Abstract
Type 2 diabetes mellitus (T2DM) drastically affects the population of Middle East countries with an ever-increasing number of overweight and obese individuals. The precise links between T2DM and gut microbiome composition remain elusive in these populations. Here, we performed 16 S rRNA and ITS2- gene based microbial profiling of 50 stool samples from Emirati adults with or without T2DM. The four major enterotypes initially described in westernized cohorts were retrieved in this Emirati population. T2DM and non-T2DM healthy controls had different microbiome compositions, with an enrichment in Prevotella enterotype in non-T2DM controls whereas T2DM individuals had a higher proportion of the dysbiotic Bacteroides 2 enterotype. No significant differences in microbial diversity were observed in T2DM individuals after controlling for cofounding factors, contrasting with reports from westernized cohorts. Interestingly, fungal diversity was significantly decreased in Bacteroides 2 enterotype. Functional profiling from 16 S rRNA gene data showed marked differences between T2DM and non-T2DM controls, with an enrichment in amino acid degradation and LPS-related modules in T2DM individuals, whereas non-T2DM controls had increased abundance of carbohydrate degradation modules in concordance with enterotype composition. These differences provide an insight into gut microbiome composition in Emirati population and its potential role in the development of diabetes mellitus.
Subject terms: Microbial communities, Microbial genetics, Endocrine system and metabolic diseases
Introduction
The gut microbiome is a critical reservoir of microbial species and their genes and genomes present in the human gastrointestinal tract. Host genetics, environment, diet, the immune system, and many other lifestyle factors interact with the gut microbiome to regulate their composition and function1. Data are bringing convincing evidence that gut microbiome plays an important role in human health and diseases2. Studies have indeed linked gut microbiome richness and composition with a spectrum of cardiometabolic and neurodegenerative disorders including obesity, diabetes, cancer, depression, and schizophrenia amongst others3–5. Especially the pathogenic association between gut microbiome and type 2 diabetes is quickly gaining momentum in the world through many reports. This is also due to the availability of technological advancements in metagenomics, which enable the dissection of the complex relationship between gut microbiome and diabetes.
These reports suggested that T2DM is associated with dysbiosis, a reduction in microbiome richness, altered bacterial composition and functional properties6. Among these, were reported a lowered abundance of butyrate-producing microbes, an altered firmicutes / bacteroidetes ratio, and an increase in opportunistic pathogens, such as Bacteroides caccae, Clostridium hathewayi, Clostridium ramosum, Clostridium symbiosum, Eggerthella lenta and E. coli3,7–10. These changes may induce disturbances in host gut barrier, in metabolic homeostasis and low-grade inflammation, in short chain fatty acid synthesis and fat deposition as well as hormonal regulation for involving glucagon-like peptide-1 synthesis. These factors contribute to glucose metabolism alteration, insulin resistance and dyslipidemia in patients with diabetes11–14. While the interaction between gut microbiome and metabolic health has been studied in several populations, exploring these interactions in Middle East countries is of particular interest considering the very high prevalence of diabetes in this region of the world15. Researchers have mostly focused on examining the bacterial members of the gut microbiome, but very little is known about the fungal communities which are non-negligible components in the gut. Mycobiota have been described as members of the normal gut flora in 196716. Fungal populations comprise less than 1% of the total gut microbiome. However, recent studies have indicated that these fungi have relevant effects on dampening inflammatory responses in the gut, especially in inflammatory bowel diseases despite their small amount17,18. Others have reported their impact on bacterial community composition19–21. Fungi may represents a key part of the microbial community with significant impact on the gut ecosystem, and possibly the host health21. However, the potential role of fungi and their interaction with the host and with other members of the gut community and metabolic health needs further understanding.
Research groups have demonstrated a significant impact of T2DM on gut microbial richness and relative abundance4,22,23 and underscored significant contribution of gut microbiome in T2DM phenotypes as insulin resistance and low-grade inflammation24. However, little is known about the relationships between T2DM on gut microbiome in UAE population. Here, we examined bacterial and fungal microbiome composition and possible functional consequences in T2DM individuals from an Emirati population. We performed 16 S rRNA gene and ITS2-based microbial profiling analysis of 50 stool samples from 25 T2DM and 25 non-T2DM individuals. We conducted a phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) functional analyses based on 16 S rRNA gene abundance profiles to gain deeper insight on potential functional impact on the host in T2DM from this Emirati population.
Materials and methods
Patient inclusion and ethical statement
The study was performed after receiving the necessary ethical approval from University Hospital Sharjah Ethics Research Committee (UHS-HERC-021-0702). The study was performed in accordance with relevant research guidelines and regulations of the committe. We randomly identified 25 native Emirati subjects with diagnosis of T2DM attending the endocrinology clinic. We also identified 25 otherwise healthy Emirati individuals and had HbA1c level < 6% as controls. All volonteers were provided with information sheet and explanation of study objectives, design, and confidentiality. We obtained written informed consents. We provided to all subjects a sterile stool specimen container with integrated collection spoon and collection instructions. A total of 50 stool specimens, 2 to 4 grams of freshly passed stool was collected in sterile containers. The specimens were stored immediately in liquid nitrogen and transferred to −80°C for storage until further analysis. Liquid (diarrheal) stools and use of antibiotics in the last 3 months were the exclusion criteria for this study.
DNA extraction
Faecal samples were subjected to DNA extraction using QIAamp PowerFecal DNA Kit (Qiagen Ltd, GmbH, Germany) following the manufacturer’s instruction (Qiagen Ltd). The extracted DNA was stored at −80°C for further analysis.
Bacterial and fungal PCR, sequencing, and sequence analysis and Taxonomic composition
Bacterial 16 S rRNA genes were amplified using polymerase chain reaction (PCR) targeting the V4 region with dual-barcoded, as per procedure as described in25. Next, amplicons sequenced with an Illumina MiSeq using the 250-bp paired-end kit (v.2). Sequences were denoised, taxonomically classified using Greengenes (v. 13_8) as the reference database, and clustered into 97% similarity operational taxonomic units (OTUs) with the mothur software package (v. 1.39.5) previously described26, following the recommended procedure (https://www.mothur.org/wiki/MiSeq_SOP; accessed August 2018). The resulting dataset had 21257 OTUs (including those occurring once with a count of 1, or singletons). An average of 18383 quality-filtered reads generated per sample. Sequencing quality for R1 and R2 was determined using FastQC 0.11.5.
ITS2 region were sequenced on an Illumina MiSeq (v. 2 chemistry) using the dual barcoding protocol as described25. Primers and PCR conditions used for 16 S rRNA gene and ITS2 sequencing were identical to those previously described27. Bacterial sequences were processed and clustered into operational taxonomic units (OTUs) with the mothur software package (v. 1.39.5)26, following the recommended mothur SOP. Paired-end reads were merged and curated to reduce sequencing error as described in28. The resulting dataset had 3171 OTUs (including those occurring once with a count of 1, or singletons). An average of 9581 quality-filtered reads were generated per sample. Sequencing quality for R1 and R2 was determined using FastQC 0.11.5. Fungal processing pipeline was identical as the one used for bacteria, except for the following differences: (1) paired-end reads were trimmed at the non-overlapping ends, and (2) high quality reads were classified using UNITE (v. 7.1) as described before as the reference database29. A consensus taxonomy for each OTU obtained and the OTU abundances then aggregated into genera. OTU table was rarified to 10000 reads per sample to correct for differences in sequencing depth with rarefy_even_depth function of phyloseq R package30, and alpha diversity indexes (Observed species, Shannon, ACE) were computed from rarified OTU table estimate_richness function of phyloseq R package. The R package vegan was used to compute Beta-diversity matrix from rarified OTU table collapsed at genus level (vegdist function) and to visualize microbiome similaritires with principle coordinate analysis (PCoA) (cmdscale function)31. Enterotype classification was performed from the same genus abundance matrix used for PCoA analyses following two different approaches. First, samples were clustered using Jensen-Shannon divergence (JSD) distance and the Partition Around Medoids (PAM) clustering algorithm as described in Aurumugam et al32. Second, samples were clustered from genus abundance data using the Dirichlet Multinomial Mixture (DMM) method of Holmes et al33. The DMM approach groups samples if their taxon abundances can be modeled by the same Dirichlet-Multinomial (DM) distribution.
Quality control
The possibility for contamination examined by co-sequencing DNA amplified from samples and from four each of template-free controls and extraction kit reagents treated the same way as the samples. Two positive controls, consisting of cloned SUP05 DNA, were also added (number of copies = 2*10^6). Operational taxonomic units were considered putative contaminants (and were removed) if their mean abundance in controls reached or surpassed 25% of their mean abundance in samples as described before34.
Functional profiling from 16 S rRNA gene data
Gene family abundances from Kegg Orthology (KO) functional space were computed from rarified 16 S rRNA gene OTU abundance matrix and GreenGenes taxonomic annotations with PICRUSt-1.1.335. This includes correction of OTU abundances by 16 copy number of reference GreenGenes taxons with normalize_by_copy_number.py script, compute KO abundance matrix from 16 S rRNA gene copy number-corrected 16 S rRNA gene OTU abundance matrix with predict_metagenomes.py script, and determine OTU contributions to each KO abundance vector with metagenome_contributions.py script. Gut Metabolic Modules (GMMs) were quantified from the PICRUSt KO abundance matrix with GOmixer R package36.
Statistical analysis
Linear regression analyses was used to evaluate the impact of different clinical variables (age, BMI, weight, diet and gender) and disease state over alpha diversity distribution. The significance of diversity changes after excluding the variability explained by age cofounder was tested with non-parametric Wilcoxon test over the residuals of linear regression analyses of alpha diversity (dependent variable) vs. age (independent variable). To evaluate beta diversity across samples, we excluded genus occurring in fewer than 10% of the samples with a count of less than three and calculated Bray-Curtis indices. Environmental fitting of clinical variables (age, BMI, weight, diet and gender) and disease state over Principal coordinates analyses ordination from Bray-Curtis inter-sample dissimilarity matrix was computed with envfit and cmdscale functions of vegan R package37. Dissimilarity in community structure by disease state was assessed with permutational multivariate analyses of variance (PERMANOVA) with non-T2DM v.s T2DM groups as the main fixed factor and using 4,999 permutations for significance testing with adonis function of vegan R package.
To identify taxonomic and functional features associated to disease state while accounting for cofounding effect of age generalized linear models (GLM) with negative binomial distribution were fitted with feature abundance as dependent variable and disease state and age as dependent variables with DESEq. 238 and Phyloseq.30 R packages. Functional enrichment analyses of KEGG modules were carried out to identify high-order functional features associated to T2DM transition from KO adjusted P-values and log2 fold changes between health controls and T2DM as effect sizes using the Reporter Feature algorithm as implemented in the Piano R package39. The null distribution was used as significance method and P-values were adjusted for multiple comparisons with the Benjamini-Hochberg method40. All analyses were conducted in the R environment.
Results
Gut microbiome profile of T2DM Emirati subjects: compositional differences between non-T2DM and T2DM subjects
We evaluated the intra- and inter-individual variability of gut microbiome among 25 T2DM and 25 non-T2DM subjects, all from Emirati origin. Their clinical characteristics are shown in S1 Table. T2DM subjects were significantly older, had higher BMI and were more sedentary than non-T2DM subjects were (P value < 0.05; Table S1). Further, based on short food frequency questionnaire (DFI-FFQ)41, we found higher percentage of T2DM individuals with a high fiber diet compared to non-T2DM individuals (P value < 0.05; Table S1). All T2DM individuals were under Dipeptidyl peptidase-4 inhibitors (DPP4i) and metformin treatment.
S1 Table: Clinical characteristics of the study groups
Median and quartiles 1 and 3 are shown for continuous variables. Number and percentage of samples are shown for categorical variables. P values are computed from Wilcoxon rank-sum test for continuous variables and chi-squared or exact Fisher test when the expected frequencies is less than 5 in some cell. False discover rate (FDR) were computed with Benjamini-Hochberg method.
Linear regression analyses of individual covariates (age, diet, BMI, weight, and gender) and disease state over alpha diversity (observed species) shows that age has an important effect over microbiome diversity (p value < 0.05; R2 = 0.16), with alpha diversity levels significantly increasing with age (Spearman Rho = 0.4; P value < 0.05) (Fig. 1A). When we take out the variability explained by age no significant differences in microbial diversity were observed between non-T2DM and T2DM individuals (Fig. 1B; Wicoxon rank-sum test on the residuals of linear regression analyses of observed species by age; P value = 0.66), with a wider variability in microbiome diversity observed in the T2DM group. Similar results were observed with other alpha diversity indexes (ACE, Shannon; Supplemental Fig. 1A–D).
Figure 1.
Prokaryotic profiling of gut microbiome. (A) Effect sizes of clinical covariates and disease state over Alpha diversity distribution (observed species) based on linear regression analyses (** = FDR < 0.05; * = P value < 0.05, FDR > 0.05) (B) Differences in residuals of linear-regression between alpha diversity (Observed species, dependent variable) and age (independent variable) between study groups. (C) Enterotype composition in non-T2DM and T2DM individuals by PAM clustering over JSD distance matrix computed from genus abundance data. (D) Enterotype composition in non-T2DM and T2DM individuals by DMM approach from genus abundance data. (E) Effect sizes of environmental fitting of clinical variables and disease state over PCoA ordination (** = P value < 0.05; * = P value < 0.1; permutation test) (F) Principal coordinates analyses of inter-individual differences (genus-level Bray-Curtis beta-diversity) with samples colored by disease state (non-T2DM, T2DM). Arrows represents effect sizes of the significant variables identified by environmental fitting analyses of panel E. (G) Barplot of log2 fold changes in taxonomic feature abundances between health controls and T2DM (P value < 0.05 in GLM model with negative binomial distribution of feature abundance by disease state adjusted by age).
We further examined the gut microbiota characteristics in terms of community composition. Sample clustering based on genus-level 16 S rRNA gene abundance data shows the presence of microbial enterotypes that characterize gut microbiome composition in European, Asian and American cohorts42. PAM clustering of samples from JSD beta diversity matrix at k = 3 shows the presence of Bacteroides, Ruminococcus and Prevotella enterotypes according to the abundance distribution of these prokaryotic genera (Supplemental Fig. 2A,D–F). DMM clustering with genus abundance matrix splits Bacteroides enterotype into two subgroups (Supplemental Fig. 2B) as previously described43 (Bacteroides_1 and Bacteroides_243, after additional re-assignments of Prevotella samples to Ruminococcus (n = 3) and Bacteroides_1 (n = 2) and Ruminococcus samples to Bacteroides_1 enterotype (n = 7) (Supplemental Fig. 2C).
Diversity distributions across these enterotypes confirm in this Emirati population with the high diversity profile associated with Ruminococcus enterotype and the low diversity profile associated with Bacteroides 2 enterotype (Supplemental Fig. 3). Further, T2D and non-T2D groups show significant differences in microbiome composition according to different enterotyping methods. PAM clustering over JSD beta diversity matrix shows that the non-T2D group is enriched in Prevotella enterotype, whereas the T2D group is enriched in Ruminococcus enterotype (Fig. 1C, Fisher’s exact test < 0.05). When enterotyping is carried out with the Dirichlet Multinomial Mixture method, we still observe that non-T2D controls are enriched in Prevotella enterotype, whereas an enrichment of the low-diversity Bacteroides2 enterotypes is observed in the T2D group (Fig. 1D, Fisher’s exact test < 0.05). We also observed that 7 Ruminococcus samples with PAM clustering has been re-assigned to Bacteroides1 enterotype with the DMM method (Supplemental Fig. 2C), a dysbiotic microbiome composition associated to low microbial cell density and enriched in Crohn and IBD43,44. Environmental fitting of disease and other covariates over PCoA ordination space from genus abundance matrix shows a significant impact of disease over microbiome composition (R2 = 0.12; P value = 0.001) together with age (R2 = 0.34, P value = 0.001) and BMI (R2 = 0.13, P value = 0.037) (Fig. 1E,F).
Finally, we search for taxonomic features significantly different between non-T2DM and T2DM groups while accounting for cofounding variables detected in environmental fitting analyses by fitting generalized linear models of genus abundance by disease, age and BMI with negative binomial distribution from raw abundance feature counts with DESeq238. Six bacterial genus were significantly associated to disease state (P value < 0.05), four of them increased in T2DM group (Phascolarctobacterium, Mogibacterium, Acidaminococcus and Unclassified Victivallaceae; log2 fold change Health vs. T2DM < 0), whereas two of them were decreased in T2DM group (Odoribacter and Lactococcus; log2 fold change non-T2DM vs. T2DM > 0) (Fig. 1G). The association with Unclassified Victivallaceae is reproduced at higher taxonomic levels (from family to phylum; Fig. 1G). None of these features resist P value adjustment by multiple comparisons (FDR > 0.05).
Fungal composition is different between T2DM and non-T2DM subjects
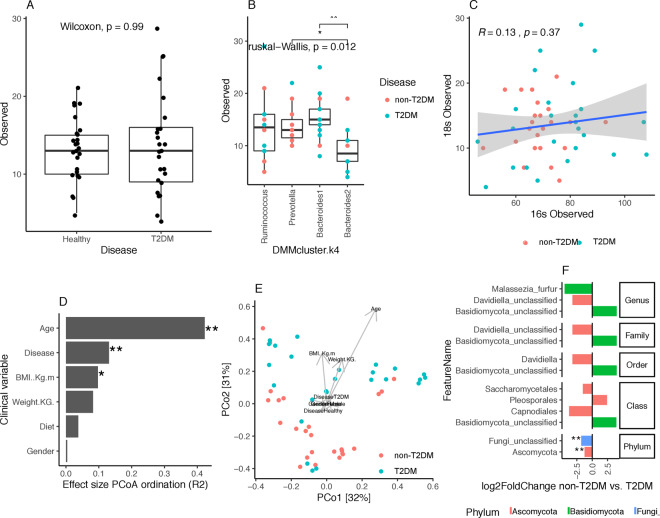
Fungi comprise a small percentage of the gut microbiome16, but reports have indicated that fungi have surprisingly strong effects on dampening inflammatory responses in the gut17,18. Others reported fungi impact on bacterial community composition19,20. Here, using ITS profiling we observed no significant difference in fungal diversity between T2DM and non-T2DM controls (P-value > 0.05 Wilcoxon test, Fig. 2A). In contrast with what we observed with prokaryotic diversity, linear regression analyses of individual covariates (age, diet, BMI, weight and gender) shows no significant associations of any of them with fungal diversity (P value > 0.05; Supplemental Fig. 4). We found no significant association between fungal and prokaryotic diversity (rho = 0.13; p value > 0.05, Fig. 2C). However, relating fungal diversity with enterotype composition, we found significant differences in fungal diversity across DMM enterotypes (P-value < 0.05 Kruskal-Wallis test; Fig. 2B), with Bacteroides 2 enterotype showing significant lower levels of fungal diversity in comparison with Bacteroides 1 and Prevotella groups (Fig. 2B).
Figure 2.
Fungal profiling of gut microbiome. (A) Alpha diversity distributions (observed species) between non-T2DM and T2DM groups. (B) Fungal diversity distributions (observed species) across DMM enterotypes (** = P value < 0.001; * = P value < 0.05; Wilcoxon rank-sum test). (C) Correlation between fungal and prokaryotic diversity (observed species). R and p corresponds to Spearman Rho and p-value of Spearman correlation test. (D) Effect sizes of environmental fitting of clinical variables and disease state over Principal coordinates ordination from panel E (** = P value < 0.05; * = P value < 0.1; permutation test). (E) Principal coordinates analyses of inter-individual differences (genus-level Bray-Curtis beta-diversity) with samples colored by disease state (non-T2DM, T2DM). Arrows represents effect sizes of the significant variables identified by environmental fitting analyses of panel D. (F) Bar plot of log2 fold changes in taxonomic feature abundance between non-T2DM controls and T2DM (P value < 0.05 in GLM model with negative binomial distribution of feature abundance by disease state adjusted by age).
Next, we examined the fungal microbiome composition as previously performed for bacterial composition. Environmental fitting of disease and other covariates over PCoA ordination space from fungal genus abundance matrix shows age (R2 = 0.42, P value = 0.001) and disease (R2 = 0.13, P value = 0.001) as the main variables with significant impact over fungal microbiome composition (Fig. 2D–E). In order to find fungal features associated to disease state while taking into account the confounding effect of age detected by environmental fitting, we follow the same approach as described above for 16 S rRNA gene data (fit generalized linear models of fungal feature abundance by disease and age with negative binomial distribution from raw feature counts). We observe a significant association of three fungal genenera with disease state (P value < 0.05), two of them (Malessezia firfur and Unclassified Davidiella) increased in the T2DM group (log2 fold change non-T2DM vs. T2DM < 0) and one (Unclassified Basidiomycota) decreased in the T2DM group (log2 fold change non-T2DM vs. T2DM > 0) (Fig. 2F). At higher taxonomic levels, T2DM groups seems to be characterized by an increase of Ascomycota lineages and a decrease of unclassified Basidiomycota lineages (Fig. 2D).
Functional profiling of T2DM and non-T2DM groups microbiomes based on 16 S rRNA gene profiles
We used the PICRUSt tool to project the functional content of the prokaryotic microbiome in the studied samples from 16 S rRNA gene OTU abundance data. In agreement with taxonomy findings, linear regression analyses of individual covariates (age, diet, BMI, weight, and gender) and disease state over functional diversity (observed KO groups) shows that disease (R2 = 0.16, P value < 0.05) and age (R2 = 0.26, P value < 0.001) have a significant impact over functional diversity (Fig. 3A). Functional diversity levels significantly increase with age (Spearman Rho = 0.51; P value < 0.001). When we excluded the variability explained by age no significant differences in functional diversity were observed between non-T2DM and T2DM individuals (Fig. 3B; P value = 0.94; Wicoxon rank-sum test on the residuals of linear regression analyses of observed species by age). Environmental fitting of disease and other covariates over PCoA ordination space from KO abundance matrix shows weight (R2 = 0.35, P value = 0.002), age (R2 = 0.29, p value = 0.001), BMI (R2 = 0.24, p value = 0.001), disease (R2 = 0.17, p value = 0.002) and diet (R2 = 0.07, p value = 0.033) as the variables with significant impact over functional prokaryotic content of the gut microbiome (Supplemental Fig. 5).
Figure 3.
Functional profiling based on PICRUS analyses of 16 S data. (A) Effect sizes of clinical covariates and disease state over functional diversity distribution (KEGG orthology (KO) groups identified in PICRUSt analyses) based on linear regression analyses (** = FDR < 0.05; * = P value < 0.05, FDR > 0.05). (B) Differences in residuals of linear-regression between functional diversity (Observed KOs, dependent variable) and age (independent variable) between study groups. (C) KEGG modules significantly enriched in differentially abundant KO groups between non-T2DM and T2DM group (** FDR < 0.05, * = P value < 0.05; Gene Set Enrichment Analyses). The mean log2 fold changes of module KOs abundances between non-T2DM controls and T2DM is represented as indicator of enrichment direction (all modules enriched in the T2DM group; mean log2 fold changes non-T2DM controls vs. T2DM < 0). (D) Bar plot of log2 fold changes in Gut metabolic modules (GMMs) abundances between health controls and T2DM (P value < 0.05 in GLM model with negative binomial distribution of GMM abundance by disease state adjusted by age).
Generalized linear models with negative binomial distribution of KO raw count data by disease state adjusted by age (4129 KOs with at least 10 counts in >20% of the samples) showed 210 KO groups significantly associated to disease state (FDR < 0.05), 32 decreased in the T2DM group (log2 fold change non-T2DM vs. T2DM group > 0) and 178 increased in the T2DM group (log2 fold change non-T2DM vs. T2DM group < 0). In order to find higher-level functional associations, we used gene set enrichment analyses of KEGG functional modules with adjusted p-values from age-adjusted GLM models and log2 fold changes of KO abundances of non-T2DM vs. T2DM as indicators of effect size. Four KEGG modules were significantly enriched in differentially abundant KOs (p value < 0.05), all of them enriched in KOs significantly increased in T2DM group (mean module KO log2 fold changes health vs. T2DM < 0). Among these we found M00064 (ADP-L-glycero-D-manno-heptose biosynthesis), a module representing the biosynthesis of glycero-manno-heptoses found in the lipopolysaccharides (LPS) of most Gram-negative bacteria, capsules and O-antigens of some Gram-negatives, and in the S-layer of certain Gram-positive bacteria45. Also we observed an enrichment of M00176 (assimilatory sulfate reduction), which was previously identified as signature of T2DM9, and an enrichment of pyruvate oxidation module (M00307) representing the pyruvate dehydrogenase complex, a key enzymatic complex linking glycolysis to TCA cycle in central metabolism during aerobic respiration46. Finally, quantification of Gut Metabolic Modules (GMM)47 based on KO abundance data shows 14 GMMs associated to disease state (Fig. 3D; FDR < 0.05; GLM models based on negative binomial distribution of module abundance by disease state adjusted by age). This analyses shows marked differences in the functional profile of gut microbiome of T2DM and non-T2DM controls, with non-T2DM controls showing significantly increases in different carbohydrate degradation modules (arabinoxylan, pectine and melibiose degradation modules, log2 fold change non-T2DM vs. T2DM > 0), whereas T2DM group showing significantly increases in several aminoacid degradation modules (isoleucine, proline, valine, cysteine, glutamine and aminobutyrate; log2 fold change non-T2DM vs. T2DM < 0), confirming also the increases in pyruvate dehydrogenase compex in T2DM group observed in the KEGG module enrichment analyses (Fig. 3C,D).
Discussion
In this study, we characterized for the first time, the prokaryotic and fungal microbiome profiles associated with T2DM and non-T2DM controls in an Emirati population where the study population was unmatched for age, BMI, and diet. When we evaluated the impact of these covariates together with disease state on microbiome diversity and composition, we observed that age had an important effect over microbiome diversity and composition. However, when we adjusted for age, there were no significant differences in microbial diversity between non-T2DM and T2DM controls. Remarkably and in contrasts with results of previous studies in westernized populations, where several factors impact gut microbiome composition and can be seen as confounders such as dietary habits, lifestyle and age48–53. One explanation can be related to dietary factors that are known to strongly impact gut microbiome composition54. For example, an Australian group demonstrated a significant effects of nutritional counseling on gut microbiome abundance and diversity among T2DM and obese individuals55. In our study, all T2DM individuals were subjected to rigorous dietary counselling as part of their clinical follow-up with a nutritionist. Furher, dietary aspects may contribute to some genera enrichment. For example, it is well known that fibers impact on Prevotella abundance which aids in polysaccharide breakdown56,57. In our study, we noticed an enrichment in Prevotella in the non-T2DM controls despite lower fiber intake based on the DFI-FFQ evaluation (Table S1). This observation is consistent with significant increase in carbohydrate degradation modules observed in the GMM modules analyses. Further, we detected an increase in aminoacid degradation modules in the T2DM group, which is in line with the observed enrichment of Bacteroides 2 enterotype and the proteolytic character of Bacteroides group58. Moreover, among the taxonomic features that resist age adjustment, we reported an increase of Victivallaceae lineage belonging to Lentisphaera phylum in the T2DM group and was notably identified from genus to phylum level. This lineage has been associated with gestational diabetes melitus in children59 and has been described to significantly increase in individuals consuming gluten-free diet60, again suggesting a potential association with the dietary counseling among T2DM group. The genus Phascolarctobacterium has also been associated both positively61–63 and negatively64 with markers of insulin sensitivity, whereas the genus Odoribacter, which includes butyrate producing bacteria that has been described negatively associated with hypertension in obese pregnant woman65. This genus also decreases in response to pre-natal metformin exposure in mice experiments66. Acidaminococcus genera has been also associated with modestly lower risk of T2DM in a mendelian randomization study67. However, the particularities of our study cohort in terms of ethnicity, and age and nutritional counseling between groups makes it difficult to extrapolate additional conclusions without further experimental evidences. All together, these findings underscores an important contribution of dietary counselling in driving these compositional changes68.
Another explanation to the observed difference from previous studies in westernized populations can be related to metformin administration among all T2DM subjects. We observed increased releative abundance of Escherichia, Akkermansia muciniphila and other unclassified Enterobacteriales lineage in T2DM subjects receiving metformin treatment. However, these differences do not resist adjustment by age. The increase in Escherichia coli and A.muciniphila in T2DM have been repeatedly reported in literature, and often associated with metformin intake69,70.
Next, we determined the presence of enterotypes that characterize microbiome composition. Prevotella enterotype is enriched in non-T2DM control group and Ruminococcus and Bacteroides 2 enterotypes is enriched in T2DM group. The compositional profile of T2DM group was also found to be heterogeneous, with enrichment of Ruminococcus enterotype that is usually associated with a more diverse microbiome profile32 and Bacteroides 2 enterotype, which generally shows an opposite association, being characterized by low microbial diversity and microbial loads and enriched in Crohn’s disease and ulcerative colitis patients43. This is also reflected in the wider range of prokaryotic diversity observed in the T2DM group in comparison with non-T2DM controls indicating a more heterogeneous microbiome profile in T2DM group, that could be attributed again to lifestyle habits as well as differences in T2DM severity.
The definition of discrete community types is a challenging task given the complexity in the landscape of community composition existing in the gut microbiome and the wide within and between-individual diversity existing in the human’s gut, which makes difficult extrapolation of conclusions based on discrete clusters to individuals in the boundary of different groups71,72. Also, and more importantly, sample clustering is strongly dependent of the other samples analyzed at the same time, which makes discretization dependent of the compositional landscape of the analyzed cohort, difficulty comparisons across studies. However, multiple studies have reproduced the presence of enterotypes with similar compositional properties across large datasets from different origins42, and the split of Bacteroides groups into two subgroups with the DMM method and the dysbiotic profile of the Bacteroides 2 group has been reproduced also in different studies and cohorts43,44,73,74. Thereby, a larger cohorts would be necessary to evaluate the strength of these community types across the Emirati population or if alternative community types could be defined.
Finally, we explored the gut microbiome functional contribution. Interestingly and in spite of the cofounding effects of age, we still observed signals at the functional level that have been identified in other quantitative metagenomic studies of T2DM, suggesting a more inflammatory profile in T2DM individuals53. For example, we noted an enrichment of ADP-L-glycero-D-manno-heptose biosynthesis module in T2DM group, a component of the bacterial LPS, associated with T2DM individuals and in agreement with other studies69. This molecule corresponds to one of the most antigenic part of the LPS, associated with low-grade inflammation that usually take place in obesity and T2DM69,75. In addition, it has been recently demonstrated as a potent pathogen-associated molecular pattern (PAMP) recognized by ALPK1 receptor and iducing NF-κB activation and cytokine expression76. Additionally, the formate conversion GMM significantly increased in the T2DM group (Fig. 3D) corresponding to the formate dehydrogenase complex responsible for formate oxidation, a metabolic signature of a dysbiosis-induced intestinal inflammation77.
Regarding fungal microbiome effect, we observed no significant differences in fungal diversity between T2DM and non-T2DM subjects. However, we detected a significant impact of disease state over fungal microbiome composition, even after normalizing the confounding impact of age. Remarkably, we found that Bacteroides 2 enterotype was associated with decreased levels of fungal diversity, in addition to its known dysbiotic phenotype, in terms of microbial diversity and loads in different pathologies like IBD and UC43. This observation extends previous findings showing that the deleterious B2 enterotype also associates with a decrease in fungal diversity. Thus, fungal diversity might been seen as an additional and novel signature of this dysbiotic microbiome composition that would need further validation in larger cohorts with fungal metagenomic data. Furthmore, we observed a shift from Candida albicans (known opportunistic) to Candida glabrata in the T2DM patients. Presence of C. glabrata has been linked to supressing genes involved in mannan biosynthesis, an important component of fungal cell wall with known protective benefits to the host78,79. Whether, this compositional shift from known commensal fungi to their virulent counterparts and the dissimilarity in mannan biosynthesis, significantly alters the intestinal barrier is yet to be explored.
In conclusion, we report a shift in gut microbiome composition and function among individuals affected by T2DM as compared to non-T2DM controls in a pilot study of Emirati people. The study population was distinctively unmatched for age, BMI, and diet, thereby providing a unique pattern and more challenging approach. Gut microbiome peculiarities have been linked to T2DM across the globe based on variation in diet, medication and ethnicity among other factors. Remarkably, our study revealed no significant differences in taxonomic and functional diversity among T2DM group, in contrast to what has been reported elsewhere, but we observed significant differences in microbiome composition (enterotypes) and functional content between study groups despite the added complexity by the unmatched confounders. We recognize that our results can be influenced by the divergence in mean age, diet intervention and highly individualized gut microbiome composition. We attributed these differences to dietary counselling provided to T2DM patients. Further, we showed that the enterotype B2 appears linked to fungal diversity that could be an additional and novel signature of this dysbiotic microbiome. We acknowledge potential limitations of this study, including relatively small sample size, detailed information regarding lifestyle and more advanced functional analysis. However, despite these limitations, this study provides meaningful insight into links between gut microbiome and its fungal community in T2DM subjects in native Emirati people. These aspects will be important to understand functional role of gut microbiome and its alterations to support host-homeostasis against metabolic and inflammatory disorders.
Supplementary information
Acknowledgements
Microbiome sequence data were provided by microbiomeinsights, Canada. This work was supported by Research Institute of Medical and Health Sciences at University of Sharjah grant P1701090226 (M.A.B), Boehringer Ingelheim grant 2016-17 (N.R.D), EU litmus grant (K.C), Le Ducq foundation (K.C. and P.B.L), and JPI-HDHL MICRODIET consortium grant (K.C and P.B.L). The funders stated above had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
All authors have approved the submitted version and have agreed both to be personally accountable for their’ s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature. M.T.B. - Conception, data acquisition, software used in the work, data analysis, manuscript preparation, revision. N.R.D.- Conception, data acquisition, software used in the work, data analysis, manuscript preparation, revision. P.B.L.- Data analysis, software used in the work, manuscript preparation, revision. B.H.B.- Data analysis, software used in the work, manuscript preparation, revision. A.M.N.- Data acquisition, manuscript preparation, revision. E.B.- Data analysis, software used in the work, manuscript preparation, revision. K.C.- Data analysis, software used in the work, manuscript preparation, revision.
Data availability
Sequencing data have been deposited in the European Bioinformatics Institute (EBI) European Nucleotide Archive (ENA) under accession number XXXX (Private access until paper acceptance). All other data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original version of this Article an affiliation for author Aml Mohamed Nada was omitted. The additional affiliation is “Mansoura University, Mansoura, Egypt.” As a result, Affiliation 7 was incorrectly listed as Affiliations 6.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/11/2021
A Correction to this paper has been published: 10.1038/s41598-021-85464-3
Change history
7/19/2021
A Correction to this paper has been published: 10.1038/s41598-021-94439-3
Contributor Information
Mohammad Tahseen Al Bataineh, Email: malbataineh@sharjah.ac.ae.
Karine Clément, Email: karine.clement2@gmail.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-66598-2.
References
- 1.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Structure, function and diversity of the healthy human microbiome. Nature486, 207–214, 10.1038/nature11234 (2012). [DOI] [PMC free article] [PubMed]
- 3.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 4.Aw W, Fukuda S. Understanding the role of the gut ecosystem in diabetes mellitus. Journal of diabetes investigation. 2018;9:5–12. doi: 10.1111/jdi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng P, et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Science advances. 2019;5:eaau8317. doi: 10.1126/sciadv.aau8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes D. Gut microbiota: Antidiabetic drug treatment confounds gut dysbiosis associated with type 2 diabetes mellitus. Nature reviews. Endocrinology. 2016;12:61. doi: 10.1038/nrendo.2015.222. [DOI] [PubMed] [Google Scholar]
- 7.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends in microbiology. 2011;19:427–434. doi: 10.1016/j.tim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS one. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 12.Wei X, et al. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell host & microbe. 2012;11:140–152. doi: 10.1016/j.chom.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nature reviews. Endocrinology. 2019;15:261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 14.Ramos-Romero S, et al. Mechanistically different effects of fat and sugar on insulin resistance, hypertension, and gut microbiota in rats. American journal of physiology. Endocrinology and metabolism. 2018;314:E552–E563. doi: 10.1152/ajpendo.00323.2017. [DOI] [PubMed] [Google Scholar]
- 15.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes research and clinical practice. 2010;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Gorbach SL, et al. Studies of intestinal microflora. II. Microorganisms of the small intestine and their relations to oral and fecal flora. Gastroenterology. 1967;53:856–867. doi: 10.1016/S0016-5085(19)34122-8. [DOI] [PubMed] [Google Scholar]
- 17.Wheeler ML, et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell host & microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol H, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo T, et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nature communications. 2018;9:3663. doi: 10.1038/s41467-018-06103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limon JJ, Skalski JH, Underhill DM. Commensal Fungi in Health and Disease. Cell host & microbe. 2017;22:156–165. doi: 10.1016/j.chom.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chehoud C, et al. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflammatory bowel diseases. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 23.Lambeth SM, et al. Composition, Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. Journal of diabetes and obesity. 2015;2:1–7. doi: 10.15436/2376-0949.15.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrema H. RG, I. J. & Nieuwdorp, M. Emerging role of intestinal microbiota and microbial metabolites in metabolic control. Diabetologia. 2017;60:613–617. doi: 10.1007/s00125-016-4192-0. [DOI] [PubMed] [Google Scholar]
- 25.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gweon HS, et al. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods in ecology and evolution. 2015;6:973–980. doi: 10.1111/2041-210x.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental microbiology. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koljalg U, et al. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. The New phytologist. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 30.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oksanen, J. et al. Vegan: Community Ecology Package. R Package Version. 2.0-10. CRAN (2013).
- 32.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PloS one. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dash NR, Khoder G, Nada AM, Al Bataineh MT. Exploring the impact of Helicobacter pylori on gut microbiome composition. PloS one. 2019;14:e0218274. doi: 10.1371/journal.pone.0218274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langille MG, et al. Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darzi Y, Falony G, Vieira-Silva S, Raes J. Towards biome-specific analysis of meta-omics data. The ISME journal. 2016;10:1025–1028. doi: 10.1038/ismej.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oksanen, J. et al. Vegan: community ecology package. R package vegan, vers. 2.2-1. R package version 2.2-1https://cran.rproject.org/web/packages/vegan/index.html (2015).
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic acids research. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling The False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J. Royal Statist. Soc., Series B. 1995;57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- 41.Healey, G. et al. Validity and Reproducibility of a Habitual Dietary Fibre Intake Short Food Frequency Questionnaire. Nutrients8, 10.3390/nu8090558 (2016). [DOI] [PMC free article] [PubMed]
- 42.Costea PI, et al. Enterotypes in the landscape of gut microbial community composition. Nature microbiology. 2018;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandeputte D, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 44.Vieira-Silva S, et al. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nature microbiology. 2019;4:1826–1831. doi: 10.1038/s41564-019-0483-9. [DOI] [PubMed] [Google Scholar]
- 45.Valvano MA, Messner P, Kosma P. Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology. 2002;148:1979–1989. doi: 10.1099/00221287-148-7-1979. [DOI] [PubMed] [Google Scholar]
- 46.Trotter EW, et al. Reprogramming of Escherichia coli K-12 metabolism during the initial phase of transition from an anaerobic to a micro-aerobic environment. PloS one. 2011;6:e25501. doi: 10.1371/journal.pone.0025501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vieira-Silva S, et al. Species-function relationships shape ecological properties of the human gut microbiome. Nature microbiology. 2016;1:16088. doi: 10.1038/nmicrobiol.2016.88. [DOI] [PubMed] [Google Scholar]
- 48.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 50.Zhernakova A, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 52.Voreades N, Kozil A, Weir TL. Diet and the development of the human intestinal microbiome. Frontiers in microbiology. 2014;5:494. doi: 10.3389/fmicb.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nature reviews. Gastroenterology & hepatology. 2019;16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 55.Marlene RSD, et al. Abundance and Diversity of Microbiota in Type 2 Diabetes and Obesity. J Diabetes Metab. 2013;4:253. doi: 10.4172/2155-6156.1000253. [DOI] [Google Scholar]
- 56.Precup, G. & Vodnar, D. C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles-A comprehensive literature review. The British journal of nutrition, 1–24, 10.1017/S0007114519000680 (2019). [DOI] [PubMed]
- 57.De Filippis F, et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell host & microbe. 2019;25(444–453):e443. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 58.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasan S, et al. Gut microbiome in gestational diabetes: a cross-sectional study of mothers and offspring 5 years postpartum. Acta obstetricia et gynecologica Scandinavica. 2018;97:38–46. doi: 10.1111/aogs.13252. [DOI] [PubMed] [Google Scholar]
- 60.Bonder MJ, et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome medicine. 2016;8:45. doi: 10.1186/s13073-016-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-Indias I, et al. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. American journal of translational research. 2016;8:5672–5684. [PMC free article] [PubMed] [Google Scholar]
- 62.Cross TL, et al. Soy Improves Cardiometabolic Health and Cecal Microbiota in Female Low-Fit Rats. Scientific reports. 2017;7:9261. doi: 10.1038/s41598-017-08965-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naderpoor, N. et al. Faecal Microbiota Are Related to Insulin Sensitivity and Secretion in Overweight or Obese Adults. Journal of clinical medicine8, 10.3390/jcm8040452 (2019). [DOI] [PMC free article] [PubMed]
- 64.Kuang YS, et al. Connections between the human gut microbiome and gestational diabetes mellitus. GigaScience. 2017;6:1–12. doi: 10.1093/gigascience/gix058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomez-Arango LF, et al. Increased Systolic and Diastolic Blood Pressure Is Associated With Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension. 2016;68:974–981. doi: 10.1161/HYPERTENSIONAHA.116.07910. [DOI] [PubMed] [Google Scholar]
- 66.Salomaki-Myftari H, et al. Neuropeptide Y Overexpressing Female and Male Mice Show Divergent Metabolic but Not Gut Microbial Responses to Prenatal Metformin Exposure. PloS one. 2016;11:e0163805. doi: 10.1371/journal.pone.0163805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Q, Lin SL, Kwok MK, Leung GM, Schooling CM. The Roles of 27 Genera of Human Gut Microbiota in Ischemic Heart Disease, Type 2 Diabetes Mellitus, and Their Risk Factors: A Mendelian Randomization Study. American journal of epidemiology. 2018;187:1916–1922. doi: 10.1093/aje/kwy096. [DOI] [PubMed] [Google Scholar]
- 68.Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nature reviews. Microbiology. 2019;17:742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- 69.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de la Cuesta-Zuluaga J, et al. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes care. 2017;40:54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 71.Knights D, et al. Rethinking “enterotypes”. Cell host & microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng M, Ning K. Stereotypes About Enterotype: the Old and New Ideas. Genomics, proteomics & bioinformatics. 2019;17:4–12. doi: 10.1016/j.gpb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aron-Wisnewsky J, et al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut. 2019;68:70–82. doi: 10.1136/gutjnl-2018-316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dao MC, et al. Akkermansia muciniphila abundance is lower in severe obesity, but its increased level after bariatric surgery is not associated with metabolic health improvement. American journal of physiology. Endocrinology and metabolism. 2019;317:E446–E459. doi: 10.1152/ajpendo.00140.2019. [DOI] [PubMed] [Google Scholar]
- 75.Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutrition in clinical practice: official publication of the American Society for Parenteral and Enteral Nutrition. 2012;27:201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou P, et al. Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature. 2018;561:122–126. doi: 10.1038/s41586-018-0433-3. [DOI] [PubMed] [Google Scholar]
- 77.Hughes ER, et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell host & microbe. 2017;21:208–219. doi: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang TT, et al. Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell host & microbe. 2017;22:809–816 e804. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West L, et al. Differential virulence of Candida glabrata glycosylation mutants. The Journal of biological chemistry. 2013;288:22006–22018. doi: 10.1074/jbc.M113.478743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the European Bioinformatics Institute (EBI) European Nucleotide Archive (ENA) under accession number XXXX (Private access until paper acceptance). All other data generated or analyzed during this study are included in this published article (and its Supplementary Information files).