Abstract
The study of biodiversity, growth, development, and metabolism of cultivated microorganisms is an integral part of modern microbiological, biotechnological, and medical research. Such studies require the development of new methods of isolation, cultivation, manipulation, and study of individual bacterial cells and their consortia. To this end, in recent years, there has been an active development of different isolation and three-dimensional cell positioning methods. In this review, the optical tweezers, surface heterogeneous functionalization, multiphoton lithography, microfluidic techniques, and laser printing are reviewed. Laser printing is considered as one of the most promising techniques and is discussed in detail.
Keywords: Laser printing, bacteria isolation, soil, unculturable microorganisms, biodiversity
1. Introduction
According to current evaluations, more than 99% of the prokaryotes biodiversity remain unculturable when using traditional cultivation methods[1-4]. To investigate the biodiversity and functions of unculturable microorganisms in situ, different molecular biological methods are used[5-7]. At the same time, cultivation is still necessary for a detailed investigation of the growth and metabolism of microorganisms, genetic manipulations, and introduction of promising strains in biotechnological processes[8-11]. The study of processes carried out by microorganisms in nature is necessary for understanding of functioning principles of ecosystems, assessment of biogeochemical flows of matter, modeling, and forecasting of various biospheric processes, which are associated with the problems of climate change and anthropogenic influence on the biosphere[12,13]. It is equally important that new species and strains of microorganisms can be valuable producers of bioactive substances, such as enzymes and antibiotics[14,15].
The impossibility of complete reproduction of the conditions of the natural habitat of microorganisms in the laboratory is considered the main reason for their unculturability[4,8]. In natural heterogeneous environments, such as lands, soils, and bottom sediments, physical-chemical conditions of the existence of microorganisms change on a microscale[16-18]. Such natural environments are combinations of microzones, different in concentration and composition of organic matter (including microbial metabolites), salts, gases, mineral composition, and other parameters. It is obvious that the reproduction of the entire set of environmental parameters in vitro is impossible. Intra- and inter-population interactions play an important role in the functioning of microorganisms within natural ecosystems[19-23]. In particular, such interactions include the formation of biofilms, production of growth regulators and signal metabolites, and production of antibiotics. Sample preparation procedures for traditional cultivation methods, including desorption of cells from mineral particles, disrupt communications between the microorganisms, which is especially important for strongly associated groups such as symbionts and pathogens[24].
2. New Isolation Methods
Nowadays, a wide variety of methods is used to expose new, previously uncultivated microorganisms[8]. In particular, new nutrient media containing specific substances are used; cultivation is carried out under different physical and chemical conditions of the environment, such as atmospheric composition, temperature, and pH.[8,25] Furthermore, applications of diluted nutrient media in combination with long incubation periods[25,26], as well as, joint cultivation of microorganisms of different species, have been studied[1,27]. In recent years, some fundamentally new methods of cultivation based on the placement of microorganisms in the natural environment without the usage of nutrient media have been developed. Such methods utilize diffusion chambers[28,29] and polymer coatings[30] in which microbial cells are placed. In this case, microorganisms receive all necessary nutrients from the natural environment, remaining isolated from it. The allocation of a new antibiotic producer (teixobactin) using such methods is considered as an important achievement[14]. Another approach includes simultaneous cultivation and screening of tens and hundreds of thousands of bacterial microcolonies on porous polymer or ceramic isolation chips[31,32]. For the cultivation of “unculturable” microorganisms, methods allowing isolation of single cells from natural environments can also be used. Among these methods, the most popular is based on the dilution of microorganism suspension, flow cytometry and cell sorting, laser microdissection, compartmentalization, and application of micromanipulators[33,34]. In addition to the cultivation of previously uncultivated species of microorganisms, the isolation of single microbial cells is necessary for the study of cell physiology, interactions between cells, as well as for the search of new metabolites, such as antibiotics and enzymes[34].
Modern scientific and technological progress provides many opportunities in terms of the development of novel methods for cultivation, isolation, manipulation, and study of individual bacterial cells and their consortia. New approaches may speed up the process of working with microorganisms significantly and allow carrying out their more complete and comprehensive studies. It is necessary to note that so far very few methods have been proposed for positioning of bacteria arrays with micrometer accuracy. In Akselrod et al.[35], three-dimensional (3D) networks of living cells in hydrogel were formed without loss of their viability using arrays of multiplexed holographic optical traps (tweezers) with unprecedented accuracy (<400 nm). To form optical traps, two lasers were used: Ar+ laser (20 W, 514 nm wavelength) and continuous wave Ti: Sapphire laser, tunable in the range of λ = 850–900 nm, as well as a combination of two diffraction elements, combined with different lenses in an inverted optical microscope. Networks of 3T3 fibroblasts surrounded by a ring of bacteria were formed. The ability to manipulate hundreds of Pseudomonas aeruginosa bacteria simultaneously in two-dimensional (2D) and 3D arrays was also demonstrated. The method of holographic optical trapping is very accurate but technically difficult to perform.
In the study of Rowan et al.[36], the method of heterogeneous functionalization of surfaces is proposed, which is a four-step lithographic process based on microcontact printing of organic monolayers, implantation of hyperbranched polymer, and its further functionalization. As a result, structures, on which the directed inoculation of bacterial cells is carried out, are obtained. The investigations of cell survival have shown that cells remain viable on the obtained structured surfaces. Large isolates of bacteria containing 18 ± 5 bacteria and small isolates containing 2 ± 1 bacteria were obtained. According to this paper, the demonstrated method can be used for high-throughput screening and biosensing. However, it is difficult to combine the heterogeneous functionalization of surfaces using this method[36] with routine biological research and conditions of microorganism cultivation (temperature, pH, nutrients, etc.). It is necessary to note that the task of finding simple ways to provide high resolution of living bacteria arrays with the opportunity of various biological studies was solved in some publications. The approaches proposed in these works are, in fact, the harbingers of 3D printing.
In a study of Weibel et al.[37], the technique of living bacteria stamping on agarose plates was proposed. Bacteria arrays were printed (the size of a single spot with bacteria >200 µm) in the area of up to 50 cm2. Polydimethylsiloxane (PDMS) stamps were produced with the help of photolithographic technique. The achieved minimum size of the print protrusion was 190 µm at the height of 140 µm, which, however, is far from the size required to separate bacteria. This method is fast, reproducible, and convenient and can be used to control the pattern, spacing, and orientation between colonies of different bacteria. In Xu et al. study[38], living bacteria arrays with cellular resolution were printed on agarose substrate using elastomeric (PDMS) stamps with a high aspect ratio, obtained by the reverse in situ lithography (RISL) method. Figure 1 shows the advantages of the RISL method over the standard ultraviolet photolithography. The only limitation of the RISL technology is the protrusion diameter, which can hardly be <1 µm due to the optical diffraction limit.
Figure 1.

The advantages of the RISL method over the standard ultraviolet photolithography. “Reprinted with permission from (Xu L, Robert L., Ouyang Q., Taddei F., Chen Y., Lindner A. B., Baigl D. Microcontact printing of living bacteria arrays with cellular resolution//Nano Lett. -2007. Vol. 7 - № 7. - P. 2068–2072). Copyright (2007) American Chemical Society.”
The method of microcontact printing of bacteria arrays works as follows: The drop of Escherichia coli in the culture medium LB is deposited on agarose gel (3 weight [wt]% in LB) and, on the agarose substrate, a monolayer of bacteria is formed (the liquid is absorbed by agarose gel). Then, the PDMS stamp contacts with a monolayer of bacteria covering the agarose gel. The stamp is removed, and the part of bacteria remains on it. Then, on the contact of the stamp with the layer of agarose gel (4 wt% in LB, thickness 200 µm), bacteria are transferred to the agarose layer. Thus, in a few seconds, arrays of E. coli bacteria can be printed directly on the agarose substrate with micron resolution, up to single bacteria, on a large area (cm2). It was shown that, after the pattern is printed, bacteria continue to grow and divide, as in the conditions of mass culture, i.e., the bacteria retain their normal physiological behavior after printing. It was also shown that the agarose concentration is crucial for good printing performance. Too little concentration leads to a distortion of the printed pattern, while too high concentration is not suitable for bacteria cultivation. To obtain arrays of single bacteria, the effect of reduction of the initial bacteria concentration in a drop of the culture medium LB was studied. For the initial concentrations of 109 and 108 cells/ml, the average number of bacteria per spot was measured at 12.1 and 1.4, respectively. A very narrow distribution was obtained at the low concentration of bacteria: 44.6% of spots had only one E. coli cell and 40.1% of spots had 0 or 2 cells. These results demonstrated that the microcontact printing allows the production of regular arrays of single bacteria. Moreover, it was shown that this approach to the separation of bacterial arrays allows analyzing the growth rates of individual lines of bacteria. This methodology provides a simple way for any desired spatial 2D distribution of bacteria and can be used for both screening and studies of bacterial phenotypic variation, population dynamics, and evolution of ecosystems.
A fast-developing area - sociomicrobiology - has identified the mechanisms with the help of which bacteria participate in collaborative and competitive relationships by influencing nearby neighbors through physical contact and modification of the chemical composition of their common microenvironment. To explore the behavior of small microbial aggregates, different technologies of microprocessing that limit bacteria in microfluidic devices, microresonators, and ultra-low volume liquid droplets were developed[39-45]. The ability to integrate analytical systems with microfluidics has made these isolation platforms attractive for high-performance screening for antibiotic resistance and enzymatic activity analysis. For example, in Eun et al.[39], a high-performance analysis and isolation of bacterial cells encapsulated in agarose microparticles with the use of fluorescent-activated cell sorting (FACS) are described. Flow-focusing microfluidic systems were used to create monodisperse microparticles with a diameter of ≈30 µm. The sizes of these particles made them compatible with flow cytometry and FACS, and the sensitivity of these methods reduced the incubation time for cell replication before carrying out of the analyses. The small volume of microparticles (≈1–50 picoliter [pl]) minimized the number of reagents required for bacterial studies. This platform made it possible to allocate and isolate bacteria effectively, as well as to use the combination of methods for quick identification of targets for biologically active small molecules. As an experimental demonstration of this method, E. coli cells were encapsulated in agarose microparticles, incubated in the presence of different concentrations of rifampicin, and analyzed with the use of FACS. The minimum inhibitory concentration of rifampicin was determined, and spontaneous mutants that had antibiotic resistance were isolated with the help of FACS and characterized by DNA sequencing. Using this approach, the time and amount of antibiotics needed to isolate mutants were reduced by 8 and 150 times, respectively, compared to traditional microbiological methods using nutrient agar plates. Thus, this method is important in the fields of chemical biology, chemistry of natural products, as well as for the discovery and characterization of biologically active secondary metabolites.
The approaches mentioned above have been useful for limiting the size, shape, and physical attributes (microhabitat); however, none of them have provided the opportunity to determine the 3D geometry of bacterial aggregates or the orientation of multiple populations arbitrarily. In addition, the process of cell encapsulation in ultra-low volume cavities often limits mass transport, leading to conditions that are incompatible with growth and signaling between physically isolated populations. A growing number of proofs highlight the importance of microcolonies in bacterial reproduction[27]; however, the lack of tools for systematic assessment of cell behavior in such communities is observed. New strategies for the creation of a 3D cultural environment on a microscopic scale can play a crucial role in identifying how bacteria manage antibiotic resistance and other social behaviors in small dense aggregates.
In Connell et al. and Connell et al.[46,47], the method of laser formation of microscopic 3D chambers based on multiphoton lithography (MPL)[48-51] is described. The MPL method has high-throughput capacity and the ability to produce arbitrary patterns. It offers opportunities for the industrial production of 3D microdevices such as micro-optical components, scaffolds for tissue engineering, and microfluidic chips. In Connell et al. study[52], bovine serum albumin (BSA), a highly soluble protein that can be crosslinked into porous, durable, and biocompatible hydrogels, was used to create microscopic 3D bacterial chambers. Some separate bacteria were enclosed in the BSA chambers with the volume of 1 pl and were then grown in clonal populations. Due to the diffusion of biologically significant molecules and antibiotics through the walls of BSA, as well as the exchange of quorum-sensitive signals, the social behavior of bacterial communities was studied in relation to the size and density of the population, the shape of the container, and the flow rate of the environment.
Within the human body, bacteria usually exist in structured 3D communities consisting of several bacterial species. To get detailed information about the effect of geometry on pathogenicity, a 3D printing strategy is described in Connell et al.[52] for bacterial communities, in which physically distinct but chemically interactive populations of a certain size, shape, and density can be organized in any form essentially (Figure 2). Using this approach, it was shown that the stability of a single pathogenic species of bacteria to the antibiotic might enhance the resistance of the second species because of their 3D relations. With the help of laser lithographic technique, microscopic containers of up to 1 pl volume with a container wall thickness of up to 2 µm around selected bacteria suspended in gelatin were formed by cross-linking of the polypeptide molecules in the focal region due to non-linear absorption of laser radiation by photosensitizer molecules. The result of this multiphoton absorption is the formation of singlet oxygen, which stimulates intramolecular and intermolecular covalent cross-linking reactions between BSA and gelatin. The unique physical and chemical properties of gelatin have motivated the interest in its use for a variety of applications, including storage, immobilization, and 3D cultivation of bacteria[53-55]. After removal of the excess reagent, the bacteria are localized in sealed cavities formed by cross-linked gelatin, which is a highly porous material and supports a rapid growth of fully enclosed cell populations. It is easily permeable for polypeptides, antibiotics, and to the physical and chemical signals with the help of which interaction between bacteria occurs. The isolation of cells in microcontainers provides the opportunities for embedding of different types/densities of containers into each other, as well as for dynamic changes in the orientation of the entire populations of bacteria within the community.
Figure 2.

Gelatin-based micro-three-dimensional printing in the presence of bacteria. (Left) engineering polymicrobial communities (right) “Reprinted from (Connell J.L., Ritschdorff E.T., Whiteley M., Shear J.B. 3D printing of microscopic bacterial communities // Proc. Natl. Acad. Sci. - 2013. - Vol. 110 - № 46. - P. 18380–18385).”
The authors have shown that spatially localized interactions of the Gram-positive Staphylococcus aureus and the Gram-negative P. aeruginosa bacteria (two human pathogens that often form persistent coinfections inside wounds, catheters, and lung of patients with mucoviscidosis) may increase Staphylococcus survival in the treatment with the β-lactam antibiotics.
Micro-3D cell printing fundamentally expands the possibilities for probing of antibiotic resistance when a single bacterial microgroup can affect the antibiotic susceptibility of adjacent surrounding or embedded populations - a matter of particular relevance for in vivo infections (e.g., wounds, oral cavity, and cystic fibrosis of the lungs) where tissues are often colonized by several species of bacteria simultaneously. The true power of 3D cell printing lies in the ability to organize the microbial communities in the unlimited range of geometries. Micro-3D cell printing can also be a valuable tool for the investigations of mechanisms and dynamics of the adaptive responses to environmental conditions.
3. Laser Bioprinting
Over the past two decades, bioprinting, including the printing of mammalian and bacterial cells, has become an extensive field of research. Printers, starting with modified inkjet printers, extrusion pens, electrospinning, and laser systems, have demonstrated the ability to create submillimeter resolution samples of biomaterials. Tests for viability, genetic damages, cell differentiation, and stress tests were performed after printing to demonstrate that each of these tools can form patterns and 3D structures of intact living cells directly without the aid of surface functionalization or patterns (lithography, masks, etc.)[56,57]. Currently, bioprinting is used in laboratories all over the world to print living cells ranging from stem cells, bacteria, and viruses to create microchips and 3D tissue engineering constructs in vitro [57-60].
Most methods, such as inkjet printers and extrusion pens, require a nozzle or print head to print microdrops of “bio-ink”[60]. These nozzles are unable to print solid particles without clogging up. The modified method of laser-induced forward transfer (LIFT), such as biological laser printing, does not require the nozzles or holes of any type because it is based on a focused laser beam. Laser bioprinting based on LIFT (Figure 3) is a relatively new bioprocessing technique[61] for placing biological materials or living cells in well-defined positions on samples (Figure 4). This method allows fast transfer of ultra-small amounts of biological material to different substrates with spatial accuracy better than 5 µm at the deposition rate up to 100 pixels of biological material per second. With the help of laser bioprinting, one can successfully create print arrays and samples of biological materials from liquid and solid-phase “bio-inks,” including proteins, viruses, mammalian cells, and bacteria[56,57,59,61,62].
Figure 3.
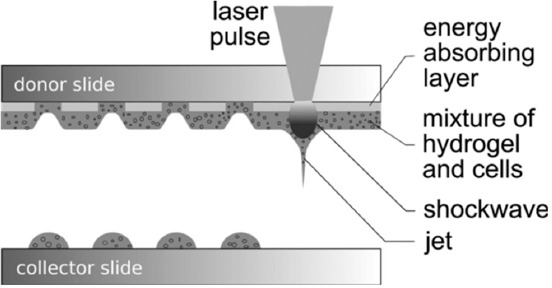
Schematic sketch of the laser-assisted bioprinting.
Figure 4.

Gel/soil microdroplets on an acceptor plate (A), soil microparticles distribution in microdroplets (B), and colonies as the result of microbial growth after gel/soil printing of gel/soil microdroplets onto agar plates (C) with E = 20 µJ.
Laser bioprinting of microorganisms opens the door for the development of new and accurate methods that could be used for the study of: (i) The development of microorganisms in solid matrices in the presence of nutrient gradients, (ii) interactions of the same and different organic colonies next to each other, (iii) response to the stress and resistance to inhibitors, and (iv) cellular communication or quorum determination. This method provides a relatively simple way to perform experiments with a large number of replicas and can even be used for the selection of strains in the future. Laser printing can also serve as a means for carrying out multifactor experiments[63].
The printing process provides an unprecedented level of accuracy. Using traditional methods, cell suspensions can be diluted in sterile environments and manually placed as droplets in certain positions on the growth matrix; realistically, the volume of the droplet cannot be <1 µl, and the accuracy of the human hand will require that the droplets were not located closer than 2–3 mm from each other. Laser printing of microdroplets of cell suspensions is carried out with micron accuracy and drop volumes of <10 pl.
The principle of laser printing is as follows: The donor slide is covered with a layer that absorbs laser radiation and a layer of biomaterial that needs to be transferred; usually, it is hydrogel with cells. Laser pulses are focused through the upper glass slide in the absorbing layer (Figure 3). The evaporation of this layer creates a high gas pressure that transfers the biomaterial to the bottom slide. The vapor bubble reaches its maximum volume in a few microseconds and collapses when its internal pressure drops below the atmospheric pressure[64]. However, accelerated biomaterial continues to move by inertia to the receiving slide and forms a thin jet at the front of the bubble, which lasts several hundred milliseconds. As a result, the volume from several pl to several nanoliters (nl) is transferred to the surface of the receiving slide (a collector) in the form of a drop (Figure 4). Biomaterial droplets can be arranged in 2D models by moving the donor and collector slides relatively to each other. The volume of printed droplets depends on the laser pulse energy, the thickness of the biomaterial layer, as well as the viscosity of the biomaterial layer on the donor slide[65]. The number of cells in each droplet usually depends on the initial cell density in the biomaterial layer and the volume of the printed droplet.
In a study of Taidi et al.[63], laser bioprinting was used for the precise placement of eukaryotic microorganisms in certain patterns. Saccharomyces cerevisiae var . bayanus and Chlorella vulgaris were the first used as the model organisms for this purpose. The authors used laser pulses with a wavelength of 1064 nm, pulse duration of 10 ns, and pulse energy of about 20 μJ corresponding to laser energy density from 1 up to 2 J/cm2 at the focal point, which was focused through the donor slide on the absorbent surface (60 nm of gold). The conditions used for printing of S. bayanus and C. vulgaris were droplet volume (180 pl) and the cell concentration of 200 cells per a droplet. The growth and development of microcolonies were studied by confocal microscopy, and the growth rates of colonies were determined by the image analysis. The developed protocols for printing of microorganisms and determining the growth rate of microcolonies are very promising for future studies of the growth and development of colonies.
In Koch et al. study[66], skin cell lines (fibroblasts/keratinocytes) and human mesenchymal stem cells (hMSCs) were selected for laser printing experiments because of their high potential in human skin regeneration and new applications of stem cell therapy. The effect of laser printing on cell survival, proliferation, apoptotic activity, and DNA damage has been investigated. Approximately 98% of skin cells and about 90% of hMSC cells survived after the laser printing procedure. All used cell types kept their ability to reproduce after laser printing. In addition, skin cells and hMSC showed no increase in apoptosis or the DNA fragmentation. The hMSCs also have maintained their phenotype, as confirmed by the analysis of sorting with the help of FACS. This study declares laser printing as a suitable method for computer positioning of different cell types and a promising tool for future applications in ex vivo tissue generation.
In Deng et al. study[67], the influences of laser pulse energy, laser spot size, distance to the acceptor substrate on the number, size, and proliferation of laser printed HELA cells are analyzed. It is shown that the laser power and the thickness of the titanium film are the main factors affecting the survival of the isolated cells. To provide a sufficient working distance and increase the viscosity of the culture medium, glycerin was used. To soften the landing of the cell on the acceptor plate a layer of alginate was used. It was found that the optimal parameters to obtain a viable cell are pulse energy - 9 µJ, spot size - 60 µm, the thickness of the titanium film - 12 nm, working distance - 700 µm, the concentration of glycerin in the culture medium 2–4%, and the thickness of the alginate is more than 1 µm. To avoid contamination and increase humidity, the process of cell shooting was carried out in a special chamber made from PDMS.
It should be noted that very few works devoted to the isolation of microorganisms from complex heterogeneous systems with the use of laser printing are existing in the literature. Nowadays, two modifications of the laser printing method - biological laser printing[68,69] and laser engineering of microbial systems (LEMS) - have been proposed for the cultivation of microorganisms from natural environments[70-72].
As it was noted above, traditional methods to isolate microorganisms from environmental samples, such as soil or sediment, require pretreatment to remove living cells from their solid-phase carrier, creating a liquid phase sample. This process destroys close relationships that can be crucial for the cultivation and study of the isolated microorganisms. In Ringeisen et al. study[68], a high-performance automated method based on laser printing that isolates pure microbial cultures and spatially bound microbial consortia directly from solid-phase complex environmental samples is described. A mixture of soil with water or water and glycerol was applied to a quartz tape coated with titanium dioxide, 85 nm thick, producing a donor slide. Adjustable amounts of soil were transferred to different substrates using a pulsed excimer laser (wavelength 248 nm, pulse duration in the range of 2–10 ns, and pulse energy varied from 7 to 23 μJ), including 96-well plates filled with broth at the rate exceeding 20 microparticles per second or more than a thousand microparticles per minute. After printing, the viability of microbial cultures, culture value, and significant morphological diversity have been demonstrated. However, it is not clear whether it exceeds the diversity obtained by cultivation with the use of traditional methods. Nevertheless, the results showed that single-stage soil printing could be used to (a) produce pure microbial cultures (isolates) and (b) isolate consortia from the micro-ecological system. The study, described here, is the first extension of bioprinting to solid-phase environmental samples for the isolation and cultivation of individual microorganisms or consortia.
The LEMS method uses 8 ns, 24 μJ, 1.06 µm laser pulses[70-72]. The donor plate is a glass coated with a 50–100 nm layer of gold, titanium, or chromium. To print biological objects, bacterial cells or soil are mixed with a gel (2% hyaluronic acid), which prevents rapid drying of the sample and spraying of microdroplets during laser printing. This technology allows obtaining a large number of separate bacterial colonies. It was also demonstrated that the LEMS technology allows cultivating a significantly higher bacterial diversity in comparison with traditional methods of cultivation (Figures 5 and 6). In particular, with the use of this method, a strain of a rare Nonomuraea [70,71] genus was isolated from the soil (Figure 5), while the isolation of bacteria of this kind by traditional methods requires the use of a number of special techniques, for example, the addition of antibiotics and vitamins to the nutrient media[73,74]. When using the LEMS method, in addition to increase of the microorganisms diversity, an increase in the number of cultivated microorganisms was observed[70,72]. This effect was demonstrated both for natural samples and for pure cultures of bacteria.
Figure 5.

Cultivated and identified groups of G+ and G− bacteria from the mollisol soil using the standard method and laser engineering of microbial systems technology.
Figure 6.

A diagram illustrating the main differences between the laser engineering of microbial systems and standard method, leading to an increase in biodiversity in the isolation of microorganisms from soil. The numbers indicate microbes that, with the standard cultivation method: 1 - easy to flush out of their microenvironment, 2 - most actively multiply, 3 - separate from those with which they exist in symbiosis, and 4 - remain in the “sleeping” state.
The reasons for the high efficiency of laser printing methods in the isolation of bacteria from heterogeneous environments are not yet sufficiently investigated. Probably, one of the reasons is that, unlike traditional cultivation methods, these methods do not use the desorption of microorganisms from mineral particles[68,70,71]. The direct printing techniques are likely to isolate neighboring microorganisms while they are still attached to soil particles (Figure 6). In this case, the natural micro-ecological environment and relationships between microorganisms in the process of isolation and screening are maintained. In the case of preserving the relationships between microorganisms, it is possible to cultivate a wider microbial diversity[1,24,68]. Changes in microbial diversity and activation of bacterial growth may also be caused by laser radiation. In the LEMS method, even though most of the radiation is absorbed by the metal film, a small part of the radiation reaches the gel-containing microbial cells[75]. At the same time, it is known that laser radiation can activate the growth and metabolism of bacteria[76-78]. Another factor that can affect the state of microorganisms during laser printing is the formation of nanoparticles, generated from the absorbing metal film on the donor glass slide[71,72,75]. Nanoparticles, in relation to their shape, size, they material and some other factors, can be both toxic to bacteria or stimulate their growth[79-82]. However, in general, the processes that cause changes in microbial diversity and growth during and after laser printing are practically not studied and require further investigations.
In addition to the isolation of microorganisms from complex substrates, it is assumed to use laser printing methods to create biochips and bioelectronic interfaces. It is shown that various modifications of laser printing allow transferring cells of pure microorganism cultures to various acceptor surfaces with high accuracy (in the zone <50 × 50 µm)[61,83-85]. At the same time, very high survival of microorganisms (up to 100%) is observed. As it was mentioned before, laser printing can also be used to isolate single microorganism cells[67,68], which is necessary for the study of growth, metabolism, the genome of individual cells, as well as for the study of interactions between microbial cells[27,68,86,87]. One of the relevant tasks of modern biotechnology is the creation of multilayer structures consisting of microorganisms to produce some new materials, in particular, bioplastics, adhesives, bio-based electrical switches, etc.[88-91] Nowadays, the use of 3D printers can help to solve these biotechnology problems[91]. Application of laser printing in this field is very promising due to the high accuracy of microbial transfer and high cell survival.
In conclusion, at present, there are only a few publications on the use of laser printing for the transfer of microorganisms. However, it has been shown that laser printing is effective for isolating microorganisms from complex natural substrates, as well as for transferring microbial cells to various acceptor surfaces with high speed and accuracy. Due to this fact, it is expected that this technology will be actively used for the studies of microbial diversity of different ecosystems, for the creation of biochips and bioelectronic interfaces, isolation of single cells of microorganisms, and printing of multilayer cell structures and biofilm studies.
Acknowledgments
This work was supported by the Ministry of Science and Higher Education within the Russian State assignment FSRC Crystallography and Photonics RAS. BNC also acknowledges financial support from deutsche forschungsgemeinschaft, the Cluster of Excellence REBIRTH, and biofabrication for NIFE project (Land Niedersachsen/Volkswagenstiftung).
References
- 1.Vartoukian S R, Palmer R M, Wade W G. 2010, Strategies for culture of “unculturable”bacteria. FEMS Microbiol Lett. 309(1):1–7. doi: 10.1111/j.1574-6968.2010.02000.x. https://doi.org/10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 2.Whitman W B, Coleman D C, Wiebe W J. 1998, Prokaryotes:The unseen majority. Proc Natl Acad Sci. 95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. https://doi.org/10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Head I M, Saunders J R, Pickup R W. 1998, Microbial evolution, diversity, and ecology:A decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 35(1):1–21. doi: 10.1007/s002489900056. https://doi.org/10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 4.Alain K, Querellou J. 2009, Cultivating the uncultured:Limits, advances and future challenges. Extremophiles. 13(4):583–594. doi: 10.1007/s00792-009-0261-3. https://doi.org/10.1007/s00792-009-0261-3. [DOI] [PubMed] [Google Scholar]
- 5.Sogin M L, Morrison H G, Huber J A, et al. 2011, Microbial diversity in the deep sea and the underexplored “rare biosphere”. Handb Mol Microb Ecol II Metagenomics Differ Habitats. 2011(30):243–252. [Google Scholar]
- 6.Hultman J, Waldrop M P, Mackelprang R, et al. 2015, Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 521(7551):208–212. doi: 10.1038/nature14238. https://doi.org/10.1038/natur.e14238. [DOI] [PubMed] [Google Scholar]
- 7.Jansson J K, Taş N. 2014, The microbial ecology of permafrost. Nat Rev Microbiol. 12(6):414–425. doi: 10.1038/nrmicro3262. https://doi.org/10.1038/nrmicro3262. [DOI] [PubMed] [Google Scholar]
- 8.Pham V H T, Kim J. 2012, Cultivation of unculturable soil bacteria. Trends Biotechnol. 30(9):475–484. doi: 10.1016/j.tibtech.2012.05.007. https://doi.org/10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Shao X, Mugler A, Kim J, et al. 2017, Nemenman I Growth of bacteria in 3-d colonies. PLoS Comput Biol. 13(7):1–19. doi: 10.1371/journal.pcbi.1005679. https://doi.org/10.1371/journal.pcbi.1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohanim Y K, Levi D, Jona G, et al. 2018, A bacterial growth law out of steady state. Cell Rep. 23(10):2891–2900. doi: 10.1016/j.celrep.2018.05.007. https://doi.org/10.1016/j.celrep.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Scott M, Hwa T. 2011, Bacterial growth laws and their applications. Curr Opin Biotechnol. 22(4):559–565. doi: 10.1016/j.copbio.2011.04.014. https://doi.org/10.1016/j.copbio.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman D C, Callaham M A, Crossley D A., Jr . 2017, Fundamentals of Soil Ecology. London: Academic Press; [Google Scholar]
- 13.Singh B K, Bardgett R D, Smith P, et al. 2010, Microorganisms and climate change:Terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 8(11):779–790. doi: 10.1038/nrmicro2439. https://doi.org/10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- 14.Ling L L, Schneider T, Peoples A J, et al. 2015, A new antibiotic kills pathogens without detectable resistance. Nature. 517(7535):455–459. doi: 10.1038/nature14098. https://doi.org/10.1038/natur.e14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodfellow M, Nouioui I, Sanderson R, et al. 2018, Rare taxa and dark microbial matter:Novel bioactive actinobacteria abound in Atacama desert soils. Antonie van Leeuwenhoek. 111(8):1315–1332. doi: 10.1007/s10482-018-1088-7. https://doi.org/10.1007/s10482-018-1088-7. [DOI] [PubMed] [Google Scholar]
- 16.Hattori T, Hattori R. 1976, The physical environemnt in soil microbiology:An attempt to extend priciples of microbiology to soil microorgansims. CRC Crit Rev Microbiol. 4:423–461. doi: 10.3109/10408417609102305. https://doi.org/10.3109/10408417609102305. [DOI] [PubMed] [Google Scholar]
- 17.Stotzky G. 1986, Interactions of soil minerals with natural organics and microbes. In: Huang P M, Schnitzer M, editors. Influence of Soil Mineral Colloids on Metabolic Processes, Growth, Adhesion, and Ecology of Microbes and Viruses. Madison, WI, USA: Soil Science Society of America; pp. p305–428. [Google Scholar]
- 18.Torsvik V, Øvreås L. 2008, Microbial diversity, life strategies, and adaptation to life in extreme soils. Microbiol Extrem Soils. 13:373. https://doi.org/10.1007/978-3-540-74231-9_2. [Google Scholar]
- 19.Fajardo A, Martínez J L. 2008, Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 11(2):161–167. doi: 10.1016/j.mib.2008.02.006. https://doi.org/10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 20.García-Contreras R, Nuñez-López L, Jasso-Chávez R, et al. 2015, Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 9(1):115–125. doi: 10.1038/ismej.2014.98. https://doi.org/10.1038/ismej.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Registan G I, Mulyukin A L, Nikolaev Y A, et al. 2005, The role of microbial low-molecular-weight autoregulatory factors (alkylhydroxybenzenes) in resistance of microorganisms to radiation and heat shock. Adv Space Res. 36(9):1718–1728. https://doi.org/10.1016/j.asr.2005.02.070. [Google Scholar]
- 22.El-Registan G I, Mulyukin A L, Nikolaev Y A, et al. 2006, Adaptogenic functions of extracellular autoregulators of microorganisms. Microbiology. 75(4):380–389. https://doi.org/10.1134/S0026261706040035. [PubMed] [Google Scholar]
- 23.Decho A W. 2000, Microbial biofilms in intertidal systems:An overview. Cont Shelf Res. 20(10-11):1257–1273. https://doi.org/10.1016/S0278-4343(00)00022-4. [Google Scholar]
- 24.Joint I, Mühling M, Querellou J. 2010, Culturing marine bacteria-an essential prerequisite for biodiscovery:Minireview. Microb Biotechnol. 3(5):564–575. doi: 10.1111/j.1751-7915.2010.00188.x. https://doi.org/10.1111/j.1751-7915.2010.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis K E R, Joseph S J, Peter H, et al. 2005, Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol. 71(2):826–834. doi: 10.1128/AEM.71.2.826-834.2005. https://doi.org/10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulschen A A, Bendia A G, Fricker A D, et al. 2017, Isolation of uncultured bacteria from antarctica using long incubation periods and low nutritional media. Front Microbiol. 8:1–12. doi: 10.3389/fmicb.2017.01346. https://doi.org/10.3389/fmicb.2017.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes W F, Chacón J, Segrè D, et al. 2018, Bioprinting microbial communities to examine interspecies interactions in time and space. Biomed Phys Eng Express. 4(5) [Google Scholar]
- 28.Bollmann A, Lewis K, Epstein S S. 2007, Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol. 6386;73(20) doi: 10.1128/AEM.01309-07. https://doi.org/10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrari B C, Winsley T, Gillings M, et al. 2008, Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat Protoc. 3(8):1261–1269. doi: 10.1038/nprot.2008.102. https://doi.org/10.1038/nprot.2008.102. [DOI] [PubMed] [Google Scholar]
- 30.Kushmaro A, Geresh S. 2012. U.S. Patent No. 8,158,401. Washington, DC: U.S. Patent and Trademark Office; [Google Scholar]
- 31.Nichols D, Cahoon N, Trakhtenberg E M, et al. 2010, Use of ichip for high-throughput in situ cultivation of “uncultivable”microbial species. Appl Environ Microbiol. 76(8):2445–2450. doi: 10.1128/AEM.01754-09. https://doi.org/10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingham C J, Sprenkels A, Bomer J, et al. 2007, The micro-petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc Natl Acad Sci. 104(46):18217–18222. doi: 10.1073/pnas.0701693104. https://doi.org/10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fröhlich J, König H. 2000, New techniques for isolation of single prokaryotic cells. FEMS Microbiol Rev. 24(5):567–572. doi: 10.1111/j.1574-6976.2000.tb00558.x. https://doi.org/10.1016/S0168-6445(00)00045-0;https://doi.org/10.1111/j.1574-6976.2000.tb00558.x. [DOI] [PubMed] [Google Scholar]
- 34.Ishii S, Tago K, Senoo K. 2010, Single-cell analysis and isolation for microbiology and biotechnology:Methods and applications. Appl Microbiol Biotechnol. 86(5):1281–1292. doi: 10.1007/s00253-010-2524-4. https://doi.org/10.1007/s00253-010-2524-4. [DOI] [PubMed] [Google Scholar]
- 35.Akselrod G M, Timp W, Mirsaidov U, et al. 2006, Laser-guided assembly of heterotypic three-dimensional living cell microarrays. Biophys J. 91(9):3465–3473. doi: 10.1529/biophysj.106.084079. https://doi.org/10.1529/biophysj.106.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowan B, Wheeler M A, Crooks R M. 2002, Patterning bacteria within hyperbranched polymer film templates. Langmuir. 18(25):9914–9917. https://doi.org/10.1021/la020664h. [Google Scholar]
- 37.Weibel D B, Lee A, Mayer M, et al. 2005, Bacterial printing press that regenerates its ink:Contact-printing bacteria using hydrogel stamps. Langmuir. 21(14):6436–6442. doi: 10.1021/la047173c. https://doi.org/10.1021/la047173c. [DOI] [PubMed] [Google Scholar]
- 38.Xu L, Robert L, Ouyang Q, et al. 2007, Microcontact printing of living bacteria arrays with cellular resolution. Nano Lett. 7(7):2068–2072. doi: 10.1021/nl070983z. https://doi.org/10.1021/nl070983z. [DOI] [PubMed] [Google Scholar]
- 39.Eun Y J, Utada A S, Copeland M F, et al. 2011, Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem Biol. 6(3):260–266. doi: 10.1021/cb100336p. https://doi.org/10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volfson D, Cookson S, Hasty J, et al. 2008, Biomechanical ordering of dense cell populations. Proc Natl Acad Sci. 105(40):15346–15351. doi: 10.1073/pnas.0706805105. https://doi.org/10.1073/pnas.0706805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boedicker J Q, Vincent M E, Ismagilov R F. 2009, Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chemie Int Ed. 48(32):5908–5911. doi: 10.1002/anie.200901550. https://doi.org/10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flickinger S T, Copeland M F, Downes E M, et al. 2011, Quorum sensing between Pseudomonas aeruginosa biofilms accelerates cell growth. J Am Chem Soc. 133:5966–5975. doi: 10.1021/ja111131f. https://doi.org/10.1021/ja111131f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carnes E C, Lopez D M, Donegan N P, et al. 2010, Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat Chem Biol. 6(1):41–45. doi: 10.1038/nchembio.264. https://doi.org/10.1038/nchembio.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaguchi T, Dwidar M, Byun C K, et al. 2012, Aqueous two-phase system-derived biofilms for bacterial interaction studies. Biomacromolecules. 13(9):2655–2661. doi: 10.1021/bm300500y. https://doi.org/10.1021/bm300500y. [DOI] [PubMed] [Google Scholar]
- 45.Cho H J, Jönsson H, Campbell K, et al. 2007, Self-organization in high-density bacterial colonies:Efficient crowd control. PLoS Biol. 5(11):2614–2623. doi: 10.1371/journal.pbio.0050302. https://doi.org/10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connell J L, Whiteley M, Shear J B. 2012, Sociomicrobiology in engineered landscapes. Nat Chem Biol. 8(1):10–13. doi: 10.1038/nchembio.749. https://doi.org/10.1038/nchembio.749. [DOI] [PubMed] [Google Scholar]
- 47.Connell J L, Wessel A K, Parsek M R. 2010, Probing prokaryotic social behaviors with bacterial “lobster traps”. Mbio. 1(4):1–8. doi: 10.1128/mBio.00202-10. https://doi.org/10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farsari M, Chichkov B N. 2009, Two-photon fabrication. Nat Photonics. 3:450–452. https://doi.org/10.1038/nphoton.2009.131. [Google Scholar]
- 49.Nielson R, Kaehr B, Shear J B. 2009, Microreplication and design of biological architectures using dynamic-mask multiphoton lithography. Small. 5(1):120–125. doi: 10.1002/smll.200801084. https://doi.org/10.1002/smll.200801084. [DOI] [PubMed] [Google Scholar]
- 50.Kawata S, Sun H B, Tanaka T, et al. 2001, Finer features for functional microdevices. Nature. 412(6848):697–698. doi: 10.1038/35089130. https://doi.org/10.1038/35089130. [DOI] [PubMed] [Google Scholar]
- 51.Kim D, So P T C. 2010, High-throughput three-dimensional lithographic microfabrication. Opt Lett. 1602;35(10) doi: 10.1364/OL.35.001602. https://doi.org/10.1364/OL.35.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connell J L, Ritschdorff E T, Whiteley M, et al. 2013, 3D printing of microscopic bacterial communities. Proc Natl Acad Sci. 110(46):18380–18385. doi: 10.1073/pnas.1309729110. https://doi.org/10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obara Y, Yamai S, Nikkawa T, et al. 1981, Preservation and transportation of bacteria by a simple gelatin disk method. J Clin Microbiol. 14(1):61–66. doi: 10.1128/jcm.14.1.61-66.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kailas L, Ratcliffe E C, Hayhurst E J, et al. 2009, Immobilizing live bacteria for AFM imaging of cellular processes. Ultramicroscopy. 109(7):775–780. doi: 10.1016/j.ultramic.2009.01.012. https://doi.org/10.1016/j.ultramic.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 55.Jordan P, Leach J, Padgett M, et al. 2005, Creating permanent 3D arrangements of isolated cells using holographic optical tweezers. Lab Chip. 5(11):1224–1228. doi: 10.1039/b509218c. https://doi.org/10.1039/b509218c. [DOI] [PubMed] [Google Scholar]
- 56.Barron J A, Krizman D B, Ringeisen B R. 2005, Laser printing of single cells:Statistical analysis, cell viability, and stress. Ann Biomed Eng. https://doi.org/10.1007/s10439-005-8971-x. 33(2):121–130. doi: 10.1007/s10439-005-8971-x. [DOI] [PubMed] [Google Scholar]
- 57.Barron J A, Wu P, Ladouceur H D, et al. 2004, Biological laser printing:A novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed Microdevices. 6(2):139–147. doi: 10.1023/b:bmmd.0000031751.67267.9f. https://doi.org/10.1023/B:BMMD.0000031751.67267.9f. [DOI] [PubMed] [Google Scholar]
- 58.Mironov V, Kasyanov V, Drake C, et al. 2008, Organ printing:Promises and challenges. Regen Med. 3(1):93–103. doi: 10.2217/17460751.3.1.93. https://doi.org/10.2217/17460751.3.1.93. [DOI] [PubMed] [Google Scholar]
- 59.Fitzgerald L A, Wu P K, Gurnon J R, et al. 2010, Isolation of the phycodnavirus PBCV-1 by biological laser printing. J Virol Methods. 167(2):223–225. doi: 10.1016/j.jviromet.2010.04.005. https://doi.org/10.1016/j.jviromet.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 60.Ringeisen B R, Othon C M, Barron J A, et al. 2006, Jet-based methods to print living cells. Biotechnol J. 1(9):930–948. doi: 10.1002/biot.200600058. https://doi.org/10.1002/biot.200600058. [DOI] [PubMed] [Google Scholar]
- 61.Barron J A, Rosen R, Jones-Meehan J, et al. 2004, Biological laser printing of genetically modified Escherichia coli for biosensor applications. Biosens Bioelectron. 20(2):246–252. doi: 10.1016/j.bios.2004.01.011. https://doi.org/10.1016/j.bios.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 62.Barron J A, Young H D, Dlott D D, et al. 2005, Printing of protein microarrays via a capillary-free fluid jetting mechanism. Proteomics. 5(16):4138–4144. doi: 10.1002/pmic.200401294. https://doi.org/10.1002/pmic.200401294. [DOI] [PubMed] [Google Scholar]
- 63.Taidi B, Lebernede G, Koch L, et al. 2016, Colony development of laser printed eukaryotic (yeast and microalga) microorganisms in co-culture. Int J Bioprinting. 2(2):37–43. https://doi.org/10.18063/IJB.2016.02.001. [Google Scholar]
- 64.Unger C, Gruene M, Koch L, et al. 2011, Time-resolved imaging of hydrogel printing via laser-induced forward transfer. Appl Phys A Mater Sci Process. 103(2):271–277. https://doi.org/10.1007/s00339-010-6030-4. [Google Scholar]
- 65.Gruene M, Unger C, Koch L, et al. 2011, Dispensing pico to nanolitre of a natural hydrogel by laser-assisted bioprinting. Biomed Eng Online. 10(3):9–12. doi: 10.1186/1475-925X-10-19. https://doi.org/10.1186/1475-925X-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koch L, Kuhn S, Sorg H, et al. 2010, Laser printing of skin cells and human stem cells. Tissue Eng Part C Methods. 16(5):847–854. doi: 10.1089/ten.TEC.2009.0397. https://doi.org/10.1089/ten.tec.2009.0397. [DOI] [PubMed] [Google Scholar]
- 67.Deng Y, Renaud P, Guo Z, et al. 2017, Single cell isolation process with laser induced forward transfer. J Biol Eng. 11(1):2. doi: 10.1186/s13036-016-0045-0. https://doi.org/10.1186/s13036-016-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ringeisen B R, Rincon K, Fitzgerald L A, et al. 2015, Printing soil:A single-step, high-throughput method to isolate micro-organisms and near-neighbour microbial consortia from a complex environmental sample. Methods Ecol Evol. 6(2):209–217. https://doi.org/10.1111/2041-210X.12303. [Google Scholar]
- 69.Ringeisen B R, Lizewski S E, Fitzgerald L A, et al. 2010, Single cell isolation of bacteria from microbial fuel cells and potomac river sediment. Electroanalysis. 22(7-8):875–882. https://doi.org/10.1002/elan.200880012. [Google Scholar]
- 70.Yusupov V I, Gorlenko M V, Cheptsov V S, et al. 2018, Laser engineering of microbial systems. Laser Phys Lett. 15(6):065604. https://doi.org/10.1088/1612-202X/aab5ef. [Google Scholar]
- 71.Gorlenko M, Chutko E, Churbanova E, et al. 2018, Laser microsampling of soil microbial community. J Biol Eng Rev. 12:27. doi: 10.1186/s13036-018-0117-4. https://doi.org/10.1186/s13036-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheptsov V S, Churbanova E S, Yusupov V I, et al. 2018, Laser printing of microbial systems:Effect of absorbing metal film. Lett Appl Microbiol Rev. 67:544–549. doi: 10.1111/lam.13074. https://doi.org/10.1111/lam.13074. [DOI] [PubMed] [Google Scholar]
- 73.Ara I, Kudo T, Matsumoto A, et al. 2007 Nonomuraea bangladeshensis Sp Nov. and Nonomuraea coxensis Sp. Nov. Int J Syst Evol Microbiol. 57(7):1504–1509. doi: 10.1099/ijs.0.65054-0. https://doi.org/10.1099/ijs.0.65054-0. [DOI] [PubMed] [Google Scholar]
- 74.Sungthong R, Nakaew N. 2015, The genus Nonomuraea :A review of a rare actinomycete taxon for novel metabolites. J Basic Microbiol. 55(5):554–565. doi: 10.1002/jobm.201300691. https://doi.org/10.1002/jobm.201300691. [DOI] [PubMed] [Google Scholar]
- 75.Yusupov V I, Zhigarkov V S, Churbanova E S, et al. 2017, Laser-induced transfer of gel microdroplets for cell printing. Quantum Electron. 47(12):1158–1165. https://doi.org/10.1070/QEL16512. [Google Scholar]
- 76.Kohli R, Bose B, Gupta P K. 2001, Induction of phr gene expression in E coli strain KY706/pPL-1 by He-Ne laser (632.8 nm) irradiation. J Photochem Photobiol B Biol. 60(2-3):136–142. doi: 10.1016/s1011-1344(01)00139-7. https://doi.org/10.1016/S1011-1344(01)00139-7. [DOI] [PubMed] [Google Scholar]
- 77.Nussbaum E, Lilge L L, Mazzulli T. 2002, Delivering radiant exposure of 1-50 J/cm 2 on three species of bacteria in vitro. Surgery. 20(6):325–333. doi: 10.1089/104454702320901116. [DOI] [PubMed] [Google Scholar]
- 78.Nussbaum E L, Lilge L, Mazzulli T. 2003, Effects of low-level laser therapy (LLLT) of 810 nm upon in vitro growth of bacteria:Relevance of irradiance and radiant exposure. J Clin Laser Med Surg. 21(5):283–290. doi: 10.1089/104454703322564497. https://doi.org/10.1089/104454703322564497. [DOI] [PubMed] [Google Scholar]
- 79.Chen Z, Lu J, Gao S H, et al. 2018, Silver nanoparticles stimulate the proliferation of sulfate reducing bacterium Desulfovibrio vulgaris. Water Res Pergamon. 129:163–171. doi: 10.1016/j.watres.2017.11.021. https://doi.org/10.1016/j.watres.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 80.Miazek K, Iwanek W, Remacle C, et al. 2015, Effect of metals, metalloids and metallic nanoparticles on microalgae growth and industrial product biosynthesis:A review. Int J Mol Sci. 16(10):23929–23969. doi: 10.3390/ijms161023929. https://doi.org/10.3390/ijms161023929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slavin Y N, Asnis J, Häfeli U O, et al. 2017, Metal nanoparticles:Understanding the mechanisms behind antibacterial activity. J Nanobiotechnol. 15(1):1–20. doi: 10.1186/s12951-017-0308-z. https://doi.org/10.1186/s12951-017-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schacht V J, Neumann L V, Sandhi S K, et al. 2013, Effects of silver nanoparticles on microbial growth dynamics. J Appl Microbiol. 114(1):25–35. doi: 10.1111/jam.12000. https://doi.org/10.1111/jam.12000. [DOI] [PubMed] [Google Scholar]
- 83.Haider A J, Haider M J, Majed M D, et al. 2017, Effect of laser fluence on a microarray droplets micro-organisms cells by LIFT technique. Energy Procedia. 119:256–263. https://doi.org/10.1016/j.egypro.2017.07.078. [Google Scholar]
- 84.Ringeisen B R, Chrisey D B, Piqué A, et al. 2002, Generation of mesoscopic patterns of viable Escherichia coli by ambient laser transfer. Biomaterials. 23(1):161–166. doi: 10.1016/s0142-9612(01)00091-6. https://doi.org/10.1016/S0142-9612(01)00091-6. [DOI] [PubMed] [Google Scholar]
- 85.Hopp B, Smausz T, Antal Z, et al. 2004, Absorbing film assisted laser induced forward transfer of fungi (Trichoderma conidia ) J Appl Phys. 96(6):3478–3481. https://doi.org/10.1063/1.1782275. [Google Scholar]
- 86.Francois K, Devlieghere F, Standaert A R, et al. 2003, Modelling the individual cell lag phase Isolating single cells:Protocol development. Lett Appl Microbiol. 37(1):26–30. doi: 10.1046/j.1472-765x.2003.01340.x. https://doi.org/10.1046/j.1472-765X.2003.01340.x. [DOI] [PubMed] [Google Scholar]
- 87.Gross A, Schoendube J, Zimmermann S, et al. 2015, Technologies for single-cell isolation. Int J Mol Sci. 16(8):16897–16919. doi: 10.3390/ijms160816897. https://doi.org/10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Höfer P, Vermette P, Groleau D. 2011, Introducing a new bioengineered bug Methylobacterium extorquens tuned as a microbial bioplastic factory. Bioeng Bugs. 2(2):71–79. doi: 10.4161/bbug.2.2.15009. https://doi.org/10.4161/bbug.2.2.15009. [DOI] [PubMed] [Google Scholar]
- 89.Zhong C, Gurry T, Cheng A A, et al. 2014, Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat Nanotechnol. 9(10):858–866. doi: 10.1038/nnano.2014.199. https://doi.org/10.1038/nna no.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen A Y, Deng Z, Billings A N, et al. 2014, Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater. 13(5):515–523. doi: 10.1038/nmat3912. https://doi.org/10.1016/j.matdes.2013.08.029;https://doi.org/10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lehner B A E, Schmieden D T, Meyer A S. 2017, A straightforward approach for 3D bacterial printing. ACS Synth Biol. 6(7):1124–1130. doi: 10.1021/acssynbio.6b00395. https://doi.org/10.1021/acssynbio.6b00395. [DOI] [PMC free article] [PubMed] [Google Scholar]


