Abstract
Background: Colorectal adenocarcinoma with mucinous component (AWMC) is a special entity of colorectal cancer. The study is aimed at analyzing the clinicopathological characteristics, mutation spectrum, and prognosis of AWMC and comparing it with classical adenocarcinoma (AC) in a Chinese cohort.
Methods: One hundred eight AMWC and 204 AC patients were included. Targeted next-generation sequencing (NGS) was performed on formalin-fixed paraffin-embedded (FFPE) tissues. AWMC was further divided into two groups: AWMC with signet ring cell component and AWMC without signet ring cell component. Clinicopathological features, mismatch repair protein (MMR) status, genetic alterations, and survival outcomes were analyzed after tumor location was taken into consideration.
Results: AWMC had larger tumor size (p = 0.014) and showed predilection for proximal colon (p < 0.001) compared with AC. Regardless of primary sites, AWMC was associated with less metastasis (p < 0.001) and earlier AJCC stage (p < 0.001). Mismatch repair protein deficiency (dMMR) was more commonly detected in AWMC than in AC for right-sided colon (p < 0.001), but the difference was not significant for left-sided colon (p = 0.081). The five most commonly mutated genes in AWMC were KRAS (45.4%), TP53 (39.8%), APC (22.2%), PIK3CA (22.2%), and SMAD4 (10.2%). AWMC showed a significantly lower mutation rate of TP53 than AC, both in right-sided colon and in left-sided colon (p < 0.001 and p = 0.033, respectively). In left-sided colon, AWMC with signet ring cell component had a significantly smaller size than tumors with signet ring cell component (p = 0.034). No dMMR cases were detected in AWMC with signet ring cell component (n = 7). Moreover, AWMC with signet ring cell component had a significantly lower KRAS mutation rate than AWMC without signet ring cell component, both in right-sided colon and in left-sided colon (p = 0.036 and p = 0.012, respectively). The disease-specific survival (DSS) for AWMC and AC were not statistically different (p = 0.0587). Multivariate analysis showed that AWMC was not an independent predictor of prognosis.
Conclusion: Regardless of primary sites, AWMC demonstrates less metastasis, earlier stages, more frequent dMMR, and lower TP53 mutation rate than AC. Our results indicate that different molecular pathogenesis might underlie mucinous morphology in colorectal carcinoma. Mucinous component is not an independent factor of outcome.
Keywords: AWMC, AC, clinicopathological characteristics, MMR, mutation spectrum, primary site
Introduction
Colorectal cancer (CRC) is the fourth most deadly cancer globally and has become a public health problem due to its rising incidence (1). It is characterized by high heterogeneity and varied and outcomes (2). Based on histological subtypes, most CRCs belong to classical adenocarcinomas (AC), with several histological variants associated with specific molecular characteristics (3, 4). Previous research suggested that mucinous histology is related to the proximal colon, microsatellite instability, and numerous upregulated or downregulated genes involved with differentiation and mucin production (5–7). Although the importance of histological appearance has been highlighted, there is a persistent debate regarding its clinical behavior (8–10). It is also unclear whether the existence of signet ring cells is related to clinicopathological characteristics or prognosis (11, 12). In recent years, the location of the tumors has been indicated to be an important predictor factor, which has added to the difficulty of discussing its clinicopathological features and outcomes (13). In this study, we emphasized on colorectal adenocarcinoma with mucinous component (AWMC) after considering the impact of location and assessed the potential differences between AWMC and AC through an institution-based cohort.
Materials and Methods
Patients
A total of 312 patients diagnosed with CRC between July 2010 and September 2018 from Peking Union Medical College Hospital (PUMCH), Chinese Academy of Medical Sciences were identified, including 108 AWMC patients and 204 AC patients. Approval for this study was obtained from PUMCH Institutional Review Board. Written informed consents were obtained from all the patients.
Pathological Evaluations
Tumor sections from formalin-fixed paraffin-embedded (FFPE) samples were stained with hematoxylin-eosin (HE) and reviewed by two experienced pathologists independently. AWMC was defined as adenocarcinoma that shows intra or extracellular mucin secretion. AWMC with signet ring cell component was defined as AWMC with signet ring cells which typically show displacement and molding of the nucleus.
Clinicopathological Features
Clinicopathological parameters were obtained from medical records and pathological reports. Tumors arising from the cecum, ascending colon, and the right 2/3 of transverse colon were considered to be right-sided and those arising in the left 1/3 of transverse colon, descending colon, sigmoid colon, and rectum were considered to be left-sided. Tumor stage was decided based on the American Joint Committee on Cancer (AJCC) tumor staging system (the eighth edition, 2017).
Mismatch Repair (MMR) Status
Immunohistochemistry (IHC) for the four MMR proteins (MLH1, PMS2, MSH2 and MSH6) was performed on FFPE slides from more recent specimens, including 33 AWMC cases and 88 AC cases. The antibodies used were: MLH1 (clone ES05, Leica), MSH2 (clone 25D12, Leica), MSH6 (clone PU29, Leica) and PMS2 (clone M0R4G, Leica). Normal colon epithelium and infiltrating lymphocytes were used as internal positive controls. dMMR was defined as complete nuclear loss of expression of one or more of these proteins.
Target Next-Generation Sequencing (NGS)
DNA of FFPE samples was isolated using a QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer's protocol. 10 ng of DNA were used as template to generate an amplicon library for sequencing for AC samples. Libraries were prepared using an Ion AmpliSeq Library Kit 2.0 and an Ion AmpliSeq Cancer Hotspot Panel v2 (Life Technologies) and quantified using a Qubit dsDNA HS Assay Kit and a Qubit 2.0 fluorometer (Life Technologies). Sequenceing was performed with an Ion Torrent PGM system (Life Technologies). The readings were mapped to the reference genome (hg19) using the Torrent Mapping Alignment Program. Variants were identified using Torrent Variant Caller (3.6.6; Life Technologies). The SGI OncoAim™DNAPanel (Singlera Genomics, Shanghai, China) was used for preparing DNA libraries of AWMC samples and the Qubit® dsDNA HS Assay kit and a Qubit 3.0 fluorimeter was used for quantification. Each library was quantified with KAPASYBR® FAST universal qPCR Kits. Libraries were pooled in equimolar amount and sequenced with an Illumina Miseq sequencer (Illumina, Hayward, CA, USA). The output data were uploaded for quality control, sequence alignment, and variant calling using a vendor-supplied bioinformatics pipeline.
Statistical Analysis
Statistical analysis was performed with IBM SPSS statistics Version 23.0 (IBM Corporation, USA). Ages were expressed as means ± s.d. and differences between groups were compared using independent sample T-test. χ2-Square test or Fisher's exact test was used for nominal scaled variables to compare the clinical features and mutation spectrum. The distribution of ordinal scaled variables (T, N, M, and AJCC stage) was performed by Mann-Whitney U-test. Cochran-Mantel-Haenszel (CMH) test was used for analyzing stratified categorical data. Disease-specific survival (DSS) was defined as the interval from the date of treatment to death specifically from colorectal cancer. Log-rank test was used to compare Kaplan–Meier survival curves. The covariated factors with a borderline significance (p < 0.2) were included in multivariate analysis, which was performed with a Cox proportional hazards regression model. A p < 0.05 was considered to be statistically significant.
Results
Clinicopathological Characteristics of AWMC and AC
A total of 108 cases diagnosed with AWMC and 204 cases diagnosed with AC were collected. Their clinicopathological features were shown in Table 1.
Table 1.
Clinicopathological features of AWMC and AC.
| AWMC (n = 108) | AC (n = 204) n/% | P (AWMC vs. AC) | P (AWMC without SRC vs. AC) | |||
|---|---|---|---|---|---|---|
| Without SRC (n = 90) n/% | With SRC (n = 18) n/% | P | ||||
| Sex | ||||||
| Male | 55 (61.1) | 9 (50.0) | 0.381 | 122 (59.8) | 0.926 | 0.833 |
| Female | 35 (38.9) | 9 (50.0) | 82 (40.2) | |||
| Age (yr), median | 61 (20-84) | 44.5 (25-79) | 0.026 | 61 (30-91) | 0.084 | 0.462 |
| Tumor size (cm) | ||||||
| ≤ 5 | 39 (43.3) | 13 (72.2) | 0.007 | 97 (47.6) | 0.014 | 0.002 |
| >5 | 50 (55.6) | 3 (16.7) | 50 (24.5) | |||
| Unknown | 1 (1.1) | 2 (11.1) | 57 (27.9) | |||
| Tumor location | ||||||
| Right-sided | 39 (43.3) | 11 (61.1) | 0.180 | 40 (19.6) | <0.001 | <0.001 |
| Left-sided | 50 (55.6) | 7(38.9) | 158 (77.4) | |||
| Multiple | 1 (1.1) | 0 (0) | 5 (2.5) | |||
| Unknown | 0 (0) | 0 (0) | 1 (0.5) | |||
| T | ||||||
| Tis | 1 (1.1) | 0 (0) | 0.488 | 0 (0) | 0.028 | 0.033 |
| T1 | 2 (2.2) | 1 (5.6) | 2 (1.0) | |||
| T2 | 12 (13.3) | 1 (5.6) | 11 (5.4) | |||
| T3 | 62 (69.0) | 13 (72.1) | 117 (57.3) | |||
| T4 | 12 (13.3) | 2 (11.1) | 32 (15.7) | |||
| Tx | 1 (1.1) | 1 (5.6) | 42 (20.6) | |||
| N | ||||||
| N0 | 39 (43.4) | 6 (33.3) | 0.111 | 44 (21.6) | 0.087 | 0.026 |
| N1 | 29 (32.2) | 0 (0) | 65 (31.9) | |||
| N2 | 20 (22.2) | 9 (50.0) | 46 (22.5) | |||
| Nx | 2 (2.2) | 3 (16.7) | 49 (24.0) | |||
| M | ||||||
| M0 | 81 (90.0) | 15 (83.3) | 0.721 | 88 (43.2) | <0.001 | <0.001 |
| M1 | 8 (8.9) | 2 (11.1) | 109 (53.4) | |||
| Mx | 1 (1.1) | 1 (5.6) | 7 (3.4) | |||
| AJCC Stage | ||||||
| I | 9 (10.0) | 1 (5.6) | 0.614 | 5 (2.5) | <0.001 | <0.001 |
| II | 28 (31.1) | 5 (27.8) | 20 (9.8) | |||
| III | 43 (47.8) | 8 (44.4) | 64 (31.4) | |||
| IV | 8 (8.9) | 2 (11.1) | 109 (53.4) | |||
| Unknown | 2 (2.2) | 2 (11.1) | 6 (2.9) | |||
SRC, signet ring cell component. Bold values were statistically significant.
In both AWMC and AC, patients were predominantly males (59% and 60%). The median ages of AWMC and AC were 60 years (range: 20–84 years) and 61 years (range: 30–91 years), respectively (p = 0.084). Compared with AC, AWMC had a significantly larger tumor size (p = 0.014), was more frequently right-sided (p < 0.001), and presented at an earlier AJCC stage (p < 0.001). After excluding cases of AWMC with signet ring cell component (n = 18), similarly, AWMC without signet ring cell component was also related to larger tumor size, right-sided colon, and earlier stages compared with AC.
Tumors with signet ring cell component was associated with younger age compared with tumors without signet ring cell component (p = 0.026). Tumor size was also shown to be different between the two subgroups (p = 0.007): the existence of signet ring cell component was associated with a smaller size, with 72.2% of the tumors no larger than 5 cm. There were no significant differences in gender, tumor location and staging (p = 0.381, 0.180, 0.614, respectively).
Since there was obvious distinction in primary tumor sites between AWMC and AC, their clinicopathological characteristics were analyzed after stratification (Table 2, Supplementary Tables 1, 2). Despite the primary sites, AWMC developed less metastasis and presented with an earlier AJCC stage. Nevertheless, other clinical characteristics were not completely the same in subgroups: In right-sided colon, sex, age and tumor size did not differ significantly between AWMC and AC, whereas the existence of signet ring cell component was associated with younger age (p = 0.022). In left-sided colon, the median age of AWMC was significantly younger than AC (55 vs. 61, p = 0.005). There was no obvious difference in tumor size between AWMC and AC (p = 0.103), while AWMC without signet ring cell component showed a larger size than AWMC with signet ring cell component (p = 0.034).
Table 2.
P-value comparing clinicopathological features between AWMC without SRC, AWMC with SRC, and AC.
| Sex | Age | Tumor size | T | N | M | Stage | |
|---|---|---|---|---|---|---|---|
| Right-sided | |||||||
| AWMC vs. AC | 0.703 | 0.422 | 0.881 | 0.145 | 0.005 | <0.001 | <0.001 |
| AWMC without SRC vs. AC | 0.746 | 0.895 | 0.471 | 0.138 | 0.002 | <0.001 | <0.001 |
| AWMC without SRC vs. AWMC with SRC | 1.000 | 0.022 | 0.068 | 0.001 | 0.371 | 0.790 | 0.662 |
| Left-sided | |||||||
| AWMC vs. AC | 0.750 | 0.005 | 0.103 | 0.004 | 0.537 | <0.001 | <0.001 |
| AWMC without SRC vs. AC | 0.506 | 0.030 | 0.025 | 0.011 | 0.978 | <0.001 | <0.001 |
| AWMC without SRC vs. AWMC with SRC | 0.404 | 0.164 | 0.034 | 0.488 | 0.054 | 0.426 | 0.400 |
| Left-sided (excluding rectal tumors) | |||||||
| AWMC vs. AC | 0.895 | 0.059 | 0.149 | 0.099 | 1.000 | <0.001 | <0.001 |
| AWMC without SRC vs. AC | 0.942 | 0.266 | 0.046 | 0.211 | 0.471 | <0.001 | <0.001 |
| Rectal | |||||||
| AWMC vs. AC | 0.586 | 0.040 | 0.381 | 0.020 | 0.403 | <0.001 | <0.001 |
| AWMC without SRC vs. AC | 0.405 | 0.059 | 0.225 | 0.026 | 0.501 | <0.001 | <0.001 |
In left-sided tumors (excluding rectal tumors) and rectal tumors, AWMC without SRC was not compared with AWMC with SRC due to the limited number of cases. Bold values were statistically significant.
For further details, the rectal tumors were separated from left-sided tumors (Table 2, Supplementary Table 2). In rectum, AWMC occurred at a younger age than AC (p = 0.040). In the other sites of the left-sided colon, no difference in sex or age was observed, whereas AWMC without signet ring cell component tended to be larger than AC (p = 0.046).
MMR Status
Deficient MMR (dMMR) was found in 14 of 33 cases (42%) of AWMC and 5 of 88 cases (6%) of AC (Figures 1, 2). AWMC demonstrated a significantly higher frequency of dMMR compared with AC (p < 0.001) (Table 3). In dMMR AWMC cases, there were 8 MLH1/PMS2 paired negative, 4 MSH2/MSH6 paired negative, and 2 PMS2 negative. In dMMR AC cases, there were 2 MLH1/PMS2 paired negative, 1 MSH2/MSH6 paired negative, 1 PMS2 negative, and 1 case with all four proteins negative. In AWMC with signet ring cell component, all the 7 tested cases were MMR-proficient (pMMR). Signet ring cell component was associated with less dMMR (p = 0.013).
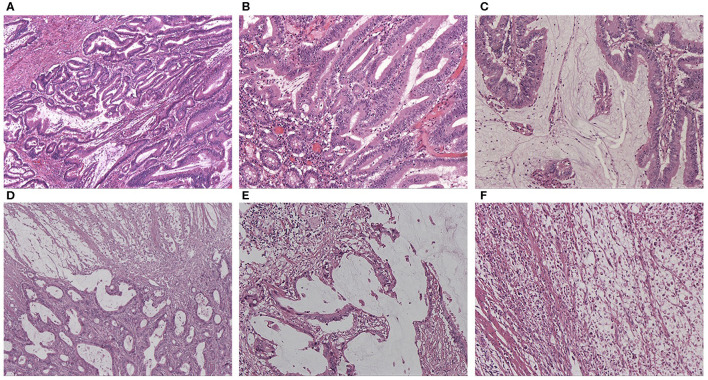
Figure 1.
(A) HE staining of AWMC showing both classical adenocarcinoma component and mucinous component (HEx40). (B) HE staining of AWMC showing classical adenocarcinoma component (HEx100). (C) HE staining of AWMC showing mucinous component (HEx100). (D) HE staining of AWMC with signet ring cell component (HEx40). (E) HE staining of AWMC with signet ring cell component showing mucin pool (HEx100). (F) HE staining showing scattered signet ring cells in the same case with D and E (HEx100).
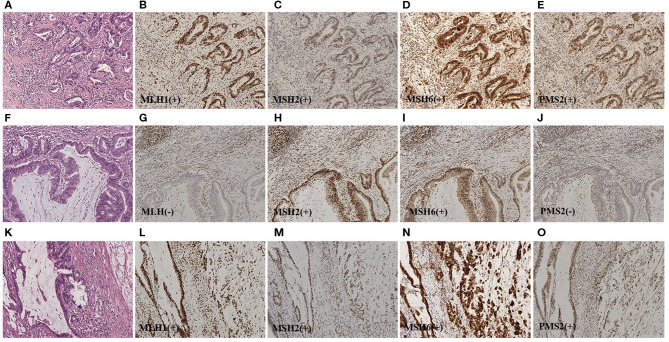
Figure 2.
(A) Representative HE staining of classical adenocarcinoma (HEx100). For (B–E), representative IHC staining for the four MMR proteins of serial sections from pMMR classical adenocarcinoma (IHCx100). Stromal cells and infiltrating lymphocytes served as internal positive controls. (F) Representative HE staining of dMMR AWMC (HEx100). For (G–J), representative IHC staining for the four MMR proteins of serial sections from dMMR AWMC (IHCx100). Stromal cells and infiltrating lymphocytes served as internal positive controls. In this case, MLH1 and PMS2 were negative. (K) Representative HE staining of AWMC with signet ring cell component (HEx100). For (L–O), representative IHC staining for the four MMR proteins of serial sections from pMMR AWMC with signet ring cell component (IHCx100). Stromal cells and infiltrating lymphocytes served as internal positive controls.
Table 3.
MMR status and mutation spectrum of AWMC and AC.
| AWMC | AC | P | P | |||
|---|---|---|---|---|---|---|
| Without SRC | With SRC | P | (AWMC vs. AC) | (AWMC without SRC vs. AC) | ||
| MMR status | n/% (n = 26) | n/% (n = 7) | 0.013 | n/% (n = 88) | <0.001 | <0.001 |
| pMMR | 12 (46.2) | 7 (100.0) | 0.023* (R) | 83 (94.3) | 0.001* (R) | <0.001* (R) |
| dMMR | 14 (53.8) | 0 (0) | 1.000* (L) | 5 (5.7) | 0.081* (L) | 0.048* (L) |
| Mutated gene | n/% (n = 90) | n/% (n = 18) | n/% (n = 204) | |||
| AKT1 | 1 (1.1) | 0 (0) | 1.000 | 2 (1.0) | 1.000 | 1.000 |
| ALK | 0 (0) | 1 (5.6) | 0.167 | 0 (0) | 0.346 | – |
| APC | 19 (21.1) | 5 (27.8) | 0.543 | 50 (24.5) | 0.651 | 0.526 |
| 0.093* (R) | 0.596* (R) | 0.260* (R) | ||||
| 0.662* (L) | 0.882* (L) | 0.706* (L) | ||||
| ATM | 2 (2.2) | 2 (11.1) | 0.129 | 1 (0.5) | 0.050 | 0.223 |
| BRAF | 9 (10.0) | 1 (5.6) | 1.000 | 10 (4.9) | 0.135 | 0.101 |
| 1.000* (R) | 1.000* (R) | 0.737* (R) | ||||
| 1.000* (L) | 0.702* (L) | 0.451* (L) | ||||
| EGFR | 1 (1.1) | 1 (5.6) | 0.307 | 1 (0.5) | 0.276 | 0.519 |
| ERBB2 | 4 (4.4) | 1 (5.6) | 1.000 | 1 (0.5) | 0.020 | 0.032 |
| 1.000* (R) | 1.000* (R) | 0.494* (R) | ||||
| 0.417* (L) | 0.018* (L) | 0.044* (L) | ||||
| FBXW7 | 4 (4.4) | 0 (0) | 1.000 | 17 (8.3) | 0.120 | 0.233 |
| –* (R) | 0.036* (R) | 0.116* (R) | ||||
| 1.000* (L) | 1.000* (L) | 1.000* (L) | ||||
| HRAS | 0 (0) | 1 (5.6) | 0.167 | 0 (0) | 0.346 | - |
| KIT | 2 (2.2) | 0 (0) | 1.000 | 1 (0.5) | 0.276 | 0.223 |
| KRAS | 47 (52.2) | 2 (11.1) | 0.001 | 96 (47.1) | 0.776 | 0.414 |
| 0.036* (R) | 0.119* (R) | 0.436* (R) | ||||
| 0.012* (L) | 0.675* (L) | 0.234* (L) | ||||
| NRAS | 5 (5.6) | 2 (11.1) | 0.330 | 8 (3.9) | 0.316 | 0.546 |
| 0.601* (R) | 0.127* (R) | 0.201* (R) | ||||
| –* (L) | 0.345* (L) | 0.339* (L) | ||||
| PIK3CA | 20 (22.2) | 4 (22.2) | 1.000 | 28 (13.7) | 0.055 | 0.069 |
| 1.000* (R) | 0.263* (R) | 0.292* (R) | ||||
| 1.000* (L) | 0.961* (L) | 0.807* (L) | ||||
| PTEN | 7 (7.8) | 1 (5.6) | 1.000 | 7 (3.4) | 0.118 | 0.137 |
| SMAD4 | 9 (10.0) | 2 (11.1) | 1.000 | 13 (6.4) | 0.229 | 0.276 |
| 1.000* (R) | 0.689* (R) | 0.675* (R) | ||||
| 1.000* (L) | 0.264* (L) | 0.250* (L) | ||||
| SMARCB1 | 1 (1.1) | 0 (0) | 1.000 | 0 (0) | 0.346 | 0.306 |
| TP53 | 33 (36.7) | 10 (55.6) | 0.135 | 135 (66.2) | <0.001 | <0.001 |
| 0.065* (R) | <0.001* (R) | <0.001* (R) | ||||
| 0.706* (L) | 0.033* (L) | 0.030* (L) | ||||
P-value after stratified analysis with CMH test. (R), right-sided colon. (L), left-sided colon. Bold values were statistically significant.
The data were analyzed according to tumors' primary sites. In right-sided colon, AWMC showed more frequent dMMR than AC (p < 0.001), and AWMC without signet ring cell component showed more dMMR than AWMC with with signet ring cell component (p = 0.023). In left-sided colon, there was no significant difference for MMR status between AWMC and AC (p = 0.081). Due to the limited number of cases, only AWMC and AC were compared when classifying left-sided colon into two subgroups (rectum and left-sided colon excluding rectum). There was no statistically difference between AWMC and AC in both groups (data not shown).
Mutation Spectrum
The five most commonly mutated genes in AWMC were KRAS (n = 49, 45.4%), TP53 (n = 43, 39.8%), APC (n = 24, 22.2%), PIK3CA (n = 24, 22.2%), and SMAD4 (n = 11, 10.2%), while those in AC were TP53 (n = 135, 66.2%), KRAS (n = 96, 47.1%), APC (n = 50, 24.5%), PIK3CA (n = 28, 13.7%), and FBXW7 (n = 17, 8.3%) (Table 3). Compared with AC, AWMC was associated with lower TP53 mutation rate (39.8 vs. 66.2%, p < 0.001) and higher ERBB2 mutation rate (4.6 vs. 0.5%, p = 0.020). The five ERBB2 mutated spots in AWMC were R678Q (3 cases), V754M, and V842I, and the ERBB2 mutated spot in AC was V842I. AWMC without signet ring cell component demonstrated similar characteristics to AWMC when compared with AC, with lower TP53 mutation rate (36.7%) and more frequent ERBB2 mutation (4.4%). Within AWMC, signet ring cell component was associated with lower KRAS mutation rate (p = 0.001) (Table 3).
Several frequently mutated genes were selected for analyzing their mutation status in different sites of the colon (Table 3). The mutation spectrum of APC, BRAF, KRAS, NRAS, PIK3CA, SMAD4, and TP53 for AWMC and AC was similar regardless of primary sites. Nonetheless, only in left-sided colon did AWMC demonstrate a higher mutation rate of ERBB2 than AC (p = 0.018). In addition, AWMC showed more frequent mutation of FBXW7 than AC in right-sided tumors (p = 0.036), although their overall mutation rates were not statistically different (p = 0.120).
The mutation spectrum of the selected genes was also analyzed in left-sided colon (excluding rectum) and rectum, respectively. There was no obvious difference between AWMC and AC in both sites for mutation rate of APC, BRAF, FBXW7, KRAS, NRAS, PIK3CA, and SMAD4. For TP53, AWMC without SRC demonstrated a lower mutation rate than AC in left-sided colon (excluding rectum) (p = 0.042), whereas no obvious difference was observed between AWMC and AC (p = 0.083). In rectum, there was no significant difference for TP53 between AWMC without signet ring cell component, AWMC with signet ring cell component, and AC. For ERBB2, all of the mutated cases arose from the left-sided colon (excluding rectum), and the existence of mucinous component was related to a higher mutation rate (AWMC without SRC vs. AC, p = 0.042; AWMC vs. AC, p = 0.018; AWMC without SRC vs. AWMC with SRC, p = 0.495).
RAS mutational status. KRAS mutation was detected in 49/108 AWMC patients (45%), whose rate is slightly higher than that of CRC previously reported. Forty-eight of the mutations were point mutation (13 of G12D, 10 of G12V, 9 of G13D, 8 of A146T, 2 of Q61R, 1 of A146V, 1 of G12C, 1 of G12S, 1 of Q61H, 1 of A146T and G13D, 1 of T20M and G13D), with only one insertion (G12_G13insA). Comparison of subgroups indicated that the rate of KRAS mutation in AWMC without signet ring cell component was significantly higher than in AWMC with signet ring cell component (52.2 vs. 11.1%, p = 0.001) (Table 3). Only one HRAS mutation was detected in AMWC. The mutation rates of NRAS were low in both groups, with 6.5% in AWMC and 3.9% in AC (p = 0.316). Two patients harbored concomitant NRAS and KRAS mutations, one of whom was KRAS (G13D) plus NRAS (G12D) and the other was KRAS (G12D) plus NRAS (G12D).
BRAF mutational status. The mutation rates of BRAF were 9.3% in AWMC and 4.8% in AC (p = 0.135). The hotspot was the typical V600E mutation (7 of V600E, 1 of G466E, 1of G469R, and 1 of R726H).
Several cases with rare concomitant mutations were identified in AWMC without signet ring cell component: one case with NRAS and BRAF mutation (Q61R and R726H, respectively), one case with concomitant KRAS and BRAF mutation (A146T and G466E, respectively), and three cases with BRAF and PIK3CA mutations (E545K and V600E, R108H and V600E, G118D and R726H).
Correlation of Mutational Status With Clinicopathological Features
The association between KRAS or BRAF mutation and clinicopathological features was analyzed in AWMC (Table 4, Supplementary Table 3). There was no correlation of sex, age, tumor size, location, or staging with mutation status of KRAS or BRAF.
Table 4.
Correlation of KRAS and BRAF status with clinicopathological features in AWMC.
| Clinicopathological characteristics | KRAS status | P | BRAF status | P | ||
|---|---|---|---|---|---|---|
| Wild (n = 59) n/% | Mutant (n = 49) n/% | Wild (n = 98) n/% | Mutant (n = 10) n/% | |||
| Sex | ||||||
| Male | 37 (62.7) | 27 (55.1) | 0.423 | 60 (61.2) | 4 (40.0) | 0.311 |
| Female | 22 (37.3) | 22 (44.9) | 38 (38.8) | 6 (60.0) | ||
| Age (yr), median | 62 (20-84) | 60 (25-81) | 0.979 | 58 (20-84) | 72 (40-79) | 0.070 |
| Tumor size (cm) | ||||||
| ≤ 5 | 31 (52.5) | 21 (42.9) | 0.278 | 46 (46.9) | 6 (60.0) | 0.526 |
| >5 | 26 (44.1) | 27(55.1) | 49 (50.0) | 4 (40.0) | ||
| Unknown | 2 (3.4) | 1 (2.0) | 3 (3.1) | 0 (0) | ||
| Tumor location | ||||||
| Right-sided | 27 (45.8) | 23 (46.9) | 0.968 | 44 (44.9) | 6 (60.0) | 0.299 |
| Left-sided | 31 (52.5) | 26 (53.1) | 54 (55.1) | 3 (30.0) | ||
| Multiple | 1 (1.7) | 0 (0) | 0 (0) | 1 (10.0) | ||
| T | ||||||
| Tis | 1 (1.7) | 0 (0) | 0.635 | 0 (0) | 1 (10.0) | 0.535 |
| T1 | 1 (1.7) | 2 (4.1) | 2 (2.0) | 1 (10.0) | ||
| T2 | 7 (11.9) | 6 (12.2) | 12 (12.3) | 1 (10.0) | ||
| T3 | 43 (72.8) | 32 (65.4) | 70 (71.4) | 5 (50.0) | ||
| T4 | 6 (10.2) | 8 (16.3) | 12 (12.3) | 2 (20.0) | ||
| Tx | 1 (1.7) | 1 (2.0) | 2 (2.0) | 0 (0) | ||
| N | ||||||
| N0 | 24 (40.7) | 21 (42.9) | 0.254 | 40 (40.8) | 5 (50.0) | 0.492 |
| N1 | 10 (16.9) | 19 (38.8) | 27 (27.6) | 2 (20.0) | ||
| N2 | 21(35.6) | 8 (16.3) | 27 (27.6) | 2 (20.0) | ||
| Nx | 4 (6.8) | 1 (2.0) | 4 (4.0) | 1 (10.0) | ||
| M | ||||||
| M0 | 54 (91.5) | 42 (85.7) | 0.328 | 86 (87.8) | 10 (100.0) | 0.286 |
| M1 | 4 (6.8) | 6 (12.3) | 10 (10.2) | 0 (0) | ||
| Mx | 1 (1.7) | 1 (2.0) | 2 (2.0) | 0 (0) | ||
| Stage | ||||||
| I | 5 (8.5) | 5 (10.2) | 0.623 | 9 (9.2) | 1 (10.0) | 0.298 |
| II | 19 (32.2) | 14 (28.6) | 29 (29.6) | 4 (40.0) | ||
| III | 28 (47.4) | 23 (46.9) | 47 (47.9) | 4 (40.0) | ||
| IV | 4 (6.8) | 6 (12.3) | 10 (10.2) | 0 (0) | ||
| Unknown | 3 (5.1) | 1 (2.0) | 3 (3.1) | 1 (10.0) | ||
Survival Analysis
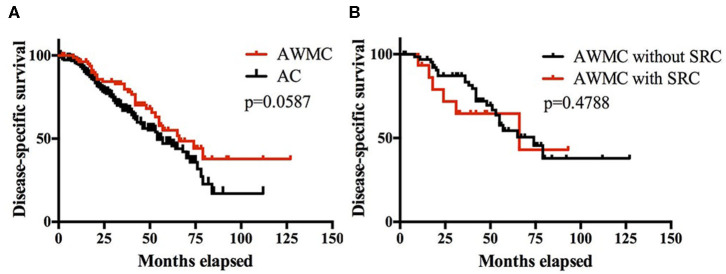
A total of 269 patients (86%) had clinical follow-up, including 188 AC, 66 AWMC without signet ring cell component, and 15 AWMC with signet ring cell component. The median follow-up time was 36 months (range: 1–127 months). The median DSS for AWMC and AC was 66 months and 54 months, respectively. The cumulative survival rates for AWMC at 1, 3, and 5 years were 96, 80, and 55%, respectively. The cumulative OS rates for AC at 1, 3, and 5 years were 93, 67, and 47%, respectively. The DSS of AWMC was not statistically different from AC (p = 0.0587) (Figure 3A). Within AWMC, the median DSS was 74 months for patients without signet ring cell component, and 66 months for patients with signet ring cell component. There was no difference between the two subgroups (p = 0.4788) (Figure 3B).
Figure 3.
(A) Kaplan-Meier survival curves showing DSS of patients of AWMC and AC. (B) Kaplan-Meier survival curves showing DSS of patients of AWMC with SRC and AWMC without SRC.
To further assess the potential impact of mucinous component on survival, univariate and multivariate analysis of CRC was performed. In univariate analysis, absence of lymph node metastasis (p = 0.037) and an earlier AJCC stage (p = 0.006) presented a favorable impact on survival (Table 5). Age, histopathology, lymph node status, and stage were included into multivariate analysis. Age over 55 years (HR 1.84, 95% CI 1.12–3.04) and AJCC stage III-IV (HR 1.82, 95% CI 1.08–3.06) were found to be independent predictors of poor outcome (p = 0.017 and p = 0.025). Nevertheless, histopathology or lymph node metastasis was not a significant predictor of survival (p = 0.446 and p = 0.543).
Table 5.
Univariate analysis of DSS of CRC patients.
| Variables | HR | 95% CI | P |
|---|---|---|---|
| Sex | |||
| Male | 1 | 0.884 | |
| Female | 1.03 | 0.71–1.49 | |
| Age (yr) | |||
| ≤ 55 | 1 | 0.054 | |
| >55 | 1.50 | 1.00–2.12 | |
| Tumor size | |||
| ≤ 5 | 1 | 0.796 | |
| >5 | 0.95 | 0.62–1.44 | |
| Tumor location | |||
| Right-sided | 1 | 0.782 | |
| Left-sided | 0.94 | 0.62–1.43 | |
| Histopathology | |||
| AWMC | 1 | 0.059 | |
| AC | 1.48 | 0.99–2.14 | |
| T | |||
| T1-T2 | 1 | 0.211 | |
| T3-T4 | 1.5 | 0.82–2.5 | |
| Lymph node metastasis | |||
| No | 1 | 0.037 | |
| Yes | 1.61 | 1.03–2.42 | |
| AJCC stage | |||
| I-II | 1 | 0.006 | |
| III-IV | 1.99 | 1.19–2.74 | |
| KRAS status | |||
| Wild | 1 | 0.496 | |
| Mutant | 1.13 | 0.79–1.65 | |
| BRAF status | |||
| Wild | 1 | 0.242 | |
| Mutant | 1.49 | 0.73–3.66 | |
HR, hazard ratio. CI, confidence interval. Bold values were statistically significant.
Discussion
Our study identifies the clinicopathological features, molecular spectrum, and clinical outcomes of AWMC and compare with AC to reveal the potential differences underlie different histology.
Previous studies suggest that special histology subtypes of CRC differ from AC in tumor biology and outcomes (10). Mucinous adenocarcinoma present homologous clinicopathological manifestations such as affecting younger age, arising from proximal colon, and larger tumor size, and signet ring cells indicate poorer outcome (11, 14, 15). However, their clinical features, especially survival outcomes, have been largely controversial (16). In recent years, more researchers have been focusing on the specific impact of site of origin in the pathogenesis of CRC (16–19). In a study of 2,413 colorectal tumors in 2017, Salem et al. reported that colorectal tumors with different primary sites present with distinct clinical features and molecular features (13). Similar conclusions have been illustrated in other studies (18, 20). The findings regarding variations between right-sided and left-sided colon should certainly be considered when exploring the potential differences between tumors with specific histology and AC. In addition, although it is common to include rectum into left-sided colon, some researchers have separated them and found variations (13, 16, 21–23). Nonetheless, this confounding variable was not considered in most of the previous studies emphasizing on histology (11, 24). Therefore, one of the strengths of our research is to compare AWMC with AC after clear stratification.
Firstly, our retrospective study illustrates that AWMC is associated with larger tumor size and proximal colon. Contrary to previous studies, AWMC is more diagnosed at T2-T3 stage, whereas AC demonstrates more metastasis, probably due to earlier symptoms of AWMC (25). We do not find significant difference of gender, tumor location, or staging between AMWC with signet ring cell component and AWMC without signet ring cell component, which is consistent with previous research (8). Nevertheless, AWMC with signet ring cell component appears to occur in younger patients and the tumor size tends to be smaller at diagnosis. Next, AWMC is compared with AC after classifying tumor location into left-sided and right-sided. We find that AWMC is always associated with less metastasis and earlier stage regardless of primary site, which highlights the influence of specific mucinous histology. There is not significant variation of metastatic status or AJCC stage among the two subgroups of AWMC. Interestingly, only in left-sided colon did AWMC show a younger age than AC. After further classifying left-sided colon, we find that the different age distribution is seemingly caused by rectal AWMC rather than other sites of left-sided colon.
In MMR IHC analysis, our results indicate that AWMC is associated with dMMR (p < 0.001 in all cases; p = 0.001 in right-sided cases). In left-sided cases, the MMR status between AWMC and AC is not significantly different. However, due to the limitation of study population (12 cases arising from left-sided colon), there is still a trend for AWMC to present more dMMR (p = 0.081). Notably, none of the AWMC with signet ring cell component (7 cases) were dMMR, which to some extent reveals its unique molecular pathogenesis.
The pathogenesis of CRC involves a series of genetic and epigenetic modifications regulating cell proliferation, apoptosis, and angiogenesis (26). RAS and BRAF are two well-known proto-oncogenes located downstream of the epidermal growth factor receptor (EGFR) signaling cascade and their mutation results in constitutive activation of EGFR pathway and colorectal tumorigenesis. Mutations of multiple genes such as KRAS and NRAS confer resistance to anti-EGFR therapy. BRAF gene encodes a protein which is part of the Ras-Raf-MEK-ERK (MAPK) signaling pathway. BRAF mutation is associated with poor prognosis, and is more frequent in signet ring cell carcinoma than AC (27). The mutation rates of KRAS and BRAF in overall CRC in different ethnic groups and different studies vary a lot, and hotspot genes are important for deciding treatments and predicting outcomes (28). However, the mutational status of CRC in Chinese population is relatively lacking. One of the largest Chinese cohort collecting 1,110 patients has illustrated the molecular spectrum of CRC, but the histology subtypes were not stratified (29). Our data for both KRAS and BRAF were similar to previously reported rates in mucinous adenocarcinoma, and further stratification indicates similar characteristics of AWMC and AC in each specific location (27).
Previous research suggests that TP53 is the most commonly mutated gene in CRC. In our study, AC has frequent TP53 mutation (66.2%), whereas a significantly lower TP53 mutation rate in AWMC is observed (39.8%, p < 0.001). Instead, KRAS is the most commonly mutated gene in AWMC (45.4%). The majority of KRAS mutations occur in codon 12 or 13, with G12D the most common, followed by the G12V, G13D, and A146T. These data differ slightly from that of western populations and oriental AC patients, suggesting that race and histological subtype might play a role in mutation patterns. Notably, AWMC shows distinct mutation rate of ERBB2 from AC (4.6 vs. 0.5%, p = 0.020), especially in left-sided colon (p = 0.018). The mutation spots in 4 AWMC cases were c.2033G>A (p.R678Q) (2 cases), c.2260G>A (p.V754M), and c.2524G>A (p.V842I), respectively. According to previous literature, the functional effect of R678Q is controversial; V754M has never been reported; V842M has been proved to be a functioning mutation in other carcinomas (30, 31). Within AWMC, tumors with signet ring cell component has a significantly lower KRAS mutation rate than tumors without signet ring cell component despite the site of origin, which is consistent with our expectations and previous reports (32). In summary, the mutation status of genes of major concern (APC, BRAF, KRAS, and TP53) do not seem to be influenced by primary sites, and AWMC shows distinct mutation spectrum from AC, mainly attributed to their histology. ERBB2 and FBXW7 are two genes with different mutation pattern in different sites, however, more investigation is needed considering the small number of cases in our cohort.
We also notice that despite the low mutation rates of HRAS (0.3%) and NRAS (4.8%) in CRC overall, AWMC with signet ring cell component has more frequent HRAS and NRAS mutations than AWMC without signet ring cell component. The trend is likely masked by the small sample size, but it indicates that the existence of signet ring cell component is the marker of specific molecular changes.
Mutation rates of BRAF are reported to be higher in signet ring cell carcinoma than in AC (32, 33). Interestingly, we detect only one BRAF mutation in 18 cases of AMWC with signet ring cell component, likely due to the ethnics and relative small sample size.
RAS and BRAF mutations are traditionally thought to be nearly mutually exclusive (34). Concomitant KRAS and BRAF mutant CRCs are extremely rare (0.001%) are often associated with more advanced stage, therefore it is recommended that KRAS-mutated patients not be tested for BRAF mutation (35). Nonetheless, recent cases indicate that the occurrence of concomitant KRAS and BRAF mutations in surgical cohorts maybe higher than hypothesized, and present variable survival outcomes (35). It still needs studying which detailed gene profiling pattern of concomitant mutated tumors is, and which tumor gene signature it imitates more. In our study, the case where both KRAS and BRAF mutations are identified is a 75-year-old female with a 3 cm tumor in rectum. Both mutations are in the less frequent sites (A146T in KRAS and G466E in BRAF) and this combination is completely new. Her tumor staging is T1N0M0. However, she was found to have vagina metastases 1 year after the rectal surgery (Miles) and was given radiotherapy both prior to and after a second resection in vagina. It is notable since most previous studies regarding KRAS/BRAF concomitant mutations involve KRAS codon 12 or codon 13 and BRAF V600E, and there is a heterogeneity in the biological features of these mutations (34–38). Some cases show that concomitant KRAS/BRAF mutant patients had poorer prognosis (36). Thus, such cases should be kept in mind to clarify the type of concomitant mutations and elucidate their biological behavior.
KRAS and BRAF mutated CRCs are associated with distinct clinicopathological features according to a large cohort of CRC in Western entity, where RAS mutation is associated with male gender and classical adenocarcinoma subtype (39). In Chinese group, previous studies mention that KRAS-mutated or BRAF-mutated CRCs tend to occur in the proximal colon but have no specific trend in sex, age, lymph node metastasis, or TNM stages (29). Our study focuses on AWMC and AWMC without signet ring cell component and found no distinct features between KRAS-mutant and wild-type patients. The prognostic value of KRAS and BRAF in AWMC requires more research. Due to the small number of cases, we do not manage to compare KRAS- or BRAF- mutated cases with wild-type cases in AWMC with signet ring cell component.
A set of other somatic mutations is summarized in previous reports but never completely evaluated in Chinese group (27, 28). Since the response of wild-type CRC to anti-EGFR therapy is not as effective as expected, it is important to identify other potential markers to evaluate the outcome. Our research includes mutational profiling of those hotspot mutations as listed in Table 3 (partially). Among the 16 PIK3CA mutant AWMC cases, 11 are accompanied with KRAS mutations, while there is no significant correlation between KRAS and PIK3CA mutation (p = 0.145), which is inconsistent with previous studies in CRCs and suggests the difference in molecular spectrum between AWMC and other types of CRC (29). Four cases of AWMC with signet ring cell component are found to harbor PIK3CA mutations, and one of them has concomitant KRAS mutation.
Mucinous histology is reported to be an independent adverse prognostic predictor in some studies, but not in others (11, 40). In our study, AWMC tends to have a favorable prognosis compared with AC, but not significantly. Univariate analysis of DSS indicates that an earlier stage and absence of lymph node metastasis present a better outcome. Multivariate analysis reveals that age and stage are independent prognostic factors. We infer that mucinous histology itself may not influence the survival outcome, but the close association between AWMC and earlier stages are having an impact on prognosis. There is no difference in DSS between AWMC with signet ring cell component and AWMC without signet ring cell component. However, considering the relatively small population, especially for AWMC with signet ring cell component, larger and multi-center studies might be a further direction.
Several limitations are of concern in our study. First, the data are gathered from a single center, and some is incomplete. Second, the majority of the patients coming to our hospital are in earlier stages, which might bring bias when analyzing the relationship between molecular profiling and biological behavior. Third, the population is relatively small with loss to follow up. Nevertheless, our research manages to focus on histology after taking primary sites into consideration and highlights the clinical significance of the unique entity of AWMC. We emphasize that this topic requires more exploration in fields of molecular biology, which will certainly provide clues for treatment. Our study also opens up avenues for larger, multi-center studies with more follow up.
Conclusion
Regardless of primary sites, AWMC demonstrates less metastasis, earlier stages, more frequent dMMR, and lower TP53 mutation rate compared with AC. Within AWMC, AWMC with signet ring cell component is associated with lower KRAS mutation rate. Mucinous histology does not have an obvious effect in DSS of CRC. Our results indicate the unique molecular pathogenesis underlying AWMC.
Data Availability Statement
The raw data of the article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The institutional review board of Peking Union Medical College Hospital approved the study. Written informed consent was obtained from all patients.
Author Contributions
JC, LZ, and HW collected the cases, analyzed the data, and wrote the manuscript. JG, TL, and JW performed the NGS sequencing and analyzed the results. HW and ZL contributed to the concept and design for the study. All authors contributed to critical interpretation of data and the final draft of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the CAMS Central Public Welfare Scientific Research Institute Basal Research Funds (2019XK320045) and CAMS Innovation Fund for Medical Sciences (2019-I2M-2-002).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00917/full#supplementary-material
References
- 1.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. (2019) 394:1467–80. 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- 2.Fakhri B, Lim KH. Molecular landscape and sub-classification of gastrointestinal cancers: a review of literature. J Gastrointest Oncol. (2017) 8:379–86. 10.21037/jgo.2016.11.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds IS, Furney SJ, Kay EW, McNamara DA, Prehn JHM, Burke JP. Meta-analysis of the molecular associations of mucinous colorectal cancer. Br J Surg. (2019) 106:682–91. 10.1002/bjs.11142 [DOI] [PubMed] [Google Scholar]
- 4.Nguyen LH, Goel A, Chung DC. Pathways of colorectal carcinogenesis. Gastroenterology. (2020) 158:291–302. 10.1053/j.gastro.2019.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hugen N, Simons M, Halilovic A, van der Post RS, Bogers AJ, Marijnissen-van Zanten MA, et al. The molecular background of mucinous carcinoma beyond MUC2. J Pathol Clin Res. (2015) 1:3–17. 10.1002/cjp2.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liddell C, Droy-Dupre L, Metairie S, Airaud F, Volteau C, Bezieau S, et al. Mapping clinicopathological entities within colorectal mucinous adenocarcinomas: a hierarchical clustering approach. Modern Pathol. (2017) 30:1177–89. 10.1038/modpathol.2017.18 [DOI] [PubMed] [Google Scholar]
- 7.Melis M, Hernandez J, Siegel EM, McLoughlin JM, Ly QP, Nair RM, et al. Gene expression profiling of colorectal mucinous adenocarcinomas. Dis Colon Rectum. (2010) 53:936–43. 10.1007/DCR.0b013e3181d320c4 [DOI] [PubMed] [Google Scholar]
- 8.Hyngstrom JR, Hu CY, Xing Y, You YN, Feig BW, Skibber JM, et al. Clinicopathology and outcomes for mucinous and signet ring colorectal adenocarcinoma: analysis from the National cancer data base. Ann Surg Oncol. (2012) 19:2814–21. 10.1245/s10434-012-2321-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan J, Burke JP, Samaha G, Condon E, Waldron D, Faul P, et al. Overall survival is improved in mucinous adenocarcinoma of the colon. Int J Colorectal Dis. (2014) 29:563–9. 10.1007/s00384-013-1826-2 [DOI] [PubMed] [Google Scholar]
- 10.Warschkow R, Tarantino I, Huttner FJ, Schmied BM, Guller U, Diener MK, et al. Predictive value of mucinous histology in colon cancer: a population-based, propensity score matched analysis. Br J Cancer. (2016) 114:1027–32. 10.1038/bjc.2016.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung CO, Seo JW, Kim KM, Do IG, Kim SW, Park CK. Clinical significance of signet-ring cells in colorectal mucinous adenocarcinoma. Mod Pathol. (2008) 21:1533–41. 10.1038/modpathol.2008.170 [DOI] [PubMed] [Google Scholar]
- 12.Song IH, Hong SM, Yu E, Yoon YS, Park IJ, Lim SB, et al. Signet ring cell component predicts aggressive behaviour in colorectal mucinous adenocarcinoma. Pathology. (2019) 51:384–91. 10.1016/j.pathol.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 13.Salem ME, Weinberg BA, Xiu J, El-Deiry WS, Hwang JJ, Gatalica Z, et al. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget. (2017) 8:86356–68. 10.18632/oncotarget.21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitsche U, Zimmermann A, Spath C, Muller T, Maak M, Schuster T, et al. Mucinous and signet-ring cell colorectal cancers differ from classical adenocarcinomas in tumor biology and prognosis. Ann Surg. (2013) 258:775–82. 10.1097/SLA.0b013e3182a69f7e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun SO, Cho YB, Lee WY, Kim HC, Yun SH, Park YA, et al. Clinical significance of signet-ring-cell colorectal cancer as a prognostic factor. Ann Coloproctol. (2017) 33:232–8. 10.3393/ac.2017.33.6.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo C, Cen S, Ding G, Wu W. Mucinous colorectal adenocarcinoma: clinical pathology and treatment options. Cancer Commun. (2019) 39:13. 10.1186/s40880-019-0361-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang SY, Cho MS, Kim NK. Difference between right-sided and left-sided colorectal cancers: from embryology to molecular subtype. Expert Rev Anticanc. (2018) 18:351–8. 10.1080/14737140.2018.1442217 [DOI] [PubMed] [Google Scholar]
- 18.Natsume S, Yamaguchi T, Takao M, Iijima T, Wakaume R, Takahashi K, et al. Clinicopathological and molecular differences between right-sided and left-sided colorectal cancer in Japanese patients. Jpn J Clin Oncol. (2018) 48:609–18. 10.1093/jjco/hyy069 [DOI] [PubMed] [Google Scholar]
- 19.Stintzing S, Tejpar S, Gibbs P, Thiebach L, Lenz HJ. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer. (2017) 84:69–80. 10.1016/j.ejca.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ, Chen G. Distinguishable prognostic signatures of left- and right-sided colon cancer: a study based on sequencing data. Cell Physiol Biochem. (2018) 48:475–90. 10.1159/000491778 [DOI] [PubMed] [Google Scholar]
- 21.McCawley N, Clancy C, O'Neill BDP, Deasy J, McNamara DA, Burke JP. Mucinous rectal adenocarcinoma is associated with a poor response to neoadjuvant chemoradiotherapy: a systematic review and meta-analysis. Dis Colon Rectum. (2016) 59:1200–8. 10.1097/DCR.0000000000000635 [DOI] [PubMed] [Google Scholar]
- 22.Imperial R, Ahmed Z, Toor OM, Erdogan C, Khaliq A, Case P, et al. Comparative proteogenomic analysis of right-sided colon cancer, left-sided colon cancer and rectal cancer reveals distinct mutational profiles. Mol Cancer. (2018) 17:177. 10.1186/s12943-018-0923-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao XH, Yu GY, Gong HF, Liu LJ, Xu Y, Hao Q, et al. Differences of protein expression profiles, KRAS and BRAF mutation, and prognosis in right-sided colon, left-sided colon and rectal cancer. Sci Rep. (2017) 7:7882. 10.1038/s41598-017-08413-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kermanshahi TR, Magge D, Choudry H, Ramalingam L, Zhu B, Pingpank J, et al. Mucinous and signet ring cell differentiation affect patterns of metastasis in colorectal carcinoma and influence survival. Int J Surg Pathol. (2017) 25:108–17. 10.1177/1066896916664990 [DOI] [PubMed] [Google Scholar]
- 25.Ott C, Gerken M, Hirsch D, Fest P, Fichtner-Feigl S, Munker S, et al. Advanced mucinous colorectal cancer: epidemiology, prognosis and efficacy of chemotherapeutic treatment. Digestion. (2018) 98:143–52. 10.1159/000487710 [DOI] [PubMed] [Google Scholar]
- 26.Gonsalves WI, Mahoney MR, Sargent DJ, Nelson GD, Alberts SR, Sinicrope FA, et al. Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/Alliance N0147. J Natl Cancer Inst. (2014) 106:dju106. 10.1093/jnci/dju106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Togashi Y, Yoshioka Y, Chikugo T, Terashima M, Mizukami T, Hayashi H, et al. Clinicopathological and genetic differences between low-grade and high-grade colorectal mucinous adenocarcinoma. Eur J Cancer. (2015) 51:S390. 10.1016/S0959-8049(16)31089-9 [DOI] [PubMed] [Google Scholar]
- 28.Al-Shamsi HO, Jones J, Fahmawi Y, Dahbour I, Tabash A, Abdel-Wahab R, et al. Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern. J Gastrointest Oncol. (2016) 7:882–902. 10.21037/jgo.2016.11.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu T, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep. (2015) 5:18678. 10.1038/srep18678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, et al. Actionable activating oncogenic ERBB2/HER2 transmembrane and juxtamembrane domain mutations. Cancer Cell. (2018) 34:792–806.e795. 10.1016/j.ccell.2018.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang L, Jiang W, Ye S, He T, Pei X, Li J, et al. ERBB2 mutation: a promising target in non-squamous cervical cancer. Gynecol Oncol. (2018) 148:311–6. 10.1016/j.ygyno.2017.12.023 [DOI] [PubMed] [Google Scholar]
- 32.Korphaisarn K, Morris V, Davis JS, Overman MJ, Fogelman DR, Kee BK, et al. Signet ring cell colorectal cancer: genomic insights into a rare subpopulation of colorectal adenocarcinoma. Br J Cancer. (2019) 121:505–10. 10.1038/s41416-019-0548-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yalcin S, Onguru O. BRAF mutation in colorectal carcinomas with signet ring cell component. Cancer Biol Med. (2017) 14:287–92. 10.20892/j.issn.2095-3941.2017.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin IH, Kazmi SM, Yorio JT, Bhadkamkar NA, Kee BK, Garrett CR. Rare though not mutually exclusive: a report of three cases of concomitant KRAS and BRAF mutation and a review of the literature. J Cancer. (2013) 4:320–2. 10.7150/jca.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshwar A, Margonis GA, Andreatos N, Barbon C, Wang J, Buettner S, et al. Double KRAS and BRAF mutations in surgically treated colorectal cancer liver metastases: an international, multi-institutional case series. Anticancer Res. (2018) 38:2891–5. 10.21873/anticanres.12535 [DOI] [PubMed] [Google Scholar]
- 36.Larki P, Gharib E, Yaghoob Taleghani M, Khorshidi F, Nazemalhosseini-Mojarad E, Asadzadeh Aghdaei H. Coexistence of KRAS and BRAF mutations in colorectal cancer: a case report supporting the concept of tumoral heterogeneity. Cell J. (2017) 19:113–7. 10.22074/cellj.2017.5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vittal A, Middinti A, Kasi Loknath Kumar A. Are all mutations the same? A rare case report of coexisting mutually exclusive KRAS and BRAF mutations in a patient with metastatic colon adenocarcinoma. Case Rep Oncol Med. (2017) 2017:2321052. 10.1155/2017/2321052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, et al. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. (2007) 26:158–63. 10.1038/sj.onc.1209758 [DOI] [PubMed] [Google Scholar]
- 39.Rimbert J, Tachon G, Junca A, Villalva C, Karayan-Tapon L, Tougeron D. Association between clinicopathological characteristics and RAS mutation in colorectal cancer. Mod Pathol. (2018) 31:517–26. 10.1038/modpathol.2017.119 [DOI] [PubMed] [Google Scholar]
- 40.Kang H, O'Connell JB, Maggard MA, Sack J, Ko CY. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. (2005) 48:1161–8. 10.1007/s10350-004-0932-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of the article will be made available by the authors, without undue reservation, to any qualified researcher.