Summary
Induction of trained immunity by Bacille-Calmette-Guérin (BCG) vaccination mediates beneficial heterologous effects, but the mechanisms underlying its persistence and magnitude remain elusive. In this study, we show that BCG vaccination in healthy human volunteers induces a persistent transcriptional program connected to myeloid cell development and function within the hematopoietic stem and progenitor cell (HSPC) compartment in the bone marrow. We identify hepatic nuclear factor (HNF) family members 1a and b as crucial regulators of this transcriptional shift. These findings are corroborated by higher granulocyte numbers in BCG-vaccinated infants, HNF1 SNP variants that correlate with trained immunity, and elevated serum concentrations of the HNF1 target alpha-1 antitrypsin. Additionally, transcriptomic HSPC remodeling was epigenetically conveyed to peripheral CD14+ monocytes, displaying an activated transcriptional signature three months after BCG vaccination. Taken together, transcriptomic, epigenomic, and functional reprogramming of HSPCs and peripheral monocytes is a hallmark of BCG-induced trained immunity in humans.
Keywords: BCG, vaccination, trained immunity, myelopoiesis, hematopoietic stem cells, monocyte, myeloid cell, epigenetic imprinting
Graphical Abstract
Highlights
-
•
Human BCG vaccination induces a persistent innate immune training of CD14+ monocytes
-
•
BCG vaccination imprints a persistent transcriptomic myeloid bias on human HSPCs
-
•
Hepatic nuclear factors are regulators of BCG-induced trained immunity in HSPCs
-
•
BCG induces persistent epigenetic changes in peripheral CD14+ monocytes
Cirovic and de Bree et al. investigate the effects of BCG vaccination in humans and reveal the induction of a transcriptomic rewiring of human stem and progenitor cells toward the myeloid cell lineage, instructed by hepatic nuclear factors, resulting in epigenetic and functional changes within CD14+ peripheral monocytes.
Introduction
Recent studies have shown that the ability to functionally adapt immune responses after an initial insult is not an exclusive property of adaptive immune cells, such as B and T cells, but is also found in a variety of innate immune cells, such as myeloid cells and natural killer (NK) cells (Netea et al., 2016, Sun et al., 2011). The functional adaptation of innate immunity after a primary insult, such as an infection or vaccination, represents a de facto innate immune memory, also termed trained immunity. This leads to a more effective non-specific (heterologous) response to a secondary insult, independently of the initial antigen (Netea et al., 2011). Epidemiological studies have shown that many vaccines, especially those employing attenuated live microorganisms, such as Bacille Calmette-Guérin (BCG), measles, or oral polio vaccine, lead to a decrease in overall childhood mortality that cannot be solely attributed to protection against the target disease alone (Aaby et al., 2011, Benn et al., 2013, Biering-Sørensen et al., 2017).
Among these vaccines, BCG has been shown to reduce neonatal mortality by 38% (17%–54%) in high infectious pressure environments (Aaby et al., 2011, Biering-Sørensen et al., 2017). Furthermore, BCG has been associated with sustained beneficial effects in older children (Storgaard et al., 2015) and adults (Rieckmann et al., 2017, Villumsen et al., 2009). In experimental models, BCG vaccination provides heterologous protection against a broad range of infections such as C. albicans or S. aureus infections (Clark et al., 1976, Freyne et al., 2015, Kleinnijenhuis et al., 2012, Murphy, 1981). It is suggested that this protective effect is mediated by a BCG-induced increase in the function of innate immune cells, including higher pro-inflammatory cytokine responses to secondary unrelated pathogens. This process is mediated by transcriptomic and epigenetic changes of myeloid cells, such as monocytes and macrophages (Arts et al., 2018, Kleinnijenhuis et al., 2012). BCG-mediated innate immune memory has been shown to be conveyed by monocytes; however, how the persistent memory function of myeloid cells, which has been shown to last months and even years after initial vaccination, is established remained elusive (Kleinnijenhuis et al., 2014, Kleinnijenhuis et al., 2012), especially considering the limited lifetime of myeloid cells in the circulation. Recently, two studies have addressed this conundrum in the mouse, by showing that systemic BCG vaccination or injection of the fungal cell wall component β-glucan induces a myeloid differentiation bias at the level of the hematopoietic stem cell within the bone marrow (BM), resulting in an increased release of monocytes and their enhanced ability to secrete cytokines and kill pathogens (Cheng et al., 2014, Kaufmann et al., 2018, Mitroulis et al., 2018). However, whether similar processes are induced by intradermal BCG vaccination in humans is unknown.
In this study, we used a human in vivo vaccination model to assess whether BCG vaccination of healthy individuals leads to epigenetic, transcriptional, and functional changes in BM hematopoietic stem and progenitor cells (HSPCs) and circulating monocytes. Furthermore, we investigated the molecular consequences of innate immune memory induction by BCG vaccination, identifying hepatic nuclear factor (HNF) 1a and b as crucial regulators of innate immune memory formation induced by BCG vaccination in humans in vivo.
Results
Peripheral Blood Mononuclear Cells Release More Pro-inflammatory Cytokines 3 Months after Intradermal BCG Vaccination upon C. albicans Restimulation
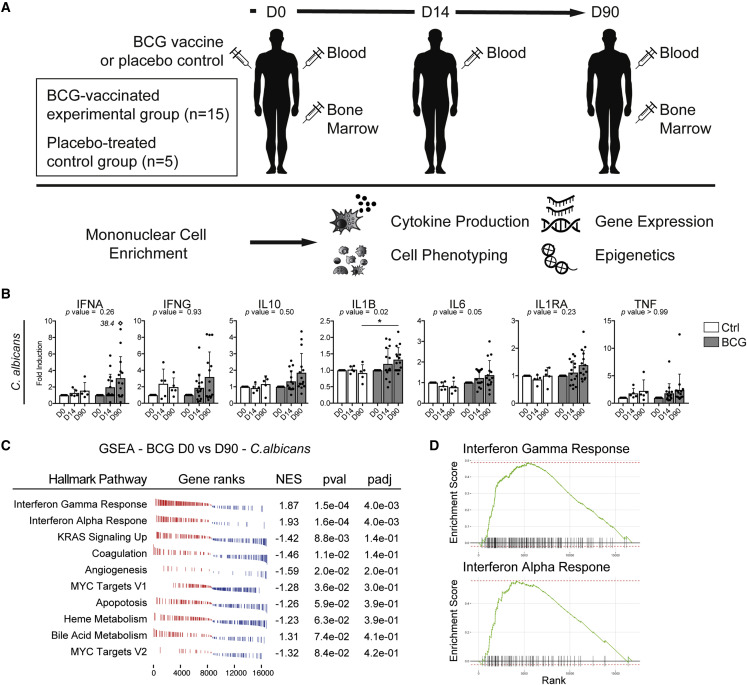
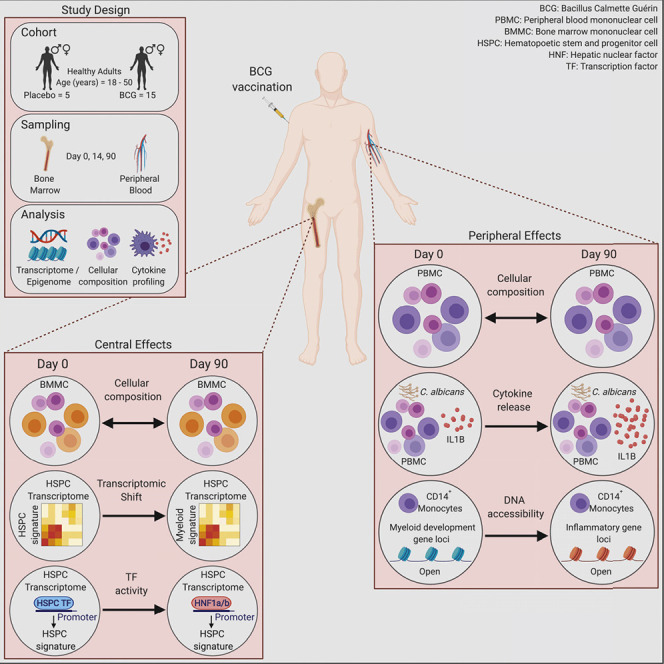
In order to fully understand the mechanisms of induction and the longevity of the enhanced heterologous innate immune response observed after BCG vaccination, we vaccinated 15 healthy BCG-naive volunteers with BCG, while 5 volunteers received placebo. On day 0, 14, and 90 (D0, D14, and D90) after vaccination (or placebo) we prepared peripheral blood mononuclear cells (PBMC). BM aspirates were collected and enriched for the mononuclear cell fraction (MNC) on D0 and D90 post-vaccination (Figure 1 A). All vaccinated individuals developed a local scar, and none had complications of vaccination (Table 1 ). No complication related to the vaccination was observed (e.g., lymphadenitis or abscess formation). Volunteers tested negative for the presence of M. bovis (the mycobacterial species from which BCG is derived) in the BM, 90 days after vaccination (data not shown). In order to validate the robustness of our in vivo BCG vaccination study approach we restimulated PBMCs isolated on D0, D14, and D90 post-BCG or placebo vaccination with C. albicans and measured the release of interferon alpha (IFNA), gamma (IFNG), interleukin 10 (IL10), 1B (IL1B), 6 (IL6), interleukin 1 receptor antagonist (IL1RA), and tumor necrosis factor (TNF) (Figure 1B). As previously demonstrated in several studies (Arts et al., 2018, Clark et al., 1976, Jensen et al., 2015, Kleinnijenhuis et al., 2012, Kleinnijenhuis et al., 2014, Murphy, 1981), BCG vaccination induced functional changes in PBMCs characterized by increased cytokine production after 24 h, compared with D0 and the placebo-treated group, specifically the functional and mechanistic hallmark cytokine IL1B (Netea et al., 2011) was markedly upregulated upon restimulation with C. albicans (Figure 1B; Table S1), thus validating our study setup in regard to the induction of heterologous innate immunity by BCG vaccination. Furthermore, also IL6 showed an increase however missed significance.
Figure 1.
BCG Vaccination Elicits Trained Innate Immunity in Healthy Individuals
(A) Study design including the experimental arm with BCG-vaccinated healthy individuals (n = 15) and placebo-treated control arm (diluent-treated, n = 5). Blood and BM aspirations were analyzed before (D0), two weeks (D14, blood only), and three months (D90) after vaccination.
(B) Cytokine measurement in supernatants of PBMC challenged ex vivo with C. albicans for 24 h. Data are presented as mean and SD. Fold changes relative to baseline (D0) are shown. Mann-Whitney test was used to compare fold induction at D90 in BCG versus controls (∗p < 0.05). Rectangular symbol indicates a single data point exceeding axis limits and the actual value next to it.
(C) Hallmark GSEA of transcriptomic data derived from the adherent PBMC fraction prepared before (D0) and after (D90) vaccination from the same individuals and treated ex vivo with C. albicans for 24 h (n = 5 per group). NES, normalized enrichment score; pval, p value; padj, adjusted p value. Top ten hallmark pathways enriched after vaccination are listed.
(D) Enrichment plots from GSEA for the two significantly enriched terms after BCG vaccination (see C).
Table 1.
Metadata of Healthy Volunteers Included in the Study
| Parameters | Controls (n = 5) | BCG-Vaccinated (n = 15) | Total (n = 20) |
|---|---|---|---|
| Age in years | 21.8 (SD 1.8, SEM 0.8) | 23.7 (SD 7.3, SEM 1.8) | 23.3 (SD 6.5, SEM 1.4) |
| Sex (% females) | 2/5 (40%) | 5/15 (33%) | 7/20 (35%) |
| Blister (90 days p.i.) | 0% | 100% | N/A |
| Blister size (90 days p.i., mm) | N/A | 4.8 (SD 1.4, SEM 0.3) | N/A |
| VAS (pain) score BM aspiration (0–10) | 3.5 (SD1.2, SEM 0.5) | 4 (SD 1.9, SEM 0.5) | 3.9 (SD 1.8, SEM 0.4) |
Summary of metadata of healthy volunteers included in the study. Presented parameters include age, sex, blister presence and size, and pain score after BM aspiration. Post-injection, p.i.; standard deviation, SD; standard error of the mean, SEM; not applicable, N/A; visual analog scale, VAS.
To further reveal whether these functional differences are also reflected within the transcriptome of the complete fraction of the PBMC, we investigated the pathways which were differentially regulated after BCG vaccination of individuals upon restimulation, using mRNA sequencing and subsequent gene set enrichment analysis (GSEA). First, we assessed the changes in the placebo group: They have not been found to be significant between D0 and D90 in placebo-treated individuals. Therefore, we conclude that one BM aspiration does not induce long-term changes in transcriptional program of PBMCs. In contrast, GSEA of C. albicans-restimulated PBMCs from BCG-vaccinated individuals before (D0) and after (D90) vaccination showed a significant enrichment of pathways related to IFNA and IFNG response signaling (Figures 1C and 1D), implying a heightened activation status of cells contained within the PBMC fraction, consistent with the significant increase in IL1B release and the trend toward a higher production of IFNα and G. These unbiased global transcriptomic and functional data suggest a transcriptomic basis for the enhanced innate immune response elicited by BCG vaccination.
BCG Vaccination Does Not Elicit Cellular Changes within the BM or Peripheral Blood in Healthy Volunteers 3 Months after Vaccination
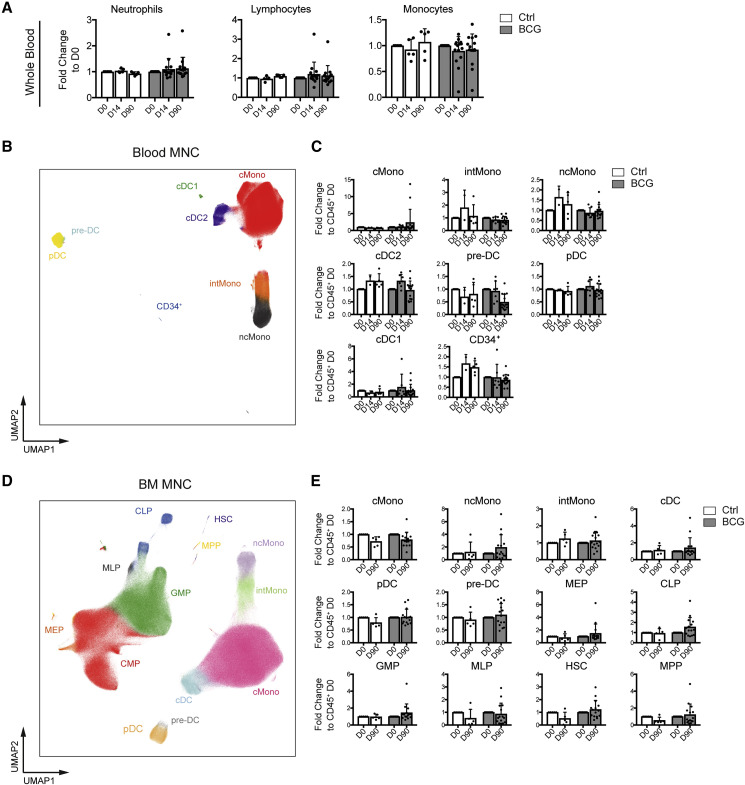
The BCG-induced increase in IL1B from the PBMC suggests the possibility of a remodeled myeloid cell and progenitor compartment as the cellular basis for trained immunity. To address this, we investigated the composition of the PBMC and the BM MNC populations of BCG-vaccinated and BCG-naive individuals on D0, D14, and D90 post-vaccination using differential white blood cell counts and multi-color flow cytometry. Differential blood count analysis did not reveal significant differences between the BCG-vaccinated and BCG-naive group within neutrophils, total lymphocytes or monocyte counts (Figure 2 A). As differential blood counts do not segregate between different mononuclear phagocyte subsets within the PBMCs, we utilized flow cytometry to investigate classical monocytes (cMono), intermediate monocytes (intMono), non-classical monocytes (ncMono), conventional dendritic cells (cDC) 1 and 2 alongside pre-DCs, pDCs and the peripheral blood resident CD34+ progenitor fraction (Figures 2B, 2C, S1A, and S1B). However, no significant alteration of the cellular frequencies between D0 and D90 in BCG-vaccinated or placebo-treated individuals could be detected over the time of the study period within the PBMC fraction, recapitulating the results of the differential blood counts. Similarly, we monitored the evolution of the myeloid mature cell and progenitor compartment in MNCs of the BM by tracking cMono, ncMono, intMono, total cDC, pDC, pre-DC, myeloid erythroid progenitors (MEPs), common lymphoid progenitors (CLPs), granulocyte macrophage progenitors (GMPs), multi-lymphoid progenitors (MLPs), multi-potent progenitors (MPPs), and hematopoietic stem cells (HSCs) on D0 and D90 post-vaccination (Figures 2D, 2E, S1C, and S1D). Again, no significant changes could be detected in the frequencies of any of these populations between D0 and D90 in vaccinated or placebo-treated individuals. Taken together, these data indicate that human trained immunity induced by intradermal BCG vaccination is not accompanied by changes in the frequencies of the mature myeloid or myeloid progenitor populations within the peripheral blood or BM MNC compartment of adult healthy volunteers during steady state.
Figure 2.
BCG Does Not Alter the Composition of Immune Cells and Progenitors in Blood and BM
(A) Whole blood counts of neutrophils, lymphocytes, and monocytes as fold change relative to D0.
(B and D) Uniform manifold approximation and projection (UMAP) representation of analyzed cell lineages of the alive Lin (CD3, CD7, CD10, CD15, CD19, and CD20)− CD45+ compartment in PBMC (B) and Lin (CD3, CD7, CD15, CD19, and CD20)− CD45+ BM MNC (D) (representative donor, 5 × 105 sampled cells).
(C and E) Quantification of cell types depicted in (B) and (D) as proportion of Lin−CD45+ relative to D0 for all time points (mean and SD; D0/D90, BCG, n = 15, Ctrl, n = 5; D14, BCG, n = 7, Ctrl, n = 2).
Unpaired t test was used in (A), (C), and (E) to compare BCG versus Ctrl D90 (all p values > 0.05). See also Figure S1.
BCG Vaccination Induces a Myeloid-Associated Transcriptomic Signature 90 Days after BCG Vaccination in BM HSPCs of Healthy Volunteers
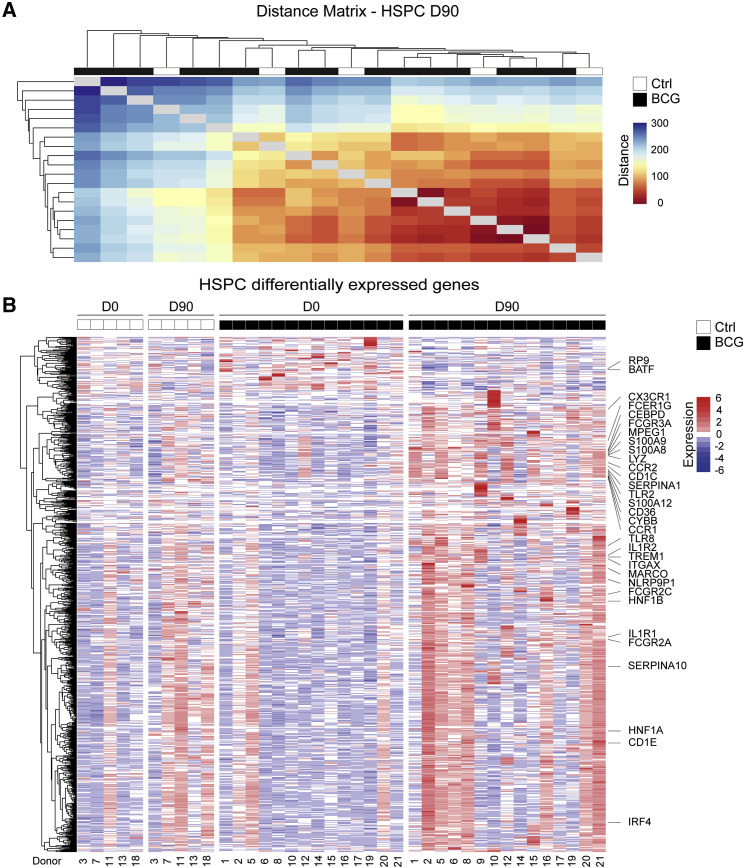
Recent murine studies have shown that systemic BCG vaccination induced transcriptional and functional changes at the level of the HSPCs (Kaufmann et al., 2018, Mitroulis et al., 2018). Therefore, we hypothesized that similar mechanisms might play a role in inducing human innate immune training in circulating monocytes 90 days post-intradermal BCG vaccination in human healthy volunteers. To investigate whether intradermal BCG vaccination-induced systemic innate immune memory is indeed mediated by transcriptional and functional remodeling of the human BM progenitor compartment, we purified HSPCs from BM aspirates using flow cytometry on day 0 and 90 post-vaccination from BCG-vaccinated and placebo-treated individuals and performed a global transcriptome analysis (Figures 3A and S1C for sorting scheme; Table S2). Global distance measurement of transcript abundance between samples at D90 detected a subset of BCG-vaccinated individuals with a different transcriptomic makeup than the placebo-treated group, thus identifying overall transcriptomic differences between BCG-vaccinated and placebo-treated individuals 90 days after vaccination (Figure 3A). In order to further understand these differences, we directly compared the transcriptome of HSPCs isolated from the same individuals on D0 and D90 post-BCG vaccination (Figure 3B). This analysis revealed an upregulation of genes associated to myeloid and granulocytic cell lineage priming (such as C-X3-C motif chemokine receptor 1 [CX3CR1], macrophage-expressed 1 [MPEG1], interferon regulatory factor 4 [IRF4], and CCAAT enhancer-binding protein delta [CEBPD]; Merad et al., 2013, Varol et al., 2015), and function (such as macrophage receptor with collagenous structure [MARCO], IL 1 receptor type 1 [IL1R1], type 2 [IL1R2], S100 calcium-binding protein A8 [S100A8], A9 [S100A9], A12 [S100A12], and serpin family A member 1 [SERPINA1]; Arts et al., 2018, Christ et al., 2018, Mitroulis et al., 2018, Sander et al., 2017) (Figure 3B; Table S3). These data indicated that BCG vaccination induces the upregulation of myeloid and granulocytic lineage-associated transcripts at the level of the HSPC, indicating the potential for a skewed myeloid development bias within healthy vaccinated individuals 90 days post-vaccination.
Figure 3.
BCG Vaccination Induces Persistent Changes in BM HSPCs
(A) Heatmap visualizing Euclidean distance measurement of transcript abundance between HSPC transcriptomes from D90 (Ctrl, n = 5; BCG, n = 15).
(B) Expression heatmap of DEGs in HSPCs (BCG, D0, n = 14 versus D90, n = 15). Samples are separated in columns based on time point and experimental arm. Pearson correlation is used as distance measure to cluster genes in rows. Names of a subset of genes previously associated with trained immunity or myeloid biology are shown. For the statistical details, see STAR Methods.
BCG Vaccination Upregulates Transcriptomic Networks Associated with Neutrophil and Mononuclear Phagocyte Development and Function 90 Days after Vaccination
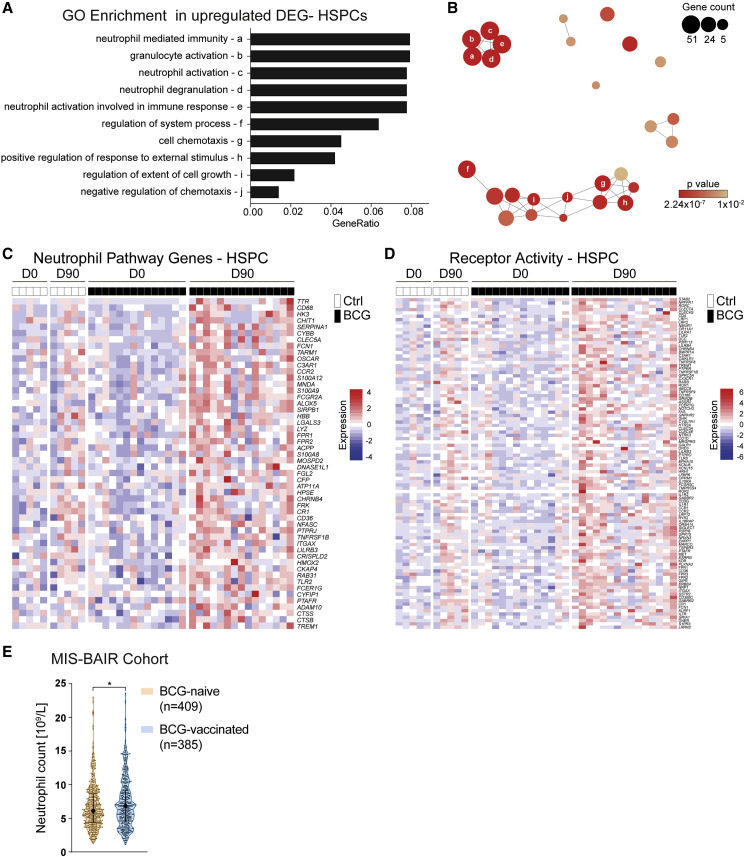
To validate a myeloid-skewed signature within HSPCs as found by inspection of the differentially expressed genes (DEGs) (Figure 3B), we performed an unbiased Gene Ontology (GO) enrichment analysis (GOEA) on the upregulated DEGs within HSPCs before and 90 days after BCG vaccination (Figure 4 A).
Figure 4.
BCG Vaccination Elicits Myeloid Cell Fate Priming
(A) GOEA of upregulated genes in HSPCs. Top ten enriched terms (designated to letters “a” to “j”) are listed (BCG, D0, n = 14 versus D90, n = 15).
(B) Network representation of GO terms significantly enriched in upregulated DEGs in HSPCs (BCG, D0 versus D90, p < 0.01). Node size and color indicate contained gene numbers and p value for term nodes, respectively. Position and edges visualize relation of nodes to each other based on involved gene lists. Annotation of nodes by lowercase letters corresponds to (A).
(C) Unified genes from neutrophil pathways-associated genes from terms (ontology terms a–e, as indicated in A) (GO:0036230, GO:0002446, GO:0002283, GO:0043312, and GO:0042119) were extracted to generate an expression heatmap of HSPC transcriptomes.
(D) Expression heatmap of genes overlapping upregulated DEGs in HSPCs and genes associated with the term for the molecular function of receptor activity (GO: 0004872).
(E) Neutrophil counts (×109/L) of infants in the MIS BAIR study (BCG-vaccinated, n=385; BCG-naive, n = 409; ∗p < 0.05).
See also Tables S4–S7.
This analysis identified pathways, such as “granulocyte activation,” “neutrophil mediated immunity,” and “positive regulation of response to external stimulus,” among others, to be significantly enriched (Figures 4A, 4B, and S2A; Table S4). Network-level analysis of the enriched pathways found by GOEA revealed a dense topological clustering of the neutrophil-associated pathways (ontology terms a–e) and the pathways associated with regulation of immune responses (ontology terms f–j), indicating overlap in the genes contributing to the enrichment of these GO terms within the D90 HSPC transcriptome of BCG-vaccinated individuals (Figure 4B).
Gene-level analysis of the upregulated “neutrophil pathway genes” as revealed by GOEA indicated upregulation of important genes involved in the development and function of neutrophils, such as SERPINA1, S100A12, S100A9, galectin 3 (LGALS3), and lysozyme (LYZ) (Figure 4C). Additionally, the pathway “positive regulation of response to external stimulus” within the day 90 BCG-vaccinated HSPC transcriptome revealed significant upregulation of transcripts associated with myeloid cell function, such as TNF, complement C3a receptor 1 (C3AR1), C-C motif chemokine receptor 2 (CCR2), and the gamma chain of the Fc epsilon receptor (FCER1G) (Figure S2A). Furthermore, inspection of DEG related to the term “signaling receptor activity” (GO:0004872) showed upregulation of genes important for myeloid cell activation and function in HSPC in individuals 90 days after BCG vaccination, including the C-type lectin domain containing 7A (CLEC7A) and C-type lectin domain family 4 member G (CLEC4G) and the cytokine receptors IL1R1, interleukin 7 receptor (IL7R), interleukin 10 receptor subunit alpha (IL10RA), and interleukin 18 receptor accessory protein (IL18RAP) (Figure 4D). Filtering of DEG according to the biological process term “DNA-binding transcription factor (TF) activity” (GO:0003700) revealed several upregulated myeloid cell-associated TFs, such as HNF1 homeobox A and HNF1 homeobox B (HNF1A/B), IRF4, and CEBPD, that serve as potential candidates driving the process of trained immunity on a gene-regulatory level (Figure S2B).
To decipher the functional consequences of the transcriptional reprogramming of the HSPC, we investigated whether there is an effect of BCG vaccination on granulocyte numbers in a significantly larger cohort of vaccinated children. The relatively small number of volunteers from whom we could collect BM aspirates, and the biological variation in cell counts in the current trial, did not permit to reach statistical significance for the differences in cell populations after BCG vaccination (see above). Therefore, we compared neutrophil numbers in the MIS BAIR cohort of 1,200 infants randomized to receive either BCG or no BCG shortly after birth (clinical trial registration NCT01906853). As shown in Figure 4E, within days of randomization, BCG-vaccinated infants had significantly clinically relevant higher numbers of neutrophils in the circulation compared with BCG-naive infants, thereby adding weight to the hypothesis that BCG vaccination skews the myeloid lineage toward granulopoeisis (Figure 4E; Tables S5–S7).
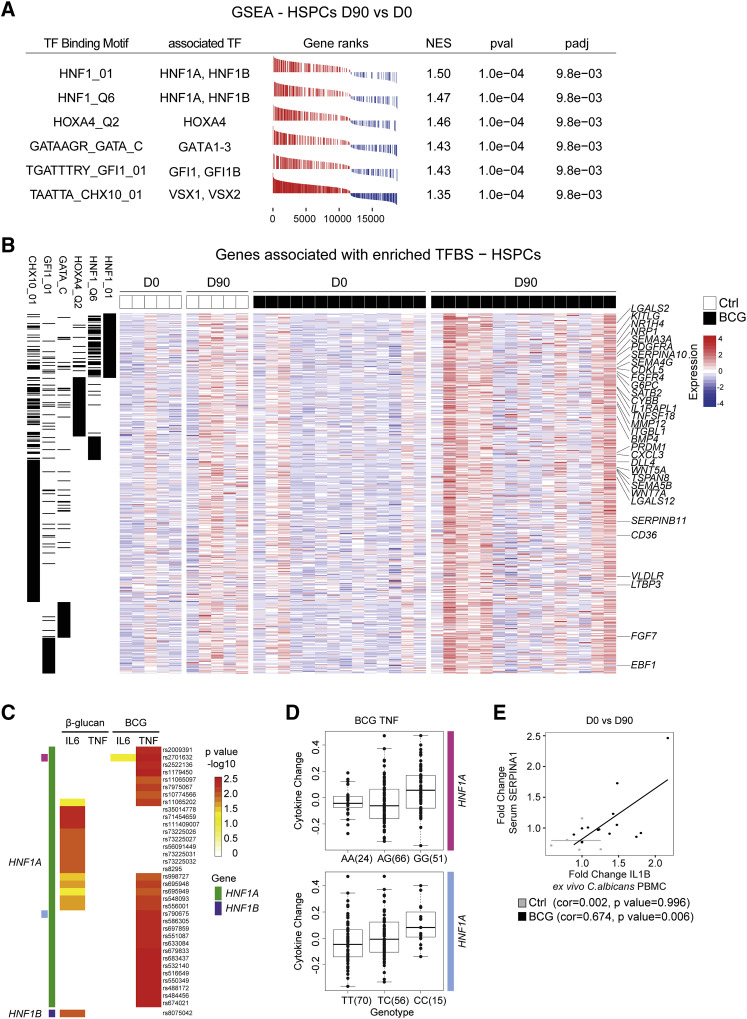
HNF1A and HNF1B Regulate the Induction of BCG-Elicited Innate Immune Training in Human HSPCs 90 Days Post-Vaccination
To reveal factors that guard the induction of the regulatory gene networks leading to the expression of genes associated with human innate immune memory, myeloid cell fate, and the increase in circulating neutrophils, we used gene sets associated with distinct cis-regulatory TF-binding motifs on ranked gene lists according to expression fold change in HSPCs from D0 to D90 and applied GSEA (Figure 5 A). This analysis indicated a significant enrichment of a set of genes associated with TF-binding motifs related to HNF1, homeobox A4 (HOXA4), GATA-binding proteins, growth factor independent 1 transcriptional repressor (GFI1), and visual system homeobox 2 (VSX2), defined by the CHX10 motif, TFs (Figure 5A). Visualizing the leading-edge genes matching significant genes with binding motifs for the TFs identified in Figure 5A indicated important factors involved in myeloid cell development, such as LGALS2, glucose-6-phosphatase catalytic subunit (G6PC), interleukin 1 receptor accessory protein like 1 (IL1RAPL1), bone morphogenetic protein 1 (BMP4), and C-X-C motif chemokine ligand 3 (CXCL3) (Figure 5B).
Figure 5.
HNF1 TFs Represent Regulatory Factors in Trained Immunity
(A) GSEA of enriched signatures associated with distinct TF-binding motifs in genes ranked according to expression fold change in HSPCs at D0 versus D90. TFs associated with the binding motifs are listed. NES, normalized enrichment score; pval, p value; padj, adjusted p value.
(B) Expression heatmap of all leading-edge genes derived from TF-binding site gene sets as found in (A). Black lines in the first six columns indicate association of genes to the respective TF signature gene sets listed in the column header.
(C) Ex vivo training of PBMCs with β-glucan or BCG and IL6/TNF cytokine measurements after LPS challenge. SNPs ± 250 kb within the genomic locus of HNF1A (green) or HNF1B (purple) and variants thereof significantly leading to a change in cytokine production for any of the conditions are listed. Threshold for inclusion, p < 0.01. Top two expression quantitative trait loci (eQTLs) are annotated as pink and bright blue.
(D) Detailed TNF cytokine level after BCG training and LPS challenge for the top two eQTLs (annotated in C as pink and bright blue). Number of individuals displaying distinct SNP variants are shown in brackets. Analysis in (C) and (D) is based on 141 individuals in total.
(E) Fold changes of IL1B after ex vivo C. albicans restimulation and serum SERPINA1 (D0 versus D90) correlated in the same individuals. Data points from BCG-vaccinated or Ctrl-treated individuals are shown in black or gray, respectively.
The two TFs with the highest significance predicted by GSEA, alongside a robust upregulation within the transcriptome of HSPCs day 90 post-vaccination, were HNF1A and HNF1B (Figures 3B and 5A), which are known to be expressed in the liver, but also in myeloid cells (Armendariz and Krauss, 2009). Furthermore, HNF1A/B and their target genes SERPINA10 and SERPIN1A were found to be upregulated 90 post-BCG vaccination in HSPCs of BCG-vaccinated individuals (Figures 3B and 4C). We therefore hypothesized that HNF1A/B may be involved in the induction of trained immunity, and we initiated genetic validation in a cohort of 141 healthy individuals (200FG cohort from the human functional genomics project) in which an ex vivo trained immunity model was applied using either BCG or β-glucan as inducers (Ter Horst et al., 2016). Indeed, polymorphisms in both HNF1A and HNF1B significantly influenced the induction of trained immunity as indicated by significant differences in the potential to release IL6 and/or TNF after restimulation with LPS in vitro (Figures 5C and 5D; Table S8). Furthermore, looking at genetic variants present at specific sites within the HNF1A locus (marked pink and bright blue in Figure 5C) revealed that specific SNP variants are associated with more or less TNF production after BCG-induced training ex vivo (Figure 5D). Furthermore, also polymorphisms in the other indicated TFs associated with motifs identified earlier (Figure 5A), such as GATA2/3, GFI1/1B alongside HOXA4, and visual system homeobox 1 (VSX1) were shown to influence the β-glucan or BCG-mediated induction of innate immune memory (Figure S3A).
One of the target genes of HNF1A is SERPINA1, coding for the acute phase protein α-1-antitrypsin (AAT) (Armendariz and Krauss, 2009), which was found to be upregulated within the transcriptome of the HSPC 90 days after BCG vaccination (Figures 3B and 4C). To validate the functional consequences of this finding, we tested whether there is a correlation between circulating AAT concentrations and the strength of induction of innate immune memory using enhanced IL1B production as an indicator of successful innate immune memory induction. We measured serum AAT levels at D0 and D90 post-vaccination and correlated this concentration with the fold changes of IL1B induction in PBMCs restimulated ex vivo with C. albicans in BCG-vaccinated and non-vaccinated individuals after 90 days. As shown in Figure 5E, the serum concentration of AAT correlated with enhanced IL1B production within restimulated PBMCs (Figure 1B), further supporting the hypothesis that a HNF1-controlled transcriptomic network is implicated in the induction of human innate immune memory. To directly test the effect of AAT on BCG-induced innate immune training on the cellular level, we isolated healthy PBMC-derived monocytes and incubated these cells with BCG in the presence of either AAT or serum-derived albumin (as control protein) (Figures S3B and S3C). After training for 24 h, the stimuli were washed off, and monocytes were rested for 5 days. On day 6 after training with BCG (with or without AAT), we restimulated the cells with E. coli-derived LPS and measured IL6 and TNF secretion 24 h later. AAT decreased trained immunity induced by BCG, especially IL6 production (Figure S3C). These data establish a regulatory role of AAT on the development of BCG-induced innate immune training ex vivo and point toward a negative regulatory role of AAT in vivo, balancing the development of innate immune training in humans.
To identify molecules which would allow prediction of individuals with the potential to mount an efficient innate immune training response, we performed an Olink-targeted proteomics analysis investigating the expression of 92 plasma proteins prior BCG vaccination and correlated it with the ex vivo PBMC cytokine release for IL1B and IL6 in response to C. albicans D14 or D90 post-BCG vaccination. Here, increased baseline (D0) plasma levels of CCL23 significantly correlated with trained immunity responses, as assessed by the fold increase of IL1B production in PBMC restimulated ex vivo with C. albicans 14 days post-BCG vaccination (Figure S3D). Furthermore, these analyses revealed that baseline plasma concentrations of CCL20 (a predicted target of HNF1A [TRANSFAC]) positively correlated with the ability to release increased amounts of IL6 in PBMCs restimulated ex vivo with C. albicans 14 days post-BCG vaccination (Figure S3E). Prior analysis has shown that neutrophil-mediated gene programs are activated 90 days post-vaccination with BCG and an increase in neutrophil counts in children days after BCG vaccination. This notion of increased granulopoesis was further supported by the fact that plasma levels of S100A12, a crucial neutrophil derived anti-microbial effector, significantly correlated with the increase in PBMC-derived IL6 release 90 days post ex vivo C. albicans challenge (Figure S3F). No other correlations were found between the rest of the inflammatory mediators and the induction of trained immunity. Thus, these data reveal HNF1A and HNF1B as crucial regulators of BCG-induced innate immune memory formation in human resting HSPCs 90 days post-vaccination.
BCG Vaccination Induces Persistent Epigenetic Changes Associated with Inflammation in Peripheral CD14+ Monocytes
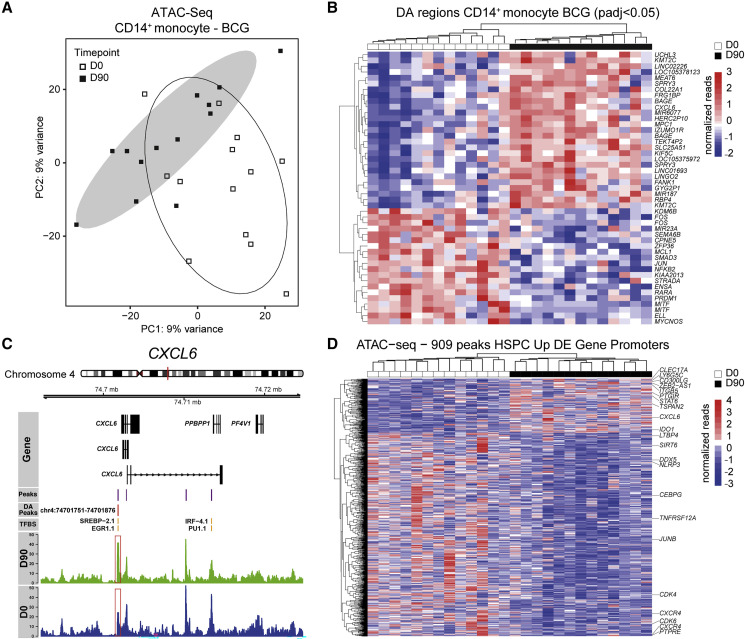
Functional investigation of C. albicans-restimulated PBMCs revealed a higher capacity of blood borne cells to produce and release IL1B 90 days after BCG vaccination (Kleinnijenhuis et al., 2014) (Figure 1B). Therefore, we hypothesized that innate immune memory induction is reflected within the epigenome of the major effector cell type within the PBMC, i.e., CD14+ monocytes, during homeostasis. To investigate this, we assessed the differential chromatin accessibility of CD14+ monocytes from BCG-vaccinated individuals before and 90 days after vaccination using the assay for transposase accessible chromatin by ATAC sequencing (ATAC-seq) (Figures 4A and 4B; Table S9). PCA of all present peaks segregated CD14+ monocytes isolated at day 0 and day 90 post-BCG vaccination indicating a clear role for regulation at the level of DNA accessibility (Figures 6A and S4C). Analysis of the genes associated to the core set of the top 47 differentially accessible peaks (robust to multiple testing correction) between day 0 and 90 post-BCG vaccination showed increased DNA accessibility in the proximity of various inflammation-associated genes, such as collagen type XXII alpha 1 chain (COL22A1) (Linge et al., 2008, Rasmussen et al., 2012), C-X-C motif chemokine ligand 6 (CXCL6) (Linge et al., 2008, Jovic et al., 2016) and retinol-binding protein 4 (RBP4) (Moraes-Vieira et al., 2014) in homeostatic CD14+ monocytes 90 days post-vaccination (Figures 6B, 6C, and S4D; Tables S10 and S11). On the other hand, peaks associated to genes involved in retinoic acid-induced immune tolerance (retinoic acid receptor alpha [RARA]) (Coombes et al., 2007, Jaensson et al., 2008, Siddiqui and Powrie, 2008), lymphoid development (PR/SET domain 1 [PRDM1]) (Bankoti et al., 2017), and inactive nuclear factor kappa B subunit 2 (NFKB2) (Baldwin, 1996, Lawrence, 2009) showed a diminished signal 90 days post-BCG vaccination in CD14+ monocytes (Figures 6B, S4E, and S4F). We then asked which HSPC-associated transcriptomic changes can be found in the epigenome of CD14+ monocytes and intersected the genes found to be differentially regulated within the HSPC of BCG-vaccinated individuals 90 days post-vaccination with ATAC-seq peaks within promoter regions in CD14+ monocytes. Interestingly, among the promoter regions associated with more open regions of genes upregulated within the transcriptome of HSPCs, we identified again CXCL6, CLEC17A, PTGIR, and STAT6 to be conserved from the HSPC transcriptome to the CD14+ monocyte epigenome (Figure 6D). Vice versa, loci associated to myeloid development and lineage priming, such as CEBPG, TNFRSF12A, and CXCR4 and to cell proliferation, such as CDK4 and CDK6 were closed down in the epigenome of CD14+ monocytes. Furthermore, hierarchical clustering of the normalized peak reads falling within promoters of genes that were transcriptionally upregulated in HSPCs revealed a clear stratification of the epigenetic makeup in monocytes before and after vaccination (Figure 6D). In conclusion, these analyses show that transcriptomic changes induced by BCG vaccination within the HSPC are epigenetically perpetrated toward the major PBMC-derived effector cells, i.e., CD14+ monocytes, at least in part through regulation of the DNA accessibility of certain inflammation-associated loci. This allows for a persistent induction of a trained immunity phenotype of CD14+ monocytes, beyond the individual cell’s lifespan.
Figure 6.
Epigenetic Changes in Blood Monocytes Are a Feature of BCG-Induced Training
(A) PCA based on normalized peak counts of open regions derived from ATAC-seq in peripheral blood CD14+ monocytes. Open or filled symbols indicate D0 or D90 time points of BCG-vaccinated individuals, respectively. Oval shapes denote confidence intervals at 0.85 for the two groups.
(B) Heatmap of normalized ATAC-seq reads focusing on the core set of 47 differentially accessible regions annotated according to the closest gene. Peaks are included reaching an adjusted p value (padj) < 0.05.
(C) Example genome track surrounding the CXCL6 region. Called peaks are visualized in violet, differentially accessible (DA) peaks in red and identified TF-binding sites (TFBS) within called peaks in yellow. Summarized reads from CD14+ monocytes before and after vaccination are shown in blue and green, respectively.
(D) ATAC-seq heatmap of 909 peaks within promoters closest to genes transcriptionally upregulated in HSPCs after 3 months from the vaccinated group (n = 13 per group).
Discussion
In this study, we demonstrate that human in vivo BCG vaccination induces sustained transcriptomic changes in HSPCs of healthy individuals, changes that are accompanied by persistent increase of innate immune cells responsiveness to heterologous stimuli. BCG vaccination was associated with a rewiring of transcriptional programs of HSPCs toward myelopoiesis. The physiological relevance of this shift was validated by the higher neutrophil counts within days of BCG vaccination observed in a randomized clinical trial of BCG vaccination in newborn infants and by the increased responsiveness of PBMCs to stimulation with unrelated bacterial and fungal stimuli ex vivo.
These findings are important at several levels. First, the demonstration of these effects of BCG vaccination in vivo at the level of human BM-resident HSPC provides the mechanistic explanation for the persistent effects of BCG vaccination on circulating innate immune cells and for the many epidemiological observations of beneficial non-specific effects of BCG years after vaccination (Biering-Sørensen et al., 2017, Rieckmann et al., 2017, Villumsen et al., 2009, Zimmermann et al., 2018). These effects on HSPCs are accompanied by a clinically relevant and significant increase in the number of neutrophils in infants vaccinated with BCG, constituting a part of the functional basis for the non-specific clinical effects of BCG vaccination (Biering-Sørensen et al., 2017, Zimmermann et al., 2018). The conclusion of a myeloid differentiation bias within the HSPCs induced by BCG vaccination is supported by recent studies in murine models in which trained immunity was induced by systemic BCG administration (Kaufmann et al., 2018) or β-glucan administration (Mitroulis et al., 2018), in which similar changes within the murine myeloid precursor cascade were observed.
Second, important insights have been obtained on the mechanisms responsible for the induction of trained immunity in humans. Thus, we demonstrate that BCG vaccination is accompanied by important changes at the level of chromatin accessibility in CD14+ monocytes, and that CD14+ monocytes before and 90 days after vaccination can be segregated on the basis of the patterns of DNA accessibility. The epigenetic changes within CD14+ monocytes relate to those found at the level of the transcriptome within HSPCs, revealing a crucial mechanism of innate immune memory imprinting across cell lineages (Arts et al., 2018, Saeed et al., 2014).
Third, an important focus of this study was to identify pathways that mediate induction of a trained immunity phenotype. By intersecting the list of TFs that were upregulated in HSPCs after vaccination, with that of gene sets associated with specific TF motifs significantly enriched after vaccination, we identified HNF1A and HNF1B as master regulators of trained immunity. In earlier studies HNF1A/B have been shown to directly transactivate both the gene activity and chromatin structure of SERPINA1 either by rearrangement of histone modifications or the recruitment of other co-factors (Gong et al., 2009, Rollini and Fournier, 1999). HNF1A/B are highly expressed in the liver and myeloid cells, especially in response to acute inflammation (Armendariz and Krauss, 2009). The hypothesis that HNF1 family TFs are important for the induction of trained immunity was subsequently validated in the 200FG cohort of healthy volunteers from the human functional genomics project, in which trained immunity was induced in an ex vivo model by either β-glucan or BCG: polymorphisms in both HNF1A and HNF1B modulated the induction of trained immunity. HNF TFs regulate transcription of numerous genes important for host defense, including several acute phase proteins such as α-1-antitrypsin (AAT, SERPINA1) (Armendariz and Krauss, 2009). In an independent validation effort, we measured AAT-circulating concentrations and demonstrate a strong correlation between AAT-circulating concentrations and the induction of trained immunity. Subsequently, using the OLink plasma screening technology we identified the baseline plasma levels of S100A12, CCL23, and the putative HNF1A target gene CCL20 as predictive biomarkers of “trained immunity induction.” These observations further strengthen the role of the HNF1A/B axis in regulating the induction of human trained immunity.
Finally, these data provide a resource for potential diagnostic markers and therapeutic targets based on trained immunity. It would be important to study whether other live vaccines that have been associated with beneficial non-specific effects induce similar changes (Arts et al., 2018). Current efforts are ongoing to identify potential trained immunity correlates of protection during BCG vaccination, which could lead to an acceleration of TB vaccine development and to its deployment against other pathogens. In this respect, we have earlier demonstrated that BCG vaccination can protect against human experimental infections with viruses (Arts et al., 2018) and parasites (Walk et al., 2019). Currently, several clinical trials in the Netherlands, Australia, and Greece investigate the capacity of BCG vaccination to prevent infection with the new coronavirus SARS-CoV-2 (NCT04328441, NCT04327206, and NCT04339712 [www.clinicaltrials.gov]). Similarly, trained immunity has been proposed to represent a new avenue for immunotherapy in cancer (Netea et al., 2017) and to contribute to the pathophysiology of inflammatory and autoimmune diseases (Bekkering et al., 2018, Crișan et al., 2016). Our study reveals HNF-mediated processes as such a potential target but also provides a catalog of genes that characterize trained BM progenitors that can be targeted for diagnostic and/or therapeutic purposes.
However, our study raises additional questions to be answered by future studies. How long does the HSPC reprogramming induced by BCG vaccination persist? Can this process be reversed? Indeed, epidemiological studies suggest that the beneficial effects of BCG vaccine on overall health can be offset by subsequent vaccination with the non-live diphtheria-tetanus-pertussis vaccine (Higgins et al., 2016). Another important aspect to be answered by future studies refers to the mediators that induce the trained immunity phenotype at the level of BM. One possibility is that release of peptidoglycans from the site of the vaccination would mediate through circulation the stimulation of BM progenitors. Initial data from another study suggest indeed that muramyl-dipeptide (a peptidoglycan component) concentrations after BCG vaccination increase (V. Mourits, personal communication), but whether this is associated with changes within the BM remains to be investigated. Another possibility is the release of endogenous mediators (e.g., cytokines) from the site of infection: Their assessment in the circulation in the first hours and days after vaccination should be investigated in future studies, as these early time points were not available in the current study.
In conclusion, we provide evidence that in vivo BCG-induced trained innate immunity induces epigenetic, transcriptional, and functional changes in human BM-derived HSPCs and peripheral CD14+ monocytes. These effects at least partly explain the beneficial and persistent heterologous effects of BCG vaccination against infections as trained HSPCs can respond faster and more efficiently toward stress signals delivered e.g., by infections. These findings should lead to a strong impetus for further exploration of this process in order to identify targets for clinical vaccine development and therapeutic targeting of trained immunity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Biotin anti-human CADM1 | MBL | Cat#CM004-6 |
| APC anti-human CD10 | BioLegend | Cat#312210; RRID: AB_314921 |
| APC/Cyanine7 anti-human CD10 | BioLegend | Cat#312212; RRID: AB_2146550 |
| PE-CF594 anti-human CD110 | BD Biosciences | Cat#562416; RRID: AB_11154044 |
| PE/Cyanine7 anti-human CD115 | Thermo Fisher Scientific | Cat#12-1159-42; RRID: AB_10717675 |
| BV421 anti-human CD116 | BD Biosciences | Cat#564045; RRID: AB_2738561 |
| BV785 anti-human CD123 | BioLegend | Cat#306032; RRID: AB_2566448 |
| PE anti-human CD135 | BD Biosciences | Cat#558996; RRID: AB_397175 |
| BV711 anti-human CD14 | BioLegend | Cat#301838; RRID: AB_2562909 |
| BV421 anti-human CD14 | BioLegend | Cat#325628; RRID: AB_2563296 |
| APC/Fire750 anti-human CD15 | BioLegend | Cat#323042; RRID: AB_2572103 |
| BV510 anti-human CD16 | BioLegend | Cat#302048; RRID: AB_2562085 |
| PE/Cyanine7 anti-human CD16 | BioLegend | Cat#302016; RRID: AB_314216 |
| APC/Cyanine7 anti-human CD19 | BioLegend | Cat# 363010; RRID: AB_2564193 |
| PE/Dazzle anti-human CD1c | BioLegend | Cat#331532; RRID: AB_2565293 |
| APC/Cyanine7 anti-human CD20 | BioLegend | Cat#302314; RRID: AB_314262 |
| APC/Cyanine7 anti-human CD3 | BioLegend | Cat#317342; RRID: AB_2563410 |
| BV650 anti-human CD33 | BioLegend | Cat#303430; RRID: AB_2650934 |
| AF700 anti-human CD34 | BioLegend | Cat#343526; RRID: AB_2561495 |
| BV510 anti-human CD38 | BioLegend | Cat#356612; RRID: AB_2563875 |
| PerCP anti-human CD45 | BioLegend | Cat#304026; RRID: AB_893337 |
| FITC anti-human CD45RA | BioLegend | Cat#304106; RRID: AB_314410 |
| APC-eFluor anti-human CD7 | Thermo Fisher Scientific | Cat#47-0079-42; RRID: AB_2573942 |
| BV711 anti-human CD90 | BD Biosciences | Cat#740786; RRID: AB_2740449 |
| APC anti-human CX3CR1 | BioLegend | Cat#341610; RRID: AB_2087424 |
| BV570 anti-human HLA-DR | BioLegend | Cat#307637; RRID: AB_10895753 |
| Bacterial and Virus Strains | ||
| BCG vaccine (Bulgaria strain) | Intervax | N/A |
| BCG vaccine (Danish strain) | Statens Serum Institute | N/A |
| Heat-inactivated Candida albicans (UC820 strain) | In house | N/A |
| Escherichia coli lipopolysaccharide | Sigma-Aldrich | N/A |
| β-glucan | Prof. David Williams, University of East Tennessee | N/A |
| Biological Samples | ||
| Human mononuclear cell fraction (peripheral and bone marrow derived, Ficoll gradient separated) | Healthy volunteers | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DRAQ7 | BioLegend | Cat#424001 |
| PE Streptavidin | BioLegend | Cat#405204 |
| Critical Commercial Assays | ||
| miRNeasy Micro Kit | QIAGEN | Cat#217084 |
| QIAseq FX Single Cell RNA Library Kit | QIAGEN | Cat#180733 |
| KAPA Library Quantification Kits | Kapa Biosystems | Cat#07-KK4852-01 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q32851 |
| Pan Monocyte Isolation Kit | Miltenyi Biotec | Cat#130-096-537 |
| ProcartaPlex luminex customized multiplex assay | ThermoFisher | N/A |
| Gentra Pure Gene Blood kit | QIAGEN | Cat#158389 |
| MinElute kit | QIAGEN | Cat#28004 |
| Human Serpin A1 DuoSet ELISA | R&D systems | Cat#DY1268 |
| Proseek Multiplex Inflammation I | OLink Proteomics | Cat#95302 |
| Deposited Data | ||
| RNA sequencing/ATAC sequencing data | This Paper | GEO: GSE124220 |
| Human functional genomics data | Ter Horst et al., 2016 | https://hfgp.bbmri.nl/ |
| Software and Algorithms | ||
| FlowJo | Tree Star | RRID: SCR_008520 |
| R statistical programming | N/A | RRID: SCR_001905 |
| DESeq2 | N/A | RRID: SCR_015687 |
| Bioconductor | N/A | RRID: SCR_006442 |
| ClusterProfiler | N/A | RRID: SCR_016884 |
| Pheatmap | N/A | RRID: SCR_016418 |
| ggplot2 | N/A | RRID: SCR_014601 |
| Fgsea | Bioconductor | https://doi.org/10.18129/B9.bioc.fgsea |
| Tximport | N/A | RRID: SCR_016752 |
| flowCore | N/A | RRID: SCR_002205 |
Resource Availability
Lead Contact
Further information and requests for reagents can be addressed to and will be fulfilled by the Lead Contact Andreas Schlitzer (andreas.schlitzer@uni-bonn.de).
Materials Availability
This study did not generate new unique reagents
Data and Code Availability
RNA sequencing and ATAC data is available under GEO: GSE124220. Human functional Genomics data is available through https://hfgp.bbmri.nl (Ter Horst et al., 2016). Scripts used to analyze these data will be shared upon request.
Experimental Model and Subject Details
Experimental Design
This single-center randomized controlled trial was conducted at the Radboud university medical center (Nijmegen, The Netherlands) from January 2017 to July 2017. Prior to inclusion, volunteers were medically screened and provided written informed consent. The trial was approved by the Arnhem-Nijmegen Ethical Committee (approval number NL55825.091.15), and performed according to the Declaration of Helsinki and Good Clinical Practice. Twenty healthy, male and female, BCG-naive volunteers (age 18–50 years, no active infection, no detectable inflammatory signs, negative test result for the Quantiferon-TB Gold test) were included and randomly assigned to two groups: 15 subjects received standard dose (0.1ml of the reconstituted vaccine) of intradermal BCG vaccination (BCG Bulgaria, Intervax), and 5 received 0.1ml of vaccine diluent as a placebo control. One volunteer (assigned to the BCG-vaccinated group) has been replaced due to technical reasons during bone marrow aspiration. The 5 placebo controls were used to calibrate the cohort and establish baseline measurements and the effect of bone marrow aspiration, whereas the experimental cohort of 15 vaccinated volunteers was assessed and analyzed before and after vaccination. Blood was drawn before, two weeks after and three months after vaccination. At baseline and after three months, bone marrow was extracted by aspiration from the iliac crest by an experienced physician assistant after local anesthetics with lidocaine. 30 mL of bone marrow was aspirated in total and collected into two 20 mL syringes, prefilled with 5 ml (150 IE/ml) sodium heparin per syringe.
Method Details
Whole Blood Counts
Complete blood counts were determined in EDTA whole blood using a Sysmex analyzer (XN-450)
Mononuclear Cell Isolation and PBMC Stimulation
Mononuclear cells from peripheral EDTA whole blood (PBMCs) and heparinized bone marrow (BM-MNCs) were isolated with Ficoll-Paque (GE healthcare, UK) density gradient separation. Cells were washed twice with phosphate buffered saline (PBS), counted in a Coulter counter (Beckman Coulter, USA), and brought to concentration in Dutch modified RPMI medium (Roswell Park Memorial Institute; Invitrogen, CA, USA), supplemented with 50μg/ml gentamycine, 2 mM Glutamax (Thermo Fisher Scientific, USA) and 1 mM pyruvate (Thermo Fisher Scientific, USA). 5 x 105 PBMCs were cultured in a final volume of 200 μl / well in round bottom 96-well plates (Greiner Bio-one, Austria) and stimulated with RPMI, heat-killed Candida albicans (1 × 106 / ml, strain UC820). After 24 hours supernatants were collected and stored at -80°C until measurements were performed.
Ex Vivo BCG Induced Innate Immune Training
To investigate the effects of AAT on BCG-induced trained immunity in vitro, a trained immunity model was used as previously described (Bekkering et al., 2016). Buffy coats from healthy donors were obtained after written informed consent (Sanquin blood bank, Nijmegen, The Netherlands). PBMCs were isolated using density-gradient separation over Ficoll-Pacque as described above. PBMC-derived monocytes were obtained by density-gradient separation over Percoll, followed by one hour of adherence to 96-well flat bottom culture plates (Corning), after which cells were washed once and cultured for 24 hours with either BCG 10 μg/ml (SSI, Denmark), or culture medium (RPMI 1640 supplemented with gentamycin, GlutaMAX and pyruvate) in an end-volume of 200 μl. Either AAT or albumin (both at a concentration of 100 μg/ml) were added to the BCG preparation. After 24 hours of stimulation at 37°C and 5% CO2, cells were washed once with warm PBS and RPMI supplemented with 10% human pool serum was added to an end-volume of 200 μl. At day 6, cells were cultured with LPS (10 ng/ml; Sigma-Aldrich, St. Louis, MO, USA) for 24 hours at 37°C and 5% CO2 after which plates were centrifuged and supernatants were stored at -80°C until cytokines were measured.
Cytokine Measurements
Cytokines were determined in PBMC culture supernatants by a multiplex Procartaplex assay, Thermo Fisher Scientific, USA) and acquired on a Luminex 200 System (Thermo Fisher Scientific, USA) according to manufacturer’s protocol.
AAT Concentration Measurement
Circulating α-1-antitrypsin concentrations were determined in serum using a R&D Duoset ELISA kit (R&D Systems, Minneapolis, USA).
Flow Cytometric Analysis and Sorting
PBMC (1 x 107) or BM MNC (4 x 107) were washed in FACS-buffer (0.5% BSA, 2 mM EDTA, PBS), resuspended in blocking-buffer (1 % rat serum (R9759-10ML; Sigma-Aldrich, USA), 1 % mouse serum (M5905-5ML; Sigma-Aldrich, USA), 5 % human serum (H4522-100ML; Sigma-Aldrich, USA) in FACS-buffer) and incubated with the respective antibody cocktails for two hours on ice in the dark (Table S12). After a washing step, secondary staining was performed for PBMCs in FACS-buffer using Streptavidin-PE (1:200) for further 15 min. PBMC and BM MNC were washed, resuspended in FACS-buffer and incubated with the Live/dead marker DRAQ7 (1:1000, BioLegend, USA) for 5 min at room temperature. Samples were acquired using BD FACS ARIAIII (BD Biosciences) and the FACS Diva software (BD Biosciences, USA). Data was analyzed using FlowJo (Tree Star, USA) and is presented as percentage of live CD45+Lin- relative to baseline (D0) for each individual. For dimensionality reduction, 5 x 105 CD45+Lin- cells were sampled for PBMC or 2.5 x 105 CD45+Lin- and 2.5 x 105 CD34+ cells for BM MNC and imported into R using the flowCore R package (v1.46.1). Imported PBMC or BM MNC populations were subjected to UMAP (Becht et al., 2019, McInnes and Healy, 1802) using the parameter values FSC-A and SSC-A (linear normalization, min=0, max=4.5) and in addition CD34, CD16, HLA-DR, CD33, CD14, CD123, CD45RA, CADM1, CD1c for PBMC or CD10, CD34, CD14, CD38, HLA-DR, CD33, CD90, CD123, CD45RA, CD135, CD16, CD110 for BM MNC. Fluorescent data points were auto-logicle transformed before dimensionality reduction. Cells in the reduced dimensionality space were visualized with the ggplot2 (v3.0.0) package in R. Expression values are indicated in the UMAP space after transformation using the flowJo_biexp_trans function in the flowWorkspace package (v3.28.1) in R. For RNA isolation from HSPCs (HSC+MPP), cells were first sorted into cooled 1.5 ml reaction tubes containing FACS-buffer. Cells were pelleted at 2200 rpm for 7 min at 4°C and vigorously resuspended in 500 μl Qiazol. Tubes were stored at -80°C until further processing.
Generation and Sequencing of cDNA Libraries for Transcriptome Analysis
Total RNA was isolated using the miRNeasy Micro Kit (QIAGEN GmbH, Germany) according to the manufacturer’s guidelines. RNA concentration and quality was assed using the Tapestation 2200 system and high sensitivity reagents (Agilent Technologies, USA). Sequencing libraries of FACS-enriched populations were prepared following the QIAseq FX Single Cell RNA Library Kit protocol (QIAGEN GmbH, Germany) and libraries from cultured PBMC were generated according to the Smart-seq2 protocol (Picelli et al., 2014). For QIAseq FX Single Cell RNA Libraries, concentration of cDNA was assessed by KAPA Library Quantification Kits (Kapa Biosystems, USA) before sequencing single read 75bp on a HiSeq1500 instrument using TruSeq SBS v3-HS chemistry. For Smart-seq2 libraries, concentrations were determined using a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, USA) and average library size was determined using an High Sensitivity D5000 assay on a Tapestation 2200 system (Agilent Technologies, USA) before pooling and sequencing single read 75bp on a NextSeq500 system using High Output v2 chemistry.
Generation of ATAC-sequencing Data
Samples for ATAC-sequencing analysis were prepared from CD14+-MACS-enriched monocytes from fresh PBMCs (Pan Monocyte Isolation Kit, Miltenyi Biotec, Germany). 50,000 cells were washed in cold PBS and resuspended in cold lysis buffer (10 mM NaCL, 3 mM MgCl2, 0.1 % IGEPAL, 10mM Tris-HCl pH7.4). After centrifugation, pellets were incubated in primer-loaded transposase Tn5 in reaction buffer (10 mM TAPS-NaOH (pH 8.5) at 25°C, 5 mM MgCl2, 10% DMF) for 30 min at 37°C. Samples were subsequently immediately purified using the MinElute kit (QIAGEN GmbH, Germany) according to the manufacturer’s guidelines, eluted in 10 μl H2O and stored at -20°C until further processing. Libraries were quantified using KAPA library quantification kits (Kapa Biosystems, USA).
PBMC Isolation and In Vitro Training Experiments
After obtaining informed consent, venous blood was drawn into 10 mL EDTA tubes. PBMC isolation was performed by density centrifugation of EDTA blood diluted 1:1 in PBS over Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden). Cells were washed twice with PBS and suspended in medium (RPMI 1640) supplemented with gentamicin (10 mg/ml), L-glutamine (10 mM) and pyruvate (10 mM). The cells were counted in a Coulter counter (Beckman Coulter, Pasadena, USA). A total of 5 x 105 PBMCs per well were incubated at 37°C for 60 minutes in flat bottom 96-well plates. After 60 minutes of adherence, the wells were washed three times with 200 μl warm PBS to wash away non-adherent cells. 200 μl of stimuli in 10% human pooled serum were added (RPMI as a negative control, 10 μg / ml BCG (BCG Denmark, SSI) or 2 μg / ml β-glucan (kindly provided by Prof. David Williams, University of East Tennessee). After 24 hours incubation, stimuli were washed away with 200 μl warm PBS and fresh medium containing 10% human pooled serum was added. Plates were incubated for another 5 days and medium was refreshed once. At day 6, restimulation was performed with RPMI (negative control) or 10 ng / ml LPS. After another 24 hours incubation, supernatants were collected and stored at -20°C until cytokine concentrations were determined.
DNA Isolation and Genotyping
DNA was isolated from EDTA venous blood using the Gentra Pure Gene Blood kit, in accordance with the manufacturer’s instructions (Qiagen, Venlo, the Netherlands). DNA samples of 141 individuals were genotyped using the commercially available SNP chip, Illumina HumanOmniExpressExome-8 v1.0. Opticall 0.7.0 with default settings was used for genotype calling. Samples with a call rate ≤ 0.99 were excluded, as were variants with a Hardy-Weinburg equilibrium (HWE) ≤ 0.0001, and minor allele frequency (MAF) ≤ 0.1.
Olink Cytokine Measurements
Circulating plasma inflammatory markers were measured before and after BCG vaccination using the commercially available Olink Proteomics AB (Uppsala Sweden) Inflammation Panel (92 inflammatory proteins), using a Proceek Multiplex proximity extension assay (Assarsson et al., 2014). In this assay proteins are recognized by pairs of antibodies coupled to cDNA strands, which bind when they are in close proximity and extend by a polymerase reaction. Detected proteins are normalized and measured on a log2-scale as normalized protein expression values (NPX).
Microbiological Assessment of Mycobacterium bovis Load
To assess presence of M. bovis BCG DNA in bone marrow cell fractions and bone marrow-derived plasma samples of study subjects, we performed a real-time PCR targeting the IS6110 insertion sequence, using the primers and probes (set 2) first described by Savelkoul et al. (2006), on the LightCycler480 platform (Roche, Basel, Switzerland).
Quantification and Statistical Analysis
RNA-sequencing Pre-processing and Data Analysis
For pre-processing, data was demultiplexed using bcl2fastq2 (v.2.20). Quality controls were performed by FastQC, reads aligned to human reference transcriptome hg38 from UCSC and transcript abundance quantified using kallisto (v0.440) with default parameters. Further analysis was based on at least 5 million sequenced reads. One sample (derived from donor 9 HSPC, D0) was omitted due to prominent de-clustering of the sample in the PCA based on normalized reads of the top 5000 most variable transcripts. Samples were imported with tximport (v1.8.0) and transcripts were filtered to remove those not corresponding to HGNC gene symbols, read sum lower than 10 across all samples or matching antisense, LINC or pseudogenes. Core analysis of the cleaned RNA-sequencing data set is based on the DESeq2 package (v1.20.0) in R and potential confounding effects are taken into account by incorporating gender and batch of library preparation into the DESeq2 object design matrix. Genes were called differentially expressed if reaching an adjusted p value (padj) < 0.05 after multi-testing correction after Benjamini-Hochberg. Functional annotation of genes and enrichment of ontology terms associated with biological processes was performed within the clusterProfiler (v3.8.1) package. Enrichment analysis of gene sets derived from the Molecular Signatures Database v6.2 using the hallmark gene set collection (“H”) or gene sets based on conserved cis-regulatory motifs (“C3”) was performed with the fGSEA (v1.6) package. Gene sets containing more than 15 and less than 700 genes were included and 10,000 permutations were performed. PCA using the top 3000 most variable genes were visualized with ggplot2 (v3.0). Expression heatmaps are visualized with pheatmap (v1.0.10). The analysis assesses differences between the moment before (D0, n=14) and 3 months after BCG vaccination (D90, n=15) within the vaccinated individuals. The data from the placebo-treated group were used for calibration of the data, to exclude any effects induced in the bone marrow by the first aspiration.
ATAC-sequencing Data Preprocessing and Analysis
Sequencing was performed paired end 2∗50 bases on a HiSeq1500 instrument using TruSeq SBS v3-HS chemistry and data was demultiplexed using bcl2fastq2 v2.20. For pre-processing, reads were aligned to human hg19 index with bowtie1 and duplicates were filtered by applying the Picard tool. Removal of adapter offset and creation of bam files was done with samtools. Open chromatin peaks were called using MACS2, blacklisted regions (ENCFF001TDO_hg19_blacklist.bed.gz) were excluded and peaks were annotated using HOMER (applying annotatePeaks.pl). Downstream analysis was performed within the DESeq2 (v1.20.0) package environment. PCA was performed including all detected peaks and differentially accessible (DA) peaks were defined as baseMean > 5 and p value < 0.05. For detection of a core set of DA peaks multiple testing correction was included (Benjamini Hochberg) and FC ≥ 2 required. Coverage of open chromatin peaks were visualized with Gviz in R.
Training Quantitative Trait Loci (QTL) Mapping In Vitro Trained Immunity - 200FG Cohort
The study was performed in a cohort of ∼200 healthy individuals of Western European ancestry from the 200FG Human Functional Genomics Project (77% males and 23% females, age range 23-73 years old). The length of the box in the box plot is interquartile range (Q3–Q1). The whiskers indicate the range of 1.5 × the length of the box from either end of the box. P values were obtained using linear regression analysis of cytokine on genotype data.
Training QTL Mapping
Both genotype and data on trained immunity responses was obtained for a total of 141 individuals. Age and gender were included as co-variables in the linear model to correct for the cytokine distributions for QTL mapping. First, raw cytokine levels were log-transformed and ratios of cytokine production in primed (BCG or B-glucan) vs. nonprimed cells (RPMI control) were taken as the change of cytokine levels. The cytokine changes were mapped to genotype data using a linear regression model with age and gender as covariates. SNPs in the vicinity of the genes of interest (±250kb) were investigated.
Comparison of Neutrophil Count in BCG-Vaccinated vs BCG-Naïve Infants in the Melbourne Infant Study: BCG for Infection & Allergy Reduction
Participants were a subset of neonates from The Melbourne Infant Study: BCG for Allergy and Infection Reduction (MIS BAIR) (clinical trials registration NCT01906853). The trial was approved by the human ethics research committees of the Royal Children’s Hospital Melbourne (RCH) (HREC 33025) and Mercy Hospital for Women (HREC R12/28). This randomised controlled trial comprises 1244 healthy participants recruited antenatally between 2013 and 2016 to investigate whether BCG vaccination (0.05mL intradermal) BCG-Denmark, SSI) at birth protects against childhood infection, allergy and asthma. All participants received hepatitis B vaccine at birth according to Australian guidelines. Inclusion criteria in MIS BAIR were: greater than 32 weeks gestation, birth weight greater than 1500 grams, mother not HIV positive, absence of symptoms or signs of illness, and no known contact with TB. From the subset of participants whose parent/guardian provided consent, a blood sample was obtained by venapuncture for a full blood examination (FBE) in the neonatal period. Blood samples were analysed on an Advia 2120i (Siemens Healthineers, Germany). Those participants who had a FBE taken after randomisation were included in the study.
Statistical Analysis of Data from MIS BAIR Study
Hemoglobin, mean cell volume, white cell count, neutrophils, lymphocytes, monocytes, neutrophil-to-lymphocyte ratio and platelets were compared between the BCG-vaccinated and BCG-naïve groups using a univariate regression analysis. To identify variables to include in a multivariate regression analysis, the effect of sex, gestational age, birth method, birth weight, age at FBE, and time between randomization and FBE as potential confounders on each FBE parameter was assessed using bivariate regressions. Age at FBE, and time between randomization and FBE were found to be related to all outcomes. As these two variables are highly correlated, only age at FBE was included in the multivariate analysis to adjust for this potential confounder. In the multivariate analysis, only the mean neutrophil count differed between the groups. The adjusted difference in the mean neutrophil count and 9 5% confidence interval (CI) between the BCG-vaccinated and BCG-naïve groups were calculated. This was repeated with the participants stratified into the following groups: 0-12 hours, 12-24 hours, 24-36 hours, 36-48 hours, and greater than 48 hours between randomization and FBE.
Additional Resources
The clinical trial registration number for the Melbourne Infant Study: BCG for Allergy and Infection Reduction (MIS BAIR) is NCT01906853 (ClinicalTrials.gov).
Acknowledgments
This study was supported by an ERC advanced grant (#833247) and a Spinoza grant of the Netherlands Organization for Scientific Research (to M.G.N.). Additionally, this study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC2151 – 390873048 (to M.G.N., A.S., and J.L.S.), and an Emmy Noether research grant (SCHL2116/1 to A.S.). CVIVA is established with a grant from the Danish National Research Foundation (DNRF108). Additionally, this project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no 733100 (to J.L.S.).
Author Contributions
Conceptualization, M.G.N., A.S., L.C.J.d.B., B.C., and B.A.B.; Formal Analysis and Investigation, B.C., L.C.J.d.B., L.G., M.E.J.B., S.P., K.H., K.K., M.O., and Y.L.; Writing, M.G.N., A.S., B.C., and L.C.J.d.B.; Supervision, M.G.N., A.S., C.S.B., W.J.F.M.v.d.V., R.v.C., J.L.S., L.A.B.J., and N.C.
Declaration of Interests
M.G.N. and L.A.B.J. are scientific founders of Trained Therapeutics Discovery. All other authors declare no competing interests.
Published: June 15, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.05.014.
Contributor Information
Mihai G. Netea, Email: mihai.netea@radboudumc.nl.
Andreas Schlitzer, Email: andreas.schlitzer@uni-bonn.de.
Supplemental Information
References
- Aaby P., Roth A., Ravn H., Napirna B.M., Rodrigues A., Lisse I.M., Stensballe L., Diness B.R., Lausch K.R., Lund N. Randomized trial of BCG vaccination at birth to low-birth-weight children: beneficial nonspecific effects in the neonatal period? J. Infect. Dis. 2011;204:245–252. doi: 10.1093/infdis/jir240. [DOI] [PubMed] [Google Scholar]
- Armendariz A.D., Krauss R.M. Hepatic nuclear factor 1-alpha: inflammation, genetics, and atherosclerosis. Curr. Opin. Lipidol. 2009;20:106–111. doi: 10.1097/mol.0b013e3283295ee9. [DOI] [PubMed] [Google Scholar]
- Arts R.J.W., Moorlag S.J.C.F.M., Novakovic B., Li Y., Wang S.Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100.e5. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S.B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A.S. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Bankoti R., Ogawa C., Nguyen T., Emadi L., Couse M., Salehi S., Fan X., Dhall D., Wang Y., Brown J. Differential regulation of effector and regulatory T cell function by Blimp1. Sci. Rep. 2017;7:12078. doi: 10.1038/s41598-017-12171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bekkering S., Blok B.A., Joosten L.A., Riksen N.P., van Crevel R., Netea M.G. In vitro experimental model of trained innate immunity in human primary monocytes. Clin. Vaccine Immunol. 2016;23:926–933. doi: 10.1128/CVI.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., Arts R.J.W., Novakovic B., Kourtzelis I., van der Heijden C.D.C.C., Li Y., Popa C.D., Ter Horst R., van Tuijl J., Netea-Maier R.T. Metabolic induction of trained immunity through the mevalonate pathway. Cell. 2018;172:135–146.e9. doi: 10.1016/j.cell.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Benn C.S., Netea M.G., Selin L.K., Aaby P. A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends Immunol. 2013;34:431–439. doi: 10.1016/j.it.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Biering-Sørensen S., Aaby P., Lund N., Monteiro I., Jensen K.J., Eriksen H.B., Schaltz-Buchholzer F., Jørgensen A.S.P., Rodrigues A., Fisker A.B., Benn C.S. Early BCG-Denmark and neonatal mortality among infants weighing <2500 g: a randomized controlled trial. Clin. Infect. Dis. 2017;65:1183–1190’. doi: 10.1093/cid/cix525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A., Günther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Baßler K. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172:162–175.e14. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I.A., Allison A.C., Cox F.E. Protection of mice against Babesia and Plasmodium with BCG. Nature. 1976;259:309–311. doi: 10.1038/259309a0. [DOI] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crișan T.O., Netea M.G., Joosten L.A. Innate immune memory: implications for host responses to damage-associated molecular patterns. Eur. J. Immunol. 2016;46:817–828. doi: 10.1002/eji.201545497. [DOI] [PubMed] [Google Scholar]
- Freyne B., Marchant A., Curtis N. BCG-associated heterologous immunity, a historical perspective: intervention studies in animal models of infectious diseases. Trans. R. Soc. Trop. Med. Hyg. 2015;109:52–61. doi: 10.1093/trstmh/tru197. [DOI] [PubMed] [Google Scholar]
- Gong Y., Ma Z., Patel V., Fischer E., Hiesberger T., Pontoglio M., Igarashi P. HNF-1beta regulates transcription of the PKD modifier gene Kif12. J. Am. Soc. Nephrol. 2009;20:41–47. doi: 10.1681/ASN.2008020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.P.T., Soares-Weiser K., López-López J.A., Kakourou A., Chaplin K., Christensen H., Martin N.K., Sterne J.A.C., Reingold A.L. Association of BCG, DTP, and measles containing vaccines with childhood mortality: systematic review. BMJ (Clin. Res. Ed.) 2016;355:i5170. doi: 10.1136/bmj.i5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaensson E., Uronen-Hansson H., Pabst O., Eksteen B., Tian J., Coombes J.L., Berg P.L., Davidsson T., Powrie F., Johansson-Lindbom B., Agace W.W. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med. 2008;205:2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.J., Larsen N., Biering-Sørensen S., Andersen A., Eriksen H.B., Monteiro I., Hougaard D., Aaby P., Netea M.G., Flanagan K.L., Benn C.S. Heterologous immunological effects of early BCG vaccination in low-birth-weight infants in Guinea-Bissau: a randomized-controlled trial. J. Infect. Dis. 2015;211:956–967. doi: 10.1093/infdis/jiu508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic S., Linge H.M., Shikhagaie M.M., Olin A.I., Lannefors L., Erjefält J.S., Mörgelin M., Egesten A. The neutrophil-recruiting chemokine GCP-2/CXCL6 is expressed in cystic fibrosis airways and retains its functional properties after binding to extracellular DNA. Mucosal Immunol. 2016;9:112–123. doi: 10.1038/mi.2015.43. [DOI] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonça L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172:176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A.B., Ifrim D.C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H.G. Bacille calmette-guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Benn C.S., Joosten L.A.B., Jacobs C., van Loenhout J., Xavier R.J., Aaby P., van der Meer J.W.M. Long-lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J. Innate Immun. 2014;6:152–158. doi: 10.1159/000355628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge H.M., Collin M., Nordenfelt P., Mörgelin M., Malmsten M., Egesten A. The human CXC chemokine granulocyte chemotactic protein 2 (GCP-2)/CXCL6 possesses membrane-disrupting properties and is antibacterial. Antimicrob. Agents Chemother. 2008;52:2599–2607. doi: 10.1128/AAC.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv. 1802 arXiv:1802.03426v2. [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 2018;172:147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes-Vieira P.M., Yore M.M., Dwyer P.M., Syed I., Aryal P., Kahn B.B. RBP4 activates antigen-presenting cells, leading to adipose tissue inflammation and systemic insulin resistance. Cell Metab. 2014;19:512–526. doi: 10.1016/j.cmet.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.R. Host defenses in murine malaria: nonspecific resistance to Plasmodium berghei generated in response to Mycobacterium bovis infection or Corynebacterium parvum stimulation. Infect. Immun. 1981;33:199–211. doi: 10.1128/iai.33.1.199-211.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Quintin J., van der Meer J.W.M. Trained immunity: A memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G., O’Neill L.A., Xavier R.J. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., van der Meer J.W.M. Hypothesis: stimulation of trained immunity as adjunctive immunotherapy in cancer. J. Leukoc. Biol. 2017;102:1323–1332. doi: 10.1189/jlb.5RI0217-064RR. [DOI] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Rasmussen A.L., Wang I.M., Shuhart M.C., Proll S.C., He Y., Cristescu R., Roberts C., Carter V.S., Williams C.M., Diamond D.L. Chronic immune activation is a distinguishing feature of liver and PBMC gene signatures from HCV/HIV coinfected patients and may contribute to hepatic fibrogenesis. Virology. 2012;430:43–52. doi: 10.1016/j.virol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann A., Villumsen M., Sørup S., Haugaard L.K., Ravn H., Roth A., Baker J.L., Benn C.S., Aaby P. Vaccinations against smallpox and tuberculosis are associated with better long-term survival: a Danish case-cohort study 1971-2010. Int. J. Epidemiol. 2017;46:695–705. doi: 10.1093/ije/dyw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollini P., Fournier R.E. The HNF-4/HNF-1alpha transactivation cascade regulates gene activity and chromatin structure of the human serine protease inhibitor gene cluster at 14q32.1. Proc. Natl. Acad. Sci. USA. 1999;96:10308–10313. doi: 10.1073/pnas.96.18.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S., Quintin J., Kerstens H.H.D., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.C., Ratter J., Berentsen K., van der Ent M.A. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J., Schmidt S.V., Cirovic B., McGovern N., Papantonopoulou O., Hardt A.L., Aschenbrenner A.C., Kreer C., Quast T., Xu A.M. Cellular differentiation of human monocytes is regulated by time-dependent interleukin-4 signaling and the transcriptional regulator NCOR2. Immunity. 2017;47:1051–1066.e12. doi: 10.1016/j.immuni.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelkoul P.H., Catsburg A., Mulder S., Oostendorp L., Schirm J., Wilke H., van der Zanden A.G., Noordhoek G.T. Detection of Mycobacterium tuberculosis complex with real time PCR: comparison of different primer-probe sets based on the IS6110 element. J. Microbiol. Methods. 2006;66:177–180. doi: 10.1016/j.mimet.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Siddiqui K.R.R., Powrie F. CD103+ GALT DCs promote Foxp3+ regulatory T cells. Mucosal Immunol. 2008;1:S34–S38. doi: 10.1038/mi.2008.43. [DOI] [PubMed] [Google Scholar]
- Storgaard L., Rodrigues A., Martins C., Nielsen B.U., Ravn H., Benn C.S., Aaby P., Fisker A.B. Development of BCG scar and subsequent morbidity and mortality in rural Guinea-Bissau. Clin. Infect. Dis. 2015;61:950–959. doi: 10.1093/cid/civ452. [DOI] [PubMed] [Google Scholar]
- Sun J.C., Lopez-Verges S., Kim C.C., DeRisi J.L., Lanier L.L. NK cells and immune “memory”. J. Immunol. 2011;186:1891–1897. doi: 10.4049/jimmunol.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Horst R., Jaeger M., Smeekens S.P., Oosting M., Swertz M.A., Li Y., Kumar V., Diavatopoulos D.A., Jansen A.F.M., Lemmers H. Host and environmental factors influencing individual human cytokine responses. Cell. 2016;167:1111–1124.e13. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Mildner A., Jung S. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- Villumsen M., Sørup S., Jess T., Ravn H., Relander T., Baker J.L., Benn C.S., Sørensen T.I.A., Aaby P., Roth A. Risk of lymphoma and leukaemia after Bacille Calmette-Guérin and smallpox vaccination: a Danish case-cohort study. Vaccine. 2009;27:6950–6958. doi: 10.1016/j.vaccine.2009.08.103. [DOI] [PubMed] [Google Scholar]
- Walk J., de Bree L.C.J., Graumans W., Stoter R., van Gemert G.J., van de Vegte-Bolmer M., Teelen K., Hermsen C.C., Arts R.J.W., Behet M.C. Outcomes of controlled human malaria infection after BCG vaccination. Nat. Commun. 2019;10:874. doi: 10.1038/s41467-019-08659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Finn A., Curtis N. Does BCG vaccination protect against nontuberculous mycobacterial infection? A systematic review and meta-analysis. J. Infect. Dis. 2018;218:679–687. doi: 10.1093/infdis/jiy207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing and ATAC data is available under GEO: GSE124220. Human functional Genomics data is available through https://hfgp.bbmri.nl (Ter Horst et al., 2016). Scripts used to analyze these data will be shared upon request.