Abstract
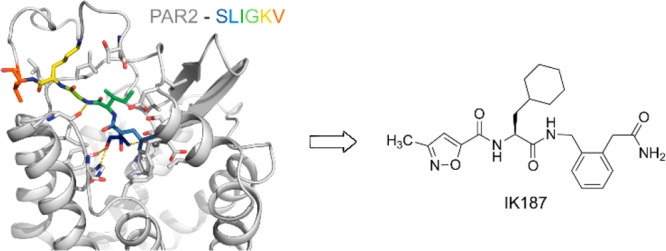
Proteinase-activated receptor 2 (PAR2) is a class A G protein-coupled receptor whose activation has been associated with inflammatory diseases and cancer, thus representing a valuable therapeutic target. Pathophysiological roles of PAR2 are often characterized using peptidic PAR2 agonists. Peptidic ligands are frequently unstable in vivo and show poor bioavailability, and only a few approaches toward drug-like nonpeptidic PAR2 ligands have been described. The herein-described ligand 5a (IK187) is a nonpeptidic PAR2 agonist with submicromolar potency in a functional assay reflecting G protein activation. The ligand also showed substantial β-arrestin recruitment. The development of the compound was guided by the crystal structure of PAR2, when the C-terminal end of peptidic agonists was replaced by a small molecule based on a disubstituted phenylene scaffold. IK187 shows preferable metabolic stability and may serve as a lead compound for the development of nonpeptidic drugs addressing PAR2.
Keywords: Proteinase-activated receptor 2, agonist, nonpeptidic ligands, structure−activity relationships
Proteinase-activated receptors (PARs) are a four-member family of class A G protein-coupled receptors (GPCRs) that share a unique mechanism of activation. While the vast majority of GPCRs are activated by diffusible ligands,1 PARs are activated by proteolytic modification of their N-terminus by serine proteinases like trypsin or thrombin, revealing a new epitope that subsequently acts as a tethered ligand activating the receptor.2 PARs, especially PAR2, are associated with diseases accompanied by an increased release of these proteinases, such as inflammatory events.2,3 PAR2 signaling has been shown to contribute to arthritis,4 colitis and inflammatory bowel diseases,5,6 airway inflammation and asthma,7,8 and cancer.9−12 Nevertheless, the relationship between individual PAR2 activation profiles and pathophysiological outcomes is not yet fully understood. Despite the value of PAR2 as a therapeutic target, drugs addressing PAR2 have not been approved. Apart from the pro-inflammatory effects, anti-inflammatory and protective effects following PAR2 activation have also been described.3,13
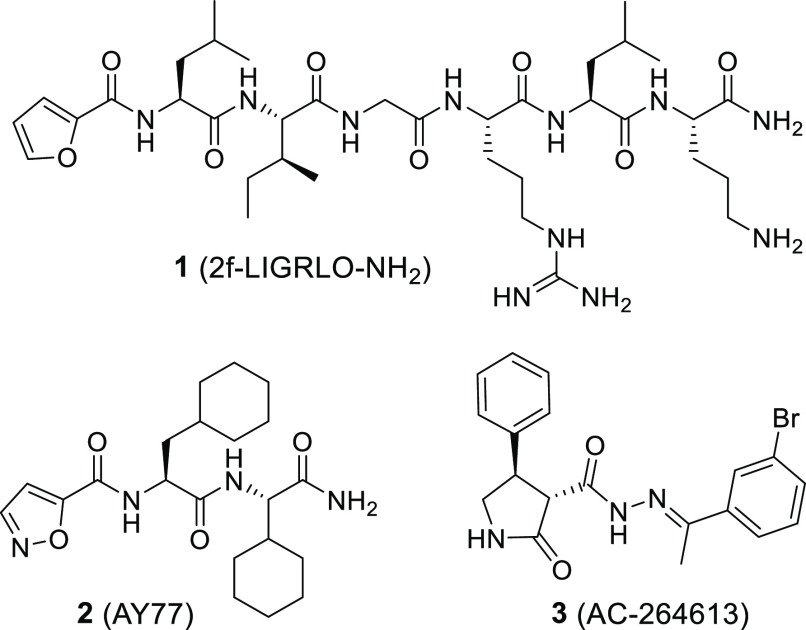
Synthetic PAR2 ligands are valuable tools to study the individual signaling patterns resulting from PAR2 activation and their impact on pathophysiological conditions. Many of the PAR2 ligands described are peptidic and derived from the sequence of the tethered ligand, S37LIGKV (human) and S37LIGRL (rodent).14 The corresponding hexapeptidic fragments were found to activate PAR2 as diffusible ligands, but with potencies in the micromolar range.15 Bioisosteric replacement of the serine residue by a 2-furoyl substituent and further structural evolution led to 1 (2f-LIGRLO-NH2; Figure 1), a potent PAR2 ligand with enhanced potency (>100-fold relative to SLIGRL-NH2).161 has been widely used as a molecular tool and reference agonist for pharmacological experiments investigating PAR2 activation.
Figure 1.
Apart from furoic acid, the terminal serine can also be replaced by other small heteroaromatic entities, including isoxazole-5-carboxylic acid and 2-aminothiazol-4-carboxylic acid.20,21 The minimum activating sequence of SLIGRL-NH2, consisting of leucine and isoleucine in addition to serine or a heterocyclic serine bioisostere, was confirmed employing a lipid-tethering approach.22 Exchanging the leucine with cyclohexylalanine improves the ligand potency and additionally confers selectivity for PAR2 over PAR1.20,23 A combination of isoxazole-5-carboxylic acid with cyclohexylalanine and isoleucine connected to a 4-(aminomethyl)piperidine substituent via a benzylamine linker yielded the PAR2 agonist GB110,20 which showed activity in a PAR2 rodent model.24 For the same class of molecules, replacement of the C-terminal amine by bulky hydrophobic residues resulted in antagonists like GB88.20,23 Further truncation experiments showed that a mimicry of the N-terminal tripeptide of the activating sequence is sufficient for PAR2-mediated Gq activation, leading to the discovery of the small peptide analogue 2 (AY77).17 To overcome the poor bioavailability of peptides and peptide analogues, a few approaches toward nonpeptidic PAR2 ligands have been pursued.25 Hence, 3 (AC-264613) (Figure 1) and analogues have been discovered and exhibit potencies ranging from 30 nM to 6 μM.18,19 However, these compounds have poor solubility, limiting their suitability as drug candidates.
The combination of ligand- and structure-based approaches has proved to be useful for the discovery of GPCR-targeted ligands.26−30 Starting from the PAR2 agonist 2, our goal was to develop novel PAR2 ligands by further reducing the peptidic nature of the lead compound. In the course of our work, we identified novel scaffolds. Potent candidates were optimized by taking advantage of a receptor–ligand model based on the recently described crystal structure of PAR2.31
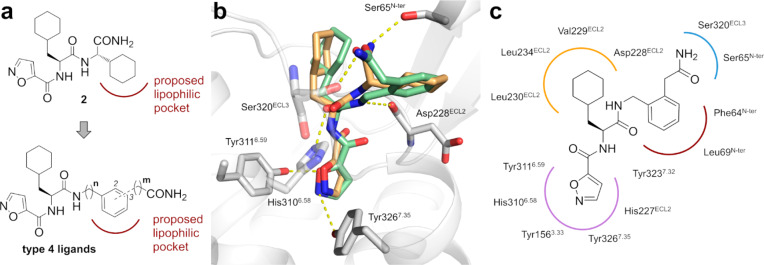
Design. Previous work by Yau and coworkes described the potential relevance of a lipophilic pocket in the orthosteric binding site of PAR2, which may be addressed by the C-terminal cyclohexyl residue of 2.17 Our strategy depended on whether we could replace the cyclohexylglycyl substructure of 2 by a more artificial disubstituted phenylene unit. To investigate whether the nonpeptidic phenylene entity could address the lipophilic pocket, we designed a series of 1,2- and 1,3-disubstituted ligands of type 4 (Figure 2a). In order to maintain a similar distance to the backbone of the ligand and to introduce a certain flexibility at the lipophilic moiety, a small set of homologues (n, m = 0 or 1) were considered.
Figure 2.
(a) Design of novel nonpeptidic PAR2 ligands. (b) Overlay of the best poses for 2 (orange) and 4b (green) in the PAR2 model (gray). Residues showing polar interactions with the ligands are depicted in sticks. (c) Schematic 2D representation of 4b and the amino acid side chains addressed by the ligand within the orthosteric binding pocket.
The designed compounds were supposed to be evaluated by molecular docking. Previous computational studies of PAR2, published before the PAR2 crystal structure31 was released, applied homology models based on the crystal structure of PAR1 or the nociceptin/orphanin FQ receptor.17 We envisioned using the recently published crystal structure of PAR2. As the receptor was crystallized in complex with an allosteric antagonist (AZ8838), which led to a partial collapse of the potential orthosteric pocket formed by the antagonist vorapaxar at PAR1, and as the receptor was stabilized in the inactive state by nine thermostabilizing mutants, which abolished agonist binding, the crystal structure construct is presumably unsuited for structure-based agonist design. To create an agonist-bound model of PAR2 applicable for docking of multiple agonistic structures, we investigated the binding of the trypsin-activated N-terminus to the transmembrane segment of wild-type PAR2. This was achieved by means of metadynamics simulations, applying the parameters of a recently published protocol for predicting the binding modes of ligands at class A GPCRs.32 The simulations were started from the above-described crystal structure with all mutations reverted to their wild-type sequence and N-terminal residues added up to Ser37 mimicking receptor activation by trypsin. This produced a PAR2 model with the trypsin-activated N-terminus (tethered ligand) bound to the extracellular side of the helix bundle of the receptor that represents an agonist bound conformation of the PAR2 (Supplementary Figure S1).
Subsequent docking of 2 and 4b (1,2-disubstituted, m = 1, n = 1), a representative example of the type 4 ligands, into the pocket formed by the N-terminus of the PAR2 model revealed that the compounds address an identical subset of residues as the tethered ligand but do not show an exact overlap. The phenylene and cyclohexylglycyl moieties of 4b and 2, respectively, form lipophilic interactions with the side chains of Phe64 and Leu69. Interestingly, this lipophilic site is not identical to the pocket that was proposed by Yau.17 The isoxazole may replace the N-terminal Ser of the signaling peptide sequence engaging a polar network within the binding pocket (Figure 2). Furthermore, the pose of 4b indicated the opportunity to introduce a substituent on the isoxazole, and we therefore included in our design ligands 5a (IK187) and 5b bearing a methyl or ethyl substituent, respectively, at position 3 of the isoxazole-5-carboxylic acid. Indeed, docking studies with our PAR2 model indicated very similar binding poses for 4b and 5a, with only a slightly different conformation of the isoxazole conferring an ideal fit into a predominantly hydrophobic pocket (Figure 3).
Figure 3.
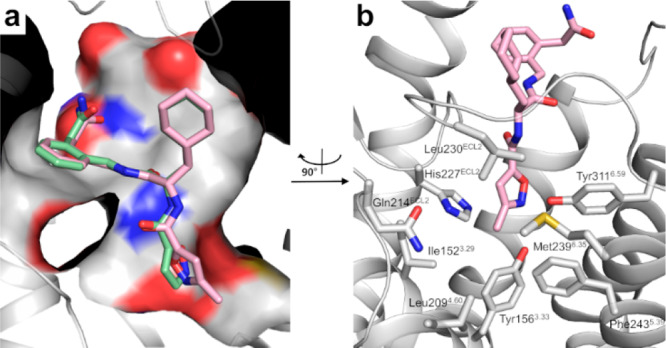
(a) Overlay of the best-scored docking poses of 5a (IK187) (pink) and 4b (green) within the orthosteric pocket of the receptor model. For clarity reasons, ECL2 is not shown. (b) Side chains of residues constituting the binding pocket of the methyl group of 5a are shown as gray sticks. Compared with (a), the view is rotated 90° clockwise.
While our study was underway, Kennedy et al. used a combination of docking with flexible side chains and insights from mutagenesis studies to develop a PAR2 model starting from the same PAR2 crystal structure.33 Docking of 4b to this model revealed two distinct orientations of the ligand (Supplementary Figure S2). While one of the poses was quite similar to the result obtained with our metadynamics-derived model, the isoxazole moiety of 4b extended deeper into the transmembrane helix bundle in the second orientation. Nevertheless, both poses revealed sufficient space for the introduction of substituents on the isoxazole (Supplementary Figure S2).
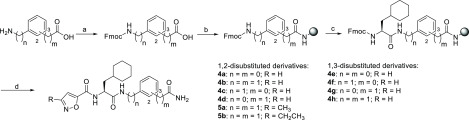
Chemical Synthesis. Ligands 4a–h, 5a, and 5b were synthesized by standard Fmoc-based and microwave-assisted methods using Rink amide resin (Scheme 1). The corresponding Fmoc-protected building blocks were either commercially available or obtained by Fmoc protection of the amine in the first step, in analogy to previously described protocols.34 After it was loaded on the resin, the protecting group was cleaved by application of piperidine and microwave irradiation. For the synthesis of 4d, we loaded the resin directly with unprotected 2-(2-aminophenyl)acetic acid. Subsequently, Fmoc-3-cyclohexylalanine was attached to form an amide bond in a microwave-supported reaction employing PyBOP and HOBT (for n = 1) as coupling reagents in the presence of DIPEA. In case of aromatic amines (n = 0), HATU and DIPEA were used as coupling reagents, and 30 instead of 20 cycles of microwave irradiation were performed since coupling in the presence of PyBOP, HOBt, and DIPEA was found to be insufficient. Consecutive Fmoc deprotection, coupling of isoxazole-5-carboxylic acid or its 3-methyl- or 3-ethyl-substituted derivative, and cleavage from the resin with a TFA/TIS/phenol/water mixture yielded the desired products.
Scheme 1. Synthesis of the Target Compounds 4a–h, 5a, and 5b.
Reagents and conditions: (a) Fmoc-Cl, Na2CO3, water/1,4-dioxane, rt, 3 h (69–95%). (b) Microwave-assisted loading on unprotected Rink resin, then PyBOP, HOBT, DIPEA, DMF, 30 cycles at 50 W, 10 s, intermittent cooling. (c) (i) 25% v/v piperidine in DMF, 5–15 cycles at 100 W, 5 s; (ii) for n = 0, Fmoc-Cha-OH, HATU, DIPEA, 30 cycles at 50 W, 10 s; for n = 1, Fmoc-Cha-OH, PYBOP, HOBT, DIPEA, 20 cycles at 50 W, 10 s. (d) (i) 25% v/v piperidine in DMF, five cycles at 100 W, 5 s; (ii) isoxazole-5-carboxylic acid, 3-methylisoxazole-5-carboxylic acid, or 3-ethylisoxazole-5-carboxylic acid, PyBOP, HOBT, DIPEA, 20 cycles at 50 W, 10 s; (iii) TFA, TIS, phenol, water, rt, 2 h.
Biological Investigations and SARs. The agonist activities of the ligands were biologically evaluated in comparison with the reference agents 1 and 2. We investigated PAR2-mediated Gq activation and β-arrestin recruitment35 in HEK293 cells using a FRET-based IP1 accumulation assay (IP-One) to measure Gq activation and an enzyme complementation assay (PathHunter) to assess recruitment of β-arrestin-2.
All of the 1,2-disubstituted (4a–d) and 1,3-disubstituted (4e–h) phenylene derivatives activated PAR2 and Gq signaling with potencies in the range of 0.59–11 μM. With exception of 4a (Emax = 70%), the compounds showed ligand efficacies greater than 90% (Table 1). In contrast, recruitment of β-arrestin-2 required high ligand concentration, with efficacies ranging between 6 and 73% at 30 μM, precluding the determination of an EC50 value.
Table 1. PAR2 Agonist Activitiesa.
| Gq activation |
β-arrestin-2
recruitment |
|||
|---|---|---|---|---|
| EC50 [μM] | Emax [%]b | EC50 [μM] | Emax [%]b | |
| 4a | 3.1 ± 1.1 | 70 ± 7 | n.c.d | 6 ± 3e |
| 4b | 0.59 ± 0.10 | 93 ± 2 | 6.0 ± 0.3 | 65 ± 6 |
| 4c | 1.0 ± 0.1 | 95 ± 1 | n.c.d | 55 ± 7e |
| 4d | 11 ± 1 | 108 ± 4 | n.c.d | 8 ± 1 |
| 4e | 2.2 ± 0.3 | 105 ± 8 | n.c.d | 53 ± 10e |
| 4f | 3.3 ± 0.4 | 93 ± 1 | n.c.d | 42 ± 4e |
| 4g | 1.3 ± 0.2 | 97 ± 2 | n.c.d | 73 ± 6e |
| 4h | 2.9 ± 0.4 | 92 ± 3 | n.c.d | 41 ± 5 |
| 5a | 0.64 ± 0.19 | 105 ± 2 | 8.9 ± 0.4 | 95 ± 3 |
| 5b | 0.96 ± 0.05 | 104 ± 4 | 18 ± 2 | 93 ± 4 |
| 6 | n.a.c | <5 | n.a.c | <5 |
| 7 | 1.9 ± 0.1 | 93 ± 1 | n.c.d | 32 ± 4e |
| 8a | n.c.d | 85 ± 2e | n.a.c | <5 |
| 8b | n.c.d | 44 ± 2e | n.a.c | <5 |
| 9 | n.c.d | 9 ± 1e | n.a.c | <5 |
| 10a | 1.8 ± 0.3 | 97 ± 3 | n.c.d | 60 ± 2e |
| 10b | 0.64 ± 0.08 | 100 ± 1 | 7.4 ± 0.3 | 87 ± 6 |
| 11 | 2.4 ± 0.2 | 98 ± 5 | n.c.d | 93 ± 5e |
| 12a | 4.2 ± 0.5 | 97 ± 4 | n.c.d | 32 ± 4e |
| 12b | 4.9 ± 0.8 | 98 ± 4 | n.c.d | 33 ± 5e |
| 13 | n.c.d | 12 ± 5e | n.a.c | <5 |
| 1 | 0.15 ± 0.03 | 100 | 0.43 ± 0.05 | 100 |
| 2 | 0.17 ± 0.03 | 110 ± 4 | 2.0 ± 0.2 | 94 ± 5 |
Data are reported as mean ± SEM for at least three independent experiments performed in duplicate.
Maximum efficacy determined relative to reference 1 (2f-LIGRLO-NH2).
n.a. indicates that no activity was observed.
n.c. indicates that the dose–response curve did not converge to a plateau and an exact EC50 was not calculated.
The dose–response curve did not converge to a plateau, and the reported value is the efficacy at a ligand concentration of 30 μM.
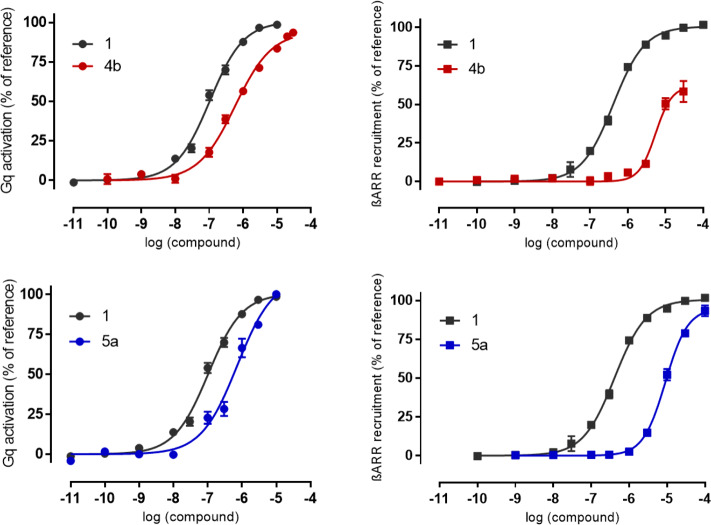
Only small differences between the 1,3-disubstituted homologues 4e–h were observed. The ligands were agonists in both assay systems and displayed low-micromolar potencies (EC50 = 1.3–3.3 μM) in the IP1 accumulation assay. Within the series of 1,2-disubstituted homologues 4a–d, noticeable differences were observed in both assays. An additional methylene group between the amine and phenyl moieties (n = 1) was clearly beneficial for ligand activity, as 4b and 4c (EC50 = 0.59 and 1.0 μM, respectively) were more potent than their n = 0 analogues 4a and 4d (EC50 = 3.1 and 11 μM, respectively) in the IP1 accumulation assay. In the β-arrestin-2 recruitment assay, this effect was even more obvious, as PAR2 activation by 4b and 4c led to recruitment of β-arrestin-2, while recruitment was barely observed for the corresponding homologues 4a and 4d. This indicates that conformational flexibility allowing the aromatic moiety to adapt to the lipophilic binding pocket may be required. The nonpeptidic PAR2 agonist 4b was identified as the most potent type 4 ligand. When tested under the same conditions in the IP1 assay, peptidomimetic 2 showed only slightly higher potency (∼3.5-fold, EC50 = 0.17 μM), proving the success of our design strategy. Interestingly, while fully activating PAR2-mediated Gq signaling, 4b was 10-fold less potent and showed only partial agonist properties for the recruitment of β-arrestin-2 (EC50 = 6 μM, Emax = 65%; Figure 4). Although slight differences in potency in the two assays were also observed for the reference peptide 1 (∼3-fold, EC50 = 0.15 and 0.43 μM, respectively), these results point toward a G protein-biased mechanism of action for 4b. Similar differences were also observed for 2 (12-fold higher potency for Gαq activation), which is in agreement with previous reports showing that mimicry of the three N-terminal amino acids of 1 leads to bias toward PAR2-mediated G protein activation.36 A preference for G protein activation over β-arrestin recruitment may have beneficial effects, since PAR2-mediated β-arrestin activation has been associated with proinflammatory effects in asthma,37,38 whereas PAR2-mediated G protein signaling can lead to relaxation of airway smooth muscles.38 Therefore, G protein-biased ligands may be useful tools to further understand the underlying effects and to develop new drugs for asthma therapy in the future.
Figure 4.
Functional activities of (top) 4b and (bottom) 5a (IK187) in the Gq pathway (left) and recruitment of β-arrestin-2 (right). Data are reported as mean ± SEM of at least three independent experiments performed in duplicate.
In agreement with the docking predictions, a methyl substituent at position 3 of the isoxazole was well-tolerated. Compound 5a showed full agonist properties in both assay systems, with EC50 values of 0.64 and 8.9 μM in the IP1 and β-arrestin-2 recruitment assays, respectively, indicating a 14-fold preference for the Gαq pathway (Figure 4). The introduction of the larger ethyl substituent did not result in a further increase in potency for 5b, indicating that the available space in the binding pocket is limited.
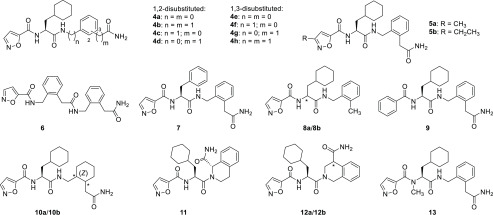
To learn more about the importance of the individual entities of the type 4 ligands, further structure–activity relationship studies were performed. Ligands 6–13 were synthesized in analogy to the procedures described above (see Supplementary Methods).20 Replacement of cyclohexylalanine by phenylalanine (7) attenuated the activity, and the phenylene analogue 6 was found to be inactive. Consistent with recent findings,17 our docking results indicated that hydrogen-bond acceptors and donors as contained in the isoxazole and terminal carboxamide moieties are crucial for ligand activity because of polar interactions (Figure 2b,c). In our model, the isoxazole forms hydrogen bonds to Tyr3116.58 and Tyr3267.35. The carboxamide of 2 and 4b form hydrogen bonds to Ser65N-ter or to Ser320ECL3 and Ser65N-ter, respectively. In fact, their removal reduced the ligand potency significantly for 8a and abolished the agonist activity almost completely for 8b and 9. Further, the N-methyl derivative 13 was found to be inactive, highlighting the importance of hydrogen-bond acceptors and donors for the activation of PAR2. Saturation of the C-terminal phenylene moiety was tolerated (10a and 10b), but no increase in ligand potency was observed. In agreement with the putative bioactive conformation of 4b, conformational restriction did not enhance the agonist activity for analogues 11, 12a, and 12b.
Compounds 6, 9, and 13 that were devoid of activity in the IP1 accumulation assay were also examined for antagonist properties. However, none of them could inhibit the effect of 300 nM 2f-LIGRLO-NH2 (Supplementary Figure S3), indicating that the structural modifications abolished not only the intrinsic activity but also binding of ligands to PAR2.
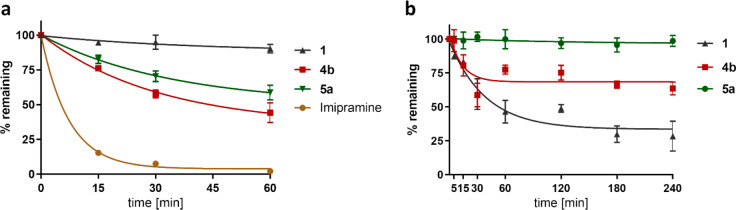
Metabolic and Plasma Stability. One major advantage of nonpeptidic drugs is their generally improved plasma stability. As a preliminary evaluation of the stability of our novel PAR2 ligands, we explored the propensity of our most potent PAR2 agonists, 4b and 5a, to undergo phase I transformations in rat liver microsomes and their stability in rat plasma. The approved antidepressant imipramine was used as a positive control that is significantly metabolized39 in rat liver microsomes (Figure 5a). As expected, the peptidic PAR2 agonist 1 was stable against metabolization via CYP enzymes (Figure 5a) but rapidly degraded within 1 h in rat plasma (Figure 5b). Ligands 4b and 5a were significantly less sensitive to CYP metabolism compared with the clinically used drug imipramine (Figure 5a) and, more importantly, significantly more stable in rat plasma than the peptidic PAR2 agonist 1 (Figure 5b). Especially the 3-methylisoxazole derivative 5a displayed very high stability, as degradation in rat plasma within 4 h could not be observed. 5a is also more stable against metabolism via CYP enzymes than its analogue 4b and did not show signs of cytotoxicity or diminished cell viability as determined by a trypan blue exclusion test in HEK 293T cells after incubation of the cells with the ligand for 24 h (93 ± 1% and 92 ± 1% cell viability at ligand concentrations of 1 or 10 μM, respectively, vs 93 ± 1% for the DMSO control). Thus, 5a represents a promising PAR2 agonist for further in vivo studies.
Figure 5.
(a) Metabolic stability in rat liver microsomes. 1 was barely metabolized by microsomes. 4b and 5a were more stable than imipramine. (b) Plasma stability assay (rat). 5a and 4b were more stable than 1 in rat plasma. Samples were analyzed by LC–MS at the time points indicated. Plotted data represent standard corrected % peak areas of samples at t = 0 min and are given as mean ± SEM (n ≥ 3).
Conclusion. Herein we have described promising nonpeptidic PAR2 ligands. Employing a combination of ligand- and structure-based approaches, we identified the potent PAR2 agonist 5a (IK187), which shows high metabolic stability regarding phase I transformations and enzymatic degradation in plasma and represents not only a valuable tool that may be useful to study PAR2 activation in vivo but also a promising starting point for further drug discovery efforts.
Acknowledgments
This work was supported by the Research Training Group GRK 1910 (DFG) and the International Doctorate Program “Receptor Dynamics” of the Elite Network of Bavaria (ENB). We thank Prof. Korbmacher (Cellular and Molecular Physiology, FAU) for generously providing the plasmids for human PAR2 and PAR2-PK1S. We gratefully acknowledge the computing resources and support provided by the Erlangen Regional Computing Center (RRZE).
Glossary
Abbreviations
- 2f
2-furoyl
- Cha
cyclohexylalanine
- DIPEA
diisopropylethylamine
- EDA
ethylenediamine
- ERK
extracellular signal-regulated kinase
- GPCR
G protein-coupled receptor
- HATU
O-(7-azobenzotriazol-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- HEK
human embryonic kidney
- HOBT
1-hydroxybenzotriazole
- IP1
inositol monophosphate
- PAR
proteinase-activated receptor
- PyBOP
benzotriazol-1-yloxytripyrrolidinophosphonium hexafluorophosphate
- SPPS
solid-phase peptide synthesis
- TIS
triisopropylsilane.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00154.
The authors declare no competing financial interest.
Supplementary Material
References
- Rosenbaum D. M.; Rasmussen S. G. F.; Kobilka B. K. The Structure and Function of G-Protein-Coupled Receptors. Nature 2009, 459, 356–363. 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossovskaya V. S.; Bunnett N. W. Protease-Activated Receptors: Contribution to Physiology and Disease. Physiol. Rev. 2004, 84, 579–621. 10.1152/physrev.00028.2003. [DOI] [PubMed] [Google Scholar]
- Adams M. N.; Ramachandran R.; Yau M. K.; Suen J. Y.; Fairlie D. P.; Hollenberg M. D.; Hooper J. D. Structure, Function and Pathophysiology of Protease Activated Receptors. Pharmacol. Ther. 2011, 130, 248–282. 10.1016/j.pharmthera.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Ferrell W. R.; Lockhart J. C.; Kelso E. B.; Dunning L.; Plevin R.; Meek S. E.; Smith A. J.; Hunter G. D.; McLean J. S.; McGarry F.; Ramage R.; Jiang L.; Kanke T.; Kawagoe J. Essential Role for Proteinase-Activated Receptor-2 in Arthritis. J. Clin. Invest. 2003, 111, 35–41. 10.1172/JCI16913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vargas N. N.; Pattison L. A.; Zhao P.; Lieu T.; Latorre R.; Jensen D. D.; Castro J.; Aurelio L.; Le G. T.; Flynn B.; Herenbrink C. K.; Yeatman H. R.; Edgington-Mitchell L.; Porter C. J. H.; Halls M. L.; Canals M.; Veldhuis N. A.; Poole D. P.; McLean P.; Hicks G. A.; Scheff N.; Chen E.; Bhattacharya A.; Schmidt B. L.; Brierley S. M.; Vanner S. J.; Bunnett N. W. Protease-Activated Receptor-2 in Endosomes Signals Persistent Pain of Irritable Bowel Syndrome. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E7438–E7447. 10.1073/pnas.1721891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenac N.; Coelho A.-M.; Nguyen C.; Compton S.; Andrade-Gordon P.; MacNaughton W. K.; Wallace J. L.; Hollenberg M. D.; Bunnett N. W.; Garcia-Villar R.; Bueno L.; Vergnolle N. Induction of Intestinal Inflammation in Mouse by Activation of Proteinase-Activated Receptor-2. Am. J. Pathol. 2002, 161, 1903–1915. 10.1016/S0002-9440(10)64466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokananthan N.; Graham P. T.; Fink J.; Knight D. A.; Bakker A. J.; McWilliam A. S.; Thompson P. J.; Stewart G. A. Activation of Protease-Activated Receptor (PAR)-1, PAR-2, and PAR-4 Stimulates IL-6, IL-8, and Prostaglandin E2 Release from Human Respiratory Epithelial Cells. J. Immunol. 2002, 168, 3577–3585. 10.4049/jimmunol.168.7.3577. [DOI] [PubMed] [Google Scholar]
- Adam E.; Hansen K. K.; Astudillo O. F.; Coulon L.; Bex F.; Duhant X.; Jaumotte E.; Hollenberg M. D.; Jacquet A. The House Dust Mite Allergen Der p 1, Unlike Der p 3, Stimulates the Expression of Interleukin-8 in Human Airway Epithelial Cells via a Proteinase-Activated Receptor-2-Independent Mechanism. J. Biol. Chem. 2006, 281, 6910–6923. 10.1074/jbc.M507140200. [DOI] [PubMed] [Google Scholar]
- Ge L.; Shenoy S. K.; Lefkowitz R. J.; DeFea K. Constitutive Protease-Activated Receptor-2-Mediated Migration of MDA MB-231 Breast Cancer Cells Requires Both Beta-Arrestin-1 and −2. J. Biol. Chem. 2004, 279, 55419–55424. 10.1074/jbc.M410312200. [DOI] [PubMed] [Google Scholar]
- Chung H.; Hamza M.; Oikonomopoulou K.; Gratio V.; Saifeddine M.; Virca G. D.; Diamandis E. P.; Hollenberg M. D.; Darmoul D. Kallikrein-Related Peptidase Signaling in Colon Carcinoma Cells: Targeting Proteinase-Activated Receptors. Biol. Chem. 2012, 393, 413–420. 10.1515/bc-2011-231. [DOI] [PubMed] [Google Scholar]
- Schaffner F.; Versteeg H. H.; Schillert A.; Yokota N.; Petersen L. C.; Mueller B. M.; Ruf W. Cooperation of Tissue Factor Cytoplasmic Domain and PAR2 Signaling in Breast Cancer Development. Blood 2010, 116, 6106–6113. 10.1182/blood-2010-06-289314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R.; Ding Y.; Ricks T. K.; Gullapalli A.; Wolfe B. L.; Trejo J. Protease-Activated Receptor-2 is Essential for Factor VIIa and Xa-Induced Signaling, Migration, and Invasion of Breast Cancer Cells. Cancer Res. 2006, 66, 307–314. 10.1158/0008-5472.CAN-05-1735. [DOI] [PubMed] [Google Scholar]
- Hollenberg M. D.; Mihara K.; Polley D.; Suen J. Y.; Han A.; Fairlie D. P.; Ramachandran R. Biased Signalling and Proteinase-Activated Receptors (PARs): Targeting Inflammatory Disease. Br. J. Pharmacol. 2014, 171, 1180–1194. 10.1111/bph.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau M. K.; Liu L.; Fairlie D. P. Toward Drugs for Protease-Activated Receptor 2 (PAR2). J. Med. Chem. 2013, 56, 7477–7497. 10.1021/jm400638v. [DOI] [PubMed] [Google Scholar]
- Maryanoff B. E.; Santulli R. J.; McComsey D. F.; Hoekstra W. J.; Hoey K.; Smith C. E.; Addo M.; Darrow A. L.; Andrade-Gordon P. Protease-Activated Receptor-2 (PAR-2): Structure-Function Study of Receptor Activation by Diverse Peptides Related to Tethered-Ligand Epitopes. Arch. Biochem. Biophys. 2001, 386, 195–204. 10.1006/abbi.2000.2207. [DOI] [PubMed] [Google Scholar]
- McGuire J. J.; Saifeddine M.; Triggle C. R.; Sun K.; Hollenberg M. D. 2-Furoyl-LIGRLO-Amide: A Potent and Selective Proteinase-Activated Receptor 2 Agonist. J. Pharmacol. Exp. Ther. 2004, 309, 1124–1131. 10.1124/jpet.103.064584. [DOI] [PubMed] [Google Scholar]
- Yau M. K.; Suen J. Y.; Xu W.; Lim J.; Liu L.; Adams M. N.; He Y.; Hooper J. D.; Reid R. C.; Fairlie D. P. Potent Small Agonists of Protease Activated Receptor 2. ACS Med. Chem. Lett. 2016, 7, 105–110. 10.1021/acsmedchemlett.5b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardell L. R.; Ma J. N.; Seitzberg J. G.; Knapp A. E.; Schiffer H. H.; Tabatabaei A.; Davis C. N.; Owens M.; Clemons B.; Wong K. K.; Lund B.; Nash N. R.; Gao Y.; Lameh J.; Schmelzer K.; Olsson R.; Burstein E. S. Identification and Characterization of Novel Small-Molecule Protease-Activated Receptor 2 Agonists. J. Pharmacol. Exp. Ther. 2008, 327, 799–808. 10.1124/jpet.108.142570. [DOI] [PubMed] [Google Scholar]
- Seitzberg J. G.; Knapp A. E.; Lund B. W.; Mandrup Bertozzi S.; Currier E. A.; Ma J.-N.; Sherbukhin V.; Burstein E. S.; Olsson R. Discovery of Potent and Selective Small-Molecule PAR-2 Agonists. J. Med. Chem. 2008, 51, 5490–5493. 10.1021/jm800754r. [DOI] [PubMed] [Google Scholar]
- Barry G. D.; Suen J. Y.; Le G. T.; Cotterell A.; Reid R. C.; Fairlie D. P. Novel Agonists and Antagonists for Human Protease Activated Receptor 2. J. Med. Chem. 2010, 53, 7428–7440. 10.1021/jm100984y. [DOI] [PubMed] [Google Scholar]
- Flynn A. N.; Tillu D. V.; Asiedu M. N.; Hoffman J.; Vagner J.; Price T. J.; Boitano S. The Protease-Activated Receptor-2-Specific Agonists 2-Aminothiazol-4-yl-LIGRL-NH2 and 6-Aminonicotinyl-LIGRL-NH2 Stimulate Multiple Signaling Pathways to Induce Physiological Responses in Vitro and in Vivo. J. Biol. Chem. 2011, 286, 19076–19088. 10.1074/jbc.M110.185264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano S.; Hoffman J.; Tillu D. V.; Asiedu M. N.; Zhang Z.; Sherwood C. L.; Wang Y.; Dong X.; Price T. J.; Vagner J. Development and Evaluation of Small Peptidomimetic Ligands to Protease-Activated Receptor-2 (PAR2) Through the Use of Lipid Tethering. PLoS One 2014, 9, e99140 10.1371/journal.pone.0099140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau M. K.; Liu L.; Suen J. Y.; Lim J.; Lohman R. J.; Jiang Y.; Cotterell A. J.; Barry G. D.; Mak J. Y.; Vesey D. A.; Reid R. C.; Fairlie D. P. PAR2Modulators Derived from GB88. ACS Med. Chem. Lett. 2016, 7, 1179–1184. 10.1021/acsmedchemlett.6b00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen J. Y.; Barry G. D.; Lohman R. J.; Halili M. A.; Cotterell A. J.; Le G. T.; Fairlie D. P. Modulating Human Proteinase Activated Receptor 2 with a Novel Antagonist (GB88) and Agonist (GB110). Br. J. Pharmacol. 2012, 165, 1413–1423. 10.1111/j.1476-5381.2011.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau M. K.; Liu L.; Lim J.; Lohman R. J.; Cotterell A. J.; Suen J. Y.; Vesey D. A.; Reid R. C.; Fairlie D. P. Benzylamide Antagonists of Protease Activated Receptor 2 with Anti-Inflammatory Activity. Bioorg. Med. Chem. Lett. 2016, 26, 986–991. 10.1016/j.bmcl.2015.12.048. [DOI] [PubMed] [Google Scholar]
- Manglik A.; Lin H.; Aryal D. K.; McCorvy J. D.; Dengler D.; Corder G.; Levit A.; Kling R. C.; Bernat V.; Hübner H.; Huang X.-P.; Sassano M. F.; Giguère P. M.; Löber S.; Da D.; Scherrer G.; Kobilka B. K.; Gmeiner P.; Roth B. L.; Shoichet B. K. Structure-Based Discovery of Opioid Analgesics with Reduced Side Effects. Nature 2016, 537, 185–190. 10.1038/nature19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Hofmann J.; Fish I.; Schaake B.; Eitel K.; Bartuschat A.; Kaindl J.; Rampp H.; Banerjee A.; Hübner H.; Clark M. J.; Vincent S. G.; Fisher J. T.; Heinrich M. R.; Hirata K.; Liu X.; Sunahara R. K.; Shoichet B. K.; Kobilka B. K.; Gmeiner P. Structure-Guided Development of Selective M3Muscarinic Acetylcholine Receptor Antagonists. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 12046–12050. 10.1073/pnas.1813988115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet B. K.; Kobilka B. K. Structure-Based Drug Screening for G-Protein-Coupled Receptors. Trends Pharmacol. Sci. 2012, 33, 268–272. 10.1016/j.tips.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M.; de Graaf C.; Swain N. A.; Tate C. G. Impact of GPCR Structures on Drug Discovery. Cell 2020, 181, 81–91. 10.1016/j.cell.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Lee Y.; Basith S.; Choi S. Recent Advances in Structure-Based Drug Design Targeting Class A G Protein-Coupled Receptors Utilizing Crystal Structures and Computational Simulations. J. Med. Chem. 2018, 61, 1–46. 10.1021/acs.jmedchem.6b01453. [DOI] [PubMed] [Google Scholar]
- Cheng R. K. Y.; Fiez-Vandal C.; Schlenker O.; Edman K.; Aggeler B.; Brown D. G.; Brown G. A.; Cooke R. M.; Dumelin C. E.; Doré A. S.; Geschwindner S.; Grebner C.; Hermansson N.-O.; Jazayeri A.; Johansson P.; Leong L.; Prihandoko R.; Rappas M.; Soutter H.; Snijder A.; Sundström L.; Tehan B.; Thornton P.; Troast D.; Wiggin G.; Zhukov A.; Marshall F. H.; Dekker N. Structural insight into allosteric modulation of protease-activated receptor 2. Nature 2017, 545, 112–115. 10.1038/nature22309. [DOI] [PubMed] [Google Scholar]
- Saleh N.; Ibrahim P.; Saladino G.; Gervasio F. L.; Clark T. An efficient metadynamics-based protocol to model the binding affinity and the transition state ensemble of G-protein-coupled receptor ligands. J. Chem. Inf. Model. 2017, 57, 1210–1217. 10.1021/acs.jcim.6b00772. [DOI] [PubMed] [Google Scholar]
- Kennedy A. J.; Ballante F.; Johansson J. R.; Milligan G.; Sundström L.; Nordqvist A.; Carlsson J. Structural Characterization of Agonist Binding to Protease-Activated Receptor 2 Through Mutagenesis and Computational Modeling. ACS Pharmacol. Transl. Sci. 2018, 1, 119–133. 10.1021/acsptsci.8b00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnam M. A. M.; Nitsche C.; Vechi S. M.; Klein C. D. C-Terminal Residue Optimization and Fragment Merging: Discovery of a Potent Peptide-Hybrid Inhibitor of Dengue Protease. ACS Med. Chem. Lett. 2014, 5, 1037–1042. 10.1021/ml500245v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; Metcalf M.; Bunnett N. W. Biased Signaling of Protease-Activated Receptors. Front. Endocrinol. 2014, 5, 67. 10.3389/fendo.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Yau M.-K.; Kok W. M.; Lim J.; Wu K.-C.; Liu L.; Hill T. A.; Suen J. Y.; Fairlie D. P. Biased Signaling by Agonists of Protease Activated Receptor 2. ACS Chem. Biol. 2017, 12, 1217–1226. 10.1021/acschembio.6b01088. [DOI] [PubMed] [Google Scholar]
- Nichols H. L.; Saffeddine M.; Theriot B. S.; Hegde A.; Polley D.; El-Mays T.; Vliagoftis H.; Hollenberg M. D.; Wilson E. H.; Walker J. K. L.; DeFea K. A. β-Arrestin-2 Mediates the Proinflammatory Effects of Proteinase-Activated Receptor-2 in the Airway. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 16660–16665. 10.1073/pnas.1208881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. K. L.; DeFea K. A. Role for β-Arrestin in Mediating Paradoxical β2AR and PAR2 Signaling in Asthma. Curr. Opin. Pharmacol. 2014, 16, 142–147. 10.1016/j.coph.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel M. H.; Baggiolini M. The Metabolism of Imipramine and its Metabolites by Rat Liver Microsomes. Biochem. Pharmacol. 1966, 15, 1155–1169. 10.1016/0006-2952(66)90281-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.