Abstract
Xyloglucan is a major hemicellulose in plant cell walls and exists in two distinct types, XXXG and XXGG. While the XXXG-type xyloglucan from dicot species only contains O-acetyl groups on side-chain galactose (Gal) residues, the XXGG-type xyloglucan from Poaceae (grasses) and Solanaceae bears O-acetyl groups on backbone glucosyl (Glc) residues. Although O-acetyltransferases responsible for xyloglucan Gal acetylation have been characterized, the biochemical mechanism underlying xyloglucan backbone acetylation remains to be elucidated. In this study, we showed that recombinant proteins of a group of DUF231 members from rice and tomato were capable of transferring acetyl groups onto O-6 of Glc residues in cello-oligomer acceptors, indicating that they are xyloglucan backbone 6-O-acetyltransferases (XyBATs). We further demonstrated that XyBAT-acetylated cellohexaose oligomers could be readily xylosylated by AtXXT1 (Arabidopsis xyloglucan xylosyltransferase 1) to generate acetylated, xylosylated cello-oligomers, whereas AtXXT1-xylosylated cellohexaose oligomers were much less effectively acetylated by XyBATs. Heterologous expression of a rice XyBAT in Arabidopsis led to a severe reduction in cell expansion and plant growth and a drastic alteration in xyloglucan xylosylation pattern with the formation of acetylated XXGG-type units, including XGG, XGGG, XXGG, XXGG,XXGGG and XXGGG (G denotes acetylated Glc). In addition, recombinant proteins of two Arabidopsis XyBAT homologs also exhibited O-acetyltransferase activity toward cellohexaose, suggesting their possible role in mediating xyloglucan backbone acetylation in vivo. Our findings provide new insights into the biochemical mechanism underlying xyloglucan backbone acetylation and indicate the importance of maintaining the regular xyloglucan xylosylation pattern in cell wall function.
Keywords: Acetylation, Acetyltransferase, Cell wall, DUF231, Rice, Tomato, Xyloglucan
Accession number: The GenBank accession numbers of the genes investigated in this study are MN996513 (OsXyBAT1), MN996514 (OsXyBAT2), MN996515 (OsXyBAT3), MN996516 (OsXyBAT4), MN996517 (OsXyBAT5), MN996518 (OsXyBAT6), MN996519 (OsXyBAT7), MN996520 (SlXyBAT1), MN996521 (SlXyBAT2), MN996522 (SlXyBAT3), MN996523 (SlXyBAT4), MN996524 (SlXyBAT5), MN996525 (SlXyBAT6), MN996526 (SlXyBAT7), MT174529 (AtXyBAT1/TBL19) and MT174530 (AtXyBAT2/TBL21).
Introduction
Xyloglucan is a hemicellulosic polysaccharide found in the cell walls of all taxa of land plants. It consists of a linear chain of β-1,4-linked d-glucosyl (Glc) residues that are highly substituted at O-6 with α-d-xylosyl (Xyl) residues. In dicot plants, the Xyl residues in xyloglucan may be further substituted at O-2 with a β-d-galactosyl (Gal) residue to form a Gal-Xyl disaccharide side chain. The Gal residues, in turn, can be linked at O-2 with an α-l-fucosyl (Fuc) residue to produce a Fuc-Gal-Xyl trisaccharide side chain. Besides Gal and Fuc, other sugars, such as l-arabinopyranose (Arap), l-arabinofuranose (Araf), d-galacturonic acid (GalA) and Xyl, may also be attached to Xyl residues to generate various di- and trisaccharide side chains in xyloglucan, depending on plant species and specific tissue types (Pauly and Keegstra 2016). A one-letter nomenclature was developed to depict the various xyloglucan side-chain structures. For example, an unsubstituted Glc is designated as G, a xylose-substituted Glc as X, a galactosyl-xylose-substituted Glc as L and a fucosyl-galactosyl-xylose-substituted Glc as F (Fry et al. 1993). Most of the genes involved in the biosynthesis of xyloglucan backbone and side chains have been uncovered by genetic and/or biochemical analyses. These include cellulose synthase-like C subfamily members as β-1,4-glucan synthases responsible for xyloglucan backbone synthesis (Cocuron et al. 2007), xyloglucan α-1,6-xylosyltransferases (XXTs) catalyzing the addition of Xyl side chains onto the glucan backbone (Cavalier and Keegstra 2006, Cavalier et al. 2008), two xyloglucan β-1,2-galactosyltransferases, MUR3 (MURUS3) and XLT2 (xyloglucan l-side-chain galactosyltransferase position 2), mediating the position-specific galactosylation of Xyl residues (Madson et al. 2003, Jensen et al. 2012), and xyloglucan α-1,2-fucosyltransferase catalyzing the transfer of Fuc onto Gal residues (Perrin et al. 1999, Vanzin et al. 2002). Furthermore, it was found that a xyloglucan arabinopyranosyltransferase from the moss Physcomitrella patens and two xyloglucan arabinofuranosyltransferases from tomato were involved in the addition of the respective Arap and Araf onto Xyl residues (Schultink et al. 2013, Zhu et al. 2018) and a xyloglucan galacturonosyltransferase from Arabidopsis was required for GalA addition onto Xyl residues (Peña et al. 2012).
In addition to glycosyl substitutions, xyloglucan may also bear acetyl moieties. Based on its xylosylation patterns, xyloglucan is generally categorized into two types, XXXG and XXGG, which differ in their acetyl distribution. The XXXG-type xyloglucan has Xyl decoration on three consecutive Glc out of every four backbone Glc residues and is present in vascular plants and hornworts (Pauly and Keegstra 2016). Its acetylation occurs on side-chain Gal residues predominantly at O-6, although a low level of 3-O- and 4-O-monoacetylation and 3,4-di-O- and 4,6-di-O-acetylation of Gal was also observed (York et al. 1988). The XXGG-type xyloglucan has Xyl substitutions on two consecutive Glc out of every four or more backbone Glc residues and is found in Poaceae (grasses), Solanaceae, ferns, lycophytes, mosses and liverworts. The XXGG-type xyloglucan from grasses and Solanaceae has acetyl moieties attached at O-6 of backbone Glc residues (Pauly and Keegstra 2016). In addition to the backbone Glc residues, xyloglucan from tomato also has acetyl substituents at O-6 of Gal and O-5 of Arap residues (Jia et al. 2005). Xyloglucan backbone acetylation has not been reported in the XXXG-type xyloglucan.
The O-acetylation of side-chain Gal residues in xyloglucan is mediated by members of the DUF231 family, including AXY4 (altered xyloglucan4)/XGOAT1 (xyloglucan O-acetyltransferase1) and AXY4L/XGOAT2 from Arabidopsis and PtrXGOATs from poplar. Mutations of the AXY4 and AXY4L genes cause defects in xyloglucan acetylation in vegetative tissues and seeds, respectively (Gille et al. 2011). Biochemical studies have revealed that XGOATs from both Arabidopsis and poplar catalyze the acetylation of Gal residues predominantly at O-6 and to a less extent at O-3, O-4 and O-4,6. It was demonstrated that fucosylated Gal residues in xyloglucan were the preferred acceptor for XGOATs in vitro, indicating that the acetylated terminal Gal residues present in Arabidopsis xyloglucan is most likely generated by the removal of Fuc residues by fucosidase from the acetylated, fucosylated Gal residues (Zhong et al. 2018b). This is in agreement with an early genetic study showing that the Arabidopsis Atfut1 and mur2 mutants defective in xyloglucan fucosylation had a loss of acetylation on Gal residues in xyloglucan (Perrin et al. 2003). Little is known about the O-acetyltransferases responsible for the O-acetylation of xyloglucan backbone. Although a DUF231 gene from Brachypodium distachyon has been implicated in xyloglucan backbone acetylation as its mutation caused a reduction in xyloglucan acetylation and its heterologous expression in Arabidopsis resulted in the occurrence of some acetylated XXGG units in xyloglucan, its enzymatic activity and acceptor substrate identity remain unknown (Liu et al. 2016).
In this report, we have identified and biochemically characterized a group of DUF231 proteins from rice and tomato and demonstrated that they are xyloglucan backbone O-acetyltransferases (XyBATs) capable of transferring acetyl groups onto O-6 of Glc residues in cello-oligomer acceptors. Sequential enzymatic reactions catalyzed by XyBATs and AtXXT1, an Arabidopsis xyloglucan xylosyltransferase, revealed that AtXXT1 could efficiently xylosylate acetylated cellohexaose and two rice XyBATs were able to acetylate xylosylated cellohexaose, generating acetylated, xylosylated cellohexaose. Furthermore, we present genetic evidence demonstrating that heterologous expression of a rice XyBAT in Arabidopsis leads to an alteration in xyloglucan xylosylation pattern and a concomitant detrimental effect on cell expansion and plant growth. These findings provide biochemical evidence establishing that these DUF231 members are XyBATs and suggest that maintaining the regular substitution pattern in xyloglucan is essential for normal cell expansion and plant growth.
Results
Identification and biochemical characterization of rice and tomato DUF231 proteins involved in xyloglucan backbone acetylation
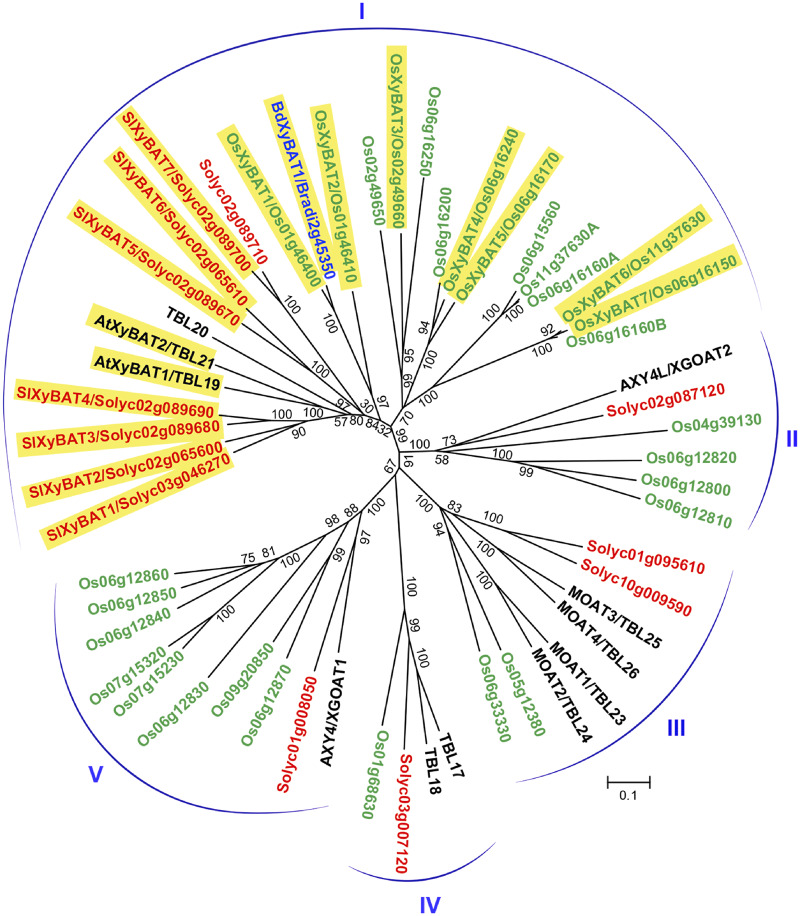
Xyloglucans from grasses and Solanaceae bear acetyl moieties at O-6 of backbone Glc residues (Pauly and Keegstra 2016). To search for genes involved in xyloglucan backbone acetylation, we first identified all DUF231 genes in rice (Oryza sativa; Os) and tomato (Solanum lycopersicum; Sl), two representative species in the grass and the Solanaceae families, respectively, by basic local alignment search tool (BLAST)-searching their genome sequence databases using the amino acid sequences of all 46 Arabidopsis DUF231 genes. A total of 66 and 69 DUF231 genes were identified in rice and tomato, respectively, and phylogenetic analysis revealed that they fell into seven distinct groups together with those from Arabidopsis (Supplementary Fig. S1). A DUF231 gene from B. distachyon, BdXyBAT1, which was previously implicated in xyloglucan backbone acetylation (Liu et al. 2016), resided in group I, which harbors AXY4/4L and MOATs (mannan O-acetyltransferases) catalyzing the acetylation of xyloglucan side-chain Gal residues and glucomannan, respectively (Gille et al. 2011, Zhong et al. 2018a, Zhong et al. 2018b). A close examination of members in group I showed that they could be divided into five subgroups (Fig. 1), which is further supported by the phylogenetic analysis of their homologs from additional dicot and grass species (Supplementary Fig. S2).
Fig. 1.
Phylogenetic relationship of group I DUF231 proteins from rice (Oryza sativa; Os), tomato (Solanum lycopersicum; Sl or Solyc) and Arabidopsis. BdXyBAT1 from Brachypodium distachyon was also included in the analysis (see Supplementary Fig. S1 for the phylogenetic tree of all DUF231 genes from these species). The tree was constructed with the neighbor-joining algorithm, and the 0.1 scale denotes a 10% change. Bootstrap values from 1,000 replicates are shown as percentages at the nodes. The DUF231 genes from rice and tomato are shown with their names and locus identifiers in green and red, respectively. The Arabidopsis DUF231 genes are shown with their names in black. The proteins characterized in this report are highlighted in yellow.
We next set out to study the possible role of subgroup I rice and tomato DUF231 members in xyloglucan backbone acetylation. Based on gene expression data from the Rice Expression Profile Database (Sato et al. 2013) and the Tomato Functional Genomics Database (Sato et al. 2012), these DUF231 genes exhibited differential expression in different organs (Supplementary Fig. S3). cDNA cloning and sequence analysis revealed that the rice Os06g16160 and Os11g37630 genes, which were annotated as single DUF231 gene in the O. sativa v7_JGI database, consisted of two DUF231 genes arranged in tandem, and thus, they were named Os06g16160A/Os06g16160B and Os11g37630A/Os11g37630B (Supplementary Fig. S1). Os06g15560 has a premature stop codon in its cDNA after the 49th codon, and Solyc02g089710 has a stop codon right after the DXXH motif and its predicted protein lacks the conserved C-terminal sequence. Therefore, they are most likely pseudogenes and not included in this study. Prediction of transmembrane helices in subgroup I rice and tomato DUF231 proteins showed that they were type II membrane proteins with an N-terminal transmembrane helix followed by a large putative catalytic domain with the conserved trichome birefringence-like (TBL) and DUF231 domains (Supplementary Fig. S4). The GDS motif located in the TBL domain and the DXXH motif in the DUF231 domain, which were previously demonstrated to be essential for the acetyltransferase activities of XGOATs, XOATs (xylan O-acetyltransferases) and MOATs (Zhong et al. 2017a, Zhong et al. 2018a, Zhong et al. 2018b), were completely conserved in these rice and tomato DUF231 proteins (Supplementary Fig. S4).
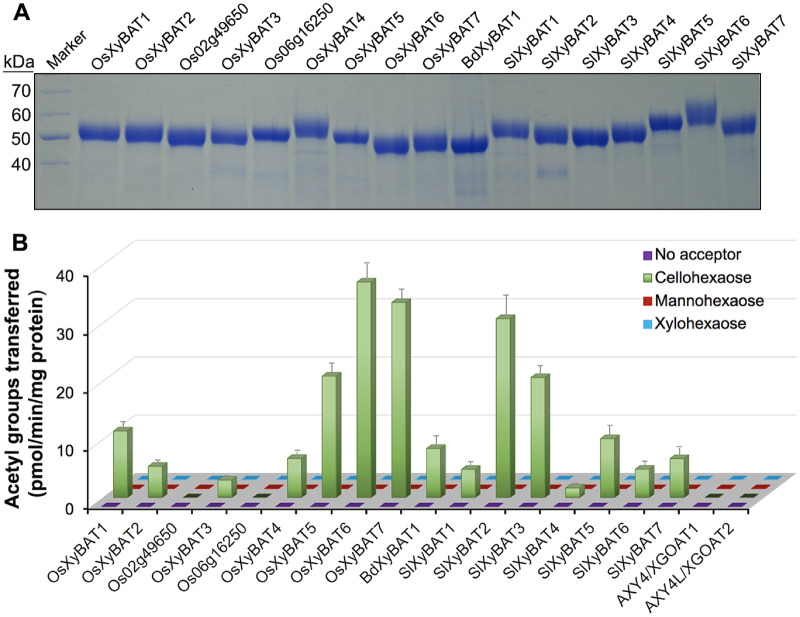
To investigate their biochemical functions, we expressed the putative catalytic domains of these rice and tomato DUF231 proteins as well as BdXyBAT1 as secreted, His-tagged proteins in the human embryonic kidney (HEK) 293 cells and assayed the purified recombinant proteins for acetyltransferase activities toward various oligosaccharide acceptors, including cellohexaose (G6), mannohexaose and xylohexaose. Out of 21 DUF231 proteins attempted, we successfully generated nine rice recombinant proteins and seven tomato ones along with BdXyBAT1 (Fig. 2A). When incubated with 14C-labeled acetyl-CoA and oligosaccharide acceptors, seven out of nine rice DUF231 proteins, all seven tomato ones and BdXyBAT1 were able to transfer radiolabeled acetyl groups onto the G6 acceptor albeit at various levels (Fig. 2B). By contrast, no acetyl transfer onto mannohexaose and xylohexaose was detected. These results suggest that these rice and tomato DUF231 proteins are O-acetyltransferases capable of transferring acetyl groups onto Glc residues in the G6 glucan acceptor and hence were named after BdXyBAT1 as OsXyBATs and SlXyBATs. The recombinant proteins of AXY4/XGOAT1 and AXY4L/XGOAT2 (Zhong et al. 2018a), two Arabidopsis XGOATs catalyzing the acetylation of side-chain Gal residues, were unable to transfer acetyl groups onto the G6 acceptor (Fig. 2B).
Fig. 2.
Production and activity assays of rice and tomato recombinant DUF231 proteins. (A) SDS–PAGE and Coomassie Blue staining of recombinant DUF231 proteins generated by heterologous expression in the mammalian HEK293 cells. The molecular masses of the protein markers are indicated at the left. (B) O-Acetyltransferase activity assays of recombinant DUF231 proteins. The proteins were incubated with 14C-labeled acetyl-CoA and various acceptors (cellohexaose, mannohexaose and xylohexaose), and their O-acetyltransferase activities were measured by counting the radioactivity of the acetylated acceptor products. Note that a number of rice and tomato DUF231 proteins, as well as BdXyBAT1, were able to transfer radiolabeled acetyl groups onto the cellohexaose acceptor. Error bars denote the SE of three biological replicates.
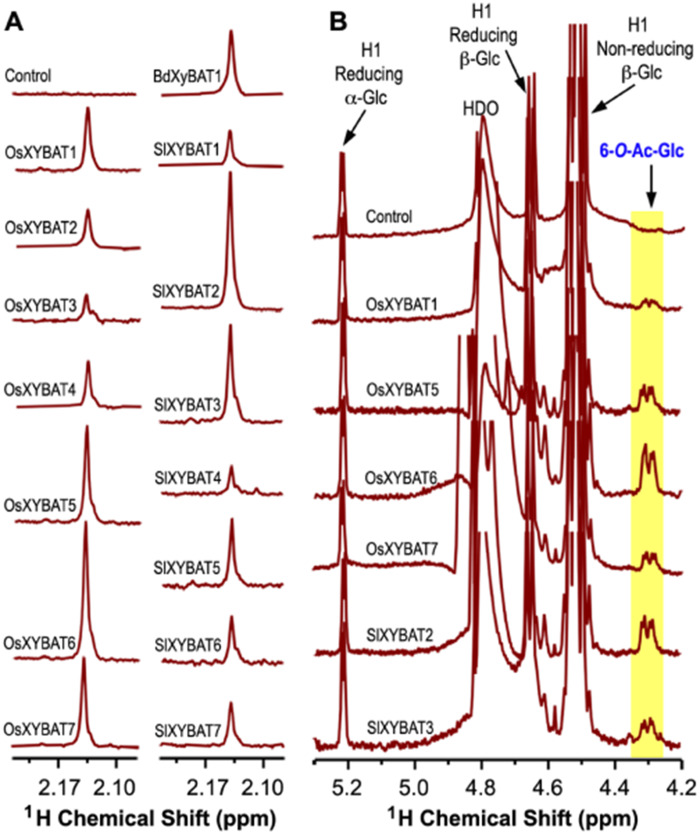
Examination of the XyBAT-catalyzed reaction products by 1H nuclear magnetic resonance (NMR) spectroscopy revealed a prominent resonance peak at 2.14 ppm in the region characteristic of resonance signals of methyl protons of acetyl groups (between 2.0 and 2.3 ppm), whereas the control reaction incubated with denatured OsXyBAT1 did not display any resonance signals in this region (Fig. 3A). Further analysis of the anomeric region of NMR spectra showed that the reaction products of several OsXyBATs and SlXyBATs with high activities exhibited a resonance doublet around 4.3 ppm (Fig. 3B) attributed to one of the methylene protons of 6-O-acetylated Glc residues based on the published NMR assignments of tomato O-acetylated xyloglucan oligosaccharides (Jia et al. 2005). These results demonstrated that these XyBATs transferred acetyl groups onto O-6 of Glc residues in the G6 acceptor, indicating that they are xyloglucan backbone 6-O-acetyltransferases.
Fig. 3.
1H NMR spectroscopic analysis of the acetylated cellohexaose products generated by the incubation of cellohexaose and acetyl-CoA with recombinant OsXyBATs, SlXyBATs and BdXyBAT1. The control was the reaction product of cellohexaose and acetyl-CoA incubated with heat-denatured OsXyBAT1. (A) The signature region of 1H NMR spectra showing the resonances corresponding to the methyl protons of acetyl groups. Note the predominant proton resonance peak at 2.14 ppm in all of the XyBAT-catalyzed reaction products. (B) The anomeric region of 1H NMR spectra showing the resonances attributed to Glc, including H1 of reducing α-Glc and β-Glc and H1 of nonreducing β-Glc, and the resonances attributed to acetylated Glc (6-O-Ac-Glc), which were shown as a doublet corresponding to one of the methylene protons of Glc acetylated at O-6. HDO, hydrogen-deuterium oxide.
Acetyl distribution pattern in XyBAT-catalyzed reaction products
To determine the distribution pattern of acetyl groups in the acetylated G6 products catalyzed by XyBATs, we first applied matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) to examine the mass spectra of the reaction products. While the control reaction incubated with denatured OsXyBAT1 displayed only the ion species at m/z 1,013 corresponding to the G6 acceptor, XyBAT-catalyzed reactions had up to three new ion species at m/z 1,055, 1,097 and 1,139 (Fig. 4). These new ion species had a successive increment of 42 Da in mass over that of G6. Because the mass of one acetyl group is 42 Da (after the loss of water), the new ion species at m/z 1,055, 1,097 and 1,139 corresponded to G6 substituted with one (G6Ac1), two (G6Ac2) and three (G6Ac3) acetyl groups, respectively. The high ratio of acetylated G6 to the G6 acceptor in the reaction products of OsXyBAT5, OsXyBAT6, OsXyBAT7, SlXyBAT2 and SlXyBAT3 (Fig. 4) is consistent with their high specific activities detected by the radiolabeling method (Fig. 2B).
Fig. 4.
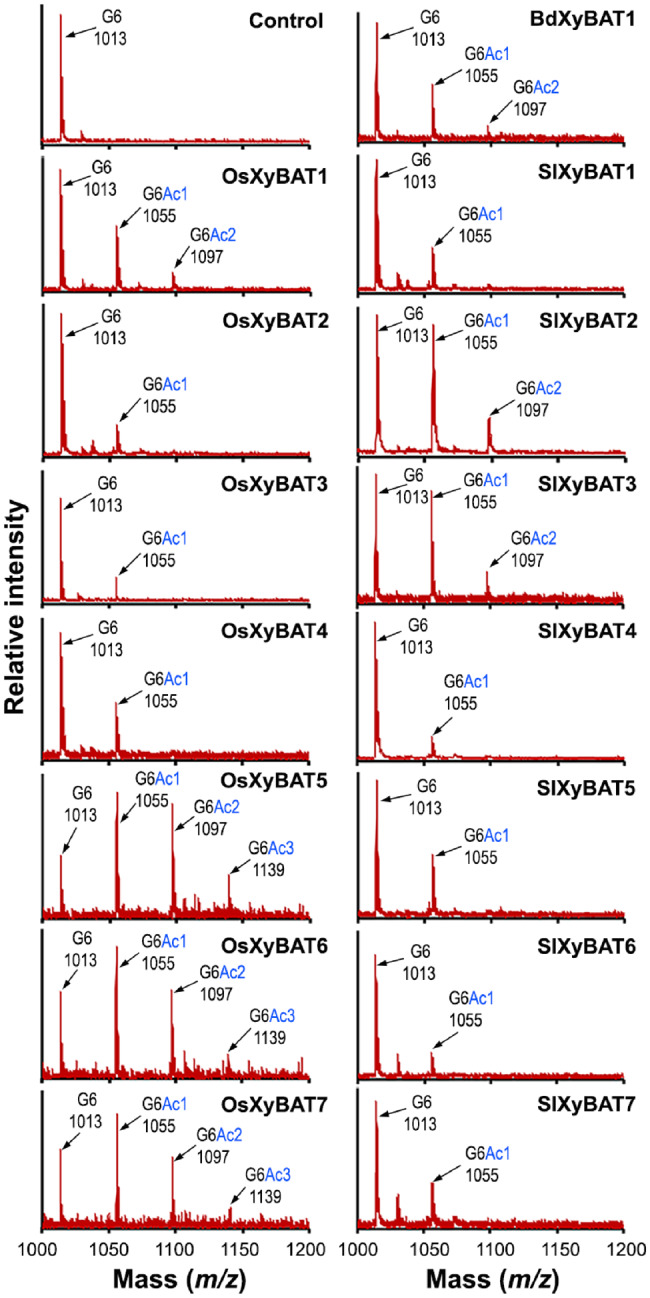
MALDI-TOF MS analysis of XyBAT-catalyzed reaction products. Recombinant OsXyBATs, SlXyBATs and BdXyBAT1 were incubated with cellohexaose and acetyl-CoA, and the reaction products were examined by MALDI-TOF MS. The control was the reaction products of cellohexaose and acetyl-CoA incubated with heat-denatured OsXyBAT1. Each ion peak is marked with its mass ([M+Na]+) and structural identity. The ion peak at m/z 1,013 corresponds to the cellohexaose (G6) acceptor. The ion peaks at m/z 1,055, 1,097 and 1,139 have a successive mass increment of 42 Da (corresponding to the mass of an acetyl group) over the mass of G6 and are attributed to G6 decorated with one (G6Ac1), two (G6Ac2) and three (G6Ac3) acetyl groups, respectively. The data shown are representatives of three biological replicates.
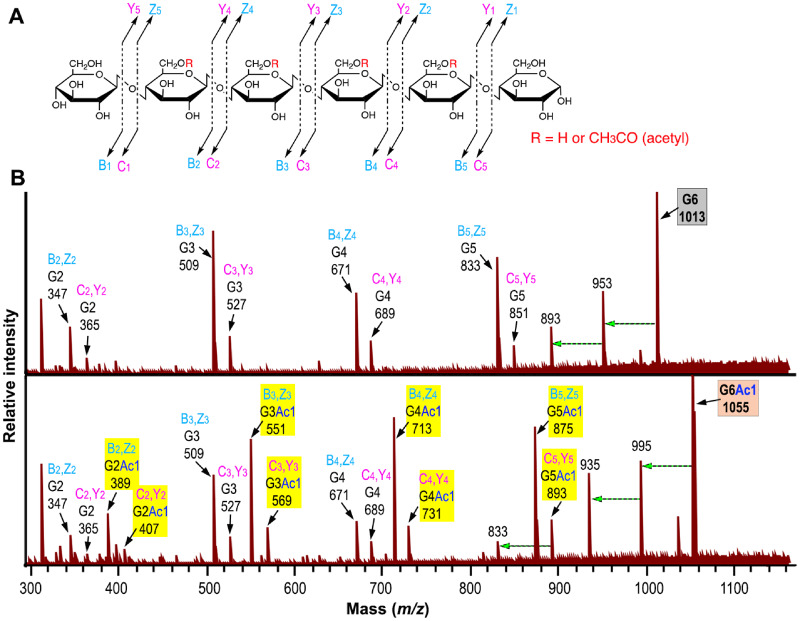
We next employed MALDI tandem MS (MS/MS) to decipher whether there was any positional preference for acetylation in the G6 acceptor by XyBATs. Since the reaction products of OsXyBAT5 exhibited abundant signals in all three acetylated ion species including G6Ac1, G6Ac2 and G6Ac3 (Fig. 4), each of these ion species as well as that of G6 was fragmented by collision-induced dissociation (CID) and the resulting fragments were detected by MS (Fig. 5A, B; Supplementary Fig. S5; Supplementary Table S1). Fragmentation of G6 (m/z 1,013) generated ion species corresponding to cellobiose (G2), cellotriose (G3), cellotetraose (G4) and cellopentaose (G5) (Fig. 5B). It was apparent that the signal intensities of the B,Z-type ion species were 3- to 4-fold higher than those of the C,Y-type ion species. The two ion species at m/z 893 and 953 with a sequential loss of 60 in mass from G6 (m/z 1,013) were most likely generated by cross-ring cleavages (An and Lebrilla 2011). Fragmentation of monoacetylated G6 (G6Ac1 at m/z 1,055) produced ion species corresponding to monoacetylated and non-acetylated fragments (Fig. 5B), which was expected since cleavage of a glycosidic bond in monoacetylated G6 would generate one fragment with the acetyl group and one without. Although both B,Z- and C,Y-type ion species for non-acetylated G2, G3 and G4 were obvious, no C,Y-type ion species for non-acetylated G5 (m/z 851) was detected and the low level of ion species at m/z 833 was most likely resulted from the ion species at m/z 893 by a loss of 60 in mass through cross-ring cleavages (Fig. 5B). Since fragmentation of monoacetylated G6 with the acetyl group on the reducing or nonreducing end Glc residue would generate non-acetylated G5, these results suggest that little acetylation occurred on the reducing and nonreducing end Glc residues, whereas an acetyl group was readily added onto any of the four internal Glc residues in G6. The presence of diacetylated G3 (G3Ac2) in the fragmentation products of diacetylated G6 and that of triacetylated G4 (G4Ac3) in those of triacetylated G6 (Supplementary Fig. S5) indicate the occurrence of two or three consecutive, acetylated Glc residues in the OsXyBAT5-catalyzed reaction products.
Fig. 5.
MALDI tandem mass spectrometry (MS/MS) analysis of the acetylation pattern of monoacetylated cellohexaose. The ion species at m/z 1,055 corresponding to monoacetylated cellohexaose (G6Ac1) and that at m/z 1,013 corresponding to the cellohexaose (G6) acceptor in the OsXyBAT5-catalyzed reaction products (Fig. 4) were fragmented by collision-induced dissociation and then detected by MS. (A) Diagram of the possible fragment ions generated by the fragmentation of cellohexaose. Bn (cyan) and Cn (pink) denote fragments containing the nonreducing terminus, and Yn (pink) and Zn (cyan) designate fragments containing the reducing terminus. The subscripts in Bn, Cn, Yn and Zn represent the number of monomeric units toward the terminus. (B) MALDI MS/MS spectrum of monoacetylated cellohexaose (G6Ac1 at m/z 1,055) compared with that of the cellohexaose acceptor (G6 at m/z 1,013). Each ion species is marked with its fragmentation type (Bn, Zn or Cn, Yn), mass ([M+Na]+) and oligomer composition (GnAcn; an oligomer containing n number of Glc decorated with n number of acetyl groups), all of which are also listed in Supplementary Table S1. A dashed horizontal arrow indicates an ion species generated from the ion species at its right end by a loss of 60 in mass through cross-ring cleavages (An and Lebrilla 2011). Ion species of acetylated cello-oligomers are highlighted in yellow. Note that while the ion species of both non-acetylated and monoacetylated G2, G3 and G4 and those of monoacetylated G5 were observed, the ones corresponding to non-acetylated G5 were absent.
Minimal length of cello-oligomers required for XyBAT-catalyzed acetylation
To find out the minimal length of cello-oligomers required by XyBATs, five representative XyBATs (OsXyBAT1/5/6 and SlXyBAT2/3) were assayed for their acetyltransferase activities toward cello-oligomers with various degrees of polymerization (G1–G6) (Supplementary Fig. S6A). Although all five XyBATs could acetylate G5 in addition to G6, only OsXyBAT5, OsXyBAT6 and SlXyBAT2 exhibited acetyltransferase activities toward G4. While none of them could use G1 or G2 as an acceptor, OsXyBAT6 still displayed a significant activity toward G3. MALDI-TOF MS of the OsXyBAT6-catalyzed reaction products revealed that up to two acetyl groups were added onto G4 and up to three acetyl groups were added onto G5 and G6 (Supplementary Fig. S6B). These results indicate that XyBATs have the differential requirement of the minimal length of cello-oligomer acceptors for their activities.
Sequential acetylation and xylosylation of cello-oligomers by XyBATs and AtXXT1
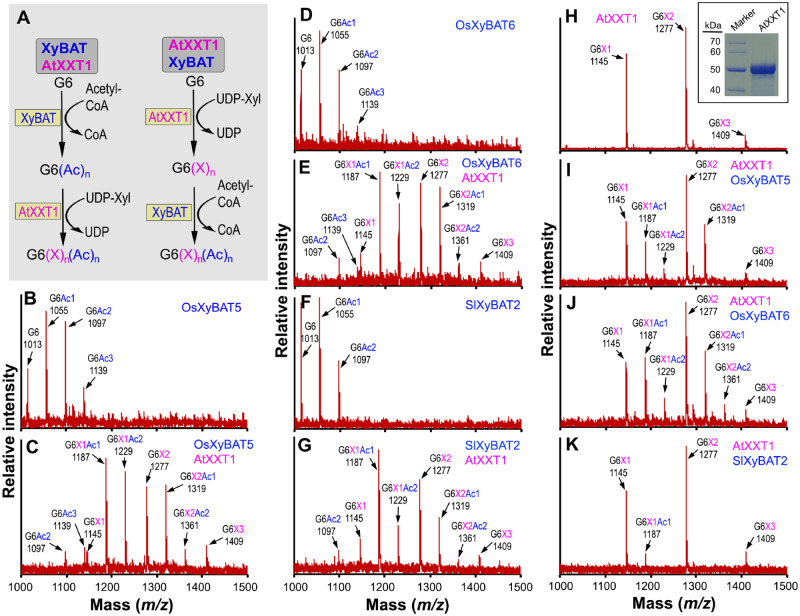
The aforementioned results established that XyBATs could catalyze the transfer of acetyl groups onto unsubstituted glucan acceptors. However, xyloglucan is a xylosylated glucan and that from rice and tomato typically has acetylated Glc residues adjacent to two consecutive xylosylated Glc residues (Pauly and Keegstra 2016). Because both acetylation and xylosylation of the xyloglucan backbone occur at O-6 of Glc residues, acetylation of a Glc residue would block its xylosylation and vice versa. It is currently unknown how xyloglucan backbone acetylation and xylosylation are coordinated during its biosynthesis and whether acetylation of a Glc residue occurs before or after xylosylation of the adjacent Glc residues. To probe these questions, we performed sequential reactions by first acetylating the G6 acceptor and then xylosylating the acetylated G6 or vice versa (Fig. 6A). The G6 acceptor was first incubated with OsXyBAT5, OsXyBAT6 or SlXyBAT2 to generate acetylated G6 oligomers (Fig. 6B, D, F; Supplementary Table S2), which were then used as acceptors for recombinant Arabidopsis AtXXT1 (Fig. 6H, inset). MALDI-TOF MS of the reaction products revealed a number of acetylated, xylosylated G6 ion species, including three abundant ones corresponding to G6X1Ac1 (m/z 1,187), G6X1Ac2 (m/z 1,229) and G6X2Ac1 (m/z 1,319) and a minor one to G6X2Ac2 (m/z 1,361) (Fig. 6C, E, G; Supplementary Table S2). These results demonstrated that AtXXT1 was able to efficiently add one Xyl residue onto both mono- and diacetylated G6 and two Xyl onto monoacetylated G6. Based on the findings that little acetylation occurs on the reducing or nonreducing end Glc residues in the XyBAT-mediated reactions (Fig. 5B) and that AtXXT1-catalyzed dixylosylation occurs predominantly on the third and fourth Glc residues from the reducing end of G6 (Cavalier and Keegstra 2006), G6X2Ac1 was most likely generated from monoacetylated G6 with an acetyl group on the second or fifth Glc residue and G6X2Ac2 from diacetylated G6 with an acetyl group on both the second and fifth Glc residues, suggesting that AtXXT1 is capable of xylosylating Glc residues adjacent to an acetylated Glc residue. This proposition was further substantiated by MALDI MS/MS analysis of the ion species corresponding to G6X2Ac1 and G6X2Ac2 showing the generation of a fragment corresponding to G4X2Ac1 (Supplementary Fig. S7).
Fig. 6.
MALDI-TOF MS detection of cellohexaose acetylation and xylosylation mediated by XyBATs and AtXXT1. (A) Schematic diagram of the sequential reactions for cellohexaose acetylation and xylosylation. XyBAT AtXXT1 denotes a reaction in which cellohexaose was first incubated with XyBAT and the reaction products were then incubated with AtXXT1. AtXXT1 XyBAT denotes a reaction in which cellohexaose was first incubated with AtXXT1, and the reaction products were then incubated with XyBAT. (B-G) MALDI-TOF mass spectra of the reaction products catalyzed by XyBATs (B, D and F) or XyBATs followed by AtXXT1 (C, E and G). Each ion peak is labeled with its mass ([M+Na]+) and oligomer composition. (H-K) MALDI-TOF mass spectra of the reaction products catalyzed by AtXXT1 (H) or AtXXT1 followed by XyBATs (I–K). The inset image in (H) shows the SDS–PAGE and Coomassie Blue staining of recombinant AtXXT1 protein used for activity assay. The molecular masses (kDa) of the markers are shown at the left. G6, cellohexaose; G6(Ac)n, cellohexaose with n number of acetyl groups (Ac); G6(X)n, cellohexaose with n number of xylosyl groups (X); G6(X)n(Ac)n, cellohexaose with n number of xylosyl groups and n number of acetyl groups.
To investigate whether XyBATs are able to use xylosylated G6 as acceptors, we first incubated G6 with AtXXT1 to generate xylosylated G6, which was then incubated with OsXyBAT5, OsXyBAT6 or SlXyBAT2. As previously reported (Cavalier and Keegstra 2006), AtXXT1 converted G6 into predominantly monoxylosylated and dixylosylated G6 (Fig. 6H). Incubation of the AtXXT1-reaction products with OsXyBAT5 or OsXyBAT6 generated a number of acetylated, xylosylated G6 ion species, including two major ones corresponding to G6X1Ac1 (m/z 1,187) and G6X2Ac1 (m/z 1,319) (Fig. 6I, J; Supplementary Table S2), a minor one to G6X1Ac2 (m/z 1,229) and, in the case of OsXyBAT6, another minor one to G6X2Ac2 (m/z 1,361). By contrast, SlXyBAT2 was only able to add one acetyl group to a small amount of monoxylosylated G6 to generate G6X1Ac1 (m/z 1,187) (Fig. 6K;Supplementary Table S2). These results demonstrated that OsXyBAT5 and OsXyBAT6 were able to use xyloxylated G6 as acceptors, whereas SlXyBAT2 could barely acetylate xylosylated G6, indicating that XyBATs exhibit differential activities toward non-xylosylated and xylosylated glucan acceptor substrates.
It is important to note that although the sequential reactions of AtXXT1 followed by SlXyBAT2 only produced a minute amount of monoacetylated, monoxylosylated G6 (Fig. 6K), those of SlXyBAT2 followed by AtXXT1 effectively converted most of the G6 acceptor to acetylated, xylosylated G6 (Fig. 6G). In addition, the reaction products of OsXyBAT5 or OsXyBAT6 followed by AtXXT1 had much higher proportions of acetylated, xylosylated G6 than those of AtXXT1 followed by OsXyBAT5 or OsXyBAT6 (Fig. 6C, E, I, J). These findings suggest that AtXXT1 could xylosylate pre-acetylated G6 more efficiently than XyBATs acetylate pre-xylosylated G6.
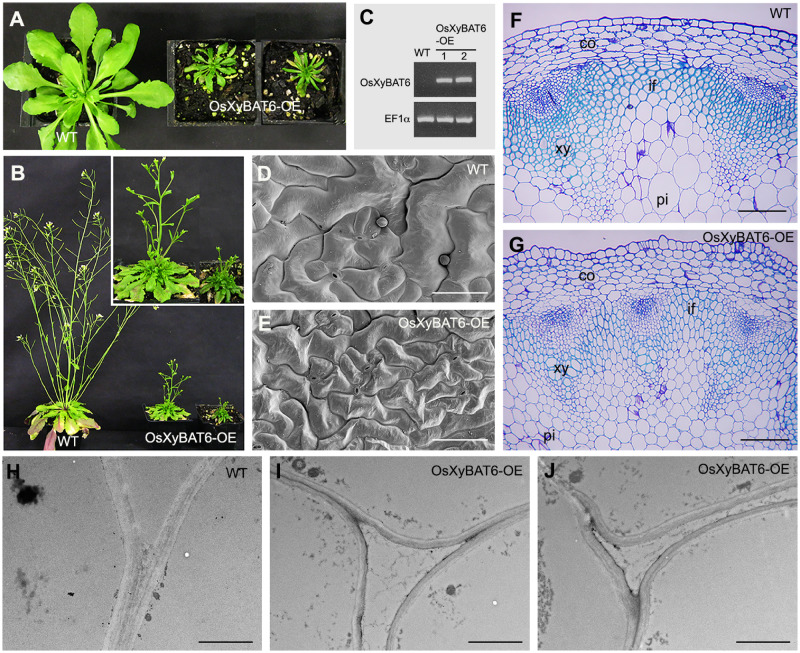
Effects of heterologous expression of OsXyBAT6 in Arabidopsis on xyloglucan structure and plant growth
To investigate the effects of xyloglucan backbone acetylation on its xylosylation pattern in planta, we expressed a representative XyBAT, OsXyBAT6, in Arabidopsis, which has XXXG-type xyloglucan without backbone acetylation. It was found that OsXyBAT6 expression resulted in a severe retardation in plant growth, leading to a much smaller rosette size and a diminished plant height compared with the wild type (Fig. 7A–C). Scanning electron microscopy showed that the leaf epidermal cells of OsXyBAT6 expressors (OsXyBAT6-OE) were smaller in size and had less sinuous contours than those of the wild type (Fig. 7D, E). The cortex and pith cells in the stems of OsXyBAT6-OE were also noticeably smaller in size (Fig. 7F, G). Furthermore, the walls of pith cells appeared to be thinner than those of the wild type (Fig. 7H–J). Consistent with the stunted plant growth, cell wall composition analysis showed a drastic reduction in the amounts of glucose and xylose, the main components of cellulose and xylan, respectively, in OsXyBAT6-OE compared with the wild type (Supplementary Fig. S8). We next examined whether there was any intracellular aggregation of xyloglucan in OsXyBAT6-OE since it has previously been reported that the dwarf phenotype of the Arabidopsis mur3-3 mutant is associated with intracellular accumulation of cell wall components in abnormal endomembrane aggregates (Kong et al. 2015). Immunocytochemical staining using two xyloglucan antibodies, LM15 (Marcus et al. 2008) and LM25 (Pedersen et al. 2012), did not reveal any intracellular aggregation of xyloglucan in OsXyBAT6-OE (Supplementary Fig. S9).
Fig. 7.
Effects of heterologous expression of OsXyBAT6 on plant growth and cell expansion in Arabidopsis. (A and B) Morphology of 4-week-old (A) and 7-week-old (B) WT and OsXyBAT6-expressing (OsXyBAT6-OE) plants. Inset in (B) shows a higher magnification of OsXyBAT6-OE plants. (C) Reverse transcription-PCR analysis showing the presence of OsXyBAT6 transcripts in two representative OsXyBAT6-OE lines. The EF1α gene was used as an internal control. (D and E) Scanning electron micrographs showing the morphology of adaxial epidermal cells in the leaves of 4-week-old WT (D) and OsXyBAT6-OE (E). Bars = 100 μm. (F and G) Cross sections of the bottom internodes of the inflorescence stems of 7-week-old WT (F) and 8-week-old OsXyBAT6-OE (G). Diameters of pith cells in the wild type and OsXyBAT6-OE are 55 ± 18 and 33 ± 11 μm, respectively. Bars = 120 μm. (H-J) Transmission electron micrographs showing the primary walls of pith cells in the inflorescence stems of WT (H) and two representative OsXyBAT6-OE lines (I, J). Bars = 1.6 μm. WT, wild type; co, cortex; if, interfascicular fiber; pi, pith; xy, xylem.
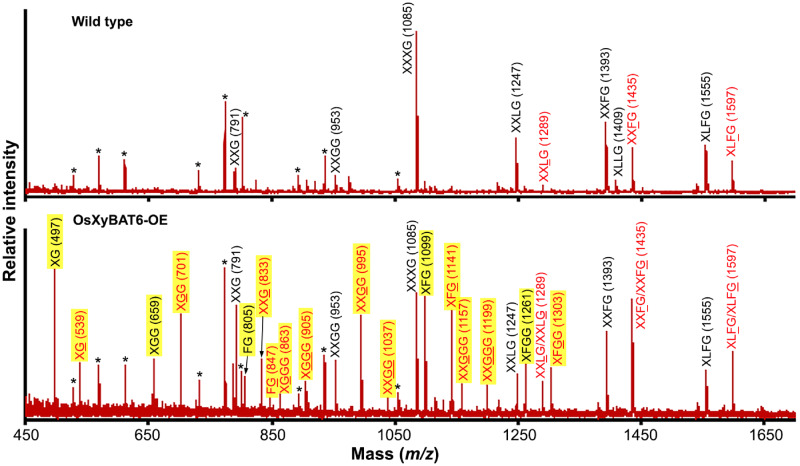
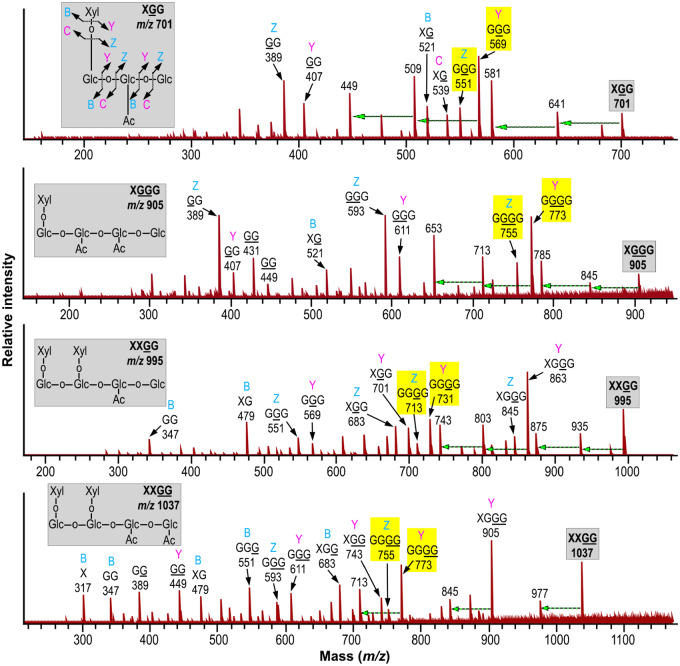
To discern whether the severe plant growth defects caused by OsXyBAT6 expression were accompanied by an alteration in xyloglucan structure, we digested the cell walls with endo-β-1,4-glucanase to release xyloglucan oligomers for structural analysis. MALDI-TOF MS analysis of xyloglucan oligomers from the cell walls of wild-type Arabidopsis revealed the typical ion species at m/z 791 (corresponding to XXG), 953 (XXGG), 1,085 (XXXG), 1,247 (XXLG), 1,289 (XXLG; L denotes that Gal in the Gal-Xyl disaccharide side chain is acetylated), 1,393 (XXFG), 1,409 (XLLG), 1,435 (XXFG; F denotes that Gal in the Fuc-Gal-Xyl trisaccharide side chain is acetylated), 1,555 (XLFG) and 1,597 (XLFG) (Fig. 8) (Günl et al. 2011). By contrast, the MALDI-TOF mass spectrum of xyloglucan oligomers from OsXyBAT6-OE displayed a large number of new ion species in addition to those observed in wild-type xyloglucan oligomers (Fig. 8). These new ion species were most likely attributed to fragments with reduced xylosyl decoration and their corresponding acetylated forms, including XG (m/z 497) and XG (m/z 539; G denotes acetylated Glc), XGG (m/z 659) and XGG (m/z 701), FG (m/z 805) and FG (m/z 847), XGGG (m/z 863) and XGGG (m/z 905), XFG (m/z 1,099) and XFG (m/z 1,141), XXGGG (m/z 1,157) and XXGGG (m/z 1,199) and XFGG (m/z 1,261) and XFGG (m/z 1,303). Other noticeable new ion species were m/z 833 corresponding to XXG and m/z 995 and m/z 1,037 corresponding to XXGG and XXGG, respectively, none of which was detected in the wild type, although their non-acetylated forms were present in both the wild type and OsXyBAT6-OE. The structural identities of a number of these ion species in OsXyBAT6-OE were verified by MALDI MS/MS. Fragmentations of ion species at m/z 659 and 701 produced fragments attributed to GGG and GGG (loss of Xyl), respectively, confirming the identities of XGG (m/z 659) and XGG (m/z 701) (Fig. 9; Supplementary Fig. S10). Likewise, the identities of XGGG (m/z 905), XXGG (m/z 953), XXGG (m/z 995), XXGG (m/z 1,037), XXGGG (m/z 1,157) and XXGGG (m/z 1,199) were proven by their fragmentation generating ion species corresponding to their respective backbone cello-oligomer (loss of Xyl) (Fig. 9; Supplementary Fig. S10). It was also observed that the relative ratios of ion species for the acetylated over the non-acetylated XXFG and XLFG were elevated in OsXyBAT6-OE compared with the wild type (Fig. 8), implying that OsXyBAT6 also acetylated Glc in these structural units. The release of XXGG-type fragments by the endoglucanase digestion of cell walls of OsXyBAT6-OE indicates that xyloglucan backbone acetylation alters its regular xylosylation pattern, resulting in a disruption of the XXXG core motifs. These results demonstrated that the expression of OsXyBAT6 in Arabidopsis had a profound effect on xyloglucan structure and severely affected cell expansion and plant growth.
Fig. 8.
MALDI-TOF MS analysis of xyloglucan oligomers released by endo-1,4-β-glucanase digestion of cell walls of wild-type Arabidopsis (top panel) and OsXyBAT6-OE (bottom panel) plants. Each ion peak is labeled with its mass ([M+Na]+) and predicted xyloglucan oligomer identity. Other possible variants of xyloglucan oligomers for each ion species are listed in Supplementary Table S3. Xyloglucan oligomers without acetylation are shown in black, and the ones decorated with acetyl groups (G, L and F) are shown in red. Ion peaks unique in OsXyBAT6-OE are highlighted in yellow. Asterisks indicate additional ion species corresponding to glucomannan or xylan oligomers generated by the endo-1,4-β-glucanase used, which contains low activities toward glucomannan and xylan.
Fig. 9.
MALDI MS/MS spectra of ion species at m/z 701, 905, 995 and 1,037 of glucanase-released xyloglucan fragments from OsXyBAT6-OE. Inset in each panel depicts the structure of the corresponding mother ion species, and the one in the top panel also indicates the types of fragmentation generating ions containing the nonreducing terminus (B, C) and those containing the reducing terminus (Y, Z). Ion species generated by the cleavage of glycosidic bonds in the mother ion species were denoted by their fragmentation types, structural identities and masses ([M+Na]+). Highlighted in yellow are the backbone cello-oligomer fragments derived by the loss of all Xyl residues from the mother ion. A dashed horizontal arrow indicates an ion species generated from the one at its right end by a loss of 60 in mass through cross-ring cleavages.
Two Arabidopsis XyBAT close homologs, TBL19 and TBL21, exhibit O-acetyltransferase activity toward cellohexaose
It is interesting that there exist three close homologs of XyBATs in Arabidopsis, namely TBL19, TBL20 and TBL21 (Fig. 1), since Arabidopsis xyloglucan is the XXXG type without backbone acetylation. To discern whether they possess the same biochemical activity as XyBATs, we produced recombinant proteins of TBL19, TBL20 and TBL21 in HEK293 cells (Fig. 10A) and assayed their O-acetyltransferase activities. It was found that, while no acetyl transfer onto mannohexaose and xylohexaose was detected, TBL19 and TBL21 were able to transfer acetyl groups onto the cellohexaose acceptor (Fig. 10B), which was confirmed by MALDI-TOF MS (Fig. 10C). NMR spectroscopy further revealed the transfer of acetyl groups onto O-6 of Glc residues in cellohexaose by TBL19 and TBL21 (Fig. 10D), indicating that they are also XyBATs and thus named AtXyBAT1 and AtXyBAT2, respectively.
Fig. 10.
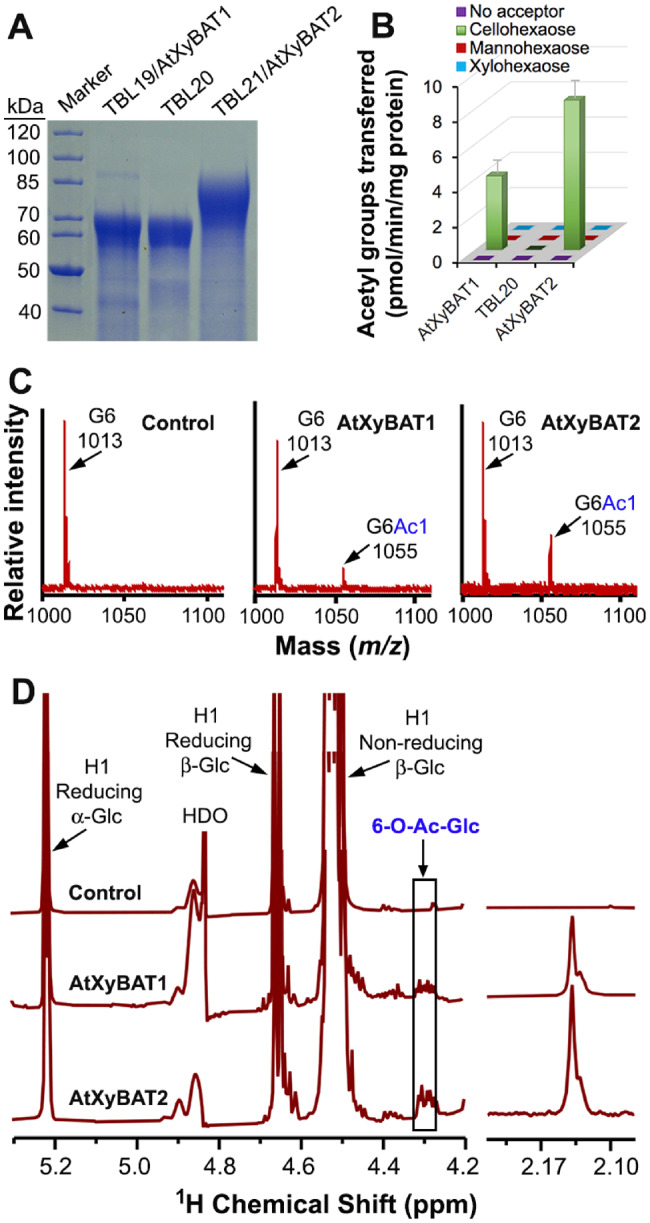
Analysis of O-acetyltransferase activities of Arabidopsis TBL19/AtXyBAT1, TBL20 and TBL21/AtXyBAT2 toward the cellohexaose acceptor. (A) SDS–PAGE and Coomassie Blue staining of recombinant proteins of TBL19, TBL20 and TBL21. Shown at the left are the molecular masses of protein markers. (B) Assay of O-acetyltransferase activities of AtXyBAT1, TBL20 and AtXyBAT2 by incubating their recombinant proteins with 14C-labeled acetyl-CoA and various acceptors, including cellohexaose, mannohexaose and xylohexaose. Error bars denote the SE of three biological replicates. (C) MALDI-TOF mass spectra of the reaction products of AtXyBAT1 and AtXyBAT2 incubated with acetyl-CoA and cellohexoase. The control is heat-denatured AtXyBAT2 incubated with acetyl-CoA and cellohexoase. Each ion peak is marked with its mass ([M+Na]+) and structural identity. Note the occurrence of an additional ion peak at m/z 1,055 corresponding to monoacetylated cellohexaose (G6Ac1) compared with the control having the cellohexaose acceptor (G6) at m/z 1,013 only. (D) 1H NMR spectra of AtXyBAT1- and AtXyBAT2-catalyzed reaction products. Note the presence of resonances attributed to acetylated Glc (6-O-Ac-Glc) in the anomeric region (left panel) and resonances corresponding to acetyl groups in the acetyl signature region (right panel) in the AtXyBAT1- and AtXyBAT2-catalyzed reaction products. HDO, hydrogen-deuterium oxide.
Discussion
A group of rice and tomato DUF231 proteins is XyBATs
Xyloglucan from rice and tomato is the XXGG type and has acetyl moieties attached at O-6 of the unsubstituted backbone Glc residues. In this report, we have provided unequivocal biochemical evidence establishing that a group of DUF231 proteins from rice and tomato are XyBATs. These XyBATs are able to transfer up to three acetyl groups onto the cellohexaose acceptor, and the acetyl moieties were shown to be attached at O-6 of the backbone Glc residues. While some XyBATs could only effectively acetylate cello-oligomers with a degree of polymerization (DP) of 5 and 6, OsXyBAT5 and SlXyBAT2 could act on cello-oligomers with a DP of as short as 4 and OsXyBAT6 as short as 3. This is in contrast to the requirement of cello-oligomers with a DP of 6 by xyloglucan xylosyltransferases for efficient xylosylation (Cavalier and Keegstra 2006).
The XyBATs from rice, tomato and Arabidopsis phylogenetically reside in group I in the DUF231 family, which includes XGOATs and MOATs (Gille et al. 2011, Zhong et al. 2018a, Zhong et al. 2018b). However, XyBATs apparently fall into a subgroup distinct from the XGOAT and MOAT subgroups. It is unclear why both rice and tomato have so many XyBATs, but considering that they are differentially expressed in different organs, it is conceivable that they may be involved in mediating xyloglucan backbone acetylation in specific organs/cell types and/or at different developmental stages.
Although xyloglucan from rice and tomato is predominantly the XXGG type with acetylation on the backbone Glc residues, XXXG-type xyloglucan has been found in the cell walls of some specialized cell types, such as rice root hairs and anthers (Liu et al. 2015) and tomato pollen tubes (Dardelle et al. 2015). It is currently unknown whether the XXXG-type xyloglucan from rice and tomato bears acetyl moieties on side-chain Gal residues, but their genomes harbor close homologs of AXY4/XGOAT1 and AXY4L/XGOAT2, including one of each in tomato and an unexpectedly large number of them in rice and switchgrass (Supplementary Fig. S2), the biochemical functions of which await to be investigated. It is intriguing that two Arabidopsis DUF231 members, TBL19/AtXyBAT1 and TBL21/AtXyBAT2, possess XyBAT activity since xyloglucan from Arabidopsis is the XXXG type and has not been reported to have backbone Glc acetylation. This finding indicates that Arabidopsis might have XXGG-type xyloglucan in some specialized cell types, a scenario similar to rice and tomato having cell-type-specific XXXG-type xyloglucan. In line with this proposition, TBL19/AtXyBAT1 and TBL21/AtXyBAT2 are preferentially expressed in trichomes and root hairs, respectively, based on the Arabidopsis transcriptome data (Supplementary Figs. S11–S13).
In addition to grasses and Solanaceae, a number of other taxonomic groups of plants, including ferns, lycophytes, mosses and liverworts, have XXGG-type xyloglucan (Pauly and Keegstra 2016). It is not known whether xyloglucan from these plants is also acetylated on the backbone Glc residues. Since the moss genome lacks close homologs of XyBATs (Zhong et al. 2019), if moss xyloglucan is acetylated on the backbone Glc residues, it would be most likely catalyzed by other members of the DUF231 family or other unknown proteins. Such a scenario was observed for moss xylan; although moss xylan is acetylated (Haghighat et al. 2016), its genome lacks close homologs of Arabidopsis xylan O-acetyltransferases (Zhong et al. 2019).
Coordination of 6-O-acetylation and 6-O-xylosylation of xyloglucan backbone Glc residues
The XXGG-type xyloglucan from grasses and Solanaceae typically has repeating units of two xylosylated Glc residues followed by two or more non-xylosylated ones that may be acetylated (XXGG or XXGGG) (Pauly and Keegstra 2016). Because both xylosylation and acetylation occur at O-6 of the backbone Glc residues, xylosylaton of a Glc residue prevents its acetylation and vice versa. Therefore, xylosylation and acetylation of the xyloglucan backbone must be precisely coordinated to achieve the regular pattern observed in the XXGG-type xyloglucan.
Our enzymatic activity assays revealed that AtXXT1 could efficiently xylosylate acetylated cellohexaose generated by XyBATs, producing a number of acetylated, xylosylated cello-oligomers, including those with two Xyl residues and one or two acetyl moieties that might reflect the xyloglucan acetylation patterns (XXGG) seen in rice cell walls. These data are further corroborated by in planta studies showing that OsXyBAT6 expression in Arabidopsis resulted in the generation of xyloglucan structural units with similar acetylation patterns as those in rice, including XXGG, XXGG, XXGGG and XXGGG. In addition, OsXyBAT5 and OsXyBAT6 were shown to be able to act on xylosylated cellohexaose generated by AtXXT1, resulting in the production of acetylated, xylosylated cello-oligomers. However, the efficiency was much lower than that of AtXXT1 on acetylated cellohexaose. Furthermore, SlXyBAT2 could barely act on xylosylated cellohexaose although its activity on non-xylosylated cellohexaose was comparable to those of OsXyBAT5 and OsXyBAT6. These findings indicate that in planta, the xyloglucan backbone might be first acetylated and then xylosylated to efficiently achieve its specific substitution patterns. It has been proposed that in Arabidopsis, the three xyloglucan xylosyltransferases XXT1, XXT2 and XXT5 form multiprotein complexes with xyloglucan backbone synthases in such a way that they xylosylate the first, second and third Glc residues, respectively, thus generating the XXXG repeating units of xyloglucan (Culbertson et al. 2018). By analogy, it is likely that in grasses and Solanaceae, XyBATs are assembled into multiprotein complexes with XXTs and xyloglucan backbone synthases in such a manner that together they synthesize the XXGG-type xyloglucan with specific backbone acetylation patterns. The exact mechanisms underlying the specific xyloglucan backbone xylosylation and acetylation patterns remain to be elucidated.
Maintaining the regular xyloglucan xylosylation pattern is crucial for normal cell expansion and plant growth
We have demonstrated that heterologous expression of OsXyBAT6 in Arabidopsis leads to a drastic alteration in the xylosylation pattern of xyloglucan. Instead of the XXXG-type fragments seen in wild-type Arabidopsis cell walls, endoglucanase digestion of cell walls of OsXyBAT6-OE generated a number of XXGG-type fragments, including XGGG, XGGG, XXGG, XXGG, XXGGG, XXGGG and XFGG, as well as some shorter fragments corresponding to XG, FG, XGG, XXG and XFG and their acetylated forms. This finding suggests that OsXyBAT6 competed with XXTs for substitutions at O-6 of the backbone Glc residues and thus disrupted the regular xylosylation of three consecutive Glc residues, resulting in XXGG-type and shorter repeating units in xyloglucan. Unusual xyloglucan oligomers, such as LG, FG, GLG, GFG, XFG, GXLG or GLLG, have also been observed in the endoglucanase-digestion products of cell walls of several Arabidopsis mutants defective in apoplastic glycosidases that modify xyloglucan structure, including axy8, xyl1 (α-xylosidase1), bgal10 (β-galactosidase10) and bglc1 (β-glucosidase active against xyloglucan1) (Günl et al. 2011. Sampedro et al. 2010, 2012, 2017a). Expression of BdXyBAT1 in Arabidopsis has previously been shown to result in the occurrence of some acetylated XXGG units in xyloglucan, indicating a reduction in xyloglucan xylosylation (Liu et al. 2016). The observed variations in the composition of endoglucanase-released xyloglucan fragments between OsXyBAT6-OE and BdXyBAT1-OE could be due to differences in the specific activities of XyBATs, their expression levels and/or the specificities of the endoglucanases used.
It is intriguing to find that although a lack of xyloglucan in the xxt1 xxt2 double mutant did not significantly affect plant growth except root hair development (Cavalier et al. 2008), an alteration in xyloglucan structure by the expression of OsXyBAT6 in Arabidopsis resulted in reduced cell expansion and severely stunted plant growth. A strong retardation in plant growth was also observed in the mur3 single and the mur3 xlt2 double mutants with Gal-depleted xyloglucan, which could be partially attributed to aggregation of the dysfunctional xyloglucan in the endomembrane system that may obstruct membrane organization and the delivery of vesicles containing cell wall polysaccharides (Schultink et al. 2013, Kong et al. 2015, Pauly and Keegstra 2016). In addition, reduced growth of some specific organs, such as siliques and flowers, was seen in the xyl1 and bgal10 mutants with altered xyloglucan xylosylation pattern (Sampedro et al. 2010, 2012). Immunolabeling of xyloglucan in OsXyBAT6-OE did not reveal any intracellular aggregation of xyloglucan, indicating that the secretion of cell wall polysaccharides was not affected. It is tempting to propose that the drastic alteration in xyloglucan xylosylation pattern most likely accounts for the severe phenotypes observed in OsXyBAT6-OE. The lack of any observable phenotypes in BdXyBAT1-OE (Liu et al. 2016) could be due to the lesser degree of alteration in xyloglucan xylosylation.
It is unclear how the altered xyloglucan structure in XyBAT6-OE leads to a severe reduction in cell expansion and plant growth. Xyloglucan crosslinks cellulose to form a cellulose/hemicellulose network, which is considered to be important for the mechanical properties of primary cell walls and plant growth (Whitney et al. 1999). It was proposed that cellulose microfibrils are connected by xyloglucan through limited regions of load-bearing junctions (Park and Cosgrove 2012a). It is likely that the altered xyloglucan structure in OsXyBAT6-OE may hinder its normal interaction with cellulose microfibrils, thus hampering cell expansion and plant growth. Xyloglucan has been demonstrated to be important for cell wall loosening by α-expansins (Park and Cosgrove 2012b, Schultink et al. 2013), and Arabidopsis xyloglucan endotransglycosylase/hydrolases (XTHs) have been shown to be involved in cell wall extension and exhibit a strong preference for XXXG instead of XXG as an acceptor (Maris et al. 2009). Therefore, it is also possible that the altered xyloglucan structure may reduce α-expansin-mediated cell wall creep and/or impede XTH-mediated cell wall extension, thus leading to stunted plant growth. Furthermore, xyloglucan oligosaccharides released by glucosidases have been shown to regulate cell expansion (McDougall and Fry 1990, Takeda et al. 2002). The altered xyloglucan structure in OsXyBAT6-OE might also affect the generation of such cell expansion-promoting xyloglucan oligosaccharides. Further investigation of the mechanistic basis of the stunted plant growth caused by the altered xyloglucan structure in XyBAT6-OE will further our understanding of the roles of xyloglucan in plant growth.
Materials and Methods
Phylogenetic analysis
The amino acid sequences of all 46 Arabidopsis DUF231 proteins were used to BLAST search for their rice and tomato homologs from the rice (O. sativa v7_JGI at Phytozome v12) and the tomato (S. lycopersicum iTAG2.4 at Phytozome v12) genomic databases, respectively. The genomes of poplar, Brachypodium, maize, sorghum and switchgrass in Phytozome v12 were BLAST-searched for close homologs of group I DUF231 proteins. The MEGA (v6.0) software with the neighbor-joining algorithm was employed to evaluate the phylogenetic relationships of DUF231 proteins.
Expression of recombinant proteins in HEK293 cells
The putative catalytic domains of DUF231 proteins and AtXXT1 were expressed as secreted His-tagged recombinant proteins in HEK293 cells (Zhong et al. 2017a). The amino acids used for recombinant protein production were from numbers 28 to 404 for OsXyBAT1, 26 to 393 for OsXyBAT2, 36 to 415 for OsXyBAT3, 30 to 406 for OsXyBAT4, 36 to 438 for OsXyBAT5, 40 to 420 for OsXyBAT6, 40 to 411 for OsXyBAT7, 36 to 416 for Os02g49650, 43 to 439 for Os06g16250, 28 to 405 for BdXyBAT1, 40 to 434 for SlXyBAT1, 40 to 430 for SlXyBAT2, 42 to 427 for SlXyBAT3, 40 to 425 for SlXyBAT4, 44 to 531 for SlXyBAT5, 43 to 455 for SlXyBAT6, 34 to 432 for SlXyBAT7, 41 to 460 for AtXXT1, 36 to 426 for TBL19/AtXyBAT1, 35 to 478 for TBL20 and 36 to 526 for TBL21/AtXyBAT2. The cDNA sequences corresponding to the catalytic domains of DUF231 proteins and AtXXT1 were PCR-amplified, cloned in-frame between the murine Igκ chain leader sequence (for protein secretion) and the c-myc epitope and six-tandem histidine tag in the pSecTag2 mammalian expression vector (Invitrogen, Waltham, MA, USA) and confirmed by sequencing. HEK293 cells were transfected with the expression constructs using the Invitrogen FreeStyle 293 Expression System. After culturing the transfected cells for 5 d, the culture media were collected and passed through nickel resin columns for the purification of His-tagged recombinant proteins. The purified proteins were examined by SDS–PAGE and Coomassie Blue staining.
Assay of O-acetyltransferase activities
Acetyltransferase activity assays for radiodetection and MALDI-TOF MS were carried out in 40-μl reaction mixtures by incubating purified recombinant proteins (20 μg) with acetyl-1-14C CoA (0.1 μCi; American Radiolabeled Chemicals, St Louis, MO, USA) or non-radiolabeled acetyl-CoA (1 mM) and various oligosaccharide acceptors (30 μg), including cello-oligomers, xylohexaose and mannohexaose, in 50 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] buffer (pH 7.0). For NMR analysis, 100 μg of recombinant proteins and 200 μg of acceptors were used in a 200-μl reaction mixture. After incubation at 37°C for 4 h (for radiodetection and MALDI-TOF MS) or 20 h (for NMR analysis), the reaction products were separated from acetyl-CoA by passing through Dowex 1X4 anion exchange resin and then counted for radioactivity with a PerkinElmer scintillation counter (Waltham, MA, USA) or subjected to MALDI-TOF MS and NMR analyses. The amount of radioactivity incorporated into oligosaccharide acceptors by the recombinant proteins was calculated by subtracting the background radioactivity present in the reaction mixture containing boiled proteins. Cello-oligomers, xylohexaose and mannohexaose were purchased from Megazyme (Bray, Co. Wicklow, Ireland). For each recombinant protein, the reaction products from three biological replicates were used for subsequent analyses.
Sequential acetyltransferase and xylosyltransferase reactions
To examine whether XyBATs could use xylosylated cellohexaose as acceptors, cellohexaose (30 μg) was first incubated with recombinant AtXXT1 (20 μg) and UDP-Xyl (2.5 mM) in 50 mM HEPES buffer (pH 7.0) containing MnCl2 (2.5 mM) in a volume of 40 μl. After 20-h incubation at 37°C, the reaction mixture was boiled to denature AtXXT1 and the xylosylated cellohexaose products were then incubated with XyBAT (20 μg) and acetyl-CoA (1 mM) in 50 mM HEPES buffer (pH 7.0). The reaction was proceeded at 37°C for 20 h, and the reaction mixtures were desalted with BioRex MSZ 501 beads (Bio-Rad, Hercules, CA, USA). The reaction products were then examined for their masses by MALDI-TOF MS. To test whether AtXXT1 could use acetylated cellohexaose as acceptors, cellohexaose was first incubated with XyBATs and then with AtXXT1 as described above. The reaction products from three replicates were analyzed, and representative spectra were shown.
MALDI-TOF MS
The reaction products were inspected for masses corresponding to acetylated and/or xylosylated cello-oligomers by MALDI-TOF MS using a Burker Autoflex TOF mass spectrometer (Billerica, MA, USA) in reflection mode (Zhong et al. 2005). The spectra were the averages of at least 100 laser shots. The reaction products from three replicates were analyzed, and representative spectra were shown.
The MALDI MS/MS spectra were acquired on a Solarix XR 12T Fourier-transform ion cyclotron resonance mass spectrometer (Bruker Daltonics, Billerica, MA, USA). Five hundred laser shots were used to produce one spectrum with the use of a 2 million data point transient size. Fragmentation was done by CID in the hexapole collision cell, with the settings of 55 V collision energy, 2,000 Vpp radio frequency amplitude and a parent ion isolation window width set to 10 m/z.
1H NMR spectroscopy
The XyBAT-catalyzed reaction products were subjected to NMR analysis using a Varian Inova 500 MHz spectrometer (Varian Inc., Palo Alto, CA, USA). A total of 512 transients were recorded for each NMR spectrum. The proton positions and residue identities in the NMR spectra were assigned based on the NMR spectral data for xyloglucan oligomers from tomato (Jia et al. 2005).
Generation of transgenic Arabidopsis plants expressing XyBAT
The full-length cDNA of OsXyBAT6 was PCR-amplified, ligated between the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator in the pBI121 binary vector and confirmed by sequencing. The construct was introduced into wild-type Arabidopsis by the agrobacterium-mediated transformation. Transgenic plants were selected by growing seeds on the Murashige–Skoog medium containing kanamycin and further grown in soil in a climate-controlled greenhouse. The expression of OsXyBAT6 in transgenic plants was examined by reverse transcription-PCR analysis. The first generation of transgenic plants was chosen for phenotypic and cell wall analyses. Cell walls from three separate pools of transgenic plants with 4–6 plants in each pool were used as biological replicates for subsequent analyses.
Histology
The bottom parts of Arabidopsis stems were fixed and embedded in LR White Resin as previously described (Burk et al. 2006). For light microscopy, stems were cut into 1-μm thickness and stained with toluidine blue for observation. For transmission electron microscopy, stems were sectioned into 60-nm thickness and post-stained with lead citrate and uranyl acetate before examination under the JEOL JEM1011 transmission electron microscope (JEOL Inc., Peabody, MA, USA). For the immunodetection of xyloglucan, 1-μm-thick stem sections were incubated with the xyloglucan monoclonal antibodies LM15 and LM25 (Plantprobes) and then with fluorescein isothiocyanate-conjugated secondary antibody as previously described (Zhong et al. 2017b). The immunolabeled sections were observed for fluorescence signals under a Zeiss LSM710 confocal microscope. For the visualization of leaf epidermis, leaves were fixed in 2% glutaraldehyde and then post-fixed in 1% osmium tetroxide. After dehydration through a gradient of ethanol from 20% to 100%, the leaves were dried in a Samdri model 780-A critical point dryer (Tousimis, Rockville, MD, USA), mounted on aluminum stubs, sputter-coated with 30 nm of gold–palladium using a Leica EM ACE600 (Leica Microsystems Inc. Buffalo Grove, IL, USA) and observed under the FEI Teneo scanning electron microscope (Thermo Fisher Scientific, Hillsboro, OR, USA). Stems and leaves from four independent transgenic plants were examined, and representative images were shown.
Cell wall composition analysis and digestion of cell walls with endoglucanase
Arabidopsis stems were ground into powder in liquid N2 and then homogenized sequentially in 70% ethanol, 100% ethanol and 100% acetone (Zhong et al. 2005). For cell wall sugar composition analysis, the alcohol-insoluble cell walls were hydrolyzed by sulfuric acid and the released monosaccharides from polysaccharides (including cellulose) were derivatized to alditol acetates, which were separated on a silica capillary column (30 m × 0.25 mm) attached to a Hewlett Packard gas chromatography (GC)-MS (Hoebler et al. 1989). For xyloglucan structural analysis, the alcohol-insoluble cell walls were digested with endo-1,4-β-glucanase from Trichoderma longibrachiatum (E-CELTR; Megazyme) and the released xyloglucan oligomers were examined by MALDI-TOF MS and MALDI MS/MS.
Supplementary Data
Supplementary data are available at PCP online.
Supplementary Material
Acknowledgments
We thank Dr. J.N. Glushka at the Complex Carbohydrate Research Center and Dr. M.K. Kandasamy at the UGA Biomedical Microscopy Core for their assistance with NMR and confocal microscope imaging, respectively, and two anonymous reviewers for their constructive comments and suggestions.
Funding
U.S. Department of Energy, Office of Science, Basic Energy Sciences (DE-FG02-03ER15415). The authors are grateful for the support of the National Institutes of Health (S10 OD025118) for funding the acquisition of the Solarix XR mass spectrometer.
Disclosures
The authors have no conflicts of interest to declare.
References
- An H.J., Lebrilla C, B. (2011) Structure elucidation of native N- and O-linked glycans by tandem mass spectrometry (tutorial). Mass Spectrom. Rev. 30: 560–578. [DOI] [PubMed] [Google Scholar]
- Burk D.H., Zhong R., Morrison W.H. III and Ye Z.-H. (2006) Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J. Integr. Plant Biol. 48: 85–98. [Google Scholar]
- Cavalier D.M., Keegstra K. (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J. Biol. Chem. 281: 34197–34207. [DOI] [PubMed] [Google Scholar]
- Cavalier D.M., Lerouxel O., Neumetzler L., Yamauchi K., Reinecke A., Freshour G., et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J.C., Lerouxel O., Drakakaki G., Alonso A.P., Liepman A.H., Keegstra K., et al. (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 104: 8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson A.T., Ehrlich J.J., Choe J.Y., Honzatko R.B., Zabotina O.A. (2018) Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers. Proc. Natl. Acad. Sci. USA 115: 6064–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F., Le Mauff F., Lehner A., Loutelier-Bourhis C., Bardor M., Rihouey C., et al. (2015) Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann. Bot. 115: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C., York W.S., Albersheim P., Darvill A., Hayashi T., Joseleau J.-P., et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89: 1–3. [Google Scholar]
- Gille S., de Souza A., Xiong G., Benz M., Cheng K., Schultink A., et al. (2011) O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M., Neumetzler L., Kraemer F., de Souza A., Schultink A., Pena M., et al. (2011) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23: 4025–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat M., Teng Q., Zhong R., Ye Z.-H. (2016) Evolutionary conservation of xylan biosynthetic genes in Selaginella moellendorffii and Physcomitrella patens. Plant Cell Physiol. 57: 1707–1719. [DOI] [PubMed] [Google Scholar]
- Hoebler C., Barry J.L., David A., Delort-Laval J. (1989) Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J. Agr. Food Chem. 37: 360–367. [Google Scholar]
- Jensen J.K., Schultink A., Keegstra K., Wilkerson C.G., Pauly M. (2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol. Plant 5: 984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Cash M., Darvill A.G., York W.S. (2005) NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr. Res. 340: 1818–1825. [DOI] [PubMed] [Google Scholar]
- Kong Y., Peña M.J., Renna L., Avci U., Pattathil S., Tuomivaara S.T., et al. (2015) Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 167: 1296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Hsia M.M., Dama M., Vogel J., Pauly M. (2016) A xyloglucan backbone 6-O-acetyltransferase from Brachypodium distachyon modulates xyloglucan xylosylation. Mol. Plant 9: 615–617. [DOI] [PubMed] [Google Scholar]
- Liu L., Paulitz J., Pauly M. (2015) The presence of fucogalactoxyloglucan and its synthesis in rice indicates conserved functional importance in plants. Plant Physiol. 168: 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall G.J., Fry S.C. (1990) Xyloglucan oligosaccharides promote growth and activate cellulase: evidence for a role of cellulase in cell expansion. Plant Physiol. 93: 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M., Dunand C., Li X., Verma R., Vanzin G.F., Caplan J., et al. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J.J., Farkas V., Pedersen H.L., et al. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A., Suslov D., Fry S.C., Verbelen J.P., Vissenberg K. (2009) Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 60: 3959–3972. [DOI] [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J. (2012. a) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158: 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J. (2012. b) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 158: 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M., Keegstra K. (2016) Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 67: 235–259. [DOI] [PubMed] [Google Scholar]
- Pedersen H.L., Fangel J.U., McCleary B., Ruzanski C., Rydahl M.G., Ralet M.C., et al. (2012) Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 287: 39429–39438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M.J., Kong Y., York W.S., O’Neill M.A. (2012) A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24: 4511–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R.M., DeRocher A.E., Bar-Peled M., Zeng W., Norambuena L., Orellana A., et al. (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979. [DOI] [PubMed] [Google Scholar]
- Perrin R.M., Jia Z., Wagner T.A., O’Neill M.A., Sarria R., York W.S., et al. (2003) Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol. 132: 768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J., Gianzo C., Iglesias N., Guitián E., Revilla G., Zarra I. (2012) AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 158: 1146–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J., Pardo B., Gianzo C., Guitián E., Revilla G., Zarra I. (2010) Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol. 154: 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J., Valdivia E.R., Fraga P., Iglesias N., Revilla G., Zarra I. (2017) Soluble and membrane-bound β-glucosidases are involved in trimming the xyloglucan backbone. Plant Physiol. 173: 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Tabata S., Hirakawa H., Asamizu E., Shirasawa K., Isobe S., et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Takehisa H., Kamatsuki K., Minami H., Namiki N., Ikawa H., et al. (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41: D1206–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A., Cheng K., Park Y.B., Cosgrove D.J., Pauly M. (2013) The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol. 163: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Furuta Y., Awano T., Mizuno K., Mitsuishi Y., Hayashi T. (2002) Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc. Natl. Acad. Sci. USA 99: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin G.F., Madson M., Carpita N.C., Raikhel N.V., Keegstra K., Reiter W.D. (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc. Natl. Acad. Sci. USA 99: 3340–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney S.E., Gothard M.G., Mitchell J.T., Gidley M.J. (1999) Roles of cellulose and xyloglucan in determining the mechanical properties of primary plant cell walls. Plant Physiol. 121: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W.S., Oates J.E., van Halbeek H., Darvill A.G., Albersheim P., Tiller P.R., et al. (1988) Location of the O-acetyl substituents on a nonasaccharide repeating unit of sycamore extracellular xyloglucan. Carbohydr. Res. 173: 113–132. [DOI] [PubMed] [Google Scholar]
- Zhong R., Cui D., Ye Z.-H. (2017. a) Regiospecific acetylation of xylan is mediated by a group of DUF231-containing O-acetyltransferases. Plant Cell Physiol. 58: 2126–2138. [DOI] [PubMed] [Google Scholar]
- Zhong R., Cui D., Ye Z.-H. (2018. a) Members of the DUF231 family are O-acetyltransferases catalyzing 2-O- and 3-O-acetylation of mannan. Plant Cell Physiol. 59: 2339–2349. [DOI] [PubMed] [Google Scholar]
- Zhong R., Cui D., Ye Z.-H. (2018. b) Xyloglucan O-acetyltransferases from Arabidopsis thaliana and Populus trichocarpa catalyze acetylation of fucosylated galactose residues on xyloglucan side chains. Planta 248: 1159–1171. [DOI] [PubMed] [Google Scholar]
- Zhong R., Cui D., Ye Z.-H. (2019) Evolutionary origin of O-acetyltransferases responsible for glucomannan acetylation in land plants. New Phytol. 224: 466–479. [DOI] [PubMed] [Google Scholar]
- Zhong R., Peña M.J., Zhou G.-K., Nairn C.J., Wood-Jones A., Richardson E.A., et al. (2005) Arabidopsis Fragile Fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Teng Q., Haghighat M., Yuan Y., Furey S.T., Dasher R.L., et al. (2017. b) Cytosol-localized UDP-xylose synthases provide the major source of UDP-xylose for the biosynthesis of xylan and xyloglucan. Plant Cell Physiol. 58: 156–174. [DOI] [PubMed] [Google Scholar]
- Zhu L., Dama M., Pauly M. (2018) Identification of an arabinopyranosyltransferase from Physcomitrella patens involved in the synthesis of the hemicellulose xyloglucan. Plant Direct 2: e00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.