Abstract
The Nonsense-mediated mRNA Decay (NMD) pathway is an RNA quality control pathway conserved among eukaryotic cells. While historically thought to predominantly recognize transcripts with premature termination codons, it is now known that the NMD pathway plays a variety of roles, from homeostatic events to control of viral pathogens. In this review we highlight the reciprocal interactions between the host NMD pathway and viral pathogens, which have shaped both the host antiviral defense and viral pathogenesis.
Keywords: Viruses, RNA biology, Nonsense-mediated mRNA decay, Anti-viral response
Abbreviations: NMD, Nonsense-mediated mRNA Decay; UPF, Up-Frameshift Protein; SFV, Semliki Forest Virus; SINV, Sindbis Virus; HCV, Hepatitis C Virus; ZIKV, Zika Virus; WNV, West Nile Virus; DENV, Dengue Virus; CoV, Coronavirus; MHV, Mouse Hepatitis Virus; PVX, Potato Virus X; TCV, Turnip Crinkle Virus; PEMV2, Pea Enation Mosaic Virus 2; HTLV, Human T Lymphotropic Virus; HIV, Human Immunodeficiency Virus; RSV, Rous Sarcoma Virus; CaMV, Cauliflower Mosaic Virus
1. Introduction
Transcripts with premature termination codons (PTCs) lead to the production of truncated proteins, which is potentially harmful for the cell [1,2]. To address this problem, the cell employs the NMD pathway, which is a translation-dependent process that targets PTC -containing transcripts with a set of factors conserved from yeast to man [3]. However, recent research has revealed that NMD plays a broader role in biology. This includes surveillance of non-PTC containing messenger RNAs (mRNAs) implicated in a wide swath of biological functions, including development, stem cell differentiation, stress response, and protection from viral infections [[4], [5], [6], [7], [8]].
While our understanding of the NMD pathway is still evolving, it can be divided into at least two branches: one which is dependent on the Exon-Junction Complex (EJC-dependent or EJC-enhanced) and one which operates in an EJC-independent manner. The current understanding of these models is summarized below. After transcription occurs, mRNAs are spliced and components of the exon-junction complex (EJC) are loaded onto mRNAs. The EJC consists of several proteins, including MAGOH, RBM8A, BTZ and eIF4A3 [7]. Transcripts are then transported out of the nucleus, where translation begins, displacing EJCs positioned along the transcript [9]. If the transcript contains a PTC, translation will terminate with one or more downstream EJCs non-displaced, which triggers initiation of “EJC-dependent” NMD (reviewed in [3]) (Fig. 1 A).
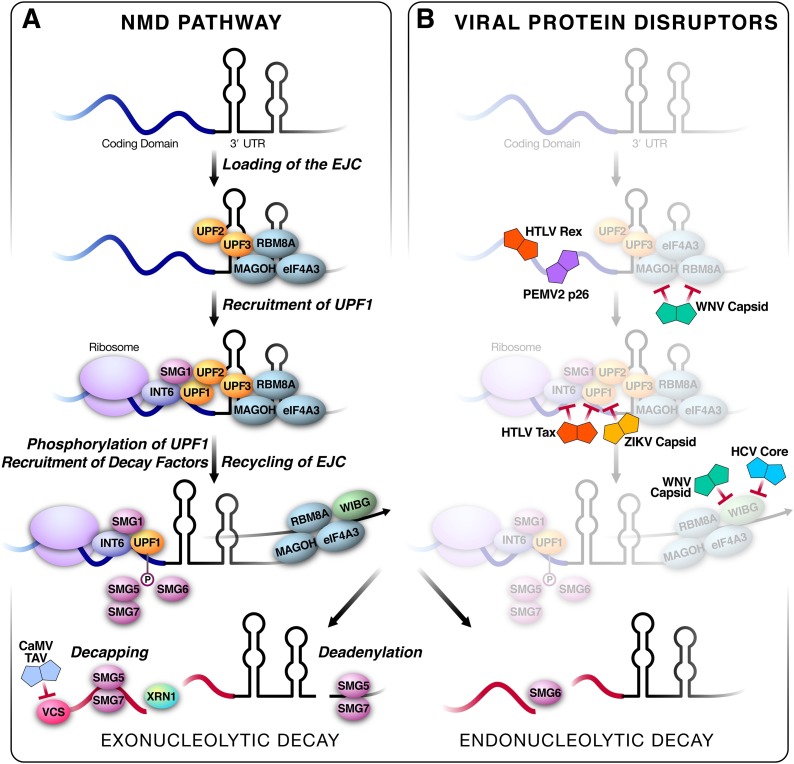
Fig. 1.
A) Schematic of the NMD pathway: Initially, a spliced mRNA has the exon junction complex (EJC) loaded on to the mRNA. This includes the core members: RBM8A, eIF4A3, and MAGOH. When there is a PTC upstream of a retained EJC, UPF2 and UPF3 act as a bridge to recruit the UPF1/SMG1 complex and initiate the NMD pathway. UPF1 is then phosphorylated by SMG1, leading to the recruitment of the decapping and deadenylating complex created by SMG5/SMG7, resulting in exonucleolytic decay. SMG6 is recruited for a separate endonucleolytic decay pathway. In plants, the complex VARICOSE (VCS) is responsible for decapping mRNAs. B) Viral interactors overlaid on the NMD pathway: HTLV Rex and PEMVp62 protects mRNA from degradation by blocking NMD initiation on transcripts. WNV capsid interacts and interferes with RBM8A and MAGOH. HTLV Tax interferes with INT6 and UPF1 function, whereas ZIKV capsid blocks UPF1 function. WNV capsid and HCV core interfere with WIBG recycling of EJC factors. The CaMV TAV protein blocks decapping by interfering with the complex VCS.
The ribosome and retained EJC recruit core NMD proteins: the Up-Frameshift (UPF) proteins, which recognize and bind to the transcript, and Suppressors with Morphological Effects on Genitalia (SMG) proteins, which induce decay [6]. The translation termination complex, which contains the helicase UPF1, the kinase SMG1, and the eukaryotic peptide chain release factor eRF1-3, provides the first signal for the NMD pathway. The remaining downstream EJC provides the second signal by binding to the UPF3 (or the partially redundant UPF3X) and UPF2 complex. The UPF2/3 complex acts as a bridge linking the EJC to the UPF1/SMG1 complex, which then activates SMG1 to phosphorylate UPF1 [1]. Phosphorylation of UPF1 activates the Decay-Inducing Complex (DECID) pathway, which consists of decapping and deadenylation of the transcript by the deadenylase CCR4-NOT via the SMG5/7 heterodimer [10,11], as well as endonucleolytic cleavage by SMG6 [12,13]. The resulting cleavage products are then further degraded, likely by the exoribonuclease XRN1 [14].
Recent systems-wide approaches uncovered that NMD not only targets “faulty” transcripts, but also a large number of “normal” transcripts, potentially 10-20% of the human transcriptome [[15], [16], [17], [18], [19], [20]]. Certain transcripts, especially those with long 3’ untranslated regions (UTRs) and GC-rich regions, are particularly susceptible to NMD degradation [[21], [22], [23]]. In addition, NMD triggered by events other than EJCs deposited downstream of stop codons (EJC-independent) is particularly relevant in interactions of NMD with positive-sense RNA viruses.
This target range and preference for “unusual” RNAs form the basis for the ability of the NMD pathway to also degrade incoming viral RNAs or viral transcripts [24]. In turn, viruses have evolved mechanisms to circumvent this innate anti-viral defense mechanism, either to protect the viral RNA from degradation or to turn members of the NMD pathway in enabling factors to enhance viral replication [5]. Below, we summarize examples of how many different kinds of viruses interact with the NMD system, and the implications that these mechanisms may have for the development of anti-viral therapies.
2. Positive Sense RNA Viruses
2.1. Alphavirus
Alphaviruses are small, enveloped viruses with a single strand positive-sense RNA genome. One of the first reports indicating a role for the NMD pathway in RNA virus infection came from the study of two alphaviruses, Semliki Forest Virus (SFV) and Sindbis Virus (SINV) [25]. In a genome-wide screen in HeLa cells, UPF1 was identified as a restriction factor of SFV infection, and this function was confirmed in a model of SINV infection. UPF1 was found to act early in infection, and knockdown of UPF1 led to increased viral RNA synthesis and production of viral progeny. Downstream effectors of the NMD pathway, SMG5 and SMG7, were also found to restrict alphavirus infection. The study’s conclusion was that UPF1, SMG5, and SMG7 target the incoming viral RNA, leading to a decrease of all downstream viral events. Interestingly, the NMD pathway is known to target host transcripts with long 3’ UTRs, but deletion of the viral 3′ UTR did not appear to impact UPF1-mediated viral RNA degradation [25]. However, alphavirus proteins are translated from two different open reading frames: the nonstructural proteins from the genomic RNA and the structural proteins from shorter so-called subgenomic RNAs. Thus, when nonstructural proteins are translated, the remaining sequence encoding the structural proteins forms a long 3′ sequence including the template for the structural proteins, potentially allowing for recognition by the NMD despite the deletion of the shorter genomic 3′ UTR [26]. UPF1 also acted as a viral restriction factor of SINV in a fly model system [27]. These data indicate that the NMD pathway, relying on UPF1 and the SMG proteins, can target incoming viral RNA, serving as an intrinsic immune system against RNA viruses.
2.2. Flavivirus
Flaviviridae also contain a positive-sense single-stranded RNA genome as a possible target for NMD. Flaviviridae were first linked to the antiviral activity of NMD in a screen for protein-protein interactions between host proteins and proteins encoded by the Hepatitis C Virus (HCV)[28]. HCV chronically infects liver cells, in most cases leading to life-long progressive, and ultimately terminal, liver disease [29]. Our group found that one NMD-associated protein, WIBG (also called partner of Y14/RBM8A and Magoh, PYM), interacted robustly with the HCV capsid protein called core. A parallel RNAi screen found that knockdown of WIBG led to decreased HCV replication. WIBG is known to interact with two members of the exon-junction complex, RBM8A and MAGOH [9,30,31], and is thought to recycle these proteins back into the nucleus [9] (Fig. 1B, Fig. 2 A). Binding of HCV core to WIBG disrupts this interaction, leading to an overall decrease in cellular NMD activity as shown by the enhanced RNA levels of select NMD targets [28]. Thus, HCV evolved to disrupt the antiviral activity of NMD. However, unlike with alphaviruses, the knockdown of UPF1 did not enhance viral replication. In contrast, our data support a model wherein HCV disrupts WIBG and the NMD pathway to support its own life cycle.
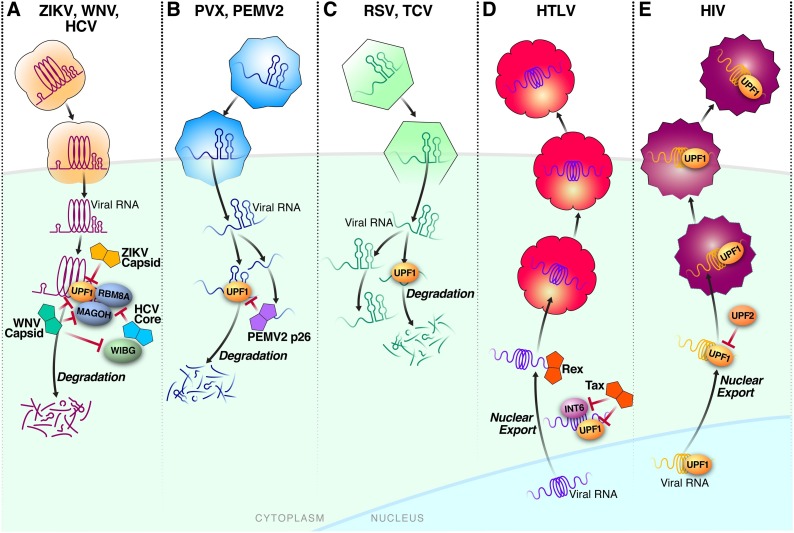
Fig. 2.
A) Zika Virus (ZIKV), West Nile Virus (WNV) and Hepatitis C Virus (HCV): Flavivirus structural proteins protect viral mRNA. ZIKV and WNV capsid degrade UPF1. HCV core and WNV capsid interact with WIBG to block interaction between WIBG and RBM8A/MAGOH. WNV capsid also relocalizes RBM8A and MAGOH. B) Potato Virus X (PVX) and Pea Enation Mosaic Virus 2 (PEMV2): Elements of the viral mRNA lead to NMD degradation. The 3’UTR contains elements responsible for NMD degradation, where removal of the 3’UTR protects viral RNA from the NMD. PEMV2 p62 interacts with RNA to protect it from degradation. C) Rous Sarcoma Virus (RSV) and Turnip Crinkle Virus (TCV): Elements of the viral mRNA protect from NMD degradation. A segment of the 3’ UTR confers protection to viral mRNAs from the NMD and protect from RNA decay machinery. D) Human T-cell Lymphotropic Virus (HTLV): Two different HTLV proteins protect viral RNA from the NMD pathway. While Rex interaction with viral RNA confers protection, Tax plays a more active role by both inhibiting the ability of UPF1 to interact with RNA and properly translocate across bound RNA molecules. Furthermore, Tax inhibits INT6 interactions with UPF1. E) Human Immunodeficiency Virus (HIV): UPF1 is involved in the RNA export of HIV genomes from the nucleus into the cytoplasm and then loaded into infectious virions. The host protein UPF2 inhibits UPF1 interaction with viral RNA and inhibits viral replication.
Zika virus (ZIKV) is a mosquito-borne flavivirus that causes microcephaly in fetuses and a dengue-like fever in adults [32]. Our group found that ZIKV has also evolved mechanisms to disrupt the NMD pathway, one of them being that the ZIKV capsid interacts with and degrades UPF1 in the nucleus [33] (Fig. 1B, Fig. 2A). Depletion of UPF1 results in a marked increase in the permissiveness of neural progenitor cells to viral infection, indicating that NMD is active in tissue-specific progenitor cells, possibly as an added protection of these cells from viral infection [33].
WIBG, MAGOH, and UPF1 were all identified as restriction factors for Dengue Virus (DENV), ZIKV and West Nile Virus (WNV) infections in human cell lines [34], emphasizing that NMD is a common host response to many flaviviruses. Furthermore, MAGOH and RBM8A proteins re-localize from the cytoplasm to host membranes when WNV capsid protein is overexpressed [34], which may explain why NMD is disrupted in WNV-infected cells (Fig. 1B, Fig. 2A). RBM8A was found to interact with viral RNA, dependent on the expression of WIBG or MAGOH proteins [34]. Interaction of RBM8A with viral RNA is surprising because EJC deposition typically occurs in the nucleus, whereas flavivirus RNA replication occurs in the cytoplasm. Relocalization of EJC members from the nucleus to the cytoplasm during WNV infection could expose not only the viral RNA but also other cytoplasmic RNAs to members of the EJC. In addition, other members of the NMD pathway, such as UPF1, which is found in the nucleus and cytoplasm, could potentially recruit EJC family members to viral RNA. Flaviviruses are vector-borne, and the interactions between NMD proteins and viral proteins likely occur in both the insect vectors and human hosts. Collectively, these data uncover multiple interactions of flaviviruses with the NMD pathway, from acute suppression of NMD in self-limiting diseases like ZIKV, DENV and WNV infections to chronic disruption of individual NMD-associated proteins, like WIBG in HCV infection.
2.3. Coronavirus
Coronaviruses (CoV) comprise a group of large positive-stranded RNA viruses (∼30 kb) that include SARS-CoV, MERS-CoV and the new SARS-CoV-2 causing the COVID-19 outbreak [35]. Mouse Hepatitis Virus (MHV), a mammalian coronavirus, has been shown to be the target of the NMD pathway and replicate more efficiently when UPF1, UPF2, SMG5 or SMG6 proteins are individually knocked down [36]. Furthermore, viral RNA transfected into cells that were rendered NMD-incompetent gained a significantly increased half-life, arguing that CoV RNA, like flavivirus and alphavirus RNA, may be a substrate for NMD-mediated degradation. Interestingly, the nucleocapsid of MHV, the N protein, which protects viral RNA from degradation, impairs the NMD pathway when co-transfected with an NMD reporter system [36], indicating an adaptation to actively antagonize the host response. Most recently, the SARS-CoV-2 nucleoprotein has also been shown to interact with UPF1 [37], highlighting that interaction of coronaviruses with the NMD pathway is conserved in both mice and humans.
2.4. Alphaflexiviridae and Tombusviridae
NMD is highly conserved among eukaryotes including plants [38]. Potato virus X (PVX) is a positive-sense single-stranded RNA virus in the Alphaflexiviridae family that typically infects solanaceous crops, which include potatoes, tomatoes and peppers. PVX is also capable of infecting Nicotiana benthamiana, a permissive laboratory model and close relative of tobacco indigenous to Australia [39,40]. Using a standard Arabidopsis plant model system artificially expressing PVX, a screen was performed to identify restricting and enabling host factors. This screen identified UPF1, SMG7, and UPF3 as restriction factors, that when inactivated led to enhanced viral replication [41]. In N. benthamiana, PVX replicated more efficiently when plants expressed a dominant negative mutant of UPF1. Furthermore, removal of the long 3’UTR from the PVX genome also conferred protection from NMD-mediated degradation (Fig. 2B). Interestingly, PVX infection also impaired NMD activity and stabilized known NMD substrates in the Arabidposis model, although this function could not be confirmed in the Nicotiana system due to the lack of known NMD targets in these plants [41]. This indicates that PVX may actively hamper NMD, as seen in other viral species.
Similar enhancement of viral replication by dominant negative UPF1 was observed when N. benthamiana was infected with Turnip crinkle virus (TCV). This virus belongs to the family of tombusviridae, and unlike PVX lacks both a 5’ cap and a 3’ polyA tail [41]. Interestingly, tombusviridae showed a spectrum of susceptibility to the antiviral activity of NMD, with Pea Enation Mosaic Virus 2 (PEMV2) being more sensitive than TCV (Fig. 2B). While the 3’UTR of TCV conferred protection against NMD degradation, the 3’UTR of PEMV2 did not appear to provide the same protection [42] (Fig. 2C). Furthermore, the PEMV2 long distance movement protein p26 protects viral and host mRNAs (Fig. 1B, 2 B) against NMD in a N. benthamiana system. The authors found that PEMV2 infection can protect nearly half of the NMD-sensitive target RNAs, which consist of host transcripts with long, GC-rich 3’UTRs. P26 expression alone increased transcript abundance for about a third of the NMD-sensitive target RNAs (Fig. 1B) [43]. This indicates that PEMV2 likely has multiples ways of perturbing the NMD pathway.
3. Retroviruses
3.1. HTLV (Human T lymphotropic virus)
The human T lymphotropic virus (HTLV) is a human oncogenic retrovirus that contains two single-stranded RNA copies in its virion [44]. Similar to the other viruses discussed above, HTLV has an RNA genome, but fundamental differences in the HTLV lifecycle, including reverse transcription of the viral genome and subsequent integration into the host, provide different avenues by which NMD proteins could target and interact with the virus. HTLV-1 infected cells exhibit suppressed NMD function [45], and viral RNA co-immunoprecipitates with UPF1 [46]. The central role of UPF1 in virus-targeted NMD was demonstrated by the finding that HTLV-1 mRNAs are sensitive to NMD degradation, which can be rescued by UPF1 knockdown [45]. In contrast, ectopic UPF1 expression decreases the presence of viral RNA, whereas inhibition of NMD either with wortmannin (a PI3K inhibitor that targets SMG1 and is known to disrupt UPF1 function [47]) or siRNA knockdown leads to enhanced levels of viral RNA [46]. In addition, two HTLV-1 proteins, the transactivator protein Tax and the viral RNA binding protein Rex, directly influence the NMD pathway [45,46,48,49](Fig. 2D).
Functional analysis showed that a reporter construct containing a PTC was significantly enriched in HTLV-1 infected cells, but only if those cell lines expressed Tax [45]. Tax alone could stabilize PTC-containing reporter mRNAs, but also stabilized both NMD-susceptible host and HTLV-1 mRNAs [45]. HTLV Tax protein interacts with three NMD proteins: UPF1, UPF2, and INT6 [45] (Fig. 1B, Fig. 2D). Tax interaction with UPF1 reduces the ability of UPF1 to interact with nucleic acids, which is thought to occur by steric hindrance of the RNA binding site. Tax also prevents translocation of UPF1 across nucleic acid substrates inducing substrate dissociation [49]. Tax expression also reduces the interaction between INT6, a member of the eIF3 translation initiation factor, and UPF1. While UPF1 is known to interact with eIF3 to inhibit translation [50], the role of the INT6-UPF1 interaction is not fully understood. However, INT6 appears to play an important role in the NMD, as Tax mutants unable to bind to INT6 do not disrupt NMD [45]. Interestingly, Tax expression induces UPF1 re-localization to P-bodies, cytoplasmic foci of untranslated mRNAs and proteins involved with mRNA decay and translation inhibition [51], and causes P-body enlargement, indicating inhibition of mRNA decay.
Similar to Tax, the HTLV Rex protein, responsible for stabilizing and exporting viral mRNAs from the nucleus [52] (Fig. 1B), can significantly hamper normal cellular NMD function. This effect was seen both for the PTC reporter system and for endogenous substrates of the NMD pathway [46]. Rex overexpression stabilizes viral RNA, possibly due to NMD inhibition, which leads to increased viral RNA and consequently protein production [46].
Collectively, these data on both Tax and Rex indicate multiple pathways inhibit NMD, both on viral RNAs and on host RNAs. This leads to the hypothesis that not only are these viral proteins protecting viral RNA, but that disruption of the NMD pathway may result in a favorable environment for HTLV to replicate.
3.2. HIV
Like HTLV discussed above, HIV is a retrovirus with a single-stranded RNA genome that gets incorporated into the host genome. The first indication that UPF1 interacts with HIV was based on known links between HIV and the Staufen protein [53,54]. Staufen is a dsRNA-binding protein known to recruit UPF1 to specific RNAs [55]. Staufen is also incorporated into HIV virions [53,54], indicating that HIV could interact with UPF1. Indeed, UPF1 is incorporated into the HIV virion, and HIV viral RNA is destabilized when UPF1 is knocked down [56] (Fig. 2E). Furthermore, UPF1 overexpression enhances HIV replication and protein production, although interestingly, this effect is dependent on UPF1 ATPase activity, and independent of its role in NMD [56]. Thus UPF1, separate from its role in the NMD pathway, appears to play a uniquely enabling role in the HIV life cycle. The necessity of UPF1 for HIV replication was independently confirmed by a group of researchers who found that HIV virions produced in UPF1-depleted cells showed reduced infectivity. Furthermore, the UPF1 ATPase region was also found to be responsible for proper virion infectivity [57].
UPF1 controls HIV replication at multiple levels, including nucleo-cytoplasmic shuttling of HIV RNA (Fig. 2E). Indeed, in the absence of the viral Rev protein, which exports unspliced viral RNAs from the nucleus [58], overexpression of UPF1 is sufficient to support viral export, and this requires the UPF1 nuclear localization sequence and nuclear export signal [59]. In the same study, UPF2 and UPF3 were shown to be negative regulators of HIV, and UPF2 was excluded from HIV RNPs [59]. This indicates that the canonical, NMD-related function of UPF1, including its interaction with UPF2 and UPF3, actually inhibits HIV replication by interfering with UPF1 recruitment to the HIV genome. Blocking UPF2 interaction with UPF1 is thought to be particularly important for the production of infectious HIV virions.
Multiple NMD factors also control HIV latency, wherein viral gene expression is transcriptionally silenced. Specifically, UPF1 can reverse latency by enhancing viral RNA levels and viral gene expression, whereas UPF2 and SMG6 promote latency by binding to UPF1 and hindering its proviral activities. UPF2 sequesters UPF1 and prevents association with viral RNAs, while SMG6, an endonuclease, is recruited to RNA bound by UPF1 and destabilizes the viral RNA [60]. The same study showed that SMG6 and UPF2 negatively impact HIV reactivation from latency in both primary CD4+ T-cells and macrophages [60,61]. In turn, SMG6 and UPF2 protein levels were decreased in infected macrophages. This underscores that the effects of NMD factors on HIV replication are complex, and point to UPF1 as a unique enabling factor for HIV independent of its role in NMD.
3.3. Rous Sarcoma Virus (RSV)
Rous Sarcoma virus (RSV), which causes sarcoma in fowl, was the first virus discovered to have oncogenic properties [62]. While early studies on RSV found that certain RNA elements of the genome determined stability of the viral RNA [[63], [64], [65]], it was only after NMD was better understood that it became clear that these elements were linked to UPF1 and the NMD pathway [[66], [67], [68], [69]]. It was found that RSV RNAs containing PTCs were degraded at a higher rate than wild type, and co-transfection of a dominant negative UPF1 prevented degradation of the PTC-containing RSV RNAs, highlighting their status as NMD targets [66]. Subsequent studies found that a 155-nucleotide element, known as the RNA Stability Element (RSE), in the 3’UTR of RSV RNAs conferred protection from NMD [67,68] (Fig. 2C). The RSE contains polypyrimidine tracts that bind PTBP1, which excludes UPF1 from the target RNA [70]. This evasion strategy is also relevant for several NMD-resistant host mRNAs [70]. Furthermore, the RSE inhibited deadenylation and decay by XRN1 when inserted into 3’UTR of canonical NMD transcripts [69]. This indicates that RSV has evolved a mechanism to escape the restriction imposed by the cellular NMD pathway and to successfully protect its genome and mRNAs from NMD attacks.
3.4. Pararetroviruses
Cauliflower Mosaic Virus (CaMV), a plant pararetrovirus in the family Caulimoviridae that infects the Cruciferae family, including Arabidopsis thaliana, also disrupts cellular NMD processes [71,72]. Pararetroviruses are similar to mammalian retroviruses, in that reverse transcription of an RNA intermediate is required to replicate, but pararetroviruses have a DNA genome that does not integrate into the host genome but instead remains in nuclear episomes [73]. The CaMV viral transactivator protein (TAV) binds and destabilizes the RNA decapping complex VARICOSE (VCS), which then leads to the accumulation of NMD substrates in the cell (Fig. 1A). Specifically, transcripts with PTCs, normally targeted and eliminated by the NMD pathway, are stabilized by TAV overexpression. The proposed model is that by inhibiting NMD, TAV increases viral RNA levels present within the cell [72]. Even though the pararetrovirus has a DNA genome, the production of a significant number of RNA intermediates for replication produces NMD substrates. Thus, protection of viral RNA from the NMD pathway remains an important aspect of infection.
4. Conclusions
In the last several years, our understanding of the importance of NMD as a cell-intrinsic antiviral restriction pathway in animals and plants has expanded rapidly. With this, we have also come to recognize that viruses have evolved multiple mechanisms to evade the antiviral activity of NMD (Fig. 1, Fig. 2). These range from hijacking UPF1 to inhibiting the deadenylation step of NMD. Remarkably, viruses such as HIV and HCV have co-opted components of the NMD pathway, making the NMD factors required for viral replication. Understanding NMD and its nuances in response to viral infections can inform diverse fields. For example, Cas9 activity generates nonsense mutations that are subsequently depleted as transcripts by the NMD pathway [74], which is an important consideration for the therapeutic applications of the CRISPR/Cas9 technology.
Understanding how viruses perturb NMD could also have implications for nonviral diseases. For example, in Duchenne’s Muscular Dystrophy, mutations in the dystrophin gene lead to truncated transcripts that are subject to NMD. Preservation of those transcripts could result in partially functional proteins, thus taking lessons from viral pathogens on how to inhibit NMD could lead to novel therapeutic approaches [75].
In other cases, there may be therapeutic benefit to amplifying NMD. For example, microcephaly is caused by haploinsufficiency of members of the EJC, which are required for EJC-dependent NMD [76,77]. In the context of ZIKV infection, the capsid protein interferes with NMD by degrading UPF1 in the nucleus [33] or binding WIBG [34]. Thus, small-molecule therapeutics that could correct the UPF1 downregulation and prevent capsid:WIBG interactions could exert anti-viral and anti-pathogenesis effects, as active NMD would be restored to prevent the establishment of viral infection and maintenance of transcript homeostasis.
As with many emerging fields, plenty of open questions remain. For example, is the interaction restricted to RNA viruses, or do DNA viruses such as herpesviruses also interact with NMD? In addition, little is known about the recognition of viral RNAs by NMD. What aspects of the NMD pathway are required to recognize “foreign” RNA, and what defines specificity towards aberrant RNA molecules? Spatial aspects are also important to clarify: many of the upstream factors of the canonical NMD pathway act in the nucleus, but many of the viruses described above, such as the flaviviruses, replicate in virally-induced compartments in the cytoplasm. Where does NMD of viral RNAs occur, and how do nuclear NMD factors interact with viral RNA?
Much more work is required to fully understand the impact of viruses on NMD and vice versa. Until then, the findings that many viruses have evolved elaborate mechanisms to impair NMD or to subvert select NMD members into enabling factors promoting viral infection remain the strongest argument for a critical role of the pathway in the innate anti-viral response.
Acknowledgements
We would like to thank John CW Carroll for figure design and Kathryn Claiborn, PhD for editorial support. This work was supported by the UCSF Medical Scientist Training Program, the UCSF Biomedical Sciences Program, the NINDS/NIH under award F31NS113432, the NIAID/NIH under award 5R01AI097552-03 and the NIDA/NIH under award 5DP1DA038043-05 UCSF Discovery Fellows (Otellini Family).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.semcdb.2020.05.018.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Kurosaki T., Maquat L.E. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129(3):461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Research. 2016;44(4):1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurosaki T., Popp M.W., Maquat L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20(7):406–420. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han X., Wei Y., Wang H., Wang F., Ju Z., Li T. Nonsense-mediated mRNA decay: a’ nonsense’ pathway makes sense in stem cell biology. Nucleic Acids Res. 2018;46(3):1038–1051. doi: 10.1093/nar/gkx1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balistreri G., Bognanni C., Muhlemann O. Virus Escape and Manipulation of Cellular Nonsense-Mediated mRNA Decay. Viruses. 2017;9(1) doi: 10.3390/v9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 7.Popp M.W., Maquat L.E. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith J.E., Baker K.E. Nonsense-mediated RNA decay--a switch and dial for regulating gene expression. Bioessays. 2015;37(6):612–623. doi: 10.1002/bies.201500007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehring N.H., Lamprinaki S., Kulozik A.E., Hentze M.W. Disassembly of exon junction complexes by PYM. Cell. 2009;137(3):536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 10.Unterholzner L., Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16(4):587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Loh B., Jonas S., Izaurralde E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013;27(19):2125–2138. doi: 10.1101/gad.226951.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lykke-Andersen S., Chen Y., Ardal B.R., Lilje B., Waage J., Sandelin A., Jensen T.H. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014;28(22):2498–2517. doi: 10.1101/gad.246538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franks T.M., Singh G., Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143(6):938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44(4):1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 16.Tani H., Imamachi N., Salam K.A., Mizutani R., Ijiri K., Irie T., Yada T., Suzuki Y., Akimitsu N. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 2012;9(11):1370–1379. doi: 10.4161/rna.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weischenfeldt J., Damgaard I., Bryder D., Theilgaard-Monch K., Thoren L.A., Nielsen F.C., Jacobsen S.E., Nerlov C., Porse B.T. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22(10):1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittmann J., Hol E.M., Jack H.M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26(4):1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17(12):2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurt J.A., Robertson A.D., Burge C.B. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23(10):1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toma K.G., Rebbapragada I., Durand S., Lykke-Andersen J. Identification of elements in human long 3’ UTRs that inhibit nonsense-mediated decay. Rna. 2015;21(5):887–897. doi: 10.1261/rna.048637.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kebaara B.W., Atkin A.L. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamachi N., Salam K.A., Suzuki Y., Akimitsu N. A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2017;27(3):407–418. doi: 10.1101/gr.206060.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contu L., Steiner S., Thiel V., Muhlemann O. The Role of Stress Granules and the Nonsense-mediated mRNA Decay Pathway in Antiviral Defence. Chimia (Aarau) 2019;73(6):374–379. doi: 10.2533/chimia.2019.374. [DOI] [PubMed] [Google Scholar]
- 25.Balistreri G., Horvath P., Schweingruber C., Zund D., McInerney G., Merits A., Muhlemann O., Azzalin C., Helenius A. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe. 2014;16(3):403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Hyde J.L., Chen R., Trobaugh D.W., Diamond M.S., Weaver S.C., Klimstra W.B., Wilusz J. The 5’ and 3’ ends of alphavirus RNAs--Non-coding is not non-functional. Virus Res. 2015;206:99–107. doi: 10.1016/j.virusres.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wernet M.F., Klovstad M., Clandinin T.R. Generation of infectious virus particles from inducible transgenic genomes. Curr Biol. 2014;24(3):R107–8. doi: 10.1016/j.cub.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramage H.R., Kumar G.R., Verschueren E., Johnson J.R., Von Dollen J., Johnson T., Newton B., Shah P., Horner J., Krogan N.J., Ott M. A Combined Proteomics/Genomics Approach Links Hepatitis C Virus Infection with Nonsense-Mediated mRNA Decay. Mol Cell. 2015;57(2):329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houghton M. Hepatitis C Virus: 30 Years after Its Discovery. Cold Spring Harb Perspect Med. 2019;9(12) doi: 10.1101/cshperspect.a037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diem M.D., Chan C.C., Younis I., Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14(12):1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- 31.Bono F., Ebert J., Unterholzner L., Guttler T., Izaurralde E., Conti E. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004;5(3):304–310. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierson T.C., Diamond M.S. The emergence of Zika virus and its new clinical syndromes. Nature. 2018;560(7720):573–581. doi: 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 33.Fontaine K.A., Leon K.E., Khalid M.M., Tomar S., Jimenez-Morales D., Dunlap M., Kaye J.A., Shah P.S., Finkbeiner S., Krogan N.J., Ott M. The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. MBio. 2018;9(6) doi: 10.1128/mBio.02126-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M., Johnson J.R., Truong B., Kim G., Weinbren N., Dittmar M., Shah P.S., Von Dollen J., Newton B.W., Jang G.M., Krogan N.J., Cherry S., Ramage H. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol. 2019;4(6):985–995. doi: 10.1038/s41564-019-0375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wada M., Lokugamage K.G., Nakagawa K., Narayanan K., Makino S. Interplay between coronavirus, a cytoplasmic RNA virus, and nonsense-mediated mRNA decay pathway. Proc Natl Acad Sci U S A. 2018;115(43):E10157–66. doi: 10.1073/pnas.1811675115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloyd J.P.B. The evolution and diversity of the nonsense-mediated mRNA decay pathway. F1000Res. 2018;7:1299. doi: 10.12688/f1000research.15872.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lico C., Benvenuto E., Baschieri S. The Two-Faced Potato Virus X: From Plant Pathogen to Smart Nanoparticle. Front Plant Sci. 2015;6:1009. doi: 10.3389/fpls.2015.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact. 2008;21(8):1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 41.Garcia D., Garcia S., Voinnet O. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe. 2014;16(3):391–402. doi: 10.1016/j.chom.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.May J.P., Yuan X., Sawicki E., Simon A.E. RNA virus evasion of nonsense-mediated decay. PLoS Pathog. 2018;14(11):e1007459. doi: 10.1371/journal.ppat.1007459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.May J.P., Johnson P.Z., Ilyas M., Gao F., Simon A.E. The Multifunctional Long-Distance Movement Protein of Pea Enation Mosaic Virus 2 Protects Viral and Host Transcripts from Nonsense-Mediated Decay. mBio. 2020;11(2) doi: 10.1128/mBio.00204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Futsch N., Mahieux R., Dutartre H. HTLV-1, the Other Pathogenic Yet Neglected Human Retrovirus: From Transmission to Therapeutic Treatment. Viruses. 2017;10(1) doi: 10.3390/v10010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mocquet V., Neusiedler J., Rende F., Cluet D., Robin J.P., Terme J.M., Duc Dodon M., Wittmann J., Morris C., Le Hir H., Ciminale V., Jalinot P. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol. 2012;86(14):7530–7543. doi: 10.1128/JVI.07021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano K., Ando T., Yamagishi M., Yokoyama K., Ishida T., Ohsugi T., Tanaka Y., Brighty D.W., Watanabe T. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect. 2013;15(6-7):491–505. doi: 10.1016/j.micinf.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Pal M., Ishigaki Y., Nagy E., Maquat L.E. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. Rna. 2001;7(1):5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mocquet V., Neusiedler J., Rende F., Cluet D., Robin J.P., Terme J.M., Duc-Dodon M., Morris C., Le Hir H., Ciminale V., Jalinot P. Nonsense-mediated mRNA decay inhibition by HTLV-1 Tax protein. Retrovirology. 2014:O61. [Google Scholar]
- 49.Fiorini F., Robin J.P., Kanaan J., Borowiak M., Croquette V., Le Hir H., Jalinot P., Mocquet V. HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition. Nat Commun. 2018;9(1):431. doi: 10.1038/s41467-017-02793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isken O., Kim Y.K., Hosoda N., Mayeur G.L., Hershey J.W., Maquat L.E. Upf1 Phosphorylation Triggers Translational Repression during Nonsense-Mediated mRNA Decay. Cell. 2008;133(2):314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Decker C.J., Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4(9):a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakano K., Watanabe T. HTLV-1 Rex Tunes the Cellular Environment Favorable for Viral Replication. Viruses. 2016;8(3):58. doi: 10.3390/v8030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatel-Chaix L., Clement J.F., Martel C., Beriault V., Gatignol A., DesGroseillers L., Mouland A.J. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24(7):2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mouland A.J., Mercier J., Luo M., Bernier L., DesGroseillers L., Cohen E.A. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74(12):5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim Y.K., Furic L., Desgroseillers L., Maquat L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell. 2005;120(2):195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 56.Ajamian L., Abrahamyan L., Milev M., Ivanov P.V., Kulozik A.E., Gehring N.H., Mouland A.J. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA. 2008;14(5):914–927. doi: 10.1261/rna.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serquina A.K., Das S.R., Popova E., Ojelabi O.A., Roy C.K., Gottlinger H.G. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J Virol. 2013;87(16):8853–8861. doi: 10.1128/JVI.00925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blissenbach M., Grewe B., Hoffmann B., Brandt S., Uberla K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J Virol. 2010;84(13):6598–6604. doi: 10.1128/JVI.02264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ajamian L., Abel K., Rao S., Vyboh K., Garcia-de-Gracia F., Soto-Rifo R., Kulozik A.E., Gehring N.H., Mouland A.J. HIV-1 Recruits UPF1 but Excludes UPF2 to Promote Nucleocytoplasmic Export of the Genomic RNA. Biomolecules. 2015;5(4):2808–2839. doi: 10.3390/biom5042808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao S., Amorim R., Niu M., Temzi A., Mouland A.J. The RNA surveillance proteins UPF1, UPF2 and SMG6 affect HIV-1 reactivation at a post-transcriptional level. Retrovirology. 2018;15(1):42. doi: 10.1186/s12977-018-0425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao S., Amorim R., Niu M., Breton Y., Tremblay M.J., Mouland A.J. Host mRNA decay proteins influence HIV-1 replication and viral gene expression in primary monocyte-derived macrophages. Retrovirology. 2019;16(1):3. doi: 10.1186/s12977-019-0465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss R.A., Vogt P.K. 100 years of Rous sarcoma virus. J Exp Med. 2011;208(12):2351–2355. doi: 10.1084/jem.20112160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barker G.F., Beemon K. Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol Cell Biol. 1994;14(3):1986–1996. doi: 10.1128/mcb.14.3.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barker G.F., Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991;11(5):2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arrigo S., Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8(11):4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LeBlanc J.J., Beemon K.L. Unspliced Rous Sarcoma Virus Genomic RNAs Are Translated and Subjected to Nonsense-Mediated mRNA Decay before Packaging. J Virol. 2004;78(10):5139–5146. doi: 10.1128/JVI.78.10.5139-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weil J.E., Beemon K.L. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA. 2006;12(1):102–110. doi: 10.1261/rna.2129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Withers J.B., Beemon K.L. Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon. Retrovirology. 2010;7:65. doi: 10.1186/1742-4690-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balagopal V., Beemon K.L. Rous Sarcoma Virus RNA Stability Element Inhibits Deadenylation of mRNAs with Long 3′UTRs. Viruses. 2017;9(8) doi: 10.3390/v9080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ge Z., Quek B.L., Beemon K.L., Hogg J.R. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife. 2016;5 doi: 10.7554/eLife.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haas M., Bureau M., Geldreich A., Yot P., Keller M. Cauliflower mosaic virus: still in the news. Mol Plant Pathol. 2002;3(6):419–429. doi: 10.1046/j.1364-3703.2002.00136.x. [DOI] [PubMed] [Google Scholar]
- 72.Lukhovitskaya N., Ryabova L.A. Cauliflower mosaic virus transactivator protein (TAV) can suppress nonsense-mediated decay by targeting VARICOSE, a scaffold protein of the decapping complex. Sci Rep. 2019;9(1):7042. doi: 10.1038/s41598-019-43414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hohn T., Rothnie H. Plant pararetroviruses: replication and expression. Curr Opin Virol. 2013;3(6):621–628. doi: 10.1016/j.coviro.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 74.Popp M.W., Maquat L.E. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165(6):1319–1322. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller J.N., Pearce D.A. Nonsense-Mediated Decay in Genetic Disease: Friend or Foe? Mutat Res Rev Mutat Res. 2014:52–64. doi: 10.1016/j.mrrev.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMahon J.J., Miller E.E., Silver D.L. The exon junction complex in neural development and neurodevelopmental disease. Int J Dev Neurosci. 2016;55:117–123. doi: 10.1016/j.ijdevneu.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao H., McMahon J.J., Tsai Y.H., Wang Z., Silver D.L. Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes p53-Mediated Microcephaly. PLoS Genet. 2016;12(9):e1006282. doi: 10.1371/journal.pgen.1006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.