Abstract
The signal transducer and activator of transcription 3 (STAT3) protein is constitutively activated in several cancers. STAT3 activity can be blocked by inhibiting its Src Homology 2 (SH2) domain, but phosphotyrosine and its isosteres have poor bioavailability. In this work, we develop peptide-based inhibitors of STAT3-SH2 by combining chemical strategies that have proven effective for targeting other SH2 domains. These strategies include a STAT3-specific selectivity sequence, non-hydrolyzable phosphotyrosine isosteres, and a high-efficiency cell-penetrating peptide. Peptides that combined these three strategies had substantial biological stability and cytosolic delivery, as measured using highly quantitative cell-based assays. However, these peptides did not inhibit STAT3 activity in cells. By comparing in vitro binding affinity, cell penetration, and proteolytic stability, this work explores the delicate balance of factors that contribute to biological activity for peptidic inhibitors of STAT3.
Keywords: STAT3, SH2 domain, phosphonates, cell-penetrating peptide, chloroalkane penetration assay, transcription factor
1. Introduction
The signal transducer and activator of transcription 3 (STAT3) protein is a transcription factor frequently dysregulated in hematological malignancies such as leukemias and lymphomas, as well as in solid tumors such as breast cancers, prostate cancers, and glioblastoma multiforme.1–3 Cytokine stimulation with IL-6 or oncostatin M leads to phosphorylation of gp130, which recruits STAT3 to the cell membrane via its Src Homology 2 (SH2) domain. Once localized to the plasma membrane, STAT3 is phosphorylated by Janus kinases. Phosphorylated STAT3 dimerizes via its SH2 domains, and the dimer translocates to the nucleus where it acts as a transcription factor.1,4 STAT3 upregulates numerous genes critical for cancer cell survival, including: survivin, Bcl-2, and Bcl-xL, which allow evasion of apoptosis; VEGF, which promotes angiogenesis; MMPs, which enhance cell motility; and PD-L1, which allows for immune evasion.5–9 Importantly, STAT3 was shown to be necessary for malignant transformation of mouse fibroblasts, but not for normal fibroblast growth, suggesting a potential therapeutic window as a cancer target.10
Membrane localization and dimerization are both critical steps in STAT3 activity, and both require the function of the STAT3 SH2 domain. This has prompted the hypothesis that inhibitors of the STAT3 SH2 domain could block STAT3 activity.11,12 However, SH2 domains are challenging targets for drug discovery. SH2 domains recognize phosphotyrosine (pTyr)-containing sequences, yet pTyr is rapidly hydrolyzed in the cytosol by protein tyrosine phosphatases. Additionally, pharmacological agents with pTyr have poor cytosolic penetration, which is typically ascribed to their multiple negative charges.13 Numerous pTyr isosteres have been developed over the last several decades in order to address these issues. Phosphonates, including phosphonomethyl phenylalanine (Pmp) and difluorophosphonomethyl phenylalanine (F2Pmp), have been applied as phosphatase-stable SH2 domain inhibitors.14–16 However, these and other pTyr isosteres still have multiple negative charges, and typically still suffer from poor cytosolic delivery.13
Significant efforts have been undertaken to develop more cell-penetrant small molecule inhibitors containing diverse pTyr isosteres including phosphonates, salicylates, benzoates, sulfonamides, benzothiophenes and others.17–27 These efforts have generated inhibitors with nanomolar binding affinities and potent STAT3 inhibition in cells. However, to date, no STAT3 small molecule inhibitor (or any other STAT3-targeted therapy) has attained FDA approval, so there is an ongoing need for STAT3 inhibitor development. Other SH2 domains constitute promising drug targets, but even small molecule ligands (developed using traditional medicinal chemistry efforts involving high-throughput screening and extensive analog testing) present challenges of balancing affinity, selectivity and cell penetration. Given these difficulties for developing small molecule inhibitors, we and others have continued to explore peptides as potential starting points for SH2 domain inhibitors.13 In this work, we revisited an earlier approach to inhibiting STAT3-SH2 by starting with a selective phosphopeptide ligand and addressing the major challenges of biological degradation and cell penetration.
Cell-penetrating peptides (CPPs) offer a promising solution for the cytosolic delivery of peptidomimetics containing pTyr or pTyr isosteres.28–30 CPPs including Tat, polyarginine, and penetratin have been used to deliver peptides containing pTyr and pTyr isosteres, resulting in inhibition of various SH2 domains and protein tyrosine phosphatases.31–35 Most of these prior efforts delivered phosphopeptides, which are prone to dephosphorylation in serum (vide infra) and in the cytosol. A handful of prior efforts have incorporated pTyr isosteres to prevent phosphate hydrolysis.34,35 For example, Watson and colleagues developed a bicyclic peptide inhibitor of the Grb7 SH2 domain containing a carboxyphenylalanine (cF) residue, and conjugated it to penetratin to promote cell uptake. This peptide inhibited Grb7 binding to FAK, HER2, and SHC in SKBR3 breast cancer cells when assessed by coimmunoprecipitation.34 Additionally, Lian and colleagues incorporated F2Pmp into a bicyclic peptide targeting the phosphatase PTP1B, in which one macrocycle incorporated a pseudosubstrate motif and the other incorporated a cyclic CPP, cyclo(FφR4) where φ is 2-naphthylalanine. This peptide produced a two-fold increase in insulin receptor phosphorylation in HepG2 liver cells when applied at 200 nM.35 These studies demonstrated that combining non-hydrolyzable pTyr isosteres, macrocyclization, and CPPs, one can develop relatively potent and cell-penetrant inhibitors of SH2 domains and protein tyrosine phosphatases.
Based on this prior work, we sought to apply a similar strategy to the inhibition of STAT3. To take advantage of recent advances in CPP development,36–38 we chose to apply an improved version of cyclo(FφR4), CPP12, to deliver STAT3-targeted peptides to the cytosol. CPP12 was demonstrated by Qian and colleagues to improve cytosolic delivery efficiency by 6-fold compared to cyclo(FφR4), and 30 to 60-fold compared to polyarginine and Tat, respectively.36 We found that STAT3-selective binding sequences from gp130 could be substituted with F2Pmp and maintain micromolar binding affinity for STAT3. Since micromolar inhibitors of STAT3 have been shown to be effective in cellular models of STAT3 activation, we tested CPP12-fused peptides to measure their effects on STAT3 activity in relevant STAT3-driven cancer cell lines. When the peptides did not act as effective STAT3 antagonists, we followed up by quantitatively measuring the serum stability, cell lysate stability, and cytosolic penetration of selected CPP12 fusions, producing a rich data set that addresses the delicate balance that must be achieved between target affinity, resistance to degradation, and cytosolic penetration for pTyr-mimetic SH2 domain inhibitors.
2. Results
2.1. Substituting pTyr with isosteres Pmp and F2Pmp
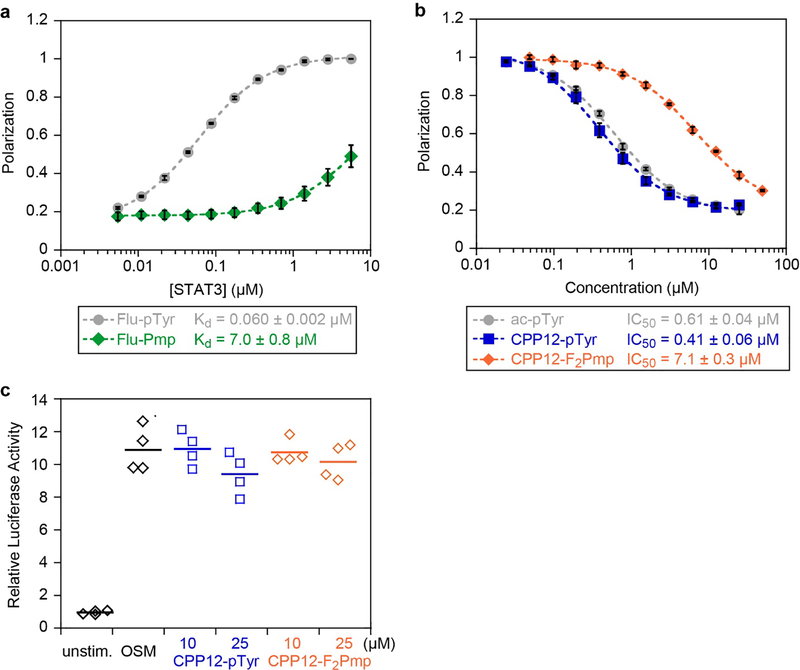
We began by directly substituting pTyr with phosphonomethyl phenylalanine (Pmp) in a previously described, high-affinity STAT3-binding peptide derived from phospho-gp130 (Ac-G(pTyr)LPQTV-NH2).39,40 While many phosphopeptide ligands of the STAT3 SH2 domain have been identified, the vast majority of these ligands have micromolar affinity for STAT3-SH2.40 This gp130-derived phosphopeptide demonstrated a 150 nM IC50 in an electrophoretic mobility shift assay measuring STAT3:DNA binding.40 In that study, phosphopeptides that incorporated a proline two residues C-terminal to the phosphotyrosine had the highest-affinity binding, likely due to optimal positioning of the glutamine residue to interact with the specificity pocket. We chose the gp130-derived phosphopeptide because it is the highest-affinity phosphopepetide ligand known for STAT3-SH2, and because it continues to be used as a scaffold for designing small molecule STAT3 inhibitors.27,41 We prepared fluorescein-labeled versions of the native pTyr peptide (flu-pTyr) and the Pmp-containing analog (flu-Pmp) to permit direct fluorescence polarization (FP) binding assays. flu-pTyr bound recombinant STAT3 with a Kd of 60 nM (Fig 1a, Fig S1a–c), consistent with previously observed binding affinities.39 However, flu-Pmp showed poor binding even at micromolar protein concentrations; this represented over a 100-fold loss in binding affinity (Fig 1a). We concluded that Pmp was not a suitable replacement for pTyr in the context of this STAT3-SH2 ligand.
Figure 1.
Binding affinities and cellular STAT3 inhibition of selected peptides. a) Fluorescence polarization binding data for selected peptides with recombinant STAT3. Fluorescein-labeled peptides were incubated at 10 nM with serial dilutions of STAT3 at room temperature for 45 min. Data points represent the averages of three biological replicates, each performed with three technical replicates, normalized to the maximum observed polarization (raw data shown in Fig. S1). Error bars show standard error of the mean for the three biological replicates. Kd values are the mean and standard error of the mean for three independent Kd curve fits to the three biological replicates. b) Competition FP for selected peptides with STAT3. flu-pTyr was incubated at 10 nM with 300 nM STAT3 protein and serial dilutions of peptide inhibitors at room temperature for 45 min. Data points represent the averages of three biological replicates, each performed with three technical replicates, normalized to the no-inhibitor control (raw data shown in Figs. S2,S3). Error bars show standard error of the mean for the three biological replicates. IC50 values are the mean and standard error of the mean for three independent curve fits to the three biological replicates. c) STAT3 transcription inhibition. STAT3-luc U3A fibrosarcoma cells were pretreated with 10 or 25 μM of CPP12-pTyr and CPP12-F2Pmp for 24 hours at 37 °C in DMEM supplemented with 10% FBS, followed by 6 hour stimulation with OSM (10 ng/mL). Controls included OSM-treated cells without peptide, and unstimulated cells. This experiment was performed with two biological replicates, each with two technical replicates (all four values displayed).
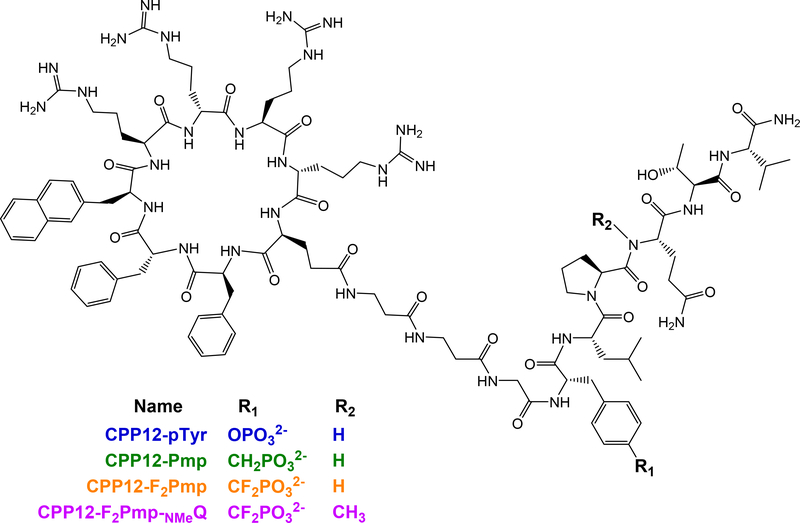
We anticipated that fusion to a CPP might alter binding affinity, so we prepared CPP12-pTyr, a pTyr-containing g130 sequence fused to CPP12 (Fig 2). CPP12 was synthesized N-terminal to the gp130-derived sequence with a linker of two β-alanines.36 Because CPP12 uses the N-terminus for cyclization, we did not attach fluorescein to these peptides, and instead used the competition FP assay to measure competitive inhibition of the STAT3-gp130 phosphopeptide interaction. CPP12-pTyr inhibited this interaction slightly better than ac-pTyr, with IC50 values of 410 nM and 610 nM, respectively (Fig 1b). This demonstrated that fusion to CPP12 did not greatly impact the inhibitory potency. Next, we substituted pTyr with difluorophosphonomethyl phenylalanine (F2Pmp), a pTyr isostere that has shown improved properties over Pmp for SH2 domain binding.15,16,42 CPP12-F2Pmp had an IC50 of 7.12 μM, which was 17-fold less potent than CPP12-pTyr (Fig 1b).
Figure 2.
Structures of CPP12-conjugated STAT3 SH2-targeting peptides.
While CPP12-F2Pmp was less potent than CPP12-pTyr, this was a significant improvement over the Pmp-containing peptide and merited additional characterization. Next, we tested CPP12-pTyr and CPP12-F2Pmp in a STAT3 transcriptional reporter assay in a U3A fibrosarcoma cell line.43 In previous work, we used this robust assay to test the effects of small molecules that affect the STAT3 pathway,43,44 and nearly identical assays have been used by others to monitor cellular activities of direct STAT3 SH2 domain inhibitors, often showing effects in the low micromolar range.24,26,45,46 However, even at 25 μM peptide incubated for 24 hours, neither CPP12-pTyr nor CPP12-F2Pmp inhibited OSM-stimulated STAT3 activity (Fig 1c). There are multiple examples in the literature of small molecule STAT3 SH2 domain inhibitors with single- to double-digit micromolar IC50 values when tested in vitro under the same conditions as our STAT3 fluorescence polarization assay, yet display potent cell-based phenotypes.19,20 As such, the lack of an observable phenotype with our peptides led us to investigate what barriers were responsible.
2.2. Cell penetration studies of CPP12-conjugated gp130 peptide fusions
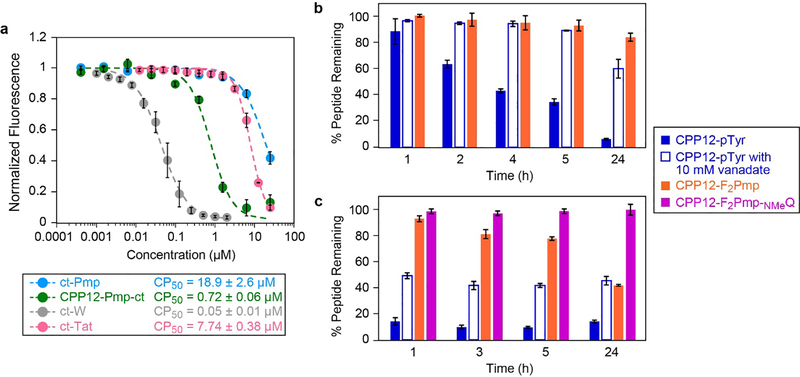
One possibility was that CPP12 was not sufficiently delivering the peptide into the cytosol. The Kritzer lab previously reported an assay for quantitatively comparing the cytosolic delivery of peptides and other biomolecules.47 This assay, called the Chloroalkane Penetration Assay or CAPA, uses a HeLa cell line that stably expresses HaloTag in the cytosol to measure the relative cytosolic penetration of molecules labeled with a small chloroalkane tag. We prepared a representative CPP12-linked peptide with a chloroalkane tag (CPP12-Pmp-ct, Fig. S5a) as well as a non-CPP-linked analog ct-Pmp. The cytosolic delivery of these peptides were compared to control molecules ct-W, a small-molecule with excellent cytosolic penetration, and ct-Tat, a molecule with moderate cytosolic penetration.47 ct-Pmp showed little cytosolic delivery except at the highest concentration tested (25 μM), while CPP12-Pmp-ct showed substantial cytosolic delivery at concentrations above 750 nM. By fitting IC50 curves to the dose-dependence data, we have derived “CP50 values” which allow direct, quantitative comparison of the extent of cytosolic delivery. CPP12-Pmp-ct had a CP50 value of 720 nM, while the Pmp-containing peptide without CPP12 (ct-Pmp) had a CP50 value of 18.9 μM (Fig 3a). A chloroalkane-linked version of the commonly used CPP Tat (ct-Tat) had a CP50 value of 7.74 μM under these conditions (Fig 3a). These data indicated that fusion to CPP12 improved cytosolic delivery of the Pmp-containing peptide by roughly 20-fold, and the CPP12-linked anionic peptide was roughly 10-fold more cytosolically penetrant than cargo-free Tat peptide. Because this assay was performed in serum-free medium and the STAT3 reporter assay was performed in DMEM with 10% FBS, we hypothesized that serum proteins may be restricting access to the cytosol, either directly through sequestering our peptide inhibitors, or indirectly through modulation of processes like endosomal uptake. We repeated CAPA in DMEM with 10% FBS and observed no difference in cytosolic penetration (Fig. S5e). Overall, the CAPA data indicated that CPP12-gp130 fusions with pTyr isosteres effectively access the cytosol when incubated at high nanomolar to low micromolar concentrations for 4 hours or longer.
Figure 3.
Cytosolic penetration, serum stability, and cell lysate stability of selected CPP12-gp130 peptide fusions. a) Chloroalkane penetration assay with CPP12-Pmp-ct (structure shown in Fig S5a), non-CPP-linked peptide ct-Pmp, and control molecules ct-Tat and ct-W. The figure shows data normalized to the no-molecule control (100% fluorescence), which indicates signal when no molecule accessed the cytosol, and no-dye control (0% fluorescence), which indicates signal if 100% of cytosolic HaloTag was blocked by chloroalkane-tagged molecules. Data show averages of three biological replicates (four biological replicates for CPP12-Pmp-ct), and within each biological replicate each data point represents the mean red fluorescence of 5,000 cells. CP50 values are reported as the mean and standard error of the mean for three separate curve fits to the three biological replicates (individual replicates shown in Fig. S5b–d). b and c) Serum and lysate stability assay for selected CPP12-gp130 peptides. Peptides were incubated for various time points in DMEM supplemented with 10% FBS (b) or HeLa cell lysate (c) at 37 °C, with and without 10 mM sodium orthovanadate. Areas under each peptide chromatogram peak were normalized to the area under the zero timepoint chromatogram peak.
2.3. Serum and lysate stability studies of CPP12-gp130 peptide fusions
Another potential barrier to cellular activity is degradation, either in serum or within cells. We investigated the first possibility by measuring degradation of selected CPP-gp130 peptides in serum-containing media. We incubated CPP12-pTyr and CPP12-F2Pmp at 150 μM in DMEM with 10% FBS, took aliquots at various time points over the course of 24 hours, and analyzed those samples via reverse-phase HPLC. In serum, the CPP12-pTyr peptide was degraded by almost 50% at 4 hours, and it was almost completely absent at 24 hours (Fig 3b, Fig S6a). Mass spectrometry analysis revealed that the primary product was the dephosphorylated peptide (Fig S7a,b). When the serum stability assay was repeated in the presence of 10 mM sodium orthovanadate, a general inhibitor of protein tyrosine phosphatases, dephosphorylation was blocked, with nearly 60% of CPP12-pTyr still intact after 24 hours (Fig 3b, Fig S6b). Consistent with these results, CPP12-F2Pmp showed little degradation, even after 24 hours in serum (Fig 3b, Fig S6c, S7c,d).
Testing peptide stability in a cell lysate was recently reported as a particularly demanding assay for benchmarking the cellular stability of peptide therapeutics.48 When tested in HeLa cell lysate, CPP12-pTyr was degraded much more quickly than in serum-containing media, with less than 20% of the original peptide remaining after just 1 hour (Fig 3c). This process could be slowed but not completely prevented with the addition of sodium orthovanadate (Fig 3c). Mass spectrometry revealed that, in the first hour of incubation, CPP12-pTyr is rapidly dephosphorylated. Following dephosphorylation, the peptide undergoes proteolysis at multiple positions including at the tyrosine generated by pTyr dephosphorylation (Fig S8a, S9a). CPP12-F2Pmp was more stable in cell lysate, with about 80% of the peptide remaining after 5 hours and 50% remaining after 24 hours of incubation (Fig 3c, Fig S8b). Interestingly, while CPP12-pTyr had numerous end products consistent with proteolysis at multiple positions, the mass spectrometry data revealed that CPP12-F2Pmp was proteolyzed at a single site. Specifically, this peptide was cleaved just N-terminal to the glutamine residue (Fig S8b).
In order to slow the intracellular degradation of CPP12-F2Pmp, we substituted the glutamine residue with N-methyl glutamine. This peptide, CPP12-F2Pmp-NMeQ, was completely resistant to degradation in cell lysate at 24 hours (Fig 3c, Fig S8c, S9c). Therefore, substitution of just two artificial amino acids (the pTyr isostere F2Pmp and a single N-methyl glutamine) produced a cell-permeable peptide with considerable stability to degradation by intracellular enzymes. Finally, we tested CPP12-F2Pmp-NMeQ in the STAT3 luciferase reporter assay (Fig S4a,b), as well as a 72-hour viability assay in the STAT3-dependent MDA-MB-468 breast cancer cell line.49 However, at 25 μM no activity was observed in either assay (Fig S10). The cause of this was revealed when we tested CPP12-F2Pmp-NMeQ in the competition FP assay. CPP12-F2Pmp-NMeQ had very poor STAT3 binding affinity (Fig S3a–c), so despite its high stability, it was unsuitable for further development as a STAT3 inhibitor.
3. Discussion and Conclusions
In this work, we sought to use CPP12 to enhance cellular delivery of peptides containing pTyr and pTyr isosteres, with the goal of producing more effective inhibitors of the STAT3 SH2 domain. Despite extensive development of STAT3 SH2 inhibitors, and prior applications of CPPs to deliver pTyr-mimicking peptides, to the best of our knowledge there are no reported efforts to develop CPP-peptide fusions to inhibit STAT3-SH2 in living cells. We quantitated the STAT3 affinity and cytosolic delivery of these CPP12 conjugates, evaluated their stability in cell lysate and serum, and measured their activity in cell culture. The medicinal chemistry of peptidomimetic pTyr isosteres has always required a difficult balance among cellular stability, cytosolic penetration, and target affinity.13 In this work, we directly measure each of these properties for CPP-linked peptides containing pTyr and pTyr isosteres.
We observed that the substitution of the pTyr with F2Pmp within the native gp130 sequence led to a 17-fold loss in STAT3 affinity, while substitution with Pmp led to over 100-fold loss in affinity. This generally matches previous reports examining Pmp and F2Pmp isosteres in other peptide-SH2 interactions, which showed large losses in affinity with Pmp but anywhere from 5-fold losses to 5-fold gains in affinity with F2Pmp.16 While Mandal and colleagues reported cellular effects on STAT3 activity using prodrug-functionalized 4-phosphonodifluoromethylcinnamate analogs of the gp130 sequence,17,18 this is the first reported application of F2Pmp to STAT3 inhibitors. It is possible that a similar prodrug approach could promote greater cytosolic penetration for gp130-derived peptides. Even more recently, Bai and colleagues utilized a phosphonomethyl indole derivative to generate high affinity STAT3-targeted peptidomimetics.27 These inhibitors were then incorporated into proteolysis-targeting chimeras (PRTACs) for the induction of STAT3 proteasomal degradation. This work highlights the continued relevance of pTyr isosteres, underscoring the need for careful evaluation of factors that can improve their cytosolic delivery.
We chose to apply CPP12 because it was shown to be more efficient than older CPPs at delivering peptides and peptidomimetics, including negatively charged cargoes.36–38,47,50,51 Since the delivery efficiency of CPP12 has been shown to vary depending on the cargo attached and the cell line employed, we used the chloroalkane penetration assay to conclusively demonstrate cytosolic delivery of a CPP12-gp130 fusion containing a pTyr isostere. These data must be interpreted carefully due to possible perturbations from the CPP, the linker between the CPP and the gp130-derived sequence, the pTyr isostere, and the chloroalkane group. Still, the data clearly show that cytosolic delivery can be achieved for pTyr-isostere-containing peptides at concentrations as low as 720 nM. Considering that the small molecule control, ct-W, had a CP50 value of 50 nM under these conditions,47,52 this is an impressive degree of cytosolic delivery for an anionic peptide. For comparison, the CPP12-fused peptide showed cytosolic localization at over 10-fold lower concentrations than Tat without cargo, and over 20-fold lower concentrations than a Pmp-containing control peptide without CPP12. The 18.9 μM CP50 of the non-CPP12-linked anionic peptide may reflect a moderate amount of cell penetration at 25 μM, or a small amount of degradation at this high concentration point; artifacts due to degradation, and additional caveats for CAPA, are described in detail elsewhere.47,53 Interestingly, while Song and colleagues observed significantly poorer uptake of CPP12 in media containing 10% FBS,38 we observed no difference in uptake of CPP12-conjugated anionic peptides between serum-free media and media with 10% FBS (Fig S5e). Additionally, while these cell penetration studies were only performed in HeLa cells and penetration may vary between different cell types, multiple other publications have demonstrated efficient cargo delivery with CPP12 across other cell types including HMLE, HEK-293, and U87MG glioblastoma cells.50,51 Still, additional penetration studies in other cell lines, including STAT3-dependent cancer cells, would be of great utility. Ultimately, while results can vary depending on the nature of the cargo and chemical tags, our results suggest that CPP12 is a viable delivery system for delivering small anionic peptides such as those containing pTyr and pTyr isosteres.
Serum and intracellular stability are also important considerations for peptidomimetic design, since biological degradation can severely limit efficacy.48 We measured extent of degradation over 24 hours in serum and in cell lysates, including analysis of degradation products by mass spectrometry. Our results suggest that CPP12 itself is completely stable in serum and in the presence of cellular enzymes including lysosomal proteases. Additionally, these results highlight that phosphopeptides are dephosphorylated rapidly in serum, and even more rapidly by intracellular enzymes. Interestingly, protection from dephosphorylation provided a significant boost to proteolytic stability, implying that dephosphorylation likely precedes proteolysis in many cases. Further, we were able to completely block proteolysis of CPP12-F2Pmp by incorporating a single N-methylated residue. These modifications allowed a 17-residue peptide, which was almost entirely degraded in cell lysate after 1 hour, to become completely stable under these conditions for 24 hours. Unfortunately, the N-methylation greatly disrupted binding to STAT3. Given published structural evidence that this glutamine residue engages a fairly narrow pocket on the STAT3 surface, it is possible that this N-methylation prevents optimal engagement of the glutamine with STAT3.27,54 Another glutamine derivative, or perhaps a non-natural proline derivative N-terminal to the cleavage site, might prevent degradation with minimal effects on binding affinity. Ultimately, these results highlight the careful balance that must be achieved between target affinity and bioavailability.
Ultimately, the most difficult challenges for peptidomimetics containing pTyr isosteres continue to be cell permeability and proteolytic stability. Our findings highlight the importance of contextualizing cell-based activity assays with not just target affinity, but quantitative assessment of cell penetration and proteolytic stability. While limited STAT3 affinity is the most likely reason for the lack of cellular activity, there are other potential causes including nonspecific binding to other proteins within the cell. If affinity is indeed the limiting factor, our data demonstrate that degradation-resistant, anionic SH2 domain inhibitors can be optimized for cytosolic delivery using advanced CPPs. Investigation of additional pTyr isosteres that may better engage the STAT3 pTyr binding pocket may provide sufficiently potent STAT3 binders which, coupled to CPP12, could more effectively inhibit STAT3 in cells. We also anticipate application of this strategy to other SH2 domains that better tolerate F2Pmp, such as PI-3K and Src, as well as cancer-relevant phosphotyrosine phosphatases.16,45,55
4. Methods
4.1. Peptide Synthesis
All peptides were synthesized via standard Fmoc solid phase peptide synthesis on an automated Tribute Peptide Synthesizer (Gyros Protein Technologies). Peptides were synthesized on low-loading Rink amide resin (substitution 0.36mmol/g) with deprotection in 20% piperidine in DMF, and coupling with 5 equiv. Each of amino acid, HBTU, and HOBt, and 10 equiv. of DIPEA. DMF washes were performed between each step. F2Pmp was allowed to couple overnight to ensure complete coupling. For on-resin CPP12 synthesis and cyclization, C-terminally allyl-protected glutamate was coupled via its sidechain to the growing peptide. When synthesis of the linear CPP was complete, the allyl group was removed with three 15-minute incubations with 0.1 equiv. palladium tetrakis and 10 equiv. of phenylsilane in anhydrous DCM. After N-terminal Fmoc deprotection, the peptide was cyclized overnight in DMF with 5 equiv. PyBOP, 5 equiv. HOBt, and 10 equiv. DIPEA.36 To prepare chloroalkane-tagged peptides, an MTT-protected lysine was deprotected with two 10-minute incubations with 1% TFA in DCM. After DMF washes, 2.5 equiv. of chloroalkane tag was coupled with 2.5 equiv. of PyBOP and 6.5 equiv. of DIPEA in DMF for 1.5 hours. To prepare fluorescein-labeled peptides, 5 equiv. of 5,6-carboxyfluorescein were coupled overnight with 10 equiv. DIPEA in DMF. To couple N-methyl glutamine, 5 equiv. each of amino acid, HATU and 10 equiv. DIPEA were coupled for 30 minutes, followed by a second coupling with the same reagents. The subsequent deprotection in 20% piperidine was followed by a second deprotection with 2% piperidine and 2% DBU in DMF. The amino acid following the N-methyl glutamine was double-coupled as well. All peptides were globally deprotected and cleaved from resin using a TFA cleavage cocktail (95:2:2:1, TFA:H2O:EDT:TIPS) for 3 hours. Following cleavage, peptides were diethyl ether precipitated and pelleted, followed by an additional diethyl ether wash and centrifugation. Peptides were then dried before resuspending in water/acetonitrile for reverse-phase HPLC purification on a preparative-scale C8 column (Agilent Technologies) at a 5 – 100% acetonitrile in 30 min gradient. Masses were determined using MALDI-TOF mass spectrometry (Bruker Microflex). MALDI Matrix used was 10mg/mL α-cyano-4-hydroxycinnamate in 50/50 water acetonitrile with 0.1% TFA. Peptides were at least 95% pure as determined via analytical HPLC on a C18 column at a 5 – 100% acetonitrile in 20 min gradient. Following purification, peptides were lyophilized and resuspended in DMSO for working solutions, which were quantified based on absorbance at 280 nm (Thermo Scientific Nanodrop 1000). Observed masses of final products are given in supplemental figure S8.
4.2. Protein Expression
STAT3 protein was expressed and purified as described.56 Rosetta BL21 E. coli were transfected with an expression plasmid for His6-STAT3 (codons 127–711). Transfected cells were grown on kanamycin agar plates and colonies were selected and grown in LB culture medium. At an optical density of 0.6 to 0.8, 1 mM IPTG was added and cells were incubated for 3 hours at 37 °C. Cells were pelleted, resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 5 mM imidazole, 0.2% lysozyme, 1 protease inhibitor cocktail pellet (Roche), and 2.5U/mL universal nuclease (Pierce), sonicated and lysed, and the lysate was centrifuged to pellet debris. The protein was purified from clarified lysate using Ni-NTA resin by incubating protein with resin for 45 min at 4 °C, rinsing with wash buffer (50 mM Tris pH 8.0, 300 mM NaCl, 5 mM imidazole), and then eluting in 50 mM Tris pH 8.0, 300 mM NaCl, 250 mM imidazole at 4°C.
The eluate was further purified by size exclusion chromatography using an automated FPLC system (AKTA, GE) on a Superdex S200 prep column in 10 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, and 2 mM DTT. Protein fractions were analyzed via SDS-PAGE, and pure fractions were pooled together. Concentration of protein was quantified via absorbance at 280 nm and confirmed via BCA assay. Protein was stored in frozen aliquots at −80°C.
4.3. Fluorescence Polarization Assays
FP assays were performed as described.39 For direct FP assay, fluorescent peptide was mixed at a final concentration of 10 nM with a serial dilution of STAT3 protein [5.6 μM to 0.0055 μM] in a final reaction volume of 50 μL in a black, flat-bottomed 384-well polypropylene plate (Greiner Bio-One). The buffer composition was 10 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, and 2 mM DTT. The plate was incubated in the dark for 45 min at room temperature, then read at 494 nm excitation and 519 nm emission. Data was normalized to the maximum polarization observed (raw data shown in supplement). For competition FP assays, fluorescent probe at a final concentration of 10 nM was mixed with STAT3 protein at a final concentration of 300 nM and a serial dilution of inhibitor peptide [25 or 50 μM to 0.025 μM]. DMSO was normalized across the plate to a final concentration not exceeding 0.5%. The final reaction volume was 50 μL in a black 384-well plate. The buffer composition was 10 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, and 2 mM DTT. The plate was incubated in the dark for 45 min at room temperature before reading at 494 nm excitation and 519 nm emission. Data was normalized to the no inhibitor (maximum polarization) control. Kd and IC50 values were obtained from curve fits using KaleidaGraph graphic software as described.57
4.4. STAT3 Luciferase Reporter Assay
Luciferase reporter assay was performed as described.43 STAT3-luc/U3A fibrosarcoma reporter cells were seeded in a 96-well plate at 104 cells per well overnight. Then, cells were treated with selected concentrations of peptide or vehicle [10 or 25 μM] for 1 or 24 hours at 37 °C, and then stimulated with OSM at 10 ng/mL for 6 hours at 37 °C. Luciferase activity was quantified using the Bright-Glo Luciferase Assay system (Promega) and a Luminoskan Ascent Luminometer (Labsystems).
4.5. Chloroalkane Penetration Assay
CAPA assay was performed as described.47,52 Halo-GFP-Mito HeLa cells were cultured using DMEM + 10% FBS + 1% Pen/Strep + 1μg/mL puromycin. Cells were seeded in a 96-well plate at 1.0 × 105 cells/well and incubated overnight. Cells were then rinsed with Opti-MEM and treated with a serial dilution of peptides [25 μM to 0.0004 μM] or ct-W control [2 μM to 0.001 μM] in Opti-MEM for 4 hours. Media was aspirated and cells were washed with Opti-MEM. Cells were then treated with 5 μM ct-TAMRA for 30 minutes before washing and trypsinizing cells. Cells were pelleted and resuspended in PBS twice before resuspending in a final volume of 250 μL of PBS and transferring them to a 96-well plate for flow cytometry analysis (Guava EasyCyte 6HT-2L benchtop flow cytometer), gating for live cells and measuring 5000 cells per sample. Fluorescence was normalized to the ct-TAMRA treated (100% fluorescence) and untreated (0% fluorescence) cells. ct-W and ct-TAMRA were prepared and characterized as described.47
4.6. Peptide Stability Assays
Serum stability assays were performed as described.58 10% FBS in DMEM was warmed to 37 °C prior to adding peptide. For lysate stability assays, HeLa cells were trypsinized, washed in PBS, and pelleted before treating with lysis buffer (50 mM Tris, 250 mM NaCl, 0.5% IGEPAL CA-630 detergent, pH 8.0) on ice for 15 min. Then, lysates were centrifuged for 10 min at 4 °C at 14,800 rpm and the clarified lysate was collected. Peptides were added to either the serum or the lysate to a concentration of 150 μM and were incubated at 37 °C. Aliquots of 40 μL were taken at each time point and quenched in 160 μL of ice-cold methanol. Samples were spun down for 10 min at 14,800 rpm prior to analysis via reverse-phase analytical HPLC on a C18 column (Agilent Technologies). Peptide chromatogram peaks were integrated to determine area under each curve. Areas for each timepoint were normalized to the zero-hour timepoint to determine percentage of peptide remaining. Data presented is the average of three biological replicates performed on different days. Peptide masses present in each sample were determined using MALDI-TOF mass spectrometry (Bruker Microflex).
4.7. Cell Viability Assay
Cell viability assay was performed as described.49 MDA-MB-468 breast cancer cells were seeded 3 × 103 cells/well in white opaque 96-well plates overnight. Peptide or vehicle [10 or 25 μM] was added to a final volume of 100 μL and incubated at 37 °C for the specified time. Cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability kit (Promega).
Supplementary Material
Acknowledgments
This work was supported by a Ruth L. Kirschstein Individual Predoctoral NRSA Fellowship F30CA220678 (NCI, NIH) and by NSF CHE-1507456. The STAT3 expression plasmid was kindly provided by Dr. Yuan Chen at the Beckman Research Institute, City of Hope Comprehensive Cancer Center.
Footnotes
Declaration of Competing Interests
There are no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2(10):740–749. doi: 10.1038/nrc906 [DOI] [PubMed] [Google Scholar]
- 3.Bromberg J Stat proteins and oncogenesis. J Clin Invest. 2002;109(9):1139–1142. doi: 10.1172/JCI15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi: 10.1038/nrclinonc.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei D, Le X, Zheng L, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22(3):319–329. doi: 10.1038/sj.onc.1206122 [DOI] [PubMed] [Google Scholar]
- 6.Xie T, Wei D, Liu M, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23(20):3550–3560. doi: 10.1038/sj.onc.1207383 [DOI] [PubMed] [Google Scholar]
- 7.Lee T-L, Yeh J, Waes CV, Chen Z. Bcl-xL is regulated by NF-KappaB and STAT3 through p53-dependent control in head and neck squamous cell carcinoma. Cancer Res. 2004;64(7 Supplement):1115–1115. [Google Scholar]
- 8.Gritsko T Persistent Activation of Stat3 Signaling Induces Survivin Gene Expression and Confers Resistance to Apoptosis in Human Breast Cancer Cells. Clin Cancer Res. 2006;12(1):11–19. doi: 10.1158/1078-0432.CCR-04-1752 [DOI] [PubMed] [Google Scholar]
- 9.Bu LL, Yu GT, Wu L, et al. STAT3 Induces Immunosuppression by Upregulating PD-1/PD-L1 in HNSCC. J Dent Res. 2017;96(9):1027–1034. doi: 10.1177/0022034517712435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlessinger K, Levy DE. Malignant Transformation but not Normal Cell Growth Depends on Signal Transducer and Activator of Transcription 3. Cancer Res. 2005;65(13):5828–5834. doi: 10.1158/0008-5472.CAN-05-0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraskouskaya D, Duodu E, Arpin CC, Gunning PT. Progress towards the development of SH2 domain inhibitors. Chem Soc Rev. 2013;42(8):3337–3370. doi: 10.1039/C3CS35449K [DOI] [PubMed] [Google Scholar]
- 12.Furtek SL, Backos DS, Matheson CJ, Reigan P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem Biol. 2016;11(2):308–318. doi: 10.1021/acschembio.5b00945 [DOI] [PubMed] [Google Scholar]
- 13.Cerulli RA, Kritzer JA. Phosphotyrosine isosteres: past, present and future. Org Biomol Chem. November 2019. doi: 10.1039/C9OB01998G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domchek SM, Auger KR, Chatterjee S, Burke TR, Shoelson SE. Inhibition of SH2 domain/phosphoprotein association by a nonhydrolyzable phosphonopeptide. Biochemistry. 1992;31(41):9865–9870. [DOI] [PubMed] [Google Scholar]
- 15.Burke TR, Smyth MS, Nomizu M, Otaka A, Roller PR. Preparation of fluoro- and hydroxy-4-(phosphonomethyl)-D,L-phenylalanine suitably protected for solid-phase synthesis of peptides containing hydrolytically stable analogs of O-phosphotyrosine. J Org Chem. 1993;58(6):1336–1340. doi: 10.1021/jo00058a009 [DOI] [Google Scholar]
- 16.Burke TR Jr, Smyth MS, Otaka A, et al. Nonhydrolyzable Phosphotyrosyl Mimetics for the Preparation Of Phosphatase-Resistant SH2 Domain Inhibitors. Biochemistry. 1994;33(21):6490–6494. doi: 10.1021/bi00187a015 [DOI] [PubMed] [Google Scholar]
- 17.Mandal PK, Liao WS-L, McMurray JS. Synthesis of Phosphatase-Stable, Cell-Permeable Peptidomimetic Prodrugs That Target the SH2 Domain of Stat3. Org Lett. 2009;11(15):3394–3397. doi: 10.1021/ol9012662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal PK, Gao F, Lu Z, et al. Potent and Selective Phosphopeptide Mimetic Prodrugs Targeted to the Src Homology 2 (SH2) Domain of Signal Transducer and Activator of Transcription 3. J Med Chem. 2011;54(10):3549–3563. doi: 10.1021/jm2000882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Yue P, Page BDG, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci. 2012;109(24):9623–9628. doi: 10.1073/pnas.1121606109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haftchenary S, Luchman HA, Jouk AO, et al. Potent Targeting of the STAT3 Protein in Brain Cancer Stem Cells: A Promising Route for Treating Glioblastoma. ACS Med Chem Lett. 2013;4(11):1102–1107. doi: 10.1021/ml4003138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiquee K, Zhang S, Guida WC, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci. 2007;104(18):7391–7396. doi: 10.1073/pnas.0609757104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bharadwaj U, Eckols TK, Xu X, et al. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget. 2016;7(18):26307–26330. doi: 10.18632/oncotarget.8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu W, Li C, Zhang W, et al. Discovery of an Orally Selective Inhibitor of Signal Transducer and Activator of Transcription 3 Using Advanced Multiple Ligand Simultaneous Docking. J Med Chem. 2017;60(7):2718–2731. doi: 10.1021/acs.jmedchem.6b01489 [DOI] [PubMed] [Google Scholar]
- 24.Lin L, Benson DM, DeAngelis S, et al. A small molecule, LLL12 inhibits constitutive STAT3 and IL-6-induced STAT3 signaling and exhibits potent growth suppressive activity in human multiple myeloma cells. Int J Cancer. 2012;130(6):1459–1469. doi: 10.1002/ijc.26152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: A Small-Molecule Inhibitor of STAT3 Activation and Dimerization. Chem Biol. 2006;13(11):1235–1242. doi: 10.1016/j.chembiol.2006.09.018 [DOI] [PubMed] [Google Scholar]
- 26.Li X, Ma H, Li L, et al. Novel synthetic bisindolylmaleimide alkaloids inhibit STAT3 activation by binding to the SH2 domain and suppress breast xenograft tumor growth. Oncogene. 2018;37(18):2469–2480. doi: 10.1038/s41388-017-0076-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai L, Zhou H, Xu R, et al. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo. Cancer Cell. 2019;36(5):498–511.e17. doi: 10.1016/j.ccell.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peraro L, Kritzer JA. Emerging Methods and Design Principles for Cell-Penetrant Peptides. Angew Chem-Int Ed. 2018;57(37):11868–11881. doi: 10.1002/anie.201801361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157(2):195–206. doi: 10.1111/j.1476-5381.2009.00057.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guidotti G, Brambilla L, Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol Sci. 2017;38(4):406–424. doi: 10.1016/j.tips.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 31.Kertész Á, Váradi G, Tóth GK, Fajka-Boja R, Monostori É, Sármay G. Optimization of the cellular import of functionally active SH2-domain-interacting phosphopeptides. Cell Mol Life Sci CMLS. 2006;63(22):2682–2693. doi: 10.1007/s00018-006-6346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wavreille A-S, Pei D. A Chemical Approach to the Identification of Tensin-Binding Proteins. ACS Chem Biol. 2007;2(2):109–118. doi: 10.1021/cb600433g [DOI] [PubMed] [Google Scholar]
- 33.Kuil J, Fischer MJE, de Mol NJ, Liskamp RMJ. Cell permeable ITAM constructs for the modulation of mediator release in mast cells. Org Biomol Chem. 2011;9(3):820–833. doi: 10.1039/C0OB00441C [DOI] [PubMed] [Google Scholar]
- 34.Watson GM, Kulkarni K, Sang J, et al. Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target. J Med Chem. 2017;60(22):9349–9359. doi: 10.1021/acs.jmedchem.7b01320 [DOI] [PubMed] [Google Scholar]
- 35.Lian W, Jiang B, Qian Z, Pei D. Cell-Permeable Bicyclic Peptide Inhibitors against Intracellular Proteins. J Am Chem Soc. 2014;136(28):9830–9833. doi: 10.1021/ja503710n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Z, Martyna A, Hard RL, et al. Discovery and Mechanism of Highly Efficient Cyclic Cell-Penetrating Peptides. Biochemistry. 2016;55(18):2601–2612. doi: 10.1021/acs.biochem.6b00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wissner RF, Steinauer A, Knox SL, Thompson AD, Schepartz A. Fluorescence Correlation Spectroscopy Reveals Efficient Cytosolic Delivery of Protein Cargo by Cell-Permeant Miniature Proteins. ACS Cent Sci. 2018;4(10):1379–1393. doi: 10.1021/acscentsci.8b00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J, Qian Z, Sahni A, Chen K, Pei D. Cyclic Cell-Penetrating Peptides with Single Hydrophobic Groups. ChemBioChem. 2019;20(16):2085–2088. doi: 10.1002/cbic.201900370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schust J, Berg T. A high-throughput fluorescence polarization assay for signal transducer and activator of transcription 3. Anal Biochem. 2004;330(1):114–118. doi: 10.1016/j.ab.2004.03.024 [DOI] [PubMed] [Google Scholar]
- 40.Ren Z, Cabell LA, Schaefer TS, McMurray JS. Identification of a High-Affinity Phosphopeptide Inhibitor of Stat3. Bioorg Med Chem Lett. 2003;13(4):633–636. doi: 10.1016/S0960-894X(02)01050-8 [DOI] [PubMed] [Google Scholar]
- 41.Mandal PK, Morlacchi P, Knight JM, et al. Targeting the Src Homology 2 (SH2) Domain of Signal Transducer and Activator of Transcription 6 (STAT6) with Cell-Permeable, Phosphatase-Stable Phosphopeptide Mimics Potently Inhibits Tyr641 Phosphorylation and Transcriptional Activity. J Med Chem. 2015;58(22):8970–8984. doi: 10.1021/acs.jmedchem.5b01321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burke TR, Kole HK, Roller PP. Potent Inhibition of Insulin Receptor Dephosphorylation by a Hexamer Peptide Containing the Phosphotyrosyl Mimetic F2Pmp. Biochem Biophys Res Commun. 1994;204(1):129–134. doi: 10.1006/bbrc.1994.2435 [DOI] [PubMed] [Google Scholar]
- 43.Nelson EA, Walker SR, Kepich A, et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112(13):5095–5102. doi: 10.1182/blood-2007-12-129718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang M, Kim H, Ho VT, et al. Gene expression–based discovery of atovaquone as a STAT3 inhibitor and anticancer agent. Blood. 2016;128(14):1845–1853. doi: 10.1182/blood-2015-07-660506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, He Y, Liu S, et al. Salicylic Acid Based Small Molecule Inhibitor for the Oncogenic Src Homology-2 Domain Containing Protein Tyrosine Phosphatase-2 (SHP2). J Med Chem. 2010;53(6):2482–2493. doi: 10.1021/jm901645u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci. 2005;102(13):4700–4705. doi: 10.1073/pnas.0409894102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peraro L, Deprey KL, Moser MK, et al. Cell Penetration Profiling Using the Chloroalkane Penetration Assay. J Am Chem Soc. 2018;140(36):11360–11369. doi: 10.1021/jacs.8b06144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Partridge AW, Kaan HYK, Juang Y-C, et al. Incorporation of Putative Helix-Breaking Amino Acids in the Design of Novel Stapled Peptides: Exploring Biophysical and Cellular Permeability Properties. Molecules. 2019;24(12):2292. doi: 10.3390/molecules24122292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker SR, Chaudhury M, Nelson EA, Frank DA. Microtubule-Targeted Chemotherapeutic Agents Inhibit Signal Transducer and Activator of Transcription 3 (STAT3) Signaling. Mol Pharmacol. 2010;78(5):903–908. doi: 10.1124/mol.110.066316 [DOI] [PubMed] [Google Scholar]
- 50.Soudah T, Khawaled S, Aqeilan RI, Yavin E. AntimiR-155 Cyclic Peptide–PNA Conjugate: Synthesis, Cellular Uptake, and Biological Activity. ACS Omega. 2019;4(9):13954–13961. doi: 10.1021/acsomega.9b01697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai B, Kim D, Akhand S, et al. Selection of DNA-Encoded Libraries to Protein Targets within and on Living Cells. J Am Chem Soc. 2019;141(43):17057–17061. doi: 10.1021/jacs.9b08085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peraro L, Zou Z, Makwana KM, et al. Diversity-Oriented Stapling Yields Intrinsically Cell-Penetrant Inducers of Autophagy. J Am Chem Soc. 2017;139(23):7792–7802. doi: 10.1021/jacs.7b01698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deprey K, Kritzer JA. Quantitative measurement of cytosolic penetration using the chloroalkane penetration assay In: Methods in Enzymology. Academic Press; 2020. doi: 10.1016/bs.mie.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMurray JS. Structural basis for the binding of high affinity phosphopeptides to Stat3. Biopolymers. 2008;90(1):69–79. doi: 10.1002/bip.20901 [DOI] [PubMed] [Google Scholar]
- 55.Frankson R, Yu Z-H, Bai Y, Li Q, Zhang R-Y, Zhang Z-Y. Therapeutic Targeting of Oncogenic Tyrosine Phosphatases. Cancer Res. 2017;77(21):5701–5705. doi: 10.1158/0008-5472.CAN-17-1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Namanja AT, Wang J, Buettner R, Colson L, Chen Y. Allosteric Communication Across STAT3 Domains Associated with STAT3 Function and Disease-causing Mutation. J Mol Biol. 2016;428(3):579–589. doi: 10.1016/j.jmb.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siegert TR, Bird MJ, Makwana KM, Kritzer JA. Analysis of Loops that Mediate Protein-Protein Interactions and Translation into Submicromolar Inhibitors. J Am Chem Soc. 2016;138(39):12876–12884. doi: 10.1021/jacs.6b05656 [DOI] [PubMed] [Google Scholar]
- 58.Quartararo JS, Wu P, Kritzer JA. Peptide Bicycles that Inhibit the Grb2 SH2 Domain. ChemBioChem. 2012;13(10):1490–1496. doi: 10.1002/cbic.201200175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.