Abstract
The improvement of the yield and quality of oil palm via precise genome editing has been indispensable goal for oil palm breeders. Genome editing via the CRISPR/Cas9 (CRISPR-associated protein 9) system, ZFN (zinc finger nucleases) and TALEN (transcription activator-like effector nucleases) has flourished as an efficient technology for precise target modifications in the genomes of various crops. Among the genome editing technologies, base editing approach has emerged as novel technology that could generate single base changes i.e. irreversible conversion of one target base in to other in a programmable manner. A base editor (adenine or cytosine) is a fusion of catalytically inactive CRISPR–Cas9 domain (Cas9 variants) and cytosine or adenosine deaminase domain that introduces desired point mutations. However, till date no such genetic modifications have ever been developed in oil palm via base editing technology. Precise genome editing via base editing approach can be a challenging task in oil palm due to its complex genome as well as difficulties in tissue culture and genetic transformation methods. However, availability of whole genome sequencing data in oil palm provides a platform for developing the base editing technology. Here, we briefly review the potential application and future implications of base editing technology for the genetic improvement of oil palm
Keywords: CRISPR/Cas, Base editors, Oil palm
Introduction
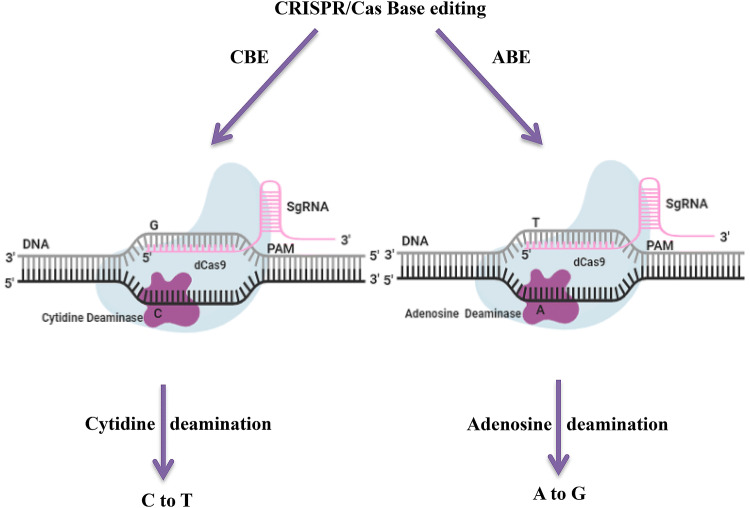
Oil palm (Elaeis guineensis, Jacq.) is the most productive oil crop that alone contribute higher proportion of vegetable oil production globally (Barcelos et al. 2015; Corley 2009). To the emerging needs of vegetable oil in the food and fuel industries, it’s mandatory to produce high oil yielding varieties of oil palm with tailored oil composition. Genetic improvement of oil palm with a tailored oil composition is possible with the help of modern genomic tools (Yarra et al. 2019; Masani et al. 2018). Availability of oil palm genome sequencing enabling us for the genetic improvement of oil palm (Singh et al. 2013). The genome editing tools such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats (CRISPR) along with Cas9 and base editors have revolutionized in precise editing of various plant genomes (Qin et al. 2019; Veillet et al. 2019; Ren et al. 2018; Zong et al. 2017; Shimatani et al. 2017). Among the GE tools, base editing tool has emerged as novel platform for efficient and precise genome modifications at target sites in plant genome (Mishra et al. 2020; Bharat et al. 2019). It represents new aspect of CRISPR/Cas mediated precise genome editing at targeted sites by generating single nucleotide changes in DNA or RNA without creating double strand breaks (DSBs) and homology directed mediated repair (HDR) (Molla et al. 2019; Eid et al. 2018; Komor et al. 2016). Additionally, very low rates of indel formation with high editing efficiency can be achieved through base editing technology (Mishra et al. 2020; Bharat et al. 2019; Chen et al. 2019; Eid et al. 2018). Base editing platforms mainly consisting of a modified Cas9 variants such as dCas9 or Cas9 nickase fused with a cytosine or adenosine deaminases for precise gene editing (Fig. 1). Two types of base editors (adenine BE or cytidine BE) are widely used in base editing approach. The base editors are chimeric proteins composed of a DNA targeting module and a catalytic domain, capable of deaminating a cytidine or adenine base. Adenine base editors (ABE) converts A into G, or A–T into G–C, where as cytidine base editors (CBE) converts cytosine into uracil (Fig. 1). These two base editing systems (ABEs and CBEs) are widely employed in precise genome editing in various plants (Mishra et al. 2020; Bharat et al. 2019; Eid et al. 2018). Thus single guide RNA (sgRNA) associated with the ncas9 or dCas9/deaminase fusion targets to the desired region and then permits the deamination on the non-complementary strand (Eid et al. 2018). This programmable genome modification via base editing in oil palm holds great promise in crop improvement.
Fig. 1.
CRISPR/Cas mediated base editing mechanism. A catalytically inactive Cas9 (dCas9) fused to adenosine or cytidine deaminase domain, guided by a sgRNA to generate single‐base substitutions without double‐stranded breaks (DSBs) in the DNA. The cytidine deaminase catalyzes the deamination of cytosine (C) and converts cytosine (C) to thymine (T) at the target site. The adenosine deaminase catalyzes the deamination of adenine (A) and converts adenine (A) to guanine (G) at the target site. ABE, adenosine base editor; CBE, cytidine base editor
Though base-editing approach is an advanced genome editing tool with high sensitivity and precision, there are still many aspects and challenges that should be overcome before accepting its efficacy in genome editing. The major limitations of this technology are concerned with specific PAM sequence requirement, size of catalytic window and off-target editing (Mishra et al. 2020). To extend the compatibility of PAM sequence, several base editors (ABEs and CBEs) with Cas9 variants have been developed to recognize different PAM motifs (Mishra et al. 2020). The wide window (~ 4–5 nucleotides) activity of base editors is a major limitation for obtaining high editing efficiency. Though attempts have been made to generate base editors with narrow catalytic window, still more efforts are needed for generating high precision base editors that can edit precisely with in the catalytic window (Mishra et al. 2020). Off-target editing is also one of the constraints in base editing approach. Off target activities are mostly caused by CBEs and occurs when additional cytosines proximal to the target base gets edited. Off-targets can be minimized by optimizing the deaminase domain, UGI components and with improved variants of base editors (Mishra et al. 2020).
Till date, no genome editing tools have been exploited or reported for the genetic improvement of oil palm. Though tissue culture and genetic transformation methodologies were well established for oil palm, little progress has been achieved for the genetic improvement. For the sustainability of oil palm with high yield varieties, it is imperative to apply the novel genomic tools for genome editing of oil palm. This can be achieved with the help of genome editing technologies such as CRISPR/Cas9 or base editing approaches. Especially, base editing application for the genetic improvement of oil palm will pave the way for genome editing of other palm crops.
Proposed base editing strategy in oil palm
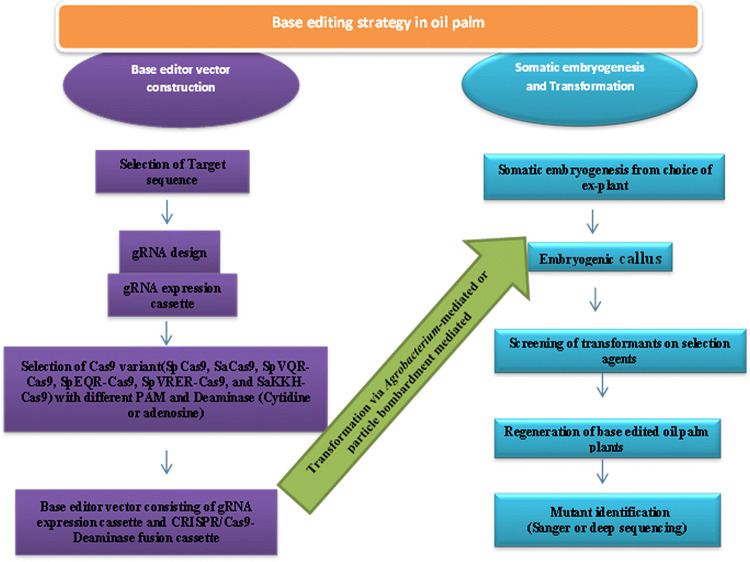
The success of base editing technology is associated with the different platforms such as well-established transformation methods and construction of base editor vectors with sgRNA, appropriate Cas9 variants that recognize different PAM sequence and deaminases(cytidine deaminase or adenosine deaminase) (Fig. 2). Here we propose the strategy for base editing in oil palm.
Fig. 2.
Base editing technology strategy in oil palm. Suitable base editor vector is constructed by cloning both gRNA expression cassette and CRISPR/Cas9 adenosine or cytidine deaminase fusion cassette. Base editor construct will be further transformed to embryogenic calli of oil palm followed by selection, regeneration, and mutant identification by sanger or deep sequencing
Somatic embryogenesis
A reliable, amenable and efficient genetic transformation methods are essential for genome editing of plants. Somatic embryogenesis is the extensively used and well established method for genetic transformation of oil palm (Yarra et al. 2019; Masani et al. 2018). Embryogenic callus induced from choice of explants can be used as a target tissue in base editing of oil palm.
Construction of base editor vector sgRNA design and online tools
Primary requirement for the successful base editing is the suitable sgRNA sequences for targeting site in the genome. For designing guide RNAs, particular requirements should be taken in to consideration such as off target activity, optimal activities for DNA modifying and PAM sequences. Various online tools such as BE-Designer (https://www.rgenome.net/be-designer/), beeditor (https://pypi.org/project/beeditor/) and benchling (https://benchling.com) are currently employing to design sgRNAs for CRISPR base editors (Dandage et al. 2019; Hwang et al. 2018). These tools provide a list of potential sgRNAs from a given input target DNA sequence along with editable sequences in a target window, relative target positions, GC content, and potential off-target sites.
Selection of Cas9 variant with different PAM and deaminase
Selection of Cas9 variant with specific PAM is required for efficient and precise base editing at targeted site. Currently available Cas9 variants with specific PAM sequences are SpCas9 (5′-NGG-3′), SpCas9-VQR (5′-NGAN-3′), SpCas9-EQR (5′-NGAG-3′), SpCas9-VRER (5′-NGCG-3′), xCas9 3.7 (TLIKDIV SpCas9; 5′-NGR-3′ and 5′-NG-3′), StCas9 (5′-NNAGAAW-3′), CjCas9 (5′-NNNVRYAC-3′), SaCas9 (5′-NNGRRT-'3) and, SaCas9-KKH (5′-NNNRRT-'3). Proper selection and fusion of either cytidine deaminase or adenine deaminase with Cas9 variants is important to convert one base to another base at targeted region.
CRISPR/Cas9 deaminase vector construction and delivery
Designed guide RNA should be cloned into the gRNA expression vector with Cas9 protein compatible to our host system. Then the base editor vector can be constructed by cloning sgRNA expression cassette and CRISPR/Cas9 -deaminase fusion (cytidine or adenine deaminases) cassette into a suitable vector. Till date, various cytidine deaminases (APOBEC, APOBEC1, pmCDA1 and human AID) or adenine deaminases (ecTadA) have been successfully utilized in base editing of various plants. These deaminases can be used in oil palm base editing. Subsequently, the base editor vector consisting of gRNA expression cassette and CRISPR/Cas9-deaminase fusion cassette will be transformed to suitable target tissue (Embryogenic callus) via Agrobacterium or particle bombardment mediated method as genetic transformation methods were well established in oil palm (Masani et al. 2018; Yarra et al. 2019; Parveez 2000; Parveez et al. 2015; Izawati et al. 2012, 2015; Bahariah et al. 2013; Masli et al. 2009). Transformed embryogenic calli can be further cultured on selection, regeneration medium to regenerate the base edited plants.
Detection of on-target mutations
Base edited oil palm plants with single base conversions at targeted regions can be analyzed by T7E1, PCR-RE assay, sanger sequencing and deep sequencing. As T7E1 and PCR-RE assays are depending on the endonucleases and PCR technology, it’s difficult to detect single base changes at targeted region. Due to high sensitivity and precision, sanger and deep sequencing methods are the reliable to analyze the results of base editing. However, deep sequencing is very reliable with high throughput and inexpensive compared to sanger sequencing method. Base conversion ratios can be calculated by uploading the targeted deep sequencing data into the BE-analyzer tool (https://www.rgenome.net/be-analyzer/).
Advantages and prospective genes for base editing in oil palm
Employing traditional breeding approaches to introduce desired trait in the crops with polyploidy nature of genomes like oil palm is time consuming and laborious. Moreover, conventional breeding approach takes 10–12 years to release an improved oil palm variety. Enhancing the palm oil production via modifying the metabolic pathways is impossible through conventional breeding strategy. But it can be achieved to some extent by over-expressing the genes through transgenic approach. Now with the genome editing technology like base editing, these hindrances can be overcome with fruitful progress. Base editing (BE) is a potent genome editing tool that harnesses Cas9-mediated gene targeting to induce specific point mutations in DNA or RNA. Various important traits in plants are mostly conferred by SNPs in their genome and base editing tool plays an essential function in correcting those point mutations for trait improvement. Many SNP trait associations have been identified in oil palm (Astorkia et al. 2020; Babu et al. 2019a, b; Xia et al. 2018, 2019; Teh et al. 2016) and cytosine, adenine base editors can be employed to edit specific genes conferred by SNPs in oil palm plants. The base editing approach can be utilized to develop disease resistant plants against various pathogens by modifying the target regions in genome (Mushtaq et al. 2019).The major disease that affects significant losses in oil palm yield is the basal stem rot caused by the fungal pathogen Ganoderma boninense (Barcelos et al. 2015; Rees et al. 2009). This novel genome editing technology solves this problem by editing single bases in genes or transcription factors that negatively regulating resistance against this fungal pathogen.
Abiotic stresses such as cold, high temperature are the major constraints for edible oil production and expected to exacerbate with anticipated climate change (Xiao et al. 2017; Murugesan et al. 2017). Especially, low temperature causes yellowing and withering of young leaves and flowers in oil palm. Various EgWRKY, DREB1, EgRBP42, EgEREBP and EgNAC genes were identified and characterized in oil palm under different abiotic stresses (Xiao et al. 2017; Azzeme et al. 2017; Yeap et al. 2012, 2019; Omidvar et al. 2013). Base editing tool can be applied to edit endogenous WRKY DREB1, RBP42, EREBP and NAC genes for developing stress tolerant oil palm plants.
Being oil palm is the most oil yielding crop, it is important to elucidate the genes that are responsible the biosynthesis of oil. Till date, very few genes related to oil yield have been characterized in oil palm (Nakkaew et al. 2013; Zheng et al. 2018). Among them, wrinkled1(wrl1) transcriptional factor is essential for the transcriptional control of plant oil biosynthetic pathways and its function has been validated in many plants including oil palm(Kong et al. 2019; Yeap et al. 2017; Bourgis et al. 2011). Yeap et al. (2017) reported that novel regulators such as EgNF-YA3, EgNF-YC2, and EgABI5 specifically bind to the EgWRI1 promoter and activate EgWRI1 expression. With the help of base editing technology, it’s possible to change the precise base changes at the binding sites to elucidate the exact function and unlock the oil yield related expression at transcription level in oil palm. Another important gene EgGDSL, associated with the oil content has been characterized (Zhang et al. 2018) and will be a potent gene to target through base editing technology.
The complexities in a polyploid genome and challenges ahead
The oil palm has huge genome size about 1.8 GB with haploid chromosome number of 16. Though various biotechnological tools have been employed for the genetic improvement of oil palm, still many efforts and novel technologies are needed for improving yield related traits in oil palm. Oil palm is recalcitrant for both forward and reverse genetic studies due to its large complex genome, high ploidy and high proportion of repetitive DNA.
One of the vital necessities for base editing technology is the ease availability of functional genomic resources for designing gRNAs for precise editing at target regions. Especially, scientists working on this important oil crop require sound knowledge in designing sgRNAs and use of specific base editors for precise and efficient editing. Moreover, the transcriptomic and EST data is not available or limited and many of the important agronomic trait genes yet to be identified (Chan et al. 2017). Base editing technology requires specific Cas9 variants regulated by highly efficient promoters. Its also a big challenge to select the promoters for highly efficient genome editing in oil palm. Evaluating the efficacy of base editor for precise editing in oil palm is also one of the major hurdles to achieve the base editing in oil palm.
Base editing technology utilizes the range of plant transformation methods such as protoplast transformation, agro-infiltration and stable transformation. Transient expression methods are not well established and unsuccessful in oil palm. Though, Agrobacterium and particle bombardment mediated method of transformations are well established, they are laborious and time consuming. The frequency of obtaining base edited plants is very low and needs more transgenics to analyze the edited events. Obtaining more number of oil palm transgenics is difficult. For achieving high regeneration efficiency in base editing technology, oil palm plants transformed with EgSERK1 (Lee et al. 2019) can be used as a mother plants for base editing technology.
Due to the complexities in polyploidy genome of oil palm, multiple gRNAs can be designed to edit the genome via base editing technology. Therefore, experts in bioinformatics are needed to develop the web based tools for targeting the specific copy or homologous copies. Another challenge ahead is to screen the base edited plants with edited events at target regions. Oil palm mutant identification by T7E1, PCR-RE assays, sanger sequencing is expensive and sensitive. More methods for screening base edited plants are needed to be explored in future.
However, scientist should also think about the GM regulations before producing the base edited plants. This genome editing technology also raise the serious concerns regarding ecological imbalance due to introduction of genome edited plants. Nevertheless, base editing approach is well established in various important food crops like rice, wheat and tomato and can be useful to obtain high yield varieties of oil palm. Though, none of this genetically modified food plants is accepted for general consumption in food chain.
Conclusion and future prospectives
Base editing is a novel technology in biological research that converts one base to another base. However base editing has been successfully applied for rice and wheat crop improvement, still its application to various crops is in infancy stage. Various Cas9 variants and base editors generated so far can be utilized to apply this technology to the important crops like oil palm. Further improvement of base editing technology with various base editors will show a path to edit the genome of oil palm with improved traits in near future. This GE platform paves the way for remarkable changes in oil palm industry.
Author contributions
R.Y. drafted and edited the manuscript. R.Y., C.H., J.L., Y.M., and Z.L collected the background information. All authors read and approved the final manuscript.
Funding
This work was supported by the Central public-interest scientific institution basal research fund for the Chinese Academy of Tropical Agricultural Sciences (No.1630152017008 and No.1630152019002), the fund for the construction of experimental agricultural station of the Chinese Academy of Tropical Agricultural Sciences (No.SYZ2019-12),the fund for The fund for species and varieties conservation of sector project of the Ministry of Agriculture and Rural Affairs (125163015000160004) and the Coconut Research Institute, Chinese Academy of Tropical Agricultural Sciences, Wenchang, Hainan, People’s Republic of China.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Astorkia M, Hernández M, Bocs S, et al. Detection of significant SNP associated with production and oil quality traits in interspecific oil palm hybrids using RARSeq. Plant Sci. 2020;291:110366. doi: 10.1016/j.plantsci.2019.110366. [DOI] [PubMed] [Google Scholar]
- Azzeme AM, Abdullah SNA, Aziz MA, Wahab PEM. Oil palm drought inducible DREB1 induced expression of DRE/CRT- and non-DRE/CRT-containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol Biochem. 2017;112:129–151. doi: 10.1016/j.plaphy.2016.12.025. [DOI] [PubMed] [Google Scholar]
- Babu BK, Mathur RK, Ravichandran G, et al. Genome-wide association study (GWAS) for stem height increment in oil palm (Elaeis guineensis) germplasm using SNP markers. Tree Gen Genom. 2019;15:40. [Google Scholar]
- Babu BK, Mathur RK, Ravichandran G, Anitha P, Venu MVB. Genome-wide association study for leaf area, rachis length and total dry weight in oil palm (Eleaeisguineensis) using genotyping by sequencing. PLoS ONE. 2019;14(8):e0220626. doi: 10.1371/journal.pone.0220626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahariah B, Parveez GKA, Masani MYA, Masura SS, Khalid N, Othman RY. Biolistic transformation of oil palm using the phosphomannose isomerase (pmi) gene as a positive selectable marker. Biocatal Agric Biotechnol. 2013;2:295–304. [Google Scholar]
- Barcelos E, Rios SA, Cunha RNV, Lopes R, Motoike S, Babiychuk E, Skirycz A, Kushnir S. Oil palm natural diversity and the potential for yield improvement. Front Plant Sci. 2015;6:190. doi: 10.3389/fpls.2015.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat SS, Li S, Li J, Yan L, Xia L. Base editing in plants: current status and challenges. Crop J. 2019 doi: 10.1016/j.cj.2019.10.002. [DOI] [Google Scholar]
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci USA. 2011;108:12527–12532. doi: 10.1073/pnas.1106502108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Tatarinova TV, Rosli R, et al. Evidence-based gene models for structural and functional annotations of the oil palm genome. Biol Direct. 2017;12:21. doi: 10.1186/s13062-017-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Ann Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Corley RHV. How much palm oil do we need? Environ Sci Policy. 2009;12:134–139. [Google Scholar]
- Dandage R, Després PC, Yachie N, Landry CR. beditor: A computational workflow for designing libraries of guide RNAs for CRISPR-mediated base editing. Genetics. 2019;212(2):377–385. doi: 10.1534/genetics.119.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid A, Alshareef S, Mahfouz MM. CRISPR base editors: genome editing without double-stranded breaks. Biochem J. 2018;475(11):1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang GH, Park J, Lim K, Kim S, Yu J, Yu E, Kim ST, Eils R, Kim JS, Bae S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinform. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawati AMD, Masani MYA, Ismanizan I, Parveez GKA. Evaluation on the effectiveness of 2-deoxyglucose-6-phosphate phosphatase (DOGR1) gene as a selectable marker for oil palm (Elaeis guineensis Jacq.) embryogenic calli transformation mediated by Agrobacterium tumefaciens. Front Plant Sci. 2015;6:727. doi: 10.3389/fpls.2015.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawati AMD, Parveez GKA, Masani MYA. Transformation of oil palm using Agrobacterium tumefaciens. Methods Mol Biol. 2012;847:177–188. doi: 10.1007/978-1-61779-558-9_15. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Yuan L, Ma W. WRINKLED1, a “Master Regulator” in transcriptional control of plant oil biosynthesis. Plants. 2019;8(7):238. doi: 10.3390/plants8070238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FC, Ong-Abdullah M, Ooi SE, Ho CL, Namasivayam P. Cloning and characterization of Somatic Embryogenesis Receptor Kinase I (EgSERK I) and its association with callus initiation in oil palm. Vitro Cell Dev Biol Plant. 2019;55:153. [Google Scholar]
- Masani MYA, Izawati AMD, Rasid OA, Parveez GKA. Biotechnology of oil palm: current status of oil palm genetic transformation. Biocat Agric Biotechnol. 2018;15:335–347. [Google Scholar]
- Masli DIA, Parveez GKA, Yunus AMM. Transformation of oil palm using Agrobacterium tumefaciens. J Oil Palm Res. 2009;21:643–652. [Google Scholar]
- Mishra R, Joshi RK, Zhao K. Base editing in crops: current advances, limitations and future implications. Plant Biotechnol J. 2020;18(1):20–31. doi: 10.1111/pbi.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla KA, Yang Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;S0167–7799(19):30053–30058. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Murugesan P, Aswathy GM, Kumar SK, Masilamani P, Kumar V, Ravi V. Oil palm (Elaeis guineensis) genetic resources for abiotic stress tolerance: a review. Ind J Agricul Sci. 2017;87(5):571–579. [Google Scholar]
- Mushtaq M, Sakina A, Wani SH, Shikari AB, Tripathi P, Zaid A, Galla A, Abdelrahman M, Sharma M, Singh AK, Salgotra RK. Harnessing genome editing techniques to engineer disease resistance in plants. Front Plant Sci. 2019;10:550. doi: 10.3389/fpls.2019.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakkaew A, Thitichai N, Nualkaew S, Chotigeat W, Phongdara A, Altpeter F. Cloning, characterization and overexpression of a 14-3-3 ω protein from oil palm (E laeis guineensis) Plant Breed. 2013;132(6):701–710. [Google Scholar]
- Omidvar V, Abdullah SNA, Ho CL, Mahmood M. Isolation and characterization of an ethylene-responsive element binding protein (EgEREBP) from oil palm (Elaeis guineensis) Aust J Crop Sci. 2013;7:219–226. [Google Scholar]
- Parveez GKA. Production of transgenic oil palm (Elaeis guineensis Jacq.) using biolistic techniques. In: Jain SN, Minocha SC, editors. Molecular Biology of Woody Plants 2. Tranbjerg Denmark: Kluwer Academic Publishers; 2000. pp. 327–350. [Google Scholar]
- Parveez GKA, Bahariah B, Ayub NH, Masani MYA, Rasid OA, Tarmizi AH, Ishak Z. Production of polyhydroxybutyrate in oil palm (Elaeis guineensis Jacq.) mediated by microprojectile bombardment of PHB biosynthesis genes into embryogenic calli. Front Plant Sci. 2015;6:598. doi: 10.3389/fpls.2015.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Li J, Wang Q, Xu Z, Sun L, Alariqi M, Manghwar H, Wang G, Li B, Ding X, Rui H, Huang H, Lu T, Lindsey K, Daniell H, Zhang X, Jin S. High efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2019;18(1):45–56. doi: 10.1111/pbi.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees RW, Flood J, Hasan Y, Potter U, Cooper RM. Basal stem rot of oil palm (Elaeis guineensis); mode of root infection and lower stem invasion by Ganoderma boninense. Plant Pathol. 2009;58:982–989. [Google Scholar]
- Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol Plant. 2018;11:623–626. doi: 10.1016/j.molp.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Singh R, Ong-Abdullah M, Low ET, Manaf MA, Rosli R, Nookiah R, Ooi LC, Ooi SE, Chan KL, Halim MA, Azizi N, Nagappan J, Bacher B, Lakey N, Smith SW, He D, Hogan M, Budiman MA, Lee EK, DeSalle R, Kudrna D, Goicoechea JL, Wing RA, Wilson RK, Fulton RS, Ordway JM, Martienssen RA, Sambanthamurthi R. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature. 2013 doi: 10.1038/nature12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh CK, Ong AL, Kwong QB, Apparow S, Chew FT, Mayes S, et al. Genome-wide association study identifies three key loci for high mesocarp oil content in perennial crop oil palm. Sci Rep. 2016;6:19075. doi: 10.1038/srep19075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet F, Perrot L, Chauvin L, Kermarrec MP, Guyon-Debast A, Chauvin JE, Nogué F, Mazier M. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Luo T, Dou Y, Zhang W, Mason AS, Huang D, Huang X, Tang W, Wang J, Zhang C, Xiao Y. Identification and validation of candidate genes involved in fatty acid content in oil palm by genomewide association analysis. Front Plant Sci. 2019;10:1263. doi: 10.3389/fpls.2019.01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Luo T, Dou Y, Zhang W, Mason AS, Huang D, Huang X, Tang W, Dou Y, Zhang C, Xiao Y, et al. Identification of genes affecting saturated fat acid content in Elaeis guineensis by genome-wide association analysis. BioRxiv. 2018;34:1347. [Google Scholar]
- Xiao Y, Zhou L, Lei X, Cao H, Wang Y, et al. Genome-wide identification of WRKY genes and their expression profiles under different abiotic stresses in Elaeis guineensis. PLoS ONE. 2017;12(12):e0189224. doi: 10.1371/journal.pone.0189224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarra R, Jin L, Zhao Z, Cao H. Progress in tissue culture and genetic transformation of oil palm: an overview. Int J Mol Sci. 2019;20(21):5353. doi: 10.3390/ijms20215353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap WC, Lee FC, Shabari Shan DK, Musa H, Appleton DR, Kulaveerasingam H. WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 2017;91:97–113. doi: 10.1111/tpj.13549. [DOI] [PubMed] [Google Scholar]
- Yeap WC, Namasivayam P, Ooi TE, Appleton DR, Kulaveerasingam H, Ho CL. EgRBP42 from oil palm enhances adaptation to stress in Arabidopsis through regulation of nucleocytoplasmic transport of stress-responsive mRNAs. Plant Cell Environ. 2019;42(5):1657–1673. doi: 10.1111/pce.13503. [DOI] [PubMed] [Google Scholar]
- Yeap WC, Ooi TE, Namasivayam P, Kulaveerasingam H, Ho CL. EgRBP42 encoding an hnRNP-like RNA-binding protein from Elaeis guineensis Jacq. is responsive to abiotic stresses. Plant Cell Rep. 2012;31(10):1829–1843. doi: 10.1007/s00299-012-1297-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bai B, Lee M, Alfiko Y, Suwanto A, Yue GH. Cloning and characterization of EgGDSL, a gene associated with oil content in oil palm. Sci Rep. 2018;8:11406. doi: 10.1038/s41598-018-29492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YS, Chen H, Yuan Y, Wang Y, Chen L, Liu X, Li DD. Cloning and functional characterization of long-chain acyl-CoA synthetase 1 from the mesocarp of African oil palm (Elaeis guineensis Jacq.) Ind crops products. 2018;122:252–260. [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]