Abstract
Microglial cells play essential volume-related actions in the brain that contribute to the maturation and plasticity of neural circuits that ultimately shape behavior. Microglia can thus be expected to have similar cell sizes and even distribution both across brain structures and across species with different brain sizes. To test this hypothesis, we determined microglial cell densities (the inverse of cell size) using immunocytochemistry to Iba1 in samples of free cell nuclei prepared with the isotropic fractionator from brain structures of 33 mammalian species belonging to males and females of five different clades. We found that microglial cells constitute ∼7% of non-neuronal cells in different brain structures as well as in the whole brain of all mammalian species examined. Further, they vary little in cell density compared with neuronal cell densities within the cerebral cortex, across brain structures, across species within the same clade, and across mammalian clades. As a consequence, we find that one microglial cell services as few as one and as many as 100 neurons in different brain regions and species, depending on the local neuronal density. We thus conclude that the addition of microglial cells to mammalian brains is governed by mechanisms that constrain the size of these cells and have remained conserved over 200 million years of mammalian evolution. We discuss the probable consequences of such constrained size for brain function in health and disease.
SIGNIFICANCE STATEMENT Microglial cells are resident macrophages of the CNS, with key functions in recycling synapses and maintaining the local environment in health and disease. We find that microglial cells occur in similar densities in the brains of different species and in the different structures of each individual brain, which indicates that these cells maintain a similar average size in mammalian evolution, suggesting in turn that the volume monitored by each microglial cell remains constant across mammals. Because the density of neurons is highly variable across the same brain structures and species, our finding implies that microglia-dependent functional recovery may be particularly difficult in those brain structures and species with high neuronal densities and therefore fewer microglial cells per neuron.
Keywords: cell density, cell numbers, comparative, evolution, microglia
Introduction
Microglial cells are mesoderm-derived macrophages that reside in the CNS; and together with neuroectoderm-derived astrocytes and oligodendrocytes, as well as endoderm-derived endothelial cells, they compose the non-neuronal population of brain cells. Microglia play key surveillance and housekeeping roles as sensors of their local environment, responding to injury, phagocytosing apoptotic cells, pathogens, and cellular debris, and releasing inflammatory modulators and cytokines (Wake et al., 2013; Hickman et al., 2018). During development, they engulf both seemingly healthy and apoptotic neural precursors and immature neurons, thus contributing to establishing the final number of neurons in brain structures (and consequently tissue volume) (Cunningham et al., 2013; Kabba et al., 2018). In adult neurogenesis, microglia support survival, proliferation, and migration of neuronal precursors from the subventricular zone to the olfactory bulbs through the rostral migratory system (Xavier et al., 2015); and in neural plasticity, they regulate synaptic formation, maturation, and elimination (Wake et al., 2009, 2013; Tremblay et al., 2010; Kettenmann et al., 2011; Paolicelli et al., 2011; Schafer et al., 2012; Parkhurst et al., 2013).
All of the above functions concern formation and maintenance of brain tissue volume (Nimmerjahn et al., 2005; Kettenmann et al., 2011). As macrophages that they are, individual microglia can be construed to be monitors of their local environment, each in charge of its respective volume in the parenchyma, a view that is compatible with their typically homogeneous distribution in all adult cerebral cortices examined to date (Nimmerjahn et al., 2005; Cunningham et al., 2013; but for noncortical structures, see Lawson et al., 1990; Mittelbronn et al., 2001). Given that their functions are volume-based, a question of fundamental importance for understanding healthy and diseased brain function is how the volume of brain tissue monitored by the average individual microglial cell compares across species.
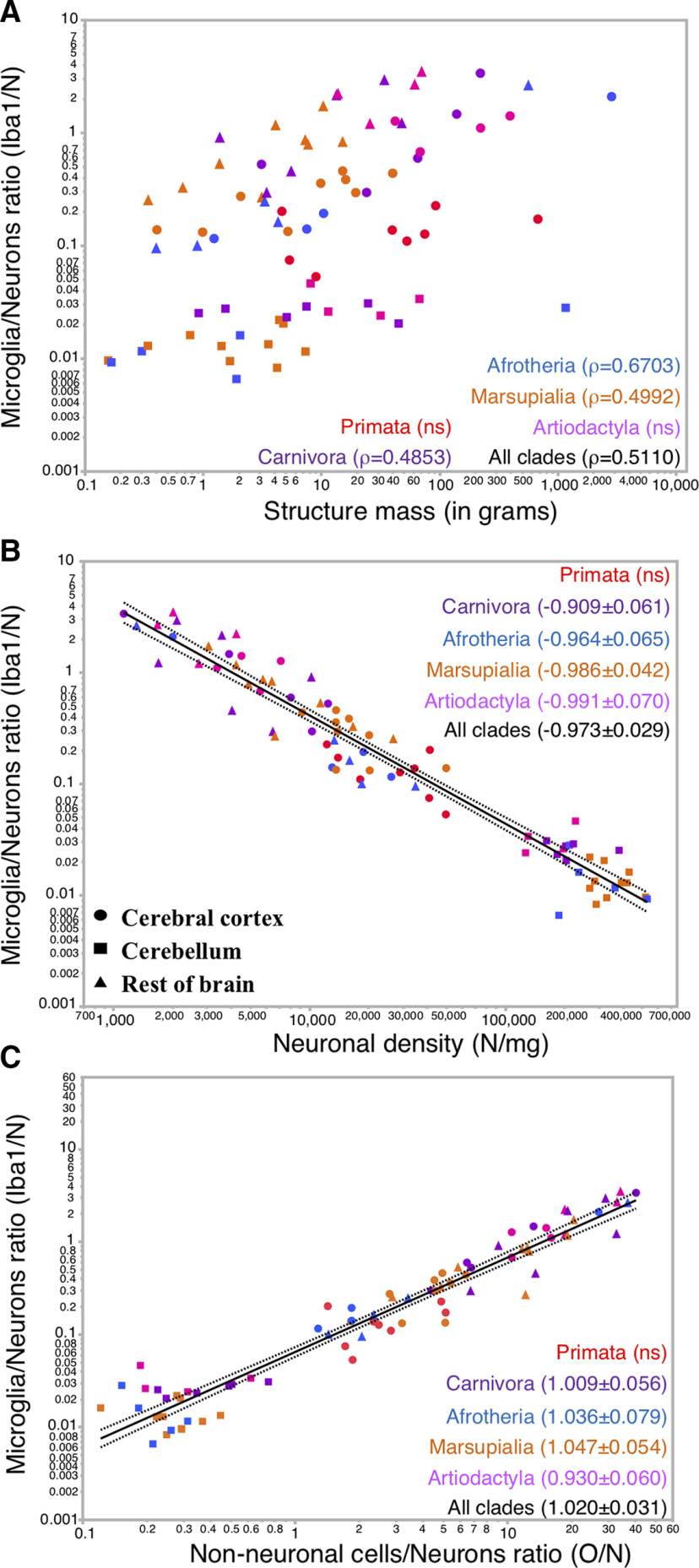
We have previously found that, while neuronal densities vary by 1000-fold across brain structures in >60 mammalian species, densities of non-neuronal cells as a whole vary only by 10-fold in the same sample, and are thus comparatively restricted, which is compatible with volume-related functions of non-neuronal cells (Herculano-Houzel and Dos Santos, 2018). As a consequence, the lower the neuronal density (i.e., the larger the average size of neurons), the larger the ratio of non-neuronal cells per neuron found in a tissue (Herculano-Houzel, 2014; Mota and Herculano-Houzel, 2014).
Our finding of constant densities of non-neuronal cells across brain structures and species does not translate automatically into constant densities of each non-neuronal cell type (Herculano-Houzel and Dos Santos, 2018). In the few instances where microglia have been quantified, they constitute a very small minority of all brain cells, amounting to 5% of cells in the cerebral cortex (Cx) or corpus callosum and 12% of cells in the substantia nigra of the mouse brain (Lawson et al., 1990), from 0.3% of cells in the gray matter of the cerebellum (Cb) to 11% of cells in the medulla oblongata of the human brain (Mittelbronn et al., 2001), and to 5% of glial cells in the human cortical gray matter (Pelvig et al., 2008). These small percentages imply that, while the restricted densities of non-neuronal cells found in our previous studies may apply to the different non-neuronal cell types separately, they might also mask marked variations in microglial size and distribution (Herculano-Houzel and Dos Santos, 2018).
Here we undertake a systematic examination of microglial densities across a wide range of brain structures and species of mammals to determine the extent of their variation. Our analysis is directed to determine how microglial densities vary at four levels: (1) across different sites within a same structure (gray and white matter of the Cx); (2) across brain structures; (3) across species in the same clade; and (4) across mammalian clades.
Materials and Methods
We used 2069 samples of free nuclei prepared with the isotropic fractionator technique (Herculano-Houzel and Lent, 2005) from previous studies that determined total numbers of cells and the fraction of neurons in different brain structures from 33 mammalian species of five clades: marsupials, afrotherians, primates, carnivorans, and artiodactyls (Ribeiro et al., 2013; Herculano-Houzel et al., 2014; Kazu et al., 2014; Neves et al., 2014; Gabi et al., 2016; Dos Santos et al., 2017; Jardim-Messeder et al., 2017; compiled in Herculano-Houzel et al., 2015a). These clades encompass over 218 million years of evolution since their divergence from the last common ancestor shared exclusively by all of them (Murphy et al., 2001, 2004; Bininda-Emonds et al., 2007).
To determine microglial densities and infer total numbers of these cells in each brain structure, we subjected free nuclei in each frozen stored sample to immunocytochemistry against the universal microglial marker Iba1 (ionized calcium binding adapter molecule 1), which is structurally and functionally conserved among Metazoa (Imai et al., 1996; Imai and Kohsaka, 2002; Janušonis, 2018). Total numbers of cells in the structures examined as well as structure mass are available for each species in the original publications, as listed above. Applying the percentage of nuclei found to be Iba1+ to total numbers of cells/mg reported in the same structure gives us the microglial density in each structure analyzed.
Sample
The 33 species analyzed here consist of 10 species of marsupials (Dos Santos et al., 2017), 4 species of artiodactyls (Kazu et al., 2014), 8 species of primates (Ribeiro et al., 2013; Gabi et al., 2016), five species of afrotherians (Herculano-Houzel et al., 2014; Neves et al., 2014), and 6 species of carnivorans (Jardim-Messeder et al., 2017) for which we had samples of suspensions of free nuclei in storage (see below). This excluded all rodent and primate species of our initial studies. All were adults of either sex, except for the giraffe, which was a juvenile (Kazu et al., 2014), with no signs of extreme aging, neurologic diseases, or brain abnormalities. With the exception of a cat and two dog brains, which were not perfused, and the human brain, which was perfused several hours postmortem, almost all specimens were perfused shortly after death. The majority of freshly perfused specimens combined with the similarity of results obtained in the few species processed differently suggests that postmortem interval and any infiltration of the brain by blood macrophages are not significant issues for the present study. For 5 of the 33 species (Petrodromus tetradactylus, Elephantulus myurus, Procavia capensis, Canis lupus familiaris, and Mustela putorius furo), two specimens of each were available; thus, data reported for those species correspond to the average data of both specimens each, so as to avoid duplication and bias. For the other 28 species, we examined only one specimen of each. While a single specimen per condition would generally be deemed insufficient for biological comparisons across conditions, we consider that for a study of scaling of cell numbers and densities across multiple orders of magnitude, intraspecific variation is mathematically irrelevant, while remaining an interesting issue of its own, which we have addressed previously (Herculano-Houzel et al., 2015b).
For all species, only one hemisphere of the brain was analyzed, as reported previously. Each brain was separated into different structures of interest: Cb, Cx, and what we report as the rest of the brain (RoB), constituted by the remaining hindbrain, midbrain, diencephalon, striatum, and hypothalamus (Dos Santos et al., 2017; Watson et al., 2017). Only cerebral cortical structures were available from primates. For all species, the Cx was sectioned into coronal sections (1 mm thick for Didelphis aurita; 12.8 mm for Loxodonta africana; 2 mm for all other species), and gray and white matter dissected from each coronal section were processed separately for all species but the four smallest marsupials and afrotherians (Metachirus nudicaudatus, Marmosops incanus, Petrodomus tetradactylus, and Elephantulus myurus). The hippocampus was separated from the rest of the cortical gray matter and counted separately. Where a single Cx value is reported in a species, it corresponds to the ensemble of cortical gray matter, white matter, and hippocampus; whenever values apply exclusively to gray or white matter, that is so indicated. “Cortical sites” refer either to the various slabs taken from coronal sections in the original studies (L. africana, C. familiaris, F. catus, P. leo, U. arctos, D. goodfellowi, D. aurita, M. fuliginosus, M. parma, M. rufogriseus, S. harrisii, W. bicolor, and all primate cortices) (Ribeiro et al., 2013; Herculano-Houzel et al., 2014; Gabi et al., 2016; Dos Santos et al., 2017; Jardim-Messeder et al., 2017) or to each of the whole coronal sections, as in Neves et al. (2014) and Kazu et al. (2014).
Immunostaining
All samples were isotropic suspensions of free isolated nuclei that had been stored in an antifreeze solution (30% glycerol, 30% ethylene glycol, 30% distilled water and 10% 0.244 M phosphate buffer 2xPO4) (Manger et al., 2009) at −20°C (Ribeiro et al., 2013; Herculano-Houzel et al., 2014; Kazu et al., 2014; Neves et al., 2014; Gabi et al., 2016; Dos Santos et al., 2017; Jardim-Messeder et al., 2017). The proportion of microglial cell nuclei in each sample was determined by immunocytochemical detection of the antigen Iba1, specifically expressed in both the nucleus and cytoplasm of active and resting microglia (Imai et al., 1996; Imai and Kohsaka, 2002), an antibody commonly used to label microglia both in vivo and in vitro (Sousa et al., 2017). We first incubated the samples of free nuclei in 0.2 m boric acid, pH 9.0, for 2 h at 70°C for antigen retrieval. To maximize the chances of compatibility across species, we used a rabbit polyclonal primary antibody against Iba1 (Wako, 019-19 741) diluted 1:100 in a buffer made of 0.1 M PBS, pH 7.4, 3% NGS and 2% BSA, incubated for 2 d at 4°C. All samples were further incubated with AlexaFluor-488-conjugated goat anti-rabbit secondary antibody (Abcam, ab150077) diluted 1:500 in a similar buffer as above and incubated for 2 d at 4°C, and finally labeled in 0.1 M PBS, pH 7.4, with the fluorescent DNA marker DAPI (Invitrogen) at 2 mg/L for 30 min at room temperature. The fraction of DAPI+ cell nuclei that were Iba1+ was determined by unbiased systematic counting of contiguous squares in a modified Neubauer chamber, under a Carl Zeiss AxioImager A2 fluorescent microscope with a 40× objective (Carl Zeiss EC-Plan-NEOFLUAR 40×/0,75 420360-9900), until at least 500 DAPI+ nuclei had been scored. Depending on the percentage of Iba1+ nuclei, typically the latter were typically 10-30 of all nuclei scored for each brain structure analyzed.
To confirm that Iba1 is conserved enough that the same polyclonal antibody labels nuclei with anatomic characteristics of microglia in all the mammalian species analyzed, we conducted immunohistochemistry on 50 μm coronal tissue sections of the Cx of 2 species of each clade (primates, Northern greater galago and owl monkey; artiodactyls, springbok and pig; marsupials, big-eared opossum and Bennett's wallaby; carnivorans, ferret and cat; afrotherians, African elephant and elephant shrew; Fig. 1). We incubated the tissue sections in 0.2 m boric acid, pH 9.0, for 2 h at 70°C for antigen retrieval and applied to them the same 1:100 dilution of anti-Iba1 antibody used on the respective free nuclei in the presence of goat and bovine sera (5% NGS; 3% BSA in 0.1 M PB/0.25% Triton X-100) for 2 d at 4°C followed by incubation with AlexaFluor-488-conjugated goat anti-rabbit secondary antibody (Abcam, ab150077) diluted at 1:500 in an incubation buffer made of 0.1 M PB, pH 7.4, 5% NGS, and 3% BSA for 2 d at 4°C and ultimately in 0.1 M PB, pH 7.4, with DAPI at 2 mg/L for 1 h at room temperature. As shown in Figure 1, Iba1+ cells have the typical shape of microglial cells with extensive thin processes, and their distribution seems homogeneous across different cortical and brain regions, as has been previously reported for the mouse Cx, where minimal distances across neighboring microglial cells were fairly constant across cortical layers (Nimmerjahn et al., 2005). Two-dimensional photomontages were acquired on a Carl Zeiss AxioImager M2 fluorescent microscope under 5×, 20×, 40×, or 63× objective magnification using StereoInvestigator software (MicroBrightField). Colabeling with antibodies against Iba1 and other glial and neuronal markers showed no overlap in mouse brain sections (Herculano-Houzel and Dos Santos, 2018), confirming the specificity of this marker for microglial cells only, in agreement with a recent report (Valério-Gomes et al., 2018). Heretofore, Iba1+ cells and cell nuclei will be referred to as microglial cells (or I, for Iba1+ cells or nuclei).
Figure 1.
Immunostaining of the microglial Iba1 marker on mammalian cortical tissue sections. Coronal 50-μm-thick sections of the Cx of two marsupial species (D. aurita in A and M. rufogriseus in B), two primate species (O. garnettii in C and A. trivirgatus in D), two afrotherian species (L. africana in E and E. myurus in F), two carnivoran species (M. putorius furo in G and F. catus in H), and two artiodactyl species (A. marsupialis in I and S. scrofa domesticus in J) are shown at 63× magnification with all cell nuclei stained with DAPI (blue) and microglial cells stained for Iba1 (green). Scale bar, 50 μm.
Data analysis
All statistical analyses were performed in JMP Pro 14.0 (SAS Institute). Regressions to power functions were performed to find the best fit for each distribution by applying linear fits to automatically log-transformed data. All averages and exponents are reported ± SE and with the corresponding p value; 95% CI are not reported explicitly as they can be easily calculated as average or exponent ± 2 SE. To evaluate the statistical dependence between two variables, we do not assume normality in the data and use a nonparametric Spearman's rank correlation coefficient instead. We consider a cutoff p value of p < 0.01 throughout the manuscript to estimate significance of our statistical analyses. Because we focus directly on the question of whether microglial cell densities are universal across species, we chose to analyze the raw data directly without introducing any inferences about phylogenetic relationships about the species analyzed. All data are presented in the tables for future use in other studies where accounting for phylogenetic relationships may be of interest.
Data reported in Table 1 represent the sums of masses, total microglial and neuronal cells, and their densities in each structure analyzed in the present study. Total numbers of microglial cells in each structure were estimated by applying the percentages of Iba1+ nuclei in the samples to their respective recorded original total number of cell nuclei in the sample. Microglial cell density is the estimated total number of microglial cells obtained this way divided by the original sample mass, in cells/mg of tissue. Where a brain structure had been subdivided into multiple samples, we first calculated the absolute number of Iba1+ cells in each sample, then the total tally for the whole structure, and only then calculated the average microglial cell density for the whole structure as above. That is, the averages reported in Table 1 represent the real average when sampling the entire tissue, not an average of averages in multiple samples.
Table 1.
Numbers and densities of total, neuronal, and microglial cellsa
| Species name | Structure | Structure mass (g) | C | N | I | N/mg | I/mg | I/N | No. of samples |
|---|---|---|---|---|---|---|---|---|---|
| Dendrohyrax dorsalis | Cx | 7.556 | 282 500 000 | 98 960 000 | 13 842 895 | 13 098 | 1832 | 0.140 | 3 |
| Dendrohyrax dorsalis | Cb | 1.918 | 438 500 000 | 360 929 350 | 2367 900 | 188 180 | 1235 | 0.007 | 1 |
| Dendrohyrax dorsalis | RoB | 3.328 | 197 138 000 | 44 671 471 | 10 982 209 | 13 423 | 3299 | 0.246 | 3 |
| Dendrohyrax dorsalis | Ctx | 6.830 | 257 310 000 | 90 644 478 | 12 325 149 | 13 272 | 1805 | 0.136 | 2 |
| Dendrohyrax dorsalis | Hp | 0.726 | 25 190 000 | 8312 700 | 1516 438 | 11 450 | 2089 | 0.182 | 1 |
| Dendrohyrax dorsalis | Br | 12.802 | 918 138 000 | 504 551 401 | 27 176 885 | 39 412 | 10 129 | 0.052 | 7 |
| Elephantulus myurus | Cx* | 0.450 | 46 970 000 | 21 847 255 | 1052 128 | 48 549 | 2338 | 0.048 | 3 |
| Elephantulus myurus | Cb | 0.168 | 112 681 000 | 89 312 372 | 822 571 | 531 494 | 4896 | 0.009 | 2 |
| Elephantulus myurus | RoB | 0.401 | 43 010 000 | 14 012 533 | 1324 708 | 34 520 | 3295 | 0.095 | 2 |
| Elephantulus myurus | Ctx | 0.356 | 34 554 000 | 16 422 814 | 580 507 | 46 132 | 1631 | 0.035 | 2 |
| Elephantulus myurus | Hp* | 0.086 | 12 080 000 | 5159 368 | 353 944 | 59 993 | 4116 | 0.069 | 1 |
| Elephantulus myurus | Br* | 0.958 | 195 628 000 | 124 766 317 | 2699 666 | 130 236 | 2818 | 0.022 | 7 |
| Loxodonta africana | Cx | 2847.954 | 160 509 115 478 | 5593 241 033 | 11 667 281 742 | 1964 | 4192 | 2.086 | 264 |
| Loxodonta africana | Cb | 1171.63 | 287 226 757 500 | 250 708 527 536 | 7025 615 044 | 213 983 | 5982 | 0.028 | 80 |
| Loxodonta africana | RoB | 564.674 | 28 148 254 366 | 741 704 844 | 1947 975 176 | 1314 | 3450 | 2.626 | 44 |
| Loxodonta africana | Ctx | 2823.530 | 157 990 215 478 | 5556 610 521 | 11 462 555 196 | 1968 | 4149 | 2.063 | 263 |
| Loxodonta africana | Hp | 24.426 | 1763 900 000 | 36 630 512 | 199 354 517 | 1500 | 8175 | 5.442 | 1 |
| Loxodonta africana | Br | 4168.620 | 477 844 127 344 | 257 043 473 412 | 20 919 937 679 | 56 071 | 4562 | 0.081 | 388 |
| Petrodromus tetradactylus | Cx | 1.239 | 74 433 000 | 33 947 000 | 3908 787 | 27 236 | 3019 | 0.115 | 4 |
| Petrodromus tetradactylus | Cb | 0.304 | 145 311 000 | 110 653 150 | 1278 737 | 362 537 | 4206 | 0.012 | 2 |
| Petrodromus tetradactylus | RoB | 0.894 | 40 436 000 | 12 231 811 | 1217 665 | 13 696 | 1845 | 0.100 | 2 |
| Petrodromus tetradactylus | Ctx | 0.968 | 60 480 000 | 27 651 699 | 2921 184 | 28 566 | 3018 | 0.106 | 2 |
| Petrodromus tetradactylus | Hp | 0.272 | 13 953 000 | 6294 000 | 1067 062 | 23 164 | 3032 | 0.170 | 2 |
| Petrodromus tetradactylus | Br | 2.440 | 260 180 000 | 156 830 795 | 6564 788 | 64 104 | 2743 | 0.042 | 8 |
| Procavia capensis | Cx | 10.478 | 564 554 000 | 197 933 000 | 38 108 306 | 19 134 | 3637 | 0.193 | 6 |
| Procavia capensis | Cb | 2.058 | 579 378 000 | 488 3703 000 | 7806 491 | 242 415 | 3801 | 0.016 | 2 |
| Procavia capensis | RoB | 4.317 | 232 343 000 | 69 358 240 | 11 199 666 | 16 436 | 2594 | 0.161 | 4 |
| Procavia capensis | Ctx | 9.184 | 488 188 000 | 171 217 186 | 33 099 146 | 18 643 | 3604 | 0.193 | 4 |
| Procavia capensis | Hp | 1.294 | 76 311 438 | 26 712 953 | 4994 695 | 21 712 | 3860 | 0.187 | 2 |
| Procavia capensis | Br | 16.853 | 1376 275 000 | 755 653 256 | 57 043 095 | 45 576 | 3389 | 0.075 | 12 |
| Antidorcas marsupialis | Cx | 68.806 | 4443 685 152 | 396 896 159 | 268 018 858 | 5768 | 3791 | 0.675 | 29 |
| Antidorcas marsupialis | Cb | 11.458 | 2724 458 750 | 2257 214 074 | 58 539 299 | 196 999 | 5147 | 0.026 | 2 |
| Antidorcas marsupialis | RoB | 25.810 | 1389 765 624 | 70 485 000 | 84 309 976 | 2731 | 3267 | 1.196 | 3 |
| Antidorcas marsupialis | Ctx | 65.372 | 4216 083 590 | 376 412 018 | 254 974 497 | 5758 | 3790 | 0.677 | 28 |
| Antidorcas marsupialis | Hp | 3.434 | 227 601 562 | 20 484 141 | 12 973 289 | 5965 | 3778 | 0.633 | 1 |
| Antidorcas marsupialis | Br | 106.074 | 8557 909 526 | 2724 595 233 | 402 984 165 | 25 686 | 3810 | 0.148 | 34 |
| Giraffa camelopardalis | Cx | 398.808 | 29 244 220 002 | 1730 513 460 | 2430 910 509 | 4339 | 6330 | 1.405 | 45 |
| Giraffa camelopardalis | Cb | 67.730 | 14 398 437 500 | 8878 076 563 | 298 047 656 | 131 080 | 4401 | 0.034 | 1 |
| Giraffa camelopardalis | RoB | 70.680 | 5021 562 500 | 142 697 625 | 495 014 837 | 2019 | 7005 | 3.469 | 5 |
| Giraffa camelopardalis | Ctx | 391.322 | 28 341 393 752 | 1671 925 960 | 2365 907 262 | 4272 | 6278 | 1.415 | 44 |
| Giraffa camelopardalis | Hp | 7.486 | 681 250 000 | 58 587 500 | 64 786 875 | 8435 | 9327 | 1.106 | 1 |
| Giraffa camelopardalis | Br | 537.218 | 48 442 643 752 | 10 751 287 650 | 3335 932 264 | 19 678 | 6278 | 0.310 | 51 |
| Sus scrofa domesticus | Cx | 42.202 | 3476 643 442 | 307 082 404 | 388 217 638 | 7276 | 9048 | 1.264 | 24 |
| Sus scrofa domesticus | Cb | 8.128 | 2207 031 250 | 1858 320 313 | 85 632 813 | 228 632 | 10 536 | 0.046 | 1 |
| Sus scrofa domesticus | RoB | 13.850 | 1155 531 250 | 58 709 836 | 129 924 626 | 4238 | 9386 | 2.213 | 3 |
| Sus scrofa domesticus | Ctx | 40.274 | 3300 354 380 | 289 142 456 | 359 715 940 | 7179 | 8932 | 1.244 | 23 |
| Sus scrofa domesticus | Hp | 1.928 | 146 289 062 | 12 907 084 | 22 141 064 | 6692 | 11 480 | 1.715 | 1 |
| Sus scrofa domesticus | Br | 64.180 | 6839 205 942 | 2224 112 553 | 598 831 830 | 34 654 | 9310 | 0.269 | 28 |
| Tragelaphus strepsiceros | Cx | 213.370 | 13 064 871 626 | 762 567 178 | 837 474 821 | 3574 | 3718 | 1.098 | 37 |
| Tragelaphus strepsiceros | Cb | 31.776 | 5308 593 750 | 4042 494 141 | 96 616 406 | 127 218 | 3041 | 0.024 | 1 |
| Tragelaphus strepsiceros | RoB | 61.716 | 3515 369 752 | 106 590 230 | 284 487 989 | 1727 | 4485 | 2.669 | 5 |
| Tragelaphus strepsiceros | Ctx | 202.434 | 12 392 809 376 | 726 411 196 | 799 590 074 | 3588 | 3772 | 1.101 | 36 |
| Tragelaphus strepsiceros | Hp | 10.936 | 535 156 250 | 28 363 281 | 29 326 563 | 2594 | 4710 | 3.946 | 1 |
| Tragelaphus strepsiceros | Br | 306.860 | 21 751 929 | 4911 651 549 | 1204 929 478 | 16 006 | 3800 | 0.245 | 43 |
| Canis familiaris | Cx | 65.491 | 3944 367 750 | 527 911 882 | 313 577 236 | 8061 | 4788 | 0.594 | 86 |
| Canis familiaris | Cb | 7.512 | 2525 312 500 | 1676 633 212 | 48 233 469 | 223 194 | 6421 | 0.029 | 2 |
| Canis familiaris | RoB | 13.459 | 976 243 750 | 48 149 310 | 103 384 213 | 3577 | 7681 | 2.147 | 6 |
| Canis familiaris | Ctx | 63.481 | 3801 711 500 | 519 305 298 | 302 616 235 | 8180 | 4767 | 0.583 | 84 |
| Canis familiaris | Hp | 2.010 | 142 656 250 | 8606 584 | 11 141 453 | 4282 | 5543 | 1.295 | 2 |
| Canis familiaris | Br | 86.462 | 7445 924 000 | 2252 694 404 | 465 370 250 | 26 054 | 5382 | 0.207 | 94 |
| Felis catus | Cx | 24.176 | 1346 938 750 | 249 834 196 | 73 677 550 | 10 334 | 3048 | 0.295 | 44 |
| Felis catus | Cb | 5.110 | 1269 000 000 | 942 902 390 | 21 826 800 | 184 521 | 4271 | 0.023 | 1 |
| Felis catus | RoB | 5.572 | 328 062 500 | 22 472 558 | 10 202 744 | 4033 | 1831 | 0.454 | 3 |
| Felis catus | Ctx | 23.300 | 1282 938 750 | 241 786 292 | 71 716 276 | 10 377 | 3078 | 0.297 | 43 |
| Felis catus | Hp | 0.876 | 64 000 000 | 8047 904 | 1952 000 | 9187 | 2228 | 0.243 | 1 |
| Felis catus | Br | 34.858 | 2944 001 250 | 1215 209 144 | 105 689 645 | 34 862 | 3032 | 0.087 | 48 |
| Mungos mungo | Cx** | NA | NA | NA | NA | NA | NA | NA | 20 |
| Mungos mungo | Cb | 1.548 | 470 000 000 | 315 738 964 | 8695 000 | 203 966 | 5617 | 0.028 | 1 |
| Mungos mungo | RoB | 3.454 | 174 060 000 | 22 514 320 | 6614 280 | 6518 | 1.915 | 0.294 | 3 |
| Mungos mungo | Ctx** | NA | NA | NA | NA | NA | NA | NA | 19 |
| Mungos mungo | Hp | 0.428 | 25 625 000 | 2664 224 | 863 563 | 6225 | 2018 | 0.324 | 1 |
| Mungos mungo | Br | NA | NA | NA | NA | NA | NA | NA | 24 |
| Mustela putorius furo | Cx | 3.123 | 303 008 200 | 38 952 070 | 20 392 452 | 12 473 | 6530 | 0.524 | 24 |
| Mustela putorius furo | Cb | 0.920 | 430 860 000 | 351 264 942 | 8832 630 | 381 810 | 9601 | 0.025 | 2 |
| Mustela putorius furo | RoB | 1.386 | 143 542 500 | 14 210 998 | 12 847 054 | 10 253 | 9269 | 0.904 | 4 |
| Mustela putorius furo | Ctx | 2.794 | 270 508 200 | 35 760 238 | 17 528 931 | 12 799 | 6274 | 0.490 | 22 |
| Mustela putorius furo | Hp | 0.329 | 32 500 000 | 3191 834 | 2856 750 | 9702 | 8683 | 0.895 | 2 |
| Mustela putorius furo | Br | 5.429 | 877 410 700 | 404 428 012 | 42 115 714 | 74 494 | 7758 | 0.104 | 30 |
| Panthera leo | Cx | 139.902 | 7806 210 250 | 545 238 686 | 795 452 825 | 3897 | 5686 | 1.459 | 107 |
| Panthera leo | Cb | 24.882 | 7093 750 000 | 4049 750 938 | 124 140 625 | 162 758 | 4989 | 0.031 | 1 |
| Panthera leo | RoB | 34.162 | 2165 910 000 | 72 045 082 | 210 743 043 | 2109 | 6169 | 2.925 | 2 |
| Panthera leo | Ctx | 136.254 | 7615 560 250 | 531 700 630 | 784 402 706 | 3902 | 5757 | 1.475 | 106 |
| Panthera leo | Hp | 3.648 | 190 650 000 | 13 538 056 | 10 886 115 | 3711 | 2984 | 0.804 | 1 |
| Panthera leo | Br | 198.946 | 17 065 870 250 | 4667 034 704 | 1129 760 611 | 23 459 | 5679 | 0.242 | 110 |
| Ursus arctos | Cx | 222.000 | 10 404 967 500 | 250 969 270 | 843 842 866 | 1130 | 3801 | 3.362 | 116 |
| Ursus arctos | Cb | 45.106 | 11 550 000 000 | 9253 250 478 | 188 265 000 | 205 145 | 4174 | 0.020 | 1 |
| Ursus arctos | RoB | 47.898 | 2743 825 000 | 81 384 332 | 98 777 700 | 1699 | 2062 | 1.214 | 4 |
| Ursus arctos | Ctx | 217.870 | 10 147 192 500 | 242 359 888 | 835 113 943 | 1112 | 3833 | 3.446 | 115 |
| Ursus arctos | Hp | 4.130 | 257 775 000 | 8609 382 | 8815 905 | 2085 | 2135 | 1.024 | 1 |
| Ursus arctos | Br | 315.004 | 24 698 792 500 | 9585 604 080 | 1131 204 697 | 30 430 | 3591 | 0.118 | 121 |
| Dendrolagus goodfellowi | Cx | 19.570 | 1532 188 750 | 273 434 984 | 80 439 909 | 13 972 | 4110 | 0.294 | 46 |
| Dendrolagus goodfellowi | Cb | 4.236 | 1545 500 000 | 1237 018 200 | 10 200 300 | 292 025 | 2408 | 0.008 | 1 |
| Dendrolagus goodfellowi | RoB | 7.742 | 518 100 000 | 37 858 500 | 29 687 130 | 4890 | 3835 | 0.784 | 2 |
| Dendrolagus goodfellowi | Ctx | 18.820 | 1431 388 750 | 255 280 904 | 74 289 076 | 13 965 | 4064 | 0.291 | 45 |
| Dendrolagus goodfellowi | Hp | 1.290 | 100 800 000 | 18 154 080 | 6189 120 | 14 073 | 4798 | 0.341 | 1 |
| Dendrolagus goodfellowi | Br | 31.548 | 3595 788 750 | 1548 311 684 | 120 458 923 | 49 078 | 3818 | 0.078 | 49 |
| Didelphis aurita | Cx | 2.136 | 163 477 500 | 42 499 454 | 11 581 340 | 19 897 | 5511 | 0.273 | 31 |
| Didelphis aurita | Cb | 0.778 | 375 031 250 | 334 265 353 | 5362 947 | 429 647 | 6893 | 0.016 | 1 |
| Didelphis aurita | RoB | 1.378 | 108 137 500 | 15 732 651 | 8326 588 | 11 417 | 6043 | 0.529 | 3 |
| Didelphis aurita | Ctx | 1.750 | 133 775 000 | 35 269 454 | 9740 554 | 20 154 | 5679 | 0.276 | 30 |
| Didelphis aurita | Hp | 0.386 | 30 000 000 | 7230 000 | 1848 000 | 18 731 | 4788 | 0.256 | 1 |
| Didelphis aurita | Br | 4.292 | 643 138 750 | 392 497 458 | 25 297 939 | 91 449 | 5961 | 0.064 | 35 |
| Macropus fuliginosus | Cx | 40.194 | 2740 637 500 | 370 171 727 | 161 697 613 | 9210 | 4023 | 0.437 | 54 |
| Macropus fuliginosus | Cb | 7.358 | 2721 875 000 | 1989 962 813 | 22 863 750 | 270 449 | 3107 | 0.011 | 1 |
| Macropus fuliginosus | RoB | 15.172 | 1245 625 000 | 97 741 950 | 80 965 625 | 6442 | 5337 | 0.828 | 2 |
| Macropus fuliginosus | Ctx | 37.256 | 2513 387 500 | 325 403 477 | 145 776 475 | 8734 | 3913 | 0.448 | 53 |
| Macropus fuliginosus | Hp | 2.938 | 227 250 000 | 44 768 250 | 15 975 675 | 15 238 | 5438 | 0.357 | 1 |
| Macropus fuliginosus | Br | 62.724 | 6708 137 500 | 2457 876 490 | 265 642 245 | 39 186 | 4235 | 0.108 | 57 |
| Macropus parma | Cx | 9.914 | 869 532 500 | 135 625 473 | 48 259 054 | 13 680 | 4868 | 0.356 | 35 |
| Macropus parma | Cb | 1.690 | 721 687 500 | 558 080 944 | 5268 319 | 330 225 | 3117 | 0.009 | 1 |
| Macropus parma | RoB | 4.110 | 352 250 000 | 17 420 050 | 20 254 375 | 4238 | 4928 | 1.163 | 2 |
| Macropus parma | Ctx | 8.676 | 750 782 500 | 127 146 723 | 40 467 177 | 14 655 | 4664 | 0.318 | 34 |
| Macropus parma | Hp | 1.238 | 118 750 000 | 8478 750 | 7801 875 | 6849 | 6302 | 0.920 | 1 |
| Macropus parma | Br | 15.714 | 1943 470 000 | 711 126 467 | 73 851 860 | 45 254 | 4700 | 0.104 | 38 |
| Macropus rufogriseus | Cx | 16.102 | 1423 953 750 | 256 784 370 | 98 537 600 | 15 947 | 6120 | 0.384 | 41 |
| Macropus rufogriseus | Cb | 3.572 | 1479 625 000 | 1022 568 838 | 13 612 550 | 286 273 | 3811 | 0.013 | 1 |
| Macropus rufogriseus | RoB | 7.320 | 571 812 500 | 42 476 569 | 36 538 819 | 5803 | 4992 | 0.860 | 2 |
| Macropus rufogriseus | Ctx | 14.306 | 1259 953 750 | 246 009 570 | 84 038 915 | 17 196 | 5874 | 0.342 | 40 |
| Macropus rufogriseus | Hp | 1.796 | 164 000 000 | 10 774 069 | 14 514 000 | 5999 | 8081 | 1.347 | 1 |
| Macropus rufogriseus | Br | 26.994 | 3475 391 250 | 1321 829 777 | 148 746 746 | 48 968 | 5510 | 0.113 | 44 |
| Macropus rufus | Cb | 4.786 | 1976 624 000 | 1532 871 912 | 31 230 659 | 320 282 | 6525 | 0.020 | 1 |
| Macropus rufus | P + M | 2.886 | 227 500 000 | 19 337 500 | 17 881 500 | 6700 | 6196 | 0.925 | 1 |
| Marmosops incanus | Cx | 0.408 | 57 975 000 | 20 413 493 | 2811 788 | 50 033 | 6892 | 0.138 | 2 |
| Marmosops incanus | Cb | 0.158 | 106 500 000 | 82 399 050 | 788 100 | 521 513 | 4988 | 0.010 | 1 |
| Marmosops incanus | RoB | 0.344 | 35 825 000 | 9238 790 | 2332 208 | 26 857 | 6780 | 0.252 | 3 |
| Marmosops incanus | Ctx | 0.348 | 51 700 000 | 17 831 330 | 2553 980 | 51 239 | 7339 | 0.143 | 1 |
| Marmosops incanus | Hp | 0.060 | 6275 000 | 2582 163 | 258 530 | 43 036 | 4309 | 0.100 | 1 |
| Marmosops incanus | Br | 0.910 | 200 300 000 | 112 051 333 | 5928 880 | 123 133 | 6515 | 0.053 | 6 |
| Metachirus nudicaudatus | Cx | 0.992 | 84 775 000 | 20 191 038 | 2653 458 | 20 354 | 2675 | 0.131 | 2 |
| Metachirus nudicaudatus | Cb | 0.342 | 166 312 500 | 134 131 031 | 1729 650 | 392 196 | 5057 | 0.013 | 1 |
| Metachirus nudicaudatus | RoB | 0.674 | 66 256 250 | 11 272 337 | 3670 596 | 16 725 | 5446 | 0.326 | 3 |
| Metachirus nudicaudatus | Ctx | 0.848 | 72 625 000 | 15 999 288 | 2222 325 | 18 867 | 2621 | 0.139 | 1 |
| Metachirus nudicaudatus | Hp | 0.144 | 12 150 000 | 4191 750 | 428 895 | 29 109 | 2978 | 0.102 | 1 |
| Metachirus nudicaudatus | Br | 2.008 | 317 343 750 | 166 594 406 | 8060 531 | 82 467 | 4014 | 0.049 | 6 |
| Sarcophilus harrisii | Cx | 5.228 | 437 941 000 | 71 655 547 | 9547 114 | 13 706 | 1826 | 0.133 | 26 |
| Sarcophilus harrisii | Cb | 1.434 | 735 437 500 | 601 073 069 | 7722 094 | 419 158 | 5385 | 0.013 | 1 |
| Sarcophilus harrisii | RoB | 3.140 | 276 752 500 | 20 966 121 | 5590 401 | 6677 | 1780 | 0.267 | 3 |
| Sarcophilus harrisii | Ctx | 4.594 | 367 941 000 | 59 013 547 | 7248 438 | 12 846 | 1578 | 0.123 | 25 |
| Sarcophilus harrisii | Hp | 0.634 | 70 000 000 | 12 642 000 | 2275 000 | 19 940 | 3588 | 0.180 | 1 |
| Sarcophilus harrisii | Br | 9.802 | 1450 131 000 | 693 694 737 | 22 912 070 | 70 771 | 2337 | 0.033 | 30 |
| Wallabia bicolor | Cx | 15.182 | 1239 254 750 | 208 113 314 | 95 174 765 | 13 708 | 6269 | 0.457 | 45 |
| Wallabia bicolor | Cb | 4.420 | 1528 750 000 | 1195 941 125 | 26 141 625 | 270 575 | 5914 | 0.022 | 1 |
| Wallabia bicolor | RoB | 10.410 | 687 750 000 | 31 865 550 | 54 676 125 | 3061 | 5252 | 1.716 | 2 |
| Wallabia bicolor | Ctx | 13.616 | 1115 954 750 | 195 142 154 | 88 272 021 | 14 332 | 6483 | 0.452 | 44 |
| Wallabia bicolor | Hp | 1.566 | 123 300 000 | 12 971 160 | 6892 470 | 8283 | 4401 | 0.531 | 1 |
| Wallabia bicolor | Br | 30.012 | 3455 754 750 | 1435 919 989 | 175 897 917 | 47 845 | 5861 | 0.122 | 48 |
| Aotus trivirgatus | Cx | 9.124 | 1303 931 000 | 451 671 982 | 23 875 354 | 49 504 | 2635 | 0.053 | 43 |
| Cebus apella | Cx | 53.784 | 3035 947 220 | 970 159 664 | 106 083 459 | 18 038 | 1991 | 0.109 | 78 |
| Homo sapiens sapiens | Cx | 696.708 | 58 983 401 216 | 9547 198 728 | 1634 984 719 | 13 703 | 2403 | 0.171 | 290 |
| Homo sapiens sapiens | Ctx | 693.422 | 58 732 501 216 | 9513 780 748 | 1612 228 351 | 13 720 | 2381 | 0.169 | 271 |
| Homo sapiens sapiens | Hp | 3.286 | 250 900 000 | 33 417 980 | 10 136 360 | 10 170 | 3085 | 0.303 | 19 |
| Macaca fascicularis | Cx | 40.000 | 4755 230 252 | 1404 683 000 | 192 394 153 | 35 117 | 4727 | 0.137 | 79 |
| Macaca nemestrina | Cx | 76.964 | 7787 044 390 | 2194 785 386 | 277 022 786 | 28 517 | 3668 | 0.126 | 106 |
| Otolemur garnettii | Cx | 4.876 | 494 053 510 | 196 070 964 | 39 378 618 | 40 211 | 8294 | 0.201 | 23 |
| Papio anubis cynocephalus | Cx | 94.744 | 4934 116 014 | 1144 837 998 | 257 818 766 | 12 083 | 2773 | 0.225 | 102 |
| Saguinus midas | Cx | 5.030 | 603 175 002 | 220 924 134 | 16 398 445 | 43 921 | 3041 | 0.074 | 28 |
aCx includes gray matter, white matter, and hippocampus; RoB includes pons, medulla oblongata, mesencephalon, diencephalon, and striatum; P + M, Pons and medulla oblongata; Ctx includes gray and white matter, excluding hippocampus; Hp, hippocampus; Br, whole brain (corresponding to the joint value of the respective Cx, Cb, and RoB); C, total number of cells in the structure; N, total number of neurons in the structure; M, total number of microglia in the structure; N/mg, number of neuronal cells per milligram of structure; I/mg, number of microglia per milligram of structure; I/N, microglia/neuron ratio. Structure mass, C, N, and N/mg list the values published previously for each species and for each structure in its entirety (Ribeiro et al., 2013; Herculano-Houzel et al., 2014; Kazu et al., 2014; Neves et al., 2014; Gabi et al., 2016; Dos Santos et al., 2017; Jardim-Messeder et al., 2017). I/mg and I/N list the values measured from the samples available for each species and structure (for some structures, some samples were no longer available for the present analysis). Absolute numbers of microglial cells (I) are estimated by applying the product of the I/N ratios measured in this study by the previously published number of neurons N in each structure and species (I/N × N). All values refer to the two hemispheres together, calculated as 2× the values obtained for one hemisphere, as in the original reports.
*Data available for only one specimen of the species.
**Samples were only available for some cortical sites, so total values were not calculated (NA). In both, however, microglial densities and ratios between numbers of neurons and microglial cells from the samples available are included in analyses across sites of the Cx. For M. rufus, only two structures were available: Cb and Pons and Medulla (P + M).
Results
Microglial cells are ∼6% of all cells in the mammalian Cx
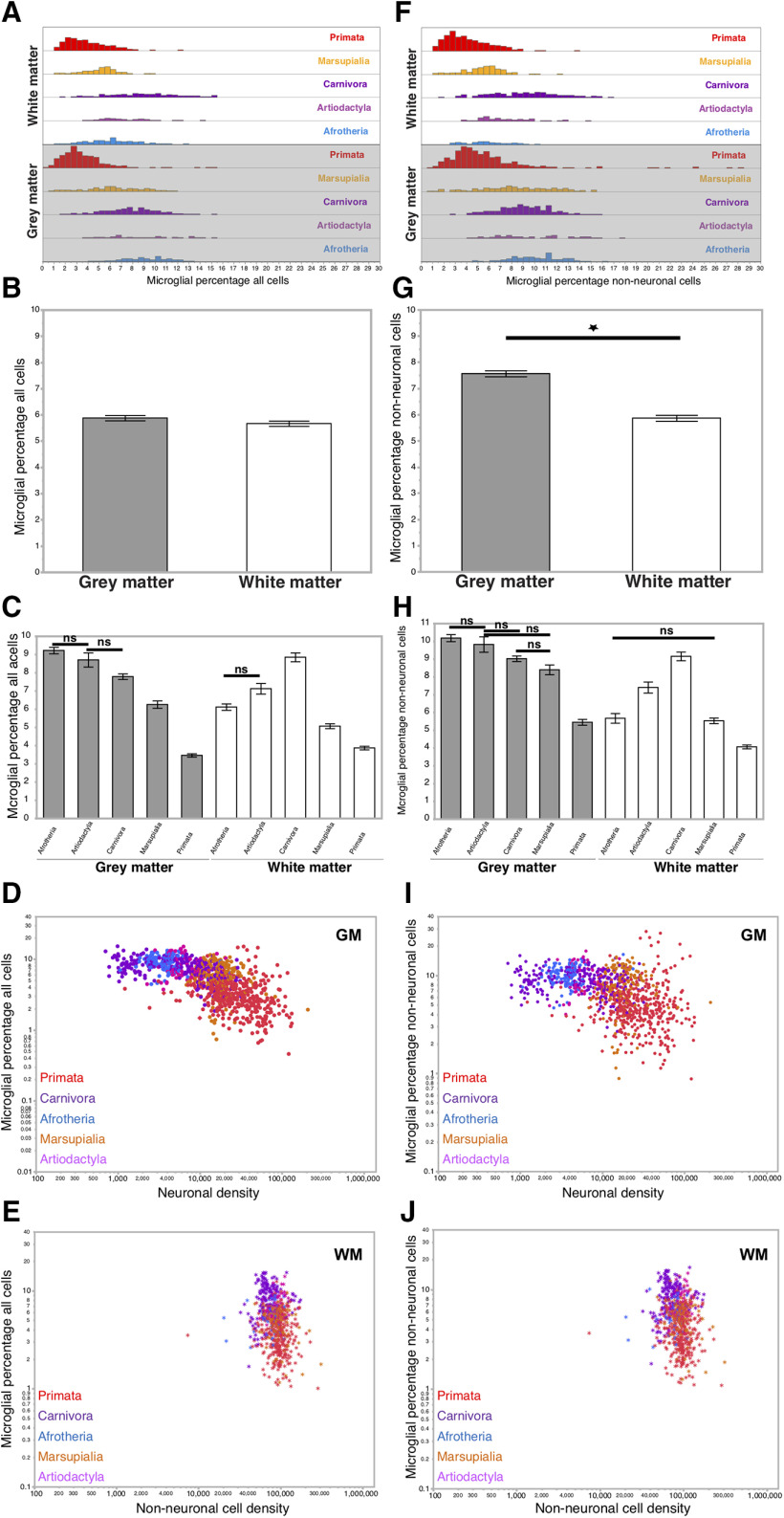
We first focus on the percentage of microglia among all cells across the 1776 cerebral cortical samples of gray and white matter analyzed across 30 species (Table 2). There is an overall 33-fold variation in the percentage of cells that are microglia across cortical sites, from 0.46% in the red-handed tamarin gray matter to 15.37% in the ferret white matter, but these percentages span overlapping ranges across the 5 mammalian clades examined (Table 2; Fig. 2A). When all species and clades are considered together, microglia represent similar, small percentages of all cortical cells in gray and white matter (5.88 ± 0.10% and 5.67 ± 0.11%, respectively; Wilcoxon p = 0.2650, Table 2; Fig. 2B). Compared across clades, the average percentage of microglia among all cortical cells is lowest in primate gray and white matter, and highest in afrotherian gray matter and carnivoran white matter (Fig. 2C; “all clades” in Table 2). Within each clade, we find significant differences in the percentage of cells that are microglial between gray and white matter (Wilcoxon p values all <0.01), but percentages were not systematically higher in either one (Fig. 2C; “per clade” in Table 2). Within an individual cortex, 12 of the 26 species with multiple cortical sites available have significantly different percentages of cells that are microglia between gray and white matter (Wilcoxon p values <0.01; “per species” in Table 2). In these cases, percentages of microglial cells were higher in the gray matter samples for all species, except carnivorans.
Table 2.
Microglia, % of all cells in different cortical sitesa
| Gray matter |
White matter |
Wilcoxon, %I in gray versus white matter | |||||
|---|---|---|---|---|---|---|---|
| Range | Mean ± SE | % versus N/mg, Spearman ρ and p value | Range | Mean ± SE | % versus O/mg, Spearman ρ and p | ||
| All clades | [0.46–15.25%] | 5.88 ± 0.10% | ρ = −0.6836 | [1.01–15.37%] | 5.67 ± 0.11% | ρ = −0.2625 | p = 0.2650 |
| p < 0.0001 | p < 0.0001 | ||||||
| Per clade | |||||||
| Afrotheria | [4.45–13.25%] | 9.22 ± 0.18% | ρ = −0.0467 | [1.08–12.50%] | 6.11 ± 0.18% | ρ = 0.1829 | p < 0.0001 |
| p = 0.6123 | p = 0.2185 | ||||||
| Artiodactyla | [3.74–15.10%] | 8.70 ± 0.39% | ρ = 0.1455 | [3.54–14.03%] | 7.12 ± 0.29% | ρ = 0.3189 | p = 0.0037 |
| p = 0.2758 | p = 0.0156 | ||||||
| Carnivora | [1.86–15.25%] | 7.79 ± 0.15% | ρ = −0.4731 | [1.69–15.37%] | 8.85 ± 0.24% | ρ = 0.2088 | p = 0.0004 |
| p < 0.0001 | p = 0.0080 | ||||||
| Marsupialia | [0.74–11.52%] | 6.25 ± 0.20% | ρ = −0.0797 | [1.16–9.56%] | 5.06 ± 0.13% | ρ = −0.1453 | p < 0.0001 |
| p = 0.3390 | p = 0.1058 | ||||||
| Primata | [0.46–15.11%] | 3.46 ± 0.09% | ρ = −0.3786 | [1.01–12.46%] | 3.87 ± 0.10% | ρ = −0.0876 | p = 0.0004 |
| p < 0.0001 | p = 0.1221 | ||||||
| Per species | |||||||
| Afrotheria* | |||||||
| L. africana | [4.45–13.25%] | 9.28 ± 0.18% | ρ = −0.0029 | [1.08–12.50%] | 6.14 ± 0.18% | ρ = 0.1916 | p < 0.0001 |
| p = 0.9752 | p = 0.2127 | ||||||
| Artiodactyla | |||||||
| G. camelopardalis | [5.70–15.10%] | 11.07 ± 0.51% | ρ = −0.2827 | [3.56–10.28%] | 7.24 ± 0.41% | ρ = −0.1000 | p < 0.0001 |
| p = 0.2272 | p = 0.6748 | ||||||
| T. strepsiceros | [4.45–8.56%] | 6.31 ± 0.30% | ρ = 0.3147 | [4.26–8.67%] | 6.26 ± 0.31% | ρ = −0.2912 | p = 0.8358 |
| p = 0.2352 | p = 0.2739 | ||||||
| S. scrofa domesticus | [8.41–14.01%] | 10.92 ± 0.59% | ρ = −0.7173 | [8.56–14.03%] | 10.56 ± 0.62% | ρ = −0.0500 | p = 0.6532 |
| p = 0.0195 | p = 0.8984 | ||||||
| A. marsupialis | [3.74–7.28%] | 6.11 ± 0.32% | ρ = 0.3328 | [3.54–6.78%] | 5.50 ± 0.26% | ρ = 0.4685 | p = 0.1058 |
| p = 0.2906 | p = 0.1245 | ||||||
| Carnivora | |||||||
| M. mongo | [1.86–3.44%] | 2.76 ± 0.20% | ρ = 0.2143 | [1.69–5.20%] | 3.58 ± 0.29% | ρ = −0.0909 | p = 0.0832 |
| p = 0.6445 | p = 0.7787 | ||||||
| U. arctos | [4.85–15.25%] | 8.59 ± 0.27% | ρ = −0.1508 | [5.06–13.88%] | 8.33 ± 0.31% | ρ = 0.0509 | p = 0.4334 |
| p = 0.2267 | p = 0.7282 | ||||||
| F. catus | [3.37–7.83%] | 5.42 ± 0.24% | ρ = 0.3102 | [3.54–8.60%] | 5.58 ± 0.34% | ρ = −0.0912 | p = 0.8113 |
| p = 0.1153 | p = 0.7370 | ||||||
| C. familiaris | [3.66–12.26%] | 7.38 ± 0.26% | ρ = −0.4702 | [5.27–13.52%] | 9.30 ± 0.30% | ρ = −0.2102 | p < 0.0001 |
| p = 0.0006 | p = 0.2403 | ||||||
| M. putorius furo | [4.78–9.80%] | 8.07 ± 0.48% | ρ = −0.1364 | [6.63–15.37%] | 8.98 ± 0.84% | ρ = −0.6000 | p = 1.0000 |
| p = 0.6893 | p = 0.0876 | ||||||
| P. leo | [4.72–12.19%] | 8.76 ± 0.21% | ρ = −0.5220 | [8.15–15.36%] | 11.90 ± 0.30% | ρ = −0.5297 | p < 0.0001 |
| p < 0.0001 | p = 0.0004 | ||||||
| Marsupialia | |||||||
| M. rufrogriseus | [1.94–10.23%] | 7.30 ± 0.40% | ρ = −0.3403 | [4.74–7.10%] | 6.04 ± 0.15% | ρ = −0.4684 | p = 0.0015 |
| p = 0.1312 | p = 0.0431 | ||||||
| D. aurita | [4.87–11.49%] | 7.93 ± 0.47% | ρ = −0.2879 | [4.04–7.36%] | 5.74 ± 0.32% | ρ = −0.6643 | p = 0.0043 |
| p = 0.2466 | p = 0.0185 | ||||||
| D. goodfellowi | [2.81–8.10%] | 5.04 ± 0.27% | ρ = −0.3313 | [2.70–7.72%] | 5.33 ± 0.28% | ρ = −0.2989 | p = 0.3568 |
| p = 0.1138 | p = 0.1881 | ||||||
| M. parma | [2.81–8.36%] | 5.68 ± 0.36% | ρ = −0.0158 | [2.94–6.39%] | 4.61 ± 0.26% | ρ = −0.2377 | p = 0.0480 |
| p = 0.9488 | p = 0.3936 | ||||||
| W. bicolor | [1.28–11.52%] | 8.24 ± 0.51% | ρ = −0.7087 | [1.78–9.56%] | 5.84 ± 0.37% | ρ = −0.1624 | p = 0.0002 |
| p = 0.0002 | p = 0.4819 | ||||||
| S. harrisii | [0.74–3.02%] | 1.76 ± 0.18% | ρ = 0.2967 | [1.16–4.14%] | 2.55 ± 0.29% | ρ = −0.0629 | p = 0.0535 |
| p = 0.3249 | p = 0.8459 | ||||||
| M. fuliginosus | [3.73–9.30%] | 6.26 ± 0.29% | ρ = 0.1845 | [2.01–5.87%] | 4.60 ± 0.18% | ρ = −0.2859 | p = 0.0001 |
| p = 0.3474 | p = 0.1660 | ||||||
| Primata | |||||||
| P. anubis cynocephalus | [1.45–7.29%] | 3.88 ± 0.21% | ρ = −0.5007 | [2.25–8.43%] | 4.87 ± 0.21% | ρ = 0.0075 | p = 0.0014 |
| p = 0.0001 | p = 0.9609 | ||||||
| M. fascicularis | [0.98–5.76%] | 3.25 ± 0.18% | ρ = −0.1461 | [1.01–9.58%] | 5.06 ± 0.34% | ρ = −0.4542 | p < 0.0001 |
| p = 0.3749 | p = 0.0070 | ||||||
| O. garnettii | [3.64–15.11%] | 9.15 ± 1.05% | ρ = −0.6703 | [4.58–12.46%] | 7.36 ± 0.84% | ρ = 0.2167 | p = 0.2703 |
| p = 0.0087 | p = 0.5755 | ||||||
| H. sapiens sapiens | [0.66–11.13%] | 3.34 ± 0.12% | ρ = −0.5234 | [1.06–8.06%] | 2.89 ± 0.11% | ρ = 0.1011 | p = 0.0041 |
| p < 0.0001 | p = 0.2865 | ||||||
| A. trivirgatus | [0.93–3.14%] | 1.82 ± 0.14% | ρ = −0.4979 | [1.41–3.01%] | 2.22 ± 0.11% | ρ = −0.2012 | p = 0.0332 |
| p = 0.0113 | p = 0.4233 | ||||||
| S. midas | [0.46–3.80%] | 2.60 ± 0.20% | ρ = −0.0518 | [1.79–5.67%] | 3.53 ± 0.32% | ρ = 0.1678 | p = 0.0303 |
| p = 0.8544 | p = 0.5837 | ||||||
| M. nemestrina | [1.44–7.07%] | 3.50 ± 0.17% | ρ = −0.3023 | [1.83–7.68%] | 4.57 ± 0.19% | ρ = 0.1054 | p < 0.0001 |
| p = 0.0235 | p = 0.4807 | ||||||
| C. apella | [1.19–6.01%] | 2.94 ± 0.19% | ρ = −0.6219 | [1.10–6.47%] | 3.62 ± 0.23% | ρ = −0.2720 | p = 0.0371 |
| p<0.0001 | p=0.1197 | ||||||
a%I, Microglial percentage of all cells; N/mg, number of neuronal cells per milligram of structure; O/mg: number of non-neuronal (other) cells per milligram of structure.
*For Afrotheria, analyses were done, including all the species of the clade (D. dorsalis, E. myurus, L. africana, P. tetradactylus, and P. capensis). Analyses within afrotherian species cortices were only possible in the elephant (L. africana), where multiple data points were available.
Figure 2.
Average percentages of PI microglial cells represent 6% of all cortical cells in gray and white matter and correlate only slightly with neuronal and non-neuronal cell densities. A, Distribution of the percentage of all cortical cells that are microglial cells in the gray and white matter per clade. The ranges of distribution and means of the percentage of all cortical cells that are microglia are similar across clades within the cortical gray matter (values in Table 2). B, Average percentages of total cells that are microglia across clades and species represent a constant ∼6% in both cortical gray and white matter. Wilcoxon p = 0.2650 between gray and white matter in all mammalian species. C, Average percentages of total cells that are microglia in gray and white matter across clades and species. In the gray matter, significant differences are found across all pairs of clades (Wilcoxon, p < 0.0001), except between Artiodactyla and both Afrotheria (Wilcoxon, p = 0.1000; nonsignificant) and Carnivora (Wilcoxon, p = 0.0963). In the white matter, significant differences are found across all pairs of clades (p < 0.01 for all pairs), except between Artiodactyla and Afrotheria (p = 0.0121). For each clade individually, we find significant differences in the percentage of all cells that are microglial between white and gray matter samples (Wilcoxon p < 0.0001 for Afrotheria and Marsupialia; p = 0.0037 for Artiodactyla; p = 0.0004 for Carnivora and Primata). D, Percentages of microglial cells among all cells in the gray matter fail to correlate systematically with the neuronal cell density in the sample. Spearman ρ and p values for each clade are given in Table 2. E, Percentages of microglial cells among all cells in the white matter fail to correlate systematically with the non-neuronal cell density in the sample. Spearman ρ and p values for each clade are given in Table 2. F, Distribution of the percentage of cortical non-neuronal cells that are microglial cells in the gray and white matter per clade. Ranges and means are listed in Table 3. G, Microglia are a larger average percentage of all non-neuronal cells in cortical gray matter (7.56 ± 0.11%) than in the white matter (5.86 ± 0.13%, Wilcoxon p < 0.0001). H, Average percentages of non-neuronal cells that are microglia in gray and white matter across clades and species. In the gray matter, significant differences are found across all pairs of clades (Wilcoxon, p < 0.01 for all clades), except between Artiodactyla and Afrotheria, Marsupialia, and Carnivora (p = 0.2645, p = 0.0240, and 0.2106, respectively) and between Marsupialia and Carnivora (p = 0.0837). In the white matter, significant differences are found across all pairs of clades (Wilcoxon, p = 0.0002 between Artiodactyla and Afrotheria; and p < 0.0001 for all other pairs), except between Afrotheria and Marsupialia (p = 0.8582). I, Percentages of microglial cells among non-neuronal cells in the gray matter do not correlate systematically with the non-neuronal cell density in the sample. Spearman correlation ρ and p values are given in Table 3. J, Percentages of microglial cells among non-neuronal cells in the white matter do not correlate systematically with the neuronal cell density in the sample. Spearman correlation ρ and p values are given in Table 3. D, E, I, J, Each data point represents the values found per cortical site examined for each species colored by clade as in the key. Filled circles represent cortical gray matter sites of the Cx, including the hippocampus. *White matter sites. G, *Significance.
We next determined whether the smaller and larger percentages of cells that are microglial in primate and afrotherian cortical gray matter, respectively, are related to the high neuronal densities found in primate versus the low neuronal densities found in the elephant cortex (Herculano-Houzel et al., 2014; Gabi et al., 2016). When all data are combined, we find a strong correlation between local neuronal density and the percentage of cells that are microglial in gray matter cortical sites across species and clades, as shown in Figure 2D (Spearman correlation for all clades together: ρ = −0.6836, p < 0.0001). This correlation indicates that microglial cells are an up to 10-fold higher percentage of all gray matter cells where neuronal densities are lower. However, across cortical sites within each clade, this correlation only applies for carnivorans and primates; and within each species, only within C. familiaris, P. leo, W. bicolor, P. anubis cynocephalus, O. garnettii, H. sapiens sapiens, and C. apella.
A slight but detectable correlation between the local non-neuronal cell density and the percentage of microglial cells among all cells in each white matter cortical site across species and clades is observed, as shown in Figure 2E (Spearman correlation for all clades together: ρ = −0.2625, p < 0.0001). However, across cortical sites within a clade, this correlation only applies for carnivorans; and within each species, only in P. leo and M. fascicularis. It thus appears more likely that any apparent correlation between local non-neuronal cell density and percentage of microglial cells in the gray or white matter of the Cx is related to clade-specific (evolutionary) differences, and not to biological scaling.
Microglial cells are a variable percentage of all cortical non-neuronal cells across cortical sites and clades
To avoid the confound of variable percentages of cortical cells that are neurons across mammalian species, depending on cortical mass and clade (Herculano-Houzel, 2017), we next analyzed the representation of microglial cells among only the non-neuronal cells in each sample. In this case, microglia are found to represent a slightly larger average percentage of non-neuronal cells in gray matter sites (7.56 ± 0.11%) than in white matter sites (5.86 ± 0.11%, Wilcoxon p < 0.0001; Fig. 2F,G; Table 3, “all clades”). This is the case in each clade separately, except in carnivorans (Table 3, “per clade”). Again, microglial cells are a higher percentage of non-neuronal cells in the gray matter in afrotherians (10.18 ± 0.20%) and lower in primates (5.44 ± 0.16%). Across white matter sites, a higher percentage of microglia is found in carnivorans (9.15 ± 0.24% of all non-neuronal cells), and the lowest percentage in primates (4.06 ± 0.11% of all non-neuronal cells; Fig. 2F,H). Within each clade, with the exception of carnivorans, microglial cells are a significantly higher percentage of non-neuronal cells in the gray matter than in the white matter (Fig. 2H; Table 3, “per clade”). Within each individual cortex, 12 of the 26 species with multiple data points available have significantly different percentages of microglia among other cells between gray and white matter (Wilcoxon p values <0.01; Table 3, “per species”) with percentages of microglial cells higher in the gray matter than in the white matter for all 12 species, except carnivorans.
Table 3.
Microglia percentage of non-neuronal cells in different cortical sitesa
| Gray matter |
White matter |
Wilcoxon, I%O in gray versus white matter | |||||
|---|---|---|---|---|---|---|---|
| Range | Mean ± SE | I% versus N/mg, Spearman r, and p value | Range | Mean ± SE | % versus O/mg, Spearman r, and p value | ||
| All clades | [0.88–28.15%] | 7.56 ± 0.11% | ρ = −0.4805 | [1.09–16.73%] | 5.86 ± 0.11% | ρ = −0.2613 | p < 0.0001 |
| p < 0.0001 | p < 0.0001 | ||||||
| Per clade | |||||||
| Afrotheria | [4.67–16.39%] | 10.18 ± 0.20% | ρ = 0.1082 | [2.64–10.12%] | 5.67 ± 0.27% | ρ = 0.1285 | p < 0.0001 |
| p = 0.2393 | p = 0.3895 | ||||||
| Artiodactyla | [4.02–17.76%] | 9.82 ± 0.43% | ρ = 0.2185 | [3.60–14.64%] | 7.40 ± 0.31% | ρ = 0.2997 | p < 0.0001 |
| p = 0.0994 | p = 0.0235 | ||||||
| Carnivora | [2.69–15.95%] | 9.02 ± 0.16% | ρ = −0.1793 | [1.80–16.73%] | 9.15 ± 0.24% | ρ = 0.2002 | p = 0.6766 |
| p = 0.0069 | p = 0.0112 | ||||||
| Marsupialia | [0.89–15.45%] | 8.40 ± 0.27% | ρ = 0.0954 | [1.26–12.18%] | 5.53 ± 0.16% | ρ = −0.1915 | p < 0.0001 |
| p = 0.2521 | p = 0.0324 | ||||||
| Primata | [0.88–28.15%] | 5.44 ± 0.16% | ρ = −0.1184 | [1.09–13.65%] | 4.06 ± 0.11% | ρ = −0.0805 | p < 0.0001 |
| p = 0.0148 | p = 0.1554 | ||||||
| Per species | |||||||
| Afrotheria* | |||||||
| L. africana | [4.67–14.73%] | 10.12 ± 0.20% | ρ = 0.0834 | [2.64–9.13%] | 5.60 ± 0.27% | ρ = 0.1787 | p < 0.0001 |
| p = 0.3710 | p = 0.2458 | ||||||
| Artiodactyla | |||||||
| G. camelopardalis | [6.81–17.76%] | 12.47 ± 0.57% | ρ = −0.1083 | [3.60–10.64%] | 7.43 ± 0.42% | ρ = −0.1293 | p < 0.0001 |
| p = 0.6496 | p = 0.5869 | ||||||
| T. strepsiceros | [4.91–9.79%] | 7.06 ± 0.34% | ρ = 0.3147 | [4.45–8.85%] | 6.41 ± 0.30% | ρ = −0.3294 | p = 0.1468 |
| p = 0.2352 | p = 0.2128 | ||||||
| S. scrofa domesticus | [9.18–15.19%] | 12.25 ± 0.60% | ρ = −0.6121 | [9.22–14.64%] | 11.25 ± 0.63% | ρ = 0.0333 | p = 0.3913 |
| p = 0.0600 | p = 0.9322 | ||||||
| A. marsupialis | [4.02–8.56%] | 7.06 ± 0.41% | ρ = 0.4895 | [3.82–6.96%] | 5.79 ± 0.24% | ρ = 0.3636 | p = 0.0166 |
| p = 0.1063 | p = 0.2453 | ||||||
| Carnivora | |||||||
| M. mongo | [2.69–5.62%] | 4.18 ± 0.41% | ρ = 0.5357 | [1.80–5.60%] | 3.93 ± 0.31% | ρ = −0.1748 | p = 0.6420 |
| p = 0.2152 | p = 0.5868 | ||||||
| U. arctos | [4.98–15.95%] | 9.02 ± 0.28% | ρ = −0.0980 | [5.13–13.93%] | 8.40 ± 0.31% | ρ = 0.0486 | p = 0.1126 |
| p = 0.4335 | p = 0.7403 | ||||||
| F. catus | [4.52–10.35%] | 7.44 ± 0.35% | ρ = 0.5275 | [3.64–9.68%] | 6.10 ± 0.40% | ρ = −0.1265 | p = 0.0317 |
| p = 0.0047 | p = 0.6407 | ||||||
| C. familiaris | [4.77–15.05%] | 9.24 ± 0.31% | ρ = −0.2504 | [5.51–14.23%] | 9.87 ± 0.32% | ρ = −0.2209 | p = 0.1064 |
| p = 0.0794 | p = 0.2166 | ||||||
| M. putorius furo | [5.31–13.20%] | 9.70 ± 0.64% | ρ = 0.3909 | [6.76–16.73%] | 9.42 ± 0.96% | ρ = −0.6667 | p = 0.0946 |
| p = 0.2345 | p = 0.0499 | ||||||
| P. leo | [5.88–13.74%] | 9.91 ± 0.22% | ρ = −0.3891 | [8.40–15.54%] | 12.13 ± 0.30% | ρ = −0.5305 | p < 0.0001 |
| p = 0.0014 | p = 0.0004 | ||||||
| Marsupialia | |||||||
| M. rufrogriseus | [5.31–13.38%] | 10.08 ± 0.45% | ρ = −0.0870 | [4.92–12.18%] | 6.72 ± 0.35% | ρ = −0.3404 | p < 0.0001 |
| p = 0.7076 | p = 0.1539 | ||||||
| D. aurita | [6.68–15.45%] | 11.24 ± 0.63% | ρ = −0.1909 | [4.63–8.45%] | 6.57 ± 0.37% | ρ = −0.7413 | p < 0.0001 |
| p = 0.4479 | p = 0.0058 | ||||||
| D. goodfellowi | [3.63–10.43%] | 7.03 ± 0.36% | ρ = −0.0400 | [2.94–8.18%] | 6.07 ± 0.29% | ρ = −0.5727 | p = 0.0670 |
| p = 0.8528 | p = 0.0067 | ||||||
| M. parma | [3.40–11.19%] | 7.43 ± 0.47% | ρ = 0.1561 | [3.20–6.51%] | 4.84 ± 0.26% | ρ = −0.1893 | p = 0.0001 |
| p = 0.5233 | p = 0.4993 | ||||||
| W. bicolor | [1.81–14.31%] | 10.91 ± 0.63% | ρ = −0.4674 | [1.86–10.05%] | 6.25 ± 0.39% | ρ = −0.1922 | p < 0.0001 |
| p = 0.0245 | p = 0.4039 | ||||||
| S. harrisii | [0.89–3.85%] | 2.30 ± 0.25% | ρ = 0.4231 | [1.26–4.38%] | 2.72 ± 0.30% | ρ = −0.0699 | p = 0.3412 |
| p = 0.1497 | p = 0.8290 | ||||||
| M. fuliginosus | [4.68–12.13%] | 7.90 ± 0.39% | ρ = 0.3914 | [2.44–6.07%] | 4.84 ± 0.18% | ρ = −0.3038 | p < 0.0001 |
| p = 0.0395 | p = 0.1398 | ||||||
| Primata | |||||||
| P. anubis cynocephalus | [1.98–10.77%] | 5.33 ± 0.31% | ρ = −0.3339 | [2.27–8.88%] | 5.03 ± 0.22% | ρ = 0.0117 | p = 0.9779 |
| p = 0.0127 | p = 0.9391 | ||||||
| M. fascicularis | [1.29–11.00%] | 5.45 ± 0.34% | ρ = 0.0935 | [1.09–10.06%] | 5.47 ± 0.38% | ρ = −0.4674 | p = 0.8726 |
| p = 0.5712 | p = 0.0053 | ||||||
| O. garnettii | [10.60–28.15%] | 18.60 ± 1.59% | ρ = −0.4462 | [4.88–13.65%] | 8.04 ± 0.94% | ρ = 0.2000 | p = 0.0003 |
| p = 0.1098 | p = 0.6059 | ||||||
| H. sapiens sapiens | [1.13–15.94%] | 4.71 ± 0.15% | ρ = −0.3137 | [1.11–8.86%] | 2.98 ± 0.12% | ρ = 0.0787 | p < 0.0001 |
| p < 0.0001 | p = 0.4075 | ||||||
| A. trivirgatus | [1.56–5.12%] | 3.14 ± 0.23% | ρ = −0.1377 | [1.70–3.24%] | 2.42 ± 0.11% | ρ = −0.1476 | p = 0.0503 |
| p = 0.5116 | p = 0.5590 | ||||||
| S. midas | [0.88–6.75%] | 4.77 ± 0.36% | ρ = 0.1464p = 0.6025 | [2.02–6.40%] | 4.04 ± 0.37% | ρ = 0.1484 | p = 0.0972 |
| p = 0.6286 | |||||||
| M. nemestrina | [2.75–12.55%] | 6.11 ± 0.30% | ρ = −0.1228 | [1.92–8.40%] | 4.84 ± 0.21% | ρ = 0.1164 | p = 0.0048 |
| p = 0.3671 | p = 0.4357 | ||||||
| C. apella | [2.31–10.75%] | 5.07 ± 0.30% | ρ = −0.4000 | [1.16–6.70%] | 3.73 ± 0.25% | ρ = −0.2834 | p = 0.0022 |
| p = 0.0087 | p = 0.1043 | ||||||
aI%O, Microglial percentage of non-neuronal cells; N/mg, number of neuronal cells per milligram of structure; O/mg, number of non-neuronal (other) cells per milligram of structure.
*For Afrotheria, analyses were done including all the species of the clade (D. dorsalis, E. myurus, L. africana, P. tetradactylus and P. capensis); Analyses within afrotherian species cortices were only possible where at least three data points were available (L. africana).
When data are combined across all species and clades, we find a detectable correlation between local neuronal density and the percentage of microglial cells among non-neuronal cells in each gray matter cortical site across species and clades, as shown in Figure 2I (Spearman correlation for all clades together: ρ = −0.4805, p < 0.0001). However, this correlation only applies across cortical sites within carnivorans, and only applies within the species F. catus, P. leo, H. sapiens sapiens, and C. apella.
In the white matter, a slight but detectable correlation is observed between local non-neuronal cell density and the percentage of microglial cells among non-neuronal cells in each cortical site across species and clades, as shown in Figure 2I (Spearman correlation for all clades together: ρ = −0.2613, p < 0.0001). However, this correlation does not apply across cortical sites within each clade, and only applies within each species in P. leo, D. aurita, D. goodfellowi, and M. fascicularis.
It thus again appears more likely that any apparent correlation between local non-neuronal cell density and percentage of non-neuronal cells that are microglial in the gray or white matter of the Cx is related to clade-specific (evolutionary) differences, and not to biological scaling. Overall, we find that microglial cells constitute systematically <10% of all non-neuronal cells in both gray and white matter of the Cx across all species examined, with an exact percentage that is smaller in primates than in other clades, and only slightly and nonsystematically variable across cortical sites in most species, independently of local neuronal densities.
No systematic variation of microglial density within the Cx
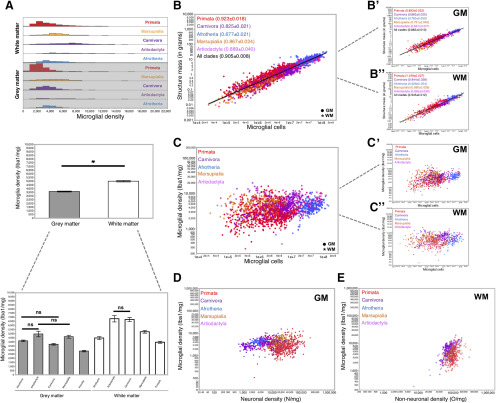
Although microglial cells are a slightly higher percentage of non-neuronal cells in the gray matter than in the white matter (Fig. 2G), we find that the average absolute density of microglial cells across all species and clades examined is actually higher in the white matter (5016 ± 93 microglia/mg) than in the gray matter (3620 ± 63 microglia/mg, Wilcoxon p < 0.0001; Fig. 3A, middle; Table 4, “all clades”), due to the overall slightly higher densities of non-neuronal cells in the former than in the latter (Herculano-Houzel and Dos Santos, 2018). The vast majority of individual gray matter sites examined has microglial densities between 1000 and 10,000 microglia/mg, whereas neuronal densities at the same sites vary between 1000 and 100,000 neurons/mg (Fig. 3D). Average microglial cell densities in gray and white matter by clade are shown in Figure 3A (bottom). Across clades, the lowest average microglial cell densities are found in primates, both in gray (2890 ± 92 microglia/mg) and white matter (3965 ± 123 microglia/mg; Table 4, “per clade”), although the broad overall ranges of microglial cell densities encompass largely overlapping distributions across clades and cortical sites (Fig. 3A, top, bottom). Within each clade, there is not much variation in microglial densities across species (Table 4, “per species”). Within an individual cortex, microglial densities are significantly higher in white matter sites compared with those in gray matter in 12 of the species sampled, including the human cortex (Table 4).
Figure 3.
Little variation of microglial density in cortical gray and white matter. A, Distribution of microglial density in the cortical gray and white matter at individual sites per clade (top), average across all samples (middle), and average per clade (bottom). Values are given in Table 4. Average microglial densities in gray matter cortical sites differ significantly across clades at the p < 0.01 level, except between Artiodactyla and Afrotheria (Wilcoxon, p = 0.1877; not significant), Afrotheria and Marsupialia (p = 0.0442), and Artiodactyla and Marsupialia (p = 0.7454), and in the white matter, between Artiodactyla and Carnivora (p = 0.8260). Differences in microglial cell density between white and gray matter are significant (Wilcoxon p < 0.01) in all clades (see Table 4), except Afrotheria (p = 0.2835) and Marsupialia (p = 0.0151). B, Relationship between structure mass and number of microglial cells across cortical sites and mammalian species. C, Function plotted applies to all species and gray and white matter together with exponent (0.905 ± 0.008, p < 0.0001, r2 = 0.878). B′, B′′, This same relationship for gray (B′) and white (B′′) matter sites separately. Spearman correlations, p values, and other fit values in each clade and species are listed in Table 4. C, Relationship between the density of microglial cells and the number of microglial cells across cortical sites and mammalian species. C′, C′′, This same relationship for gray (C′) and white (C′′) matter sites separately. Spearman correlation ρ and p values are given in Table 4. D, Relationship between microglial density and neuronal cell density across cortical gray matter sites and mammalian species. Spearman coefficients and p values as well as exponents and p values for each clade and species are listed in Table 4. E, Relationship between microglial density and non-neuronal density across cortical white matter sites and mammalian species. Spearman coefficients and p values as well as exponents and p values for each clade and species are listed in Table 4. B, B′, B′′, C, C′, C′′, D, E, Each data point represents the values found per cortical site examined for each species colored by clade as in the key. Filled circles represent cortical gray matter sites of the Cx, including the hippocampus (GM). *White matter sites (WM). A, *Significance.
Table 4.
Microglial density and microglia/neuron ratio in different cortical sitesa
| Gray matter (microglia/mg) |
White matter (microglia/mg) |
Wilcoxon I/mg versus white matter | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I/mg |
M vs. I, (exponent and p value) | I/N |
I/mg |
M vs. I, (exponent and p value) | |||||||||
| (range and mean ± SE) | (I/mg vs I, Spearman ρ and p value) | (I/mg vs N/mg, Spearman ρ andp value) | (I/mg vs N/mg, exponent and p value) | (I/mg vs N/mg, exponent, and p value) | (I vs I/mg, exponent, and p value) | (range and mean ± SE) | (I/mg vs I, Spearman ρ, andp value) | (I/mg vs O/mg, Spearman ρ, andp value) | (I/mg vs O/mg exponent, and p value) | ||||
| All clades | [106-16 061] 3620 ± 63 |
ρ=0.2942 p < 0.0001 |
ρ=−0.1108 p = 0.0005 |
−0.025 ± 0.016 p = 0.1165 |
0.882 ± 0.010 p < 0.0001 |
−1.020 ± 0.016 p < 0.0001 |
1.104 ± 0.063 p < 0.0001 |
[271-19 827] 5016 ± 93 |
ρ=0.1614 p < 0.0001 |
ρ=0.2172 p < 0.0001 |
0.596 ± 0.064 p < 0.0001 |
0.945 ± 0.012 p < 0.0001 |
p < 0.0001 |
| Per clade | |||||||||||||
| Afrotheria | [930-7971] 4140 ± 97 |
ρ=0.3005 p = 0.0009 |
ρ=0.1538 p = 0.0936 |
0.083 ± 0.066 p = 0.2094 |
0.760 ± 0.050 p < 0.0001 |
−0.917 ± 0.066 p < 0.0001 |
0.840 ± 0.126 p < 0.0001 |
[371-10 673] 4482 ± 163 |
ρ=0.4252 p < 0.0001 |
ρ=0.5710 p < 0.0001 |
1.144 ± 0.149 p < 0.0001 |
0.628 ± 0.054 p < 0.0001 |
p = 0.2835 |
| Artiodactyla | [1075-15 647] 4975 ± 345 |
ρ=0.4382 p = 0.0006 |
ρ=0.5049 p = 0.0001 |
0.715 ± 0.136 p < 0.0001 |
0.847 ± 0.071 p < 0.0001 |
−0.285 ± 0.136 p = 0.0397 |
0.536 ± 0.088 p < 0.0001 |
[2576-16 518] 6833 ± 387 |
ρ=0.2862 p = 0.0309 |
ρ=0.7154 p < 0.0001 |
1.472 ± 0.202 p < 0.0001 |
0.882 ± 0.039 p < 0.0001 |
p = 0.0002 |
| Carnivora | [919-10 728] 3722 ± 98 |
ρ=0.2386 p = 0.0003 |
ρ=0.2954 p < 0.0001 |
0.122 ± 0.026 p < 0.0001 |
0.863 ± 0.025 p < 0.0001 |
−0.878 ± 0.026 p < 0.0001 |
0.276 ± 0.156 p = 0.0775 |
[741-19 827] 6740 ± 242 |
ρ=0.3512 p < 0.0001 |
ρ=0.7023 p < 0.0001 |
1.323 ± 0.103 p < 0.0001 |
0.844 ± 0.028 p < 0.0001 |
p < 0.0001 |
| Marsupialia | [600-15 098] 4648 ± 175 |
ρ=0.2954 p = 0.0003 |
ρ=0.3813 p < 0.0001 |
0.368 ± 0.092 p = 0.0001 |
0.791 ± 0.040 p < 0.0001 |
−0.632 ± 0.092 p < 0.0001 |
0.730 ± 0.068 p < 0.0001 |
[1220-12 132] 5242 ± 162 |
ρ=0.2362 p = 0.0080 |
ρ=0.4062 p < 0.0001 |
0.474 ± 0.128 p = 0.0003 |
0.890 ± 0.026 p < 0.0001 |
p = 0.0151 |
| Primata | [106-16 061] 2890 ± 92 |
ρ=0.1876 p = 0.0001 |
ρ=0.3226 p < 0.0001 |
0.301 ± 0.036 p < 0.0001 |
0.893 ± 0.022 p < 0.0001 |
−0.657 ± 0.037 p < 0.0001 |
0.537 ± 0.054 p < 0.0001 |
[271-12 511] 3965 ± 123 |
ρ=−0.0689 p = 0.2197 |
ρ=0.3998 p < 0.0001 |
0.864 ± 0.088 p < 0.0001 |
1.019 ± 0.027 p < 0.0001 |
p < 0.0001 |
| Per species | |||||||||||||
| Afrotheria* | |||||||||||||
| L. africana | [930-7971] 4170 ± 97 |
ρ=0.2704 p = 0.0032 |
ρ=0.1538 p = 0.0936 |
0.280 ± 0.080 p = 0.0006 |
0.791 ± 0.054 p < 0.0001 |
−0.719 ± 0.080 p < 0.0001 |
0.652 ± 0.099 p < 0.0001 |
[371-10 673] 4511 ± 166 |
ρ=0.4081 p < 0.0001 |
ρ=0.5491 p = 0.0001 |
1.240 ± 0.154 p < 0.0001 |
0.604 ± 0.058 p < 0.0001 |
p = 0.2732 |
| Artiodactyla | |||||||||||||
| G. camelopardalis | [2764-9975] 5825 ± 352 |
ρ=0.6361 p = 0.0026 |
ρ=0.2060 p = 0.3835 |
0.228 ± 0.170 p = 0.1969 |
0.604 ± 0.141 p = 0.0004 |
−0.772 ± 0.170 p = 0.0003 |
0.602 ± 0.297 p = 0.0576 |
[3769-10 746] 7197 ± 466 |
ρ=0.5789 p = 0.0075 |
ρ=0.4226 p = 0.0634 |
0.827 ± 0.365 p = 0.0361 |
0.876 ± 0.101 p < 0.0001 |
p = 0.0223 |
| T. strepsiceros | [1075-4298] 2847 ± 214 |
ρ=0.6118 p = 0.0118 |
ρ=0.6824 p = 0.0036 |
1.066 ± 0.211 p = 0.0002 |
0.658 ± 0.133 p = 0.0002 |
0.066 ± 0.211 p = 0.7581 |
0.394 ± 0.120 p = 0.0054 |
[3502-6804] 5070 ± 231 |
ρ=0.5353 p = 0.0326 |
ρ=0.4471 p = 0.0825 |
0.432 ± 0.373 p = 0.2663 |
0.933 ± 0.032 p < 0.0001 |
p < 0.0001 |
| S. scrofa domesticus | [5584-15 647] 8481 ± 939 |
ρ=0.1879 p = 0.6032 |
ρ=−0.1758 p = 0.6272 |
0.400 ± 0.250 p = 0.1478 |
1.170 ± 0.089 p < 0.0001 |
−0.600 ± 0.250 p = 0.0430 |
0.393 ± 0.379 p = 0.3305 |
[8595-16 518] 11 742 ± 803 |
ρ=0.5333 p = 0.1392 |
ρ=0.5000 p = 0.1705 |
0.983 ± 0.525 p = 0.1032 |
0.907 ± 0.139 p = 0.0003 |
p = 0.0101 |
| A. marsupialis | [1941-6161] 3476 ± 377 |
ρ=−0.1189 p = 0.7129 |
ρ=0.8531 p = 0.0004 |
0.625 ± 0.114 p = 0.0003 |
1.138 ± 0.164 p < 0.0001 |
−0.375 ± 0.114 p = 0.0080 |
−0.203 ± 0.218 p = 0.3754 |
[2576-7398] 4894 ± 399 |
ρ=0.4685 p = 0.1245 |
ρ=0.9231 p < 0.0001 |
1.285 ± 0.220 p = 0.0002 |
0.831 ± 0.041 p < 0.0001 |
p = 0.0194 |
| Carnivora | |||||||||||||
| M. mongo | [919-2130] 1400 ± 172 |
ρ=0.3214 p = 0.4821 |
ρ=0.7857 p = 0.0362 |
0.698 ± 0.201 p = 0.0177 |
0.398 ± 0.511 p = 0.4715 |
−0.302 ± 0.201 p = 0.1926 |
−0.014 ± 0.292 p = 0.9642 |
[740-2918] 1926 ± 156 |
ρ=0.7343 p = 0.0065 |
ρ=0.1678 p = 0.6021 |
1.391 ± 0.916 p = 0.1600 |
0.250 ± 0.193 p = 0.2235 |
p = 0.0572 |
| U. arctos | [1432-5774] 2801 ± 94 |
ρ=0.1660 p = 0.1828 |
ρ=−0.0423 p = 0.7359 |
0.022 ± 0.088 p = 0.8050 |
0.953 ± 0.032 p < 0.0001 |
−0.978 ± 0.088 p < 0.0001 |
0.956 ± 0.178 p < 0.0001 |
[1970-11 965] 5738 ± 281 |
ρ=0.3883 p = 0.0058 |
ρ=0.5573 p < 0.0001 |
1.178 ± 0.184 p < 0.0001 |
0.953 ± 0.047 p < 0.0001 |
p < 0.0001 |
| F. catus | [1536-4755] 2902 ± 186 |
ρ=0.3510 p = 0.0726 |
ρ=0.4829 p = 0.0107 |
0.588 ± 0.211 p = 0.0101 |
0.846 ± 0.078 p < 0.0001 |
−0.412 ± 0.211 p = 0.0622 |
0.598 ± 0.145 p = 0.0004 |
[2276-5590] 3406 ± 227 |
ρ=0.6941 p = 0.0029 |
ρ=0.3824 p = 0.1439 |
0.492 ± 0.376 p = 0.2117 |
0.850 ± 0.070 p < 0.0001 |
p = 0.0899 |
| C. familiaris | [1867-6866] 4170 ± 132 |
ρ=0.2634 p = 0.0646 |
ρ=0.1200 p = 0.4065 |
0.085 ± 0.076 p = 0.2666 |
0.948 ± 0.041 p < 0.0001 |
−0.915 ± 0.076 p < 0.0001 |
0.699 ± 0.268 p = 0.0121 |
[4846-15 717] 8094 ± 358 |
ρ=0.2406 p = 0.1773 |
ρ=0.6086 p = 0.0002 |
0.668 ± 0.142 p < 0.0001 |
1.018 ± 0.048 p < 0.0001 |
p < 0.0001 |
| M. putorius furo | [4523-10 728] 7366 ± 525 |
ρ=0.1636 p = 0.6307 |
ρ=0.4455 p = 0.1697 |
0.260 ± 0.160 p = 0.1396 |
0.892 ± 0.190 p = 0.0011 |
−0.740 ± 0.160 p = 0.0013 |
0.131 ± 0.536 p = 0.8119 |
[10 348-19 827] 13 278 ± 909 |
ρ=−0.0833 p = 0.8312 |
ρ=−0.1333 p = 0.7324 |
−0.583 ± 0.562 p = 0.3343 |
1.039 ± 0.144 p = 0.0002 |
p = 0.0003 |
| P. leo | [2192-8166] 4285 ± 126 |
ρ=0.2756 p = 0.0263 |
ρ=0.0137 p = 0.9139 |
0.081 ± 0.070 p = 0.2482 |
0.895 ± 0.051 p < 0.0001 |
−0.919 ± 0.070 p < 0.0001 |
0.740 ± 0.223 p = 0.0015 |
[5816-10 588] 8123 ± 199 |
ρ=0.4866 p = 0.0013 |
ρ=0.3747 p = 0.0158 |
0.472 ± 0.132 p = 0.0010 |
0.943 ± 0.026 p < 0.0001 |
p < 0.0001 |
| Marsupialia | |||||||||||||
| M. rufogriseus | [4090-7414] 5753 ± 225 |
ρ=0.0610 p = 0.7927 |
ρ=0.1117 p = 0.6298 |
0.058 ± 0.075 p = 0.4500 |
1.053 ± 0.087 p < 0.0001 |
−0.942 ± 0.075 p < 0.0001 |
0.476 ± 0.679 p = 0.4919 |
[5303-9640] 6775 ± 257 |
ρ=−0.0561 p = 0.8194 |
ρ=0.2614 p = 0.2797 |
0.224 ± 0.247 p = 0.3770 |
1.053 ± 0.053 p < 0.0001 |
p = 0.0127 |
| D. aurita | [3290-8565] 5801 ± 425 |
ρ=0.8576 p < 0.0001 |
ρ=0.6182 p = 0.0063 |
0.485 ± 0.173 p = 0.0126 |
0.461 ± 0.100 p = 0.0003 |
−0.515 ± 0.173 p = 0.0088 |
0.319 ± 0.242 p = 0.2069 |
[4113-8611] 5506 ± 402 |
ρ=0.1678 p = 0.6021 |
ρ=0.6573 p = 0.0202 |
0.553 ± 0.131 p = 0.0018 |
0.884 ± 0.166 p = 0.0003 |
p = 0.5677 |
| D. goodfellowi | [1780-7000] 3465 ± 239 |
ρ=0.2591 p = 0.2214 |
ρ=0.2417 p = 0.2551 |
0.275 ± 0.168 p = 0.1174 |
0.991 ± 0.076 p < 0.0001 |
−0.725 ± 0.168 p = 0.0003 |
0.608 ± 0.241 p = 0.0192 |
[3536-7873] 5382 ± 246 |
ρ=0.4442 p = 0.0437 |
ρ=0.1558 p = 0.4999 |
0.259 ± 0.272 p = 0.3529 |
0.826 ± 0.068 p < 0.0001 |
p < 0.0001 |
| M. parma | [2351-15 098] 5547 ± 640 |
ρ=−0.0912 p = 0.7103 |
ρ=0.7123 p = 0.0006 |
0.510 ± 0.164 p = 0.0063 |
1.117 ± 0.174 p < 0.0001 |
−0.490 ± 0.164 p = 0.0082 |
0.287 ± 0.228 p = 0.2257 |
[2671-5518] 4316 ± 222 |
ρ=0.4071 p = 0.1320 |
ρ=0.5179 p = 0.0480 |
0.484 ± 0.248 p = 0.0730 |
0.831 ± 0.067 p < 0.0001 |
p = 0.1270 |
| W. bicolor | [1163-8680] 6191 ± 330 |
ρ=0.3468 p = 0.1049 |
ρ=−0.0524 p = 0.8124 |
−0.136 ± 0.209 p = 0.5223 |
0.736 ± 0.076 p < 0.0001 |
−1.136 ± 0.209 p < 0.0001 |
1.145 ± 0.223 p = 0.000044 |
[4194-12 132] 6966 ± 429 |
ρ=0.2403 p = 0.2942 |
ρ=0.5026 p = 0.0202 |
0.238 ± 0.179 p = 0.1987 |
0.937 ± 0.066 p < 0.0001 |
p = 0.6052 |
| S. harrisii | [600-2571] 1394 ± 156 |
ρ=0.3187 p = 0.2886 |
ρ=0.5495 p = 0.0518 |
0.945 ± 0.411 p = 0.0422 |
0.807 ± 0.226 p = 0.0045 |
−0.055 ± 0.411 p = 0.8956 |
0.657 ± 0.149 p = 0.0011 |
[1220-4633] 2971 ± 324 |
ρ=0.7273 p = 0.0074 |
ρ=0.3007 p = 0.3423 |
0.483 ± 0.634 p = 0.4638 |
0.765 ± 0.146 p = 0.0004 |
p = 0.0006 |
| M. fuliginosus | [2113-5441] 3725 ± 197 |
ρ=0.5047 p = 0.0062 |
ρ=0.2906 p = 0.1335 |
0.328 ± 0.171 p = 0.0657 |
0.916 ± 0.060 p < 0.0001 |
−0.672 ± 0.171 p = 0.0006 |
0.621 ± 0.197 p = 0.0041 |
[1945-5161] 4029 ± 166 |
ρ=0.2538 p = 0.2208 |
ρ=0.5646 p = 0.0033 |
0.881 ± 0.301 p = 0.0076 |
0.832 ± 0.026 p < 0.0001 |
p = 0.1277 |
| Primata | |||||||||||||
| P. anubiscynocephalus | [856-8907] 2402 ± 154 |
ρ=0.0651 p = 0.6337 |
ρ=0.2333 p = 0.0865 |
0.200 ± 0.087 p = 0.0254 |
1.023 ± 0.050 p < 0.0001 |
−0.800 ± 0.087 p < 0.0001 |
0.546 ± 0.197 p = 0.0077 |
[1983-11 992] 4385 ± 320 |
ρ=0.2047 p = 0.1724 |
ρ=0.6150 p < 0.0001 |
1.071 ± 0.169 p < 0.0001 |
0.960 ± 0.103 p < 0.0001 |
p < 0.0001 |
| M. fascicularis | [989-11 057] 3718 ± 257 |
ρ=0.6443 p < 0.0001 |
ρ=0.4089 p = 0.0097 |
0.482 ± 0.178 p = 0.0102 |
0.756 ± 0.048 p < 0.0001 |
−0.518 ± 0.178 p = 0.0061 |
0.657 ± 0.127 p < 0.0001 |
[1620-10 255] 5997 ± 348 |
ρ=0.3559 p = 0.0306 |
ρ=0.1566 p = 0.3764 |
0.095 ± 0.220 p = 0.6700 |
0.873 ± 0.059 p < 0.0001 |
p < 0.0001 |
| O. garnettii | [3384-16 061] 9259 ± 909 |
ρ=0.2264 p = 0.4364 |
ρ=0.0637 p = 0.8286 |
0.021 ± 0.291 p = 0.9434 |
0.472 ± 0.265 p = 0.1001 |
−0.979 ± 0.291 p = 0.0057 |
0.979 ± 0.286 p = 0.0050 |
[2226-12 240] 7912 ± 1172 |
ρ=0.8833 p = 0.0016 |
ρ=0.5000 p = 0.1705 |
1.251 ± 0.341 p = 0.0080 |
−0.063 ± 0.221 p = 0.7835 |
p = 0.3951 |
| H. sapiens sapiens | [664-7812] 2379 ± 81 |
ρ=0.3915 p < 0.0001 |
ρ=−0.1368 p = 0.0695 |
−0.138 ± 0.058 p = 0.0192 |
0.889 ± 0.023 p < 0.0001 |
−0.993 ± 0.065 p < 0.0001 |
1.222 ± 0.087 p < 0.0001 |
[915-9079] 2891 ± 133 |
ρ=0.1703 p = 0.0713 |
ρ=0.6150 p < 0.0001 |
1.244 ± 0.201 p < 0.0001 |
0.999 ± 0.050 p < 0.0001 |
p = 0.0005 |
| A. trivirgatus | [1155-4145] 2612 ± 180 |
ρ=0.4946 p = 0.0120 |
ρ=0.2215 p = 0.2872 |
0.099 ± 0.121 p = 0.4215 |
0.815 ± 0.090 p < 0.0001 |
−0.901 ± 0.121 p < 0.0001 |
0.714 ± 0.350 p = 0.0530 |
[1782-4366] 2863 ± 193 |
ρ=0.1909 p = 0.4479 |
ρ=0.5872 p = 0.0104 |
0.991 ± 0.254 p = 0.0013 |
0.966 ± 0.108 p < 0.0001 |
p = 0.4381 |
| S. midas | [691-7771] 3304 ± 532 |
ρ=0.2750 p = 0.3212 |
ρ=0.5821 p = 0.0228 |
0.705 ± 0.230 p = 0.0089 |
0.491 ± 0.268 p = 0.0898 |
−0.295 ± 0.230 p = 0.2206 |
0.404 ± 0.194 p = 0.0582 |
[1987-8209] 4437 ± 550 |
ρ=0.2912 p = 0.3344 |
ρ=0.6648 p = 0.0132 |
1.194 ± 0.389 p = 0.0107 |
0.715 ± 0.305 p = 0.0390 |
p = 0.1173 |
| M. nemestrina | [1413-8300] 3732 ± 223 |
ρ=0.0196 p = 0.8839 |
ρ=0.4969 p = 0.0001 |
0.469 ± 0.110 p < 0.0001 |
1.045 ± 0.062 p < 0.0001 |
−0.531 ± 0.110 p < 0.0001 |
0.467 ± 0.126 p = 0.0005 |
[1510-12 511] 4782 ± 295 |
ρ=0.2114 p = 0.1493 |
ρ=0.6303 p < 0.0001 |
1.112 ± 0.172 p < 0.0001 |
0.902 ± 0.080 p < 0.0001 |
p = 0.0030 |
| C. apella | [106-2901] 1626 ± 96 |
ρ=0.4959 p = 0.0007 |
ρ=0.2610 p = 0.0950 |
0.505 ± 0.106 p < 0.0001 |
0.633 ± 0.081 p < 0.0001 |
−0.495 ± 0.106 p < 0.0001 |
0.282 ± 0.150 p = 0.0684 |
[271-7371] 2985 ± 244 |
ρ=0.5126 p = 0.0016 |
ρ=0.3580 p = 0.0376 |
0.856 ± 0.164 p < 0.0001 |
0.400 ± 0.146 p = 0.0100 |
p < 0.0001 |
aI, Number of microglial cells; I/mg, number of microglial cells per milligram of structure; N/mg, number of neuronal cells per milligram of structure; O/mg, number of non-neuronal (other) cells per milligram of structure; M, structure mass; I/N, microglia/neuron ratio.
*For Afrotheria, analyses were done, including all the species of the clade (D. dorsalis, E. myurus, L. africana, P. tetradactylus, and P. capensis). Analyses within afrotherian species cortices were only possible where at least three data points were available (L. africana).
Across all cortical gray and white matter sites analyzed (n = 980 and n = 796, respectively), we find that the mass of each cortical site varies as a power function of the number of microglial cells found in it that is significantly different from linearity whether gray and white matter sites are analyzed together (exponent, 0.905 ± 0.008, p < 0.0001) or separately (exponents 0.882 ± 0.010, p < 0.0001 and 0.945 ± 0.012, p < 0.0001 for gray and white matter, respectively; Fig. 3B,B′,B′′). These strong correlations still hold within individual cortices, with significant sublinear or linear relationships between mass and number of microglial cells, except for four species (M. mungo gray and white matter, and gray matter in S. scrofa, F. catus, and S. harrisii; Table 4). With exponents that include unity, a linear relationship cannot be excluded for gray and/or white matter in 21 of the 26 species analyzed, which suggests that microglial density does not really vary systematically within the Cx gray and white matter in these species. Indeed, across all cortical gray and white matter sites, the local density of microglial cells increases only weakly with their number across species and clades, with a significant but weak correlation across gray and white matter sites together (Spearman ρ = 0.2373, p < 0.0001) or separately (Spearman ρ = 0.2942, p < 0.0001 and Spearman ρ = 0.1614, p < 0.0001 for gray and white matter, respectively; Figure 3C,C′,C′′). Within individual cortices, weak correlations, most of the times nonsignificant, within both gray and white mater sites or at least one cortical region are observed in 23 of the 26 species (Table 4). Thus, microglial cells appear to be added in highly variable numbers but fairly similar densities to different cortical sites.
The slight but significant variation in microglial cell density across cortical sites could reflect local variations in neuronal density, which is known to be significant even within an individual Cx (Ribeiro et al., 2013; Herculano-Houzel et al., 2015b; Gabi et al., 2016). However, variations in local microglial densities in the cortical gray matter are only poorly correlated with local neuronal densities when data points from all sites are pooled together across species and clades (Spearman ρ = −0.1108, p = 0.0005; Fig. 3D), and the fit to a power function is not significant (p = 0.1165; Fig. 3D).
Within each clade, pooling cortical sites across species reveals in all clades, except Afrotheria, a significant positive relationship between local microglial density and neuronal density in the cortical gray matter (Fig. 3D; Table 4). However, most individual cortices do not present significant relationships between local microglial and neuronal densities (Table 4). Thus, any apparent systematic variation in microglial cell density seen here is most likely the result of pooling data across clades, and not of a systematic, biological relationship between local microglial and neuronal densities.
Similarly, local microglial densities in the cortical white matter sites are slightly correlated with local densities of non-neuronal cells when data points from all sites are pooled together across species and clades (Spearman ρ = 0.2172, p < 0.0001; Fig. 3E), with a significant fit (exponent, 0.596 ± 0.010, p < 0.0001) but with a very low r2 of 0.112. As for the gray matter, pooling cortical sites across species within each clade reveals a significant positive relationship between local microglial density and non-neuronal cell density for all clades (Fig. 3E; Table 4). However, only half of the species analyzed present significant relationships between local microglial and non-neuronal cell densities within an individual Cx (Fig. 3E; Table 4), again suggesting that any apparent relationship in the overall dataset is the result of clade-specific differences, with slightly lower densities in primate cortices.
Microglia/neuron ratio in the cortical gray matter is a universal function of neuronal density across sites, species, and clades
We find that there are typically fewer microglial cells than neurons in the cortical gray matter: that was the case in 973 of all 1776 sites analyzed in the 30 species. More microglial cells than neurons in the cortical gray matter as a whole were only found in 5 species, all of them the largest-brained nonprimate species of carnivorans, artiodactyls, or the African elephant. Even in those cases, there were at most 2-3 microglial cells per neuron in the cortical gray matter. Generally, we find that there are 1-100 neurons per microglial cell in the gray matter of most mammalian species (Table 1, I/N).
As reported previously (Herculano-Houzel, 2014; Herculano-Houzel and Dos Santos, 2018), we find that the number of non-neuronal cells per neuron varies inversely with neuronal density in our current dataset pooled across all cortical sites, species, and clades as a sublinear power function of local neuronal density (exponent –0.793 ± 0.009, p < 0.0001, r2 = 0.888; Fig. 4A). There is a similar trend of fewer microglial cells per neuron (i.e., a smaller I/N ratio) where neuronal densities are higher: the I/N ratio varies as a power function of neuronal density across all gray matter cortical sites and across species and clades with an exponent that is not significantly different from inverse linearity (−1.020 ± 0.016, p < 0.0001, r2 = 0.799; Fig. 4B). This correlation holds within each clade, except for Artiodactyla (Table 4). Within the cortex of each species, strong correlations, with significant sublinear or linear relationships between I/N and neuronal density, are found for all but six species, and are present in the only afrotherian species included, the African elephant (Table 4). In contrast with neuronal density, there is no single scaling relationship that applies between the I/N ratio and microglial density across all cortical sites, clades, and species (Fig. 4C; Table 4). These findings strongly indicate that variations in I/N across cortical sites and species are due to variations in neuronal density in the presence of only slight variations in microglial cell density.
Figure 4.
Microglia/neuron ratio varies uniformly with neuronal density, but not with microglial density across cortical gray matter sites. A, Non-neuronal cells/neuron ratio (O/N) is plotted as a function of neuronal density in the gray matter sites across species and clades. p value for Spearman correlation for all clades together is p < 0.0001. Exponent, p value, and r2 for the relationship plotted in A (−0.793 ± 0.009, p < 0.0001, 0.888). B, Microglia/neuron ratio is plotted as a function of neuronal density in the gray matter sites across species and clades. p values for Spearman correlations are all p < 0.0001, except for Artiodactyla (p = 0.1819). The power function plotted in B applies to all gray matter sites together and mammalian species with exponent (−1.020 ± 0.016, p < 0.0001, r2 = 0.799). Exponents and p values for each clade and species are listed in Table 4. C, Microglia/neuron ratio is plotted as a function of microglial density in the gray matter sites across species and clades. p values for Spearman correlations are as follows: p < 0.0001 for all clades together and each clade separately except for Carnivora (p = 0.8414). Exponents and p values for each clade and species are listed in Table 4. Each data point represents the values found per cortical gray matter site examined for each species colored by clade as in the key.
Similar microglial proportions and densities across brain structures and species
We next turn to the question of how microglial cell density compares across brain structures in different mammalian species. Relative to the total number of cells in the structure, microglial cells amount to 5.77 ± 0.43% in the Cx as a whole, 1.50 ± 0.15% in the Cb, and 6.39 ± 0.51% in the RoB (Fig. 5A,B; Table 5). These average percentages, calculated among all species and clades, are significantly higher in both Cx and RoB compared with Cb (Wilcoxon, p < 0.0001), but not significantly different between the former two structures (Wilcoxon p = 0.4283) (Fig. 5B; Table 5). The percentage of all cells that are microglial in each major brain division varies both across structures in the same clade and within the same structure across clades (Fig. 5C). Variation in the percentage of all cells that are microglial correlates strongly with the neuronal density in the respective structure, as shown in Figure 5D (Spearman correlation: ρ = −0.8058, p < 0.0001 for all clades): the higher the neuronal density, the lower the percentage of all cells that is microglial.
Figure 5.
Average percentages of microglial cells vary across brain structures but correlate with numbers of non-neuronal cells of which they represent a constant ∼7%. A, Distribution of the percentage of all cells that are microglial cells in the three main brain structures per clade. The ranges of distribution and means of the percentage of all cells that are microglia differ across clades and brain structures (values in Table 5). B, Average percentages of total cells that are microglia differ across brain structures. Wilcoxon p < 0.0001 between Cb and both Cx and RoB in all mammalian species. Wilcoxon p = 0.4283 between Cx and RoB in all mammalian species. C, Average percentages of total cells that are microglia in brain structures across clades and species. In the Cb, significant differences are found between Marsupialia and both Artiodactyla and Carnivora (Wilcoxon, p = 0.0058 and p = 0.0020, respectively). In the Cx and RoB, no significant differences are found between clades, Wilcoxon p values of all pairs being ≥0.01. Within clades, significant differences are also found between the Cb and the Cx and RoB for each clade, except Afrotheria and Artiodactyla: Carnivora, between Cb and Cx Wilcoxon p = 0.0081 and between Cb and RoB Wilcoxon p = 0.0051; and Marsupialia, between Cb and Cx Wilcoxon p = 0.0003 and between Cb and RoB Wilcoxon p = 0.0003. D, Percentages of microglial cells among all cells across all brain structures and species correlate strongly with the neuronal cell density in the sample. The power function plotted in D applies to brain structures together and mammalian species with exponent (−0.378 ± 0.028, p < 0.0001, r2 = 0.709). Spearman ρ values are indicated between brackets for each clade when significant (otherwise, indicated as nonsignificant) with respective p values falling to <0.01, except for Artiodactyla (p = 0.0386; nonsignificant) and Primata (p = 0.6932). E, Distribution of the percentage of non-neuronal cells that are microglial cells in the three main brain structures per clade. The ranges of distribution and means of the percentage of non-neuronal cells that are microglia are similar across clades and brain structures (values in Table 5). F, Average percentages of non-neuronal cells that are microglia represent a constant ∼7% across brain structures and clades in all mammalian species. Wilcoxon p values >0.01 for all pairs of structures. G, Average percentages of non-neuronal cells that are microglia in brain structures across clades and species. Across each structure, all Wilcoxon, p > 0.01. Within each structure across all clades, all Wilcoxon, p > 0.01. H, Percentages of microglial cells among non-neuronal cells across all brain structures and species fail to correlate significantly with the neuronal cell density in the sample. Spearman, all p values >0.01. D, H, Each data point represents the values found per brain structure examined for each species colored by clade as in the key. Filled circles represent the Cx (including the hippocampus). Squares represent the Cb. Triangles represent the RoB. B, C, *Significance.
Table 5.
Microglial percentages and densities in different brain structuresa
| Cx |
Cb |
RoB |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I% all cells | I% O cells | I/mg | I/N | I% all cells | I% O cells | I/mg | I/N | I% all cells | I% O cells | I/mg | I/N | |
| (range and mean ± SE) | (range and mean ± SE) | (range and mean ± SE) | (range and I/mg vs N/mg, exponent. andp value) | (range and mean ± SE) | (range and mean ± SE) | (range and mean ± SE) | (range and I/mg vs N/mg, exponent andp value) | (range and mean ± SE) | (range and mean ± SE) | (range and mean ± SE) | (range and I/mg vs N/mg, exponent, andp value) | |
| All clades | [1.84-10.98%] 5.77 ± 0.43% |
[2.61-14.03%] 7.24 ± 0.50% |
[1826-9048] 4375 ± 341 |
[0.053-3.362] −1.038 ± 0.088 p < 0.0001 |
[0.54-3.88%] 1.50 ± 0.15% |
[2.98-24.56%] 7.37 ± 1.05% |
[1234-10 536] 5021 ± 406 |
[0.007-0.046] −0.720 ± 0.224 p = 0.0038 |
[2.02-11.25%] 6.39 ± 0.51% |
[2.18-11.85%] 7.20 ± 0.50% |
[1780-9386] 4664 ± 464 |
[0.094-3.469] −1.033 ± 0.121 p < 0.0001 |
| Per clade | ||||||||||||
| Afrotheria | [4.90-7.46%] 6.04 ± 0.64% |
[7.54-10.39%] 8.65 ± 1.31% |
[1832-4192] 3170 ± 506 |
[0.115-2.086] −1.137 ± 0.200 p = 0.0296 |
[0.54-2.43%] 1.19 ± 0.34% |
[3.05-18.29%] 7.45 ± 2.90% |
[1234-5982] 4024 ± 789 |
[0.007-0.028] −0.298 ± 0.725 p = 0.7084 |
[3.08-6.92%] 4.89 ± 0.65% |
[4.57-7.20%] 6.53 ± 0.50% |
[1845-3450] 2897 ± 302 |
[0.094-2.626] −1.075 ± 0.113 p = 0.0025 |
| Artiodactyla | [5.86-10.98%] 7.97 ± 1.17% |
[6.42-12.03%] 8.61 ± 1.29% |
[3718-9048] 5722 ± 1264 |
[0.675-1.405] −0.104 ± 0.714 p = 0.8971 |
[1.82-3.88%] 2.48 ± 0.47% |
[5.40-24.56%] 12.67 ± 4.28% |
[3040-10 536] 5781 ± 1644 |
[0.024-0.046] 0.557 ± 0.590 p = 0.4444 |
[6.07-11.25%] 8.76 ± 1.13% |
[6.39-11.85%] 9.13 ± 1.19% |
[3267-9386] 6036 ± 1361 |
[1.196-3.469] −0.458 ± 0.723 p = 0.5910 |
| Carnivora | [5.47-10.19%] 7.69 ± 0.78% |
[6.72-10.96%] 8.58 ± 0.72% |
[3048-6530] 4770 ± 626 |
[0.295-3.362] −0.929 ± 0.176 p = 0.0132 |
[1.63-2.05%] 1.82 ± 0.06% |
[4.08-11.10%] 6.90 ± 1.01% |
[4174-9601] 5845 ± 826 |
[0.020-0.031] −0.078 ± 0.253 p = 0.7732 |
[3.11-10.59%] 6.63 ± 1.42% |
[3.34-11.14%] 7.09 ± 1.49% |
[1831-9269] 4821 ± 1351 |
[0.294-2.925] −0.721 ± 0.547 p = 0.2580 |
| Marsupialia | [2.18-7.68%] 5.40 ± 0.61% |
[2.61-9.75%] 6.82 ± 0.77% |
[1826-6892] 4699 ± 567 |
[0.131-0.457] −0.680 ± 0.328 p = 0.0771 |
[0.66-1.71%] 1.07 ± 0.12% |
[2.98-13.16%] 5.51 ± 1.02% |
[2408-6893] 4721 ± 487 |
[0.008-0.022] −0.278 ± 0.494 p = 0.5894 |
[2.02-7.95%] 6.01 ± 0.57% |
[2.18-9.01%] 6.80 ± 0.68% |
[1780-6780] 4932 ± 476 |
[0.252-1.716] −0.811 ± 0.202 p = 0.0050 |
| Primata | [1.84-8.26%] 3.75 ± 0.69% |
[2.82-14.03%] 5.48 ± 1.27% |
[1991-8294] 3692 ± 722 |
[0.053-0.225] −0.569 ± 0.298 p = 0.1044 |
NA | NA | NA | NA | NA | NA | NA | NA |
aI% all cells, Microglial percentage of all cells; I% O cells, microglial percentage of non-neuronal (other) cells; I/mg, number of microglial cells per milligram of structure; I/N, microglia/neuron ratio.
However, within the non-neuronal cell population in each structure, microglial cells are a much less variable subpopulation in all brain structures, species, and clades examined, representing ∼7% of all non-neuronal cells in any structure (Table 5; Fig. 5F). Even if the exact percentage per structure varies from 2.18% in the Tasmanian devil's RoB to 24.56% in the pig Cb (Fig. 5E), the distribution of the percentages of non-neuronal cells that are microglial spans a very similar range across different structures and clades (Fig. 5E), and average percentages do not vary much across structures and clades (Fig. 5F,G). Moreover, the percentage of microglial cells among non-neuronal cells does not correlate significantly with the density of neurons in each structure as shown in Figure 5H (Spearman ρ = −0.2190, p = 0.0525). Thus, as brain structures vary in size, number of neurons, and neuronal density across mammalian species and clades, microglial cells appear to remain a fairly constant percentage of all non-neuronal cells.
Similar densities of microglial cells across brain structures and mammalian species
Like the percentage of non-neuronal cells that are microglial, average microglial cell densities are remarkably similar across brain structures and mammalian species of all clades: 4374 ± 341 microglia/mg in the Cx (gray and white matter combined, including hippocampus), 5021 ± 406 microglia/mg in the Cb (which also combines gray and white matter), and 4664 ± 464 microglia/mg in the RoB, with an overall average of 4667 ± 230 microglia/mg (all pairwise comparisons, Wilcoxon p > 0.1; Fig. 6A, middle, bottom; Table 5). In each of these structures, the coefficient of variation of microglial cell density is <0.10, both across species and across clades (Table 5). Whereas neuronal densities vary by >1000-fold across brain structures and mammalian species combined, microglial cell density varies by only 5-fold to 8-fold (Fig. 6E; Tables 1, 5), similar to densities of non-neuronal cells as a whole (which include astrocytes, oligodendrocytes, and vasculature-associated cells) (Herculano-Houzel and Dos Santos, 2018). When all brain structures, species, and clades are pooled together, there is no systematic variation in the density of microglial cells in the major brain divisions analyzed with variation in the total number of microglial cells in them (Spearman ρ = 0.1435, p = 0.2071; Fig. 6B). Similarly, no correlation is observed between those variables across species and clades when each structure is analyzed individually (Spearman, p = 0.3649 for Cx, p = 0.3076 for Cb, and p = 0.1351 for RoB). Thus, microglial cells appear to be added in highly variable numbers but fairly similar densities to all major brain structures, within and across brains and species.
Figure 6.
Constant average microglial density across brain structures, mammalian clades, and species. A, Distribution of microglial density in the different brain structures per clade (top), average across all samples (middle), and average per clade (bottom). Values are given in Table 5. Densities vary 8.5-, 5.0-, and 5.3-fold across species in Cb, Cx, and RoB, respectively (top; values in Table 1). Invariant average microglial density across brain structures in all mammalian species together (middle; Wilcoxon p values >0.01 for each pair) and across mammalian clades (bottom; Wilcoxon p values across clades are >0.01 across brain structures and clades). B, Relationship between the density of microglial cells and the number of microglial cells across brain structures and mammalian species. p values for Spearman correlations when all structures are analyzed together, except for Primata, Cx only, are all >0.01. C, Relationship between structure mass and number of microglial cells across brain structures and mammalian species. Function plotted in C applies to all structures and species with exponent (0.962 ± 0.024, p < 0.0001, r2 = 0.952). p values for each clade separately are all p < 0.0001. p values for Spearman correlations all fall to <0.01. D, Relationship between microglial density and structure mass across brain structures and mammalian species. p values for Spearman correlations when all structures are analyzed together, except for Primata, Cx only, are all superior to 0.01. E, Relationship between microglial density and neuronal density across brain structures and mammalian species. p values for Spearman correlations when all structures are analyzed together, except for Primata, Cx only, are all nonsignificant (p > 0.01). D, E, Each data point represents the values found per brain structure examined for each species colored by clade as in the key. Filled circles represent the Cx (including the hippocampus). Squares represent the Cb. Triangles represent the RoB. C, Values are exponent ± SE.
Further support of no significant variation in microglial density across brain structures and species is the linear relationship we find between structure mass and the number of microglial cells (Fig. 6C; exponent, 0.962 ± 0.024, p < 0.0001). This linear relationship approximates that of the distribution of non-neuronal cells as a whole for all mammalian species analyzed previously (exponent, 1.056 ± 0.013, p < 0.0001) (Dos Santos et al., 2017; Jardim-Messeder et al., 2017). The linearity of this relationship implies that brain structures of similar size are composed of similar numbers of microglial cells across different mammalian brains, whatever the structure identity, species, and even clade. Similar linear relationships between structure mass and number of Iba+ cells are also found across structures and species within each clade (p < 0.0001; Fig. 6C).
Similarly, there is no significant correlation between microglial cell density and structure mass across species and clades for any particular brain structure (Spearman, p = 0.9117, p = 0.8753, and p = 0.6103 for Cx, Cb, and RoB, respectively), and also no correlation across species within or across clades when all structures are considered together (Fig. 6D). Despite the >1000-fold variation in neuronal density in the dataset, microglial cell density in each major brain structure is not correlated with neuronal density across structures and species neither within nor across clades (Fig. 6E). Thus, as brain structures vary in size, number of neurons, and neuronal density across mammalian species and clades, microglial cells both remain a fairly constant percentage of all non-neuronal cells and occur at fairly constant densities in each brain and its various structures (Fig. 6D,E).
Microglia/neuron ratio scales universally with neuronal density across structures and species
As a consequence of the very large variation in neuronal densities with little variation in microglial density across brain structures in the different species, the number of microglial cells found per neuron in each structure (the I/N ratio) is enormously variable, from >100 neurons per microglial cell in the Cb of several species to ∼3 microglial cells per neuron in the Cx or RoB of large nonprimate brains (Fig. 7A; Tables 1, 5). Typically, there are more neurons than microglial cells in most brain structures, with an exact I/N ratio that correlates strongly with the local density of neurons (Fig. 7B). In contrast, I/N ratios vary only loosely with structure mass across species and clades taken together (Fig. 7A). Finally, and as expected, the I/N ratio is highly correlated with the total numbers of non-neuronal cells per neuron (O/N; Spearman ρ = 0.9479, p < 0.0001), and consistently with the finding that microglial cells are a fairly constant percentage of all non-neuronal cells in any brain structure and species, the ratio I/N can be described as a power function of O/N with an exponent that is indistinguishable from linearity (1.020 ± 0.031, p < 0.0001, r2 = 0.932; Fig. 7C). These findings thus demonstrate that, with a relatively constant density of microglial cells, brain structures with low neuronal densities (and thus large neurons) (Mota and Herculano-Houzel, 2014) have more microglial cells per neuron, whereas brain structures with high neuronal densities (and thus small neurons) have few microglial cells per neuron.
Figure 7.
Microglial cell/neuron ratio varies uniformly with neuronal density but not with structure mass across brain structures and mammalian species. Microglial cell/neuron ratio is plotted as a function of the brain structure mass (A), neuronal density (B), or non-neuronal cell/neuron ratio (C). A, p values for Spearman correlations when all structures are analyzed together, except for Primata, Cx only, fall below p < 0.01 for all clades together and individually, except for Carnivora (p = 0.0483; not significant), Primata (p = 0.3851), and Artiodactyla (p = 0.2453). Spearman p values when Cb is analyzed individually are nonsignificant, except when all clades are analyzed together (p = 0.0006). Spearman p values when Cx is analyzed individually are nonsignificant, except when all clades are analyzed together (p = 0. 0015) and for Afrotheria (p < 0.0001). Spearman p values when RoB is analyzed individually are nonsignificant, except when all clades are analyzed together (p < 0.0001). B, p values for Spearman correlations when all structures are analyzed together, except for Primata, Cx only, fall below p < 0.01 for all clades together and individually, except for Primata (p = 0.1600) and Artiodactyla (p = 0.0016). The power function plotted in B has an exponent of −0.973 ± 0.029 (p < 0.0001; r2 = 0.936) and applies to all mammalian species and brain structures. Exponents and p values for each clade individually are listed in Table 5. C, p values for Spearman correlations when all structures are analyzed together, except for Primata, Cx only, fall all below 0.01, except for Primata (p = 0.4198). The power function plotted in C has an exponent of 1.020 ± 0.031 (p < 0.0001; r2 = 0.932) and applies to all brain structures and mammalian species. All clades have a significant relationship, except Primata (p = 0.2252). Each data point represents the values found per brain structure examined for each species colored by clade as in the key. Filled circles represent the Cx, including the hippocampus. Squares represent the Cb. Triangles represent the RoB. B, C, Values are exponent ± SE.
Microglial cell numbers and densities in the whole brain
Across the 23 species examined for which all brain structures were available, total brain mass varies by 5038-fold, and the total estimated number of microglial cells in the brain varies by a fairly similar 7749-fold (Table 1). In contrast, and as previously reported, numbers of brain neurons vary by only 2300-fold across the same species. Overall, microglial cells amount to 4.42 ± 1.65% of all cells and to 7.60 ± 2.06% of non-neuronal cells in the whole brain.
Microglial cell densities in the brain as a whole vary <3-fold across species of all clades (Table 1), with an overall average of 4658 ± 371 microglia/mg. We find no differences in average microglial cell density across clades nor across species within each clade (Wilcoxon, all p > 0.05). However, because neuronal densities are enormously variable, the I/N ratio calculated for the whole brain varies in our dataset between 3 neurons per microglial cell in the giraffe brain and 45 neurons for every microglial cell in the elephant shrew brain (Table 1).
Discussion
Here we find that microglial cells occur in fairly similar and low densities across brain structures as different as multiple sites along the anteroposterior axis of the cerebral cortical gray and white matter; the Cb; the remaining structures in the hindbrain, midbrain, and forebrain; and across a wide sample of mammalian species that diverged as much as 218 million years ago (Murphy et al., 2004; Meredith et al., 2011). Finding consistent microglial cell densities across brain structures in such a wide range of mammalian species is indicative of little variation in the average size of these cells, as found previously for macrophages across species (Haley et al., 1991; Krombach et al., 1997). The unlikelihood that such consistent microglial cell densities occur by chance across so many brain structures and species indicates that microglial cell size and distribution have been conserved in mammalian evolution, whether due to biological, physiological, or physical reasons. An example of biological constraint would be the existence of fundamental morphologic, genetic, or regulatory mechanisms that constrain cell size, which may have been lifted only in some cell types, such as neurons (Savage et al., 2007; Herculano-Houzel and Dos Santos, 2018). There might also be physiological constraints in place, for instance, due to the volume-based macrophage-like functions of microglial cells, that render unviable any major variation in their size and/or distribution. Additionally, if microglial cell size is constrained and microglial cell proliferation is self-controlled such that it is arrested by contact inhibition, when confluence is reached in any tissue during development, a physical (mechanical) constraint to microglial cell distribution will follow, resulting in fairly constant microglial cell densities, as observed here. In support of such paired biological-physical constraints to microglial cell densities, contact inhibition has been reported during microglial progenitor cell proliferation (Gomez-Nicola and Perry, 2015; Askew et al., 2017) as well as during active scanning of the parenchyma, maintaining the mosaic distribution of microglial cells in the tissue (Nimmerjahn et al., 2005). Such a constant mosaic (tiled) distribution of microglial cells is also consistent with our finding of fairly constant densities of microglial cells across brain structures and species. Consequently, each microglial cell seems to take care of a territory of fairly similar volume across all brain structures and mammalian species.
Importantly, the remarkable constancy in microglial density reported here does not imply that microglial cells are functionally similar or equivalent across structures, and there is evidence to the effect that they are not (Xavier et al., 2015). It remains to bedetermined whether microglial cell densities are also constant across brain structures and species during development. While developmental trajectories appear to differ across mammalian species (Ueno and Yamashita, 2014; Matcovitch-Natan et al., 2016; Hanamsagar et al., 2017), the process of embryonic colonization by microglia is highly conserved across vertebrate species (Perry et al., 1985; Ashwell, 1991; Herbomel et al., 2001; Verney et al., 2010; Schlegelmilch et al., 2011; Swinnen et al., 2013). In mice, total numbers of microglial cells were found to peak at 3 weeks after birth and to decrease thereafter, showing similar densities at 1 and 9 months of age (Nikodemova et al., 2015). Aging has been shown in several species to affect microglial cell morphology and soma size, but not microglial cell density (Davies et al., 2017; Chan et al., 2018; Ali et al., 2019), which is consistent with our findings of relatively similar microglial densities across a wide range of species whose chronological age we cannot ascertain due to the opportunistic nature of our specimen collection.
Because microglial densities are similar while neurons are distributed unevenly and variably across brain structures and species, it follows that the number of neurons in the territory of each microglial cell is widely dissimilar across structures and species. Here we find that there is only rarely more than one microglial cell to each neuron in the brain; typically, there are one or more neurons per microglial cell in any brain tissue, in any mammalian species. As a consequence, to the extent that the physiological load on microglial function depends on the number of neurons served or monitored, microglial function should be more strained where there are more neurons per microglial cell (i.e., where neuronal density is higher), for instance, in the granular layers of the hippocampus and Cb, and in the densest regions of the Cx, such as entorhinal cortex. It is thus possible that the high vulnerability of these regions to neurodegenerative diseases, such as Alzheimer's disease (Braak et al., 1993; Larner, 1997; Mu and Gage, 2011; Jacobs et al., 2018), is at least partly associated with a high susceptibility to inflammation given their heavy neuronal load per microglial cell. Given the evidence that aging affects microglial function and morphology but not densities, as mentioned above, compensatory alterations are unlikely, such that different neuron/microglia ratios across brain sites probably have a lasting role in establishing susceptibility of different brain regions to insults over the lifespan. Similarly, one could expect the Cx to be more susceptible to inflammation in mammalian species with higher neuronal densities, such as mice and rats, compared with the human Cx. In that case, the laboratory rabbit, with neuronal densities closer to those found in humans (McMullen et al., 1994), would be a better model to study inflammation in the human cortex.
We report microglial percentages among non-neuronal cells in mammalian cortices that are comparable with the 5% found previously in the human cortical gray matter (Pelvig et al., 2008). Strikingly, not only the density of microglial cells is fairly constant across brain structures and species, but they also represent a fairly constant 7% of all non-neuronal cells. The latter include vasculature-related cells, which are possibly also a fairly constant (and small) percentage of all non-neuronal cells, given that capillary density appears to be consistently low across the few species sampled so far (Lauwers et al., 2008; Ventura-Antunes and S.H.-H., manuscript in preparation). A constant microglial percentage of all non-neuronal cells is consistent with neural tissue being composed of fixed percentages of astrocytes and oligodendrocytes as well (Herculano-Houzel and Dos Santos, 2018). Importantly, constant densities of each non-neuronal cell type in any brain structure and species would follow naturally if the average cell size of each cell type were invariable (Savage et al., 2007). Alternatively, the lack of much variation in cell density might be particular to microglial cells, since they do not originate from the same precursors that give rise to neurons and macroglial cells (astrocytes and oligodendrocytes) during development (for review, see Herculano-Houzel and Dos Santos, 2018). Our ongoing studies estimating directly the densities of astrocytes and oligodendrocytes will address whether or not macroglial cells also occur in similar densities across brain structures and species. Importantly, microglial cells are no exception in occurring at invariant densities and thus presumably constant average cell size: this seems to be the case of most somatic cell types other than neurons (with the possible exception of alveolar macrophages and adipocytes) (Savage et al., 2007). Neurons might thus be one of the rare cell types that escape the limitations that constrain cell size that have applied to other cell types over 200 million years of mammalian evolution.
We found that microglial densities do vary somewhat between cortical gray and white matter, with higher densities found in the latter, which might be simply a direct consequence of the scarcity of neuronal cell bodies in the cortical white matter. A higher density of microglial cells in white than in gray matter agrees with previous results in the adult rat (Savchenko et al., 2000), mouse (Askew et al., 2017), and human brain (Mittelbronn et al., 2001; Askew et al., 2017). Our finding that similar proportions of all cells are microglial between the gray and white matter is in conflict with previous results where authors found either higher proportions of microglia in the gray matter in the mouse Cx (Lawson et al., 1990), or in the white matter in the human Cx (Mittelbronn et al., 2001). While these differences in proportions might be due to the different technique applied in our study (the isotropic fractionator, instead of stereological methods) (see von Bartheld et al., 2016), they are most likely explained by the use in previous studies of antibodies that are now known to not stain all the microglial cell population (F4/80) (Lawson et al., 1990) or CD68, LCA, LCF, and MHC-II (Mittelbronn et al., 2001).
The highly variable ratio of neurons served per microglial cell that results from the fairly constant microglial cell densities uncovered here might have a very fundamental consequence for brain tissue function due to the role of microglial cells in synaptic pruning (Paolicelli et al., 2011; Schafer et al., 2012; Ueno et al., 2013; Li and Barres, 2018). While it is possible that constant microglial densities might be imposed by constant synaptic densities (which has been largely assumed to be the case, but for which solid, systematic evidence is still lacking) (O'Kusky and Colonnier, 1982; Miki et al., 1997), we find it much more likely that it is the fairly constant microglial densities reported here that contribute to imposing fairly constant synaptic densities across brain structures and species. One reason is the very early invasion of the developing parenchyma by microglial progenitors, well before formation of the first synapses (Ginhoux et al., 2010). Similarly, we expect that fairly constant densities of astrocytes would also induce fairly constant densities of synapses across brain structures and species. To date, the only systematic survey of synaptic densities was described recently for the adult mouse brain (Zhu et al., 2018). Only a systematic analysis of the distribution and densities of synapses in a large set of mammalian species will allow these fundamental questions about brain tissue organization to be addressed.
Footnotes
The authors declare no competing financial interests.
This work was supported by the Carlos Chagas Filho Foundation for Research Support of the State of Rio de Janeiro-FAPERJ (SDS fellowship), generous crowdfunding support by Brazilian supporters to the former S.H.-H. laboratory at Universidade Federal do Rio de Janeiro, the James S. McDonnell Foundation Grant 220020232 to C.C.S. and Grant 220020293 to S.H.-H., and Vanderbilt University laboratory startup funds to S.H.-H. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Anderson Miller and Louise Botelho for help in experiments; Mary Ann Raghanti for providing marsupial brains; and Laura Trice for sectioning the brains used for immunohistochemistry.
References
- Ali S, Liu X, Queen NS, Patel RS, Wilkins RK, Mo X, Cao L (2019) Long-term environmental enrichment affects microglial morphology in middle age mice. Aging 11:2388–2402. 10.18632/aging.101923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell K. (1991) The distribution of microglia and cell death in the fetal rat forebrain. Dev Brain Res 58:1–12. 10.1016/0165-3806(91)90231-7 [DOI] [PubMed] [Google Scholar]
- Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, Tipton T, Chapman MA, Riecken K, Beccari S, Sierra A, Molnár Z, Cragg MS, Garaschuk O, Perry HV, Gomez-Nicola D (2017) Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep 18:391–405. 10.1016/j.celrep.2016.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck R, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A (2007) The delayed rise of present-day mammals. Nature 446:507–512. 10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, Bohl J (1993) Staging of Alzheimer-related cortical destruction. Eur Neurol 33:403–408. 10.1159/000116984 [DOI] [PubMed] [Google Scholar]
- Chan TE, Grossman YS, Bloss EB, Janssen WG, Lou W, McEwen BS, Dumitriu D, Morrison JH (2018) Cell-type specific changes in glial morphology and glucocorticoid expression during stress and aging in the medial prefrontal cortex. Front Aging Neurosci 10:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, Noctor SC (2013) Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci 33:4216–4233. 10.1523/JNEUROSCI.3441-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DS, Ma J, Jegathees T, Goldsbury C (2017) Microglia show altered morphology and reduced arborization in human brain during aging and Alzheimer's disease. Brain Pathol 27:795–808. 10.1111/bpa.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos SE, Porfirio J, da Cunha FB, Manger PR, Tavares W, Pessoa L, Raghanti M, Sherwood CC, Herculano-Houzel S (2017) Cellular scaling rules for the brains of marsupials: not as “primitive” as expected. Brain Behav Evol 89:48–63. 10.1159/000452856 [DOI] [PubMed] [Google Scholar]
- Gabi M, Neves K, Masseron C, Ribeiro PF, Ventura-Antunes L, Torres L, Mota B, Kaas JH, Herculano-Houzel S (2016) No relative expansion of the number of prefrontal neurons in primate and human evolution. Proc Natl Acad Sci USA 113:9617–9622. 10.1073/pnas.1610178113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng L, Stanley RE, Samokhvalov IM, Merad M (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Nicola D, Perry HV (2015) Microglial dynamics and role in the healthy and diseased brain. Neuroscientist 21:169–184. 10.1177/1073858414530512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley P, Muggenburg B, Weissman D, Bice D (1991) Comparative morphology and morphometry of alveolar macrophages from six species. Am J Anat 191:401–407. 10.1002/aja.1001910407 [DOI] [PubMed] [Google Scholar]
- Hanamsagar R, Alter MD, Block CS, Sullivan H, Bolton JL, Bilbo SD (2017) Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65:1504–1520. 10.1002/glia.23176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C (2001) Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol 238:274–288. 10.1006/dbio.2001.0393 [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S. (2014) The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 62:1377–1391. 10.1002/glia.22683 [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S. (2017) 2153–2180 What modern mammals teach us about the cellular composition of early brains and mechanisms of brain evolution. In: Evolution of nervous systems (Kaas JH, Herculano-Houzel S, eds), pp 2153–2180. New York: Elsevier. [Google Scholar]
- Herculano-Houzel S, Dos Santos SE (2018) You do not mess with the glia. Neuroglia 1:14. [Google Scholar]
- Herculano-Houzel S, Lent R (2005) Isotropic fractionator: a simple, rapid method for the quantification of total cell and neuron numbers in the brain. J Neurosci 25:2518–2521. 10.1523/JNEUROSCI.4526-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Avelino-de-Souza K, Neves K, Porfírio J, Messeder D, Feijó L, Maldonado J, Manger PR (2014) The elephant brain in numbers. Front Neuroanat 8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Catania K, Manger PR, Kaas JH (2015a) Mammalian brains are made of these: a dataset of the numbers and densities of neuronal and nonneuronal cells in the brain of Glires, primates, Scandentia, Eulipotyphlans, Afrotherians and Artiodactyls, and their relationship with body mass. Brain Behav Evol 86:145–163. 10.1159/000437413 [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S, Messeder DJ, Fonseca-Azevedo K, Pantoja NA (2015b) When larger brains do not have more neurons: increased numbers of cells are compensated by decreased average cell size across mouse individuals. Front Neuroanat 9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, Khoury J (2018) Microglia in neurodegeneration. Nat Neurosci 21:1359–1369. 10.1038/s41593-018-0242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Ibata I, Ito D, Ohsawa K, Kohsaka S (1996) A novel geneiba1in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem Biophys Res Commun 224:855–862. 10.1006/bbrc.1996.1112 [DOI] [PubMed] [Google Scholar]
- Imai Y, Kohsaka S (2002) Intracellular signaling in M‐CSF‐induced microglia activation: role of Iba1. Glia 40:164–174. 10.1002/glia.10149 [DOI] [PubMed] [Google Scholar]
- Jacobs HI, Hopkins DA, Mayrhofer HC, Bruner E, van Leeuwen FW, Raaijmakers W, Schmahmann JD (2018) The cerebellum in Alzheimer's disease: evaluating its role in cognitive decline. Brain 141:37–47. 10.1093/brain/awx194 [DOI] [PubMed] [Google Scholar]
- Janušonis S. (2018) Some galeomorph sharks express a mammalian microglia-specific protein in radial ependymoglia of the telencephalon. Brain Behav Evol 91:17–30. 10.1159/000484196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim-Messeder D, Lambert K, Noctor S, Pestana FM, de Leal ME, Bertelsen MF, Alagaili AN, Mohammad OB, Manger PR, Herculano-Houzel S (2017) Dogs have the most neurons, though not the largest brain: trade-off between body mass and number of neurons in the cerebral cortex of large carnivoran species. Front Neuroanat 11:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabba J, Xu Y, Christian H, Ruan W, Chenai K, Xiang Y, Zhang L, Saavedra JM, Pang T (2018) Microglia: housekeeper of the central nervous system. Cell Mol Neurobiol 38:53–71. 10.1007/s10571-017-0504-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazu RS, Maldonado J, Mota B, Manger PR, Herculano-Houzel S (2014) Cellular scaling rules for the brain of Artiodactyla include a highly folded cortex with few neurons. Front Neuroanat 8:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Hanisch UK, Noda M, Verkhratsky A (2011) Physiology of microglia. Physiol Rev 91:461–553. 10.1152/physrev.00011.2010 [DOI] [PubMed] [Google Scholar]
- Krombach F, Münzing S, Allmeling A, Gerlach J, Behr J, Dörger M (1997) Cell size of alveolar macrophages: an interspecies comparison. Environ Health Perspect 105:1261–1263. 10.2307/3433544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner AJ. (1997) The cerebellum in Alzheimer's disease. Dement Geriatr Cogn Disord 8:203–209. 10.1159/000106632 [DOI] [PubMed] [Google Scholar]
- Lauwers F, Cassot F, Lauwers-Cances V, Puwanarajah P, Duvernoy H (2008) Morphometry of the human cerebral cortex microcirculation: general characteristics and space-related profiles. Neuroimage 39:936–948. 10.1016/j.neuroimage.2007.09.024 [DOI] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P, Gordon S (1990) Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39:151–170. 10.1016/0306-4522(90)90229-W [DOI] [PubMed] [Google Scholar]
- Li Q, Barres BA (2018) Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 18:225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Manger PR, Pillay P, Maseko BC, Bhagwandin A, Gravett N, Moon DJ, Jillani N, Hemingway J (2009) Acquisition of brains from the African elephant (Loxodonta africana): perfusion-fixation and dissection. J Neurosci Methods 179:16–21. 10.1016/j.jneumeth.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, et al. (2016) Microglia development follows a stepwise program to regulate brain homeostasis. Science 353:8670 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- McMullen NT, Smelser CB, de Venecia RK (1994) A quantitative analysis of parvalbumin neurons in rabbit auditory neocortex. J Comp Neurol 349:493–511. 10.1002/cne.903490402 [DOI] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, et al. (2011) Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334:521–524. 10.1126/science.1211028 [DOI] [PubMed] [Google Scholar]
- Miki T, Fukui Y, Itoh M, Hisano S, Xie Q, Takeuchi Y (1997) Estimation of the numerical densities of neurons and synapses in cerebral cortex. Brain Res Protoc 2:9–16. 10.1016/S1385-299X(97)00021-4 [DOI] [PubMed] [Google Scholar]
- Mittelbronn M, Dietz K, Schluesener H, Meyermann R (2001) Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol 101:249–255. 10.1007/s004010000284 [DOI] [PubMed] [Google Scholar]
- Mota B, Herculano-Houzel S (2014) All brains are made of this: a fundamental building block of brain matter with matching neuronal and glial masses. Front Neuroanat 8:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Gage FH (2011) Adult hippocampal neurogenesis and its role in Alzheimer's disease. Mol Neurodegeneration 6:85 10.1186/1750-1326-6-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ (2001) Molecular phylogenetics and the origins of placental mammals. Nature 409:614–618. 10.1038/35054550 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pevzner PA, O'Brien SJ (2004) Mammalian phylogenomics comes of age. Trends Genet 20:631–639. 10.1016/j.tig.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Neves K, Ferreira FM, Tovar-Moll F, Gravett N, Bennett NC, Kaswera C, Gilissen E, Manger PR, Herculano-Houzel S (2014) Cellular scaling rules for the brain of afrotherians. Front Neuroanat 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodemova M, Kimyon RS, De I, Small AL, Collier LS, Watters JJ (2015) Microglia numbers attain adult levels after undergoing a rapid decrease in cell number in the third postnatal week. J Neuroimmunol 278:280–288. 10.1016/j.jneuroim.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F (2005) Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308:1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- O'Kusky J, Colonnier M (1982) A laminar analysis of the number of neurons, glia, and synapses in the visual cortex (area 17) of adult macaque monkeys. J Comp Neurol 210:278–290. 10.1002/cne.902100307 [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira T, Guiducci E, Dumas L, Ragozzino D, Gross CT (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333:1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, Hempstead BL, Littman DR, Gan WB (2013) Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155:1596–1609. 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelvig DP, Pakkenberg H, Stark AK, Pakkenberg B (2008) Neocortical glial cell numbers in human brains. Neurobiol Aging 29:1754–1762. 10.1016/j.neurobiolaging.2007.04.013 [DOI] [PubMed] [Google Scholar]
- Perry VH, Hume DA, Gordon S (1985) Immunohistochemical localization of macrophages and microglia in the adult and developing mouse brain. Neuroscience 15:313–326. 10.1016/0306-4522(85)90215-5 [DOI] [PubMed] [Google Scholar]
- Ribeiro PF, Ventura-Antunes L, Gabi M, Mota B, Grinberg LT, Farfel JM, Ferretti-Rebustini RE, Leite RE, Filho WJ, Herculano-Houzel S (2013) The human cerebral cortex is neither one nor many: neuronal distribution reveals two quantitatively different zones in the gray matter, three in the white matter, and explains local variations in cortical folding. Front Neuroanat 7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH, West GB (2007) Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc Natl Acad Sci USA 104:4718–4723. 10.1073/pnas.0611235104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko VL, McKanna JA, Nikonenko IR, Skibo GG (2000) Microglia and astrocytes in the adult rat brain: comparative immunocytochemical analysis demonstrates the efficacy of lipocortin 1 immunoreactivity. Neuroscience 96:195–203. 10.1016/S0306-4522(99)00538-2 [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B (2012) Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74:691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch T, Henke K, Peri F (2011) Microglia in the developing brain: from immunity to behaviour. Curr Opin Neurobiol 21:5–10. 10.1016/j.conb.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Sousa C, Biber K, Michelucci A (2017) Cellular and molecular characterization of microglia: a unique immune cell population. Front Immunol 8:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen N, Smolders S, Avila A, Notelaers K, Paesen R, Ameloot M, Brône B, Legendre P, Rigo J (2013) Complex invasion pattern of the cerebral cortex by microglial cells during development of the mouse embryo. Glia 61:150–163. 10.1002/glia.22421 [DOI] [PubMed] [Google Scholar]
- Tremblay MÈ, Lowery RL, Majewska AK (2010) Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8:e1000527 10.1371/journal.pbio.1000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T (2013) Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci 16:543–551. 10.1038/nn.3358 [DOI] [PubMed] [Google Scholar]
- Ueno M, Yamashita T (2014) Bidirectional tuning of microglia in the developing brain: from neurogenesis to neural circuit formation. Curr Opin Neurobiol 27:8–15. 10.1016/j.conb.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Valério-Gomes B, Guimaraes DM, Szczupak D, Lent R (2018) The absolute number of oligodendrocytes in the adult mouse brain. Front Neuroanat 12:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verney C, Monier A, Fallet‐Bianco C, Gressens P (2010) Early microglial colonization of the human forebrain and possible involvement in periventricular white‐matter injury of preterm infants. J Anat 217:436–448. 10.1111/j.1469-7580.2010.01245.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bartheld CS, Bahney J, Herculano‐Houzel S (2016) The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J Comp Neurol 524:3865–3895. 10.1002/cne.24040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J (2009) Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci 29:3974–3980. 10.1523/JNEUROSCI.4363-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Miyamoto A, Nabekura J (2013) Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci 36:209–217. 10.1016/j.tins.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Watson C, Mitchelle A, Puelles L (2017) A new mammalian brain ontology based on developmental gene expression. In: Evolution of nervous systems (Kaas JH, Herculano-Houzel S, eds), pp 253–275. New York: Elsevier. [Google Scholar]
- Xavier AL, Kress BT, Goldman SA, de Menezes JR, Nedergaard M (2015) A distinct population of microglia supports adult neurogenesis in the subventricular zone. J Neurosci 35:11848–11861. 10.1523/JNEUROSCI.1217-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Cizeron M, Qiu Z, Benavides-Piccione R, Kopanitsa MV, Skene NG, Koniaris B, DeFelipe J, Fransén E, Komiyama NH, Grant S (2018) Architecture of the mouse brain synaptome. Neuron 99:781–799.e10. 10.1016/j.neuron.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]