Highlights
-
•
For IgG detection >14 days after symptoms onset was 100.0 % for all assays.
-
•
Specificity for IgG was greater than 98 % for CLIA and LFIA compared to ELISA.
-
•
LFIA (NG-Test®) is reliable and accurate to diagnose SARS-CoV-2 infection.
-
•
Best agreement was observed between CLIA and LFIA assays (97 %; k = 0.936).
Keywords: SARS-CoV-2, COVID-19, Performance, Automated immunoassays, Lateral flow immunoassay
Abstract
Background
The emergence of new SARS-CoV-2 has promoted the development of new serological tests that could be complementary to RT-PCR. Nevertheless, the assessment of clinical performances of available tests is urgently required as their use has just been initiated for diagnose.
Objectives
The aim of this study was to assess the performance of three immunoassays for the detection of SARS-CoV-2 antibodies.
Methods
Two automated immunoassays (Abbott SARS-CoV-2 CLIA IgG and Euroimmun Anti-SARS-CoV-2 ELISA IgG/IgA assays) and one lateral flow immunoassay (LFIA NG-Test® IgG-IgM COVID-19) were tested. 293 specimens were analyzed from patients with a positive RT-PCR response, from patients with symptoms consistent with COVID-19 but exhibiting a negative response to the RT-PCR detection test, and from control group specimens. Days since symptoms onset were collected from clinical information sheet associated with respiratory tract samples.
Results
Overall sensitivity for IgG was equivalent (around 80 %) for CLIA, ELISA and LFIA. Sensitivity for IgG detection, >14 days after onset of symptoms, was 100.0 % for all assays. Overall specificity for IgG was greater for CLIA and LFIA (more than 98 %) compared to ELISA (95.8 %). Specificity was significantly different between IgA ELISA (78.9 %) and IgM LFIA (95.8 %) (p < 0.05). The best agreement was observed between CLIA and LFIA assays (97 %; k = 0.936).
Conclusion
Excellent sensitivity for IgG detection was obtained >14 days after onset of symptoms for all immunoassays. Specificity was also excellent for IgG CLIA and IgG LFIA. Our study shows that NG-Test® is reliable and accurate for routine use in clinical laboratories.
1. Background
A new acute respiratory syndrome named coronavirus disease 2019 (COVID-19) has emerged from the region of Wuhan in China in December 2019. This infection, widespread all over the world, is caused by a novel Sarbecovirus designated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), associated with severe morbidity and mortality [[1], [2], [3]]. The detection of viral RNA by real time reverse transcriptase-Polymerase chain reaction (RT-PCR) in respiratory tract samples is considered as the gold standard method for screening and diagnosis in the early phase of infection. However, sensitivity is variable depending on sample types, suitable sampling technique, the anatomic site, time of infection and viral load [[4], [5], [6]]. Chest computed tomography (CT) may be helpful for the diagnosis, complementary to RT-PCR, but it remains unspecific [7]. Development of new serological tests [8,9], readily available and easier to perform compared to requirements of molecular assays in laboratories [10], could be helpful as a complementary diagnostic tool and to increase the sensitivity of tests especially in patients with late complications i.e. severe pneumonia. Different assays have recently been commercialized: automated tests (enzyme-linked immunosorbent assays [ELISA] or chemiluminescence enzyme immunoassays [CLIA]) or rapid detection test (lateral flow immunoassays, LFIA). LFIA seems to be very attractive for large seroprevalence studies because these tests can be used easily as point of care tests or in the laboratory, with a result in less than 15 min. Serological tests can be used for symptomatic individuals for which RT-PCR testing was either not performed at the time of acute illness or for which nasopharyngeal swab result was found to be negative, and also for epidemiological studies (close contacts screening, screening of health care workers …) [11,12]. However, the relevance of serological tests is highly related to their clinical performance, hence antibody (Ab) assays with good sensitivity and specificity are needed. Despite a growing number of available assays, related clinical performances are still scarce [[13], [14], [15], [16], [17]] or unknown and individual studies are usually inconclusive. Moreover, the quality and diagnostic performance of rapid tests have already been questioned in Spain and United Kingdom [18,19].
2. Objectives
The aim of the study was to assess the clinical performance of CE marked assays available in Europe to detect SARS-CoV-2 antibodies: two automated immunoassays (Euroimmun and Abbott assays) targeting two different proteins and also one lateral flow immunoassay (NG Biotech).
3. Methods
3.1. Specimens
This retrospective study included 293 residual sera from patients with RT-PCR confirmed SARS-CoV-2 infection, patients with symptoms consistent with COVID-19 but with a negative RT-PCR result (clinical diagnosis of pneumonia of unknown etiology), and control individuals (presumed negative). These samples were collected in the virology laboratory of Angers University Hospital, France. Serum samples (n = 141) obtained from 82 patients (median age: 67 years) with confirmed COVID-19 by RT-PCR, performed in our laboratory [20], were tested. 57 serum specimens obtained from 52 patients (median age: 64 years) with symptoms consistent with COVID-19, but with negative RT-PCR results were analyzed. Information about days since symptoms onset was determined by clinical information sheet associated with respiratory tract samples. 50 residual serum samples presumed negative collected before the emergence of SARS-CoV-2, in March 2019 and stored at −80 °C were used as control specimens.
Then, 25 serum samples with a potential cross-reaction to the SARS-CoV-2 immunoassays were investigated (Table 1 ). Samples from 10 pregnant women and 10 sera from patients with positive rheumatoid factor (RF) were also tested.
Table 1.
Selected specimens potentially containing cross-reacting antibodies with SARS-CoV-2.
| Pathogen potentially cross-reactive with SARS-CoV-2 | Number of specimens |
|---|---|
| Seasonal coronaviruses (HKU1, NL63, 229E, OC43) | 2 |
| Influenza A virus | 3 |
| Respiratory Syncitial Virus | 3 |
| Rhinovirus | 3 |
| Parainfluenzae virus | 1 |
| Acute EBV infection (positive for EBV VCA IgM and EBV VCA IgG) | 7 |
| Acute CMV infection (positive for CMV IgM) | 1 |
| M. pneumonia infection | 2 |
| Acute Hepatitis A infection | 1 |
| Acute hepatitis E infection | 2 |
The study was approved by the Institutional Board of the Angers University Hospital.
3.2. Serological assays
3.2.1. ELISA assay
The Euroimmun Anti-SARS-CoV-2 ELISA IgG and IgA assays (Euroimmun, Lüebeck, Germany) were performed according to the manufacturer’s guidelines on the DS2® system, an automated microplate technology (Dynex Technologies GmbH, Denkendorf, Germany). The microplate wells are coated with recombinant S1 structural protein and the assay detects anti-SARS-CoV-2 IgG and IgA against the viral spike protein (Sp).
3.2.2. CLIA assay
The Abbott SARS-CoV-2 IgG (Abbott Diagnostics, IL, USA) was performed according to the manufacturer’s instructions on the automated Abbott ARCHITECT i2000SR Instrument.
The assay is a CLIA for qualitative detection of IgG antibodies against the SARS-CoV-2 nucleoprotein (Np) in serum or plasma.
3.2.3. Lateral flow test
NG-Test® IgG-IgM COVID-19 (NG Biotech Laboratoires, Guipry-Messac, France) is an immune colloidal technique intended for the qualitative detection of IgG and IgM antibodies against the SARS-CoV-2 nucleoprotein in serum or plasma. 10 μL of specimen, were added onto the sample loading area followed by two drops of sample dilution solution. The results were read and interpreted 15 min after testing.
3.3. Statistical analysis
All statistical analyses were performed using IBM® SPSS® 15.0 Statistics software (Statistical Package for Social Sciences, IBM Corp., Chicago, IL). To assess the sensitivity and specificity, we choose the RT-PCR method as gold standard. Time from onset symptoms was used to determine sensitivity and specificity. Grey zone was considered positive for the statistical analyses. A p value <0.05 was considered statistically significant. The Cohen’s Kappa value was determined for agreement between assays.
4. Results
Sensitivities and specificities obtained with three immunoassays are summarized in Table 2 . The sensitivity of IgG ELISA at ≤7 days of symptoms was 28.1 %, at 8–14 days 72.4 %, and >14 days was 100.0 %. The sensitivity of IgG CLIA at ≤7 days of symptoms was 46.9 %, at 8–14 days 69 %, and was 100.0 % >14 days. Sensitivity of IgG LFIA at ≤7 days of symptoms was 31.3 %, at 8–14 days 69.0 %, and was 100.0 % >14 days. Overall sensitivity for IgG was equivalent (around 80 %) for CLIA, ELISA and LFIA. Overall specificity for IgG was greater than 98 % for CLIA and LFIA compared to ELISA (95.8 %). Comparison of the sensitivity of IgA ELISA (59.4 %) and IgM LFIA (43.8 %), during the first seven days after onset of symptoms, was not significant (p > 0.05). By contrast, specificity was significantly different between IgA ELISA and IgM LFIA (p < 0.05).
Table 2.
Sensitivities of immunoassays for SARS-CoV-2 according to the onset of COVID-19 symptoms and specificities data. CI: confidence interval.
| Overall % (CI 95%) | Time from the symptom onset |
|||
|---|---|---|---|---|
| 0 to 7 days % (CI 95%) | 8 to 14 days % (CI 95%) | 15 or more days % (CI 95%) | ||
| ELISA assay | ||||
| IgG or IgA | Se: 87.4 (81.0-91.9%) | Se: 59.4 (42.3-74.5%) | Se: 82.8 (65.5-92.4%) | Se: 100.0 (95.5-100.0%) |
| Sp: 82.0 (75.1-87.3%) | ||||
| IgG | Se: 78.3 (70.9-84.3%) | Se: 28.1 (42.3-74.5%) | Se: 72.4 (54.3-85.3%) | Se:100.0 (95.5-100.0%) |
| Sp: 96.7 (92.4-98.6% | ||||
| IgA | Se: 86.7 (80.2-91.3%) | Se: 59.4 (15.6-45.4%) | Se: 79.3 (61.6-90.2%) | Se: 100.0 (95.5-100.0%) |
| Sp: 82.7 (75.8-87.9%) | ||||
| CLIA assay | ||||
| IgG | Se: 81.8 (74.7-87.3%) | Se: 46.9 | Se: 69.0 | Se: 100.0 |
| Sp: 99.3 (96.3-99.9%) | (30.9-63.6%) | (50.8-82.7%) | (95.5-100.0%) | |
| Lateral flow immunoassay | ||||
| IgG or IgM | Se: 81.8 (74.7-87.3%) | Se: 43.8 (28.2-60.7%) | Se: 72.4 (54.3-85.3%) | Se: 100.0 (95.5-100.0%) |
| Sp: 95.3 (90.7-97.7%) | ||||
| IgG | Se: 78.3 (70.9-84.3%) | Se: 31.3 (18.0-48.6%) | Se: 69.0 (50.8-82.7%) | Se: 100.0 (95.5-100.0%) |
| Sp: 98.0 (94.3-99.3%) | ||||
| IgM | Se: 81.8 (74.7-87.3%) | Se: 43.8 (28.2-60.7%) | Se: 72.4 (54.3-85.3%) | Se: 100.0 (95.5-100.0%) |
| Sp: 95.3 (90.7-97.7%) | ||||
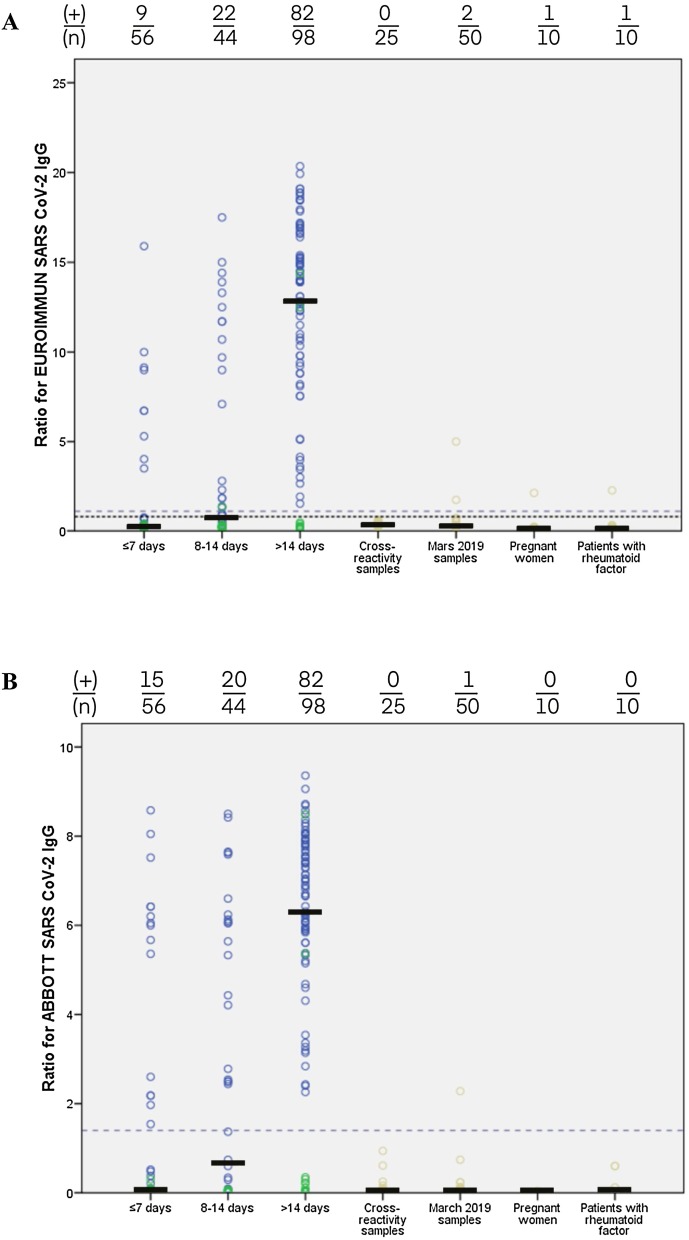
Among the control samples and the group of patients with negative RT-PCR, 26 false positives were observed with IgA ELISA (17.3 %): seven specimens from the cross-reactivity study; seven from pre-epidemic specimens (March 2019); two from pregnant women; four from patients with RF; six from patients with negative SARS-CoV-2 RT-PCR and symptoms of pneumonia/dyspnea without a chest CT argument for COVID-19 or seroconversion (median time between symptom onset and sera: 9.5 days). Fewer false positives were observed with IgM LFIA: three specimens from the cross-reactivity study; one from pre-epidemic sera; three from patients with negative RT-PCR result and symptoms of pneumonia/dyspnea without a chest CT argument (including two specimens from the same patient). Five false positives were observed with IgG ELISA (Fig. 1 ): two pre-pandemic specimens, one sample from pregnant woman, one sample from a patient with RF and one with negative RT-PCR result (negative result with other assays). Only one false positive result was observed with IgG CLIA and corresponded to a pre-pandemic specimen (Fig. 1). Using IgG LFIA, three false positives were observed; two were from a patient (negative RT-PCR) for whom the etiology of pneumonia was undetermined.
Fig. 1.
Seropositivity of tested specimens with ELISA Euroimmun and CLIA Abbott assays. Seropositivity analysis in 95 presumed negatives control samples (cross-reactivity samples, march 2019 samples, pregnant women samples, patients with RF samples), 57 samples from 52 patients with RT-PCR negative relative to days from symptom onset (≤7 days; 8-14 days; >14 days) and 141 samples from 82 patients with RT-PCR positive relative to days from symptom onset. Blue circles correspond to sera from patients exhibiting a positive RT-PCR result. Green circles correspond to sera from patients with negative RT-PCR result. Ochre circles correspond to sera from individuals for whom RT-PCR detection has not been performed. The black line represents the median of ratio. (+): number of seropositive sera; (n): total number of specimens tested. A) Seropositivity with ELISA Euroimmun assay. Dashed grey line represents cutoff for positivity (ratio ≥1.1). Dotted purple line corresponds to cutoff for negativity (ratio <0.8). B) Seropositivity with CLIA Abbott assay. Dashed grey line represents cutoff for positivity (ratio ≥1.4). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
All patients with positive RT-PCR were positive for serological assays. Two discordant results between serologic assays and molecular method were reported: two patients clinically adjudicated as COVID-19 but with negative RT-PCR. These two samples were tested positive for all immunoassays (Fig. 1). To determine the specificity for IgG of the three assays, we excluded two specimens positive for serological assays but negative for RT-PCR because the symptoms were strongly compatible with the COVID-19 and RT-PCR was performed 17–24 days after symptom onset.
Among patients with RT-PCR confirmed SARS-CoV-2 infection, there were only two individuals without COVID-19 symptoms, but with a notion of contact with infected patients. Both SARS-CoV-2 RT-PCR and serological assays were positives for these patients.
Table 3 summarized overall agreement and agreement relative to the time of symptoms onset between three immunoassays. Overall, excellent agreement was observed between the three assays. The best agreement was observed between CLIA and LFIA (97 %; Cohen kappa index of 0.936). Even for the first week of symptoms onset, an excellent agreement was observed between ELISA and LFIA assays (95 %; k = 0.810). However, poor agreement was observed between ELISA and CLIA (89 %; k = 0.687). Overall agreement between IgG/IgA ELISA and IgG/IgM LFIA was excellent (96 %; k = 0.914).
Table 3.
Agreement between IgG serological assays.
| Euroimmun % (Kappa) | NG Biotech % (Kappa) | ||
|---|---|---|---|
| Overall | Abbott | 96 % (0.908) | 97 % (0.936) |
| NG Biotech | 96 % (0.914) | ||
| 0 to 7 days | Abbott | 89 % (0.687) | 91 % (0.745) |
| NG Biotech | 95 % (0.810) | ||
| 8 to 14 days | Abbott | 95 % (0.909) | 98 % (0.954) |
| NG Biotech | 93 % (0.864) | ||
| 15 or more days | Abbott | 100 % (1.000) | 99 % (0.962) |
| NG Biotech | 99 % (0.962) |
The IgA, IgM and IgG Ab kinetics were studied using specimens from seven patients (positive RT-PCR) with serial results and interesting kinetics (Fig. 2 ). Then, five patients presented an earlier IgG seroconversion using CLIA compared to ELISA, the first week of symptom onset. Among these patients, we observed in three patients an IgM line with LFIA and IgA ELISA was positive for four patients.
Fig. 2.
Anti-SARS-CoV-2 antibodies seroconversion profiles for seven individuals. X-axis: time from symptoms onset. Y-axis: interpretation ratio of two semi-quantitative immunoassays. Dotted black line represents the day of positivity of LFIA NG-Test® IgG-IgM COVID-19. The cutoff for positivity with ELISA Euroimmun assay is ≥1.1 (dotted blue line) and the cutoff for positivity with CLIA Abbott assay is ≥1.4 (dotted red line). Patients 1, 2, 4 and 6 developed a prolonged immune response one month after symptoms onset and up 64 days for patient 4. Patients 1, 2 and 7 had early seroconversion in the second week after symptoms onset and patient 5 had already seroconversion in the first week. Patient 3 had a seroconversion in the third week after symptoms onset. Patient 5 produced fewer antibodies compared to other patients and notably IgA production is close to the threshold of positivity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Using LFIA, results were more easily interpretable for IgG line than for IgM line. IgM line was difficult for reading, notably for seven sera.
5. Discussion
A strong clinical performance of assays in diagnosis and management of COVID-19 is essential to quickly contain the COVID outbreak worldwide. Therefore, the development of serological assays, routinely used in clinical laboratories to determine recent infection or previous contact with viruses, is a good option complementary to RT-PCR method [21]. On May 2020, the French Health Authority (Haute Autorité de Santé) and Infectious Diseases Society of America recommended that patients with symptoms consistent with COVID-19 but having a negative result for SARS-CoV-2 by RT-PCR may be diagnosed by serological tests [22,23]. Various immunoassays are available on the European market [24,25] and subjected to European regulations with the mandatory CE marked for sales. Nevertheless, the European Commission, in its April 2020 recommendations, allowed exceptionally the marketing of tests that do not have the CE marked, in the interest of public health [22].
Here, we evaluated three different CE marked commercial immunoassays for detection of SARS-CoV-2 antibodies in human serum and plasma. ELISA assay was performed on a semi-automated microplate technology requiring high handling and with a limited capacity of tests per day (90 tests per 4 h). In contrast, CLIA assay is a fully automated random-access test and that can perform over 4000 tests per 24 h. These two assays are used in clinical laboratories, unlike LFIA, which can be used as a point of care test or in clinical laboratories and provides a result within 15 min.
Performance of Euroimmun assay has been evaluated in some studies [13,14,[26], [27], [28], [29]], showing sensitivity for IgG between 85 % and 95 % >14 days after symptoms onset and specificity between 95 % and 100 %. Few studies reported clinical performance of Abbott assay [14,16,26,28]. Sensitivity for IgG was between 94 % and 100 % more than 14 days post symptom onset and specificity between 99 % and 100 %. In our study, we showed a sensitivity for IgG of 100 % for CLIA Abbott and ELISA Euroimmun assays >14 days after symptoms onset and an overall specificity for IgG of 78.3 % and 81.8 % with ELISA and CLIA respectively. We carried out a large cross-reactivity study and more false positives results were observed using ELISA than CLIA as previously described [14].
Recently, many CE marked LFIA became available. Two studies showed that sensitivity and specificity were similar to those of Euroimmun assay [13,29]. However, to our knowledge, only one study compared clinical performance between CLIA Abbott and LFIA [30] and no study described diagnostic performance of NG-Test®. Here, we observed an excellent agreement for IgG between CLIA and LFIA 15 days after onset symptoms (k = 0.810), and an excellent sensitivity and specificity for both assays. LFIA advantages are the ability to reach larger population groups, when used in point-of-care, and to evaluate the herd immunity without saturating the capacity of laboratories. However, these devices must be used with caution. Trained staff or automated reader devices are needed for good interpretation of result. Traceability of results may be at fault in case of use at the point-of-care and results may not be reported to the health authorities for seroprevalence studies.
To evaluate sensitivity, some manufacturers or authors used the time from positive RT-PCR rather than the time from symptom onset. However, there is a risk of misestimating sensitivity as some patients presented late after the onset of symptoms with disease progression at time of the first PCR testing. Then, sensitivity and specificity must be interpreted with caution. The use of RT-PCR as gold standard may decrease the real number of patients infected by SARS-CoV-2 due to false negative results.
In our study, we observed false positive results with IgA ELISA and few with IgM LFIA. No false positive with IgM LFIA were observed with RF specimens whereas interferences were described with some other immunoassays [31]. Elslande et al. pointed out that the ELISA IgA should not be used for the screening of asymptomatic persons. It might be better not to measure IgM or IgA since it may result in a significant number of false-positive results without improving diagnostic performance. [29]. It would appear here that IgM detection with the LFIA provides a gain in diagnostic performance.
Developed immunoassays target either the Sp or the Np of SARS-CoV-2 [32], involving different immune Ab responses. However, related studies are controversial. Some studies described that early antibody response was targeted against Np and then Sp inducing an earlier positivity of the tests targeting Np [14,33]. By contrast, another study revealed that the Sp-based ELISA was more sensitive than the Np-based one in the detection of IgM [34]. Here, we did not observe any significant difference between sensitivity of IgA ELISA and IgM LFIA which target two different proteins. Positive results with serological tests do not indicate the presence of neutralizing antibodies, i.e. possible protective immunity to SARS-CoV-2 [35], but are only indicative of a contact with SARS-CoV-2.
One of the strong points of our study is the use of large number of samples for cross reactivities study with other pathogens and other causes of false positive results i.e. pregnancy and individuals with RF. Among symptomatic patients with positive RT-PCR, most of patients were elderly with a potential risk of low immune response.
Some limitations of the study are the subjectivity in the perception of symptoms by patients, in particular for elderly patient. Our study included few patients with asymptomatic infections and positive RT-PCR because specific indications of tests in France were, at the time of the study, mainly limited to patients who were symptomatic, due in particular to the shortage of tests.
In conclusion, our study showed equivalent clinical performance for IgG of three immunoassays (ELISA, CLIA and LFIA) >14 days after symptoms onset. The three assays had, as expected, a poor sensitivity during first days of symptom onset. Therefore, serological tests can be useful to confirm past COVID-19, to do epidemiologic studies 15 days after symptoms onset [36] or to identify people who could return to the workplace, even if its use is still widely discussed [37]. For asymptomatic patients with RT-PCR negative, a higher threshold must be used [16]. A lower threshold (8–14 days) should be used for symptomatic patients >7 days with negative RT-PCR and clinical presentation consistent with COVID-19. Currently, it is not clear whether IgG antibodies are protective against reinfection [38]. Finally, even if the LFIA is reliable on serum or plasma, studies should be conducted to evaluate the performance on fingerstick; a process commonly used for seroprevalence studies.
Funding
This research did not receive any specific grant from funding agencies. NG-Test® IgG-IgM COVID-19 rapid test cassettes (NG Biotech Laboratoires) were kindly provided by the manufacturer.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors thank the laboratory technicians who helped us. The authors thank Pr Thomas Guillemette for proofreading the English manuscript.
References
- 1.2020. World Health Organization, Coronavirus Disease 2019.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (Accessed 23 May 2020) [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. China novel coronavirus investigating and research team, a novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vashist S.K. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. 2020;10:202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J., Peng L., Wong G., Zheng H., Liao M., Feng K., Li J., Yang Q., Zhao J., Zhang Z., Liu L., Liu Y. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020 doi: 10.1101/2020.02.11.20021493. [DOI] [Google Scholar]
- 7.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;395:1101–1102. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerging Infect. Dis. 2020;26 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . 2020. Laboratory Biosafety Guidance Related to the Novel Coronavirus (2019-nCoV)https://www.who.int/docs/default-source/coronaviruse/laboratory-biosafety-novel-coronavirus-version-1-1.pdf?sfvrsn=912a9847_2 (Accessed 27 May 2020) [Google Scholar]
- 11.Xu Y., Xiao M., Liu X., Xu S., Du T., Xu J., Yang Q., Xu Y., Han Y., Li T., Zhu H., Wang M. Significance of serology testing to assist timely diagnosis of SARS-CoV-2 infections: implication from a family cluster. Emerg. Microbes Infect. 2020;9:924–927. doi: 10.1080/22221751.2020.1752610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montesinos I., Gruson D., Kabamba B., Dahma H., Van den Wijngaert S., Reza S., Carbone V., Vandenberg O., Gulbis B., Wolff F., Rodriguez-Villalobos H. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., Farnsworth C.W. Clinical performance of two SARS-CoV-2 serologic assays. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., Long X., Guo S., Zhao Z., Liu Y., Hu H., Xue H., Li Y. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A., Jerome K.R., Mathias P.C., Greninger A.L. Performance characteristics of the abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zainol Rashid Z., Othman S.N., Abdul Samat M.N., Ali U.K., Wong K.K. Diagnostic performance of COVID-19 serology assays. Malays. J. Pathol. 2020;42:13–21. [PubMed] [Google Scholar]
- 18.Sevillano E. 2020. Covid-19 In Spain: Unreliability of New Tests Delays Effort to Slow Coronavirus Spread in Spain | Society | EL PAÍS in English.https://english.elpais.com/society/2020-03-27/unreliability-of-new-tests-delays-effort-to-slow-coronavirus-spread-in-spain.html (Accessed 9 June 2020) [Google Scholar]
- 19.Brennan D. 2020. U.K. Says Millions of Coronavirus Test Kits Bought From China Are Unreliable for Most Patients, Newsweek.https://www.newsweek.com/uk-says-millions-coronavirus-test-kits-bought-china-unreliable-most-patients-1496506 (Accessed 9 June 2020) [Google Scholar]
- 20.Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.-C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.-F., Lina B., van-der-Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., Chen Y., Gu B. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microbes Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Autorité de Santé Haute. 2020. Place Des Tests Sérologiques Dans La Stratégie De Prise En Charge De La Maladie COVID-19; p. 37. [Google Scholar]
- 23.Infectious Diseases Society of America . 2020. IDSA COVID-19 Antibody Primer.https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-covid-19-antibody-testing-primer.pdf (Accessed 10 June 2020) [Google Scholar]
- 24.Ministère des Solidarités et de la Santé . 2020. Plateforme COVID-19.https://covid-19.sante.gouv.fr/tests (Accessed 25 May 2020) [Google Scholar]
- 25.2020. SARS-CoV-2 Diagnostic Pipeline.https://www.finddx.org/testing-matters/ (Accessed 9 June 2020) [Google Scholar]
- 26.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J. Med. Virol. 2020 doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theel E.S., Harring J., Hilgart H., Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E., Van Ranst M., Lagrou K., Vermeersch P. Diagnostic performance of 7 rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dellière S., Salmona M., Minier M., Gabassi A., Alanio A., Le Goff J., Delaugerre C., Chaix M.-L. Evaluation of COVID-19 IgG/IgM rapid test from orient gene biotech. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01233-20. JCM.01233-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q., Du Q., Guo B., Mu D., Lu X., Ma Q., Guo Y., Fang L., Zhang B., Zhang G., Guo X. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 33.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of SARS-CoV-2 IgG antibody tests and potential protective immunity. Microbiology. 2020 doi: 10.1101/2020.05.08.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter A.K., Hegde S.T. The important role of serology for COVID-19 control. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstein M.C., Freedberg K.A., Hyle E.P., Paltiel A.D. Waiting for certainty on Covid-19 antibody tests — at what cost? N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2017739. 2017739. [DOI] [PubMed] [Google Scholar]
- 38.Melgaço J.G., Azamor T., Ano Bom A.P.D. Protective immunity after COVID-19 has been questioned: what can we do without SARS-CoV-2-IgG detection? Cell. Immunol. 2020;353 doi: 10.1016/j.cellimm.2020.104114. [DOI] [PMC free article] [PubMed] [Google Scholar]