Abstract
B-cell lymphoma 6 (BCL6) is a transcriptional repressor frequently deregulated in lymphoid malignancies. BCL6 engages with number of corepressors, and these protein–protein interactions are being explored as a strategy for drug development. Here, we report the development of an irreversible BCL6 inhibitor TMX-2164 that uses a sulfonyl fluoride to covalently react with the hydroxyl group of Tyrosine 58 located in the lateral groove. TMX-2164 exhibits significantly improved inhibitory activity compared to that of its reversible parental compound and displays sustained target engagement and antiproliferative activity in cells. TMX-2164 therefore represents an example of a tyrosine-directed covalent inhibitor of BCL6 which demonstrates advantages relative to reversible targeting.
Keywords: BCL6, Protein−protein interactions, Covalent inhibitor, Sulfonyl fluoride
B-cell lymphoma 6 (BCL6) is an essential protein for the formation and maintenance of the germinal center (GC) during the humoral immune response1−3 and is therefore important for B-cell development. GCs are transient and dynamic substructures of lymph nodes that are dedicated to the selection of B-cells expressing high-affinity antibodies in response to T-cell-dependent antigen stimulation.4,5 Within GCs, BCL6 represses the expression of genes that are required to sustain mutagenic activity without activating the DNA damage response or apoptosis,6−8 ensuring that GC B-cells undergo immunoglobulin affinity maturation.6,9 BCL6 also represses genes required for exit from the GC cycle, ensuring that GC B-cells have sufficient time to acquire somatic hypermutation of their immunoglobulin genes.6 The activity of BCL6 must be switched off once GC B-cells acquire appropriate affinity for the inciting antigen, which allows their differentiation into memory B-cells and plasma cells.10 However, deregulation of BCL6 results in a highly proliferative GC phenotype with accumulating DNA damage, eventually leading to malignant transformation of B-cells.11,12 Overexpression of BCL6 has been found in diffuse large B-cell lymphoma (DLBCL), nodular lymphocyte predominant Hodgkin lymphomas (NLPHL), and follicular lymphoma (FL). Transgenic IμHA-BCL6 mice developed DLBCL-like tumors,11 and genetic knockdown of BCL6 with shRNA induced lethality in DLBCL cell lines,13 suggesting that BCL6 represents a therapeutic target for cancer treatment.
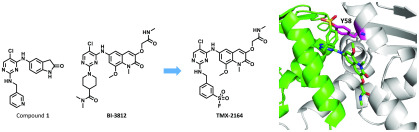
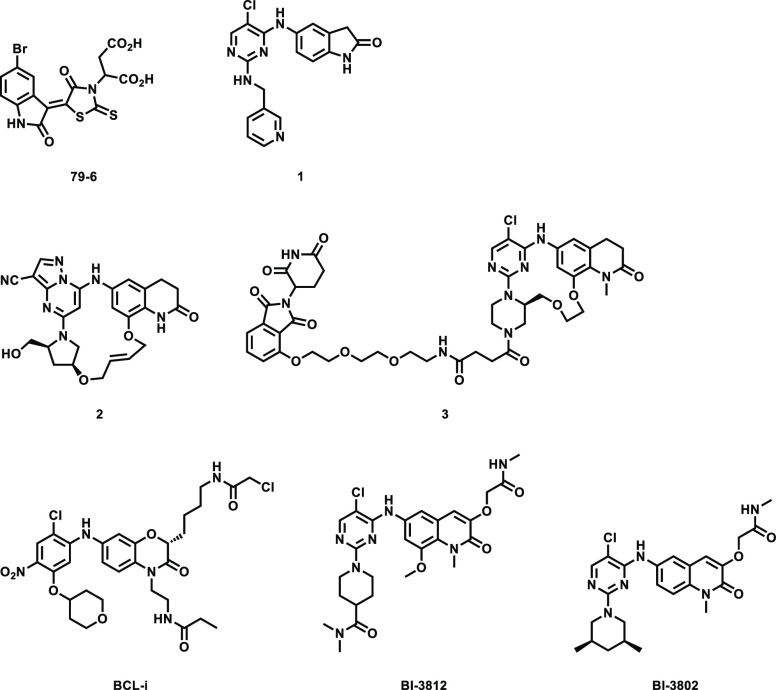
Mechanistically, BCL6 regulates gene transcription through recruiting corepressor proteins such as BCL6 corepressor (BCOR). BCL6 functions as an obligatory dimer, and this dimerization, which is mediated by the BTB domain, creates two identical lateral grooves that engage binding partners.14,15 The lateral groove also is a hotspot for small molecule inhibitor binding, such as compound 79-6 which was the first reported small molecule inhibitor of BCL6.16 Further optimization efforts resulted in inhibitors with improved activity, including compound 1,17BI-3812,18 and macrocyclic compound 2.19 All of these compounds employ a reversible mode of binding. Recently, Sameshima et al. reported an irreversible BCL6 inhibitor BCL-i that targets cysteine 53 (Cys53) located within a cavity found in the BTB domain.20 In addition to these compounds that bind BCL6 and inhibit protein–protein interactions, more recently, several compounds that induce BCL6 degradation have also been reported. For example, BI-3802 was serendipitously identified as a BCL6 degrader,18 whereas heterobifunctional molecule 3 that hijacks E3 ligase cereblon (CRBN) to induce proteasome-mediated degradation of BCL6 was rationally designed21 (Figure 1). Here, we report the rational design of TMX-2164, a covalent inhibitor that targets tyrosine 58 (Tyr58) located in the lateral groove of BCL6. We validate that TMX-2164 covalently binds Tyr58 using a range of in vitro assays and confirm that it engages BCL6 in cells. TMX-2164 exhibits single digit micromolar antiproliferative activity in DLBCL cells and outperforms the reversible parental compound.
Figure 1.
Chemical structures of published BCL6 inhibitors and BCL6 degraders.
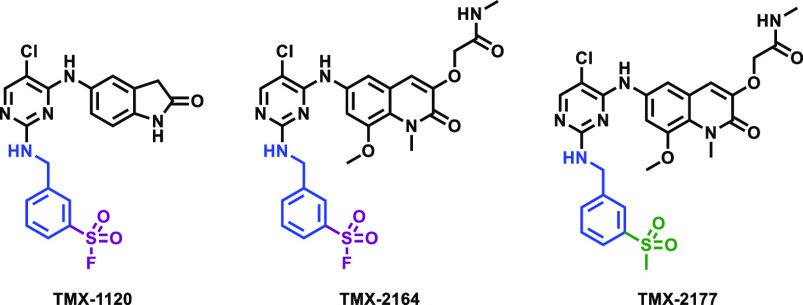
The cocrystal structure of compound 1 with BCL6 (PDB: 5X4Q) (Figure 2) provided the basis for the design of a covalent inhibitor. Tyr58 of BCL6 is favorably located within 4.2 Å as measured by the distance between Tyr58 −OH group and meta-carbon of pyridine ring. We also noted that the guanidinium group of Arg28 is within 2.4 Å of the Tyr58 −OH and likely to facilitate deprotonation of the Tyr58 −OH, as was previously observed in reports that describe covalent targeting of tyrosines.22−24 Additionally, we observed that the pyridine moiety was solvent-exposed and disordered, suggesting that there should be no steric hindrance to reach the phenol group. Taken together, we reasoned that introducing sulfonyl fluoride, a previously used electrophilic warhead for targeting tyrosines22,25−27 to compound 1, would potentially yield a covalent inhibitor for BCL6. Thus, compound TMX-1120 (Figure 3) was designed and docked into the BCL6 crystal structure. As shown in Figure 2, TMX-1120 is predicted to bind to BCL6BTB in a binding mode similar to that of compound 1. The carbonyl oxygen of the cyclic amide moiety was observed to interact with Glu115; the linker nitrogen formed a hydrogen bond with the main-chain oxygen of Met51, and one of the pyrimidine nitrogen atoms interacted with Arg28. Importantly, the original interaction of Tyr58 −OH with Arg28 was replaced by the oxygen of the sulfonyl moiety, strongly suggesting the potential for covalent bond formation.
Figure 2.
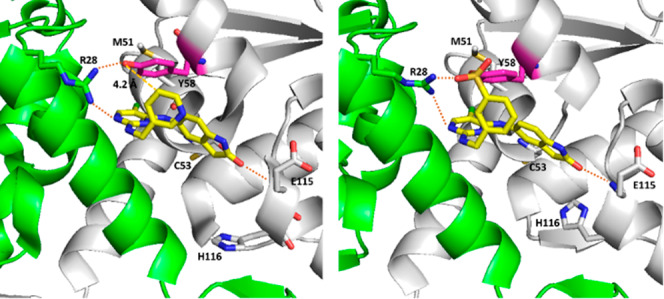
(Left) Co-crystal structure of compound 1 in complex with BCL6BTB (PDB code: 5X4Q). The side chain of Tyr58 (colored pink) is located near the 5-position of the pyridine ring (4.2 Å). (Right) Docking-based structure of TMX-1120 bound to BCL6BTB.
Figure 3.
Chemical structures of covalent BCL6 inhibitors and nonreactive methylsulfonyl-bearing control compound used in this study.
To validate that TMX-1120 binds and inhibits BCL6, we performed a series of in vitro experiments. To assess the binding, we used a TR-FRET-based biochemical assay where a BodipyFL-labeled BCOR peptide was displaced from BCL6 upon titration with increasing concentrations of TMX-1120. At a fixed time point of 30 min, TMX-1120 exhibited an IC50 of 251 nM and, in contrast, the reversible compound 1 demonstrated an IC50 of only 2699 nM (Figure 4). To examine whether the improved activity was due to the covalent bond formation with Tyr58, we analyzed recombinant expressed BCL6 protein by LC-MS after incubation with a 10-fold molar excess of TMX-1120 for 2 h at room temperature. We observed a mass shift consistent with the stoichiometric modification of the protein by TMX-1120 (accompanied by the loss of HF as expected for the reaction between Tyr −OH with the sulfonyl fluoride warhead, Figure 5A). To determine the site of modification, the labeled protein was digested with trypsin, and peptides were analyzed by capillary electrophoresis-mass spectrometry (CE-MS). Database search revealed exclusive modification of BCL6 Tyr58 (Figure 5B). Taken together, our mass spectrometry-based analysis confirmed that TMX-1120 forms a covalent adduct with BCL6 by reacting with Tyr58. Next, we took advantage of the well-established structure–activity relationship (SAR) for this scaffold to hybridize the sulfonyl fluoride warhead with BI-3812, which is a more potent reversible BCL6 inhibitor compared to compound 1. This resulted in the design of compound TMX-2164 and its reversible counterpart compound TMX-2177 (Figure 3). After confirming the covalent binding of TMX-2164 with BCL6 by mass spectrometry (Figure 5C and D), we measured the affinity of TMX-2164 using the TR-FRET-based displacement assay described above. As shown in Figure 4, TMX-2164 displayed an IC50 of 152 nM. The reversible counterpart, TMX-2177, showed a comparable activity, in agreement with a stronger reversible binding with BCL6 compared to compound 1.
Figure 4.
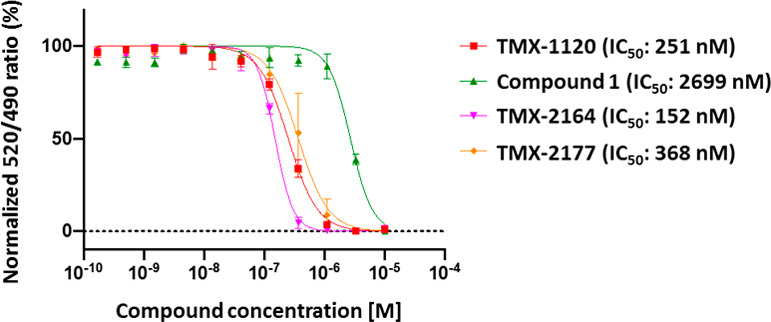
BCL-6 corepressor peptide displacement assay.
Figure 5.
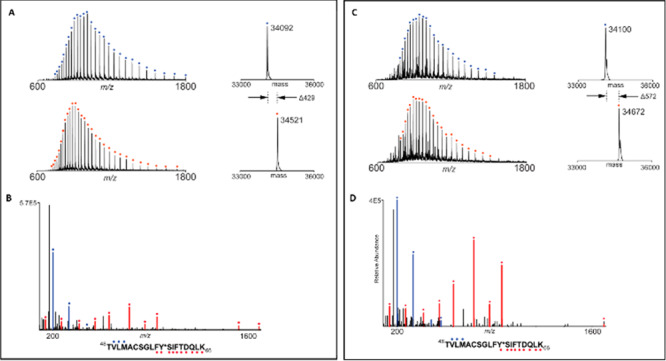
Mass spectrometry analysis reveals that TMX-1120 and TMX-2164 react with BCL6 protein at Tyr58. (A) Mass spectra (left) and zero-charge mass spectra (right) of BCL6 protein treated with DMSO (top) or 10-fold molar excess of TMX-1120 (bottom) for 2 h at room temperature. The observed mass shift of 429 Da is consistent with covalent addition of a single molecule of TMX-1120 (with loss of HF). (B) CE-MS/MS spectrum of tryptic BCL6 peptide (residues 48–66) with Y58 modified by TMX-1120. Ions of type b and y are indicated with blue and red glyphs, respectively. Y*, TMX-1120-modified tyrosine. Panels C and D are the same as panels A and B except with TMX-2164 (mass shift of 572 Da is consistent with TMX-2164 with loss of HF).
To determine whether our compounds indeed function as covalent inhibitors in cells, we developed a fluorescence-activated cell sorting (FACS)-based reporter assay in HEK293T-cells, which allows quantification of BCL6 levels by monitoring the eGFP to mCherry ratio (Figure 6A).28 Cells were treated with either covalent inhibitors TMX-1120 and TMX-2164 or reversible compounds 1 and TMX-2177 at a fixed concentration of 5 μM for 30 h, and excess compound was then washed away. The cells were then exposed to BCL6 degrader BI-3802 at various concentrations, and BCL6 protein levels were monitored using the ratio of eGFP over mCherry. As shown in Figure 6B, while TMX-1120 and TMX-2164 rescued BCL6 from BI-3802-induced degradation, compound 1 and TMX-2177 did not, demonstrating a prolonged occupancy on BCL6 protein by a covalent inhibitor. Furthermore, we evaluated the antiproliferative activity of these covalent inhibitors in SU-DHL-4 cells (Figure S1), a DLBCL model system. With 5-day treatment, TMX-2164 showed the most effective cell growth inhibition with single digit micromolar GI50 (Figure 6C). However, all the reversible compounds, including TMX-2177 that had a biochemical activity comparable to that of TMX-2164, significantly lost ability to protect BCL6 from degradation as well as antiproliferative activity, suggesting that a covalent inhibitor may be superior to a reversible one under the conditions tested.
Figure 6.
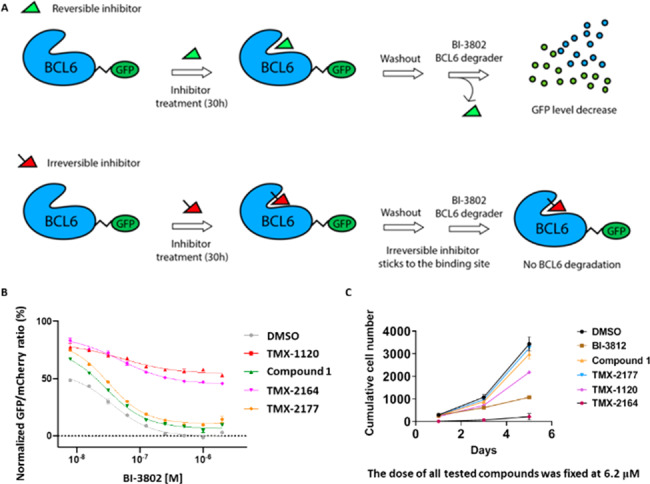
(A) A schematic cartoon describing the washout assay in BCL6-eGFP reporter cells. (B) The BCL6 target engagement for the covalent (irreversible) and reversible BCL6 inhibitors in HEK293T-cells. (C) Antiproliferative effects of the covalent and reversible BCL6 inhibitors after 1-day, 3-day, and 5-day treatment in SU-DHL-4 cells at a fixed dose of 6.2 μM. See also Figure S1 for additional information.
Experimental Section
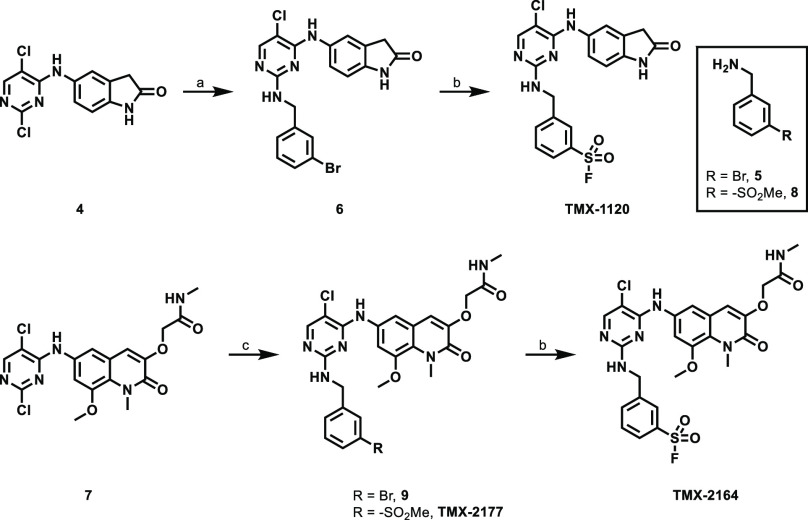
Synthetic methods for compounds TMX-1120, TMX-2164, and TMX-2177 are shown in Scheme 1. 5-((2,5-Dichloropyrimidin-4-yl)amino)indolin-2-one (4) and 2-[(6-((2,5-dichloropyrimidin-4-yl)amino)-8-methoxy-1-methyl-2-oxo-1,2-dihydroquinolin-3-yl)oxy]-N-methylacetamide (7) were used as staring materials. N-(3-Bromobenzyl)pyrimidin-2-amines 6 and 9 were then generated through aromatic nucleophilic substitution of 2-chloropyrimidine 4 or 7, respectively, by using benzylamine as the nucleophile. Next, a one-pot palladium-catalyzed synthesis of sulfonyl fluoride from aryl bromide, an elegant methodology recently developed by the Willis’s group,29 was applied to successfully generate both TMX-1120 and TMX-2164, albeit in low yields.
Scheme 1. Synthesis of Compounds TMX-1120, TMX-2164, and TMX-2177.
Reagents and conditions: (a) 5, TEA, DMSO, 95 °C, 67%; (b) DABSO, PdCl2(AmPhos)2, TEA, i-PrOH, 75 °C; then NFSI, rt, 6% (for TMX-1120) or 7% (for TMX-2164); (c) 5 or 8, TEA, DMF, MeOH, 70 °C, 59% (for 9) or 9% (for TMX-2177).
In summary, here we report the development of a covalent BCL6 inhibitor, TMX-1120. We used a structure-guided rational design strategy that introduced a tyrosine-directed covalent warhead, sulfonyl fluoride, to a reversible BCL6 inhibitor 1. We validated that TMX-1120 forms a covalent bond with Tyr58 on BCL6 and sustainably inhibits BCL6 in cellular context after washout, whereas the reversible inhibitors required continuous exposure. Further optimization resulted in the development of TMX-2164, which showed an improved antiproliferation activity in SU-DHL-4 cells, a DLBCL model system. Thus, the covalent inhibitor TMX-2164 represents a new example of tyrosine-directed covalent targeting strategy applied to BCL6 with advantages over reversible targeting.
Acknowledgments
We thank Dr. Milka Kostic (Twitter: @MilkaKostic) for assistance with critical reading and polishing the manuscript. J.A.M. acknowledges generous support from the NIH (R01-CA233800).
Glossary
Abbreviations
- BCL6
B-cell lymphoma 6
- TEA
triethylamine
- DMSO
dimethyl sulfoxide
- DABSO
1,4-diazabicyclo[2.2.2]octane bis(sulfur dioxide) adduct
- AmPhos
ditert-butyl(4-dimethylaminophenyl)phosphine
- NFSI
N-fluorobenzenesulfonimide
- i-PrOH
isopropanol
- rt
room temperature
- DMF
dimethylformamide
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00111.
Details of biological assays, synthetic procedures, and analytical data (PDF)
Author Contributions
M.T., T.Z., and N.S.G. generated the concept. M.T. designed and synthesized compounds, analyzed and interpreted data, and prepared the manuscript. S.B.F. and J.A.M. carried out mass spectrum labeling and trypsin digestion. H.Y. and E.S.F. expressed and purified protein constructs and performed TR-FRET-based biochemical assay and washout assay. J.C. built the docking model. J.Z. performed cell proliferation assay. T.Z. and N.S.G. provided strategic direction and coordinated manuscript preparation. All authors discussed the results and commented on the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): Eric S. Fischer is a founder, science advisory board (SAB) member, and equity holder in Civetta Therapeutics and an equity holder and SAB member in C4 Therapeutics. The Fischer lab receives or has received research funding from Astellas, Novartis, Voronoi, and Deerfield. Jarrod A. Marto serves on the SAB of 908 Devices and has received sponsored research support from AstraZeneca and Vertex. Nathanael S. Gray is a founder, SAB member, and equity holder in Gatekeeper, Syros, Petra, C4, B2S, Aduro, and Soltego. The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voronoi, Her2llc, Deerfield, and Sanofi.
Supplementary Material
References
- Ye B. H.; Cattoretti G.; Shen Q.; Zhang J.; Hawe N.; Waard R. d.; Leung C.; Nouri-Shirazi M.; Orazi A.; Chaganti R.S.K.; Rothman P.; Stall A. M.; Pandolfi P.-P.; Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997, 16 (2), 161–170. 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Dent A. L.; Shaffer A. L.; Yu X.; Allman D.; Staudt L. M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 1997, 276 (5312), 589–592. 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Fukuda T.; Yoshida T.; Okada S.; Hatano M.; Miki T.; Ishibashi K.; Okabe S.; Koseki H.; Hirosawa S.; Taniguchi M.; Miyasaka N.; Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 1997, 186 (3), 439–448. 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K.; Dalla-Favera R. Germinal centres and B-cell lymphomagenesis. Nat. Rev. Immunol. 2015, 15 (3), 172–184. 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- Mesin L.; Ersching J.; Victora G. D. Germinal Center B Cell Dynamics. Immunity 2016, 45 (3), 471–482. 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K.; Melnick A. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014, 20 (6), 343–352. 10.1016/j.molmed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K.; Saito M.; Sumazin P.; Margolin A. A.; Wang K.; Lim W. K.; Kitagawa Y.; Schneider C.; Alvarez M. J.; Califano A.; Dalla-Favera R. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B-cells. Blood 2010, 115 (5), 975–984. 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W.; Polo J. M.; Cerchietti L.; Shaknovich R.; Wang L.; Yang S. N.; Ye K.; Farinha P.; Horsman D. E.; Gascoyne R. D.; Elemento O.; Melnick A. The BCL6 transcriptional program features repression of multiple oncogenes in primary B-cells and is deregulated in DLBCL. Blood 2009, 113 (22), 5536–5548. 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U.; Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008, 8 (1), 22–33. 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Saito M.; Gao J.; Basso K.; Kitagawa Y.; Smith P. M.; Bhagat G.; Pernis A.; Pasqualucci L.; Dalla-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B-cells is blocked by BCL6 gene alterations in B-cell lymphoma. Cancer Cell 2007, 12 (3), 280–292. 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Cattoretti G.; Pasqualucci L.; Ballon G.; Tam W.; Nandula S. V.; Shen Q.; Mo T.; Murty V. V.; Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B-cell lymphomas in mice. Cancer Cell 2005, 7 (5), 445–455. 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Baron B. W.; Anastasi J.; Montag A.; Huo D.; Baron R. M.; Karrison T.; Thirman M. J.; Subudhi S. K.; Chin R. K.; Felsher D. W.; Fu Y. X.; McKeithan T. W.; Baron J. M. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (39), 14198–14203. 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranuncolo S. M.; Polo J. M.; Dierov J.; Singer M.; Kuo T.; Greally J.; Green R.; Carroll M.; Melnick A. Bcl-6 mediates the germinal center B-cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 2007, 8 (7), 705–714. 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- Ahmad K. F.; Engel C. K.; Prive G. G. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. U. S. A. 1998, 95 (21), 12123–12128. 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetu A. F.; Corcoran C. M.; Cerchietti L.; Bardwell V. J.; Melnick A.; Prive G. G. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell 2008, 29 (3), 384–391. 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti L. C.; Ghetu A. F.; Zhu X.; Da Silva G. F.; Zhong S.; Matthews M.; Bunting K. L.; Polo J. M.; Fares C.; Arrowsmith C. H.; Yang S. N.; Garcia M.; Coop A.; Mackerell A. D. Jr.; Prive G. G.; Melnick A. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 2010, 17 (4), 400–411. 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y.; Sakai N.; Sogabe S.; Ida K.; Oki H.; Sakamoto K.; Lane W.; Snell G.; Iida M.; Imaeda Y.; Sakamoto J.; Matsui J. Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach. J. Med. Chem. 2017, 60 (10), 4358–4368. 10.1021/acs.jmedchem.7b00313. [DOI] [PubMed] [Google Scholar]
- Kerres N.; Steurer S.; Schlager S.; Bader G.; Berger H.; Caligiuri M.; Dank C.; Engen J. R.; Ettmayer P.; Fischerauer B.; Flotzinger G.; Gerlach D.; Gerstberger T.; Gmaschitz T.; Greb P.; Han B.; Heyes E.; Iacob R. E.; Kessler D.; Kolle H.; Lamarre L.; Lancia D. R.; Lucas S.; Mayer M.; Mayr K.; Mischerikow N.; Muck K.; Peinsipp C.; Petermann O.; Reiser U.; Rudolph D.; Rumpel K.; Salomon C.; Scharn D.; Schnitzer R.; Schrenk A.; Schweifer N.; Thompson D.; Traxler E.; Varecka R.; Voss T.; Weiss-Puxbaum A.; Winkler S.; Zheng X.; Zoephel A.; Kraut N.; McConnell D.; Pearson M.; Koegl M. Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6. Cell Rep. 2017, 20 (12), 2860–2875. 10.1016/j.celrep.2017.08.081. [DOI] [PubMed] [Google Scholar]
- McCoull W.; Abrams R. D.; Anderson E.; Blades K.; Barton P.; Box M.; Burgess J.; Byth K.; Cao Q.; Chuaqui C.; Carbajo R. J.; Cheung T.; Code E.; Ferguson A. D.; Fillery S.; Fuller N. O.; Gangl E.; Gao N.; Grist M.; Hargreaves D.; Howard M. R.; Hu J.; Kemmitt P. D.; Nelson J. E.; O’Connell N.; Prince D. B.; Raubo P.; Rawlins P. B.; Robb G. R.; Shi J.; Waring M. J.; Whittaker D.; Wylot M.; Zhu X. Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors. J. Med. Chem. 2017, 60 (10), 4386–4402. 10.1021/acs.jmedchem.7b00359. [DOI] [PubMed] [Google Scholar]
- Sameshima T.; Yamamoto T.; Sano O.; Sogabe S.; Igaki S.; Sakamoto K.; Ida K.; Gotou M.; Imaeda Y.; Sakamoto J.; Miyahisa I. Discovery of an Irreversible and Cell-Active BCL6 Inhibitor Selectively Targeting Cys53 Located at the Protein-Protein Interaction Interface. Biochemistry 2018, 57 (8), 1369–1379. 10.1021/acs.biochem.7b00732. [DOI] [PubMed] [Google Scholar]
- McCoull W.; Cheung T.; Anderson E.; Barton P.; Burgess J.; Byth K.; Cao Q.; Castaldi M. P.; Chen H.; Chiarparin E.; Carbajo R. J.; Code E.; Cowan S.; Davey P. R.; Ferguson A. D.; Fillery S.; Fuller N. O.; Gao N.; Hargreaves D.; Howard M. R.; Hu J.; Kawatkar A.; Kemmitt P. D.; Leo E.; Molina D. M.; O’Connell N.; Petteruti P.; Rasmusson T.; Raubo P.; Rawlins P. B.; Ricchiuto P.; Robb G. R.; Schenone M.; Waring M. J.; Zinda M.; Fawell S.; Wilson D. M. Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6. ACS Chem. Biol. 2018, 13 (11), 3131–3141. 10.1021/acschembio.8b00698. [DOI] [PubMed] [Google Scholar]
- Hett E. C.; Xu H.; Geoghegan K. F.; Gopalsamy A.; Kyne R. E. Jr.; Menard C. A.; Narayanan A.; Parikh M. D.; Liu S.; Roberts L.; Robinson R. P.; Tones M. A.; Jones L. H. Rational targeting of active-site tyrosine residues using sulfonyl fluoride probes. ACS Chem. Biol. 2015, 10 (4), 1094–1098. 10.1021/cb5009475. [DOI] [PubMed] [Google Scholar]
- Hanoulle X.; van Damme J.; Staes A.; Martens L.; Goethals M.; Vandekerckhove J.; Gevaert K. A new functional, chemical proteomics technology to identify purine nucleotide binding sites in complex proteomes. J. Proteome Res. 2006, 5 (12), 3438–3445. 10.1021/pr060313e. [DOI] [PubMed] [Google Scholar]
- Gu C.; Shannon D. A.; Colby T.; Wang Z.; Shabab M.; Kumari S.; Villamor J. G.; McLaughlin C. J.; Weerapana E.; Kaiser M.; Cravatt B. F.; van der Hoorn R. A. L. Chemical proteomics with sulfonyl fluoride probes reveals selective labeling of functional tyrosines in glutathione transferases. Chem. Biol. 2013, 20 (4), 541–548. 10.1016/j.chembiol.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Dong J.; Krasnova L.; Finn M. G.; Sharpless K. B. Sulfur(VI) fluoride exchange (SuFEx): another good reaction for click chemistry. Angew. Chem., Int. Ed. 2014, 53 (36), 9430–9448. 10.1002/anie.201309399. [DOI] [PubMed] [Google Scholar]
- Hatcher J. M.; Wu G.; Zeng C.; Zhu J.; Meng F.; Patel S.; Wang W.; Ficarro S. B.; Leggett A. L.; Powell C. E.; Marto J. A.; Zhang K.; Ki Ngo J. C.; Fu X. D.; Zhang T.; Gray N. S. SRPKIN-1: A Covalent SRPK1/2 Inhibitor that Potently Converts VEGF from Pro-angiogenic to Anti-angiogenic Isoform. Cell Chem. Biol. 2018, 25 (4), 460–470. 10.1016/j.chembiol.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee H.; Su N.; Belmonte M. A.; Hargreaves D.; Patel J.; Tentarelli S.; Aquila B.; Grimster N. P. Discovery and optimization of covalent Bcl-xL antagonists. Bioorg. Med. Chem. Lett. 2019, 29 (23), 126682. 10.1016/j.bmcl.2019.126682. [DOI] [PubMed] [Google Scholar]
- Sievers Q. L.; Petzold G.; Bunker R. D.; Renneville A.; Slabicki M.; Liddicoat B. J.; Abdulrahman W.; Mikkelsen T.; Ebert B. L.; Thoma N. H. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 2018, 362 (6414), eaat0572 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A. T.; Curto J. M.; Bagley S. W.; Willis M. C. One-pot palladium-catalyzed synthesis of sulfonyl fluorides from aryl bromides. Chem. Sci. 2017, 8 (2), 1233–1237. 10.1039/C6SC03924C. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.