Abstract
Objective
Numerous studies have reported that patients with cancer seem to be more likely to be diagnosed with COVID-19. However, it is still unknown the prevalence of different types of cancer in patients with COVID-19. Therefore, this study will explore which type of cancer patients are more susceptible to the SARS-COX-2.
Methods
Eligible studies were identified by searching several electronic databases for relevant studies published before April 28, 2020. The language was restricted to English or Chinese. The meta-analysis was performed using Stata 12.0. The GEPIA database evaluated the expression of SARS-COX-2 infection key genes in different types of normal and tumor samples.
Results
A total of 6 studies, including 205 patients, were identified to be eligible for this meta-analysis. Among the cancer patients infected by SARS-COX-2, the proportion of patients with the lung, colorectal, breast, esophagus, bladder, pancreatic and cervical cancer were 24.7 %, 20.5 %, 13.0 %, 7.6 %,7.3 %,6.1 %,and 6.0 %, respectively. These findings were also corroborated by the results of the GEPIA database.
Conclusion
Compared with other types of cancer, lung cancer and colorectal cancer are more susceptible to SARS-COX-2 infection.
Keywords: COVID-19, Cancer, SARS-CoV-2, Meta-analysis
1. Introduction
Coronavirus disease 2019(COVID-19), a new and highly infectious disease that is caused by a previously undescribed coronavirus in humans, has created a significant public health threat in the world (Chen et al., 2020; Wang et al., 2020a,b). According to the most recent World Health Organization (WHO) estimate, more than 3.3 million people worldwide are infected with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). There is a broad consensus that patients with comorbid diseases may be more susceptible to SARS-CoV-2 (Ali et al., 2020; Guan et al., 2020; Hu et al., 2020). Cancer is a common comorbidity in COVID-19 patients. Previous studies mentioned that patients with cancer seemed to be more likely to be diagnosed with COVID-19 and have a higher risk of a severe event (Sidaway, 2020). The prevalence of different types of cancer in patients with COVID-19 is inconsistent and inconclusive due to limited sample sizes (Wang and Zhang, 2020; Xia et al., 2020). Given the rapid spread and high mortality rate of COVID-19, it is necessary to evaluate the prevalence of different types of cancer.
The coronavirus receptor angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) have been recognized play key functions in SARS-CoV-2-infected cells (Hoffmann et al., 2020). Analysis of ACE2 and TMPRSS2 expression levels in different tumor cells could help to investigate cancer susceptibility. Until now, several ongoing clinical trials are now exploring the epidemiology of SARS-CoV-2 in cancer patients (NCT04344002, NCT04352556, NCT04341207). Given this situation, use available data to understanding the prevalence of different types of cancer in patients with COVID-19 can be valuable to clinicians in their individualized approach to prevention.
As far as we know, this is the first meta-analysis to explore the prevalence of different types of cancer in patients with Covid-19. Furthermore, we also evaluated the level of ACE2 and TMPRSS2 expression in tumor cells to investigate the susceptibility of different types of cancer.
2. Methods
2.1. Registration
This study was not registered. This study was performed based on the PRISMA checklist.
2.2. Search strategy
Relevant literature was extracted by systematic retrieval of PubMed(Medline), EMBASE, Springer, Web of Science, Cochrane Library, China National Knowledge Infrastructure (CNKI), and Wanfang databases up to date to April 28, 2020. Our search strategy included terms for: “2019-nCoV” or “Coronavirus” or“COVID-19” or “SARS-CoV-2” or“2019-nCoV” or “Wuhan Coronavirus” and “cancer” or “tumor” or “carcinoma” or “malignancy.” Besides, we manually screened out the relevant potential article in the references selected. The above process was performed independently by two participants.
2.3. Inclusion and exclusion criteria
Inclusion criteria are as follows: (1) published studies focused on SARS-CoV-2 infection in patients cancer; (2) reporting the prevalence of different types of cancer; (3) the language was restricted to English or Chinese.
The exclusion criteria: (1) reviews, summaries of meetings or discussions; (2) insufficient data information provided; (3) duplicate publications; (4) the sample size was less than 5; (5) in vitro experiments or animal studies.
2.4. Data extraction and quality assessment
Two participants separately conducted literature screening, data extraction, literature quality evaluation, and any differences that could be resolved through discussion or a third analyst. Information extracted from the included literature: first author surname, year of publication, country of the population, sample size, data on the prevalence of different types of cancer in patients with COVID-19, etc.
The Newcastle-Ottawa scale (NOS) was adopted to evaluate the process in terms of queue selection, comparability of queues, and evaluation of results (Stang, 2010). The quality of the included studies was assessed independently by two participants.NOS scores of at least six were considered high-quality literature. Higher NOS scores showed higher literature quality.
2.5. Statistical analysis
The prevalence rate and the 95 % confidence interval (CI) were estimated for each of all the included studies using the STATA 12.0. Proportions were first transformed using the Freeman-Tukey double arc-sin transformation before being combined (Barendregt et al., 2013; Freeman and Tukey, 1950). After that, the heterogeneity test was conducted. When P ≥ 0.05 or I2<50 % was performed, it indicated that there was no obvious heterogeneity, and the fixed-effect model should be applied for a merger. Otherwise, the random-effect model was applied. Publication bias was assessed using Begg funnel plot and Egger test linear regression test (where at least five studies were available). If P < 0.05 indicates obvious publication bias.
Gene Expression Profiling Interactive Analysis (GEPIA) was established using gene expression data via RNA sequencing from Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx). The expression levels of ACE2 and TMPRSS2 in tumor tissues and normal samples were validated by the GEPIA platform, which is a free online database (http://gepia.cancer-pku.cn/).
3. Results
3.1. Process of literature selection and description of qualified studies
The flow diagram (Fig. 1 ) showed detailed literature search steps. The initial 538 articles screened through databases of PubMed(Medline), EMBASE, Springer, Web of Science, Cochrane Library, CNKI, and Wanfang databases and one additional record identified through other sources. After the exclusion of duplicate references, 261 articles were considered for the meta-analysis. Of the remaining 261 articles, we excluded 251 articles after reading the title and the abstract. After careful review of the full texts, four studies were excluded, as the sample size was less than five or comment.
Fig. 1.
Flow diagram of the literature search and selection process in the meta-analysis.
Six studies with a total of 205 cancer patients were included in our meta-analysis (Dai et al., 2020; Liang et al., 2020; Ma et al., 2020; Yu et al., 2020; Zhang et al., 2020a, b). Participants’ demographic information is shown in Table 1 . All six studies were performed in China. Six studies reported lung cancer and breast cancer, respectively. Five of the included studies covered colorectal cancer, esophagus cancer, and cervical cancer, and four involved bladder cancer and pancreatic cancer. The NOS score of eligible articles ranged from 6 to 8, which indicated that all included studies were of high quality.
Table 1.
Main characteristics of the included studies in meta-analysis.
| Study | J.Yu | W.Liang | J.Ma | L.Zhang | Y.Zhang | M.Dai |
|---|---|---|---|---|---|---|
| Year | 2020 | 2020 | 2020 | 2020 | 2020 | 2020 |
| Country | China | China | China | China | China | China |
| Sample | 12 | 18 | 37 | 28 | 5 | 105 |
| Median(IQR)/Mean Age (years) | 66(30.0) | 63.1 ± 12.1 | 62(11) | 65.0(56.0−70.0) | 67. 6 ± 16. 7 | 64(14.0) |
| Male | 10 | 12 | 20 | 17 | 4 | 57 |
| With anti-tumor therapy | 6 | 6 | 13 | 18 | NA | 48 |
| Cancer type | ||||||
| Lung cancer | 7 | 5 | 8 | 7 | 1 | 22 |
| Breast cancer | 1 | 3 | 7 | 3 | 0 | 11 |
| Colorectal cancer | 2 | 4 | 11 | 2 | 1 | NA |
| Bladder cancer | 0 | 2 | NA | 0 | 1 | NA |
| Esophagus cancer | 0 | 0 | NA | 4 | 1 | 6 |
| Pancreatic cancer | 1 | 0 | NA | 0 | 1 | NA |
| NOS | 7 | 8 | 8 | 7 | 6 | 8 |
3.2. The prevalence of lung cancer, colorectal cancer, and breast cancer
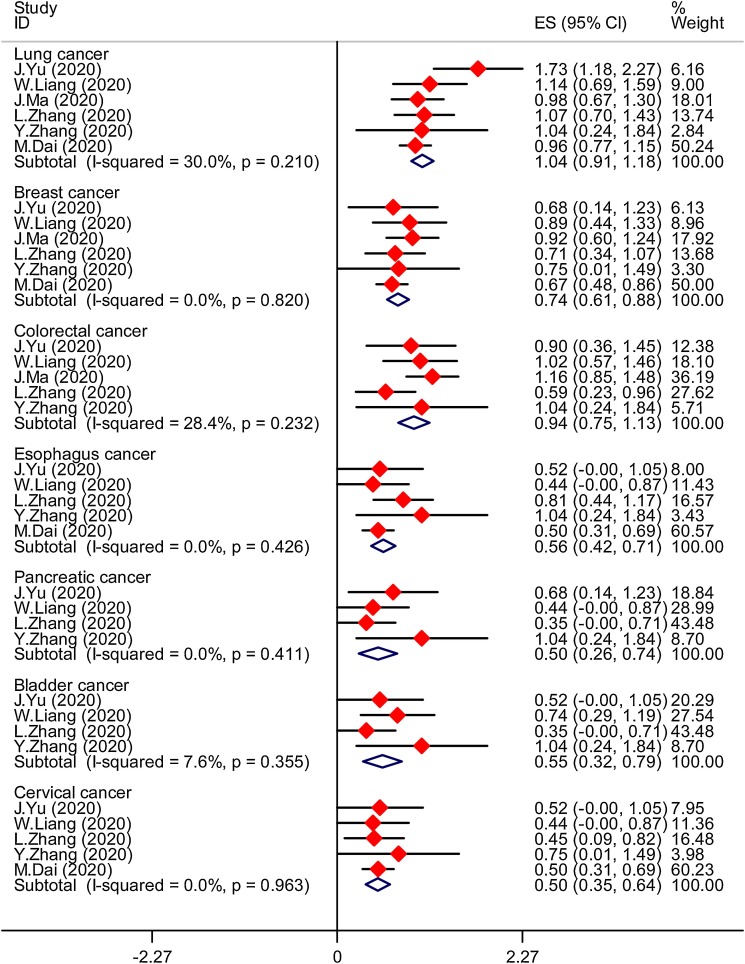
Six studies provided the data in terms of lung cancer and breast cancer (Dai et al., 2020; Liang et al., 2020; Ma et al., 2020; Yu et al., 2020; Zhang et al., 2020a, b). With no obvious heterogeneity (lung cancer:I2 = 30.0 %, P = 0.210;breast cancer: I2 = 0.0 %, P = 0.820) among these studies, so a fixed-effect pattern was used for assessment. The results find that the prevalence of lung cancer and breast cancer were 24.7 % (95 %CI, 0.19−0.30) and 13.0 % (95 %CI,0.09−0.18), respectively. Five studies were evaluated for determining the prevalence of colorectal cancer. Pooled prevalence of colorectal cancer was 20.5 % (95 %CI, 0.13−0.29), with no obvious heterogeneity between estimates (I2 = 28.4 %, P = 0.232) (Fig. 2 )(Liang et al., 2020; Ma et al., 2020; Yu et al., 2020; Zhang et al., 2020a, b).
Fig. 2.
The prevalence of different types of cancer in patients with COVID-19.
3.3. The prevalence of esophagus cancer, bladder cancer, pancreatic cancer and cervical cancer
Five included studies reported esophagus cancer and cervical cancer (Dai et al., 2020; Liang et al., 2020; Yu et al., 2020; Zhang et al., 2020a, b). A fixed-effects model was used since the heterogeneity test suggested that there was no heterogeneity(esophagus cancer:I2 = 0.0 %, P = 0.426; cervical cancer: I2 = 0.0 %, P = 0.963). The prevalence of esophagus cancer and cervical cancer were 7.6 % (95 %CI, 0.04−0.12) and 6.0 % (95 %CI, 0.03−0.10). Among the included studies, four of the studies covered data on bladder cancer and pancreatic cancer. Prevalence of bladder cancer and pancreatic cancer were 7.3 % (95 % CI:0.03−0.14) and 6.1 % (0.02−0.13), which indicates no obvious heterogeneity (Fig. 2) (Liang et al., 2020; Yu et al., 2020; Zhang et al., 2020a, b).
3.4. Publication bias
The risk of publication bias was analyzed in the following cancers: lung cancer, colorectal cancer, breast cancer, esophagus cancer, and cervical cancer. Table 2 shows the results of publication bias, which were evaluated by funnel plots and Eggers test. Begg’s test (All P > 0.05) and Egger’s regression test (All P > 0.05) suggest no significant publication bias.
Table 2.
The results of meta-analysis.
| No.of studies | Prevalence% (95 %CI) | P-Value | Heterogeneity |
Model used | Begg's Test | Egger's test | ||
|---|---|---|---|---|---|---|---|---|
| I² | Ph | |||||||
| Lung cancer | 6 | 24.7 % (0.19−0.30) | <0.001 | 30.0 % | 0.210 | Fixed | 0.452 | 0.622 |
| Breast cancer | 6 | 13.0 % (0.09−0.18) | <0.001 | 0.0 % | 0.820 | Fixed | 1.000 | 0.877 |
| Colorectal cancer | 5 | 20.5 % (0.13−0.29) | <0.001 | 28.4 % | 0.232 | Fixed | 0.462 | 0.190 |
| Bladder cancer | 4 | 7.3 % (0.03−0.14) | <0.001 | 7.6 % | 0.355 | Fixed | NA | NA |
| Esophagus cancer | 5 | 7.6 % (0.04−0.12) | <0.001 | 0.0 % | 0.426 | Fixed | 1.000 | 0.481 |
| Pancreatic cancer | 4 | 6.1 % (0.02−0.13) | <0.001 | 0.0 % | 0.411 | Fixed | NA | NA |
| Cervical cancer | 5 | 6.0 % (0.03−0.10) | <0.001 | 0.0 % | 0.963 | Fixed | 1.000 | 0.709 |
3.4.1. Analysis of core genes by the GEPIA
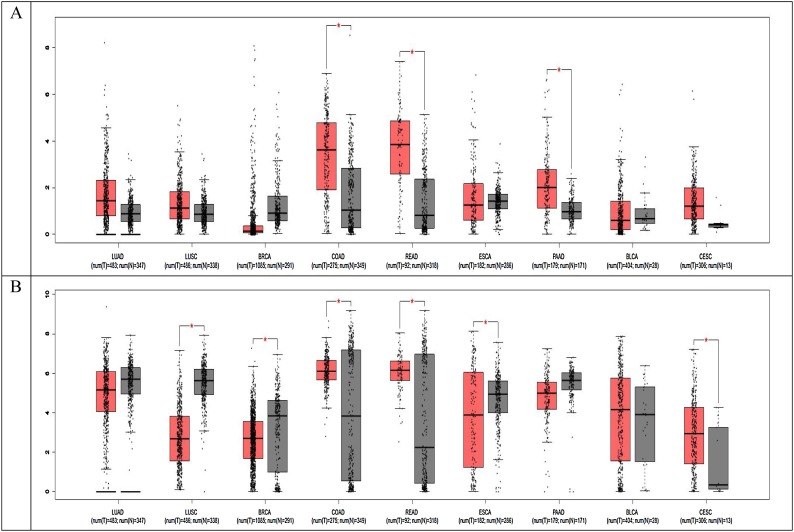
To further explore different types of cancer patients SARS-COX-2 susceptibility, we evaluated the expression of infection key genes, including ACE2 and TMPRSS2 in normal and tumor samples. The mRNA expression level of ACE2 and TMPRSS2 in seven common cancers, analyzed by GEPIA. Taken together, the results of GEPIA analysis suggested that the mRNA expression of ACE2 and TMPRSS2 was higher in lung cancer and colorectal cancer than other types of cancer (Fig.3 ).
Fig. 3.
The expression of ACE2 and TMPRss2 in cancer and normal tissue. LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; BRCA: Breast invasive carcinoma ;COAD: Colon adenocarcinoma; READ: Rectum adenocarcinoma; ESCA: Esophageal carcinoma; PAAD: Pancreatic adenocarcinoma; BLCA: Bladder Urothelial Carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; T: Cancer tissue; N: Normal tissue.
4. Discussion
SARS-CoV-2 is the most recent viral pathogen to cause a global public health emergency. Currently, there are still significant knowledge gaps in the cancer management of COVID-19 patients (Hanna et al., 2020; Moujaess et al., 2020; Wang et al., 2020a,b). A better understanding of the prevalence of different types of cancer in COVID-19 patients allows the development of more specific and more efficient ways of prevention and therapy. Previous studies mentioned that lung cancer patients are more susceptible to SARS-CoV-2 infection (Yu et al., 2020). However, the conclusions of previous studies are not entirely consistent. Therefore, we used meta-analysis to analyze which type of cancer patients are more susceptible to SARS-COX-2.
In our meta-analysis, including 205 patients from 6 retrospective studies, found the prevalence of different types of cancer in patients with COVID-19. We analyzed the susceptibility of seven cancers, including lung cancer, colorectal cancer, breast cancer, esophagus cancer, bladder cancer, pancreatic cancer, and cervical cancer. The prevalence of lung cancer was 24.5 % among cancer patients infected by SARS-COX-2, which demonstrates a high presence of this infection. We also found that colorectal cancer patients with a prevalence of 20.5 % are also susceptible to SARS-COX-2.
Furthermore, we evaluated the expression of infection core genes in different types of cancer by the GEPIA database. Numerous studies have reported that ACE2 and TMPRSS2 are the critical proteins for SARS-CoV-2 entry into host cells.SARS-CoV-2 uses the SARS-CoV receptor (ACE2) for entry and the serine protease TMPRSS2 for S protein priming (Hoffmann et al., 2020). Previous studies study explored the existence of host cell receptor ACE2 in various cell types using published single-cell RNA-seq data to track the potential SARS-CoV-2 infection in human organs and constructed a risk map indicating the vulnerability of different organs to SARS-CoV-2 infection (Xu et al., 2020; Zou et al., 2020). Therefore,the cells with ACE2 and TMPRSS2 expression may act as target cells and are susceptible to SARS-CoV-2 infection. In our results of the GEPIA database, we found that the mRNA expression of ACE2 and TMPRSS2 was higher in lung cancer and colorectal cancer than other types of cancer, which further support the finding of this meta-analysis.
However, this conclusion needs to be interpreted with some caution. Firstly, the results provided in this article come from six retrospective studies, and the pooled results of some types of cancer only come from four studies. Secondly, the small sample size could have affected the significance of the results. Thirdly, some patients may have more than one type of cancer. Finally, the quality of different studies was different, which might lead to bias.
5. Conclusion
The meta-analysis and bioinformatics analysis identified lung cancer and colorectal cancer are two types of cancer susceptible to SARS-CoV-2 infection. Knowledge of the prevalence in different types of cancer can better define those COVID-19 patients at higher risk, and thus allow a more targeted and specific approach to preventing SARS-CoV-2 infection.
Funding
None.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
References
- Ali H., Daoud A., Mohamed M.M., Salim S.A., Yessayan L., Baharani J., Murtaza A., Rao V., Soliman K.M. Survival rate in acute kidney injury superimposed COVID-19 patients: a systematic review and meta-analysis. Ren. Fail. 2020;42(1):393–397. doi: 10.1080/0886022X.2020.1756323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendregt J.J., Doi S.A., Lee Y.Y., Norman R.E., Vos T. Meta-analysis of prevalence. J. Epidemiol. Community Health. 2013;67(11):974–978. doi: 10.1136/jech-2013-203104. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Liu D., Liu M., Zhou F., Li G., Chen Z., Zhang Z., You H., Wu M., Zheng Q., Xiong Y., Xiong H., Wang C., Chen C., Xiong F., Zhang Y., Peng Y., Ge S., Zhen B., Yu T., Wang L., Wang H., Liu Y., Chen Y., Mei J., Gao X., Li Z., Gan L., He C., Li Z., Shi Y., Qi Y., Yang J., Tenen D.G., Chai L., Mucci L.A., Santillana M., Cai H. 2020. Patients with Cancer Appear More Vulnerable to SARS-COV-2: a Multi-center Study During the COVID-19 Outbreak. Cancer discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M.F., Tukey J.W. Transformations related to the angular and the square root. Ann. Math. Stat. 1950;21(4):607–611. [Google Scholar]
- Guan W.J., Liang W.H., Zhao Y., Liang H.R., Chen Z.S., Li Y.M., Liu X.Q., Chen R.C., Tang C.L., Wang T., Ou C.Q., Li L., Chen P.Y., Sang L., Wang W., Li J.F., Li C.C., Ou L.M., Cheng B., Xiong S., Ni Z.Y., Xiang J., Hu Y., Liu L., Shan H., Lei C.L., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Cheng L.L., Ye F., Li S.Y., Zheng J.P., Zhang N.F., Zhong N.S., He J.X., China Medical Treatment Expert Group for, C Comorbidity and its impact on 1590 patients with Covid-19 in China: a Nationwide Analysis. Eur. Respir. J. 2020 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna T.P., Evans G.A., Booth C.M. Cancer, COVID-19 and the precautionary principle: prioritizing treatment during a global pandemic. Nature Rev. Clin. Oncol. 2020;17(5):268–270. doi: 10.1038/s41571-020-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Sun J., Dai Z., Deng H., Li X., Huang Q., Wu Y., Sun L., Xu Y. Prevalence and severity of corona virus disease 2019 (COVID-19): a systematic review and meta-analysis. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., Li S., He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21(3):335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Yin J., Qian Y., Wu Y. Clinical characteristics and prognosis in cancer patients with COVID-19: a single center’s retrospective study. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moujaess E., Kourie H.R., Ghosn M. Cancer patients and research during COVID-19 pandemic: a systematic review of current evidence. Crit. Rev. Oncol. Hematol. 2020;150 doi: 10.1016/j.critrevonc.2020.102972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidaway P. COVID-19 and cancer: what we know so far. Nat. Rev. Clin. Oncol. 2020 doi: 10.1038/s41571-020-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- Wang B., Li R., Lu Z., Huang Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging. 2020;12(7):6049–6057. doi: 10.18632/aging.103000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. 2020. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang L. Risk of COVID-19 for patients with cancer. Lancet Oncol. 2020;21(4):e181. doi: 10.1016/S1470-2045(20)30149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Jin R., Zhao J., Li W., Shen H. Risk of COVID-19 for patients with cancer. Lancet Oncol. 2020;21(4):e180. doi: 10.1016/S1470-2045(20)30150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Ouyang W., Chua M.L.K., Xie C. 2020. SARS-CoV-2 Transmission in Patients with Cancer at a Tertiary Care Hospital in Wuhan, China. JAMA Oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhu F., Xie L., Wang C., Wang J., Chen R., Jia P., Guan H.Q., Peng L., Chen Y., Peng P., Zhang P., Chu Q., Shen Q., Wang Y., Xu S.Y., Zhao J.P., Zhou M. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann. Oncol. 2020 doi: 10.1016/j.annonc.2020.03.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li M., Gan L., Li B. Analysis of clinical characteristics of 5 tumor patients with coronavirus disease 2019. Guangdong Med. J. 2020:41. [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020 doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]