Abstract
Purpose
Survivin (BIRC5) is a promising tumor biomarker. Conflicting data exist on its prognostic effect in breast cancer. These data may at least be partly due to the manual interpretation of immunohistochemical staining, especially as survivin can be located in both the nucleus and cytoplasm. Quantitative determination of survivin expression using image analysis offers the opportunity to develop alternative scoring models for survivin immunohistochemistry. Here, we present such a model.
Experimental Design
A breast cancer tissue microarray containing 102 tumors was stained with an anti-survivin antibody Whole-slide scanning was used to capture high-resolution images. These images were analyzed using automated algorithms to quantify the staining.
Results
Increased nuclear, but not cytoplasmic, survivin was associated with a reduced overall survival (OS; P = 0.038) and disease-specific survival (P = 0.0015). A high cytoplasmic-to-nuclear ratio (CNR) of survivin was associated with improved OS (P = 0.005) and disease-specific survival (P = 0.05). Multivariate analysis revealed that the survivin CNR was an independent predictor of OS (hazard ratio, 0.09; 95% confidence interval, 0.01–0.76; P = 0.027). A survivin CNR of >5 correlated positively with estrogen receptor (P = 0.019) and progesterone receptor (P = 0.033) levels, whereas it was negatively associated with Ki-67 expression (P = 0.04), p53 status (P = 0.005), and c-myc amplification (P = 0.016).
Conclusion
Different prognostic information is supplied by nuclear and cytoplasmic survivin in breast cancer. Nuclear survivin is a poor prognostic marker in breast cancer. Moreover, CNR of survivin, as determined by image analysis, is an independent prognostic factor.
Survivin (BIRC5) is a member of the inhibitor of apoptosis protein family. It is a multifunctional protein that inhibits apoptosis, regulates cell division, and enhances angiogenesis (1). Early work using serial analysis of gene expression revealed that survivin was the fourth most highly expressed transcript in several common cancers but was rarely present in normal terminally differentiated tissues (2). Consequently, surviving has been the subject of intense investigation about its potential role as a tumor biomarker (2, 3). Multiple studies in several different cancers have investigated the prognostic value of survivin (see ref. 1 for review).
Studies in breast cancer have produced conflicting data about the prognostic influence of survivin. In breast cancer, its prognostic value has been reported to be nonexistent (4–6), associated with improved outcome (7), or associated with adverse outcome (8–10). Despite these conflicting findings, survivin is one of the 16 cancer-related genes represented in the Oncotype DX assay (11). Additionally, following reanalysis of gene expression microarray data published by van ‘t Veer et al. (12), we identified that survivin gene expression was specifically up-regulated in breast tumors with poor prognosis. Survivin ranked 18th of the top 100 genes associated with poor prognosis in the supervised reanalysis (13).
Immunohistochemical-based studies have failed to reach a consensus about how survivin staining should be interpreted (i.e., does examination of cytoplasmic fraction, nuclear fraction, or both provide more useful information?). In a review of the literature, Li et al. (14) identified 19 publications that measured nuclear survivin in different cancer types and showed that 9 studies concluded that nuclear survivin was associated with an unfavorable prognosis, whereas 5 showed an association with a favorable prognosis. Manual interpretation of survivin is challenging because of the presence of signal within both the cytoplasm and nucleus. This dilemma can be addressed by image analysis, whereby one can accurately identify the nuclear and cytoplasmic fractions of the stain and investigate any link between these fractions and survival.
In this study, we used high-throughput image analysis to fulfill two objectives: (a) the development of an automated quantitative scoring model for survivin and (b) the identification of new prognostic subgroups that were not evident following manual analysis.
Materials and Methods
Patients and tumor samples
The tissue microarray (TMA) used in this study was constructed from a consecutive cohort of 102 patients diagnosed with breast cancer at Umeå University Hospital (Umeå, Sweden) with a median follow-up time of 77 months and has been described previously (15, 16). The study was approved by the ethics committee at Umeå University. The only exclusion criteria for this study were tumors that were too small to snap freeze for ELISA (see below).
The median age at diagnosis was 60 years (range, 30–87 years). Patients did not receive neoadjuvant treatment and were treated with modified radical mastectomy or wide local excision and axillary lymph node dissection. All patients who had breast-conserving surgery received adjuvant radiotherapy to the remaining breast tissue. Forty-eight percent of patients were lymph node positive. Node-negative patients did not receive adjuvant therapy; however, node-positive premenopausal patients received adjuvant chemotherapy and node-positive postmenopausal patients received adjuvant tamoxifen, concurrent with local recommendations at the time. Eighty-eight percent of tumors were classified as ductal, 6% as lobular, 3% as medullary, and 3% as mucinous. The median tumor size was 2.2 cm (0.8–10 cm).
Following surgical resection, a piece of fresh tumor was immediately snap frozen in liquid nitrogen and stored at −80°C until protein extraction took place. The hormone receptor content was measured by ELISA, with 72% of tumors being estrogen receptor (ER) positive and 62% being progesterone receptor (PR) positive. Diagnostic specimens were all formalin fixed and paraffin embedded in the Department of Pathology in Umeå University Hospital. All tissue blocks were stored in this department before construction of the TMA.
Cell lines
All cell lines (MDA-MB-231, MCF-7, T47D, SKBR3, Hs578T, and BT474) were obtained from the European Collection of Cell Cultures and maintained as previously described (13, 17).
Western blot analysis
Western blot analysis was done as previously described (17). Lysates were separated by SDS-PAGE, and survivin epression was analyzed using a mouse monoclonal anti-human antibody D8 (Santa Cruz Biotechnology, Inc.) at a dilution of 1:50. An anti-β-actin antibody (Abcam) at a dilution of 1:5,000 was used as a loading control.
TMAs and immunohistochemistry
Breast cancer TMAs were constructed as previously described (16) and the blocks were stored in the Department of Pathology in Malmö University Hospital. TMA sections were cut immediately before staining. Sections (4 μm) were dried, deparaffinized, and rehydrated through descending concentrations of ethanol. Heat-mediated antigen retrieval was done using microwave treatment for 2 × 5 min in a citrate buffer before being processed either in the Ventana Benchmark system (Ventana Medical Systems, Inc.) using a prediluted antibody to Her2 (Pathway CB-USA, 760–2694) or in the Dako Techmate 500 system (Dako) for Ki-67 (1:200, M7240; Dako) and survivin (1:50, D8; Santa Cruz Biotechnology).
Manual evaluation of immunohistochemical staining
Survivin expression levels in tumor specimens were evaluated by two pathologists and scored for both nuclear and cytoplasmic staining. All discordant cases were reevaluated and a consensus was reached. Nuclear survivin was scored using the following categories for percent positive cells: 0, 1 to 5 (1+), 6 to 15 (2+), and 16 to 100 (3+). Nuclear and cytoplasmic survivin intensity was divided into four categories: negative (0), mild (1+), moderate (2+), and high staining intensity (3+; Fig. 1). The interpretation of Her2, Ki-67, and p53 immunohistochemistry, as well as c-myc amplification, has been previously described (18, 19).
Fig. 1.
Western blotting and immunohistochemical analysis using anti-survivin antibody.Western blot of survivin in a panel of breast cancer cell lines showing the specificity of the antibody. A, β-actin was used as a loading control. Survivin immunohistochemistry and H&E-stained cores. B, cytoplasmic survivin was scored manually.
Image acquisition, management, and analysis
The Aperio ScanScope CS Slide Scanner (Aperio Technologies) system was used to capture whole-slide digital images with a 20× objective. A positive pixel count algorithm (Aperio Technologies) was used to develop a qualitative scoring model for both nuclear and cytoplasmic survivin expression (discussed below).
Statistical analysis
Spearman’s Rho correlation was used to estimate the relationship between duplicate cores from individual tumors, automated and manual analysis, as well as nuclear and cytoplasmic staining intensity. Differences in distribution of clinical data and tumor characteristics between samples with a high and low cytoplasmic-to-nuclear ratio (CNR; described below) were evaluated using the χ2 test. Kaplan-Meier analysis and the log-rank test were used to illustrate differences between overall survival (OS), metastases-free survival, and breast cancer-specific survival (BCSS) according to survivin expression. Cox regression proportional hazards models were used to estimate the relationship to OS and BCSS of survivin, Her2, lymph node status, tumor grade, Ki-67, p53, PR, and ER in the patient cohort. Multivariate models included any variable that displayed a significant association with outcome following univariate analysis. All calculations were done using Statistical Package for the Social Sciences version 11.0 (SPSS, Inc.). A P value of <0.05 was considered statistically significant. Random forest clustering (RFC) was performed using R software.8 Hierarchical clustering of random forest data was done in MatLab version 7.4 (The MathWorks, Inc.).
Results
Quantitative determination of survivin expression as determined by image analysis
As previously indicated, immunohistochemical-based studies of survivin in breast cancer have produced conflicting results. We, therefore, used image analysis to develop a quantitative scoring model for both nuclear and cytoplasmic survivin expression, as assessed by immunohistochemistry. The specificity of the anti-survivin antibody was first confirmed by Western blot analysis. The anti-survivin antibody recognized a discrete band at 17 kDa in all six of a panel of human breast cancer cell lines examined (Fig. 1A). Following optimization of the staining procedure, it was possible to evaluate survivin protein expression in 96 (91.4%) of the 102 tumors represented on the breast cancer TMA. The pattern of staining is shown in Fig. 1B. Manual analysis revealed a weak association between nuclear survivin and BCSS (P = 0.05) and no relationship between overall or cytoplasmic survivin and outcome (Supplementary Fig. S1).
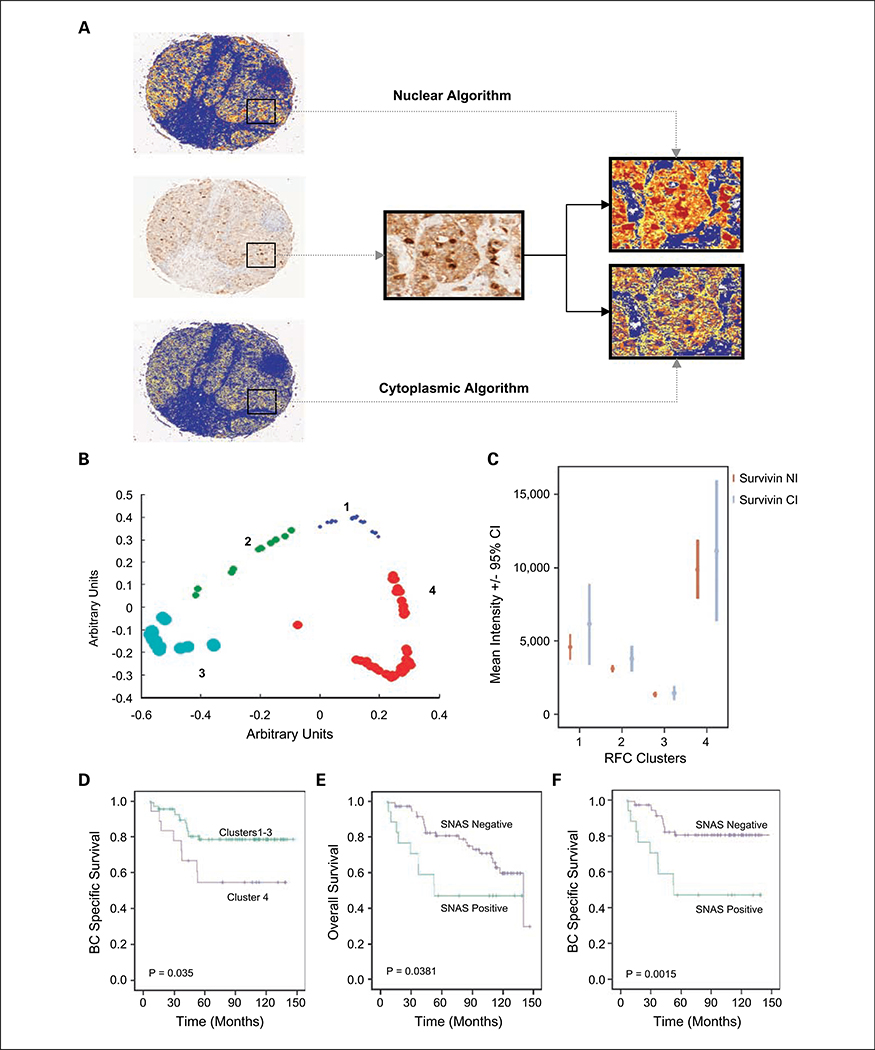
Quantitative determination of survivin expression was then ascertained using an image analysis approach, particularly via the use of a commercial positive pixel count algorithm (Aperio Technologies). A pseudocolor “markup” image was generated as an algorithm result, thus allowing the user to confirm that the algorithm was accurately identifying nuclear and cytoplasmic pixels (Fig. 2A). A full description of the algorithm is available in the Supplementary Data.
Fig. 2.
Automated analysis of survivin immunohistochemistry reveals a relationship between nuclear survivin and poor prognosis. Immunohistochemistry of survivin with corresponding markup images of nuclear and cytoplasmic algorithms. High-power image showing nuclear and cytoplasmic algorithm markup images. A, note that nuclei are red in nuclear markup but blue in the cytoplasmic markup. The algorithm outputs were nuclear and cytoplasmic intensity, which were of a linear nature (histograms of outputs are available; Supplementary Fig. S2). B, RFC of nuclear and cytoplasmic intensity revealing four distinct clusters. C, error bar showing mean survivin cytoplasmic intensity and nuclear intensity, based on random forest clusters, shows greater separation of nuclear intensity compared with cytoplasmic intensity. Kaplan-Meier estimate of BCSS comparing clusters 1 to 3 (n = 66) with cluster 4 (n = 30; D) and OS and BCSS based on survivin nuclear autoscore (SNAS; E and F).
The algorithm was used to calculate the total intensity of survivin for each core, as well as survivin cytoplasmic intensity and survivin nuclear intensity. There was a strong correlation between duplicate cores from individual tumors for both nuclear intensity (Spearman’s Rho = 0.853; P < 0.001) and cytoplasmic intensity (Spearman’s Rho = 0.85; P < 0.001). As tumors were arrayed in duplicate, the maximum value for each tumor was taken for further analysis. The algorithm accurately distinguished between nuclear and cytoplasmic staining in all 96 cores as confirmed by a histopathologist (S.M.H.). There was a strong correlation between manual and automated scoring (Spearman’s Rho = 0.713; P < 0.0001), indicating that image analysis can be used to analyze survivin immunohistochemistry. Additionally, there was also a strong correlation between nuclear and cytoplasmic staining intensity (Spearman’s Rho = 0.842; P < 0.001).
Associations between nuclear survivin as determined by automated image analysis and survival
We were initially unable to establish any link between survivin nuclear intensity, cytoplasmic intensity or total intensity, and survival, despite analyzing all functions as continuous variables. We, therefore, proceeded to use RFC in an attempt to identify new prognostic subgroups. RFC is an unsupervised strategy that has been used to profile tumors based on TMA data (20, 21). It is attractive for TMA data as it handles highly skewed nonparametric data well (Supplementary Fig. S2).
RFC was done on survivin nuclear intensity and cytoplasmic intensity data, which revealed four distinct clusters (Fig. 2B). The clusters contained the following number of tumors: cluster 1 had 14, cluster 2 contained 32, cluster 3 contained 20, and cluster 4 contained 30. We then determined the mean survivin nuclear intensity and cytoplasmic intensity in all four clusters. As displayed in Fig. 2C, there was a more distinct separation of survivin nuclear intensity in the clusters than survivin cytoplasmic intensity. We performed Kaplan-Meier analysis on the four clusters, which revealed a trend toward worse survival in cluster 4 compared with clusters 1, 2, and 3. When clusters 1 to 3 (n = 66) were combined, they displayed an improved BCSS compared with cluster 4 (n =30; P = 0.035; Fig. 2D). Given that the significant difference between the clusters was based around nuclear survivin, we proceeded to examine the relationship between nuclear intensity and survival.
One of the issues to consider when examining staining intensity alone is that even a small fraction of staining artifact can significantly alter intensity values. Therefore, to reduce noise secondary to staining artifact, we examined the relationship between nuclear pixel ratio and survival. By using a cutoff of 25th percentile (8%), we also showed a relationship between a high nuclear pixel ratio and decreased BCSS (P = 0.0051; Supplementary Fig. S1E). A survivin nuclear autoscore (SNAS) was devised based on tumors in cluster 4 (n = 30) and staining in >8% of pixels. Twenty-four percent of tumors (n = 23) met these criteria. A positive SNAS (a marker of nuclear expression) was associated with a decreased BCSS (P = 0.0015) and OS (P = 0.0381; Fig. 2E and F). Given the highly significant univariate analysis of SNAS, we proceeded to conduct a Cox multivariate regression analysis of BCSS (Table 1) in relation to SNAS. Multivariate analysis revealed that the SNAS was a significant predictor of BCSS in this cohort [hazard ratio (HR), 4.70; 95% confidence interval (95% CI), 1.45–15.26; P = 0.010], along with lymph node status (HR, 3.90; 95% CI, 0.17–2.37; P = 0.027), tumor size (HR, 1.04; 95% CI, 1.01–1.07; P = 0.019), and Her2 status (HR, 3.94; 95% CI, 1.09–13.01; P = 0.025). The relationship between SNAS and other clinicopathologic variables was also examined, with the only significant relationship being between positive SNAS and high grade (P = 0.034; data not shown).
Table 1.
Cox regression analysis of BCSS in entire cohort (n = 96)
| BCSS in entire cohort |
||||
|---|---|---|---|---|
| Univariate |
Multivariate* |
|||
| HR (95% C) | P | HR (95% CI) | P | |
| SNAS (positive vs negative) | 3.61 (1.54–8.46) | 0.003 | 4.70 (1.45–15.26) | 0.010 |
| Nodal status (positive vs negative) | 3.42 (1.36–8.62) | 0.009 | 3.90 (1.17–12.96) | 0.027 |
| Grade (1 and 2 vs 3) | 2.89 (1.29–6.49) | 0.010 | 0.63 (0.17–2.37) | 0.489 |
| ER status (positive vs negative) | 0.21 (0.09–0.46) | 0.003 | 0.40 (0.10–1.61) | 0.195 |
| Her2 (1 and 2+ vs 3+) | 3.08 (1.37–6.94) | 0.006 | 3.94 (1.19–13.01) | 0.025 |
| PR status (positive vs negative) | 0.3 (0.13–0.69) | 0.004 | 0.66 (0.18–2.41) | 0.529 |
| Tumor size (continuous) | 1.04 (1.02–1.06) | 0.001 | 1.04 (1.01–1.07) | 0.019 |
| Ki-67 (0–10% vs 11–100%) | 10.46 (1.14–7.37) | 0.020 | 1.84 (0.02–16.49) | 0.583 |
| p53 (0–10% vs 11–100%) | 2.55 (1.18–5.51) | 0.018 | 0.69 (0.19–2.54) | 0.579 |
Abbreviations: Her2, Her2 immunohistochemistry; p53, p53 immunohistochemistry; SNAS, survivin nuclear autoscore.
Adjusted for all other variables in the table.
CNR of survivin expression
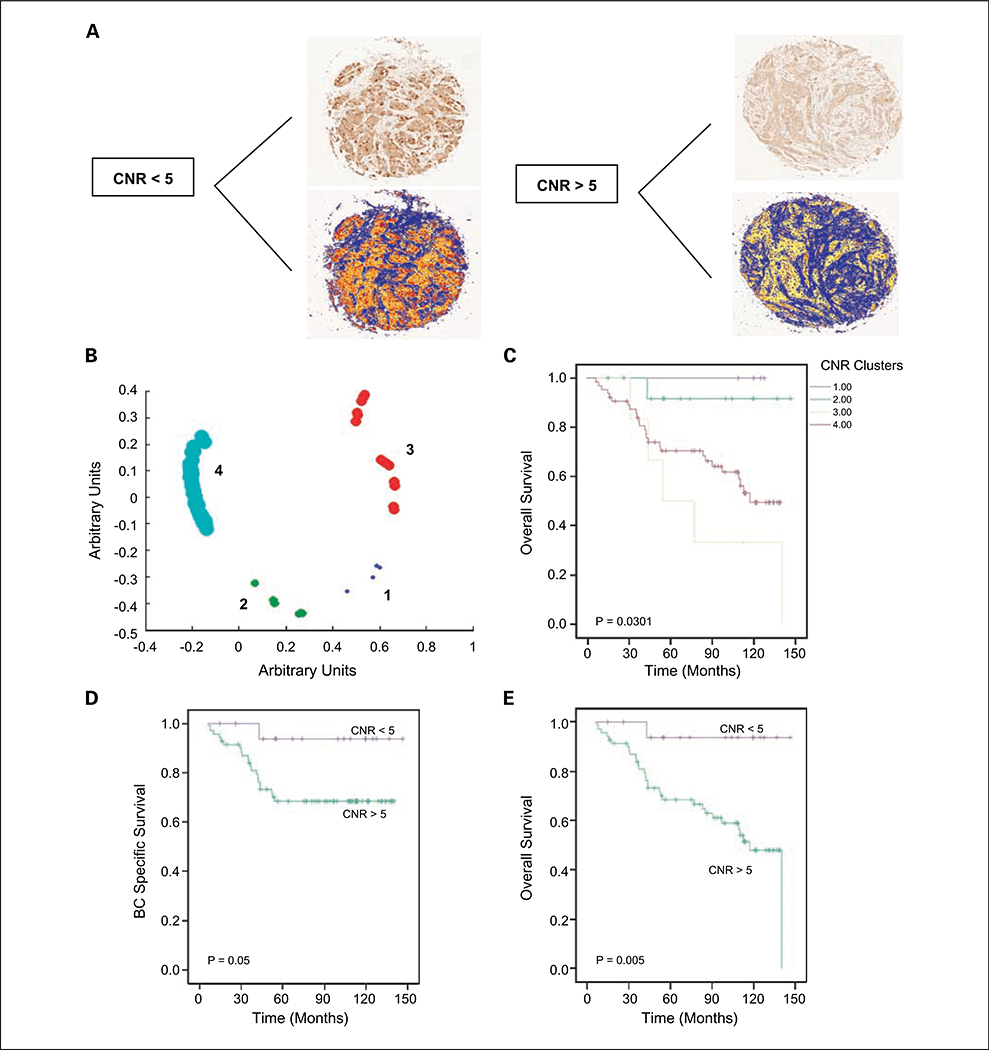
Having shown a significant relationship between nuclear survivin and survival, we proceeded to use image analysis to examine the CNR of survivin. Tumor cores with low and high survivin CNRs are illustrated in Fig. 3A. Univariate Cox regression analysis of survivin CNR as a continuous variable revealed an association between survivin CNR and OS (HR, 0.78; 95% CI, 0.63–0.97; P = 0.025). We proceeded to do RFC analysis on survivin CNR values for all samples, which revealed four distinct clusters (Fig. 3B). Kaplan-Meier analysis of these clusters revealed a significantly decreased OS (P = 0.038), with greatest difference seen when clusters 1 (n = 6) and 2 (n = 12) were grouped together and compared with clusters 3 (n = 14) and 4 (n = 64; Fig. 3C). Based on this analysis, survivin CNR values were thus dichotomized using a CNR cutoff of 5.
Fig. 3.
Survivin CNR is a prognostic factor in breast cancer. A, tumor cores with markup images showing a low and high CNR. Note marked difference in number of red pixels (identifying nuclear survivin) in the CNR < 5 core compared with the CNR > 5 core, which shows minimal nuclear staining. B, RFC of survivin CNR reveals four clusters. Kaplan-Meier estimate of OS based on RFC of survivin CNR (C) and OS and BCSS using a CNR of5 as a cutoff (D and E).
Using this cutoff, 19% (n = 18) of tumors had a survivin CNR of >5. Kaplan-Meier analysis of OS and BCSS revealed that a survivin CNR of <5 was associated with a reduced OS (P = 0.0052) and BCSS (P = 0.05; Fig. 3C and D). We proceeded to do a Cox multivariate regression analysis of OS in the entire cohort (n = 96), in relation to survivin CNR values. Multivariate analysis (Table 2) revealed that survivin CNR was a significant predictor of OS in this cohort (HR, 0.09; 95% CI, 0.01–0.76; P = 0.027), along with tumor size (HR, 1.05; 95% CI, 1.02–1.08; P = 0.002) and lymph node status (HR, 2.71; 95% CI, 1.18–6.24; P = 0.019).
Table 2.
Cox regression analysis of OS in entire cohort (n = 96)
| OS in entire cohort |
||||
|---|---|---|---|---|
| Univariate |
Multivariate* |
|||
| HR (95% CI) | P | HR (95% CI) | P | |
| CNR (<5 vs >5) | 0.1 (0.01–0.73) | 0.023 | 0.09 (0.01–0.76) | 0.027 |
| Nodal status (positive vs negative) | 3.03 (1.48–6.20) | 0.002 | 2.71 (1.18–6.24) | 0.019 |
| Grade (1 and 2 vs 3) | 2.52 (1.32–4.81) | 0.005 | 0.65 (0.27–1.55) | 0.330 |
| ER status (positive vs negative) | 0.38 (0.20–0.73) | 0.004 | 0.58 (0.20–1.72) | 0.338 |
| Her2 (1 and 2+ vs 3+) | 2.19 (1.06–4.52) | 0.034 | 2.04 (0.81–5.15) | 0.129 |
| PR status (positive vs negative) | 0.41 (0.21–0.80) | 0.009 | 0.85 (0.35–2.07) | 0.727 |
| Tumor size (continuous) | 1.04 (1.02–1.06) | 0.001 | 1.05 (1.02–1.08) | 0.002 |
| Ki-67 (0–10% vs 11–100%) | 2.60 (1.01–6.67) | 0.047 | 0.98 (0.31–3.10) | 0.975 |
| p53 (0–10% vs 11–100%) | 2.19 (1.15–4.17) | 0.017 | 0.93 (0.34–2.51) | 0.882 |
Adjusted for all other variables in the table.
The relationship between survivin CNR and other well-documented clinicopathologic variables was then examined (Table 3). A survivin CNR of >5 correlated positively with ER and PR, as well as low and intermediate grade. A survivin CNR of <5 correlated positively with Ki-67, p53 status, and c-myc amplification. There was no significant association between survivin CNR and age, tumor size, nodal status, or Her2 overexpression.
Table 3.
Clinical and tumor characteristics of evaluated cohort (n = 96) stratified according to CNR of survivin protein expression
| CNR < 5 (n = 78), n (%) | CNR > 5 (n = 18), n (%) | P | |
|---|---|---|---|
| Age | |||
| Mean (SD) | 58 (14.7) | 62.3 (13.25) | 0.245* |
| Tumor size (mm) | |||
| Median (range) | 22 (10–100) | 24 (11–60) | |
| 0–20 | 33 (42) | 6 (33) | 0.601† |
| >21 | 45 (58) | 12 (67) | 0.019§ |
| ER status ‡ | |||
| ER− | 26 (33) | 1 (6) | |
| ER+ | 52 (67) | 17 (94) | |
| PR status ‡ | 0.033§ | ||
| PR− | 35 (45) | 3 (17) | |
| PR+ | 43 (55) | 15 (83) | |
| Nodal status | |||
| Negative | 38 (49) | 12 (67) | 0.83† |
| Positive | 40 (51) | 6 (33) | |
| NHG | |||
| NHG I and II | 37 (47) | 18 (100) | || |
| NHG III | 41 (53) | 0 | |
| Her2 status¶ | 0.512§ | ||
| 0–2+ | 61 (78) | 15 (83) | |
| 3+ | 16 (21) | 2(11) | |
| Missing | 2 | 0.005§ | |
| p53 status¶ | |||
| P53− | 53 (68) | 17 (94) | |
| P53+ | 25 (42) | 1 (6) | |
| Myc amplification** | 0.016§ | ||
| Low | 43 (55) | 17 (94) | |
| Intermediate/high | 16(21) | 1 (6) | |
| Missing | 22 | ||
| Ki-67¶ | |||
| 0–10% | 12(17) | 7 (39) | 0.04 |
| 11–100% | 57 (83) | 11 (61) | |
| Missing | 11 |
Abbreviation: NHG, Nottingham Harris Grade.
Independent t test.
χ2 test.
Measured by ELISA.
Fisher’s exact test.
No test done as one cell empty.
Measured by immunohistochemistry.
Measured by fluorescence in situ hybridization.
Discussion
Advances in high-throughput molecular profiling methodologies, such as DNA microarrays, have revolutionized the scientific approach to highly complex diseases such as breast cancer (22). Despite these developments, histopathology remains the gold standard for diagnostic and therapeutic decisions. Histopathology has traditionally been a low-throughput, labor-intensive technique relying on observation, description, and experience. Automated scoring systems for immunohistochemistry offer the opportunity to further advance this well-established and clinically used assay, leveraging the capacity of digital imaging instrumentation to accurately and reproducibly quantify staining intensity.
The manual interpretation of immunohistochemical staining remains a time-consuming, subjective process, to which only limited statistical confidence can be assigned due to inherent interobserver and intraobserver variability and the semiquantitative nature of the data (23). The introduction of digital imaging devices and computer-assisted image analysis has provided a major advance toward quantitative description of biological systems. It is anticipated that quantitative scoring systems will, in time, evolve toward integrated environments where the interface between tissue staining, image acquisition, data analysis, and visualization of results will be totally transparent to the end user (24).
This study is the first to use automated quantitative algorithms to analyze survivin immunohistochemical data. Given the fact that survivin expression is associated with the inhibition of apoptosis and the promotion of angiogenesis and proliferation, there is a rationale that increased survivin levels would be associated with an aggressive phenotype (1). Indeed, this has borne out in multiple studies as indicated previously. In breast cancer, however, there have been several conflicting reports about the prognostic value of survivin. Two relatively large studies of 275 patients (9) and 420 patients (8) using cell-free extracts showed that quantitative measurement of survivin expression was a poor prognostic marker associated with high-grade and hormone receptor negativity (8, 9). In general, our data agree with these studies.
By applying image analysis, we showed that increased expression of nuclear survivin, as opposed to cytoplasmic survivin, was associated with a decreased OS and BCSS. Additionally, we applied two novel scoring models (i.e., the SNAS and CNR). We found a significant relationship between grade and SNAS (P = 0.034) and survivin CNR (P < 0.001). Moreover, we showed a significant relationship between low survivin CNR (which is a marker of increased nuclear staining) and ER (P = 0.019) and PR (P = 0.013) negativity. This agrees with the findings of Span et al. (25) who showed that high levels of survivin protein were associated with a poor response to endocrine therapy.
To exclude any confounding variables, our new scoring models were compared with other well-established breast cancer prognostic factors (Table 1). A Cox multivariate regression analysis revealed that SNAS was a significant predictor of BCSS (HR, 4.70; 95% CI, 1.45–15.26; P = 0.010), along with lymph node status (HR, 3.90; 95% CI, 0.17–2.37; P = 0.027), tumor size (HR, 1.04; 95% CI, 1.01–1.07; P = 0.019), and Her2 status (HR, 3.94; 95% CI, 1.09–13.01; P = 0.025).
Likewise, a second Cox multivariate analysis revealed that CNR was a significant predictor of OS (HR, 0.09; 95% CI, 0.01–0.76; P = 0.027), along with tumor size (HR, 1.05; 95% CI, 1.02–1.08; P = 0.002) and lymph node status (HR, 2.71; 95% CI, 1.18–6.24; P = 0.019). These findings would suggest that increased levels of nuclear survivin as determined by image analysis is a predictor of outcome even when controlling for other well-recognized prognostic markers.
As mentioned previously, the prognostic relevance of survivin as measured by immunohistochemistry in breast cancer is a controversial issue and several smaller qualitative studies have produced conflicting results. It is possible that the quantitative measurement of survivin (either by ELISA or image analysis) is necessary for its use as a biomarker in breast cancer. Interestingly, our study and those of Span et al. (25) and Ryan et al. (8) all used quantitative methods to evaluate survivin expression and found similar results. Our study has an added benefit in that it used formalin-fixed, paraffin-embedded materials as opposed to frozen tissue specimens. The use of a TMA as a biomarker discovery and validation platform raises issues of tumor heterogeneity, and definitive utility of a biomarker requires replication on whole patient samples. The fact that we saw an excellent correlation between nuclear intensity (Spearman’s Rho = 0.852; P < 0.001) and cytoplasmic intensity (Spearman’s Rho = 0.85; P < 0.001) for duplicate cores from individual tumors would suggest that tumor heterogeneity is not a significant contributor to survivin expression. However, this question could only be answered by doing a study of SNAS and survivin CNR on full sections.
There is much debate in the literature about the nuclear-cytoplasmic transport of survivin and its implications for tumorigenesis (26, 27). This study is the first to examine the relationship between the ratio of cytoplasmic to nuclear survivin and outcome. Our results are consistent with the hypothesis that the nuclear and cytoplasmic fractions of survivin have different biological roles (28).
It is well recognized that nuclear-cytoplasmic shuttling of survivin is controlled by an evolutionary conserved Crm1-dependent nuclear export signal (NES; refs. 26, 29, 30). Knauer et al. (26) used small interfering RNA-mediated Crm1 depletion to show that an active NES is necessary to promote the antiapoptotic functions of survivin and inhibition of the NES makes cells more susceptible to chemotherapy- or radiotherapy-induced apoptosis. Colnaghi et al. (30) showed that point mutations in survivin NES abrogated the antiapoptotic effect of survivin. However, they also showed that the mitotic effect of survivin is maintained in NES mutants (30). It could, therefore, be argued that increased levels of nuclear survivin could lead to a proliferative aggressive phenotype.
The role of survivin as a mitotic regulator is well described. Expression of survivin peaks in the G2-M phase of the cell cycle (31) and it is a key member of the chromosomal passenger complex during mitosis, where it regulates levels of the spindle checkpoint protein BubRl (32–34). One could, therefore, argue that increased levels of nuclear survivin (possibly secondary to a deficient NES) are associated with dysregulation of the mitotic apparatus due to increased levels of BubR1 and a sustained checkpoint response. Our data support this hypothesis, as we saw a strong correlation between low survivin CNR and c-myc amplification, p53 expression, and Ki-67, all of which are associated with a proliferative phenotype (35–37). Furthermore, by applying two novel scoring models, we showed that increased nuclear survivin was associated with poor prognosis. Based on these data, it is not surprising that immunohistochemical-based studies of survivin have produced conflicting results. This may relate to varying specificity of the antibodies used or possibly due to inherent interobserver and intraobserver variability in the manual interpretation of survivin immunohistochemistry. As mentioned previously, it may also be necessary to quantitatively determine survivin levels to fully unearth its prognostic potential in breast cancer. It is in this field that automated image analysis of survivin immunohistochemistry may play an important role and allow for further stratification of patients.
In conclusion, we have presented an alternative method of quantitatively determining the expression of survivin via immunohistochemistry. In more detail, we applied a combination of automated image analysis of survivin immunohistochemistry and RFC to identify new prognostic subgroups. Our data support the hypothesis that the different subcellular pools of survivin have distinct functions and increased levels of nuclear survivin are associated with a proliferative phenotype. Such an approach may be helpful in further dissecting the debate surrounding the role of survivin as a prognostic marker in breast cancer and also as a putative therapeutic target.
Supplementary Material
Acknowledgments
Grant support: Enterprise Ireland, Cancer Research Ireland (for support, in part, of Dr. Brennan’s postgraduate studies), British Association for Cancer Research, and Health Research Board of Ireland (under the auspices of the “Breast Cancer Metastasis: Biomarkers and Functional Mediators” research programme). This study has also been supported by grants from the Swedish Cancer Society, Swegene/Wallenberg Consortium North, Gunnar, Arvid and Elisabeth Nilsson Cancer Foundation, Per-Eric and Ulla Schyberg Foundation, Lund University Research Funds, and Malmö University Hospital Research and Cancer Funds. Finally, the cross-national component of the project was facilitated by the Marie Curie Transfer of Knowledge Industry-Academia Partnership research programme, TargetBreast (http://www.targetbreast.com). The UCD Conway Institute is funded by the Programme for Third Level Institutions, as administered by the Higher Education Authority of Ireland. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Duffy MJ, O’Donovan N, Brennan DJ, Gallagher WM, Ryan BM. Survivin: a promising tumor biomarker. Cancer Lett 2007;249:49–60. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu VE, Madden SL, Zhang L, et al. Analysis of human transcriptomes. Nat Genet 1999;23:387–8. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosini G, Adida C, Altieri DC. A novel antiapoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997;3:917–21. [DOI] [PubMed] [Google Scholar]
- 4.Chu JS, Shew JY, Huang CS. Immunohistochemical analysis of survivin expression in primary breast cancers. J Formos Med Assoc 2004;103:925–31. [PubMed] [Google Scholar]
- 5.O’Driscoll L, Linehan R, Kennedy SM, et al. Lack of prognostic significance of survivin, survivin-ΔEx3, survivin-2B, galectin-3, bag-1, bax-α and MRP-1 mRNAs in breast cancer. Cancer Lett 2003;201: 225–36. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka K, Iwamoto S, Gon G, et al. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res 2000;6:127–34. [PubMed] [Google Scholar]
- 7.Kennedy SM, O’Driscoll L, Purcell R, et al. Prognostic importance of survivin in breast cancer. Br J Cancer 2003;88:1077–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan BM, Konecny GE, Kahlert S, et al. Survivin expression in breast cancer predicts clinical outcome and is associated with HER2,VEGF, urokinase plasminogen activator and PAI-1. Ann Oncol 2006;17: 597–604. [DOI] [PubMed] [Google Scholar]
- 9.Span PN, Sweep FC,Wiegerinck ET, et al. Survivin is an independent prognostic marker for risk stratification of breast cancer patients. Clin Chem 2004;50: 1986–93. [DOI] [PubMed] [Google Scholar]
- 10.Hinnis AR, Luckett JC, Walker RA. Survivin is an independent predictor of short-term survival in poor prognostic breast cancer patients. Br J Cancer 2007; 96:639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paik S, Shak S,Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26. [DOI] [PubMed] [Google Scholar]
- 12.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530–6. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien SL, Fagan A, Fox EJ, et al. CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer. Int J Cancer2007;120:1426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F, Yang J, Ramnath N, Javle MM, Tan D. Nuclear or cytoplasmic expression of survivin: what is the significance? Int J Cancer 2005;114:509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svensson S, Jirstrom K, Ryden L, et al. ERK phosphorylation is linked to VEGFR2 expression and Ets-2 phosphorylation in breast cancer and is associated with tamoxifen treatment resistance and small tumours with good prognosis. Oncogene 2005;24: 4370–9. [DOI] [PubMed] [Google Scholar]
- 16.Ryden L, Linderholm B, Nielsen NH, et al. Tumor specific VEGF-A and VEGFR2/KDR protein are coexpressed in breast cancer. Breast Cancer Res Treat 2003;82:147–54. [DOI] [PubMed] [Google Scholar]
- 17.Brennan DJ, Jirstrom K, Kronblad A, et al. CA IX is an independent prognostic marker in premenopausal breast cancer patients with one to three positive lymph nodes and a putative marker of radiation resistance. Clin Cancer Res 2006;12:6421–31. [DOI] [PubMed] [Google Scholar]
- 18.Roos G, Nilsson P, Cajander S, et al. Telomerase activity in relation to p53 status and clinico-pathological parameters in breast cancer. Int J Cancer 1998;79:343–8. [DOI] [PubMed] [Google Scholar]
- 19.Loden M, Stighall M, Nielsen NH, et al. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene 2002;21:4680–90. [DOI] [PubMed] [Google Scholar]
- 20.Shi T, Seligson D, Belldegrun AS, Palotie A, Horvath S. Tumor classification by tissue microarray profiling: random forest clustering applied to renal cell carcinoma. Mod Pathol 2005;18:547–57. [DOI] [PubMed] [Google Scholar]
- 21.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005;435:1262–6. [DOI] [PubMed] [Google Scholar]
- 22.Brennan DJ, O’Brien SL, Fagan A, et al. Application of DNA microarray technology in determining breast cancer prognosis and therapeutic response. Expert Opin Biol Ther 2005;5:1069–83. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez DC, Bhargava R, Hewitt SM, Levin IW. Infrared spectroscopic imaging for histopathologic recognition. Nat Biotechnol 2005;23:469–74. [DOI] [PubMed] [Google Scholar]
- 24.Brennan DJ, Kelly C, Rexhepaj E, et al. Contribution of DNA and tissue microarray technology to the identification and validation of biomarkers and personalised medicine in breast cancer. Cancer Genomics Proteomics 2007;4:3–16. [PubMed] [Google Scholar]
- 25.Span PN, Tjan-Heijnen VC, Manders P, et al. High survivin predicts a poor response to endocrine therapy, but a good response to chemotherapy in advanced breast cancer. Breast Cancer Res Treat 2006;98:223–30. [DOI] [PubMed] [Google Scholar]
- 26.Knauer SK, Kramer OH, Knosel T, et al. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J 2007;21:207–16. [DOI] [PubMed] [Google Scholar]
- 27.Knauer SK, Mann W, Stauber RH. Survivin’s dual role: an export’s view. Cell Cycle 2007;6: 518–21. [DOI] [PubMed] [Google Scholar]
- 28.Fortugno P,Wall NR, Giodini A, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci 2002;115:575–85. [DOI] [PubMed] [Google Scholar]
- 29.Knauer SK, Bier C, Habtemichael N, Stauber RH. The survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep 2006;7:1259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colnaghi R, Connell CM, Barrett RM, Wheatley SP. Separating the anti-apoptotic and mitotic roles of survivin. J Biol Chem 2006;281:33450–6. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 1998;396:580–4. [DOI] [PubMed] [Google Scholar]
- 32.Sampath SC, Ohi R, Leismann O, et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 2004;118:187–202. [DOI] [PubMed] [Google Scholar]
- 33.Gassmann R, Carvalho A, Henzing AJ, et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol 2004; 166:179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolton MA, Lan W, Powers SE, et al. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol Biol Cell 2002;13: 3064–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappellen D, SchlangeT, Bauer M, Maurer F, Hynes NE. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep 2007;8:70–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isola J,Visakorpi T, Holli K, Kallioniemi OP. Association of overexpression of tumor suppressor protein p53 with rapid cell proliferation and poor prognosis in node-negative breast cancer patients. J Natl Cancer Inst 1992;84:1109–14. [DOI] [PubMed] [Google Scholar]
- 37.Barbareschi M, Leonardi E, Mauri FA, Serio G, Dalla Palma P. p53 and c-erbB-2 protein expression in breast carcinomas. An immunohistochemical study including correlations with receptor status, proliferation markers, and clinical stage in human breast cancer. Am J Clin Pathol 1992;98:408–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.