Abstract
Injured skeletal muscles which lose more than 20% of their volume, known as volumetric muscle loss, can no longer regenerate cells through self-healing. The traditional solution for recovery is through regenerative therapy. As the technology of three-dimensional (3D) bioprinting continues to advance, a new approach for tissue transplantation is using biocompatible materials arranged in 3D scaffolds for muscle repair. Ultrashort self-assembling peptide hydrogels compete as a potential biomaterial for muscle tissue formation due to their biocompatibility. In this study, two sequences of ultrashort peptides were analyzed with muscle myoblast cells (C2C12) for cell viability, cell proliferation, and differentiation in 3D cell culture. The peptides were then extruded through a custom-designed robotic 3D bioprinter to create cell-laden 3D structures. These constructs were also analyzed for cell viability through live/dead assay. Results showed that 3D bioprinted structures of peptide hydrogels could be used as tissue platforms for myotube formation – a process necessary for muscle repair.
Keywords: Three-dimensional bioprinting, Peptide, Biomaterials, Bioinks, Tissue engineering, Myoblasts
1. Introduction
Drastic muscle loss resulting from injury, birth defect, or cancer ablation restrains muscles’ ability to reconstruct through self-healing, consequently requiring regenerative treatments through engineered tissues[1]. Around 45 million cases of reconstructive surgeries are reported yearly in the USA[2]. Autologous tissue transfer is the present treatment for massive tissue loss. However, patients undergo complications and functional restrictions resulted from harvesting tissues from a donor[3]. Moreover, the access to sufficient tissues and organs for all patients is nearly impossible, and many patients die waiting for available organs due to long transplant waiting lists. Furthermore, implanting compatible foreign biomaterials can cause dislodgment, fracture, and infection. These challenges have simulated a need to pursue and develop innovative approaches to deliver required tissue[4].
Skeletal muscle is a soft tissue that constitutes approximately half of the human adult body mass[5]. Muscles mass is profoundly affected by many factors such as nutritional level, hormonal status, physical activity, and illness or injury, which influence the balance of protein synthesis and degradation[6]. Skeletal muscle is a voluntary moveable tissue that has the ability to convert chemical energy into mechanical energy and then transfer it to tendon tissue. It also supports soft tissue and maintains body posture[7]. In addition, this tissue is responsible for different functions of the body such as respiration and protection of abdominal viscera, and also controls the movement of limbs[8]. Skeletal muscle tissue exhibits the native capability to regenerate and repair through the activation of local satellite cells[8,9].
However, this ability declines with age as well as in clinical conditions such as tumor resection and traumatic sport injuries including concussions and strains, and muscular dystrophy that may result in volumetric muscle loss (VML). In these injuries, approximately 20% or more of the muscle mass is lost[10,11] and, as a result, tissues lose the ability to signal each other and become unable to repair themselves through natural physiological processes. Thus, surgical intervention is needed[12-15] to restore normal function and prevent the formation of scar tissue[13], which may lead to muscle atrophy and prevent muscle regeneration[16]. Around the world, millions of people are affected by these clinical conditions which cause significant social and economic problems[17,18]. As such, alternative technologies are urgently needed for the reconstruction of skeletal muscle tissues that have experienced VML and need to regenerate new functional tissue[10,19].
An alternative approach for VML treatment and organ fabrication is tissue engineering through the use of biological scaffolds[20]. The process of muscle tissue engineering is the same as that of skin tissue engineering: The cells are grown in a three-dimensional (3D) environment, similar to how they would grow in vivo using biomaterial scaffolds. Particular interest is taken in scaffolds made from self- assembling peptides for 3D culture and bioprinting because of their synthetic, yet natural background. They have been used as biomaterials and matrices to deliver encapsulated bioactive molecules in therapeutic applications and regenerative medicine[21-25]. Many hydrogels have been used and assessed for their mechanical properties, cellular activity, and myogenic potential. However, a need is still present to develop the most appropriate material that is efficient in maintaining mechanical stability and promoting myotube formation[26].
The principle of 3D bioprinting allows the capability of fabricating constructs of a fully customized muscle. This technology depends on forming a complex biological construct by dispensing cells and bionics in a layer-by-layer fashion. Due to these excellent features, 3D bioprinting has become the ultimate solution for tissue engineering, especially when reconstructing skeletal muscles. Inspired by this emerging technology, we aim to study the printability of our custom-designed robotic 3D bioprinting system[27,28] to fabricate 3D scaffolds for the differentiation of myoblast cells. The process of 3D bioprinting is believed to enhance the arrangement of homogeneous cellular scaffolds and improve cell proliferation and adhesion for myotube formation. Two sequences of self-assembling peptides are tested and analyzed for cell viability, proliferation, and differentiation. The promising results indicate that 3D bioprinting of self-assembling ultrashort peptides may valuably improve the process of muscle tissue engineering.
2. Materials and Methods
Two tetrameric self-assembling peptides CH-01 and CH-02 were custom-synthesized in our Laboratory for Nanomedicine. Mouse myoblast cells (C2C12) were obtained from ATCC, USA. The following materials were ordered from Gibco, USA: Dulbecco’s modified eagle medium (DMEM), fetal bovine serum (FBS), heat-inactivated horse serum, Dulbecco’s phosphate-buffered saline (PBS) solution, and penicillin-streptomycin (P/S) antibiotics. An 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay kit and a LIVE/DEAD Viability/Cytotoxicity kit were purchased from Promega, USA. Immunostaining antibody myosin heavy chain (MHC) was purchased from Abcam. Cell culture flasks and 96-well plates were ordered from Corning, USA.
2.1. Preparation of Peptide Hydrogel
CH-01 and CH-02 peptide powders were dissolved in Milli-Q water. Then, 10× PBS was mixed into the peptide solution. Gelation of both peptides occurred within a few minutes at a minimum concentration of 4 mg/mL and 3 mg/mL for CH-01 and CH-02, respectively, as shown in Figure 1. The final volume ratio of peptide solution and 10× PBS was 9:1.
Figure 1.
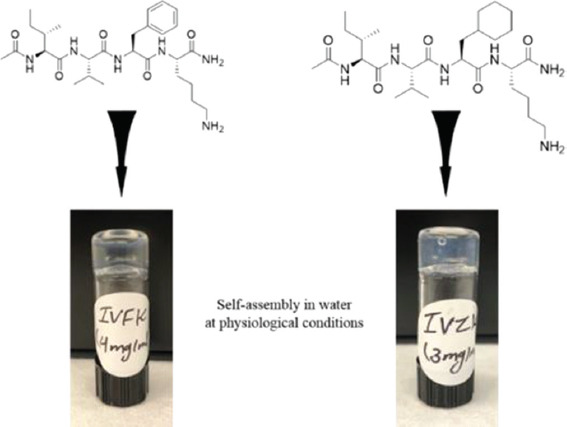
The self-assembling peptides CH-01 (4 mg/ml) and CH-02 (3 mg/ml) generate macromolecular nanofibrous hydrogels in an aqueous solution, the gelation was enhanced using phosphate buffer saline.
2.2. Characterization of the Topography and Morphology of Peptide Hydrogels
2.2.1. Evaluation of Fiber Structures by Field-emission Scanning Electron Microscopy (SEM)
The peptide nanogels were dehydrated by gradually increasing concentrations of 30%, 50%, 70%, 90%, and 100% (v/v) ethanol solutions for 15 min in each solution. Further dehydration in 100% ethanol solution was continued by changing the absolute ethanol solution with a fresh one twice for 15 min each. The dehydrated samples were subsequently kept in 1:2 ratio of hexamethyldisilazane (HMDS) and ethanol for 20 min, followed by 20 min of incubation in a fresh solution of 2:1 ratio of HMDS and ethanol and then in 100% HMDS, performed twice for 20 min. Finally, the samples were stored overnight in a fume hood to allow HMDS to evaporate. Before imaging, the samples were mounted onto SEM grids using conductive carbon tape, and then sputter-coated with a 5 nm thick coating of iridium and a 3 nm thick coating of gold/palladium. Images were taken of the coated samples with a field emission SEM system (FEI Nova Nano630 SEM, Oregon, USA).
2.3. Cell Culture and Growth Conditions
2.3.1. Mouse Myoblast Cells (C2C12)
Mouse myoblast cells (C2C12) were cultured either in a T175 or T75 culture flask in complete DMEM media (10% FBS and 1% P/S). The cells were placed in a humidified incubator with 95% air and 5% CO2 at 37°C. Then they were subcultured using trypsin at approximately 80% confluence. Fresh culture media was added every 48 h.
2.3.2. 3D Culture of Myoblast Cells in Peptide Hydrogels
In a 96-well plate, mouse myoblast cells were encapsulated in peptide hydrogels, as previously described[1]. Briefly, peptide solutions CH-01 (4 mg/ mL) and CH-02 (3 mg/mL) were added at 40 μL/well. Mouse myoblast cells (30,000 cells/well) that were re-suspended in 2xPBS were mixed gently with the peptide solutions. The gelation time was 3-5 min. Subsequently, the culture medium was added to the wells.
2.3.3. Differentiation of Myoblast Cells within 3D Culture Construct
6 days of culturing myoblast cells inside the 3D environment in the growth medium, the culture conditions were then changed to differentiated mode to study differentiation behavior for 8 days. The differentiation medium contained DMEM supplemented with 2% horse serum and 1% P/S.
2.4. Biocompatibility Evaluation of Tetrameric Ultrashort Self-assembling Peptides in Two-dimensional (2D) Culture
2.4.1. Cell Viability Assay (MTT Assay)
All biocompatibility studies were performed in a 96-well plate. C2C12 (10,000 cells/well) were seeded in a complete medium. After 2 days, the medium was discarded, and the cells were incubated for 48 h with different concentrations of peptide solution, at 37°C, 95% air, and 5% CO2, Matrigel was used as a control. A colorimetric MTT assay was used to determine cell viability as advised in the manufacturer’s protocol. Briefly, the phenol-free fresh medium was mixed with 10% MTT reagent. Each well was incubated for 4 h with 100 μL MTT reagent including the positive control wells. Insoluble crystals of formazan were dissolved by adding 100 μL of dimethyl sulfoxide to each well. Finally, the absorption of individual wells was recorded at 540 nm using a plate reader (PHERAstar FS, Germany).
2.5. 3D Bioprinting of Myoblast Cells
2.5.1. 3D Bioprinting
In two vials, 15 mg of CH-01 and CH-02 peptide powder each were weighed out and placed under UV for 30 min sterilization. The peptide powder was dissolved in 1mL of MilliQ water and the peptide solution was then vortexed and sonicated to obtain a homogenous solution. The vials were placed in an incubator at 37°C and 5% CO2. The incubation time for the CH-01 pre-gel bioink solution was 3.5 h, but was 2 h when using the CH-02 pre-gel bioink solution.
A custom-designed robotic 3D bioprinter[27,28] was set up with commercial microfluidic pumps. A homemade two-inlet nozzle was used for extrusion. A heatbed was set to 37°C to create a suitable environment for the cells once extruded within the peptide bioink. Two commercial microfluidic pumps were loaded for extrusion. A simple gcode file was used to create a structure of 8 layers.
Pump 1 was loaded with the peptide pre-gel solution and set to a flow rate of 60 µL/min. Pump 2 was loaded with myoblast cells containing serum-free DMEM culture media. The same procedure was conducted for both peptides. 17-18 samples were printed for each peptide with a height of 7-8 layers for each sample.
2.5.2. Live/Dead Assay
A two-color fluorescence assay was used to assess the cell viability within the printed constructs. Calcein was used as a marker for living cells and ethidium homodimer for dead cells. The bioprinted tissues were washed in PBS 3 times and treated with calcein AM (green) and ethidium homodimer-1 (red) at 1:2 ratio in PBS. The samples were then placed for 20 min in a dark incubator at 37°C and 5% CO2. After staining, they were washed again 3 times in PBS. A confocal microscope (Leica SP8) was used for image acquisition.
2.6. Immunofluorescence Staining of Differentiated Myoblasts
The differentiation of mouse myoblast cells within both hydrogels was studied in a glass confocal dish (12mm) by immunofluorescence analysis. C2C12 (30,000 cells/plate) were embedded in different hydrogels. After 8 days of differentiation, 4% paraformaldehyde solution was used for cells fixation. After 20 min incubation at room temperature, the cells were permeabilized and labeled with primary anti-MHC (1:300 PBS) for 1 h followed by 1 h incubation with secondary anti-mouse IgG-fluorescein isothiocyanate and DAPI. The myotube formation was observed with fluorescence confocal microscopy (Zeiss LSM 710 Inverted Confocal Microscope, Germany).
2.7. Statistical Analysis
All results are presented as a mean±standard deviation. Each type of test was repeated in three similar experiments. Statistical differences among the experimental groups were determined with one-way analysis of variance. When P<0.05, the results were considered to be statistically significant.
3. Results
3.1. The Nanofibrous Morphology of Self-assembling Peptides
The nanofibrous morphology of the self-assembling peptides was observed through SEM imaging. It was then compared to the morphology observed in bovine collagen (Figure 2A), which is comprised by a unique triple-helical structure[23]. SEM results confirmed that the fibrous structures of these peptides resemble the fibrous structure of collagen in terms of architecture. The detailed assessment of CH-01 (Figure 2B) and CH-02 (Figure 2C) showed that the fibrous structures of these peptides resemble the fibrous structure of collagen in terms of architecture. This nanofibrous structure was produced from the antiparallel pairing of two peptide monomers (Figure 1). Subsequently, the assembly of the peptide pairs by stacking facilitated the formation of the fibers. The hydrogel was formed by the condensation of these fibers.
Figure 2.
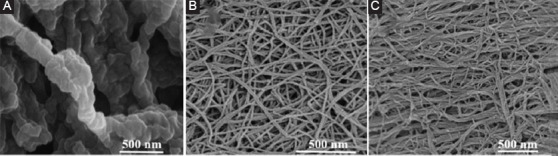
Field emission scanning electron microscopy images of nanofibrous structure of 2.5 mg/mL bovine collage type I (A), 4 mg/mL CH-01 (B), and 3 mg/mL CH-02 (C).
3.2. Cell Viability Results (MTT assay)
After 24 h of incubation, cell proliferation was tested with different peptide concentrations to evaluate biocompatibility. The MTT assay was used to quantify the number of viable cells. This was done by plotting a standard curve for a known number of cells (Figure 3B). Test results indicated that the differences between both peptides CH-01 (Figure 3C), CH-02 (Figure 3D), and positive control, Matrigel were non-significant, indicating that both scaffolds were suitable and biocompatible on muscle myoblast cells.
Figure 3.
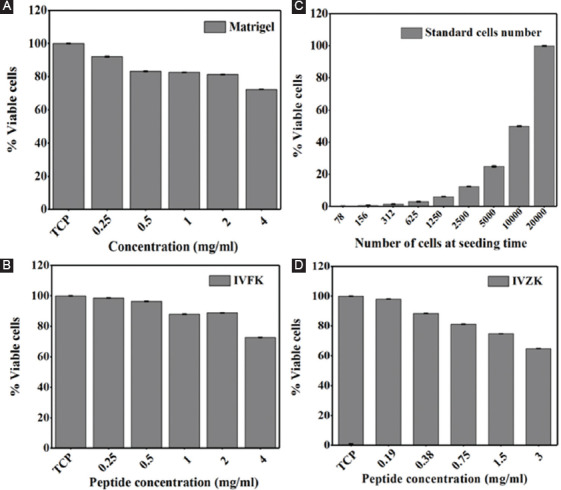
Graphical representation of 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay of mouse myoblast cells incubated with different peptide concentrations for 24 h, CH-01 (C), CH-02 (D), and positive controls, Matrigel (A) was used. A standard curve for a known number of cells (B).
3.3. Differentiation of Muscle Myoblasts
To confirm whether these scaffolds induce differentiation of C2C12 myoblasts, the expression of MHC, which is a late-stage differentiation marker of myogenesis, was observed through immunostaining. After inducing differentiation of the cells in differentiation media for 8 days, MHC expression was observed from myoblasts cultured on both scaffolds and was found to be similar to the positive control Matrigel, as shown in Figure 4A. These findings indicate that both scaffolds promote muscle cell differentiation, thus suggesting that these materials may prove to be beneficial in increasing muscle mass. The fusion index was calculated from MHC stained cells, which is defined as the number of nuclei present in myotubes in comparison to the total number of nuclei present in the observed field. Statistical analysis revealed a significant increase in the number of myotubes containing four or more nuclei in cells encapsulated within CH-01, when compared to other tested materials (Figure 4B). In addition, quantitative investigation of cell elongation within the scaffolds was estimated by the cell aspect ratio, which is defined as the proportion between the length of the longest line and the length of the shortest line across the nuclei. The results demonstrated a slight increase in the cell aspect ratio in the 3D cultures using peptide hydrogels and Matrigel as the 3D control, different to the 2D culture. However, these increases did not reach statistical significance (Figure 4C).
Figure 4.
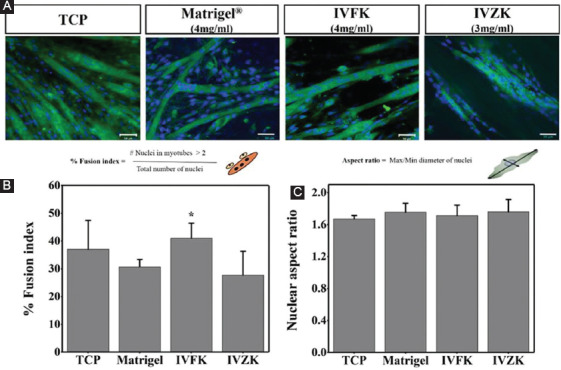
Overlaid confocal fluorescent images of differentiated mouse myoblast cells encapsulated in the peptide (4 mg/ml CH-01 and 3 mg/ml CH-02) and 4 mg/ml Matrigel. The encapsulated cells were cultured for 8 days in differentiation medium. Nucleus showed in blue and myosin heavy chain shown in green, (A) percentage of fusion index after 8 days, (B) and nuclear aspect ratio of differentiated muscle cells, (C) scale bar is 50 µm.
3.4. Cell Viability Results of 3D Bioprinted Structures
The intensity of green fluorescence of the 3D printed cell-laden constructs shown in Figure 5 revealed that most of the cells remained viable in both peptide hydrogels throughout 5 days indicating that the diffusion of nutrients and removal of waste products were sufficient to maintain cell viability. There were only very few dead cells visible within the matrix. It is worth mentioning that the reduction in cell viability with 4 mg/ml (Figure 3C) and 3 mg/ml (Figure 3D) is not due to the toxicity of the hydrogels and cell death, but due to a change in the local cellular microenvironment and diffusion barrier.
Figure 5.
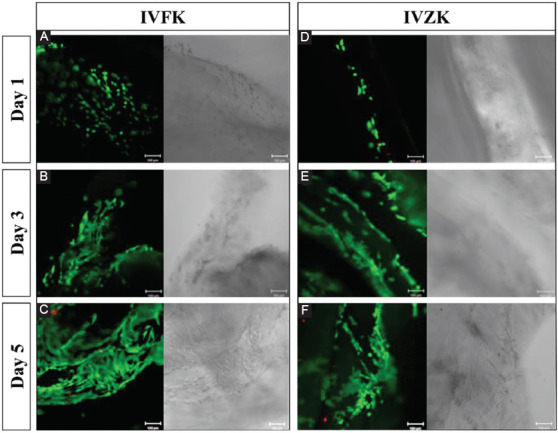
Overlaid confocal fluorescent images of three-dimensional bioprinted mouse myoblast cells in peptide hydrogels; CH-01 and CH-02 and cultured for different time points (Live cells shown in green and dead cells in red) CH-01 (A-C) and CH-02 (D-F) at Days 1, 3, and 5, respectively. Scale bars 100 μm.
4. Discussion and Conclusion
Myotube formation plays a key role in repairing muscular functions. The enhancement of differentiation of myoblast cells into myotubes using different biomaterials is a valuable area of interest. Conventionally, skeletal muscle tissue is engineered by fabricating muscle tissues in vitro using myoblast cells and modified scaffolds. Key factors including biocompatibility, biodegradability, and formation of polar parallel myotubes determine the success of tissue-repaired transplantation. Studies have shown that orderly arranged 3D scaffolds can promote cell adhesion and proliferation[29]. Ideal scaffolds should create environments that are suitable for cell proliferation, differentiation, alignment, orientation, and migration during the reparation of tissue damages[30]. This study used 3D printed structures to promote myogenesis, a process necessary for muscle repair. The structures were 3D bioprinted from biocompatible and biodegradable materials that simulate highly complex structures of extracellular matrix (ECM), and their effects on differentiation in 3D culture myoblast cells were observed.
In this study, we used previously designed tetrameric peptides for the following purposes: The first purpose aimed to test the ability of these materials to be used as scaffolds to facilitate myotube formation in a 3D culture, which is needed in muscle repair. The second purpose was to test the efficacy of our designed peptide nanogels to maintain the viability of skeletal muscle cells after 3D bioprinting. These purposes focus on the aim to assess the biocompatibility of the tetrameric peptides on skeletal muscle cell proliferation and differentiation as well as to fabricate a 3D muscle model.
The outcome of the nanofiber network formed from the self-assembling of ultrashort peptides CH-01 and CH-02 was confirmed by SEM, with an average diameter of peptide nanofibers of around 10-20 nm, where the fibers structurally resemble collagen fibers with respect to topography[31]. The diameter of these nanoscale fibers ranges within the diametric scope found in the natural ECM (5-300 nm)[32]. In our previous study[33], the mechanical stiffness and stability of both peptide nanogels were determined using oscillatory rheology based on measuring the storage modulus (G’) and loss modulus (G”). The G” values of CH-01 and CH-02 were found to be less than their G’ values indicating the gel state of both samples[34].
Cellular proliferation, adhesion and the formation of 3D cellular networks play a key role for tissue repair and regeneration. Thus, the cytocompatibility of the peptide nanogels was evaluated using mouse myoblast cells (C2C12). The in vitro investigation demonstrated that exposure of C2C12 to different concentrations of peptide nanogels did not affect cell growth when compared to cell growth in tissue culture plates and positive control, Matrigel®. The results demonstrated that the cells were metabolically active in response to different concentrations.
Based on this observation, we could confirm that the peptide nanogels are promising materials for the fabrication of muscle substitutes as well as 3D muscle graft models, particularly in the context of VML. In summary, our studies show that newly developed peptide nanogels provide native cues to mouse myoblast cells as most cells were found to be alive with very few dead cells.
In our previous paper[33], we had shown that both peptides have good printability, which opens the possibility of 3D bioprinting different cell types. In this work, the 3D bioprinted scaffolds, which simulate highly complex structures of ECM, were engineered by our custom-designed robotic 3D bioprinter. The cells were infused into the 3D constructs during printing through a custom extrusion method. The two-inlet nozzle, fabricated in-house, allowed the gelation of the peptide and even distribution of the cells within each layer of the construct. The results showed that the 3D printed scaffolds could enhance adhesion and proliferation for at least 5 days as can be seen in the results of the live-dead assay. Moreover, they could promote myotube formation and hence induce the myogenic differentiation of C2C12 myoblast cells in 3D culture. This confirms the biocompatibility of the 3D bioprinted structures and suggests that they can potentially be used as cell culture platforms for skeletal tissue engineering and regeneration.
Various studies argue that improved adhesion or proliferation of myoblasts promotes differentiation due to the confluence effect[35]. Our findings show that the 3D culture system not only enhances cell adhesion and proliferation but also helps in myogenic differentiation, as shown by the expression levels of MHC in C2C12 cells cultured within 3D scaffolds. Cell proliferation and migration can be further enhanced by forming a 3D scaffold of cell-laden layers. These scaffolds can strongly influence the polarity of cells through a process called “contact guidance”[35]. The proliferation and differentiation of the cells can only be facilitated if the cells can penetrate into the scaffolds, and hence form skeletal muscle tissues. Although the 3D bioprinted constructs could not completely mimic the structure and functions of a native cell microenvironment, their transplantation into the injured or punctured skeletal muscle in future in vivo studies may contribute to improved muscle repair. Overall, our results demonstrate that the 3D bioprinted constructs are biocompatible and may be used as biomimetic platforms to promote cell differentiation, adhesion, and proliferation.
Further, in vivo studies should be performed to assess how the 3D peptide scaffolds work when seeded together with autologous myoblast cells. Follow-up studies are critically needed as they will allow for a more precise evaluation of the injuries’ fate post-grafting. We believe that the described results represent an advancement in the context of skeletal muscle tissue engineering, opening up opportunities for tissue replacement and repair.
Acknowledgments
The research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST). The authors would like to acknowledge Dr. Dana Alhattab and Francesca Melle for their contribution to the optimization of the 3D bioprinting process.
Authors’ Contributions
CAEH supervised the project. WA designed and conducted the experiments. KK and ZK handled the 3D bioprinting experiments. All authors wrote the manuscript.
Conflicts of Interest
The authors declare that they do not have any competing interest.
References
- 1.Choi YJ, Jun YJ, Kim DY, et al. A 3D Cell Printed Muscle Construct with Tissue-derived Bioink for the Treatment of Volumetric Muscle Loss. Biomaterials. 2019;206:160–9. doi: 10.1016/j.biomaterials.2019.03.036. DOI 10.1016/j.biomaterials.2019.03.036. [DOI] [PubMed] [Google Scholar]
- 2.Kim JH, Seol YJ, Ko IK, et al. 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Scientific Reports. 2018;8:12307. doi: 10.1038/s41598-018-29968-5. DOI 10.1038/s41598-018-29968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.VanDusen KW, Syverud BC, Williams ML, et al. Engineered Skeletal Muscle Units for Repair of Volumetric Muscle Loss in the Tibialis Anterior Muscle of a Rat. Tissue Engineering Part A. 2014;20(21-22):2920–30. doi: 10.1089/ten.tea.2014.0060. DOI 10.1089/ten.tea.2014.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua CK, Yeong WY. Bioprinting:Principles and Applications. Vol. 1. Singapore: World Scientific Publishing Co, Inc; 2014. [Google Scholar]
- 5.Kwee BJ, Mooney DJ. Biomaterials for Skeletal Muscle Tissue Engineering. Current Opinion in Biotechnology. 2017;47:16–22. doi: 10.1016/j.copbio.2017.05.003. DOI 10.1016/j.copbio.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frontera WR, Ochala J. Skeletal muscle:A Brief Review of Structure and Function. Calcified Tissue International. 2015;96(3):183–95. doi: 10.1007/s00223-014-9915-y. DOI 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 7.Beldjilali-Labro M, Garcia AG, Farhat F, et al. Biomaterials in Tendon and Skeletal Muscle Tissue Engineering:Current Trends and Challenges. Materials. 2018;11(7):1116. doi: 10.3390/ma11071116. DOI 10.3390/ma11071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Relaix F, Zammit PS. Satellite Cells are Essential for Skeletal Muscle Regeneration:The Cell on the Edge Returns Centre Stage. Development. 2012;139(16):2845–56. doi: 10.1242/dev.069088. DOI 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- 9.Brack AS, Rando TA. Tissue-specific Stem Cells:Lessons from the Skeletal Muscle Satellite Cell. Cell Stem Cell. 2012;10(5):504–14. doi: 10.1016/j.stem.2012.04.001. DOI 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grogan BF, Hsu JR, Consortium STR. Volumetric Muscle Loss. JAAOS Journal of the American Academy of Orthopaedic Surgeons. 2011;19:S35–7. doi: 10.5435/00124635-201102001-00007. DOI 10.5435/00124635-201102001-00007. [DOI] [PubMed] [Google Scholar]
- 11.Turner NJ, Badylak SF. Regeneration of Skeletal Muscle. Cell and Tissue Research. 347(3):759–74. doi: 10.1007/s00441-011-1185-7. DOI 10.1007/s00441-011-1185-7. [DOI] [PubMed] [Google Scholar]
- 12.Lynch GS, Schertzer JD, Ryall JG. Anabolic Agents for Improving Muscle Regeneration and Function After Injury. Clinical and Experimental Pharmacology and Physiology. 2008;35(7):852–58. doi: 10.1111/j.1440-1681.2008.04955.x. DOI 10.1111/j.1440-1681.2008.04955.x. [DOI] [PubMed] [Google Scholar]
- 13.Järvinen TA, Järvinen TL, Kääriäinen M, et al. Muscle Injuries:Optimising Recovery. Best Practice and Research Clinical Rheumatology. 2007;21(2):317–31. doi: 10.1016/j.berh.2006.12.004. DOI 10.1016/j.berh.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Järvinen TA, Järvinen TL, Kääriäinen M, et al. Muscle Injuries:Biology and Treatment. The American Journal of Sports Medicine. 2005;33(5):745–64. doi: 10.1177/0363546505274714. DOI 10.1177/0363546505274714. [DOI] [PubMed] [Google Scholar]
- 15.Järvinen TA, Kääriäinen M, Järvinen M, Kalimo H. Muscle Strain Injuries. Current Opinion in Rheumatology. 2000;12(2):155–61. doi: 10.1097/00002281-200003000-00010. DOI 10.1097/00002281-200003000-00010. [DOI] [PubMed] [Google Scholar]
- 16.Corona BT, Wu X, Ward CL, et al. The Promotion of a Functional Fibrosis in Skeletal Muscle with Volumetric Muscle Loss Injury following the Transplantation of muscle-ECM. Biomaterials. 2013;34(13):3324–35. doi: 10.1016/j.biomaterials.2013.01.061. DOI 10.1016/j.biomaterials.2013.01.061. [DOI] [PubMed] [Google Scholar]
- 17.Manring H, Abreu E, Brotto L, et al. Novel Excitation-contraction Coupling Related Genes Reveal Aspects of Muscle Weakness Beyond Atrophy new Hopes for Treatment of Musculoskeletal Diseases. Frontiers in Physiology. 2014;5:37. doi: 10.3389/fphys.2014.00037. DOI 10.3389/fphys.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasman JM, Zayas MJ, Page RL, et al. Biomimetic Scaffolds for Regeneration of Volumetric Muscle Loss in Skeletal Muscle Injuries. Acta Biomaterialia. 2015;25:2–15. doi: 10.1016/j.actbio.2015.07.038. DOI 10.1016/j.actbio.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mase VJ, Hsu JR, Wolf SE, et al. Clinical Application of an Acellular Biologic Scaffold for Surgical Repair of a Large, Traumatic Quadriceps Femoris Muscle Defect. Orthopedics. 2010;33(7):511. doi: 10.3928/01477447-20100526-24. DOI 10.3928/01477447-20100526-24. [DOI] [PubMed] [Google Scholar]
- 20.Zorlutuna P, Annabi N, Camci-Unal G, et al. Microfabricated Biomaterials for Engineering 3D Tissues. Advanced Materials. 2012;24(14):1782–804. doi: 10.1002/adma.201104631. DOI 10.1002/adma.201104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser CA, Zhang S. Designer Self-assembling Peptide Nanofiber Biological Materials. Chemical Society Reviews. 2010;39(8):2780–90. doi: 10.1039/b921448h. DOI 10.1039/b921448h. [DOI] [PubMed] [Google Scholar]
- 22.Loo Y, Zhang S, Hauser CA. From Short Peptides to Nanofibers to Macromolecular Assemblies in Biomedicine. Biotechnology Advances. 2012;30(3):593–603. doi: 10.1016/j.biotechadv.2011.10.004. DOI 10.1016/j.biotechadv.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Wu EC, Zhang S, Hauser CA. Self-assembling Peptides as Cell-interactive Scaffolds. Advanced Functional Materials. 2012;22(3):456–68. DOI 10.1002/adfm.201101905. [Google Scholar]
- 24.Hauser CA, Deng R, Mishra A, et al. Natural Tri-to Hexapeptides Self-assemble in Water to Amyloid β-type Fiber Aggregates by Unexpected α-helical Intermediate Structures. Proceedings of the National Academy of Sciences. 2011;108(4):1361–6. doi: 10.1073/pnas.1014796108. DOI 10.1073/pnas.1014796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra A, Loo Y, Deng R, et al. Ultrasmall Natural Peptides Self-assemble to Strong Temperature-resistant Helical Fibers in Scaffolds Suitable for Tissue Engineering. Nano Today. 2011;6(3):232–9. DOI 10.1016/j.nantod.2011.06.010. [Google Scholar]
- 26.Pollot BE, Rathbone CR, Wenke JC, et al. Natural Polymeric Hydrogel Evaluation for Skeletal Muscle Tissue Engineering. Journal of Biomedical Materials Research Part B:Applied Biomaterials. 2018;106(2):672–9. doi: 10.1002/jbm.b.33859. DOI 10.1002/jbm.b.33859. [DOI] [PubMed] [Google Scholar]
- 27.Kahin K, Khan Z, Albagami M, et al. Development of a Robotic 3D Bioprinting and Microfluidic Pumping System for Tissue and Organ Engineering. Microfluidics, Biomems, and Medical Microsystems. 2019 Doi 10.1117/12.2507237. [Google Scholar]
- 28.Khan Z, Kahin K, Rauf S, et al. Optimization of a 3D Bioprinting Process Using Ultrashort Peptide Bioinks. International Journal of Bioprinting. 2018;5(1):1–3. doi: 10.18063/ijb.v5i1.173. DOI 10.18063/ijb.v5i1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenneisen P, Blaudschun R, Gille J, et al. Essential role of an Activator Protein-2 (AP-2)/Specificity Protein 1 (Sp1) Cluster in the UVB-mediated Induction of the Human Vascular Endothelial Growth Factor in HaCaT Keratinocytes. Biochemical Journal. 2003;369(2):341–9. doi: 10.1042/BJ20021032. DOI 10.1042/bj20021032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendriks J, Riesle J, van Blitterswijk CA. Co-culture in Cartilage Tissue Engineering. Journal of Tissue Engineering and Regenerative Medicine. 2007;1(3):170–8. doi: 10.1002/term.19. DOI 10.1002/term.19. [DOI] [PubMed] [Google Scholar]
- 31.Loo Y, Lakshmanan A, Ni M, et al. Peptide Bioink:Self-assembling Nanofibrous Scaffolds for Three-dimensional Organotypic Cultures. Nano Letters. 2015;15(10):6919–25. doi: 10.1021/acs.nanolett.5b02859. DOI 10.1021/acs.nanolett.5b02859. [DOI] [PubMed] [Google Scholar]
- 32.Jayawarna V, Ali M, Jowitt TA, et al. Nanostructured Hydrogels for Three-dimensional Cell Culture Through Self-assembly of Fluorenylmethoxycarbonyl Dipeptides. Advanced Materials. 2006;18(5):611–4. DOI 10.1002/adma.200501522. [Google Scholar]
- 33.Arab W, Rauf S, Al-Harbi O, et al. Novel Ultrashort Self-Assembling Peptide Bioinks for 3D Culture of Muscle Myoblast Cells. International Journal of Bioprinting. 2018;4(2):129. doi: 10.18063/IJB.v4i2.129. DOI 10.18063/ijb.v4i1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lakshmanan A, Cheong DW, Accardo A, et al. Aliphatic Peptides Show Similar Self-assembly to Amyloid Core Sequences, Challenging the Importance of Aromatic Interactions in Amyloidosis. Proceedings of the National Academy of Sciences. 2013;110(2):519–24. doi: 10.1073/pnas.1217742110. DOI 10.1073/pnas.1217742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Zhong J, Wang J, et al. Enhanced Growth and Differentiation of Myoblast Cells Grown on E-jet 3D Printed Platforms. International Journal of Nanomedicine. 2019;14:937–50. doi: 10.2147/IJN.S193624. DOI 10.2147/ijn.s193624. [DOI] [PMC free article] [PubMed] [Google Scholar]


