Graphical abstract
Keywords: Salt intake, Food intake, FGF21, United Arab Emirates, Food Frequency Questionnaire (FFQ), Multiple correspondence analysis
Abstract
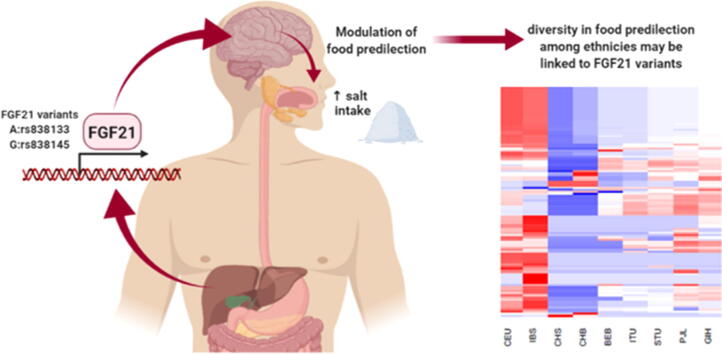
Food predilection is linked to variants in the hepatokine “Fibroblast Growth Factor-21” gene (FGF21); with rs838133 linked to the sweet tooth in Caucasians. The effect of FGF21 variants on food intake is still unclear in other populations. A cohort of 196 healthy Emirati subjects was investigated [age: 30.34 ± 9.75yrs (44.4% males)]. The FGF21 rs838133 and rs838145 were genotyped. The daily intake was calculated based on a 61-item food frequency questionnaire. Multivariate analysis was performed using in house R script that implements two-way unsupervised hierarchical clustering to detect the association of the studied single-nucleotide polymorphisms (SNPs) and related SNPs in linkage disequilibrium, using data from the 1000 genome project. Both SNPs were in Hardy-Weinberg Equilaribium (HWE). BMI positively correlated with age (p = 0.002), but not with caloric intake. Salt intake was significantly higher in subjects homozygous (A: rs838133) and (G:rs838145),(p = 0.03 and 0.01, respectively). An interaction was observed between both SNPs; significantly associated with high salt intake. Using publicly available data, both SNPs fall within a region transmitted in Iberians which has a profile closely similar to Caucasians, but far from Chinese population. In conclusion, the minor alleles of FGF21 rs838145 and rs838133 are associated with high salt intake in Emiratis and may suggest neuro-metabolic link to dietary preference across different populations.
Introduction
Liver hormones play an essential role in the gut-brain link. A good example is the fibroblast growth factor-21 (FGF21); a hepatokine with a myriad of functions including central mechanisms of appetite control [1] as well as insulin-sensitizing properties [2]. Appetite is controlled by neural and hormonal mechanisms. The neural mechanism involves the hypothalamic arcuate nucleus, the lateral hypothalamus, and calcitonin gene-related peptide neurons located in the parabrachial nucleus [3]. Gut hormones control appetite by many modulators; including ghrelin, a unique orexigenic hormone, and others [4]. The β‐klotho receptors of FGF21 are present in the paraventricular nucleus of the hypothalamus and may mediate the effect of FGF21 on decreasing the appetite predilection to sweet food [5]. FGF21 may be linked to the preference of specific food item intake, and possibly, to dependence; it was recently shown to reduce the preference for morphine in the mouse conditioned place preference assay [6].
An interesting example of gene-environment interaction is the effect of gene polymorphisms on preference of certain types of food. The “A” allele of FGF21 at rs838133 is associated with higher carbohydrate intake, despite insignificant change of total calorie intake [7], [8], with predeliction to sugary food [9]. Recently, Frayling et al. showed that the rs838133 A allele is associated with hypertension and a high waist-hip ratio, but lower total body-fat percentage [10]. The studies investigated such links in Caucasians, but no studies have investigated other ethnicities yet.
The therapeutic utility of FGF21 is being actively investigated in clinical trials for the treatment of type 2 diabetes and non-alcoholic steatohepatitis (NASH), [11]. Several studies are currently testing FGF21-class molecules, including modified proteins, through phase 1 or phase 2 clinical studies [12]. Information derived from human genetic investigations may be crucial to inform further drug development [12].
The FGF family of proteins display a wide range of mitogenic and cell survival activities and are involved in a variety of biological processes. The FGF21 gene is located on chromosome 19q13.33 [13]. Diseases associated with FGF21 include acquired lipodystrophy and overnutrition. Among a myriad of pathways are Ribosomal protein S6 kinase beta-1 (p70S6K) signaling and activation of cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (PKA). According to “Gene Ontology” annotations, an important paralog of this gene is FGF19. FGF21 facilitates glucose uptake in monophthalate-treated adipocytes. It also stimulates glucose uptake in differentiated adipocytes via inducing the expression of glucose transporter 1 (GLUT1; also known as solute carrier family 2 facilitated glucose transporter member 1, SLC2A1) in presence of β‐klotho receptors [14], [15].
The Glutamate Ionotropic Receptor NMDA Type Subunit 2D (GRIN2D) gene is located close to the FGF21 gene (19q13.33), [13]. The gene encodes a subunit of the N-methyl-D-aspartate (NMDA) receptor [16]. The NMDA receptors mediate afferent taste signals to cortical taste neurons in rats. In addition, they are involved in auditory, visual, and somatosensory transmission, as well as memory encoding [17], [18].
The UAE is one of the countries of high obesity prevalence [19], with remarkable changes in lifestyle and eating habits during the last few decades [20]. The current study investigates the potential effect of two single-nucleotide polymorphisms (SNPs) of the FGF21 gene; rs838133 and rs838145, on the food intake pattern in an Emirati cohort of healthy adults. We hypothesize that those SNPs are affecting food predilection.
Subjects and methods
Subjects
This study is a cross-sectional study of two FGF21 SNPs is conducted across a total of 196 subjects from the Emirati population. The formula S = [(1.96)2p (1 − p)]/d2 was used to calculate the sample size; where ‘p’ is the expected prevalence in the population-based on previous/pilot studies and ‘d’ is the absolute error or precision (i.e. d = calculated prevalence-true prevalence). This formula considers a type I error of 5% where p < 0.05 is considered as statistically significant.
The study included adult healthy Emirati subjects, who can consent for participation and complete the questionnaire. We excluded subject with a body mass index (BMI) of < 16 or above 40 kg/m2, chronic diseases (e.g. hypertension and diabetes mellitus), or following strict dietary changes.
The study protocol was approved by the Research and Ethics Committee at the University of Sharjah (REC-15-11-P001) and all participants consented to the study’s protocol. 213 healthy adult Emiratis, aging 18–73 years old, were recruited from the University of Sharjah as well as primary health care centers. Amongst these 213 subjects, 196 were selected after excluding subjects with missing data or who provide extreme values for food intake. Two milliliters of saliva (phlegm-free) were collected from the subjects after abstaining from eating for 30 min. The samples were preserved for a maximum of 7 days at −20 °C for DNA extraction, performed using QIAamp extraction kit (cat# 51306).
Anthropometry
Using tandardized techniques and calibrated equipments were used to the anthropometric parameters were measured for all participants: (1) weight (to the nearest 0.1 kg in light indoor clothing) and (2) height (to the nearest 0.5 cm using a stadiometer)). The BMI, calculated using the equation BMI = weight/height2 (kg/m2), was categorized as per the classification of the WHO[21]
Dietary survey
A sixty-one-item FFQ was used to assess dietary intake over the past year [22]. The subjects were asked to report the frequency of consumption of a list of commonly consumed food items and beverages in UAE. A standard portion and a reference portion was identified for each food item, using a visual aid as previously described [23]. The reported frequencies were converted to daily portion intake from which consumption was calculated by NUTRITIONIST PROTM diet analysis software (Axxya Systems LLC., USA, version 5.1.0, 2014, First Fata Bank, Nutritionist Pro, San Bruno, CA).
Genotyping
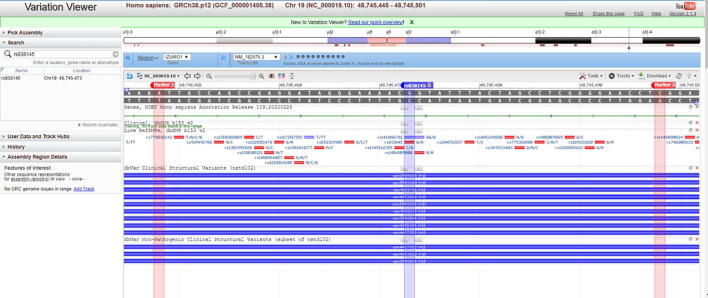
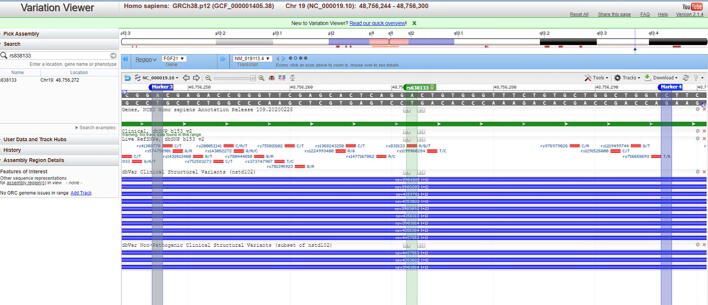
Genotyping for chromosome 19 (19q13.33) FGF21 rs838145 (A > G) and rs838133 (A > G) was performed as described in our previous study [23]. The genotyping was conducted using the TaqMan® DMGAssay (Applied Biosystems, USA) and Real-Time PCR StepOne Systems (Thermo Fischer Scientific, USA). Box 1 shows the context sequence for both SNPs. Each sequence is aligned to the FGF21 gene and the position of each SNP is visualized through the “Variation Viewer” from UCSC (Supplementary Fig. S1 -A&B). The location of each SNP is mapped on chromosome 19 (S1-C). The wild type allele binds to VIC, whereas mutant allele binds to FAM. The genotyping data were used to calculate the Chi-square p-value for the HWE and the distribution of haplotypes and alleles of interest [24].
Box 1. Context Sequence of FGF21 rs838145 (A>G) and rs838133 (A>G).
| NCBI SNP reference. | Context sequence. |
| rs838145. | ATTGCCAGCCGAGGATAGGGAAAAC[A/G]GTATTTACTAGCCTCGGGGAACCTC. |
| rs838133. | ACGAGACCGGGTTCGAGCACTCAGG[A/G]CTGTGGGTTTCTGTGCTGGCTGGTC. |
Statistical analysis
Frequencies (number of cases) and percentages were used as appropriate. We described data in terms of mean ± standard deviation (SD) when normally distributed. Categorical data were compared using Chi-square (X2). A Shapiro-Wilk’s test (p < 0.05), and a visual inspection of histograms, Q-Q plots showed that the body mass index (BMI) was non-parametric for both males and females. There was a skewness of 0.68 (SE = 0.23) and a kurtosis of 0.314 (SE = 0.46) for females and a skewness of 1.02 (SE = 0.26) and kurtosis of 1.89 (SE = 0.51) for males [25], with z- value for all > 1.96, mandating the use of non-parametric statistical tests [26].
In addition, we performed the same tests for Caloric intake, total protein, carbohydrate and fat intake as dependent variables, genotypes and age groups as independent variables. All data were not normally distributed. We expressed our data by “median” and “mean rank”, using non-parametric tests to determine significance; Mann Witney U test when comparing 2 groups and Kruskal Wallis if>2 groups are compared, with significance set at a p-value of ≤ 0.05. We calculated the effect size by deriving “r” from the equation r = z/ √N, where N is the total sample size; r = 0.1 is small, r = 0.3 is medium and r = 0.5 is large effect [27].
All statistical calculations were done using computer program SPSS (Statistical Package for the Social Science; SPSS Inc., Chicago, IL, USA] version 26 for Microsoft Windows.
Multivariate analysis was carried out using in house R script that implements two-way unsupervised hierarchical clustering [28] used to examine the association of the two FGF21 SNPs; rs838145 and rs838133 with other SNPs reported to be in linkage disequilibrium in different subpopulations. The results were visualized using heatmaps.
Results
Subject characteristics
A sample of 196 Emiratis was recruited (mean age 30.34 ± 9.75, Median 29.00, range 18–73 years, 44.4% males). The mean BMI of the population was 26.95 ± 6.42 kg/m2, median 26.32 indicating overweight. Male participants had a non-significant higher BMI than females (mean 27.58 and 26.44 kg/m2, median respectively, p = 0.28). Table 1 shows further baseline characteristics of the study group. The BMI positively correlated with age (p = 0.002, Pearson correlation = 0.22, but R2 linear = 0.04), but not with total caloric intake. The BMI did not differ according to the genotype of both SNPs.
Table 1.
Participant characteristics.
| [Median/number(%)] | (Mean ± SD) | ||
|---|---|---|---|
| Age (years) | 29 | 30.3 ± 9.8 | |
| Gender | Male Female |
87 (44%) 109 (55.6%) |
|
| Total caloric intake (Kcal) | 3228.9 | 3483.3 ± 1186.0 | |
| Total carbohydrate intake (g/d) | 390.8 | 405.7 ± 149.7 | |
| Total protein intake (g/d) | 144.5 | 155.2 ± 63.1 | |
| Total fat intake (g/d) | 118.7 | 131.2 ± 62.4 | |
| Total mono-unsaturated fat intake (g/d) | 45.7 | 52.7 ± 26.9 | |
| Total sodium intake (g/d) | 3561.0 | 3450.1 ± 1344.1 | |
| BMI (Kg/m2) | 26.3 | 27 ± 6.4 | |
| FGF21 rs838145 | A/G | 79 (40.3%) | X2 = 0.047 p-value = 0.83 MAF = 0.27 |
| G/G | 14 (7.1%) | ||
| A/A | 103 (52.6%) | ||
| FGF21 rs838133 | A/G | 72 (36.7%) | X2 = 2.7 p-value = 0.09 MAF = 0.3 |
| G/G | 102 (52.0%) | ||
| A/A | 22 (11.2%) | ||
196 subjects were genotyped for both FGF21 SNPs, BMI = body mass index, MAF = minor allele frequency, X2 = Chi-square.
Both SNPs were in Hardy-Weinberg equilibrium (Table 1). Twelve subjects carried homozygous the minor variant of both SNPs. Vitamin D intake is far below recommended.
Effect of genotype on food intake:
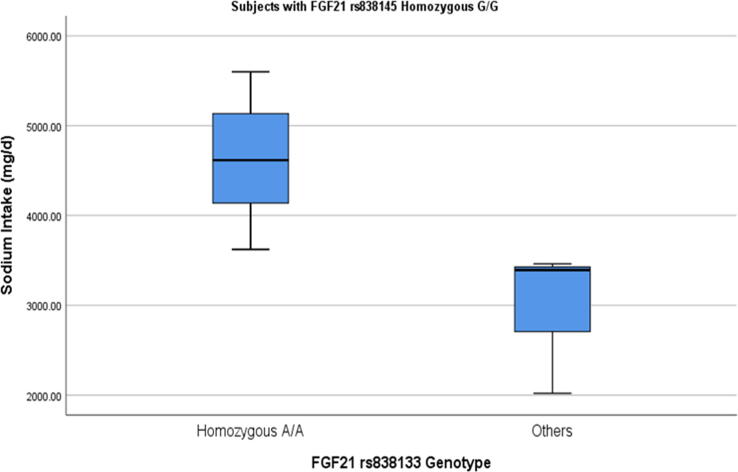
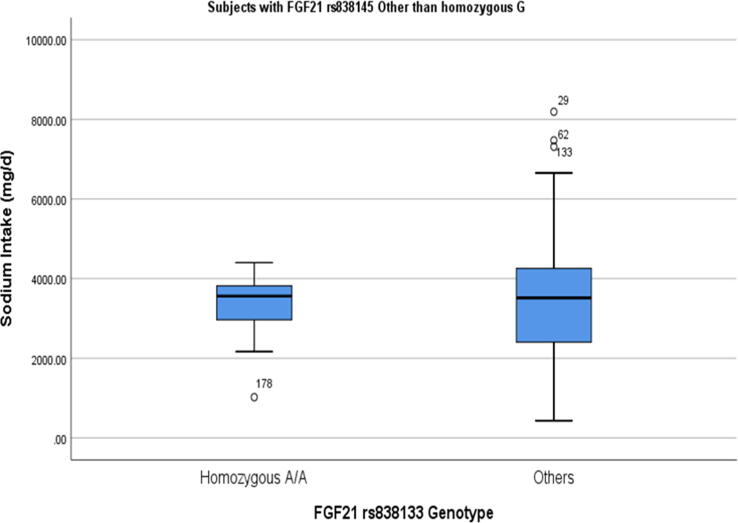
Sodium intake was significantly higher in subjects homozygous for minor alleles of both SNPs (A: rs838133) and (G:rs838145), (Table 2). Males homozygous for the minor G allele of rs838145 showed a significantly higher intake of total sodium (p = 0.021), whereas females with the same genotype had a higher intake of vitamin D (p = 0.015), (Table 3). Compared to other genotypes, females homozygous for minor A allele of rs838133 had a significantly higher intake of vitamin D. This effect was not observed in males. Sodium intake positively correlated with the intake of vegetables and fruits in rs838133 genotypes other than the homozygous (G:rs838145), (p < 0.001, Pearson's = 0.550**). Similarly, sodium intake positively correlated with the intake of vegetables and fruits in rs838145 genotypes other than the homozygous (A:rs838133), (p < 0.001, Pearson's = 0.682**). We explored the potential effect of co-occurrence of homozygous for (A: rs838133) and (G:rs838145). In subjects homozygous for (A: rs838133), the presence of homozygous minor allele (G:rs838145), is significantly associated with high salt intake, but not in other genotypes of rs838133 (Fig. 2).
Table 2.
The effect of FGF21 rs838145 and rs838133 genotypes on nutrient intake.
| Food category Intake (Median, Mean Rank) | FGF21 rs838145 |
U | p-value (two-tailed) | Z | Effect size (r) | FGF21 rs838133 |
U | p-value (two-tailed) | z | Effect size (r) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G/G (n = 14) | Others (n = 182) | A/A (n = 22) | Others (n = 174) | |||||||||
| Caloric intake (Kcal) | 3411.8, 50.7 | 3219.2, 46.4 | 501.5 | 0.58 | −0.55 | 0 | 3584.3, 112.4 | 3200.4, 96.7 | 1608 | 0.22 | −1.22 | −0.1 |
| Carbohydrate (g/d) | 444.6, 52.8 | 389.2, 46 | 472 | 0.38 | −0.87 | −0.1 | 466.1, 118.8 | 381.6, 95.9 | 1467 | 0.08 | −1.78 | −0.1 |
| Protein (g/d) | 149.5, 47.9 | 144.3, 46.9 | 541 | 0.89 | −0.13 | 0 | 157.1, 105.8 | 143.5, 97.6 | 1753 | 0.52 | −0.64 | 0 |
| Fat (g/d) | 111.6, 51.4 | 121.1, 46.2 | 491 | 0.51 | −0.67 | 0 | 111.2, 95.9 | 122, 98.8 | 1856 | 0.82 | −0.23 | 0 |
| Mono-unsaturated fat (g/d) | 44.3, 52 | 46.2, 46.1 | 483 | 0.45 | −0.75 | −0.1 | 39.4, 91.6 | 47.7, 99.4 | 1761 | 0.54 | −0.61 | 0 |
| Vitamin D (IU/d) | 171.2, 57.7 | 146.4, 45.1 | 403 | 0.11 | −1.61 | −0.1 | 160, 111.5 | 147.6, 96.9 | 1629 | 0.26 | −1.14 | −0.1 |
| Calcium (mg/d) | 1,844, 56.4 | 1,560, 45.3 | 421 | 0.16 | −1.42 | −0.1 | 1,844, 112.7 | 1560, 96.7 | 1602 | 0.21 | −1.25 | −0.1 |
| Potassium(mg/d) | 1,085, 42.5 | 1,216, 47.8 | 490 | 0.49 | −0.68 | 0 | 1,224, 99.6 | 12128.5, 98.4 | 1891 | 0.93 | −0.09 | 0 |
| Sodium (mg/d) | 4,371, 63.6 | 3,529, 44.1 | 320 | 0.01* | −2.50 | −0.2 | 3,895, 123.2 | 3,475, 95.4 | 1371 | 0.03* | −2.17 | −0.2 |
| Added sugar | 521.5, 116.9 | 349, 97.1 | 1016.5 | 0.21 | −1.26 | −0.1 | 486.8, 114.1 | 347.7, 96.5 | 1572 | 0.17 | −1.36 | −0.1 |
| Total Fruits and vegetables (g/d) | 722.5, 125.5 | 451.6, 96.4 | 896 | 0.03* | −2.16 | −0.2 | 642.6, 125 | 444.4, 95.2 | 1340 | 0.02* | −2.29 | −0.2 |
| High Fiber food (g/d) | 1003.9, 142.9 | 669.2, 95.1 | 653 | 0.002* | −3.04 | 0.2 | 820.3, 117.1 | 679, 96.2 | 1506 | 0.10 | −1.63 | −0.1 |
Mann Witney U test was used to compare subjects homozygous for minor allele versus other genotypes. The effect size “r” was derived from the equation r = z/ √N, where N is the total sample size (=196); r = 0.1 is small, r = 0.3 is medium and r = 0.5 is large effect. Vitamin D was calculated in mcg (as per the food frequency questionnaire) then in IU (1 IU = 0.025 mcg). *P < 0.05.
Table 3.
Gender differences in nutrient intake.
| Food category Intake (Median, Mean Rank) | Female (Median, Mean Rank, n = 109) | Male (Median, Mean Rank, n = 87) | U | p-value (two-tailed) | z | Effect size (r) |
|---|---|---|---|---|---|---|
| Caloric intake (Kcal) | 3063.7, 88 | 3418.3, 111.6 | 3598 | 0.004* | −2.89 | −0.2 |
| Carbohydrate (g/d) | 364.9, 90.2 | 406.1, 108.9 | 3841 | 0.022* | −2.28 | −0.2 |
| Protein (g/d) | 129.2, 85.3 | 156.7, 115.1 | 3302 | <0.001* | −3.65 | −0.3 |
| Fat (g/d) | 107.7, 91.3 | 134, 107.5 | 3960 | 0.048* | −1.98 | −0.1 |
| Mono-unsaturated fat intake (g/d) | 41.3, 91.4 | 53.5, 107.4 | 3970 | 0.051 | −1.96 | −0.1 |
| Vitamin D (IU/d) | 141.2, 93.4 | 162.4, 104.9 | 4181 | 0.155 | −1.42 | −0.1 |
| Calcium (mg/d) | 1529, 92.5 | 1785, 106 | 4087 | 0.097 | −1.66 | −0.1 |
| Potassium(mg/d) | 1183, 92.3 | 1,255, 106.3 | 4067 | 0.087 | −1.71 | −0.1 |
| Sodium (mg/d) | 3470, 94.2 | 3,652, 103.9 | 4268 | 0.230 | −1.20 | −0.1 |
| Added sugar(g/d) | 347.4, 96.8 | 366, 100.7 | 4551 | 0.629 | −0.48 | 0.0 |
| Total Fruits and vegetables(g/d) | 401, 91 | 502, 108 | 3918.5 | 0.015* | −2.44 | −0.2 |
| High Fiber food(g/d) | 648.9, 90.8 | 757.9, 108.1 | 39.2 | 0.033* | −2.13 | −0.2 |
Mann Witney U test was used to compare male and female subjects. The effect size “r” was derived from the equation r = z/ √N, where N is the total sample size (=196); r = 0.1 is small, r = 0.3 is medium and r = 0.5 is large effect. Vitamin D was calculated in mcg (as per the food frequency questionnaire) then in IU (1 IU = 0.025 mcg). *P < 0.05.
Fig. 2.
Interaction of FGF21 rs838133 and rs838145 on sodium intake. In the group of subjects homozygous for A allele (risk allele) in rs838133, there is an additive effect of rs838145 risk allele G (A). This was not noticed in other FGF21 rs838133 genotypes (B). The two groups were compared using Mann Witney U non-parametric test. There were 12 subjects homozygous for both minor alleles.
Vitamin C intake was significantly higher in the homozygous minor allele of both SNPs. Vitamin A intake was significantly higher in subjects with AA genotype of rs838133. Compared to other genotypes, females homozygous for minor A allele of rs838133 had a significantly higher intake of carbohydrates, vitamin C, vitamin D, vitamin B1, B2, and B6. The age group I with the homozygous minor variant of either SNP had significantly higher vitamin B1, B2, B6, and selenium intake compared to genotypes. In addition, homozygous A: rs838133 had also significantly higher vitamin C and vitamin D intake. Zinc intake significantly decreased with age (p = 0.01, Pearson’s = −0.237**, R2 linear = 0.056). Further results are presented in Supplementary material (Table S1–S4).
Effect of gender and age on food intake
The caloric and macronutrient intake was significantly higher in males compared with females (Table 3). The caloric and carbohydrate intake negatively correlated with age, despite the increase in BMI (total caloric intake; p = 0.01, Pearson's = -0.184* and carbohydrate; p = 0.006, Pearson's = -0.194**). However, the correlation was week R2 linear = 0.034 and 0.0.38 respectively. Sodium intake did not differ with age. The recruited subjects were subdivided into 3 age groups; group-I aged 18–24, group-II 25–34, group-III ≥ 35 years. Total sugar intake was significantly lower in the age group-III compared to the other 2 groups (p = 0.005 by non-parametric Kruskal Wallis test). The effect was more prominent in females (p = 0.046) than in males (p = 0.08). Further results are available in Supplementary material (Table S5).
Carbohydrate intake significantly correlated with sweet intake in males and females (Pearson correlation 0.315, 0.361, p = 0.013& 0.001, respectively) at all age groups. However, the correlation is weak (R2 is below 0.2 in all). Group I with the homozygous minor variant of either SNP had significantly higher total caloric and carbohydrate intake. In addition, homozygous A: rs838133 had also significantly higher vitamin D intake. The intake of some micronutrients (iron, zinc, and selenium) was significantly higher in males.
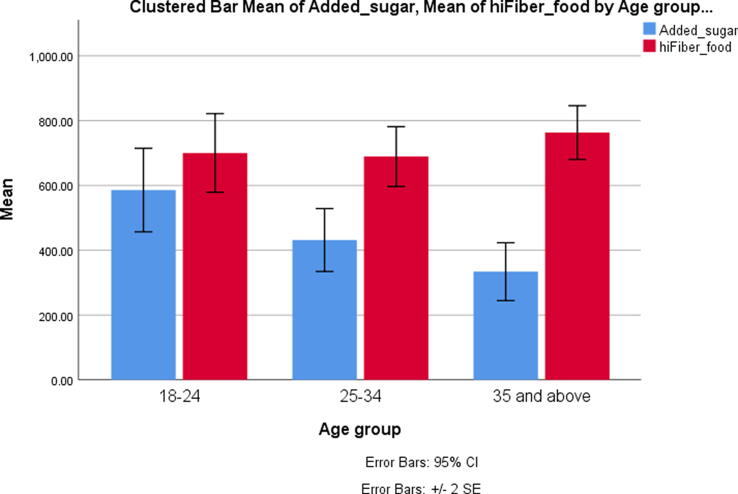
Added Sugar, high-fiber diet, vegetables, and fruits
Intake of added sugars negatively correlated with age (p = 0.004, Pearson = -0.20, but R2 = 0.04), with a significant difference among age groups (Fig. 1). There was no difference in the added sugar intake according to gender or genotypes of both SNPs. The high-fiber diet was higher in males (p = 0.33). It was also higher in subjects homozygous for G: rs838145 compared to others (p = 0.002), but not in homozygous A: rs838133. Fiber intake showed a trend to increase with age (Fig. 1, Fig. 2). Subjects with homozygous A: rs838133 had a significantly higher intake of fruits and vegetables compared with other genotypes (p = 0.022). Similarly, subjects homozygous for the minor G allele of rs838145 had higher fruit and vegetable intake (p = 0.009).
Fig. 1.
Daily intake of added sugar and high-fiber diet according to the age group. The recruited subjects (n = 196) were subdivided into 3 age groups; group-I aged 18–24 (n = 71), group-II 25–34 (n = 57), group-III ≥ 35 years (n = 68). There was a signficiant difference in added sugar among the three age groups (p = 0.003) by Kruskal Wallis test. High fiber intake showed non-significant increase with age (p = 0.15).
Correlation of sodium intake with different food items
The salt-rich food items were further analyzed for a significant correlation to total sodium intake according to age and sex. The participants aged between 18 and 24 had a significantly strong correlation between sodium and salad seasoning intake (p = 0.001, r = 0.828). While it was significant but weakly correlated with cheese, falafel, pizza, roasted and salted nuts, and white bread. Similarly, frequent consumption of salad dressing was highly correlated with the high sodium intake of participants aged 24–35 (p = 0.001, r = 865) and participants whose age > 35 years old (p = 0.001, r = 0.818) and weekly correlated with cheese, falafel, pizza, labneh luncheon meat, and cheese pie. Sodium intake was highly significant and strongly correlated with salad seasoning among females and males (p = 0.001, r = 0.855; p = 0.001, r = 0.786). Moreover, females' consumption of falafel, shawarma, pizza, roasted and salted nuts, labneh, bread, fried potatoes was significant but weakly correlated with total sodium intake. Among males, cheese, falafel, pizza, luncheon meat, bread and manakeesh (thyme and cheese) consumption was significant but weakly correlated with total sodium intake.
Analysis of linkage disequilibrium in subpopulations:
Analysis of linkage disequilibrium was carried out using publicly available data through the 1000 genome project (HG19 build). All SNPs with r2 ≥ 0.05 were extracted, using a phyton script, for each of the two FGF21 SNPs; rs838145 and rs838133; r2 is the correlation between a pair of loci. An r2 value of 0 indicates that the two loci are in complete linkage equilibrium and a value of 1 denotes the two loci are in complete linkage disequilibrium and coinherited The prevalence of the two FGF21 SNPs was compiled (Supplementary Table S6, S7). Both fall within a region transmitted in Iberians (n = 107) which represents a Caucasian subpopulation, but very far from Chinese as shown in Fig. 3. Interestingly, we included five South Asian subpopulations as they are geographically close to the Emirati population [41]. https://pubmed.ncbi.nlm.nih.gov/31604968/?from_term=al+safar+h&from_sort=date&from_pos=2. The Utah residents of European ancestry were added for comparison. The coverage/depth was chosen to be by around x40 for rs838133 and x30 for rs838145 using the Genome Aggregation Database (gnomAD) [http://gnomad.broadinstitute.org/].
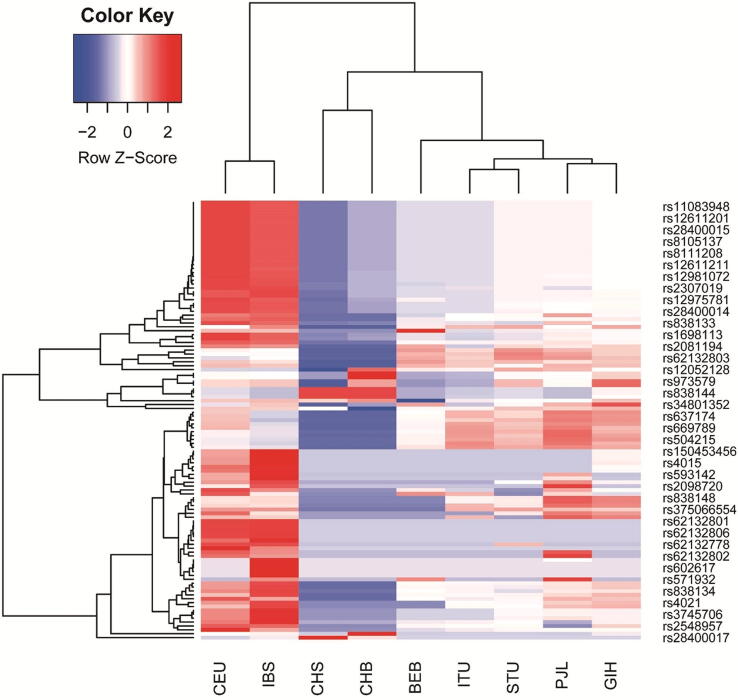
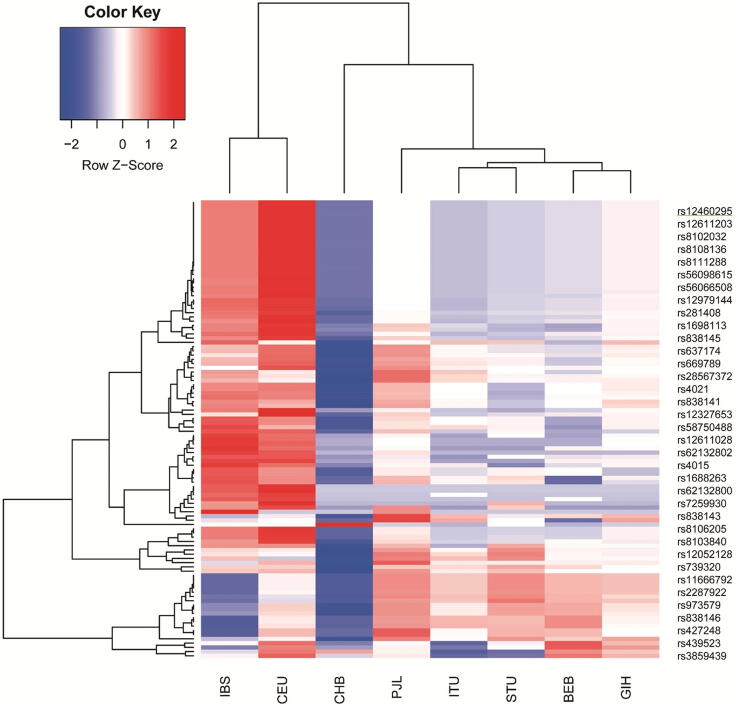
Fig. 3.
Unsupervised hierarchical clustering and heatmaps of the identified SNPs in linkage disequilibrium with FGF21 rs838133 and rs838145 (columns) across different subpopulations (columns). Cells are color-coded according to the log10 of p-value, which tests the enrichment/depletion of an effect allele in a population, compared with the average of all. The red color represents allele enrichment, and the blue represents allele depletion. Cluster dendrograms are added as supplementary figures. (A) Heatmap of FGF21 rs838133 and SNPs in linkage disequilibrium (B) Heatmap of FGF21 rs838145 and SNPs in linkage disequilibrium. CEU: Utah Residents (CEPH) with Northern and Western European Ancestry (n = 99) CHS: Southern Han Chinese (n = 105) CHB: Han Chinese in Bijing, China (n = 103), BEB: Bengali from Bangladesh (n = 86), IBS: Iberian Population in Spain (n = 107), ITU: Indian Telugu from the UK (n = 102), STU: Sri Lankan Tamil from the UK (n = 102), PJL: Punjabi from Lahore, Pakistan (n = 96), GIH: Gujarati Indian from Houston, Texas (n = 103). No data were available on CHS data for rs838145 in the 1000Genome database.
Thirty-six loci were in LD with rs838145, and 29 with rs838133. As expected, apart from a few SNPs (7/36 in LD with rs838145), Iberians (IBS) and Utah residents (CEU) subpopulations showed similar allele enrichment/ depletion patterns. Similarly, the south Asian subpopulations show similar patterns (within the same continental group). However, Han Chinese (CHB and CHS) showed a distinguished allele enrichment/depletion patterns closer to the Asians. In addition, Punjabi in Lahore, Pakistan (PJL) showed a pattern closer to the CEU that to south Asians. All alleles in the 1000 Genomes Project were reported on the forward strand Fig. 3, Supplementary Fig. S2, Supplementary Fig. S2). Analysing the allele frequency for both alleles show relative closeness between different populations for the two alleles; rs838145 and rs838133 showed allele frequency of around 0.4 and 0.5 for IBS, CEU and other European populations, respectively.
Discussion
The current study explored the effect of FGF21 gene variants on food predilection in the Emirati population. The most striking finding in our study is the significant increase in sodium intake in subjects homozygous for minor alleles of both FGF21 SNPs (regardless of age and gender), whereas those genotypes were previously linked to the sweet tooth in Caucasians [9]. We showed an interaction between the two SNPs with significantly increased salt intake in subjects homozygous for the minor allele of both. Interestingly, a recent GWAS showed the rs838133 as the strongest locus for urinary sodium [29]. We assume that this SNP is linked to an adaptive mechanism regulating the sodium balance. High salt intake, in addition to its effect on blood pressure, is also an independent risk factor for other cardiovascular and kidney diseases [29]. A marked association also exists between high salt intake, obesity and asthma, osteoporosis, and stomach cancer. At the cellular and molecular levels, high sodium intake leads to hypertrophy of vascular smooth muscles, increases oxidative stress, and reduces nitric oxide, decreases aortic hyaluronan content and large artery compliance [30]. High salt intake also contributes to resistance to antihypertensive therapy. Interestingly, adult sodium intake exceeds physiological needs worldwide, particularly in Asian countries [31].
In our study, The BMI did not differ according to the genotype of both SNPs. This is in contrast to Frayling’s study that showed a correlation between A allele of FGF 21 rs838133 and total body fat [10]. Noteworthy, added sugar was not increased in the minor alleles of both SNPs in contrast to a previous study in Caucasians [9], denoting a significant ethnic variability in food predilection as a result of gene-environmentt interaction. Importantly, vitamin D intake is far lower than the recommended 400–800 IU/d, with even lower intake in females than males.
Recent studies have shown that FGF21 affects the balance of consumed macronutrients and results in variations in nutritional status [32]. In the current study, there was no difference in macronutrient intake among the two FGF21 SNP’s. However, it was noted that participants consumed high fiber intake as evident by increase intake of fruits and vegetables. A high-fiber diet is one factor that has received considerable attention in obesity prevention, with some studies reporting that increasing dietary fiber can reduce weight gain and obesity [33]. In our study, males consumed more high-fiber diet as compared to females among subjects homozygous for G: rs838145 compared to others. It was noted that subjects with homozygous A: rs838133 and homozygous for the minor G allele of rs838145 had a significantly higher intake of fruits and vegetables compared with other genotypes. So, targeting diet composition is one potential approach for decreasing bodyweight since certain foods, such as fiber‐rich vegetables, promote health by increasing satiety and replacing energy‐dense food options.
Considering the possible link to NMDA receptor encoding gene GRIN2D [13]; the FGF21 gene may be implicated in neurally-mediated predilection to certain food items. In addition, FGF21 receptors in the paraventricular nucleus of the hypothalamus are other possible mediators of the FGF21 effect on appetite [5]. However, the effect on appetite differs according to ethnicity and is directed towards the most required food item according to the diverse environmental factors (sweet or high caloric food in Caucasians versus salty food in hot countries, where high sodium loss may prevail). At the level of taste buds, the sodium/glucose cotransporter 1 (SGLT1) may provide a plausible explanation of the paradox. the SGLT1 transports glucose into sweet-sensing taste buds, leading to membrane depolarization through blocking ATP-inhibited K+ channels. The involvement of sodium-dependent transporter (SGLT1) may also explain the potentiation of sweet taste by sodium salts [32]
Multivariate analysis using two-way unsupervised hierarchical clustering showed that the prevalence of the two FGF21 SNPs rs838145 and rs838133 in our Emirati cohort is close to the European subpopulations, but far from the Chinese population.
We explored the SNPs in LD with the FGF21 (A: rs838133) and (G:rs838145) in five South Asians, Iberians, Utah residents of European ancestry and Han Chinese. A hierarchical model was used to assess if the SNP was significantly enriched or depleted in each of the 7 populations compared with the overall average. The p-values were then used to generate an enrichment/depletion heatmap (Fig. 3). The SNPs of the Caucasian subpopulations are markedly distinguished from the South Asians and are closer to Iberians forming strong cluster that is independent of the other populations. The Chinese have another cluster pattern.
Based on the previous studies relating (A: rs838133) and (G:rs838145) to sweet intake [9] and our results that link both to sodium intake in the Emirati population, we speculate that genetic variant frequency in a given population may determine the food predilection, and at least in part, shape the food culture and its diversity all over the world. This may highlight the fact that the “cultural” difference in food predilection may be dictated, at least in part, by genomics.
Analysis of data derived from the 1000 genome project showed Linkage disequilibrium (LD) of FGF21 rs838133 with several SNPs in the Izumo Sperm-Egg Fusion 1 (IZUMO1) and Fucosyltransferase 1 (FUT1) genes. Similarly, LD analysis of rs838145 showed LD with several SNPs in Ras Interacting Protein 1 (RASIP1) and IZUMO1 genes. FUT1 (H blood group) gene encodes a Golgi stack membrane protein, involved in the formation of the H antigen, and mutations lead to the H-Bombay blood group. Associated diseases include Bombay Phenotype (autosomal recessive; related to tetralogy of Fallot and endocardial fibroelastosis) and Chronic Purulent Otitis Media [13]. The IZUMO1 is essential for sperm-egg plasma membrane binding and fusion. Associated diseases include normal-tension glaucoma and Hajdu-Cheney Syndrome (acroosteolysis with skeletal changes and distinctive facial features and potential spine fractures), [33]. RASIP1 is required for the proper formation of vascular structures and angiogenesis. It is a negative regulator of amino acid starvation-induced autophagy. RASIP1 may be associated with enamel Erosion of teeth [34].
All ethnic groups at all ages and of both genders are at an increasing prevalence of obesity. Aging is linked to increased abdominal adiposity and skeletal muscle fat deposition. Lifestyle changes in older adults may cause a long-term positive energy balance state. Interestingly, shorter telomeres have been associated with increased BMI and waist to hip ratio and accumulation of excess visceral fat [35].In our study, intake of added sugars has a significant negative correlation with age, whereas the high-fiber diet positively, but non-significantly, correlated with age.
Gender differences have been reported for dietary intakes in this study. This sex difference has been observed in all populations although its magnitude is influenced by ethnic, genetic, and environmental factors. In the current study, males had significantly higher total daily caloric and protein intake than females. Men usually need more calories and have higher energy intake than women due to different body composition. Men have more muscle mass as compared to women. Similarly, in other studies, men reported higher energy intake as compared to women consequently there was an increased intake of macronutrients [36]. The impact of the dietary intake on metabolic variables measured can be influenced by sex-related factors such as sex hormones [37]. In our study, females homozygous for minor A allele of rs838133 had a significantly higher intake of vitamin D. A few studies explored the association between different SNPs and vitamin D and calcium levels in the blood. Zhang et al screened 15 key genes in the vitamin D metabolic pathway. The results highlighted several SNPs that might contribute to variability in the serum 25(OH)D levels in a healthy Chinese population [38].
There may be fluctuations in the expression of certain genes related to immune and/or inflammatory response according to the menstrual cycle phases [39]. Previous studies have noticed major differences in gene expression profiles between men and women [40].
The genetic variants of FGF21 are likely associated with ethnic-dependent food predilection. Up to our knowledge, only the link to the sweet tooth was studied in Caucasians. In view of the well recognized “food culture” diversity across different ethnicities, studies are required to further investigate gene-environment interaction effect on food predilection.
Conclusion
The current study investigated the role of two FGF21 gene polymorphisms; rs838133 and rs838145 in defining preferences to certain types of food. The minor alleles of both studied FGF21 variants are associated with high salt intake in the Emirati population. Through the bioinformatics analysis, a potential effect of those SNPs on the diet and taste in different populations can be speculated. Linkage disequilibrium analysis showed FGF21 SNPs diversity amongst subpopulations. In addition, our study emphasizes that the interpretation of genotype-phenotype correlations may vary significantly according to ethnicity.
Statement of authors’ contributions to the manuscript
M.S. conceived the idea of the study, developed the overall plan, conducted the analysis and wrote the manuscript. S.M. and S.H. carried out the genotyping. H.R. supervised the administration, performed the analysis of the FFQ and reviewed the manuscript. R.W., H.J and A.A. administered FFQ and collected the saliva samples. P.H. and S.M. carried out the basic bioinformatics part. R.H. was consulted for the statistical analysis and carried out bioinformatics meta-analysis.
Funding
The study was funded by Al-Jalila Foundation seed grant (AJF 201768). M.S-A is funded by University of Sharjah Targeted grant (1801090141-P) and the MBRU-AlMahmeed Research Award 2019.
Compliance with Ethics Requirements
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We would like to acknowledge the kind help of Dr. Hayat Hassan to recruit subjects for the study (Al Qarain Health Center, Sharjah).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.05.020.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary Fig. S1.
Alignment of the sequence used for genotyping on the FGF21 gene. (A) The sequence used for genotyping the samples fro FGF21 rs838145 is marked (marker 1 and 2), the FGF21 rs838145 G/A SNP location is shown. The SNP is located close to the Izumo sperm-egg fusion 1 (IZUMO1) protein-coding region. (B) The sequence used for genotyping for the FGF21 rs838133 is marked (marker 3 and 4), the FGF21 rs838133 A/G SNP location is shown. The SNP is located on exon 2 of the FGF-21 gene. (C) Mapping of the FGF21 rs838145 and rs838133 location on chromosome 19.
Supplementary Fig. S1.
Supplementary Fig. S2.
Dendrograms of the heatmap of the identified SNPs in linkage disequilibrium with FGF21 rs838133 and rs838145 showing subpopulation clusters.
Supplementary Fig. S2.
References
- 1.von Holstein-Rathlou S, BonDurant LD, Peltekian L, Naber MC, Yin TC, Claflin KE, et al. FGF21 Mediates Endocrine Control of Simple Sugar Intake and Sweet Taste Preference by the Liver. Cell Metab [Internet]. 2016 Feb; 23(2): 335–43. Available from: https://linkinghub.elsevier.com/retrieve/pii/S155041311500618X. [DOI] [PMC free article] [PubMed]
- 2.BonDurant L.D., Ameka M., Naber M.C., Markan K.R., Idiga S.O., Acevedo M.R. FGF21 regulates metabolism through adipose-dependent and -independent mechanisms. Cell Metab [Internet]. 2017;25(4):935–944.e4. doi: 10.1016/j.cmet.2017.03.005. https://linkinghub.elsevier.com/retrieve/pii/S1550413117301560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sternson SM, Eiselt A-K. Three pillars for the neural control of appetite. Annu Rev Physiol [Internet]. 2017 Feb 10; 79(1): 401–23. Available from: https://doi.org/10.1146/annurev-physiol-021115-104948. [DOI] [PubMed]
- 4.Perry B., Wang Y. Appetite regulation and weight control: the role of gut hormones. Nutr Diabetes [Internet]. 2012;2(1):e26. doi: 10.1038/nutd.2011.21. http://www.nature.com/articles/nutd201121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talukdar S., Owen B.M., Song P., Hernandez G., Zhang Y., Zhou Y. FGF21 regulates sweet and alcohol preference. Cell Metab [Internet]. 2016;23(2):344–349. doi: 10.1016/j.cmet.2015.12.008. https://linkinghub.elsevier.com/retrieve/pii/S1550413115006233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorval L, B. I. Knapp JMB. Morphine preference in mice is decreased by fibroblast growth factor 21 (FGF21). In: Program No 24014 2019 Neuroscience Meeting Planner Society for Neuroscience; Chicago, IL. 2019. p. Online.
- 7.Chu A.Y., Workalemahu T., Paynter N.P., Rose L.M., Giulianini F., Tanaka T. Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet [Internet]. 2013;22(9):1895–1902. doi: 10.1093/hmg/ddt032. http://www.ncbi.nlm.nih.gov/pubmed/23372041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T., Ngwa J.S., van Rooij F.J., Zillikens M.C., Wojczynski M.K., Frazier-Wood A.C. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr [Internet]. 2013;97(6):1395–1402. doi: 10.3945/ajcn.112.052183. https://academic.oup.com/ajcn/article/97/6/1395/4576919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Søberg S., Sandholt C.H., Jespersen N.Z., Toft U., Madsen A.L., von Holstein-Rathlou S. FGF21 Is a Sugar-Induced Hormone Associated with Sweet Intake and Preference in Humans. Cell Metab [Internet]. 2017;25(5):1045–1053.e6. doi: 10.1016/j.cmet.2017.04.009. https://linkinghub.elsevier.com/retrieve/pii/S1550413117302140 [DOI] [PubMed] [Google Scholar]
- 10.Frayling T.M., Beaumont R.N., Jones S.E., Yaghootkar H., Tuke M.A., Ruth K.S. A Common Allele in FGF21 Associated with Sugar Intake Is Associated with Body Shape, Lower Total Body-Fat Percentage, and Higher Blood Pressure. Cell Rep [Internet]. 2018;23(2):327–336. doi: 10.1016/j.celrep.2018.03.070. https://linkinghub.elsevier.com/retrieve/pii/S2211124718304315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolumam G., Chen M.Z., Tong R., Zavala-Solorio J., Kates L., van Bruggen N. Sustained Brown Fat Stimulation and Insulin Sensitization by a Humanized Bispecific Antibody Agonist for Fibroblast Growth Factor Receptor 1/βKlotho Complex. EBioMedicine. 2015;2(7):730–743. doi: 10.1016/j.ebiom.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda J, Chen MZ, Baruch A. FGF21-receptor agonists: an emerging therapeutic class for obesity-related diseases. Horm Mol Biol Clin Investig [Internet]. 2017 May 24;30(2). Available from: http://www.degruyter.com/view/j/hmbci.2017.30.issue-2/hmbci-2017-0002/hmbci-2017-0002.xml. [DOI] [PubMed]
- 13.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol Cell Proteomics [Internet]. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. http://www.mcponline.org/lookup/doi/10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S., Choi J., Mohanty J., Sousa L.P., Tome F., Pardon E. Structures of $β$-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature [Internet]. 2018;553(7689):501–505. doi: 10.1038/nature25010. http://www.nature.com/articles/nature25010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu J.-W., Yeh S.-C., Tsai F.-Y., Chen H.-W., Tsou T.-C. Fibroblast growth factor 21 secretion enhances glucose uptake in mono(2-ethylhexyl)phthalate-treated adipocytes. Toxicol Vitr [Internet]. 2019;59:246–254. doi: 10.1016/j.tiv.2019.04.021. https://linkinghub.elsevier.com/retrieve/pii/S0887233318307082 [DOI] [PubMed] [Google Scholar]
- 16.Li D., Yuan H., Ortiz-Gonzalez X.R., Marsh E.D., Tian L., McCormick E.M. GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am J Hum Genet [Internet]. 2016;99(4):802–816. doi: 10.1016/j.ajhg.2016.07.013. https://linkinghub.elsevier.com/retrieve/pii/S0002929716302877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Núñez-Jaramillo L., Rangel-Hernández J.A., Burgueño-Zúñiga B., Miranda M.I. Activation of nucleus accumbens NMDA receptors differentially affects appetitive or aversive taste learning and memory. Front Behav Neurosci. 2012 doi: 10.3389/fnbeh.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris R.G.M. NMDA receptors and memory encoding. Neuropharmacology [Internet]. 2013;74:32–40. doi: 10.1016/j.neuropharm.2013.04.014. https://linkinghub.elsevier.com/retrieve/pii/S0028390813001512 [DOI] [PubMed] [Google Scholar]
- 19.Radwan H., Ballout R.A., Hasan H., Lessan N., Karavetian M., Rizk R. The Epidemiology and Economic Burden of Obesity and Related Cardiometabolic Disorders in the United Arab Emirates: A Systematic Review and Qualitative Synthesis. J Obes [Internet]. 2018;3(2018):1–23. doi: 10.1155/2018/2185942. https://www.hindawi.com/journals/jobe/2018/2185942/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montagnese C, Santarpia L, Iavarone F, Strangio F, Sangiovanni B, Buonifacio M, et al. Food-Based Dietary Guidelines around the World: Eastern Mediterranean and Middle Eastern Countries. Nutrients [Internet]. 2019 Jun 13; 11(6): 1325. Available from: https://www.mdpi.com/2072-6643/11/6/1325. [DOI] [PMC free article] [PubMed]
- 21.Phillips CM, Tierney AC, Perez-Martinez P, Defoort C, Blaak EE, Gjelstad IMF, et al. Obesity and Body Fat Classification in the Metabolic Syndrome: Impact on Cardiometabolic Risk Metabotype. Obesity [Internet]. 2012 Jun 28; Available from: http://doi.wiley.com/10.1038/oby.2012.188. [DOI] [PubMed]
- 22.Naja F., Nasreddine L., Itani L., Chamieh M.C., Adra N., Sibai A.M. Dietary patterns and their association with obesity and sociodemographic factors in a national sample of Lebanese adults. Public Health Nutr [Internet]. 2011;14(09):1570–1578. doi: 10.1017/S136898001100070X. http://www.journals.cambridge.org/abstract_S136898001100070X [DOI] [PubMed] [Google Scholar]
- 23.Saber-Ayad M., Manzoor S., El Serafi A., Mahmoud I., Hammoudeh S., Rani A. The FTO rs9939609 “A” allele is associated with impaired fasting glucose and insulin resistance in Emirati population. Gene [Internet]. 2019;681:93–98. doi: 10.1016/j.gene.2018.09.053. https://linkinghub.elsevier.com/retrieve/pii/S0378111918310175 [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez S., Gaunt T.R., Day I.N.M. Hardy-Weinberg Equilibrium Testing of Biological Ascertainment for Mendelian Randomization Studies. Am J Epidemiol [Internet]. 2009;169(4):505–514. doi: 10.1093/aje/kwn359. https://academic.oup.com/aje/article-lookup/doi/10.1093/aje/kwn359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanusz Z., Tarasińska J. Normalization of the Kolmogorov-Smirnov and Shapiro-Wilk tests of normality. Biometrical Lett [Internet]. 2015;52(2):85–93. http://content.sciendo.com/view/journals/bile/52/2/article-p85.xml [Google Scholar]
- 26.Doane DP, Seward LE. Measuring Skewness: A Forgotten Statistic? J Stat Educ [Internet]. 2011 Jul 29; 19(2). Available from: https://www.tandfonline.com/doi/full/10.1080/10691898.2011.11889611.
- 27.Greenland S., Senn S.J., Rothman K.J., Carlin J.B., Poole C., Goodman S.N. Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol [Internet]. 2016;31(4):337–350. doi: 10.1007/s10654-016-0149-3. http://link.springer.com/10.1007/s10654-016-0149-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semreen MH, Alniss HY, Grgic SR, El-Awady RA, Almehdi AH, Mousa MK, et al. Comparative metabolomics of MCF-7 breast cancer cells using different extraction solvents assessed by mass spectroscopy. Sci Rep [Internet]. 2019 Dec 11; 9(1): 13126. Available from: http://www.nature.com/articles/s41598-019-49509-y. [DOI] [PMC free article] [PubMed]
- 29.Pazoki R, Evangelou E, Mosen-Ansorena D, Pinto RC, Karaman I, Blakeley P, et al. GWAS for urinary sodium and potassium excretion highlights pathways shared with cardiovascular traits. Nat Commun [Internet]. 2019 Dec 13; 10(1): 3653. Available from: http://www.nature.com/articles/s41467-019-11451-y. [DOI] [PMC free article] [PubMed]
- 30.Sanada H., Jones J.E., Jose P.A. Genetics of salt-sensitive hypertension. Curr Hypertens Rep. 2011;13(1):55–66. doi: 10.1007/s11906-010-0167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He F.J., MacGregor G.A. Reducing Population Salt Intake Worldwide: From Evidence to Implementation. Prog Cardiovasc Dis [Internet]. 2010;52(5):363–382. doi: 10.1016/j.pcad.2009.12.006. https://linkinghub.elsevier.com/retrieve/pii/S0033062009001273 [DOI] [PubMed] [Google Scholar]
- 32.Hill C.M., Qualls-Creekmore E., Berthoud H.-R., Soto P., Yu S., McDougal D.H. FGF21 and the physiological regulation of macronutrient preference. Endocrinology. 2020;163(3) doi: 10.1210/endocr/bqaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosseini-Esfahani F, Koochakpoor G, Daneshpour MS, Mirmiran P, Sedaghati-khayat B, Azizi F. The interaction of fat mass and obesity associated gene polymorphisms and dietary fiber intake in relation to obesity phenotypes. Sci Rep [Internet]. 2017 Dec 22; 7(1): 18057. Available from: http://www.nature.com/articles/s41598-017-18386-8. [DOI] [PMC free article] [PubMed]
- 34.Roper S.D., Chaudhari N. Taste buds: cells, signals and synapses. Nat Rev Neurosci [Internet]. 2017;18(8):485–497. doi: 10.1038/nrn.2017.68. http://www.nature.com/articles/nrn.2017.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singaravelu G., Rahimi S., Krauchunas A., Rizvi A., Dharia S., Shakes D. Forward Genetics Identifies a Requirement for the Izumo-like Immunoglobulin Superfamily spe-45 Gene in Caenorhabditis elegans Fertilization. Curr Biol [Internet]. 2015;25(24):3220–3224. doi: 10.1016/j.cub.2015.10.055. https://linkinghub.elsevier.com/retrieve/pii/S0960982215013494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Kreuk B.-J., Gingras A.R., Knight J.D., Liu J.J., Gingras A.-C., Ginsberg M.H. Heart of glass anchors Rasip1 at endothelial cell-cell junctions to support vascular integrity. Elife [Internet]. 2016 Jan;19:5. doi: 10.7554/eLife.11394. https://elifesciences.org/articles/11394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzanetakou I.P., Katsilambros N.L., Benetos A., Mikhailidis D.P., Perrea D.N. Is obesity linked to aging? Ageing Res Rev [Internet]. 2012;11(2):220–229. doi: 10.1016/j.arr.2011.12.003. https://linkinghub.elsevier.com/retrieve/pii/S1568163711000754 [DOI] [PubMed] [Google Scholar]
- 38.Archer E, Hand GA, Blair SN. Validity of U.S. Nutritional Surveillance: National Health and Nutrition Examination Survey Caloric Energy Intake Data, 1971–2010. Johannsen D, editor. PLoS One [Internet]. 2013 Oct 9; 8(10): e76632. Available from: https://dx.plos.org/10.1371/journal.pone.0076632. [DOI] [PMC free article] [PubMed]
- 39.Bédard A., Tchernof A., Lamarche B., Corneau L., Dodin S., Lemieux S. Effects of the traditional Mediterranean diet on adiponectin and leptin concentrations in men and premenopausal women: do sex differences exist? Eur J Clin Nutr [Internet]. 2014;68(5):561–566. doi: 10.1038/ejcn.2014.27. http://www.nature.com/articles/ejcn201427 [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z., He J.-W., Fu W.-Z., Zhang C.-Q., Zhang Z.-L. An analysis of the association between the vitamin D pathway and serum 25-hydroxyvitamin D levels in a healthy Chinese population. J Bone Miner Res [Internet]. 2013;28(8):1784–1792. doi: 10.1002/jbmr.1926. http://doi.wiley.com/10.1002/jbmr.1926 [DOI] [PubMed] [Google Scholar]
- 41.AlSafar HS, Al-Ali M, Elbait GD, et al. Introducing the first whole genomes of nationals from the United Arab Emirates. Sci Rep. 2019;9(1):14725. Published 2019 Oct 11. doi:10.1038/s41598-019-50876-9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.