, HGF, SEMA3D, BMP7, IL33, and PTHLH (asterisks in the models denote ligands made by 4-fold more of the indicated cell relative to all other cell types). Moreover, 33 of these were highest in the endoneurial mesenchymal cells. Another 22 ligands were highest in Schwann cells, including ARTN, BTC, DHH, FGF5, GDNF, SEMA3B, SHH, and UCN2. The remaining 43 predicted interactions were split almost equally between endothelial cells (17), VSM/pericyte cells (14), and immune cells (12), and included well-characterized nerve ligands such as NGF, NT3 (Ntf3), and IGF2. Notably, while some of these ligands were predicted to signal via dedicated receptors (Fig. 6A,B; Extended Data Fig. 6-1A,B; Table 9), many others shared receptors or coreceptors, suggesting the potential for convergent signaling. As examples, CLCF1, CRLF1, and LIF were all predicted to share the receptor components LIFR and gp130, and ANGPT1, ANGPT2, and ANGPT4 were all predicted to act by interacting with TEK.
Figure 6.
Predicted unidirectional ligand-receptor interactions between injured sciatic nerve Schwann cells or endoneurial mesenchymal cells and sympathetic neurons (see also Extended Data Fig. 6-1). Models showing predicted unidirectional interactions between the ligands most highly expressed by injured nerve Schwann cells (A) or endoneurial mesenchymal cells (B) and their receptors on cultured sympathetic neurons (SCGs). Ligands are shown in the central columns in A, B and are color coded as in Figure 5 (Schwann cell ligands in gray and endoneurial mesenchymal cell ligands in yellow). Receptors are shown on either side of the ligand column and also include coreceptors that are well-characterized components of receptor complexes. Receptors that were observed at both the transcriptomic and proteomic levels are colored green while those defined only at the transcriptomic level are colored blue. Arrows indicate directionality of interactions. Note that many ligands interact with multiple receptors and, conversely, that multiple ligands are sometimes predicted to share receptors.
Modeling predicts that many nerve ligands have the capacity to act on both PNS and CNS neurons
This analysis predicts that peripheral nerve cells produce ligands that could act on at least two populations of peripheral neurons. To test the idea that this might reflect a ligand environment that is generally supportive of axonal growth, we asked about motor neurons, which also project axons via the sciatic nerve, and RGCs, CNS neurons that normally do not regenerate in the CNS, but will regenerate into peripheral nerve grafts (Politis and Spencer, 1986; for review, see Benowitz et al., 2017).
For motor neurons, we analyzed previously published microarray data from microdissected P7 mouse lumbar motor neurons (Kaplan et al., 2014), identifying receptor mRNAs using the same thresholding cutoff as for the sensory neurons (top 87% of mRNAs). Of 322 receptor mRNAs defined in the motor neuron dataset using this approach, 272 were also expressed by sensory and sympathetic neurons (Table 10). Computational modeling with the motor neuron receptors and the 143 injured nerve ligands showed that of the 122 shared sympathetic and sensory neuron interactions, 121 were also predicted for motor neurons, with the endoneurial ligand EDA the only exception (Fig. 7A,C; Table 9). There were also two predicted nerve to motor neuron interactions involving GRP-GRPR and GNRH1-GNRHR that were not shared with both sympathetic and sensory neurons.
Table 10.
Receptors identified on motor neurons and RGCs using microarrays and RNA-seq
| MNs (322) | DRGs. SCGs,and MNs (272) | RGCs (320) | DRGs, SCGs,MNs, andRGCs (258) |
|---|---|---|---|
| Acvr1 | Acvr1 | Acvr1 | Acvr1 |
| Acvr1b | Acvr1b | Acvr1b | Acvr1b |
| Acvr1c | Acvr2a | Acvr1c | Acvr2a |
| Acvr2a | Acvr2b | Acvr2a | Acvr2b |
| Acvr2b | Acvrl1 | Acvr2b | Acvrl1 |
| Acvrl1 | Adcyap1r1 | Acvrl1 | Adcyap1r1 |
| Adcyap1r1 | Adipor1 | Adcyap1r1 | Adipor1 |
| Adipor1 | Adipor2 | Adipor1 | Adipor2 |
| Adipor2 | Adora2a | Adipor2 | Adora2a |
| Adora2a | Adra1b | Adora2a | Adra1b |
| Adra1a | Adrb2 | Adra1a | Adrb2 |
| Adra1b | Ager | Adra1b | Ager |
| Adrb2 | Alk | Adrb2 | Alk |
| Ager | Amfr | Ager | Amfr |
| Alk | Aplnr | Alk | Aplnr |
| Amfr | Avpr1a | Amfr | Avpr1a |
| Amhr2 | Axl | Amhr2 | Axl |
| Aplnr | Bdkrb2 | Aplnr | Bdkrb2 |
| Ar | Bmpr1a | Ar | Bmpr1a |
| Avpr1a | Bmpr1b | Avpr1a | Bmpr1b |
| Avpr1b | Bmpr2 | Avpr2 | Bmpr2 |
| Avpr2 | C3ar1 | Axl | C3ar1 |
| Axl | C5ar1 | Bdkrb2 | C5ar1 |
| Bdkrb2 | Calcrl | Bmpr1a | Calcrl |
| Bmpr1a | Cckar | Bmpr1b | Cckar |
| Bmpr1b | Cckbr | Bmpr2 | Cckbr |
| Bmpr2 | Ccr10 | Btn1a1 | Ccr10 |
| Btn1a1 | Cd14 | C3ar1 | Cd14 |
| C3ar1 | Cd4 | C5ar1 | Cd4 |
| C5ar1 | Cd44 | Calcr | Cd44 |
| Calcr | Cd5l | Calcrl | Cd5l |
| Calcrl | Cd7 | Cckar | Cd74 |
| Cckar | Cd74 | Cckbr | Cntfr |
| Cckbr | Cntfr | Ccr1 | Crhr1 |
| Ccr1 | Crhr1 | Ccr10 | Crhr2 |
| Ccr10 | Crhr2 | Ccr2 | Crlf1 |
| Ccr2 | Crlf1 | Ccr4 | Crlf2 |
| Ccr3 | Crlf2 | Ccr5 | Csf1r |
| Ccr4 | Csf1r | Ccr9 | Csf2ra |
| Ccr5 | Csf2ra | Cd14 | Csf2rb |
| Ccr6 | Csf2rb | Cd27 | Csf3r |
| Ccr7 | Csf3r | Cd33 | Ctf1 |
| Ccr8 | Ctf1 | Cd4 | Cx3cr1 |
| Ccr9 | Cx3cr1 | Cd40 | Cxcr3 |
| Cd14 | Cxcr3 | Cd44 | Cxcr4 |
| Cd27 | Cxcr4 | Cd5l | Dcc |
| Cd33 | Dcc | Cd74 | Ddr1 |
| Cd4 | Ddr1 | Cntfr | Derl1 |
| Cd40 | Derl1 | Cr2 | Dip2a |
| Cd44 | Dip2a | Crhr1 | Ednra |
| Cd5l | Ednra | Crhr2 | Ednrb |
| Cd7 | Ednrb | Crlf1 | Egfr |
| Cd74 | Egfr | Crlf2 | Eng |
| Cntfr | Eng | Csf1r | Epha1 |
| Cr2 | Epha1 | Csf2ra | Epha2 |
| Crhr1 | Epha2 | Csf2rb | Epha3 |
| Crhr2 | Epha3 | Csf3r | Epha4 |
| Crlf1 | Epha4 | Ctf1 | Epha5 |
| Crlf2 | Epha5 | Cx3cr1 | Epha7 |
| Csf1r | Epha7 | Cxcr2 | Ephb1 |
| Csf2ra | Ephb1 | Cxcr3 | Ephb2 |
| Csf2rb | Ephb2 | Cxcr4 | Ephb3 |
| Csf3r | Ephb3 | Cxcr5 | Ephb4 |
| Ctf1 | Ephb4 | Cxcr6 | Epor |
| Cx3cr1 | Epor | Darc | Eps15l1 |
| Cxcr2 | Eps15l1 | Dcc | Erbb2 |
| Cxcr3 | Erbb2 | Ddr1 | Erbb3 |
| Cxcr4 | Erbb3 | Derl1 | Esr2 |
| Cxcr5 | Esr2 | Dip2a | F2r |
| Cxcr6 | F2r | Dpp4 | Fgfr1 |
| Dcc | Fas | Edar | Fgfr2 |
| Ddr1 | Fgfr1 | Ednra | Fgfr3 |
| Derl1 | Fgfr2 | Ednrb | Fgfr4 |
| Dip2a | Fgfr3 | Egfr | Fgfrl1 |
| Dpp4 | Fgfr4 | Eng | Flt1 |
| Edar | Fgfrl1 | Epha1 | Flt3 |
| Ednra | Flt1 | Epha2 | Flt4 |
| Ednrb | Flt3 | Epha3 | Folr1 |
| Egfr | Flt4 | Epha4 | Fzd1 |
| Eng | Folr1 | Epha5 | Fzd2 |
| Epha1 | Fshr | Epha7 | Fzd4 |
| Epha2 | Fzd1 | Ephb1 | Fzd5 |
| Epha3 | Fzd2 | Ephb2 | Fzd9 |
| Epha5 | Fzd4 | Ephb3 | Gabbr1 |
| Ephb1 | Fzd5 | Ephb4 | Galr2 |
| Ephb2 | Fzd9 | Epor | Gcgr |
| Ephb3 | Gabbr1 | Eps15l1 | Gfra2 |
| Epor | Galr2 | Erbb2 | Gfra3 |
| Eps15l1 | Gcgr | Erbb3 | Gfra4 |
| Erbb2 | Gfra2 | Erbb4 | Ghr |
| Erbb3 | Gfra3 | Esr1 | Gipr |
| Erbb4 | Gfra4 | Esr2 | Gosr1 |
| Esr1 | Ghr | F2r | Grik5 |
| Esr2 | Ghrhr | F2rl1 | Grin2a |
| F2r | Gipr | F2rl3 | Grin2b |
| F2rl1 | Glp1r | Fgfr1 | Grin2c |
| F2rl2 | Gosr1 | Fgfr2 | Grin2d |
| F2rl3 | Grik5 | Fgfr3 | Gucy2c |
| Fas | Grin2a | Fgfr4 | Hcrtr1 |
| Fgfr1 | Grin2b | Fgfrl1 | Hcrtr2 |
| Fgfr2 | Grin2c | Flt1 | Hnf4a |
| Fgfr3 | Grin2d | Flt3 | Hpn |
| Fgfr4 | Gucy2c | Flt4 | Ifnar1 |
| Fgfrl1 | Hcrtr1 | Folr1 | Ifnar2 |
| Flt1 | Hcrtr2 | Fzd1 | Ifngr1 |
| Flt3 | Hnf4a | Fzd2 | Ifngr2 |
| Flt4 | Hpn | Fzd4 | Igf1r |
| Folr1 | Ifnar1 | Fzd5 | Igf2r |
| Fshr | Ifnar2 | Fzd8 | Igfbp1 |
| Fzd1 | Ifngr1 | Fzd9 | Igfbp2 |
| Fzd2 | Ifngr2 | Gabbr1 | Igfbp3 |
| Fzd4 | Igf1r | Galr1 | Igfbp4 |
| Fzd5 | Igf2r | Galr2 | Igfbp5 |
| Fzd8 | Igfbp1 | Gcgr | Igfbp6 |
| Fzd9 | Igfbp2 | Gfra1 | Igfbp7 |
| Gabbr1 | Igfbp3 | Gfra2 | Il10ra |
| Galr1 | Igfbp4 | Gfra3 | Il10rb |
| Galr2 | Igfbp5 | Gfra4 | Il12rb1 |
| Gcgr | Igfbp6 | Ghr | Il15ra |
| Gfra1 | Igfbp7 | Ghsr | Il17ra |
| Gfra2 | Il10ra | Gipr | Il17rc |
| Gfra3 | Il10rb | Glp2r | Il18rap |
| Gfra4 | Il12rb1 | Gosr1 | Il1r1 |
| Ghr | Il15ra | Gpr151 | Il1rap |
| Ghrhr | Il17ra | Grik5 | Il1rl1 |
| Ghsr | Il17rc | Grin2a | Il1rl2 |
| Gipr | Il18r1 | Grin2b | Il21r |
| Glp1r | Il18rap | Grin2c | Il27ra |
| Gnrhr | Il1r1 | Grin2d | Il2rb |
| Gosr1 | Il1r2 | Gucy2c | Il2rg |
| Gpr151 | Il1rap | Hcrtr1 | Il3ra |
| Grik5 | Il1rl1 | Hcrtr2 | Il6st |
| Grin2a | Il1rl2 | Hnf4a | Il7r |
| Grin2b | Il21r | Hpn | Insr |
| Grin2c | Il22ra1 | Ifnar1 | Irs1 |
| Grin2d | Il27ra | Ifnar2 | Itga2 |
| Grpr | Il2ra | Ifngr1 | Itga2b |
| Gucy2c | Il2rb | Ifngr2 | Itga5 |
| Hcrtr1 | Il2rg | Igf1r | Itga9 |
| Hcrtr2 | Il3ra | Igf2r | Itgal |
| Hnf4a | Il6st | Igfbp1 | Itgav |
| Hpn | Il7r | Igfbp2 | Itgb1 |
| Hrh4 | Insr | Igfbp3 | Itgb3 |
| Ifnar1 | Irs1 | Igfbp4 | Itgb5 |
| Ifnar2 | Itga2 | Igfbp5 | Itgb6 |
| Ifngr1 | Itga2b | Igfbp6 | Itgb8 |
| Ifngr2 | Itga5 | Igfbp7 | Itpr3 |
| Igf1r | Itga9 | Il10ra | Kit |
| Igf2r | Itgal | Il10rb | Ldlr |
| Igfbp1 | Itgav | Il12rb1 | Lgals3bp |
| Igfbp2 | Itgb1 | Il12rb2 | Lgr5 |
| Igfbp3 | Itgb3 | Il13ra1 | Lifr |
| Igfbp4 | Itgb5 | Il15ra | Lingo1 |
| Igfbp5 | Itgb6 | Il17ra | Lrp1 |
| Igfbp6 | Itgb8 | Il17rb | Lrp5 |
| Igfbp7 | Itpr3 | Il17rc | Lrp6 |
| Il10ra | Kit | Il18rap | Lsr |
| Il10rb | Ldlr | Il1r1 | Ltbr |
| Il12rb1 | Lgals3bp | Il1rap | Mc1r |
| Il12rb2 | Lgr5 | Il1rl1 | Mc3r |
| Il13ra1 | Lifr | Il1rl2 | Mc5r |
| Il13ra2 | Lingo1 | Il20ra | Mchr1 |
| Il15ra | Lrp1 | Il20rb | Met |
| Il17ra | Lrp5 | Il21r | Mst1r |
| Il17rb | Lrp6 | Il27ra | Ncoa3 |
| Il17rc | Lsr | Il2rb | Ncor1 |
| Il18r1 | Ltbr | Il2rg | Neo1 |
| Il18rap | Marco | Il3ra | Ngfr |
| Il1r1 | Mc1r | Il6st | Notch1 |
| Il1r2 | Mc2r | Il7r | Notch2 |
| Il1rap | Mc3r | Il9r | Notch3 |
| Il1rl1 | Mc5r | Insr | Npr1 |
| Il1rl2 | Mchr1 | Irs1 | Npr2 |
| Il20ra | Met | Itga2 | Npr3 |
| Il20rb | Mpl | Itga2b | Npy1r |
| Il21r | Mst1r | Itga5 | Npy5r |
| Il22ra1 | Ncoa3 | Itga9 | Nr3c1 |
| Il22ra2 | Ncor1 | Itgal | Nrp1 |
| Il27ra | Neo1 | Itgav | Nrp2 |
| Il2ra | Ngfr | Itgb1 | Ntng1 |
| Il2rb | Notch1 | Itgb2 | Ntng2 |
| Il2rg | Notch2 | Itgb3 | Ntrk1 |
| Il3ra | Notch3 | Itgb5 | Ntrk2 |
| Il5ra | Npffr2 | Itgb6 | Ntrk3 |
| Il6st | Npr1 | Itgb8 | Ntsr1 |
| Il7r | Npr2 | Itpr3 | Oprl1 |
| Il9r | Npr3 | Kdr | Osmr |
| Insr | Npy1r | Kit | Oxtr |
| Irs1 | Npy2r | Ldlr | Pdgfa |
| Itga2 | Npy5r | Lepr | Pdgfra |
| Itga2b | Nr3c1 | Lgals3bp | Pdgfrb |
| Itga5 | Nrp1 | Lgr5 | Pgr |
| Itga9 | Nrp2 | Lhcgr | Plaur |
| Itgal | Ntng1 | Lifr | Plgrkt |
| Itgav | Ntng2 | Lingo1 | Plxna1 |
| Itgb1 | Ntrk1 | Loxl2 | Plxna2 |
| Itgb2 | Ntrk2 | Lrp1 | Plxna3 |
| Itgb3 | Ntrk3 | Lrp2 | Plxna4 |
| Itgb5 | Ntsr1 | Lrp5 | Plxnb1 |
| Itgb6 | Oprl1 | Lrp6 | Plxnc1 |
| Itgb8 | Osmr | Lsr | Plxnd1 |
| Itpr3 | Oxtr | Ltbr | Procr |
| Kdr | Pdgfa | Mc1r | Prokr1 |
| Kit | Pdgfra | Mc3r | Prokr2 |
| Ldlr | Pdgfrb | Mc4r | Ptch1 |
| Lepr | Pgr | Mc5r | Ptch2 |
| Lgals3bp | Plaur | Mchr1 | Pth1r |
| Lgr5 | Plgrkt | Met | Ptprk |
| Lhcgr | Plxna1 | Mst1r | Ptprs |
| Lifr | Plxna2 | Ncoa3 | Ptprz1 |
| Lrp1 | Plxna3 | Ncor1 | Ret |
| Lrp2 | Plxna4 | Neo1 | Robo3 |
| Lrp5 | Plxnb1 | Ngfr | Ror1 |
| Lrp6 | Plxnc1 | Nmbr | Ror2 |
| Lsr | Plxnd1 | Nmur2 | Rorb |
| Ltbr | Procr | Notch1 | Rtn4r |
| Marco | Prokr1 | Notch2 | Rtn4rl1 |
| Mc1r | Prokr2 | Notch3 | Rxrg |
| Mc2r | Ptch1 | Npffr1 | Ryr1 |
| Mc3r | Ptch2 | Npr1 | Ryr2 |
| Mc5r | Pth1r | Npr2 | Sctr |
| Mchr1 | Ptprk | Npr3 | Sdc4 |
| Met | Ptprs | Npy1r | Sfrp1 |
| Mpl | Ptprz1 | Npy5r | Sfrp2 |
| Ncoa3 | Ret | Nr3c1 | Slc1a5 |
| Ncor1 | Robo3 | Nrp1 | Sorcs3 |
| Ngfr | Ror1 | Nrp2 | Sort1 |
| Nmbr | Ror2 | Ntng1 | Sstr1 |
| Nmur2 | Rorb | Ntng2 | Sstr2 |
| Notch1 | Rtn4r | Ntrk1 | Sstr3 |
| Notch2 | Rtn4rl1 | Ntrk2 | Sstr4 |
| Notch3 | Rxrg | Ntrk3 | Sstr5 |
| Npffr2 | Ryr1 | Ntsr1 | Tek |
| Npr1 | Ryr2 | Oprl1 | Tgfbr2 |
| Npr2 | Sctr | Osmr | Tgfbr3 |
| Npr3 | Sdc4 | Oxtr | Thbd |
| Npy1r | Sfrp1 | Pdgfa | Thra |
| Npy2r | Sfrp2 | Pdgfra | Thrap3 |
| Npy5r | Slc1a5 | Pdgfrb | Tnfrsf10b |
| Nr3c1 | Sorcs3 | Pgr | Tnfrsf11a |
| Nrp1 | Sort1 | Plat | Tnfrsf11b |
| Nrp2 | Sstr1 | Plaur | Tnfrsf12a |
| Ntrk1 | Sstr2 | Plgrkt | Tnfrsf13c |
| Ntrk2 | Sstr3 | Plxna1 | Tnfrsf14 |
| Ntrk3 | Sstr4 | Plxna2 | Tnfrsf17 |
| Ntsr1 | Sstr5 | Plxna3 | Tnfrsf18 |
| Oprl1 | Tek | Plxna4 | Tnfrsf1a |
| Osmr | Tgfbr2 | Plxnb1 | Tnfrsf1b |
| Oxtr | Tgfbr3 | Plxnb2 | Tnfrsf25 |
| Pdgfa | Thbd | Plxnc1 | Tnfrsf8 |
| Pdgfra | Thra | Plxnd1 | Tnfrsf9 |
| Pdgfrb | Thrap3 | Prlhr | Tshr |
| Pgr | Tnfrsf10b | Prlr | Unc5b |
| Plat | Tnfrsf11a | Procr | Unc5c |
| Plaur | Tnfrsf11b | Prokr1 | Uts2r |
| Plgrkt | Tnfrsf12a | Prokr2 | Vipr1 |
| Plxnb2 | Tnfrsf13c | Ptch1 | Vldlr |
| Prlr | Tnfrsf14 | Ptch2 | Vtn |
| Procr | Tnfrsf17 | Pth1r | |
| Prokr1 | Tnfrsf18 | Pth2r | |
| Prokr2 | Tnfrsf1a | Ptprh | |
| Ptch1 | Tnfrsf1b | Ptprk | |
| Ptch2 | Tnfrsf25 | Ptprs | |
| Pth1r | Tnfrsf8 | Ptprz1 | |
| Pth2r | Tnfrsf9 | Ret | |
| Ptprk | Tshr | Robo3 | |
| Ptprs | Unc5b | Ror1 | |
| Ptprz1 | Unc5c | Ror2 | |
| Ret | Uts2r | Rorb | |
| Robo3 | Vipr1 | Rtn4r | |
| Ror1 | Vldlr | Rtn4rl1 | |
| Ror2 | Vtn | Rxfp2 | |
| Rorb | Rxfp4 | ||
| Rtn4r | Rxrg | ||
| Rxfp1 | Ryr1 | ||
| Rxfp2 | Ryr2 | ||
| Rxrg | Sctr | ||
| Ryr1 | Sdc4 | ||
| Ryr2 | Sfrp1 | ||
| Sctr | Sfrp2 | ||
| Sdc4 | Slc1a5 | ||
| Sfrp1 | Sorcs3 | ||
| Sfrp2 | Sort1 | ||
| Slc1a5 | Sstr1 | ||
| Sorcs3 | Sstr2 | ||
| Sort1 | Sstr3 | ||
| Sstr1 | Sstr4 | ||
| Sstr2 | Sstr5 | ||
| Sstr3 | Tek | ||
| Sstr4 | Tgfbr1 | ||
| Sstr5 | Tgfbr2 | ||
| Tek | Tgfbr3 | ||
| Tgfbr1 | Thbd | ||
| Tgfbr2 | Thra | ||
| Tgfbr3 | Thrap3 | ||
| Thbd | Tnfrsf10b | ||
| Thra | Tnfrsf11a | ||
| Thrap3 | Tnfrsf11b | ||
| Tnfrsf10b | Tnfrsf12a | ||
| Tnfrsf11a | Tnfrsf13b | ||
| Tnfrsf11b | Tnfrsf13c | ||
| Tnfrsf12a | Tnfrsf14 | ||
| Tnfrsf13b | Tnfrsf17 | ||
| Tnfrsf13c | Tnfrsf18 | ||
| Tnfrsf14 | Tnfrsf1a | ||
| Tnfrsf17 | Tnfrsf1b | ||
| Tnfrsf18 | Tnfrsf25 | ||
| Tnfrsf1a | Tnfrsf4 | ||
| Tnfrsf1b | Tnfrsf8 | ||
| Tnfrsf25 | Tnfrsf9 | ||
| Tnfrsf4 | Trhr | ||
| Tnfrsf8 | Tshr | ||
| Tnfrsf9 | Unc5b | ||
| Trhr | Unc5c | ||
| Tshr | Uts2r | ||
| Unc5c | Vipr1 | ||
| Uts2r | Vipr2 | ||
| Vipr1 | Vldlr | ||
| Vipr2 | Vtn | ||
| Vldlr | Xcr1 | ||
| Vtn | |||
| Xcr1 |
Receptor mRNAs identified by microarray for motor neurons (MNs; column 1) and by RNA-seq for RGCs (column 3), as defined using the updated ligand-receptor database (modified from Yuzwa et al., 2016). Only included are receptor mRNAs that had expression levels exceeding a cutoff of the top 87% of mRNAs for motor neurons and FPKM of 1 for RGCs. Also shown are receptor mRNAs that overlap between sensory, sympathetic and motor neurons (column 2) or between all four populations of neurons (column 4). The sensory and sympathetic neuron receptor mRNAs are shown in Table 8. The total numbers of receptor mRNAs in each column are indicated.
We obtained similar findings when we analyzed RGCs using a bulk RNA-seq dataset generated from P5–P7 rat RGCs that were cultured for 12 h (Blanco-Suarez et al., 2018). Using an FPKM of 1 as a threshold for expression, we found 320 receptors expressed by RGCs, with 258 of them also expressed by the three peripheral neuron types (Table 10). Modeling of the unidirectional paracrine interactions between the injured nerve and RGCs identified 126 predicted interactions that included all 122 shared sensory and sympathetic neuron interactions (Fig. 7B, shared interactions are outlined in purple; Table 9). There were also four interactions that were unique to the RGC model involving CCL9, CXCL1, CXCL2, and CXCL16. Thus, the injured peripheral nerve and in particular the Schwann cells and endoneurial mesenchymal cells are predicted to provide a ligand environment that acts on multiple populations of neurons.
The communication networks identify mesenchymal-derived ligands that regulate peripheral axon growth
These models predict that, in addition to Schwann cells, nerve mesenchymal cells are key growth factor sources in the injured nerve. To validate this concept, we focused on two predicted endoneurial ligands that have not previously been explored within a nerve context, ANGPT1 and CCL11 (Fig. 8A,B). We also analyzed VEGFC, which is expressed at a low level in injured nerve mesenchymal cells (Fig. 8A), but that had a coreceptor (NPR1) that was validated at the protein level in both sensory and sympathetic neurons.
Figure 8.
Identification and characterization of ligands expressed in nerve mesenchymal cells that locally promote sympathetic axon growth. A, tSNE gene expression overlays of Angpt1 and Vegfc on the combined 3 and 9 DPI nerve dataset shown in Figure 1E. Cells that detectably express the ligand are colored blue, and the numbers correspond to the clusters. B, Heatmap showing expression of Ccl11 mRNA in single cells within clusters of the combined injured nerve scRNA-seq dataset shown in Figure 1E. Each column line represents the level of expression in a single cell. Gene expression represents scaled expression values using Seurat’s scaling function and is color coded as per the adjacent color key, where yellow indicates the highest expression. Cluster numbers are on the bottom, and the cell types in that cluster are annotated on the top. Mes. = mesenchymal, Endoth. = endothelial, Endo Mes. = endoneurial mesenchymal, SC = Schwann cell. C–E, Neonatal (P4) rat sciatic nerve mesenchymal cells were sorted for cell-surface PDGFRα using FACS, cultured in defined growth medium, and medium was collected after 1–4 d of conditioning. C, Schematic of the experiments. D, Representative image of the mesenchymal cell cultures immunostained for PDGFRα and for the Schwann cell protein S100β to indicate the relative purity of the cultures. Scale bar = 50 μm. E, Quantitative ELISA analysis of the nerve mesenchymal cell conditioned medium for ANGPT1, CCL11, and VEGFC (CM). Control growth medium was used as a negative control in each experiment (Con). Shown are the mean ± SEM from three independent experiments; **p < 0.01 (CCL11, p = 0.0055; VEGFC, p = 0.0045) and *p < 0.05 (ANGPT1, p = 0.031), two-tailed unpaired Student’s t test. F, Images of the 9-d injured distal sciatic nerve of an adult PdgfraEgfp/+ mouse analyzed by FISH for Angpt1, Ccl11, and Vegfc mRNAs. Hatched white lines outline EGFP-positive cells (green nuclei) that were also positive for the mRNA of interest (red dots). Also shown is Hoechst 33258 counterstaining (white/gray) to highlight cell nuclei. The arrows indicate cells that are shown at higher magnification in the insets. Scale bars = 20 μm. Scale bars in insets = 8.75 μm. G, Schematic of the compartmented culture axon outgrowth experiments. Neonatal SCG sympathetic neurons were established in compartmented cultures in the presence of 10 ng/ml NGF. When their axons had crossed into the side compartments, the side compartment medium was replaced with medium containing 0.5 ng/ml NGF plus 100 ng/ml of ANGPT1, CCL11, or VEGFC for three additional days. As a positive control, 50 ng/ml NGF was added into side compartments and as a baseline control, axons were maintained in 0.5 ng/ml NGF alone. H, Brightfield images of sympathetic axons in side compartments grown as described in G, located within 1 mm of the furthest extent of axonal outgrowth. Positive control (Max NGF) = 50 ng/ml, negative control (Min NGF) = 0.5 ng/ml. Scale bar = 100 μm. I, Scatter plots showing the density of outgrowth in response to the different ligands. A vertical line was drawn within the farthest 1 mm of axonal outgrowth, and the number of axons crossing the line was quantified. A maximum of eight separate lanes was scored per technical replicate, with three to four technical replicate cultures (i.e., cultures generated from sympathetic ganglia harvested from the same litter) scored per biological replicate (n values in plot). Individual points show mean ± SEM of individual biological replicates; *p < 0.05 (p = 0.041), ****p < 0.0001, one-way ANOVA with Holm–Sidak’s multiple comparison’s test (n = 5 for all treatments except VEGFC, where n = 3).
We first asked whether these three ligands were secreted by nerve mesenchymal cells. To do this, we isolated mesenchymal cells from rat sciatic nerves using antibody-based flow sorting for cell-surface PDGFRα. We cultured and expanded these sorted mesenchymal cells for 2.5–4 weeks, at which point the cultures were comprised of over 90% PDGFRα-positive mesenchymal cells, with the remaining cells being S100β-positive Schwann cells (Fig. 8C,D). After three further days in culture, we added defined, serum-free medium for 24–96 h, collected this conditioned medium, and performed ELISAs. ANGPT1, CCL11, and VEGFC were all detected in three independent preparations of nerve mesenchymal cell conditioned medium (Fig. 8E).
Having confirmed that these ligands were secreted by nerve mesenchymal cells in culture, we asked whether Angpt1, Ccl11, and Vegfc mRNAs were expressed in endoneurial cells of the injured nerve. To do this, we analyzed injured distal nerve sections from the PdgfraEgfp/+mice. We resected sciatic nerves and at 9 DPI performed single molecule FISH. Angpt1 and Ccl11 mRNAs were detectable in many Pdgfra-EGFP-positive mesenchymal cells within the injured nerve endoneurium (Fig. 8F). Consistent with the scRNA-seq data, many but not all EGFP-positive cells were positive for these mRNAs (Fig. 8A,B; 24% and 87% of endoneurial cells express detectable Angpt1 and Ccl11 mRNAs, respectively). There were also some Angpt1 or Ccl11-positive cells that were Pdgfra-EGFP-negative. For Angpt1 mRNA these are likely VSM/pericyte cells, while for Ccl11 mRNA, they could be VSM/pericytes or immune cells (Fig. 8B). Vegfc mRNA was also detected in scattered endoneurial Pdgfra-EGFP-positive cells but consistent with the scRNA-seq analysis (Fig. 8A), there were fewer double-labeled cells and the FISH signal was low (Fig. 8F).
Having validated their expression, we asked whether ANGPT1, CCL11, or VEGFC could promote growth of peripheral axons. To do this, we used compartmented cultures of neonatal SCG sympathetic neurons (Campenot et al., 1991; Park et al., 2010). In these cultures, cell bodies are physically separated from axons so that ligands can be applied just to the axons, as would occur in the peripheral nerve (Fig. 8G). We established sympathetic neurons in these compartmented cultures with 10 ng/ml NGF, their obligate survival factor, in all compartments. Three days later, when axons had crossed into the side compartments, we replaced the side compartment medium with medium containing 0.5 ng/ml NGF plus 100 ng/ml of ANGPT1, CCL11, or VEGFC for three additional days. As a positive control, we added 50 ng/ml NGF into side compartments, and as a baseline control, we maintained axons in 0.5 ng/ml NGF alone. To quantify the density of axonal growth in these compartments, we drew a line perpendicular to the axis of the outgrowth within the furthest 1 mm of outgrowth where axons were maximally defasciculated. This analysis showed that 50 ng/ml NGF caused a robust increase in the density of sympathetic axons relative to the 0.5 ng/ml NGF baseline control (Fig. 8H,I). Notably, all three mesenchymal ligands modestly but significantly enhanced axonal density, although to a lesser degree than maximal NGF (Fig. 8H,I). Thus, at least some of the mesenchymally derived ligands predicted in our models were bioactive on sympathetic axons.
Discussion
In the present study, we have characterized the ligand environment of the uninjured, injured, and developing sciatic nerves using bulk and single-cell transcriptional profiling. We have identified receptor proteins on the surface of two types of peripheral neurons (sensory and sympathetic) and made predictions of ligand-receptor paracrine interactions between the injured nerve and peripheral neurons. We then go on to show, based on these predictions, that mesenchymal cells express factors that are capable of augmenting growth of peripheral axons in vitro, indicating that at least some of these ligands may directly contribute to the positive axonal growth environment of the developing and regenerating peripheral nerves.
Peripheral nerves provide a highly supportive environment for axonal growth during development and following injury (Chen et al., 2007; Cattin and Lloyd, 2016; Fledrich et al., 2019) and promote the repair and regeneration of innervated tissues (Kumar and Brockes, 2012; Johnston et al., 2013, 2016; Mahmoud et al., 2015). Many nerve-derived growth factors have already been well studied, including NGF, BDNF, NTF3, GDNF, and cytokines of the LIF/CNTF family (for review, see Terenghi, 1999). These factors are generally assumed to be Schwann cell derived, although macrophages express factors like GAS6 that promote proper function of Schwann cells in the regenerating nerve (Stratton et al., 2018). To identify other important factors that might be involved in providing a supportive peripheral nerve environment, we used a modeling strategy based on transcriptomic analysis and cell-surface mass spectrometry, as has been previously done for embryonic cortical development and digit tip regeneration (Johnston et al., 2016; Yuzwa et al., 2016). However, while bulk transcriptomic profiling was used in these earlier studies, here we added single-cell transcriptional profiling, thereby providing a level of cellular resolution previously not possible for complex tissues. This approach allowed us to define a previously unappreciated role for mesenchymal cells in establishing the nerve paracrine environment, to identify new nerve ligands, and to predict that many nerve ligands will act on both PNS and CNS neurons, thereby potentially providing an explanation for why peripheral nerves can promote growth of CNS axons.
Our study defined many growth factor mRNAs induced in Schwann cells following nerve injury. Some of these encoded previously studied factors like Artn, Bdnf, Gdnf, Pdgfa, Shh, and Lif, while others encoded factors not well studied in this regard, including Ucn2, Fgf5, and the CNTF-like cytokines Clcf1 and Crlf1. Previous studies have proposed that this ligand induction is part of a unique Schwann cell “repair” phenotype (Jessen and Mirsky, 2019) that is important for axonal regeneration in the case of ligands like BDNF, GDNF, and LIF, and for tissue repair, in the case of PDGFA (Johnston et al., 2016). What is this repair phenotype? Previous work has shown that following injury Schwann cells display altered morphology and gene expression that is thought to be conducive to promoting axonal regeneration (Gomez-Sanchez et al., 2017; Jessen and Mirsky, 2019). Repair Schwann cells have also been reported to have enhanced epithelial to mesenchymal transition gene expression and TGFβ signaling that contributes to the establishment of an invasive, “mesenchymal-like” phenotype (Arthur-Farraj et al., 2017; Clements et al., 2017). Our findings also show that following nerve injury, Schwann cells induce ligand genes that are not expressed at detectable levels in the uninjured neonatal or adult nerves. Moreover, our transcriptional comparisons expand on previous work and show that repair Schwann cells are more similar to neonatal than to adult uninjured nerve Schwann cells but that they are nonetheless distinct. In this regard, our developmental comparison was limited to the neonatal nerve when myelination is ongoing, and it would be interesting to use single-cell transcriptional profiling to see how similar repair Schwann cells are to embryonic nerve Schwann cells before myelination has commenced.
An important finding of this work is that Pdgfra-positive mesenchymal cells are a major source of ligands in the developing, adult, and injured nerves and that they express well-characterized nerve growth factors like NGF, HGF, and LIF. Of particular interest is the high ligand expression by endoneurial mesenchymal cells, which are neural crest derived (Joseph et al., 2004) and are scattered throughout the endoneurial space in close apposition to Schwann cells and axons. These endoneurial mesenchymal cells are thus ideally positioned to regulate axon and Schwann cell biology, and, like Schwann cells, they display increased expression of many ligand mRNAs following injury, including well-studied ligands like Crlf1, Ngf, and Lif and less-studied ligands such as Angpt1, Ccl9, and Sema7a. Equally intriguing was the observation that endoneurial cells express many little-studied ligands under homeostatic conditions, including Adm, Bmp7, Il33, Pthlh, and Wnt5a. Since mesenchymally derived ligands include both well-studied nerve growth factors such as NGF as well as many ligands with unknown roles in nerve biology, it is likely that mesenchymal cells express ligands that are critical components of the regenerative response of the injured nerve and/or the growth program of the developing nerve. Our validation studies indicate that at least some of these endoneurial cell ligands are active on axons, but they might be equally important for other nerve cell types and/or for the tissues they innervate. As one example, PTHLH and nerve innervation are both important for bone homeostasis and repair (Elefteriou et al., 2014; Ansari et al., 2018), and endoneurial cells, which express Pthlh, migrate into the injured bone where they directly contribute to bone repair (Carr et al., 2019). As a second example, the vasodilator peptide adrenomedullin (ADM) was previously shown to stimulate cAMP accumulation in endothelial cells and Schwann cells (Dumont et al., 2002), suggesting that endoneurial cell-derived ADM might be important for nerve vasculature and/or Schwann cell biology. As a final example, BMP7 inhibits myelin gene expression in Schwann cells (Liu et al., 2016) and promotes mammalian digit tip regeneration (Yu et al., 2010), suggesting that endoneurial cell-derived BMP7 might play multiple important roles.
Another important finding is that most injured nerve ligands are predicted to act on all three populations of PNS neurons as well as RGCs. In this regard, the nerve could promote axonal growth and regeneration in two somewhat disparate ways. In one model, Schwann cells and endoneurial mesenchymal cells would produce different ligands depending on the axons that they are currently or have previously interacted with, thereby tailoring the nerve environment to the axons that need to grow or regenerate. Support for this model comes from studies showing that denervated Schwann cells of motor versus sensory nerves provide ligands specific to different types of axons (Höke et al., 2006; Brushart et al., 2013). In a second model, during development or following nerve damage Schwann cells and mesenchymal cells could express a broad repertoire of ligands, thereby ensuring that growth of all types of PNS axons would be supported. Our findings support this latter model, since we find broad injury-induced ligand expression and a relatively broad repertoire of receptor expression on four different types of neurons, culminating in many similar predicted paracrine interactions. Such a mechanism would provide maximum flexibility and would explain why peripheral nerve grafts promote regeneration of multiple types of injured CNS neurons, which do not normally project in peripheral nerves. Nonetheless, our findings are still consistent with the finding of differential ligand expression in different nerve subtypes (Höke et al., 2006; Brushart et al., 2013), since we have only defined the ligand landscape in a mixed nerve.
How predictive are these models? Previous studies using bulk transcriptomics and/or cell-surface mass spectroscopy predicted three factors important for embryonic cortical neurogenesis (IFNγ, Neurturin, and GDNF; Yuzwa et al., 2016) and one for oligodendrogenesis (Fractalkine; Voronova et al., 2017), and two factors important for digit tip regeneration (PDGFA and OSM; Johnston et al., 2016). In those studies, the cell of origin for each ligand was identified either by isolating cells or by performing single-cell resolution morphologic approaches. Here, we have instead used single-cell transcriptional profiling to provide the necessary resolution, an approach with much broader applicability. The validity of the resultant models is attested to by our finding that almost all ligands previously shown to be important for peripheral nerve regeneration were independently assigned in our models, including many ligands known to be expressed in Schwann cells. Nonetheless, to ensure the validity of these models, we also examined three ligands that were predicted to be made by nerve mesenchymal cells, ANGPT1, CCL11, and VEGFC. Two of these factors, ANGPT1 and VEGFC, are well-known angiogenesis factors, while the third, CCL11 or eotaxin, is a chemokine involved in eosinophil recruitment (Jose et al., 1994). None of the three has been studied as a positive factor within the context of the injured nerve, although ANGPT1 has been shown to promote growth of cultured sensory neurons (Kosacka et al., 2006), VEGFC promotes growth of developing motor neurons in zebrafish (Kwon et al., 2013), and CCL11 inhibits Schwann cell differentiation (Büttner et al., 2018). Based on our predictive models, we tested these ligands and found that all three (1) were expressed by endoneurial mesenchymal cells in the injured nerve, as shown by both scRNA-seq and FISH analyses; (2) were synthesized and secreted by cultured nerve-derived PDGFRα-positive mesenchymal cells; and (3) enhanced sympathetic axon outgrowth when applied locally in the presence of minimal NGF. While we recognize that additional studies will be required to show that these three ligands are secreted by mesenchymal cells in vivo, our data indicate that they are highly expressed following nerve injury, raising the possibility that they are important for nerve repair. In this regard, ANGPT1, CCL11, and VEGFC were not as potent as NGF in promoting growth of sympathetic axons in culture, but they did enhance growth and thus could be important factors for peripheral axon growth in a regenerating nerve context. We therefore feel that our studies validate the predictive value of the modeling approach and provide support for the idea that mesenchymal cells within the nerve are important ligand sources, particularly within the context of nerve repair and regeneration.
The data presented here reinforce the importance of Schwann cells as sources of growth promoting factors and provide evidence that mesenchymal cells also play an important role in determining the ligand environment of the developing and injured nerve. Notably, some well-known nerve regeneration ligands such as GDNF and BDNF were expressed at the highest levels in Schwann cells, while others, such as NGF and HGF, were instead highest in mesenchymal cells. In addition, both cell types express ligands that are not well-characterized as nerve injury ligands, including BTC and UCN2 for Schwann cells, and ANGPT1, CCL11, and VEGFC for mesenchymal cells. While the relative contributions of growth factors from these two cell types to nerve growth and repair remain unknown, our study does highlight mesenchymal cells as a previously overlooked reservoir of growth factors for axon growth and potentially for nerve tissue regeneration, an area we are only now starting to understand (for examples, see Parrinello et al., 2010; Cattin et al., 2015). The data presented here thus provide an important step toward defining nerve paracrine interactions not only with regard to axon growth and peripheral nerve regeneration, but also with regard to the paracrine roles of the nerve during repair and regeneration of target tissues.
Acknowledgments
Acknowledgements: We thank Dennis Aquino, Jon Krieger, Troy Ketela, and Konstantin Feinberg for advice and technical assistance.
Synthesis
Reviewing Editor: Jeffery Twiss, University of South Carolina
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Ahmet Hoke, Moses Chao.
I apologize for the delay in getting this review process completed. Normally I would put together a synthesis statement that incorporates both reviewers comments and an online discussion. The extraordinary circumstances that we are under now has everyone distracted, so I did not get to conduct the online discussion to the extent that would be needed for my usual synthesis. I am appending the direct comments from each reviewer below my summary of their comments and my suggestions for revision.
Both reviewers encouraged resubmission. Both reviewers and the reviewing editor fully recognize the amount of work that went into this study. I think that critiques give clear guidance on points to address for revision. In particular, you should reorganize the text to make sure that the main message of the manuscript is not lost in the single cell RNA data - shortening the manuscript may help as well as being selective on what extended data needs to be included. The statistical analyses are difficult to follow, so more clearly outlining the tests used and rationale for those would help. With the newness of scRNA-Seq, you will do the reader a service by carefully justifying the statistical measures.
For the comments from reviewer 1 on no data showing the ligands play a role in regeneration and issue of Schwann cells outnumbering mesenchymal cells many fold, I think this could be addressed by text changes. Specifically backing off on conclusions and referring to these ligands as potential growth modulators. I fully agree with the reviewer that showing in vitro activity in SCGs is not sufficient to make a conclusion on regeneration. The conclusions would of course be strengthened by in vivo data, but I do not think that is necessary if you can appropriately qualify your conclusions.
For the comment from reviewer 2 on Vegfc, Ccl11 and Angpt1 secretion, I agree with the reviewer that UniProt database designations are not sufficient. Your experimental system could certainly behave differently than the studies that UniProt draws from. Data on secretion in peripheral nerve would certainly strengthen the manuscript. Alternatively, I think that you could suggest this for future studies, acknowledging the limitations of public databases (also I recognize the difficulty in showing secretion in vivo in the PNS, and I think that would unfortunately be the necessary data).
Reviewer 1:
This is an interesting paper that examines potentially secreted ligands that can act on neurons from cells in the injured nerve and concludes that about half of potential ligands are coming from mesenchymal cells. Although potentially a very important observation, the work presented is very preliminary and does not justify the conclusions, including the title. There is absolutely no data shown that these molecules actually play a role in regeneration. Demonstrating that some of the candidate molecules have in vitro activity in SCG neurons is not sufficient to prove that they have a role in regeneration. The authors fail to take into consideration that the number of mesenchymal cells in the distal nerve are dwarfed by Schwann cells and macrophages and that any potentially secreted ligand may or may not have any substantial effect on regeneration.
I think the authors need to be commended for performing this scRNA-Seq analysis as it will be a resource for the whole scientific community. However, the references to regeneration need to be removed from the paper as data is not there to justify such conclusions.
Reviewer 2:
The study covers a topic of interest-transcriptional changes in the sciatic nerve after injury. The goal is to identify ligand-receptor expression in neighboring cells with the goal of identifying molecules involved in regeneration. The manuscript covers the results in a very extensive manner. Unfortunately, the description of the results is very lengthy, unwieldly and drawn out. The results and the conclusions should be edited and shortened to cover the main points in a more succinct manner.
This study is carefully performed with appropriate programs and algorithms (Seurat, Gene Ontology, Panther). The manuscript needs to be strengthened by considering several issues and questions. They include the following questions, which should be addressed and clarified. -
1) The Materials and Methods section has a thorough description of the procedures; the isolation of single cells; the scRNA-seq and computational analysis. However, it is interspersed with commentary and citations about the results. To organize the paper better, the description of the results and Figures should be integrated within the Results section.
2) A major finding is that Vegfc, Ccl11 and Angpt1 are expressed in endoneurial cells after injury. Do the authors have direct evidence the three ligands are secreted? Do they all give a similar receptor signaling response downstream of their receptors? The UniProtKB database is cited as the source of secreted ligands, but the manuscript would be stronger if more data were included.
3) In this regard, the presented facts can be condensed by placing relevant results in separate themes and biological contexts, instead of listings of enriched genes.
4) Several descriptions of the Results should be better explained and more comprehensible. For example, “This analysis identified seven Sox10-positive clusters containing 5331 Schwann cells” does not convey the significance of this finding.
5) The effect of injury upon ligand mRNAs needs to be clarified, as it is unclear if the levels of Ccl1 are greatly affected (Fig. 4G). Also, Ccl11 appears to be expressed in many cell types-Schwann, vascular, immune cells (Fig. 8). The cell specificity of ligand expression should be emphasized from the analysis.
References
- Ansari N, Ho PW, Crimeen-Irwin B, Poulton IJ, Brunt AR, Forwood MR, Divieti Pajevic P, Gooi JH, Martin TJ, Sims NA (2018) Autocrine and paracrine regulation of the murine skeleton by osteocyte-derived parathyroid hormone-related protein. J Bone Miner Res 33:137–153. 10.1002/jbmr.3291 [DOI] [PubMed] [Google Scholar]
- Arthur-Farraj PJ, Morgan CC, Adamowicz M, Gomez-Sanchez JA, Fazal SV, Beucher A, Razzaghi B, Mirsky R, Jessen KR, Aitman TJ (2017) Changes in the coding and non-coding transcriptome and DNA methylome that define the Schwann cell repair phenotype after nerve injury. Cell Rep 20:2719–2734. 10.1016/j.celrep.2017.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz LI, He Z, Goldberg JL (2017) Reaching the brain: advances in optic nerve regeneration. Exp Neurol 287:365–373. 10.1016/j.expneurol.2015.12.015 [DOI] [PubMed] [Google Scholar]
- Blanco-Suarez E, Liu TF, Kopelevich A, Allen NJ (2018) Astrocyte-secreted chordin-like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron 100:1116–1132. 10.1016/j.neuron.2018.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL (2011) Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 8:552–565. 10.1016/j.stem.2011.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushart TM, Aspalter M, Griffin JW, Redett R, Hameed H, Zhou C, Wright M, Vyas A, Höke A (2013) Schwann cell phenotype is regulated by axon modality and central-peripheral location, and persists in vitro. Exp Neurol 247:272–281. 10.1016/j.expneurol.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge MB (2016) Efficacy of Schwann cell transplantation for spinal cord repair is improved with combinatorial strategies. J Physiol 594:3533–3538. 10.1113/JP271531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner R, Schulz A, Reuter M, Akula AK, Mindos T, Carlstedt A, Riecken LB, Baader SL, Bauer R, Morrison H (2018) Inflammaging impairs peripheral nerve maintenance and regeneration. Aging Cell 17:e12833. 10.1111/acel.12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenot RB, Walji AH, Draker DD (1991) Effects of sphingosine, staurosporine, and phorbol ester on neurites of rat sympathetic neurons growing in compartmented cultures. J Neurosci 11:1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr MJ, Toma JS, Johnston APW, Steadman PE, Yuzwa SA, Mahmud N, Frankland PW, Kaplan DR, Miller FD (2019) Mesenchymal precursor cells in adult nerves contribute to mammalian tissue repair and regeneration. Cell Stem Cell 24:240–256. 10.1016/j.stem.2018.10.024 [DOI] [PubMed] [Google Scholar]
- Cattin AL, Burden JJ, Van Emmenis L, Mackenzie FE, Hoving JJ, Garcia Calavia N, Guo Y, McLaughlin M, Rosenberg LH, Quereda V, Jamecna D, Napoli I, Parrinello S, Enver T, Ruhrberg C, Lloyd AC (2015) Macrophage-induced blood vessels guide Schwann cell-mediated regeneration of peripheral nerves. Cell 162:1127–1139. 10.1016/j.cell.2015.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin AL, Lloyd AC (2016) The multicellular complexity of peripheral nerve regeneration. Curr Opin Neurobiol 39:38–46. 10.1016/j.conb.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Chen ZL, Yu WM, Strickland S (2007) Peripheral regeneration. Annu Rev Neurosci 30:209–233. 10.1146/annurev.neuro.30.051606.094337 [DOI] [PubMed] [Google Scholar]
- Clements MP, Byrne E, Camarillo Guerrero LF, Cattin AL, Zakka L, Ashraf A, Burden JJ, Khadayate S, Lloyd AC, Marguerat S, Parrinello S (2017) The wound microenvironment reprograms Schwann cells to invasive mesenchymal-like cells to drive peripheral nerve regeneration. Neuron 96:98–114. 10.1016/j.neuron.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S, Aguayo AJ (1981) Axonal elongation into peripheral nervous system “bridges” after central nervous system injury in adult rats. Science 214:931–933. 10.1126/science.6171034 [DOI] [PubMed] [Google Scholar]
- Dumont CE, Muff R, Flühmann B, Fischer JA, Born W (2002) Paracrine/autocrine function of adrenomedullin in peripheral nerves of rats. Brain Res 955:64–71. 10.1016/s0006-8993(02)03365-6 [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Campbell P, Ma Y (2014) Control of bone remodeling by the peripheral sympathetic nervous system. Calcif Tissue Int 94:140–151. 10.1007/s00223-013-9752-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg K, Eshed-Eisenbach Y, Frechter S, Amor V, Salomon D, Sabanay H, Dupree JL, Grumet M, Brophy PJ, Shrager P, Peles E (2010) A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron 65:490–502. 10.1016/j.neuron.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg K, Kolaj A, Wu C, Grinshtein N, Krieger JR, Moran MF, Rubin LL, Miller FD, Kaplan DR (2017) A neuroprotective agent that inactivates prodegenerative TrkA and preserves mitochondria. J Cell Biol 216:3655–3675. 10.1083/jcb.201705085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fledrich R, Kungl T, Nave KA, Stassart RM (2019) Axo-glial interdependence in peripheral nerve development. Development 146:dev151704 10.1242/dev.151704 [DOI] [PubMed] [Google Scholar]
- Gardiner NJ (2011) Integrins and the extracellular matrix: key mediators of development and regeneration of the sensory nervous system. Dev Neurobiol 71:1054–1072. 10.1002/dneu.20950 [DOI] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac-Santel M, Nowoshilow S, Kageyama J, Khattak S, Currie JD, Camp JG, Tanaka EM, Treutlein B (2018) Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362:eaaq0681 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanchez JA, Pilch KS, van der Lans M, Fazal SV, Benito C, Wagstaff LJ, Mirsky R, Jessen KR (2017) After nerve injury, lineage tracing shows that myelin and remak Schwann cells elongate extensively and branch to form repair Schwann cells, which shorten radically on remyelination. J Neurosci 37:9086–9099. 10.1523/JNEUROSCI.1453-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P (2003) Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23:4013–4025. 10.1128/mcb.23.11.4013-4025.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höke A, Redett R, Hameed H, Jari R, Zhou C, Li ZB, Griffin JW, Brushart TM (2006) Schwann cells express motor and sensory phenotypes that regulate axon regeneration. J Neurosci 26:9646–9655. 10.1523/JNEUROSCI.1620-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes BT, Bader GD (2019) scClustViz – Single-cell RNAseq cluster assessment and visualization. F1000Res 7:1522 10.12688/f1000research.16198.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R (2019) The success and failure of the Schwann cell response to nerve injury. Front Cell Neurosci 13:33. 10.3389/fncel.2019.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston APW, Naska S, Jones K, Jinno H, Kaplan DR, Miller FD (2013) Sox2-mediated regulation of adult neural crest precursors and skin repair. Stem Cell Reports 1:38–45. 10.1016/j.stemcr.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AP, Yuzwa SA, Carr MJ, Mahmud N, Storer MA, Krause MP, Jones K, Paul S, Kaplan DR, Miller FD (2016) Dedifferentiated Schwann cell precursors secreting paracrine factors are required for regeneration of the mammalian digit tip. Cell Stem Cell 19:433–448. 10.1016/j.stem.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Jose PJ, Griffiths-Johnson DA, Collins PD, Walsh DT, Moqbel R, Totty NF, Truong O, Hsuan JJ, Williams TJ (1994) Eotaxin: a potent eosinophil chemoattractant cytokine detected in a guinea pig model of allergic airways inflammation. J Exp Med 179:881–887. 10.1084/jem.179.3.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph NM, Mukouyama YS, Mosher JT, Jaegle M, Crone SA, Dormand EL, Lee KF, Meijer D, Anderson DJ, Morrison SJ (2004) Neural crest stem cells undergo multilineage differentiation in developing peripheral nerves to generate endoneurial fibroblasts in addition to Schwann cells. Development 131:5599–5612. 10.1242/dev.01429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A, Spiller KJ, Towne C, Kanning KC, Choe GT, Geber A, Akay T, Aebischer P, Henderson CE (2014) Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron 81:333–348. 10.1016/j.neuron.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Bablaenko Y, Brenner M, Loh PR, Raychaudhuri S (2019) Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16:1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosacka J, Nowicki M, Kacza J, Borlak J, Engele J, Spanel-Borowski K (2006) Adipocyte-derived angiopoietin-1 supports neurite outgrowth and synaptogenesis of sensory neurons. J Neurosci Res 83:1160–1169. 10.1002/jnr.20811 [DOI] [PubMed] [Google Scholar]
- Kumar A, Brockes JP (2012) Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci 35:691–699. 10.1016/j.tins.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP (2007) Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318:772–777. 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Fukuhara S, Asakawa K, Ando K, Kashiwada T, Kawakami K, Hibi M, Kwon YG, Kim KW, Alitalo K, Mochizuki N (2013) The parallel growth of motoneuron axons with the dorsal aorta depends on Vegfc/Vegfr3 signaling in zebrafish. Development 140:4081–4090. 10.1242/dev.091702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibrock J, Lottspeich F, Hohn A, Hofer M, Hengerer B, Masiakowski P, Thoenen H, Barde YA (1989) Molecular cloning and expression of brain-derived neurotrophic factor. Nature 341:149–152. 10.1038/341149a0 [DOI] [PubMed] [Google Scholar]
- Lindsay RM (1988) Nerve growth factors (NGF, BDNF) enhance axonal regeneration but are not required for survival of adult sensory neurons. J Neurosci 8:2394–2405. 10.1523/JNEUROSCI.08-07-02394.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhao Y, Peng S, Zhang S, Wang M, Chen Y, Zhang S, Yang Y, Sun C (2016) BMP7 retards peripheral myelination by activating p38 MAPK in Schwann cells. Sci Rep 6:31049. 10.1038/srep31049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, McCarthy DJ, Marioni JC (2016) A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5:2122. 10.12688/f1000research.9501.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161:1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud AI, O'Meara CC, Gemberling M, Zhao L, Bryant DM, Zheng R, Gannon JB, Cai L, Choi W-Y, Egnaczyk GF, Burns CE, Burns CG, MacRae CA, Poss KD, Lee RT (2015) Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev Cell 34:387–399. 10.1016/j.devcel.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CA, Yang JY, Marathe V, Yen TY, Macher BA (2009) Combining results from lectin affinity chromatography and glycocapture approaches substantially improves the coverage of the glycoproteome. Mol Cell Proteomics 8:287–301. 10.1074/mcp.M800272-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef L, Rodgers P (2014) eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One 9:e101717. 10.1371/journal.pone.0101717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir D (2010) The potentiation of peripheral nerve sheaths in regeneration and repair. Exp Neurol 223:102–111. 10.1016/j.expneurol.2009.05.038 [DOI] [PubMed] [Google Scholar]
- Naveilhan P, ElShamy WM, Ernfors P (1997) Differential regulation of mRNAs for GDNF and its receptors Ret and GDNFR alpha after sciatic nerve lesion in the mouse. Eur J Neurosci 9:1450–1460. 10.1111/j.1460-9568.1997.tb01499.x [DOI] [PubMed] [Google Scholar]
- Park KJ, Grosso CA, Aubert I, Kaplan DR, Miller FD (2010) p75NTR-dependent, myelin-mediated axonal degeneration regulates neural connectivity in the adult brain. Nat Neurosci 13:559-566. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Napoli I, Ribeiro S, Wingfield Digby P, Fedorova M, Parkinson DB, Doddrell RD, Nakayama M, Adams RH, Lloyd AC (2010) EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell 143:145–155. 10.1016/j.cell.2010.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis MJ, Spencer PS (1986) Regeneration of rat optic axons into peripheral nerve grafts. Exp Neurol 91:52–59. 10.1016/0014-4886(86)90025-7 [DOI] [PubMed] [Google Scholar]
- Schiess R, Mueller LN, Schmidt A, Mueller M, Wollscheid B, Aebersold R (2009) Analysis of cell surface proteome changes via label-free, quantitative mass spectrometry. Mol Cell Proteomics 8:624–638. 10.1074/mcp.M800172-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer MA, Mahmud N, Karamboulas K, Borrett MJ, Yuzwa SA, Gont A, Androschuk A, Sefton MV, Kaplan DR, Miller FD (2020) Acquisition of a unique mesenchymal precursor-like blastema state underlies successful adult mammalian digit tip regeneration. Dev Cell 52:509–524. 10.1016/j.devcel.2019.12.004 [DOI] [PubMed] [Google Scholar]
- Stratton JA, Holmes A, Rosin NL, Sinha S, Vohra M, Burma NE, Trang T, Midha R, Biernaskie J (2018) Macrophages regulate Schwann cell maturation after nerve injury. Cell Rep 24:2561–2572. 10.1016/j.celrep.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Terenghi G (1999) Peripheral nerve regeneration and neurotrophic factors. J Anat 194:1–14. 10.1046/j.1469-7580.1999.19410001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupp M, Rydén M, Jörnvall H, Funakoshi H, Timmusk T, Arenas E, Ibáñez CF (1995) Peripheral expression and biological activities of GDNF, a new neurotrophic factor for avian and mammalian peripheral neurons. J Cell Biol 130:137–148. 10.1083/jcb.130.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A, Yuzwa SA, Wang BS, Zahr S, Syal C, Wang J, Kaplan DR, Miller FD (2017) Migrating interneurons secrete fractalkine to promote oligodendrocyte formation in the developing mammalian brain. Neuron 94:500–516. 10.1016/j.neuron.2017.04.018 [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, Taketo MM, Karlsson S, Iwama A, Nakauchi H (2011) Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 147:1146–1158. 10.1016/j.cell.2011.09.053 [DOI] [PubMed] [Google Scholar]
- Yu L, Han M, Yan M, Lee EC, Lee J, Muneoka K (2010) BMP signaling induces digit regeneration in neonatal mice. Development 137:551–559. 10.1242/dev.042424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa SA, Yang G, Borrett MJ, Clarke G, Cancino GI, Zahr SK, Zandstra PW, Kaplan DR, Miller FD (2016) Proneurogenic ligands defined by modeling developing cortex growth factor communication networks. Neuron 91:988–1004. 10.1016/j.neuron.2016.07.037 [DOI] [PubMed] [Google Scholar]
- Yuzwa SA, Borrett MJ, Innes BT, Voronova A, Ketela T, Kaplan DR, Bader GD, Miller FD (2017) Developmental emergence of adult neural stem cells as revealed by single-cell transcriptional profiling. Cell Rep 21:3970–3986. 10.1016/j.celrep.2017.12.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the 3- and 9-d injured sciatic nerve scRNA-seq datasets. A, t-SNE gene expression overlays on the 3 DPI total cell dataset (shown in Fig. 1C and in the adjacent legend) for the endothelial cell marker Pecam1, the immune cell marker Trbc2, the VSM/pericyte cell marker Rgs5, and the Pdgfra-positive mesenchymal epineurial and endoneurial cell markers Dpp4 and Wif1. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. B, t-SNE gene expression overlays on the combined 3 and 9 DPI total cell datasets (shown in Figure 1E and in the adjacent legend) for the endothelial cell marker Pecam1, the immune cell marker Aif1, the VSM/pericyte cell marker Rgs5, the mesenchymal marker Pdgfra, the Schwann cell marker Sox10, and the B cell marker Cd19. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. C, t-SNE gene expression overlays on the combined 3 and 9 DPI total cell dataset (shown in Fig. 1E and in the adjacent legend) for markers for the different types of Pdgfra-positive mesenchymal cells, including Etv1-positive endoneurial cells, Pcolce2-positive epineurial cells, Msln-positive perineurial cells, and Dlk1-positive differentiating mesenchymal cells. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. D, t-SNE gene expression overlays of the combined 3 and 9 DPI total cell dataset for Fgf10, Adm, Pthlh, and Ntn1. Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Specific cell types are circled and annotated, including mesenchymal (Mes.), endoneurial (Endo.), and epineurial/perineurial (Epi/Perineurial) cells. Download Figure 1-1, TIF file (2.5MB, tif)
Characterization of the uninjured and neonatal sciatic nerve scRNA-seq datasets. A, t-SNE gene expression overlays of the combined 3 and 9 DPI total cell dataset (shown in Fig. 1E and the adjacent legend) for Bdnf, Dhh, and Shh. Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Schwann cells are circled and annotated (SC). B, t-SNE gene expression overlays on the uninjured sciatic nerve total cell dataset (shown in Fig. 2C and in the adjacent legend) for the VSM/pericyte cell marker Acta2, the endothelial cell marker Pecam1, the immune cell marker Cd52, and the proliferating cell marker Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. C, t-SNE cluster visualization of neonatal sciatic nerve single-cell transcriptomes (as in Fig. 2E and the adjacent legend) showing dataset of origin. Set 1 (red) refers to the neonatal nerve cells isolated by FACS and Set 2 (blue) to the neonatal nerve cells isolated by treatment with the myelin removal beads. The right t-SNE cluster visualization indicates the datasets of origin following Harmony data integration batch correction of the combined datasets. D, t-SNE gene expression overlays on the neonatal sciatic nerve total cell dataset (shown in Fig. 2E and in the adjacent legend) for Osr2, which marks endoneurial mesenchymal cells, Casq2, which is expressed in perineurial cells, the endothelial cell marker Pecam1, the VSM/pericyte cell marker Acta2, and the proliferating cell marker Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. Download Figure 2-1, TIF file (3.1MB, tif) .
Characterization of the combined Schwann cell sciatic nerve scRNA-seq dataset. A, t-SNE gene expression overlays on the combined and batch-corrected neonatal, injured adult and uninjured adult Schwann cell data (shown in Fig. 3A and the adjacent legend) for Sox10, the myelination gene Mag, the pre-myelinating Schwann cell marker Pou3f1, the non-myelinating Schwann cell gene Emp1, and the proliferation marker Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. B, Plots show correlation analyses of average transcript expression in the in the combined injured nerve dataset (Fig. 1E) showing Schwann cell cluster 6 compared to endoneurial cell cluster 5 (left plot) and to epineurial cell cluster 3 (right plot). Outlier transcripts expressed in the mesenchymal cell clusters are highlighted red and labelled. C, D, t-SNE gene expression overlays of the combined Schwann cell data (shown in Fig. 3A and the adjacent legend) for Ccl3, Crlf1, Lif, Shh, and Tgfb1 (C) and Bmp1, Fgf7, Mdk, and Pdgfa (D). Cells that detectably express the ligand are colored blue and the numbers correspond to the clusters. Injured Schwann cell cluster 3 is circled and annotated (C, Inj.). Download Figure 3-1, TIF file (2.7MB, tif) .
Characterization of the combined Pdgfra-positive mesenchymal cell sciatic nerve scRNA-seq dataset. t-SNE gene expression overlays on the combined and batch-corrected neonatal, injured adult and uninjured adult Pdgfra-positive mesenchymal cell data (shown in Fig. 4A and the adjacent legend) for Pdgfra, the epineurial markers Pcolce2 and Dpp4, the perineurial gene Slc2a1, the endoneurial gene Meox1, the differentiating injured cell genes Mest and Dlk1, and the proliferation gene Top2a. Relative transcript expression levels are color coded as per the adjacent color keys and numbers correspond to clusters. Download Figure 4-1, TIF file (1.5MB, tif) .
Identification of cell-surface proteins on sensory and sympathetic neurons. A, Bar graphs showing the percentage of cells expressing the neuronal protein βIII-tubulin (Tubb3), the Schwann cell protein S100β, or the fibroblast protein Fibronectin in cultures of DRG sensory neurons or SCG sympathetic neurons as shown in Figure 5A. The total number of cells in the cultures was determined by counterstaining with Hoechst 33258. Values: mean ± SEM, n = 6 for DRGs, n = 4 for SCGs except for cultures immunostained for Fibronectin where n = 2. B, Venn diagram showing the overlap of cell-surface proteins detected by mass spectrometry in sensory neurons and sympathetic neurons. All proteins included were annotated by the terms “cell membrane” and/or “secreted” by the UniProtKB database (http://uniprot.org). C, Bar graphs showing classification of the proteins detected by cell-surface capture mass spectrometry on sensory neurons (DRGs, blue) and sympathetic neurons (SCGs, red). Proteins were classified as receptors based on the ligand-receptor database, GO terms in the PANTHER classification system, as well as by manual curation, and were further classified into receptor types as shown in Figure 5B. The remainder of the graph includes proteins classified using PANTHER (http://pantherdb.org). D, Graphs showing the distribution of proteins detected by cell-surface capture mass-spectrometry relative to their transcript expression levels (based on microarray analyses described in the text) in sensory neurons (DRGs, left) and sympathetic neurons (SCGs, right). The cutoffs used to define receptor expression in the microarray data were based on the receptors detected by mass spectrometry analysis that had the lowest levels of mRNA expression. This was Itgam for sensory neurons (DRGs) and Sorcs3 for sympathetic neurons (SCGs, shown in red). Download Figure 5-1, TIF file (573.7KB, tif) .
Predicted unidirectional ligand-receptor interactions between injured sciatic nerve Schwann cells or endoneurial mesenchymal cells and sensory neurons. Models showing predicted unidirectional interactions between the ligands most highly expressed by injured nerve Schwann cells (A) or endoneurial mesenchymal cells (B) and their receptors on cultured sensory neurons (DRGs). Ligands are shown in the central columns in A, B and are color coded as in Figure 5 (Schwann cell ligands in grey and endoneurial mesenchymal cell ligands in yellow). Receptors are shown on either side of the ligand column and also include coreceptors that are well-characterized components of receptor complexes. Receptors that were observed at both the transcriptomic and proteomic levels are colored green while those defined only at the transcriptomic level are colored blue. Arrows indicate directionality of interactions. Note that many ligands interact with multiple receptors and, conversely, that multiple ligands are sometimes predicted to share receptors. Download Figure 6-1, TIF file (1.4MB, tif) .