Highlights
-
•
Microneutralisation test (MNT) was carried out for 62 COVID-19 patients.
-
•
Results from six commercial SARS-CoV-2 immunoassays were compared to MNT.
-
•
Performance of different immunoassays were variable.
Keywords: SARS-CoV-2, COVID-19, Serology, IgG, IgA, Neutralisation
Abstract
There is an urgent need for reliable high-throughput serological assays for the management of the ongoing COVID-19 pandemic. Preferably, the performance of serological tests for a novel virus should be determined with clinical specimens against a gold standard, i.e. virus neutralisation. We compared the performance of six commercial immunoassays for the detection of SARS-COV-2 IgG, IgA and IgM antibodies, including four automated assays [Abbott SARS-COV-2 IgG (CE marked), Diasorin Liaison® SARS-COV-2 S1/S2 IgG (research use only, RUO), and Euroimmun SARS-COV-2 IgG and IgA (CE marked)], and two rapid lateral flow (immunocromatographic) tests [Acro Biotech 2019-nCoV IgG/IgM (CE marked) and Xiamen Biotime Biotechnology SARS-COV-2 IgG/IgM (CE marked)] with a microneutralisation test (MNT). Two specimen panels from serum samples sent to Helsinki University Hospital Laboratory (HUSLAB) were compiled: the patient panel (N=70) included sera from PCR confirmed COVID-19 patients, and the negative panel (N=81) included sera sent for screening of autoimmune diseases and respiratory virus antibodies in 2018 and 2019. The MNT was carried out for all COVID-19 samples (70 serum samples, 62 individuals) and for 53 samples from the negative panel. Forty-one out of
62 COVID-19 patients showed neutralising antibodies.The specificity and sensitivity values of the commercial tests against MNT, respectively, were as follows: 95.1 %/80.5 % (Abbott Architect SARS-CoV-2 IgG), 94.9 %/43.8 % (Diasorin Liaison SARS-CoV-2 IgG; RUO), 68.3 %/87.8 % (Euroimmun SARS-CoV-2 IgA), 86.6 %/70.7 % (Euroimmun SARS-CoV-2 IgG), 74.4 %/56.1 % (Acro 2019-nCoV IgG), 69.5 %/46.3 % (Acro 2019-nCoV IgM), 97.5 %/71.9 % (Xiamen Biotime SARS-CoV-2 IgG), and 88.8 %/81.3 % (Xiamen Biotime SARS-CoV-2 IgM). This study shows variable performance values. Laboratories should carefully consider their testing process, such as a two-tier approach, in order to optimize the overall performance of SARS- CoV-2 serodiagnostics.
1. Introduction
Serosurveys are considered essential for creating timely snapshots for global and regional public health management of the ongoing COVID-19 pandemic [1]. Thus, there is an urgent need for the development of high-throughput serological assays, which enable population screening, as well as other epidemiological investigations.
Setting up a serological assay for a completely novel pathogen is challenging in many respects. At present, there is inadequate knowledge as to when and what kind of immune response follows SARS-CoV-2 infection [2]. We are also yet to learn about factors that may disturb reliable serology, such as potential cross reaction from seasonal coronaviruses.
The aim of this study was to compare the performance of four automated immunoassays [Abbott SARS-COV-2 IgG (chemiluminescent microparticle immunoassay (CMIA); CE marked), Diasorin Liaison® SARS-CoV-2 S1/S2 IgG (chemiluminescent assay (CLIA); research use only, RUO), Euroimmun SARS-CoV-2 IgG (enzyme linked immunoassay (ELISA); CE marked), and Euroimmun SARS-CoV-2 IgA (enzyme linked immunoassay (ELISA); CE marked)], and two rapid lateral flow (immunocromatographic) tests [Acro Biotech 2019-nCoV IgG/IgM (CE marked) and Xiamen Biotime Biotechnology SARS-CoV-2 IgG/IgM (CE marked)] with a SARS-CoV-2 microneutralisation test (MNT) by using clinical serum specimens.
2. Materials and methods
The patient samples consisted of serum specimens sent to the Department of Virology and Immunology, Helsinki University Hospital Laboratory, Finland for diagnostic purposes. A subset of these specimens has been included in a previous publication evaluating the Euroimmun SARS-CoV-2 IgG and IgA assays, and are included here for comparison [3].
3. Serum samples comprising the negative panel
The negative panel consisted of 81 serum samples (from 81 individuals) (median age 64 years, range 2–89 years; 33 males, 48 females) (Table 1 ). All of these samples originated from 2018−2019, i.e. before the circulation of SARS-CoV-2 in Europe.
Table 1.
Negative serum sample panel consisting of samples collected retrospectively during years 2018-2019, prior the SARS-CoV-2 epidemic.
| Number and type of samples (serum) |
aAbbott, IgG, nucleoprotein antigen (INDEX) | bEuroimmun, IgA, S1 antigen (ratio) | bEuroimmun, IgG, S1 antigen (ratio) | cLiaison, IgG, S1/S2 antigen (AU/mL) | dAcro IgG/IgM (x/x), pos or neg | eXiamen Biotime IgG/IgM (x/x), pos or neg | fMNT (titer) | |
|---|---|---|---|---|---|---|---|---|
| Nuclear Ab, pattern (titer)1 Rf (+/-)1 | ||||||||
| 1 | homogeneous (1280), Rf(-) | NEG (0.03) | NEG (0.59) | NEG (0.35) | NEG (0.95) | pos/pos | neg/neg | <40 |
| 2 | homogeneous (1280,) Rf(-) | NEG (0.07) | NEG (0.20) | NEG (0.43) | NEG (2.38) | pos/pos | neg/neg | <40 |
| 3 | homogeneous (>5000), Rf(-) | NEG (0.09) | INCONC.(1.05) | NEG (0.31) | INCONC.(13.2) | pos/pos | neg/neg | <40 |
| 4 | homogeneous (1280), Rf(-) | NEG (0.31) | NEG (0.54) | NEG (0.58) | NEG (3.02) | pos/neg | neg/neg | <40 |
| 5 | homogeneous (>5000), Rf(-) | NEG (0.06) | NEG (0.15) | INCONC.(0.93) | NEG (5.25) | pos/neg | neg/neg | <40 |
| 6 | homogeneous (1280,) Rf(-) | NEG (0.04) | INCONC.(0.99) | NEG (0.44) | NEG (2.56) | neg/neg | neg/neg | <40 |
| 7 | homogeneous (1280), Rf(-) | NEG (0.03) | POS (2.45) | POS (1.13) | Invalid result | neg/neg | pos/pos | <40 |
| 8 | speckled (>5000), Rf(-) | NEG (0.13) | NEG (0.39) | NEG (0.31) | NEG (2.25) | neg/neg | neg/neg | <40 |
| 9 | speckled (1280), Rf(-) | NEG (0.09) | NEG (0.55) | NEG (0.28) | NEG (3.38) | neg/neg | neg/neg | <40 |
| 10 | speckled (>5000), Rf(-) | NEG (0.03) | POS (1.12) | NEG (0.41) | NEG (2.56) | neg/neg | neg/neg | <40 |
| 11 | speckled (1280), Rf(+) | NEG (0.06) | NEG (0.69) | NEG (0.61) | NEG (6.91) | neg/neg | neg/neg | <40 |
| 12 | speckled (1280), Rf(-) | POS (1.82) | NEG (0.21) | NEG (0.38) | NEG (2.28) | neg/neg | neg/neg | <40 |
| 13 | speckled (1280), Rf(-) | NEG (0.04) | INCONC.(0.96) | NEG (0.61) | NEG (3.01) | neg/neg | neg/neg | <40 |
| 14 | speckled (>5000), Rf(+) | NEG (0.02) | NEG (0.31) | NEG (0.33) | NEG (4.30) | neg/neg | neg/neg | <40 |
| 15 | Centromere + AMA (1280), Rf(-) | NEG (0.02) | NEG (0.15) | NEG (0.29) | NEG (2.06) | neg/neg | neg/neg | <40 |
| 16 | centromere (1280), Rf(-) | NEG (0.07) | POS (9.42) | NEG (0.64) | NEG (1.50) | neg/neg | neg/neg | <40 |
| 17 | centromere (1280), Rf(-) | NEG (0.01) | INCONC.(1.01) | NEG (0.68) | NEG (3.12) | neg/neg | neg/neg | <40 |
| 18 | centromere (1280), Rf(-) | NEG (0.01) | NEG (0.16) | NEG (0.24) | NEG (3.22) | neg/neg | neg/neg | <40 |
| 19 | centromere (1280), Rf(-) | NEG (0.02) | NEG (0.07) | NEG (0.23) | POS (35.5) | neg/neg | neg/neg | <40 |
| 20 | nucleolar. (80), Rf(-) | NEG (0.02) | NEG (0.47) | NEG (0.28v | NEG (1.28) | pos/neg | neg/neg | <40 |
| 21 | speckled (5000) and nuclear dots (1280), Rf(-) | NEG (0.39) | NEG (0.20) | NEG (0.32) | NEG (4.31) | neg/pos | neg/neg | <40 |
| Phospolipase receptor 2A pos (titer)1, Rf(+/-)1 | 20/21 neg | 14/21 neg | 19/21 neg | 18/21 neg | 14/21 neg | 20/21 neg | ||
| 1 | 50, Rf(-) | NEG (0.04) | NEG (0.13) | NEG (0.20 | NEG (1.49) | neg/neg | neg/neg | <40 |
| 2 | 50, Rf(-) | NEG (0.01) | NEG (0.17) | NEG (0.19 | NEG (2.43) | pos/pos | neg/neg | <40 |
| 3 | 50, Rf(-) | NEG (0.06) | NEG (0.11) | NEG (0.22 | NEG (0.96) | neg/neg | neg/neg | <40 |
| 4 | 50, Rf(-) | NEG (0.03) | NEG (0.42) | NEG (0.21 | NEG (2.11) | neg/pos | neg/pos | <40 |
| 5 | 50, Rf(-) | NEG (0.03) | POS (2.06) | NEG (0.37 | NEG (3.51) | pos/pos | neg/pos | <40 |
| 6 | 50, Rf(-) | NEG (0.05) | NEG (0.46) | NEG (0.31 | NEG (1.64) | neg/pos | neg/neg | <40 |
| 7 | 250, Rf(-) | NEG (0.02) | NEG (0.21) | NEG (0.21 | NEG (1.49) | neg/neg | neg/neg | <40 |
| 8 | 50, Rf(-) | NEG (0.01) | NEG (0.30) | NEG (0.41 | NEG (0.93) | pos/neg | neg/neg | <40 |
| 9 | 50, Rf(-) | NEG (0.01) | NEG (0.11 | NEG (0.16 | NEG (0.27) | pos/neg | neg/neg | <40 |
| 10 | 50, Rf(+) | NEG (0.15) | NEG (0.26 | NEG (0.32 | NEG (1.69) | neg/neg | neg/neg | <40 |
| GBM Ab pos (titer)1, Rf(+/-)1 | 10/10 neg | 9/10 neg | 10/10 neg | 10/10 neg | 4/10 neg | 8/10 neg | ||
| 1 | 250, Rf(+) | NEG (0.04) | NEG (0.18) | NEG (0.24) | NEG (3.02) | neg/neg | neg/neg | <40 |
| 2 | 250, Rf(-) | NEG (0.04) | NEG (0.19) | NEG (0.32) | Invalid result | neg/neg | neg/neg | <40 |
| 3 | 50, Rf(-) | NEG (0.14) | NEG (0.35) | NEG (0.23) | NEG (2.50) | neg/pos | neg/neg | <40 |
| ANCA Ab pos (titer)1, Rf(+/-)1 | 3/3 neg | 3/3 neg | 3/3 neg | 2/3 neg | 2/3 neg | 3/3 neg | ||
| 1 | atypical C-ANCA (50), Rf(-) | NEG (0.10) | NEG (0.69) | NEG (0.45) | NEG (3.73) | neg/neg | neg/neg | <40 |
| 2 | C-ANCA (1280), Rf(-) | NEG (0.12) | NEG (0.44) | NEG (0.30) | NEG (4.34) | neg/neg | neg/neg | <40 |
| 3 | P-ANCA (200), Rf(-) | NEG (0.03) | NEG (0.28) | NEG (0.24) | Invalid result | neg/neg | neg/neg | <40 |
| 4 | C-ANCA (50), P-ANCA (1280), Rf(-) | NEG (0.07) | NEG (0.28) | NEG (0.31) | NEG (2.20) | neg/neg | neg/neg | <40 |
| 5 | P-ANCA (200), Rf(+) | NEG (0.07) | NEG (0.13) | NEG (0.23) | NEG (3.02) | pos/pos | neg/neg | <40 |
| Primary EBV infection (IgG, IgM, AVI)2 | 5/5 neg | 5/5 neg | 5/5 neg | 4/5 neg | 4/5 neg | 5/5 neg | ||
| 1 | POS, POS, LOW | NEG (0.07) | NEG (0.73) | INCONC.(1.09) | NEG (3.71) | pos/pos | neg/pos | <40 |
| 2 | POS, POS, LOW | NEG (0.02) | NEG (0.32) | NEG (0.33) | NEG (1.83) | neg/pos | neg/pos | <40 |
| 3 | POS, POS, LOW | NEG (0.09) | INCONC.(0.82) | INCONC.(0.96) | NEG (1.86) | neg/pos | neg/neg | <40 |
| HCoV samples3 | 3/3 neg | 2/3 neg | 1/3 neg | 3/3 neg | 0/3 neg | 1/3 neg | ||
| 1 | HCoV OC43 | NEG (0.05) | INCONC.(0.97) | NEG (0.13) | NEG (3.97) | neg/neg | neg/neg | <40 |
| 2 | HCoV OC43 | NEG (0.02) | NEG (0.09) | NEG (013) | NEG (2.33) | pos/neg | neg/neg | <40 |
| 3 | HCoV OC43 | NEG (0.05) | NEG (0.25) | NEG (0.17) | NEG (2.39) | neg/neg | neg/neg | <40 |
| 4 | HCoV OC43 | NEG (0.08) | POS (1.22) | POS (2.54) | NEG (2.20) | neg/neg | neg/neg | <40 |
| Samples from year 20194 | 4/4 neg | 2/4 neg | 3/4 neg | 4/4 neg | 4/4 neg | 4/4 neg | ||
| 1 | NEG (0.04) | POS (5.12) | INCONC.(1.07) | NEG (2.19) | neg/neg | neg/neg | <40 | |
| 2 | NEG (0.02) | NEG (0.23) | NEG (0.16) | POS (17.8) | neg/neg | neg/neg | <40 | |
| 3 | NEG (0.15) | NEG (0.43) | NEG (0.30) | POS (16.0) | neg/pos | neg/neg | <40 | |
| 4 | NEG (0.07) | INCONC.(1.07) | INCONC.(0.96) | NEG (3.27) | neg/neg | neg/neg | <40 | |
| 5 | NEG (0.02) | POS (1.73) | POS (5.71) | NEG (2.44) | neg/pos | neg/pos | <40 | |
| 6 | NEG (0.03) | POS (1.25) | POS (2.42) | NEG (2.47) | pos/neg | nd/nd | <40 | |
| 7 | NEG (0.02) | POS (4.51) | POS (1.70) | NEG (1.70) | pos/pos | nd/nd | <40 | |
| 8 | NEG (0.04) | POS (1.52) | NEG (0.28) | NEG (1.89) | nd/nd | nd/nd | nd | |
| 9 | NEG (0.11) | NEG (0.23) | NEG (0.35) | NEG (3.73) | nd/nd | nd/nd | nd | |
| 10 | NEG (0.07) | NEG (0.28) | NEG (0.29) | NEG (3.25) | nd/nd | nd/nd | nd | |
| 11 | NEG (0.03) | NEG (0.48) | NEG (0.76) | NEG (5.76) | nd/nd | nd/nd | nd | |
| 12 | NEG (0.01) | NEG (0.25) | NEG (0.29) | NEG (2.11) | nd/nd | nd/nd | nd | |
| 13 | NEG (0.04) | NEG (0.18) | NEG (0.14) | NEG (1.59) | nd/nd | nd/nd | nd | |
| 14 | NEG (0.11) | POS (7.96) | NEG (0.60) | NEG (3.02) | nd/nd | nd/nd | nd | |
| 15 | NEG (0.03) | INCONC.(1.02) | NEG (0.31) | NEG (2.72) | nd/nd | nd/nd | nd | |
| 16 | NEG (0.02) | NEG (0.09) | NEG (0.25) | NEG (5.60) | nd/nd | nd/nd | nd | |
| 17 | NEG (0.02) | NEG (0.42) | NEG (0.27) | NEG (1.48) | nd/nd | nd/nd | nd | |
| 18 | NEG (0.02) | NEG (0.17) | NEG (0.22) | NEG (1.32) | nd/nd | nd/nd | nd | |
| 19 | NEG (0.02) | NEG (0.56) | NEG (0.23) | NEG (2.13) | nd/nd | nd/nd | nd | |
| 20 | NEG (0.02) | NEG (0.39) | NEG (0.20) | NEG (2.86) | nd/nd | nd/nd | nd | |
| 21 | NEG (0.02) | NEG (0.26) | NEG (0.18) | NEG (1.15) | nd/nd | nd/nd | nd | |
| 22 | NEG (0.02) | NEG (0.77) | NEG (0.18) | NEG (1.00) | nd/nd | nd/nd | nd | |
| 23 | NEG (0.01) | NEG (0.73) | NEG (0.13) | NEG (1.04) | nd/nd | nd/nd | nd | |
| 24 | NEG (0.03) | NEG (0.43) | NEG (0.32) | NEG (2.36) | nd/nd | nd/nd | nd | |
| 25 | POS (2.09) | NEG (0.35) | NEG (0.21) | NEG (1.64) | nd/nd | nd/nd | nd | |
| 26 | NEG (0.01) | NEG (0.21) | NEG (0.15) | NEG (2.04) | nd/nd | nd/nd | nd | |
| 27 | NEG (0.10) | POS (6.82) | NEG (0.44) | NEG (1.54) | nd/nd | nd/nd | nd | |
| 28 | NEG (0.02) | POS (1.52) | NEG (0.71) | NEG (5.75) | nd/nd | nd/nd | nd | |
| 29 | NEG (0.04) | POS (1.80) | NEG (0.21) | NEG (5.41) | nd/nd | nd/nd | nd | |
| 30 | NEG (0.05) | NEG (0.61) | NEG (0.30) | NEG (2.69) | nd/nd | nd/nd | nd | |
| 31 | NEG (0.01) | NEG (0.75) | NEG (0.30) | NEG (2.26) | nd/nd | nd/nd | nd | |
| 32 | NEG (0.01) | NEG (0.16) | NEG (0.21) | NEG (1.28) | nd/nd | nd/nd | nd | |
| 33 | NEG (0.03) | NEG (0.77) | NEG (0.29) | NEG (2.87) | nd/nd | nd/nd | nd | |
| 34 | NEG (0.01) | NEG (0.08) | NEG (0.14) | NEG (0.73) | nd/nd | nd/nd | nd | |
| 35 | NEG (0.04) | NEG (0.23) | NEG (0.29) | NEG (1.70) | nd/nd | nd/nd | nd | |
| 34/35 neg | 24/35 neg | 30/35 neg | 33/35 neg | 3/7 neg | 4/5 neg | |||
| Specificity % | 97.5 % | 75.3 % | 87.7 % | 91.4 % | 30/53 neg | 45/51 neg | ||
| Assay process successful % of samples | 100 % | 100 % | 100 % | 96.3 % | ||||
Neg, negative; pos, positive; inconc., inconclusive; nd, not determined; Rf (-), rheumatoid factor negative; Rf(+), rheumatoid factor positive; PLA2R, phospolipase A2 receptor; GBM, glomerular basement membrane; ANCA, antineutrophil cytoplasmic antibodies; EBV, Epstein-Barr virus; MNT, microneutralisation assay.
Nuclear, phospholipase A2 receptor (PLA2R), glomerular basement membrane (GBM), antineutrophil cytoplasmic (ANCA) antibodies were detected using immunofluorescence assays of NOVA Lite® DAPI ANA Kit (Inova Diagnostics, California, USA), Anti-Phospholipase A2 Receptor IIFT (IgG) (Euroimmun, Lübeck, Germany), EUROPLUS kidney (monkey) and GBM antigen IIFT (Euroimmun, Lübeck, Germany) and NOVA Lite® ANCA IFA Kit (Inova Diagnostics, California, USA), respectively. Rheumatoid factors (Rf) were determined using RapiTex® RF (Siemens Healthcare Diagnostics, Erlangen, Germany).
EBV IgG and IgM were determined using Enzygnost Anti-EBV/IgG and Anti-EBV/IgM II (Siemens Healthcare Diagnostics, Erlangen, Germany).
HCoV were detected using xTAG® Respiratory Viral panel kit (Luminex Corporation, Texas, USA) from nasopharyngeal samples and corresponding serum samples taken from the patient were used for testing antibodies.
All serum samples were sent for antibody testing of influenza A, B, respiratory syncytial virus, parainfluenza virus, enterovirus IgG antibodies (HUSLAB, Finland) in 2019.
Architect SARS-CoV-2 IgG Assay (Abbott, Illinois, USA).
Anti-SARS-CoV-2 IgA and IgG EIA (Euroimmun, Lübeck, Germany).
LIAISON® SARS-CoV-2 IgG (DiaSorin, Saluggia, Italy).
2019-nCoV IgG/IgM Rapid Test Cassette (Acro Biotech, California, USA).
SARS-CoV-2 IgG/IgM Rapid Test (Xiamen Biotime, Fujian, China).
Microneutralisation assay were carried out according protocol described by Haveri et al. (2020).
Thirty-nine out of 81 samples contained autoantibodies: 21 had anti-nuclear antibodies in Hep-2 cell IFA analysis [NOVA Lite® DAPI ANA Kit, Inova Diagnostics, California, USA; staining patterns: homogenous, 7/21; speckled 7/21; centromere 4/21; centromere + anti-mitochondrial antibodies 1/21; nucleolar 1/21; speckled and nuclear dots 1/21], 10 were positive for phospholipase receptor A2 (PLA2R) antibodies (Anti-Phospholipase A2 Receptor IIFT (IgG) (Euroimmun, Lübeck, Germany), three for glomerular basement membrane (GBM) antibodies (EUROPLUS kidney (monkey) and GBM antigen IIFT, Euroimmun, Lübeck, Germany), and five for antineutrophil cytoplasmic (ANCA) antibodies (NOVA Lite® ANCA IFA Kit (Inova Diagnostics, California, USA) (Table 1). Presence of rheumatoid factor was tested for these 39 samples using RapiTex® RF (Siemens Healthcare Diagnostics, Erlangen, Germany) agglutination assay.
Three serum samples were from patients with primary Epstein-Barr virus infection (mononucleosis; EBV IgG and IgM were determined using Enzygnost Anti-EBV/IgG and Anti-EBV/IgM II kits (Siemens Healthcare Diagnostics, Erlangen, Germany)), four were from patients who had an ongoing Human coronavirus (HCoV) OC43 infection. HCoVs were detected using xTAG® Respiratory Viral panel kit (Luminex Corporation, Texas, USA) from nasopharyngeal samples and corresponding serum samples were collected and used for testing antibodies. In addition, 35 were serum samples originally sent for testing of respiratory virus antibodies (Helsinki University Hospital Laboratory, HUSLAB, Helsinki, Finland; Table 1).
4. Serum samples comprising the COVID-19 patient panel
The patient panel consisted of serum samples from coronavirus 19 disease (COVID-19) patients, who had been diagnosed by PCR-based methods from nasopharyngeal samples in our laboratory (Table 2 ). For molecular testing, three different methods were used: cobas® SARS-CoV-2 test on the Cobas® 6800 system (Roche Diagnostics, Basel, Switzerland), Amplidiag® COVID-19 test (Mobidiag, Espoo, Finland) and a protocol based on Corman et al. [2020; [4]].
Table 2.
a and b. Days after onset of symptoms and results from PCR-confirmed COVID-19 patients using microneutralisation test and Abbott SARS-CoV-2 IgG, Euroimmun SARS-CoV-2 IgA, Euroimmun SARS-CoV-2 IgG and LiaisonSARS-CoV-2 IgG (Diasorin) automated immunoassays (a) and two lateral flow rapid tests of Acro and Xiamen Biotime aimed for detection of IgG and IgM antibodies (b).
| a | |||||||
|---|---|---|---|---|---|---|---|
| Days after onset of symptoms | ID No | Severity | MNT (titer) | Abbott IgG CMIA (N ag; index) | EIM IgA ELISA (S1 ag; ratio) | EIM IgG ELISA (S1 ag; ratio) | Liaison IgG CLIA (S1/S2 ag; AU/mL) |
| 1 - 5 | 1 | Mild | <40 | neg (0.02) | neg (0.54) | neg (0.26) | neg (5.15) |
| 2 | Mild | <40 | neg (0.03) | neg (0.36) | neg (0.34) | neg (2.16) | |
| 4 | Mild | <40 | neg (0.02) | neg (0.43) | neg (0.35) | neg (1.91) | |
| 48 | Mild | <40 | neg (0.05) | neg (0.3) | neg (0.31) | neg (1.56) | |
| 6 | Mild | <40 | neg (0.04) | neg (0.41) | neg (0.35) | neg (2.4) | |
| 46 | Severe | <40 | neg (0.02) | neg (0.73) | neg (0.19) | neg (1.75) | |
| 3−1 | Moderate | <40 | neg (0.02) | eq (0.8) | neg (0.26) | neg (1.95) | |
| 51 | Moderate | <40 | neg (0.02) | eq (0.82) | neg (0.47) | neg (2.94) | |
| 7−1 | Moderate | <40 | neg (0.06) | eq (0.98) | neg (0.3) | neg (2.42) | |
| 39 | Severe | <40 | neg (0.02) | eq (0.19) | neg (0.16) | neg (11.7) | |
| 36 | Severe | 80 | pos (5.63) | pos (4.43) | pos (2.89) | neg (11.6) | |
| 50 | Moderate | 160 | pos (5.27) | pos (3.96) | pos (6.22) | pos (22.8) | |
| 6 -14 | 10−1 | Moderate | <40 | neg (0.03) | neg (0.52) | neg (0.40) | neg (1.28) |
| 26 | Severe | <40 | neg (0.01) | neg (0.39) | neg (0.16) | neg (1.43) | |
| 24 | Severe | <40 | neg (0.05) | neg (0.19) | neg (0.19) | neg (1.83) | |
| 38 | Severe | <40 | neg (0.07) | neg (0.78) | neg (0.25) | neg (2.95) | |
| 43 | Severe | <40 | neg (0.04) | neg (0.62) | neg (0.22) | neg (3.56) | |
| 32 | Severe | <40 | neg (0.07) | neg (0.39) | neg (0.16) | neg (5.08) | |
| 3−2 | Moderate | <40 | neg (0.02) | eq (0.92) | neg (0.21) | neg (1.61) | |
| 20 | Mild | <40 | neg (0.02) | pos (4.00) | neg (0.79) | neg (1.18) | |
| 8−1 | Moderate | <40 | neg (0.09) | pos (2.29) | neg (0.56) | neg (3.27) | |
| 44−1 | Moderate | <40 | neg (0.22) | pos (1.80) | neg (0.32) | neg (4.65) | |
| 7−2 | Moderate | <40 | neg (0.07) | pos (1.24) | neg (0.26) | neg (2.63) | |
| 13 | Severe | <40 | neg (0.03) | pos (2.24) | neg (0.25) | neg (7.59) | |
| 42 | Moderate | <40 | pos (1.93) | pos (1.56) | neg (0.33) | neg (2.23) | |
| 29 | Moderate | 40 | neg (0.06) | neg (0.17) | neg (0.28) | neg (7.78) | |
| 40 | Moderate | 40 | neg (0.72) | neg (0.75) | eq (1.06) | neg (2.60) | |
| 25−1 | Moderate | 40 | neg (1.22) | pos (1.31) | neg (0.49) | neg (5.72) | |
| 28 | Severe | 40 | neg (1.11) | pos (1.74) | neg (0.32) | neg (10) | |
| 14 | Moderate | 40 | neg (0.45) | pos (4.02) | neg (0.46) | neg (7.59) | |
| 15 | Severe | 80 | pos (1.64) | neg (0.67) | neg (0.34) | neg (3.17) | |
| 49 | Severe | 80 | pos (2.48) | pos (3.85) | neg (0.78) | neg (1.97) | |
| 22 | Moderate | 80 | pos (7.48) | pos (1.88) | neg (0.64) | neg (3.43) | |
| 62 | Mild | 80 | pos (6.48) | pos (1.47) | neg (0.50) | ND | |
| 47 | Severe | 80 | pos (2.23) | pos (4.14) | pos (1.33) | neg (8.60) | |
| 16 | Moderate | 80 | pos (5.69) | pos (4.86) | pos (1.50) | neg (3.76) | |
| 8−2 | Moderate | 80 | pos (3.12) | pos (6.90) | pos (2.01) | eq (13.1) | |
| 30 | Severe | 160 | neg (0.24) | neg (0.53) | neg (0.21) | neg (2.61) | |
| 10−2 | Moderate | 160 | pos (2.04) | pos (3.42) | neg (0.55) | eq (12.4) | |
| 18 | Severe | 160 | pos (1.52) | pos (8.66) | eq (0.84) | neg (5.17) | |
| 23 | Severe | 160 | pos (5.05) | pos (3.11) | pos (1.62) | neg (11.8) | |
| 21 | Moderate | 160 | pos (5.39) | pos (3.50) | pos (1.54) | neg (5.34) | |
| 35 | Severe | 160 | pos (4.15) | pos (10.51) | pos (5.96) | eq (14.8) | |
| 34 | Moderate | 160 | pos (7.91) | pos (4.19) | pos (1.68) | pos (6.18) | |
| 44−2 | Moderate | 160 | pos (4.75) | pos (30.12) | pos (1.90) | pos (43.1) | |
| 31 | Severe | 160 | pos (7.27) | pos (5.39) | pos (1.3) | pos (20.0) | |
| 53 | Moderate | 320 | pos (8.57) | pos (28.17) | pos (10.84) | pos (51.4) | |
| 25−2 | Moderate | 320 | pos (6.03) | pos (5.73) | pos (5.24) | pos (22.8) | |
| 12 | Moderate | 640 | neg (0.97) | pos (6.37) | neg (0.44) | neg (4.29) | |
| 27 | Mild | >2560 | pos (4.74) | pos (10.9) | pos (1.84) | pos (40.2) | |
| 15 - 21 | 52 | Moderate | <40 | neg (0.17) | pos (1.43) | neg (0.42) | neg (2.92) |
| 5−1 | Mild | <40 | pos (1.74) | pos (4.26) | neg (0.75) | neg (9.91) | |
| 5−2 | Mild | <40 | pos (2.39) | pos (3.19) | neg (0.60) | neg (5.28) | |
| 37−1 | Moderate | 320 | pos (4.00) | pos (31.11) | pos (2.42) | pos (45.3) | |
| 19 | Mild | 1280 | pos (2.57) | pos (31.11) | pos (10.09) | pos (42.0) | |
| 33 | Severe | >2560 | pos (7.92) | pos (31.11) | pos (13.41) | pos (52.4) | |
| >21 | 57 | Mild | 80 | pos (4.01) | pos (8.95) | pos (6.60) | ND |
| 58 | Mild | 80 | pos (3.38) | pos (5.48) | pos (8.08) | ND | |
| 61 | Mild | 160 | pos (9.27) | pos (2.93) | pos (12.49) | ND | |
| 37−2 | Moderate | 320 | pos (6.13) | pos (31.11) | pos (9.22) | pos (78.2) | |
| 59 | Mild | 320 | pos (4.68) | pos (4.93) | pos (11.44) | ND | |
| 60 | Severe | 640 | pos (6.49) | pos (30.22) | pos (14.58) | ND | |
| 56 | Mild | 1280 | pos (8.26) | pos (30.22) | pos (15.07) | ND | |
| NA | 45 | Moderate | <40 | neg (0.01) | neg (0.67) | neg (0.15) | neg (1.79) |
| 17 | NA | <40 | neg (0.02) | neg (0.19) | neg (0.19) | neg (2.04) | |
| 9 | NA | <40 | neg (0.06) | neg (0.51) | eq (0.85) | neg (2.54) | |
| 41 | NA | 80 | pos (2.69) | pos (6.27) | eq (0.97) | pos (27.3) | |
| 55 | NA | 80 | pos (2.07) | pos (6.59) | pos (4.46) | ND | |
| 11 | NA | 160 | neg (0.81) | pos (2.49) | neg (0.47) | neg (1.97) | |
| 54 | Mild | 640 | pos (8.10) | pos (5.95) | pos (14.45) | ND | |
| b | |||||||
|---|---|---|---|---|---|---|---|
| Days after onset of symptoms | ID No | Severity | MNT (titer) | Acro IgG | Acro IgM | Xiamen Biotime IgG | Xiamen Biotime IgM |
| 1 - 5 | 1 | Mild | <40 | neg | neg | neg | neg |
| 2 | Mild | <40 | neg | neg | neg | neg | |
| 4 | Mild | <40 | pos | neg | neg | neg | |
| 48 | Mild | <40 | neg | neg | neg | neg | |
| 6 | Mild | <40 | neg | neg | neg | neg | |
| 46 | Severe | <40 | neg | pos | neg | neg | |
| 3−1* | Moderate | <40 | neg | neg | neg | neg | |
| 51 | Moderate | <40 | neg | neg | neg | neg | |
| 7−1* | Moderate | <40 | neg | neg | neg | neg | |
| 39 | Severe | <40 | neg | neg | neg | neg | |
| 36 | Severe | 80 | pos | pos | pos | pos | |
| 50 | Moderate | 160 | neg | neg | pos | pos | |
| 6 -14 | 10−1* | Moderate | <40 | neg | neg | neg | neg |
| 26 | Severe | <40 | neg | pos | neg | neg | |
| 24 | Severe | <40 | neg | neg | neg | neg | |
| 38 | Severe | <40 | neg | pos | neg | neg | |
| 43 | Severe | <40 | neg | pos | neg | neg | |
| 32 | Severe | <40 | neg | neg | neg | neg | |
| 3−2* | Moderate | <40 | neg | pos | neg | neg | |
| 20 | Mild | <40 | pos | pos | neg | pos | |
| 8−1* | Moderate | <40 | neg | neg | neg | neg | |
| 44−1* | Moderate | <40 | pos | pos | pos | pos | |
| 7−2* | Moderate | <40 | neg | neg | neg | neg | |
| 13 | Severe | <40 | neg | pos | neg | neg | |
| 42 | Moderate | <40 | pos | neg | pos | pos | |
| 29 | Moderate | 40 | neg | neg | neg | neg | |
| 40 | Moderate | 40 | neg | neg | neg | neg | |
| 25−1* | Moderate | 40 | pos | pos | neg | pos | |
| 28 | Severe | 40 | neg | neg | neg | pos | |
| 14 | Moderate | 40 | neg | pos | neg | pos | |
| 15 | Severe | 80 | pos | pos | pos | pos | |
| 49 | Severe | 80 | pos | pos | pos | pos | |
| 22 | Moderate | 80 | pos | neg | pos | pos | |
| 62 | Mild | 80 | neg | neg | ND | ND | |
| 47 | Severe | 80 | pos | pos | pos | pos | |
| 16 | Moderate | 80 | pos | pos | pos | pos | |
| 8−2* | Moderate | 80 | pos | neg | neg | neg | |
| 30 | Severe | 160 | neg | neg | neg | neg | |
| 10−2* | Moderate | 160 | pos | pos | pos | pos | |
| 18 | Severe | 160 | pos | pos | neg | pos | |
| 23 | Severe | 160 | neg | pos | pos | pos | |
| 21 | Moderate | 160 | pos | pos | pos | pos | |
| 35 | Severe | 160 | pos | pos | pos | pos | |
| 34 | Moderate | 160 | neg | neg | pos | pos | |
| 44−2* | Moderate | 160 | pos | pos | pos | pos | |
| 31 | Severe | 160 | pos | pos | pos | pos | |
| 53 | Moderate | 320 | neg | neg | pos | neg | |
| 25−2* | Moderate | 320 | pos | pos | pos | pos | |
| 12 | Moderate | 640 | pos | neg | neg | neg | |
| 27 | Mild | >2560 | neg | pos | pos | pos | |
| 15 - 21 | 52 | Moderate | <40 | neg | neg | neg | neg |
| 5−1* | Mild | <40 | pos | pos | neg | pos | |
| 5−2* | Mild | <40 | pos | pos | neg | neg | |
| 37−1* | Moderate | 320 | neg | neg | pos | pos | |
| 19 | Mild | 1280 | neg | pos | pos | pos | |
| 33 | Severe | >2560 | pos | pos | pos | pos | |
| >21 | 57 | Mild | 80 | pos | neg | ND | ND |
| 58 | Mild | 80 | pos | neg | ND | ND | |
| 61 | Mild | 160 | neg | neg | ND | ND | |
| 37−2* | Moderate | 320 | neg | neg | pos | pos | |
| 59 | Mild | 320 | neg | neg | ND | ND | |
| 60 | Severe | 640 | pos | neg | ND | ND | |
| 56 | Mild | 1280 | neg | neg | ND | ND | |
| NA | 45 | Moderate | <40 | neg | neg | neg | neg |
| 17 | NA | <40 | neg | neg | neg | neg | |
| 9 | NA | <40 | pos | neg | neg | neg | |
| 41 | NA | 80 | pos | neg | pos | pos | |
| 55 | NA | 80 | pos | neg | ND | ND | |
| 11 | NA | 160 | pos | pos | pos | pos | |
| 54 | Mild | 640 | neg | neg | ND | ND | |
ND, not determined due to the limitation of tests available; NA, not available; MNT, microneutralisation test; eq, equivocal; pos, positive; neg, negative.
a) All results were determined according to manufacturers´ instructions. In this study, the Liaison SARS-CoV-2 IgG chemiluminescent assay (CLIA; Diasorin) was research use only (RUO) kit. Abbott SARS-CoV-2 IgG chemiluminescent microparticle immunoassay (CMIA), Euroimmun (EIM) SARS-CoV-2 IgA and IgG enzyme linked immunoassay (ELISA) were all CE marked kits.
b) All results were determined according to manufacturers´ instructions. Acro IgG/IgM rapid lateral flow test and Xiamen Biotime IgG/IgM rapid lateral flow test are both CE marked kits.
Two separate samples were available from the patients. First number is the identification code for patient, second for the sample; examples, 3−1 and 3−2, 5−1 and 5−2.
In total, 70 serum samples from 62 individuals (median age 54 years, range 24–86 years; 28 males, 34 females; Table 2) were available for this study. Data were collected and samples treated according to permit HUS/32/2018 (Helsinki University Hospital, Finland).
5. Automated immunoassays for anti-SARS-CoV-2 IgG or IgA detection
The analysis of SARS-COV-2 IgG or IgA antibodies were carried out using the Architect Plus i2000sr Analyzer (Abbott, Illinois, USA) and SARS-COV-2 IgG CMIA kit (nucleoprotein based antigen; Abbott; CE marked), EUROLabworkstation (Euroimmun, Lübeck, Germany) and SARS-COV-2 IgG and IgA ELISA kits (S1-based antigen; Euroimmun) and Diasorin Liaison® XL (DiaSorin, Saluggia, Italy) and SARS-CoV-2 S1/S2 IgG CLIA kit (S1/S2 based antigen; DiaSorin; RUO) according to the manufacturers´ instructions. All samples from the negative panel (N = 81) and the patient panel (N = 70) were tested with Abbott SARS-CoV-2 IgG, and Euroimmun SARS-CoV-2 IgA and IgG. All samples from the negative panel (N = 81) and (due to limited kit supply) 61/70 samples (53/62 individuals) from the COVID-19 patient panel were tested with DiaSorin SARS-CoV-2 S1/S2 IgG.
6. Rapid lateral flow tests
2019-nCoV IgG/IgM (Acro Biotech, California, USA; CE marked)] and SARS-CoV-2 IgG/IgM (Xiamen Biotime Biotechnology, Fujian, China; CE marked) rapid lateral flow (immunocromatographic) tests were evaluated. Altogether, 53/81 samples from the negative panel and all 70 specimens from the patient panel were tested with Acro Biotech, and 51/81 samples from the negative panel and 61/70 from the patient panel were tested with Xiamen Biotime.
7. Microneutralisation test
MNT was conducted for 53/81 of the negative panel and all of the 70 specimens from the COVID-19 patient panel (Table 1, Table 2). Microneutralisation assays were carried out for 39 (39/53) samples positive for autoantibodies, three (3/53) samples from patients with primary EBV infection, four (4/53) samples from patients with acute HCoV OC43 infection, and seven (7/53) samples which had been sent for testing of respiratory virus antibodies.
MNT was performed in a BSL-3 laboratory as described previously [5] with modifications. Briefly, SARS-CoV-2/Finland/1/2020 was passaged five times in Vero E6 cells in MEM supplemented with 2% of heat-inactivated FBS, l-glutamine, penicillin and streptomycin. The infectious virus titer was determined by plaque assay in Vero E6 cells. For MNT, Vero E6 cells (50 000/well) were plated the previous day on 96-well plate in MEM with 10 % FBS. Inactivated serum samples were 2-fold serially diluted in triplicates starting from 1:40 dilution in MEM with 2% FBS. Fifty plaque forming units (PFU) of SARS-CoV-2 were added to serum dilutions and incubated for 1 h at 37 °C. The growth medium was removed and the virus–serum mixture was added to the cells and incubated for 4 days at 37 °C with 5% CO2, after which the cells were stained with crystal violet to detect cytopathic effect (CPE). The neutralisation endpoint titer was determined as the endpoint serum dilution that inhibited the SARS-CoV-2 induced CPE in at least 2 out 3 parallel wells. The MNT titer ≥40 was considered as positive.
8. Results
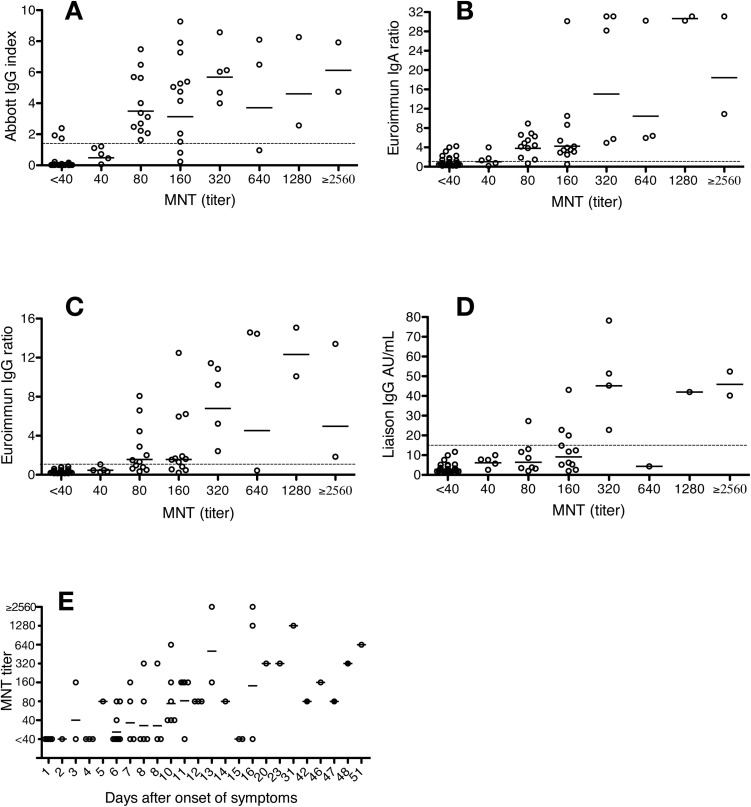
For 55 COVID-19 patients out of 62, the date of disease onset was available, and disease severity could be rated (mild, moderate or severe; based on Siddiqi et al. [2020; [6]]) (Table 2, Fig. 1 ). In the COVID-19 patients included in this study, the earliest time point for the MNT to become positive was 3 days from onset of illness (patient ID 50), while the furthest time point for a negative MNT was 16 days from onset (patient ID 5) (Fig. 1E; Table 2). Disease severity did not appear to be reflected in the MNT titers of the patients, however, the number of patients in each category was too low to assess significance (Table 2).
Fig. 1.
Comparison of microneutralisation test (MNT) and immunoassay results, and onset of illness. A) Abbott SARS-CoV-2 IgG assay (index); n = 70. B) Euroimmun SARS-CoV-2 IgA (ratio), n = 70. C) Euroimmun SARS-CoV-2 IgG (ratio), n = 70, D) Liaison SARS-CoV-2 IgG (RUO) (AU/mL), n = 62. E) Microneutralisation titers for 63 serum samples collected from 55 COVID-19 patients, organized according to the time lapse between the onset of symptoms and the sample collection. The geometric mean is marked for each titer with a solid line and immunoassay cut-off values are indicated with a dotted line.
Numeric results of Abbott Architect SARS-CoV-2 IgG, Euroimmun SARS-CoV-2 IgA, Euroimmun SARS-CoV-2 IgG and Diasorin Liaison SARS-CoV-2 IgG (RUO) were plotted against the MNT titer values (Table 2, Fig. 1). The geometric mean of the patient panel specimens exceeded the test cut-off at the following MNT titers: Abbott IgG (test cut-off 1.4 index exceeded with geometric mean 3.50 index at MNT titer 80); Euroimmun IgA (test cut-off 1.1 ratio exceeded with geometric mean 3.79 ratio at MNT titer 80), Euroimmun IgG (test cut-off 1.1 ratio exceeded with geometric mean 1.57 ratio at MNT titer 80), and Liaison IgG (test cut-off 15 AU/mL exceeded with geometric mean 45.1 AU/mL at MNT titer 320) (Fig. 1). However, the geometric mean in the Euroimmun IgG assay lingered in close proximity (geometric mean 1.57 ratio) of the cut-off (1.1 ratio) still at MNT titer of 160.
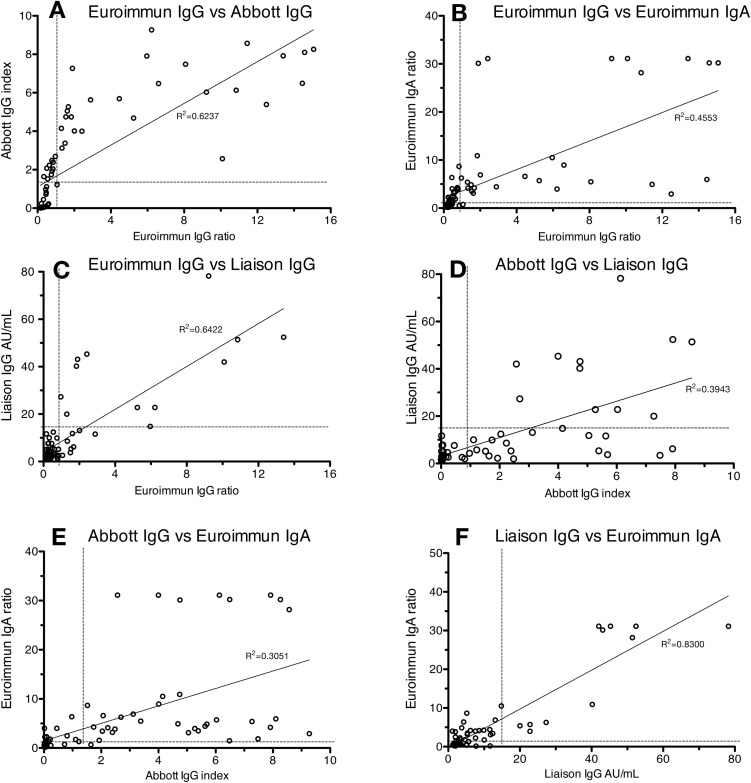
Negative and positive agreements (specificity and sensitivity) for immunoassays were calculated in comparison with MNT, in which MNT titer <40 was considered negative and ≥40 positive (Table 1, Table 2, Table 3). Equivocal results (eq in Table 1, Table 2, Table 3 ) of the commercial assays were regarded as reactive in the performance calculations. Altogether, 53 samples from the negative panel and all of the 70 samples from the COVID-19 patient panel were tested with MNT (Table 1, Table 2, Table 3). The negative and positive agreement (specificity and sensitivity) values, respectively, were as follows: 95.1 %/80.5 % (Abbott Architect SARS-CoV-2 IgG), 94.9 %/43.8 % (Diasorin Liaison SARS-CoV-2 IgG; RUO), 68.3 %/87.8 % (Euroimmun SARS-CoV-2 IgA), 86.6 %/70.7 % (Euroimmun SARS-CoV-2 IgG), 74.4 %/56.1 % (Acro Biotech 2019-nCoV IgG), 69.5 %/46.3 % (Acro Biotech 2019-nCoV IgM), 97.5 %/71.9 % (Xiamen Biotime SARS-CoV-2 IgG), and 88.8 %/81.3 % (Xiamen Biotime SARS-CoV-2 IgM). Test results from the automated immunoassays plotted against each other are shown in Fig. 2 . By using the cut-offs provided by the manufacturers, a trend was observed in which Abbott IgG yielded positive signals in specimens still negative in Euroimmun IgG and Liaison (RUO) IgG (Fig. 2). Rheumatoid factor was detected in five of negative panel specimens (Table 1). More detailed results are provided in Table 1, Table 2, Table 3.
Table 3.
Specificity and sensitivity of the six commercial immunoassays compared with MNT. MNT titer ≥40 was considered as positive. Equivocal results of the commercial assays were regarded as reactive in this analysis. The total number of specimens tested with MNT, and each of the commercial immunoassays, with their respective results are presented in Tables 1–2.
| Immunoassay, platform, antigen used, RUO/CE marked | Immunoassay qualitative test result | Number of specimens |
Specificity (compared with MNT) | Sensitivity (compared with MNT) | |
|---|---|---|---|---|---|
| MNT titer <40 | MNT titer ≥40 |
||||
| Abbott SARS-CoV-2 IgG CMIA Based on nucleoprotein antigen CE marked | pos | 4 | 33 | 95.1 % | 80.5 % |
| neg | 78 | 8 | |||
| Total | 82 | 41 | |||
| Euroimmun SARS-CoV-2 IgA ELISA Based on S1 antigen CE marked | pos | 18 | 36 | 68.3 % | 87.8 % |
| eq | 8 | 0 | |||
| neg | 56 | 5 | |||
| Total | 82 | 41 | |||
| Euroimmun SARS-CoV-2 IgG ELISA Based on S1 antigen CE marked | pos | 5 | 26 | 86.6 % | 70.7 % |
| eq | 6 | 3 | |||
| neg | 71 | 12 | |||
| Total | 82 | 41 | |||
| Liaison SARS-CoV-2 IgG CLIA Based on S1/S2 antigen Research Use Only (RUO) | pos | 3 | 11 | 94.9 % | 43.8 % |
| eq | 1 | 3 | |||
| neg | 75 | 18 | |||
| Total* | 79 | 32 | |||
| Acro Biotech, 2019-nCoV IgG, lateral flow rapid test No antigen information provided CE marked | pos | 21 | 23 | 74.4 % | 56.1 % |
| neg | 61 | 18 | |||
| Total | 82 | 41 | |||
| Acro Biotech, 2019-nCoV IgM, lateral flow rapid test No antigen information provided CE marked | pos | 25 | 19 | 69.5 % | 46.3 % |
| neg | 57 | 22 | |||
| Total | 82 | 41 | |||
| Xiamen Biotime, SARS-CoV-2 IgG, lateral flow rapid test No antigen information provided CE marked | pos | 2 | 23 | 97.5 % | 71.9 % |
| neg | 78 | 9 | |||
| Total* | 80 | 32 | |||
| Xiamen Biotime, SARS-CoV-2 IgM, lateral flow rapid test No antigen information provided CE marked | pos | 9 | 26 | 88.8 % | 81.3 % |
| neg | 71 | 6 | |||
| Total* | 80 | 32 | |||
Pos, positive; neg, negative; eq, equivocal; RUO, research use only; MNT, microneutralisation test.
Due to limited kit supply, not all specimens tested with MNT could be analysed with these commercial tests.
Fig. 2.
Test results from the automated immunoassays plotted against each other. A) Euroimmun IgG vs Abbott IgG, B) Euroimmun IgG vs. Euroimmun IgA, C) Euroimmun IgG vs. Liaison IgG (RUO), D) Abbott IgG vs Liaison IgG (RUO), E) Abbott IgG vs Euroimmun IgA, F) Liaison IgG (RUO) vs Euroimmun IgA. The immunoassay cut-off values (dotted line) and trendlines are provided.
All of the six immunoassays gave reactive results to a varying degree for the negative panel specimens (Table 1). Particularly the Acro Biotech rapid test and Euroimmun IgA assay reacted in samples retrieved from patients with autoantibodies.
As the sensitivity of the Acro Biotech rapid test was lower than the other immunoassays tested, we randomly chose an MNT positive specimen (ID 61), conducted a dilution series of 1:2 for it, and tested the specimen again with the Acro Biotech test. An evident prozone effect was detected, and the originally negative test turned IgG positive at serum dilution 1:4 up until dilution of 1:16.
9. Discussion
As serological assays for SARS-CoV-2 are now becoming available in the market in abundance [7], assessment of their analytical performance by using clinical specimens is of critical importance. In this study, we assessed the specificity and sensitivity of six commercial immunoassays for the detection of SARS-CoV-2 antibodies, including two rapid lateral flow tests, in comparison with a neutralisation test. While neutralisation assays are considered to be the gold standard in terms of specificity, they also provide evidence as to development of immunity.
Eighty-one of the specimens were retrieved in 2018 and 2019 in Finland, rendering these specimens as ascertained negative for SARS-CoV-2 antibodies, and subsequently verifying the very high specificity of the neutralisation test we used (100 % were negative in MNT). We chose serum dilution 1/40 as the limit of detection for the MNT. Failure to detect very low antibody concentrations in this setup is possible. However, four of the 62 PCR-positive individuals showed neutralising antibodies without reactivity in any of the IgG tests used, suggesting a reasonable level of sensitivity in our neutralisation assay.
RF, which is an autoantibody against the Fc portion of IgG, and a common cause of cross-reactivity in immunoassays [8], was analysed in the specimens collected in 2018 and 2019. Five out of 39 of these specimens were positive for RF; 4/5 were negative in all SARS-CoV-2 immunoassays, and 1/5 gave a positive reaction in the Acro IgG and IgM test. We conclude that the majority of positive test reactions in the six different immunoassays by using the negative serum panel from 2018−2019 were not due to RF. Of note, we observed a prozone phenomenon [9] by diluting specimen ID 61 for the Acro lateral flow assay. While we did not investigate prozone phenomenon extensively in this study, we do consider it may be an important cause for false negative test results. The prozone phenomenon has been reported for other lateral flow assays previously [10].
Of the automated assays included in this study, and by using the cut-off values set by the manufacturers, the best specificity values were observed with Abbott IgG (95.1 %). A previous report from the United States reported a 99.9 % specificity [11]. In our study, Liaison IgG (RUO) assay (94.9 %) also showed a good specificity. Euroimmun SARS-CoV-2 IgA assay had the best positive agreement (sensitivity) (87.8 %), while the positive agreement of the Liaison IgG (RUO) assay was the lowest (43.8 %). The CE marked Diasorin Liaison SARS-CoV-2 IgG assay was not available for this evaluation.
The automated assays from the three manufacturers were all based on different antigen components (S1, S2, nucleocapsid). This is noteworthy, as antibody responses against each of these antigens may develop with varying kinetics, which remains a subject for further investigation. In addition, the immunoassays may detect nonneutralizing antibodies, not detected by neutralization assays. However, the topic of interest in our study was specifically on comparability of the assays with neutralising antibodies.
When interpreting sensitivity values, the time from onset of illness in COVID-19 patients needs to be accounted for. By using the Abbott IgG assay, SARS-CoV-2 IgG seroconversion was previously reported in all patients by the day 17 post onset of illness [11]. Previous reports suggest a median seroconversion time for SARS-CoV-2 from 11 days [12] to 13 days [13]. The present study also suggests a relatively long period required for serological response to take place (Table 2, Fig. 1E). Even though extensive conclusions cannot be made from our data, Liaison IgG (RUO) appears to turn positive at a later point in time from onset of illness in comparison with the other immunoassays evaluated in our study (Table 2). Perkmann et al. ([14]; 2020) have also reported this phenomenon, and it should be investigated more thoroughly whether antibodies against SARS-CoV-2 S1/S2 antigen, in general, are detected in later time point.
Of the two rapid lateral flow assays, the Xiamen IgG/IgM showed a good specificity (97.5 % / 88.8 %) with a modest positive agreement (sensitivity) (71.9 % / 81.3 %). In line with a previous report [15], the performance of the Acro Biotech IgG/IgM rapid test appears not to be adequate for clinical use, with specificity of 74.4 % / 69.5 % and positive agreement (sensitivity) of 56.1 % / 46.3 %.
The currently very low seroprevalence of SARS-CoV-2 in most regions globally render low positive predictive values in the serological testing of individual patients. This can be somewhat improved by good targeting of groups tested. The analytical test performance can be optimised by placing several consecutive assays, with varying antigenic features, in the test workflow, ideally emphasizing sensitivity in the screening and specificity in the second-line testing. The very variable performance values observed in this study highlights the need for laboratories to carefully consider their testing process in order to optimize the overall performance of SARS-CoV-2 serodiagnostics.
Funding
Funded by Helsinki University Hospital, HUSLAB, Helsinki, Finland (KLIMIK).
Contribution
AJJ carried out the collection of retrospective samples together with HKK and MA. AJJ carried out the immunoassays (EIAs and rapid tests) and analysed the resulting data. SK carried out the microneutralisation assays. ELKE collected the data from patient records and carried out part of the rapid tests. AJJ and HJ carried out RF tests. HKK, RL, SKU and ML took part in the PCRs evaluation and setting up PCR assays in the laboratory. AJJ, SK, SKU, HJ and ML reviewed the data. HJ helped in formulating the Tables and Figures. SKU wrote the manuscript together with AJJ. AJ, SK, HKK, OV, HJ, ELKE, MA, RL, SKU and ML reviewed and modified the manuscript and approved its final version.
Declaration of Competing Interest
None of the authors have any conflict of interest.
Acknowledgements
We would like to thank Pamela Österlund (Finnish Institute for Health and Welfare, Helsinki, Finland) for providing the virus strain. We would also like to thank (in alphabetical order) Anu Jääskeläinen (Helsinki University Hospital), Pia Jokela (Helsinki University Hospital), Laura Mannonen (Helsinki University Hospital), Tarja Sironen (University of Helsinki), Satu Suuronen (Helsinki University Hospital), Anne Toivonen (Helsinki University Hospital), and Mira Utriainen (University of Helsinki).
References
- 1.Winter A.K., Hegde S.T. The important role of serology for COVID-19 control. Lancet Infect. Dis. 2020;(20):S1473–3099. doi: 10.1016/S1473-3099(20)30322-4. 30322-30324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-Specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jääskeläinen Aj, Kekäläinen E., Kallio-Kokko H., Mannonen L., Kortela E., Vapalahti O., Kurkela S., Lappalainen M. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Euro Surveill. 2020;25(18):2000603. doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu DKW Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haveri A., Smura T., Kuivanen S., Österlund P., Hepojoki J., Ikonen N., Pitkäpaasi M., Blomqvist S., Rönkkö E., Kantele A., Strandin T., Kallio-Kokko H., Mannonen L., Lappalainen M., Broas M., Jiang M., Siira L., Salminen M., Puumalainen T., Sane J., Melin M., Vapalahti O., Savolainen-Copra C. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25(11) doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqi H.K., Mehra M.R. COVID-19 illness in native and immunosuppressed states: a clinical-therapeutic staging proposal. J. Heart Lung Transplant. 2020 doi: 10.1016/j.healun.2020.03.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;4(395):1101–1102. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salonen E.M., Vaheri A., Suni J., Wager O. Rheumatoid factor in acute viral infections: interference with determination of IgM, IgG, and IgA antibodies in an enzyme immunoassay. J. Infect. Dis. 1980;142:250–255. doi: 10.1093/infdis/142.2.250. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs J.F., van der Molen R.G., Bossuyt X., Damoiseaux J. Antigen excess in modern immunoassays: to anticipate on the unexpected. Autoimmun. Rev. 2015;14:160–167. doi: 10.1016/j.autrev.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Lee G., Arthur I., Leung M. False-negative serum cryptococcal lateral flow assay result due to the prozone phenomenon. J. Clin. Microbiol. 2018;56:e01878–17. doi: 10.1128/JCM.01878-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A., Jerome K.R., Mathias P.C., Greninger A.L. Performance Characteristics of the Abbott Architect SARS-CoV-2 IgG Assay and Seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020;2020 doi: 10.1128/JCM.00941-20. pii: JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 14.Perkmann T., Perkmann-Nagele N., Breyer M.K., Breyer-Kohansal R., Burghuber O.C., Hartl S., Aletaha D., Sieghart D., Quehenberger P., Marculescu R., Mucher P., Strassl R., Wagner O.F., Binder C.J., Haslacher H. Side by side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. medRxiv. 2020 doi: 10.1101/2020.06.04.20117911. 06.04.20117911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lassaunière R., Frische A., Harboe Z.B., Nielsen ACY, Fomsgaard A, Krogfelt K.A., et al. medRxiv preprint doi: https://doi.org/10.1101/2020.04.09.20056325.