Abstract
In this review, we focused on the origins of the novel coronavirus (SARS-CoV-2), origin, pathogenesis, immune responses, genes and genetic variations, phylogenetic analyses, and potential therapeutic strategies to summarize approaches for developing broadly effective preventions and vaccines to cope COVID-19. Towards the end of 2019, SARS-CoV-2 has emerged in association with the SARS, later was named COVID-19 caused an environment of chaos worldwide and infected a massive number of lives. Since these epidemics or pandemics had spread to 210 countries and territories around the world and 2 international conveyances with 6,467,229 confirmed cases, including, 382,766 deaths, as of June 03, 2020 (https://www.worldometers.info/coronavirus/), hence the World Health Organization declared it as a global Public Health Emergency. There are no clinically approved vaccines or antiviral drugs available for either of new or old corona infections; thus, the development of effective therapeutic and preventive strategies that can be readily available to cope with these strains.
Keywords: SARS-CoV-2, Origin, Pathogenesis, Variations, Evolution, Phylogenetic analysis
Abbreviations: COVID-19, Coronavirus Disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WHO, World Health Organization; TGEV, transmissible gastroenteritis virus; BCoV, bovine coronavirus; IBV, avian infectious bronchitis viruses; ACE2, angiotensin receptor 2
1. Introduction
Before December 2019, very little was known about the coronaviruses (CoVs) because previously CoVs were not hit the global community so badly even though CoVs have been infected many species including humans and discussed for more than 70 years. Bailey et al. (1949) were reported murine virus (JHM) for the first time in 1949. Molecular mechanisms and pathogenesis of different CoVs have been intensely studied in different animal species such as porcine transmissible gastroenteritis virus (TGEV), bovine coronavirus (BCoV), and avian infectious bronchitis viruses (IBV). The CoVs having emergence and re-emergence history causing respiratory and intestinal infections in animals and humans (Masters and Perlman, 2013). This family of viruses remained unclear because they can only cause common cold symptoms in immune-competent individuals until the outbreak of severe acute respiratory syndrome (SARS) in 2003 (Weiss and Navas-Martin, 2005). Before the SARS epidemic in 2002–03, it was supposed that CoVs are not deadly pathogens to humans. Subsequently, a decade later in 2012, another Zoonotic highly pathogenic coronavirus emerged in Middle Eastern countries caused the Middle East respiratory syndrome (MERS) epidemic (Zaki et al., 2012). The ongoing SARS-CoV-2 pandemic situation got worldwide attention and became the utmost priority of the global health community due to the higher rate of human-human transmission. Previous studies have confirmed that SARS-CoV-2 adopted recombination and mutations strategies that help to adapt rapidly changing host environments through genotype adjustment via reproductive adaptability and ultimately escape from human immune reconnaissance (Rehman et al., 2020).
In a broader perspective, this review will describe the SARS-CoV-2 origins, epidemiology, pathogenesis, clinical manifestations, immune enhancement, genes and their variations, phylogenetic analyses, potential therapeutic strategies and future perspective to summarize approaches for developing broadly effective prevention of Corona Virus Disease-19 (COVID-19).
2. Origin
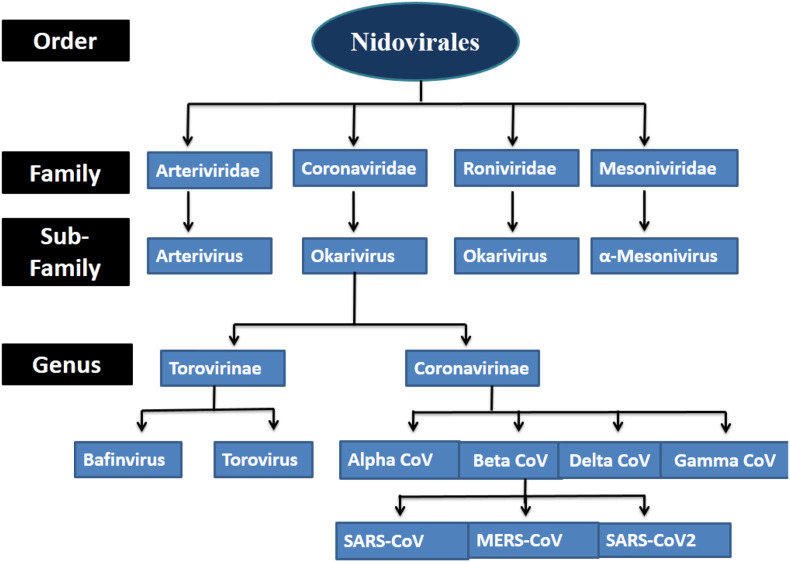
Coronaviridae family divided into two subfamilies, the coronaviruses, and the toroviruses. All identified CoVs are categorized into four different genera alpha coronaviruses, beta coronaviruses, gammacoronaviruses, and delta coronaviruses. The human infecting CoVs belongs to alpha coronaviruses and beta coronaviruses, whereas gammacoronaviruses, and delta coronaviruses showed susceptibility for fish and birds as shown in Fig. 1 .
Fig. 1.
Classification of coronaviruses.
All human infecting CoVs can cause mild to severe infection in humans and used spillover intermediate hosts (Fig. 2 ). Earlier to 2019, there were only six CoVs that were known to infect humans and cause respiratory diseases. Four out of six (HCoV-229E, HCoV-OC43, HCoV-NL63, and HKU1) human infecting CoVs can only cause mild upper respiratory disease, and in rare cases, some of them can cause severe infection in infants, young children, and elders (Fig. 2). Although, SARS-CoV and MERS-CoV which are zoonotic in origin can infect the lower respiratory tract and cause a severe respiratory syndrome in humans (Chang et al., 2006; N. Chen et al., 2020; Woo et al., 2012). Previously studied known pathogenic coronaviruses are presented in the table (Table 1 ).
Fig. 2.
Human infection-causing coronavirus and their origin.
Table 1.
Pathogenic coronaviruses, strain, host, and disease symptoms.
| Accession no. | Virus | Strain | Host | Symptoms |
|---|---|---|---|---|
| NC_002645.1 | Human CoV-229E | HCoV-229E | Human | Mild respiratory tract infections |
| NC_005831.2 | Human CoV-NL63 | HCoV-NL63 | Human | Mild respiratory tract infections |
| MH940245.1 | Human CoV-HKU1 | HCoV-HKU1 | Human | Pneumonia |
| KU131570.1 | Human coronavirus OC43 strain | Human CoV-OC43 | Human | Mild respiratory tract infections |
| AY345986 | Severe acute respiratory syndrome | SARS-CoV | Human | Severe acute respiratory syndrome,10% mortality rate |
| NC_045512.2 | Severe acute respiratory syndrome coronavirus 2 | SARS-CoV-2 | Human | Fever, Cough, Shortness of breath or difficulty breathing |
| MN988668.1 | Wuhan seafood market pneumonia virus isolate 2019-nCoV WHU01 | 2019-nCoV WHU0 | Human | Severe acute respiratory syndrome |
| MH734115.1 | Middle East respiratory syndrome coronavirus | MERS-CoV | Human | Severe acute respiratory syndrome,37% mortality rate |
| DQ811787.1 | Porcine respiratory coronavirus | PRCV/ISU-1 | Pig | Mild respiratory tract infections |
| NC_038861.1 | Transmissible gastroenteritis virus | TGEV/PUR46-MAD | Pig | Diarrhea,with100%mortalityinpiglets less than 2-week-old |
| KU558701 | Porcine epidemic diarrhea virus | PEDV/ZJU-G1-2013 | Pig | Severe watery diarrhea |
| KU558701 | Swine enteric alphacoronavirus | SeACoV-CH/GD-01 | Pig | Severe and acute diarrhea and acute vomiting |
| NTU336/F/2008 | Canine CoV | CCOV | Dog | Mild clinical signs, diarrhea |
| DQ848678.1 | Feline infectious peritonitis virus | FCoV C1Je | Cat | Fever, vasculitis, and serositis, with or without effusions |
| EF424623 | Bovine CoV/ENT | Bovine CoV/ENT | Cow | Diarrhea |
| AY394989 | Equine Severe acute respiratory syndrome | EquineCoV/Obihiro12-1 | Horse | Fever, anorexia, leucopenia |
| NC_001846.1 | Mouse hepatitis virus strain MHV-A59 C12 | MHV-A59 | Mouse | Acute pneumonia and severe lung injuries |
| NC_010646 | Beluga whale coronavirus SW1 | Beluga Whale CoV/SW1 | Whale | Pulmonary disease, terminal acute liver failure |
| M21883 | infectious bronchitis virus strain | IBV | Chicken | Severe respiratory disease |
| NC_011547.1 | Bulbul CoV HKU11-934 | HKU11-934 | Bulbul | Respiratory disease (collected from the respiratory tract of dead wild birds) |
| NC_009657 | Sparrow coronavirus HKU17 | HKU17 | Sparrow | Respiratory disease (collected from the respiratory tract of dead wild birds) |
| MG772934.1 | Bat SARS-like coronavirus | Bat-SL-CoVZXC21 | Bat | Fever, Cough, Shortness of breath or difficulty breathing |
| MG772933.1 | Bat SARS-like coronavirus | Bat-SL-CoVZC45 | Bat | Fever, Cough, Shortness of breath or difficulty breathing |
SARS and the MERSCoVs belong to beta coronaviruses have been characterized as they were transmitted from animals to humans and caused severe disease outbreaks in the past. SARS-CoV was transmitted in humans from bats via the intermediary host of palm civet cats (Fig. 2) in the Guangdong province of China and about 8422 infected cases with 916 deaths were recorded and the mortality rate was 11%. Furthermore, a decade later in 2012, a bat origin virus MERS-CoV epidemic was emerged in Saudi Arabia through the dromedary camels (Fig. 2) and caused 858 deaths out of 2494 infected people with a 34% fatality rate (Singhal, 2020).
Now, in late December 2019, a couple of patients were diagnosed with concentrated pneumonia with an unknown etiology in Wuhan, China (Bogoch et al., 2020; Hui et al., 2019; Lu et al., 2020). The newly emerged novel CoV retained 99.8–99.9% nucleotide sequence homology with beta bat CoVs that directed the reemergence of another viral strain, later entitled as SARS-CoV-2 (Ren et al., 2020) and genetic analysis of the SARS-CoV-2 represented genetic similarity 50% with MERS-CoV and 80% with SARS-CoV (Lu et al., 2020; Ren et al., 2020; Rehman et al., 2020).
3. Epidemiology
As of 03 June 2020, a total of 6,467,229 cases of COVID-19 have been confirmed worldwide including 382,766 deaths (WHO, 2020). COVID-19 as an acute respiratory infectious disease, primarily spreads by the mean of the respiratory tract, via droplets, respiratory secretions emitted from an infected person or direct contact for a low infective dose (Li et al., 2020; Lee and Hsueh, 2020). Significantly high level of viral loads was also observed in the nasal cavity as compared to the throat where the viral load was the same between symptomatic and asymptomatic people (Zou et al., 2020). Patients can be a career of infection even on clinical recovery and few people may act as a strong candidate to spread the infection, for example, an infected UK resident caused 11 people to infect by COVID-19. The incubation period of this disease ranges from 2 to 14 days (median 5 days) (Singhal, 2020). SARS-CoV-2 virus responds to the same receptor, angiotensin receptor 2 (ACE2), to enter the respiratory mucosa as the SARS-CoV entry receptor (Cheng and Shan, 2020).
4. Pathogenesis
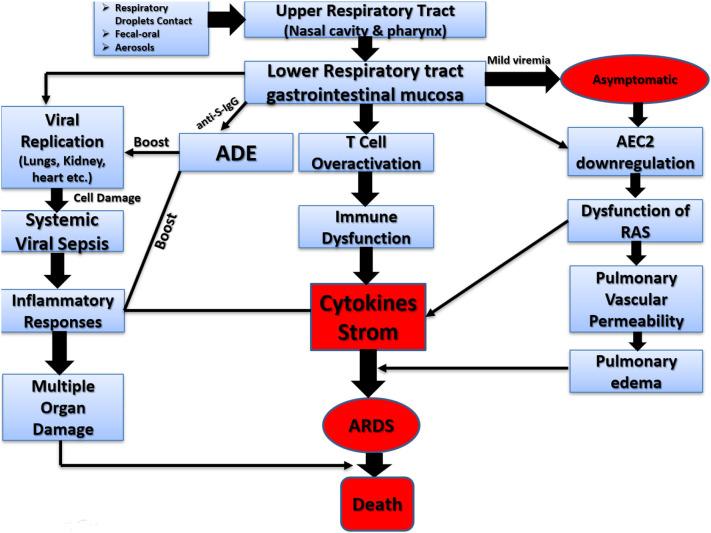
The SARS-CoV-2 is a respiratory system targeting virus therefore the prime pathogenesis of the COVID-19 severe pneumonia, RNAaemia, combined with the incidence of ground-glass opacities, and acute cardiac injury. Also, some patients were exhibited non-respiratory symptoms such as the acute liver and heart injury, kidney failure, diarrhea, implying multiple organ involvement (Y. Chen et al., 2020; G. Guan et al., 2020; C. Huang et al., 2020; P. Huang et al., 2020; Su et al., 2016; W. Wang et al., 2020). Crucially, viral replication is supposed to occur in the mucosal epithelium of the upper respiratory tract (nasal cavity and pharynx), in addition, proliferation is in the lower respiratory tract and gastrointestinal mucosa that results in the mild viremia (Xiao et al., 2020; Yuefei et al., 2020). Fig. 3 is a hypothetical explanation of the pathogenesis of SARS-CoV-2 infection.
Fig. 3.
Pathogenesis of SARS-CoV-2 infection.
5. Clinical manifestations
The clinical features of COVID-19 range from an asymptomatic condition to acute respiratory distress syndrome and multiple organ dysfunction, so it is difficult to distinguish it from other respiratory infections. Almost 5.2 days' time period of incubation is required for the appearance of COVID-19 symptoms (Li et al., 2020), and after one week of the onset of disease patients experience pneumonia, the respiratory failure that leads to death (N. Chen et al., 2020). The primary and common symptoms at the early stage of COVID-19 infection are fever, cough, and fatigue, whereas other symptoms such as sputum, headache, hemoptysis, diarrhea, dyspnoea, and lymphopenia have also been observed (Carlos et al., 2020; W.-j. Guan et al., 2020; P. Huang et al., 2020; Ozaslan et al., 2020; Guan et al., 2020; D. Wang et al., 2020). Clinical findings related to acute lung injury, ARDS, shock, kidney injury can cause death (Carlos et al., 2020; Ozaslan et al., 2020; Phan et al., 2020). The shorter period was witnessed in patients with the age of more than 70-years old (Carlos et al., 2020). Incompatible death results are more common in the elderly and those with underlying co-morbidities (50–75% of fatal cases). In the adult, aged patient's fatality rate ranges between 4 and 11% while the overall fatality rate varies between 2 and 3% (Ozaslan et al., 2020; Rothan and Byrareddy, 2020).
In another study, similar symptoms for COVID-19 and earlier betacoronavirus have been observed in chest CT scans (P. Huang et al., 2020). Although, some distinctive clinical features in COVID-19 exist including lower airway and upper respiratory tract symptoms like rhinorrhea, sneezing, and throat soring (Assiri et al., 2013; Lee et al., 2003). Similarly, chest radiography of some patients showed an infiltrate in the upper lobe of the patients (Phan et al., 2020). Whereas, gastrointestinal symptoms like diarrhea were also diagnosed in some COVID-19 patients (Gu et al., 2020), which were also found in MERS-CoV or SARS-CoV patients (Assiri et al., 2013; Lee et al., 2003).
6. Immune enhancement against COVID-19
Antibody-dependent enhancement (ADE) occurs when non-neutralizing antiviral proteins facilitate virus entry into host cells, leading to increased infectivity in the cells. Recently, ADE is proposed as a responsible for the inflammation because of SARS-CoV-2 infection (Fu et al., 2020). ADE can interact with virus–antibody complexes their receptors, and increasing the target cell infection (Takada and Kawaoka, 2003). The interaction between specific receptor and virus complex can activate both pathways such as inflammatory responses and viral replication in the patients (Gu et al., 2020). In another study, loss of ACE2 function is related to acute lung injury because the ACE2downregulation can result in RAS dysfunction, and endorse the inflammation that causes vascular permeability (Imai et al., 2008). These biomarkers propose both a molecular description and a feasible treatment for acute respiratory distress syndrome (ARDS) following SAS-CoV-2 infection. Moreover, in severe cases, CD4 and CD8 T cells exhibited hyper-activation and high level of expression of cytotoxic granules CD8 T cells and pro-inflammatory CD4 T cells suggested immune responses against viral attack (Xu et al., 2020). Additionally, lymphopenia is also reported as a common characteristic of COVID-19 which is a serious reason account for severe infection and a higher rate of mortality (P. Huang et al., 2020; Zhu et al., 2020). The virus can remain viable on surfaces in suitable conditions and can be smashed within 60 s by common disinfectants like sodium hypochlorite and hydrogen peroxide (Kampf et al., 2020).
7. Genes involved
Interestingly, the histopathologically ARDS has been identified in the patients of all type of cases such as SARS-CoV, MERS-CoV, and SARS-CoV-2 (Ding et al., 2003; Ng et al., 2016; Xu et al., 2020). Elevated levels of cytokines and chemokines in COVID-19 patients including IL1β, IL1RA, IL7, IL8, IL9, IL10, basic FGF2, GCSF, GMCSF, IFN γ, IP10, MCP1, MIP1 α, MIP1 β, PDGFB, TNF α, and VEGFA were also observed. Additionally, a few patients with a severe infection in the intensive care unit also showed increased levels of pro-inflammatory cytokines including IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1 α, and TNF α that were responsible for disease severity (P. Huang et al., 2020). After genetic variability, the cytokines were studied and the incidence of ARDS was evidenced. Many candidate genes, i.e. ACE2, IL-10, TNF, VEGF are believed to be associated with ARDS development or outcome (Meyer and Christie, 2013). In addition, the increased levels of IL-6 and IL-8 were confirmed to be associated with ARDS (Thompson et al., 2017).
8. Genomic variations in SARS-CoV-2
The genetic information of any life is preserved in its genome, and the annotation is the first step to explain the sequence. While the length of the SARS-CoV genome is above ~29 kb, it appears that only a few coding genes do not match the general characteristics of the viral genome and the group of minimal hereditary data. In previous studies, 156 variants were found in 95 samples available on NCBI databases and 116 variants were identified uniquely. In addition, these variants have 46 missense, 52 synonymous, 2 insertions, 1 deletion, and 14 non-coding alleles. The most common variant were found 8782C>T (ORF1ab) in 13 samples, 28144T>C (ORF8) in 14 samples and 29095C>T (N) in 8 samples. The 8782C>T and 28144T>C coincided. Also, 29095C>T was located in one of these subsets. Both 8782C>T and 29095C>T were synonymous; However, 28144T>C causes amino acids to replace L84S in ORF8. For 46 missense variants, ORF1ab has 24 variants, that occupies 2/3 of the entire genome. All non-coding mutations are within 3′UTR or 5′UTR regions that have C>T changes (Rozhgar et al., 2020).
In another study, Rehman et al. (2020) were compared the Envelop (E), Nucleocapsid, Membrane (M), and Spike protein of SARS-CoV-2 with SARS-CoV. It represented 7%, 8%, 7% and 19% amino acid sequence variations in E, M, N and S regions respectively (Table 2 ). Moreover, they also presented nine putative recombinants in SARS-CoV-2 and hypothesized that SARS-CoV-2 is the recombinant of SARS and SARS-like CoVs. Six out of nine recombinants are in S motif which is important for viral pathogenicity. Furthermore, SARS-CoV-2 receptor-binding domain (RBD) was analyzed and showed 73% sequence similarity with the pandemic RBD (Rehman et al., 2020). While the binding free energies that were calculated for human ACE2 and S protein binding complexes and SARS-CoV-2 free binding energy increased up to 28 kcal mol−1, representing higher binding affinity to the human ACE2 receptor (Rehman et al., 2020).
Table 2.
SARS-CoV-2 homology and genetic variations in different genomic regions with respect to SARS-CoV.
| Envelop protein |
Membrane protein |
Nucleocapsid protein |
Spike protein |
||||
|---|---|---|---|---|---|---|---|
| Homology | Genetic variations | Homology | Genetic variations | Homology | Genetic variations | Homology | Genetic variations |
| 93% | 07% | 92% | 08% | 93% | 07% | 81% | 19% |
SARS-CoV-2 (Wuhan-Hu-1-CoV) and SARS-CoV (GZ02).
9. Evolution of SARS-CoV-2
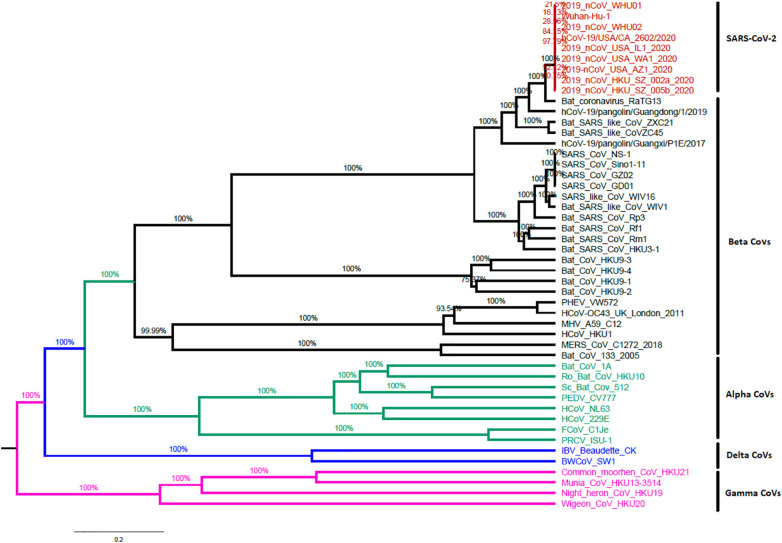
Distinguishing the origins of an emerging pathogen can be critical during the beginning periods of an outbreak as it may allow for containment measures to be precisely targeted. Before December 2019, four strains of Betacoronavirus, HKU1, MERS-CoV, OC43, and SARS-CoV, had been accounted to cause severe human infections (Paola et al., 2020). The fifth strain, a novel beta coronavirus SARS-CoV-2 causing human pneumonia, was first detailed in Wuhan, Central China (Paola et al., 2020). Coronaviruses are naturally hosted and developmentally formed by bats. To do this, it has been proposed that the majority of human coronaviruses are derived from the bat reservoir (Tang et al., 2020). Several teams have recently confirmed the genetic similarity between SARS-CoV-2 and the beta betacoronavirus of the subgenus Sarbecovirus (Tang et al., 2020). The genome sequence of the new virus is 96.2% identical to that of the bat. SARS-related coronavirus (SARSr-CoV; RaTG13) has been collected in Wuhan, China, but is not identical to the genome of SARS-CoV (about 79%) or MERS-CoV (approximately 50%) as shown in Fig. 4 . Replacement and recombination of nucleotides has been proposed a mechanism of viral development in nature (Rehman et al., 2020; Phan, 2020).
Fig. 4.
Whole genome-based phylogenetic tree of CoVs constructed with the maximum-likelihood method using BEAST with GTR + I + G as the nucleotide model of substitution.
In addition, SARS-CoV-2 uses the same receptor, angiotensin II converting enzyme (ACE2) as SARS-CoV. Although the specific pathway of transmission from natural reservoirs to humans is unclear (Peter et al., 2020; Tang et al., 2020), several studies have shown that pangolin may confer partial spike genes to SARS-CoV-2; The critical functional regions of the spike protein SAR-CoV-2 are nearly the same as those defined in viruses isolated from a pangolin (Tang et al., 2020).
10. Potential therapeutic strategies
In the early era until 1960 CoVs were thought to be simple viruses causing flu-like symptoms. In later studies, it was proved that CoV is a more serious and dangerous virus and it causes SARS epidemics (2002 to 2003) (Cascella et al., 2020; Li et al., 2020; W. Wang et al., 2020). The outbreak of MERS in Middle Eastern Countries in 2012 posed a very serious condition that proved it more dangerous (Burke et al., 2020; Cascella et al., 2020; Li et al., 2020; W. Wang et al., 2020).
To manage the COVID-19 antipyretics and analgesics are directed. Moreover, hydration is maintained and also the respiratory system is supported by mechanical ventilation or extracorporeal membrane oxygenation. In case of bacterial infection along with COVID-19 antibiotics are found beneficial. We may use chloroquine and hydroxychloroquine having synergistic effects, but further studies needed (De Wit et al., 2020; Ozaslan et al., 2020). Also, we have the option of Favipiravir and Remdesivir which were trialed by China and Japan but further studies are required (Fu et al., 2020; Hui et al., 2020).
Also, many evidences showed that convalescent plasma from patients recovering from viral infections can be used as a treatment in patients infected with SARS-CoV-2 (Chen et al., 2020). In addition, by influencing ACE2 levels, cardiovascular diseases and/or their treatment can play an important role in the infectivity and consequences of COVID - 19. Whether ACE2 treatment or upregulation caused by the disease immediately affects the COVID - 19 course (Sommerstein et al., 2020). During this global emergency of the COVID-19 pandemic, strategies are taken into account to quickly monitor the timeline for licensing a vaccine against COVID-19, especially by compressing the generally long duration of phase II–III study (Callaway, 2020; De Wit et al., 2020; Boodman, 2020; Eyal et al., 2020).
11. Conclusion
It is widely accepted that there are many viruses in their natural reservoirs for a very long time. The constant spread of viruses from natural hosts to humans and other animals is largely due to human activities. Data collected on genetic evolution, receptor binding and pathogenesis have shown that SARS-CoV is most likely caused by bats by sequential recombination of bat SARS-CoVs. The rapid proliferation of SARS-CoV-2 raises many questions, whether its development is driven by the mutation or any other mechanisms. So the Government and health authorities must develop and implement the guidelines for researchers and the public for the development of drugs and vaccines to reduce the spread rate of COVID-19.
Acknowledgments
Acknowledgments
We highly appreciate many members of the frontline medical and nursing staff who demonstrated selfless and heroic devotion to duty in the face of this outbreak.
Data availability statement
All relevant data are within the manuscript.
Funding
The authors received no specific funding for this work.
Declaration of competing interest
The authors have declared that no competing interests exist.
Ethical approval
We have followed all ethical approvals for this study.
Informed consent
All authors have read and approved the contents and manuscript.
References
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F. Epidemiological, demographic, and clinical characteristics of 47 cases of MiddleEastrespiratorysyndromecoronavirus disease from Saudi Arabia: a descriptive study. LancetInfect.Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey O.T., Pappenheimer A.M., Cheever F.S., Daniels J.B. Amurinevirus(JHM) causing disseminated encephalomyelitis with extensive destruction of myelin: II. Pathology. J. Exp. M. 1949;90(3):195–212. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U., Khan K. Pneumonia of unknown etiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel Med. 2020 doi: 10.1093/jtm/taaa1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boodman Eric. STAT; 2020. Coronavirus Vaccine Clinical Trial Starting Without Usual Animal Data. (Retrieved 19 April 2020) [Google Scholar]
- Burke R.M., Midgley C.M., Dratch A., Fenstersheib M., Haupt T., Holshue M., et al. Active monitoring of persons exposed to patients with confirmed COVID-19 — United States. MMWR Morb. Mortal. Wkly Rep. 2020 doi: 10.15585/mmwr.mm6909e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Should scientists infect healthy people with the coronavirus to test vaccines? Nature. 2020;580(7801):17. doi: 10.1038/d41586-020-00927-3. [DOI] [PubMed] [Google Scholar]
- Carlos W.G., Dela Cruz C.S., Cao B., Pasnick S., Jamil S. Novel Wuhan (2019-nCoV) coronavirus. AmJRespirCritCareMed. 2020;201(4):7–8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C. StatPearls Publishing; Treasure Island, FL: 2020. Napoli RD: Features, Evaluation and Treatment Coronavirus (COVID-19) [PubMed] [Google Scholar]
- Chang C.-k., Sue S.-C., Yu T.-h., Hsieh C.-M., Tsai C.-K., Chiang Y.-C., Lee S.-j., Hsiao H.-h., Wu W.-J., Chang W.-L. ModularorganizationofSARScoronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020 doi: 10.1016/s1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumoniainWuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Shan J. 2019 novel coronavirus: where we are and what we know. Infection. 2020;48:155–163. doi: 10.1007/s15010-020-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 2003;200(3):282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Nir, Lipsitch Marc, Smith Peter G. Human challenge studies to accelerate coronavirus vaccine licensure. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol.Sin. 2020:1–6. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Wei-jie, Zheng-yi N., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G., Gao L., Wang J., Wen X., Mao T., Peng S., Zhang T., Chen X., Lu F. Exploring the mechanism of liver enzyme abnormalities in patients with novel coronavirus- infected pneumonia. Zhonghuaganzangbingzazhi=Zhonghuaganzangbingzazhi= Chinese journal of hepatology. 2020;28(2):E002. doi: 10.3760/cma.j.issn.1007-3418.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S. 2020. Clinical Characteristics of 2019 Novel Coronavirus Infection in China. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., Hu X., Chen J., Liu B. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., Chow B.K., Lo T., Tsang O.T.Y., Ko F.W., Ng S.S., Gin T., Chan M.T.V. Exhaled air dispersion during high-flow nasal cannula therapy versus CPAP via different masks. Eur. Respir. J. 2019;53(4) doi: 10.1183/13993003.02339-2018. (Apr) [DOI] [PubMed] [Google Scholar]
- Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Penninger J.M. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp. Physiol. 2008;93(5):543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.-I., Hsueh P.-R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;20:30011–30016. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C., To K. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. In: Fields Virology. Knipe D.M., Howley P.M., editors. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. Coronaviridae; pp. 825–858. [Google Scholar]
- Meyer N.J., Christie J.D. Seminars in Respiratory and Critical Care Medicine. 2013. Genetic heterogeneity and risk of acute respiratory distress syndrome; pp. 459–474. [DOI] [PubMed] [Google Scholar]
- Ng D.L., Al Hosani F., Keating M.K., Gerber S.I., Jones T.L., Metcalfe M.G., Tong S., Tao Y., Alami N.N., Haynes L.M. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am. J. Pathol. 2016;186(3):652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaslan Mehmet, Safdar Muhamad, Halil Kilic I., Khailany Rozhgar A. Practical measures to prevent COVID-19: a mini-review. J. Biol. Sci. 2020;20:100–102. [Google Scholar]
- Paola Stefanelli, Giovanni Faggioni, Alessandra Lo Presti, Stefano Fiore, Antonella Marchi, Eleonora Benedetti, Concetta Fabiani, Anna Anselmo, Andrea Ciammaruconi, Antonella Fortunato, Riccardo De Santis, Silvia Fillo, Rosaria Capobianchi Maria, Rita Gismondo Maria, Alessandra Ciervo, Giovanni Rezza, Rita Castrucci Maria, Florigio Lista, ISS COVID-19 study group Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25(13) doi: 10.2807/1560-7917.ES.2020.25.13.2000305. (pii=2000305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter F., Lucy F., Colin R., Michael F. Phylogenetic network analysis of SARS-CoV-2 genomes. 2020. www.pnas.org/cgi/doi/10.1073/pnas.2004999117
- Phan Tung. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. (July) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q., Nguyen T.T., Cao T.M., Pham Q.D. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N. Engl. J. Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman S.U., Shafique L., Ihsan A., Liu Q. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. 2020;9(3):240. doi: 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L.L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., Jiang Y.-Z., Xiong Y., Li Y.-J., Li H. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhgar A.M., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerstein R., Kochen Michael M., Messerli Franz H., Gräni C. Coronavirus disease 2019 (COVID-19): do angiotensin-converting enzyme inhibitors/angiotensin receptor blockers have a biphasic effect? J. Am. Heart Assoc. 2020;9(7):e016509. doi: 10.1161/JAHA.120.016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev. Med. Virol. 2003;13(6):387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.T., Chambers R.C., Liu K.D. Acute respiratory distress syndrome. N. Engl. J. Med. 2017;377(6):562–572. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2020;92(4):441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2020. https://www.worldometers.info/coronavirus/
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and delta coronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Li C., He J., Hong Z., Huang S., Zhang Z., Lin X., Fang Z. 2020. Evidence for Gastrointestinal Infection of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuefei J., Haiyan Y., Wangquan J., Weidong W., Shuaiyin C., Weiguo Z., Guangcai D. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12(4):372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.