Abstract
For proteins and DNA secondary structures such as G-quadruplexes and i-motifs, nanoconfinement can facilitate their folding and increase structural stabilities. However, properties of physiologically prevalent B-DNA duplex have not been elucidated inside nanocavity. By using a 17-bp DNA duplex in the form of a hairpin stem, here, we probed folding and unfolding transitions of the hairpin DNA duplex inside a DNA origami nanocavity. Compared to the free solution, the DNA hairpin inside the nanocage with a 15×15 nm cross section showed a drastic decrease in mechanical (20→9 pN) and thermodynamic (25→6 kcal/mol) stabilities. Free energy profiles revealed that activation energy of unzipping the hairpin DNA duplex decreased dramatically (28→8 kcal/mol) whereas the transition state moved closer to the unfolded state inside nanocage. All these indicate that nanoconfinement weakens the stability of hairpin DNA duplex to an unexpected extent. In a DNA hairpin made of a stem that contains complementary telomeric G-quadruplex (GQ) and i-motif (iM) forming sequences, the formation of the Hoogsteen base pairs underlining the GQ or iM is preferred over the Watson-Crick base pairs in the DNA hairpin. These results shed light on the behavior of DNA in nanochannels, nanopores, or nanopockets of various natural or synthetic machineries. It also elucidates an alternative pathway to populate non-canonical DNA over B-DNA in cellular environment where nanocavity is abundant.
Keywords: B-DNA, nanoconfinement, water activity, hairpin, tetraplex
Graphical Abstract
INTRODUCTION
Elucidating the property of duplex DNA in nanoconfinement is of fundamental importance in many fields ranging from single-molecule biophysics to DNA sequencing. Recently, individual DNA molecules have been stretched inside nanochannels to investigate the interaction between proteins and DNA.1 Biochemical reactions such as enzymatic digestions and RNA transcriptions can also be investigated using confined DNA templates. In approaches leveraged for next generation sequencing, DNA strands are guided through nanopores or nanochannels for accurate reading of individual bases or specific DNA segments.2–3 Results from these experiments are often interpreted with the speculation that behavior of DNA inside nanoconfinement remains the same as that in free solutions. Changes in the stability of duplex DNA bring complexity to these processes. Inside cells, DNA strands are often constricted in nanocavities of DNA binding proteins or DNA processing machineries. In telomerase for example, semi-enclosed pocket exists to clasp telomere DNA template.4 In polymerases, nanometer sized reaction sites are abundant for DNA strands. Therefore, it is necessary to reveal the behavior of confined B-DNA duplex to fully understand these fundamental biochemical processes.
Nanoconfinement is known to increase the stability of proteins 5–7 and non-B DNA structures such as G-quadruplexes and i-motifs 8–11. These species share one common feature: water molecules are lost during the folding.12–13 Inside nanocavity with hydrophilic walls as those found in DNA origami nanoassemblies, water molecules become increasingly ordered when cavity gets smaller due to increased ion-dipole interactions.9 The resultant decreased water activity in the smaller cavity provides a driving force to accommodate released water molecules during folding of macromolecules, which increase stabilities of the macromolecules. In duplex DNA, although there is a net release of water molecules during DNA hybridization,14 the interaction of water molecules to the minor groove of dsDNA gets stronger whereas no significant change is observed elsewhere in the structure.15 Reduced water activity may also compromise the duplex DNA stability by weakening its base stacking.16 In addition, various reports on the stabilization or destabilization of duplex DNA in solutions of negatively charged nanoparticles or polymers suggest complex cosolute effects on DNA properties.17–19 It is therefore difficult to predict the effect of the nanoconfinement on the property of the DNA duplex.
In this work, we quantify for the first time the stability of B-DNA in the stem of DNA hairpin in nanocavity. DNA hairpin is made of a duplex DNA stem with a single-stranded loop. Upon unfolding of the hairpin, the Watson-Crick base pairs in the stem de-hybridize whereas the loop remains single-stranded. Therefore, stability of the hairpin is governed by the stem. Since the hairpin stem is stabilized by the same Watson-Crick base pairs and base stacking in double stranded B-DNA,16 it has been well accepted that DNA hairpin stem is a good mimic of B form of DNA.20 Here, we placed a DNA hairpin inside a DNA origami nanocage with 15 × 15 nm cross section. Using mechanical unfolding in an optical tweezers instrument, we found that mechanical stability of the hairpin decreases from 20.2 pN outside nanocage to 9.4 pN inside nanocage, which demonstrated that duplex DNA became weakened to an unexpectedly low level inside nanocages. Using population analyses, we retrieved unfolding free energy trajectories of free and confined DNA hairpins. We found that the energy barrier to unfold DNA hairpins is much reduced in nanocages compared to free solutions. Next, we compared the formation of the B-DNA with non-B DNA using a hairpin that contains G-quadruplex and i-motif forming sequences in the two complementary stem strands. We revealed that inside nanocage only 2% population was hairpin duplex whereas 62% was tetraplex structures. These findings shed light on the property of physiologically prevalent B-DNA inside nanochannels, nanopores, or nanopockets of natural or synthetic machineries. Given the abundance of DNA sequences with a propensity to form non-canonical DNA structures in human genome,21 our finding reveals a new physiological situation in which non-B DNA structures are preferred over B form of DNA.
MATERIALS AND METHODS
Materials
All the chemicals, unless specified, were purchased either from VWR (www.vwr.com) or Nacalai Tesque (www.nacalai.com). Bovine serum albumin (BSA, biotechnology grade) was purchased from Amresco. p8064 plasmid and all DNA staples were purchased from Eurofins Genomics. All the oligos modified with biotin, digoxigenin, photocleavable linker and PEG linker were obtained from Japan Bio Services. The pET-26b (+) plasmid for handle preparation was obtained from Novagen. The Sephacryl S-300 and the gel-filtration column were purchased from GE Healthcare and Bio-Rad Laboratories respectively. The streptavidin or anti-digoxigenin coated polystyrene beads were purchased from Spherotech.
Synthesis of the DNA origami nanocages that contain hairpin hosting DNA fragment
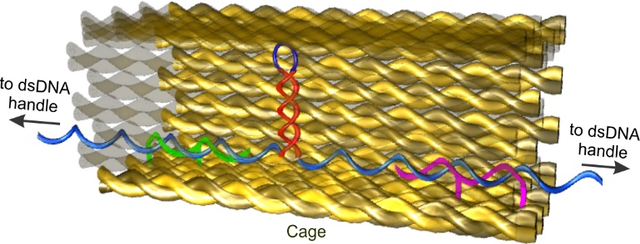
The DNA nanocage structures were designed using the protocol described elsewhere.8 In short, for the preparation of each nanocage (Figure 1), p8064 plasmid was digested with specific restriction enzymes in presence of complementary primer strands shown in Table S1. The scaffold ssDNA was purified by agarose gel followed by quantification. For the synthesis of nanocage, 25 nM of DNA scaffold was isothermally assembled at 50 °C for 1 hour with 0.2 μM of staple sequences (see Table S2) to form open nanocages. The hairpin forming sequences (see Figure S3, the stem region was taken from the sequence of bcl-2 promotor 22; and Figure S4, the stem contained human telomeric tetraplexes sequences) were placed inside the open nanocage with the help of two photocleavable guide and two capture strands (Figures S1&S2) followed by closing of the nanocage by using 4 equiv. closing staples (see Tables S2). The product was purified by hand-packed Sephacryl S-400 gel-filtration column. The purified nanocages were annealed with two double-stranded DNA handles (each 2520-bp in length) by slowly cooling the mixture from 40 to 15 °C at a rate of −1 °C per min.
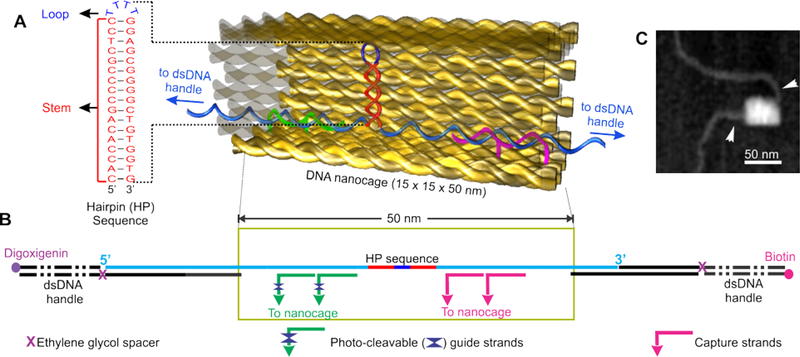
Figure 1.
Design and characterization of the hairpin@nanocage construct. (A) Schematic of the DNA construct containing hairpin forming sequence inside the DNA nanocage. Sequence of the DNA hairpin is shown to the left. (B) A hairpin (HP) forming sequence (taken from the bcl-2 promoter) inside nanocage with two dsDNA handles, which are labelled with biotin and digoxigenin at the two ends, respectively, for affinity attachments. (C) AFM image of a nanocage after annealing with dsDNA handles (arrowheads).
Characterization of the single molecular DNA nanocage constructs by AFM
AFM images (Figures 1c & S5) were obtained at scan rate of 0.2 frames per second (fps) in an AFM system (Nano Live Vision, RIBM, Tsukuba, Japan) with a silicon nitride cantilever (resonant frequency = 1.0 – 2.0 MHz, spring constant = 0.1 – 0.3 N/m, EBD Tip radius <15 nm, Olympus BLAC10EGS-A2). The sample preparation for imaging was done by adsorption of 2 μL sample onto a freshly cleaved mica plate [Φ 1.5 mm, pretreated with 0.1% 3-aminopropyl trimethoxysilane (APTES)] for 5 min at room temperature followed by several washing with 20 mM Tris buffer (pH 7.8) containing 10 mM MgCl2 and 1 mM EDTA.
Mechanical unfolding experiments in optical tweezers
First, 0.5 μL of sample was exposed to 365 nm UV for 10 min to break the photocleavable linker X in guide strands to avoid unwanted strain on the nanocage during mechanical unfolding’s. By incubation of the exposed sample with streptavidin coated polystyrene bead, the construct was immobilized on the surface of the bead via streptavidin/biotin linkage. The immobilized DNA on streptavidin coated beads and anti-digoxigenin beads were flowed into top and bottom channels of the three-channel microfluidic chamber, respectively. These beads were flowed to the middle channel via two micropipettes (id: 25 μm, King Precision Glass, Claremont, CA) connecting top and bottom channels respectively to the middle channel. Each bead was separately trapped by a 1,064 nm laser beam in a custom-made dual-trap laser tweezers. Two beads were brought closer to each other by a steerable mirror in the laser tweezers instrument to form a DNA tether by digoxigenin/anti-digoxigenin interaction between the free end of the DNA and the anti-digoxigenin coated bead. The tether was stretched and relaxed at the loading rate of ~5.5 pN/s by the same steerable mirror. The force versus extension (F-X) traces were recorded at 1,000 Hz using a Labview program. The experiments were carried out in a 20 mM Tris (pH 7.8) buffer or a 10 mM MES (pH 5.5) buffer supplemented with 10 mM MgCl2, 100 mM KCl, and 1 mM EDTA at 25 °C.
RESULTS
Preparation of DNA hairpins inside DNA origami nanocage
Mechanical unfolding and refolding experiments were performed in an optical-tweezers instrument described previously.23 First, we placed a DNA hairpin sequence (5′-CACCACAGCCCCGCTCC-TTTT-GGAGCGGGGCTGTGGTG, the stem sequence (underlined) is taken from the bcl-2 promoter) inside a DNA origami nanocage assembly8–9 (Figure 1a&b). Two ends of the hairpin stem were tethered to two duplex DNA handles, which were attached to the two optically trapped polystyrene beads by affinity linkages. Two sides of the nanocage were left open to allow the passage of each DNA handle. This design ensured that force is applied directly on the DNA hairpin for mechanical unfolding and refolding experiments. To ensure that DNA hairpin is contained inside nanocage, the nanocage is always anchored to one of the DNA pulling handles via two capture strands close to the hairpin (see Figure 1b and SI for details). Molecular simulation revealed that the DNA hairpin formed inside a 9×9 nm nanocage (which is smaller than the 15×15 nm nanocage used here) is not sterically hindered (Figure S15). AFM images have revealed successful preparation of the origami construct (Figure 1c).
Mechanical unfolding and refolding of DNA hairpins inside nanocage
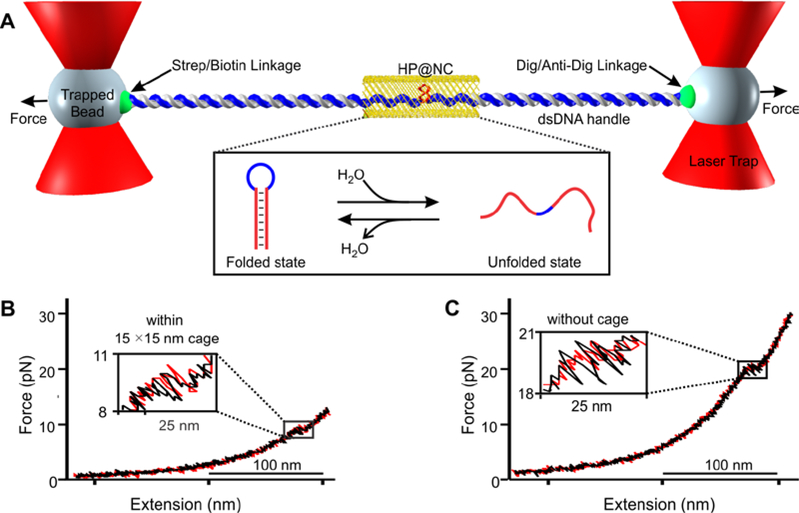
To start mechanical unfolding and refolding of individual DNA hairpins tethered between two optically trapped polystyrene particles (Figure 2a), we moved one of the trapped beads away from another using a steerable mirror at a load force of 5.5 pN/s. This increased the tension in the DNA construct until the hairpin was unfolded (Figure 2b&S10b, inset). As a control, the same experiments were carried out on the DNA construct without nanocage (Figure 2c&S10a). Compared to the unfolding force of the hairpin without nanocage (Figures 2&3, 20.2 pN), it came to our attention that unfolding force was significantly smaller for the hairpin inside nanocage (9.4 pN). The same trend was observed for the refolding forces (18.5 vs 7.4 pN (without vs within nanocage) see Figure S6).
Figure 2.
Mechanical unfolding of the bcl-2 hairpin (HP) within DNA nanocage (NC). (A) The DNA construct in Figure 1 is tethered between two optically trapped beads via affinity interactions. Inset shows unfolding and refolding transitions of the hairpin inside nanocage (HP@NC). Strep, Dig and Anti-Dig represent streptavidin, digoxigenin and anti-digoxigenin respectively. Force versus extension curve of the DNA hairpin (B) within and (C) without 15×15 nm nanocage. Red and black traces indicate stretching and relaxing curves, respectively. Zigzag features in insets depict rapid unfolding/refolding transitions. Experiments were performed in a 20 mM Tris buffer (pH 7.8) supplemented with 100 mM KCl, 10 mM MgCl2, and 1 mM EDTA at 25 °C.
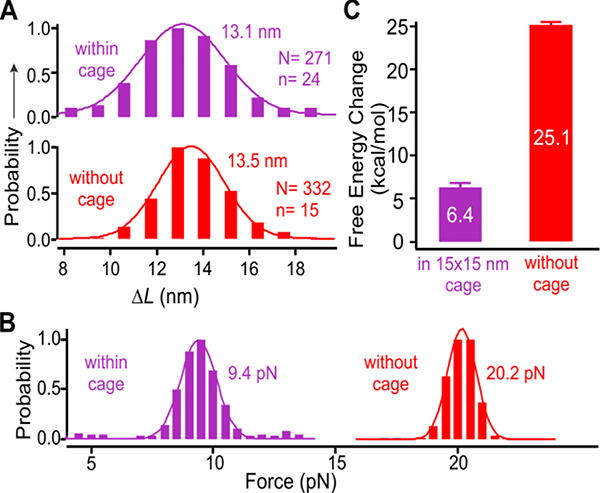
Figure 3.
Mechanical properties of the bcl-2 hairpin within (purple) and without (red) the 15 × 15 nm nanocage. (A) Change-in-contour-length (ΔL) and (B) unfolding force histograms of the bcl-2 hairpin inside and outside nanocage. (C) Change in the free energy of unzipping the bcl-2 hairpin within (purple) and without (red) nanocage. Solid curves depict Gaussian fittings. N and n depict the numbers of unfolding features and molecules, respectively.
To explain the different transition forces between these two DNA samples, we analyzed structures formed within and without nanocage. Outside nanocage, rapid and reversible folding and unfolding transitions were observed at 20.2 pN (Figures 2c&3b), which were consistent with those observed for DNA hairpins.20 When we plotted the change-in-contour-length (ΔL) histogram, we found that ΔL (13.5–14.2 nm, Figures 3a&S7) is consistent with that expected for a fully folded hairpin (expected ΔL 14.3 nm, see SI for calculation). Inside the nanocage with a 15 × 15 nm cross section, the hairpins were also fully folded as revealed by the ΔL histograms (Figures 3a&S7, ΔL ~ 13.2 nm). Therefore, the much-reduced unfolding/refolding force of the hairpin within nanocage with respect to that outside (Figures 3b & S6) must be due to different environment inside nanocage. This result demonstrated that hairpin duplex DNA in nanoconfinement has surprisingly large reduction in mechanical stability. The unexpected large reduction was also observed in the thermodynamic stability of the hairpin when the change in free energy of hairpin unfolding (ΔGunfold) was estimated from the unfolding work using the Jarzynski equality expression 24 (25.1: 6.4 kcal/mol (outside : inside nanocage), see Figure 3c and SI Figure S12 for unfolding work histograms). It is noteworthy that ΔGunfold calculated by Jarzynski equality is identical to that calculated by the mfold® method 25 (Figure S13).
Unfolding free energy profiles of the bcl-2 DNA hairpin
Next, we retrieved free energy profile of the entire unfolding trajectory of the bcl-2 hairpin using reported methods. 26–27 To this end, we collected hundreds of unfolding/refolding events of the DNA hairpins within and without nanocage. To account for baseline drifts due to different molecules, we calculated the change-in-contour-length (ΔL) between the stretching and relaxing force-extension curves (Figure 4 a&b, left panel) during the force range in which folding and unfolding transitions occur.28 Three force regions were shown in ΔL−F plots (Figure 4 a&b, middle panel). Right at the transition force, the positive and negative ΔL populations reflect the unfolding and refolding transitions, respectively, between the folded and unfolded hairpins. At the force smaller than the hairpin transition, ΔL reduces to zero, which indicates folded hairpins in both stretching and relaxing F-X curves. At the force larger than the hairpin transition, ΔL reduces to zero again, corresponding to the unfolded hairpin in both stretching and relaxing F-X curves. Change-in-free-energy (ΔG(L, F)) along the unfolding coordinate (Lunfold) at a particular force (F) can be calculated by the Boltzmann equation, ΔG(L,F) = kBT ln[P(L)], where P(L) is the population probability density.26 By grouping all unfolding transitions of the hairpins around the transition force (Figure 4 a&b, right panel, only positive ΔL was considered), we were able to deconvolute the probability density P and obtain the unfolding free energy trajectory of the bcl-2 hairpin (Figure 4c) using a point spread function (PSF) 26, which was obtained from the same DNA handles without hairpin forming sequence at a specific transition force (Figure S14).
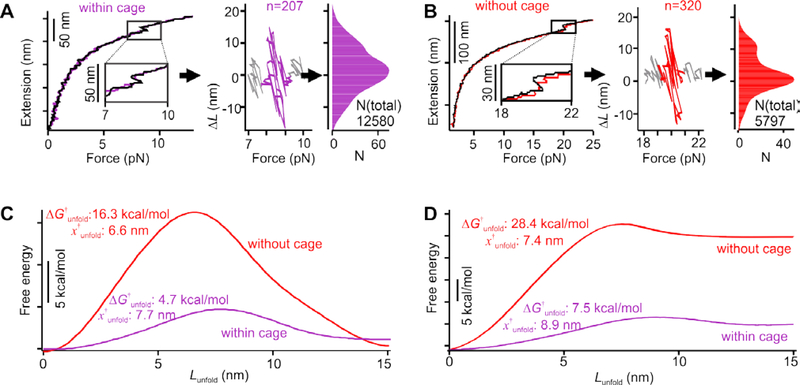
Figure 4.
Unfolding free energy profiles of the bcl-2 hairpin. Population density profiles of the bcl-2 hairpin at the transition forces within (A) and without (B) the 15 × 15 nm nanocage. Left panels, extension vs force traces of the hairpin. Colored and black traces depict stretching and relaxing processes respectively. The change-in-contour-length (ΔL) versus force (F) plots (middle panels) are calculated based on the difference in the extension between the stretching and relaxing traces at the same force around the transition events. Colors in the middle panels depict the unfolding and refolding transitions of the hairpin. Right panels show population profiles of the corresponding ΔL-F plots in the middle panels. Only populations with positive ΔL are used to obtain the unfolding energy profiles of the hairpin. The n depicts the number of transition events used for analyses. Unfolding energy profile of the bcl-2 hairpin without (red) and within (purple) nanocage at specific unfolding forces (C) and zero force (D). See text for notations.
To retrieve the unfolding free energy profile at F=0 pN (Figure 4d), we further accounted the energies stored in the dsDNA, in the unfolded hairpin, as well as in the two optical traps 26 (see SI). As these energies are either constant, which does not influence the shape of the free energy profile, or linearly proportional with respect to the reaction coordinate Lunfold, they contribute linearly to the final free energy profile. Therefore, this linear energy correction was determined by the change in the free energy between fully folded and fully unfolded hairpin, ΔGunfold (obtained by the Jarzynski equality, see above and SI). As shown in Figure 4d, we found that the energy barrier to unfold DNA hairpin (ΔGunfold†) is much reduced inside nanocage compared to that without. This observation is in contrast to the unfolding of the G-quadruplex and i-motif, which demonstrated much larger ΔG†unfold in nanoconfinement.8–9 Significantly, we also found that the transition state of the hairpin in nanoconfinement at zero force is located closer to the unfolded state (the distance between the folded to the transition states, x†unfold, is 8.9 nm, Figure 4d) than that observed in the free solution (x†unfold = 7.4 nm), indicating that hairpin structure becomes softer in confined space.29 This trend falls into the Leffler−Hammond postulate that defines the correlation between the force and the position of the transition state.30–31
Competitive formation of the B-DNA versus non-B DNA in nanoconfinement
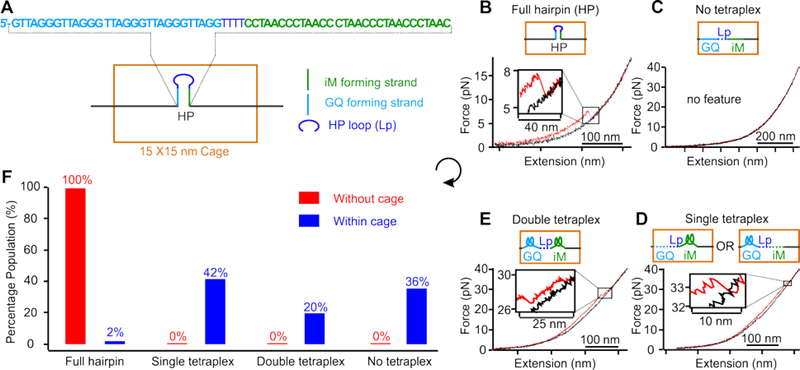
Compared to the much-increased stability of DNA G-quadruplex and i-motif structures in DNA nanocages, 8–9 the drastically decreased stability in duplex DNA suggests that in the confinement of many cellular machineries, formation of non-B DNA structures, such as G-quadruplex and i-motif tetraplexes which employ Hoogsteen base pairs, can be preferred over duplex DNA. To test this hypothesis, we compared the formation probability of the hairpin duplex DNA and the tetraplex structures using a hairpin in which G-quadruplex and i-motif forming sequences are placed in the two complementary stem strands (Figure 5a). In this design, the formations of the fully folded hairpin and any of the two tetraplexes (G-quadruplex or i-motif) are mutually exclusive. It is noteworthy that the nanocage with a 15×15 nm cross section can readily accommodate the hairpin presented in any orientation (the 30-bp stem of the hairpin has 10.2 nm in length) (Figure S15e). After the DNA construct was placed inside the 15×15 nm nanocage (Figure S5), mechanical unfolding experiments were performed in a 10 mM MES buffer (pH 5.5) supplemented with 100 mM KCl, 10 mM MgCl2, and 1 mM EDTA at 25 °C (Figures 5b–e, S8, S9, and S11). Previous studies have demonstrated that telomeric G-quadruplex and i-motif32, as well DNA hairpins33, can form under similar conditions (pH 5.5 and 100 mM KCl).
Figure 5.
Competitive formation of B-DNA versus non-B DNA in nanoconfinement. (A) Schematic of a 15×15 nm nanocage that contains a hairpin with i-motif (iM) and G-quadruplex (GQ) forming sequences in complementary strands in the hairpin stem. Typical force versus extension curves of the hairpin inside nanoconfinement showing (B) full hairpin, (C) no tetraplex, (D) single tetraplex, and (E) double tetraplex formations. (F) Percentage populations of folded species without (red) and within (blue) the nanocage. Assignment of folded structures in each hairpin is based on ΔL and unfolding force histograms (see text and Figure S8; see Figure S9 for all observed structures). Note the “No tetraplex” population contains 8% partially folded hairpins. These experiments were performed in a 10 mM MES buffer (pH 5.5) supplemented with 100 mM KCl, 10 mM MgCl2, and 1 mM EDTA at 25 °C.
Each of the two tetraplexes is expected to have higher unfolding force and lower ΔL than fully folded hairpins (Figure S8). Therefore, from the magnitude of the ΔL and the rupture force associated with each unfolding feature in individual F-X traces, we were able to identify folded structures in the hairpin (Figures 5&S9). We revealed that only 2% population was fully folded hairpin, whereas 42% and 20% were single and double tetraplex structures, respectively, inside the 15×15 nm nanocage (Figures 5f&S9). In comparison, without nanocage (Figure S8a&b), the population was exclusively fully folded hairpin. These results confirmed that the B-DNA was weaker than the non-B DNA tetraplex structures in nanoconfinement.
DISSCUSSION
At the molecular level, it has been shown that increased unfolding energy barriers for G-quadruplex and i-motif in nanoconfinement are due to the hydration of water molecules during the transition.9 In the nanocavity confined by charged walls such as those in the DNA origami nanoassembly, water molecules are well aligned with reduced activities due to increased ion-dipole interactions.8 Therefore, it is more difficult to interact with water in the nanocage during unfolding of the DNA tetraplexes, which increases the energy barrier. In duplex DNA, much-decreased stability of dsDNA was observed in molecularly crowded solution at a similar ionic strength, which was fully consistent with what we observed here.34 The decreased stability was ascribed to the cosolute-mediated hydration during the hybridization of duplex DNA. 34 Without cosolute, investigations now indicated the release of water molecules during the folding of duplex DNA.14 However, it has been found that in the minor groove of duplex DNA, binding of the water molecule becomes much tighter compared to the ssDNA, whereas no significant difference is found for the phosphate group or the major groove.15 The much-compromised stability of duplex DNA in nanocage can be attributed to this predominating enthalpic factor. Inside DNA origami nanocage, water molecules with much-reduced activities 9 are sluggish to tightly interact with the minor groove of the dsDNA, which decreases the stability of the duplex DNA in the nanocage. In other studies, similar destabilization effect on the duplex DNA from reduced water activity has been attributed to the weakening of the base stacking in duplex DNA.16, 35 Molecular simulation revealed that the stem of the hairpin inside the nanocage is located towards the walls of the nanocage (Figure S15 A&B), which presented lower water activities compared to the center of the nanocage due to increased ion-dipole interactions close to the nanocage surface. After quantification of this geometrical effect, we found reduced water activity contributed at least 87% of reduced stability of the 17-bp bcl-2 hairpin inside the 15×15 nm nanocage (see SI, Figure S16). Other than the water activity effect, it is also possible that cations interacting with negatively charged origami surface may result in reduced cation activity in nanoconfinement, which compromises the stability of duplex DNA by the reduced charge screening effect. In addition, the repulsive force between the DNA hairpin and the nanocage wall may destabilize the hairpin structure. This is because the unfolded form of the hairpin (ssDNA) is expected to maintain a longer distance to the nanocage wall due to its more flexible nature with respect to the duplex DNA in the hairpin stem. Molecular dynamics simulation can be used to better understand these different factors on the stability of DNA structures36,37 in nanoconfinement.
The surprisingly low stabilities of B-DNA in nanoconfinement give ramifications to correctly interpret results obtained from experiments where duplex DNA is imaged or analyzed in nanochannels or nanopores especially with negatively charged surfaces. This behavior is also of high physiological significance. Inside cells, DNA can be temporarily confined in many machineries employed for processes such as transcriptions and replications. Our results indicate that nanoconfinement weakens B-DNA, making it easier to unwind DNA duplex for biochemical processes such as the formation of the open complex during transcription initiation or propagation of the transcription bubble.38 At the same time, non-canonical DNA structures in the same DNA region become more stable in the confinement.8–9 As a result, population equilibrium shifts to favor the formation of non-B DNA structures. As these non-canonical structures have demonstrated regulatory roles,39 the nanoconfinement can offer a unique way to modulate cellular processes. From this perspective, the nanoconfinement bears similarity to other cellular environment such as molecular crowding 12 and torsional constrained template 40–41. Whereas the former condition often has a global effect applicable to the entire system at the steady state, the latter is quite dynamic in nature 40. The nanoconfinement, however, provides localized environment with its efficacy determined by the availability as well as the number of cellular machineries (or nanocavities) working on DNA templates.
CONCLUSIONS
By mechanical unfolding and refolding of DNA hairpins inside a DNA origami nanocage using optical tweezers, we quantified the property of B-DNA in nanoconfinement. We found both mechanical and thermodynamic stabilities of B-DNA hairpin decrease to unexpectedly low levels inside nanocavity. Direct comparison for the formation of hairpin duplex DNA versus tetraplex DNA revealed preferential formation of tetraplex structures. These surprising results shed light on many in vivo or in vitro processes in which DNA and associated components are confined inside nanochannels, nanopores, or nano reaction sites. They reveal a new pathway by which non-B DNA structures become preferred species in the context of double-stranded DNA, which justifies purported regulatory roles of non-canonical DNA structures for many biochemical processes inside cells.
Supplementary Material
ACKNOWLEDGMENT
H.M. thanks financial support from National Science Foundation [CHE-1904921] and National Institutes of Health [NIH 1R01CA236350]. This work was supported by a Grant-in-Aid for Scientific Research JSPS KAKENHI Fund for the Promotion of Joint International Research (Fostering Joint International Research (B)) (Grant Numbers 18KK0139 and 16H06356)) to H.S. and M.E. Financial supports from the Nakatani Foundation and the Uehara Memorial Foundation to M.E. were acknowledged. We thank Tomoko Emura and Kumi Hidaka for the preparation of DNA hairpin inside the nanocage.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Synthesis of the DNA origami nanocages that contain hairpin hosting DNA fragment, Synthesis strategy for the single-stranded DNA containing the bcl-2 hairpin forming sequence, Synthesis strategy for the single-stranded DNA that contains a telomeric G-quadruplex sequence and a telomeric i-motif sequence, Characterization of the single molecular DNA nanocage constructs by AFM, Unfolding/refolding force and change-in-contour-length measurements, Percentage populations of the hairpin and tetraplex structures, Unfolding force versus extension plots, Expected change-in-contour-length (ΔL), Calculation of the change in free energy of unfolding (ΔGunfold), Estimation of change in free energy of hairpin unfolding using mfold®, Energy stored in the unfolding pathway of the bcl-2 hairpin, Point spread function used to convert species populations to relative free energies, Coarse-grained molecular dynamics (MD) simulation of bcl-2 hairpin inside the nanocage, Quantitation of factors in the reduced stability of the duplex DNA, Supplemental references.
The authors declare no competing financial interests.
REFERENCES
- 1.Persson F; Tegenfeldt JO, DNA in nanochannels—directly visualizing genomic information. Chemical Society Reviews 2010, 39 (3), 985–999. [DOI] [PubMed] [Google Scholar]
- 2.Clarke J; Wu H-C; Jayasinghe L; Patel A; Reid S; Bayley H, Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotechnology 2009, 4 (4), 265–270. [DOI] [PubMed] [Google Scholar]
- 3.Jin Q; Fleming AM; Burrows CJ; White HS, Unzipping Kinetics of Duplex DNA Containing Oxidized Lesions in an α-Hemolysin Nanopore. Journal of the American Chemical Society 2012, 134 (26), 11006–11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell M; Gillis A; Futahashi M; Fujiwara H; Skordalakes E, Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat. Struct. Mol. Biol. 2010, 17 (4), 513–518. [DOI] [PubMed] [Google Scholar]
- 5.Zhou H-X; Dill KA, Stabilization of Proteins in Confined Spaces. Biochemistry 2001, 40 (38), 11289–11293. [DOI] [PubMed] [Google Scholar]
- 6.Takagi F; Koga N; Takada S, How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: Molecular simulations. Proc. Natl Acad. Sci. U.S.A 2003, 100 (20), 11367–11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinker A; Pfeifer G; Kerner MJ; Naylor DJ; Hartl FU; Hayer-Hartl M, Dual Function of Protein Confinement in Chaperonin-Assisted Protein Folding. Cell 2001, 107 (2), 223–233. [DOI] [PubMed] [Google Scholar]
- 8.Shrestha P; Jonchhe S; Emura T; Hidaka K; Endo M; Sugiyama H; Mao H, Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 2017, 12 (6), 582–588. [DOI] [PubMed] [Google Scholar]
- 9.Jonchhe S; Pandey S; Emura T; Hidaka K; Hossain MA; Shrestha P; Sugiyama H; Endo M; Mao H, Decreased water activity in nanoconfinement contributes to the folding of G-quadruplex and i-motif structures. Proc. Natl. Acad. Sci. U S A 2018, 115 (38), 9539–9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pramanik S; Nagatoishi S; Sugimoto N, DNA tetraplex structure formation from human telomeric repeat motif (TTAGGG):(CCCTAA) in nanocavity water pools of reverse micelles. Chem. Comm. 2012, 48 (40), 4815–4817. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J; Wei C; Jia G; Wang X; Feng Z; Li C, Formation and stabilization of G-quadruplex in nanosized water pools. Chem. Commun. 2010, 46 (10), 1700–1702. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi D; Karimata H; Sugimoto N, Hydration Regulates Thermodynamics of G-Quadruplex Formation under Molecular Crowding Conditions. J. Am. Chem. Soc. 2006, 128 (24), 7957–7963. [DOI] [PubMed] [Google Scholar]
- 13.Zhao C; Ren J; Qu X, Single-Walled Carbon Nanotubes Binding to Human Telomeric i-Motif DNA Under Molecular-Crowding Conditions: More Water Molecules Released. Chem. Eur. J. 2008, 14, 5435–5439. [DOI] [PubMed] [Google Scholar]
- 14.Son I; Shek YL; Dubins DN; Chalikian TV, Hydration Changes Accompanying Helix-to-Coil DNA Transitions. Journal of the American Chemical Society 2014, 136 (10), 4040–4047. [DOI] [PubMed] [Google Scholar]
- 15.Nakano M; Tateishi-Karimata H; Tanaka S; Tama F; Miyashita O; Nakano S.-i.; Sugimoto N, Local thermodynamics of the water molecules around single- and double-stranded DNA studied by grid inhomogeneous solvation theory. Chemical Physics Letters 2016, 660, 250–255. [Google Scholar]
- 16.Feng B; Sosa RP; Mårtensson AKF; Jiang K; Tong A; Dorfman KD; Takahashi M; Lincoln P; Bustamante CJ; Westerlund F; Nordén B, Hydrophobic catalysis and a potential biological role of DNA unstacking induced by environment effects. Proceedings of the National Academy of Sciences 2019, 116 (35), 17169–17174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khimji I; Shin J; Liu J, DNA duplex stabilization in crowded polyanion solutions. Chemical Communications 2013, 49 (13), 1306–1308. [DOI] [PubMed] [Google Scholar]
- 18.Moriyama R; Iwasaki Y; Miyoshi D, Stabilization of DNA Structures with Poly(ethylene sodium phosphate). The Journal of Physical Chemistry B 2015, 119 (36), 11969–11977. [DOI] [PubMed] [Google Scholar]
- 19.Zinchenko A; Tsumoto K; Murata S; Yoshikawa K, Crowding by Anionic Nanoparticles Causes DNA Double-Strand Instability and Compaction. The Journal of Physical Chemistry B 2014, 118 (5), 1256–1262. [DOI] [PubMed] [Google Scholar]
- 20.Woodside MT; Behnke-Parks WM; Larizadeh K; Travers K; Herschlag D; Block SM, Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. U S A 2006, 103 (16), 6190–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huppert JL; Balasubramanian S, Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33 (9), 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui Y; Koirala D; Kang H; Dhakal S; Yangyuoru P; Hurley LH; Mao H, Molecular Population Dynamics of DNA Structures in a Bcl-2 Promoter Sequence is Regulated by Small-molecules and the Transcription Factor hnRNP LL. Nucleic Acids Res. 2014, 42, 5755–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z; Schonhoft JD; Dhakal S; Bajracharya R; Hegde R; Basu S; Mao H, ILPR G-Quadruplexes Formed in Seconds Demonstrate High Mechanical Stabilities. J. Am. Chem. Soc. 2009, 131 (5), 1876–1882. [DOI] [PubMed] [Google Scholar]
- 24.Collin D; Ritort F; Jarzynski C; Smith SB; Tinoco IJ; Bustamante C, Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature 2005, 437, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M, Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodside MT; Anthony PC; Behnke-Parks WM; Larizadeh K; Herschlag D; Block SM, Direct Measurement of the Full, Sequence-Dependent Folding Landscape of a Nucleic Acid. Science 2006, 314, 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koirala D; Punnoose JA; Shrestha P; Mao H, Yoctoliter thermometry for single-molecule investigations: A generic bead-on-a-tip temperature-control module. Angew. Chem. Int. Ed. Engl. 2014, 53 (13), 3470–3474. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z; Gaerig V; Cui Y; Kang H; Gokhale V; Zhao Y; Hurley LH; Mao H, Tertiary DNA Structure in the Single-Stranded hTERT Promoter Fragment Unfolds and Refolds by Parallel Pathways via Cooperative or Sequential Events. J. Am. Chem. Soc. 2012, 134 (11), 5157–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alemany A; Ritort F, Force-Dependent Folding and Unfolding Kinetics in DNA Hairpins Reveals Transition-State Displacements along a Single Pathway. The Journal of Physical Chemistry Letters 2017, 8 (5), 895–900. [DOI] [PubMed] [Google Scholar]
- 30.LEFFLER JE, Parameters for the Description of Transition States. Science 1953, 117 (3039), 340–341. [DOI] [PubMed] [Google Scholar]
- 31.Hammond GS, A Correlation of Reaction Rates. Journal of the American Chemical Society 1955, 77 (2), 334–338. [Google Scholar]
- 32.Cui Y; Kong D; Ghimire C; Xu C; Mao H, Mutually Exclusive Formation of G-Quadruplex and i-Motif Is a General Phenomenon Governed by Steric Hindrance in Duplex DNA. Biochemistry 2016, 55 (15), 2291–2299. [DOI] [PubMed] [Google Scholar]
- 33.Jonchhe S; Shrestha P; Ascencio K; Mao H, A New Concentration Jump Strategy Reveals the Lifetime of i-Motif at Physiological pH without Force. Analytical Chemistry 2018, 90 (3), 1718–1724. [DOI] [PubMed] [Google Scholar]
- 34.Nakano M; Tateishi-Karimata H; Tanaka S; Tama F; Miyashita O; Nakano S -i.; Sugimoto, N., Thermodynamic properties of water molecules in the presence of cosolute depend on DNA structure: a study using grid inhomogeneous solvation theory. Nucleic Acids Research 2015, 43 (21), 10114–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hormeño S; Ibarra B; Valpuesta JM; Carrascosa JL; Ricardo Arias-Gonzalez J, Mechanical stability of low-humidity single DNA molecules. Biopolymers 2012, 97 (4), 199–208. [DOI] [PubMed] [Google Scholar]
- 36.Galindo-Murillo R; Roe DR; Cheatham TE, Convergence and reproducibility in molecular dynamics simulations of the DNA duplex d(GCACGAACGAACGAACGC). Biochimica et Biophysica Acta (BBA) - General Subjects 2015, 1850 (5), 1041–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panczyk T; Wojton P; Wolski P, Mechanism of unfolding and relative stabilities of G-quadruplex and I-motif noncanonical DNA structures analyzed in biased molecular dynamics simulations. Biophysical Chemistry 2019, 250, 106173. [DOI] [PubMed] [Google Scholar]
- 38.Djordjevic M; Bundschuh R, Formation of the open complex by bacterial RNA polymerase--a quantitative model. Biophysical Journal 2008, 94 (11), 4233–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddiqui-Jain A; Grand CL; Bearss DJ; Hurley LH, Direct Evidence for a G-quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c-MYC Transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouzine F; Liu J; Sanford S; Chung HJ; Levens D, The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 2004, 11 (11), 1092–100. [DOI] [PubMed] [Google Scholar]
- 41.Selvam S; Koirala D; Yu Z; Mao H, Quantification of Topological Coupling between DNA Superhelicity and G-quadruplex Formation. J. Am. Chem. Soc. 2014, 136, 13967–13970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.