Dear Editor,
On 31st December 2019, the World Health Organization was alerted to cases of pneumonia of an unknown aetiology in Wuhan City, China [1]. The novel virus responsible, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has subsequently caused a pandemic.
Global health surveillance was quickly implemented to identify cases of COVID-19. Although effective, a study by Deslandes et al. [2] indicated potential virus circulation prior to cases first detected via surveillance. The study detailed a Parisian patient with no recent travel history admitted to ICU, who retrospectively tested positive for SARS-CoV-2 [2]. The respiratory sample in question was taken approximately one month before it is believed the epidemic began in France [2].
The first Scottish positive case was confirmed on 1st March, in a patient who had travelled to Italy [3]. At this time sampling was recommended only for individuals who had travelled to an epidemic region and displayed symptoms. This screening criteria may have missed community cases. To investigate further, we retrospectively tested respiratory samples from Greater Glasgow and Clyde (GGC) which had been sent to the West of Scotland Specialist Virology Centre (WoSSVC).
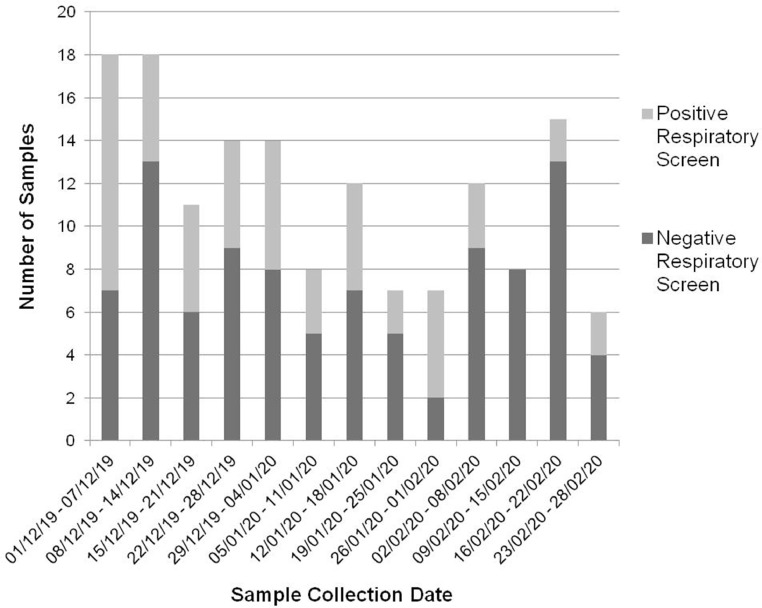
Respiratory samples were selected which had been submitted from adult ICUs, HDUs and CCUs, across three hospital sites in NHS GGC between 1st December 2019 and 28th February 2020 (none appropriate for 29th February). 206 samples were accepted for extended respiratory screening from 164 patients. 160 had not received a SARS-CoV-2 test as part of the initial respiratory screen. Four were negative. Following routine testing these samples had been stored at −80 °C.
For each adult patient (n = 160), we tested the initial sample, unless a superior sample type was received within 48 h e.g. samples from the lower respiratory tract (LRT) rather than upper respiratory tract (URT) [4], or the sample was previously inhibited or insufficient. If all samples from a patient were insufficient, previously inhibited or unavailable then the patient was excluded (n = 12). In two instances, two samples were tested from the same patients following admission to two different wards several weeks apart. This resulted in our testing 150 samples from 148 adult patients (Fig. 1 ).
Fig. 1.
Summary of the 150 respiratory samples from 148 adult ICU patients, with positive and negative respiratory screening results, which were then retrospectively tested for SARS-CoV-2.
The initial extended respiratory screen was negative for 96 samples and positive for 54 (Table 1 ). A range of respiratory samples were included – 99 from URT (gargle n = 43, nasopharyngeal aspirate n = 3, and nose/nose and throat/throat swab n = 53), and 51 from LRT (bronchial aspirate n = 11, bronchoalveolar lavage n = 5, and sputum n = 35).
Table 1.
Summary of respiratory screening results for the 150 adult samples included in the retrospective study. URT: Upper respiratory tract sample. LRT: lower respiratory tract sample.
| Extended respiratory screen target | Number of samples (% total samples) | URT sample | LRT sample |
|---|---|---|---|
| rhinovirus | 12 (8.00) | 8 | 4 |
| respiratory syncytial virus | 10 (6.67) | 3 | 7 |
| influenza A or B | 9 (6.00) | 6 | 3 |
| seasonal coronavirus | 6 (4.00) | 5 | 1 |
| parainfluenza 1,2,3,or 4 | 4 (2.67) | 1 | 3 |
| human metapneumovirus | 3 (2.00) | 3 | 0 |
| adenovirus | 2 (1.33) | 1 | 1 |
| Mycoplasma pneumonia | 1 (0.67) | 1 | 0 |
| 2 respiratory pathogens | 6 (4.00) | 3 | 3 |
| 3 respiratory pathogens | 1 (0.67) | 0 | 1 |
| Total number of samples with positive respiratory screen | 54 (36) | 31 | 23 |
| Total number of samples with negative respiratory screen | 96 (64) | 68 | 28 |
| Total number of samples tested | 150 (100) | 99 | 51 |
A sub-set of samples from paediatric ICU patients were included, which had no significant virological or microbiological results two weeks prior to sample collection. 24 samples, from 23 patients were collected between 4th December 2019 and 28th February 2020, 13 being URT and 11 being LRT samples.
Samples were extracted using the Abbott M2000sp instrument (Abbott Laboratories, Illinois) or the NUCLISENS easyMAG (Biomérieux, Marcy-l'Étoile). Sample extracts and appropriate controls underwent real-time RT-PCR, using primers and probes for RNA-dependant RNA polymerase (RdRp) and envelope (E) genes [5]. In-house verification demonstrated a limit of detection of 1000 copies/mL for RdRp gene and 200 copies/mL for E gene. Samples requiring confirmation were re-tested using the Roche cobas SARS-CoV-2 Test to detect open reading frame 1a (ORF 1a) and E genes (Roche Diagnostics, Basel) as it had similar sensitivity to that of the E gene assay.
Of the 174 samples tested, 166 were negative for SARS-COV-2 using real-time RT-PCR to detect RdRp and E genes. The remaining eight samples which were RdRp gene negative but had spurious traces on the E gene assay were re-tested using the cobas and also deemed negative.
SARS-CoV-2 was not detected in >90% of the relevant adult ICU population prior to March 2020. It is unlikely, therefore, that patients were hospitalised before March 2020 with clinically significant respiratory symptoms due to SARS-CoV-2. The use of two different PCR gene targets reduced the likelihood of false-negatives through primer/probe mismatches. Additionally, SARS-CoV-2 was absent in our sub-set of paediatric ICU samples. This is unsurprising given the sample-set size and as severe COVID-19 occurs in a low percentage of children [6].
These findings differ from those of Deslandes et al. [2] who tested 14 nasopharyngeal samples taken between 2nd December 2019 and 16th January 2020. The authors detected one positive from late December [2]. Additionally, Hogen et al. [7] retrospectively tested 2888 nasopharyngeal and bronchoalveolar lavage samples taken between January and February in San Francisco. Two were positive from late February, which overlapped with the first reported cases in the nearby area [7]. However, Hogan et al. [8] demonstrated no positives in 1700 samples retrospectively tested from November to December 2019.
The results support the initial Scottish SARS-CoV-2 testing strategy. Retrospective testing of a larger set of samples is necessary to fully rule-out early community transmission. However, the benefit must be balanced with existing limitations in reagents and current testing demands. Sample pooling could overcome this, and although less sensitive, would allow economical surveillance of a larger population to inform Scottish transmission dynamics prior to March 2020 [7,8].
There are limitations in this study. Firstly, the initial storage of the samples may have impacted detection [9]. Long-term storage at −80 °C is, however, unlikely to have been detrimental. Secondly, testing was not exhaustive. Presentation of COVID-19 in GGC ICUs prior to March 2020 cannot be ruled out without blanket testing.
Additionally, only one sample was retrospectively tested for the majority of patients included. Significantly higher rates of SARS-CoV-2 RNA detection is found in LRT samples, compared with URT [4]. Thus, with only a third of tested samples being of the LRT, false-negative results could have occurred for the remaining two thirds. Specifically, gargle samples may have been less appropriate. Gargles have, however, been demonstrated to be a suitable alternative to sputum for SARS-CoV-2 detection [10]. Finally, samples were only tested from patients hospitalised in three GGC sites. The lack of SARS-CoV-2 positivity cannot, therefore, be assumed for those in ICUs in the rest of Scotland, and we cannot rule-out community transmission.
In summary this retrospective study demonstrated no SARS-CoV-2 positivity in the 174 selected respiratory samples collected from GGC ICUs prior to the first reported Scottish case.
Declaration of Competing Interest
None.
Acknowledgments
Acknowledgement
Thanks are given to the staff at West of Scotland Specialist Virology Centre for their technical expertise during the study.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data Statement
Research data is confidential.
Ethics Approval
None required.
References
- 1.World Health Organization . World Health Organization; Geneva: 2020. Novel Coronavirus (2019-nCoV), situation report – 1.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4 [Accessed 01/06/2020] [Google Scholar]
- 2.Deslandes A., Berti V., Tandjaoui-Lambotte Y., Alloui C., Carbonnelle E., Zahar J.R. SARS-CoV-2 was already spreading in France in late December 2019. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106006. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill K.J., Russell C.D., Clifford S., Templeton K., Mackintosh C.L., Koch O. The index case of SARS-CoV-2 in Scotland: a case report. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.022. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammadi A., Esmaeilzadeh E., Li Y., Bosch R.J., Li J.Z. SARS-CoV-2 detection in different respiratory sites: a systematic review and meta-analysis. Med Rxiv. 2020 doi: 10.1101/2020.05.14.20102038. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. pii=2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L. Features of 20,133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020:369. doi: 10.1136/bmj.m1985. m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323(19):1967–1969. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogan C.A., Garamani N., Sahoo M.K., Huang C.H., Zehnder J., Pinsky B.A. Retrospective pooled screening for SARS-CoV-2 RNA in late 2019. Med Rxiv[Prepint] 2020. doi: 10.1101/2020.05.14.20102079. [DOI] [PMC free article] [PubMed]
- 9.Li L., Li X., Guo Z., Wang Z., Zhang K., Li C. Influence of storage conditions on SARS-CoV-2 nucleic acid detection in throat swabs. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa272. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W.-.D., Chang S.-.Y., j-T Wang, Tsai M.-.J., Hung C.-.C., Hsu C.-.L. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.063. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]