ABSTRACT
We previously identified the cyclin dependent kinase Cdk8 as a putative silencing factor for Xist. To investigate its role in X inactivation, we engineered a Cdk8 mutation in mouse embryonic stem cells (ESCs) carrying an inducible system for studying Xist function. We found that Xist repressed X-linked genes at half of the expression level in Cdk8 mutant cells, whereas they were almost completely silenced in the controls. Lack of Cdk8 impaired Ezh2 recruitment and the establishment of histone H3 lysine 27 tri-methylation but not PRC1 recruitment by Xist. Transgenic expression of wild-type but not catalytically inactive Cdk8 restored efficient gene repression and PRC2 recruitment. Mutation of the paralogous kinase Cdk19 did not affect Xist function, and combined mutations of Cdk8 and Cdk19 resembled the Cdk8 mutation. In mice, a Cdk8 mutation caused post-implantation lethality. We observed that homozygous Cdk8 mutant female embryos showed a greater developmental delay than males on day 10.5. Together with the inefficient repression of X-linked genes in differentiating Cdk8 mutant female ESCs, these data show a requirement for Cdk8 in the initiation of X inactivation.
KEY WORDS: Cdk8, Cyclin dependent kinase, Xist, X inactivation, Polycomb, Gene regulation
Summary: Mutagenesis studies enabled the functional characterization of the cyclin-dependent kinase Cdk8 in X chromosome inactivation, and showed that Cdk8 is required for efficient gene repression and PRC2 recruitment by Xist.
INTRODUCTION
Mammals achieve dosage compensation for the different number of X chromosomes in male and female cells by silencing of the transcription of one of the two X chromosomes (Lyon, 1962). X chromosome inactivation (XCI) is initiated by the expression of the long noncoding Xist RNA, which localizes to the future inactive X chromosome (Xi), and triggers chromatin modifications and gene repression in an almost chromosome-wide manner (Galupa and Heard, 2018). The process of chromosomal silencing has been studied in mice and mouse embryonic stem cells (ESCs). Initially, in female mouse embryos, Xist is expressed from the paternally inherited X chromosome at the four-cell stage and imprinted XCI is observed. In the cells of the inner cell mass of the blastocyst that form the epiblast lineage, imprinted XCI is reversed and two active X chromosomes are present in the female embryos. Subsequently, the inactivation of either the maternal or paternal inherited X chromosome is initiated in the embryo at day 5.5. The initiation of random XCI has also been extensively studied in female mouse ESCs, which initially possess two active X chromosomes and undergo XCI upon differentiation. Xist is expressed from and localizes to the future Xi and establishes a domain of repressive chromatin (Chaumeil et al., 2008). Initially, this repressive compartment is spatially separated from genes, which remain expressed and are silenced by a separate mechanism that requires A repeat sequences of Xist RNA, as well as the RNA-binding protein Spen (Chu et al., 2015; McHugh et al., 2015; Monfort et al., 2015; Wutz et al., 2002). The recruitment of polycomb proteins by Xist induces chromosome-wide histone modifications and this recruitment requires hnRNPK (Chu et al., 2015; Pintacuda et al., 2017), and the polycomb group proteins Pcgf3 and Pcgf5 (Almeida et al., 2017). It is thought that hnRNPK binds to Xist repeat B, and through the recruitment of Ring1b- and Ring1A-containing complexes, leads to ubiquitylation of histone H2A lysine 119 (H2AK119ub). This initial chromatin modification triggers the recruitment of additional polycomb repressive complex 1 (PRC1) and PRC2 (Almeida et al., 2017). PRC2 recruitment in turn catalyses trimethylation of histone H3 lysine 27 (H3K27me3). Multiple interactions of different polycomb group proteins with H2AK119ub and H3K27me3 are thought to cause an enrichment of a large number of polycomb proteins on the Xi, and to contribute to the establishment of a domain of repressive chromatin. Recent biochemical and genetic studies have identified a number of additional factors that contribute to the process of X inactivation (Chu et al., 2015; McHugh et al., 2015; Moindrot et al., 2015; Ridings-Figueroa et al., 2017). The function of Xist in chromosomal silencing has also been studied by forced expression of Xist in male ESCs, allowing the uncoupling of the formation of a silent chromosome from the complex regulation of the Xist gene (Chaumeil et al., 2008; Chu et al., 2015; McHugh et al., 2015; Plath et al., 2003; Wutz and Jaenisch, 2000). We have previously used an inducible Xist expression system for a genetic screen in mouse haploid ESCs for identifying mutations that prevent the repression of X-linked genes by forced Xist expression (Monfort et al., 2015). Among the candidate genes identified as required for Xist function is the atypical cyclin-dependent kinase Cdk8.
Cdk8 interacts with the mediator complex, which has a central role in the regulation of transcription. Biochemically, Cdk8 associates with a kinase module of the mediator that contains the Cdk8, CycC, Med12 and Med13 proteins (Clark et al., 2015; Jeronimo and Robert, 2017). It is thought that Cdk8 has a role in fine-tuning transcription and can exert repressive, as well as activating, functions on mediator-regulated gene expression (Gobert et al., 2010; Papadopoulou et al., 2016). Negative regulation of TFIIH activity through the phosphorylation of cyclin H by Cdk8, has been demonstrated (Akoulitchev et al., 2000). However, it is likely that Cdk8 acts through additional mechanisms that remain to be fully understood (Andrau et al., 2006; Fukasawa et al., 2015). In mouse pluripotent cells, the Med12/Med13 module associates with PRC1-binding sites and H2AK119ub (Papadopoulou et al., 2016). However, Cdk8 appears to be stably bound to only a small subset of these sites. In addition, Cdk8 binding is associated with active genes. Recent evidence from the inhibition of Cdk8 kinase activity in lymphoma cells suggests a repressive function for Cdk8 on genes that are regulated by super-enhancers (Pelish et al., 2015). The function of mediator kinases is difficult to assess, as a homologous complex containing Cdk19, CycC, Med12L and Med13L is also present in vertebrates (Bourbon 2008; Daniels et al., 2013). It is thought that Cdk8 and Cdk19 form mutually exclusive complexes that might have overlapping functions. Cdk8 and Cdk19 share 97% sequence homology in their catalytic domain and differ at their C-termini (Sato et al., 2004). Here, we investigate the function of the mediator kinases for gene repression by Xist.
RESULTS
Cdk8 is required for efficient gene repression by Xist
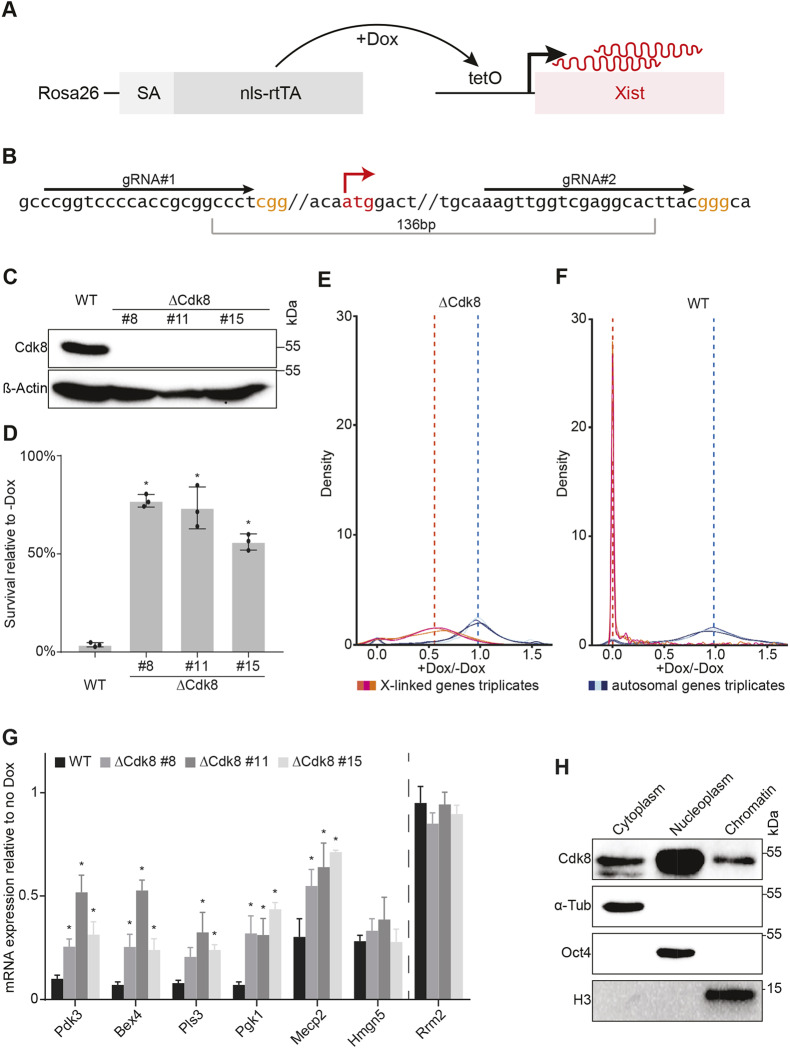
For investigating the effect of Cdk8 on Xist function, we engineered a mutation of Cdk8 in mouse HATX3 ESCs that carry a doxycycline-inducible Xist allele (Fig. 1A). HATX3 ESCs were established from haploid mouse embryos (Monfort et al., 2015) and became diploid in culture. Induction of Xist causes repression of X-linked genes and leads to cell death. A small deletion that includes the start codon was introduced into the Cdk8 locus (Fig. 1B). Several clones were identified to carry homozygous mutations and the absence of Cdk8 protein was confirmed by western blot analysis (Fig. 1C). Three independent clones were selected for further analysis. Sequencing of the genomic locus revealed that a small insertion had occurred in one clone (Fig. S1A) that might explain residual transcript in this clone (Fig. S1B). However, Cdk8 protein was undetectable by western blot analysis. Induction of Xist expression in parental HATX3 and Cdk8 mutant ESCs (ΔCdk8) resulted in cell loss. To quantify the magnitude of cell loss, we applied a single cell assay, whereby individual cells were deposited into 96-well plates and the number of colonies were counted after 14 days in the presence or absence of doxycycline. The ratio of the number of colonies obtained in the presence of doxycycline relative to the absence of doxycycline was calculated to determine the percentage of survival after Xist induction (Fig. S1C). This assay revealed a substantial increase in the survival of cells lacking Cdk8 (Fig. 1D). This suggested a potential requirement for Cdk8 in Xist function. To further assess whether this survival was caused by changes in X-linked gene repression, we performed RNAseq analysis of Cdk8 mutant and wild-type cells after 48 h of Xist induction, and in uninduced conditions. We quantified expression changes as the ratio of gene expression in induced conditions relative to uninduced conditions (Fig. 1E,F). Residual expression of X-linked genes in wild-type cells expressing Xist, was strongly reduced compared with uninduced cells (Fig. 1F). However, in cells lacking Cdk8, X-linked genes were on average expressed at half the level measured in uninduced conditions (Fig. 1E, Table S1). Overall, transcription was not affected by Xist, as shown by the unchanged expression of autosomal genes (Fig. 1E,F). Furthermore, the Cdk8 mutation did not lead to an overall change in X-linked gene expression before the induction of Xist (Fig. S1D). This observation suggested that Xist was able to repress genes in the absence of Cdk8 but the repression was incomplete. Investigation of the gene level further showed variability over different X-linked genes (Fig. S1E), which likely reflects the different half-lives of the transcripts, as well as the efficiency of silencing. The latter has been observed before and is probably caused by multiple and gene-specific repression mechanisms acting in X inactivation (Żylicz et al., 2019). To obtain an independent confirmation, we determined the expression levels of several genes by qRT-PCR. The X-linked Pdk3, Bex4, Pls3, Pgk1 and Mecp2 genes showed higher residual expression in Cdk8 mutant cells compared with wild-type control cells, when Xist was induced (Fig. 1G). In contrast, Hmgn5 was repressed to comparable levels in Cdk8 mutant and control cells, showing that the requirement of Cdk8 for efficient repression was gene specific (Fig. 1G). Induction of Xist had no measurable effect on the autosomal Rrm2 gene (Fig. 1G). In addition, we assessed the relative expression differences of the above genes in Cdk8 mutant and control cells in the absence of Xist induction. Expression levels were comparable, which confirmed that the baseline expression in control and Cdk8 mutant cells is comparable before the induction of Xist (Fig. S1F). Biochemical fractionation showed that Cdk8 is localized to the nucleus in ESCs and can be detected in the chromatin-associated fraction (Fig. 1H), which is consistent with its function in gene regulation.
Fig. 1.
Loss of Cdk8 impairs X-linked gene silencing by Xist. (A) Schematic of the Xist expression system in HATX3 ESCs. The nls-rtTA transactivator binds an inducible tetO promoter at the start site of the Xist gene. Doxycycline addition leads to Xist expression. nls-rtTA, nuclear localization signal-reverse tetracycline-controlled transactivator; tetO, tetracycline operator; Dox, doxycycline; SA, splice acceptor; Rosa26, genomic locus of nls-rtTA integration. (B) CRISPR/Cas9 strategy to engineer ΔCdk8 HATX3 ESCs; two gRNAs were designed to excise 136 bp, including the Cdk8 start codon. Yellow, PAM sequence; red, start codon; black arrows, gRNA target sequences. (C) Immunoblot confirming the absence of Cdk8 protein in ΔCdk8 ESC clones 8, 11 and 15. WT represents parental HATX3 ESCs. β-Actin, loading control. (D) Single cell survival assay for measuring Xist function. The ratio of survival of WT and ΔCdk8 after Xist induction relative to uninduced conditions is shown. The experiments were performed in triplicate. Data are mean±s.d.; asterisk indicates significant changes relative to WT (P<0.05). (E,F) RNAseq data representation of expression ratio after 48 h of Xist expression relative to untreated conditions for ΔCdk8 cells (E) and wild-type cells (F). Red curves, ratio of X-linked gene expression; blue curves, ratio of autosomal gene expression; dashed lines, median values. (G) qRT-PCR validation of differentially regulated X-linked genes in ΔCdk8 compared with WT ESCs after 48 h of Xist expression. Rrm2 serves as autosomal control. Expression levels were normalised to Gapdh and are relative to uninduced conditions. The experiments were performed in triplicate. Data are mean±s.d.; asterisk indicates significant changes relative to WT (P<0.05). (H) Western blot analysis of cell fractionation showing localisation of Cdk8, α-Tubulin (cytoplasmic marker), Oct4 (nucleoplasmic marker) and histone H3 (chromatin fraction).
The catalytic activity of Cdk8 is required for Xist function
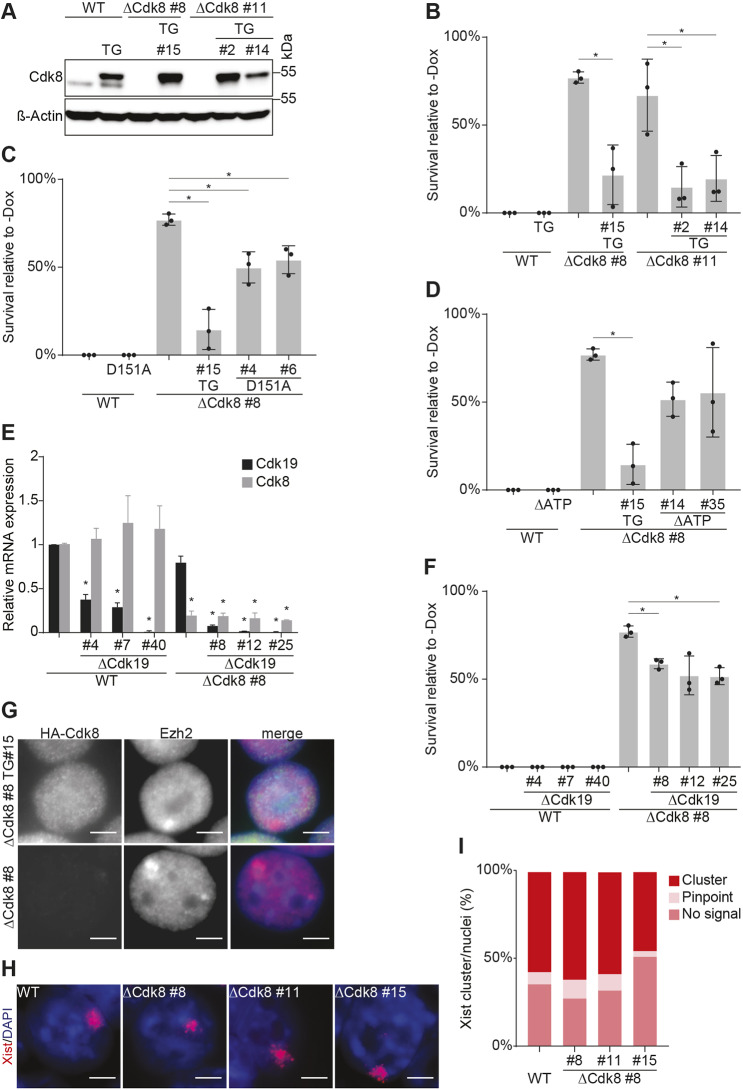
To demonstrate the specificity of the Cdk8 mutation and exclude potential off-target effects, we complemented two Cdk8 mutant cell lines with cDNA expression constructs. Several ESC clones were isolated and Cdk8 protein expression was investigated by western blot analysis (Fig. 2A). In these complemented cells, the restoration of the function of Xist was evident through reduced cell survival upon Xist induction (Fig. 2B). Taken together, these data indicate that Cdk8 contributes to Xist-induced gene repression in mouse ESCs.
Fig. 2.
Cdk8 kinase activity is required for Xist function but not Xist localization. (A) Immunoblot confirming expression of Cdk8 wild-type transgene in ΔCdk8 (clones 8 and 11) and wild-type ESCs. Transgenic HA-Cdk8 shows a higher molecular weight. β-Actin, loading control. (B-D) Survival ratio after Xist induction (as in Fig. 1D) of ESCs expressing a wild-type Cdk8 transgene (B), a D151A mutant Cdk8 transgene (C) and ΔATP mutant Cdk8 transgene (D), showing complementation of ΔCdk8 ESCs with the wild-type Cdk8 transgene. The experiments were performed in triplicate. Data are mean±s.d.; asterisk indicates statistically significant changes (P<0.05). ΔCdk8#8 and ΔCdk8#8 TG are the same in all panels. (E) qRT-PCR analysis of Cdk8 and Cdk19 expression in ΔCdk19 mutant clones, ΔCdk8#8 ΔCdk19 double mutant clones and control HATX3 (WT) ESCs, using primers in Cdk19 exons 1 and 2, and Cdk8 exons 2 and 3. Expression is normalized using Gapdh and shown relative to WT. Experiments were performed in triplicate. Data are mean±s.d.; asterisk indicates statistically significant changes relative to WT (P<0.05). (F) Survival ratio after Xist induction of ΔCdk19 and ΔCdk8#8 ΔCdk19 ESCs (as in B). Experiments were performed in triplicate; data are mean±s.d.; asterisk indicates statistically significant changes (P<0.05). (G) Immunofluorescence analysis showing localisation of HA-Cdk8 and Ezh2 (a Xi marker). (H) Xist RNA FISH (red) of wild-type and ΔCdk8 ESCs. DAPI was used to stain DNA (blue). (I) Quantification of Xist RNA FISH showing percentage of clusters (red), pinpoint signals (light pink) and no signal (dark pink) for genotypes as indicated. The experiments were performed in triplicate; >100 nuclei counted. Scale bars: 5 μm.
To further delineate the mechanism of Cdk8 in Xist function, we next investigated the requirement for Cdk8 kinase activity. We introduced two mutations into the Cdk8 cDNA that either abolish ATP binding or the function of the proton acceptor. Both mutations are predicted to lead to a loss of catalytic activity (Akoulitchev et al., 2000; Furumoto et al., 2007; Schneider et al., 2011). We introduced these mutated versions into Cdk8 mutant and wild-type control ESCs, and confirmed protein expression by western blot analysis (Fig. S2A,B). To assess the effect on Xist function, we performed cell survival assays. Whereas expression of wild-type Cdk8 restored Xist function in Cdk8 mutant cells, neither of the mutated versions of Cdk8 were able to complement Xist function in Cdk8 mutant cells (Fig. 2C,D). Furthermore, the expression of either Cdk8 mutant cDNAs in control wild-type cells did not lead to a measurable effect on Xist function. Taken together, these data show that the catalytic activity of Cdk8 is required for Xist function in our cell system.
Cdk8 but not Cdk19 contributes to Xist function in mouse ESCs
Cdk19 is a paralogue of Cdk8 that is expressed in mouse ESCs and could potentially compensate for its function in Cdk8 mutant cells. It is thought that Cdk19 complexes with Med12L and Med13L to form a submodule of the mediator complex that is similar to that formed by Cdk8. Either Cdk8 or Cdk19 submodules can associate with different core mediator complexes. To assess whether a specific requirement for Cdk8 exists or whether Cdk19 could also contribute to Xist function, we performed an analysis of the Cdk19 mutation in our ESC system. For this purpose, we engineered mutations in the Cdk19 gene in wild-type HATX3 cells and Cdk8 mutant ΔCdk8 cells. RT-PCR analysis showed a strong reduction of Cdk19 transcripts in cells carrying homozygous mutations in Cdk19 (Fig. 2E). We did not detect a measurable change in the expression of Cdk8 in Cdk19 mutant cells. Similarly, no change of Cdk19 expression in Cdk8 mutant cells could be observed (Fig. 2E). These observations show that there is no reciprocal regulation at the transcriptional level between the two homologous kinases. Importantly, Cdk8 and Cdk19 double-deficient ESCs had an appearance similar to control ESCs, demonstrating that mediator kinases are dispensable for the self-renewal of pluripotent cells. This allowed us to analyse the effect of combined mediator kinase mutations on Xist function. In contrast to the Cdk8 mutation, loss of Cdk19 did not have a measurable effect on Xist function, as determined by our single cell survival assay (Fig. 2F). In addition, the combined mutations of Cdk8 and Cdk19 resembled the Cdk8 mutation and did not further increase cell survival after Xist induction (Fig. 2F). These results strongly suggest that Cdk19 does not contribute to Xist function in mouse ESCs and demonstrate a specific requirement of Cdk8.
Cdk8 acts downstream of Xist localization
We next analysed a potential effect of the Cdk8 mutation on Xist expression and localization. Xist was detected in Cdk8 mutant cells at a level comparable with control cells (Fig. 2H,I). We counted Xist clusters and pinpoint signals 24 h after Xist induction, and did not observe a statistically significant difference between wild-type, Cdk8 mutant and complemented cells (Fig. S2C). Further quantification of total fluorescence of the Xist clusters confirmed that Xist clusters in wild-type and ΔCdk8 ESCs contained comparable amounts of Xist (Fig. S2D). The measurement of Xist transcript abundance by RT-PCR revealed a higher expression level in wild-type cells, compared with Cdk8 mutant or complemented cells. However, there was no statistically significant difference between Cdk8 mutant and complemented cells (Fig. S2E). The higher observed Xist expression in wild-type cells is probably due to the polyclonal nature of the parental HATX3 ESCs, which might have included some differentiated cells. To investigate a potential recruitment by Xist, we used ESCs expressing HA-tagged Cdk8 protein for immunofluorescence staining, together with Ezh2 antisera for identifying the X chromosome. HA-tagged Cdk8 showed a diffuse nuclear localization pattern without an enrichment over the Ezh2 cluster, whereas in cells that did not express HA-tagged Cdk8, no signal was observed (Fig. 2G). Taken together, our results show that mutation of Cdk8 does not affect the expression and localization of Xist, which suggests that Cdk8 acts downstream of Xist localization.
Cdk8 is required for the efficient recruitment of PRC2 by Xist
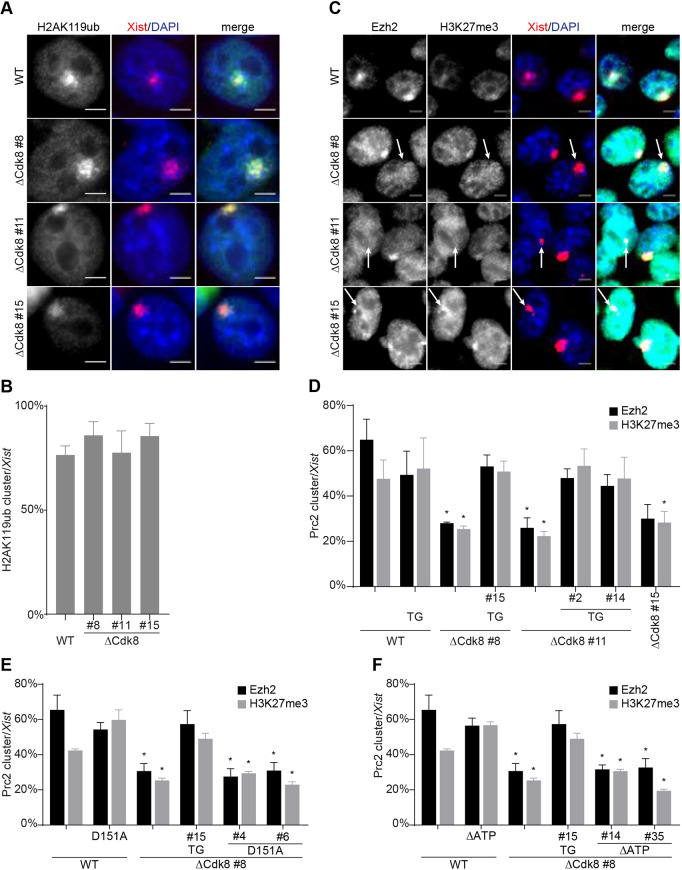
To investigate a potential role for Cdk8 in chromatin modifications of the Xi, we next measured the ability of Xist to recruit polycomb complex activity. We performed immunofluorescence staining with antisera specific for Ezh2, H3K27me3 and H2AK119ub in combination with Xist RNA fluorescent in situ hybridisation (FISH) after 24 h of Xist induction (Fig. 3, Fig. S3). We counted the number of foci relative to the number of Xist clusters (Fig. 3B-F). In ∼60% of nuclei, clear Xist clusters were detected using our combined staining technique, with little variation between different cell lines. Importantly, similar fractions of cells with Xist clusters were observed in cells mutant or wild-type for Cdk8, consistent with our earlier observation using Xist RNA FISH (Fig. S2C, Fig. S3A). The percentage of Xist clusters overlapping with H2AK119ub foci was comparable between Cdk8 mutant and control cells (Fig. 3B). In wild-type cells, 40% of Xist clusters colocalised with H3K27me3 foci using our combined immunofluorescence RNA FISH staining technique (Fig. 3D-F). In cells lacking Cdk8, a significant reduction in H3K27me3 foci was observed, with ∼25-30% of Xist clusters colocalised with clear H3K27me3 foci (Fig. 3D). A similar pattern was observed for Ezh2 foci (Fig. 3D). Importantly, the reduction in Ezh2 and H3K27me3 clusters in Cdk8 mutant cells was rescued by the expression of wild-type Cdk8 cDNAs but not by catalytically inactive versions of Cdk8 (Fig. 3D-F, Fig. S3D-G, Fig. S4A). Therefore, the kinase activity of Cdk8 is required for the efficient recruitment of Ezh2 and PRC2 activity by Xist. To further investigate a general effect of Cdk8 on polycomb histone modifications, we performed a western blot analysis using antisera specific for H3K27me3 and H2AK119ub (Fig. S3B). We observed comparable amounts of both histone modifications in control and ΔCdk8 ESCs, showing that Cdk8 was specifically required for PRC2 recruitment by Xist.
Fig. 3.
Cdk8 is required for efficient PRC2 recruitment by Xist. (A) Images of combined H2AK119ub immunofluorescence with Xist RNA FISH (red) for wild-type and ΔCdk8 cells. DAPI was used to stain DNA (blue). (B) Quantification of the percentage of Xist clusters with H2AK119ub foci after 24 h of Xist expression in ΔCdk8 and wild-type ESCs. Percentages are relative to counted Xist clusters; experiments were performed in triplicate; data are mean±s.d. (C) Combined immunofluorescence (Ezh2 and H3K27me3) with Xist-FISH (red) for wild-type and ΔCdk8 ESCs. White arrows indicate Xist clusters lacking PRC2 marks. DNA was stained with DAPI (blue). (D-F) Quantification of the percentage of Xist clusters with PRC2 (Ezh2 and H3K27me3) foci after 24 h of Xist expression in (D) ΔCdk8 ESCs and wild-type Cdk8 transgene complemented ΔCdk8 ESCs, (E) D151A and (F) ΔATP mutant Cdk8 transgene complemented ΔCdk8 ESCs. Percentages are relative to counted Xist clusters. Wild-type, ΔCdk8 #8 and ΔCdk8 #8 TG#15 samples are the same in E and F. The experiments were performed in triplicate. Data are mean±s.d.; asterisk indicates significant changes relative to WT (P<0.05). Scale bars: 5 μm.
Mutation of Cdk8 is embryonic lethal with a sex-specific phenotypic dimorphism
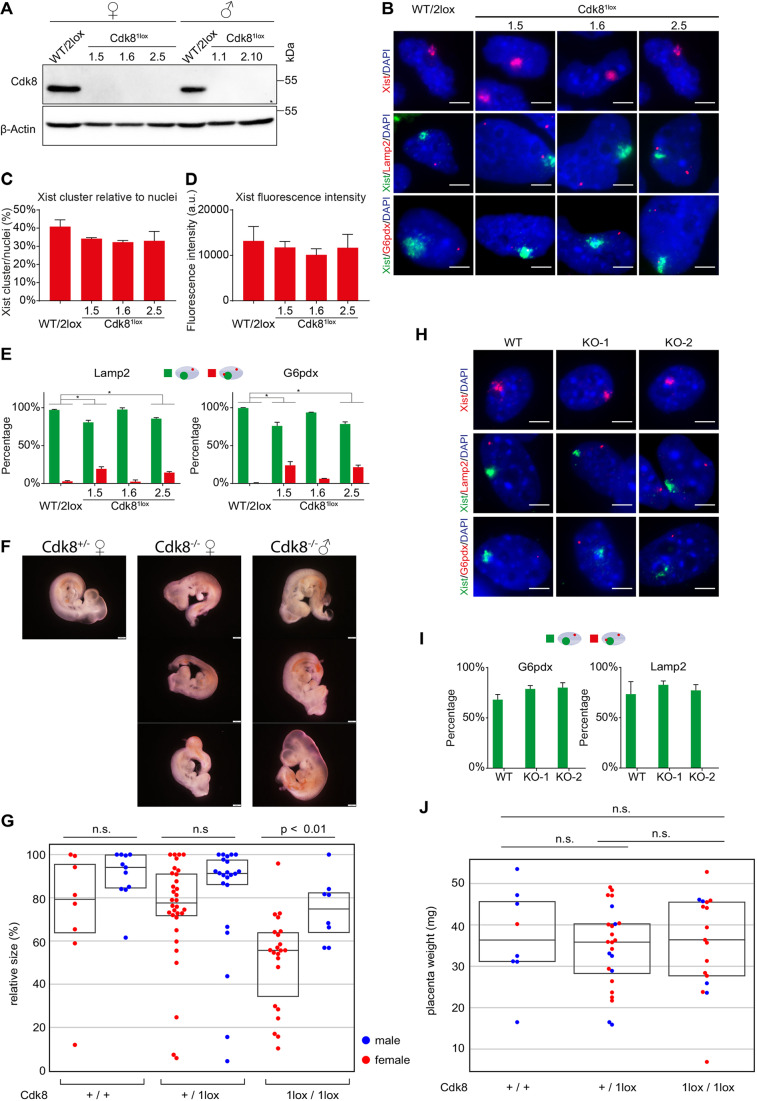
We next investigated whether there was a requirement for Cdk8 in XCI. We established ESCs from blastocysts of a cross between females homozygous for a Cdk82lox conditional mutation and heterozygous Cdk81lox/+ mutant males that also carry a Sox2-Cre transgene for epiblast-specific expression of Cre recombinase under the Sox2 regulatory region (Hayashi et al., 2002). We obtained two male and three female ESC lines that were homozygous for the Cdk81lox allele. Western blot analysis confirmed that Cdk8 protein was undetectable in these newly established ESCs (Fig. 4A). The potential for differentiation was further confirmed by analysis of Pou5f1, Pax6 and Gata6 expression before and after induction with retinoic acid (RA) for 4 days (Fig. S4B). Next, we investigated the expression of Xist and the X-linked genes Lamp2 and G6pdx by RNA FISH after 4 days of differentiation in the presence of RA (Fig. 4B-E, Fig. S5A). The percentage of cells with Xist clusters and the fluorescence intensity of the Xist clusters were comparable between Cdk8-deficient and wild-type female ESCs (Fig. 4B-D). Furthermore, no difference in Xist abundance or Pgk1 expression was observed by qRT-PCR (Fig. S5A, Fig. S4C). To assess silencing of Lamp2 and G6pdx, we counted the percentage of cells that had FISH signals overlapping the Xist cluster, as well as cells that had a single FISH signal that did not overlap with Xist (Fig. 4E). In control cells, very few cells showed two signals for Lamp2, with one apparently originating from the Xi, as inferred from an overlap with Xist. Similarly, most cells displayed a single non-overlapping G6pdx signal. In contrast, in Cdk8 mutant cells an increase in the number of cells with biallelic expression was observed for both X-linked genes, which was paralleled by an increase in the number of signals overlapping Xist (Fig. 4E). These data indicate that Xist did not silence Lamp2 or G6pdx in a significant number of Cdk8 mutant differentiating female ESCs, which is consistent with our earlier results showing that Cdk8 is required for efficient gene repression by Xist.
Fig. 4.
Cdk8 contributes to the initiation of XCI in female mouse development. (A) Western blot analysis of Cdk8 with β-Actin as loading control in ESCs established from embryos of crosses between Cdk8WT/1lox Sox2-Cre+/− males and Cdk82lox/2lox females. (B) RNA FISH expression analysis of Xist and X-linked genes Lamp2 (middle row) or G6pdx (bottom row) in homozygous female Cdk81lox and control female ESCs after 96 h RA differentiation. Top row, Xist (red); middle and bottom rows, Xist (green) and X-linked genes (red), Lamp2 (middle) and G6pdx (bottom) RNA FISH. DNA was stained with DAPI (blue). (C) Quantification of Xist RNA FISH analysis of Cdk81lox and control (WT/2LOX) female ESCs after 96 h RA differentiation. The percentage of Xist clusters is shown relative to counted nuclei (>100 counted). The experiments were performed in triplicate. Data are mean±s.d. a.u., arbitrary units. (D) Quantification of the fluorescence intensity of Xist clusters from RNA FISH (>100 clusters measured). The experiments were performed in triplicate. Data are mean±s.d. (E) Double FISH quantification of biallelic expression of the X-linked genes Lamp2 and G6pdx, and overlap with an Xist cluster. Percentage (>100 Xist clusters analysed) of Xist clusters without detectable biallelic X-linked gene expression (green bars), and Xist clusters with overlapping X-linked gene expression (red bars) are shown. The experiment was performed in triplicate. Data are mean±s.d. Asterisk indicates a significant difference from the wild-type control (P<0.05). (F) Representative images of heterozygous control female (left), and homozygous female (middle) and male Cdk8 mutant (right) embryos from one litter at E10.5. (G) Statistical analysis of embryo size as determined by image segmentation. The swarm plot shows the size normalized to largest size of the litter for each embryo with a box plot showing the mean and quartiles superimposed (n.s., not significant). (H,I) Female primary mouse embryonic fibroblasts derived from E10.5 Cdk81lox/1lox embryos. (H) RNA FISH expression analysis of Xist and X-linked genes Lamp2 (middle) or G6pdx (bottom). Top, Xist RNA FISH (red); middle and bottom, Xist (green); and X-linked genes (red) RNA FISH. DNA was stained using DAPI (blue). (I) Quantification of RNA FISH analysis of biallelic X-linked gene expression in the presence of Xist clusters (as in B). The percentage of nuclei showing monoallelic expression of the X-linked genes G6pdx and Lamp2 (green bars) in nuclei with Xist clusters are shown (>100 Xist clusters containing cells analysed). No cells with biallelic Xist expression were observed. The experiments were performed in triplicate. Data are mean±s.d. (J) Statistical analysis of placenta weights. Swarm plot showing the weight of individual placentae in mg for each genotype. Sex is indicated by the same colours as in G (n.s., not significant). Numbers above the lanes and panels in A and B, respectively, and on the x-axes in C, D and E indicate the clone numbers of independent ESC lines. Scale bars: 5 μm (B,H); 500 μm (F).
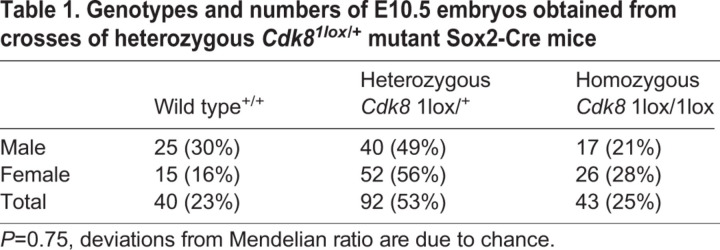
Preliminary analysis of crosses between homozygous conditional Cdk82lox mutant and heterozygous Cdk81lox/+ mutant Sox2-Cre mice indicated that the Cdk8 mutation is lethal around embryonic day (E) 10.5 (Table S3, Fig. S4D,E). To assess a potential requirement of Cdk8 for XCI in the embryo, we crossed mice that carried a heterozygous Cdk81lox/+ mutation. We obtained a total of 175 embryos, which included 43 homozygous Cdk81lox/1lox mutant embryos (Table 1). The sex of these embryos was established by Sry- and Zfy-specific PCR to identify the Y chromosome. All female homozygous Cdk8 mutant embryos that were obtained showed a more pronounced developmental delay and smaller size compared with males (Fig. 4F, Fig. S5B-D). We carried out the segmentation of microscopy images to determine the size of 103 embryos and analysed the statistical significance of the size difference between male and female embryos for all genotypes (Fig. 4G, Fig. S6, Table S4, Data S1; Wutz et al., 2020). The size difference between female and male homozygous Cdk8 mutant embryos was statistically significant, whereas the sex-specific differences for other genotypes were not significant. The more severe phenotype of female Cdk8 mutant embryos is consistent with a function of Cdk8 in XCI.
Table 1.
Genotypes and numbers of E10.5 embryos obtained from crosses of heterozygous Cdk81lox/+ mutant Sox2-Cre mice
We established fibroblast cultures from wild-type and Cdk8 mutant female embryos. We noted an impaired proliferation of Cdk8 mutant fibroblast and could ultimately obtain two cultures (Fig. S5E) in which the expression of Xist (Fig. S5F) and X-linked genes could be examined. We detected no statistically significant difference in the percentage of cells with Xist clusters between wild-type and Cdk8 mutant cells (Fig. 4H, Fig. S5G). In addition, the total fluorescence intensity of Xist clusters in Cdk8 mutant cells was comparable with wild-type controls (Fig. S5H), showing that Xist was expressed and formed clusters in the absence of Cdk8. We then examined the expression of the X-linked genes Lamp2 and G6pdx using RNA FISH (Fig. 4I). Both genes appeared efficiently silenced in wild-type and Cdk8 mutant fibroblast cells. This finding indicated that the absence of Cdk8 did not impair dosage compensation in somatic cells and suggests that Cdk8 is a factor that contributes to the initiation of XCI.
To investigate whether a potential defect in the placenta could contribute to the phenotype of the Cdk8 mutation, we determined the weight of placentae from E10.5 embryos of our heterozygous cross. We obtained weights for 51 placentae, which included 17 homozygous mutants (Fig. 4J, Fig. S7, Table S4, Data S1; Wutz et al., 2020). The weight difference between wild-type, homozygous and heterozygous, or male and female homozygous mutant placentae was not statistically significant, showing that placental weight was largely unaffected by the Cdk8 mutation. This observation suggests that the phenotype of the Cdk8 mutation is caused by embryonic defects.
Loss of Cdk8 leads to the deregulation of gene expression associated with Notch signalling
To further investigate the lethality of the Cdk8 mutation, we identified genes that are differentially expressed between wild-type and Cdk8 mutant ΔCdk8 ESCs (Fig. S5I). Our RNAseq datasets revealed that 210 genes were significantly upregulated and 105 genes were downregulated in Cdk8 mutant compared with wild-type control ESCs (Table S2). Among the upregulated genes were members of the Zscan4 family that are associated with cleavage-stage transcriptional profiles and the two-cell-like state of mouse ESCs. Previous studies have shown that Cdk8 regulates the Notch intracellular domain (Nicd). Consistent with this finding, we observed the upregulation of the Notch target gene Hes1 in Cdk8 mutant cells. We confirmed a 2.5-fold upregulation of Hes1 in Cdk8 mutant cells by RT-PCR (Fig. S5J). Investigation of the Nicd by western blot analysis indicated a higher level of the cleaved Nicd fragment in ΔCdk8 cells (Fig. S5K). Taken together, these findings indicate a deregulation of Notch signalling in the absence of Cdk8, which is consistent with the proposed role of Cdk8 in the regulation of the Nicd. Although an increase in Notch activity was detectable, it did not affect the growth of Cdk8 mutant ESCs, which is consistent with previous observations on overexpressing Nicd in ESCs (Lowell et al., 2006).
Cdk8 has also been implicated in the activation of Stat1 and Stat3 by phosphorylation. Stat3 activation was of particular interest as it contributes to the stabilisation of pluripotent mouse ESCs (Ying et al., 2008). However, western blot analysis did not detect changes in Stat1 or in Stat3 phosphorylation (Fig. S5K). Importantly, we did not observe any changes in the expression of chromatin regulators, including polycomb group genes in Cdk8 mutant cells. Overall, the mutation of Cdk8 induced relatively few changes in the transcriptome of ESCs, suggesting it plays only a minor role in gene regulation. This observation indicates that the contribution of Cdk8 to Xist function is not caused by transcriptome changes in ESCs.
DISCUSSION
Our study implicates Cdk8 as a new factor for X inactivation in mice. We found that Cdk8 is required for efficient gene silencing by Xist and recruitment of PRC2 but is dispensable for Xist expression and localization. In the absence of Cdk8, PRC1 activity is recruited by Xist, which suggests there is a specific requirement for PRC2 recruitment. This is consistent with the current model of polycomb complex recruitment in X inactivation, which implies PRC1 activity as an initial signal for recruiting PRC2 (Almeida et al., 2017; Pintacuda et al., 2017).
The absence of Cdk8 had a modest effect on gene expression in mouse ESCs. Notably, we did not detect changes in polycomb gene expression or in the expression of chromatin regulatory proteins among the top regulated genes. The effect of the Cdk8 mutation was most dramatic on the repressive effect of Xist on X-linked genes. In the absence of Cdk8, X-linked genes remained, on average, expressed at half the level of the active X chromosome. In contrast, in control cells with an intact Cdk8, gene repression was almost complete. This effect on gene repression is consistent with an earlier observation of a repressive effect of Cdk8 on super-enhancers in lymphoma cells (Pelish et al., 2015). This study showed that Cdk8 localizes to sites of mediator binding and acts to downregulate expression of associated genes. The remaining activity of Xist in Cdk8 mutant cells is probably due to the activity of pathways that act in parallel.
We found that the paralogous kinase Cdk19 is not required for Xist function. Considering the high level of sequence identity between the Cdk8 and Cdk19 proteins, this might appear surprising. Nonetheless, this observation is consistent with previous studies that have shown that Cdk8 and Cdk19 form distinct biochemical complexes that can act independently (Bourbon, 2008; Daniels et al., 2013). In our cell system, the Cdk19 mutation did not have a measurable effect on Xist function and a combined mutation of Cdk8 and Cdk19 resembled the effect of the Cdk8 mutation. Furthermore, we did not detect evidence of compensatory gene regulation between Cdk8 and Cdk19, and neither mutation affected the expression of the respective other kinase gene. Therefore, we conclude that gene repression by Xist specifically requires Cdk8. Notably, the absence of both mediator kinases Cdk8 and Cdk19 does not impair ESC self-renewal. Homozygous Cdk8 and Cdk19 mutant ESCs are a resource for future studies of mediator kinase function in signalling and gene expression.
Complementation experiments with mutant versions of Cdk8 that are predicted to lack kinase activity, demonstrate that the catalytic activity of Cdk8 is required for Xist function. Interaction of mediator kinases with PRC2 subunits Ezh2 and Suz12, and phosphorylation of Ezh2, has been reported (Fukasawa et al., 2015), consistent with a direct role of Cdk8 kinase for PRC2 recruitment in X inactivation. We observed Cdk8 as a nuclear localized and chromatin-associated protein but we did not detect an enrichment over the X chromosome using HA-tagged Cdk8 expressing ESCs. Although the failure to detect an enrichment might be due to technical limitations, it is conceivable that Cdk8 kinase activity is locally activated on Xi by factors targeted by Xist. Cdk8 also does not contain a discernible RNA interaction motif. Notably, the cyclin-binding protein Ciz1 was previously observed on the Xi (Ridings-Figueroa et al., 2017). Ciz1 shows enrichment over the Xist domain at the initiation of XCI in ESCs but is not required until somatic cell fates are generated. It is enticing to speculate that Ciz1 could contribute to locally activate among other kinases, as well as Cdk8 (Andrau et al., 2006). However, this aspect would need further exploration in future studies.
Consistent with a requirement for X inactivation, we found that the Cdk8 mutation causes a sex-specific dimorphic phenotype with a female-specific increased developmental delay at E10.5. Homozygous Cdk8 mutant female embryos were smaller and had a more pronounced developmental delay compared with males. These observations are consistent with a defect in dosage compensation in female Cdk8 mutant embryos. Although female embryo development is impaired more strongly than in males, lethality at E10.5 does not suggest a complete abrogation of dosage compensation. This observation is consistent with a partial defect of gene repression by Xist that is also gene specific in our ESC system. Our analysis further indicates the presence of an Xi in somatic cells in female Cdk8 mutant embryos. These cells are probably selected in embryonic development as cells with dosage compensation defects are eliminated.
Our observation of a postimplantation lethality of Cdk8 mutant embryos is inconsistent with an earlier study of a Cdk8 gene trap mutation, which reported a developmental arrest in preimplantation embryos (Westerling et al., 2007). We did not detect Cdk8 protein in homozygous mutants of the Cdk81lox allele, which suggests a loss-of-function mutation. Statistical analysis shows that at E10.5 Cdk8 mutant embryos are not under-represented and are observed at their expected Mendelian ratios. The discrepancy between our study and the earlier study can be reconciled by considering the different genetic backgrounds of the mouse strains and the differences in the structure of the Cdk8 mutant alleles. In our hands, the Cdk8 mutation is lethal around E10.5, with a more severe developmental delay in the female embryos that were recovered. From our placental weight measurements, we suggest that the Cdk8 mutation predominantly affects the embryo and does not reduce the weight of the placenta at E10.5. We observed reduced embryo size and malformations with a striking defect in head development. This phenotype was associated with a delay or defect in neural tube closure. Taken together, our data demonstrate that Cdk8 contributes to gene repression and PRC2 recruitment during the initiation of X inactivation, and is an essential gene for the post-implantation development of the mouse embryo.
MATERIALS AND METHODS
Cell lines
Diploidised HATX3 cells (Monfort et al., 2015), mutant derivatives and established ESC lines derived from mice were cultured as described previously (Wutz and Jaenisch, 2000). Briefly, cells were plated on gelatine-coated dishes containing high glucose Dulbecco's modified eagle medium (DMEM) (Life Technologies, 41965039) supplemented with 15% foetal bovine serum (Pan, P140402), 1% each of non-essential amino acids (Life Technologies, 11140035), sodium pyruvate (Life Technologies, 11360070) and L-glutamine (Life Technologies, 25030081), 8 μl/l β-mercaptoethanol (Sigma-Aldrich, M7522) and 1000 units/ml leukaemia inhibitory factor (LIF) (homemade). For maintenance culture, 3 μM Gsk3β inhibitor (Chir99021, Axon Medchem) and 1 μM Mek1/2 inhibitor (PD035901, Axon Medchem) were added (Ying et al., 2008). To induce Xist expression (Monfort et al., 2015) in HATX3 and mutant derivatives, 1 μg/ml doxycycline (Sigma-Aldrich, 324385) was administered. ESCs were differentiated for 96 h in the presence of 200 nM RA (Sigma-Aldrich, R2625). Primary mouse embryonic fibroblasts were cultured in a medium composed of high glucose DMEM (Life Technologies, 41965039) supplemented with 10% foetal bovine serum (Pan, P140402), 1% each of non-essential amino acids (Life Technologies, 11140035), sodium pyruvate (Life Technologies, 11360070) and L-glutamine (Life Technologies, 25030081).
Cdk8 and Cdk19 loss-of-function mutations were generated using a CRISPR/Cas9 strategy described previously (Monfort et al., 2015). Two guide RNAs targeting the region around the start codon were designed using the Massachusetts Institute of Technology algorithm (crispr.mit.edu) (Cdk8: gRNA1, 5′-ccggtccccaccgcggccct-3′ and gRNA2, 5′-aagttggtcgaggcacttac-3′; Cdk19: gRNA1, 5′-caccgtttcaaggcgaagctggcgg-3′ and gRNA2: 5′-caccgtaagagcgcgagcggggagt-3′) and inserted into PX330 vector (Addgene, 422300). Plasmids were sequenced for correct integration using a primer targeting the U6 promoter (5′-gactatcatatgcttaccgt-3′). The corresponding vectors, Cdk8 gRNA1/gRNA2 or Cdk19 gRNA1/gRNA2, were lipofected (Lipofectamine 3000, Thermo Fisher Scientific, L3000015) into HATX3 cells, together with tdTomato-N1 (Addgene, 54642), and after 48 h the cells were sorted for red fluorescence (MoFlo Astrios EQ, Beckman Coulter) and plated at limiting dilution in order to obtain clonal populations. Deletions were detected using PCR with genomic DNA (Cdk8: 5′-tctctcggaggagctaccggctgt-3′ and 5′-caaaactgagtgtcaccagccataggtttg-3′; Cdk19: 5′-ccaggttccaaaacaaggaa-3′ and 5′-acccctaaactccacctcca-3′) and validated by Sanger sequencing. Rescue and kinase-dead constructs were generated using the PiggyBac transposase system. Wild-type Cdk8 cDNA, reverse transcribed from RNA, was inserted into EcoRI-digested PB-EF1α-MCS-IRES-Neo vector (PB-EF1α-MCS-IRES-Neo cDNA cloning and expression vector, System Biosciences, PB533A-2) using a directional seamless cloning kit (In-Fusion HD Cloning Plus, Takara Bio, 638911) (primer: 5′-gcggccgatgactatgactttaaagtgaagctgagcag-3′ and 5′-ccgatttaaattcgaatttcagtaccgatgtgtctgatgtgagtac-3′). Additionally, DNA coding for an HA-Strep-tagII tag (primer: 5′-ctctagagctagcgaattatgtacccatacgatgttcccgac-3′ and 5′-gtcatagtcatcggccgctttttcgaac-3′) was cloned upstream of the Cdk8 cDNA. Site-directed mutagenesis to generate mutant Cdk8 cDNA vectors was achieved by an inverse PCR following the manufacturer's instructions (In-Fusion HD Cloning Plus, Takara Bio, 638911) (D151A: 5′-agggctttgaaacctgctaatattttagttatggg-3′ and 5′-gtttcaaagccctgtgcaacacc-3′; ΔATP: 5′-aaagactacgctttacaaatagaaggaactggaatttctatgtcgg-3′ and 5′-agttccttctatttgtaaagcgtagtctttatcgt-3′). Sequence-validated plasmids were lipofected into corresponding cells together with hyperactive PiggyBac transposase and a tdTomato fluorescent reporter in a ratio of 10:10:1, and sorted for tdTomato expression after 48 h. Independent clones were derived, selected for plasmid integration by the addition of 5 mg/ml G418 Sulphate (Life Technologies, 11811031), PCR screened for plasmid insertion (5′-gaccctgcttgctcaactct-3′ and 5′-tatagacaaacgcacaccg-3′) and validated by Sanger sequencing using the PCR screening primer. All cell lines tested negative for mycoplasma.
Single cell survival assay
Single cells were sorted (MoFlo Astrios EQ, Beckman Coulter) into 96-well tissue culture plates containing ESC medium without Gsk3 and Mek1/2 inhibitors, which – within a plate – alternately contains 1 μg/ml Dox. The medium was changed after 5 days and emerging colonies were quantified after 12 to 14 days. The experiments were performed in technical duplicates and in biological triplicates.
RNA extraction, cDNA synthesis and qPCR
RNA was extracted using the RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer's protocol; including an on-column DNA digest using RNase-free DNase (Qiagen, 79254). RNA concentration was determined using a NanoDrop spectrophotometer. Equal amounts of RNA were deployed to reverse transcription, using the SuperScript IV reverse transcriptase kit (Thermo Fisher Scientific, 18090200). Oligo(dT)15 primer (Promega, C1101) was used to specifically reverse transcribe polyadenylated transcripts. qPCR experiments were performed in technical duplicates and biological triplicates using a 384-well format on a Roche 480 Lightcycler instrument using the SYBR Green method (KAPA SYBR FAST qPCR Kit, Kapa Biosystems, KK4611). Fold change expression was calculated by the ΔΔct method. Gapdh was used for normalisation.
Primer sequences for gene expression analysis
The following primer sequences were used for gene expression analysis: Bex4, 5′-gataggcccaggagtgatg-3′, 5′-gggttcttcttcactttgtttg-3′; Cdk19, 5′-ggatctgtttgagtacgaaggg-3′, 5′-acaagccgacatagatattcctg-3′; Cdk8, 5′-agaggaaagatgggaaggac-3′, 5′-gctctcggagtaatgctatctc-3′; Gapdh, 5′-cgaaggtggaagagtgggag-3′, 5′-tgaagcaggcatctgaggg-3′; Hes1, 5′-caccggacaaaccaaagacg-3′, 5′-ggaatgccgggagctatctt-3′; Hmgn5, 5′-aaagaaaggctgcaggtg-3′, 5′-ggtttcaactccggtgtaaag-3′; Lef1, 5′-ccctgatgaaggaaagcatc-3′, 5′-gggtcgctgttcatattgg-3′; Mecp2: 5′-ccggggacctatgtatgatg-3′, 5′-aggaggtgtctcccaccttt-3′; Pdk3, 5′-gttcagagctggtacatgcag-3′, 5′-ggccattgtaggaacaacatc-3′; Pgk1, 5′-cccttcctggctatcttggg-3′, 5′-gatgtgccaatctccatgttgt-3′; Pls3, 5′-gcaggaatgaagcactgg-3′, 5′-cccatctcagcagaagctc-3′; Rrm2, 5′-cgccgagctggaaagtaaagcg-3′, 5′-tcgatgggaaagacaacgaagcg-3′; Xist, 5′-tgccatcctccctacctcagaa-3′, 5′-cctgacattgttttccccctaacaacc-3′; Pou5f1, 5′-caactcccgaggagtccca-3′, 5′- ctgggtgtaccccaaggtga-3′; Pax6, 5′-taacggagaagactcggatgaagc-3′, 5′-cgggcaaacacatctggataatgg-3′; Gata6, 5′gaagcgcgtgccttcatc-3′, 5′-gtagtggttgtggtgtgacagttg-3′; Atrx, 5′-cagtggatgatgacgacgac-3′, 5′-cccatcctcatcagagaaa-3′; Uba1, 5′-tttcctcctgaccagctc-3′, 5′-tttgggtccagaccagaa-3′; Armcx1, 5′-gggcagggtgcctgtatc-3′, 5′-ccttccctgcttcttggtttag-3′; and Huwe1, 5′-tgactacccccacaactg-3′, 5′-caccaacctttgctggag-3′.
Western blot
Whole-cell lysates were prepared by resuspending cells in TNTE buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 0.5% Triton X-100 and 1 mM EDTA] supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM MgCl2, 5 mM CaCl2 and 3000 U/ml DNase I, and incubation on a rotating wheel for 30 min at 4°C followed by 20 min centrifugation at 18,000 g. Cellular fractions were prepared as described previously (Graumann et al., 2008) with minor modifications. Briefly, collected cells were incubated for 10 min in ice-cold buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2 and 0.2% Igepal-C630, supplemented with 1 mM PMSF. The suspension was homogenised using a glass douncer and a loose pestle. After centrifugation for 15 min at 400 g, the supernatant containing cytoplasmic proteins was collected and stored on ice. The pellet was washed in ice-cold PBS and resuspended in buffer, containing 420 mM NaCl, 20 mM HEPES (pH 7.9), 20% glycerol, 2 mM MgCl2, 0.2 mM EDTA, supplemented with 1 mM PMSF, and incubated on ice for 60 min. After centrifugation for 15 min at 18,000 g, the nucleoplasmic protein-containing supernatant was collected and stored on ice. The pellet was washed in ice-cold PBS and resuspended in PBS supplemented with 600 mM NaCl, 1% Igepal-C630, 10 mM MgCl2, 5 mM CaCl2 and 3000 U/ml DNase I, and incubated on a shaker at 10°C at 600 rpm for 30 min. After centrifugation for 15 min at 18,000 g, the chromatin-bound protein containing supernatant was collected and stored on ice. Histones were extracted by pre-extraction using 1 ml 0.5% Triton-X100 in PBS+2 mM PMSF per 107 cells followed by 10 min centrifugation at 400 g at 4°C. Resulting pellets were incubated in 0.2 M HCl (1 ml/4×107 cells) overnight at 4°C followed by 10 min centrifugation at 400 g at 4°C. Protein concentrations were determined using the Bio-Rad DC Protein Assay (DC Protein Assay Kit II, Bio-Rad, 5000112). Equal protein concentrations for total cell lysates and equal volumes for cellular fractions were subjected to SDS-PAGE, and blotted on polyvinylidene difluoride membranes. The membranes were blocked in TBS+0.1% Tween 20 (TBST)+5% non-fat dried milk. Membrane washes were performed in TBST and antibody incubations were performed in TBST+5% non-fat dry milk. Blots were developed using the enhanced chemiluminiscence detection method on a Bio-Rad ChemiDoc system. Clarity Western ECL substrate (Bio-Rad, 1705060) was used for abundant proteins and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, 34095) was applied for proteins expressed at low levels.
Primary antibodies used for immunoblotting were as follows: goat anti-Cdk8 (Santa Cruz Biotechnology, SC-1521, dilution 1:400), anti-β-Actin-HRP (Sigma-Aldrich, A3854, dilution 1:20,000), rabbit anti-Notch1 (Cell Signaling Technology, 3608S, dilution 1:500), rabbit anti-phospho-Stat1(S727) (Cell Signaling Technology, 9177S, dilution: 1:200), mouse anti-Stat1 (BD Biosciences, 610186, dilution: 1:1000), rabbit anti-phospho-Stat3(S727) (Cell Signaling Technology, 9134T, dilution: 1:500), mouse anti-Stat3 (Cell Signaling Technology, 9139T, dilution: 1:1000), rabbit anti-Oct4 (Santa Cruz Biotechnology, sc-9081, dilution: 1:250), mouse anti-α-Tubulin (GenScript, A01410-40, dilution: 1:30,000), mouse anti-H3 (Cell Signaling Technology, 3638T, dilution 1:1000) and rabbit anti-H2AK119ub (Cell Signaling Technology, 8240S, dilution 1:2000). Secondary antibodies, purchased from Jackson ImmunoResearch and used at a dilution of 1:20,000, were as follows: peroxidase AffiniPure bovine anti-goat IgG (H+L) (805-035-180), peroxidase AffiniPure donkey anti-mouse IgG (H+L) (715-035-180) and peroxidase AffiniPure donkey anti-rabbit IgG (H+L) (711-035-152).
RNA FISH
RNA FISH was performed as described previously (Wutz and Jaenisch, 2000). Cells were seeded on laminin- (Sigma-Aldrich, L2020-1MG) coated (5 μg/ml in PBS) Roboz slides (CellPoint Scientific) in appropriate medium. Cells were rinsed in PBS, washed in cytoskeletal (CSK) buffer [10 mM PIPES (pH 6.8), 100 mM NaCl, 300 mM sucrose and 3 mM MgCl2] and proteins were extracted by incubation in CSK buffer, supplemented with 0.5% Triton X-100, for 7 min. After one wash in CSK buffer, cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Slides were dehydrated in an ethanol series of 70%, 80%, 95% and 100%. FISH probe was applied to the cells and incubated overnight at 37°C in a light-protected humidified chamber. Slides were washed three times each for 5 min at 39°C with 50% formamide/2× SSC [300 mM NaCl, 30 mM sodium citrate (pH 7.0)] and 2× SSC, respectively, followed by one wash with 1× SSC. Cellular DNA was stained with DAPI and slides were mounted with mounting medium (Vectashield H-1000, Vectorlabs, H-1000). Xist FISH probes were prepared as described previously (Karen and Wutz, 2007) using the random primer labelling technique (Prime-It II Random Primer Labelling Kit, Stratagene, 300385) and Cy3-dCTP using a Xist cDNA template [blueprint vector containing Xist cDNA, pBP5.6 (Wutz and Jaenisch, 2000)]. For double FISH experiments, Xist probes were prepared using Fluorescein-High Prime kit (Sigma-Aldrich, 11585622910) and X-linked gene probes were prepared using the random primer labelling technique and Cy3-dCTP using genomic bacterial artificial chromosome templates (Lamp2: RP24173A8, G6pdx: RP2313D21).
Combined immunofluorescence and RNA-FISH
This experiment was performed as described previously (Chaumeil et al., 2008) with minor modifications. Cells grown as mentioned for RNA FISH, were rinsed with ice-cold PBS and fixed for 10 min in 4% paraformaldehyde in PBS at room temperature. After two washes in PBS, cells were permeabilised in 0.1% sodium citrate/0.5% Triton X-100 in PBS supplemented with 2 mM ribonuleoside vanadyl complex (RVC) (NEB, S1402S) for 10 min on ice. Slides were washed two times in ice-cold PBS+0.1% Tween 20 (PBS-T) and blocked for 45 min at room temperature in 2.5% BSA in PBS-T supplemented with 2 mM RVC. Primary antibodies [mouse anti-Ezh2 (AC22) (dilution 1:100, Cell Signaling Technology, 3147); rabbit anti-H3K27me3 (dilution 1:200, Active Motif, 39155); rabbit anti-HA (C29F4) (dilution 1:1000, Cell Signaling Technology, 3724); rabbit anti-H2AK119ub (dilution 1:500, Cell Signaling Technology, 8240)] diluted in blocking solution supplemented with 2 mM RVC and 1 U/μl RiboLock RNase inhibitor (Thermo Fisher Scientific, EO0381), were added to the cells for 60 min at room temperature. Following three washes with PBS-T, fluorophore-labelled secondary antibodies [Alexa Fluor 488 AffiniPure donkey α-mouse IgG (H+L), Jackson ImmunoResearch, 715-545-150; and Alexa Fluor 647 AffiniPure Donkey α-Mouse IgG(H+L), Jackson ImmunoResearch, 715-605-150], in blocking solution/RVC/RNase inhibitor, were applied at a dilution of 1:1000 and incubated for 60 min at room temperature in the dark. Slides were washed twice with PBS-T and once with PBS. Slides were post-fixed for 10 min with 4% paraformaldehyde in PBS at room temperature, washed once in PBS followed by a wash in 2× SSC. After air drying of the samples, the FISH protocol applied from the point of adding the probe to the slides.
Microscopy
Samples were analysed with a Zeiss Axio Observer Z.1 fluorescence microscope equipped with a Hamamatsu OrcaFlash 4.0 camera and a Plan Apochromat 100×/1.46 oil DIC objective. Images were processed using Zeiss Zen Pro 2.0 software and figures were prepared using ImageJ/Fiji and Adobe Photoshop to crop pictures and adjust brightness and contrast. Fluorescence intensity measurements were taken by measuring the fluorescence of a defined area with a fixed exposure time, and intensity was calculated by subtracting background fluorescence. Experiments were performed in triplicates and more than 100 measurements or counts were taken for each experiment. Embryo images were acquired using an Olympus MVX10 Stereo-Zoom microscope equipped with an Olympus DP73 camera using the cellSens software. Image segmentation was performed using a custom Python script (Data S1; Wutz et al., 2020) from the scikit-image package. Statistical analysis was performed using a custom Python script based on the scikit-stats package (see supplementary Materials and Methods).
Mouse experiments
All in vivo experiments were performed under the licence ZH152/17 in accordance with the standards and regulations of the Kantonale Ethikkommission Zürich. Breeding, maintenance, timed matings and plug checks were carried out at the ETH Phenomics Center Mouse Facility. B6.CDK8tm1(fl/fl)Eucomm mice were a kind gift from Professor Markus Stoffel (ETH Zurich, Switzerland) and B6.Cg-Tg(Sox2-cre)1Amc/J (Hayashi et al., 2002) mice were kindly provided by Professor Jennifer Nichols (University of Cambridge, UK). Embryos were collected in accordance with Institute of Molecular Health Sciences regulations. Pictures were taken at the same magnification using an Olympus MVX10 stereo microscope mounted with an Olympus DP73 camera. Pictures were prepared using Adobe Photoshop to crop and adjust brightness and contrast. ESCs were derived in accordance with published procedures (Nichols and Jones, 2017). Briefly, eight-cell-stage embryos were flushed from oviducts at 2.5 dpc and incubated in KSOM medium (Millipore, MR-020P-5D), supplemented with 3 μM Gsk3β inhibitor (Chir99021, Axon Medchem) and 1 μM Mek1/2 inhibitor (PD035901, Axon Medchem) overnight, at 37°C/5% CO2. The next day, the medium was replaced with N2B27 (Takara Bio, Y400002), supplemented with Chir99021+PD0335901, and incubated for another 2 days. Inner cell masses were isolated by immunosurgery consisting of incubation in N2B27 with 20% rabbit α-mouse serum (Sigma-Aldrich M5774) for 1 h at 37°C, 5% CO2, followed by incubation in N2B27 with 20% guinea pig serum (Sigma-Aldrich, G9774) for 10 min. Isolated epiblasts were placed in tissue culture plates containing N2B27 supplemented with Chir99021, PD035901 and LIF. Primary mouse embryonic fibroblasts were derived from E10.5 embryos by pushing the embryos through a 27-gauge needle. Expanded cell lines were genotyped using the following primers: Cdk8 deletion, 5′-cttccctcttcccagaggac-3′, 5′-caaccccttttgaggttgaa-3′; Xist: 5′-gtagatatggctgttgtca-3′, 5′-ctccatccaagttctttctg-3′; Sry: 5′-tcttaaactctgaagaagagac-3′, 5′-gtcttgcctgtatgtgatgg-3′; and Zfy: 5′-aagataagcttacataatcacatgga-3′, 5′-cctatgaaatcctttgctgcacatgt-3′. Embryos were genotyped for Cdk8 deletion using the following primers: 5′-ggtgctggaggattaagtgc-3′, 5′-cacagaggacagcacagagc-3′.
Transcriptomic analysis
RNA was extracted as described above and sequenced by the Functional Genomics Center Zürich. Polyadenylated RNA was enriched using Oligo(dT) beads and libraries were prepared using an Illumina TruSeq Stranded mRNA kit. Libraries were sequenced on an Illumina HiSeq 4000 machine, creating 125 base single-end reads. CLC Genomics Workbench 11 (Qiagen), licensed by ETH, was used for analysis. Adapters were not trimmed and the reads were aligned to the Ensembl reference genome (GRCm38.94). Reads were normalised using the transcripts per kilobase million method. Statistically significant differentially expressed genes were obtained by comparing the data set of interest with a control group, meaning treated against non-treated for differentially regulated X-linked genes or ΔCdk8 non-treated against wild type non-treated for general transcriptional changes using a Wald test. Confounding factors were not taken into consideration. The threshold for significance was set to an absolute fold change greater than two and a false discovery rate P-value less than 0.01.
Statistics
Paired parametric two-tailed t-tests were conducted to determine significance with a P-value threshold less than 0.05. Calculations were performed using Prism GraphPad Version 7.
Supplementary Material
Acknowledgements
We thank J. Nichols and M. Stoffel for kindly providing the Sox2-Cre and Cdk8 mouse lines; S. Sting for embryo analysis; R. Freimann for flow cytometry; and members of the group for critical discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.W.; Methodology: A.P., C.E.D.; Software: A.P.; Validation: A.P., C.E.D.; Formal analysis: A.P.; Investigation: A.P., C.E.D.; Resources: A.P.; Data curation: A.P., C.E.D.; Writing - original draft: A.P., A.W.; Visualization: A.P.; Supervision: A.W.; Project administration: A.W.; Funding acquisition: A.W.
Funding
This work was supported by the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (31003A_152814/1 and 31003A_175643/1 to A.W.); and the Eidgenössische Technische Hochschule Zürich (ETH-38 16-1 to A.W.). Deposited in PMC for immediate release.
Data availability
Raw data and processed information from the RNAseq experiment were deposited in GEO under the accession number GSE129338. Image segmentation for size calculation of Cdk8 mutant embryos, microscopy images, segmentation summary and embryo sizes for all litters analysed are available from the Dryad Digital Repository (Wutz et al., 2020): doi:10.5061/dryad.tqjq2bvw0.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.175141.supplemental
References
- Akoulitchev S., Chuikov S. and Reinberg D. (2000). TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407, 102-106. 10.1038/35024111 [DOI] [PubMed] [Google Scholar]
- Almeida M., Pintacuda G., Masui O., Koseki Y., Gdula M., Cerase A., Brown D., Mould A., Innocent C., Nakayama M. et al. (2017). PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081-1084. 10.1126/science.aal2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrau J.-C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M. G., van de Peppel J., Werner M. and Holstege F. C. P. (2006). Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22, 179-192. 10.1016/j.molcel.2006.03.023 [DOI] [PubMed] [Google Scholar]
- Bourbon H.-M. (2008). Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993-4008. 10.1093/nar/gkn349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J., Augui S., Chow J. C. and Heard E. (2008). Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol. Biol. 463, 297-308. 10.1007/978-1-59745-406-3_18 [DOI] [PubMed] [Google Scholar]
- Chu C., Zhang Q. C., da Rocha S. T., Flynn R. A., Bharadwaj M., Calabrese J. M., Magnuson T., Heard E. and Chang H. Y. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404-416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. D., Oldenbroek M. and Boyer T. G. (2015). Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 50, 393-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Ford M., Schwinn M. K.; Benink H., M.D. G., Amunugama R., Jones R., Okazaki N., Yamakawa H., Miki F. et al. (2013). Mutual Exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK-mediator kinase module. J. Proteomic Bioinf. S2, 004 10.4172/jpb.S2-004 [DOI] [Google Scholar]
- Fukasawa R., Iida S., Tsutsui T., Hirose Y. and Ohkuma Y. (2015). Mediator complex cooperatively regulates transcription of retinoic acid target genes with Polycomb Repressive Complex 2 during neuronal differentiation. J. Biochem. 158, 373-384. 10.1093/jb/mvv055 [DOI] [PubMed] [Google Scholar]
- Furumoto T., Tanaka A., Ito M., Malik S., Hirose Y., Hanaoka F. and Ohkuma Y. (2007). A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells 12, 119-132. 10.1111/j.1365-2443.2007.01036.x [DOI] [PubMed] [Google Scholar]
- Galupa R. and Heard E. (2018). X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535-566. 10.1146/annurev-genet-120116-024611 [DOI] [PubMed] [Google Scholar]
- Gobert V., Osman D., Bras S., Augé B., Boube M., Bourbon H.-M., Horn T., Boutros M., Haenlin M. and Waltzer L. (2010). A genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/ RUNX-activated transcription in Drosophila. Mol. Cell. Biol. 30, 2837-2848. 10.1128/MCB.01625-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H. and Mann M. (2008). Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672-683. 10.1074/mcp.M700460-MCP200 [DOI] [PubMed] [Google Scholar]
- Hayashi S., Lewis P., Pevny L. and McMahon A. P. (2002). Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119 Suppl. 1, S97-S101. 10.1016/S0925-4773(03)00099-6 [DOI] [PubMed] [Google Scholar]
- Jeronimo C. and Robert F. (2017). The mediator complex: at the nexus of RNA polymerase II transcription. Trends Cell Biol. 27, 765-783. 10.1016/j.tcb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Karen N. and Wutz A. (2007). RNA FISH on cultured cells in interphase. CSH Protoc. 2007, pdb prot4763 10.1101/pdb.prot4763 [DOI] [PubMed] [Google Scholar]
- Lowell S., Benchoua A., Heavey B. and Smith A. G. (2006). Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 4, e121 10.1371/journal.pbio.0040121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F. (1962). Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 14, 135-148. [PMC free article] [PubMed] [Google Scholar]
- McHugh C. A., Chen C.-K., Chow A., Surka C. F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A. et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232-236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., Schermelleh L., Nesterova T. B. and Brockdorff N. (2015). A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562-572. 10.1016/j.celrep.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A., Di Minin G., Postlmayr A., Freimann R., Arieti F., Thore S. and Wutz A. (2015). Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554-561. 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. and Jones K. (2017). Derivation of mouse Embryonic Stem (ES) cell lines using small-molecule inhibitors of Erk and Gsk3 signaling (2i). Cold Spring Harb. Protoc. 2017, pdb.prot094086. 10.1101/pdb.prot094086 [DOI] [PubMed] [Google Scholar]
- Papadopoulou T., Kaymak A., Sayols S. and Richly H. (2016). Dual role of Med12 in PRC1-dependent gene repression and ncRNA-mediated transcriptional activation. Cell Cycle 15, 1479-1493. 10.1080/15384101.2016.1175797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelish H. E., Liau B. B., Nitulescu I. I., Tangpeerachaikul A., Poss Z. C., Da Silva D. H., Caruso B. T., Arefolov A., Fadeyi O., Christie A. L. et al. (2015). Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526, 273-276. 10.1038/nature14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G., Wei G., Roustan C., Kirmizitas B. A., Solcan N., Cerase A., Castello A., Mohammed S., Moindrot B., Nesterova T. B. et al. (2017). hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955-969.e910. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K., Fang J., Mlynarczyk-Evans S. K., Cao R., Worringer K. A., Wang H., De la Cruz C. C., Otte A. P., Panning B. and Zhang Y. (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131-135. 10.1126/science.1084274 [DOI] [PubMed] [Google Scholar]
- Ridings-Figueroa R., Stewart E. R., Nesterova T. B., Coker H., Pintacuda G., Godwin J., Wilson R., Haslam A., Lilley F., Ruigrok R. et al. (2017). The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 31, 876-888. 10.1101/gad.295907.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A. S., Jin J., Cai Y., Washburn M. P. et al. (2004). A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685-691. 10.1016/j.molcel.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Schneider E. V., Böttcher J., Blaesse M., Neumann L., Huber R. and Maskos K. (2011). The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J. Mol. Biol. 412, 251-266. 10.1016/j.jmb.2011.07.020 [DOI] [PubMed] [Google Scholar]
- Westerling T., Kuuluvainen E. and Mäkelä T. P. (2007). Cdk8 is essential for preimplantation mouse development. Mol. Cell. Biol. 27, 6177-6182. 10.1128/MCB.01302-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. and Jaenisch R. (2000). A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5, 695-705. 10.1016/S1097-2765(00)80248-8 [DOI] [PubMed] [Google Scholar]
- Wutz A., Postlmayr A. and Dumeau C. (2020). Data from: Cdk8 is required for establishment of H3K27me3 and gene repression by Xist and mouse development, v2. Dryad Digital Repository. 10.5061/dryad.tqjq2bvw0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A., Rasmussen T. P. and Jaenisch R. (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167-174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519-523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żylicz J. J., Bousard A., Žumer K., Dossin F., Mohammad E., da Rocha S. T., Schwalb B., Syx L., Dingli F., Loew D. et al. (2019). The implication of early chromatin changes in X chromosome inactivation. Cell 176, 182-197.e123. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.