Abstract
The complex signaling dynamics of transcription factors can encode both qualitative and quantitative information about the extracellular environment, which increases the information transfer capacity and potentially supports accurate cellular decision-making. An important question is how these signaling dynamics patterns are translated into functionally appropriate gene regulation programs. To address this question for transcription factors of the nuclear factor κB (NF-κB) family, we profiled the single-cell dynamics of two major NF-κB subunits, RelA and c-Rel, induced by a panel of pathogen-derived stimuli in immune and nonimmune cellular contexts. Diverse NF-κB–activating ligands produced different patterns of RelA and c-Rel signaling dynamic features, such as variations in duration or time-integrated activity. Analysis of nascent transcripts delineated putative direct targets of NF-κB as compared to genes controlled by other transcriptional and posttranscriptional mechanisms and showed that the transcription of more than half of the induced genes was tightly linked to specific dynamic features of NF-κB signaling. Fibroblast and macrophage cell lines shared a cluster of such “NF-κB dynamics–decoding” genes, as well as cell type–specific decoding genes. Dissecting the subunit specificity of dynamics-decoding genes suggested that target genes were most often linked to both RelA and c-Rel or to RelA alone. Thus, our analysis reveals the cell type–specific interpretation of pathogenic information through the signaling dynamics of NF-κB.
Introduction
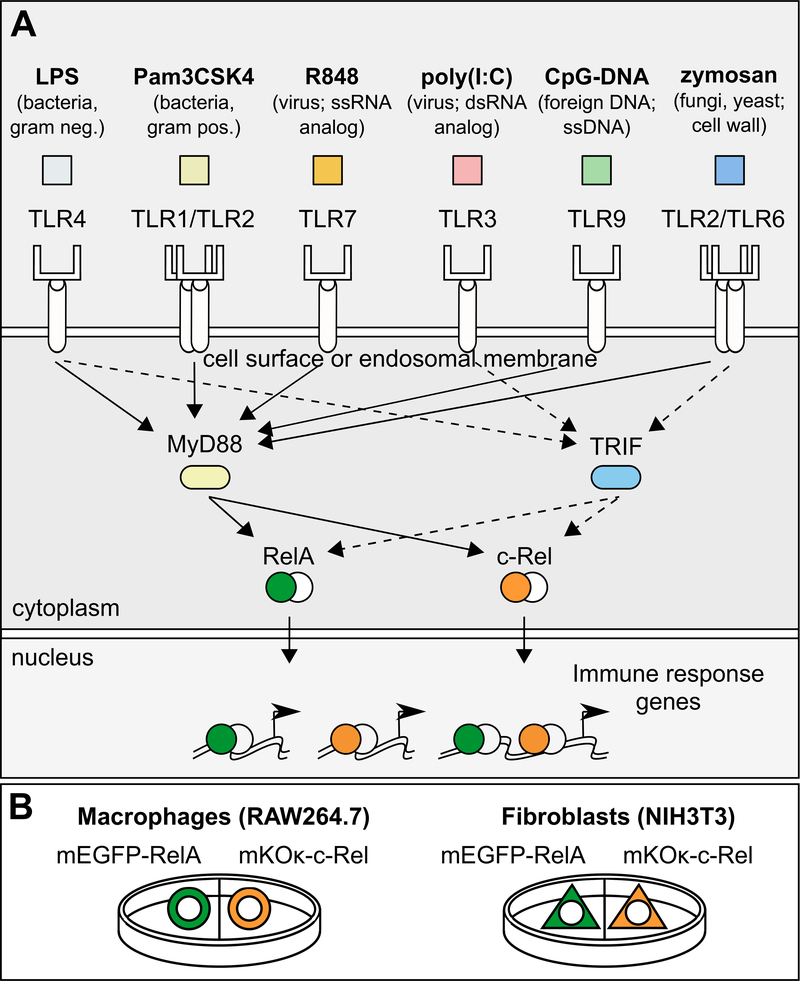
Innate immune cells can sense diverse pathogen-associated molecular patterns (PAMPs) as ligands binding to a panel of Toll-like receptors (TLRs) found either on the cell surface or in endosomes (1, 2). Whereas the PAMP-TLR interactions have evolved to be specialized for each receptor, the downstream signaling adaptors and transcription factors (TFs) are extensively shared across the PAMPs (Fig. 1A). Because innate immune cells respond to each PAMP with a functionally tailored gene expression program, the shared usage of TFs, such as nuclear factor-κB (NF-κB), suggests that biochemical specificity alone cannot adequately explain the basis of PAMP-specific responses. The activity dynamics of TFs can have profound effects in gene regulation and cell fate decisions (3). Therefore, it is possible that the signaling dynamics of TLR-activated TFs may be used to distinguish among the different PAMP stimuli and achieve functional specificity.
Fig. 1. Experimental design for imaging multiple NF-κB subunits in two cell types.
(A) Macrophages (RAW264.7 cells) and fibroblasts (NIH3T3 cells) were treated with an array of TLR ligands representing bacterial, viral, and fungal pathogens. Portrayed are the TLR ligands that produced detectable NF-κB activation in this study, as well as the targeted TLRs. LPS activates TLR4 (48); Pam3CSK4 activates TLR1/TLR2 heterodimers (49); R848 activates TLR7 (TLR7 and TLR8 in humans) (50, 51); zymosan activates TLR2/TLR6 heterodimers (52) and dectin-1 (53) (not depicted); poly(I:C) activates TLR3 (54); CpG-DNA activates TLR9 (55, 56); not shown: flagellin activates TLR5 (57) and profilin activates TLR11/TLR12 heterodimers (58, 59). (B) Macrophages or fibroblasts stably transfected with mEGFP-RelA or mKOκ-c-Rel were cultured in the same dish, treated with each of the TLR ligands, and then subjected to live-cell imaging. ssRNA, single-stranded RNA; dsRNA, double-stranded RNA; ssDNA, single-stranded DNA.
NF-κB dimers are activated by the TLR-MyD88 (myeloid differentiation marker 88) or TLR-TRIF (TIR domain–containing adaptor protein–inducing interferon-β) signaling axes. Stimulus-dependent degradation of inhibitor of NF-κB proteins (IκBs) induces the release of NF-κB dimers from IκBs and the accumulation of NF-κB in the nucleus for gene regulation. NF-κB heterodimers or homodimers containing RelA (also known as p65) or c-Rel subunits are critical mediators of the rapid transcriptional response in TLR-stimulated cells. These subunits contain a transactivation domain that mediates the recruitment of transcriptional coactivators, such as p300 and CBP (4). Given that NF-κB signaling dynamics (the temporal profile of nuclear abundance) can encode information about the dose and identity of extracellular stimuli (5–8), an important question is whether the signaling dynamics of NF-κB may also be used to decode the upstream PAMP information in terms of gene regulation. Pioneering single-cell imaging studies of RelA coupled with gene expression analysis have established a convincing link between RelA signaling dynamics and the expression of individual target genes (5, 7, 9–17). However, these findings were limited to RelA dynamics induced by a few cytokines or a single TLR ligand, lipopolysaccharide (LPS). Moreover, the exclusive focus on RelA is difficult to justify, because c-Rel is a highly abundant subunit in many immune cells and may exert a more potent functional role than RelA in certain situations (18).
Understanding the overall contribution of NF-κB in the functional decoding of signaling information requires data about the dynamics of all of the relevant NF-κB dimers, in different cell contexts, in response to diverse stimuli. The examination of diverse stimuli is important, especially because there are no precise and rapid pharmacological methods of perturbing specific features (for example, duration and amplitude) of NF-κB signaling dynamics. Widely used perturbation methods, such as small-molecule inhibitors, siRNA-mediated knockdown, and CRISPR-based knockout systems entail nonspecific effects and long-term compensatory mechanisms that hinder our ability to manipulate specific features of NF-κB dynamics. Therefore, more insight will be gained about the functional importance of NF-κB signaling dynamics from a greater number of naturally occurring distinct dynamic patterns.
Here, we used a comprehensive profiling approach to investigate the link between NF-κB dynamics, as reflected in RelA and c-Rel, and gene expression in two distinct cell types. Macrophages are immune cells specialized in detecting and responding to diverse pathogens and pathogen-derived substances. We contrasted the responses of a macrophage-like cell line with those of a non-immune cell type, NIH3T3 fibroblasts, in our integrative analysis and identified shared and tissue-specific decoding of signaling dynamics induced by an array of TLR ligands representing bacterial, viral, and fungal pathogens (Fig. 1). The results suggest that different cell types decode TF signaling dynamics differentially to interpret the stimuli and produce cell type–specific gene expression responses.
Results
Both RelA and c-Rel are activated by TLR ligands in RAW264.7 and NIH3T3 cell lines
To monitor the real-time dynamics of NF-κB dimers containing RelA or c-Rel by live-cell imaging, we stably transfected pools of macrophage-like cells (RAW264.7 cells) (herein referred to as macrophages) with plasmids encoding mEGFP-RelA or mKOκ-c-Rel (fig. S1). RelB, p50, and p52 subunits were not considered for imaging analysis because RelB is not robustly activated by canonical stimuli such as TLR ligands in most cell types, and the other two subunits form transcriptionally repressive homodimers (4). We also generated stable cell lines of transfected NIH3T3 cells (hereafter referred to as fibroblasts) using the same expression plasmids (fig. S1). We observed a wide range of fusion protein abundance in individual cells, and excluded single cells overexpressing the fluorescent fusion protein in our microscopy analysis to avoid effects from unphysiologically high amounts of RelA or c-Rel. The cells stably expressing mEGFP-RelA or mKOκ-c-Rel were treated with TLR ligands that represent widely different types of pathogens, spanning several major TLRs and the downstream intracellular signaling pathways (Fig. 1). Flagellin and profilin were not included in our comprehensive analysis because they did not visibly activate RelA or c-Rel in either cell line. Live-cell, time-lapse imaging was performed to quantify the single-cell time course of nuclear abundance (referred to as signaling dynamics) of RelA and c-Rel induced by each TLR ligand. Custom-written MATLAB code was used for cell-tracking and quantification of nuclear and total cellular intensities of each fusion protein.
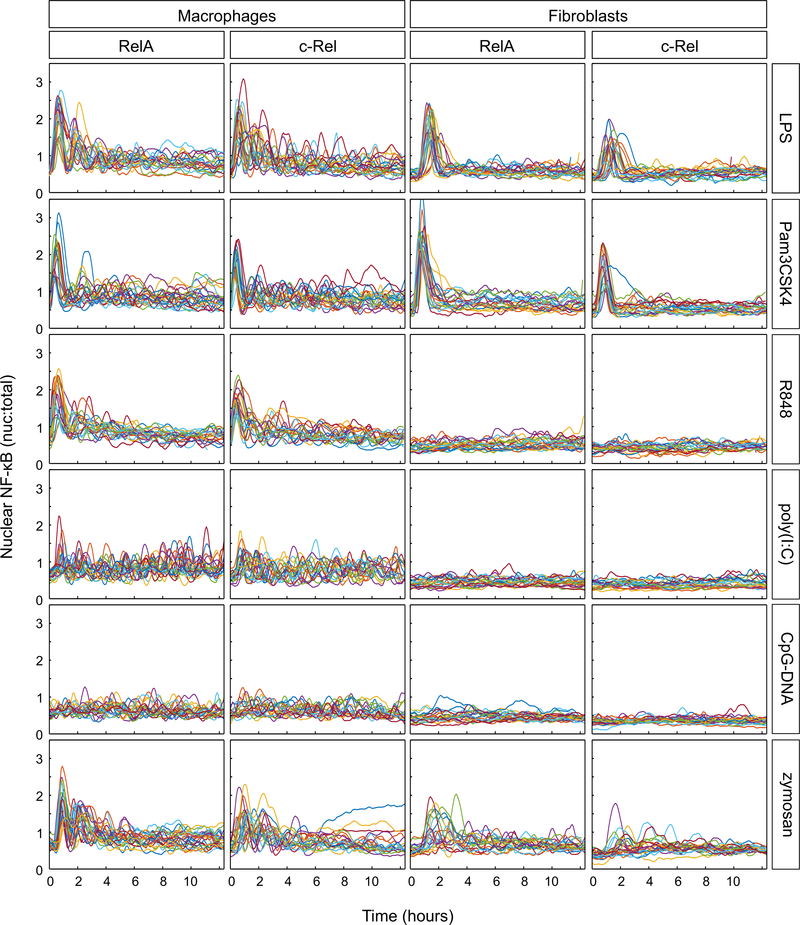
LPS, the prototypical immune stimulant, induced RelA responses in macrophages, consisting of a high-amplitude initial peak followed by two or more peaks (Fig. 2), as previously reported (7, 19). LPS-induced c-Rel signaling dynamics closely mirrored those of RelA (Fig. 2). Poly(I:C) and CpG-DNA induced lower-amplitude and variable RelA and c-Rel dynamics (Fig. 2). In both cell types, c-Rel signaling dynamics in response to the various TLR ligands were generally similar to those of RelA, with somewhat reduced magnitudes (Fig. 2). Only half of the TLR ligands (LPS, Pam3CSK4, and zymosan) induced appreciable RelA and c-Rel responses in fibroblasts (Fig. 2). The lack of response to poly(I:C), R848, and CpG-DNA in fibroblasts was presumably due to little or no expression of the cognate TLRs (fig. S2). The ligands that activated both macrophages and fibroblasts elicited cell type–specific signaling dynamics. The LPS and Pam3CSK4 responses in macrophages were characterized by marked oscillations, whereas the fibroblast responses largely consisted of a single peak of nuclear activity which quickly returned to baseline with minor fluctuations thereafter (Fig. 2). Fibroblast responses to zymosan were delayed and heterogenous, in comparison to those of macrophages. Note that we chose ligand concentrations that were sufficiently high to induce robust responses but were considered non-saturating for both cell types. A higher concentration of zymosan could not be used because its autofluorescence interferes with signal quantification upon cell binding and internalization.
Fig. 2. TLR ligand–induced NF-κB dynamics in single cells.
Macrophages and fibroblasts stably expressing fluorescent RelA or c-Rel fusion proteins were treated with the indicated TLR ligands and imaged for approximately 12.5 hours. Ratios of nuclear-to-total (nuc:total) cellular fluorescence of RelA and c-Rel were calculated and plotted. Ligand concentrations were carefully chosen from the reference literature, where available. The concentrations of TLR ligands used were as follows: LPS (10 ng/ml); Pam3CSK4 (40 ng/ml); R848 (1 μM; 350 ng/ml); poly(I:C) (20 μg/ml); CpG-DNA (25 μg/ml); zymosan (5 μg/ml); flagellin and profilin (250 ng/ml). n ≥ 20 cells for each ligand and NF-κB subunit, in each cell type. Results were obtained from at least two independent experiments per ligand, NF-κB subunit, and cell type.
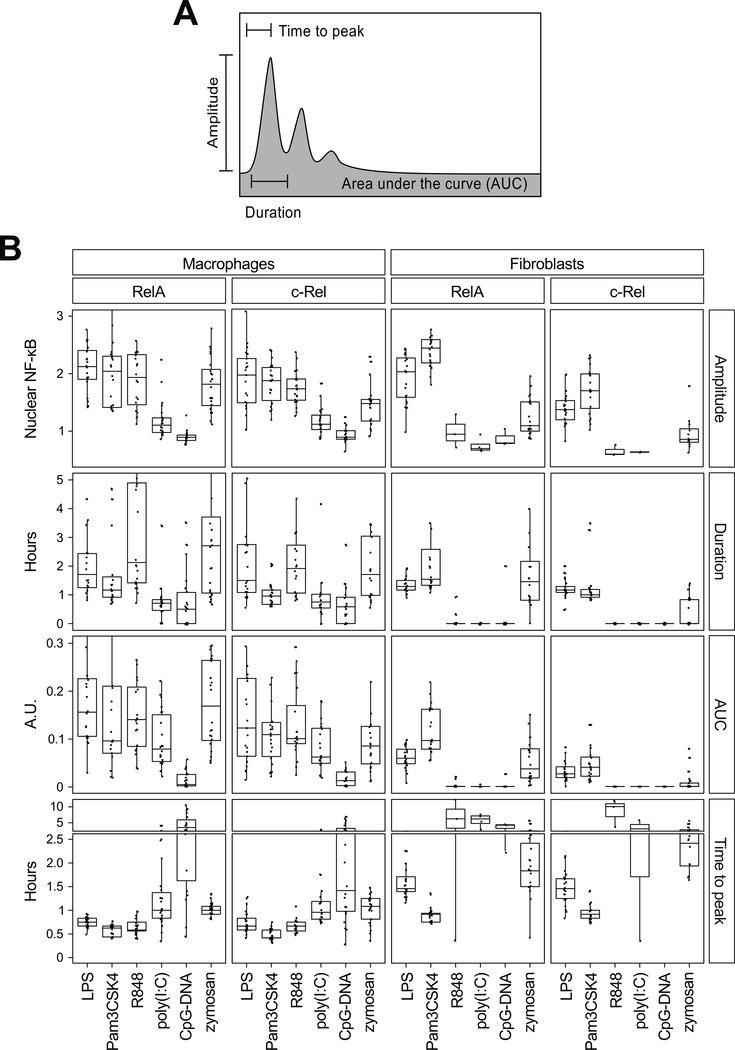
To capture quantitative features of NF-κB signaling dynamics, we focused on the following four measures, which essentially define a single-cell RelA or c-Rel response: (i) the delay until the nuclear signal reached a maximum amplitude (time to peak); (ii) the amplitude of the initial peak (peak amplitude); (iii) the duration of the initial peak (duration); and (iv) the time-integrated nuclear occupancy [area under the (nuclear:total ratio) curve, AUC] (Fig. 3A). We also prepared a complete summary of the quantified results including statistical differences and similarities amongst the dynamics (table S1). The quantitative analysis showed that some dynamic features were ligand-specific or cell type–specific (Fig. 3B). First, the time to RelA or c-Rel peaks was ligand-specific, with the shortest time observed for Pam3CSK4. Among the three ligands that activated both macrophages and fibroblasts, the rank order of the time to peak was preserved between the two cell types, with a general delay in fibroblasts. However, the rank orders of the other dynamic features across the ligands were not the same between the cell types. For example, the relative strengths of the responses to LPS and Pam3CSK4 were reversed in the two cell types (LPS and Pam3CSK4 resulted in higher values in macrophages and fibroblasts, respectively). Regarding c-Rel, the amplitude, duration, and AUC were slightly weaker than those of RelA, particularly in fibroblasts. Furthermore, each dynamic feature had distinct profiles across the ligands, particularly in macrophages, which suggests that no single feature can hold the information content of NF-κB responses (Fig. 3B). Each feature of NF-κB dynamics may have a differential effect in the transcriptional control of a specific subset of target genes, providing cells with multiple channels to decode the signaling information into functional cell type–specific gene expression responses.
Fig. 3. Quantification of TLR ligand–induced NF-κB dynamics.
(A) Features of NF-κB signaling dynamics that were analyzed. (B) Quantification of the single-cell RelA and c-Rel dynamics induced by the six TLR ligands that induced responses in either cell type (from the data in Fig. 2). The median value of each dynamic feature is depicted with quartile ranges. One-way ANOVA was performed to measure statistical significance amongst groups, whereas the Mann–Whitney test was performed for pairwise comparisons. Statistical test results can be found in table S1. AUC, area under the curve; A.U., arbitrary units.
TLR ligands induce cell type–specific patterns of gene expression
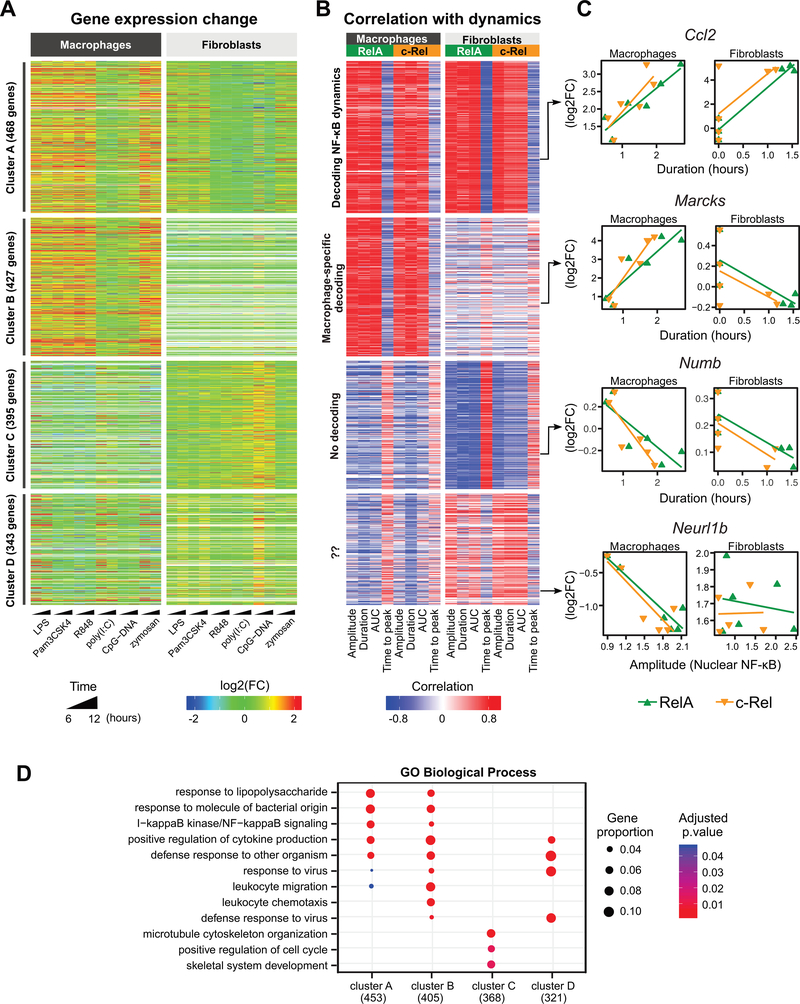
To explore whether distinct NF-κB dynamics translated into functional genome regulation, we next investigated the transcriptome-wide changes induced by each of the TLR ligands as assessed by total RNA-seq. Parental lines of macrophages and fibroblasts were treated with the six TLR ligands for 6 or 12 hours (Fig. 4A). Because NF-κB is primarily a transcriptional activator, we focused our analysis on 1633 genes whose expression was induced (log2 fold-change ≥ 1) by any of the TLR ligands in either cell type. We chose 6 and 12 hours of ligand stimulation as time points that correspond to the outcome of the gene regulatory responses. Even though these are not early time points, a substantial proportion (46 to 65%) of the differentially expressed genes were previously reported to be immediate early genes (20, 21). The gene induction by TLR ligands largely reflected the strengths of the RelA and c-Rel responses from the respective ligands. In macrophages, LPS, Pam3CSK4, R848, and zymosan generated the greatest increases in gene expression, whereas poly(I:C) and CpG-DNA induced only moderate changes (Fig. 4A). In fibroblasts, LPS, Pam3CSK4, and zymosan also induced robust increases in gene expression, with overall fold changes that were not as large as those observed in macrophages.
Fig. 4. Dynamic features of NF-κB signaling are linked to gene expression.
(A) Heatmap of the 1633 differentially expressed genes [log2 fold-change (log2FC) ≥ 1 and false discovery rate (FDR) < 0.05 in at least one treatment] obtained from whole-transcriptome analysis of TLR ligand–induced changes in macrophage and fibroblast gene expression. Displayed are the log2FC values of the treated cells relative to unstimulated cells. Each row represents one gene. (B) Pearson correlation values were determined for the log2FC value (mean of the 6- and 12-hour treatments) of each gene with respect to the medians of the single-cell NF-κB dynamics in each cell type and for each NF-κB subunit. Clustering was performed based on the correlation values using partitions around medoids. Values beyond the legend scales were set to extremes. The ordering of the genes is the same as that in (A). (C) Correlations for representative genes derived from each of the four clusters in (A) and (B). Shown are the genes’ expression level changes relative to the median of the single-cell dynamics features for each of the NF-κB subunits in each cell type. The dynamics feature that most strongly correlated with gene expression for each example gene is depicted. (D) Gene Ontology Biological Process (GOBP) enrichment analysis of dynamics-correlated genes in each of the clusters presented in (A) and (B). The three most statistically significant GOBP terms for each cluster were merged and the combined GOBP terms are shown. The number of genes with GOBP annotations are indicated in parentheses. Significant terms are marked as dots. Dot size represents the proportion of cluster genes annotated for each term. P values were adjusted using the default Benjamini-Hochberg method.
As expected, the TLR ligands induced numerous NF-κB–regulated immune response genes. Genes whose expression was increased in both cell types included Nfκbia, Tnfaip3, and Ccl2. Other genes, including Il1b and Cd40, were only increased in expression in macrophages, whereas genes such as Cxcl1 and Cxcl5 were only increased in expression in fibroblasts. The expression of Nfκbiε, which encodes a feedback inhibitor of NF-κB (IκBε) (22, 23), was induced by TLR ligands to a greater degree in fibroblasts than in macrophages (table S2), despite macrophages exhibiting stronger NF-κB signaling (Figs. 2 and 3). The higher induction of Nfκbiε expression and possibly that of other feedback genes in fibroblasts may contribute to the shorter-lived NF-κB dynamics that were observed fibroblasts in comparison to the sustained dynamics in macrophages. Our transcriptome-wide profiles show the extent of the cell-type specificity in gene induction patterns (Fig. 4A and table S2).
Features of NF-κB dynamics relate to some, but not all, gene clusters
We sought to discover genes potentially involved in the decoding of NF-κB signaling dynamics, by integrating the live-cell imaging data with the total RNA-seq data. We calculated the correlations between dynamic features of RelA and c-Rel signaling and gene induction across the TLR ligand treatments (Fig. 4, B and C; see Materials and Methods). If the correlation was strong, the gene was operationally termed as “decoding” a dynamic feature of the NF-κB subunit. The analysis revealed a large number of genes whose expression was strongly correlated with the RelA and c-Rel dynamics across the TLR ligands (Fig. 4B). Because short response times are generally associated with stronger responses, time to peak was often anti-correlated with gene induction. We prepared a complete list of correlation coefficients and gene expression values (table S2).
Using an unsupervised clustering analysis, we identified four distinct gene clusters based on how the gene expression was correlated to NF-κB dynamic features (Fig. 4B). Genes in cluster A were distinguished by strong correlations with RelA and c-Rel dynamics in both macrophages and fibroblasts (Fig. 4, A and B; “Decoding NF-κB dynamics”), with Ccl2 as an example of a duration-decoding gene. This cluster includes many genes previously established to be direct targets of NF-κB, such as Nfκbia, Nfkbie, Tnfaip3, and Myc (fig. S3). Cluster B genes had their expression changes strongly correlated to RelA and c-Rel dynamics only in macrophages (Fig. 4, A and B, “Macrophage-specific decoding”), for example, Tnf, Il1b, Cd40, Icam1, Nlrp3, and Marcks. In fibroblasts, cluster B genes either had negligible correlations or were not expressed (41% with mean absolute correlation < 0.6; 42% with undetectable expression; Fig. 4, A and B). Gene ontology (GO) analysis showed that only these dynamics-decoding clusters (A and B) were enriched for “NF-κB signaling” genes (Fig. 4D).
The potential relevance of clusters C and D was less clear. Expression of genes in cluster C was mostly anti-correlated with NF-κB dynamics in both cell types (Fig. 4, A to C), suggesting that these genes may be regulated by other transcription factors that may function in a mutually exclusive manner with respect to NF-κB. For example, despite the inability of CpG-DNA to induce detectable RelA or c-Rel responses in fibroblasts, CpG-DNA produced substantial gene induction in cluster C (Fig. 4A). Cluster D genes exhibited a mixture of heterogeneous correlation patterns. These findings raised the possibility that the linkage of NF-κB dynamics to gene regulation may not have been accurate in this integrative approach, which prompted us to seek alternative methods.
Nascent transcript-based re-clustering reveals cell type–specific decoding of NF-κB dynamics
Hypothesizing that the initial integrative analysis was potentially missing information, we wondered whether we could obtain deeper information from our total RNA-seq data. Because nascent transcripts are a more direct readout of active transcription factors, we surmised that they would produce a clearer relationship between NF-κB dynamics and gene regulation patterns. Furthermore, our bulk RNA-seq libraries had sufficient sequencing depths to enable the detection and quantification of nascent transcripts (using intron-mapped reads) that generally exist at lower abundance (Fig. 5A). Therefore, instead of using the default RNA-seq quantification (of mostly mature transcripts), we were able to quantify the expression of nascent transcripts using reads that map to introns of the mouse reference genome.
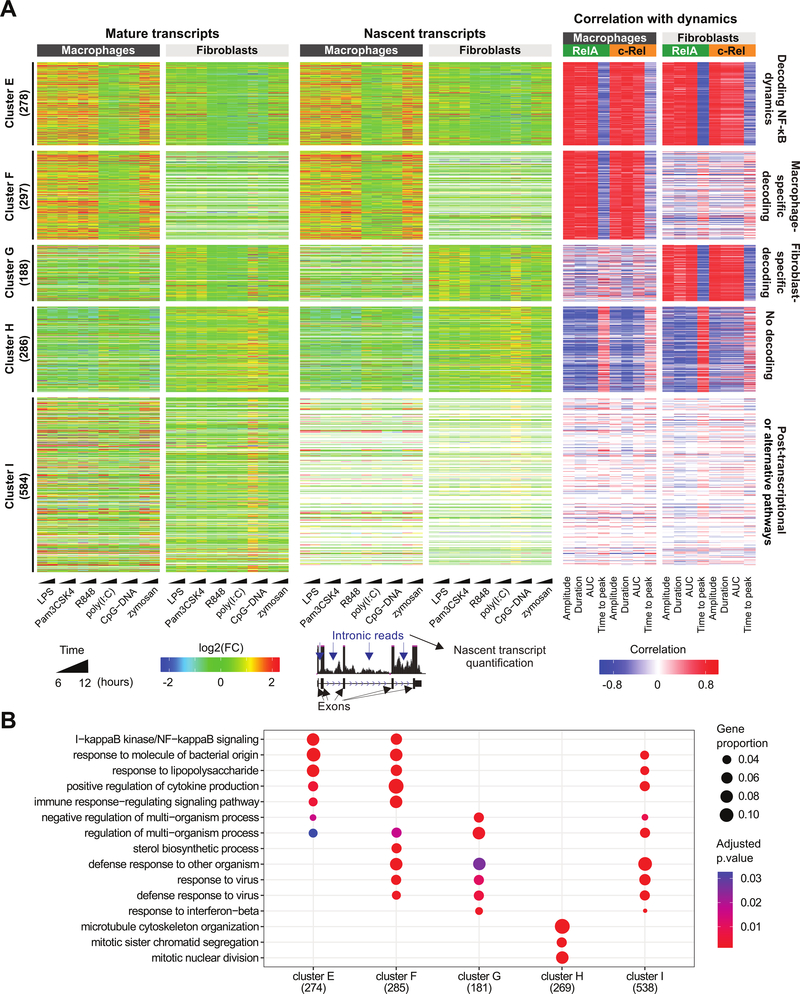
Fig. 5. NF-κB dynamics are better correlated to nascent transcription.
(A) The 1633 differentially expressed genes from Fig. 4A (left) were re-analyzed and re-clustered based on nascent-level gene expression changes (center) and correlations with NF-κB dynamics (right). Displayed are the log2FC values of cells treated with the indicated TLR ligands relative to unstimulated cells. Pearson correlation values were determined for the log2FC value (mean of the 6- and 12-hour treatments) of each gene at the nascent level with respect to the medians of single-cell NF-κB dynamics in each cell type and for each NF-κB subunit. Clustering was performed based on the correlation values using partitions around medoids. Values beyond the legend scales were set to extremes. (B) GOBP enrichment analysis of dynamics-correlated genes in each cluster presented in (A). The three most statistically significant GOBP terms for each cluster were merged and the combined GOBP terms are shown. The number of genes with GOBP annotations are indicated in parentheses. Significant terms are marked as dots. Dot size represents the proportion of cluster genes annotated for each term. P values were adjusted using the default Benjamini-Hochberg method.
Indeed, re-analysis of the RNA-seq data and re-clustering of the genes based on correlations between NF-κB dynamics and nascent transcripts delineated the genes presumably regulated by transcriptional (clusters E, F, G, and H) versus alternative mechanisms (cluster I) (Fig. 5A). The display of expression changes in mature versus nascent transcripts alongside the gene clusters highlighted that the genes in cluster I did not have a well-defined correlation with NF-κB dynamics because only their mature transcripts, but not their nascent transcripts, were increased in abundance in ligand-treated cells. Some genes within this cluster had undetectable nascent transcripts, and the correlation to NF-κB dynamics could not even be defined (generating blank entries in the heatmap, Fig. 5A). The gene Zfp36, which encodes the post-transcriptional regulator tristetraprolin (TTP), is in cluster I (24, 25). TTP promotes the degradation of its target mRNAs and is regulated by p38 mitogen-activated protein kinase (MAPK) activity (25). This cluster also helps to explain why cluster D was difficult to characterize: 51% of the genes in cluster D were assigned to cluster I (fig. S4). These results suggest that some genes in cluster I may be regulated by alternative mechanisms, including post-transcriptional control, and that analysis of mature transcript abundances may have been inappropriate for correlation with NF-κB dynamics (as was initially done to generate clusters A to D).
Clusters E, F, and H generally re-captured the patterns observed in the previous clusters A, B, and C (Figs. 4 and 5). Cluster E represents a cell type–independent “NF-κB dynamics-decoding” gene cluster, which includes Nfkbia, Nfkbib, Nfkbid, Nfkbie, Nfkbiz, Relb, Nfkb1, Nfkb2, Bcl3, Tnfaip3, Ikbke, Ccl2, Ccl7, and Myc. A macrophage-specific decoding gene cluster was also redefined as cluster F, which included inflammation-related genes, such as Tnf, Nlrp3, Cd40, Tlr1, Tlr6, Ccl5, Il1a, Il1b, Lcn2, and H2-Q6. The macrophage-specific cluster F had genes with either undetectable expression (44%) or negligible correlation (41%) in fibroblasts. The re-analysis also resulted in “NF-κB signaling” being the top enriched GO term among the NF-κB dynamics–correlated genes in clusters E and F, confirming the specificity of these clusters (Fig. 5B). A “no decoding” cluster H, consisting of cell cycle–related genes showing little correlation to NF-κB dynamics, was reduced with fewer constituent genes in comparison to its previous counterpart, cluster C. These genes may be largely influenced by other TLR-activated factors (for examples, IRFs and AP-1) that do not involve NF-κB.
Note that the new integrative analysis gave a fibroblast-specific gene cluster G, which was missing from the previous analysis. Cluster G contains Cxcl5, Vcam1, Isg20, Ifitm3, Ccl8, Ccl25, Irf1, Irf2, and Numb (mistakenly thought to be anti-correlated in the previous analysis), H2-Q4, Brd4, and Ifit3. About half of the fibroblast-specific decoding genes are from cluster D, which was of unknown relevance, but their nascent transcript abundances strongly correlated with the dynamic features of both RelA and c-Rel (Fig. 5A and fig. S4). Unlike the macrophage-specific clusters described so far, the fibroblast-specificity of cluster G was in large part due to the lack of correlation (rather than lack of expression) in macrophages: only 25% of the genes were not expressed and 69% had negligible correlation in macrophages.
The strength of correlation to the dynamic features of NF-κB may be gene-specific, and individual genes may depend more on certain features over others for their regulation. For example, some genes may better decode RelA amplitude, whereas others mostly decode c-Rel AUC. Although such tantalizing examples could be found, we examined the genome-wide influence of each dynamic feature. In macrophages, the strongest absolute correlation between NF-κB dynamics and global changes in gene expression was from the duration of nuclear c-Rel signaling (fig. S5, A and B). Correlation to RelA duration was similarly strong, which is consistent with a study of RelA dynamics and target gene expression using scRNA-seq (16). In fibroblasts, duration and time to peak of RelA displayed the highest absolute correlations. Overall, when nascent transcript expression was used for analysis, most TLR-induced genes were linked to NF-κB dynamics: 91 and 64% were correlated to at least one feature of NF-κB dynamics in macrophages and fibroblasts, respectively (absolute correlation ≥ 0.8), with RelA showing a broader and stronger effect than c-Rel (fig. S5C). These findings support the functional importance of NF-κB signaling dynamics in transcriptional responses to diverse TLR ligands, particularly in macrophages.
Transcription of dynamics-decoding genes is linked to RelA or to both RelA and c-Rel
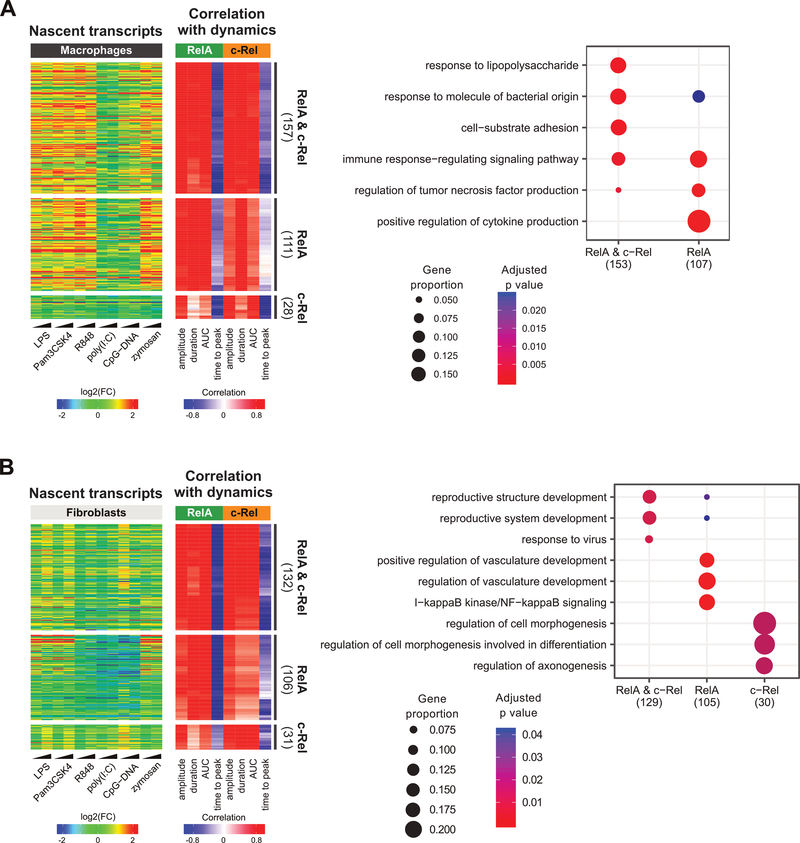
Finally, having observed a similar overall correlation of transcriptional responses to RelA versus c-Rel dynamics, we were curious to examine more closely whether there were any genes that seemed to track the dynamics of one subunit over the other. Because many NF-κB dynamics–decoding (positively correlated to the first three features and negatively correlated to time to peak) genes were cell type–specific (Fig. 5A), we performed this analysis separately for fibroblasts and macrophages with genes from clusters E through G. In each cell type, the largest set was from TLR-induced genes whose transcription was linked to dynamic features of both RelA and c-Rel (Fig. 6). Slightly smaller in numbers were genes showing transcriptional responses linked to RelA but only weakly to c-Rel. For example, the strongest fibroblast gene induction from the zymosan treatment was from the RelA dynamics-decoding cluster (Fig. 6B), which reflects the weak c-Rel responses relative to those of RelA in zymosan-treated fibroblasts (Fig. 2 and Fig. 3B). Few genes showed c-Rel–specific correlations. We note that these results should be interpreted with caution, given the unavoidable difference between the fusion protein abundances in the two reporter cell lines. Further validations will be necessary to ascertain the roles of RelA or c-Rel in specific regulation of these genes using rapid-acting tools that overcome the limitations of conventional methods such as knock-down, knock-out, or pulsing of stimuli (11, 22, 26, 27). Nevertheless, GO analysis indicated some functional categories related to LPS responses and NF-κB signaling were differentially enriched in these gene subsets (Fig. 6 and table S3).
Fig. 6. Subunit-specific correlation analysis.
(A and B) For macrophages (A) and fibroblasts (B), genes from clusters E to G were classified based on the average strength of correlation to RelA and c-Rel dynamic features (see Materials and Methods). Display schemes are the same as those used in Fig. 5. Right: GOBP enrichment analysis of dynamics-correlated genes in each gene subset in the heatmap. The three most statistically significant GOBP terms for each set were merged and the combined GOBP terms are listed in the plot. The number of genes with GOBP annotations are indicated in parentheses. Significant terms are marked as dots. Dot size represents the proportion of cluster genes annotated for each term. P values were adjusted using the default Benjamini-Hochberg method.
Discussion
Our study enabled a side-by-side comparison of an immune cell type and a non-immune cell type responding to a panel of pathogen-related stimuli and revealed the extent of cell type–specific decoding of complex NF-κB signaling dynamics. Previous studies of NF-κB dynamics in single cells have largely focused on one cell type each (6, 7, 12, 13, 15, 16, 26, 28–30). Integrative analyses of the extensive transcriptomic profiles and NF-κB signaling dynamics in matching conditions suggest that distinct features of NF-κB dynamics are decoded by different target genes in a cell type–specific manner. Furthermore, a previously uncharacterized relationship between NF-κB dynamics and gene regulation emerged when nascent transcripts were considered instead of mature transcripts. The marked difference between information gained from mature versus nascent transcript abundance also serves as a cautionary note for the utility of single cell RNA-seq methods for studying how TF signaling dynamics are decoded by target genes, because they generally measure mature transcripts.
We characterized c-Rel signaling dynamics by live-cell imaging and found that they closely resembled those of RelA (Figs. 2 and 3). The overall similarity of the responses suggests that RelA and c-Rel activities are likely regulated by the same or comparable signaling mechanisms. However, c-Rel signaling appeared to be somewhat less robust, relative to that of RelA, consistent with the expectation that RelA is the predominant NF-κB subunit in most settings. We note that the RelA and c-Rel fluorescent fusion proteins in this study were expressed from a constitutive promoter, reporting the abundance and localization of the existing pool of NF-κB containing these subunits. Therefore, the signaling dynamics of endogenous RelA and c-Rel, which are themselves induced by NF-κB, may diverge from those observed here, especially in the late phase of responses. Our findings will be useful in dissecting endogenous signaling dynamics, which are a superimposition of existing versus newly synthesized RelA or c-Rel proteins.
The cell type–specific decoding of NF-κB signaling dynamics may occur partly through gene-specific chromatin requirements for productive transcription. For example, some target genes may need the sustained presence of NF-κB at key regulatory sites, thereby decoding the duration of RelA or c-Rel signaling. Others may depend particularly on a high concentration of nuclear NF-κB, thereby decoding the amplitude of RelA or c-Rel. AUC-decoding genes may produce a transcriptional output that is generally proportional to the total NF-κB signaling activity over time. Genes decoding time to peak may diminish their transcription if the NF-κB arrives at their regulatory sites too late, such as when the enhancers are occupied or chromatin accessibility is limited by other, early-acting TFs. Such tissue-specific genomic compartmentalization in the decoding of NF-κB signaling dynamics is an interesting but difficult topic to be investigated with improved tools in the future. Such a regulatory architecture is also consistent with the finding that some target genes are co-regulated by NF-κB dynamics (16). Finally, it will be important in the future to obtain additional insight from varying the dose of activating ligands for each of the TLRs. In summary, our results highlight the need for more in-depth investigations of the potential effects of specific features of NF-κB dynamics on transcriptional control in both immune and non-immune cells.
Materials and Methods
Reagents
The plasmids mEGFP-N1 (54767), mKOκ (amplified from Chicken Mermaid S188) (53617) (31), RelA cFlag pcDNA3 (20012), and c-Rel cFlag pcDNA3 (20013) (32) were purchased from Addgene. The pSF-EF1α-Ub-Neo plasmid (OG606) was purchased from Oxford Genetics. Purchased TLR ligands included LPS (Enzo Life Sciences, ALX-581–008), Pam3CSK4 (Tocris; 4633), R848 (Invivogen; tlrl-R848), poly(I:C) (Invivogen; tlrl-picw), CpG-DNA (Invivogen; ODN 2395), zymosan (Sigma; 24250), profilin (Sigma; SRP8050), and flagellin (Sigma; SRP8029). The primary antibodies polyclonal rabbit anti-RelA (SC-372) and polyclonal rabbit anti-c-Rel (SC-71) were purchased from Santa Cruz. Monoclonal rabbit anti-Gapdh (14C10) was purchased from Cell Signaling Technology. Donkey anti-rabbit antibody (GE Healthcare; NA934V) was used as the secondary antibody.
Plasmid generation
Briefly, cDNA from plasmids purchased from Addgene was amplified by PCR using Phusion polymerase mix [New England BioLabs (NEB); M0532S] and the appropriate primers (table S4). PCR products were gel-purified using a QIAquick gel extraction kit (Qiagen; 28115). The constructs and the pSF-EF1α-Ub-Neo plasmid were digested with restriction enzymes from NEB (EcoR1-HF, R3101S; EcoRV-HF, R3195S; BsiW1, R0553S; BsrGI-HF, R3575S), where applicable. The plasmid was dephosphorylated with Antarctic Phosphatase (NEB; M02809S). Digested PCR products were purified with a QIAquick PCR purification kit (Qiagen, 28104) and ligated to each other and then into the pSF-EF1α-Ub-Neo plasmid using Promega LigaFast Rapid DNA Ligation kit (Fisher Scientific; PR-M8221). Portions of the ligation reactions were used to transform DH5α cells (Thermofisher; 18265017). Plasmids from single antibiotic–resistant clones were purified using Mini-prep kit(s) (Qiagen; 27104) and Maxi-prep kit(s) (Qiagen; 12362). Construction of the plasmids was verified by DNA sequencing.
Cell culture and transfections
NIH3T3 cells (CRL-1658) and RAW264.7 cells (TIB-71) were newly purchased from ATCC. All cell lines were cultured and maintained at 37ºC in a 5% CO2/95% air environment in growth medium composed of phenol-red-free Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco; 21063–029) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gemini Bioproducts; 100–500) and 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (1% PSG) (Gibco; 15140–122). Cells were passaged by brief exposure to 0.25% trypsin-EDTA (Gibco, 25200–056) (NIH3T3) or by gently scraping (RAW264.7). Cells in growth medium lacking 1% PSG were transfected with expression plasmids using Fugene HD (Promega; E2311) according to the manufacturer’s instructions. Pools of cells with stable integration of the EEF1A1-promoter–driven fusion constructs were obtained by selection with G418 sulfate solution (500 μg/mL) (Mirus, MIR5920). Stable expression and function of the fusion proteins was tested through a combination of Western blotting and microscopy.
Western blotting
Cells were lysed in ice-cold RIPA cell lysis buffer (Millipore, 20–188) containing Complete Ultra Mini protease inhibitors (Roche; 05892970001). Lysates were vortexed, centrifuged at 18,000g for 15 min, and homogenized with insulin syringes. Sample proteins concentrations were equalized using the Protein Assay Dye reagent concentrate (Biorad; 5000006) and heated at 95ºC for 5 min in LDS sample buffer (ThermoFisher; NP0007) containing 10% 2-mercaptoethanol (Gibco; 31350–010). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using 4 to 12% NuPage Bis-Tris gels (ThermoFisher; NP0322BOX) and MOPS buffer (ThermoFisher; NP0001). Proteins were transferred using transfer buffer (ThermoFisher; NP00061) onto 0.45-μm PVDF membranes (Millipore; IPVH00010). Membranes were blocked in 5% (w/v) non-fat milk for 30 min and then sequentially incubated with primary and horseradish peroxidase (HRP)-conjugated secondary antibodies. HRP activity was detected using SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher; 34580). Where indicated in the figure legends, membranes were stripped for 15 min with Restore Western Blot Stripping Buffer (21059).
Microscopy
Pools of transfected NIH3T3 and RAW264.7 cells stably expressing RelA or c-Rel fusion protein were seeded individually in Ibidi 4-well culture inserts (80466) at medium confluence on 35-mm glass-bottomed dishes (MatTek; P35G-1.5–20-C) and cultured for up to 36 hours as described earlier. The Ibidi insert was then removed and the medium was changed to growth medium containing 2% serum (1:5 dilution in serum-free growth medium). Cells were then placed in the Zeiss LSM880 AxioObserver confocal microscope incubation system and maintained at 37ºC in a humidified environment of 5% CO2 for 3 to 4 hours, at which time recording was begun. Immediately before the second frame, cells were treated with 3X concentrations of each TLR ligand, also in growth medium containing 2% serum, to achieve the indicated 1X final concentrations. Time-lapse images were acquired at 5-min intervals for a time-course of ~12.5 hours (150 frames) using laser powers of 0.12% (for NIH3T3 cells) and 0.8% (for RAW264.7 cells), and excitation wavelengths of 488 nm (for mEGFP) and 514 nm (for mKOκ). Additional imaging conditions: a 40X/1.4 NA Plan-Apochromat oil objective, a fully-open pinhole (600 μm), 16-bit, 1024×1024, LineSequential Scan Mode, ChS1 channel, 0.52 μs pixel dwell, 0.63 s frame time; 0.52 μs pixel time, 0.03 ms line time, 750 detector (master) gain, 1 digital gain, and 1.0 zoom. Acquired LSM files were exported as 16-bit TIFF files for further analysis.
Image analysis
Using custom MATLAB code that uses the Image Processing toolbox, responding cells in each time lapse field that did not divide, stayed in view and focus for at least 10 hours, had little overlaps with other cells, and exhibited no signs of cellular stress were subject to further analysis. Cellular boundaries were identified automatically by intensity-thresholding. Boundaries were manually verified, adjusted, and separated if necessary. Because no nuclear markers were used, to avoid perturbing cell physiology and potentially NF-κB regulation, nuclear boundaries were carefully identified and segmented manually in every frame. This strict analysis precluded analyzing many more cells. Background-subtracted intensity values were used to calculate the ratios of mean nuclear fluorescence intensity to mean total cellular fluorescence intensity (nuc:total). Where necessary, fibroblasts that did not visibly respond to TLR ligands [R848, poly(I:C), and CpG-DNA] were analyzed.
Total RNA-seq
Parental NIH3T3 and RAW264.7 cells were seeded at medium confluence on 24-well glass-bottomed plates (MatTek, P24G-1.5–13-F) and cultured overnight as described earlier. The next day, the medium was changed to growth medium containing 2% serum (1:5 dilution in serum-free growth medium) and the cells were cultured for an additional 3 to 4 hours. As described earlier, cells were then treated with 3X concentrations of each TLR ligand to achieve final 1X concentrations. For each replicate, non-treated cells, as well as cells treated with TLR ligands for 6 and 12 hours, were lysed in RLT buffer and frozen at −80ºC. RNA was later purified using QiaShredder kit(s) (Qiagen, 79656) and RNeasy kit(s) (Qiagen, 74136). RNA concentrations were determined using a NanoDrop spectrophotometer (ThermoFisher). Determination of RNA integrity and other quality control was performed by the Center for Cancer Research (CCR) Sequencing Facility at the National Cancer Institute (Frederick, MD). Only high-quality RNA samples (RIN (RNA integrity number) ≥ 9) were used for RNA-seq library preparation with TruSeq Stranded Total RNA kit(s) (Illumina). Three independent biological replicates, from three independently thawed batches of cells, were performed to obtain two QC-passing biological replicates for every sample. In total, 56 RNA-seq libraries were generated [2 cell types x 6 ligands x 2 time-points x 2 biological replicates, as well as 8 untreated control samples taken at the 0-hour (2 from each of the 2 replicates per cell type)]. The libraries were multiplexed and sequenced on two NextSeq flow cells at the CCR Sequencing Facility. From each sample, we collected from 16.1 to 55.5 million (median = 30.3 million) sequencing reads per library with 79 to 90% non-duplicate reads and at least 91% reads with 90% bases having the Phred Score quality Q30 and above. The percentage of uniquely mapped reads ranged from 72 to 86% (median = 84%).
RNA-seq analysis
The stranded paired-end 75-bp reads were mapped to the mouse reference genome [GRCm38, mm10 (33)] with the STAR v2.5.3a mapper (34) using the Gencode M14 primary assembly annotation (35). For the exonic-based mature expression, the reads were counted per gene using RSEM (36). For the intronic-based nascent expression, we first extracted unique intronic regions, as suggested previously (37), with BEDtools v2.25.0 (38). The reads were counted with the featureCounts function of the Subread package v1.4.6-p3 (39) using the following options: -p –primary -B -C -s 2. The count data was then used for differential expression analysis with the edgeR v3.18.1 (40), R v3.4.2. Before the analysis, we filtered out the following genes: (i) lowly expressed genes (< 1 FPKM (Fragments Per Kilobase of transcript per Million mapped reads) for exonic, < 0.5 FPKM for intronic in > 60% of samples in each cell type separately); (ii) short genes (< 200bp); (iii) ribosomal, transporter RNA genes, and other small RNA genes; (iv) genes from unplaced chromosomes. Read counts were normalized for the library size and common, trended, and tagwise negative binomial dispersions by weighted likelihood empirical Bayes method, using the calcNormFactors and estimateDisp functions of the edgeR package. The counts were then fitted into gene-wise negative binomial generalized linear models with the glmFit function. Differential expression was determined pairwise for each ligand and time point compared to untreated samples by likelihood ratio tests using the glmLRT function with contrasts accordingly. A detectable batch effect was caused by using multiple flow cells in the sequencing and therefore this factor was included in our gene-wise negative binomial general linear model fit (~ flow cell + group) before testing for differential expression. For each cell type, the data were analyzed separately but the P value correction was performed only once for all differential expression results. Amongst the differentially increased genes, we excluded 334 that were not annotated as protein-coding genes per Gencode M14 gene biotype. A gene was considered to be differentially expressed if the |log2FC| ≥ 1 and false discovery rate (FDR) < 0.05.
Correlation analysis
We first filtered genes whose expression was statistically significantly induced by any of the ligands at any time point and by any expression measure (mature or nascent transcripts). We then used the mean of the edgeR-calculated log2 fold-change (FC) values from the two time points (6 and 12 hours) for each of the ligands and calculated the Pearson’s correlations between gene expression (measured by log2FC upon stimulation compared to untreated cells) and each of the four dynamic features: the amplitude of the first peak (amplitude), the duration of the first peak (duration), the time-integrated area under the curve (AUC), and time to the first peak (time to peak) for each of the NF-κB subunits (RelA and c-Rel) and each cell type (macrophages and fibroblasts) separately. We used the median of measurable dynamics values across the number of single cells analyzed (table S1). We calculated the correlations using the corAndPvalue function of the WGCNA R package (41) for both mature and nascent expression (measured as log2FC relative to untreated cells). We then performed a cluster analysis with the partition around medoids (PAM) algorithm (42) based on the correlation values calculated between the expression changes and all four dynamics features for both cell types and both NF-κB subunits. For the purpose of clustering, missing values were set to zero and later reverted to missing values before plotting or any other analyses. The identified clusters were then used to generate a heatmap of correlated and anti-correlated genes with the ComplexHeatmap R package (43). The number of clusters was increased until no new patterns were detected. Genes within clusters were ordered by hierarchical clustering. The correlation heatmap was plotted next to the expression heatmap with the same gene order (based on PAM clustering of the correlation values). For the subunit-specific correlation, we considered genes to be correlated if the average of the absolute correlation coefficients was at least 0.7, whereas genes were considered to be uncorrelated if the average of the absolute correlation coefficients was below 0.6. Despite the panel of TLR ligands analyzed, our sample size was not large enough to support stringent statistical filtering. Hence, we did not correct the correlation coefficients for any multiple testing and instead used their absolute value as a measure of confidence.
Gene set enrichment analysis
The Ensembl gene identifiers for each of the clusters of genes were used for the gene set enrichment analysis using the ClusterProfiler R package (44) using all mouse genes (Gencode MV14) as a reference set. The tool uses a hypergeometrical test with the Benjamini-Hochberg correction for multiple testing (adjusted P values) (45). We used GO Biological Process terms (46, 47) (GO.db R package version 3.5.0) and performed compareCluster analysis with enrichGO function. The statistical significance threshold for the adjusted P value was set to 0.05. Only the three most significantly enriched terms were included in Figs. 4D and 5B.
Statistical analysis
For the single-cell dynamics, statistically significant differences among groups were calculated using one-way ANOVA and Mann–Whitney tests (pairwise comparisons). P values < 0.05 were considered to be statistically significant. For RNA-seq and related analyses, statistical methods are described in the relevant sections of the Materials and Methods.
Supplementary Material
Table S1. Single-cell dynamics characteristics.
Table S2. Complete list of differentially expressed genes and correlations.
Table S3. List of RelA versus c-Rel specificity in correlations.
Fig. S1. Expression of NF-κB fusion proteins in stably transfected cells.
Fig. S2. TLR expression in macrophages and fibroblasts.
Fig. S3. TLR ligand–induced expression of NF-κB target genes and correlations with NF-κB dynamics.
Fig. S4. Reclassification of TLR-induced genes based on the correlation of changes in mature versus nascent transcripts to NF-κB dynamics.
Fig. S5. Correlation of TLR ligand–induced transcriptional changes with NF-κB dynamics.
Table S4. Primers used to generate expression plasmids containing mEGFP-RelA and mKOκ-c-Rel coding sequences.
Acknowledgments
We thank M. Davidson for providing mEGFP-N1, V. Pieribone for Chicken Mermaid S188, and S. Smale for RelA cFlag pcDNA3 and c-Rel cFlag pcDNA3. The authors thank the National Cancer Institute CCR Sequencing Facility for RNA-seq QC, library preparation, and sequencing. This work used the computational resources of NIH High Performance Computing Systems, including the Biowulf cluster.
Funding: This study was entirely funded by the NIA Intramural Research Program at the National Institutes of Health.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The materials and data in this study are available upon reasonable request or through appropriate MTAs. Raw RNA-seq data from the 56 samples were deposited at the European Nucleotide Archive (ENA) and are available at https://www.ebi.ac.uk/ena/data/view/PRJEB24055.
References and Notes
- 1.Akira S, Uematsu S, Takeuchi O, Pathogen recognition and innate immunity. Cell 124, 783–801 (2006); published online EpubFeb 24 ( 10.1016/j.cell.2006.02.015). [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki T, Kawai T, Toll-like receptor signaling pathways. Front Immunol 5, 461 (2014) 10.3389/fimmu.2014.00461). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G, p53 dynamics control cell fate. Science 336, 1440–1444 (2012); published online EpubJun 15 ( 10.1126/science.1218351). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden MS, Ghosh S, NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26, 203–234 (2012); published online EpubFeb 1 ( 10.1101/gad.183434.111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW, Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 (2010); published online EpubJul 8 ( 10.1038/nature09145). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner DA, Paszek P, Woodcock DJ, Nelson DE, Horton CA, Wang Y, Spiller DG, Rand DA, White MR, Harper CV, Physiological levels of TNFalpha stimulation induce stochastic dynamics of NF-kappaB responses in single living cells. J Cell Sci 123, 2834–2843 (2010); published online EpubAug 15 ( 10.1242/jcs.069641). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung MH, Li N, Lao Q, Gottschalk RA, Hager GL, Fraser ID, Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-kappaB signaling. Sci Signal 7, ra6 (2014); published online EpubJan 14 ( 10.1126/scisignal.2004764). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, Covert MW, A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Sci Signal 2, ra65 (2009); published online EpubOct 20 ( 10.1126/scisignal.2000599). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR, Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708 (2004); published online EpubOct 22 ( 10.1126/science.1099962). [DOI] [PubMed] [Google Scholar]
- 10.Zambrano S, De Toma I, Piffer A, Bianchi ME, Agresti A, NF-kappaB oscillations translate into functionally related patterns of gene expression. Elife 5, e09100 (2016); published online EpubJan 14 ( 10.7554/eLife.09100). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellogg RA, Tay S, Noise facilitates transcriptional control under dynamic inputs. Cell 160, 381–392 (2015); published online EpubJan 29 ( 10.1016/j.cell.2015.01.013). [DOI] [PubMed] [Google Scholar]
- 12.Sung MH, Salvatore L, De Lorenzi R, Indrawan A, Pasparakis M, Hager GL, Bianchi ME, Agresti A, Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS One 4, e7163 (2009); published online EpubSep 29 ( 10.1371/journal.pone.0007163). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamson A, Boddington C, Downton P, Rowe W, Bagnall J, Lam C, Maya-Mendoza A, Schmidt L, Harper CV, Spiller DG, Rand DA, Jackson DA, White MR, Paszek P, Signal transduction controls heterogeneous NF-kappaB dynamics and target gene expression through cytokine-specific refractory states. Nat Commun 7, 12057 (2016); published online EpubJul 6 ( 10.1038/ncomms12057). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee RE, Walker SR, Savery K, Frank DA, Gaudet S, Fold change of nuclear NF-kappaB determines TNF-induced transcription in single cells. Mol Cell 53, 867–879 (2014); published online EpubMar 20 ( 10.1016/j.molcel.2014.01.026). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong VC, Bass VL, Bullock ME, Chavali AK, Lee REC, Mothes W, Gaudet S, Miller-Jensen K, NF-kappaB-Chromatin Interactions Drive Diverse Phenotypes by Modulating Transcriptional Noise. Cell Rep 22, 585–599 (2018); published online EpubJan 16 ( 10.1016/j.celrep.2017.12.080). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane K, Van Valen D, DeFelice MM, Macklin DN, Kudo T, Jaimovich A, Carr A, Meyer T, Pe’er D, Boutet SC, Covert MW, Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-kappaB Activation. Cell Syst 4, 458–469 e455 (2017); published online EpubApr 26 ( 10.1016/j.cels.2017.03.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rand U, Rinas M, Schwerk J, Nohren G, Linnes M, Kroger A, Flossdorf M, Kaly-Kullai K, Hauser H, Hofer T, Koster M, Multi-layered stochasticity and paracrine signal propagation shape the type-I interferon response. Mol Syst Biol 8, 584 (2012); published online EpubMay 22 ( 10.1038/msb.2012.17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shokhirev MN, Almaden J, Davis-Turak J, Birnbaum HA, Russell TM, Vargas JA, Hoffmann A, A multi-scale approach reveals that NF-kappaB cRel enforces a B-cell decision to divide. Mol Syst Biol 11, 783 (2015); published online EpubFeb 13 ( 10.15252/msb.20145554). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagnall J, Boddington C, England H, Brignall R, Downton P, Alsoufi Z, Boyd J, Rowe W, Bennett A, Walker C, Adamson A, Patel NMX, O’Cualain R, Schmidt L, Spiller DG, Jackson DA, Muller W, Muldoon M, White MRH, Paszek P, Quantitative analysis of competitive cytokine signaling predicts tissue thresholds for the propagation of macrophage activation. Sci Signal 11, (2018); published online EpubJul 24 ( 10.1126/scisignal.aaf3998). [DOI] [PubMed] [Google Scholar]
- 20.Oh KS, Patel H, Gottschalk RA, Lee WS, Baek S, Fraser IDC, Hager GL, Sung MH, Anti-Inflammatory Chromatinscape Suggests Alternative Mechanisms of Glucocorticoid Receptor Action. Immunity 47, 298–309 e295 (2017); published online EpubAug 15 ( 10.1016/j.immuni.2017.07.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST, Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012); published online EpubJul 20 ( 10.1016/j.cell.2012.05.043). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann A, Levchenko A, Scott ML, Baltimore D, The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 (2002); published online EpubNov 8 ( 10.1126/science.1071914). [DOI] [PubMed] [Google Scholar]
- 23.Basak S, Hoffmann A, Crosstalk via the NF-kappaB signaling system. Cytokine Growth Factor Rev 19, 187–197 (2008); published online EpubJun-Aug ( 10.1016/j.cytogfr.2008.04.005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao S, Baltimore D, The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10, 281–288 (2009); published online EpubMar ( 10.1038/ni.1699). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, Hartweger H, Vielnascher R, Karaghiosoff M, Rulicke T, Muller M, Hofacker I, Lang R, Kovarik P, Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol Syst Biol 7, 560 (2011); published online EpubDec 20 ( 10.1038/msb.2011.93). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee RE, Qasaimeh MA, Xia X, Juncker D, Gaudet S, NF-kappaB signalling and cell fate decisions in response to a short pulse of tumour necrosis factor. Sci Rep 6, 39519 (2016); published online EpubDec 22 ( 10.1038/srep39519). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand DA, See V, White MR, Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science 324, 242–246 (2009); published online EpubApr 10 ( 10.1126/science.1164860). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutschow MV, Mason JC, Lane KM, Maayan I, Hughey JJ, Bajar BT, Amatya DN, Valle SD, Covert MW, Combinatorial processing of bacterial and host-derived innate immune stimuli at the single-cell level. Mol Biol Cell 30, 282–292 (2019); published online EpubJan 15 ( 10.1091/mbc.E18-07-0423). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellogg RA, Tian C, Etzrodt M, Tay S, Cellular Decision Making by Non-Integrative Processing of TLR Inputs. Cell Rep 19, 125–135 (2017); published online EpubApr 4 ( 10.1016/j.celrep.2017.03.027). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellogg RA, Tian C, Lipniacki T, Quake SR, Tay S, Digital signaling decouples activation probability and population heterogeneity. Elife 4, e08931 (2015); published online EpubOct 21 ( 10.7554/eLife.08931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Z, Jin L, Platisa J, Cohen LB, Baker BJ, Pieribone VA, Fluorescent protein voltage probes derived from ArcLight that respond to membrane voltage changes with fast kinetics. PLoS One 8, e81295 (2013) 10.1371/journal.pone.0081295). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, Smale ST, A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev 19, 2138–2151 (2005); published online EpubSep 15 ( 10.1101/gad.1329805). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, Albracht D, Kremitzki M, Rock S, Kotkiewicz H, Kremitzki C, Wollam A, Trani L, Fulton L, Fulton R, Matthews L, Whitehead S, Chow W, Torrance J, Dunn M, Harden G, Threadgold G, Wood J, Collins J, Heath P, Griffiths G, Pelan S, Grafham D, Eichler EE, Weinstock G, Mardis ER, Wilson RK, Howe K, Flicek P, Hubbard T, Modernizing reference genome assemblies. PLoS Biol 9, e1001091 (2011); published online EpubJul ( 10.1371/journal.pbio.1001091). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013); published online EpubJan 1 ( 10.1093/bioinformatics/bts635). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudge JM, Harrow J, Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome 26, 366–378 (2015); published online EpubOct ( 10.1007/s00335-015-9583-x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011); published online EpubAug 4 ( 10.1186/1471-2105-12-323). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madsen JG, Schmidt SF, Larsen BD, Loft A, Nielsen R, Mandrup S, iRNA-seq: computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res 43, e40 (2015); published online EpubMar 31 ( 10.1093/nar/gku1365). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010); published online EpubMar 15 ( 10.1093/bioinformatics/btq033). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014); published online EpubApr 1 ( 10.1093/bioinformatics/btt656). [DOI] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010); published online EpubJan 1 ( 10.1093/bioinformatics/btp616). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008); published online EpubDec 29 ( 10.1186/1471-2105-9-559). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds AP, Richards G, de la Iglesia B, Rayward-Smith V. J. J. J. o. M. M., Algorithms, Clustering Rules: A Comparison of Partitioning and Hierarchical Clustering Algorithms. 5, 475–504 (2006); published online EpubDecember 01 ( 10.1007/s10852-005-9022-1). [DOI] [Google Scholar]
- 43.Gu Z, Eils R, Schlesner M, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016); published online EpubSep 15 ( 10.1093/bioinformatics/btw313). [DOI] [PubMed] [Google Scholar]
- 44.Yu G, Wang LG, Han Y, He QY, clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012); published online EpubMay ( 10.1089/omi.2011.0118). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29 (2000); published online EpubMay ( 10.1038/75556). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. The Gene Ontology, Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45, D331–D338 (2017); published online EpubJan 4 ( 10.1093/nar/gkw1108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B, Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 (1998); published online EpubDec 11 ( [DOI] [PubMed] [Google Scholar]
- 49.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO, Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 (2007); published online EpubSep 21 ( 10.1016/j.cell.2007.09.008). [DOI] [PubMed] [Google Scholar]
- 50.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S, Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3, 196–200 (2002); published online EpubFeb ( 10.1038/ni758). [DOI] [PubMed] [Google Scholar]
- 51.Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, Lipford G, Bauer S, Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol 3, 499 (2002); published online EpubJun ( 10.1038/ni0602-499). [DOI] [PubMed] [Google Scholar]
- 52.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A, The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A 97, 13766–13771 (2000); published online EpubDec 5 ( 10.1073/pnas.250476497). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM, Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med 197, 1107–1117 (2003); published online EpubMay 5 ( 10.1084/jem.20021787). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA, Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001); published online EpubOct 18 ( 10.1038/35099560). [DOI] [PubMed] [Google Scholar]
- 55.Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB, Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A 98, 9237–9242 (2001); published online EpubJul 31 ( 10.1073/pnas.161293498). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurk M, Schulte B, Kritzler A, Noll B, Uhlmann E, Wader T, Schetter C, Krieg AM, Vollmer J, C-Class CpG ODN: sequence requirements and characterization of immunostimulatory activities on mRNA level. Immunobiology 209, 141–154 (2004) 10.1016/j.imbio.2004.02.006). [DOI] [PubMed] [Google Scholar]
- 57.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A, The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001); published online EpubApr 26 ( 10.1038/35074106). [DOI] [PubMed] [Google Scholar]
- 58.Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, Sher A, TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308, 1626–1629 (2005); published online EpubJun 10 ( 10.1126/science.1109893). [DOI] [PubMed] [Google Scholar]
- 59.Koblansky AA, Jankovic D, Oh H, Hieny S, Sungnak W, Mathur R, Hayden MS, Akira S, Sher A, Ghosh S, Recognition of profilin by Toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity 38, 119–130 (2013); published online EpubJan 24 ( 10.1016/j.immuni.2012.09.016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Single-cell dynamics characteristics.
Table S2. Complete list of differentially expressed genes and correlations.
Table S3. List of RelA versus c-Rel specificity in correlations.
Fig. S1. Expression of NF-κB fusion proteins in stably transfected cells.
Fig. S2. TLR expression in macrophages and fibroblasts.
Fig. S3. TLR ligand–induced expression of NF-κB target genes and correlations with NF-κB dynamics.
Fig. S4. Reclassification of TLR-induced genes based on the correlation of changes in mature versus nascent transcripts to NF-κB dynamics.
Fig. S5. Correlation of TLR ligand–induced transcriptional changes with NF-κB dynamics.
Table S4. Primers used to generate expression plasmids containing mEGFP-RelA and mKOκ-c-Rel coding sequences.