Abstract
Emerging technologies are set to play an important role in our response to the COVID-19 pandemic. This paper explores three prominent initiatives: COVID-19 focused datasets (e.g., CORD-19); Artificial intelligence-powered search tools (e.g., WellAI, SciSight); and contact tracing based on mobile communication technology. We believe that increasing awareness of these tools will be important in future research into the disease, COVID-19, and the virus, SARS-CoV-2.
Key words: COVID-19, SARS-CoV-2, artificial intelligence, machine learning, contact tracing
INTRODUCTION
The COVID-19 pandemic has created unprecedented challenges for the medical and clinical diagnostic community. The fight against COVID-19 is being supported by a number of databases and artificial intelligence (AI)-based initiatives aimed at assessing dissemination of the disease [1], aiding in detection and diagnosis, minimizing the spread of the disease, and facilitating and accelerating research globally [2-7].
Prominent among these initiatives are: the COVID-19 Open Research Dataset (CORD-19) [8-10], and databases curated by the CDC [11,12], NLM [13], and the WHO [14]; AI-powered tools such as those from WellAI [15,16] and the Allen Institute for AI (SciSight) [17-19]; and contact tracing based on mobile communication technology [20,21].
CORD-19
The CORD-19 Dataset has resulted from a partnership between the Semantic Scholar team at the Allen Institute for AI and leading research groups (Chan Zuckerberg Initiative, Georgetown University’s Center for Security and Emerging Technology, Microsoft Research, the Kaggle AI platform (Google), and the National Library of Medicine–National Institutes of Health) in coordination with the White House Office of Science and Technology Policy.
Publications in the collection are sourced from PubMed Central, the bioRxiv and medRxiv preprint servers, and the WHO COVID-19 Database. CORD-19 is freely available, downloadable and it is updated weekly. The collection currently contains over 128,000 publications (with over 59,000 full text as of 26 May 2020) on the disease, COVID-19, and the virus, SARS-CoV-2, and related coronaviruses. It is part of a call to action to the AI community to develop AI techniques in order to generate new insights to assist in the fight against COVID-19 [9]. This call to action has been informed by a series of tasks described in the form of a series of questions that are listed in Table 1 [22].
Table 1.
COVID-19 Open Research Dataset Challenge (CORD-19) – tasks
| What is known about transmission, incubation, and environmental stability? |
| What do we know about COVID-19 risk factors? |
| What do we know about virus genetics, origin, and evolution? |
| What do we know about vaccines and therapeutics? |
| What has been published about medical care? |
| What do we know about non-pharmaceutical interventions? |
| What do we know about diagnostics and surveillance? |
| What has been published about ethical and social science considerations? |
| What has been published about information sharing and inter-sectoral collaboration? |
AI-POWERED SEARCH TOOLS
Analysis of the vast amount of COVID-19 data that has already accumulated (e.g., CORD-19 Dataset, COVID-19 cases data, Hospital Data and case statistics) [23] is a daunting challenge, however, this big data type of problem is amenable to AI-based search tools [24] such as those from WellAI and the Allen Institute for AI (SciSight). There are several advantages of AI-powered tools that exploit natural language processing (NLP) compared to a conventional search engine, e.g., unlocking buried information [25-27], and these are summarized in Table 2.
Table 2.
Comparison of machine learning tools based on NLP and a conventional search engine
| AI-powered search tool based on NLP (e.g., WellAI) |
A publication search engine (e.g., PubMed) |
|
|---|---|---|
| General objective | Neural networks summarize, generalize and predict relationships | Searches for key words and phrases in an article. Cannot make conclusions about relationships. |
| Synonyms (correlated concepts) | Understands synonyms and correlated concepts. For example, understands that “hypertension” is a synonym for “high blood pressure” and “elevated blood pressure”. This knowledge helps build more accurate relationships between concepts. | The results produced match the search words or phrases, without knowledge of synonyms and related concepts. |
| Result aggregated and summarized? | Yes. Every single concept suggestion is based on a large number of articles. | No. The result is a list of articles that contain the key words or phrases. |
| Output & next step | A structured list of concepts with ranked probabilities. This narrows the scope of work and results in greater efficiency. Focus on concepts of interest and exploration of relationships - not only between concepts (e.g., COVID-19 and Diagnostics Radiology), but between clusters of concepts (e.g., COVID-19 + Diagnosis, Clinical + Diagnostic Tests and Diagnostics Radiology) |
A list of every single occurrence (i.e., every article) of a word or a phrase. Read the articles (time consuming), summarize, and make generalizations. |
| Example | Starting with “COVID-19” as the preselected concept, selecting “READ ARTICLES” corresponding to “Diagnosis, Clinical” produces a list of articles in which the machine learning models have determined there is a relationship between COVID-19 and clinical diagnosis, and not just the whole list of articles that mentions both COVID-19 and clinical diagnosis. In addition, the models know there is a difference between clinical diagnosis and diagnosis. | The result for search terms “COVID-19” and “clinical diagnosis”, is a list of all articles that mention “COVID-19” and “clinical diagnosis” irrespective of whether there is a relationship between the two phrases mentioned in the article. For example, hypothetically speaking, the article may not be about clinical diagnosis at all, the phrase “Clinical diagnosis” may be just mentioned in the References section. |
WellAI
WellAI has developed a Machine Learning (ML) search and analytics tool, based on four neural networks and incorporating the complete list of NIH medical categories [Unified Medical Language System (UMLS)] semantic types, for interrogation of the CORD-19 Dataset and this is available at https://wellai.health/covid/ [16]. It is now widely agreed that ML has significant applications in the physical and biological sciences [28]. In the WellAI COVID-19 application, a subset of ML -- i.e. neural networks – is being used. Neural networks facilitate discovery of highly complex and nonlinear relationships between sets of variables without having to search for a closed form mathematical solution. Neural networks can contain tens of thousands to millions of variables, and this is the basis of their power. The complexity of relationships neural networks can uncover is difficult to fathom but is enabled by an ever-increasing computing power. Somewhat surprisingly, one of the biggest trends of the past 10 years is the increasing scientific role of neural network models of a language. At first glance it seems counterintuitive that something so qualitative and subjective as language, plays a role in learning about physical or biological sciences, which by their nature strive for precision. However, NLP is set to play a major role in scientific learning over the coming decades, because arguably the biggest ‘problem’ for scientists today is an ever-growing body of data, which defies any traditional tools of comprehension [29]. For example, the CORD-19 dataset already contains >128,000 articles. Digesting such a vast amount of information quickly can only be done by the NLP methods and can extend beyond capturing “known knowledge” and reveal new information and hidden connections [27].
The WellAI COVID-19 application uses NLP neural networks to ‘learn’ from the CORD-19 dataset in order to summarize existing knowledge. It can also be used to make discoveries in an unsupervised manner. This application is based on unsupervised learning [19, 20], but its main goal is to enable a researcher to generate ideas for the next set of concepts that are relevant to the discovery. The UMLS concepts are used as variables in the model and these concepts provide a vast terminology. Crucially, they deal with synonymy, and by including all of the synonyms, the number of UMLS concepts increased to 4,224,512! Only 60,892 concepts are used in the WellAI COVID-19 model, grouped into 69 categories (or UMLS semantic types). Broader WellAI models are based on >25 million medical articles and use millions of concepts.
These concepts are a helpful starting point. However, they had to be altered for WellAI models because they are somewhat outdated, specifically when it comes to the terminology surrounding the novel coronavirus. The altered concepts were applied to the CORD-19 dataset. This whole process was not trivial because application of concepts requires context. Different words can mean different things in different contexts. Complex ML models sensitive to the context of an article needed to be developed. A series of WellAI neural network models have been utilized to learn relationships between medical concepts. Relationships of any single concept to a set of concepts along with probabilities (strength of the relationship) is routine. However, it is more difficult to work with a group of concepts as inputs, especially if the number of variables is not constant. A researcher may use any number of concepts as a starting point of their research and a model was developed that can accept any number of concepts as inputs and update predicted related concepts, along with actionable probabilities.
At a practical level, searching the CORD-19 Data-set using the WellAI tool begins with the results of the initial analysis, based on COVID-19 and SARS Coronavirus as the preloaded concepts, and this produces a list of 69 concept categories. Each concept category has an associated list of concepts, ranked according to their significance in relation to COVID-19 based on log probability (or negative log likelihood loss) [30] of the strength of the concept relationship to COVID-19, according to the WellAI neural networks.
For clinical diagnostics there are several relevant major concept categories in the list, including: “Diagnostic Procedure”; “Laboratory Procedure”; “Laboratory or Test Result”.
Associated with each major concept category is a list of related concepts, each linked to relevant publications (“READ ARTICLES”). The search can be refined by adding any of the concepts to the “Selected Concepts” list. A rerun of the search (“Find by selected concepts” option) produces the new lists of concepts that are most related to the new list of “Selected Concepts” (Figure. 1).
Figure 1.
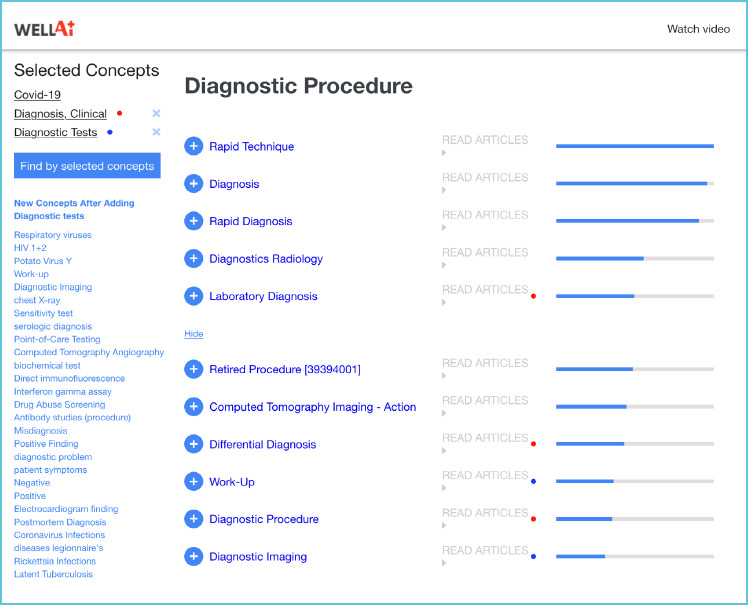
Example of WellAI search results for the combination of three concepts - “Covid-19” and “Diagnosis, Clinical” and “Diagnostic tests”
Underlying this AI-powered tool is a network of servers that make the searching quick and seemingly effortless. Significantly, most of the questions in Table 1 could be answered by the WellAI COVID-19 tool by entering a concept (e.g., transmission mode) or looking at the relevant concept category (e.g., “Gene or Genome” for virus genetics and virus origin question).
SciSight
SciSight is an AI-powered visualization tool for exploring associations between concepts appearing in the CORD-19 Dataset and visualizing the emerging literature network around COVID-19 [17-19,31]. It is available at: https://SciSight.apps.allenai.org/ [17]. SciSight is based on SciBERT, a pretrained language model, trained on a large corpus of scientific publications, to provide improved performance in natural language processing [32]. Initially, SciSight provides four different search options, namely, two scientific concepts that are important to the study of the virus, “Proteins/genes/cells” and “Diseases/chemicals”, and a “Network of Science” search and a “Faceted search”.
The user can explore associations between either of two preselected scientific concepts – “Proteins/genes/cells” or “Diseases/chemicals” in the CORD-19 Dataset as follows. Selection of one of the preselected concepts displays the “Try:” list below the search box, and this lists salient keywords with high relevance to SARS-CoV-2. There is also a graphical display of the network of associations between the preselected scientific concept and the top related terms mined from the Dataset. The thickness of the edges signifies that terms are co-mentioned more often in close proximity to each other in publications in the database. Clicking on an edge reveals the list of linked full text papers and hovering over a term reveals co-mentioned terms. This is illustrated in Figure 2 for the associations between the preselected concept “diseases/chemicals” and the key words “virus infection” selected from the “Try:” list. Alternatively, one of two preselected scientific concepts can be chosen, and a search term entered. This generates and displays a list of autocompleted search suggestions. Selecting one of these suggestions again displays the network of top associations in the dataset.
Figure 2.
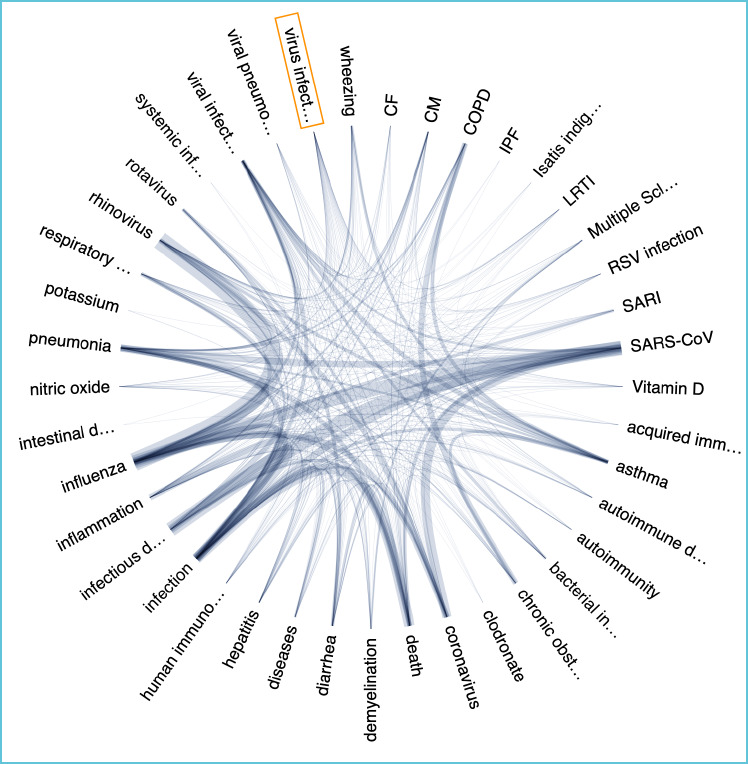
Example of SciSight search results for combination of “diseases/chemicals” associated with “virus infection”
A “Network of science” search option allows the user to visualize research groups and their ties in the context of COVID-19. Searches can be by “Topics”, “Affiliation” or “Authors” or by the seven preloaded topics in the “Try:” list. Multiple combinations of “Topics” “Affiliation” or “Authors” can be selected. Results are shown as a network of boxes that are color coded from high to low relevance. Each box shows top authors, top affiliations and top topics in a group, and the color-coded links between boxes reveal shared authors or topics. Selection of a box provides a list of publications relating to the contents of that particular box. Also, results are ranked within each topic category (e.g., “Author”) by means of a shaded bar.
Another search option is “Faceted search”. This reveals how authors and topics interact over time in the context of COVID-19. Searches can be made by selecting combinations of Author, Co-author, Characteristic, Intervention, Outcome, Journal, License or Source, and/or by selecting one of seven preloaded topics in the “Try:” list. Multiple combinations of Topics, Affiliation, or Authors can be selected. Results are ranked within each topic category (e.g., Author) by means of a shaded bar and a list of relevant publications and a graphic shows the number of papers per year.
DIGITAL CONTACT TRACING
Population-wide datasets are now emerging that show the response of society to COVID-19. The data includes commonly used terms in internet search engines, satellite mapping data of human activity and the emerging interactive data from digital contact tracing. Contact tracing is an essential monitoring process for combating the spread of an infectious disease [19-21]. It comprises three basic steps: 1) Contact identification; 2) Contact listing; and, 3) Contact follow-up - and it forms one part of the “Test, Trace and Quarantine” mantra. Conventionally, contact tracing is a manual process relying on finding individuals who have tested positive, and then interviewing those individuals to identify all individuals who need to be quarantined. The widespread availability of mobile communication technology (e.g., smartphones) is providing new ways of enabling contact tracing by using Bluetooth to track nearby phones, keep logs of those contacts, and to warn people about others with whom they have been in contact. In the digital age, contact tracing can be passively achieved and integrated with diagnostic testing results. On an individual level, the actions can be bi-directional. An individual can test positive and then initiate a cascade of notifications of all recent contacts. Alternatively, an individual can be notified that they were in Bluetooth proximity to an anonymous person who has tested positive. Public health authorities empowered with digital tracing can quickly identify positive contacts with a minimal workforce.
In the US, Apple and Google are collaborating on tracking technology for iOS and Android smartphones [33]. Elsewhere in the world, an example of a contact tracing app is Trace Together which has been deployed in Singapore [34,35]. If a person is found to be positive for COVID-19, then the app uses a smartphone’s Bluetooth network to notify every participating Trace Together user that person was within 2 meters of for more than 30 minutes.
In China, the Alipay Health Code on the Alipay app dictates freedom of travel based on three categories: green for unrestricted travel, yellow for a seven-day quarantine, and red for a two-week quarantine [36]. In South Korea, people receive location-based emergency text messages from the government to inform them if they have been in the vicinity of a confirmed case of COVID-19 [37]. In Italy the app “Immuni” [38,39] combines a personal clinical diary and contact tracing. Anonymous identification codes are generated by the user’s app rather than a central server in order to improve privacy. By placing identification on the individual user’s device, the contact tracing information is separate from identification. The App complies with the European model outlined by the PEPP-PT (Pan European Privacy-Preserving Proximity Tracing) consortium [40]. It is delivered for free and on a voluntary basis. There has been resistance to app-based monitoring [39], but the Italian government expect 60-70% of people will download the app. In the UK, a contact tracing app (NHS COVID-19) is currently being trialed in a limited geographical area with a population of ~140,000 [41]. This app registers duration and distance between devices and the data is fed into a centralized system where a risk algorithm estimates infection risk and triggers notifications.
Other examples of pandemic data infrastructures include the Google tool, COVID Near You, to identify patterns and hot spots by location (zip-code) [42]; COVID Trace [43] that warns of exposure to COVID-19 by comparing your locations over the previous 3 weeks against the time and locations of reported exposures; CoronApp, which provides localized, real-time data about COVID-19 based on the geographic location of their smartphones [44]; and a hashtag tracking tool for the evolution of COVID hashtags on Twitter (>628 million tweets about COVID-19) [45]. Twitter is also being used to understand the impact of COVID-19 (e.g., psychological impact) [46]. One significant concern over digital contact tracing has been ethical issues (e.g., privacy) and the consequent impact on the rate of adoption of the apps [47,48]. Some technology developers are focused on developing tracing apps that ensure privacy protection [49].
Currently, in response to COVID-19, clinical laboratories and the IVD industry are grappling with test development, test validation, fast-track clearance (e.g., Emergency Use Authorization) [50], availability of analyzers, tests and related supplies, and testing capacity for both molecular tests for SARS-CoV-2 and tests for IgM/IgG antibodies against this virus [51,52]. Once these issues have been resolved, the next major hurdle will be contact tracing to reduce the risk of future outbreaks. AI-powered tools will be valuable to identify trends and associations between digital contact tracing, tests and outbreaks of disease.
CONCLUSIONS
Easily accessible AI-powered tools and databases are valuable in all types of research, but especially so, in the context of the urgent diagnostic and therapeutic challenges presented by the COVID-19 pandemic. It is hoped that the new AI-powered search tools will accelerate research and development in COVID-19 as the world strives to develop efficient and timely testing and effective therapies to combat this disastrous pandemic. Another important part of our fight against COVID-19 will be efficient digital contact tracing enabled by mobile communication technology linked with massively scaled-up testing as outlined in the recent “Roadmap to Pandemic Resilience” [53].
REFERENCES
- 1.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med 2020;1–3 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaisha R, Javid M, Khan IH, Haleem A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab Syndr Clin Res Rev 2020;14:337-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alimadadi A, Aryal S, Manandhar I, Monroe PB, Joe B, Cheng X. Artificial intelligence and machine learning to fight COVID-19. Physiol Genom 2020;52:200-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting DSW, Carin L, Dzau V, Wong TY. I. Digital technology and COVID-19. Nat Med 2020;26:458-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mashamba-Thompson TP, Crayton EB. Blockchain and artificial intelligence technology for novel coronavirus disease 2019 self-testing. Diagn 2020;10:198 doi:10.3390/diagnostics10040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allam Z, Jones DS. On the coronavirus (COVID-19) outbreak and the smart city network: Universal data sharing standards coupled with artificial intelligence (AI) to benefit urban health monitoring and management. Healthcare 2020;8:46 Doi:10.3390/healthcare8010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao ASR, Vazquez JA. Identification of COVID-19 can be quicker through artificial intelligence framework using a mobile phone-based survey in the populations when cities/towns are under quarantine. Infect Control Hosp Epidemiol 2020:March 3;1-5. DOI: 10.1017/ice.2020.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen Institute for AI. COVID-19 Open Research Dataset (CORD-19). https://pages.semanticscholar.org/coronavirus-research. [Accessed May 26, 2020].
- 9.Office of Science and Technology Policy. Call to action to the tech community on new machine readable COVID-19 dataset. March 16, 2020. https://www.whitehouse.gov/briefings-statements/call-action-tech-community-new-machine-readable-covid-19-dataset/. [Accessed May 26, 2020].
- 10.Wang LL, Lo K, Chandrasekhar Y, Reas R, Yang J, Eide D, Funk K, Kinney R, Liu Z, Merrill W, Mooney P, Murdick D, Rishi D, Sheehan J, Shen Z, Stilson B, Wade AD, Wang K, Wilhelm C, Xie B, Raymond D, Weld D S, Etzioni O, Kohlmeier S. CORD-19: The COVID-19 Open Research Dataset. arXiv:2004.10706 [cs.DL]. [Google Scholar]
- 11.Stephen B. Thacker CDC Library. COVID-19 research articles downloadable data base, https://www.cdc.gov/library/researchguides/2019novelcoronavirus/research-articles.html. [Accessed May 26, 2020].
- 12.CDC. COVIDView. A weekly surveillance summary of U.S. COVID-19 activity, https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html, [Accessed May 26, 2020].
- 13.NIH. NLM. LITCOVID. General Info. https://www.ncbi.nlm.nih.gov/research/coronavirus/docsum?filters=topics.General%20Info. [Accessed May 26, 2020].
- 14.WHO. Global research on coronavirus disease (COVID-19), https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov. [Accessed May 26, 2020].
- 15.WellAI develops COVID-19 research tool in response to White House’s call to action. PR Newswire. April 8, 2020. [Accessed May 26, 2020]. [Google Scholar]
- 16.COVID-19 machine learning analytics for researchers. https://wellai.health/covid/. [Accessed May 26, 2020].
- 17.Allen Institute for AI. SciSight is a tool for exploring the evolving network of science in the COVID-19 Open Research Dataset, from Semantic Scholar at the Allen Institute for AI. https://scisight.apps.allenai.org/. [Accessed May 26, 2020].
- 18.Hope T, Borchardt J, Portenoy J, Vasan K, West J. Exploring the COVID-19 network of scientific research with SciSight. https://medium.com/ai2-blog/exploring-the-covid-19-network-of-scientific-research-with-scisight-f75373320a8c. [Accessed May 26, 2020].
- 19.Hope T, Portenoy J, Vasan K, Borchardt J, Horvitz E, Weld DS, Hearst MA, West J. SciSight: Combining faceted navigation and research group detection for COVID-19 exploratory scientific search. bioRxiv 2020May23 112284 10.1101/2020.05.23.112284. [Accessed May 26, 2020]. [DOI]
- 20.CDC. Contact tracing. https://www.cdc.gov/corona-virus/2019-ncov/php/open-america/contact-tracing.html. [Accessed May 26, 2020].
- 21.WHO. Operational planning guidelines to support country preparedness and response. WHO, 2020. [Google Scholar]
- 22.Allen Institute for AI and 8 collaborators. COVID-19 open research dataset challenge (CORD-19). Kaggle. https://www.kaggle.com/allen-institute-for-ai/CORD-19-re-search-challenge/tasks [Accessed May 26, 2020].
- 23.Razzaq A. List of COVID-19 resources for machine learning and data science research. Marktechpost. https://www.marktechpost.com/2020/04/12/list-of-covid-19-resources-for-machine-learning-and-data-science-research/ [Accessed May 26, 2020].
- 24.Schmitt M. How to fight COVID-19 with machine learning. Towards Data Science. April 7, 2020. https://towards-datascience.com/fight-covid-19-with-machine-learning-1d1106192d84. [Accessed May 26, 2020].
- 25.Badal VD, Kundrotas PJ, Vakser IA. Natural language processing in text mining for structural modeling of protein complexes. BMC Bioinformatics 2018;19:84 10.1186/s12859-018-2079-4. [Accessed May 26, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spasic I, Nenadic G. Clinical text data in machine learning: Systematic review. JMIR Med Inform. 2020;8(3): e17984 doi:10.2196/17984. [Accessed May 26, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tshitoyan V, Dagdelen J, Weston L, Dunn A, Rong Z, Kononova O, Persson KA, Ceder G, Jain A. Unsupervised word embeddings capture latent knowledge from materials science literature. Nature 2019;571(7763):95-98. [DOI] [PubMed] [Google Scholar]
- 28.Dey A. Machine learning algorithms: A review. Intl J Comput Sci Info Technol. 2016;7:1174-1179. [Google Scholar]
- 29.Young T, Hazarika D, Poria S, Cambria E. Recent trends in deep learning based natural language processing. IEEE Comp Intell Mag 2018;13:55-75. https://arxiv.org/abs/1708.02709. [Accessed May 26, 2020]. [Google Scholar]
- 30.Yao H, Zhu D-L, Jiang B, Yu P. Negative log likelihood ratio loss for deep neural network classification. ArXiv 2019;Corpus ID: 14031265. https://arxiv.org/pdf/1804.10690.pdf. [Accessed May 26, 2020].
- 31.Hope T, Borchardt J. SciSight: Helping scientists visualize and explore COVID-19 literature with AI. Ai2. https://medium.com/ai2-blog/coviz-helping-scientists-visualize-and-explore-covid-19-literature-with-ai-9359559368e5. [Accessed May 26, 2020].
- 32.Beltagy I, Lo K, Cohan A. SCIBERT: A pretrained language model for scientific text. Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing, Hong Kong, China, November 3–7, 2019:3615–20. https://www.aclweb.org/anthology/D19-1371.pdf. [Accessed May 26, 2020]. [Google Scholar]
- 33.Greenberg A. Does Covid-19 contact tracing pose a privacy risk? Your questions, answered. Wired. April 17, 2020. https://www.wired.com/story/apple-google-contact-tracing-strengths-weaknesses/ [Accessed May 26, 2020].
- 34.Yu E. Singapore introduces contact tracing app to slow coronavirus spread. ZDNet. March 21, 2020. https://www.zdnet.com/article/singapore-introduces-contact-tracing-app-to-slow-coronavirus-spread/. [Accessed May 26, 2020].
- 35.How TraceTogether works. https://www.tracetogeth-er.gov.sg/. [Accessed May 26, 2020]
- 36.Mozur P, Zhong R, Krolik A. In coronavirus fight, China gives citizens a color code, with red flags. New York Times March 1, 2020. https://www.nytimes.com/2020/03/01/business/china-coronavirus-surveillance.html. [Accessed May 26, 2020].
- 37.Kwon S. COVID-19: Lessons from South Korea. Health System Global. 31 March 2020. https://www.health-systemsglobal.org/blog/406/COVID-19-Lessons-from-South-Korea.html. [Accessed May 26, 2020]
- 38.Immuni: Italy’s Coronavirus-tracking App. Leaders League. 21-April-2020. https://www.leadersleague.com/en/news/immuni-italy-s-coronavirus-tracking-app. [Accessed May 26, 2020]
- 39.Pollina E. Italy tests contact-tracing app to speed lockdown exit. Reuters April 17, 2020. https://www.reuters.com/article/us-health-coronavirus-italy-technology/italy-tests-contact-tracing-app-to-speed-lockdown-exit-idUSKBN21Z0VJ. [Accessed May 26, 2020].
- 40.PEPP-PT. Pan-European Privacy-Preserving Proximity Tracing. https://www.pepp-pt.org/. [Accessed May 26, 2020].
- 41.Lomas N. NHS COVID-19: The UK’s coronavirus contacts-tracing app explained. Tech Crunch. May 5, 2020. https://techcrunch.com/2020/05/05/nhs-covid-19-the-uks-coronavirus-contacts-tracing-app-explained/. [Accessed May 26, 2020].
- 42.COVID Near You. https://www.covidnearyou.org/#!/ [Accessed May 26, 2020].
- 43.COVID Trace. https://covidtrace.com [Accessed May 26, 2020].
- 44.Lee T. Providing people with coronavirus-related data most useful to them. Berkeley Engineering. April 2, 2020. https://engineering.berkeley.edu/news/2020/04/providing-people-with-coronavirus-related-data-most-useful-to-them/. [Accessed May 26, 2020].
- 45.TweetBinder blog.#Covid 19-Twitter evolution. Twitter Analytics, https://www.tweetbinder.com/blog/covid-19-coronavirus-twitter/ [Accessed May 26, 2020].
- 46.Eichstaedt J, Guntuku S, Schwartz A. Using Twitter to understand the impact of COVID-19. HAI. https://hai.stanford.edu/sites/g/files/sbivbi10986/f/16johanneseichstaedt_languageofcovid_results_v5_rc4.pdf. [Accessed May 26, 2020].
- 47.Palmer D. Coronavirus contact-tracing apps: What are the privacy concerns? ZDNet. April 14, 2020. https://www.zdnet.com/article/coronavirus-contact-tracing-apps-what-are-the-privacy-concerns/ [Accessed May 26, 2020].
- 48.Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dorner L, Parker M, Bonsall D, Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 2020. 10.1126/science.abb6936. [Accessed May 26, 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenberg A. Clever cryptography could protect privacy in covid-19 contact-tracing apps. Wired. 04-August-2020. https://www.wired.com/story/covid-19-contact-tracing-apps-cryptography/. [Accessed May 26, 2020].
- 50.FDA. Emergency Use Authorizations. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations. [Accessed May 26, 2020].
- 51.IFCC information guide on COVID-19. https://www.ifcc.org/ifcc-news/2020-03-26-ifcc-information-guide-on-covid-19/. [Accessed May 26, 2020].
- 52.AACC. Coronavirus resources for labs, https://www.aacc.org/global-health-outreach/how-labs-can-prepare-for-coronavirus-and-other-outbreaks. [Accessed May 26, 2020].
- 53.Allen D, Block S, Cohen J, Eckersley P, Eifler M, Gostin L, Goux D, Gruener D, Hart VI, Hitzig Z, Krein U, Langford J, Nordhaus T, Rosenthal M, Sethi R, Siddarth D, Simons J, Sitaraman G, Slaughter A-M, Stanger A, Tabarrok A, Tretikov LA, Weyl EG. Roadmap to pandemic pandemic resilience. Massive scale testing, tracing, and supported isolation (TTSI) as the path to pandemic resilience for a free society. Edmond J Safra Center for Ethics. April 20 2020. https://ethics.harvard.edu/files/center-for-ethics/files/road-maptopandemicresilience_updated_4.20.20_0.pdf. [Accessed May 26, 2020].


