Heart failure (HF) is a leading clinical and public health concern because of its high prevalence and poor prognosis. It is thus critical to identify novel risk factors for developing HF. Mitochondrial DNA copy number (mtDNA-CN), an indirect biomarker of mitochondrial dysfunction, is associated with atherosclerotic cardiovascular disease endpoints, cardiovascular risk factors, all-cause mortality, and sudden cardiac death.1-3 The association between mtDNA-CN and the risk of incident HF, however, is unknown. We examined this association in the Atherosclerosis Risk in Communities (ARIC) cohort.
The ARIC study is a population-based prospective cohort of 15,792 individuals 45–65 years of age at the time of recruitment (1987–1989).1 Among 11,431 participants with DNA collected in one of the visits to generate mtDNA-CN measurements, we excluded Black participants recruited from Minnesota or Maryland (n=1), and participants without follow-up information (n=1), with prevalent HF at the time of DNA collection (n=596), or with missing data in relevant variables (n=31). The final sample included 10,802 participants (4,904 men and 5,898 women). All centers obtained approval from their institutional review boards and all participants provided written informed consent.
DNA was extracted from buffy coat collected in Visits 1–4 (429 participants in Visit 1 [1987–1989], 8,655 in Visit 2 [1990–1992], 1,654 in Visit 3 [1993–1995], and 64 in Visit 4 [1996–1998]). mtDNA-CN was calculated from probe intensities of mitochondrial single nucleotide polymorphisms on the Affymetrix Genome-Wide Human SNP Array 6.0 (www.genvisis.org).4 The mtDNA-CN metric used in this analysis was obtained as standardized residuals (mean 0, standard deviation 1) from a linear regression of raw estimates of mtDNA-CN against age, sex, center, surrogate variables used in the surrogate variable analysis, and white blood cell count. For each participant, we used the visit in which DNA for mtDNA-CN assays was obtained as the baseline visit.
Incident HF was defined as the first hospitalization for HF or HF-related death.5 Incident HF events since 2005 were adjudicated by the ARIC HF Classification Committee and further classified as HF with reduced ejection fraction (HFrEF, left ventricular ejection fraction [LVEF] <50%), or HF with preserved ejection fraction (HFpEF, LVEF ≥50%).5
Study participants were followed from the visit of DNA collection until the development of HF, death, loss to follow-up, or December 31, 2017, whichever occurred first. We used a Cox proportional hazards model to estimate hazard ratios (HR) and 95% confidence intervals (CI) for the association between mtDNA-CN and incident HF, adjusting for age, sex, race/ethnicity, center, body mass index, smoking, alcohol intake, total and HDL cholesterol, cholesterol medication, hypertension, diabetes, and prevalent coronary heart disease. As the proportional hazards assumption was not met (P<0.001), we used a parametric survival model with separate spline variables for baseline hazard and for time-dependent effects. All statistical analyses were performed using Stata version 15.0 (StataCorp LP, College Station, TX, USA).
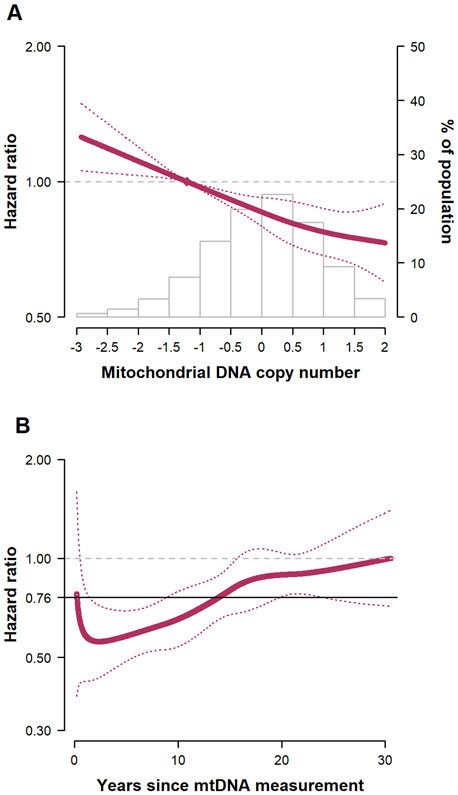
The median age (interquartile range) of study participants at baseline was 57 (52–62) years. During a median follow-up of 23.1 years, we identified 2,227 new cases of HF (incidence rate 10.3 per 1000 person-years). The adjusted HRs (95% CI) for HF comparing the 2nd through 5th quintiles of mtDNA-CN to the 1st quintile were 0.91 (0.80–1.04), 0.82 (0.72–0.93), 0.81 (0.71–0.92), and 0.74 (0.65–0.85), respectively (P for trend<0.001; Figure, A). When we compared the 90th to the 10th percentiles of mtDNA-CN (mtDNA-CN levels 1.16 and −1.21, respectively) as a continuous variable, the adjusted HR for HF was 0.76 (0.69–0.84). The results were similar when we used hospital discharge codes for HF instead of adjudicated outcomes. The association between mtDNA-CN and HF was progressively attenuated from the time of mtDNA-CN measurement (Figure, B). The adjusted HRs for HFpEF (564 events) and HFrEF (504 events) comparing the 90th to the 10th percentile of mtDNA-CN were 1.06 (0.87–1.28) and 0.89 (0.73–1.09), respectively.
Figure. Hazard ratios for incident heart failure associated with mitochondrial DNA copy number levels.
Results were adjusted for age, sex, race/ethnicity, body mass index, smoking, alcohol intake, total and HDL cholesterol, cholesterol medication, hypertension, and prevalent coronary heart disease. A, Hazard ratios by levels of mitochondrial DNA copy number as a continuous variable. The curves represent adjusted hazard ratios (solid line) and their 95% confidence intervals (dotted lines) based on restricted quadratic splines for mtDNA copy number with knots at 5th, 50th, and 95th percentiles of its distribution, under the proportional hazards assumption. The mtDNA-CN values used in the figure were unitless and were calculated as the standardized residuals (mean 0, standard deviation 1) from a linear regression of raw estimates of mtDNA-CN against age, sex, center, surrogate variables used in the surrogate variable analysis, and white blood cell count. The reference value (diamond dot) was set at the 10th percentile of the distribution. The inverse association of mtDNA-CN with incident heart failure was approximately linear (P-value for non-linear spline terms 0.74). The adjusted HR for HF comparing the 90th to the 10th percentile of mtDNA-CN (mtDNA-CN levels of 1.16 and −1.21, respectively) as a continuous variable was 0.76 (0.69–0.84). Histograms represent the frequency distribution of mtDNA copy number at baseline.
B, Time-dependent hazard ratios for incident heart failure comparing the 90th to the 10th percentile of mitochondrial DNA copy number. To allow the association of mtDNA copy number with incident HF to vary by time, we used a parametric survival model with restricted cubic splines with 4 degrees of freedom for log-time, restricted cubic splines with 4 degrees of freedom for mtDNA-CN, and time by mtDNA-CN interactions. The selection of degrees of freedom was based on the lowest AIC. The analyses were performed using the stpm2 command in Stata with the tvc option. The curve represents time-dependent adjusted hazard ratios (solid red line) and the dotted red lines represent its 95% confidence interval comparing the 90th to the 10th percentile of mtDNA copy number. The fully adjusted HRs for HF comparing the 90th to the 10th percentile of mtDNA-CN at 10, 20, and 30 years since mtDNA-CN measurement were 0.65 (0.54–0.79), 0.89 (0.76–1.04), and 0.99 (0.72–1.37), respectively. The solid black line represents the time-fixed adjusted hazard ratio comparing the 90th to the 10th percentile of mtDNA copy number (0.76, 95% CI 0.69– 0.84).
There are several limitations in our data. First, mtDNA-CN measurements were derived from the buffy coat of peripheral blood and were not measured directly in cardiomyocytes. Second, mtDNA-CN was measured only once and we could not evaluate changes after the baseline visit. Third, systematic adjudication of HF events occurred only after 2005. However, our analyses were consistent when we used only information from discharge codes. Finally, the analysis for subtypes of HF was restricted to events occurring after 2005, and we had limited power to identify differences between HFpEF and HFrEF.
In conclusion, mtDNA-CN was inversely associated with the risk of incident HF suggesting that reduced levels of mtDNA-CN, a biomarker of mitochondrial dysfunction, was a risk factor for HF. The association was approximately linear, but it was strongest early after measurement of mtDNA-CN and was progressively attenuated over 30 years of follow-up. Further studies are needed to better understand the underlying mechanisms and to characterize the association of mtDNA-CN with different types of HF.
ACKNOWLEDGEMENTS
The authors thank the staff and participants of the ARIC study for their important contributions. A previous version of this manuscript is available in the preprint server medRxiv (doi: https://doi.org/10.1101/19012013).
SOURCES OF FUNDING
US National Institutes of Health grants (R01HL131573 to E.G., R.J.L., C.A.C., D.E.A.; and R01HL111267 to R.J.L., D.E.A.). The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL059367, R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
All authors report no conflict of interest.
Data is available upon request from the ARIC Coordinating Center, following established procedures as laid out by the ARIC study (https://sites.cscc.unc.edu/aric/).
REFERENCES
- 1.Zhang Y, Guallar E, Ashar FN, Longchamps RJ, Castellani CA, Lane J, Grove ML, Coresh J, Sotoodehnia N, Ilkhanoff L, Boerwinkle E, Pankratz N and Arking DE. Association between mitochondrial DNA copy number and sudden cardiac death: findings from the Atherosclerosis Risk in Communities study (ARIC). Eur Heart J. 2017;38:3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashar FN, Zhang Y, Longchamps RJ, Lane J, Moes A, Grove ML, Mychaleckyj JC, Taylor KD, Coresh J, Rotter JI, Boerwinkle E, Pankratz N, Guallar E and Arking DE. Association of Mitochondrial DNA Copy Number With Cardiovascular Disease. JAMA Cardiol. 2017;2:1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashar FN, Moes A, Moore AZ, Grove ML, Chaves PHM, Coresh J, Newman AB, Matteini AM, Bandeen-Roche K, Boerwinkle E, Walston JD and Arking DE. Association of mitochondrial DNA levels with frailty and all-cause mortality. J Mol Med (Berl). 2015;93:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longchamps R, Castellani C, Newcomb C, Sumpter J, Lane J, Grove M, Guallar E, Pankratz N, Taylor K, Rotter J, Boerwinkle E and Arking D. Evaluation of mitochondrial DNA copy number estimation techniques. bioRxiv. 2019:610238. 610238; doi: 10.1101/610238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosamond WD, Chang PP, Baggett C, Johnson A, Bertoni AG, Shahar E, Deswal A, Heiss G and Chambless LE. Classification of heart failure in the atherosclerosis risk in communities (ARIC) study: a comparison of diagnostic criteria. Circ Heart fail. 2012;5:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]