Abstract
Purpose
The threat of Staphylococcus aureus antimicrobial resistance is increasing worldwide. Niosomes are a new drug delivery system that enhances the antimicrobial potential of antibiotics. We hereby aim to evaluate the antimicrobial and antibiofilm activity of ciprofloxacin-loaded niosomes.
Methods
The antimicrobial susceptibility of clinical S. aureus isolates (n=59) was determined by Kirby–Bauer disk diffusion method. Their biofilm formation activity was tested by Christensen’s method. Two ciprofloxacin-loaded niosomal formulations were prepared by thin-film hydration method, and their minimum inhibitory concentrations (MIC) were determined by agar dilution method, against ciprofloxacin-resistant and biofilm-forming isolates (n=24). Their ability to inhibit biofilm formation and eradicate already formed biofilms was evaluated and further confirmed by scanning electron microscope images. Non-synonymous mutations, in a quinolone resistance-determining regions of S. aureus isolates, were detected by polymerase chain reaction.
Results
Most of the isolates were methicillin- (47/59) and ciprofloxacin-resistant (45/59). All except two isolates were capable of biofilm production. Niosomal preparation I reduced ciprofloxacin MIC by twofold in four isolates, whereas preparation II reduced ciprofloxacin MIC of most isolates by 8- to 32-fold, with three isolates that became ciprofloxacin-susceptible. Non-synonymous mutations were detected in isolates that maintained phenotypic ciprofloxacin resistance against ciprofloxacin-loaded niosomal preparation II. Ciprofloxacin-loaded niosomes reduced the minimum biofilm inhibitory concentration and the minimum biofilm eradication concentration in 58% and 62% of the tested isolates, respectively.
Conclusion
Ciprofloxacin-loaded niosomes can restore ciprofloxacin activity against resistant S. aureus isolates. To our knowledge, this is the first report on the inhibition of biofilm formation and eradication of formed biofilms by ciprofloxacin-loaded niosomes.
Keywords: biofilm, ciprofloxacin, niosomes, Staphylococcus aureus, resistance
Introduction
Staphylococcus aureus is a human pathogen responsible for a wide range of critical infections, such as infective endocarditis as well as bloodstream, bone, skin, and soft tissue infections.1 It has the ability to attach to different surfaces, forming a biofilm; which accounts for its adherence to different medical devices in the hospital environment.2 In addition, biofilm formation, in vivo, causes a delay in wound healing, leading to chronic infection, where a high percentage (43–88%) of S. aureus isolates are from ulcers.3
Several challenges are facing the treatment of S. aureus infections. S. aureus is resistant to a wide variety of available antibiotics. Methicillin resistant S. aureus (MRSA) was listed as a “serious threat” by a report from the Center of Disease Control and Prevention, where there were 323,700 reported cases of invasive MRSA infections with about 10,600 related deaths that occurred between 2012 and 2017.4 The incidence of MRSA infections was found to be about 55% among burn patients admitted into intensive care units in different areas.5 MRSA is resistant to all currently available beta-lactam agents and most non-beta-lactam compounds. Vancomycin is the drug of choice for treatment of MRSA infections. However, S. aureus with reduced susceptibility to vancomycin has emerged, threatening its future use.6 Another challenge in the treatment of S. aureus infection is its ability to form biofilms, where bacteria in biofilms are more resistant to antibiotics compared to planktonic cells.7 They are tightly packed in an extracellular polysaccharide matrix which helps them escape the immune response and antimicrobials in the environment.1 This matrix hinders the penetration of many antibiotics, resulting in a significant decrease in antibiotic efficacy. Also, bacterial cells in deep layers of biofilm have a slow rate of metabolism and growth due to limited nutrient access.7
Development of new antimicrobials against S. aureus infection is urgently needed. This is tackled by the obstacles facing the process of drug discovery, involving the long time expended in developing new agents with the low success rate and uncertain safety.8 An alternative strategy to new drug development is to enhance the efficacy of existing drugs. Recently, the advances in nanotechnology field provided a promising tool for enhancing the activity and safety of available antimicrobial agents. The formulations of antimicrobials in nanoforms and different vesicular systems have enhanced their efficacy and selective delivery to both extracellular and intracellular infections as well as their antibiofilm activity, compared to the free drug.9,10
Liposomes are an example of such a nano-system. They are small vesicles that are made of phospholipid bilayer(s) enclosing an internal aqueous environment. They can encapsulate antimicrobial compounds and enhance their efficacy by fusion with the microbial cell membrane. Liposomes are effective in delivering antibiotics and other therapeutics to various bacterial cells and biofilms. However, their physical and chemical instability represents a major problem hindering their widespread application.9
Similarly, noisomes are bilayered structures, made from non-ionic surfactants that behave like the liposomes, in vivo. However, they are more stable, biodegradable, biocompatible, nonimmunogenic with low toxicity, and no special conditions are required for handling and storage. Thus, they overcome the problems associated with liposomal preparations.11 Niosomal formulation has enhanced the activity of several antimicrobial agents. Formulation of ciprofloxacin as niosomes has enhanced its activity against several resistant strains of S. aureus, Escherichia coli, and Pseudomonas aeruginosa, as well as in targeting intracellular and pulmonary infections.12,14 The effect of niosomes in the enhancement of vancomycin and norfloxacin antibiofilm activity, against S. aureus and P. aeruginosa, was reported previously.15,16
This study aimed at preparing ciprofloxacin-loaded niosomes and testing their potential antimicrobial effect against ciprofloxacin-resistant S. aureus clinical isolates as well as the possible inhibition of biofilm formation or eradication of already formed biofilms. Two different formulations of ciprofloxacin-loaded niosomes were prepared. The formulation containing span 60 and equimolar ratio of surfactants and cholesterol was able to highly reduce ciprofloxacin MIC, against ciprofloxacin-resistant and biofilm-forming strains of S. aureus, by 8- to 32-fold. Also, this formulation was able to inhibit biofilm formation and eradicate already formed biofilms.
Materials and Methods
Bacterial Strains
S. aureus ATCC 25923 was used as a reference strain. S. aureus clinical isolates (n=59) were collected from El-Kasr EL-Aini hospital in Egypt, during the period from September 2016 to January 2017. The collected isolates were from wounds (n=20), pus (n=13), sputum (n=7), blood (n=15), burn (n=2), bronchoalveolar lavage (n=1), and ascitic fluid (n=1). Oral informed consent was obtained from patients, since samples were collected during routine diagnosis and no special interventions were made. They were identified, using a Matrix-assisted laser desorption ionization/time-of-flight (MALDI/TOF) detector (Ultraflextreme, Bruker, Germany), at the Faculty of Medicine, Alexandria University.
Antimicrobial Susceptibility Testing
The antimicrobial susceptibility profile, of S. aureus isolates, was determined using Kirby-Bauer disk diffusion method following the Clinical and Laboratory Standards Institute (CLSI) guidelines.17 The following antibiotics were tested; penicillin (10 units), gentamicin (10 µg), amikacin (30 µg), tobramycin (10 µg), azithromycin (15 µg), clarithromycin (15 µg), erythromycin (15 µg), tetracycline (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), clindamycin (2 µg), sulfamethoxazole/trimethoprim (1.25/23.75 µg), chloramphenicol (30 µg), rifampin (5 µg), and linezolid (30 µg). MRSA were identified by determination of zone diameter against cefoxitin disk (30 µg). All antibiotic discs were purchased from Oxoid, UK. After 18–24 hours of incubation at 37°C, the plates were examined, and inhibition zone diameters around antibiotic disks were measured. Susceptibility of isolates, to vancomycin, was determined by agar dilution method.18 The antimicrobial susceptibility of tested isolates was interpreted according to CLSI breakpoints. Isolates with a zone diameter ≤21 mm against cefoxitin disk were categorized as MRSA.19 Determination of multidrug resistant isolates was done following Magiorakos et al's definition.20 S. aureus ATCC 25923 was used as a quality control strain.
Detection of Biofilm Formation
The detection of biofilm formation was carried out using the method of Christensen et al,21 with minor modifications according to Manandhar et al.22 Briefly, an overnight bacterial culture was grown at 37°C in Tryptic Soy broth (TSB) supplemented with 1% (w/v) glucose followed by 1:100 dilution using fresh media. The diluted culture (200 µL) was transferred to a 96-well microtiter plate. Uninoculated Tryptic Soy broth was used as negative control. The plates were incubated aerobically for 48 hours at 37°C. After incubation, the content of each well was removed and wells were washed three times with 200 µL of sterile distilled water, to remove planktonic cells. Adherent biofilms were fixed with 95% ethanol for 15 minutes followed by staining with 100 µL of 1% crystal violet for 5 minutes. Unbound stain was removed and the wells were washed with 200 µL of sterile distilled water. The water was removed, and the plate was air dried; then 100 µL of ethanol was added to dissolve the formed biofilm. The optical density (OD) of the stained biofilms was measured at 570 nm, using a micro plate reader (BioTek, USA). All isolates were tested in triplicate. The average OD values were calculated, for each tested strain and negative controls. The cut-off value (ODc) was established, according to Nasr et al,23 to be equal to average OD of negative control+(3*SD of negative control) and the test strains were categorized according to their biofilm formation into:
Non biofilm producer: OD≤ODc
Weak biofilm producer: ODc<OD≤2*ODc
Moderate biofilm producer: 2*ODc<OD≤4*ODc
Strong biofilm producer: 4*ODc<OD
Isolates that were strong or moderate biofilm-forming and ciprofloxacin-resistant (24 isolates) were selected for further testing.
Preparation of Ciprofloxacin-Loaded Niosomes
Ciprofloxacin (Global pharmaceutical industries, Egypt) niosomes were prepared by thin-film hydration method. Two preparations were tested, that differed in composition. Preparation I was composed of span 40 (Loba Chemie co., India) and tween 40 (Qualikems, India) mixed with cholesterol (El Nasr pharmaceutical industries, Egypt) at a molar ratio of 3:3:4, respectively, and dissolved in 10 mL chloroform (Fisher Scientific, USA).14 Preparation II was composed of span 60 (Loba Chemie co., India) and tween 40 mixed with cholesterol at a molar ratio of 1:1:2, respectively, and dissolved in 3 mL chloroform.24 Glass beads were added to both mixtures and the chloroform was evaporated under reduced pressure, using a rotary evaporator (Heidolph, Germany) at 60°C and at 45°C for preparations I and II, respectively. The lipid film formed on the flask wall was hydrated with 5 mL of ciprofloxacin solution (4 mg/mL), or with 5 mL of sterile distilled water in the case of blank niosomes, followed by shaking in a water bath for 1 hour at 110 rpm. The obtained niosomes were sonicated for 5 minutes to reduce the particle size.
Determination of Minimum Inhibitory Concentration (MIC)
The MIC of free ciprofloxacin solution and ciprofloxacin-loaded niosomes was determined by agar dilution method18 at a concentration range from 2–1,024 µg/mL of the original solution. Ciprofloxacin-free niosomes were tested as a blank, using the same dilution procedure used with ciprofloxacin-loaded niosomes. MIC was determined as the lowest concentration of the agent that completely inhibits the growth of the microorganisms. The niosomal preparation showing the better antimicrobial activity was selected for further study.
Characterization of Ciprofloxacin-Loaded Noisomes
Size Analysis, Size Distribution, and Zeta Potential Measurement
The mean size, size distribution, and zeta potential of the selected ciprofloxacin-loaded niosomal preparation were determined by Dynamic Light Scattering technique in a Malvern Nano ZS light scattering apparatus (Malvern Instruments Ltd., UK). Freshly prepared niosomal dispersions were diluted (1:20) with deionized water, to avoid multiple scattering phenomena due to interparticle interactions, and size analysis was carried out at 25°C. The z-average diameter and the polydispersity index of the niosomes were determined and their zeta potential was measured.
Entrapment Efficiency (EE)
The entrapment efficiency was calculated by determining the amount of ciprofloxacin that had not been entrapped in the formed niosomes (free ciprofloxacin). The formed niosomes were separated by ultracentrifugation (RemiLab., India) at 14,000 rpm for 1 hour at 4°C. Ciprofloxacin, in the supernatant, was assayed spectrophotometrically at 276 nm. Ciprofloxacin concentration was determined using a constructed standard curve. The EE% was calculated with reference to total amount of ciprofloxacin added during the preparation.13
Detection of Possible Mutations in Quinolone Resistance-Determining Regions (QRDRs)
Four isolates were selected for testing (5, 21, 31, and 57) in which ciprofloxacin-loaded niosomes have reduced the MIC by ≥8-fold, compared to that of free ciprofloxacin; however, only isolates 5 and 21 have reverted to the susceptible phenotype. DNA was extracted by boiling method,25 and polymerase chain reaction was performed to amplify QRDRs of the gyrA, gyrB, grlA, and grlB genes, according to Horii et al26 with the following primers; grlA (F: 5ʹ-GATGAGGAGGAAATCTAG and R: 5ʹ-GTTGGAAAATCGGACCTT), grlB (F: 5ʹ-GACAATTGTCTAAATCACTTGTG and R: 5ʹ-CATCAGTCATAATAATTACAC), gyrA (F: 5ʹ-GCGATGAGTGTTATCGTTGCT and R: 5ʹ-CAGGACCTTCAATATCCTCC), and gyrB (F: 5ʹ-CAGCGTTAGATGTAGCAAGC and R: 5ʹ-CGATTTTGTGATATCTTGCTTTCG). Polymerase chain reaction products were purified using GeneJet PCR purification kit (Thermo Fisher Scientific, USA) and sequenced by ABITM3500 Genetic Analyzer DNA sequencer (Applied Biosystems, Foster City, CA). Mutations were detected by translation and alignment, of the resulting peptide sequences, to that of the corresponding translated region of S. aureus ATCC 25923, using the Clustal Omega program (www.ebi.ac.uk/Tools/msa/clustalo) with the default settings.
Effect of Niosomal Encapsulation on Anti-Biofilm Activity of Ciprofloxacin
Determination of Minimum Biofilm Inhibitory Concentration (MBIC)
Anti-biofilm activity of the selected ciprofloxacin-loaded niosomes was compared with that of the free drug, through determination of MBICs. Biofilms were allowed to form, as described previously (Detection of Biofilm Formation section), in the presence of different concentrations of free ciprofloxacin and ciprofloxacin-loaded niosomes (1–512 µg/mL). Blank niosomes were diluted and tested similarly. The MBIC was determined as the lowest concentration of the test agent with a mean biofilm OD less than or equal to the optical density of negative control, at 570 nm. Uninoculated Tryptic Soy broth (Oxoid, USA) was used as a negative control, while plain S. aureus culture was used as a positive control.15 Experiments were run in duplicate.
Determination of Minimum Biofilm Eradication Concentration (MBEC)
MBEC measures the ability of the test agents to disturb already formed biofilms. The tested strains were allowed to form biofilms, as under determination of biofilm formation (Detection of Biofilm Formation section). Wells were then washed by sterile distilled water, and 200 μL of each tested concentration (1–512 µg/mL) of free ciprofloxacin solution and ciprofloxacin-loaded niosomes were added. Similar dilutions of blank niosomes were also tested. Plates were incubated for 24 hours aerobically at 37°C and the OD of biofilms was measured, at 570 nm. Uninoculated Tryptic Soy broth (Oxoid, USA) was used as a negative control, while plain S. aureus culture was used as a positive control. The MBEC was determined as the lowest concentration giving a mean biofilm OD less than or equal to the OD of negative control.15 Experiments were run in duplicate.
Biofilm Visualization by Scanning Electron Microscopy
Isolate 34 (highly ciprofloxacin resistant isolate with MIC=1,024 µg/mL) was selected for visualization of effect of ciprofloxacin and ciprofloxacin-loaded niosomes on biofilm formation. The isolate was incubated overnight with sub-inhibitory concentration (1/8 MIC) of either free ciprofloxacin or ciprofloxacin-loaded niosomes; where biofilms were allowed to form on the surface of sterile cover slips mounted in the bottom of the 6-well tissue culture plates. At the end of the incubation period, the medium was removed, the wells were gently washed twice with phosphate buffered saline (PH=7.4), and biofilms were fixed with 99% methanol for 3 minutes. Cover slips were gently removed from the wells, affixed to the scanning electron microscope (SEM) stubs with double-sided carbon tape, and coated with gold in a SPI-Module. They were photographed using scanning electron microscope (JEOL, JSM-5200 LV SEM, Japan) with the voltage set to 25 kV at 5,000x magnification power.
Results
Antimicrobial Susceptibility of Collected S. aureus Isolates
Among tested S. aureus isolates, 45 (76.27%) were resistant to fluoroquinolone class (ciprofloxacin and levofloxacin). In addition, 47 (79.6%) of the isolates were displaying MRSA phenotype, while 53 isolates (89.83 %) were found to be multidrug resistant isolates. Vancomycin resistance was detected in 33 (56%) of the collected isolates (Table S1).
Biofilm Production
All except two isolates were capable of biofilm production to variable degrees. About 15% of the isolates were strong biofilm producers (n=9), while 25% were moderate biofilm producers (n=15). The remaining biofilm forming isolates were weak biofilm producer (33 isolates). Isolates that were strong or moderate biofilm-forming and ciprofloxacin-resistant (24 isolates) were selected for further testing.
Antimicrobial Activity of Ciprofloxacin-Loaded Niosomal Preparations
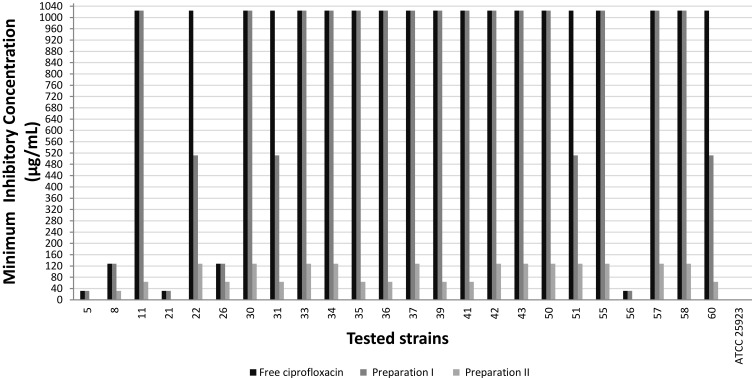
The MIC of the two ciprofloxacin-loaded niosomal preparations was compared to that of the free ciprofloxacin, against S. aureus ATCC 25923 and the selected S. aureus clinical isolates (n=24). S. aureus ATCC 25923 had MIC≤1 µg/mL, against free ciprofloxacin and against both ciprofloxacin-loaded niosomal preparations. Niosomal preparation I reduced the MIC by 2-fold in only four clinical isolates, while preparation II reduced the MIC of most clinical isolates by 8–32-fold; with three isolates having lost their resistance phenotype (MIC≤1 µg/mL; Figure 1). Therefore, ciprofloxacin-loaded preparation II was selected for further testing and evaluation. The blank noisome preparations lacked any antimicrobial activity.
Figure 1.
Minimum inhibitory concentration of free ciprofloxacin and ciprofloxacin-loaded niosomal preparations against tested S. aureus strains.
Characterization of Ciprofloxacin-Loaded Niosomes
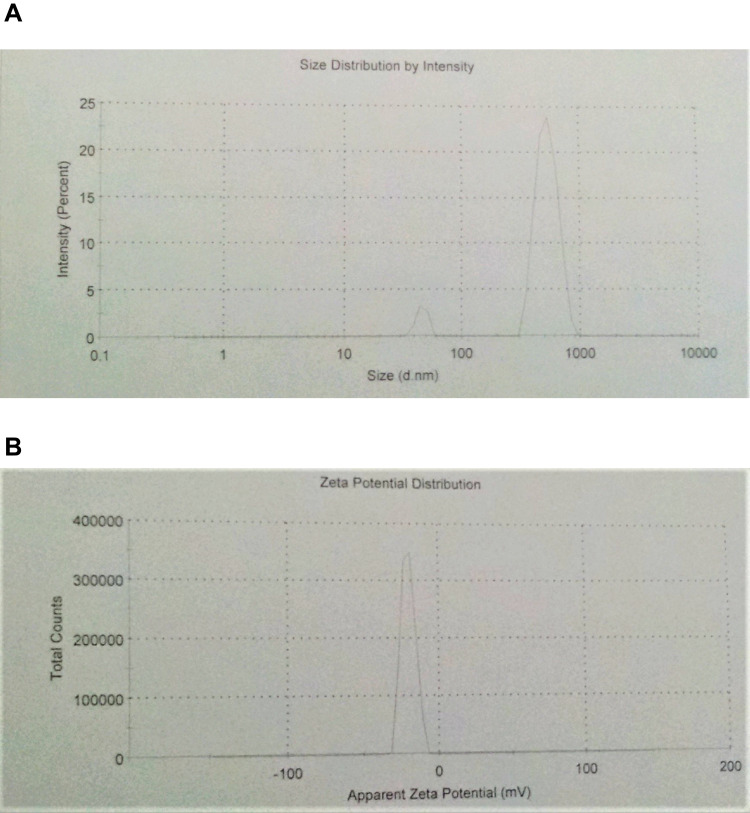
The size, size distribution, and zeta potential of ciprofloxacin-loaded niosomal preparation II were determined by dynamic light scattering. The number-averaged hydrodynamic diameter was 494.4 nm with 93% of the particles exhibiting a hydrodynamic diameter of 525.6 nm (SD=108.6). The polydispersity index was 0.337 and the zeta-potential value was −20.6mV (Figure 2).
Figure 2.
Characterization of the prepared ciprofloxacin-loaded niosomes using dynamic light scattering technique. (A) Size distribution histogram, (B) zeta potential distribution.
Entrapment efficiency of the ciprofloxacin in niosomes was 77.45%.
Detection of Mutation in QRDRs of Selected Isolates
A polymerase chain reaction was carried out to amplify QRDRs in selected isolates followed by sequencing. The obtained sequences were translated and aligned to the corresponding regions of S. aureus ATCC 25923. Results of alignment revealed mutations in isolates 57 and 31 but not isolates 5 or 21. One mutation was detected in the grlB gene of isolate 57 (Pro451Ser) and three mutations were detected in isolate 31; two mutations in grlA gene (Ser80Phe and Ser143Pro) and one mutation in gyrA gene (Ser84Leu).
Anti-Biofilm Activity of the Formulated Niosomes
The anti-biofilm activity of ciprofloxacin-loaded niosomes was compared with that of the free ciprofloxacin, through determination of MBICs and MBECs. Ciprofloxacin-loaded niosomes reduced the MBIC by 2–4-fold in 14 out of 24 of the tested isolates, compared to free ciprofloxacin. They also reduced the MBEC of 15 isolates by 2–4-fold compared to free ciprofloxacin (Table 1). Blank niosomes failed to inhibit biofilm formation or eradicate already formed biofilms at the tested concentrations.
Table 1.
Minimum Biofilm Inhibitory Concentration and Minimum Biofilm Eradication Concentration of Free Ciprofloxacin Solution and Ciprofloxacin-Loaded Noisomal Preparation II against S. aureus Isolates
| Isolates No. | Minimum Biofilm Inhibitory Concentration (µg/mL) | Minimum Biofilm Eradication Concentration (µg/mL) | ||
|---|---|---|---|---|
| Free Ciprofloxacin | Niosomal Ciprofloxacin | Free Ciprofloxacin | Niosomal Ciprofloxacin | |
| 5 | 8 | 8 | 32 | 32 |
| 8 | 128 | 32 | 64 | 32 |
| 11 | 64 | 32 | 256 | 128 |
| 21 | 32 | 16 | 32 | 8 |
| 22 | 128 | 32 | 512 | 256 |
| 26 | 4 | 4 | 256 | 256 |
| 30 | 16 | 16 | 32 | 16 |
| 31 | 16 | 16 | 32 | 32 |
| 33 | 16 | 8 | 256 | 128 |
| 34 | 32 | 8 | 256 | 128 |
| 35 | 128 | 128 | 256 | 256 |
| 36 | 32 | 16 | 512 | 256 |
| 37 | 64 | 32 | >512 | >512 |
| 39 | 32 | 16 | 128 | 32 |
| 41 | 32 | 16 | 256 | 16 |
| 42 | 128 | 128 | 32 | 16 |
| 43 | 32 | 16 | 128 | 128 |
| 50 | 64 | 64 | 128 | 128 |
| 51 | 32 | 16 | 128 | 128 |
| 55 | 16 | 16 | 512 | 256 |
| 56 | 32 | 16 | 256 | 64 |
| 57 | 32 | 32 | 128 | 128 |
| 58 | 128 | 32 | 256 | 64 |
| 60 | 16 | 16 | 128 | 16 |
| Staphylococcus aureus ATCC 25923 | 32 | 16 | 512 | 128 |
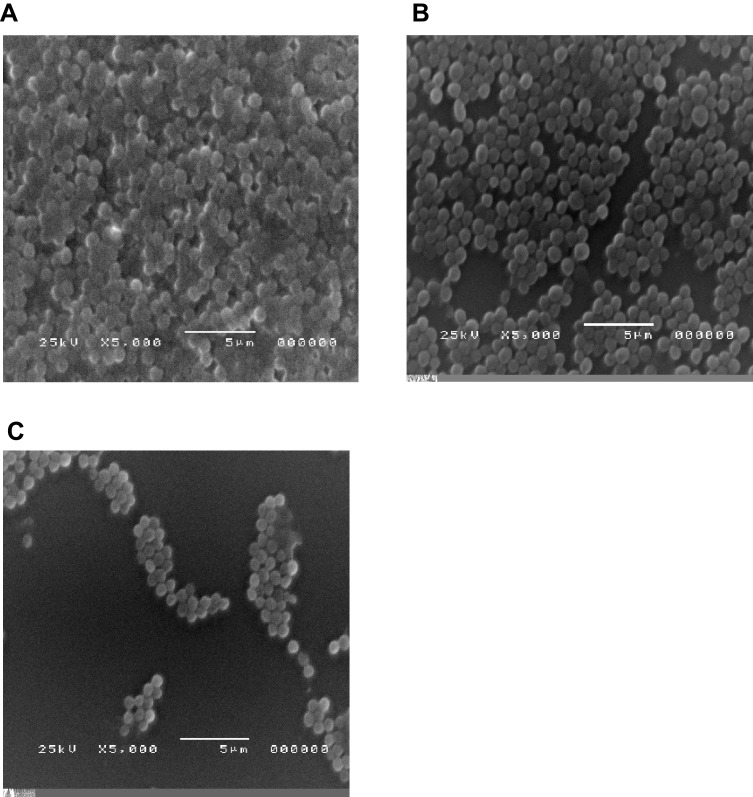
This effect of ciprofloxacin-loaded niosomes on inhibition of biofilm formation was further confirmed by the electron micrographs of biofilms formed in the presence of either free ciprofloxacin or ciprofloxacin-loaded niosomes compared to those formed in the absence of ciprofloxacin (Figure 3). It was clear that the density of the formed biofilms was greatly reduced by the presence of 1/8 MIC of ciprofloxacin-loaded niosomes compared to that formed in untreated culture or in the presence of 1/8 MIC of free ciprofloxacin. Free ciprofloxacin slightly reduced the formed biofilm compared to the untreated culture.
Figure 3.
Visualization of S. aureus biofilms formed in the presence of subinhibitory concentration of free ciprofloxacin and ciprofloxacin-loaded niosomes, using an electron microscope. (A) Untreated culture (positive control), (B) culture in the presence of 1/8 MIC of ciprofloxacin, (C) culture in the presence of 1/8 MIC of ciprofloxacin-loaded niosomes.
Discussion
Microbial resistance, to available antibiotics, is a major concern.4 The use of antibiotics in animal feeding has helped the widespread emergence of resistant strains.27 Treating infections caused by microbial resistant organisms is hampered by the increasing rate of multidrug resistant isolates, limiting the available therapeutic options.28 This was the case with S. aureus isolates collected in this study, where nearly all isolates were multidrug resistant, in addition to the high percentage of detected MRSA (79.6%) and the few drug options available for MRSA treatment. What worsen the condition is the detected high rate of vancomycin resistance (56%), where vancomycin is the drug of choice for MRSA treatment.6 Recently, there has been a reported rise in vancomycin resistance rates where a similar rate was recorded previously in Egypt.29,30 Also, most of the collected isolates were biofilm forming, which further complicates the treatment problem.31
Niosomal formulations of antimicrobial agents represent a promising tool for enhancement of their activity.32 Two ciprofloxacin-loaded niosomal preparations were tested; preparation II had an enhanced antimicrobial activity against ciprofloxacin resistant clinical isolates compared to preparation I. Encapsulation of ciprofloxacin, in niosomal preparation II, reduced its MIC by 8–32-fold in most isolates, and caused reversal of resistance phenotype in three isolates. A previous study by Satish et al13 has recorded a reduction of only 2–4-fold in ciprofloxacin MIC, by niosomal formulations tested on ciprofloxacin resistant isolates.
The enhanced activity of niosomes was attributed to either vesicle adsorption to the microbial surface with subsequent drug release near the bacterial cell or adsorption followed by uptake of the drug enclosed within the lipophilic system by endocytosis.33 Another important factor in enhanced activity of niosomes is their nanosize, which facilitates their transport. These proposed mechanisms of enhancement were confirmed by the lack of any antimicrobial activity of blank niosomes, which was in agreement with the findings of other studies.15,16
The difference in the antimicrobial effect, between the two niosomal preparations or the difference from that reported in Satish et al's study, may have resulted from the effect of the surfactant used. Span 60 (used in preparation II) has a longer saturated alkyl chain than that of span 40, where the length of alkyl chain of used surfactants is an important factor affecting the permeability of the prepared niosomes. Long chain products cause the niosomal preparations to be less leaky and more retaining of encapsulated drug.34 In addition, the lower hydrophile-lipophile balance of the span used (span 60=4.7, span 40=6.7) may be responsible for further reduction of the size of the produced vesicle and higher contact with the outer surface of the microorganism, enhancing its antimicrobial activity.35 Cholesterol is one of the common additives included for stabilization of the niosomal formulations. It abolishes the gel-to-lipid phase transition of niosomal systems which prevents the leakage of drug out of the niosomes. Formulation of the niosomes with a molar ratio of 1:1 for the surfactants and cholesterol was reported to be the most beneficial in making the niosomal membrane more compact and well organized.34
The results of the characterization tests of preparation II confirmed their small size (≈500 nm). Akbari et al14 have tested different formulations with variable compositions, where the diameter of the produced niosomes ranged from 163–1,100 nm. Other studies have reported a higher diameter for niosomal preparations that reached 15 µm.13,15 In addition, the negative zeta potential values recorded (−20.6 mV) are highly advantageous in terms of high stability of the dispersed niosomes resulting from the electrostatic repulsion between the particles. The negative values recorded for zeta potential have been reported previously for niosomal preparations; however, with smaller and higher absolute values.14,15 This negative values may be attributed to the effect of the hydroxyl group in cholesterol molecules or the physicochemical effect of the loaded drug, as reported by Manosroi et al36 and Akbari et al,14 respectively.
To further elucidate the reason for the variable antimicrobial effect of ciprofloxacin-loaded niosomes on different tested strains, we tested the possible presence of mutations in QRDRs of selected isolates. Non-synonymous mutations were detected in isolates 31 and 57. Similar mutations were detected previously in quinolone resistant isolates.26
Isolates 31 and 57 had a high level of ciprofloxacin resistance (MIC=1,024 µg/mL). Ciprofloxacin-loaded niosomal preparation II reduced their MIC by 8–16-fold only but could not revert it to the susceptible phenotype. However, isolates 5 and 21 showed low level ciprofloxacin resistance (MIC=32 µg/mL) and reverted to susceptible phenotype (MIC˂2 μg/mL) on using ciprofloxacin-loaded niosomes. Low level ciprofloxacin resistance usually arises from over-expression of efflux pumps rather than mutations in QRDRs.37 Therefore, the effect of niosomes in enhancing ciprofloxacin action, in these two isolates, may have arisen from the small size of niosomes that enhanced their penetration into microbial cells, overcoming the effect of efflux pumps.
Ciprofloxacin-loaded niosomes were also able to reduce MBIC by 2–4-fold in 58% of tested isolates, compared to free ciprofloxacin. Additionally, they reduced MBEC in 15 of the tested isolates (62.5%). This antibiofilm activity was further confirmed by the images obtained by electron microscopy. The antibiofilm activity of niosomal formulations of other antimicrobials, against a wide variety of pathogens was reported previously.15,16,36 Manandhar et al38 reported that clinical isolates of S. aureus tend to develop biofilms on medical devices, where S. aureus in biofilm displays high rates of antimicrobial resistance, multidrug resistance and methicillin resistance compared to non-biofilm-forming bacteria.39 Therefore, treatment of S. aureus infection usually requires inhibition and eradication of biofilm formation in addition to bacterial inhibition.
Conclusion
Ciprofloxacin-loaded niosomes offer an alternative approach to restore the efficacy of ciprofloxacin, using a stable low-cost method and preserving the newer effective agents for treatment of more resistant infection. To our knowledge, this is the first report on the effect of ciprofloxacin-loaded niosomes in inhibition of biofilm formation and eradication of formed biofilms, in addition to their pronounced antimicrobial activity against ciprofloxacin-resistant S. aureus clinical isolates.
Ethical Approval
The study was approved by the ethical committee of the Faculty of Pharmacy, Cairo University with approval number: MI (1767).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khatoon Z, McTiernan CD, Suuronen EJ, Mah TF, Alarcon EI. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4(12):e01067. doi: 10.1016/j.heliyon.2018.e01067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao G, Hochwalt PC, Usui ML, et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 2010;18(5):467–477. doi: 10.1111/j.1524-475X.2010.00608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S.: Department of Health and Human Services, CDC; 2019. [Google Scholar]
- 5.Khan TM, Kok YL, Bukhsh A, et al. Incidence of methicillin resistant Staphylococcus aureus (MRSA) in burn intensive care unit: a systematic review. Germs. 2018;8(3):113–125. doi: 10.18683/germs.2018.1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boswihi SS, Udo EE. Methicillin-resistant Staphylococcus aureus: an update on the epidemiology, treatment options and infection control. Curr Med Res Pract. 2018;8(1):18–24. doi: 10.1016/j.cmrp.2018.01.001 [DOI] [Google Scholar]
- 7.Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8(1):76. doi: 10.1186/s13756-019-0533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore DM, Blaser M, Carrs O. Discovery research: the scientific challenge of finding new antibiotics. J Antimicrob Chemother. 2011;66(9):1941–1944. doi: 10.1093/jac/dkr262 [DOI] [PubMed] [Google Scholar]
- 9.Pinto RM, Lopes-de-Campos D, Martins MCL, Van Dijck P, Nunes C, Reis S. Impact of nanosystems in Staphylococcus aureus biofilms treatment. FEMS Microbiol Rev. 2019;43(6):622–641. doi: 10.1093/femsre/fuz021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugazhendhi A, Kumar SS, Manikandan M, Saravanan M. Photocatalytic properties and antimicrobial efficacy of Fe doped CuO nanoparticles against the pathogenic bacteria and fungi. Microb Pathog. 2018;122:84–89. doi: 10.1016/j.micpath.2018.06.016 [DOI] [PubMed] [Google Scholar]
- 11.Gharbavi M, Amani J, Kheiri-Manjili H, Danafar H, Sharafi A. Niosome: a promising nanocarrier for natural drug delivery through blood-brain barrier. Adv Pharmacol Sci. 2018;2018:6847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moazeni E, Gilani K, Sotoudegan F, et al. Formulation and in vitro evaluation of ciprofloxacin containing niosomes for pulmonary delivery. J Microencapsul. 2010;27(7):618–627. doi: 10.3109/02652048.2010.506579 [DOI] [PubMed] [Google Scholar]
- 13.Satish J, Amusa AS, Gopalakrishna P. In vitro activities of fluoroquinolones entrapped in non-ionic surfactant vesicles against ciprofloxacin-resistant bacteria strains. J Pharm Technol Drug Resr. 2012;1(1):5. doi: 10.7243/2050-120X-1-5 [DOI] [Google Scholar]
- 14.Akbari V, Abedi D, Pardakhty A, Sadeghi-Aliabadi H. Ciprofloxacin nano-niosomes for targeting intracellular infections: an in vitro evaluation. J Nanopart Res. 2013;15(4):1556. [Google Scholar]
- 15.Barakat HS, Kassem MA, El-Khordagui LK, Khalafallah NM. Vancomycin-eluting niosomes: a new approach to the inhibition of staphylococcal biofilm on abiotic surfaces. AAPS PharmSciTech. 2014;15(5):1263–1274. doi: 10.1208/s12249-014-0141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelaziz AA, Elbanna TE, Sonbol FI, Gamaleldin NM, El Maghraby GM. Optimization of niosomes for enhanced antibacterial activity and reduced bacterial resistance: in vitro and in vivo evaluation. Expert Opin Drug Deliv. 2014;12(2):163–80. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial disk susceptibility tests; approved standard 11th In: CLSI Document M02-A11. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 18.CLSI. Methods for dilution antimicrobial susceptibility tests f or bacteria that grow aerobically; approved standard Ninth In: CLSI Document M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing 27th In: CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 20.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 21.Christensen GD, Simpson WA, Younger JJ, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(6):996–1006. doi: 10.1128/JCM.22.6.996-1006.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Evaluation of methods to detect in vitro biofilm formation by staphylococcal clinical isolates. BMC Res Notes. 2018;11(1):714. doi: 10.1186/s13104-018-3820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasr RA, AbuShady HM, Hussein HS. Biofilm formation and presence of icaAD gene in clinical isolates of staphylococci. Egypt J Med Hum Genet. 2012;13(3):269–274. doi: 10.1016/j.ejmhg.2012.04.007 [DOI] [Google Scholar]
- 24.Mohamed HB, El-Shanawany SM, Hamad MA, Elsabahy M. Niosomes: a strategy toward prevention of clinically significant drug incompatibilities. Sci Rep. 2017;7(1):6340. doi: 10.1038/s41598-017-06955-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green MR, Sambrook J. Molecular cloning: A laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2014. [Google Scholar]
- 26.Horii T, Suzuki Y, Monji A, et al. Detection of mutations in quinolone resistance-determining regions in levofloxacin- and methicillin-resistant Staphylococcus aureus: effects of the mutations on fluoroquinolone MICs. Diagn Microbiol Infect Dis. 2003;46(2):139–145. doi: 10.1016/S0732-8893(03)00037-3 [DOI] [PubMed] [Google Scholar]
- 27.Boovaragamoorthy GM, Anbazhagan M, Piruthiviraj P, et al. Clinically important microbial diversity and its antibiotic resistance pattern towards various drugs. J Infect Public Health. 2019;12(6):783–788. doi: 10.1016/j.jiph.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 28.van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am. 2016;30(2):377–390. doi: 10.1016/j.idc.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghasemian A, Najar Peerayeh S, Bakhshi B, Mirzaee M. Several virulence factors of multidrug-resistant Staphylococcus aureus isolates from hospitalized patients in Tehran. Int J Enteric Pathog. 2015;3(2). doi: 10.17795/ijep25196 [DOI] [Google Scholar]
- 30.Al-Amery K, Elhariri M, Elsayed A, et al. Vancomycin-resistant Staphylococcus aureus isolated from camel meat and slaughterhouse workers in Egypt. Antimicrob Resist Infect Control. 2019;8(1):129. doi: 10.1186/s13756-019-0585-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeed A, Ahsan F, Nawaz M, Iqbal K, Rehman KU, Ijaz T. Incidence of vancomycin resistant phenotype of the methicillin resistant Staphylococcus aureus isolated from a tertiary care hospital in Lahore. Antibiotics. 2019;9(1):3. doi: 10.3390/antibiotics9010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo P, Lim C, Chye S, Kiong Ling A, Koh R. Niosomes: a review of their structure, properties, methods of preparation, and medical applications. Asian Biomed. 2018;11(4):301–314. doi: 10.1515/abm-2018-0002 [DOI] [Google Scholar]
- 33.Daraee H, Etemadi A, Kouhi M, Alimirzalu S, Akbarzadeh A. Application of liposomes in medicine and drug delivery. Artif Cells Nanomed Biotechnol. 2016;44(1):381–391. doi: 10.3109/21691401.2014.953633 [DOI] [PubMed] [Google Scholar]
- 34.Hao Y, Zhao F, Li N, Yang Y, Li K. Studies on a high encapsulation of colchicine by a niosome system. Int J Pharm. 2002;244(1–2):73–80. doi: 10.1016/S0378-5173(02)00301-0 [DOI] [PubMed] [Google Scholar]
- 35.Yoshioka T, Sternberg B, Florence AT. Preparation and properties of vesicles (niosomes) of sorbitan monoesters (Span 20, 40, 60 and 80) and a sorbitan triester (Span 85). Int J Pharm. 1994;105(1):1–6. doi: 10.1016/0378-5173(94)90228-3 [DOI] [Google Scholar]
- 36.Manosroi A, Khanrin P, Lohcharoenkal W, et al. Transdermal absorption enhancement through rat skin of gallidermin loaded in niosomes. Int J Pharm. 2010;392(1–2):304–310. doi: 10.1016/j.ijpharm.2010.03.064 [DOI] [PubMed] [Google Scholar]
- 37.Sulavik MC, Barg NL. Examination of methicillin-resistant and methicillin-susceptible Staphylococcus aureus mutants with low-level fluoroquinolone resistance. Antimicrob Agents Chemother. 1998;42(12):3317–3319. doi: 10.1128/AAC.42.12.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Biofilm producing clinical Staphylococcus aureus isolates augmented prevalence of antibiotic resistant cases in tertiary care hospitals of Nepal. Front Microbiol. 2018;9:2749. doi: 10.3389/fmicb.2018.02749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. 2018;11:25–32. doi: 10.2147/IJGM.S153268 [DOI] [PMC free article] [PubMed] [Google Scholar]